Abstract
Background & Aims
Vitamin D exerts regulatory roles via vitamin D receptor (VDR) in mucosal immunity, host defense, and inflammation involving host factors and microbiome. Human Vdr gene variation shapes the microbiome and VDR deletion leads to dysbiosis. Low VDR expression and diminished vitamin D/VDR signaling are observed in colon cancer. Nevertheless, how intestinal epithelial VDR is involved in tumorigenesis through gut microbiota remains unknown. We hypothesized that intestinal VDR protects mice against dysbiosis via modulating the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway in tumorigenesis.
Methods
To test our hypothesis, we used an azoxymethane/dextran sulfate sodium–induced cancer model in intestinal VDR conditional knockout (VDRΔIEC) mice, cell cultures, stem cell–derived colonoids, and human colon cancer samples.
Results
VDRΔIEC mice have higher numbers of tumors, with the location shifted from the distal to proximal colon. Fecal microbiota analysis showed that VDR deletion leads to a bacterial profile shift from normal to susceptible carcinogenesis. We found enhanced bacterial staining in mouse and human tumors. Microbial metabolites from VDRΔIEC mice showed increased secondary bile acids, consistent with observations in human CRC. We further identified that VDR protein bound to the Jak2 promoter, suggesting that VDR transcriptionally regulated Jak2. The JAK/STAT pathway is critical in intestinal and microbial homeostasis. Fecal samples from VDRΔIEC mice activate the STAT3 signaling in human and mouse organoids. Lack of VDR led to hyperfunction of Jak2 in response to intestinal dysbiosis. A JAK/STAT inhibitor abolished the microbiome-induced activation of STAT3.
Conclusions
We provide insights into the mechanism of VDR dysfunction leading to dysbiosis and tumorigenesis. It indicates a new target: microbiome and VDR for the prevention of cancer.
Keywords: Cancer, Colonoids, Dysbiosis, Host–Bacterial Interactions, Inflammation, Microbiome, Nuclear Receptor, STAT, VDR, Vitamin D
Abbreviations used in this paper: 1,25(OH)2D3, 1α,25-dihydroxy vitamin D3; AOM/DSS, azoxymethane dextran sodium sulfate; ChIP, chromatin immunoprecipitation; CRC, colon rectal cancer; ESI, electrospray ionization; IEC, intestinal epithelial cell; Lcn-2, lipocalin 2; Jak/STAT, Janus kinase/signal transducer and activator of transcription; KO, knockout; LPS, lipopolysaccharide; PCNA, proliferating cell nuclear antigen; PCR, polymerase chain reaction; SDS, sodium dodecyl sulfate; UPLC-MS/MS, ultra high-performance liquid chromatography–tandem mass spectroscopy; VDR, vitamin D receptor
Summary.
Vitamin D/vitamin D receptor (VDR) deficiency causes a high risk of colon cancer, but how VDR is involved in tumorigenesis through microbiota remains unknown. We provide insights into the mechanism of VDR dysfunction leading to dysbiosis and tumorigenesis, indicating a new target: microbiome and VDR for the prevention of cancer.
Current research has implicated vitamin D deficiency as a critical factor in the pathology and clinical outcome of colon rectal cancer (CRC).1,2 Low plasma vitamin D is associated with adverse CRC survival after surgical resection.3,4 Vitamin D receptor (VDR) is a nuclear receptor that mediates functions of 1,25-dihydroxyvitamin D (1,25[OH]2D3), the biological active form of vitamin D.5 Higher VDR expression in tumor stromal fibroblast is associated with longer survival in a large cohort of CRC patients.2 The parallel appreciation of a role for the VDR in cancer biology began approximately 3 decades ago and subsequently a remarkable increase has occurred in the understanding of its actions in normal and malignant systems.6
The VDR regulation of gut microbiome in human and animal studies represents a newly identified and highly significant activity for VDR.7, 8, 9 Human Vdr gene variation shapes the gut microbiome and Vdr deletion leads to dysbiosis.8 Our study on VDR and bacteria establishes a microorganism-induced program of epithelial cell homeostasis and repair in the intestine.10 Dysregulation of bacterial–host interaction can result in chronic inflammatory and overexuberant repair responses, and is associated with the development of various human diseases including cancers.11,12 Even though vitamin D/VDR is an active topic in cancer research, the mechanism underlying host–microbiome interactions in cancer is incompletely understood. We know little about the mechanisms for the intestinal epithelial VDR and microbiome in CRC.
In the current study, we focused on the functions of VDR in intestinal epithelial cells and the microbiome. We hypothesized that intestinal VDR protects mice against dysbiosis via modulating the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway in tumorigenesis. VDR is required for intestinal epithelium functions and microbial homeostasis. We tested our hypothesis in an azoxymethane/dextran sulfate sodium (AOM/DSS)-induced cancer model, using intestinal VDR conditional knockout VDRΔintestinal epithelial cell (IEC) mice, colonoids, and human samples. Lack of the VDR signaling pathway led to increased tumors in colon and a shift in tumor distribution in the intestinal VDR knockout (KO) mice. We investigated how the absence of intestinal VDR leads to dysfunction in epithelial cells–microbiome interactions and the mechanism through the JAK/STAT3 signaling. Emerging data have suggested that interference in the JAK/STAT3 pathway may suppress the growth of colon cancer.13,14 JAK/STAT inhibitors are clinically used in patients with inflammatory bowel diseases.15 Thus, VDR regulation of the JAK/STAT3 pathway indicates a new target: microbiome and VDR signaling in anti-inflammation and anticancer. Our study provides insights into the mechanisms of VDR in maintaining intestinal and microbial homeostasis and protecting against intestinal tumorigenesis.
Results
Intestinal Epithelial VDR KO Mice Have Higher Tumor Numbers and Shifted Tumor Location
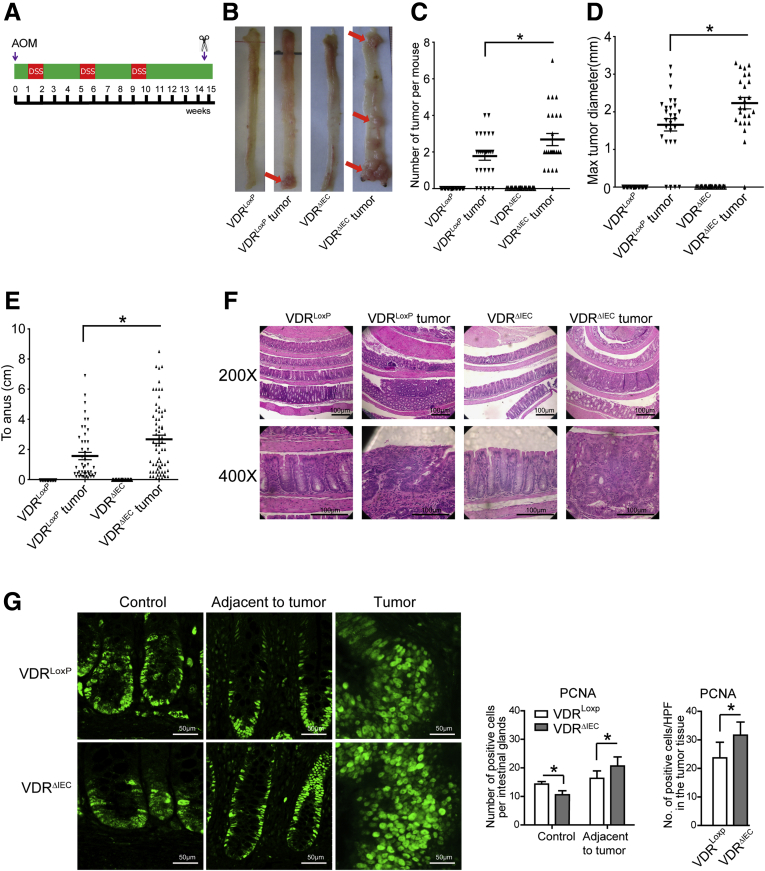
We tested our hypothesis in an AOM/DSS-induced cancer model using intestinal epithelial VDR conditional knockout VDRΔIEC mice (Figure 1A). AOM mice develop hyperproliferative colonic mucosa, aberrant crypt foci, and, eventually, carcinomas.16 AOM/DSS provides a widely used paradigm to study colitis-associated colon cancer. There was a striking difference in tumor incidence in mice with VDRLoxP and VDRΔIEC mice. We found that the VDRΔIEC mice developed more tumors (Figure 1B and C). The number and size of the tumors were significantly larger in the VDRΔIEC mice compared with the VDRLoxP mice (Figure 1C and D). Interestingly, tumor location in the VDRΔIEC mice significantly shifted from the distal to proximal colon, compared with tumors mainly in the distal colon of VDRLoxP mice (Figure 1B and E). Furthermore, the pathologic analysis of colon samples (Figure 1F) indicated a difference in tumor stage (carcinoma vs adenoma) between VDRΔIEC mice and VDRLoxP AOM/DSS experimental groups. Epithelial hyperproliferation plays a critical role in the development of colon cancer. Our immunohistochemical data of the proliferative marker proliferating cell nuclear antigen (PCNA) showed that PCNA in the colon was increased significantly in VDRΔIEC mice compared with VDRLoxP mice (Figure 1G).
Figure 1.
Intestinal epithelial cell VDR KO mice developed more tumors. (A) Schematic overview of the AOM/DSS-induced colon cancer model. AOM (10 mg/kg) was injected on day 0. On day 7, 2% DSS solution was administered to mice in drinking water. Seven days of DSS was followed by 3 weeks of drinking water. An additional 2 cycles of DSS were administered before killing. At week 15, mice were killed. (B) Colonic tumors in situ. Representative colons from different groups. Tumors are indicated by red arrows. (C) Tumor numbers in AOM/DSS-induced colon cancer model: VDRLoxP and VDRΔIEC mice. Data are expressed as means ± SD. n = 25–30, 1-way analysis of variance test. No tumors in controls for VDRLoxP and VDRΔIEC mice, therefore controls are not included for comparisons. (D) Maximum tumor size in the AOM/DSS-induced colon cancer model: VDRLoxP and VDRΔIEC mice. Data are expressed as means ± SD. n = 25–30, 1-way analysis of variance test. (E) The distance of each tumor to the anus was measured. Data are expressed as means ± SD. n = 25–30, 1-way analysis of variance test. (F) Representative H&E staining of Swiss rolls of representative colons from the indicated groups. Images are from a single experiment and are representative of 10 mice per group. (G) Quantitation of PCNA-positive cells in control mucosa/per intestinal glands or in the tumor tissue/high-power field. PCNA expression in the tumor tissue of VDRΔIEC mice was significantly higher than that in the VDRLoxP mice. Data are from a single experiment and are representative of 5 mice per group. Data are expressed as means ± SD. n = 5, Student t test. ∗P < .05.
