Highlights
-
•
We evaluated Ansh Labs SARS CoV-2 IgG and IgM ELISA’s as part of orthogonal testing.
-
•
The IgG and IgM ELISA assays showed acceptable performance.
-
•
Concordance with RT-PCR was 100% > 6 days after symptom onset.
Keywords: Serology; SARS CoV-2 antibody; Validation; IgG, IgM
Abstract
Background
While the diagnosis of SARS-CoV-2 infection is primarily based on detection of viral RNA, the detection of SARS-CoV-2 antibodies is useful for assessing past prevalence of the disease, and in corroborating a current infection in challenging cases. Sensitive and specific immunoassays provide the ability to identify exposure to SARS-CoV-2, to determine seroconversion, to confirm eligibility for donation of convalescent plasma as well as play an essential part in epidemiological studies. We report on the validation of the Ansh Laboratories SARS-CoV-2 IgG and SARS-CoV-2 IgM ELISA immunoassays. These assays were evaluated for detection of anti-SARS-CoV-2 IgG and IgM antibodies for clinical use in our hospital as part of an orthogonal testing algorithm recommended by the CDC.
Methods
Diagnostic specificity and sensitivity of the IgG and IgM ELISA assays were tested using samples confirmed to be negative or positive for COVID-19 by RT-PCR. We also evaluated precision, analytical interference, and cross-reactivity with known cases of infection with other viruses. Additionally, we validated concordance with molecular and other serological testing and evaluated seroconversion in our patient population.
Results
The IgG and IgM ELISA assays showed acceptable precision, were robust to analytical interference and did not exhibit cross reactivity with specimens positive for common respiratory viruses. Both assays exhibited 95% agreement with a primary screening serological assay utilized at our institution as well as with a reference laboratory semi-quantitative method. Concordance with RT-PCR was excellent > 6 days after symptom onset (100%).
Conclusions
The Ansh SARS-CoV-2 ELISA assays have good analytical performance suitable for clinical use.
1. Introduction
Rapid global spread of SARS-CoV-2, the causative virus of COVID-19 disease, has led to over 12 million confirmed infections and >500,000 reported deaths worldwide [1]. Timely and accurate diagnosis of the SARS-CoV-2 infection is essential to provide appropriate treatment for patients and to limit the spread of virus. Laboratory diagnosis of SARS-CoV-2 infection is primarily based on viral RNA detection via RT-PCR. However, viral loads in upper respiratory tract secretions peak early during disease course and may quickly decline below the limit of detection for patients presenting later in the course of infection [2]. Moreover, in individuals who have recovered from COVID-19, a negative RT-PCR result provides no information about prior exposure. Recent studies suggest that combining RNA and antibody testing improves the sensitivity of diagnosis in COVID-19 patients in different phases of the disease [3], and provides a way to determine a past infection.
Serological tests are routinely used for diagnosis and management of many viral diseases to verify that an individual has had exposure to a pathogen and mounted an immune response [4]. In response to the urgent need for reliable antibody detection, there has been a rapid development in serological assays for SARS-Cov-2. Currently available serological tests for SARS-CoV-2 measure IgG, IgM, IgA or a combination of this antibodies [8]. IgM antibodies are known to develop earlier in infected patients and are most useful for determining acute infection, whereas IgG may not develop until later but remain present for a longer period of time [5]. However, it remains unknown for how long IgG or IgM antibodies to SARS-CoV-2 remain present in circulation after the infection has been cleared [6], [7].
The absence of recurrent cases of COVID-19 so far, and the success of convalescent plasma treatment in many cases, suggests that patients infected with SARS-CoV-2 may produce neutralizing antibodies against the virus. Studies suggest that the average time to seroconversion for IgM and IgG antibodies is 13 days after onset of symptoms [5], however, the titer or type of antibodies that confer protection are not yet established [8].
