Abstract
Humans have limited regenerative potential of musculoskeletal tissues following limb or digit loss. The murine digit has been used to study mammalian regeneration, where stem/progenitor cells (the ‘blastema’) completely regenerate the digit tip after distal, but not proximal, amputation. However, the molecular mechanisms responsible for this response remain to be determined. Here, we evaluated the spatiotemporal formation of bone and fibrous tissues after level-dependent amputation of the murine terminal phalanx and quantified the transcriptome of the repair tissue. Distal (regenerative) and proximal (non-regenerative) amputations showed significant differences in temporal gene expression and tissue regrowth over time. Genes that direct skeletal system development and limb morphogenesis are transiently upregulated during blastema formation and differentiation, including distal Hox genes. Overall, our results suggest that digit tip regeneration is controlled by a gene regulatory network that recapitulates aspects of limb development, and that failure to activate this developmental program results in fibrotic wound healing.
Keywords: regenerative medicine, blastema, amputation, wound repair, osteogenesis, RNA-seq
Introduction
Injuries or diseases that lead to the loss of limbs or digits pose important unmet challenges to the medical community. In the United States, an estimated 185,000 people undergo limb amputations annually due to trauma or secondary to diabetes mellitus, dysvascular disease, or malignancy (1, 2). Although prostheses remain the gold standard of care, they remain costly, require extensive rehabilitation, and ultimately fail to restore the full form and function of the limb (3). Therefore, strategies that restore the biological composition and structure of the limb have potential to substantially improve patient outcomes. Unfortunately, the regenerative potential of musculoskeletal tissues is restricted, such that healing often culminates in fibrotic scarring. However, there is clinical evidence for digit tip regrowth following injury, especially in young children (4). To potentially harness this regenerative capability in humans, the cellular and molecular mechanisms driving regeneration must be better understood.
Towards this end, the murine digit tip is an attractive model to study a true regenerative process in adult mammals. After distal resection of the terminal phalanx bone (P3), the distal portion of P3 is first degraded and extruded (5, 6). Afterwards, a mass of hyperproliferative stem/progenitor cells, collectively called the ‘blastema,’ appear at the wound stump to regrow the bone and surrounding fibrous tissues (5, 7, 8). Lineage tracing studies indicate that blastema cells are germ layer-restricted and derived from local tissues (9, 10), including the bone marrow and periosteum (osteoblast lineage cells) (7, 9), blood vessels (endothelial progenitor cells) (5, 10), dermis (fibroblasts) (11, 12), and nerve (dedifferentiated Schwann cells and nerve-derived mesenchymal precursors) (13, 14). It is well established that this regenerative response depends on the level of amputation, such that removing the distal portion of P3 (1/3 length) results in the restoration of lost tissue, whereas increasingly proximal amputations (>2/3 length) lead to scar formation (8, 15–21). A similar phenomenon occurs after damage to the human fingertip (4, 8), making this finding directly relevant to clinical finger injuries.
While the molecular mechanisms responsible for level-dependent regeneration are not fully understood, regenerative competency is associated with the transient expression of developmental transcription factors such as Msx1/Msx2 and activation of the bone morphogenetic protein (BMP) signaling pathway in the blastema mesenchyme (8, 20, 21). Another regenerative feature is the preservation of the nail organ, which promotes reinnervation and subsequent bone regrowth through canonical Wnt signaling by the nail epithelium (8, 15, 17). Despite recent advances in our understanding of the pathways involved in the digit blastema, it remains to be determined how such diverse cell types and signals work in concert to restore the amputated tissues. A critical anatomical boundary that permits regeneration versus fibrotic scarring may exist, but whether this threshold is defined by the availability of competent stem/progenitor cell populations, the activation of regenerative genes, and/or the persistence of fibrotic pathways is currently unknown. The temporal dynamics of regenerative genes also requires further investigation and may shed light on the time-dependent response of cells to therapeutic intervention (18). To probe these questions, previous studies have relied on methods such as in situ hybridization, polymerase chain reaction, and microarrays for transcriptomic analysis of the regenerating tissue (13, 22, 23). While these methods provide important information about the regenerative process, they are inherently limited to a set of known transcripts. In contrast, next-generation RNA sequencing (RNA-seq) technology provides a global view of the transcriptomic landscape and facilitates the discovery of novel and rare transcripts, including non-coding RNAs, with higher specificity and sensitivity and at a wider dynamic range.
Therefore, to investigate the signaling pathways of the digit blastema, we used RNA-seq to determine the global transcriptomic response of these cells throughout the time course of regeneration. We hypothesized that successful regeneration depends on the coordinated remodeling and regrowth of bone and adjacent tissues, which is characterized by a gene regulatory network that recapitulates aspects of limb development. To test this hypothesis, we first quantified the spatiotemporal changes to bone and fibrous tissues after distal amputation (regenerative) or proximal amputation (non-regenerative) of P3 using microcomputed tomography and multiple histologic and imaging modalities. Next, we determined the differentially expressed genes of the Regen and Non-Regen repair tissue at various time points using RNA-seq. From this data, we identified key transcription factors that may be critical for regeneration and confirmed their expression in the digit tip using RNA fluorescence in situ hybridization. Our findings suggest that regeneration may depend on the transient activation of a limb-specific developmental program, whereas regenerative failure may result from reduced stem/progenitor cell populations, altered developmental signals, and/or accelerated fibrosis.
Materials & Methods
Digit amputation model
To investigate the effect of amputation level on adult murine digit regeneration, we performed bilateral resection of the digit tips of twenty female C57BL/6J mice at 10–12 weeks old (Jackson Laboratories, Bar Harbor, ME, USA). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, with all procedures approved by the Institutional Animal Care and Use Committee of Washington University in St. Louis (Protocol #20170244). Mice anesthetized with isoflurane were subjected to amputation of digits 2 and/or 4 of the hind limbs under a dissection microscope and received sustained-release buprenorphine (0.5 mg/kg subcutaneously) for analgesia. Regenerative (Regen) and Non-Regenerative (Non-Regen) states were established by resection of 20–30% (distal amputation) and 60–70% (proximal amputation) of the length of the terminal phalanx bone (P3), respectively (Fig. 1). Unamputated digit 3 of the same limb was used as a control. To establish the feasibility of multiple amputations per limb, both single (digit 4 only) and double-digit amputations (digits 2 and 4) were tested (n=2 mice/group/time point). Animals were allowed free cage activity post-operatively until euthanasia via CO2 narcosis at either 10 (wound closure), 12 (blastema formation), 14 (blastema differentiation), 21 (bone regrowth), or 56 days post-amputation (DPA) (bone restoration) in accordance with institutional policy for the humane sacrifice of animals.
Figure 1. Schematic of the murine distal digit model.
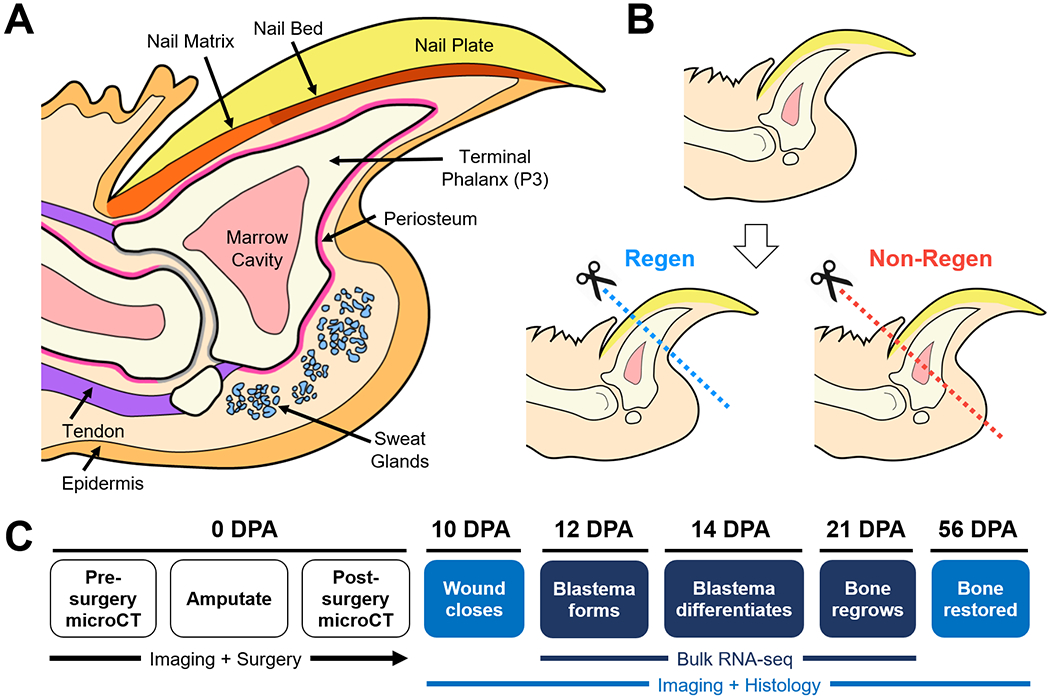
(A) Mid-sagittal section of the mouse digit tip with major anatomical structures. (B) Level-dependent amputation of the terminal phalanx bone (P3). Blue and red dashed lines indicate amputation planes for the Regenerative (Regen) and Non-Regenerative (Non-Regen) groups, respectively. (C) Experimental schematic show time course of regeneration at various days post-amputation (DPA).
