Abstract
COPD is an umbrella definition encompassing multiple disease processes. COPD heterogeneity has been described as distinct subgroups of individuals (subtypes), or as continuous measures of COPD variability (disease axes). There is little consensus on whether subtypes or disease axes are preferred, and the relative value of disease axes and subtypes for predicting COPD progression is unknown. Using a propensity score approach to learn disease axes from pairs of subtypes, we demonstrate that these disease axes predict prospective FEV1 decline and emphysema progression more accurately than the subtype pairs from which they were derived.
Introduction
The heterogeneity of chronic obstructive pulmonary disease (COPD) obscures our understanding of its natural history and molecular mechanisms. COPD heterogeneity is often represented as distinct subgroups of subjects (subtypes), but it can also be represented as continuous axes of variability, i.e. disease axes[1]. A multi-cohort study demonstrated that subtypes identified by clustering were not reproducible across cohorts, whereas disease axes from the same cohorts were more consistent[2]. There is currently no consensus on the best approach to characterize COPD heterogeneity.
We define a subtype as a single subgroup of subjects and a COPD disease axis as any continuous representation of COPD heterogeneity. We describe a method, similar in concept to propensity scores[3], where a pair of COPD subtypes can be used to define a single disease axis by using the subtype pair as the response in a logistic regression model that predicts the likelihood of subtype membership. These predictions constitute a subtype-defined disease axis. For example, in the case of chronic bronchitis, the chronic bronchitis subtype is a binary yes/no classification based on patient symptoms. Conversely, the chronic bronchitis disease axis is a continuous measure derived from a predictive model that describes the propensity of each subject to have chronic bronchitis. Using longitudinal data from the Genetic Epidemiology of COPD (COPDGene) Study, we demonstrate that subtype-defined disease axes provide better prediction of prospective COPD progression than the original subtype pairs from which they were derived.
Methods
Subjects in COPDGene with complete five-year follow-up data were analyzed (n=4,726). Four general subtype classes were selected for study: Chronic Bronchitis (CB) per the ATS-DLD definition[4], the pink puffer (PP)/blue bloater (BB) subtype[5], frequent exacerbators (>=2 COPD exacerbations over the previous 12 months)[6], and upper/lower lobe emphysema predominant subjects with a log U/L ratio >1.5 for upper lobe or <−1.5 for lower lobe predominance. We refer to a subtype pair as two subtypes that are conceptually related and therefore used to construct a disease axis. For example, the CB subtype class yields a single subtype pair (chronic bronchitis present versus absent), whereas the PP/BB subtype class yields two pairs (PP/neither and BB/neither).
For each subtype pair, we used weighted logistic regression to identify a linear combination of predictors that provide optimal classification for that pair. The beta-coefficients of this regression were used to calculate the disease axis value for each analyzed subject. This software is available at https://github.com/Chen-Jxiang/SODA. We selected the baseline values of 27 variables to serve as the predictors in the regression models (see Supplemental Materials for variables used). Disease axes were generated only from visit 1 data.
For the analyses of COPD progression, separate regression models were used to relate subtypes or disease axis scores to either five-year change in FEV1 % of predicted or change in emphysema. To formally test for whether disease axes provide incremental improvement in prediction beyond that provided by a subtype pair, we constructed nested regression models in which a disease axis was added to a base model containing the original subtype pair. Additional information is included in the Supplement.
Results
A conceptual overview of our approach is shown in Figure 1. Subtype definitions and characteristics of the subjects are shown in ST 1 and 2. One disease axis was identified for each subtype pair, resulting in a total of six disease axes (one each for frequent/non-frequent exacerbators and presence/absence of chronic bronchitis, and two each for the Pink Puffer/Blue Bloater and Upper/Lower Emphysema subtypes). To determine how well the disease axes could correctly classify their original subtypes, we examined the discrimination performance which was excellent for the pink puffer/blue bloater and upper/lower emphysema subgroups (AUC > 0.98), and reasonable for the frequent exacerbator (AUC = 0.79) and chronic bronchitis subgroups (AUC = 0.67). The predictors and beta coefficients from these models are shown in ST 3 and 4.
Figure 1. Overview of Subtype-Oriented Disease Axis Approach.