Lack of Intestinal VDR Leads to Dysbiosis and a Shift of the Bacterial Profile for a Higher Risk of CRC
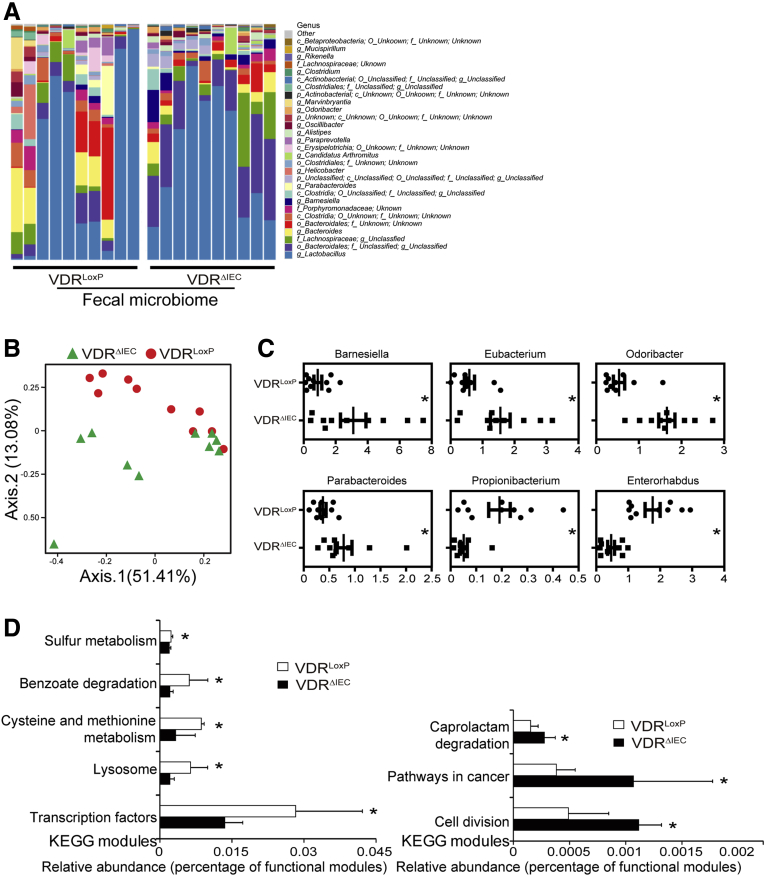
VDRΔIEC mice are known to have dysbiosis and the absence of intestinal epithelial VDR confers a transmissible risk for colitis.7 By using 16S sequencing, we showed the difference in fecal microbiome between VDRΔIEC mice and VDRLoxP mice (n = 10 each) at the genus level (n = 10) (Figure 2A). Figure 2B showed the unweighted Unique Fraction Metric distances of stool samples from VDRLoxP and VDRΔIEC mice on a principal coordinate analysis scale. We further showed the percentages of the affected genera between VDRLoxP mice and VDRΔIEC mice (Figure 2C). Functional alterations of the intestinal microbiome were detected by fecal microbiota Kyoto Encyclopedia of Genes and Genomes analysis. Lacking VDR leads to a bacterial profile shift from normal to carcinogenesis susceptibility (Figure 2D), indicating that cancer risk was significantly higher in the VDRΔIEC mice.
Figure 2.
Dysbiosis leads to high cancer risk in VDRΔIEC mice. (A) Composition of the bacterial community at the genus level in stool samples from separate cages of VDRLoxP mice (n = 10) and VDRΔIEC (n = 10) mice. (B) Unweighted Unique Fraction Metric distances of stool samples from VDRLoxP and VDRΔIEC mice on a principal coordinate analysis scale. (C) The percentages of the affected genera were compared between VDRLoxP mice and VDRΔIEC mice. Data are expressed as means ± SD. n = 10, Welch 2-sample t test. (D) Functional alterations of the intestinal microbiome related to VDR status. Data are expressed as means ± SD. n = 10, Student t-test. ∗P < .05. KEGG, Kyoto Encyclopedia of Genes and Genomes.
VDR Deletion Enhanced Bacteria in the Tumors of VDRΔIEC Mice and Impacted Bile Acid Metabolism
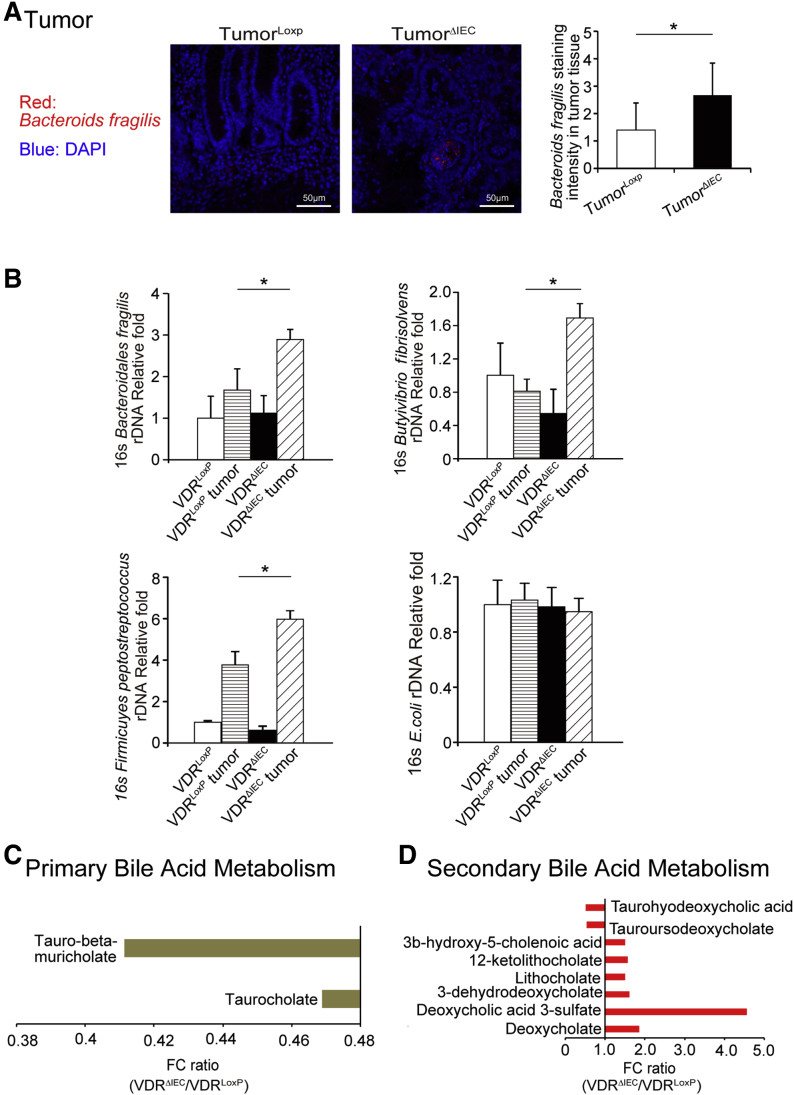
We then analyzed the relative bacteria abundance in the tumors. Bacteroides fragilis, a bacterial species enhanced in colon cancer, showed more staining in the tumors of VDRΔIEC mice compared with the VDRLoxP mice (Figure 3A). Figure 3B shows that B fragilis, Butyrivibrio fibrisolvens, and Firmicutes peptostreptococcus were enhanced in tumors in VDRΔIEC mice compared with VDRLoxP mice in tumor tissue. These bacteria are known to be associated with changes of metabolite (eg, short-chain fatty acids, bile acids) in CRC.17, 18, 19
Figure 3.
Lacking intestinal VDR leads to dysbiosis and a shift of the bacterial profile. (A) More B fragilis in tumor tissue of VDRΔIEC mice was found by fluorescent in situ hybridization. Images are from a single experiment and are representative of 5 mice per group. Data are expressed as means ± SD. n = 5, Student t test. (B) B fragilis, B fibrisolvens, and F peptostreptococcus were enhanced in tumors in VDRΔIEC mice compared with VDRLoxP mice. Data are expressed as means ± SD. n = 6, 1-way analysis of variance test. (C) The fold change ratios of the average concentrations of primary bile acid in the VDRΔIEC group was significantly lower compared with that in the control group (VDRLoxP, n = 16; VDRΔIEC, n = 17, Welch 2-sample t test, metabolite ratio < 1.00; P < .05). (D) The fold change ratios of the average concentrations of secondary bile acid in the VDRΔIEC group was significantly higher compared with that in the control group (VDRLoxP, n = 16; VDRΔIEC, n = 17, Welch 2-sample t test, metabolite ratio ≥ 1.00; P < .05). ∗P < .05. DAPI, 4′,6-diamidino-2-phenylindole; FC, fecal coliforms; rDNA, ribosomal DNA.
