Abstract
PURPOSE
Plasma genotyping may identify mutations in potentially “actionable” cancer genes, such as BRCA1/2, but their clinical significance is not well defined. We evaluated the characteristics of somatically acquired BRCA1/2 mutations in patients with MBC.
METHODS
Patients with MBC undergoing routine cell-free DNA (cfDNA) next generation sequencing (73 gene panel) before starting a new therapy were included. Somatic BRCA1/2 mutations were classified as known germline-pathogenic mutations or novel variants, and linked to clinicopathological characteristics. The effect of the PARP inhibitor olaparib was assessed in vitro, using cultured circulating tumor cells (CTCs) from a patient with a somatically acquired BRCA1 mutation and a second patient with an acquired BRCA2 mutation.
RESULTS
Among 215 MBC patients, 29 (13.5%) had somatic cfDNA BRCA1/2 mutations (nine (4%) known germline-pathogenic and rest (9%) novel variants). Known germline-pathogenic BRCA1/2 mutations were common in younger patients (p=0.008), those with triple-negative disease (p=0.022), and they were more likely to be protein-truncating alterations and be associated with TP53 mutations. Functional analysis of a CTC culture harboring a somatic BRCA1-mutation demonstrated high sensitivity to PARP inhibition, while another CTC culture harboring a somatic BRCA2 mutation showed no differential sensitivity. Across the entire cohort, APOBEC mutational signatures (COSMIC Signatures 2 and 13) and the “BRCA” mutational signature (COSMIC Signature 3) were present in BRCA1/2 mutant and wild-type cases, demonstrating the high mutational burden associated with advanced MBC.
CONCLUSION
Somatic BRCA1/2 mutations are readily detectable in MBC by cfDNA analysis, and may be present as both known germline-pathogenic and novel variants.
Keywords: Metastatic breast cancer, Cell-free DNA, Somatic BRCA1/2 mutations, Tumor genotyping, PARP inhibitor
Introduction
Tumor genotyping is the central tenet of precision oncology and is increasingly becoming part of routine clinical care in breast cancer to identify actionable mutations for potential therapeutic intervention. However, tumor tissue genotyping of the primary tumor alone does not identify clonal evolution and mutations that are acquired during the course of treatment, some of which may be therapeutically relevant. Obtaining serial tumor biopsies from metastatic sites has been applied as proof of principle to guide successive therapeutic choices, but it is limited by the risks to the patient of an invasive procedure and accessibility of the metastatic site to biopsy, as well as biased sampling of a single tumor lesion in the midst of widespread sites of disease. Consequently, cell-free DNA (cfDNA) analyses or so-called “liquid biopsies” have emerged as an important strategy to monitor acquired cancer mutations and there has been a major rise in the clinical utilization of cfDNA assays (1–6).
While cfDNA assays are widely used in the clinic, the interpretation of multiple subclonal mutations and novel variants represents a major diagnostic challenge. This is particularly important for genes that have matched therapy approved in more traditional germline or tumor genotyping contexts, such as PARP inhibitors for patients harboring germline BRCA1/2 mutations (7, 8). Cancer predisposing heterozygous germline BRCA1/2 mutations, leading to somatic BRCA-null phenotypes, have been well studied in breast cancer (7–12), but de novo somatic BRCA1/2 mutations are thought to be rare in breast cancer. An analysis of The Cancer Genome Atlas (TCGA) noted the prevalence of somatic BRCA1 mutations in primary breast cancer as 1.55%, and somatic BRCA2 mutations as 1.68% (13). However, progression to metastatic breast cancer (MBC) is associated with an increased frequency of mutations, particularly in metastatic triple-negative breast cancer (TNBC), where mutations in components of the homologous recombination pathway are more common, and the percentage of somatic BRCA1 mutations is around 6% (14). Even in such cases of definitive acquired BRCA1/2 mutations identified by traditional tumor genotyping, the functional implications for sensitivity to PARP inhibition are not established. Additionally, unlike germline BRCA1/2 genotyping, where large datasets have been curated to help interpret pathogenic and silent genetic variants within various populations, there are no such guidelines for interpreting the even more diverse potential variations in BRCA1/2 that may be acquired somatically. These challenges are further magnified in cfDNA by the variable allele fractions and subclonal tumor cell populations that they represent. Given the general availability of cfDNA genotyping, its non-invasiveness as a diagnostic tool, and the potential for identifying impactful acquired mutations, its rapidly expanding applications require careful review before they are used to trigger therapeutic interventions.
The primary objective of this study was to understand the clinical and functional characteristics of somatic BRCA1 and BRCA2 mutations detectable by cfDNA in patients with MBC.
Methods
Study population
Patients with MBC who underwent cfDNA analysis as part of routine clinical care at the Massachusetts General Hospital before starting a new therapy from February 2015 to July 2017 were identified. The subset of patients with BRCA1 and/or BRCA2 mutations detectable by cfDNA analysis (Next generation sequencing (NGS)/Guardant360®) was determined. All consecutive patients with MBC who had Guardant360® testing during the aforementioned time interval were included, and no cases were excluded. A retrospective review of medical and pathology records, based on an IRB approved institutional protocol, was conducted to identify tumor subtype, patient demographics, germline BRCA1/2 testing results via standard commercial germline testing and subsequent review of all cfDNA results with Guardant360® to verify the somatic nature of the mutations identified, as a post-hoc analysis, tumor genotyping results (NGS, institutional platform), and treatment outcomes post-cfDNA testing. This research was conducted in accordance with recognized ethical guidelines, including the Declaration of Helsinki, and the retrospective review was conducted based on an IRB approved institutional protocol.
CfDNA analysis
CfDNA analysis was performed using Guardant360® testing, an NGS based clinical assay evaluating 73 genes. Guardant360® employs massively parallel and deep sequencing, with an analytic sensitivity of 0.1% mutant allele fraction (MAF), with quoted specificity above 99.9%, and clinical sensitivity of 85.0% (compared to 80.7% tissue sensitivity) (15). The average molecule count is about 8000 molecules, and the average single read depth is approximately 15,000 molecules. For BRCA1 (chromosome 17q21), exons 2–23 are included, and for BRCA2 (chromosome 13q13), exons 2–27 are included. A retrospective chart review of the Guardant360® cfDNA reports was performed to determine the presence of somatic BRCA1 or BRCA2 mutations, to identify coexisting cfDNA mutations, and to characterize clonality. Based on the MAF of co-existing alterations, we defined BRCA1 or BRCA2 mutations as clonal (MAF ratio of BRCA1/2 mutation/gene mutation with highest MAF ≥ 0.25) or subclonal (MAF ratio of BRCA1/2 mutation/gene mutation with highest MAF < 0.25) (16).