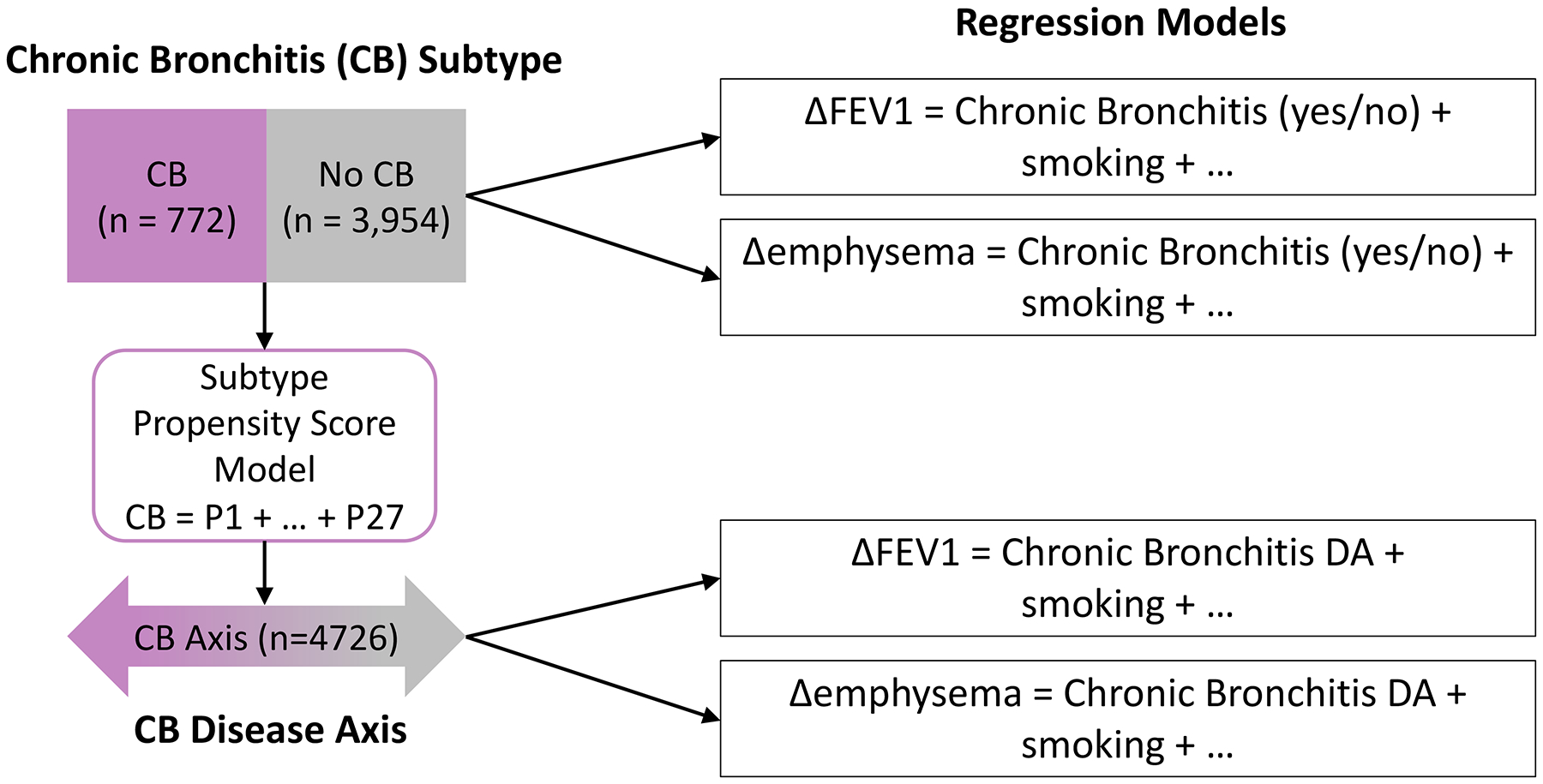
Chronic bronchitis (CB) subtypes are used as the response variable for a predictive model that uses 27 predictors (P1-P27) to classify subjects into the proper CB subtype group. The resulting predicted values constitute a continuous CB disease axis. Both subtype assignment and disease axis values are used as predictors in separate regression models in which 5-year change in FEV1 or emphysema serve as the response variables.