MicroCT imaging and analysis
P3 morphology was assessed with a micro-computed tomography (microCT) scanner designed for in vivo use (Bruker SkyScan 1176; Bruker Corporation, Billerica, MA, USA). Mice anesthetized with isoflurane were placed on the scanner bed in sternal recumbency. Scans of the hind limb digits were obtained at 9 μm resolution (200 μA, 50 kV, 1 mm Al filter, 47° rotation/step) at baseline (pre-amputation; 0 DPA), immediately post-amputation (0 DPA), and after euthanasia, with additional imaging at 28 DPA for 56 DPA animals. Scans were processed using NRecon and DataViewer (Bruker Corporation) to obtain sagittal sections of each digit. P3 volume and mid-sagittal length were quantified using CTAn (Bruker Corporation) and normalized to pre-amputation values. Volume measurements excluded the sesamoid bone and any calcified tissue that was distinctly separate from the stump bone, such as the extruded bone segment. Mid-sagittal length was defined as the average of measurements from the dorsal and ventral proximal edges of P3 to the distal tip, acquired at the volume centroid. Digits from both single and double-digit models were pooled for statistical analysis (n=4 mice, 6 digits/group/time point).
Histology and SHG imaging
Harvested digits were fixed in 10% neutral buffered formalin (NBF; VWR International, LLC, Radnor, PA, USA) at room temperature (RT) overnight. Next, samples were decalcified in 10.6% buffered formic acid at 4°C overnight (Cal-Ex II; Thermo Fisher Scientific, Waltham, MA, USA) and processed for histologic analysis. Mid-sagittal sections were cut with a paraffin microtome (8 μm thickness) and stained with Hematoxylin and Eosin (H&E) to visualize cells and extracellular matrix (ECM), and with Picrosirius Red (PSR) to visualize collagen (n=3 digits/group/time point). Samples were imaged at 10X magnification with bright field and polarized light microscopy using an inverted light microscope (Olympus VS120; Olympus America, Inc., Center Valley, PA, USA) to evaluate collagen deposition and organization. To visualize the fibrillar collagen microstructure, unstained paraffin sections were coverslipped using an aqueous mounting medium (VectaMount; Vector Laboratories, Inc., Burlingame, CA, USA) for second harmonic generation (SHG) imaging (n=3 digits/group/time point). A tunable coherent Chameleon laser produced an excitation wavelength of 820 nm and SHG signal was collected using a photomultiplier tube detecting wavelengths between 371–442 nm. Tiled z-stacks at 1 μm intervals were acquired using a Zeiss LSM 880 with an inverted microscope at 20X magnification (Carl Zeiss Microscopy GmbH, Jena, Germany), and maximum z-stack projections were generated using the open-source platform Fiji.
RNA isolation, preparation, and sequencing
To evaluate the transcriptomic landscape during digit regeneration versus scarring, bulk RNA sequencing (RNA-seq) was performed on Regen and Non-Regen digits at 12, 14, and 21 DPA (n=4 mice/group/time point). Time points were selected to match histological observations of blastema formation (12 DPA), differentiation (14 DPA), and bone regrowth (21 DPA) in Regen digits. Samples were generated using the double-digit model as described previously, where amputation level and osteogenesis were assessed by microCT. After euthanasia, the tissues distal to the distal interphalangeal joint, excluding the ventral fat pad, were collected under a dissection microscope and stored at −80°C until RNA isolation. Frozen tissues were crushed with a pestle and the tissue lysates homogenized (QIAshredder; QIAGEN, Hilden, Germany). Total RNA was isolated (RNeasy Mini Kit; QIAGEN) (n=4 digits/group/time point, biological replicates taken from different animals) following the manufacturer’s protocol, depleted of ribosomal RNA via a hybridization method (Ribo-Zero Gold rRNA Removal Kit; Illumina, Inc., San Diego, CA, USA), and purified with DNase treatment (Ambion DNA-free DNA Removal Kit; Thermo Fisher Scientific). Library preparation was performed with 1 μg of total RNA per sample (average RNA integrity number of 8), where RNA quality was assessed using an automated electrophoresis system and bioanalyzer (Agilent 4200 Tapestation; Agilent Technologies, Inc., Santa Clara, CA, USA). Reverse transcription to cDNA was accomplished using SuperScript III Reverse Transcriptase enzyme (Life Technologies Corporation, Carlsbad, CA, USA) and random hexamers, with a second strand reaction performed to yield ds-cDNA. cDNA was blunt ended, had an A base added to the 3’ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were amplified for 11 cycles using primers incorporating unique index tags and then sequenced on an Illumina HiSeq 3000 (Illumina, Inc.) using single reads extending 50 bases, generating approximately 32 million reads per sample.
Transcriptomic analysis
Reads were processed using an in-house pipeline and open-source R packages. Briefly, raw reads were first trimmed using cutadapt to remove low quality bases and reads. Trimmed reads were then aligned to the mouse genome mm10 with GENCODE annotation vM15 using STAR (v2.5.4) with default parameters. Transcript quantification was performed using featureCounts from the subread package (v1.4.6-p4). Further quality control assessments were made using RSeQC and rsem, and batch-correction was performed using edgeR, EDASeq, and RUVseq. Gene type and transcription factor (TF) annotation was performed using mouse GENCODE vM15 and AnimalTFDB, respectively. Principle component analysis (PCA) and differential expression analysis for Regen and Non-Regen groups were determined using DESeq2 in negative binomial mode using batch-corrected transcripts from featureCounts (>2-fold expression change, >1 count per million (CPM), Benjamini corrected P<0.05) (24). Pairwise comparisons were made between time points to determine differentially expressed genes (DEGs) within each group. To examine expression patterns, the average reads per kilobase million (RPKM) per time point for Regen (1513 DEGs) and Non-Regen groups (1400 DEGs) were z-score scaled and used for k-means clustering. DEGs were grouped into four clusters (Upregulation, Transient Upregulation, Transient Downregulation, and Downregulation) and their relative expression over time was plotted using ggplot2. DEGs from each distinct Regen and Non-Regen cluster were used for gene ontology (GO) enrichment analysis using DAVID (v6.8) (Benjamini corrected P<0.05) (25). Next, to compare the expression of Regen-specific DEGs between all groups and time points, the average RPKM from Non-Regen samples was z-score scaled and joined to the Regen samples in their k-means clusters and plotted using pheatmap. The expression of DEGs specific to Non-Regen samples was similarly compared to the expression of corresponding genes in Regen samples. TFs that were transiently upregulated in the Regen group were used to build a gene regulatory network (GRN) using Cytoscape (v3.0.2).
ChIP-seq data analysis
To identify potential TF binding targets, Hoxa13/Hoxd13 (Hox13) and Runx2 chromatin immunoprecipitation followed by sequencing (ChIP-seq) data from mouse E11.5 distal limb cells (26) and mouse MC3T3-E1 pre-osteoblasts and their mature osteoblast progeny (27), respectively, were downloaded from Gene Expression Omnibus (GEO; GSE81358 and GSE41955). Raw fastq files were aligned to the mouse genome mm10 assembly by the Burrows-Wheeler Alignment tool. The bam files were then processed using methylQA to generate bed and bigwig files. The MACS (v2.0.10) peak caller was used to compare the ChIP-seq signal to a corresponding input control to identify narrow regions of enrichment (peaks) of Hox13 and Runx2 that passed a q-value threshold of 0.01. ChIP-seq signal density (bigwig) and peak location (bed) were further visualized using the WashU Epigenome Browser. ChIP-seq peaks were assigned to the nearest transcription start site of genes using the annotatePeaks.pl function from HOMER2 to determine the binding targets of Hox13 and Runx2. GO enrichment analysis was performed as previously described using target genes for Hox13 only, Runx2 only, and both Hox13 and Runx2 (Benjamini corrected P<0.05).
RNA FISH and image analysis
To validate RNA-seq findings and to visualize spatiotemporal gene expression, single-molecule RNA fluorescence in situ hybridization (FISH) was performed for three genes whose levels varied by group and/or time point: Hoxa13, Runx2, and H19. Regen and Non-Regen digits from 12, 14, and 21 DPA (n=4 digits/group/time point) were harvested, fixed overnight in 10% NBF at RT, and decalcified in 10% ethylenediaminetetraacetic acid for 10 days at 4°C. Samples were processed for paraffin embedding and cut into sagittal sections (5 μm thickness). Sample pre-treatment and RNA probe hybridization, amplification, and signal development were performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, Inc., Newark, CA, USA) following the manufacturer’s protocol. Probe signals for Hoxa13, Runx2, and H19 were developed using TSA Plus fluorescein, cyanine 5, and cyanine 3 (PerkinElmer, Inc., Waltham, MA, USA), respectively, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Samples were imaged with multi-channel confocal microscopy using a Zeiss LSM 880 (Carl Zeiss Microscopy GmbH). Z-stacks were taken at 20X (tiled image) and 63X magnification to capture the entire digit tip and specific regions of interest (e.g., blastema), respectively. To quantify probe signal distribution throughout the digit, the maximum z-stack projection for each channel at 20X was converted into a binary image using Fiji. Particles larger than 20 pixels2 were excluded from analysis (autofluorescent red blood cells and nail keratin). A density heat map of each probe signal was generated using the open-source platform Bio7 and the R package spatstat, where the intensity value represents the expected number of random points per unit area. Heat maps were overlaid with schematics created using Illustrator CS2 (Adobe Inc., San Jose, CA, USA) that depict the boundaries for bone, skin/nail, and underlying connective tissue.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v8.0; GraphPad Software, Inc., San Diego, CA, USA). Significance was assessed by two-way analysis of variance (ANOVA) with Tukey’s HSD post-hoc tests to compare between groups (P<0.05). Primary metrics include bone length, bone volume, and gene expression level (RPKM) as dependent variables, with independent variables of group and time point. Results are presented as mean±s.d. unless specified otherwise.