Guardant360® can identify both germline and somatic BRCA1/2 mutations in a single test. The germline versus somatic origin of a BRCA mutation is determined using a decision tree algorithm, which relies on annotation from external databases (ExAC, COSMIC, ClinVar, etc) and the observed MAF of the BRCA variant relative to other known germline variants. All identified variants are first annotated with information from external databases to identify those variants that are known germline variants. Thereafter, variants that have an insufficient annotation to determine their origin are evaluated further based on their observed MAF relative to that of nearby known germline variants, and then a beta-binomial significance test is applied, and the variant is scored as germline or somatic. Notably, somatic BRCA1/2 mutations are usually at a variant allele fraction two orders of magnitude lower than germline BRCA1/2 mutations (17).
While germline results were initially suppressed in Guardant360® testing reports, as a post hoc analysis we worked with the Guardant360® team to verify the somatic nature of detected mutations.
Somatic BRCA1/2 mutations were further classified as either known germline-pathogenic variants or as novel/unclassified variants by two independent genetic counselors, who were blinded to the Guardant360® reports. The genetic counselors evaluated the specific DNA variants seen in cfDNA, which were specifically requested from Guardant360® for this analysis. This classification was based on review of the ClinVar database (18) to identify variants that had high classification confidence (3 and 4 star review status) as of October 2018. For variants that had moderate to low classification confidence (2 stars or fewer), additional criteria such as classification reports from CLIA-certified germline genetic testing laboratories or review by consortia was evaluated. Lastly, for variants not currently in ClinVar, the likelihood of a loss of function variant (such as nonsense mutations, frameshifts, mutations in the ±1 or 2 splice site locations) was assessed, as outlined in ACMG/AMP guidelines for DNA variant classification (19). All variants not categorized as known germline-pathogenic by this analysis were then categorized as novel/unclassified, including the majority of missense mutations.
Tumor genotyping analysis
A chart review of tumor genotyping results from archival tumor tissue was performed to identify coexisting tumor mutations. An institutional NGS assay evaluating 98 genes for mutations and 91 genes for copy number changes (SNaPshot) was utilized for tumor tissue genotyping (20). This anchored multiplex PCR assay detects gene rearrangements, insertions and deletions, single nucleotide variants, and copy number changes present at allelic frequencies at 5% or higher with 100% analytical sensitivity and 100% analytical specificity (20). BRCA1 exons 2–23 and BRCA2 exons 2–27 are included in the assay. The time interval between cfDNA collection and the tumor tissue biopsy used for tissue genotyping was determined.
Mutation signature analysis
Mutation signatures were analyzed using Non-Negative Matrix Factorization (NMF) as described previously (21–23). Mutations detected by Guardant360® in the 29 BRCA-mutant patients were combined into a single “virtual patient,” which was then analyzed together with 785 breast cancer patients from The Cancer Genome Atlas (TCGA). These mutation calls from whole-exome sequencing (WXS) were obtained from the TCGA Unified Ensemble “MC3” Call Set (24), the public, open-access dataset of somatic mutation calls produced by the MC3 calling effort (“Multi-Center Mutation Calling in Multiple Cancers”), downloaded from the following link: http://www.synapse.org/#!Synapse:syn7214402/wiki/405297 (The results here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/ as outlined in the TCGA publications guidelines (http://cancergenome.nih.gov/publications/publicationguidelines)). Joint analysis of this combined TCGA+Guardant360® dataset by NMF (k=6) revealed mutation signatures corresponding to Aging (COSMIC Signature 1), APOBEC (COSMIC Signatures 2+13), the “BRCA” signature (COSMIC Signature 3), and MSI, microsatellite instability (COSMIC Signatures 6+26). The number and fraction of mutations due to each signature was then estimated and reported.
Statistical analysis
The association between cfDNA BRCA1/2 mutation status and patient age was determined with the Wilcoxon rank sum test, and associations with cfDNA BRCA1/2 mutation status and tumor subtypes, prior treatment, and first treatment post-cfDNA testing were performed with the Pearson’s chi-squared test. The impact of cfDNA BRCA1/2 mutation status on progression-free survival (PFS) on the first treatment post-cfDNA testing, and overall survival (OS) was determined with the log-rank test. Cox regression analysis was used to determine the hazard ratio of cfDNA BRCA1/2 mutation status on PFS and OS. In addition, a multi-variate analysis correcting for age and number of prior therapies was performed to determine the impact of cfDNA BRCA1/2 mutation status on PFS and OS. For all analyses, p<0.05 was considered statistically significant.
Establishing BRCA1 mutant ex vivo circulating tumor cell (CTC) culture
A cell line (Brx401) was established from CTCs enriched from a patient with a somatic BRCA1 mutation (in this study cohort #16). This patient had a known germline-pathogenic BRCA1 mutation (splice site SNV ENST00000357654.3:c.5075–1G>C) detectable in the cfDNA, which was acquired after treatment with chemotherapy for metastatic TNBC. The patient had no known germline BRCA1/2 mutation. A second CTC cell line (Brx142) was established from a patient with hormone receptor positive (HR+) MBC. In this case an early CTC culture showed wild type BRCA2, but a subsequent culture, acquired after treatment with an oral selective estrogen receptor degrader, identified a BRCA2 mutation (missense mutation E3071Q). Again, the germline testing showed no BRCA2 mutation. Additional CTC cultures were used as controls (wild type BRCA1/2) as previously described (4). For all CTC collections, written and signed informed consent was obtained as per institutional review board approved protocol. CTCs were isolated using the microfluidic CTC i-CHIP and ex vivo cultures were established as described previously (4). CTC cultures were routinely checked for mycoplasma with the MycoAlert-Lonza Kit and authenticated against matched blood sample via STR profiling by Genetica DNA Laboratories (a LabCorp brand; Burlington, NC) using the commercially available PowerPlex®16HSamplification kit (Promega Corporation; mouse marker included) and GeneMapper ID v3.2.1 software (Applied Biosystems).
Whole-exome sequencing
For whole-exome sequencing (WES), the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) was used for extraction of genomic DNA. DNA was quantified in triplicate using a standardized PicoGreen® dsDNA Quantitation Reagent (Invitrogen, Carlsbad, CA) assay. The quality control identification check was performed using fingerprint genotyping of 95 common SNPs by Fluidigm Genotyping (Fluidigm, San Francisco, CA). Library construction was performed using the KAPA Library Prep kit, with palindromic forked adapters from Integrated DNA Technologies. All library construction, hybridization and capture steps were automated on the Agilent Bravo liquid handling system. Flowcells were sequenced utilizing Sequencing-by Synthesis chemistry for HiSeq 4000 flowcells. Each pool of whole exome libraries was sequenced on paired 76 cycle runs with two 8 cycle index reads across the number of lanes needed to meet coverage for all libraries in the pool (raw data available on request).
