Abstract
Objective:
To compare CNN models implemented using different strategies in the CT assessment of EGFR mutation status in patients with lung adenocarcinoma.
Methods:
1,010 consecutive lung adenocarcinoma patients with known EGFR mutation status were randomly divided into a training set (n=810) and a testing set (n=200). CNN models were constructed based on ResNet-101 architecture but implemented using different strategies: dimension filters (2D/3D), input sizes (small/middle/large and their fusion), slicing methods (transverse plane only and arbitrary multi-view planes), and training approaches (from scratch and fine-tuning a pre-trained CNN). The performance of the CNN models was compared using AUC.
Results:
The fusion approach yielded consistently better performance than other input sizes, although the effect often did not reach statistical significance. Multi-view slicing was significantly superior to the transverse method when fine-tuning a pre-trained 2D CNN but not a CNN trained from scratch. The 3D CNN was significantly better than the 2D transverse plane method but only marginally better than the multi-view slicing method when trained from scratch. The highest performance (AUC=0.838) was achieved for the fine-tuned 2D CNN model when built using the fusion input size and multi-view slicing method.
Conclusion:
The assessment of EGFR mutation status in patients is more accurate when CNN models use more spatial information and are fine-tuned by transfer learning. Our finding about implementation strategy of a CNN model could be a guidance to other medical 3D images applications. Compared with other published studies which used medical images to identify EGFR mutation status, our CNN model achieved the best performance in a biggest patient cohort.
Keywords: CNN, EGFR, Implementation strategy
I. Introduction
The emerging technology of deep learning, in particular convolutional neural networks (CNN) [1–4], is increasingly demonstrating its value in medical applications including detection of lung nodules [5], segmentation of liver [6] or heart [7], and diagnosis of skin cancer [8]. CNNs are an advance over classification methods using traditional machine learning techniques [9, 10] (e.g., radiomic analysis), which consist of three separate steps: 1) feature extraction, using predefined methods which are based on mathematic equations and/or representations of prior knowledge that cannot be automatically adapted from task to task; 2) feature selection; and 3) classification. CNNs are able to incorporate the feature extraction and selection processes into the classification process. More importantly, CNN’s backward propagation of errors for training purposes enables the network to self-learn novel features which are most useful for a specific application, overcoming the limitations of pre-defined features.
The impressive results achieved by CNNs have been enabled by the availability of huge datasets for training these algorithms. For example, the well-known ImageNet Large Scale Visual Recognition Challenge[11] provided about 1.4 million annotated images to train CNN architectures for analysis of 2D natural images. Far fewer images are available for most medical applications, usually less than one thousand. CNNs trained on these smaller datasets can suffer from overfitting, or learning to base its classification on random fluctuations in the training data, which limits the generalization of the algorithm to other datasets. The overfitting problem must be overcome for CNNs to succeed in medical applications. Researchers have proposed a number of strategies including data augmentation, transfer learning [12–14], and partition of input image [5, 15–17]. Augmentation can increase the amount of data available for training by creating new images through minor alterations to existing ones such as flips, translations, and/or rotations. Transfer learning means that the weights of a network model are initialized using a model pre-trained by a large dataset of natural images (ImageNet) and then fine-tuned using the smaller dataset of the specific medical application. Partition of input image trains CNNs using a region of interest (ROI) to reduce the size of the original image, thus offering fewer parameters which the algorithm might overfit.
Medical applications pose a problem for CNNs not merely because of the small number of study patients available for training the algorithm, but also because medical images acquired from CT, MR, and PET machines are three dimensional (3D). 3D images entail a vast increase in the data to be processed, causing limitations from the current capability of computer hardware (e.g., memory). As a result, there has been no 3D pre-trained CNN architecture from which a new 3D CNN model could be fine-tuned for a specific clinical application. One approach to construct a 3D clinical model is to crop volumes of interest (VOI) from full-size 3D image series [5, 18, 19] and then use the VOIs to train the 3D model from scratch. Such models may not be robust due to overfitting from the limited training sample data. Multiple approaches to VOI size have been proposed, but the effect of VOI size on a model’s performance has not been studied. A more popular approach to constructing a clinical model to handle 3D images is to adopt an existing 2D CNN architecture that are already pre-trained by natural images. To do so, a 3D image volume needs to be sliced into 2D images [16, 17, 20], either along the transverse direction or at an arbitrary direction in 3D. The sliced 2D images are then used as the input images to fine-tune the existing 2D CNN for the desired clinical application. To the best of our knowledge, to date there has been no study reporting the effects of these different implementation strategies on CNN models for any clinical application.
Our work addresses this gap in the literature by using a well-studied backbone CNN architecture, ResNet-101, for an important medical application, assessment of epithelial growth factor receptor (EGFR) mutation status of patients with lung adenocarcinoma using CT images[21]. Such assessment of EGFR mutation status is a clinical prerequisite for initiating treatment with tyrosine kinase inhibitors (TKIs) in stage IV non-small cell lung cancer (NSCLC) [22]. While EFGR mutations are usually assessed by genomic analysis of biopsy samples acquired through endoscope or fine needle aspiration (FNA), such biopsies have limitations in clinical practice. First, they are invasive, limiting the potential to perform repeated assessments during the course of treatment to monitor genetic changes. Second, because such assays are localized to the biopsy site, they poorly capture the intra- and inter-tumor heterogeneity which is a major factor in treatment success and the development of resistance to targeted therapies. Third, not all tumors are appropriate for biopsy due to their small sizes and atypical locations. Characterizing tumor phenotypes via imaging and image features (radiomic features) can overcome the limitations of molecular- and tissue-based analyses, offering the promise of a “virtual biopsy” that can depict the entire tumor, tumor metastases, and surrounding tissues at multiple body sites sequentially using non-invasive CT scan images that are routinely acquired in clinical practice and clinical trials.