We quantitively profiled metabolites derived from host–microbial co-metabolism in fecal samples using the unbiased method. We found the changes in primary bile acid metabolism and secondary bile acid metabolism in VDRΔIEC mice. The fold change ratios of the identified bile acid species were significantly higher in the VDRΔIEC group than those in the control group (Figure 3C and D). These changes are consistent with the recent observations in human CRC that bile acid metabolism is among the top biomarkers of patients.20
Increased Inflammation in VDRΔIEC Mice
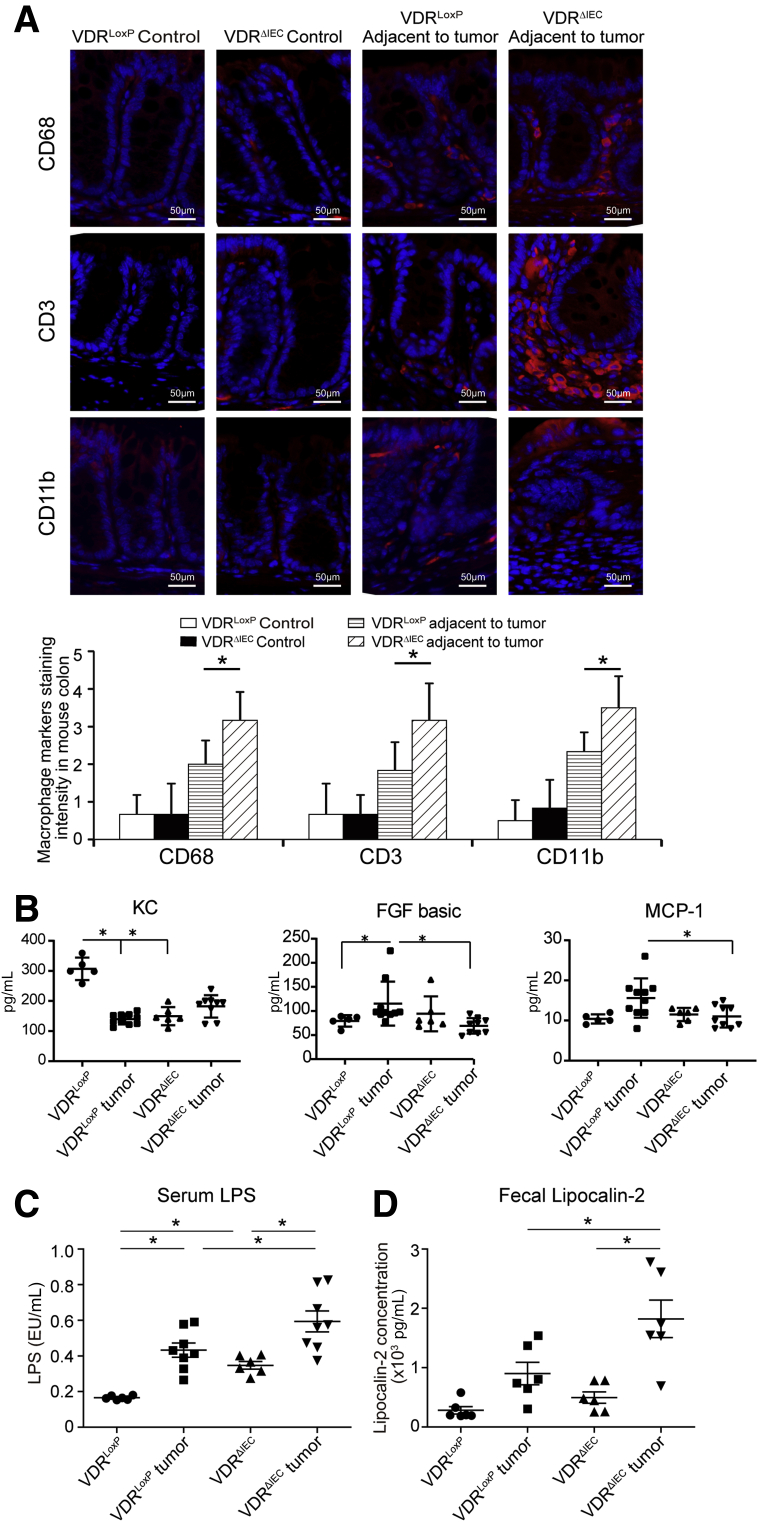
We further hypothesized that the altered intestinal epithelial and microbial functions lead to chronic inflammation, thus exacerbating colon cancer progression. We assessed several lymphocyte markers in normal colon and colonic tumors. Levels of CD68, CD3, and CD11b significantly increased in tumors, especially in VDRΔIEC mice (Figure 4A). We also detected the cytokines in serum samples from VDRLoxP and VDRΔIEC mice with or without tumor. We found that the level of fibroblast growth factor basic and monocyte chemoattractant protein-1 in the tumor tissue of VDRLoxP mice were higher than that of VDRΔIEC mice (Figure 4B). In the gastrointestinal tract, tissue barrier integrity is particularly important. Serum samples from VDRLoxP and VDRΔIEC mice were used to measure bacterial endotoxin with Limulus amebocyte lysate chromogenic end point assays. We found more bacterial endotoxin lipopolysaccharide (LPS) in VDRΔIEC mice than in VDRLoxP mice, especially in tumor groups (Figure 4C). Lipocalin-2 (Lcn-2) is used as a marker of intestinal inflammation.21 We found that the expression level of fecal Lcn-2 was significantly higher in tumor tissue of VDRΔIEC mice than that of VDRLoxP mice (Figure 4D).
Figure 4.
Altered intestinal epithelial and microbial functions may lead to chronic inflammation. (A) Several lymphocyte markers were detected in colon tissue by immunofluorescence staining. Levels of CD68, CD3, and CD11b was increased significantly in tumors, especially in VDRΔIEC mice. Data are expressed as means ± SD. n = 6, 1-way analysis of variance test. (B) Serum samples were collected from VDRLoxP and VDRΔIEC mice with or without tumor, and then cytokines were detected by the Luminex detection system. Data are expressed as means ± SD. n = 5–10, 1-way analysis of variance test. (C) Serum LPS was significantly higher in the VDRΔIEC mice. Data are expressed as means ± SD. n = 6, 1-way analysis of variance test. (D) Fecal lipocalin-2 was increased in the VDRΔIEC mice with tumors. Data are expressed as means ± SD. n = 6, 1-way analysis of variance test. ∗P < .05. FGF, fibroblast growth factor; KC, Keratinocyte Chemoattractant; MCP-1, monocyte chemoattractant protein-1.
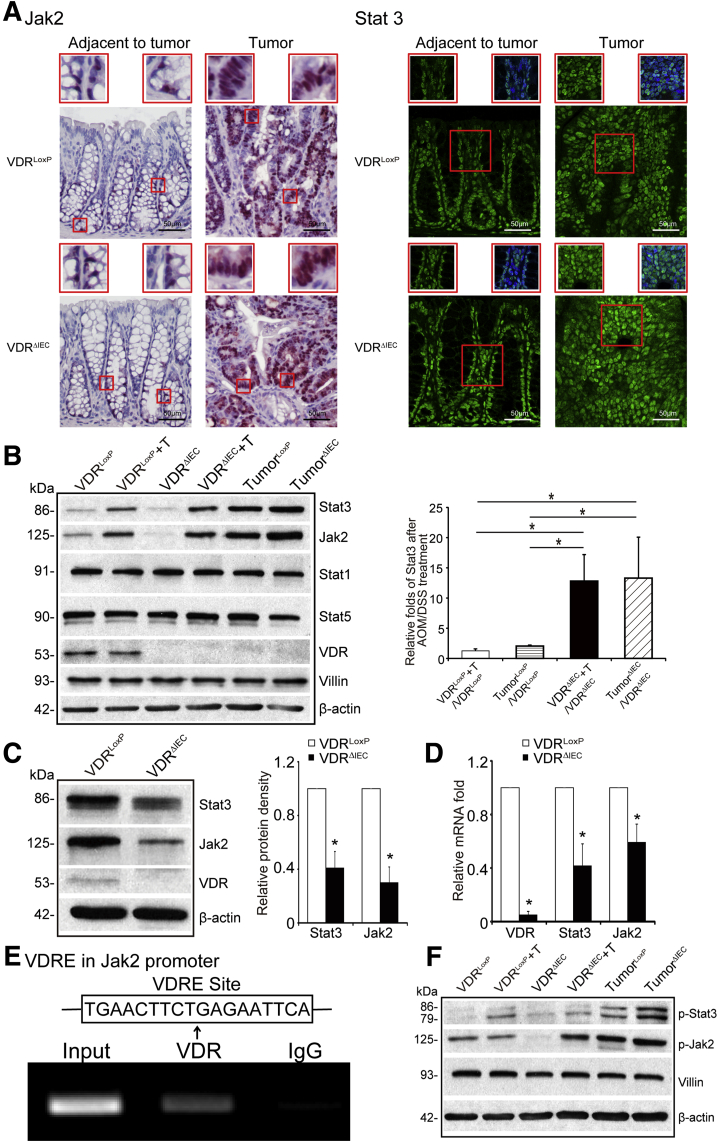
VDR Deletion Leads to Hyperfunction of the Jak2/STAT3 Signaling in Tumor Tissue
The JAK/STAT3 pathway is known to suppress the growth of colon cancer.13 After AOM/DSS treatment in VDRΔIEC mice, we observed up-regulation of total Jak2 and STAT3 protein expression in colon cancer tissue using immunostaining (Figure 5A). Furthermore, Western blot confirmed that the total protein levels of Jak2 and STAT3 were enhanced in tumors of AOM/DSS-treated VDRΔIEC mice (Figure 5B). However, VDR deletion changed the expression of total STAT1 and STAT5 in the colon tumor tissue. Interestingly, without any treatment, VDR deletion led to reduced total STAT3 and Jak2 in the basal levels of cells at the protein level and the messenger RNA level (Figure 5C and D). Furthermore, we identified VDR protein bound to the Jak2 promoter (TGAACTTCTGAGAATTCA) by chromatin immunoprecipitation (ChIP) assay (Figure 5E). Taken together, our observations show that the absence of intestinal epithelial VDR leads to the hyperfunction of JAK/STAT3 signaling in inflammation. We also examined the protein expressions of activated phosphorylated Stat3/phosphorylated Jak2 in normal tissue and tumors. We found that the expression levels of phosphorylated Stat3 and phosphorylated Jak2 were higher in the tumors in AOM/DSS–induced VDRΔIEC mice compared with those in VDRLoxp mice (Figure 5F).
Figure 5.