Somatic mutation calling from whole exome sequencing (WES) data
Exome sequencing data of CTC lines were used to identify somatic single nucleotide variations (sSNVs) and somatic small insertions and deletions (sINDELs). Output from Illumina software was processed by the Picard and GATK toolkits developed at the Broad Institute. The BAM files were generated by aligning with bwa version 0.5.9 to the NCBI Human Reference Genome Build hg19. Prior to variant calling, the impact of oxidative damage (oxoG) to DNA during sequencing was quantified as described previously (25). The cross-sample contamination was measured with ContEst (26) based on the allele fraction of homozygous SNPs, and this measurement was used in MuTect. From the aligned BAM files, somatic alterations were identified using a set of tools developed at the Broad Institute (www.broadinstitute.org/cancer/cga). The details of the sequencing data processing have been described previously (27, 28). Following our standard procedure, sSNVs were detected using MuTect (version 1.1.6) (28); sINDELs were detected using Strelka (version 1.0.11) (29). Then an allele fraction specific panel-of-normals (PoN) filter was applied to filter false positive germline variants and common artifacts from mutation calls, which compares the detected variants to a large panel of normal exomes or genomes and removes variants that were observed in the panel-of-normals. All somatic mutations, insertions and deletions were annotated using Oncotator (version 1.4.1) (30) sSNVs and sINDELS in only cancer genes (Cancer Gene Census) (31) were used for mutation status analysis.
Olaparib sensitivity studies
Three independent breast cancer CTC lines were tested for drug sensitivity: Brx401, harboring a somatically acquired known germline-pathogenic BRCA1 mutation, Brx142, harboring a somatically acquired mutation in BRCA2, and Brx07, with wild type BRCA1/2 alleles. CTC lines were seeded in 96 well ultra-low attachment plates (Corning) at 1000 cells per well. Increasing concentrations of olaparib (Selleckchem S1060) ranging from 0.01μM to 50μM were added to quadruplicate wells. Cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay per the manufacturer’s instructions at day 5.
Immunoblot
Cell pellets were lysed in 100mM Tris pH 6.8 1% SDS, sonicated for 10 seconds using a 4710 Series Ultrasonic Homogenizer (Cole-Parmer) and incubated for 3 min at 95°C. Protein lysates were then quantified and normalized using Pierce BCA Protein Assay Kit (23227, ThermoFisher Scientific). Lysates were then combined 1:1 with 2x sample buffer (100 mM Tris pH 6.8, 12% glycerol, 3.5% SDS, 0.2M DTT) and 20 μg of protein was loaded onto 4–12% Bolt Bis-Tris Plus gels (NW04122BOX, ThermoFisher Scientific) and transferred onto PVDF membranes by liquid transfer with CBS Scientific Electrophoretic Blotting System (EBX-700, 100V, 2 hours). Membranes were immunoblotted using BRCA1 (1:1000, D-9, Santa Cruz), GAPDH (1:20000, AB516, Millipore) and H3 (1:40000, ab1791, Abcam) antibodies and HRP-conjugated secondary anti mouse (1:5000, 115–035-003, Jackson Immunoresearch) and anti-rabbit (1:5000, 111–035-003, Jackson Immunoresearch) antibodies. Signals were detected using the Chemidoc imaging system (Bio Rad) with Image Lab v6.0.1 software.
Results
Patient demographics
We identified 215 patients at the Massachusetts General Hospital with MBC who had undergone cfDNA analysis before the start of a new therapy (first-line or greater) from February 2015 to July 2017. Supplemental figure S1 provides a consort diagram delineating the study population. Of the total population with MBC, 29 (13.5%) had somatic BRCA1 or BRCA2 mutations detectable by cfDNA. In nine patients (4.2%) mutations previously described as known germline-pathogenic were detected (as described in Supplemental Table S1), while in 20 (9.3%) novel variants (not previously reported in public databases) were identified (18).
Altogether, patients with cfDNA BRCA1 or BRCA2 mutations had similar age, cancer subtype distribution, and number of prior lines of chemotherapy, and went on to receive similar therapies post testing as those lacking cfDNA BRCA1/2 mutations (BRCA WT population) (supplemental Table S1). The majority of patients with somatic BRCA1/2 mutations had MBC which was recurrent (97%), rather than de-novo. The characteristics of patients with either known germline-pathogenic BRCA1/2 somatic mutations or novel variants are shown in Table 1. Interestingly, the patients with known germline-pathogenic BRCA1/2 mutations were significantly younger (median age of 48 years vs. 55 years) (p=0.008) and more often had TNBC (44% vs. 5%) (p=0.022), compared with the novel variants which were generally seen in HR+ and in some HER2+ MBC. The analyses of somatic BRCA1/2 status on patient outcomes is described in the supplemental section and supplemental Figure S2.
Table 1.
Clinical characteristics of patients with metastatic breast cancer and known germline-pathogenic BRCA1/2 mutations or novel BRCA1/2 variants.
| Clinical Variable | cfDNA BRCA1/2 mutation absent (BRCA wild-type (WT)) (N=186) | cfDNA BRCA1/2 known germline-pathogenic mutation present (BRCA known germline-pathogenic mutant)* (N=9) | cfDNA BRCA1/2 novel variant mutation present (BRCA novel variant)* (N=20) | p value for difference between BRCA WT and BRCAknown germline-pathogenic mutant+ | p value for difference between BRCA known germline-pathogenic mutant and BRCA novel variant+ |
|---|---|---|---|---|---|
| Median age at metastatic breast cancer diagnosis | 57 (48–65) | 48 (46–52) | 55 (52–67) | 0.059 | 0.008 |
| Tumor Subtype | 0.023 | 0.022 | |||
| HER2+ | 11 (5.9%) | 0 (0%) | 3 (15%) | ||
| HR+ | 134 (72%) | 4 (44.4%) | 14 (70%) | ||
| TNBC | 24 (12.9%) | 4 (44.4%) | 1 (5%) | ||
| Unknown | 17 (9.1%) | 1 (11.1%) | 2 (10%) | ||
| Number of prior lines of chemotherapy | 0.98 | 0.26 | |||
| 0–1 | 124 (66.7%) | 6 (66.7%) | 17 (85%) | ||
| ≥2 | 61 (32.8%) | 3 (33.3%) | 3 (15%) | ||
| Unknown | 1 (0.5%) | 0 (0%) | 0 (0%) | ||
| First therapy post-cfDNA testing | 0.32 | 0.42 | |||
| Endocrine | 57 (30.6%) | 3 (33.3%) | 7 (35%) | ||
| HER2 therapy | 13 (7.0%) | 0 (0.0%) | 2 (10%) | ||
| Immunotherapy | 14 (7.5%) | 2 (22.2%) | 2 (10%) | ||
| Chemotherapy | 49 (26.3%) | 3 (33.3%) | 3 (15%) | ||
| Other | 36 (19.4%) | 0 (0.0%) | 3 (15%) | ||
| None | 13 (7.0%) | 0 (0.0%) | 2 (10%) | ||
| Unknown | 4 (2.2%) | 1 (11.1%) | 1 (5%) | ||
Patients with both known germline-pathogenic and novel variants present in cfDNA were included in the known germline-pathogenic category for these analyses.
