Abstract
Background
Carotid and vagal paragangliomas (CPGLs and VPGLs) are rare neoplasms that arise from the paraganglia located at the bifurcation of carotid arteries and vagal trunk, respectively. Both tumors can occur jointly as multiple paragangliomas accounting for approximately 10 to 20% of all head and neck paragangliomas. However, molecular and genetic mechanisms underlying the pathogenesis of multiple paragangliomas remain elusive.
Case presentation
We report a case of multiple paragangliomas in a patient, manifesting as bilateral CPGL and unilateral VPGL. Tumors were revealed via computed tomography and ultrasound study and were resected in two subsequent surgeries. Both CPGLs and VPGL were subjected to immunostaining for succinate dehydrogenase (SDH) subunits and exome analysis. A likely pathogenic germline variant in the SDHD gene was indicated, while likely pathogenic somatic variants differed among the tumors.
Conclusions
The identified germline variant in the SDHD gene seems to be a driver in the development of multiple paragangliomas. However, different spectra of somatic variants identified in each tumor indicate individual molecular mechanisms underlying their pathogenesis.
Keywords: Multiple paragangliomas, Carotid and vagal paragangliomas, SDHx, Germline and somatic mutations, High-throughput exome sequencing, Immunohistochemistry, Case report
Background
Paragangliomas of the head and neck (HNPGLs) are rare neuroendocrine tumors [1]. There are several common paraganglioma localizations corresponding to the locations of paraganglia from which they are formed. Carotid paragangliomas (CPGLs) are most common, followed by middle ear paragangliomas, vagal (VPGL), and laryngeal paragangliomas [2]. These tumors are highly hereditary and associated with the germline mutations in known susceptibility genes, including SDHx, SDHAF2, TMEM127, MAX, and others [3]. Mutations in these genes predispose to different forms of paragangliomas (early, syndromic, multiple, and malignant).
HNPGLs commonly develop as single unilateral tumors, with only 1% of sporadic cases being multiple [4]. However, the number of multiple HNPGLs greatly increases in familial tumors. As multiple paragangliomas are rare, every case is important to study for a better understanding of genetics and molecular mechanisms causing their initiation and progression.
Case presentation
The case below describes multiple paragangliomas diagnosed in a Russian woman, presenting as two CPGLs at both sides of the neck and one VPGL. The aim of the study was to investigate the molecular mechanisms underlying the development of multiple paragangliomas by examining clinical and pathological characteristics along with the genetic variations of the three tumors.
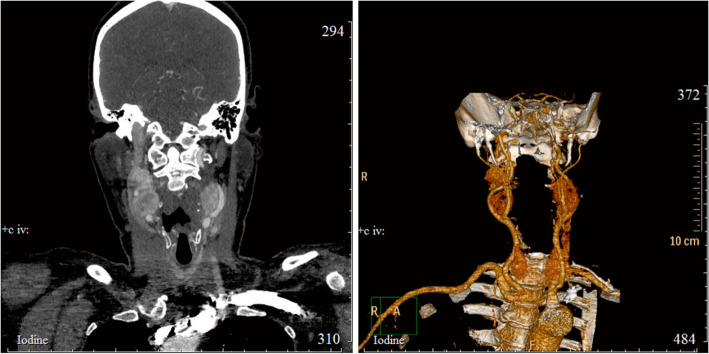
A 50-year-old female was diagnosed with extravascular compression of the carotid arteries and CPGLs on both sides of the neck. Clinical symptoms include arterial hypertension and painless rounded masses. Computed tomography (CT) and ultrasound (US) study revealed the presence of tumors in the areas of the carotid bifurcation, solid neoplasia 32 × 25 mm on the left side of the neck and two-nodal tumor 46 × 24 mm on the right side of the neck, respectively. These neck masses were heterogeneous in structure and predominantly hypoechoic and hypervascular. The CT study with contrast also revealed the presence of hypointense, highly vascularized masses at the right and left carotid bifurcations (Fig. 1).
Fig. 1.
