Abstract
ADAM metallopeptidase domain 12 (ADAM12) is a promising biomarker because of its low expression in normal tissues and high expression in a variety of human cancers. However, ADAM12 levels in ovarian cancer have not been well characterized. We previously identified ADAM12 as one of the signature genes associated with poor survival in high-grade serous ovarian carcinoma (HGSOC). Here, we sought to determine if high levels of the ADAM12 protein and/or messenger RNA (mRNA) are associated with clinical variables in HGSOC. We show that high protein levels of ADAM12 in banked preoperative sera are associated with shorter progression-free and overall survival. Tumor levels of ADAM12 mRNA were also associated with shorter progression-free and overall survival as well as with lymphatic and vascular invasion, and residual tumor volume following cytoreductive surgery. The majority of genes co-expressed with ADAM12 in HGSOC were transforming growth factor (TGF)β signaling targets that function in collagen remodeling and cell–matrix adhesion. In tumor sections, the ADAM12 protein and mRNA were expressed in epithelial cancer cells and surrounding stromal cells. In vitro data showed that ADAM12 mRNA levels can be increased by TGFβ signaling and direct contact between epithelial and stromal cells. High tumor levels of ADAM12 mRNA were characteristic of the mesenchymal/desmoplastic molecular subtype of HGSOC, which is known to have the poorest prognosis. Thus, ADAM12 may be a useful biomarker of aggressive ovarian cancer for which standard treatment is not effective.
Summary
Elevated ADAM12 is a prognostic factor associated with adverse clinical outcomes in high-grade serous ovarian carcinoma. ADAM12 is highly expressed in tumor epithelial cells and adjacent stromal cells and is associated with the mesenchymal/desmoplastic molecular subtype of ovarian cancer.
Introduction
ADAM metallopeptidase domain 12 (ADAM12) encodes a member of the ADAM (a disintegrin and metalloprotease) protein family. In humans, two isoforms of ADAM12 (also known as meltrin-α) exist as a result of alternative messenger RNA (mRNA) splicing: a long transmembrane form (ADAM12-L) and a truncated secreted form lacking the transmembrane and cytoplasmic domains (ADAM12-S). Both ADAM12-L and ADAM12-S are proteolytically processed, and the mature forms translocate to the plasma membrane and extracellular space, respectively (1), to assume their proteolytic function (2–5).
Multiple studies have demonstrated that increased levels of ADAM12 correlate with tumor progression but it is unknown if ADAM12 is an actual perpetrator in tumor progression. In mouse models of breast and prostate cancers, tumor growth and metastasis were diminished in ADAM12−/− mice in comparison with wild-type littermates, indicating that ADAM12 may be required for tumor progression (6,7). Overexpression studies also support the role of ADAM12 in tumor progression and provide mechanistic insight into the relevance of its adhesion and proteolytic functions (8–12).
ADAM12 has attracted attention as a biomarker because of its restricted expression in normal tissues and considerable activation in various disease processes. Aside from high expression in the human placenta and transient expression during embryonic morphogenesis of muscle and bone (5), postnatal ADAM12 expression in healthy and non-injured organs is low. However, levels of ADAM12 are elevated in diseases accompanied by fibrosis (13). Further, increased levels of ADAM12 have been reported in human cancers including cancers of the breast (6,14–16), liver (17–22), head and neck (11,23,24), stomach (25), bladder (26), prostate (7), lung (27), brain (28) and bone (29).
ADAM12 has not been examined as a potential biomarker in ovarian cancer. However, ADAM12 was identified in an unbiased screen as one of the transmembrane proteins expressed in ovarian tumor vasculature but not the vasculature of normal ovaries (30). In the same study, it was noted that expression of ADAM12 was highly variable among ovarian cancers, with high expression in some samples and minimal expression in others, suggesting that ADAM12 might serve as a biomarker in ovarian cancer (30). We previously identified gene signatures associated with poor survival in high-grade serous ovarian carcinoma (HGSOC) (31,32). Since ADAM12 was among the signature genes, we hypothesize that high levels of ADAM12 are associated with adverse outcome in HGSOC.
Methods
Patient samples
Studies involving human specimens were approved by the Cedars-Sinai Medical Center Institutional Review Board. All patients signed an institutional review board-approved consent for biobanking, clinical data extraction and molecular analysis. Banked frozen preoperative sera were obtained from the Women’s Cancer Program Bioepository and prepared for analysis as described in our previous publications (33,34). All patients in this study had advanced stage (FIGO III or IV), high-grade (2 or 3) serous ovarian carcinoma. Patients with other malignancies, borderline ovarian tumors and ovarian tumors of non-epithelial histology were excluded. All patients underwent initial surgical exploration with the intent of optimal cytoreduction (defined as residual disease <1cm) and were treated with at least six cycles of platinum-based chemotherapy. Patients who received intraperitoneal chemotherapy or underwent neoadjuvant chemotherapy were excluded. Immunohistochemical staining and in situ hybridization were performed on formalin-fixed, paraffin-embedded tumors surgically removed from patients and obtained from the Pathology Department archives.
Enzyme-linked immunosorbent assay
A solid-phase enzyme-linked immunosorbent assay (ELISA) was performed using the Quantikine human ADAM12 ELISA kit (R&D Systems) following the manufacturer’s instructions. Briefly, 100 μl of Assay Diluent was added to each well of the 96-well plate precoated with a monoclonal antibody specific for human ADAM12. Fifty microliters of ADAM12 standard (0–100ng/ml) or patient sera were added to each well and incubated for 2h at room temperature on a horizontal orbital microplate shaker (500 r.p.m.). The liquids were carefully discarded and the wells were washed four times with 400 μl of the Wash Buffer. Two hundred microliters of ADAM12 Conjugate, an enzyme-linked monoclonal antibody specific for human ADAM12, was added to each well and incubated for 2h at room temperature on the shaker. After washing four times with the Wash Buffer, 200 μl of Substrate Solution was added to each well and incubated for 30min at room temperature. The color development was stopped by adding 50 μl of Stop Solution to each well and the optical density at 450nm was measured by a microplate reader. ADAM12 concentration (ng/ml) in patient sera was calculated by a formula obtained from the ADAM12 standard curve.