Results
Temporal dynamics of bone and soft tissue regrowth
Pre- and post-operative microCT imaging revealed that 24.4±2.8% and 64.2±2.1% of the terminal phalanx bone (P3) length was amputated for the Regenerative (Regen) and Non-Regenerative (Non-Regen) groups, respectively (Fig. 2). Due to the tapered shape of P3, 7.7±1.0% and 37.5±3.9% of the bone volume was removed for the Regen and Non-Regen groups, respectively. Tracking the morphologic time course of P3 after amputation, we confirmed that resecting the distal portion led to a regenerative response with blastema formation and osteogenesis, whereas proximal amputations resulted in fibrotic scar formation, consistent with previous reports (8, 15–21). Both P3 length and volume were reduced for the Regen group after distal amputation at 10 days post-amputation (DPA) and at subsequent time points up to 21 DPA when compared to post-surgical values at 0 DPA, indicating histolysis and tissue remodeling (P<0.05, Fig. 2B, Fig. 2C). At 10 DPA, Hematoxylin and Eosin (H&E) staining showed incomplete re-epithelialization at the distal tip (Fig. 3A). Between 10 and 12 DPA, a distal bone fragment was extruded as the wound epidermis closed over the P3 surface (Fig. 2A). Maximum bone volume loss occurred at 12 DPA for Regen digits (48.3±5.5%). This initial stage of partial bone degradation opened the marrow cavity and was followed by a local surge in cellularity and extracellular matrix (ECM) deposition. By 14 DPA, a hypercellular mass (the blastema) extended from inside the marrow cavity to the digit tip (Fig. 3A). There was a marked periosteal reaction in Regen digits, most notably along the dorsal surface. By 21 DPA, Picrosirius Red (PSR) staining revealed collagen deposition (Fig. 3B) and mineralization (Fig. 2A) contiguous with the edges of P3, indicating new bone formation distal to the degraded bone. By 28 DPA, both P3 length and volume had increased to reach post-amputation levels at 0 DPA (P>0.05, Fig. 2B, Fig. 2C). By 56 DPA, P3 length had recovered to pre-amputated levels in Regen digits, with a 23% volume overshoot (P<0.05, Fig. 2B, Fig. 2C), and digit morphology resembled that of unamputated control digits (Fig. S1).
Fig. 2. Bone remodeling and regrowth follow distal, but not proximal, amputation.
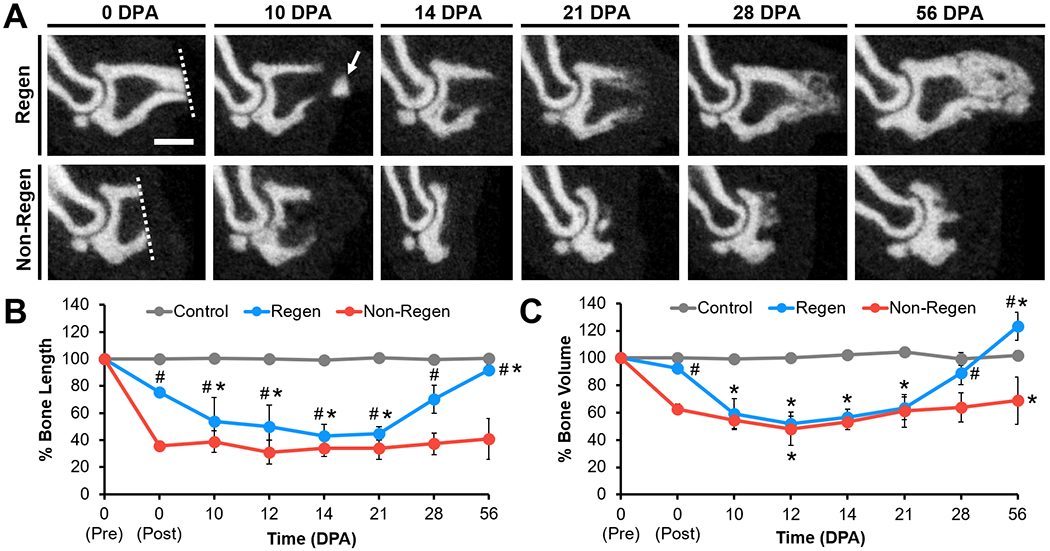
(A) Mid-sagittal microCT images of P3 at various DPA for Regen and Non-Regen groups. Dotted lines show amputation planes. Arrow points to distal fragment. Scale: 0.5 mm. (B) P3 length and (C) volume changes over time (% of pre-amputation, n=6 digits/group/time point from 4 mice, mean±s.d.). *P<0.05 vs. 0 DPA post-amputation. #P<0.05 vs. Non-Regen.
Fig. 3. Histological time course of regeneration.
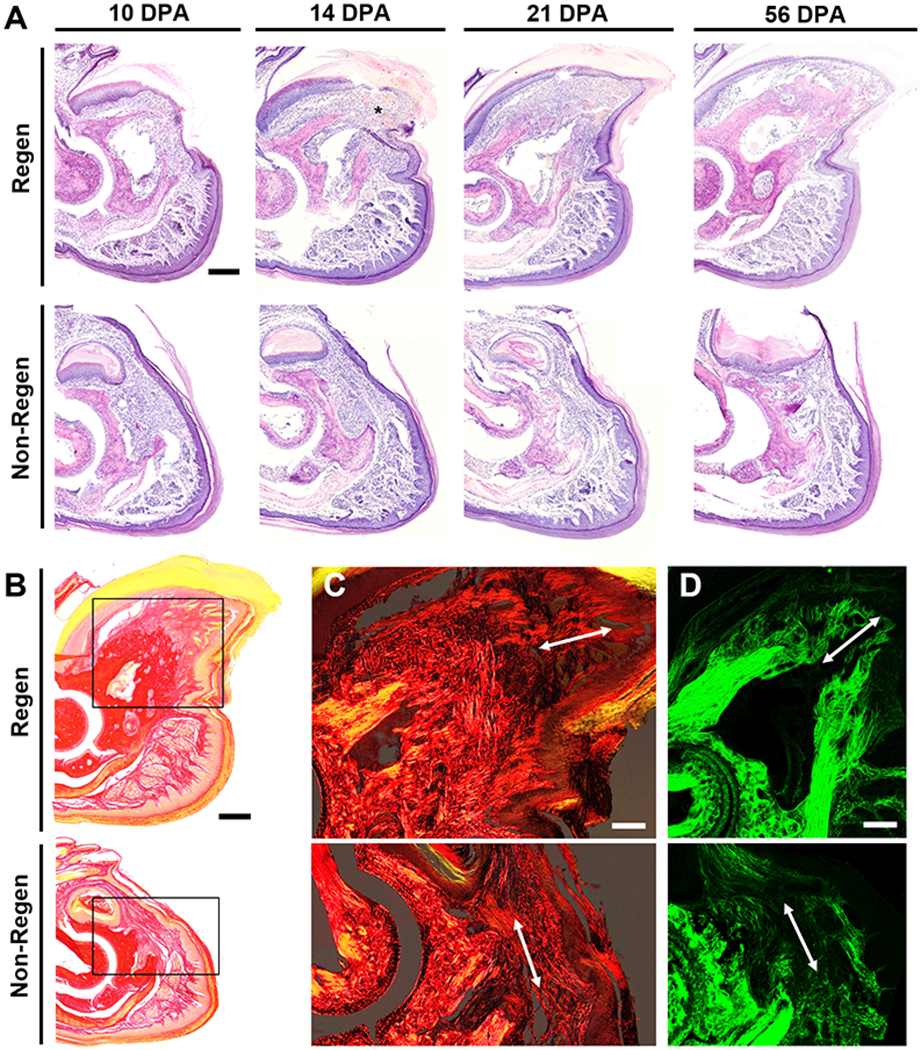
(A) Hematoxylin and Eosin (H&E) staining at various DPA for Regen and Non-Regen groups. Asterisk indicates blastema at 14 DPA. Scale: 0.2 mm. (B) Picrosirius Red (PSR) staining imaged with bright field microscopy at 21 DPA. Scale: 0.2 mm. Inset shows area imaged in (C). (C) PSR staining visualized with polarized light microscopy and (D) second harmonic generation (SHG) imaging show collagen fibers (arrow) extending proximodistally in the Regen but not the Non-Regen state at 21 DPA. Scale: 0.1 mm.