In this paper, we examined multiple approaches to building ResNet-101 CNN models in the test case of automated CT assessment of EGFR mutation status of patients with lung adenocarcinoma. Our analysis quantitatively compared the performances of models constructed using different implementation strategies, including input dimensions, input image sizes, the method used to slice 3D image series into 2D images, and training methods.
II. Materials and Methods
A. Clinical data
CT images from 1,010 consecutive patients with known EGFR status were retrospectively collected from 2013 to 2017, including 510 patients whose tumors were EGFR-mutated and 500 who were wild type [23]. Details are shown in TABLE I. The median tumor diameter was 26 mm (max: 95 mm; min: 8 mm). Patients were randomized into a training set (810 patients) and testing set (200 patients). Patient characteristics including gender, age, EGFR status, and sample type did not differ significantly between these two sets. 710 patients in the training set were used to train the model and the remaining 100 patients were used as validation set.
TABLE I.
Characteristics of the patient cohort
| Characteristic | Overall (n= 1010) | Mutation (n=510) | Wild Type (n=500) |
|---|---|---|---|
| Gender | |||
| Male | 553 | 209 (40.9%) | 344 (68.8%) |
| Female | 457 | 301 (58.9%) | 156 (31.2%) |
| Age | |||
| Median age | 63 | 62 | 61 |
| Range | 25–88 | 30–88 | 25–85 |
| Sample Type | |||
| Biopsy | 386 (38.2%) | 177 (34.7%) | 209(41.8%) |
| Surgery | 624 (61.8%) | 333 (65.3%) | 291 (58.2%) |
EGFR mutation tests were based on tissue samples acquired from surgery or biopsy via the PCR machine (Stratagene Mx3000PTM) provided by Agilent. The Human EGFR Gene Mutation Detection Kit was manufactured by Amoy Diagnostics Co., Ltd.
B. Image data
Non-contrast enhanced CT scans were performed about a week before surgery or biopsy, on two scanner types (GE Discovery and Philips Brilliance) available in the institution. The image resolutions were 5 mm slice thickness and about 1 mm in-plane resolutions. Linear interpolation was applied to the original image data to obtain isotropic image resolutions at 1 mm along the x -, y-, and z-axial directions. Regions of interest (ROI) were delineated by two experienced clinicians using the lung window (−400 HU∼1600 HU) to define a closed boundary surrounding the tumor area in each image containing the tumor.
C. Models
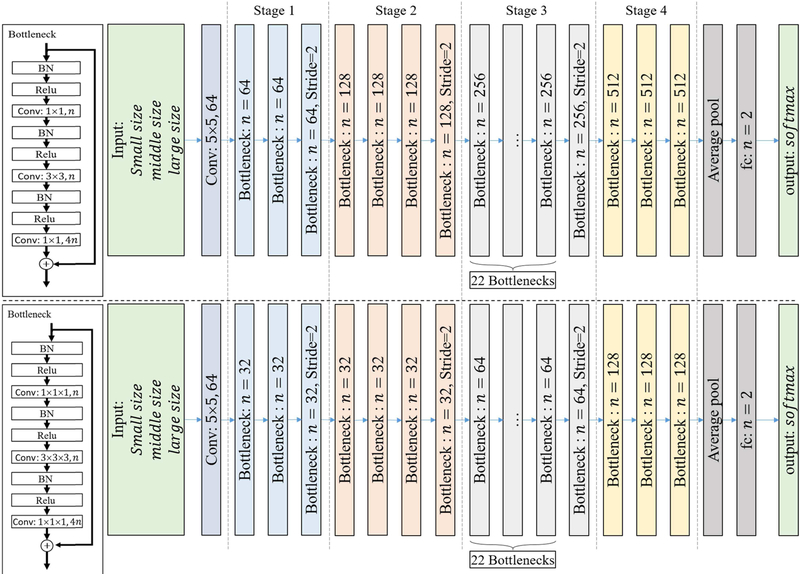
Based on a residual net with 101 layers (ResNet 101), we designed several CNN models using different implementation approaches (Fig. 1). The CNN models included 2D and 3D structures. For 2D models, we chose transverse plane (only) and arbitrary multi-view plane to slice 3D volume images into 2D images and took them as input images. For 3D models, we used multiple sizes of VOIs as 3D input images. All models, except the 3D one (no pre-trained 3D model available), were built using pre-trained as well as from scratch training methods. The detail of each method is described below.
Fig. 1.
Structure of the ResNet 101 CNN using 2D filters (top) and 3D filters (bottom).
1). Input Size:
Three different input sizes (small, middle, large) were used to capture the information of lung tumors. For a 2D CNN, the pixel resolution was 1mm∗1mm, and the input sizes were 51∗51 (small), 101∗101 (medium), or 151∗151 pixels (large). For a 3D CNN, the voxel resolution was 2mm∗2mm∗2mm, and the input sizes were 21∗21∗21 (small), 31∗31∗31 (medium), and 41∗41∗41 voxels (large). A lower resolution was used for 3D CNN due to limitations on computer memory (GPU). Each CNN (small, medium, or large input size) output a probability of EGFR gene mutation. A fourth approach, fusion, was obtained by combining the output of these three CNNs. Specifically, the output probability of EGFR for each CNN was regarded as a feature, and the three features (corresponding to small/middle/large input sizes) were fused by logistical regression whose output was the fusing prediction result.