Immunohistochemical staining and in situ hybridization
Immunohistochemical detection of ADAM12 was performed using the Vectastatin Elite ABC kit with rabbit immunoglobulin G (Vector Laboratories) following the manufacturer’s instructions. Formalin-fixed, paraffin-embedded tissue sections were deparaffinized and rehydrated in a series of xylene and diluted alcohol. Antigen retrieval was performed by boiling the slides in the Antigen Unmasking Solution (Vector Laboratories). Endogenous peroxidase was inactivated by a 30min incubation in 0.3% H2O2 solution in methanol. After blocking with goat serum, a polyclonal ADAM12 Prestige Antibody (Sigma–Aldrich) was incubated at 1:150 dilution for 30min at room temperature. Slides were washed and incubated with the biotinylated rabbit immunoglobulin G for 30min at room temperature. After washing, the slides were incubated with the ABC reagent for 30min at room temperature, then incubated in the ImmPACT DAB (Vector Laboratories) for 8min, counterstained with Harris hematoxylin (Sigma–Aldrich), dehydrated and mounted with Permount (Fisher Scientific).
ADAM12 in situ hybridization was performed using RNAscope 2.0 FFPE Assay (Advanced Cell Diagnostics) as described in Cheon et al. (31). Slides were examined using the Olympus BX43 upright microscope (Olympus).
Cell culture
The OVCAR3 ovarian cancer cell line was obtained from Dennis Slamon (University of California, Los Angeles). All other ovarian cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA). Cell line authenticity was confirmed by Laragen using the short tandem repeat method. The TRS3 normal ovarian stroma cell line was generated as described previously (31). The ovarian cancer cells and TRS3 cells were cultured in Dulbecco’s modified Eagle’s medium (Corning) and a 1:1 mixture of MCDB 105 (Sigma) and 199 (Gibco) media, respectively, supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Ovarian cancer cells were cocultured for 48h with TRS3 cells using Millicell 6-well inserts with 0.4 µm PET membrane (Merck Millipore). Alternatively, green fluorescent protein (GFP)-labeled ovarian cancer cells were cultured on a monolayer of TRS3 cells for 48h and GFP+ cells were sorted using the BD FACSAria™ III cell sorter (BD Biosciences) by the Cedars-Sinai Medical Center flow cytometry core staff. For transforming growth factor (TGF)β1 treatment, 105 cells were plated in six-well plates, serum-starved overnight, then incubated with 10ng/ml TGFβ1 (Sigma) for 48h before harvesting.
RNA isolation and quantitative real-time PCR analysis
Total RNA was extracted using the RNeasy mini kit (Qiagen) and was reverse-transcribed to complementary DNA using the QuantiTect Reverse Transcription Kit (Qiagen). A total of 50ng of complementary DNA was mixed with primers and iQ SYBR Green Supermix (Bio-Rad) in a 96-well plate format. For primers, the RT2 qPCR Primer Assay for Human ADAM12 (Qiagen; PPH07647A) and the ribosomal protein L32 (internal control) (Forward: 5′-ACAAAGCACATGCTGCCCAGTG-3′; Reverse: 5′-TTCCACGATGGCTTTGCGGTTC-3′) were used. The quantitative reverse transcription–PCR reaction was performed using a CFX96 thermo cycler (Bio-Rad) and the data were analyzed by the 2−∆CT method. Samples were in triplicate and the experiment was repeated twice.
Statistical methods
Abstracted data from medical charts included age, stage, grade, status of cytoreductive surgery and time to recurrence and death. For statistical considerations, we defined an elevated ADAM12 level as >1.0ng/ml. Differences in clinical and histopathologic factors between patients with high and low serum ADAM12 were examined with chi-square and Fisher’s exact test. The Cox regression analysis was performed to assess the significance of potential prognostic factors. Patient survival was analyzed with Kaplan–Meier curves. A P value of <0.05 was considered statistically significant.
Analyses of public databases
R2 (http://hgserver1.amc.nl/) was used to statistically analyze and graph data from public microarray data sets. The Kaplan–Meier online plotter tool (http://kmplot.com/analysis/) was used to generate survival curves by combining ADAM12 mRNA data from serous ovarian cancer patients from 13 public ovarian cancer data sets (Supplementary Table I is available at Carcinogenesis Online). cBioPortal (http://www.cbioportal.org/) was used to identify ADAM12-correlated transcripts in the ovarian cancer TCGA data set. DAVID (http://david.abcc.ncifcrf.gov/) and Ingenuity Pathway Analysis were used for functional annotation of the transcripts and identification of upstream regulators, respectively.
Results
High serum protein levels of ADAM12 are associated with poor survival in patients with HGSOC
Eighty-four patients with HGSOC met the criteria for inclusion in the study. All patients underwent initial surgical cytoreduction followed by adjuvant chemotherapy. The majority of patients had grade 3, stage III disease and were optimally resected (residual disease <1cm). ADAM12 levels in banked preoperative sera were determined by ELISA. The protein levels ranged from 0 to 5.76ng/ml with an average of 1.06ng/ml and a median of 0.83ng/ml. We arbitrarily selected 1ng/ml as a cutoff to divide the 84 patients into two groups: 48 patients with low (<1ng/ml; range 0.00–0.98ng/ml) and 36 patients with high (>1ng/ml; range 1.03–5.76ng/ml) levels of ADAM12. The distribution of cohort characteristics between patients with low and high levels of ADAM12 are shown in Supplementary Table II is available at Carcinogenesis Online.