VDR deletion leads to dysfunction of Jak2/Stat3 signaling in the tumor tissue. (A) Jak2 and Stat3 were increased in tumor tissue of VDRΔIEC mice compared with tumor tissue of VDRLoxP mice by immunofluorescence staining. Images are from a single experiment and are representative of 6 mice per group. Red boxes are higher magnification of select area. (B) VDR deletion increased the total Jak2 and Stat3 in colon tumor tissue, but not the expression levels of total Stat1 and Stat5 protein in colon. Data are expressed as means ± SD. n = 3, 1-way analysis of variance test. (C) VDR deletion decreased Jak2 and Stat3 at protein levels in colon. Data are expressed as means ± SD. n = 5, Student t test. (D) VDR deletion decreased Jak2 and Stat3 at messenger RNA (mRNA) levels in colon without any treatment. Data are expressed as mean ± SD. n = 5, Welch 2-sample t test. (E) Vitamin D response element binds to the Jak2 promoter. ChIP–PCR amplification showed binding of VDR to the promoter regions of Jak2. PCR was performed including input and negative controls. n = 3 separate experiments. (F) The expression levels of p-Stat3 and p-Jak2 were higher in the tumors in AOM/DSS-induced mice, compared with those in the VDRLoxp mice. ∗P < .05. T, tumor.
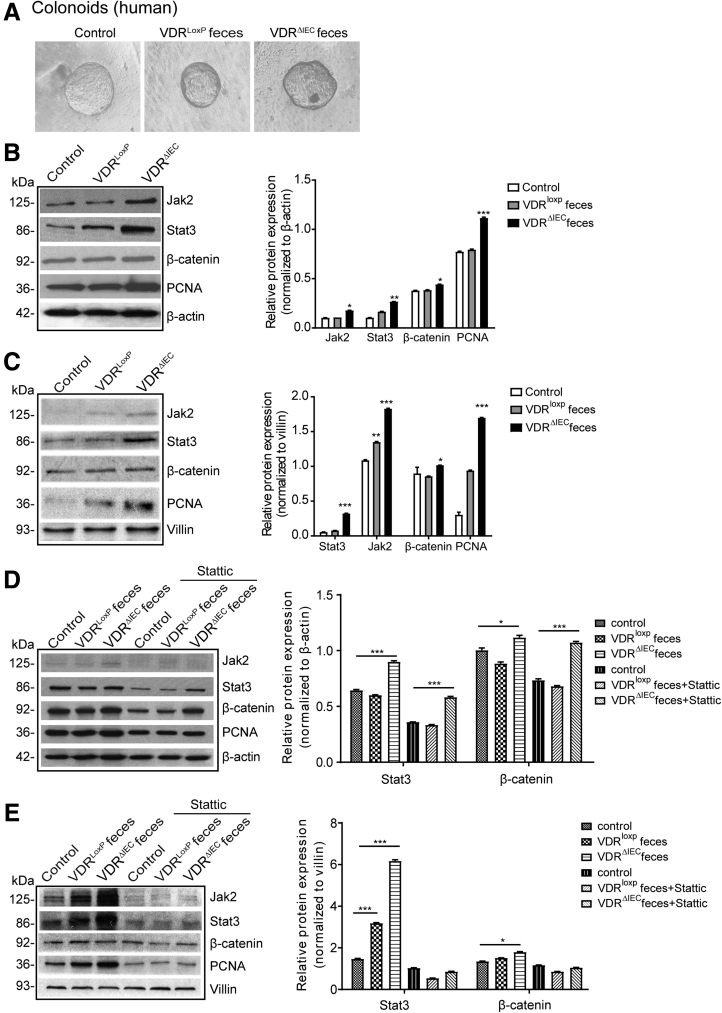
Gut Microbiome From VDRΔIEC Mice Actives JAK/STAT Signaling in Colonoids
By using the stem cell–derived colonoid systems (Figure 6A), we further investigated the influence of intestinal VDR during the activation of JAK/STAT signaling. PCNA, a proliferation marker, and proliferation regulator β-catenin, were increased in the VDRΔIEC feces-treated group followed by activation of Stat3 (human colonoids in Figure 6B). The similar hyper-regulation of STAT3 also was observed in the mouse colonoids treated with microbiome from VDRΔIEC mice (Figure 6C). We then treated the organoid with stattic, a STAT3 inhibitor. The total Stat3 was decreased compared with the no-stattic–treated mouse colonoids (Figure 6D). However, the expressions of Stat3 and β-catenin in the VDRΔIEC group still were higher than in the VDRLoxP group (Figure 6D). We observed the similar effect of stattic in inhibiting the microbiome activation of Jak2/STAT3 signaling in mouse colonoids (Figure 6E). Interestingly, stattic treatment also reduced the proliferation regulator β-catenin and the proliferation marker PCNA in colonoids.
Figure 6.
Gut microbiota from VDRΔIEC mice activates JAK/STAT signaling in human and mouse organoids. (A) Human colonoids were prepared and treated with feces from VDRloxP or VDRΔIEC mice for 2 hours. (B) The expression of Jak2 and Stat3 in human colonoids and (C) mouse colonoids was detected by Western blot. PCNA and β-catenin were increased in the VDRΔIEC feces-treated group. Data are expressed as means ± SD. n = 3, 1-way analysis of variance test. (D) Human and (E) mouse organoids were pretreated with 20 μmol/L of stattic for 2 hours, then treated with feces for 2 hours. The expression of Jak2 and was increased after stattic treatment, especially in the VDRΔIEC group. The total Stat3 was decreased compared with the no-stattic group. Data are expressed as means ± SD. n = 3, 2-way analysis of variance test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 compared with the control group.
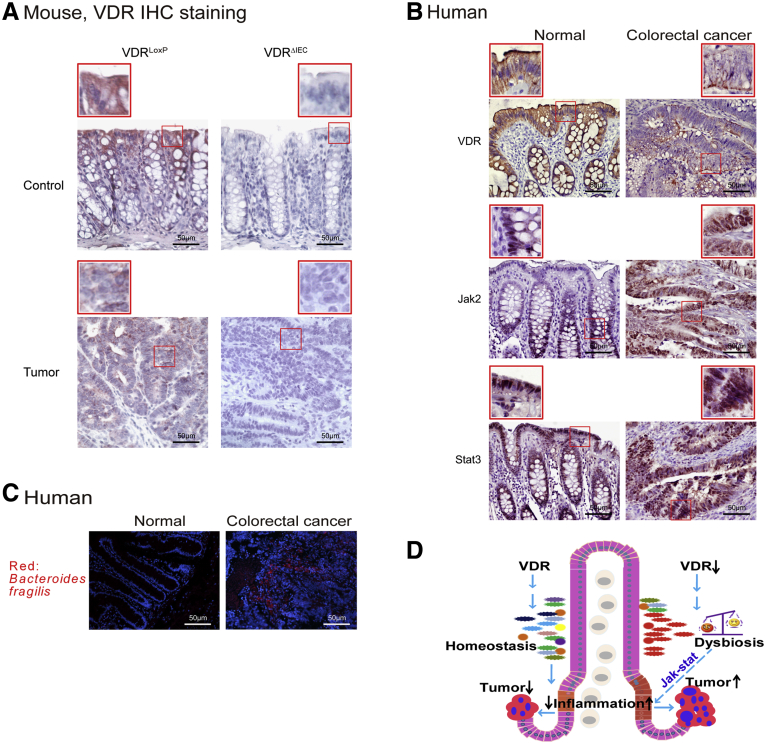
Reduced VDR and Enhanced Bacteria in Human Colon Cancer Tissue
VDR expression was decreased in the AOM/DSS–induced colon cancer model (Figure 7A). We continued to explore VDR expression in human colorectal colon samples. Our data showed that increased total Jak2 and STAT3 were associated with a reduction of intestinal VDR in human CRC intestines (Figure 7B), suggesting that JAK/STAT3 is up-regulated in human CRC with protective VDR. Interestingly, we identified bacteria in human colorectal colon samples. Fluorescent in situ hybridization data showed B fragilis in tumors from patients with CRC (Figure 7C).
Figure 7.
Enhanced bacteria, reduced VDR, and increased Jak2 and STAT3 expression was observed in human CRC patients and the AOM/DSS-induced colon cancer model. (A) Intestinal VDR expression was decreased in the AOM/DSS-induced colon cancer model. Images are from a single experiment and are representative of 6 mice per group. Red boxes are used to show the higher magnification of selected area (B) Intestinal VDR, Jak2, and STAT3 staining in human CRC samples. Compared with normal intestines, CRC patient intestines had a statistically significantly lower VDR and higher Jak2/STAT3 expression. Images are representative of experiments that we performed in triplicate; normal, n = 10; colorectal cancer, n = 10. (C) B fragilis was found in human CRC samples compared with normal tissue. Images are representative of experiments that we performed in triplicate. Normal, n = 10; colorectal cancer, n = 10. (D) A working model of VDR protects against dysbiosis and tumorigenesis via the JAK/STAT pathway in intestine. Lack of intestinal epithelial VDR causes dysbiosis and hyper-regulation of JAK/STAT3, thus leading to the overgrowth of tumors in colon.
Discussion
In the current study, we have shown that VDR deficiency in intestine leads to a bacterial profile shift from normal to susceptible carcinogenesis. VDRΔIEC mice have higher tumor numbers with their tumor location shifted from the distal to proximal colon. Enhanced bacterial staining was found in tumors. Microbial metabolites from VDRΔIEC mice showed increased secondary bile acids, which is consistent with observations in human CRC. Furthermore, our study provides the mechanism of VDR dysfunction leading to dysbiosis and tumorigenesis through the hyperfunctioning Jak2. Fecal samples from VDRΔIEC mice enhance the expression of STAT3 in human and mouse organoids. A JAK/STAT inhibitor abolished the microbiome-induced activation of STAT3. Our study fills the gaps by showing mechanisms that are important to normal intestinal homeostasis and to chronic inflammation and dysbiosis, thus suggesting new therapeutic targets for restoring VDR functions in colitis-associated colon cancer (Figure 7D, working model).
Epidemiologic and experimental studies have indicated a protective action of vitamin D against colorectal cancer.22, 23, 24, 25, 26, 27 Vitamin D3 exerts its chemopreventive activity by interrupting crosstalk between tumor epithelial cells and the tumor microenvironment in a VDR-dependent manner.23 Moreover, there is increasing interest regarding the use of vitamin D compounds for disease prevention and therapy.28 If we do not understand the mechanism of the receptor of vitamin D, vitamin D taken by people may not be used effectively and efficiently. Hence, our current study fills the gap by characterizing the precise role for intestinal epithelial VDR in colon cancer models.
