Abstract
Background
We sought to identify microRNAs (miRNAs) associated with response to anti-TNF-α or glucocorticoids in children with inflammatory bowel disease (IBD) to generate candidate pharmacodynamic and monitoring biomarkers.
Methods
Clinical response was assessed by Pediatric Crohn’s Disease Activity Index and Pediatric Ulcerative Colitis Activity Index. Quantitative real-time polymerase chain reaction via Taqman Low-Density Array cards were used to identify miRNAs in a discovery cohort of responders (n = 11) and nonresponders (n = 8). Seven serum miRNAs associated with clinical response to treatment, along with 4 previously identified (miR-146a, miR-146b, miR-320a, miR-486), were selected for further study. Candidates were assessed in a validation cohort of serum samples from IBD patients pre- and post-treatment and from healthy controls. Expression of miRNA was also analyzed in inflamed mucosal biopsies from IBD patients and non-IBD controls.
Results
Discovery cohort analysis identified 7 miRNAs associated with therapeutic response: 5 that decreased (miR-126, miR-454, miR-26b, miR-26a, let-7c) and 2 that increased (miR-636, miR-193b). In the validation cohort, 7 of 11 candidate miRNAs changed in the same direction with response to anti-TNF-α therapies, glucocorticoids, or both. In mucosal biopsies, 7 out of 11 miRNAs were significantly increased in IBD vs healthy controls.
Conclusions
Five candidate miRNAs associated with clinical response and mucosal inflammation in pediatric IBD patients were identified (miR-126, let-7c, miR-146a, miR-146b, and miR-320a). These miRNAs may be further developed as pharmacodynamic and response monitoring biomarkers for use in clinical care and trials.
Keywords: inflammatory bowel disease, microRNA, biomarker, pharmacodynamic
Discovery and validation cohort analyses were conducted to identify serum microRNAs associated with therapeutic response. Plausibility was refined by evaluation of expression in inflamed mucosal biopsies. Five serum pharmacodynamic and candidate monitoring biomarkers for pediatric IBD were identified.
INTRODUCTION
Inflammatory bowel disease (IBD) is treated with multiple therapies, both serially and concomitantly. Therapies are selected based upon stratification of disease phenotype and severity. Evaluation of response to therapy in pediatric IBD includes the calculation of clinical scores, such as the Pediatric Crohn’s Disease Activity Index (PCDAI)1 or the Pediatric Ulcerative Colitis Activity Index (PUCAI)2 and clinician-reported outcome measures, which incorporate symptoms, clinical exam findings, and laboratory results. These scores are monitored over time, along with fecal and blood biomarkers, cross-sectional imaging, and endoscopic assessments. About one third of IBD patients treated with antitumor necrosis factor (TNF)-α therapies are primary nonresponders to therapy, and about 10% develop loss of response over time.3 Objective, noninvasive approaches to monitoring progression of disease and response are expected to enable more efficient optimization of dosing and use of combination therapies, and avoidance of overescalation of therapy or using polypharmacy unnecessarily.
In clinical practice, particularly in pediatrics, the feasibility of serial assessments is difficult, as endoscopic and histologic assessments require uncomfortable, invasive, and costly procedures. Endoscopic surveillance of disease response typically requires waiting 3 to 6 months after treatment adjustment, which complicates timely optimization of therapy. Radiologic techniques, such as contrast-enhanced ultrasound, are being used to monitor disease response/progression,4, 5 though interpretation of disease improvement is subjective, and changes in transmural inflammation may occur at variable rates. A biomarker that measures disease progression or response to therapy over time is known as a monitoring biomarker.6 Objective serum-monitoring biomarkers could provide an alternative method of measuring disease activity and potentially allow for earlier assessment of response and optimization of therapy.
In clinical trials, endoscopic end points allow for objective disease activity assessment and central blinded reading to allow reproducibility. Histologic improvement is emerging as a gold-standard in disease assessment after therapy because it can be correlated with longer-term outcomes such as risk of relapse, need for surgery, and hospitalization.7–10 Monitoring biomarkers may be used as pharmacodynamic (PD) biomarkers to identify those patients who are early responders to drugs in clinical trials, thereby facilitating randomized, placebo-controlled trial enrichment. Pharmacodynamic biomarkers may also be used to demonstrate an exposure-response relationship in early phase drug trials, aiding in modeling and extrapolation to other groups of patients. Less invasive PD biomarkers indicative of response are important for pediatric IBD clinical care and drug development, as frequent serial colonoscopies are especially problematic in this population.
There are limitations to currently utilized biomarkers in IBD. Serum erythrocyte sedimentation rate and C-reactive protein may be normal in one fourth to one third of newly diagnosed pediatric IBD patients.11, 12 European consensus guidelines for management of pediatric Crohn’s Disease state that fecal calprotectin is useful for monitoring the resolution of intestinal inflammation, but the cutoff values that should trigger management changes are unknown.13
We previously sought to increase the range of serum PD/response biomarkers for disease activity in IBD and have identified serum proteins and microRNAs (miRNAs) that were associated with response to glucocorticoids (GCs), anti-TNF-α treatment, or both.14 Pharmacodynamic biomarkers may provide an early indication of efficacy. However, to be clinically useful, there must be evidence that biomarker changes are indicative of a drug’s effect on a clinical end point of interest. In this current study, we sought to identify the treatment-responsive serum miRNA biomarkers that were most correlated with clinical response using established pediatric IBD clinical scores (PUCAI and PCDAI). These PD biomarkers could also be considered candidates for further study as monitoring biomarkers. To further understand the biological plausibility, we assessed whether miRNA candidates showed variable expression in actively inflamed intestinal biopsies obtained from IBD patients compared with controls.