We then studied how well each subtype pair and its respective disease axis could predict two measures of COPD progression, change in FEV1 and quantitative CT emphysema progression. For both outcomes, we observed that regression models containing disease axes as predictors explained a greater proportion of the variance of COPD progression than similar models containing the subtype pair, with particularly marked improvement noted for emphysema progression (Table 1). To formally test for significant improvement in prediction from disease axes, for each subtype-containing model we added the corresponding disease axes and compared the two models (Table 2).
Table 1.
Regression models using either subtypes or disease axes to predict change in emphysema or change in FEV1 % of predicted.
| Progression Measure | Subtype Class | Subtype Pair | Subtype Models | Disease Axis Models | ||||
|---|---|---|---|---|---|---|---|---|
| % Variance Explained | % Variance Explained | |||||||
| Δ emphysema (Perc15) | Chronic Bronchitis | Chronic Bronchitis (No versus Yes) | 7.5 | 12.8 | ||||
| Frequent Exacerbator | Frequent Exacerbators (No versus Yes) | 7.5 | 8.0 | |||||
| Pink Puffer/Blue Bloater | PP/BB (Neither versus PP) | 7.6 | 8.5 | |||||
| Upper/Lower Emphysema | Upper/Lower Emphysema (Neither versus LLE) | 8.3 | 12.8 | |||||
| Δ FEV1 % of predicted | Chronic Bronchitis | Chronic Bronchitis (No versus Yes) | 6.0 | 6.4 | ||||
| Frequent Exacerbator | Frequent Exacerbators (No versus Yes) | 5.8 | 6.5 | |||||
| Pink Puffer/Blue Bloater | PP/BB (Neither versus PP) | 5.7 | 6.4 | |||||
| Upper/Lower Emphysema | Upper/Lower Emphysema (Neither versus LLE) | 5.7 | 5.8 | |||||
For each outcome and subtype class, two regression models were constructed including either the subtype(s) or disease axes as predictors. The COPD progression outcomes were either change in FEV1 or change in emphysema between visit 1 and 2. Values in the subtype model columns are from models that include categorical subtype assignment as a predictor, and values in the disease axis columns are from models that include the corresponding disease axes as a predictor. All models also include baseline FEV1 % predicted, baseline emphysema, and current smoking status at visit 1 and 2. PP/BB and Upper/Lower emphysema subtypes have three categories and thus 2 contrasts are included in the same model for each subtype group, respectively.
Perc15 – 15th percentile of the lung density histogram.
PP – Pink Puffer; BB – Blue Bloater; LLE = lower lobe emphysema predominant subtype; ULE – upper lobe emphysema predominant subtype.
Table 2.
Regression models using both subtypes and disease axes to predict change in emphysema or change in FEV1.
| Progression Measure | Subtype Class | Subtype Pair | Subtypes | Disease Axes | % Variance Explained | ||
|---|---|---|---|---|---|---|---|
| P | P | ||||||
| Δ emphysema (Perc15) | Chronic Bronchitis | Chronic Bronchitis (No versus Yes) | 0.78 | <0.001 | 12.8 | ||
| Frequent Exacerbator | Frequent Exacerbators (No versus Yes) | 0.27 | <0.001 | 8.0 | |||
| Pink Puffer/Blue Bloater | PP/BB (Neither versus PP) | 0.92 | <0.001 | 8.6 | |||
| Upper/Lower Emphysema | Upper/Lower Emphysema (Neither versus LLE) | 0.60 | <0.001 | 13.9 | |||
| Δ FEV1 % of predicted | Chronic Bronchitis | Chronic Bronchitis (No versus Yes) | <0.001 | <0.001 | 6.8 | ||
| Frequent Exacerbator | Frequent Exacerbators (No versus Yes) | 0.02 | <0.001 | 6.6 | |||
| Pink Puffer/Blue Bloater | PP/BB (Neither versus PP) | 0.20 | <0.001 | 6.4 | |||
| Upper/Lower Emphysema | Upper/Lower Emphysema (Neither versus LLE) | 0.63 | 0.20 | 5.9 | |||
For each outcome and subtype class, one regression model was constructed for each subtype class. This model contained the relevant categorical subtypes as well as the corresponding disease axes. The COPD progression outcomes were either change in FEV1 or change in emphysema between visit 1 and 2. All models also include baseline FEV1 % predicted, baseline emphysema, and current smoking status at visit 1 and 2. PP/BB and Upper/Lower emphysema subtypes have three categories and thus 2 contrasts are included in the same model for each subtype group, respectively.