Computed tomography of the patient's head and neck before surgery. CT scan (left); 3D reconstruction (right)
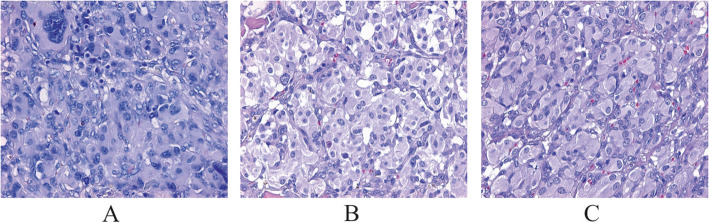
The patient was subjected to surgery for the left tumor resection. At the time of surgery, the lower pole of the hypervascularized tumor was located below the outer carotid artery (OCA) bifurcation, spreading along the carotid arteries in the proximal direction and wrapping around the posterior, anterior, and lateral surfaces. Bifurcation of carotid arteries was involved in the tumor mass. The upper pole of the tumor was associated with the vagus nerve. The tumor (25 × 2 × 17 mm) was completely removed and subjected to pathological evaluation. The patient was discharged with a planned re-hospitalization to remove the tumor on the right. Histological examination of the resected tumor confirmed carotid paraganglioma (Fig. 2). Hematoxylin-eosin (H&E) staining showed a Zellballen structure that is typical for paragangliomas. Chief tumor cells exhibited positive staining for chromogranin A, synaptophysin, and CD56 antibodies indicating a neuroendocrine tumor. S100 protein was expressed in sustentacular cells. Tumor cells were negative for cytokeratin AE1/AE3.
Fig. 2.
Hematoxylin-eosin (H&E) staining of carotid and vagal paragangliomas. a left CPGL, b right CPGL, and c right VPGL. Specific “Zellballen” growth pattern of paragangliomas (small nests of chief cells with pale eosinophilic staining, surrounded by supporting sustentacular cells) can be seen
A surgery on the right side of the neck was performed after a year. As per the US study reports, VPGL and enlarged lymph node were primarily detected. During the surgery, lymph node (15 × 5 mm) above the carotid artery bifurcation was removed and further subjected to histological examination for metastases. In addition, tumor-like mass (35 × 20 mm), laterally suppressing the internal carotid artery (ICA) was observed directly at carotid artery bifurcation and resected. After the tumor resection, one more tumor-like formation (60 × 20 mm) originating from the vagus nerve was visualized under it and removed. The vagus nerve was resected and ligated. Both the tumors were histologically examined and displayed paragangliomas with no lymph node metastases (Fig. 2).
For all the tumor samples from the patient (left and right CPGLs, VPGL), immunohistochemistry (IHC) analysis of succinate dehydrogenase (SDH) subunit expression was performed (Additional file 1). SDH complex consists of four subunits (SDHA, SDHB, SDHC, and SDHD) encoded by the corresponding genes [5, 6]. Germline and somatic mutations in the SDHx genes are commonly associated with paragangliomas/pheochromocytomas [7, 8]. Immunohistochemistry for SDH subunits is a valuable additional tool in the histopathological study of paragangliomas that is used in the clinic for assessment of SDH loss, which can be associated with the pathogenic mutations in any SDHx genes.
Immunoreactions for SDH subunits were carried out using primary antibodies from Abcam (USA) for each SDH subunit: SDHA, monoclonal, clone 2E3GC12FB2AE2; SDHB, monoclonal, clone 21A11AE7; SDHC, monoclonal, clone EPR11035(B); SDHD, polyclonal. We found weak diffuse weak SDHB staining in VPGL and both the left and right side CPGLs. According to the literature, weak diffuse staining of the SDHB subunit can reflect pathogenic mutations in any SDHx genes. We detected weak diffuse SDHB staining in all studied tumors indicating the presence of germline pathogenic mutation in one of the SDHx genes in the patient.