MATERIALS AND METHODS
Ethical Approval
Institutional approval was obtained from the Children’s National Hospital institutional review board and was in accordance with all requirements.
Patient Enrollment and Sample Collection
Patients ages 4 to 21 years cared for at Children’s National Hospital were eligible for this observational study if they were undergoing ileocolonoscopic evaluation for suspected IBD or were previously diagnosed with IBD but were not taking any medications for IBD other than mesalamine. Informed consent and/or informed assent, if applicable, were obtained from all patients or legal guardian(s) before enrollment. Pretreatment blood samples and pinch mucosal biopsies were obtained. If the patient was given a non-IBD diagnosis after ileocolonoscopy (eg, functional abdominal pain), samples were used as controls. Post-treatment blood samples were obtained before the addition of another therapy. In patients treated with ant-TNFα therapy, the post-treatment sample was collected before the 3rd or 4th infusion. All patients were characterized as “responders” or “nonresponders”; response to therapy was defined as a decrease in PCDAI by 12.5 points or a decrease in PUCAI by 20 points. Data were maintained on a password-protected Microsoft Excel file.
At each blood draw, 10 mL of whole blood was collected in a red top BD Vacutainer, and tubes were inverted about 5 times and allowed to clot at room temperature. Specimen was centrifuged for 15 minutes between 1110–1300 g. Four random mucosal biopsies (one each from the terminal ileum, ascending colon, descending colon, and rectum) were obtained and frozen by immersion into isopentane, which was precooled on liquid nitrogen. Serum and biopsies were stored in Sarstedt polypropylene tubes in a −80°C freezer. Crohn’s Disease Endoscopic Index of Severity (for Crohn’s disease patients) and Mayo endoscopic scores (for ulcerative colitis patients) were recorded by the endoscopist at the time of endoscopy. Areas of visually inflamed mucosa were confirmed histologically.
Discovery of Serum miRNAs by Taqman Low-density Array
RNA was isolated from 150 µL of serum according to ThermoFisher protocol for Trizol LS liquid extraction. Total RNA was converted to cDNA using ThermoFisher High-Capacity Reverse Transcription Kit and multiplexed RT primers. Synthesized cDNA was preamplified according to ThermoFisher preamplification protocol using PreAmp MasterMix, and multiplexed TM primers corresponding to the RT primers were used in initial cDNA reaction. MiRNA analysis was completed via a single Taqman Low-Density Array Card (Thermofisher, TaqMan Array Human MicroRNA A Cards v2.0), capable of assaying 384 miRNAs. Relative quantification (Rq) was used to determine change in expression of miRNA in serum samples pre- and post-treatment. Specifically, a value >1 indicates an increase, and a value <1 indicates a decrease in miRNA expression in post-treatment serum compared with pretreatment sample. Paired t test was used to compare miRNAs in pre- and post-treatment groups; P ≤ 0.05 was considered significant.
Validation of Serum miRNA Biomarker Candidates in an Independent Sample Set
miRNAs that were significantly correlated with response in the discovery set analysis were identified. These candidates, plus 4 additional miRNAs associated with therapeutic response that were previously identified by our group (miRNA-146a, 146b, 320a, and 486), were further assayed in another cohort of IBD patients.14
RNA was isolated from 150 µL of serum according to ThermoFisher protocol for Trizol LS liquid extraction. Total RNA was converted to cDNA using ThermoFisher High-Capacity Reverse Transcription Kit and multiplexed RT primers. Synthesized cDNA was preamplified according to ThermoFisher preamplification protocol using PreAmp MasterMix and multiplexed TM primers corresponding to the RT primers used in initial cDNA reaction. MiRNAs were quantified with individual TaqMan assays on an ABI QuantStudio 7 real-time polymerase chain reaction (PCR) machine (Applied Biosystems; Foster City, CA). Used assay IDs include hsa-miR-193b, assay ID 002367; hsa-miR-454, assay ID 002323; hsa-miR-let7c, assay ID 000379; hsa-miR-26a, assay ID 000405; hsa-miR-26b, assay ID 000407; hsa-miR-636, assay ID 002088; hsa-miR-126, assay ID 002228; hsa-miR-146b, assay ID 001097; hsa-miR-146a, assay ID 000468; hsa-miR-320, assay ID 002277; hsa-miR-486, assay ID 001278; hsa-miR-342–3p, assay ID 002260; and hsa-miR-150, assay ID 000473. Expression levels of all miRNAs were normalized to the geometric mean of multiple control genes (miR-150 and miR-342–3p) shown previously to be stable in IBD patient serum.14, 15 Pre- to post-treatment change in expression was analyzed via paired t test analysis, including assessment of directionality. P ≤ 0.05 was considered significant. Baseline anti-TNF-α cohort samples were compared with non-IBD controls using a unpaired Student t test. Data are presented as mean ± SEM unless otherwise noted.
Assessment of Candidate microRNAs in Inflamed IBD and Control Mucosal Biopsies
During diagnostic ileocolonoscopies, 4 biopsies per patient were obtained from non-IBD controls (n = 2) and IBD patients (n = 2 Crohn’s disease, n = 2 ulcerative colitis). Inflammatory bowel disease biopsies were assessed histologically for active inflammation; those with no histologic inflammation were not used. Each biopsy was used as a sample within the inflamed IBD group (n = 11) or the non-IBD control group (n = 8). RNA was isolated via ThermoFisher protocol for Trizol extraction. Total RNA was converted to cDNA using ThermoFisher High-Capacity Reverse Transcription Kit and multiplexed RT primers. Synthesized cDNA was preamplified according to ThermoFisher preamplification protocol using PreAmp MasterMix and multiplexed TM primers corresponding to the RT primers used in initial cDNA reaction. The miRNAs were then quantified using individual TaqMan assays on an ABI QuantStudio 7 real-time PCR machine (Applied Biosystems, Foster City, CA). Assay IDs match those in the validation studies. Expression levels of all miRNAs were normalized to the geometric mean of multiple control genes (sno234, U6, and RNU48). The miRNA expression in IBD biopsies was compared with controls using an unpaired t test. P ≤ 0.05 was considered significant; data are presented as the mean ± SEM unless otherwise noted.