Endogenous enteric bacteria play a crucial role in the pathogenesis of colon cancer.29 Dysregulation of bacterial–host interactions can result in chronic inflammatory and the development of cancer.30,31 Multiple mechanisms of VDR affecting cancers have been found, focusing on the host factors (eg, β-catenin pathway and inflammation).32 However, very little is known about the physiological effects and molecular mechanisms responsible for intestinal epithelial VDR regulation of the microbiome community. Our study on VDR regulation of gut bacteria has shown a microorganism-induced program of epithelial cell homeostasis and repair in the intestine.10,33 An abundance of Parabacteroides was affected by VDR signaling in both human and mouse samples.8 However, the specific relationship between the function of intestinal VDR and microbiome in tumorigenesis is not understood.34 Here, we found that VDR directly regulates host–bacterial interactions via JAK/STAT pathways and its downstream genes.
Microbial metabolites from VDRΔIEC mice showed bile acid dysregulation and increased secondary bile acids, which is consistent with the observed microbiome markers in human CRC.11,12 Loss of VDR is known to disquiet the bile acid homeostasis.35,36 Our recent study37 used intestinal epithelium-specific VDR knock out (VDRΔIEC) mice and myeloid cell–specific VDR KO (VDRΔlyz) mice to assess whether the microbiome-associated metabolic changes linked with conditional loss of VDR in a particular tissue. Among different secondary bile acids, lithocholate and deoxycholate were increased owing to loss of VDR in VDRΔIEC and in VDRΔlyz mice. We believe that host factors (eg, VDR status in specific tissues) modulate microbial metabolites and the microbiome, thus contributing to the high risk of digestive diseases.
We used colonoids and mice lacking intestinal VDR expression to confirm the physiological relevance and molecular mechanism in epithelial–microbiome interactions. Research of intestinal VDR provides a framework to understand how the intestinal epithelial cells in the gut inadvertently may promote the development of cancer as an extension of its normal role in defense and repair. These insights are important for understanding health as well as disease.
We noted a consistent link between low vitamin D/VDR signaling and high intestinal inflammation. Our previous studies suggested that cells lacking VDR are in a pre-inflammatory state,10,38,39 and overexpression of VDR substantially reduced inflammation in VDR-/- cells.38 VDR also is identified as a suppressor of IFN-α–induced signaling through the JAK/STAT pathway.40 The JAK/STAT pathway plays a critical role in intestinal and microbial homeostasis.41 The JAK/STAT inhibitors have been tested recently as novel biological therapeutic strategies in inflammatory bowel diseases.15 Because low-dose proinflammatory cytokines are sufficient to induce bacterial endocytosis by epithelial cells, subclinical or low-grade changes below the threshold may tip the balance of tolerance toward full-blown inflammation owing to subsequent intracellular microbial sensing and paracellular permeability damage. VDR expression increases epithelial integrity and attenuates inflammation. Thus, it is not surprising that the mucosal inflammation associated with VDR down-regulation in intestine contributes to the initiation and progression of colon cancer.
The current study is focused on studying the tissue-specific roles of VDR in intestinal epithelial cells in blocking intestinal inflammation and cancer development. In addition to intestinal epithelial cells, VDR also is expressed in several other cell types, including immune cells.42 We found that loss of myeloid VDR results in impaired Paneth cell differentiation and weakened host defense to pathogens.43 We investigated metabolites from mice with tissue-specific deletion of VDR in intestinal epithelial cells or myeloid cells.37 Conditional VDR deletion severely changed metabolites specifically produced from carbohydrate, protein, lipid, and bile acid metabolism.37 In the future, we will study VDR in immune cells during cancer development.
Conclusions
We provide a definitive characterization of the intestinal epithelial VDR in regulating diversity of the microbiome and colon cancer. It opens a new direction in the understanding of the microbial-VDR interactions in inflammation and cancer. It indicates a new target: microbiome and VDR for the prevention of cancer. VDR expression was decreased in the colon cancer mice after AOM/DSS treatment, which is consistent with the clinical observation in colitis-associated colon cancer patients.44 In the future, we also could consider restoring the protective role of intestinal epithelia VDR using VDR activators or probiotics in CRC. Understanding of the abnormal interactions between the host and microbiome will aid in developing novel strategies for managing chronic inflammatory diseases and cancers.
Materials and Methods
Human Tissue Samples
This study was performed in accordance with approval from the University of Rochester Ethics Committee (RSRB00037178). Colorectal tissue samples were obtained from 10 CRC patients with neoplasia and 10 patients without neoplasia (ages, 49–74 y). Human endoscopy samples from the University of Illinois at Chicago (UIC) hospital were collected for human organoid culture (Institutional Review Board: 2017-0384).
Animals
VDRLoxP mice were originally reported by Dr Geert Carmeliet.45 VDRΔIEC mice were obtained by crossing VDRLoxP mice with villin-cre mice (004586; Jackson Laboratory), as we previously reported.7 Experiments were performed on 2- to 3-month-old male and female mice. Mice were provided with water ad libitum and maintained on a 12-hour dark/light cycle. The animal work was approved by the University of Rochester (when the author’s [J.S.] laboratory was at the University of Rochester), the Rush University Animal Resources committee, and the UIC Office of Animal Care.
Induction of Colon Cancer by AOM/DSS in Mice
Mice were treated with 10 mg/kg of AOM (Sigma-Aldrich, Milwaukee, WI) by intraperitoneal injection as previously described.46 After a 7-day recovery period, mice received 3 cycles of 2% DSS in the drinking water. The initial sample size was 30 mice in the control group with no treatment and 30 mice in each experimental group. Tumor counts and measurements were performed in a blinded fashion under a stereo-dissecting microscope (SMZ1000; Nikon, Melville, NY). Microscopic analysis was performed for severity of inflammation and dysplasia on H&E-stained Swiss-rolled colons by a gastrointestinal pathologist blinded to treatment conditions. Mice were killed under anesthesia.
Cell Culture
HCT116 cells were grown in high-glucose Dulbecco’s modified Eagle medium (SH30243.01; Hyclone, Erie, PA) containing 10% (v/v) fetal bovine serum (900-108; GEMINI, West Sacramento, CA), 50 μg/mL streptomycin, and 50 U/mL penicillin (30-002CI; Mediatech, Inc, Manassas, VA), as previously described.47,48
Colonoid Cultures and Treatment With Mice Feces
C57BL/6J mice colonoids were prepared and maintained as previously described.49,50 Mini gut medium (advanced Dulbecco's modified Eagle medium/F12 supplemented with HEPES, L-glutamine, N2, and B27) was added to the culture, along with R-spondin, Noggin, epidermal growth factor, and Wnt-3a. On day 7 after passage, colonoids were colonized by indicated mice feces for 2 hours, washed, and then incubated for 2 hours in mini gut medium with gentamicin (500 μg/mL).
Human organoids were developed using endoscopy samples in the UIC hospital. Crypts were released from colon tissue by incubation for 30 minutes at 4°C in phosphate-buffered saline containing 2 mmol/L EDTA. Isolated crypts were counted and pelleted. A total of 500 crypts were mixed with 50 μL Matrigel (BD Biosciences, San Jose, CA) and plated in 24-well plates.51 The colonoids were maintained in Human IntestiCult Organoid Growth Medium (STEMCELL Technologies, Inc, Vancouver, BC).
Fresh feces were collected from 5 healthy VDRLoxP or VDRΔIEC mice (8 weeks) and then well mixed. A total of 100 mg feces was homogenized in 6 mL Hanks Balanced Salt Solution and centrifuged for 30 seconds at 300 rpm, at 4°C, to pellet the particulate matter. Organoids were treated with 250 μL feces supernatant for 2 hours, the organoids were washed 3 times with Hanks Balanced Salt Solution, and then the cells were incubated in regular organoid culture medium for 2 hours.52
Western Blot Analysis and Antibodies
Mouse colonic epithelial cells were collected by scraping the tissue from the colon of the mouse, including the proximal and distal regions.47,53 The cells were sonicated in lysis buffer (10 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, pH 8.0, 1% Triton X-100, Sigma, St. Louis, MO) with 0.2 mmol/L sodium orthovanadate and protease inhibitor cocktail. The protein concentration was measured using the Bio-Rad reagent (Bio-Rad, Hercules, CA). Cultured cells were rinsed twice with ice-cold Hank’s balanced salt solution, lysed in protein loading buffer (50 mmol/L Tris, pH 6.8, 100 mmol/L dithiothreitol, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol), and then sonicated. Equal amounts of protein were separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with primary antibodies. The following antibodies were used: anti-STAT3 (9132; Cell Signaling Technology, Danvers, MA), anti-Jak2 (3230; Cell Signaling Technology), anti-VDR (SC-13133; Santa Cruz Biotechnology, Dallas, TX), anti-villin (SC-7672; Santa Cruz Biotechnology), anti–p-β-catenin (9566; Cell Signaling Technology), anti–β-catenin (610154; BD Biosciences), anti-PCNA (SC-25280; Santa Cruz Biotechnology), anti-LC3B (2775; Cell Signaling Technology), or anti–β-actin (A5316; Sigma-Aldrich) antibodies and were visualized by ECL (32106; Thermo Fisher Scientific, Waltham, MA). Membranes that were probed with more than 1 antibody were stripped before reprobing.
Immunofluorescence
Colonic tissues were freshly isolated and embedded in paraffin wax after fixation with 10% neutral buffered formalin. Immunofluorescence was performed on paraffin-embedded sections (4 μm), after preparation of the slides as described previously,48 followed by incubation for 1 hour in blocking solution (2% bovine serum albumin and 1% goat serum in Hank’s balanced salt solution) to reduce nonspecific background. The tissue samples were incubated overnight with primary antibodies at 4°C. The following antibodies were used: anti-CD3, anti-CD11B, and anti-CD68 (Santa Cruz Biotechnology). Slides were washed 3 times for 5 minutes each at room temperature in wash buffer. Samples then were incubated with secondary antibodies (goat anti-rabbit, 1:200, Alexa Fluor 488; Molecular Probes, Eugene, OR) for 1 hour at room temperature. Tissues were mounted with the SlowFade Antifade Kit (s2828; Life Technologies, Grand Island, NY), followed by a coverslip, and the edges were sealed to prevent drying. Specimens were examined with a Zeiss laser scanning microscope LSM 710 (Carl Zeiss, Inc, Oberkochen, Germany).