To assure the quality of the available tests, as of May 4, 2020, the FDA has required commercially marketed serologic tests for SARS-CoV-2 to receive Emergency Use Authorization (EUA) [9]. Additionally, to reduce the likelihood of a false-positive result and maximize the positive predictive value of testing, the CDC Interim Guidelines for COVID-19 Antibody Testing suggests an orthogonal testing algorithm so that individuals who are positive by one antibody test are retested with a second antibody test [10]. The increase in test specificity offered by the combination of two tests rises significantly when the viral antigen targeted of the two tests are distinct [10].
Recently our laboratory successfully validated and implemented a total anti-SARS-CoV-2 antibody test (CoV2T) on the Vitros 5600 automated chemistry analyzer [11]. To minimize false positive test results from the use of a single assay, and to further adhere to CDC’s recommendation of orthogonal testing algorithm, we validated IgG and IgM ELISA immunoassays for use as confirmatory testing. The ELISA assays target different epitopes of SARS-CoV-2 and permit distinction of antibody subtypes.
2. Materials and Methods
The SARS-CoV2 IgG and SARS-CoV2 IgM ELISA Immunoassays (Ansh Laboratories) were evaluated for use on the Dynex-DS2 automated immunoassay system. The SARS-CoV2 IgG assay uses SARS-CoV-2 recombinant proteins, targeting antibodies which recognize epitopes of the nucleocapsid (N) and spike (S) proteins, whereas the SARS-CoV2 IgM ELISA uses anti-human IgM capture antibody. The IgG and IgM ELISA assays are semiquantitative, and report measurements in standard arbitrary units (AU/ml). Samples with concentration < 10 AU/ml are considered non-reactive, samples > 12 AU/ml are considered reactive and samples with AU/ml 10–12 are considered equivocal. The manufacturer provides a set of 3 calibrators and three levels of controls. Mean absorbance readings for each of the calibrators is plotted along the y-axis versus the calibrator concentrations in AU/ml along the x-axis and calibration curve is achieved using a linear regression curve-fit.
Specimens for our validation study were obtained from healthy volunteers and from confirmed COVID-19 patients, under a protocol approved by the Baylor College of Medicine Institutional Review Board. Positive patients were previously diagnosed with COVID-19 by RT-PCR or TMA (Transcription-mediated amplification) methods at our institution, or at other in the Texas Medical Center (Baylor St. Luke’s and Ben Taub Hospitals). Patient specimens were collected by venipuncture into K2EDTA tubes or serum separator tubes and processed upon receipt by the laboratory, with plasma or serum stored for up to 5 days at 4 °C until analysis.
Intra- and inter-assay precision studies were performed in accordance with CLSI EP5-A2 guidelines on negative and positive specimens. Intra-run precision was assessed by measurement of 10 replicates within one run, and inter-assay precision was assessed by measurement of 3 samples once a day for 20 days. Assay precision was expressed as coefficient of variation (%CV). Accuracy was determined by comparing twenty samples measured in parallel, in a blinded manner, by a reference laboratory (Ansh Laboratories). Results were analyzed statistically using EP Evaluator. Clinical sensitivity was determined using 38 specimens with negative SARS CoV-2 RT-PCR results and 104 patient samples that were RT-PCR positive for SARS CoV-2. Samples were tested on different days, by different operators. Accuracy was assessed as concordance with the positive or negative RT-PCR status of the specimen. A subset of these (n = 113) for which we had enough sample volume, were also tested for concordance with the CoV2T Vitros Anti-SARS-Cov-2 Total antibody assay (IgG, IgA and IgM) (Ortho Clinical Diagnostics, Raritan, NJ). The CoV2T assay is a qualitative assay and results are reported as reactive (above the signal/cutoff threshold) or nonreactive (below the threshold).
Seroconversion in our patient population was assessed by correlation of chart review of 42 patients who were repeatedly assessed in our hospital. These patients were positive for SARS-CoV-2 by RT-PCR, had a known date of symptom onset, and were evaluated for anti-SARS-CoV-2 antibodies by IgG and IgM ELISA. Specimens were grouped by the number of days elapsed since the first reported symptom per patient history. Results were analyzed and graphed using GraphPad Prism.