Additionally, we carried out the exome-sequencing of three tumors, lymph node, and blood from the patient. The DNA from tumors and lymph node was extracted with a High Pure FFPET DNA Isolation Kit (Roche, Switzerland). The DNA was isolated from blood cells using a MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche) on a MagNA Pure Compact Instrument (Roche). Exome libraries were prepared with the Rapid Capture Exome Kit (left CPGL) and TruSeq Exome Library Prep Kit (right CPGL and VPGL) from Illumina (USA). High-throughput exome sequencing was performed on a NextSeq 500 System (Illumina) under a paired-end mode of 76 × 2 bp for tumors and lymph node, and 156 × 2 bp for blood with 300x minimum coverage. The exome sequencing data of paragangliomas are available in the NCBI SRA under the accession numbers PRJNA411769 (left CPGL, Pat01), PRJNA476932 (right CPGL, Pat104), and PRJNA561073 (VPGL, Pat6). Bioinformatic analysis is described in our previous study [9]. Missense variants were considered as likely pathogenic if they were predicted by at least three prediction tools and characterized by conservation score ≥ 0.5.
Exome analysis revealed a likely pathogenic germline missense variant in the SDHD gene, NM_003002.3: c.305A > G, p.H102R (chr11: 111959726, rs104894302). Pathogenic/likely pathogenic germline variants in other genes, for which the association with paragangliomas/pheochromocytomas has been shown, were not found.
Identified likely pathogenic somatic variants were different for each tumor (Additional file 2). In left CPGL, we found missense likely pathogenic somatic variants in two genes, TENM3 [NM_001080477: c.C5082A, p.N1694K (chr4: 183696084)] and EPHA5 [NM_004439: c.G682A, p.V228I (chr4: 66467587)].
In right CPGL, a variety of likely pathogenic variants (stop-gain, frameshift, and missense) were detected. Stop-gain variants were found in NRXN3 [NM_004796: c.C1387T, p.Q463X (chr14: 79432478)] and RELN [NM_005045: c.C9052T, p.R3018X (chr7: 103137114)], missense variants were revealed in TRIP12, JAG1, ASXL1, LMBRD1, DHX9, AASS, and TP53. For the TP53 gene, we found two mutations: a pathogenic/likely pathogenic variant, NM_001126115: c.A446T, p.D149V (chr17: 7577096, rs587781525), that has been reported in ClinVar, and a previously undescribed likely pathogenic variant, NM_000546: c.A170G, p.D57G (chr17: 7579517).
In the case of VPGL, we found a pathogenic variant in mtDNA (MT: 3243, rs199474657) and likely pathogenic missense, frameshift and stop-gain variants in a number of genes (LRP1, SPEN, PPP4R1, XPO6, FBN1, C1QB, and others) (Additional file 2).
Discussion
We found a germline pathogenic variant in the SDHD gene in the patient. According to the literature, SDHD mutations are frequently associated with multiple paragangliomas. A study of 176 patients with HNPGLs divulged multiple paragangliomas in 33 individuals (18.9%) [10]. SDHx mutations were found in 34 patients, of which 22 were diagnosed with multiple paragangliomas. Moreover, 18 out of 22 patients with multiple paragangliomas had mutations in SDHD, 1 patient exhibited SDHB mutation, and 3 patients carried variations in the SDHC gene.
In a recent report, multiple synchronous or metachronous HNPGLs were found in 79 out of 147 patients studied (54%) [11]. A group of patients (98/147) were tested for the status of SDHx mutations; 74 patients carried mutations in either SDHB (10/74) or SDHD (64/74) genes. SDHB mutations were found in two cases of multiple paragangliomas, whereas 56 had mutated SDHD. Notably, p.Asp92Tyr mutation in the SDHD gene (one of the Dutch founder mutations) was the most prevalent variant identified in 50% of SDHD mutation carriers (32/64).
Another study on SDHB, SDHC, and SDHD gene mutation analysis in a large cohort of patients with a personal or family history of paragangliomas/pheochromocytomas was performed [12]. From the 1832 individuals tested, 876 patients carried mutations in either SDHB (673), SDHC (43), or SDHD (160). In summary, results from all the studies indicate a high frequency of SDHx mutations in paragangliomas. Moreover, most multiple paragangliomas are associated with SDHx variants, predominantly SDHD mutations, which are in accordance with our results.