Fluorescence In Situ Hybridization
Fluorescent in situ hybridization54 was performed using antisense single-stranded DNA probes targeting the bacterial 16S ribosomal RNA. The Bfra602 probe (5'-GAGCCGCAAACTTTCACAA-3') was used for the B fragilis group.55 Before performing the fluorescent in situ hybridization assay, 5-μm tissue sections were baked overnight at 55°C. Tissue sections were deparaffinized in xylene, dehydrated with 100% ethanol, air dried, incubated in 0.2 mol/L HCl for 20 minutes, and heated in 1 mmol/L sodium thiocyanate at 80°C for 10 minutes. Samples were pepsin-digested (4% pepsin in 0.01 N HCl) for 20 minutes at 37°C), slides were washed in wash buffer (0.3 mol/L NaCl, 0.03 mol/L sodium citrate, pH 7, and 0.1% SDS), fixed in 10% buffered formalin for 15 minutes, washed and dried, and hybridized with the probes at 5 ng/μL concentration each for 5 minutes at 96°C in hybridization buffer (0.9 mol/L NaCl, 30% formamide, 20 mmol/L Tris-HCl, pH 7.4), and 0.01% SDS and incubated at 37°C overnight. Slides were washed 4 times for 5 minutes each at 45°C in wash buffer. For visualization of the epithelial cell nuclei, the slides were counterstained with 4', 6'-diamidino-2-phenylindole/antifade solution. Slides were examined with a Zeiss laser scanning microscope LSM 710 (Carl Zeiss, Inc). Fluorescence staining was scored by a well-trained pathologist. Staining scores initially were quantified according to staining intensity (0, no staining; 1, minimal staining; 2, slight staining; 3, moderate staining; and 4, marked staining intensity). We randomly selected 3 spots for each mouse sample.
Mouse Cytokines
Mouse blood samples were collected by cardiac puncture and placed in tubes containing EDTA (10 mg/mL). Mouse cytokines were measured using a mouse cytokine 10-Plex Panel kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, beads of defined spectral properties were conjugated to protein-specific capture antibodies and added along with samples (including standards of known protein concentration, control samples, and test samples) into the wells of a filter-bottom microplate, where proteins bound to the capture antibodies over the course of a 2-hour incubation. After washing the beads, protein-specific biotinylated detector antibodies were added and incubated with the beads for 1 hour. After removal of excess biotinylated detector antibodies, the streptavidin-conjugated fluorescent protein R-phycoerythrin was added and allowed to incubate for 30 minutes. After washing to remove unbound streptavidin–R-phycoerythrin, the beads were analyzed with the Luminex detection system (CS1000 Autoplex Analyzer; PerkinElmer, Waltham, MA).
Real-Time Quantitative Polymerase Chain Reaction
Total RNA was extracted from epithelial cell monolayers or mouse colonic epithelial cells using TRIzol reagent (15596026; Thermo Fisher Scientific). The RNA integrity was verified by gel electrophoresis. RNA reverse-transcription was performed using the iScript complementary DNA synthesis kit (1708891; Bio-Rad Laboratories) according to the manufacturer's directions. The reverse-transcription complementary DNA reaction products were subjected to quantitative real-time polymerase chain reaction (PCR) using the MyiQ single-color real-time PCR detection system (Bio-Rad Laboratories) and the iTaq Universal SYBR green supermix (1725121; Bio-Rad Laboratories) according to the manufacturer's directions. All expression levels were normalized to β-actin levels of the same sample. The percentage expression was calculated as the ratio of the normalized value of each sample to that of the corresponding untreated control cells. All real-time PCR reactions were performed in triplicate. Primer sequences were designed using Primer-BLAST (Bethesda, MD) or were obtained from the Primer Bank (Cambridge, MA) primer pairs listed in Table 1.
Table 1.
Real-Time PCR Primers
| Primer name | Sequence |
|---|---|
| hJAK2F | 5’-TCTGGGGAGTATGTTGCAGAA-3’ |
| hJAK2R | 5’-AGACATGGTTGGGTGGATACC-3’ |
| hActinF | 5’-AGAGCAAGAGAGGCATCCTC-3’ |
| hActinR | 5’-CTCAAACATGATCTGGGTCA-3’ |
| mJAK2F | 5’-AGACTTCCAGAACCAGAACAAAG-3’ |
| mJAK2R | 5’-TCACAGTTTCTTCTGCCTAGCTA-3’ |
| mStat3F | 5’-CAGCAGCTTGACACACGGTA-3’ |
| mStat3R | 5’-AAACACCAAAGTGGCATGTGA-3’ |
| mActinF | 5’-GTGACGTTGACATCCGTAAAGA-3’ |
| mActinR | 5’-GCCGGACTCATCGTACTCC-3’ |
h, human; F, forward; m, mouse; R, reverse.
Real-Time PCR Measurement of Bacterial DNA
Mice feces sample DNA was extracted using the stool DNA Kit (Omega Bio-tek, Norcross, GA) according to the manufacturer's instructions. 16S ribosomal DNA PCR reactions were used the MyiQ single-color real-time PCR detection system (Bio-Rad Laboratories) and iTaq Universal SYBR green supermix (1725121; Bio-Rad Laboratories) according to the manufacturer's directions. Primers specific to 18S ribosomal RNA were used as an endogenous control to normalize loading between samples.56 The relative amount of 16S ribosomal DNA in each sample was estimated using the Delta-Delta cycle threshold. Primer sequences were designed using Primer-BLAST or were obtained from the Primer Bank primer pairs listed in Table 2.
Table 2.
Bacterial 16S Ribosomal DNA Real-Time PCR Primers
| Primer name | Sequence |
|---|---|
| B fragilis 16s forward | 5’-GGCGCACGGGTGAGTAACA-3’ |
| B fragilis 16s reverse | 5’-CAATATTCCTCACTGCTGC-3’ |
| B fibrisolvens 16s forward | 5’-CTAACACATGCAAGTCGAACG-3’ |
| B fibrisolvens 16s reverse | 5’-CCGTGTCTCAGTCCCAATG-3’ |
| F peptostreptococus 16s forward | 5’-CATTGGGACTGAGACAC-3’ |
| F peptostreptococus 16s reverse | 5’-AATCCGGATAACGCTTGC-3’ |
| E coli 16s forward | 5’-CCTACGGGAGGCAGCAGT-3’ |
| E coli 16s reverse | 5’-CGTTTACGGCGTGGACTAC-3’ |
| Univ bacteria 16s forward | 5’-TCCTACGGGAGGCAGCAGT-3’ |
| Univ bacteria 16s reverse | 5’-GGACTACCAGGGTATCTAATCCTGTT-3’ |
Univ, universal.
ChIP Assay
Binding of VDR to the Jak2 promoter was investigated using the ChIP assay as described previously.39 Briefly, HCT116 cells were treated with 1% formaldehyde for 10 minutes at 37°C. Cells were washed twice in ice-cold, phosphate-buffered saline containing protease inhibitor cocktail tablets (Roche, Indianapolis, IN). Cells were scraped into conical tubes, pelleted, and lysed in SDS lysis buffer. The lysate was sonicated to shear DNA into fragments of 200–1000 bp (4 cycles of 10-s sonication, 10-s pausing, Branson Sonifier 250, Buffalo Grove, IL). The chromatin samples were precleared with salmon sperm DNA–bovine serum albumin-sepharose beads, then incubated overnight at 4°C with VDR antibody (Santa Cruz Biotechnology). Immune complexes were precipitated with salmon sperm DNA–bovine serum albumin-sepharose beads. DNA was prepared by treatment with proteinase K, extraction with phenol and chloroform, and ethanol precipitation. By searching the mouse ATG16L1 gene, we found a similar sequence to the VDRE sequence (G/A)G(G/T)TCA. We then designed primers for ChIP. PCR was performed using the following promoter-specific primers: Jak2 forward, 5’-TGAATCCCAGGACACATTT-3’; reverse, 5’-GGTAAGCCACTGAAGGTT-3’.
Histology of Intestine
Intestines were harvested, fixed in 10% formalin (pH 7.4), processed, and paraffin-embedded. Sections (5 μm) were stained with H&E. For immunostaining, antigens were retrieved by 10-minute boiling in 10 mmol/L citrate (pH 6.0). The slides were stained with antibodies as previously described.48 Blinded histologic inflammatory scores were performed by a validated scoring system by a trained pathologist.57
LPS Detection
LPS in serum samples was measured with limulus amebocyte lysate chromogenic end point assays (HIT302; Hycult Biotech, Plymouth Meeting, PA) according to the manufacturer's indications. The samples were diluted 1:4 with endotoxin-free water and then heated at 75°C for 5 minutes on a warm plate to denature the protein before the reaction. A standard curve was generated and used to calculate the concentrations, which were expressed as EU/mL, in the blood samples.
Quantification of Fecal and Serum Lcn-2 by Enzyme-Linked Immunosorbent Assay
Freshly collected fecal samples were reconstituted in phosphate-buffered saline containing 0.1% Tween 20 (100 mg/mL) and vortexed for 20 minutes to get a homogenous fecal suspension. These samples then were centrifuged for 10 minutes at 12,000 rpm and 4°C. Clear supernatants were collected. Lcn-2 levels were estimated in the supernatants using the Duoset murine Lcn-2 enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN), as described in our previous study.58
Mucosa Microbial and Fecal 454 Pyrosequencing
The tubes for microbial sampling were autoclaved and then irradiated with UV light to destroy the environmental bacterial DNA. The mice then were anesthetized and dissected. Fecal matter was isolated freshly from the gut and placed into the specially prepared tubes, as described in our previously published reports.59,60 The samples were kept at a low temperature with dry ice and mailed to the Research and Testing Laboratory (Lubbock, TX) for 454 pyrosequencing. The V4–V6 region of the samples was amplified at the Research and Testing Laboratory for pyrosequencing using a forward and reverse fusion primer. The sequences were denoised and subjected to quality checking. Taxonomic identifications were assigned by queries against the National Center for Biotechnology Information.