Interference testing was performed by spiking viral RNA-confirmed negative or positive samples with known concentrations of hemoglobin, conjugated bilirubin, and triglyceride-rich lipid (Sun Diagnostics) and biotin (Millipore Sigma). Samples were measured in triplicates, results were considered acceptable if the difference from neat specimens was <15%.
The effect of the collection tube type was assessed by collecting specimens from five volunteers into serum separator, K2EDTA tubes or plasma-heparin tubes, and measuring the SARS-CoV2 IgG or IgM in resultant serum or plasma by IgG or IgM ELISA. Difference in antibody concentration between tube types was assessed.
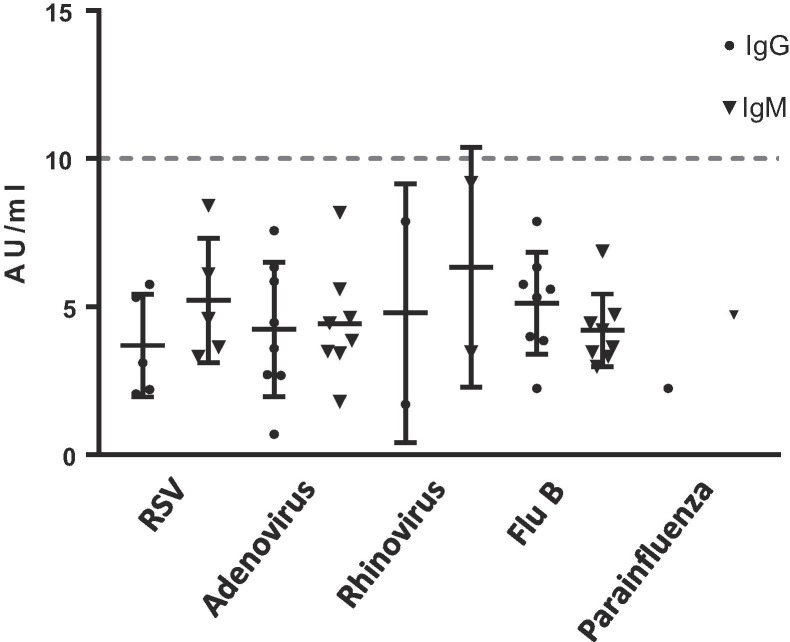
Analytical specificity was assessed by testing 19 different patient samples known to be positive for other viruses by molecular testing (including Influenza A, Influenza B, respiratory syncytial virus (RSV), adenovirus, rhinovirus), but negative for SARS-CoV-2 by RT-PCR (3 samples did not have RT-PCR result, but had no known exposure, travel history, or symptoms of COVID-19).
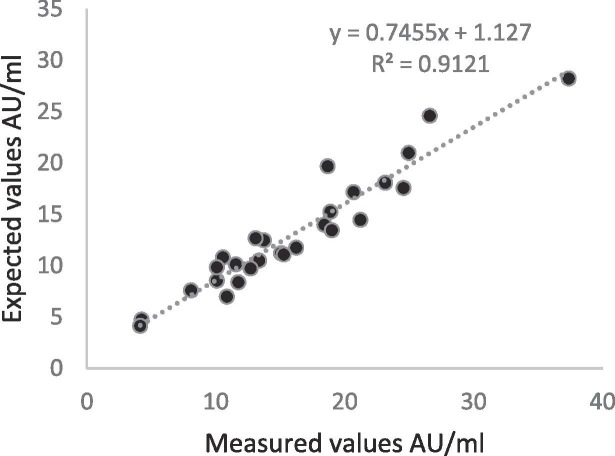
For dilution studies, positive samples were diluted with charcoal stripped human plasma in EDTA K2 (BioChemed) negative for presence of anti-SARS-CoV2 antibodies. Diluted samples of 9 positive specimens (27 dilutions from 1:200 to 1:700) were assayed as per manufacturer’s instruction for use. Expected AU/ml values were calculated including dilution factor and used to plot against measured values. In addition, because this is a semiquantitative assay, and could potentially be used in assessing the effectiveness of convalescent plasma, we examined the effect of serial dilutions of positive patient samples on the Ansh IgG ELISA assay. Statistical analysis was performed in EP Evaluator or GraphPad Prism. Results are given as mean ± SD.