2). 2D/3D CNN Structure:
The filters of a CNN can be two dimensions (2D CNN; Fig. 1(top)) or three dimensions (3D CNN; Fig. 1(bottom)). The number of filters/kernels in a 3D CNN structure is smaller than that in a 2D CNN. The input of each CNN contains the original image data and its corresponding segmented tumor mask. For 2D CNN, the input has three channels which correspond to R.G.B. of natural images: CT image, tumor mask, and CT image + tumor mask. For 3D CNN, the input has two channels, CT image and tumor mask. Both 2D slice input and 3D volume input were cropped from the original images focused at the tumor center. In the implementation, the input slices/volumes were augmented to reduce overfitting and increase the model’s accuracy. Explicitly, for each iteration, the 2D slices were generated by randomly transmuting several pixels and the 3D volumes were augmented by randomly transmuting several voxels and rotating a random angle in 3D directions. Theoretically, this random generation for each iteration meant that the CNNs could be trained on an infinite number of different images, as it is highly unlikely that the process would generate two exactly identical images.
3). Transverse/Multi-view Slicing:
For 2D CNNs, the input slice was cropped either from a fixed transverse plane or from an arbitrary multi-view plane. For transverse plane models, the 2D slice was taken along the x-y transverse plane. For multi-view plane models, the 2D slice was taken along any arbitrary direction in 3D. In both cases, the 2D slice always passed through the center of the rotated tumor.
4). Transfer Learning/Training From Scratch:
Two training methods were used for 2D CNN models: training from scratch or transfer learning. For training from scratch, training weights were initialized by Xavier filter. For transfer learning, training weights were initially adopted from the ImageNet[11] pre-trained model and then fine-tuned using the EFGR image data.
5). Constructed models:
By combining the implementation strategies described above, we constructed 5 groups of CNN models:
2D-Transverse-Scratch: 2D CNN with input of transverse plane and trained from scratch;
2D-Transverse-Fine-tune: 2D CNN with input of transverse plane and using the fine-tune (transfer learning) training method;
2D-MultiView-Scratch: 2D CNN with input of multi-view plane and trained from scratch;
2D-MultiView-Fine-tune: 2D CNN with input of multi-view plane and using the fine-tune (transfer learning) training method;
3D-Volume-Scratch: 3D CNN with input of volume using and trained from scratch.
Each model group contained four models, three of which were models having one input image size of small, middle, and large, respectively. The forth one, fusion model, fused the previous three input size models. In total, 20 (5∗4) models were constructed using the different implementation approaches for comparison of their performance.
6). Training and Testing:
Cross-entropy function (loss function) and stochastic gradient descent (SGD) were used to train all CNN models. The initial learning rate was 0.1 when training from scratch and 0.001 when fine-tuning a model. The learning rate was reduced to 0.96 for each 10 epochs, with the maximum iterative epoch set to 1000.
Given a test sample, the input slice/volume was generated 32 times and obtained 32 different prediction probabilities, and the final prediction result was computed by averaging all prediction probabilities.
D. Statistical Analysis
The area under receiver operating characteristic (ROC) curve (AUC) was used to evaluate the performance of the models. The performance of the 20 models was pairwise compared using the DeLong test [24]. P-value less than 0.05 was considered as significant. For each pairwise comparison, only one variable / implementation strategy (e.g., input size, training method) changed while the others remained the same. To guarantee the generalization, 5-folds cross-validation on the entire data was performed as well in supplementary.
III. Results
The performances (AUCs) of the 20 models are reported in TABLE II. The 2D CNN fusion model using the multi-view plane and the fine-tuning methods, 2D-MultiView-Fine-tune fusion model, showed the highest AUC value of 0.838 in the detection of EGFR mutation. The two 2D CNN models using the transverse plane and the large input size either with or without pre-training, 2D-Transverse-Fine-tuneand 2D-Transverse-Scratch, showed the lowest AUC values of 0.642 and 0.649 respectively. Notably, 2D CNN models using the multi-view plane and the fine-tuning methods had AUCs larger than 0.80 at all input sizes.
TABLE II.
Performance of each CNN model measured by AUC
| Comparison Method | Training Method | Small Input Size | Middle Input Size | Large Input Size | Fusion |
|---|---|---|---|---|---|
| 2D slice image transverse plane | scratch | 0.703 | 0.687 | 0.649 | 0.712 |
| fine-tune | 0.739 | 0.721 | 0.642 | 0.766 | |
| 2D slice image multi-view plane | scratch | 0.711 | 0.722 | 0.721 | 0.733 |
| fine-tune | 0.808 | 0.821 | 0.806 | 0.838 | |
| 3D volume image[19] | scratch | 0.753 | 0.784 | 0.774 | 0.809 |
To determine the effect of input size on CNN performance, the models using the small, middle, and large input sizes were individually compared with the fusion model within each model group. Although the fusion models showed higher AUCs compared to the other three models across the 5 model groups, the differences were statistically significant only for the 2D-Transverse-Fine-tune model using the large input size, 2D-Transverse-Scratch models using the middle and large input sizes, and the 3D-Volume-Scratch model using the small input size (TABLE III).
TABLE III.