RESULTS
Discovery of Novel Serum Candidate miRNA Biomarkers Associated With Therapeutic Response
Ten patients with CD received anti-TNF-α treatment (Remicade, n = 6; Inflectra, n = 4). All 10 patients received treatment via standard 5 mg/kg IV induction dose and schedule. Four patients with CD and 5 patients with UC were treated with intravenous or oral GCs; GC dosing for all included was 1 mg/kg daily to a maximum of 40 mg daily. Of the 19 patients, 11 were responders (average PCDAI decrease of 27.5, PUCAI of 55), and 8 were nonresponders (average decreased PCDAI of 6.5, PUCAI of 4). Demographics are shown in Table 1. Three patients, all in the nonresponder group, were taking mesalamine at the time of pre-and post-treatment sampling.
TABLE 1.
Discovery of Serum Candidate miRNA Biomarkers Associated With Treatment Response. Shown Are Demographics of the Discovery Cohort, Separated into Responder and Nonresponder Groups.
| Responders | Nonresponders | |
|---|---|---|
| N | 11 | 8 |
| Age in years (mean ± SD) | 14.8 ± 3.52 | 14.5 ± 2.67 |
| Males: Females | 5:6 | 3:5 |
| Diagnosis Crohn’s Disease Ulcerative Colitis |
9 2 |
5 3 |
| Months since Diagnosis (mean ± SD) | 1.45 ± 3.12 | 1.92 ± 3.6 |
| Paris Classification37 Crohn’s Disease |
||
| Distal Ileum ± Cecal disease | 2 | 1 |
| Distal Ileum ± Cecal disease; Upper disease proximal to Ligament of Treitz | — | 1 |
| Colonic | 2 | — |
| Colonic; Upper disease proximal to Ligament of Treitz | 1 | — |
| Ileocolonic | 4 | 3 |
| Ulcerative Colitis Left sided UC, Never severe |
2 | — |
| Left sided UC, Never severe | — | 1 |
| Extensive UC, Ever severe | — | 1 |
| Pancolitis, Never severe | — | 1 |
| Glucocorticoids (n = 4 CD, 5 UC) | ||
| Days between pre-and post-treatment sample collection (mean ± SD) | 18.25 ± 9.43 | 17.2 ± 11.48 |
| Anti-TNF-α (n = 10 CD) | ||
| Days between pre-and post-treatment sample collection (mean ± SD) | 60.57 ± 27.97 | 53.34 ± 32.52 |
| N with concurrent mesalamine at time of both pre- and post-treatment samples | 0 | 3 |
| PCDAI (mean ± SD) | ||
| Pretreatment | 40 ± 15.6 | 29.5 ± 14.40 |
| Post-treatment | 12.5 ± 11.9 | 23 ± 10.95 |
| PUCAI (mean ± SD) | ||
| Pretreatment | 62.5 ± 3.54 | 38 ± 25.63 |
| Post-treatment | 7.5 ± 10.60 | 34 ± 22.87 |
Seven miRNA biomarker candidates showed a significant change from before to after treatment in treatment responders but not in nonresponders (shown in Table 2). Three of these miRNAs showed no apparent change in nonresponders (miR-454, miR-126, and miR-26b). One miRNA changed in the opposite direction in nonresponders (let-7c). One miRNA showed a greater than 2-fold reduction in change (miR-636) in the nonresponder group compared with the responders. Two of the candidates, miR-193b and miR-26a, showed a larger change in relative quantification (Rq) in nonresponders than in responders; however, the change did not reach the threshold for significance within the nonresponder group due to variability. Thus, these 7 candidates were brought forward for downstream serum confirmation in a drug-specific cohort and for analysis in mucosal biopsy studies.
TABLE 2.
Differential Expression of 7 Serum miRNAs in the Discovery Cohort, Separated into Responder and Nonresponder Groups and Indicating the Direction of Change from Pre- to Post-treatment
| Responders | Nonresponders | |||||
|---|---|---|---|---|---|---|
| miR | ↑ or ↓ | P | Rq | ↑ or ↓ | P | Rq |
| 193b | ↑ | <0.001 | 7.81 | ↑ | Ns | 15.1 |
| 636 | ↑ | 0.01 | 6.71 | ↑ | Ns | 3.28 |
| 454 | ↓ | 0.02 | 0.11 | n.d. | Ns | 1 |
| 126 | ↓ | 0.02 | 0.32 | n.d. | Ns | 1 |
| 26b | ↓ | 0.02 | 0.11 | n.d. | Ns | 1 |
| let7c | ↓ | 0.02 | 0.14 | ↑ | Ns | 6.52 |
| 26a | ↓ | 0.04 | 0.16 | ↓ | Ns | 0.04 |
Abbreviations: Rq, relative quantification; n.d., no difference; Ns, not significant; P ≤ 0.05 significant.