Conversely, bone remodeling and regrowth were not observed after proximal amputation in the Non-Regen digits. While P3 volume was slightly reduced at 12 DPA compared to immediately post-surgery at 0 DPA (P<0.05, Fig. 2B, Fig. 2C), bone length remained constant through 56 DPA (P>0.05). Cell accumulation at the cortical bone stump and complete re-epithelialization was apparent by 10 DPA, but minimal periosteal reaction, cellular outgrowth, or bone regeneration was observed at subsequent time points (Fig. 3A). Instead, collagen deposition at the endosteal surface was apparent by 14 DPA, and by 21 DPA, the bone stump, as well as the overlying nail matrix and nail plate, was surrounded by a thin fibrous capsule that became more pronounced over time (Fig. 3B). P3 length and volume did not significantly change for control digits over the course of the study. Furthermore, the single and double-digit amputation models behaved similarly, suggesting that the healing response acts locally.
To examine the ECM structure of amputated digits, we utilized polarized light microscopy to visualize collagen distribution and alignment in PSR-stained tissue sections (Fig. 3C, Fig. S2A). Over time, collagen at the distal tip became increasingly aligned in the proximodistal direction for Regen digits and in the dorsoventral direction for Non-Regen digits. These findings were confirmed with second harmonic generation (SHG) imaging (Fig. 3D, Fig. S2B), which revealed fine collagen fibrils emanating from the bone stump of Regen digits by 14 DPA, converging to form the tip of P3 by 21 DPA. In contrast, a dense network of thick collagen bundles traversed parallel to the amputation plane in Non-Regen digits by 14 DPA. Collectively, these results suggest that the disparate healing trajectories in response to level-dependent P3 amputation are established by two weeks post-amputation in the adult mouse.
Activation of limb-specific developmental processes after distal amputation
Given these morphologic observations, we next examined the molecular signals that delineate regenerative competency (Regen) versus failure (Non-Regen) using RNA-seq at time points that reflect blastema formation (12 DPA), blastema differentiation (14 DPA), and the beginning of bone regrowth (21 DPA) (Fig. 1). Principal component analysis (PCA) revealed sample clustering by group and time point (Fig. 4A). While the Regen and Non-Regen groups remained largely separate, Non-Regen digits at 14 DPA overlapped with Regen digits at 12 DPA. Since the same time point reflects different states of healing for Regen and Non-Regen groups, we focused our analysis on the genes that were dynamically expressed within groups. Pairwise comparisons between time points within each group revealed 1513 and 1400 differentially expressed genes (DEGs) in Regen and Non-Regen digits, respectively, the majority of which were protein coding transcripts (Fig. S3). DEGs were subsequently grouped into 4 clusters based on the time course of expression using k-means clustering (Fig. 4B). Although DEGs within each cluster exhibited similar fold changes for Regen and Non-Regen groups, only a quarter of the 1513 Regen DEGs had the same expression pattern in the Non-Regen group, suggesting distinct transcriptomic differences between Regen and Non-Regen digits (Fig. 4C). Furthermore, the majority of Regen DEGs exhibited transient upregulation at 14 DPA (49.8%), whereas the majority of Non-Regen DEGs showed downregulation with time (38.9%).
Fig. 4. Temporal transcriptomic dynamics of regeneration competency versus failure.
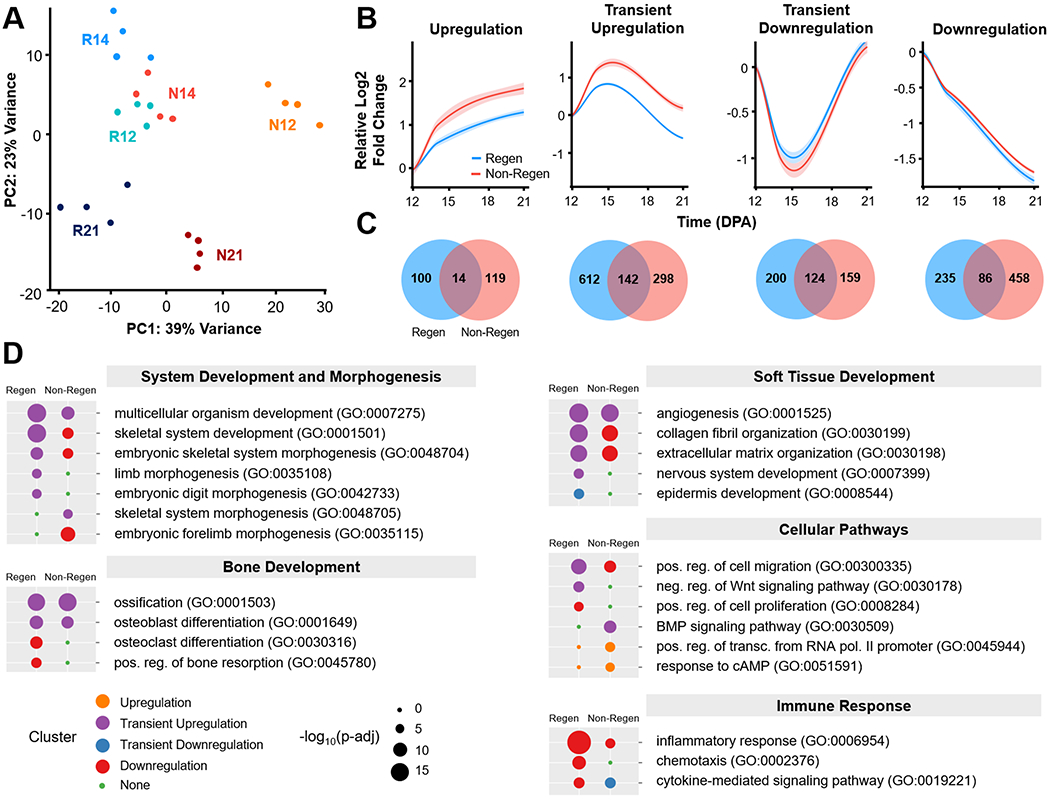
(A) Principle component analysis (PCA) illustrates the relative similarity between Regen (R) and Non-Regen (N) groups at 12, 14, and 21 DPA (n=4 mice, 4 digits/group/time point). (B) K-means clustering identified four differentially expressed gene (DEG) clusters with similar temporal expression patterns for each group. Solid lines represent the relative log2 fold change in expression level (RPKM) compared to 12 DPA. Lighter shades indicate 95% confidence interval. (C) Venn diagrams show number of DEGs present in Regen, Non-Regen, or both groups for each cluster. (D) Relevant GO terms (biological processes) for Regen and Non-Regen groups, where the circle color and size represent cluster type and Benjamini corrected p-value (p-adj), respectively.
Gene ontology (GO) enrichment analysis revealed that biological processes related to digit regeneration, including skeletal system development, embryonic skeletal system morphogenesis, limb morphogenesis, and embryonic digit morphogenesis, were transiently upregulated in Regen digits but not in Non-Regen digits (Benjamini corrected P<0.05, Fig. 4D). Genes in the Regen transient upregulation cluster also identified with pro-regenerative terms such as multicellular organism development, collagen fibril organization, ECM organization, osteoblast differentiation, ossification, angiogenesis, and nervous system development. Genes associated with wound healing (including Col3a1) and tissue remodeling (including Mmp2) were transiently upregulated. Transiently upregulated cellular pathways include positive regulation of cell migration and negative regulation of the Wnt signaling pathway. Epidermis development was transiently downregulated, most likely due to the comparative upregulation of other processes at 14 DPA. The Regen downregulation cluster contained genes related to bone remodeling (osteoclast differentiation and bone resorption), cell division (positive regulation of cell proliferation), and immune response (inflammatory response, chemotaxis, and cytokine-mediated signaling pathway), indicating that these processes had peaked by 12 DPA. There was no enrichment of GO terms for the upregulation cluster due to the relatively small number of genes (114 genes).
In contrast, while Non-Regen digits shared a subset of transiently upregulated developmental GO terms (multicellular organism development, ossification, osteoblast differentiation, and angiogenesis) (Benjamini corrected P<0.05), limb morphogenetic processes either fell in the downregulation cluster or were absent (Fig. 4D). GO terms associated with the ECM (collagen fibril organization and ECM organization) were downregulated in Non-Regen digits, whereas GO terms related to bone degradation and soft tissue development (nervous system and epidermis) were absent. Indeed, out of the 544 downregulated Non-Regen DEGs, 269 were transiently upregulated in Regen digits (Fig. S4). Furthermore, out of the 754 transiently upregulated Regen DEGs, including 60 transcription factors (TFs), only 142 of those same genes (13 TFs) were also transiently upregulated in Non-Regen digits (Fig. 5A, Fig. S5B). Interestingly, the BMP signaling pathway was transiently upregulated, indicating that there may be a pro-osteogenic response at 14 DPA at the transcriptomic level. These data suggest that although Regen and Non-Regen digits may share some similar signaling pathways, the temporal activation of a limb-specific developmental program may be necessary for a fully regenerative outcome. On the other hand, upregulated GO terms unique to Non-Regen digits (positive regulation of transcription from RNA polymerase II promoter and response to cAMP) revealed how specific genes and pathways may be suppressed during regeneration and utilized during scar formation, such as transcription factors in the Fos family (Fos, Fosb, Fosl1), which are transiently downregulated in Regen digits.