For the statistical analyses, the Wilcoxon rank-sum test (age variable) and Pearson chi-squared test (all categorical variables) were used.
Characteristics of cfDNA BRCA1/2 mutations
There was significant heterogeneity in the type of mutation and clinico-genomic characteristics of somatic BRCA1/2 mutations as depicted in Table 2. Four patients (13.8%) had polyclonal (≥2) BRCA1/2 mutations and 3 patients (10.3%) had both BRCA1 and BRCA2 cfDNA mutations.
Table 2.
Characteristics of cfDNA BRCA1/2 mutations.
| Characteristics of cfDNA BRCA1/2 mutations (N=29, overall cohort) | |
|---|---|
| Characteristic | Number of patients |
| BRCA1 or BRCA2 |
BRCA1: 15 (51.7%) BRCA2: 11 (37.9%) Both BRCA1 and BRCA2: 3 (10.3%) |
| Previously known germline-pathogenic vs. novel variants | Known germline-pathogenic: 9 (31%) Novel variants: 20 (69%) |
| Clonal vs. subclonal | Clonal: 16 (45.7%) Subclonal: 19 (54.3%) |
| Prior platinum or anthracycline treatment before cfDNA testing | Prior platinum: 4 (13.8%) Prior anthracycline: 16 (55.2%) None: 11 (37.9%) |
| Coexisting germline BRCA1/2 mutation | Germline BRCA1 mutation: 1 (3.4%) Germline BRCA2 mutation: 0 (0%) No known germline BRCA1 or BRCA2 mutation: 28 (96.6%) |
| Coexisting BRCA1/2 mutation detectable by tumor tissue genotyping |
BRCA1: 3 (15.8%) BRCA2: 0 (0%) No BRCA1 or BRCA2 mutation detected by tumor tissue genotyping: 16 (84.2%) |
| Characteristics of cfDNA previously known germline-pathogenic BRCA1/2 mutations (N=9)* | |
| BRCA1 or BRCA2 |
BRCA1: 5 (55.6%) BRCA2: 3 (33.3%) Both BRCA1 and BRCA2: 1 (11.1%) |
| Clonal vs. subclonal | Clonal: 4 (44.4%) Subclonal: 5 (55.6%) |
| Prior platinum or anthracycline treatment before cfDNA testing | Prior platinum: 4 (44.4%) Prior anthracycline: 5 (55.6%) None: 2 (22.2%) |
| Coexisting germline BRCA1/2 mutation | Germline BRCA1 mutation: 1 (11.1%) Germline BRCA2 mutation: 0 (0%) No known germline BRCA1 or BRCA2 mutation: 8 (88.9%) |
| Coexisting BRCA1/2 mutation detectable by tumor tissue genotyping |
BRCA1: 2 (40%) BRCA2: 0 (0%) No BRCA1 or BRCA2 mutation detected by tumor tissue genotyping: 3 (60%) |
| Characteristics of cfDNA novel variant BRCA1/2 mutations (N=20)* | |
| BRCA1 or BRCA2 |
BRCA1: 10 (50.0%) BRCA2: 5 (25.0%) Both BRCA1 and BRCA2: 5 (25.0%) |
| Clonal vs. subclonal | Clonal: 10 (50.0%) Subclonal: 10 (50.0%) |
| Prior platinum or anthracycline treatment before cfDNA testing | Prior platinum: 0 (0%) Prior anthracycline: 11 (55.0%) None: 9 (45.0%) |
| Coexisting germline BRCA1/2 mutation | Germline BRCA1 mutation: 0 (0%) Germline BRCA2 mutation: 0 (0%) No known germline BRCA1 or BRCA2 mutation: 20 (100%) |
| Coexisting BRCA1/2 mutation detectable by tumor tissue genotyping |
BRCA1: 1 (6.7%) BRCA2: 0 (0%) No BRCA1 or BRCA2 mutation detected by tumor tissue genotyping: 14 (93.3%) |
For these analyses, patients with both known germline-pathogenic and novel variants in cfDNA were included in the known germline-pathogenic category.
Among the various BRCA1/2 mutations detected in the cohort, 11 (29%) were protein-truncating alterations (6 frameshift insertions/deletions, 2 splice variants, and 3 nonsense mutations), all of which were predicted to be pathogenic. In contrast, 20 (53%) were missense point mutations, the majority of which were novel variants of unknown significance.
Altogether, 45.7% of the detected mutations were clonal (i.e. MAF ratio ≥25%) and 54.3% were subclonal (MAF ratio <25%). In the entire BRCA1/2 mutant population, 62% of patients with a cfDNA BRCA1/2 mutation had received prior platinum and/or anthracycline therapy, and this treatment distribution was similar in patients with known germline-pathogenic mutations. However, fewer patients with novel variants had received prior anthracycline or platinum therapy.
Of the 29 patients with somatic BRCA1 or BRCA2 mutations, 21 had archival tumor available for analysis (details in supplemental section, 52.4% on metastatic lesion at MBC diagnosis, 33.3% on a metastatic lesion post MBC diagnosis, and 14.3% on primary tumor specimen). Out of 21, only 3 patients had somatic BRCA1 mutations detectable in the archival tumor tissue, all from metastatic specimens. The BRCA1 variants in these 3 cases were identical in the blood and metastatic tumor tissue (details in supplemental section). The detailed clinical history and timing of tissue versus blood genotyping is outlined in supplemental Table S2.
One patient (patient ID #17) had a known coexisting germline BRCA1 mutation (c. 3875del4 mutation) as well as 3 additional somatic BRCA1 mutations in exon 10 which appeared to be reversion mutations, restoring the open reading frame in different ways. This patient had received platinum chemotherapy, which may have triggered the development of the BRCA1 reversion mutations that can restore BRCA1 function leading to acquired resistance to platinum and/or PARP inhibitors (32, 33).