Confirmation of miRNA Therapeutic Response in IBD Serum and Expression in Healthy Controls
Next, we performed a validation study on the 7 candidate miRNA biomarkers that were discovered, along with 4 miRNA PD biomarkers discovered previously,14 in serum from a new cohort of patients. Patients studied included those treated with anti-TNF-α (n = 8) and GCs (n = 8) and controls (n = 8; shown in Table 3). Three patients were counted in both the GC and anti-TNF-α groups; they were treated with GCs, which were tapered off, and then were subsequently started on anti-TNF-α for reinduction and maintenance therapy. Two patients in the anti-TNF-α treatment group and 1 patient in the GC treatment group were also taking mesalamine at baseline. All patients in this cohort were responders. Average decreases in PCDAI and PUCAI, respectively, were 25.83 and 37.55 in the GC treatment group and 40 and 41 in the anti-TNF-α treatment group.
TABLE 3.
Demographics of Validation Cohort Separated by IBD Treatment Groups and Controls.
| Anti-TNF-α | Glucocorticoids | Control | |
|---|---|---|---|
| N | 8 | 8 | 8 |
| Age in years (mean ± SD) | 12.38 ± 3.02 | 14.12 ± 4.88 | 13 ± 3.96 |
| Males: Females | 3:5 | 4:4 | 4:4 |
| Diagnosis Crohn’s Disease Ulcerative Colitis |
3 5 |
3 5 |
|
| Months since Diagnosis (mean ± SD) | 0.84 ± 0.84 | 9 ± 25.44 | |
| Paris Classification3 Crohn’s Disease |
|||
| Distal Ileum + Cecal disease | 0 | 1 | |
| Colonic | 1 | 2 | |
| Ileocolonic | 1 | — | |
| Ileocolonic; Upper disease proximal to Ligament of Treitza | 1 | — | |
| Ulcerative Colitis Left sided UC, Never severe |
— | 1 | |
| Extensive Colitis, Never severe | 2 | — | |
| Extensive Colitis, Ever severe | 1 | — | |
| Pancolitis, Never severe | 1 | 2 | |
| Pancolitis, Ever severe | 1 | 2 | |
| Days between pre-and post-treatment sample collection: median (mean ± SD) | 53.5 (61.3 ± 29.1) | 8.5 (15.5 ± 22.3) | |
| N with concurrent mesalamine at time of both pre- and post-treatment samples | 2 | 1 | |
| PCDAI (mean ± SD) | |||
| Pretreatment | 45 ± 13.0 | 50 ± 9.01 | |
| Post-treatment | 5 ± 8.66 | 24.17 ± 10.10 | |
| PUCAI (mean ± SD) | |||
| Pretreatment | 54 ± 22.47 | 70 ± 10.60 | |
| Post-treatment | 13 ± 10.37 | 32.5 ± 16.0 |
All IBD patients in this cohort were responders to therapy.
a13 patients with IBD were enrolled, 3 patients were treated with GCs, which were tapered, and subsequently with anti-TNF-α and included in both groups.
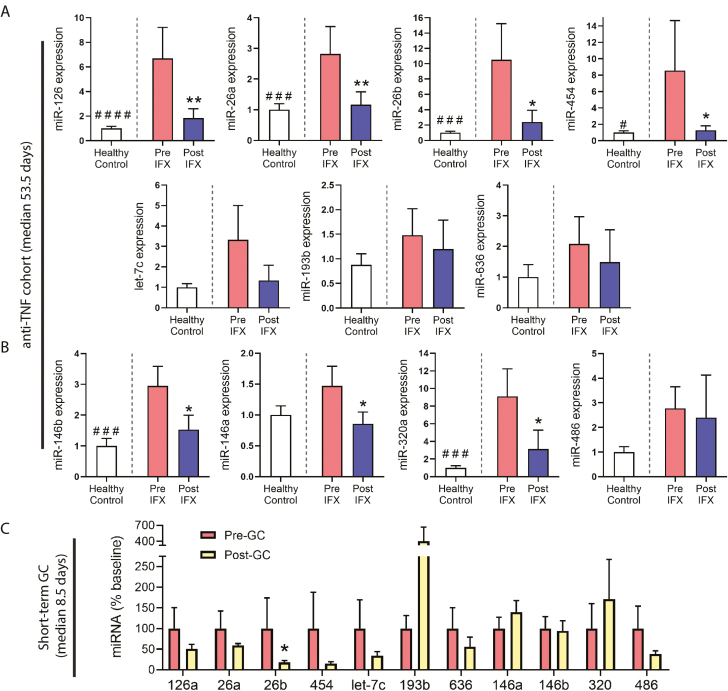
In the anti-TNF-α therapy group, 7 of the 9 miRNAs shown previously to decrease with treatment also showed a significant decrease after treatment in this validation cohort (Fig. 1A); these include miR-126, miR-26a (P ≤ 0.01), miR-26b, and miR-454 (P ≤ 0.05) from the previously mentioned discovery experiment, and miR-146a, miR-146b, and miR-320a (P ≤ 0.05) from our previous study. In this validation cohort, miR-193, miR-486, miR-454, and let-7c did not show significant changes, though let-7c trended toward significance in the same direction as the discovery set. The 2 miRNAs that increased after treatment in our discovery cohort, miR-193b and miR-636, did not show an increase in the validation cohort. In healthy control reference samples, all 11 candidate miRNAs showed at least a trend of decreased expression when compared with baseline IBD samples. Six of the miRNAs were expressed at significantly lower levels in healthy control samples, including miR-454 (P ≤ 0.05), miR-126, miR-26a, miR-26b, miR-146b, and miR-320a (P ≤ 0.005). These data validate the pharmacological response of a majority of these serum miRNA biomarkers in patients that respond to anti-TNF-α and show their dysregulation in diseased vs healthy serum at baseline.
FIGURE 1.