Fig. 5. Developmental transcriptional program is transiently activated in regenerating digits.
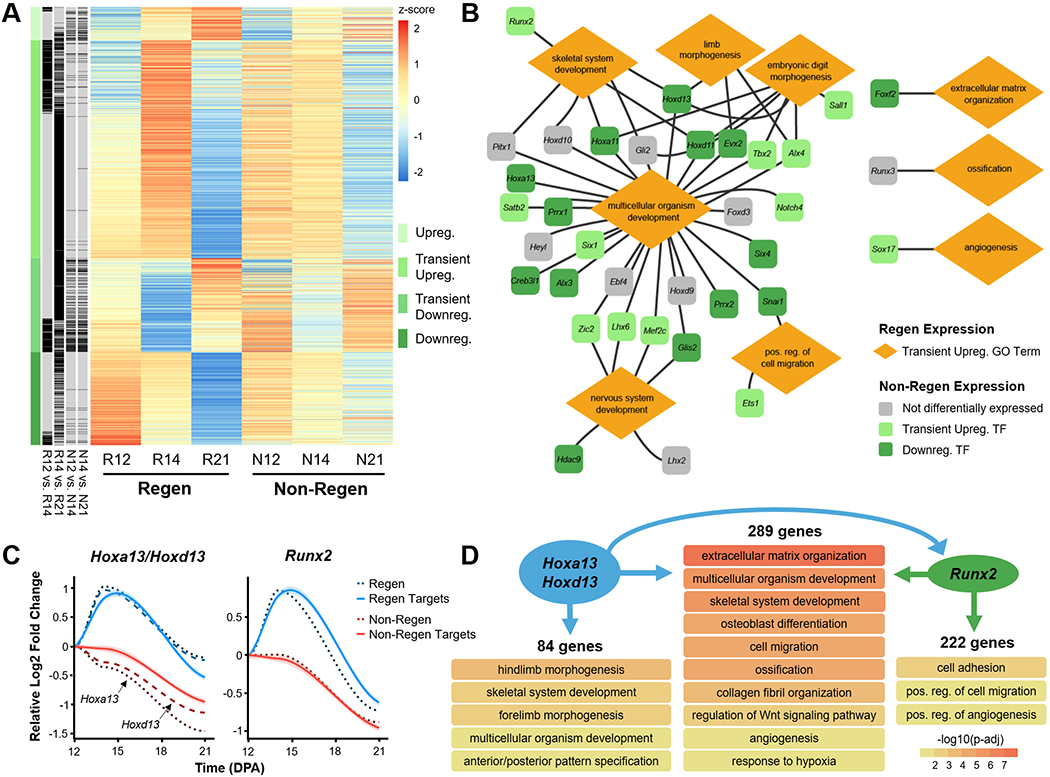
(A) Heat map shows the scaled z-score of the average RPKM of 1513 DEGs from Regen groups at 12, 14, and 21 DPA, alongside the corresponding z-score of Non-Regen groups. Red and blue cells indicate relative gene upregulation and downregulation, respectively. Green cells indicate cluster type, and black cells indicate genes that are significantly different between time points for Regen and Non-Regen groups (Benjamini corrected P<0.05). (B) Gene regulatory network illustrates various transiently upregulated Regen transcription factors (TFs) and their relationship to Regen GO terms. Lines indicate association of TF with GO term. Colors indicate the temporal expression pattern of corresponding TFs in the Non-Regen state. (C) Gene expression profiles of the TFs Hoxa13/Hoxd13 (Hox13) and Runx2 (dashed lines) and their downstream targets (solid lines), as determined by ChIP-seq analysis (n=4 mice, 4 digits/group/time point, mean). Lines represent the relative log2 fold change in expression (RPKM) compared to 12 DPA. Lighter shades indicate 95% confidence interval. (D) GO terms (biological processes) associated with gene targets of either Hox13, Runx2, or both (Benjamini corrected P<0.05). Arrows specify direction of gene regulation, including Hox13 regulation of Runx2 expression.
Gene regulatory network reveals TFs driving regeneration
To investigate how specific genes may direct regeneration, we built a gene regulatory network (GRN) using 35 transiently upregulated Regen TFs that were related to pro-regenerative GO terms, 12 of which were also transiently upregulated in Non-Regen digits (Fig. 5B, Fig. S5). Most TFs were associated with multicellular organism development and may also be linked to skeletal system development, limb morphogenesis, embryonic digit morphogenesis, nervous system development, and/or positive regulation of cell migration. TFs of note include Hox genes, which direct distal limb (Hoxa11/Hoxd11) and digit development (Hoxa13/Hoxd13) (28–30); Prrx1, which is required for early skeletogenesis and modulates limb segment length (31); Alx4 and Tbx2, which affect limb patterning (32, 33); and Runx2, which is a master regulator of osteoblast differentiation (34). Other transiently upregulated TFs in Regen digits include those involved in ECM organization (Foxf2) (35), ossification (Runx3) (36), and angiogenesis (Sox17) (37).
Next, to determine the downstream implications of transient TF upregulation, we analyzed the DNA binding targets of Hoxa13/Hoxd13 (hereafter referred to as Hox13) (26) and Runx2 (27) using publicly available chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) data. We identified 636 target genes of Hox13 and found that 373 (59%) were transiently upregulated in Regen digits (Fig. 5C). Similarly, 511 of the 879 (58%) target genes of Runx2 were also transiently upregulated in Regen digits, 289 of which were shared targets of Hox13 and Runx2. GO enrichment analysis revealed that unique targets of Hox13 were specifically related to limb morphogenesis, whereas shared targets were associated with processes involved in osteogenesis (e.g., ECM organization, osteoblast differentiation, ossification, and angiogenesis) (Fig. 5D). Furthermore, the ChIP-seq analysis revealed that Hox13 could regulate Runx2 by binding 5.2 kb upstream of a major intragenic transcription start site (Fig. S6). While the data did not show direct regulation of Hox13 by Runx2, Runx2 may potentially affect the transcription of Hoxd13, but not Hoxa13, via an upstream enhancer. The same target genes in Non-Regen digits reflected the downregulation of Hox13 and Runx2 in those samples (Fig. 5C), which is consistent with our analysis.
Spatiotemporal expression of regulatory genes
To validate RNA-seq results and to localize gene expression, we performed RNA fluorescence in situ hybridization (FISH) for two transiently upregulated TFs, Hoxa13 and Runx2, as well as for H19, a long non-coding RNA (lncRNA) that showed significantly higher expression in Non-Regen digits compared to Regen digits at 12 and 14 DPA (Fig. 6A). RNA FISH showed increased expression of both Hoxa13 and Runx2 in Regen digits compared to Non-Regen digits at 14 DPA (Fig. 6B), reflecting the transcriptomic data. In Regen digits at 14 DPA, the blastema (cell mass distal to P3), bone interface, and distal portions of the marrow cavity and endosteum showed areas of high Hoxa13 and Runx2 expression, whereas the distal periosteum was largely restricted to Runx2 expression (Fig. 6C). Hoxa13 and Runx2 were expressed at comparatively lower levels in Regen digits at 12 DPA (Fig. S7), suggesting that their tissue-level expression significantly increased between the stages of blastema formation and differentiation. The lncRNA H19 was found in Non-Regen digits at all time points, predominantly located at the bone interface and within the marrow cavity (Fig. 6, Fig. S7). H19 was minimally expressed in Regen digits at 12 and 14 DPA but was present in the region between the distal regenerating bone and epidermis at 21 DPA. H19 was also concentrated at the ventral epidermal-dermal junction in all samples, including control digits (Fig. S1). Taken together, these findings highlight the dynamic, spatiotemporal gene expression patterns that occur after digit amputation, resulting in signaling pathways that may either promote or inhibit the regenerative process.
Fig. 6. Gene expression is localized to regenerating tissues.