The variant in the SDHD gene identified in the study has been described only once in a malignant paraganglioma [13]. Thus, the mutation seems to be quite rare; although, at the same position, histidine replaced with leucine, proline, or tyrosine was found in several cases of hereditary paragangliomas [14–16].
We observed different spectra of somatic mutations in three tumors studied. In left CPGL, only two genes, TENM3 and EPHA5 with likely pathogenic somatic variants were identified. Mutations in these genes have first been found in paragangliomas/pheochromocytomas. TENM3 and EPHA5 encode for proteins with important functions in neuronal cells and have been shown to be involved in tumorigenesis [17–20].
In right CPGL, a greater number of likely pathogenic somatic variants were revealed; however, no variants were found in the genes mutated in left CPGL. Stop-gain variants were determined in NRXN3 and RELN encoding for proteins that are involved in cell adhesion and cell-cell interactions in neural cells, respectively. Both genes were shown to be associated with glioblastoma pathogenesis [21–23] and variants in these genes were also detected in a number of tumors [24–26]. We also found two variants in the TP53 gene in the right CPGL. One variant was previously reported in ClinVar as a pathogenic mutation, the other variant was first detected in ourstudy. The presence of these two variants in TP53 can enhance the deleterious effect on protein function. More missense likely pathogenic mutations and frameshift variants were identified in the genes encoding for proteins that participate in important cellular processes, such as ubiquitin fusion degradation and regulation of DNA repair (TRIP12, a tumor suppressor gene), mediation of Notch signaling (JAG1), and transcriptional regulation (DHX9, KLF12, and KAT6B). Several genes with frameshift mutations are highly expressed in neural cells and involved in the neurite growth (PLPPR5) and the brain development (KAT6B and ADGRV1).
In VPGL, we identified variants that have not been found in the left and right CPGLs in the patient. Stop-gain mutations were revealed in the genes involved in the regulation of cell cycle (LIN54) and transcription (ZNF717), and in the CCDC82 gene, encoding for protein with unknown function. Variants in ZNF717 were previously observed in gastric, colorectal, and hepatocellular cancer [27–29]. Missense mutations in the genes shown to be involved in tumorigenesis that were predicted as likely pathogenic by a majority of prediction tools (C1QB, XPO6, PPP4R1, PPIP5K2, and LRP1), were also found in VPGL [30–34]. Frameshift mutations which usually result in nonfunctional protein were observed in many genes. In addition, we detected mutation in mtDNA that was reported in ClinVar as a germline or somatic pathogenic variant associated with mitochondrial diseases.
Conclusion
We identified a likely pathogenic germline variant in the SDHD gene and likely pathogenic somatic variants in a number of genes in the patient with multiple paragangliomas (left and right CPGLs, and VPGL). Somatic variants differed amongst the tumors. Thus, we assume that the variant found in the SDHD gene is a driver mutation and its co-occurrence with other mutations in each of the three tumors can lead to the development of paraganglioma via different molecular pathways.
Supplementary information
Additional file 1. Immunohistochemical staining of SDH subunits in carotid and vagal paragangliomas.
Additional file 2. Pathogenic/likely pathogenic somatic variants in carotid and vagal paragangliomas.
Acknowledgments
The authors thank the Vishnevsky Institute of Surgery for tissue samples, the National Medical Research Center of Radiology and A. N. Severtsov Institute of Ecology and Evolution for assistance in data analysis. This work was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
About this supplement
This article has been published as part of BMC Medical Genomics Volume 13 Supplement 8, 2020: Selected Topics in “Systems Biology and Bioinformatics” - 2019: medical genomics. The full contents of the supplement are available online at https://bmcmedgenomics.biomedcentral.com/articles/supplements/volume-13-supplement-8.