Serum miRNA expression in validation cohorts. Serum was obtained from patients at baseline and after treatment with either anti-TNF-α or GCs. A, B, Patients were treated with anti-TNF-α therapy for a median of 53.5 days (post-treatment sample drawn before third or fourth dose), then serum miRNAs assayed by real-time quantitative reverse transcription PCR (qRT-PCR) pre-and post-therapy. To illustrate expression levels in a healthy baseline state, non-IBD controls are also presented as a phenotypic healthy control reference. The miRNA expression is presented as fold expression relative to healthy control levels. Seven candidate miRNAs identified in the discovery cohort were assayed (A); in addition, 4 miRNA candidates from a prior study were assayed (B). C, Patients were treated with GCs for a median of 8.5 days, then serum miRNAs assayed pre- and post-therapy by qRT-PCR. (#P ≤ 0.05, ###P ≤ 0.005, ####P ≤ 0.0005, t test of healthy control reference vs baseline pre-IFX, n = 8 per group; *P ≤ 0.05, **P ≤ 0.01, paired 1-tailed t test comparing post-treatment to baseline in direction of discovery, n = 8 per group.)
In the GC treatment group, of 11 miRNAs tested, only miR-26b showed a significant decrease after treatment (P ≤ 0.05). Several others, including miR-126a, miR-26a, miR-454, miR-486, and let-7c, showed trends of decreased expression in response to treatment. The miRNA miR-193b, which increased in responders after treatment in the discovery experiment, also showed a trend of increased expression after GC treatment in the validation cohort. Three of the 4 miRNAs we identified previously as responsive to GCs (miR-146a, miR-146b, miR-320a) did not show a significant change after GC administration in this validation cohort. However, it is important to note that the median length of GC treatment in this cohort was 8.5 days, whereas the median length of GC treatment in Heier et al was 10 weeks. Therefore, this may account for the lack of validation of these miRNAs. These data may further support a potential time effect for some miRNA responses in serum, particularly if changes are reflective of anti-inflammatory or repair processes occurring at the level of the target tissue.
Corresponding Behavior of miRNA Biomarkers in IBD Vs Control Tissue Biopsies
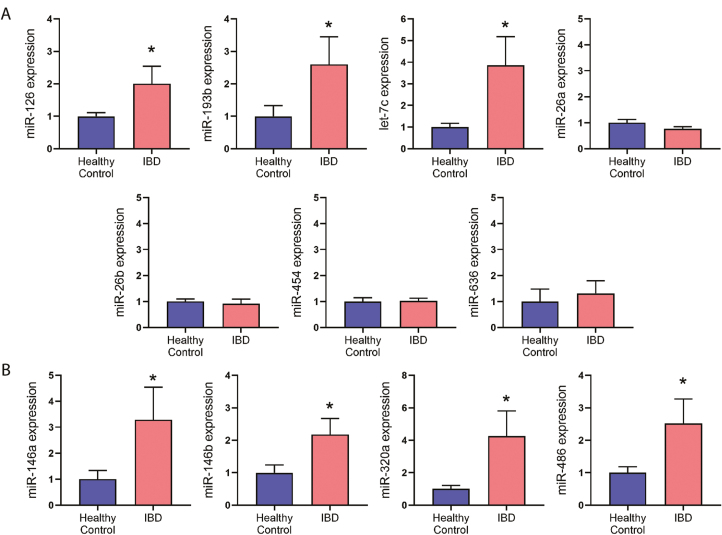
To gain insight into their potential source and behavior within diseased tissue, we analyzed expression of the 11 candidate miRNA biomarkers in mucosal biopsies from patients taken during diagnostic colonoscopies. Six patients were included in the analysis: 2 with CD, 2 with UC, and 2 controls. All IBD patients had active disease and were not on any biologics or GCs at the time of the biopsy collection (demographics are presented in Table 4). One patient had been previously exposed to anti-TNF-α therapy, which was discontinued 9 months prior (patient/parent decision). Four random biopsies were obtained from each patient, 1 each from terminal ileum, ascending colon, descending colon, and rectum. The presence of active inflammatory disease was noted by the endoscopist visually during endoscopy and confirmed histologically. For the IBD group, all biopsies confirmed to come from histologically inflamed bowel segments were grouped together for analysis. For control non-IBD patients, all biopsies were confirmed to be free of histologic inflammation, then grouped together and analyzed. Location of biopsies analyzed for each patient are shown in Table 4. Three of the 7 miRNAs identified in our previously mentioned discovery experiment showed significantly increased expression in inflamed IBD biopsies vs healthy controls (P ≤ 0.05): miR-126, miR-193b, and let-7c (Fig. 2). The remaining 4 miRNAs from this set showed no apparent change in inflamed tissue vs control tissue. Of the 4 miRNA biomarkers identified previously (miR-146a, 146b, 320a, and 486),14 all were significantly upregulated in inflamed vs control tissue biopsies (P ≤ 0.05).
TABLE 4.
Demographics from Patients Donating Colon Biopsies.
| Patients | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Diagnosis | CD | CD | UC | UC | Control | Control |
| Age (years) | 9 | 11 | 13 | 15 | 17 | 15 |
| Years since diagnosis | 0.25 | 4.9 | 3.5 | 0 | — | — |
| Disease location/Phenotype | Ileocolitis | Colonic; Upper disease proximal to Ligament of Treitz | Left sided colitis | Pancolitis | — | — |
| Biopsies analyzed in this study | TI, A, D, R | TI, A | A, Da | A, D, R | TI, A, D, R | TI, A, D, R |
| PCDAI | 37.5 | 17.5 | — | — | — | — |
| PUCAI | — | — | 60 | 75 | — | — |
Four random mucosal biopsies were obtained per patient from terminal ileum, ascending colon, descending colon and rectum. For this analysis, only tissue biopsies from endoscopically ulcerated areas of the bowel were analyzed from IBD patients, as indicated below. For controls, all biopsies were analyzed. Inflammation from inflamed areas was confirmed histologically on clinical biopsies.
aPatient 3: On colonoscopy, macroscopic evidence of left colitis involving the descending colon, sigmoid and rectum; Histologic inflammation of ascending and descending colon
Abbreviations: TI, terminal ileum; A, ascending colon; D, descending colon; R, rectum
FIGURE 2.