Sample Preparation for Metabolites
Fecal samples were prepared using the automated MicroLab STAR system from the Hamilton Company. Several recovery standards were added before the first step in the extraction process for quality check purposes. To remove protein, small molecules bound to protein or trapped in the precipitated protein matrix were dissociated, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 minutes (Glen Mills GenoGrinder 2000), followed by centrifugation. The resulting extract was divided into 5 fractions: 2 for analysis by 2 separate reverse-phase/ultra high-performance liquid chromatography–tandem mass spectroscopy (UPLC-MS/MS) methods with positive ion mode electrospray ionization (ESI), 1 for analysis by reverse-phase/UPLC-MS/MS with negative ion mode ESI, 1 for analysis by Hydrophilic Interaction Liquid Chromatography/UPLC-MS/MS with negative ion mode ESI, and 1 sample was reserved for backup. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis.
Metabolite Analysis
For the metabolite experiments, 33 mice were divided into VDRΔIEC (N = 17) and control VDRLoxP (N = 16) groups. All mice were housed in specific pathogen-free environments under a controlled condition of 12-hour light/dark cycles at 20°C–22°C and 45% ± 5% humidity, with free access to food and ultrapure water. At 16 weeks of age, the fecal contents of each mouse were collected carefully in separate Eppendorf tubes, labeled with a unique identification number, and stored at −80°C until shipped. Samples were transported to Metabolon, Inc (Morrisville, NC) in dry ice by overnight shipment for analysis.
After receipt, samples were assigned a unique identifier by the laboratory information management system and immediately stored at -80°C until processed. Samples were prepared using the automated MicroLab STAR system from Hamilton Company. First proteins and other associated small molecules were precipitated, and then diverse metabolites were recovered by grinding and centrifugation. The resulting extract was analyzed by 2 separate reverse-phase/UPLC-MS/MS methods with positive ion mode ESI, or with negative ion mode ESI, and 1 by Hydrophilic Interaction Liquid Chromatography/UPLC-MS/MS with negative ion mode ESI. Several types of controls were analyzed along with the experimental samples to ensure accurate and consistent identification. UPLC-MS/MS was used as an analyzer. Metabolon’s hardware and software were used to extract the raw data, followed by the identification of peaks and quality checks. These systems are built on a web-service platform using Microsoft’s NET technologies.
Microbiome Data Analysis
Differences in microbial communities between VDRLoxP and VDRΔIEC groups were analyzed as in previous studies.59,60 Briefly, principal coordinates analysis of unweighted Unique Fraction Metric distances plots were plotted using quantitative insights into microbial ecology (QIIME).61 To determine differences in microbiota composition between the animal groups, the analysis of similarities function in the statistical software package PRIMER 6 (PRIMER-E, Ltd, Lutton, UK) was used on the unweighted Unique Fraction Metric distance matrixes.62
Statistical Analysis
Metabolite data were expressed as a fold change ratio, all other data are expressed as the means ± SD. All statistical tests were 2-sided. P values less than .05 were considered statistically significant. For metabolite data, after log transformation and imputation of missing values, if any, with the minimum observed value for each compound, analysis of variance contrasts and the Welch 2-sample t test were used to identify biochemicals that differed significantly between experimental groups. For other data analyses, the differences between samples were analyzed using the Student t test for 2-group comparisons, and 1-way analysis of variance for comparisons of more than 2 groups, respectively.
Sequence files and metadata for all samples used in this study have been deposited in https://www.ncbi.nlm.nih.gov/bioproject/593562; submission ID: SUB6615727; BioProject ID: PRJNA593562.
Acknowledgments
The authors would like to thank Eric Xia and Jason Xia for helping with proofreading.
CRediT Authorship Contributions
Yong-Guo Zhang, PHD (Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Resources: Supporting; Validation: Lead; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Rong Lu (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Lead; Resources: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Shaoping Wu (Investigation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting); Ishita Chatterjee (Data curation: Supporting; Methodology: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting); David Zhou (Resources: Supporting; Validation: Supporting; Visualization: Supporting; Writing – review & editing: Supporting); Yinglin Xia (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Resources: Supporting; Software: Supporting; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting); Jun Sun, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Equal; Visualization: Equal; Writing – original draft: Lead; Writing – review & editing: Lead).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the University of Illinois at Chicago Cancer Center, the National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health grants R01 DK105118, R01DK114126, and Department of DefenseBC160450P1 (J.S.). The study sponsors played no role in the study design, data collection, analysis, or interpretation of data.
References
- 1.McCullough M.L., Zoltick E.S., Weinstein S.J., Fedirko V., Wang M., Cook N.R., Eliassen A.H., Zeleniuch-Jacquotte A., Agnoli C., Albanes D., Barnett M.J., Buring J.E., Campbell P.T., Clendenen T.V., Freedman N.D., Gapstur S.M., Giovannucci E.L., Goodman G.G., Haiman C.A., Ho G.Y.F., Horst R.L., Hou T., Huang W.Y., Jenab M., Jones M.E., Joshu C.E., Krogh V., Lee I.M., Lee J.E., Mannisto S., Le Marchand L., Mondul A.M., Neuhouser M.L., Platz E.A., Purdue M.P., Riboli E., Robsahm T.E., Rohan T.E., Sasazuki S., Schoemaker M.J., Sieri S., Stampfer M.J., Swerdlow A.J., Thomson C.A., Tretli S., Tsugane S., Ursin G., Visvanathan K., White K.K., Wu K., Yaun S.S., Zhang X., Willett W.C., Gail M.H., Ziegler R.G., Smith-Warner S.A. Circulating vitamin D and colorectal cancer risk: an international pooling project of 17 cohorts. J Natl Cancer Inst. 2019;111:158–169. doi: 10.1093/jnci/djy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrer-Mayorga G., Gomez-Lopez G., Barbachano A., Fernandez-Barral A., Pena C., Pisano D.G., Cantero R., Rojo F., Munoz A., Larriba M.J. Vitamin D receptor expression and associated gene signature in tumour stromal fibroblasts predict clinical outcome in colorectal cancer. Gut. 2017;66:1449–1462. doi: 10.1136/gutjnl-2015-310977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaughan-Shaw P.G., Zgaga L., Ooi L.Y., Theodoratou E., Timofeeva M., Svinti V., Walker M., O'Sullivan F., Ewing A., Johnston S., Din F.V.N., Campbell H., Farrington S.M., Dunlop M.G. Low plasma vitamin D is associated with adverse colorectal cancer survival after surgical resection, independent of systemic inflammatory response. Gut. 2020;69:103–111. doi: 10.1136/gutjnl-2018-317922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng K., Nimeiri H.S., McCleary N.J., Abrams T.A., Yurgelun M.B., Cleary J.M., Rubinson D.A., Schrag D., Miksad R., Bullock A.J., Allen J., Zuckerman D., Chan E., Chan J.A., Wolpin B.M., Constantine M., Weckstein D.J., Faggen M.A., Thomas C.A., Kournioti C., Yuan C., Ganser C., Wilkinson B., Mackintosh C., Zheng H., Hollis B.W., Meyerhardt J.A., Fuchs C.S. Effect of high-dose vs standard-dose vitamin D3 supplementation on progression-free survival among patients with advanced or metastatic colorectal cancer: the SUNSHINE randomized clinical trial. JAMA. 2019;321:1370–1379. doi: 10.1001/jama.2019.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haussler M.R., Whitfield G.K., Haussler C.A., Hsieh J.-C., Thompson P.D., Selznick S.H., Dominguez C.E., Jurutka P.W. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13:325–349. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 6.Thorne JC, Campbell JM. The Molecular Cancer Biology of the VDR. In: Trump D, Johnson C. (eds) Vitamin D and Cancer. Springer, New York, NY, 2010.