Effect of input size on CNN performance: p-values of pairwise compared ROCS between models using a single input size (small/middle/large) and their fusion
| Comparison Method | Training Method | Small Input Size | Middle Input Size | Large Input Size |
|---|---|---|---|---|
| 2D slice image transverse plane | scratch | 0.414 | 0.045 | 0.008 |
| fine-tune | 0.161 | 0.078 | <0.001 | |
| 2D slice image multi-view plane | scratch | 0.196 | 0.410 | 0.155 |
| fine-tune | 0.093 | 0.156 | 0.077 | |
| 3D volume image | scratch | 0.033 | 0.112 | 0.115 |
To determine the effect of transverse/multi-view slicing, comparisons were made within 2D CNNs using the same training method (from scratch or fine-tuning). As slicing is not required for 3D models, they are omitted from these comparisons. The model group using the transverse plane was pairwise compared with the group using the multi-view plane (2D-Transverse-Scratch vs 2D-MultiView-Scratch and 2D-Transverse-Fine-tune vs 2D-MultiView-Fine-tune). Using the fine-tuning method, the multi-view plane method significantly outperformed the transverse plane method at all input sizes, but this was not true when the models were trained from scratch (with the exception of the model using the large input size) (TABLE IV).
TABLE IV.
Effect of transverse/multi-view slicing on the performance of 2D CNN models: p-values of pairwise compared ROCS between models using the transverse plane and the multi-view plane
| Comparison Method | Training Method | Small Input Size | Middle Input Size | Large Input Size | Fusion |
|---|---|---|---|---|---|
| 2D slice image | scratch | 0.896 | 0.229 | 0.037 | 0.378 |
| fine-tune | 0.040 | 0.005 | <0.001 | 0.006 |
To determine the effect of training method, comparisons were made within 2D CNNs using the same slicing method (transverse or multi-view). As no datasets currently exist to enable fine-tuning of 3D CNNs, they are omitted from these comparisons. The model groups using the from scratch and fine-tuning training methods were pairwise compared (i.e., 2D-Transverse-Scratch vs. 2D-Transverse-Fine-tune and 2D-MultiView-Scratch vs. 2D-MultiView-Fine-tune). The performance of the 2D CNN built by fine-tuning a pre-trained model was significantly superior to that of the model trained from scratch using all input sizes when using the multi-view plane. This was not true for the transverse plane method (TABLE V), with the exception of the fusion input size.
TABLE V.
Effect of training method on the performance of 2D CNN models: p-values of pairwise compared ROCS between models trained from scratch and using the fine-tuning method
| Comparison Method | Small Input Size | Middle Input Size | Large Input Size | Fusion |
|---|---|---|---|---|
| 2D transverse plane | 0.232 | 0.374 | 0.881 | 0.046 |
| 2D multi-view plane | 0.004 | <0.001 | 0.005 | <0.001 |
To determine the effect of 2D vs. 3D CNN architecture, comparisons were made between the 3D CNN and the 2D models (transverse and multi-view). Given that no pre-trained 3D CNN architectures are available, comparisons were only made within the model groups trained from scratch (i.e., 3D-Volume-Scratch vs. 2D-Transverse-Scratch and 3D-Volume-Scratch vs. 2D-MultiView-Scratch). The 3D model performed significantly better than the 2D models at all input sizes when using the transverse plane. However, the advantage of the 3D model disappeared when the 2D models were built using multi-view planes with small or large input sizes (TABLE VI).
TABLE VI.
Effect of 2D/3D CNN architecture on the performance of models trained from scratch: p-values of pairwise compared ROCS Between the 3D CNN model group and two 2D CNN model groups
| Comparison Method | Small Input Size | Middle Input Size | Large Input Size | Fusion |
|---|---|---|---|---|
| 2D transverse plane | 0.049 | 0.006 | 0.003 | <0.001 |
| 2D multi-view plane | 0.082 | 0.023 | 0.125 | 0.006 |
IV. Discussion
Achieving the potential of deep learning methods [1–4, 25] for clinical applications requires understanding how different strategies for developing and implementing a CNN structure influence the performance of the resulting model. For clinical applications using 3D radiographic images, the dimensionality of the CNN backbone architecture represents a key decision. 3D CNNs are designed to process the spatial information contained in 3D images, but no pre-trained CNN architectures are currently available. 2D CNNs can be pre-trained through transfer learning using vast existing image databases of 2D natural images (e.g. ImageNet), which may improve their performance, but the decision to analyze 3D images using a 2D architecture requires making another choice regarding whether the source images should be sliced using transverse or multi-view planes. For both types of architecture, the input image size used presents another strategic decision.
The comparison study presented in this paper is the first effort to quantify the effect of these decisions on the performance of a CNN model for a clinical application using 3D radiographic image. In our study, we adopted the ResNet 101 CNN architecture with either 2D or 3D input filters, which is widely available and has been used in a variety of medical contexts. The clinical application we chose was assessment of EGFR mutation status in patients with lung adenocarcinoma using routinely acquired CT images. As there is no a priori reason to expect that the effect of strategic decisions on the performance of CNNs will be significantly different between this model task and other clinical applications. Our results should thus be widely applicable to other efforts to deploy CNNs for the interpretation of medical images.