MiRNA expression in mucosal biopsies from IBD patients (areas of inflammation) and non-IBD controls. Four biopsies from each patient were collected during diagnostic colonoscopy; biopsied tissues included the terminal ileum, ascending colon, descending colon, and rectum. Only inflamed biopsies were assayed for the IBD group. A, B, Expression of candidate miRNA biomarkers was assayed in healthy noninflamed control biopsies vs IBD biopsies showing active inflammation (CD and UC patients combined). MiRNA expression is presented as fold expression relative to healthy control levels. Real-time quantitative reverse transcription PCR was used to analyze (A) expression levels of miRNA biomarker candidates associated with treatment response from the serum discovery cohort and (B) expression levels of miRNA biomarker candidates associated with treatment response in a previous study.14 (n = 8 biopsies collected from 2 healthy control patients, and n = 11 biopsies collected from 4 IBD patients; * P ≤ 0.05, Student t test.)
DISCUSSION
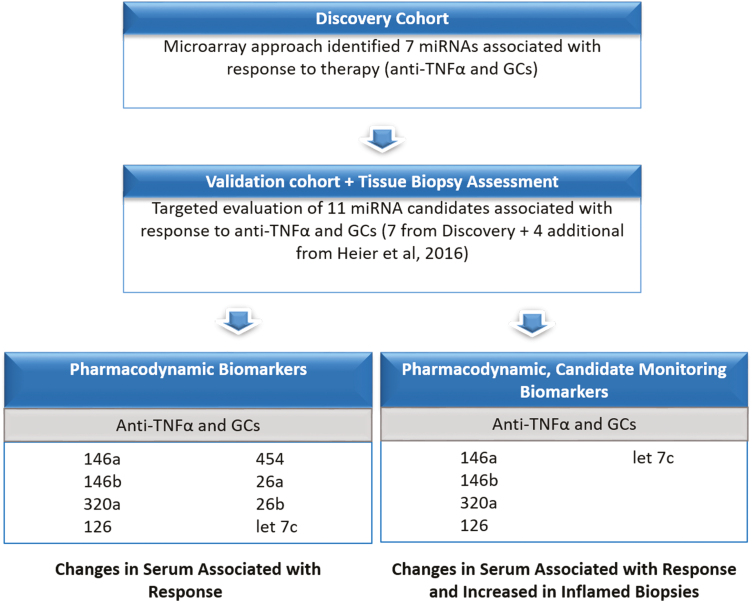
We and others have previously described the utility of miRNAs as biomarkers in IBD.14, 16–18Adding to our previous miRNA studies in pediatric IBD, here we have now identified 8 serum miRNA biomarkers that are associated with clinical response to anti-TNF-α therapy (after weeks) and GC treatment (after weeks)14 but (with the exception of miR-26b) not with shorter courses of GCs (after days). These are miR-126, miR-146a, miR-146b, miR-26a, miR-26b, miR-320a, miR-454, and let-7c. We then narrowed down this list to those biomarkers that are significantly elevated in inflamed IBD biopsies compared with controls, as this provides additional evidence that the biomarkers lie along the causal pathway of disease response: miR-146a, miR-146b, miR-320a, miR-126, and let-7c (see schematic in Fig. 3). These 5 candidate biomarkers hold potential as monitoring biomarkers in pediatric IBD and have all previously been implicated in the pathogenesis of IBD.
FIGURE 3.
This schematic shows how candidate PD monitoring biomarkers were selected. The first panel shows serum miRNAs that are associated with treatment response. These candidates were then narrowed down to 5 candidates that also showed increased expression in inflamed mucosal biopsies. These additional data provide more biological plausibility in demonstrating that biomarkers lie along the causal pathway of disease response.
In our study, serum miR-146a and miR-146b were correlated with clinical response, and both decrease with IFX treatment and longer-term GCs (weeks) but not with short term GCs (days). Expression of both miR-146a and miR-146b was increased in inflamed biopsy tissue. In studies of its acute effects, miR-146 is a negative regulator of innate immune signaling, acting as an inhibitor of NF-κB.19 However, chronic overexpression of miR-146 is associated with increased inflammation and disease pathology.20, 21 miR-146a and miR-146b are endotoxin responsive, and upregulation of miR-146a or miR-146b depends on the inflammatory stimulus.22 Inhibition of miR-146a in a murine model of ulcerative colitis alleviated disease activity through suppression of the TLR4/MyD88/NF-κB signaling pathway.23 The miRNA miR-146a has previously been shown to be more highly expressed in inflamed vs noninflamed biopsy tissue from pediatric ulcerative colitis and Crohn’s disease patients.24, 25 Circulating miR-146b has previously been described as a monitoring biomarker for IBD, correlating highly with endoscopic disease activity and with more specificity than serum C-reactive protein.26
In our study, serum miR-320a decreases in responders to infliximab (IFX) therapy and longer-term steroids (weeks)14 but not with shorter courses of GCs. The miRNA miR-320a is known to exist at higher levels in the quiescent colonic mucosa of ulcerative colitis and Crohn’s disease patients when compared with controls and may play a role in the sensitization of the quiescent mucosa to environmental factors27; thus, it is not unexpected that we found it to be more highly expressed in IBD tissue than in control tissue. However, miR-320a is also expressed more highly in noninflamed IBD tissue compared with inflamed IBD tissue27 and has been described as a negative regulator of NOD2.28 Interleukin-33 has been shown to induce epithelial-derived miR-320a, which plays an important role in intestinal epithelial restitution and recovery from acute colitis.29 Taken together, these data may imply decreased release of miRNA into blood as tissue inflammation resolves; thus, more miR-320a may be retained in the recovering intestinal mucosa when compared with serum.