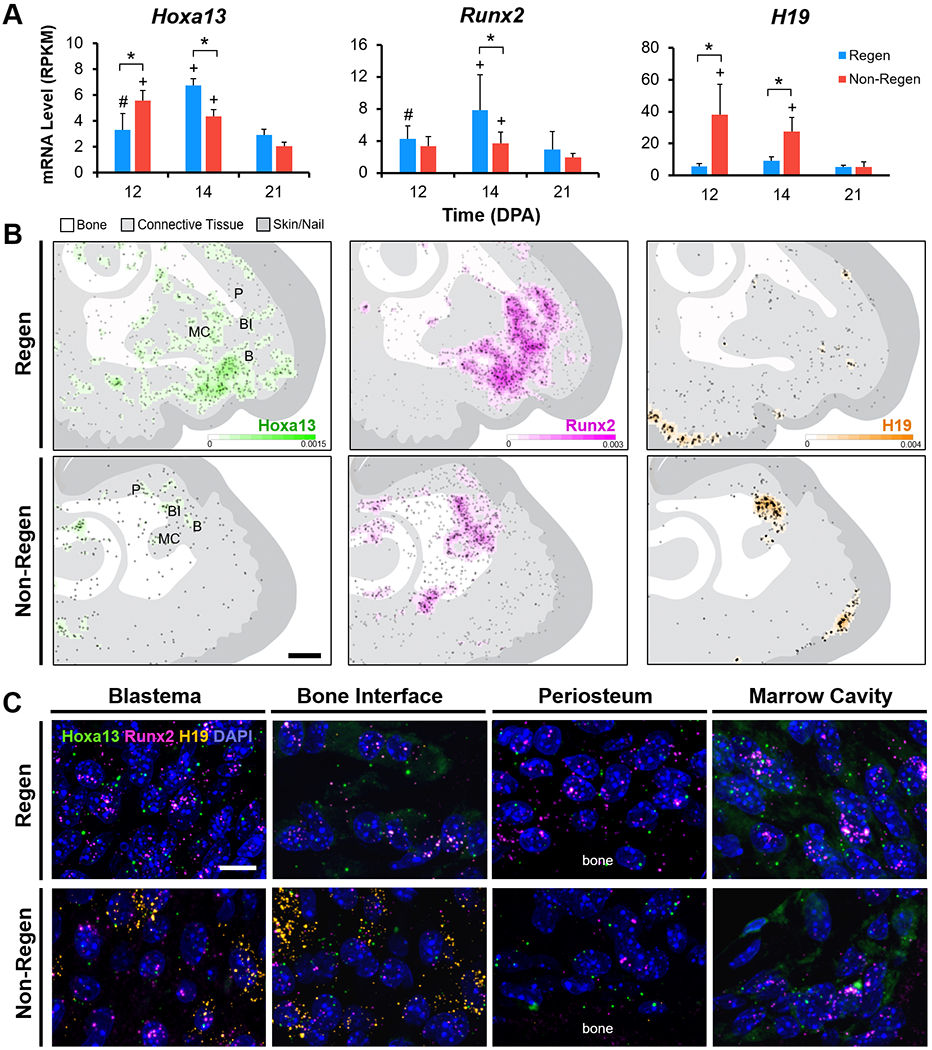
(A) Bulk RNA-seq mRNA levels (RPKM) of Hoxa13, Runx2, and H19 (n=4 mice, 4 digits/group/time point, mean±s.d.). *P<0.05 vs. Non-Regen. #P<0.05 vs. 14 DPA. +P<0.05 vs. 21 DPA. (B) Density heat maps show estimated RNA FISH probes per unit area for Hoxa13 (green), Runx2 (magenta), and H19 (orange) in Regen and Non-Regen digits at 14 DPA. Circles represent individual probe signals. Regions of interest shown in (C) include the blastema (B), bone interface (BI), periosteum (P), and marrow cavity (MC). Scale: 0.2 mm. (C) Confocal images at 63X magnification show RNA FISH probes for Hoxa13, Runx2, and H19, and cell nuclei (DAPI, blue) in various Regen and Non-Regen tissues at 14 DPA. Scale: 10 μm.
Discussion
The murine digit tip represents a unique system to investigate the factors that lead to mammalian regeneration versus scarring, as the extent of the healing process in response to amputation is dependent on the level of resection. Building upon previous studies that have extensively documented this phenomenon (8, 15–19), we established the global transcriptomic landscape of digits with differential regenerative outcomes. As expected, distal amputation (Regen) and proximal amputation (Non-Regen) groups showed significant differences in bone remodeling, ECM organization, and gene expression over time (Fig. 7). Regen digits were characterized by a limb-specific transcriptional program that result in aligned collagen deposition and ossification extending distally from the amputated cortical bone stump. Despite the presence of numerous genes related to osteogenesis, Non-Regen digits did not exhibit significant bone regrowth, but instead produced a fibrous capsule that may hinder tissue outgrowth. These findings support our hypothesis that successful digit regeneration depends on the temporal activation of genes that are implicated in limb development.
Fig. 7. Digit tip regeneration is a level-dependent and spatiotemporally controlled process.
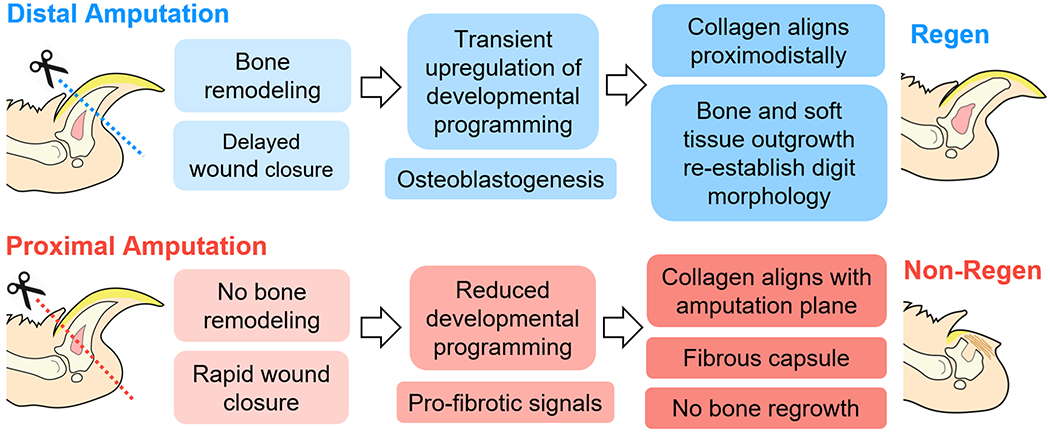
After distal amputation of the digit tip, bone degradation is followed by transient upregulation of limb-specific developmental genes and osteogenesis, where the regenerating extracellular matrix becomes aligned in the direction of tissue elongation. In contrast, proximal amputation results in minimal bone remodeling or regrowth. Regenerative failure may be due to altered developmental signals and cell populations, and/or accelerated fibrosis.
Our transcriptomic analysis, which reflects the resolution of inflammation and bone resorption with the ascension of cell migration, osteoblast differentiation, and bone formation, is consistent with previous reports (5, 7, 38). Although we did not profile the transcriptomic landscape during the entire course of regeneration, it is likely that the biological processes that occur within the first week after amputation (e.g., inflammation and histolysis) also play a significant role in shaping the regenerative outcome (38). Early regeneration prior to blastema formation is facilitated by delayed wound closure (19) and bone degradation (5), events that may promote stem/progenitor cell proliferation and migration from the marrow cavity (6, 38). In addition, localized ECM degradation releases matrix-bound growth factors, which may stimulate osteoprogenitors (39) and/or act as chemoattractants to recruit stem/progenitor cells to the wound site (40). The period spanning blastema formation and differentiation in distally amputated Regen digits is marked by the transient upregulation of genes primarily associated with skeletal/limb development and morphogenesis, as well as with nervous system development. Dawson et al. reported that the blastema is formed by a blend of periosteal and marrow/endosteal cells that differentiate in a progressive wave to form new bone distal to the P3 stump (7). Here, we demonstrate the expansion of Hoxa13+ and Runx2+ cells from the open marrow cavity and periosteum, supporting the conclusion that the blastema is comprised primarily of mesenchymal-lineage stem/progenitor cells derived from these compartments. The attenuated osteogenesis after proximal amputation is most likely due to fewer tissue-resident osteoprogenitors available to amount a regenerative response, especially those from the dorsal periosteum (7). Premature exposure of progenitor cells to a pro-inflammatory, pro-fibrotic environment may also impede blastema formation, as successful regeneration depends on the execution of a macrophage-mediated immune response (38). In addition, nerve regrowth aids the regenerative effort by delivering pro-mitotic growth factors such as fibroblast growth factor-2 (FGF-2) (17) and platelet-derived growth factor-AA (PDGF-AA) to the blastema (13), providing mesenchymal precursor cells (14), and affecting limb patterning (41). Therefore, the absence of nervous system development after proximal amputation may also hinder blastema formation and tissue outgrowth.
Appropriate spatiotemporal regulation of developmental signaling pathways is essential for the outgrowth and patterning of the limb bud. Limb formation is coordinated by the collinear expression of the HoxA and HoxD complexes (homology groups 9 to 13), a subset of evolutionarily conserved homeobox genes that determine body segment identity (28–30, 42). In adults, Hox genes are expressed by mesenchymal stem/progenitor cells in the bone marrow and periosteum, and is maintained in a regional pattern that reflects limb development (43, 44). The transient upregulation of Hox genes seen after distal amputation (Hoxa11/Hoxd11, Hoxa13/Hoxd13) suggests that this cell population is activated after injury and participates in blastema-mediated regeneration. Although we did not examine the baseline transcriptome due to the limited cellularity of uninjured digit tissues, we demonstrate that Hoxa13, a marker of the future autopod (hands and feet) during development, is abundantly expressed by stem/progenitor cells extending from the marrow cavity at 14 DPA, but is largely absent in digit tissues at homeostasis. Our analysis of transiently upregulated downstream targets of Hoxa13/Hoxd13 (Hox13) include many of the TFs identified in our GRN, including Prrx1, Alx4, Tbx2, and Runx2. Conversely, Hoxa13 expression after proximal amputation steadily declined after 12 DPA, which may reduce the expression of these downstream signals. Knocking out Hox13 leads to the loss of the autopod (28), suggesting that these genes help trigger a digit-specific program. The induction of developmentally relevant genes during digit tip regeneration brings this model closer to salamander limb regeneration, where the limb blastema cells revert to an embryonic-like state before differentiating into multiple connective tissue lineages (45, 46). However, we cannot ascertain from the bulk RNA-seq data whether these results are due to dynamic gene expression in single cells and/or fluctuations in cell populations. Similarly, it is unclear to what extent the TFs in our GRN are expressed and interact with one another within the same cell. To identify the cellular heterogeneity masked by tissue-level sequencing, single-cell RNA-seq may be utilized to evaluate which cell types are present throughout regeneration, as well as their varying stages of differentiation (14, 45–48). Another consideration in the interpretation of our findings is that the transcriptomic landscape may not correspond to protein expression due to post-transcriptional events. Therefore, an integrated analysis of transcriptomic and proteomic data, ideally with single-cell resolution, may provide additional insights into the mechanisms regulating regenerative or non-regenerative responses.