In order to determine if serum ADAM12 levels correlate with clinical outcome, we used Kaplan–Meier analyses for both time to first recurrence and time to death. Women with low serum ADAM12 levels had a longer median progression-free survival than those with high ADAM12 levels (21 months versus 14 months, P = 0.037) (Figure 1A). Similarly, women with low ADAM12 levels had a longer median overall disease-specific survival than those with high ADAM12 levels (57 months versus 45 months, P = 0.033) (Figure 1A). The significance of ADAM12 as an independent prognostic factor was evaluated by Cox regression analysis. ADAM12 levels retained statistical significance (P = 0.02, risk ratio 1.36, confidence intervals 1.06–1.75) after controlling for age, stage (III or IV), grade (2 or 3) and cytoreduction status (optimal or suboptimal) (Supplementary Table III is available at Carcinogenesis Online).
Figure 1.
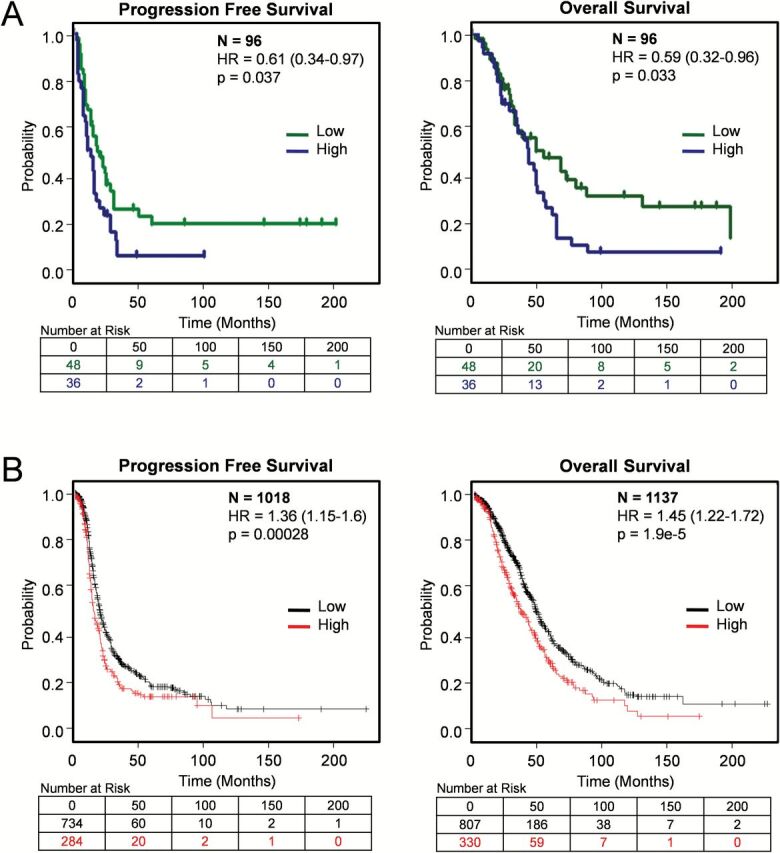
Kaplan–Meier survival curves in serous ovarian carcinoma patients with low and high levels of ADAM12. (A) Survival curves in HGSOC patients with low (<1ng/ml) and high (>1ng/ml) preoperative serum levels of ADAM12. (B) Survival curves in serous ovarian cancer patients with low and high expression levels of ADAM12 mRNA (202952_s_at) from 13 combined public ovarian cancer data sets in the Kaplan–Meier plotter database.
ADAM12 mRNA levels are associated with poor patient survival, increased tumor invasion and decreased success in surgical cytoreduction
The existence of multiple expression profile data sets for ovarian cancer facilitated the correlative analysis of ADAM12 mRNA levels with clinical parameters in a large number of patients. High levels of ADAM12 mRNA were associated with poor progression-free and overall survival in a cohort of serous ovarian cancer patients that integrated data from 13 different data sets (Figure 1B). To determine whether levels of ADAM12 correlate with various clinical parameters, we used the ovarian cancer TCGA data set (35). ADAM12 mRNA levels correlated with lymphatic invasion, venous invasion and size of residual tumor after cytoreductive surgery (Figure 2), whereas there was no statistically significant correlation with tumor stage, tumor grade, patient age at diagnosis, performance status, race or ethnicity (data not shown).
Figure 2.