Abbreviations
- HNPGL
Head and neck paraganglioma
- CPGL
Carotid paraganglioma
- VPGL
Vagal paraganglioma
- CT
Computed tomography
- US
Ultrasound study
- CCA
Common carotid artery
- ICA
Internal carotid artery
- OCA
Outer carotid artery
Authors’ contributions
DVK and ALG managed the patient and obtained the patient’s history; MSF, EAP, and ZGG carried out experimental work; DVK performed immunohistochemistry; ENL, AAD, and GSK performed bioinformatics analysis; MVS, AVL, SAS, and NVM participated in data interpretation; MVK, TBD, and ADK assisted in the manuscript editing; VSP, AVS, DVK, and AVK wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work and publication costs were financially supported by the Russian Science Foundation, grant 17–75-20105 and 19–15-00419. The design of the study and writing of the manuscript were supported by both grants. Sample collection and experimental procedure (high-throughput exome sequencing, histological and immunohistochemical examination), as well as data analysis and interpretation concerning carotid tumors of the patient were supported by the grant 17–75-20105. All procedures for vagal paraganglioma of the patient, including sample collection, exome sequencing, histological and immunohistochemical study, data analysis and interpretation, were performed with the financial assistance of the grant 19–15-00419.
The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed in this study are included in the published article. The exome sequencing data of carotid paragangliomas are available in the NCBI SRA under the accession numbers PRJNA411769 and PRJNA476932. The sequencing data of vagal paraganglioma are available in the NCBI SRA under the accession number PRJNA561073.
Ethics approval and consent to participate
The study was approved by the ethics committee from the Vishnevsky Institute of Surgery and performed according to the Declaration of Helsinki (1964). Written informed consent was obtained from the patient.
Consent for publication
Written informed consent was obtained from the patient for publication of this study and its copy is available for the journal.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vladislav S. Pavlov and Dmitry V. Kalinin contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-00789-8.
References
- 1.Lopez-Vazquez ME, Llamas-Macias FJ, Nuno-Escobar C, Gonzalez-Ojeda A, Fuentes-Orozco C, Macias-Amezcua MD. Carotid body paraganglioma in a teenager. Case report. Cir Cir. 2014;82(3):316–322. [PubMed] [Google Scholar]
- 2.El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ. Classification of head and neck tumours. Fourth ed. World Health Organ. 2017;9:348. [Google Scholar]
- 3.Buffet A, Burnichon N, Favier J, Gimenez-Roqueplo AP. An overview of 20 years of genetic studies in pheochromocytoma and paraganglioma. Best Pract Res Clin Endocrinol Metab. 2020;32:101416. doi: 10.1016/j.beem.2020.101416. [DOI] [PubMed] [Google Scholar]
- 4.Ikram A, Rehman A. StatPearls. Treasure Island (FL) 2020. Paraganglioma. [Google Scholar]
- 5.Zhikrivetskaya SO, Snezhkina AV, Zaretsky AR, Alekseev BY, Pokrovsky AV, Golovyuk AL, Melnikova NV, Stepanov OA, Kalinin DV, Moskalev AA, et al. Molecular markers of paragangliomas/pheochromocytomas. Oncotarget. 2017;8(15):25756–25782. doi: 10.18632/oncotarget.15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snezhkina AV, Lukyanova EN, Fedorova MS, Kalinin DV, Melnikova NV, Stepanov OA, Kiseleva MV, Kaprin AD, Pudova EA. Kudryavtseva AV: [Novel genes associated with the development of carotid paragangliomas] Mol Biol. 2019;53(4):613–626. doi: 10.1134/S0026893319040137. [DOI] [PubMed] [Google Scholar]