Coexisting cfDNA mutations
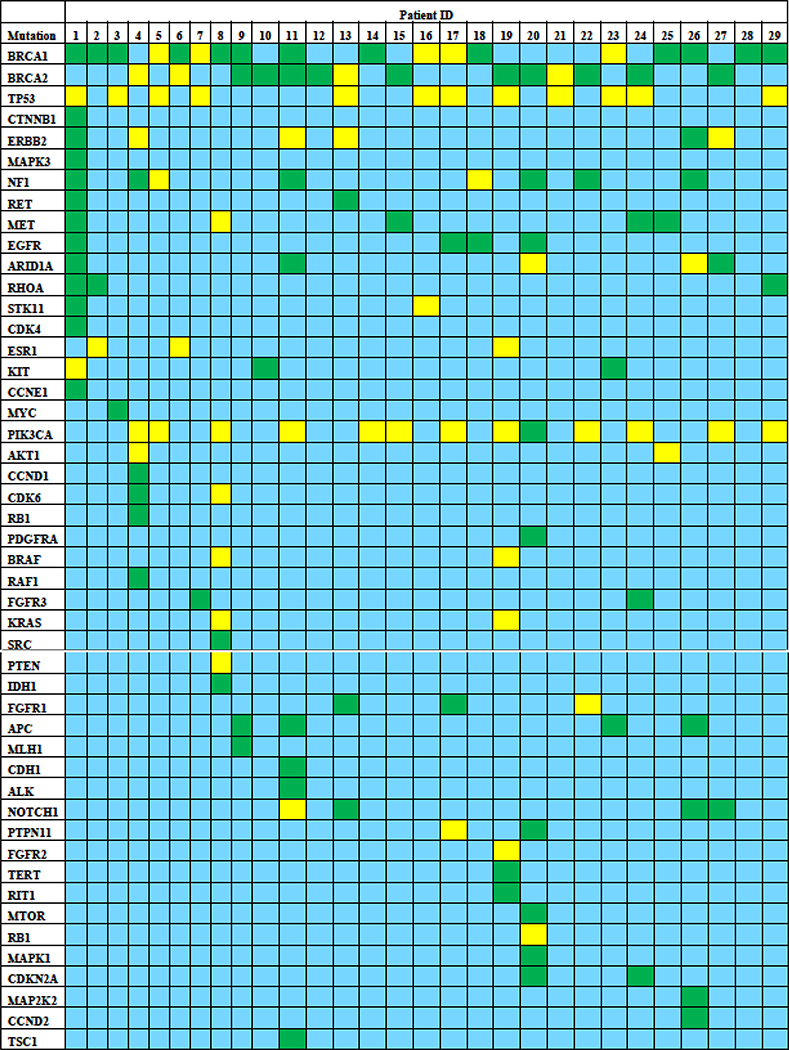
As depicted in Figure 1, a wide spectrum of coexisting mutations were detected with somatic BRCA1/2 mutations, highlighting genomic complexity and clonal heterogeneity. The most common mutations included PIK3CA (44.8%), TP53 (41.4%), NF1 (27.6%), ERBB2 (20.7%), MET (17.2%), ARID1A (17.2%), EGFR (13.8%), APC (13.8%), NOTCH1 (13.8%), RHOA (10.3%), ESR1 (10.3%), KIT (10.3%), and FGFR3 (6.9%).
Figure 1. Coexisting cfDNA mutations in patients with MBC with cfDNA BRCA1/2 mutations.
Blue depicts wild-type genes, yellow denotes known pathogenic mutations, and green signifies novel variants.
TP53 mutations were more common among patients with known germline-pathogenic BRCA1/2 mutations (77%), as compared to patients with novel BRCA1/2 variants (30%). Supplemental Figure S3 depicts the mutation spectrum by MAF for each patient in this cohort.
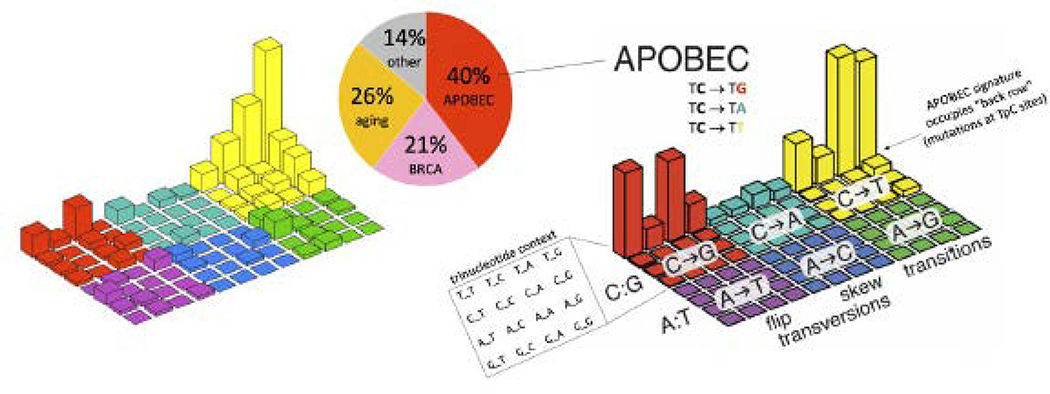
In terms of mutation signatures, comprehensive analysis revealed that both the “BRCA” signature (COSMIC Signature 3, associated with homologous recombinant deficiency) as well as APOBEC mutational signatures (COSMIC Signatures 2+13) were present in the BRCA1/2 cohort, highlighting the functional heterogeneity with somatic BRCA mutations (Figure 2).
Figure 2. APOBEC mutation signature in somatic BRCA-mutant patients.
The “Lego” plot on the left is the study data (all somatic mutations in the cohort). The “Lego” plot on the right is a “reference” for comparison: the APOBEC mutation signature. It also has the plot axes labeled. The rows are not mutational signature, but rather the whole plot is a mutation signature. The APOBEC mutation signature was clearly observed in this cohort. Approximately 40% of mutations were assignable to the APOBEC mutation signature (back row of bars in the “Lego” plot, COSMIC signatures 2+13), summing mutations across the 29 patients that were found to carry somatic BRCA mutations. Other contributors to the mutations in these BRCA-mutant patients were the “aging” signature (COSMIC Signature 1) and the “BRCA” signature (COSMIC Signature 3).