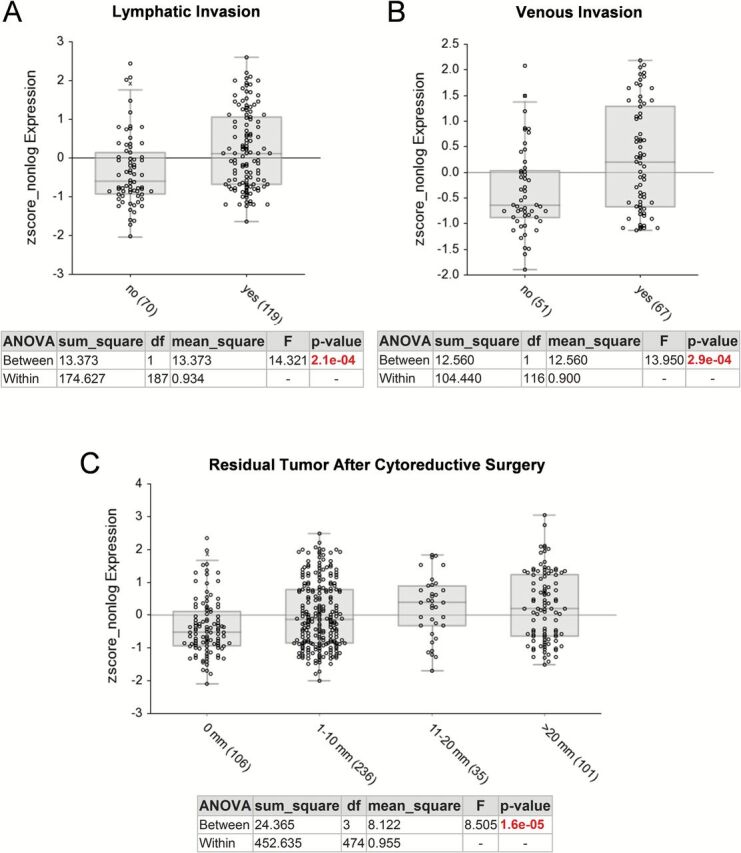
Comparison of ADAM12 expression levels and clinicopathological parameters in the ovarian cancer TCGA data set. (A) Lymphatic invasion, (B) Venous invasion, (C) Residual tumor after cytoreductive surgery. The graphs and statistical data were generated using R2 Genomics Analysis and Visualization Platform. The x-axis shows individual groups where the number of patients in each group is indicated in parentheses. The y-axis represents a relative value of ADAM12 mRNA (202952_s_at) expression.
ADAM12 mRNA levels are associated with the mesenchymal/desmoplastic molecular subtype of ovarian carcinoma
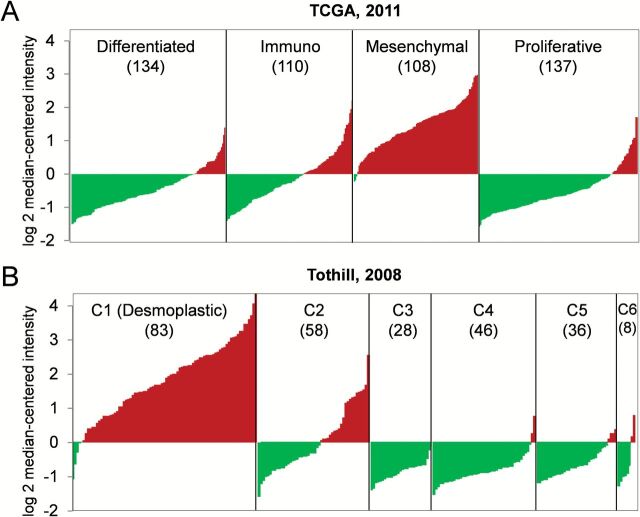
Since high serum and mRNA levels of ADAM12 were associated with worse clinical outcomes, we hypothesized that tumors in HGSOC patients with high levels of ADAM12 would exhibit aggressive biology, including increased invasion, suboptimal cytoreduction and poor survival. To determine if high levels of ADAM12 were associated with a specific molecular subtype of HGSOC, we used the ovarian cancer TCGA data set from 489 patients with HGSOC (35). Based on expression profiles, the cancer samples in this data set have been clustered into four molecular subtypes: differentiated, immunoreactive, mesenchymal and proliferative (35). Strikingly, almost all tumors of the mesenchymal subtype had elevated levels of ADAM12 (Figure 3A). Similarly in the Tothill data set, 259 serous and endometrioid ovarian carcinomas have been clustered based on their expression profile into six distinct molecular subtypes (C1–C6) (36). The C1 subtype has been characterized by a reactive stroma gene expression signature and was shown to be enriched in tumors with extensive desmoplasia (36). Almost all tumors in the C1 subtype exhibited elevated levels of ADAM12 (Figure 3B).
Figure 3.
Association of high expression levels of ADAM12 with a specific molecular subtype of ovarian carcinoma. A diagram of ADAM12 mRNA distribution in (A) 489 serous ovarian carcinomas in the ovarian cancer TCGA data set grouped into four distinct molecular subtypes and (B) 259 ovarian serous and endometrioid carcinomas in the Tothill data set grouped into six distinct molecular subtypes. The x-axis shows individual tumors that are merged into a continuous plot (the number of tumor samples in each subtype is indicated in parentheses). The y-axis represents a relative value of mRNA expression. Red indicates positive values, green indicates negative values.
ADAM12 is expressed in epithelial cancer cells and surrounding stromal cells and can be induced by epithelial–stromal interaction and TGFβ signaling
To better understand the biology of the tumors with high levels of ADAM12, we identified gene transcripts that most closely correlate with expression of ADAM12 in the ovarian cancer TCGA data set. The majority of the ADAM12-correlated genes were matricellular and extracellular matrix proteins, such as collagens and collagen-remodeling enzymes (Table 1), which we previously identified as part of a gene signature of poor survival in HGSOC (31). Gene Ontology analysis showed that ADAM12-correlated genes are primarily involved in collagen remodeling, tissue development and cell adhesion (Table 2).
Table 1.