- 7.Snezhkina AV, Lukyanova EN, Kalinin DV, Pokrovsky AV, Dmitriev AA, Koroban NV, Pudova EA, Fedorova MS, Volchenko NN, Stepanov OA, et al. Exome analysis of carotid body tumor. BMC Med Genet. 2018;11(Suppl 1):17. doi: 10.1186/s12920-018-0327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudryavtseva AV, Lukyanova EN, Kalinin DV, Zaretsky AR, Pokrovsky AV, Golovyuk AL, Fedorova MS, Pudova EA, Kharitonov SL, Pavlov VS, et al. Mutational load in carotid body tumor. BMC Med Genet. 2019;12(Suppl 2):39. doi: 10.1186/s12920-019-0483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snezhkina AV, Lukyanova EN, Zaretsky AR, Kalinin DV, Pokrovsky AV, Golovyuk AL, Krasnov GS, Fedorova MS, Pudova EA, Kharitonov SL, et al. Novel potential causative genes in carotid paragangliomas. BMC Med Genet. 2019;20(Suppl 1):48. doi: 10.1186/s12881-019-0770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papaspyrou K, Mewes T, Rossmann H, Fottner C, Schneider-Raetzke B, Bartsch O, Schreckenberger M, Lackner KJ, Amedee RG, Mann WJ. Head and neck paragangliomas: report of 175 patients (1989-2010) Head Neck. 2012;34(5):632–637. doi: 10.1002/hed.21790. [DOI] [PubMed] [Google Scholar]
- 11.Rijken JA, de Vos B, van Hest LP, Dreijerink KMA, den Heijer M, Wisselink W, Blom GJ, Hensen EF, Leemans CR. Evolving management strategies in head and neck paragangliomas: a single-centre experience with 147 patients over a 60-year period. Clin Otolaryngol. 2019;44(5):836–841. doi: 10.1111/coa.13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews KA, Ascher DB, Pires DEV, Barnes DR, Vialard L, Casey RT, Bradshaw N, Adlard J, Aylwin S, Brennan P, et al. Tumour risks and genotype-phenotype correlations associated with germline variants in succinate dehydrogenase subunit genes SDHB, SDHC and SDHD. J Med Genet. 2018;55(6):384–394. doi: 10.1136/jmedgenet-2017-105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poeppel TD, Yuece A, Boy C, Metz KA, Kaminsky E, Neumann HP, Rosenbaum SJ, Mann K, Moeller LC. Novel SDHD gene mutation (H102R) in a patient with metastatic cervical paraganglioma effectively treated by peptide receptor radionuclide therapy. J Clin Oncol. 2011;29(33):e812–e815. doi: 10.1200/JCO.2011.36.2269. [DOI] [PubMed] [Google Scholar]
- 14.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 15.Burnichon N, Rohmer V, Amar L, Herman P, Leboulleux S, Darrouzet V, Niccoli P, Gaillard D, Chabrier G, Chabolle F, et al. The succinate dehydrogenase genetic testing in a large prospective series of patients with paragangliomas. J Clin Endocrinol Metab. 2009;94(8):2817–2827. doi: 10.1210/jc.2008-2504. [DOI] [PubMed] [Google Scholar]
- 16.Piccini V, Rapizzi E, Bacca A, Di Trapani G, Pulli R, Giache V, Zampetti B, Lucci-Cordisco E, Canu L, Corsini E, et al. Head and neck paragangliomas: genetic spectrum and clinical variability in 79 consecutive patients. Endocr Relat Cancer. 2012;19(2):149–155. doi: 10.1530/ERC-11-0369. [DOI] [PubMed] [Google Scholar]
- 17.Ziegler A, Corvalan A, Roa I, Branes JA, Wollscheid B. Teneurin protein family: an emerging role in human tumorigenesis and drug resistance. Cancer Lett. 2012;326(1):1–7. doi: 10.1016/j.canlet.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Zhu Y, Ma C, Qiu Z, Zhang X, Kang Z, Wu Z, Wang H, Xu X, Zhang H, et al. Downregulation of EphA5 by promoter methylation in human prostate cancer. BMC Cancer. 2015;15:18. doi: 10.1186/s12885-015-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Wei X, Guo S, Wang J, Liu J, Wang H. Differential expression of EphA5 protein in gastric carcinoma and its clinical significance. Oncol Lett. 2019;17(6):5147–5153. doi: 10.3892/ol.2019.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Wang R, Wei B, Peng C, Wang L, Hu G, Kong D, Du C. Gene and microRNA signatures are associated with the development and survival of glioblastoma patients. DNA Cell Biol. 2019;38(7):688–699. doi: 10.1089/dna.2018.4353. [DOI] [PubMed] [Google Scholar]