BRCA protein expression and olaparib sensitivity in CTC-culture lines
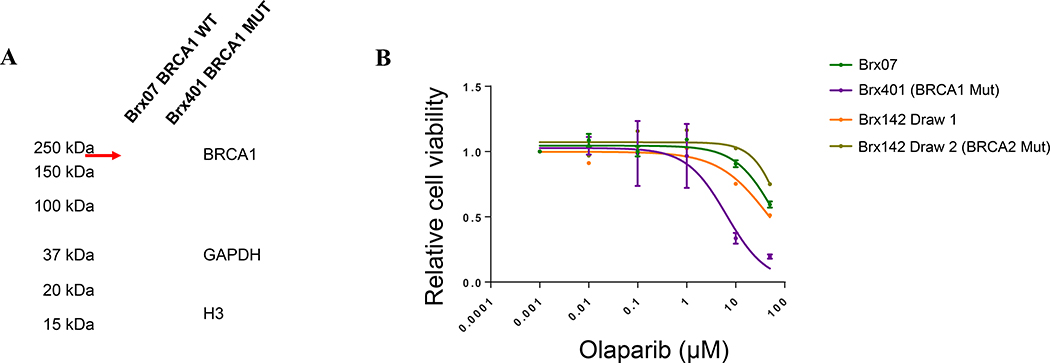
Finally, to evaluate the functional significance of somatic BRCA1 mutations, we analyzed gene expression and BRCA1 protein expression in the CTC lines Brx401 (harboring a known germline-pathogenic somatic BRCA1 mutant derived from patient ID #16 in this cohort) and Brx07 (harboring BRCA1 WT) using Western Blot (Figure 3A). Additional coexisting mutations in Brx401 included TSC2, TP53, and NOTCH2. No BRCA1 protein was seen in the cell line with a somatic BRCA1 mutation (Brx401) highlighting functional loss of BRCA1 protein, but full-length BRCA1 protein was seen in the cell line harboring BRCA1 WT (Brx07). In addition, we treated the Brx401 and Brx07 CTC cell lines with olaparib for 5 days and evaluated cell proliferation. The known germline-pathogenic somatic BRCA1 mutant line (BRx401) demonstrated increased sensitivity to olaparib (IC50 6.48μM) compared to the BRCA WT line (BRx07) (IC50 63.68μM) (Figure 3B). Indeed, the patient (patient ID #16 from whom the CTC-culture line BRx401 was developed) derived therapeutic benefit with carboplatin (PFS approximately 6 months), but not eribulin (PFS 3 months), further confirming that the somatic BRCA1 mutation was a likely driver mutation and consistent with the known platinum sensitivity of pathogenic BRCA1/2 mutations.
Figure 3. BRCA protein expression and olaparib sensitivity in CTC-culture lines.
(A) CTC lines Brx401 (acquired somatic BRCA1 mutant, MUT) and Brx07 (wild-type, WT) were analyzed by Western blot for BRCA1 protein expression. No BRCA1 protein was detected in Brx401. The red arrow indicates full-length BRCA1 protein (220kDa) detected in Brx07. (B) Brx401, Brx07, and BRx142 (novel variant BRCA2 mutant acquired (Brx142 (Draw 2) from baseline Brx142 (Draw 1) after treatment with a serum estrogen receptor degrader) CTC cell lines were treated with increasing concentrations of olaparib for 5 days and cell proliferation was evaluated. Brx401 (acquired somatic BRCA1 mutant, IC50: 6.48μM) was more sensitive to PARP inhibition compared to BRx07 (wild-type, IC50: 63.68μM) and the Brx142 lines.
Furthermore, in a third breast cancer CTC line harboring a novel somatic BRCA2 variant (Brx142) from a patient with HR+ metastatic breast cancer, we observed no increased sensitivity to olaparib compared with cells with BRCA WT. Interestingly, in this line, an APOBEC mutational signature was widely evident, and it encompassed the novel BRCA2 mutation itself. Additional coexisting mutations observed included SMARCA4 (p.1787M and p.E1606Q), CIC (p.E2258Q and p.K2423N), PI3KCA, BCLAF1, FAM135B, ALK, CSMD3, MYCN, FAT1, NF2, MUC16, MAFB, ZNF331, APC, and HIF1A, but a TP53 mutation was not present in this line. Thus, the somatic BRCA2 variant is likely a passenger mutation induced by increased APOBEC activity.
Discussion
We report that a proportion of patients with MBC harbor somatically acquired BRCA1/2 mutations in cfDNA, but that there is significant diversity in their associated clinico-genomic characteristics. While some of these mutations may be pathogenic in nature, others may not have functional significance. We were able to showcase the differences among these in selected cases for which cultured CTCs could be generated, but in general, distinguishing between cases with pathogenic BRCA1/2 mutations likely to respond to PARP inhibition and those with passenger mutations will require careful interpretation of both mutational and clinical parameters, and ultimately confirmation in prospective clinical trials. We identified that many detected mutations are subclonal. The clinical utility of using PARP inhibition to treat subclones with acquired BRCA1/2 mutations within a heterogeneous cancer is not known. The advent of PARP inhibitors as an approved therapy for germline BRCA1/2 mutant advanced breast cancer and the efficacy of DNA damaging agents in BRCA1/2 germline mutant patients makes the identification of nonfamilial cases with tumors that have BRCA-like features important, since this may help extend the application of PARP inhibitors (7, 8, 34), as has been demonstrated in ovarian cancer where germline and somatic BRCA mutant tumors have similar responses to PARP inhibition and platinum salts (35–43).
We identified that 13.5% of patients with MBC had somatic BRCA1 or BRCA2 mutations detectable by cfDNA. This mutation frequency is higher than expected based on the rates of somatic BRCA1/2 mutations in primary breast cancer (13), and possibly reflects the acquisition of mutations under therapeutic pressure (3, 4, 44). The majority of the BRCA1/2 mutations detected by cfDNA are not presently known to be pathogenic, and rather were novel variants, some of which might be passenger mutations resulting from APOBEC activity and other mutagenic conditions (14, 23, 45). A limitation of the Guardant360® assay used in these analyses is that tumor mutation burden cannot be calculated so we could not determine whether the presence of cfDNA BRCA1/2 mutations may be linked to an increased mutation rate, although more sophisticated cfDNA assays to evaluate this association could be considered in the future.