Genes co-expressed with ADAM12 in the ovarian cancer TCGA data set.
| Correlated gene | Pearson’s correlation | Spearman’s correlation |
|---|---|---|
| COL5A2 | 0.89 | 0.78 |
| COL3A1 | 0.88 | 0.9 |
| POSTN | 0.88 | 0.85 |
| COL5A1 | 0.87 | 0.84 |
| ADAMTS12 | 0.87 | 0.84 |
| THBS2 | 0.87 | 0.8 |
| COL1A1 | 0.85 | 0.86 |
| SPARC | 0.84 | 0.85 |
| VCAN | 0.84 | 0.8 |
| COL11A1 | 0.84 | 0.77 |
| FAP | 0.83 | 0.85 |
| MMP2 | 0.83 | 0.85 |
| LUM | 0.83 | 0.79 |
| ADAMTS2 | 0.83 | 0.78 |
| CRISPLD2 | 0.83 | 0.71 |
| FN1 | 0.83 | 0.7 |
| INHBA | 0.83 | 0.58 |
| OLFML2B | 0.82 | 0.81 |
| COL6A3 | 0.82 | 0.78 |
| ECM1 | 0.82 | 0.78 |
| SNAI2 | 0.82 | 0.71 |
| KCNE4 | 0.82 | 0.7 |
| MMP11 | 0.82 | 0.68 |
| COL5A3 | 0.82 | 0.67 |
| ALPK2 | 0.81 | 0.74 |
| PRRX1 | 0.81 | 0.35 |
| COL1A2 | 0.8 | 0.87 |
| LOX | 0.79 | 0.78 |
| CHSY3 | 0.79 | 0.77 |
| LRRC15 | 0.79 | 0.74 |
Agilent microarray, 489 ovarian cancer samples.
Table 2.
Functional annotation of genes co-expressed with ADAM12 in the ovarian cancer TCGA data set
| GO term | Count | % | P value | Genes |
|---|---|---|---|---|
| GO:0030199~collagen fibril organization | 7 | 23.3333 | 1.25E-11 | LUM, COL3A1, COL1A1, COL5A3, COL5A2, COL11A1, COL5A1 |
| GO:0001501~skeletal system development | 11 | 36.6667 | 1.76E-10 | INHBA, CTSK, FBN1, COL3A1, PRRX1, POSTN, SPARC, COL1A1, COL5A2, COL11A1, MMP2 |
| GO:0032963~collagen metabolic process | 5 | 16.6667 | 2.13E-07 | COL3A1, COL1A1, MMP2, COL5A1, MMP11 |
| GO:0043588~skin development | 5 | 16.6667 | 2.46E-07 | COL3A1, COL1A1, COL5A3, COL5A2, COL5A1 |
| GO:0007155~cell adhesion | 10 | 33.3333 | 3.56E-06 | COL6A3, COL3A1, ITGA11, VCAN, POSTN, COL5A3, THBS2, COL11A1, COL5A1, FN1 |
| GO:0007160~cell–matrix adhesion | 4 | 13.3333 | 6.41E-04 | COL3A1, ITGA11, COL5A3, FN1 |
GO, Gene Ontology.
To evaluate the cellular localization of ADAM12 in tumors, an ADAM12-specific polyclonal antibody and an ADAM12-specific probe were used to perform immunohistochemical staining and in situ hybridization on tumor sections from several of our patients with HGSOC. ADAM12 protein and mRNA were detected in tumor epithelial cells and adjacent stromal cells but not in distant (>1mm) stromal cells (Figure 4A). To determine if ADAM12 expression is induced by an interaction between stromal and cancer cells, SV40 large T-antigen-transformed stromal cells from a normal ovary (TRS3) were cocultured with a panel of ovarian cancer cell lines (OVCAR3, OVCAR433, HEY and SKOV3). OVCAR3 and OVCAR433 cells have a cobblestone morphology characteristic of epithelial cells (Figure 4B) and have been classified as ‘epithelial’ and ‘intermediate epithelial’ cells, respectively, based on their expression of epithelial markers and low migratory and invasive potential (37). We have demonstrated in OVCAR3 cells that their migration and invasion can be augmented by coculture with stromal TRS3 cells (J.A.Beach et al. in preparation). In contrast, HEY and SKOV3 cells are more elongated and spindle-shaped (Figure 4B) and have been classified as ‘intermediate mesenchymal’ cells based on their expression of mesenchymal markers and high migratory and invasive potential (37). We observed that the epithelial OVCAR3 and OVCAR433 cell lines had low levels of ADAM12 mRNA that could be significantly induced by both direct and indirect coculture with TRS3 stromal cells (Figure 4C). Conversely, mesenchymal HEY and SKOV3 cell lines had high levels of ADAM12 mRNA that were similar to that of TRS3 stromal cells. Further, neither direct nor indirect coculture with TRS3 cells significantly altered ADAM12 mRNA levels in HEY and SKOV3 cells (Figure 4C). Ingenuity Pathway Analysis revealed that many of the ADAM12-correlated genes in Table 1 are expressed in tumor stroma and are downstream targets of TGFβ (data not shown), indicating that ADAM12 may be regulated by TGFβ signaling. Consistent with this idea, ADAM12 levels in TRS3 cells increased ~10-fold in the presence of recombinant TGFβ1 and were further increased by direct coculture with OVCAR3 cells (Figure 4D).
Fig. 4.