- 22.Sun HT, Cheng SX, Tu Y, Li XH, Zhang S. FoxQ1 promotes glioma cells proliferation and migration by regulating NRXN3 expression. PLoS One. 2013;8(1):e55693. doi: 10.1371/journal.pone.0055693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulze M, Violonchi C, Swoboda S, Welz T, Kerkhoff E, Hoja S, Bruggemann S, Simburger J, Reinders J, Riemenschneider MJ. RELN signaling modulates glioblastoma growth and substrate-dependent migration. Brain Pathol. 2018;28(5):695–709. doi: 10.1111/bpa.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusinska R, Gorniak P, Pastorczak A, Fendler W, Potemski P, Mlynarski W, Kordek R. Influence of genomic variation in FTO at 16q12.2, MC4R at 18q22 and NRXN3 at 14q31 genes on breast cancer risk. Mol Biol Rep. 2012;39(3):2915–2919. doi: 10.1007/s11033-011-1053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J, Yin Y, Ma Q, Wang G, Olman V, Zhang Y, Chou WC, Hong CS, Zhang C, Cao S, et al. Comprehensive characterization of the genomic alterations in human gastric cancer. Int J Cancer. 2015;137(1):86–95. doi: 10.1002/ijc.29352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, Jiang L, Zhong X, Hochwald SN, Wang Y, Huang L, Nie Q, Huang H, Xu JF. Discovery of aberrant alteration of genome in colorectal cancer by exome sequencing. Am J Med Sci. 2019;358(5):340–349. doi: 10.1016/j.amjms.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Duan M, Hao J, Cui S, Worthley DL, Zhang S, Wang Z, Shi J, Liu L, Wang X, Ke A, et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28(3):359–373. doi: 10.1038/cr.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangogna A, Belmonte B, Agostinis C, Zacchi P, Iacopino DG, Martorana A, Rodolico V, Bonazza D, Zanconati F, Kishore U, et al. Prognostic implications of the complement protein C1q in gliomas. Front Immunol. 2019;10:2366. doi: 10.3389/fimmu.2019.02366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao J, Chiang YT, Gout PW, Wang Y. Elevated XPO6 expression as a potential prognostic biomarker for prostate cancer recurrence. Front Biosci (Schol Ed) 2016;8:44–55. doi: 10.2741/s445. [DOI] [PubMed] [Google Scholar]
- 32.Addou-Klouche L, Adelaide J, Finetti P, Cervera N, Ferrari A, Bekhouche I, Sircoulomb F, Sotiriou C, Viens P, Moulessehoul S, et al. Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol Cancer. 2010;9:213. doi: 10.1186/1476-4598-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Sun X, Ge W, Qian Y, Bai R, Zheng S. A seven-gene signature predicts overall survival of patients with colorectal cancer. Oncotarget. 2017;8(56):95054–95065. doi: 10.18632/oncotarget.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maletinska L, Blakely EA, Bjornstad KA, Deen DF, Knoff LJ, Forte TM. Human glioblastoma cell lines: levels of low-density lipoprotein receptor and low-density lipoprotein receptor-related protein. Cancer Res. 2000;60(8):2300–2303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Immunohistochemical staining of SDH subunits in carotid and vagal paragangliomas.
Additional file 2. Pathogenic/likely pathogenic somatic variants in carotid and vagal paragangliomas.
Data Availability Statement
All data generated or analyzed in this study are included in the published article. The exome sequencing data of carotid paragangliomas are available in the NCBI SRA under the accession numbers PRJNA411769 and PRJNA476932. The sequencing data of vagal paraganglioma are available in the NCBI SRA under the accession number PRJNA561073.