Perc15 – 15th percentile of the lung density histogram.
PP – Pink Puffer; BB – Blue Bloater; LLE = lower lobe emphysema predominant subtype; ULE – upper lobe emphysema predominant subtype.
We also examined how well baseline DA values predicted the consistency of subtype assignment over time, which is an important issue for the CB and frequent exacerbator subtypes. We classified subjects as persistent or intermittent members of these subtypes according to their status at both COPDGene Study visits, and we observed that persistent subjects had higher DA values than intermittent subjects (Supplemental Figures 1 and 2, p<0.001 for CB and p=0.007 for frequent exacerbators).
Discussion
Previous work has shown that COPD variability typically occurs along a continuum[2]. Thus, while subtypes may have intuitive appeal, disease axes are more accurate. The method presented here turns subtypes into disease axes, providing more accurate representations of COPD heterogeneity that represent a continuum defined by two COPD subtypes. These disease axes were more predictive of COPD progression than the subtypes from which they were derived; because 1) disease axes “expand” subtype information to all subjects in a dataset, and 2) disease axes extract subtype-related information from a large number of input variables and thus contain more COPD-related information than subtypes alone.
Since this method uses pre-defined subtypes to guide data-driven analysis, the strengths of this approach are the interpretability of the disease axes and the improved prediction of disease progression. However, when the sole goal is prediction, purely data-driven methods may yield superior performance. These disease axes were generated in a single cohort, so independent assessment of their generalizability is needed. These results provide proof of concept that subtype-defined disease axes provide more powerful prediction of COPD progression. In the future, it would be useful to define disease axes that can be produced from readily available variables, which would allow disease axes to be generated in a larger set of COPD studies.
In summary, relative to subtypes, disease axes provide more accurate clinical predictions, and in the future disease axes may improve our clinical characterization of COPD and enable more powerful biological discovery.
Supplementary Material
Acknowledgements:
COPDGene Phase 3
Grant Support and Disclaimer
The project described was supported by Award Number U01 HL089897 and Award Number U01 HL089856 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
COPD Foundation Funding
The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens and Sunovion.
Disclosures: PJC reports consulting fees and grant support from GSK and Novartis outside the submitted work. MHC has received grant support from GSK. In the past three years, Edwin K. Silverman received honoraria from Novartis for Continuing Medical Education Seminars and grant and travel support from GlaxoSmithKline.
References
- 1.Kinney GL, Santorico SA, Young KA, et al. Identification of Chronic Obstructive Pulmonary Disease Axes That Predict All-Cause Mortality: The COPDGene Study. American Journal of Epidemiology 2018;187:2109–16. doi: 10.1093/aje/kwy087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castaldi PJ, Benet M, Petersen H, et al. Do COPD subtypes really exist? COPD heterogeneity and clustering in 10 independent cohorts. Thorax 2017;72:998–1006. doi: 10.1136/thoraxjnl-2016-209846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ROSENBAUM PR RUBIN DB. The central role of the propensity score in observational studies for causal effects. Biometrika 1983;70:41–55. doi: 10.1093/biomet/70.1.41 [DOI] [Google Scholar]
- 4.Ferris BG. Epidemiology Standardization Project (American Thoracic Society). Am Rev Respir Dis 1978;118:1–120. [PubMed] [Google Scholar]
- 5.Hersh CP, Make BJ, Lynch DA, et al. Non-emphysematous chronic obstructive pulmonary disease is associated with diabetes mellitus. BMC Pulm Med 2014;14:164. doi: 10.1186/1471-2466-14-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst JR, Vestbo J, Anzueto A, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med 2010;363:1128–38. doi: 10.1056/NEJMoa0909883 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