Establishing a provisional matrix that acts as an organizing scaffold is also a crucial step of digit regeneration. Col3a1, which encodes type III collagen, is transiently upregulated in the in the regenerative state but downregulated in the non-regenerative state. Type III collagen facilitates cutaneous wound healing by minimizing scar tissue deposition and myofibroblast differentiation (49). Indeed, a transient, reticular fiber and type III collagen-rich scaffold is synthesized by fibroblasts during the early stages of regeneration (11), which may provide spatial instruction for stem/progenitor cell migration and differentiation prior to osteoblast differentiation and skeletal outgrowth. Therefore, the absence of a physical pro-regenerative environment may predispose the non-regenerative case to scarring. Furthermore, differences in the expression of genes regulating collagen synthesis and/or degradation between the regenerative and non-regenerative cases may drive non-regenerative digits toward fibrous encapsulation, generating a biophysical barrier to cell proliferation, migration, and subsequent tissue regeneration (50). Mmp2, which encodes matrix metalloproteinase-2 (MMP-2), a gelatinase that participates in the secondary breakdown of collagen during remodeling, is transiently upregulated in the regenerative state but downregulated in the non-regenerative state. MMP-2 deficiency exacerbates liver and kidney fibrosis, a result of impaired degradation of matrix proteins and/or enhanced deposition of type I collagen (51). We also identified upregulated genes that may contribute to scar tissue formation in the non-regenerative digits, including transcription factors in the Fos family, which can mediate fibrosis by increasing collagen production and accumulation (52). In addition, we found elevated levels of H19, a parentally imprinted lncRNA that is expressed during embryogenesis and repressed postnatally (53), at the bone interface of non-regenerative digits compared to regenerative digits at early time points. H19 can affect fibrosis by expediting fibroblast proliferation and type I collagen production (54), and may also accelerate osteoblast differentiation at the bone interface (55), thus reducing the pool of dividing osteoprogenitors and leading to precocious ossification. Taken together, our data suggest that proximally amputated digits attempt a regenerative response around 12 to 14 DPA but ultimately fail, potentially due to a combination of reduced stem/progenitor populations, insufficient activation or premature termination of developmental pathways, and/or an enhanced fibrotic response. While we did not test the signaling role of the nail matrix (17), we observed that nail regrowth was maintained in several non-regenerative cases in the absence of bone regeneration. Therefore, nail regrowth alone cannot rescue digit regeneration in the absence of other processes. Future work will investigate how the gene candidates identified in this study, as well as the cell populations that drive their dynamic expression, may contribute to these disparate but complementary mechanisms.
In conclusion, digit regeneration is a complex process that balances both pro-regenerative and pro-fibrotic signaling pathways. While the recapitulation of developmental pathways may not be unique to regenerators, the timing of their expression may need to follow other local processes, such as inflammation and histolysis. Elucidating the cellular and molecular mechanisms that regulate regeneration will open new avenues to therapies that will help restore the structure and function of the lost tissues after proximal amputation. Preventing early scar formation by targeting fibrotic signaling pathways (56) may give stem/progenitor cells an opportunity to expand and differentiate. Alternatively, reprogramming the wound microenvironment via enzymatic degradation can trigger cell proliferation and migration (57, 58) and promote the regrowth of vasculature and nerves (58). Finally, localized temporal delivery of morphogenetic signaling molecules by genetically modified cells and/or biomaterials is a promising strategy for inducing and sustaining the regenerative process, especially with regards to skeletal elongation and synovial joint formation (18, 19, 59–61).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Mr. Nicholas Benshoff for his technical assistance with surgery, Dr. Christopher Rowland and Mrs. Sara Oswald for their imaging expertise, and the Genome Technology Access Center at Washington University in St. Louis for RNA library preparation and sequencing.
Funding
This work was supported by the National Institutes of Health (T32 AR060719 and F32 AR074895 to F. Qu, R25 DA027995 to B. Zhang, AG15768 and AG46927 to F. Guilak, P30 AR073752, P30 AR074992), the Arthritis Foundation, the Nancy Taylor Foundation for Chronic Diseases, and the Shriners Hospitals for Children.
Abbreviations
- ANOVA
analysis of variance
- BMP
bone morphogenetic protein
- cDNA
complementary DNA
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput sequencing
- CPM
count per million
- DEG
differentially expressed gene
- DPA
days post-amputation
- ECM
extracellular matrix
- FISH
fluorescence in situ hybridization
- FGF
fibroblast growth factor
- GO
gene ontology
- GRN
gene regulatory network
- Hox13
Hoxa13/Hoxd13
- H&E
Hematoxylin and Eosin
- lncRNA
long non-coding RNA
- MicroCT
micro-computed tomography
- MMP
matrix metalloproteinase
- NBF
neutral buffered formalin
- P3
terminal phalanx bone
- PCA
principal component analysis
- PDGF
platelet-derived growth factor
- PSR
Picrosirius Red
- RNA-seq
next-generation RNA sequencing
- RPKM
read per kilobase million
- RT
room temperature
- SHG
second harmonic generation
- TF
transcription factor
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
High-throughput RNA sequencing results are accessible on the GEO database (GSE131078). All relevant data are available from the authors upon request.
References
- 1.Owings MF, and Kozak LJ (1998) Ambulatory and inpatient procedures in the United States, 1996. Vital Health Stat. 13, 1–119 [PubMed] [Google Scholar]
- 2.Varma P, Stineman MG, and Dillingham TR (2014) Epidemiology of limb loss. Phys. Med. Rehabil. Clin. N. Am 25, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MJ, Stevens-Lapsley J, Fields TT, Coons D, Bray-Hall S, Sullivan W, and Christiansen CL (2017) Physical activity behavior change for older veterans after dysvascular amputation. Contemp. Clin. Trials 55, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinkevich Y, Maan ZN, Walmsley GG, and Sen SK (2015) Injuries to appendage extremities and digit tips: a clinical and cellular update. Dev. Dyn 244, 641–650 [DOI] [PubMed] [Google Scholar]
- 5.Fernando WA, Leininger E, Simkin J, Li N, Malcom CA, Sathyamoorthi S, Han M, and Muneoka K (2011) Wound healing and blastema formation in regenerating digit tips of adult mice. Dev. Biol 350, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simkin J, Sammarco MC, Dawson LA, Tucker C, Taylor LJ, Van Meter K, and Muneoka K (2015) Epidermal closure regulates histolysis during mammalian (Mus) digit regeneration. Regeneration (Oxf) 2, 106–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson LA, Schanes PP, Kim P, Imholt FM, Qureshi O, Dolan CP, Yu L, Yan M, Zimmel KN, Falck AR, and Muneoka K (2018) Blastema formation and periosteal ossification in the regenerating adult mouse digit. Wound Repair Regen. 26, 263–273 [DOI] [PubMed] [Google Scholar]
- 8.Han M, Yang X, Lee J, Allan CH, and Muneoka K (2008) Development and regeneration of the neonatal digit tip in mice. Dev. Biol 315, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehoczky JA, Robert B, and Tabin CJ (2011) Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. USA 108, 20609–20614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinkevich Y, Lindau P, Ueno H, Longaker MT, and Weissman IL (2011) Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrero L, Simkin J, Sammarco M, and Muneoka K (2017) Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration (Oxf) 4, 69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Wang K, Karapetyan A, Fernando WA, Simkin J, Han M, Rugg EL, and Muneoka K (2013) Connective tissue fibroblast properties are position-dependent during mouse digit tip regeneration. PLOS ONE 8, e54764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston AP, Yuzwa SA, Carr MJ, Mahmud N, Storer MA, Krause MP, Jones K, Paul S, Kaplan DR, and Miller FD (2016) Dedifferentiated schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 19, 433–448 [DOI] [PubMed] [Google Scholar]
- 14.Carr MJ, Toma JS, Johnston APW, Steadman PE, Yuzwa SA, Mahmud N, Frankland PW, Kaplan DR, and Miller FD (2019) Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell 24, 240–256 e249 [DOI] [PubMed] [Google Scholar]
- 15.Neufeld DA, and Zhao W (1995) Bone regrowth after digit tip amputation in mice is equivalent in adults and neonates. Wound Repair Regen. 3, 461–466 [DOI] [PubMed] [Google Scholar]