Many medical images are 3D, while almost all natural images – including the vast databases such as ImageNet which have been used to pretrain CNNs – are 2D. This difference may be the most significant consideration when using CNNs in medical applications. In order to use a CNN structure developed for analysis of 2D natural images in 3D medical applications, 3D images need to be sliced into 2D images. The most common slicing method is to derive the input to the 2D CNN model by taking one or more 2D slices from the 3D volume images. A 2D slice can be taken along the transverse plane direction or in any arbitrary direction in 3D (multi-view plane). In this study, the two slicing methods were compared with the 2D CNN models built with and without transfer learning (i.e., from scratch vs. fine-tuned). We found that when a model was built using the multi-view slicing method, it yielded better performance than a model using the transverse method. The likely rationale for this finding is that more information from a tumor can be used when several 2D images are taken from multi-view planes than when only the transverse plane is used. This additional information is likely to make the multi-view CNN model more robust in the training phase, and to enable better performance when tested in the testing phase.
Due to the wide size range of lung cancer tumors, the selection of input image size is an important strategic decision. If the input image size is too small, it cannot cover the entirety of a large tumor, so that potentially useful information will not be available to guide the CNN in characterizing some of the tumors in the dataset. On the other hand, if the chosen input image size is too large, the risk of overfitting is increased by the greater availability of irrelevant information from structures surrounding a small tumor. To understand this problem, we designed three individual CNN models, each taking one size (small, medium, and large) as the input image size. Feature maps of 2D CNNs using multi-view plane inputs are shown in Fig. 2, demonstrating that when using the small input size only the CNN focused on the tumor itself while using larger input size caused extra attention to be paid to the surrounding of tumor. We also constructed a fusion model which combined the information of these three individual CNN models. We found that the fusion model achieved better performance (higher AUC) than models using the small, middle, or large input size alone. This result was remarkably consistent across all 5 model groups, although most of the pairwise comparisons between the fusion model and small, medium, or large input sizes did not reach statistical significance at p <= 0.05.
Fig. 2.
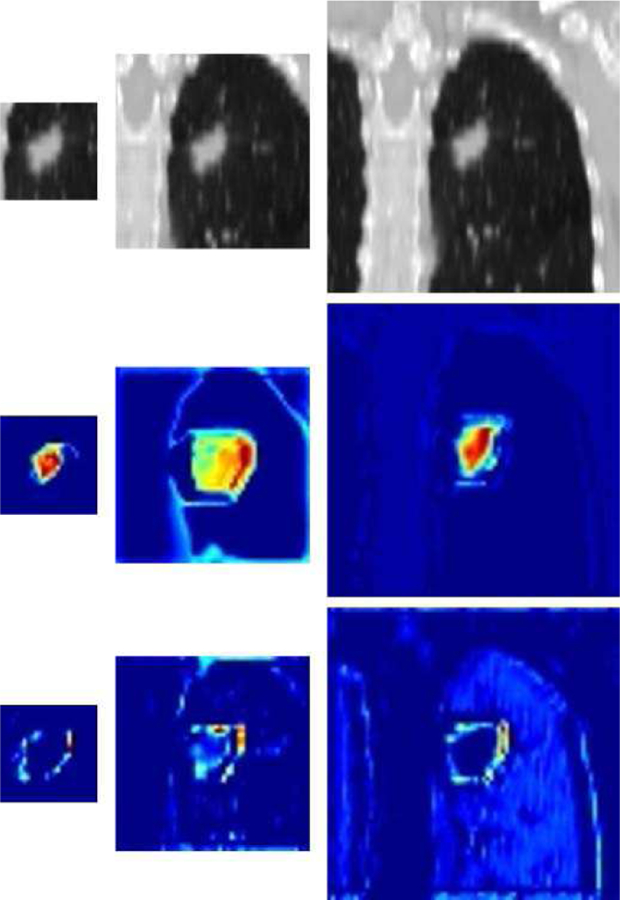
Examples of feature maps extracted from the output of 2D CNNs at the end of the first stage: original CT images (top) and their feature maps of CNN trained from scratch (middle) or fine-tuned (bottom). The left, middle, and right panels correspond to the small, middle, and large input sizes, respectively.
Transfer learning, an approach to fine-tune a pre-trained CNN model, is widely used to achieve better performance in analysis of natural images. Unfortunately, the relative shortage of 3D medical images for transfer learning means that reliable pre-trained CNN architectures are available only for 2D CNN models, not 3D. We thus compared the two training methods, trained from scratch and fine-tune training using transfer learning from a natural image dataset (ImageNet), in 2D CNN models. We found that fine-tuned models achieved significantly better performance (higher AUC) than models trained from scratch. The fine-tuned models found more information in the surface and surroundings of the tumor which correlated to EGFR mutation statue, while the model trained from scratch paid more attention to the inside of the tumor (Fig. 2).
3D CNN models can utilize the full information of a 3D medical image series, whereas sliced 2D images can only address part of the information. However, the probability of overfitting is increased by the much larger number of kernel weights used in a 3D CNN as compared to a 2D CNN structure. It is thus necessary to compare whether 2D or 3D models are more suitable for applications using 3D medical images. We thus compared 2D and 3D models, both using the training from scratch method. The 3D model achieved consistently superior performance (higher AUC) over the 2D model group, although this difference was only statistically significant for the 2D models sliced using only the transverse plane.
The overall direction of our findings can be seen clearly by focusing on the fusion models (TABLE VII), which achieved uniformly higher AUC than the other input size models. Performance consistently improved as models incorporated more spatial information (downward movement in TABLE VII). 2D multi-view models outperformed transverse models (although for the fusion input size this difference was only significant for fine-tuned models), and were in turn significantly outper-formed by 3D models (p < 0.001 for transverse 2D models and p < 0.01 for multi-view models, both trained from scratch). Likewise, fine-tuned models consistently outperformed models trained from scratch (p < 0.05 for transverse models and p < 0.001 for multi-view models). Taken together, these results strongly suggest that when a sufficiently large dataset of medical images is available to enable transfer learning for 3D CNNs models, the resulting fine-tuned 3D CNNs will offer better performance for medical applications than any of the other strategies studied in this paper.