3. Results
Both IgG and IgM ELISA assays demonstrated acceptable intra-assay precision for both negative and positive specimens. For the negative specimens, imprecision was 9.1% for IgG and 8.2% for IgM, while for the positive specimens, imprecision was 2.5% for IgG and 4.8% for IgM. Inter-assay imprecision was also acceptable, with CV for the negative specimen 13.7% for IgG and 12.4% for IgM and for the positive specimen 4.9% for IgG and 7.7% for IgM (Table 1 ).
Table 1.
Intra- and inter-assay precision study results.
| Intra-assay mean AU ± SD (%CV) |
Inter-assay mean AU ± SD (%CV) |
|||
|---|---|---|---|---|
| Sample | IgG ELISA | IgM ELISA | IgG ELISA | IgM ELISA |
| Negative | 2.06 ± 0.18 (9.1%) | 2.2 ± 1.8 (8.2%) | 2.2 ± 0.3 (13.7%) | 2.6 ± 0.3 (12.4%) |
| Positive | 64.2 ± 1.6 (2.5%) | 29 ± 1.4 (4.8%) | 63.8 ± 3.1 (4.9%) | 26.6 ± 2.05 (7.7%) |
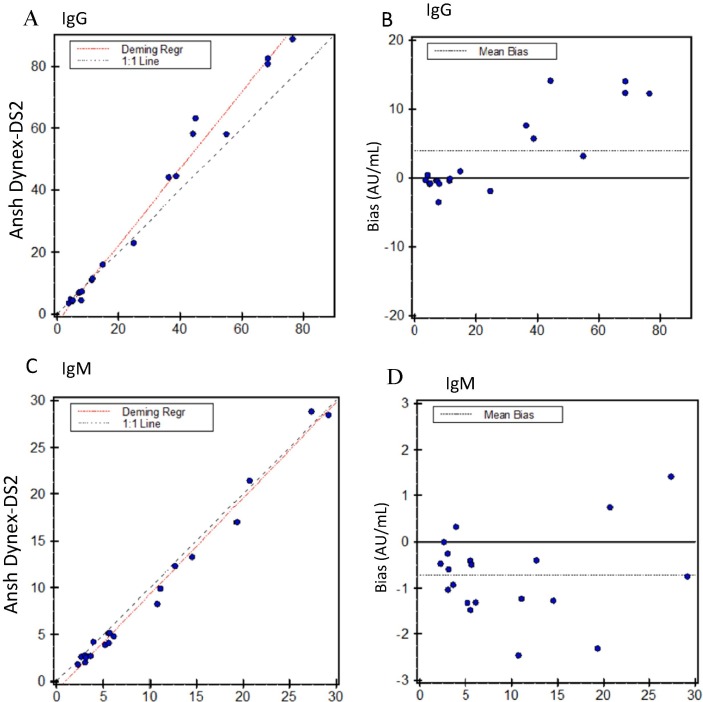
Accuracy studies with the reference laboratory indicated good agreement with the reference laboratory IgG and IgM ELISA assays, with correlation coefficients of 0.993 for the IgG, and 0.994 for the IgM assays respectively (Fig. 1 A, C). The IgG ELISA exhibited a non-significant slight positive bias relative to the reference laboratory, with a mean difference of 3.9 AU/ml (Fig. 1B). The IgM ELISA also exhibited a non-significant, negative bias relative to this assay, with a mean difference of −0.7 AU/ml (Fig. 1D).
Fig. 1.