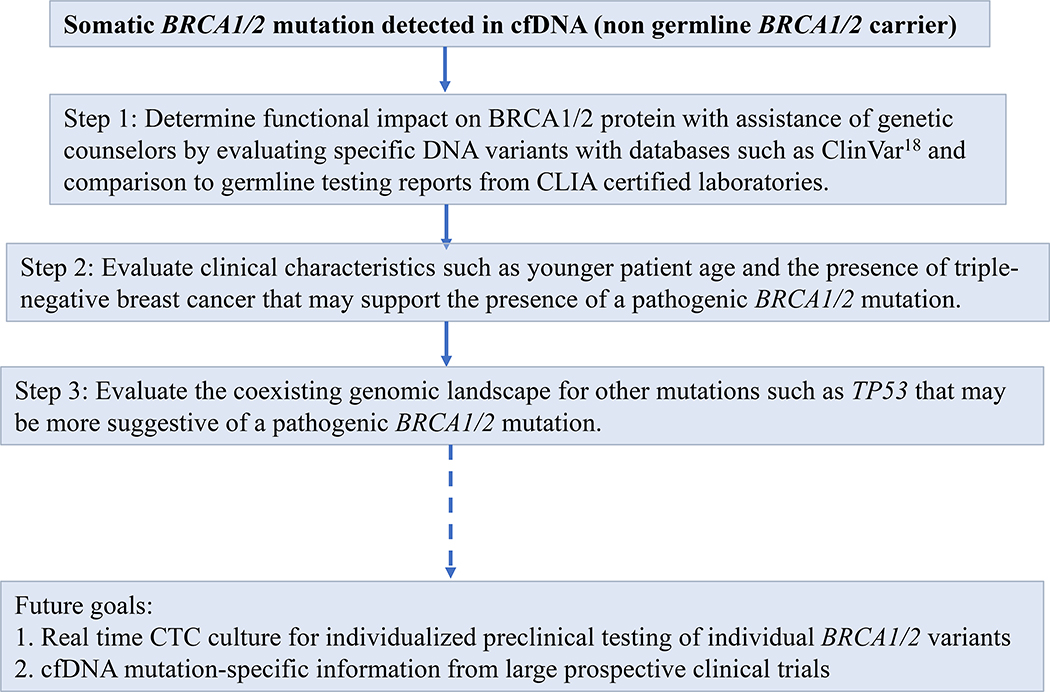
There are presently no guidelines to determine the pathogenicity of somatic BRCA1/2 mutations. Based on our work, we advocate the approach summarized in Figure 4 to determine the pathogenicity of a somatic BRCA1/2 mutation detected in cfDNA prior to consideration of PARP inhibitor therapy. We recommend initial review of detected somatic BRCA1/2 mutations by expert genetic counselors, utilizing open databases such as ClinVar (18) to understand the functional impact of genetic variants including splice site changes, frameshifts, stop codons, and missense mutations at different locations within the BRCA1/2 coding sequence. We recognize that this approach is limited by the uncertainty of extrapolating pathogenic mutation status from germline to somatic sequence analysis, and the fact that many mutations detected are likely to be novel variants whose functional significance is not known. The prospective development of large somatic genomic databases will be helpful in future classification. Clinical characteristics may help determine the potential for a somatic BRCA1/2 mutation to be pathogenic, such as TNBC histology and young age at diagnosis, criteria that are also characteristic of pathogenic germline BRCA1/2 mutations (46, 47). In contrast, most of the HR+ cases with cfDNA BRCA1/2 mutations were novel variants of uncertain significance. The coexisting genomic environment may provide clues such as coexisting TP53 mutations which we observed more commonly in patients with pathogenic BRCA1/2 mutations, similar to the association between TP53 mutations and germline BRCA1/2 mutations (48). While these criteria may provide guidance in interpreting cfDNA BRCA1/2 mutations, ultimately prospective clinical trials of PARP inhibitors in nonfamilial breast cancer are needed (49, 50).
Figure 4. Approach to establishing the pathogenic nature of a somatic BRCA1/2 mutation in cfDNA.
Based on our work, we recommend determining the functional impact on the BRCA1/2 protein (step 1), using clinical characteristics and the coexisting genomic landscape (steps 2–3) to help corroborate the presence of a pathogenic mutation. Future goals to aid in this assessment include developing real time CTC culture for individualized preclinical testing of individual BRCA1/2 variants, and obtaining data on the utility of PARP inhibition for various cfDNA BRCA1/2 mutations from large prospective clinical trials.
Supplementary Material
Statement of Translational Relevance.
Identification of somatic mutations using plasma genotyping assays in patients with metastatic breast cancer (MBC) represents an opportunity for novel therapy selection, and a challenge in distinguishing clinically impactful genetic variants. For BRCA1/2 mutations, pathogenic germline mutations are well-annotated, whereas the therapeutic significance of somatically acquired variants is not well-defined. We describe a cohort of patients with MBC, in whom we identified BRCA1/2 mutations using cell-free DNA (cfDNA) genotyping, with clinical correlates, and in selected cases conducted functional assays in cultured circulating tumor cells (CTCs). As many as 13.5% of patients with MBC harbor somatic BRCA1/2 mutations in cfDNA; 4% are known germline-pathogenic variants. In CTC-derived models, certain cell lines with somatically acquired driver variants demonstrate increased sensitivity to PARP inhibitors, while others with somatic BRCA1/2 variants resulting from increased APOBEC-mediated mutagenesis do not, behaving as passenger mutations. Detection of BRCA1/2 mutations using cfDNA requires caution before PARP inhibitor application.
Acknowledgements
We thank the patients who participated in this study.
This work was supported by grants from National Institute of Health (2O1CA129933 to D.A.H., 2U)1EB012493 to M.Toner, D.A.Haber, S.Maheswaran, 5P41EB002503 to M.Toner), the Howard Hughes Medical Institute (to D.A.Haber), National Foundation for Cancer Research (to D.A.Haber), ESSCP Breast Cancer Research Fund (to S.Maheswaran), MGH Cancer Center startup funds (to M.S.Lawrence), IBM/Broad collaboration on cancer genetics (to. G.Getz)
Relevant Disclosures
N.V. Research funding to the institution (MGH): Pfizer, Travel: Pfizer
T.D. None
M.L. MGH Cancer Center startup funds
A.S. None
A.N. none
E.B. none
B.N. Employee and stockholder at Guardant Health
W.R. none
B.C. none
B.A.R. none
G.M. none
J.L. none
S.J.I. none
D.J. none
D.M. none
S.W. Consulting/advisory board: Foundation Medicine, InfiniteMD, Eli Lilly, Puma Biotechnology; Equity: InfiniteMD
L.S. Consultant/advisory board: Novartis, Lumicell, Puma Biotechnology, Travel: Merck, Tesaro; Research Grant (Institution): Merck, Tesaro
B.M. Research funding (to MGH): PUMA Biotechnology
K.S. none
J.Y. none
R.L. Stockholder in Guardant Health, former employee at Guardant Health
M.T. Grant funding as noted above, Cofounder and equity in Tell Bio
A.J.I. None
G.G. Grant funding as noted above
L.Z. None
L.E. None
S.M. Grant funding as noted above, Cofounder and equity in Tell Bio
D.A.H. Grant funding as noted above, Cofounder and equity in Tell Bio
A.B. Consultant/advisory board: Genentech/Roche, Immunomedics, Novartis, Pfizer, Merck, Radius Health, Diiachi, Sanofi, Puma Biotechnology, Phillips
Research Grant (self): Biothernostics; Research Grant (Institution): Genentech/Roche, Immunomedics, Novartis, Pfizer, Merck, Radius Health, Sanofi, Mersana.
Footnotes
Conflict of Interest Disclosure Statement:
This study was presented in part (portion of the clinical data) at the 2017 San Antonio Breast Cancer Symposium in the Spotlight poster discussion session by N.V. (Vidula N et al. Somatic BRCA mutation detection by circulating tumor DNA analysis in patients with metastatic breast cancer: Incidence and association with tumor genotyping results, germline BRCA mutation status, and clinical outcomes [abstract]. In: Proceedings of the 2017 San Antonio Breast Cancer Symposium; 2017 Dec 5–9; San Antonio, TX. Philadelphia (PA): AACR; Cancer Res 2018; 78(4 Suppl): Abstract nr PD1-13).
Disclaimers: Massachusetts General Hospital (MGH) has applied for patents regarding the CTC-iChip technology and CTC detection signatures. D.A.H., M.T. and S.M. are cofounders and have equity in Tell Bio, which aims to commercialize the CTC-iChip technology.