TABLE VII.
Effect of 2D/3D CNN architecture on the performance of models trained from scratch: p-values of pairwise compared ROCS between the 3D CNN model group and two 2D CNN model groups
| Scratch | Fine-tune | |
|---|---|---|
| 2D slice image transverse plane | 0.712 | 0.766 |
| 2D slice image multi-view plane | 0.733 | 0.838 |
| 3D volume image[19] | 0.809 | N/A |
Training and testing models was done using the same hardware, including a NVIDIA TITAN Xp GPU and an Intel Xeon E5–2620 CPU. The average training time of one epoch and testing time of one testing sample are summarized for each model in TABLE VIII, showing that: (1) using multi-view plane input increases the computational cost of image pre-processing, requiring more time for training/testing 2D models; (2) larger input sizes increase the time for training/testing models; (3) training/testing 2D models required more time than 3D models because of the greater number of filters used in 2D CNN.
TABLE VIII.
Average processing time of training and testing
| Comparison Method | Small Input Size | Middle Input Size | Large Input Size | Fusion | |
|---|---|---|---|---|---|
| 2D slice image transverse plane | training | 53.4s | 77.3s | 160.5s | N/A |
| testing | 0.252s | 0.262s | 0.268s | 0.782s | |
| 2D slice image multi-view plane | training | 56.6s | 81.5s | 169.2s | N/A |
| testing | 0.254s | 0.263s | 0.270s | 0.788s | |
| 3D volume image | training | 35.5s | 75.3s | 134.1s | N/A |
| testing | 0.191s | 0.210s | 0.222s | 0.624s | |
The clinical utility of CNNs to medical application, and the potential impact of the methods selected for model building, can be shown by comparison of our results to other studies which similarly attempted to assess EGFR mutation status in lung cancer patients on the basis of their imaging phenotype. The studies published to date in this on-going research area [26–29] built their predictive models using a traditional machine learning method, radiomics. A comparison between the performance of their models and our optimal model, 2D-MultiView-Fine-tune, is shown in TABLE IX. The models reported by Velazquez E R[26] and Stephen SF Yip[28] achieved AUCs less than 0.7 in their testing groups. Although Ying Liu[27] and Stefania Rizzo[29] achieved relatively higher AUC scores of 0.709 and 0.82, respectively, their studies did not include independent testing groups which are essential to establishing the robustness of predictive models. Junfeng Xiong [23] and Xiaoyang Li [19] showed impressive results by using 3D deep learning model trained from scratch and achieved AUC scores of 0.776 and 0.809, respectively. Our model was trained and validated using the same patient cohort as Li’s work [19] and, in an independent testing group, achieved a higher AUC score than any previously published study in this area. Notably, the lowest-performing model in our study (a fine-tuned 2D CNN using the large input size and transverse slicing method) achieved a lower AUC (0.642) than any prior studies, highlighting the importance of optimally selecting the methods used to construct CNNs for medical applications.
TABLE IX.
Results of studies assessing EFGR mutational status using radiographic image phenotype, comparing our work to previous publications
| Method (Author) | N (mutation %) | AUC in training group | AUC in testing group |
|---|---|---|---|
| Velazquez E R[26] | 258 (45) | - | 0.67 |
| Ying Liu[27] | 298 (46) | 0.709 | - |
| Stephen SF Yip[28] | 348 (13) | - | 0.67 |
| Stefania Rizzo[29] | 285 (21) | 0.82 | - |
| Junfeng Xiong[23] | 503 (61) | - | 0.776 |
| Xiaoyang Li[19] | 1010(50) | - | 0.809 |
| Our research | 1010(50) | - | 0.838 |
Our study has some limitations. First, patient clinical features such as smoking history and gender may be associated with EGFR mutation status. Prior work has shown that incorporating these clinical features into image-based models (radiomics or deep learning) showed no significant improvement for the detection of EGFR mutation status [19, 23]. Accordingly, we did not include clinical features in the study presented here, which was focused on comparing CNN models constructed using different implementation strategies. Fusion of multimodality data promises to offer improvement in diagnosing Alzheimer’s disease [30, 31]. Hence, our future work may determine a way to combine clinical and image information to improve model performance using regression methods[32, 33]. Second, our dataset was collected from a single institution whose patient population offered limited diversity (all individuals studied were Asian). Application of our model to international patient population groups will be an important planned extension of our work. Third, due to the lack of medical image datasets available to pre-train a 3D CNN model, we were not able to extend the comparison of models trained from scratch vs. fine-tuned to include 3D CNN architectures. Building a 3D medical model pre-trained using a sufficiently large volume of medical data is a major goal for our group, and the results of the current study suggest that this method will enable still further improvements over previously studied approaches. Third, deep learning model is an end-to-end model. The input is the images and the outputs were the corresponding prediction results. We provided some example feature maps of CNN models and subjective analysis. We plan to continue performing more visualizations to help us well understand the ‘black box’.