Accuracy of the Ansh anti-SARS-CoV2 IgG and IgM ELISA. A, B. Deming regression (A) and Bland-Altman (B) analysis between Ansh Labs (reference laboratory) values and Ansh ELISA on the Dynex DS2 in our laboratory. Mean bias of the Dynex-DS2 relative to the manufacturer’s method was 3.9 AU/ml. C, D. Deming regression (C) and Bland-Altman (D) analysis between Ansh Laboratories values and the Ansh SARS-CoV2 measured on Dynex DS2 in our laboratory. Mean bias of the Dynex-DS2 relative to the manufacturer’s method was −0.7 AU/ml.
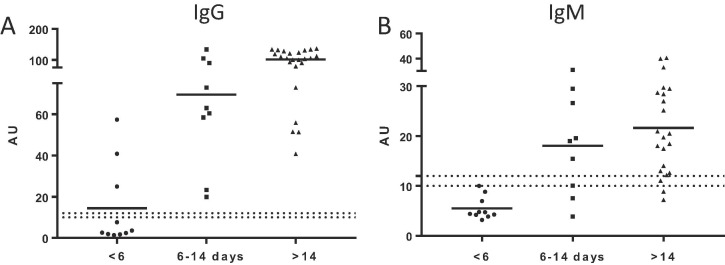
Clinical performance of IgG and IgM ELISA assays was compared with known SARS-CoV-2 RNA test results. The overall diagnostic specificity of the IgG ELISA was 94.7% (95% CI 82.7–98.5%, N = 38), and 97.4% (95% CI 86.5–99.5%, N = 38 for the IgM ELISA. Total diagnostic sensitivity is summarized in Table 2 . Anti-SARS-CoV-2 antibodies are usually detected between 7 and 14 days after symptoms onset [6]. We, therefore, correlated diagnostic specificity with days since onset of symptoms (Fig. 2 , Table 3 , and Supplementary Fig. 1). For the IgG ELISA, sensitivity at < 6 days post-symptom onset was 30% (3/10) with mean measured concentration of 14.4 AU/ml. Between day 6 and 14, sensitivity was 100% (N = 9) with mean measured concentration of 69.6 AU/ml, and for specimens collected >14 days post onset, sensitivity was 100% (N = 24) with mean measured concentration of 101.2 AU/ml. IgM ELISA sensitivity for <6d was 10% with mean measured concentration of 5.5 AU/ml, while at 6–14 days sensitivity was 77.7% with mean measured concentration of 18.1 AU/ml, and for specimens collected >14 days post symptom onset, 90.9% specificity was achieved with mean measured concentration of 21.6 AU/ml, equivocal samples were considered positive.
Table 2.
Overall clinical sensitivity of Ansh Anti-SARS-CoV-2 I3gG and IgM ELISA assays. Patients with a positive or negative RT-PCR test for SARS-CoV-2 were tested for seropositivity by IgG or IgM ELISA.
 |
Fig. 2.
Antibody reactivity by the Ansh Anti-SARS-CoV-2 IgG and IgM ELISA assays in patients positive for SARS-CoV-2 by RT-PCR, grouped by number of days since the first reported symptoms.
Table 3.
Antibody reactivity by the Ansh IgG and IgM ELISA assays in patients positive for SARS-CoV-2 by RT-PCR, grouped by number of days since the first reported symptoms (equivocal samples were considered positive).
| IgG |
IgM |
|||
|---|---|---|---|---|
| Days post-onset | % Positive | Mean AU | % Positive | Mean AU |
| < 6 | 30% (3/10) | 14.4 | 10% (1/10) | 5.5 |
| 6 to 14 | 100% (9/9) | 69.6 | 77.7% (7/9) | 18.1 |
| >14 | 100% (24/24) | 101.2 | 90.9% (20/22) | 21.6 |
There is no standard reference method for serological testing of SARS-CoV-2. We compared the Ansh IgG and IgM ELISA assays with other serological methods. We evaluated concordance with the Vitros 5600 CoV2T qualitative assay, which detects total IgG, IgA and IgM directed against SARS-Cov-2, and with a reference laboratory (Ansh Labs) ELISA for IgG and IgM assays. A total of 221 specimens were compared. Overall concordance of the Ansh IgG and IgM ELISA assays with the CoV2T assay was 0.96 (212/221). Positive percent agreement (PPA) between the IgG ELISA and the CoV2T assay was 94.4% (102/108) and negative percent agreement was 93.8% (106/113) (Table 4 ). Twenty split specimens were tested at the reference laboratory (Ansh Labs) using semi-quantitative assay for IgG and IgM antibodies separately. Positive and negative agreement were 100% for both the IgG and IgM ELISA assays.