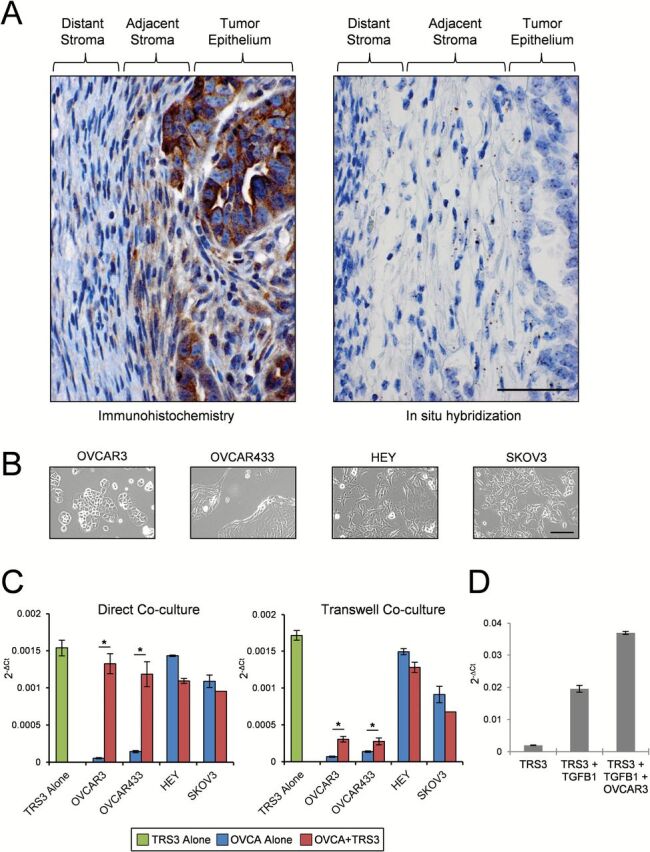
ADAM12 expression in epithelial and stromal cells in patient tumors and different culture conditions. (A) Representative localization of ADAM12 protein (immunohistochemistry; brown staining) and mRNA (in situ hybridization; brown dots) in ovarian tumor sections from HGSOC patients. Size bar for both photographs = 50 µm. (B) Bright field microscopy depicting cell morphology of ovarian cancer (OVCA) cell lines. Size bar = 100 µm. (C) Quantitative real-time PCR of ADAM12 mRNA levels in the TRS3 ovarian stromal cell line and various OVCA cell lines alone or in coculture. TRS3 cells and GFP-labeled OVCA cells were cocultured in direct contact or indirectly via transwell inserts for 48h. (D) Quantitative real-time PCR of ADAM12 mRNA in the TRS3 cells alone or in coculture with OVCAR3 epithelial cells in the presence or absence of 10ng/ml of recombinant TGFβ1. Data are normalized to ribosomal protein L32 and represent the mean ± SEM. *P < 0.05.
Discussion
Several cancer studies have demonstrated the potential utility of ADAM12 as a diagnostic and prognostic marker. For example, western blot analysis showed elevated levels of ADAM12 in urine from patients with breast cancer compared with healthy control subjects (38). In addition to detecting the presence of breast cancer, the urine levels of the ADAM12 protein also correlated with tumor stage and progressively increased from patients with in situ carcinoma to locally invasive cancer to metastatic disease (38). Similarly, urine protein levels of ADAM12 were significantly increased in patients with bladder cancer compared with healthy controls (26). In the breast cancer study and bladder cancer study, the levels of ADAM12 mRNA and protein increased as a function of cancer stage, with the highest levels found in the largest invasive tumors (26). In the small number of bladder cancer patients studied, urine protein levels of ADAM12 decreased following tumor resection and increased again upon tumor recurrence, providing further support for the diagnostic utility of ADAM12 (26).
In the current study, we identified significant differences in progression-free and overall survival between women with high and low serum ADAM12 levels in a cohort of patients with stage III/IV HGSOC. Multivariable analyses identified serum ADAM12 as an independent prognostic factor for survival. The presence of lymphovascular invasion is an important predictor of poor survival in ovarian cancer (39). We showed that ADAM12 mRNA levels correlate with lymphatic and vascular invasion in the ovarian cancer TCGA data set, supporting the hypothesis that tumors with high levels of ADAM12 are biologically aggressive. Another important predictor of survival is the extent of residual disease after primary cytoreductive surgery (40). We showed that tumor ADAM12 mRNA levels correlate with the extent of residual disease in the ovarian cancer TCGA data set, suggesting that ADAM12 may be a biomarker of unresectable ovarian cancer. However, such a biomarker would be useful only if it can stratify patients preoperatively. In cases where the extent of ovarian cancer precludes optimal resection, efforts are made to reduce tumor burden with neoadjuvant chemotherapy prior to interval cytoreductive surgery (41). Currently, there is no clinical biomarker that can be applied preoperatively to predict when optimal or suboptimal cytoreduction can be surgically accomplished. Since the majority of patients in our cohort were optimally cytoreduced, we were unable to assess the ability of preoperative serum levels of ADAM12 to predict suboptimal cytoreduction. Considering the correlation of tumor ADAM12 mRNA with residual tumor volume and the correlation of preoperative serum ADAM12 protein with poor survival, a study that directly correlates serum ADAM12 with residual disease is warranted. An effective serum biomarker of suboptimal cytoreduction would impact the management of ovarian cancer patients as they could be spared a suboptimal surgical procedure and directly triaged to neoadjuvant chemotherapy (41).
Outcome predictors based on a molecular subtype rather than surgical staging have been successfully applied in breast cancer where gene signatures are used to predict metastasis and recurrence and to identify patients who are more likely to respond to a specific therapy. In breast cancer, it has been shown that ADAM12 is predominantly upregulated in claudin-low tumors, an aggressive subtype that exhibits molecular signatures of breast tumor-initiating cells and cells undergoing epithelial to mesenchymal transition (18,19,42). In the ovarian cancer TCGA data set (35) and the Tothill data set (36), we observed that high levels of ADAM12 were associated with the mesenchymal and the C1/desmoplastic subtype, respectively. Notably, these subtypes have been associated with the poorest survival when compared with other molecular subtypes in each data set (36,43,44). A common characteristic of both the mesenchymal and C1/desmoplastic molecular subtypes is extensive desmoplasia. Consistent with this phenotype, the genes co-expressed with ADAM12 in HGSOC are known to be involved in collagen remodeling, tissue organization and cell adhesion. The mechanisms by which desmoplasia contributes to poor survival is still unclear. Possible mechanisms include the presence of a nurturing environment for cancer stem cells, formation of linear collagen tracks for efficient cancer cell migration and invasion and increased interstitial pressure that thwarts drug delivery (45). For effective and durable remission, desmoplastic tumors may require different treatment approaches that target both cancer and stromal cells. Thus, in addition to serving as a predictor of poor prognosis, ADAM12 may be a biomarker for an aggressive molecular subtype of ovarian cancer that requires aggressive treatment with current cytotoxic therapy and/or experimental therapies that target stromal cells.