V. Conclusion
Our study demonstrated that utilizing more spatial information improves the robustness and performance of CNN models for medical applications based on 3D images. Strategies which increase the spatial information available to the model include using fusion input size rather than one fixed input size; using multi-view plane 2D slices rather than transverse plane only; and using 3D CNNs rather than 2D CNNs. We also demonstrated that models fine-tuned using transfer learning were significantly more accurate than models trained from scratch. Taken together, our findings suggest that 3D CNN models will, once a large scale dataset of 3D medical images becomes available for fine-tuning them, have great potential to outperform all 2D CNN models. Given the current lack of pre-trained 3D CNN architectures, we found that of all approaches compared in this study, the fine-tuned, multi-view fusion 2D CNN is best suited to assess EGFR mutation status in patients with lung adenocarcinoma and does so better than any previous attempt.
Supplementary Material
Biography

JUNFENG XIONG received the bachelor’s degree in Biomedical Engineering from Shanghai Jiao Tong University, in 2014, where he is currently pursuing the Ph.D. degree. His research interest includes medical image analysis and deep learning.

XIAOYANG LI received the bachelor’s degree from the Department of Radiology, Anhui Medical University, Anhui, China, in 2012. He is currently pursuing the Ph.D. degree from the School of Medicine, Shanghai Jiao Tong University. His research interest includes radiation oncology and artificial intelligence in gene of lung cancer.

LIN LU received the Ph.D. degree from the Department of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, in 2013. He is currently an Associate Research Scientist with the Department of Radiology, Columbia University Medical Center. He has published over 30 research papers in a number of outstanding journals, such as the IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, Medical Physics, the Journal of Proteome Research, and the Journal of Computational Chemistry. His research interests include image processing, data mining, and machine learning

AWRENCE H SCHWARTZ is the James Picker Professor and Chairman for the Department of Radiology at Columbia University Medical Center. He also is the Chief of the Radiology Service and Attending Physician at New York-Presbyterian Hospital. He is a diplomate of the American Board of Radiology and a member of the American Roentgen Ray Society, Radiological Society of North America, International Society for Magnetic Resonance in Medicine, New York Roentgen Ray Society, Society for Computer Applications in Radiology and a Fellow at the International Cancer Imaging Society.

XIAOLONG FU received the Ph.D. degree from the School of Medicine, Fu Dan University, Shanghai, China, in 2000. He is currently a Full Professor and the Chairman for the Department of Radiation Oncology at Shanghai Chest Hospital. He is also the Vice Chairman of China Society of Therapeutic Radiation Oncology.

JUN ZHAO received the Ph.D. degree from the Department of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, in 2006. He was a Visiting Scholar with the University of Iowa, USA, from 2006 to 2007. He is currently a Full Professor and the Associate Dean of the School of Biomedical Engineering, Shanghai Jiao Tong University. He is also active in the international research groups of the IEEE EMBC, SPIE, and ISBI. He has published over 130 research papers in inter-national conferences and journals. His research interests include biomedical imaging, medical image processing, computer-aided detection, and medical applications of synchrotron radiation. He received over 10 grants from the Ministry of Science and Technology of China, the National Science Foundation of China, the Science and Technology Commission, the Education Commission of Shanghai Municipality, GE. He is recognized by a number of awards for academic and educational achievements. He serves as an Associate Editor for Computer Methods in Biomechanics and Biomedical Engineering: Imaging and Visualization, and the International Journal of Biomedical Imaging.

BINSHENG ZHAO is a Professor of Radiology (Medical Physics) and the Director of the Laboratory for Computational Image Analysis in the Department of Radiology, Columbia University Medical Center. She received her BS and MS degrees both in electronic engineering from National Institute of Technology at Changsha, China, and her DSc degree in medical informatics from University of Heidelberg, Germany. She published 90+ peer-reviewed papers. Her current research interests include quantitative imaging (QI) biomarkers for cancer diagnosis, prognosis, response prediction and assessment using radiomics and artificial intelligence approaches, and harmonization of imaging settings for reproducible and robust QI biomarkers.
Footnotes
Publisher's Disclaimer: This work was supported in part by National Natural Science Foundation of China (No. 81371634), National Key Research and Development Program (No. 2016YFC0104608), Shanghai Jiao Tong University Medical Engineering Cross Research Funds (No. YG2017ZD10, YG2014ZD05), and Grants U01 CA225431 from the National Cancer Institute (NCI).