Table 4.
Concordance between Ansh IgG ELISA assay and qualitative Vitros CoV2T assay and reference laboratory specific antibody testing.
| Vitros CoV2T |
Reference laboratory |
||
|---|---|---|---|
| IgG | IgM | ||
| (+) % Agreement | 94.4% (102/108) | 100% (10/10) | 100% (6/6) |
| (-) % Agreement | 93.8% (106/113) | 100% (7/7) | 100% (12/12) |
Interference studies were performed to examine the effect of common interferents on the Ansh IgG and IgM ELISA. Our results showed no changes in sample reactivity (CV < 15% from neat) when the positive sample was spiked with hemoglobin, conjugated bilirubin, or triglyceride-rich lipid or biotin (Table 5 ), at the concentrations tested. For the negative samples we recorded difference in interference with biotin for IgG (38.98%) and triglycerides for IgM (34.3%).
Table 5.
Summary of the interference studies. Known concentrations of interferent were spiked into a known nonreactive or reactive sample.
| Sample | Interferant | % Difference to control IgG | % Difference to control IgM |
|---|---|---|---|
| Negative sample |
Hemolysate 120 mg/dL |
3.30 | −6.50 |
| Conjugated bilirubin 30 g/dL | −8.40 | 12.70 | |
| Triglyceride-rich lipid 250 mg/dL | 4.80 | 34.30 | |
| Biotin (10,000 ng/ml) | 38.98 | 7.90 | |
| Positive sample | Hemolysate 120 mg/dL |
7.28 | −5.00 |
| Conjugated bilirubin 30 g/dL | 9.47 | 4.30 | |
| Triglyceride-rich lipid 250 mg/dL | 13.81 | 1.20 | |
| Biotin (10,000 ng/ml) | −6.31 | −5.92 |
The IgG and IgM ELISA instructions for use indicate that serum, lithium heparin plasma or K2EDTA plasma may be used for analysis. Across five specimens, we verified that all results were concordant regardless of tube type used (Table 6 ).
Table 6.
Effect of tube type on reactivity by IgG or IgM ELISA. Samples were collected into each of three tube types for the same individual (n = 10).
| Collection tube | IgG ELISA (mean AU/ml) |
IgM ELISA (mean AU/ml) |
|---|---|---|
| Serum | 10.07 | 2.6 |
| Plasma-heparin | 10.9 | 3.9 |
| K2EDTA | 11.5 | 3.4 |
Analysis of plasma from patients with other viral infections did not indicate cross-reactivity with the Ansh IgG or IgM ELISA assays. Nineteen patient specimens previously tested to be negative for SARS-CoV-2 by PCR, but positive for another respiratory viral infection by molecular analysis were nonreactive by both IgG and IgM ELISA assays (Fig. 3 ). Dilution linearity study for up to 1 in 600 dilution for IgG ELISA revealed a linear curve with R2 = 0.91 value (Fig. 4 ).
Fig. 3.
Analytical specificity studies with other respiratory virus-positive specimens. Virus-positive specimens, patient specimens previously tested positive for another virus by molecular methods were tested by Ansh Anti-SARS-CoV-2 IgG and IgM ELISA assays ay for reactivity.
Fig. 4.
Linear dilution analysis. Measured and expected values for positive sample dilution (1:200 to 1:700). Note that all samples undergo an onboard 1:100 dilution on the Dynex DS2, and the final dilution represented includes the onboard dilution. Graph represents expected AU vs measured AU.