We found that let-7c in serum decreases with anti-TNF-α therapy, decreases with GCs, and increases in IBD serum and inflamed biopsies; let-7c was also found to increase in nonresponders. Altaf-Ul-Amin et al examined the miRNA-target interactions between miRNA and genes previously implicated with IBD. Specifically, they found that let-7c may be highly related to IBD due to number of relevant miRNA-target interactions.30 The miRNA let-7c also promotes M2 macrophage polarization (anti-inflammatory) and suppresses M1 (pro-inflammatory) polarization.31
The miRNA miR-126 in serum decreases with anti-TNF-α and shows a trend of decreasing with GCs. It is higher in serum and inflamed biopsies from IBD patients than from controls. Additionally, miR-126 is an endothelial-enriched miRNA that participates in the control of leucocyte trafficking32 and an important regulator of the innate response.33, 34 Expression of miR-126 has been previously shown to be higher in IBD biopsies than controls, and miR-126 overexpression contributes to intestinal mucosal barrier dysfunction in vitro.35 Lastly, miR-126 has been shown to downregulate expression of Iκβα, an important inhibitor of NF-κB signaling pathway.36
Other miRNAs produced interesting findings but did not meet our criteria for being candidate monitoring biomarkers. For example, miR-486 is specifically responsive to GCs and shows a more variable response to anti-TNF-α therapy; it also more highly expressed in inflamed IBD biopsy tissue than in controls. Although miR-26a is a very consistent serum PD biomarker and was elevated in IBD serum compared with controls, it was not increased in inflamed tissue biopsies when compared with controls. Therefore, the PD effect of GCs on miR-486 may be drug-specific, and PD effect on miR-26a may be reflective of off-target effects, such as immunological or endocrinological dysregulation. Further studies are needed to understand these differences.
There are limitations to this study. First, due to the observational nature of this study, samples and other routine biomarker tests (fecal calprotectin, serum C-reactive protein) were obtained at variable times as part of ongoing medical treatment as provided by the primary pediatric gastroenterologist. This prohibited correlation of changes in miRNA biomarkers with changes in other biomarkers. Second, results may be affected due to small cohort size. In the discovery cohort, mean baseline PCDAI and PUCAI scores were lower in the nonresponders. This may suggest higher active inflammation in the responder group could have influenced results. The association of biomarkers to response is anchored to the PUCAI and PCDAI—and not to endoscopic findings. Biopsy tissue analysis was cross-sectional, not longitudinal. Finally, as patients in this study were treated for a much short duration with GCs before addition of other medications, these data may further support a potential time effect for some miRNA responses in serum, particularly if reflective of anti-inflammatory or repair processes occurring at the level of the target tissue. This is also supported by the finding that so many more miRNAs were found to change with the longer duration of anti-TNF-α therapy in this study.
In conclusion, pediatric IBD is a chronic autoinflammatory condition with unpredictable course and prognosis. Novel therapies need to be studied efficiently in pediatric IBD, and as more treatment options are available, there is a need for optimization and individualization of therapeutic interventions. We identified 5 candidate serum miRNAs—miRNA-146a, miRNA-146b, miRNA-320a, miRNA-126, and let-7c—which may be developed as monitoring and PD biomarkers in pediatric IBD. These candidate biomarkers may be further studied in other cohorts and in clinical trials.
ACKNOWLEDGEMENTS
Authors acknowledge the patients who participated in the study and the generous support of their division colleagues. They acknowledge Haeri Seol, Dr. Aswini Panigrahi, and Ellen Chaisson for assisting with sample processing and Dr. Eric Hoffman for suggestions on the manuscript.
Supported by: Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) T32HD087969 Postdoctoral Training in Pediatric Clinical Pharmacology (JvdA, SKB), NICHD 5U54HD090254 (JvdA, LSC, CRH), R00HL130035 (CRH), Department of Defense (grant W81XWH-17–1–047, AAF).
Conflicts of Interest: LSC and JvdA are part-time employees of ReveraGen BioPharma and own stock options in the company.