An important aspect of understanding the biomarker potential of ADAM12 in malignancy involves identification of the cells that produce and secrete ADAM12. In a variety of cell culture systems, ADAM12 expression both regulates, and can be induced by, TGFβ signaling (17,46,47). The source of ADAM12 expression within tumor tissue has been debated. Strong expression has been reported in malignant epithelial cells, stromal cells or both depending upon the cancer type and/or animal model studied (6,7,13,14,16,17,28,48,49). In a mouse model of prostate cancer where expression of SV40 large T-antigen is regulated by the prostate-specific probasin promoter, in situ hybridization demonstrated expression of ADAM12 in a subpopulation of stromal cells adjacent to prostate tumor glands (7). The ADAM12-positive stromal cells were morphologically different from adjacent spindle-shaped fibroblasts and were positive for both α-SMA and SV40 large T-antigen, indicating that they had undergone an epithelial to mesenchymal transition (7). Given the role of ADAM12 in myoblast fusion (2) and in the formation of trophoblast syncytia (50), stromal cells in the prostate that express both markers could have arisen by cell fusion. In ovarian cancer, we detected the ADAM12 mRNA and protein in tumor epithelial cells and adjacent stromal cells. In human tumor sections, it is impossible to track cells to determine if the ADAM12-positive stromal cells were derived from epithelial cells via epithelial to mesenchymal transition or cell fusion. Our in vitro coculture data support the hypothesis that ADAM12 mRNA is induced in both cell types upon direct contact. If this hypothesis is validated in other systems that effectively mimic the microenvironment in cancer, increased ADAM12 levels could be a useful readout for active epithelial-stromal signaling in cancer.
Supplementary material
Supplementary Tables I–III can be found at http://carcin.oxfordjournals.org/
Funding
The Office of the Assistant Secretary of Defense for Health Affairs through the Ovarian Cancer Research Program (Award No. W81XWH-14-1-0107 to S.O.); the American Cancer Society (RSG-10-252-01-TBG to S.O., SIOP-06-258-01-COUN to B.Y.K.); the Ovarian Cancer Research Fund Ann Schreiber Mentored Investigator Award administered by the University of California Office of the President’s Tobacco-Related Disease Research Program to D.-J.C. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
Conflict of interest statement: None declared.
Supplementary Material
Acknowledgements
We thank S.Swartwood and the Cedars-Sinai Medical Center Biobank and Translational Research Core for in situ hybridization studies and K.Daniels for assistance in the preparation of the manuscript.
Glossary
Abbreviations
- ADAM12
ADAM metallopeptidase domain 12
- ELISA
enzyme-linked immunosorbent assay
- HGSOC
high-grade serous ovarian carcinoma
- mRNA
messenger RNA
- TGF
transforming growth factor
References
- 1. Cao Y., et al. (2002) Intracellular processing of metalloprotease disintegrin ADAM12. J. Biol. Chem., 277, 26403–26411. [DOI] [PubMed] [Google Scholar]
- 2. Yagami-Hiromasa T., et al. (1995) A metalloprotease-disintegrin participating in myoblast fusion. Nature, 377, 652–656. [DOI] [PubMed] [Google Scholar]
- 3. Gilpin B.J., et al. (1998) A novel, secreted form of human ADAM 12 (meltrin alpha) provokes myogenesis in vivo. J. Biol. Chem., 273, 157–166. [DOI] [PubMed] [Google Scholar]
- 4. Hougaard S., et al. (2000) Trafficking of human ADAM 12-L: retention in the trans-Golgi network. Biochem. Biophys. Res. Commun., 275, 261–267. [DOI] [PubMed] [Google Scholar]
- 5. Kurisaki T., et al. (1998) Spatially- and temporally-restricted expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse embryo. Mech. Dev., 73, 211–215. [DOI] [PubMed] [Google Scholar]
- 6. Fröhlich C., et al. (2011) ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Mol. Cancer Res., 9, 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peduto L., et al. (2006) ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene, 25, 5462–5466. [DOI] [PubMed] [Google Scholar]
- 8. Díaz B., et al. (2013) Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J. Cell Biol., 201, 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albrechtsen R., et al. (2011) Extracellular engagement of ADAM12 induces clusters of invadopodia with localized ectodomain shedding activity. Exp. Cell Res., 317, 195–209. [DOI] [PubMed] [Google Scholar]
- 10. Roy R., et al. (2011) Potential of fluorescent metalloproteinase substrates for cancer detection. Clin. Biochem., 44, 1434–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rao V.H., et al. (2012) A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene, 31, 2888–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leyme A., et al. (2012) Identification of ILK as a new partner of the ADAM12 disintegrin and metalloprotease in cell adhesion and survival. Mol. Biol. Cell, 23, 3461–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dulauroy S., et al. (2012) Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat. Med., 18, 1262–1270. [DOI] [PubMed] [Google Scholar]
- 14. Lendeckel U., et al. (2005) Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J. Cancer Res. Clin. Oncol., 131, 41–48. [DOI] [PubMed] [Google Scholar]
- 15. Fröhlich C., et al. (2013) ADAM12 is expressed in the tumour vasculature and mediates ectodomain shedding of several membrane-anchored endothelial proteins. Biochem. J., 452, 97–109. [DOI] [PubMed] [Google Scholar]
- 16. Kveiborg M., et al. (2005) A role for ADAM12 in breast tumor progression and stromal cell apoptosis. Cancer Res., 65, 4754–4761. [DOI] [PubMed] [Google Scholar]
- 17. Le Pabic H., et al. (2003) ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology, 37, 1056–1066. [DOI] [PubMed] [Google Scholar]