4. Discussion
We evaluated the performance characteristics of the Ansh Anti-SARS-CoV2 IgG and IgM ELISA assays for isotype-specific antibody detection in our patient population. We found that both tests exhibited acceptable analytical precision, were concordant with RT-PCR positivity for SARS-CoV-2, and were concordant with other serological assays. Both assays were robust to common sample interferences, and were compatible with serum and with plasma from lithium heparin or EDTA tubes. The IgG and IgM ELISA assays also exhibited no cross-reactivity with antibodies to other common respiratory viruses. However, these studies were performed on a limited number of specimens. Considering other reports showing low but significant cross-reactivity of serological assay with other coronaviruses [12], it would be informative to evaluate specimens from patients with known infections with other coronaviruses.
We identified that the Ansh ELISA assays offered good specificity (IgG 95% and IgM 97%), which can contribute to a high positive predictive value (PPV) for these tests. However, our evaluation of this performance characteristic is limited by the lack of a standard reference method for serological testing. Additionally, the sensitivity of the test is limited by the kinetics of seroconversion in COVID-19. We identified low overall sensitivity prior to 6 days post-symptom onset, which is similar to previous reports. We found that time to seropositivity by the IgG and IgM ELISA was between 6 and 14 days. However, the small sample size (N = 9) for this time period limits our ability to accurately estimate sensitivity in this group. Presence of IgM antibodies is a marker for acute infection and IgM ELISA assays have a potential as a test for early diagnosis of patients who have been infected with SARS-CoV-2. Published data suggests that supplementary IgM test can be useful to assess subclinical patients and increases sensitivity of infection detection when compared to qPCR methods alone [13]. This test has significant value during pandemic where, rapid and correct diagnosis is essential to limit he spread of the virus.
Moreover, we relied on chart review to assess timing of symptom onset, and incomplete charting or incomplete patient history may have affected our estimation of disease onset and seropositivity kinetics. Our data are consistent with CDC recommendations, indicating that assessment of patient’s serological status before 14 days post symptoms is likely of limited value.
The performance of the Ansh SARS-CoV2 IgG and IgM ELISA immunoassays is similar to other serological assays for COVID19 [14], [15]. Antigen selection for serology testing is crucial for test specificity and sensitivity, however, the most appropriate viral antigen to use for detection of antibodies against SARS-CoV-2 has not been fully defined [16]. The Ansh SARS-Cov-2 IgG assay includes recombinant spike protein in addition to the nucleocapsid protein. The spike protein mediates entry of SARS-CoV-2 into host cells, and the nucleocapsid protein appears to be highly immunogenic.
Current FDA and CDC guidelines specify that serological testing should not to be used to diagnose an acute infection, but may be used in support of clinical assessment of patients who present late in their illness or in patients with sequelae from suspected previous infection (eg. Multisystem Inflammatory Syndrome in Children, MIS-C). Moreover, recent CDC guidelines recommend the orthogonal testing algorithm, where a second independent test is performed to confirm a positive result. Our institution has implemented a reflexive algorithm for COVID-19 serology testing, with initial screening for total antibodies using the Vitros Cov2T assay. Positive samples are reflexed to confirmatory testing by Ansh SARS-CoV2 IgG and IgM ELISA. This orthogonal testing algorithm permits both confirmation of the presence of the anti-SARS-CoV-2 antibodies, as well as identification of class of antibody (IgG or IgM). This information allows distinction between convalescence (IgG) and acute phase of illness (IgM) in our patient population. This distinction is particularly useful in patients who present with asymptomatic infection or atypical symptoms, to confirm or rule out acute infection.
The use of convalescent plasma in treatment of COVID-19 has been approved by the FDA on a compassionate use basis [17]. Serology testing provides a unique opportunity for identification of potential convalescent plasma donors. The FDA’s recently published guidance recommends a minimum neutralizing antibody titer of 1:160, and the European Commission’s guidance recommends a 1:320 titer [18], generating a growing demand for known high-titer plasma donations. Serum neutralization titer can be measured using plaque reduction neutralization test, but it is a complex and time-consuming assay not suitable for standard routine application. Therefore, results from several ELISA assays have been evaluated for their correlation with neutralization assay results [19]. Our preliminary dilution results (supplementary Fig. 2) identify positive specimens with an antibody titer up to 1:700, and highlight the possible utility of the Ansh IgG ELISA in identification of convalescent plasma donors. Being a semi quantitative assay, it appears to be useful in identifying titers in convalescent plasma donors. However, additional studies will be needed to validate the Ansh assay against the gold standard, plaque neutralization studies; the latter requires a Biosafety level 3 facilities and are not available at present.