- 7.Wu S., Zhang Y.G., Lu R., Xia Y., Zhou D., Petrof E.O., Claud E.C., Chen D., Chang E.B., Carmeliet G., Sun J. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64:1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Thingholm L.B., Skieceviciene J., Rausch P., Kummen M., Hov J.R., Degenhardt F., Heinsen F.A., Ruhlemann M.C., Szymczak S., Holm K., Esko T., Sun J., Pricop-Jeckstadt M., Al-Dury S., Bohov P., Bethune J., Sommer F., Ellinghaus D., Berge R.K., Hubenthal M., Koch M., Schwarz K., Rimbach G., Hubbe P., Pan W.H., Sheibani-Tezerji R., Hasler R., Rosenstiel P., D'Amato M., Cloppenborg-Schmidt K., Kunzel S., Laudes M., Marschall H.U., Lieb W., Nothlings U., Karlsen T.H., Baines J.F., Franke A. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy. 2016;12:1057–1058. doi: 10.1080/15548627.2015.1072670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S., Liao A.P., Xia Y., Li Y.C., Li J.D., Sartor R.B., Sun J. Vitamin D receptor negatively regulates bacterial-stimulated NF-kappaB activity in intestine. Am J Pathol. 2010;177:686–697. doi: 10.2353/ajpath.2010.090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C., Gandini S., Serrano D., Tarallo S., Francavilla A., Gallo G., Trompetto M., Ferrero G., Mizutani S., Shiroma H., Shiba S., Shibata T., Yachida S., Yamada T., Wirbel J., Schrotz-King P., Ulrich C.M., Brenner H., Arumugam M., Bork P., Zeller G., Cordero F., Dias-Neto E., Setubal J.C., Tett A., Pardini B., Rescigno M., Waldron L., Naccarati A., Segata N. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25:667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., Fleck J.S., Voigt A.Y., Palleja A., Ponnudurai R., Sunagawa S., Coelho L.P., Schrotz-King P., Vogtmann E., Habermann N., Nimeus E., Thomas A.M., Manghi P., Gandini S., Serrano D., Mizutani S., Shiroma H., Shiba S., Shibata T., Yachida S., Yamada T., Waldron L., Naccarati A., Segata N., Sinha R., Ulrich C.M., Brenner H., Arumugam M., Bork P., Zeller G. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchert M., Burns C.J., Ernst M. Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene. 2016;35:939–951. doi: 10.1038/onc.2015.150. [DOI] [PubMed] [Google Scholar]
- 14.Lu R., Zhang Y.G., Sun J. STAT3 activation in infection and infection-associated cancer. Mol Cell Endocrinol. 2017;451:80–87. doi: 10.1016/j.mce.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Amico F., Fiorino G., Furfaro F., Allocca M., Danese S. Janus kinase inhibitors for the treatment of inflammatory bowel diseases: developments from phase I and phase II clinical trials. Expert Opin Investig Drugs. 2018;27:595–599. doi: 10.1080/13543784.2018.1492547. [DOI] [PubMed] [Google Scholar]
- 16.Bird R.P., Good C.K. The significance of aberrant crypt foci in understanding the pathogenesis of colon cancer. Toxicol Lett. 2000;112–113:395–402. doi: 10.1016/s0378-4274(99)00261-1. [DOI] [PubMed] [Google Scholar]
- 17.Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagar N.M., Cree I.A., Covington J.A., Arasaradnam R.P. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterol Res Pract. 2015;2015:398585. doi: 10.1155/2015/398585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staley C., Weingarden A.R., Khoruts A., Sadowsky M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T., Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chassaing B., Srinivasan G., Delgado M.A., Young A.N., Gewirtz A.T., Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Protiva P., Cross H.S., Hopkins M.E., Kallay E., Bises G., Dreyhaupt E., Augenlicht L., Lipkin M., Lesser M., Livote E., Holt P.R. Chemoprevention of colorectal neoplasia by estrogen: potential role of vitamin D activity. Cancer Prev Res (Phila) 2009;2:43–51. doi: 10.1158/1940-6207.CAPR-08-0103. [DOI] [PubMed] [Google Scholar]
- 23.Kaler P., Augenlicht L., Klampfer L. Macrophage-derived IL-1beta stimulates Wnt signaling and growth of colon cancer cells: a crosstalk interrupted by vitamin D3. Oncogene. 2009;28:3892–3902. doi: 10.1038/onc.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fichera A., Little N., Dougherty U., Mustafi R., Cerda S., Li Y.C., Delgado J., Arora A., Campbell L.K., Joseph L., Hart J., Noffsinger A., Bissonnette M. A vitamin D analogue inhibits colonic carcinogenesis in the AOM/DSS model. J Surg Res. 2007;142:239–245. doi: 10.1016/j.jss.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 25.Nagpal S., Lu J., Boehm M.F. Vitamin D analogs: mechanism of action and therapeutic applications. Curr Med Chem. 2001;8:1661–1679. doi: 10.2174/0929867013371950. [DOI] [PubMed] [Google Scholar]
- 26.Palmer H.G., Sanchez-Carbayo M., Ordonez-Moran P., Larriba M.J., Cordon-Cardo C., Munoz A. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799–7806. [PubMed] [Google Scholar]
- 27.Chan A.T., Giovannucci E.L. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:2029–2043 e10. doi: 10.1053/j.gastro.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gombart A.F., Luong Q.T., Koeffler H.P. Vitamin D compounds: activity against microbes and cancer. Anticancer Res. 2006;26:2531–2542. [PubMed] [Google Scholar]
- 29.Wong S.H., Zhao L., Zhang X., Nakatsu G., Han J., Xu W., Xiao X., Kwong T.N.Y., Tsoi H., Wu W.K.K., Zeng B., Chan F.K.L., Sung J.J.Y., Wei H., Yu J. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153:1621–1633 e6. doi: 10.1053/j.gastro.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Terzic J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114 e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 31.Song M., Chan A.T., Jun J. Features of the gut microbiome, diet, and environment that influence risk of colorectal cancer. Gastroenterology. 2020;158:322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J. The role of vitamin D and vitamin D receptors in colon cancer. Clin Transl Gastroenterol. 2017;8:e103. doi: 10.1038/ctg.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abreu M.T. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 34.De Mattia E., Cecchin E., Montico M., Labriet A., Guillemette C., Dreussi E., Roncato R., Bignucolo A., Buonadonna A., D'Andrea M., Coppola L., Lonardi S., Levesque E., Jonker D., Couture F., Toffoli G. Association of STAT-3 rs1053004 and VDR rs11574077 with FOLFIRI-related gastrointestinal toxicity in metastatic colorectal cancer patients. Front Pharmacol. 2018;9:367. doi: 10.3389/fphar.2018.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makishima M., Lu T.T., Xie W., Whitfield G.K., Domoto H., Evans R.M., Haussler M.R., Mangelsdorf D.J. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt D.R., Holmstrom S.R., Fon Tacer K., Bookout A.L., Kliewer S.A., Mangelsdorf D.J. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem. 2010;285:14486–14494. doi: 10.1074/jbc.M110.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee I., Lu R., Zhang Y., Zhang J., Dai Y., Xia Y., Sun J. Vitamin D receptor promotes healthy microbial metabolites and microbiome. Sci Rep. 2020;10:7340. doi: 10.1038/s41598-020-64226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J., Kong J., Duan Y., Szeto F.L., Liao A., Madara J.L., Li Y.C. Increased NF-kappaB activity in fibroblasts lacking the vitamin D receptor. Am J Physiol Endocrinol Metab. 2006;291:E315–E322. doi: 10.1152/ajpendo.00590.2005. [DOI] [PubMed] [Google Scholar]
- 39.Wu S., Xia Y., Liu X., Sun J. Vitamin D receptor deletion leads to reduced level of IkappaBalpha protein through protein translation, protein-protein interaction, and post-translational modification. Int J Biochem Cell Biol. 2010;42:329–336. doi: 10.1016/j.biocel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange C.M., Gouttenoire J., Duong F.H., Morikawa K., Heim M.H., Moradpour D. Vitamin D receptor and Jak-STAT signaling crosstalk results in calcitriol-mediated increase of hepatocellular response to IFN-alpha. J Immunol. 2014;192:6037–6044. doi: 10.4049/jimmunol.1302296. [DOI] [PubMed] [Google Scholar]
- 41.Heneghan A.F., Pierre J.F., Kudsk K.A. JAK-STAT and intestinal mucosal immunology. JAKSTAT. 2013;2 doi: 10.4161/jkst.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J. Vitamin D and mucosal immune function. Curr Opin Gastroenterol. 2010;26:591–595. doi: 10.1097/MOG.0b013e32833d4b9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakke D., Agrawal A., Zhang Y., Sun J. Myeloid vitamin D receptor regulates Paneth cell function and intestinal homeostasis. Inflammatory Bowel Diseases. 2018;24:S29. [Google Scholar]
- 44.Wada K., Tanaka H., Maeda K., Inoue T., Noda E., Amano R., Kubo N., Muguruma K., Yamada N., Yashiro M., Sawada T., Nakata B., Ohira M., Hirakawa K. Vitamin D receptor expression is associated with colon cancer in ulcerative colitis. Oncol Rep. 2009;22:1021–1025. doi: 10.3892/or_00000530. [DOI] [PubMed] [Google Scholar]
- 45.Van Cromphaut S.J., Dewerchin M., Hoenderop J.G., Stockmans I., Van Herck E., Kato S., Bindels R.J., Collen D., Carmeliet P., Bouillon R., Carmeliet G. Duodenal calcium absorption in vitamin D receptor-knockout mice: functional and molecular aspects. Proc Natl Acad Sci U S A. 2001;98:13324–13329. doi: 10.1073/pnas.231474698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greten F.R., Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 47.Sun J., Hobert M.E., Duan Y., Rao A.S., He T.C., Chang E.B., Madara J.L. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am J Physiol. 2005;289:129–137. doi: 10.1152/ajpgi.00515.2004. [DOI] [PubMed] [Google Scholar]
- 48.Lu R., Wu S., Liu X., Xia Y., Zhang Y.G., Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y.G., Wu S., Xia Y., Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep. 2014;2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu R., Voigt R.M., Zhang Y., Kato I., Xia Y., Forsyth C.B., Keshavarzian A., Sun J. Alcohol injury damages intestinal stem cells. Alcohol Clin Exp Res. 2017;41:727–734. doi: 10.1111/acer.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y.G., Zhu X., Lu R., Messer J.S., Xia Y., Chang E.B., Sun J. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy. 2019;15:1935–1953. doi: 10.1080/15548627.2019.1596485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan Y., Liao A.P., Kuppireddi S., Ye Z., Ciancio M.J., Sun J. Beta-catenin activity negatively regulates bacteria-induced inflammation. Lab Invest. 2007;87:613–624. doi: 10.1038/labinvest.3700545. [DOI] [PubMed] [Google Scholar]
- 54.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., Bitton A., Dassopoulos T., Datta L.W., Green T., Griffiths A.M., Kistner E.O., Murtha M.T., Regueiro M.D., Rotter J.I., Schumm L.P., Steinhart A.H., Targan S.R., Xavier R.J., Libioulle C., Sandor C., Lathrop M., Belaiche J., Dewit O., Gut I., Heath S., Laukens D., Mni M., Rutgeerts P., Van Gossum A., Zelenika D., Franchimont D., Hugot J.P., de Vos M., Vermeire S., Louis E., Cardon L.R., Anderson C.A., Drummond H., Nimmo E., Ahmad T., Prescott N.J., Onnie C.M., Fisher S.A., Marchini J., Ghori J., Bumpstead S., Gwilliam R., Tremelling M., Deloukas P., Mansfield J., Jewell D., Satsangi J., Mathew C.G., Parkes M., Georges M., Daly M.J. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franks A.H., Harmsen H.J., Raangs G.C., Jansen G.J., Schut F., Welling G.W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan Y., Dickman K.G., Zong W.X. Akt and c-Myc differentially activate cellular metabolic programs and prime cells to bioenergetic inhibition. J Biol Chem. 2010;285:7324–7333. doi: 10.1074/jbc.M109.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sellon R.K., Tonkonogy S., Schultz M., Dieleman L.A., Grenther W., Balish E., Rennick D.M., Sartor R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu R., Zhang Y.G., Xia Y., Sun J. Imbalance of autophagy and apoptosis in intestinal epithelium lacking the vitamin D receptor. FASEB J. 2019;33:11845–11856. doi: 10.1096/fj.201900727R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Hoenig J.D., Malin K.J., Qamar S., Petrof E.O., Sun J., Antonopoulos D.A., Chang E.B., Claud E.C. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D.A., Jabri B., Chang E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Y., Sun J., Chen D.-G. Springer; Singapore: 2018. Statistical analysis of microbiome data with R. [Google Scholar]