- 18. Li H., et al. (2013) Metalloproteinase-disintegrin ADAM12 is associated with a breast tumor-initiating cell phenotype. Breast Cancer Res. Treat., 139, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H., et al. (2012) An essential role of metalloprotease-disintegrin ADAM12 in triple-negative breast cancer. Breast Cancer Res. Treat., 135, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nariţa D., et al. (2010) Molecular profiling of ADAM12 gene in breast cancers. Rom. J. Morphol. Embryol., 51, 669–676. [PubMed] [Google Scholar]
- 21. Narita D., et al. (2011) ADAM12 and ADAM17 gene expression in laser-capture microdissected and non-microdissected breast tumors. Pathol. Oncol. Res., 17, 375–385. [DOI] [PubMed] [Google Scholar]
- 22. Narita D., et al. (2012) Increased expression of ADAM12 and ADAM17 genes in laser-capture microdissected breast cancers and correlations with clinical and pathological characteristics. Acta Histochem., 114, 131–139. [DOI] [PubMed] [Google Scholar]
- 23. Uehara E., et al. (2012) Upregulated expression of ADAM12 is associated with progression of oral squamous cell carcinoma. Int. J. Oncol., 40, 1414–1422. [DOI] [PubMed] [Google Scholar]
- 24. Markowski J., et al. (2009) Metal-proteinase ADAM12, kinesin 14 and checkpoint suppressor 1 as new molecular markers of laryngeal carcinoma. Eur. Arch. Otorhinolaryngol., 266, 1501–1507. [DOI] [PubMed] [Google Scholar]
- 25. Carl-McGrath S., et al. (2005) The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int. J. Oncol., 26, 17–24. [PubMed] [Google Scholar]
- 26. Fröhlich C., et al. (2006) Molecular profiling of ADAM12 in human bladder cancer. Clin. Cancer Res., 12, 7359–7368. [DOI] [PubMed] [Google Scholar]
- 27. Mino N., et al. (2009) A disintegrin and metalloprotease 12 (ADAM12) is a prognostic factor in resected pathological stage I lung adenocarcinoma. J. Surg. Oncol., 100, 267–272. [DOI] [PubMed] [Google Scholar]
- 28. Kodama T., et al. (2004) ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am. J. Pathol., 165, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Georges S., et al. (2013) A Disintegrin And Metalloproteinase 12 produced by tumour cells accelerates osteosarcoma tumour progression and associated osteolysis. Eur. J. Cancer, 49, 2253–2263. [DOI] [PubMed] [Google Scholar]
- 30. Sasaroli D., et al. (2011) Novel surface targets and serum biomarkers from the ovarian cancer vasculature. Cancer Biol. Ther., 12, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheon D.J., et al. (2014) A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin. Cancer Res., 20, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karlan B.Y., et al. (2014) POSTN/TGFBI-associated stromal signature predicts poor prognosis in serous epithelial ovarian cancer. Gynecol. Oncol., 132, 334–342. [DOI] [PubMed] [Google Scholar]
- 33. Diaz E.S., et al. (2013) Obesity-associated adipokines correlate with survival in epithelial ovarian cancer. Gynecol. Oncol., 129, 353–357. [DOI] [PubMed] [Google Scholar]
- 34. Li A.J., et al. (2010) Serum low-density lipoprotein levels correlate with survival in advanced stage epithelial ovarian cancers. Gynecol. Oncol., 116, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature, 474, 609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tothill R.W., et al. (2008) Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res., 14, 5198–5208. [DOI] [PubMed] [Google Scholar]
- 37. Huang R.Y., et al. (2013) An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis., 4, e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy R., et al. (2004) ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J. Biol. Chem., 279, 51323–51330. [DOI] [PubMed] [Google Scholar]
- 39. Matsuo K., et al. (2012) Significance of lymphovascular space invasion in epithelial ovarian cancer. Cancer Med., 1, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang S.J., et al. (2012) Impact of complete cytoreduction leaving no gross residual disease associated with radical cytoreductive surgical procedures on survival in advanced ovarian cancer. Ann. Surg. Oncol., 19, 4059–4067. [DOI] [PubMed] [Google Scholar]
- 41. Vergote I., et al. (2010) Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N. Engl. J. Med., 363, 943–953. [DOI] [PubMed] [Google Scholar]
- 42. Prat A., et al. (2013) Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat., 142, 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Konecny G.E., et al. (2014) Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J. Natl. Cancer Inst., 106(10). pii: dju249. doi:10.1093/jnci/dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang W., et al. (2013) Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep., 4, 542–553. [DOI] [PubMed] [Google Scholar]
- 45. Egeblad M., et al. (2010) Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell, 18, 884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramdas V., et al. (2013) Canonical transforming growth factor-β signaling regulates disintegrin metalloprotease expression in experimental renal fibrosis via miR-29. Am. J. Pathol., 183, 1885–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ray A., et al. (2010) Transforming growth factor-beta1-mediated activation of NF-kappaB contributes to enhanced ADAM-12 expression in mammary carcinoma cells. Mol. Cancer Res., 8, 1261–1270. [DOI] [PubMed] [Google Scholar]
- 48. Iba K., et al. (1999) Cysteine-rich domain of human ADAM 12 (meltrin alpha) supports tumor cell adhesion. Am. J. Pathol., 154, 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bourd-Boittin K., et al. (2008) RACK1, a new ADAM12 interacting protein. Contribution to liver fibrogenesis. J. Biol. Chem., 283, 26000–26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huppertz B., et al. (2006) Trophoblast fusion: fusogenic proteins, syncytins and ADAMs, and other prerequisites for syncytial fusion. Micron, 37, 509–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.