References
- 1.Bardia A, Haber DA. Solidifying liquid biopsies: can circulating tumor cell monitoring guide treatment selection in breast cancer? J Clin Oncol 2014;32(31):3470–1. [DOI] [PubMed] [Google Scholar]
- 2.Somlo G, Lau SK, Frankel P, Hsieh HB, Liu X, Yang L, et al. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res Treat 2011;128(1):155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351(8):781–91. [DOI] [PubMed] [Google Scholar]
- 4.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science (New York, NY). 2014;345(6193):216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidula N, Juric D, Niemierko A, Spring L, Moy B, Malvarosa G, et al. Comparison of tissue genotyping (TG) vs circulating tumor DNA (ctDNA) for selection of matched therapy and impact on clinical outcomes among patients with metastatic breast cancer (MBC). J Clin Oncol 2018. May 20;36(15_suppl): 1020–1020.29380678 [Google Scholar]
- 6.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA oncology. 2019. February 1; 5(2):173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. The New England journal of medicine. 2017;377(6):523–33. [DOI] [PubMed] [Google Scholar]
- 8.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, et al. Talazoparib in patients with advanced breast cancer and germline BRCA mutation. N Engl J Med. 2018. August 23;379(8):753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campeau PM, Foulkes WD, Tischkowitz MD. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet. 2008;124(1):31–42. [DOI] [PubMed] [Google Scholar]
- 10.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33(3):244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner NC, Telli ML, Rugo HS, Mailliez A, Ettl J, Grischke E, et al. Final results of a phase II study of talazoparib (TALA) following platinum or multiple cytotoxic regimens in advanced breast cancer patients with germline BRCA 1/2 mutations (ABRAZO). Journal of Clinical Oncology. 2017;35(15_suppl):1007. [Google Scholar]
- 13.Wang X An exploration of mutation status of cancer genes in breast cancers. Next Generat Sequenc & Applic. 2014; April 26;1:103. [Google Scholar]
- 14.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–4. [DOI] [PubMed] [Google Scholar]
- 15.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taavitsainen S, Annala M, Ledet E, Beja K, Miller PJ, Moses M, et al. Evaluation of Commercial Circulating Tumor DNA Test in Metastatic Prostate Cancer. JCO Precision Oncology. 2019(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nance T, Helman E, Artieri C, Yen J, Slavin TP, Chudova D, et al. A novel approach to differentiation of somatic vs. germline variants in liquid biopsies using a betabinomial model. Proceedings of the American Association for Cancer Research Annual Meeting. 2018 Apr 14–18;Chicago, IL. Philadelphia (PA): AACR; [Google Scholar]; Cancer Res 2018; 78(13 Suppl): Abstract nr 4272. [Google Scholar]
- 18.National Center for Biotechnology Information. U.S. National Library of Medicine. ClinVar. Available online: https://wwwncbinlmnihgov/clinvar/ Accessed 2020 February.
- 19.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature medicine. 2014;20(12):1479–84. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science (New York, NY). 2019;364(6447). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell systems. 2018;6(3):271–81.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic acids research. 2013;41(6):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics (Oxford, England). 2011;27(18):2601–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics (Oxford, England). 2012;28(14):1811–7. [DOI] [PubMed] [Google Scholar]
- 30.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Human mutation. 2015;36(4):E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic acids research. 2019;47(D1):D941–d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer discovery. 2019;9(2):210–9. [DOI] [PubMed] [Google Scholar]
- 33.Weigelt B, Comino-Mendez I, de Bruijn I, Tian L, Meisel JL, Garcia-Murillas I, et al. Diverse BRCA1 and BRCA2 Reversion Mutations in Circulating Cell-Free DNA of Therapy-Resistant Breast or Ovarian Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(21):6708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilbum L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nature medicine. 2018. May;24(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science (New York, NY). 2017;355(6330):1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dougherty BA, Lai Z, Hodgson DR, Orr MCM, Hawryluk M, Sun J, et al. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget. 2017. July 4;8(27):43653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q, Yang J, Li L, Cao D, Yu M, Shen K, et al. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J Gynecol Oncol. 2017. July;28(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biranese RC, Nakamura KDM, Almeida FGDSR, Ramalho RF, Barros BDF, Ferreira ENE, et al. BRCA1 deficiency is a recurrent event in early-onset triple-negative breast cancer: a comprehensive analysis of germline mutations and somatic promoter methylation. Breast Cancer Res Treat 2018. February;167(3):803–14. [DOI] [PubMed] [Google Scholar]
- 39.Oza AM, Tinker AV, Oaknin A, Shapira-Frommer R, McNeish IA, Swisher EM, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: Integrated analysis of data from Study 10 and ARIEL2. Gynecologic oncology. 2017;147(2):267–75. [DOI] [PubMed] [Google Scholar]
- 40.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66(16):8109–15. [DOI] [PubMed] [Google Scholar]
- 41.George A, Banerjee S, Kaye S. Olaparib and somatic BRCA mutations. Oncotarget 2017. July 4;8(27):43598–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(22):3570–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lheureux S, Bruce JP, Burnier JV, Karakasis K, Shaw PA, Clarke BA, et al. Somatic BRCA1/2 Recovery as a Resistance Mechanism After Exceptional Response to Poly (ADP-ribose) Polymerase Inhibition. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2017;35(11):1240–9. [DOI] [PubMed] [Google Scholar]
- 44.Patsouris A, Tredan O, Campion L, Goncalves A, Arnedos M, Sablin MP et al. An open-label, phase Ii study of rucaparib, a PARP inhibitor, in HER2-metastatic breast cancer patients with high genomic loss of heterozygosity. J Clin Oncol. 2017; 36, no. 15_suppl:TPS1112. [Google Scholar]
- 45.Andre F, Filleron T, Ng C, Bertucci F, Letourneau C, Jacquet A, et al. Genomic characterization of metastatic breast cancer. San Antonio Breast Cancer Symposium. Abstract GS1–08. 2018; December. [Google Scholar]
- 46.Greenup R, Buchanan A, Lorizio W, Rhoads K, Chan S, Leedom T, et al. Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Annals of surgical oncology. 2013;20(10):3254–8. [DOI] [PubMed] [Google Scholar]
- 47.Copson ER, Maishman TC, Tapper WJ, Cutress RI, Greville-Heygate S, Altman DG, et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. The Lancet Oncology. 2018;19(2):169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61(10):4092–7. [PubMed] [Google Scholar]
- 49.NCT03990896 Evaluation of talazoparib, a PARP inhibitor, in patients with somatic BRCA mutant metastatic breast cancer: genotyping based clinical trial. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03990896 Accessed 2020; Feb.
- 50.NCT03344965 Olaparib in metastatic breast cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT03344965 Accessed 2020; Feb.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.