In summary, we validated the Ansh Anti-SARS-CoV-2 IgG and IgM ELISA assays and successfully integrated these tests into the COVID-19 serology testing algorithm in our institution. We anticipate this assay will be a useful method for determination of prevalence of COVID-19 infection in our population and to evaluate the immune response in patients.
CRediT authorship contribution statement
Joanna Jung: Data curation, Writing - original draft, Writing - review & editing. Emily Garnett: Data curation, Writing - original draft. Purviben Jariwala: Data curation, Writing - review & editing. Hue Pham: Data curation, Writing - original draft. Rongrong Huang: Writing - review & editing. Eduardo Benzi: . Niveen Issaq: . Martin Matzuk: Writing - review & editing. Ila Singh: Writing - review & editing. Sridevi Devaraj: Writing - review & editing, Conceptualization, Investigation.
Acknowledgement
EG and JJ were supported by the Ching Nan Ou Fellowship Endowment. Some of the validation kits used in this study were provided by Ansh Laboratories, but they did not participate in study design, validation, or data interpretation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.09.023. Supplementary Fig. 1. Antibody responses of COVID-19 patients’ cohort (n=46) in relation to duration of disease symptoms. Supplementary Fig. 2. Performance of the Ansh IgG ELISA assay in the evaluation of antibody titers of seroconverted patients. Dotted line represents the cut-off for positivity.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.https://coronavirus.jhu.edu/us-map.
- 2.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 3.La Marca A., Capuzzo M., Paglia T., Roli L., Trenti T., Nelson S.M. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online. 2020 doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farnsworth C.W., Anderson N.W. SARS-CoV-2 Serology: Much Hype Little Data. Clin. Chem. 2020;66(7):875–877. doi: 10.1093/clinchem/hvaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang B., Meng Q.H. The laboratory's role in combating COVID-19. Crit. Rev. Clin. Lab. Sci. 2020;57(6):400–414. doi: 10.1080/10408363.2020.1776675. [DOI] [PubMed] [Google Scholar]
- 8.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 9.https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas.
- 10.https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html.
- 11.Garnett E., Jung J., Tam E., Rajapakshe D., Cheney S., Brown C., Cao J., Muldrew K., Singh I., Versalovic J., Devaraj S. Clinical Validation and Performance Evaluation of the Automated Vitros Total Anti-SARS-CoV-2 Antibodies Assay for Screening of Serostatus in COVID-19. Am. J. Clin. Pathol. 2020 doi: 10.1093/ajcp/aqaa157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., Jerome K.R., Mathias P.C., Greninger A.L. Performance Characteristics of the Abbott Architect SARS-CoV-2 IgG Assay and Seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00941-20. JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q., Sharma L., Wang L., Wang J. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padoan A., Cosma C., Sciacovelli L., Faggian D., Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. 2020;58(7):1081–1088. doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- 15.Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. Clinical Performance of Two SARS-CoV-2 Serologic Assays. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P.G., Thomas D., Sullivan D.J., Paneth N., Gehrie E., Spitalnik S., Hod E.A., Pollack L., Nicholson W.T., Pirofski L.A., Bailey J.A., Tobian A.A. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020;130(6):2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C.E.G.o.t.c.a.t.o.c.C.-p.A.a. (https://ec.europa.eu/health/blood_tissues_organs/covid-19_en).
- 19.Weidner L., Gänsdorfer S., Unterweger S., Weseslindtner L., Drexler C., Farcet M., Witt V., Schistal E., Schlenke P., Kreil T.R., Jungbauer C. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.