References
- [1].LeCun Y, Bengio Y, and Hinton G, “Deep learning,” nature, vol. 521, no. 7553, p. 436, 2015. [DOI] [PubMed] [Google Scholar]
- [2].Litjens G et al. , “A survey on deep learning in medical image analysis,” Medical image analysis, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [3].Shen D, Wu G, and Suk H-I, “Deep learning in medical image analysis,” Annual review of biomedical engineering, vol. 19, pp. 221–248, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schmidhuber J, “Deep learning in neural networks: An overview,” Neural networks, vol. 61, pp. 85–117, 2015. [DOI] [PubMed] [Google Scholar]
- [5].Setio AAA et al. , “Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks,” IEEE transactions on medical imaging, vol. 35, no. 5, pp. 1160–1169, 2016. [DOI] [PubMed] [Google Scholar]
- [6].Dou Q, Chen H, Jin Y, Yu L, Qin J, and Heng P-A, “3D deeply supervised network for automatic liver segmentation from CT volumes,” 2016, pp. 149–157: Springer. [Google Scholar]
- [7].Ye C, Wang W, Zhang S, and Wang K, “Multi-Depth Fusion Network for Whole-Heart CT Image Segmentation,” IEEE Access, 2019. [Google Scholar]
- [8].Esteva A et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115 %@ 1476–4687, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jin Z, Zhou G, Gao D, and Zhang Y, “EEG classification using sparse Bayesian extreme learning machine for brain–computer interface,” Neural Computing and Applications, pp. 1–9, 2018. [Google Scholar]
- [10].Jia T-Y et al. , “Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling,” European radiology, pp. 1–9. [DOI] [PubMed] [Google Scholar]
- [11].Russakovsky O et al. , “Imagenet large scale visual recognition challenge,” International Journal of Computer Vision, vol. 115, no. 3, pp. 211–252, 2015. [Google Scholar]
- [12].Dou Q et al. , “Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks,” IEEE transactions on medical imaging, vol. 35, no. 5, pp. 1182–1195, 2016. [DOI] [PubMed] [Google Scholar]
- [13].Hoo-Chang S et al. , “Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning,” IEEE transactions on medical imaging, vol. 35, no. 5, p. 1285, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gupta A, Ayhan M, and Maida A, “Natural image bases to represent neuroimaging data,” in International conference on machine learning, 2013, pp. 987–994. [Google Scholar]
- [15].Suk H-I, Lee S-W, Shen D, and A. s. D. N. Initiative, “Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis,” NeuroImage, vol. 101, pp. 569–582, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen W, Zhou M, Yang F, Yang C, and Tian J, “Multi-scale convolutional neural networks for lung nodule classification,” in International Conference on Information Processing in Medical Imaging, 2015, pp. 588–599: Springer. [DOI] [PubMed] [Google Scholar]
- [17].Roth HR et al. , “Improving computer-aided detection using convolutional neural networks and random view aggregation,” IEEE transactions on medical imaging, vol. 35, no. 5, pp. 1170–1181, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Suk H-I, Shen D, and A. s. D. N. Initiative, “Deep learning in diagnosis of brain disorders,” in Recent Progress in Brain and Cognitive Engineering: Springer, 2015, pp. 203–213. [Google Scholar]
- [19].Li X-Y et al. , “Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks,” Journal of Thoracic Disease, vol. 10, no. 12, pp. 6624–6635 %@ 2077–6624, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ciompi F et al. , “Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box,” Medical image analysis, vol. 26, no. 1, pp. 195–202, 2015. [DOI] [PubMed] [Google Scholar]
- [21].Fukuoka M et al. , “Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non–small-cell lung cancer in Asia (IPASS),” Journal of clinical oncology, vol. 29, no. 21, pp. 2866–2874, 2011. [DOI] [PubMed] [Google Scholar]
- [22].Gridelli C et al. , “First-line erlotinib followed by second-line cisplatin-gemcitabine chemotherapy in advanced non-small-cell lung cancer: the TORCH randomized trial,” J Clin Oncol, vol. 30, no. 24, pp. 3002–3011, 2012. [DOI] [PubMed] [Google Scholar]
- [23].Xiong J-F et al. , “Identifying epidermal growth factor receptor mutation status in patients with lung adenocarcinoma by three-dimensional convolutional neural networks,” The British Journal of Radiology, vol. 91, no. 1092, p. 20180334, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].DeLong ER, DeLong DM, and Clarke-Pearson DL, “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics, pp. 837–845, 1988. [PubMed] [Google Scholar]
- [25].Liu N, Wan L, Zhang Y, Zhou T, Huo H, and Fang T, “Exploiting convolutional neural networks with deeply local description for remote sensing image classification,” IEEE Access, vol. 6, pp. 11215–11228, 2018. [Google Scholar]
- [26].Rios Velazquez E, Liu Y, Parmar C, Narayan V, Gillies R, and Aerts H, “MO - DE - 207B - 08: Radiomic CT Features Complement Semantic Annotations to Predict EGFR Mutations in Lung Adenocarcinomas,” Medical physics, vol. 43, no. 6 Part30, pp. 3706–3706, 2016. [Google Scholar]
- [27].Liu Y et al. , “Radiomic features are associated with EGFR mutation status in lung adenocarcinomas,” Clinical lung cancer, vol. 17, no. 5, pp. 441–448. e6, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yip SSF et al. , “Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non–Small Cell Lung Cancer,” Journal of Nuclear Medicine, vol. 58, no. 4, p. 569, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rizzo S et al. , “CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer,” European radiology, vol. 26, no. 1, pp. 32–42, 2016. [DOI] [PubMed] [Google Scholar]
- [30].Zhou T, Thung KH, Zhu X, and Shen D, “Effective feature learning and fusion of multimodality data using stage - wise deep neural network for dementia diagnosis,” Human brain mapping, vol. 40, no. 3, pp. 1001–1016, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhou T, Thung K-H, Liu M, and Shen D, “Brain-Wide Genome-Wide Association Study for Alzheimer’s Disease via Joint Projection Learning and Sparse Regression Model,” IEEE Transactions on Biomedical Engineering, vol. 66, no. 1, pp. 165–175, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jiao Y, Zhang Y, Wang Y, Wang B, Jin J, and Wang X, “A novel multilayer correlation maximization model for improving CCA-based frequency recognition in SSVEP brain–computer interface,” International journal of neural systems, vol. 28, no. 04, p. 1750039, 2018. [DOI] [PubMed] [Google Scholar]
- [33].Cherkassky V and Ma Y, “Practical selection of SVM parameters and noise estimation for SVM regression,” Neural networks, vol. 17, no. 1, pp. 113–126, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.