REFERENCES
- 1. Hyams JS, Ferry GD, Mandel FS, et al. Development and validation of a pediatric Crohn’s disease activity index. J Pediatr Gastroenterol Nutr. 1991;12:439–447. [PubMed] [Google Scholar]
- 2. Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423–432. [DOI] [PubMed] [Google Scholar]
- 3. Gisbert JP, Panés J. Loss of response and requirement of infliximab dose intensification in Crohn’s disease: a review. Am J Gastroenterol. 2009;104:760–767. [DOI] [PubMed] [Google Scholar]
- 4. Towbin AJ, Sullivan J, Denson LA, et al. CT and MR enterography in children and adolescents with inflammatory bowel disease. Radiographics. 2013;33:1843–1860. [DOI] [PubMed] [Google Scholar]
- 5. Anupindi SA, Podberesky DJ, Towbin AJ, et al. Pediatric inflammatory bowel disease: imaging issues with targeted solutions. Abdom Imaging. 2015;40:975–992. [DOI] [PubMed] [Google Scholar]
- 6. Dulai PS, Peyrin-Biroulet L, Danese S, et al. Approaches to integrating biomarkers into clinical trials and care pathways as targets for the treatment of inflammatory bowel diseases. Gastroenterology. 2019;157:1032–1043.e1. [DOI] [PubMed] [Google Scholar]
- 7. Chirstensen B, Erlich J, Gibson PR, et al. Histologic healing is more strongly associated with clinical outcomes in ileal Crohn’s disease than endoscopic healing. Clin Gastroenterol Hepatol. 2019; Dec 5. pii: S1542-3565(19)31396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christensen B, Hanauer SB, Erlich J, et al. Histologic normalization occurs in ulcerative colitis and is associated with improved clinical outcomes. Clin Gastroenterol Hepatol. 2017;15:1557–1564.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hefti MM, Chessin DB, Harpaz NH, et al. Severity of inflammation as a predictor of colectomy in patients with chronic ulcerative colitis. Dis Colon Rectum. 2009;52:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryant R, Burger D, Delo J, et al. Beyond endoscopic mucosal healing in UC: histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut. 2016;65(3):408–414. [DOI] [PubMed] [Google Scholar]
- 11. Turner D, Mack D, Hyams J, et al. C-reactive protein (CRP), erythrocyte sedimentation rate (ESR) or both? A systematic evaluation in pediatric ulcerative colitis. J Crohns Colitis. 2011;5:423–429. [DOI] [PubMed] [Google Scholar]
- 12. Tsampalieros A, Griffiths A, Barrowman N, et al. Use of C-reactive protein in children with newly diagnosed inflammatory bowel disease. J Pediatr. 2011;159:340–342. [DOI] [PubMed] [Google Scholar]
- 13. Ruemmele FM, Veres G, Kolho KL, et al. ; European Crohn’s and Colitis Organisation; European Society of Pediatric Gastroenterology, Hepatology and Nutrition Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J Crohns Colitis. 2014;8:1179–1207. [DOI] [PubMed] [Google Scholar]
- 14. Heier CR, Fiorillo AA, Chaisson E, et al. Identification of pathway-specific serum biomarkers of response to glucocorticoid and infliximab treatment in children with inflammatory bowel disease. Clin Transl Gastroenterol. 2016;7:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zahm AM, Thayu M, Hand NJ, et al. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr 2011;53: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soroosh AK, Pothoulakis C, Iliopoulos D. Functional role and therapeutic targeting of microRNAs in inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2018;314(2):G256–G62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaefer JA, Opekun A, Abraham B, et al. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunology. 2015;16(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neudecker VY, Bowser J, Eltzschig H. MicroRNAs in mucosal inflammation. J Mol Med. 2017;95(9):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taganov KD, Boldin MP, Chang KJ, et al. NF-κB-dependent induction of microRNA miR-146,an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma X, Zhou J, Zhong Y, et al. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci. 2014;11:810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eisenberg I, Eran A, Nishino I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paterson MR, Kriegel AJ. MiR-146a/b: a family with shared seeds and different roots. Physiol Genomics. 2017;49:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Want JP, Dong LN, Wang M, et al. MiR-146a regulates the development of ulcerative colitis via mediating the TLR4/MyD88/NF-kB signaling pathway. Eur Rev Med Pharmocol Sci. 2019;23(5):2151–2157. [DOI] [PubMed] [Google Scholar]
- 24. Szűcs D, Béres NJ, Rokonay R, et al. Increased duodenal expression of miR-146a and -155 in pediatric Crohn’s disease. World J Gastroenterol. 2016;22:6027–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Béres NJ, Szabó D, Kocsis D, et al. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:327–335. [DOI] [PubMed] [Google Scholar]
- 26. Chen P, Li Y, Li L, et al. Circulating microRNA146b-5p is superior to C-reactive protein as a novel biomarker for monitoring inflammatory bowel disease. Aliment Pharmacol Ther. 2019;49:733–743. [DOI] [PubMed] [Google Scholar]
- 27. Fasseu MT, Guichard C, Pedruzzi E, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PloS One. 2010;5(10):e13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pierdomenico MC, Cucchiara S, Vitali R, et al. NOD2 is regulated by mir-320 in physiological conditions but this control is altered in inflamed tissues of patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016;22(2):315–326. [DOI] [PubMed] [Google Scholar]
- 29. Lopetuso LR, De Salvo C, Pastorelli L, et al. IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proc Natl Acad Sci U S A. 2018;115:E9362–E9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ataf-Ul-Amin M, Karim MB, Hu P, Ono N, Kanaya S. Discovery of inflammatory bowel disease-associated miRNAs using a novel bipartite clustering approach. BMC Med Genomics. 2020; Feb 24; 13(Suppl 3):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Banerjee S, Xie N, Cui H, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190:6542–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poissonnier L, Villain G, Soncin F, Mattot VMiR126-5p repression of ALCAM and SetD5 in endothelial cells regulates leucocyte adhesion and transmigration. Cardiovasc Res. 2014;102:436–447. [DOI] [PubMed] [Google Scholar]
- 33. Agudo J, Ruzo A, Tung N, et al. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol. 2014;15:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorlacius-Ussing G, Schnack Nielsen B, Andersen V, et al. Expression and localization of miR-21 and miR-126 in mucosal tissue from patients with inflammatory bowel disease. Inflamm Bowel Dis 2017;23(5):739–752. [DOI] [PubMed] [Google Scholar]
- 35. Chen T, Xue H, Lin R, Huang Z. MiR-126 impairs the intestinal barrier function via inhibiting S1PR2 mediated activation of PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2017;494:427–432. [DOI] [PubMed] [Google Scholar]
- 36. Feng X, Wang H, Ye S, et al. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IκBα. Plos One. 2012;7:e52782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. [DOI] [PubMed] [Google Scholar]