- 16.Chamberlain CS, Jeffery JJ, Leiferman EM, Yildirim T, Sun X, Baer GS, Murphy WL, and Vanderby R (2017) Level-specific amputations and resulting regenerative outcomes in the mouse distal phalanx. Wound Repair Regen. 25, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeo M, Chou WC, Sun Q, Lee W, Rabbani P, Loomis C, Taketo MM, and Ito M (2013) Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature 499, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson LA, Yu L, Yan M, Marrero L, Schanes PP, Dolan C, Pela M, Petersen B, Han M, and Muneoka K (2017) The periosteal requirement and temporal dynamics of BMP2-induced middle phalanx regeneration in the adult mouse. Regeneration (Oxf) 4, 140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu L, Han M, Yan M, Lee E-C, Lee J, and Muneoka K (2010) BMP signaling induces digit regeneration in neonatal mice. Development 137, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han M, Yang X, Farrington JE, and Muneoka K (2003) Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development 130, 5123–5132 [DOI] [PubMed] [Google Scholar]
- 21.Reginelli AD, Wang YQ, Sassoon D, and Muneoka K (1995) Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development 121, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 22.Chadwick RB, Bu L, Yu H, Hu Y, Wergedal JE, Mohan S, and Baylink DJ (2007) Digit tip regrowth and differential gene expression in MRL/Mpj, DBA/2, and C57BL/6 mice. Wound Repair Regen. 15, 275–284 [DOI] [PubMed] [Google Scholar]
- 23.Cheng CH, Leferovich J, Zhang XM, Bedelbaeva K, Gourevitch D, Hatcher CJ, Basson CT, Heber-Katz E, and Marx KA (2013) Keratin gene expression profiles after digit amputation in C57BL/6 vs. regenerative MRL mice imply an early regenerative keratinocyte activated-like state. Physiol. Genomics 45, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 26.Sheth R, Barozzi I, Langlais D, Osterwalder M, Nemec S, Carlson HL, Stadler HS, Visel A, Drouin J, and Kmita M (2016) Distal limb patterning requires modulation of cis-regulatory activities by HOX13. Cell Rep. 17, 2913–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer MB, Benkusky NA, and Pike JW (2014) The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem 289, 16016–16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, and Chambon P (1996) Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011 [DOI] [PubMed] [Google Scholar]
- 29.Villavicencio-Lorini P, Kuss P, Friedrich J, Haupt J, Farooq M, Turkmen S, Duboule D, Hecht J, and Mundlos S (2010) Homeobox genes d11-d13 and a13 control mouse autopod cortical bone and joint formation. J. Clin. Invest 120, 1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stadler HS, Higgins KM, and Capecchi MR (2001) Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development 128, 4177–4188 [DOI] [PubMed] [Google Scholar]
- 31.ten Berge D, Brouwer A, Korving J, Martin JF, and Meijlink F (1998) Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development 125, 3831–3842 [DOI] [PubMed] [Google Scholar]
- 32.Kuijper S, Feitsma H, Sheth R, Korving J, Reijnen M, and Meijlink F (2005) Function and regulation of Alx4 in limb development: complex genetic interactions with Gli3 and Shh. Dev. Biol 285, 533–544 [DOI] [PubMed] [Google Scholar]
- 33.Sheeba CJ, and Logan MP (2017) The roles of T-box genes in vertebrate limb development. Curr. Top. Dev. Biol 122, 355–381 [DOI] [PubMed] [Google Scholar]
- 34.Komori T (2010) Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol 658, 43–49 [DOI] [PubMed] [Google Scholar]
- 35.Nik AM, Johansson JA, Ghiami M, Reyahi A, and Carlsson P (2016) Foxf2 is required for secondary palate development and Tgfβ signaling in palatal shelf mesenchyme. Dev. Biol 415, 14–23 [DOI] [PubMed] [Google Scholar]
- 36.Bauer O, Sharir A, Kimura A, Hantisteanu S, Takeda S, and Groner Y (2015) Loss of osteoblast Runx3 produces severe congenital osteopenia. Mol. Cell Biol 35, 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, Adams RH, and Dejana E (2013) Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat. Commun 4, 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simkin J, Sammarco MC, Marrero L, Dawson LA, Yan M, Tucker C, Cammack A, and Muneoka K (2017) Macrophages are required to coordinate mouse digit tip regeneration. Development 144, 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin T, Gooi JH, and Sims NA (2009) Molecular mechanisms in coupling of bone formation to resorption. Crit. Rev. Eukaryot. Gene Expr 19, 73–88 [DOI] [PubMed] [Google Scholar]
- 40.Agrawal V, Tottey S, Johnson SA, Freund JM, Siu BF, and Badylak SF (2011) Recruitment of progenitor cells by an extracellular matrix cryptic peptide in a mouse model of digit amputation. Tissue Eng. Part A 17, 2435–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinkevich Y, Montoro DT, Muhonen E, Walmsley GG, Lo D, Hasegawa M, Januszyk M, Connolly AJ, Weissman IL, and Longaker MT (2014) Clonal analysis reveals nerve-dependent and independent roles on mammalian hind limb tissue maintenance and regeneration. Proc. Natl. Acad. Sci. USA 111, 9846–9851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakany J, and Duboule D (1999) Hox genes in digit development and evolution. Cell Tissue Res. 296, 19–25 [DOI] [PubMed] [Google Scholar]
- 43.Pineault KM, Song JY, Kozloff KM, Lucas D, and Wellik DM (2019) Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life. Nat. Commun 10, 3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rux DR, Song JY, Swinehart IT, Pineault KM, Schlientz AJ, Trulik KG, Goldstein SA, Kozloff KM, Lucas D, and Wellik DM (2016) Regionally restricted Hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Dev. Cell 39, 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, Currie JD, Camp JG, Tanaka EM, and Treutlein B (2018) Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, Bryant DM, Miller BM, Ratner A, Chen A, Ye WW, Haas BJ, and Whited JL (2018) Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun 9, 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson GL, Masias EJ, and Lehoczky JA (2020) Cellular heterogeneity and lineage restriction during mouse digit tip regeneration at single-cell resolution. Dev. Cell 52, 525–540. e525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storer MA, Mahmud N, Karamboulas K, Borrett MJ, Yuzwa SA, Gont A, Androschuk A, Sefton MV, Kaplan DR, and Miller FD (2020) Acquisition of a unique mesenchymal precursor-like blastema state underlies successful adult mammalian digit tip regeneration. Dev. Cell 52, 509–524. e509 [DOI] [PubMed] [Google Scholar]
- 49.Volk SW, Wang Y, Mauldin EA, Liechty KW, and Adams SL (2011) Diminished type III collagen promotes myofibroblast differentiation and increases scar deposition in cutaneous wound healing. Cells Tissues Organs 194, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu F, Pintauro MP, Haughan JE, Henning EA, Esterhai JL, Schaer TP, Mauck RL, and Fisher MB (2015) Repair of dense connective tissues via biomaterial-mediated matrix reprogramming of the wound interface. Biomaterials 39, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giannandrea M, and Parks WC (2014) Diverse functions of matrix metalloproteinases during fibrosis. Dis. Model Mech 7, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avouac J, Palumbo K, Tomcik M, Zerr P, Dees C, Horn A, Maurer B, Akhmetshina A, Beyer C, Sadowski A, Schneider H, Shiozawa S, Distler O, Schett G, Allanore Y, and Distler JH (2012) Inhibition of activator protein 1 signaling abrogates transforming growth factor beta-mediated activation of fibroblasts and prevents experimental fibrosis. Arthritis Rheum. 64, 1642–1652 [DOI] [PubMed] [Google Scholar]
- 53.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, and Dandolo L (2009) H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136, 3413–3421 [DOI] [PubMed] [Google Scholar]
- 54.Lu Q, Guo Z, Xie W, Jin W, Zhu D, Chen S, and Ren T (2018) The lncRNA H19 mediates pulmonary fibrosis by regulating the miR-196a/COL1A1 axis. Inflammation 41, 896–903 [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Zheng Y, Jia L, and Li W (2015) Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells 33, 3481–3492 [DOI] [PubMed] [Google Scholar]
- 56.Li X, Zhu L, Wang B, Yuan M, and Zhu R (2017) Drugs and targets in fibrosis. Front. Pharmacol 8, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu F, Holloway JL, Esterhai JL, Burdick JA, and Mauck RL (2017) Programmed biomolecule delivery to enable and direct cell migration for connective tissue repair. Nat. Commun 8, 1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu X, Bellayr I, Pan H, Choi Y, and Li Y (2013) Regeneration of soft tissues is promoted by MMP1 treatment after digit amputation in mice. PLOS ONE 8, e59105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taghiyar L, Hesaraki M, Sayahpour FA, Satarian L, Hosseini S, Aghdami N, and Baghaban Eslaminejad M (2017) Msh homeobox 1 (Msx1)- and Msx2-overexpressing bone marrow-derived mesenchymal stem cells resemble blastema cells and enhance regeneration in mice. J. Biol. Chem 292, 10520–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Xu H, and Lin G (2017) Generation of iPSC-derived limb progenitor-like cells for stimulating phalange regeneration in the adult mouse. Cell Discov. 3, 17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu L, Dawson LA, Yan M, Zimmel K, Lin YL, Dolan CP, Han M, and Muneoka K (2019) BMP9 stimulates joint regeneration at digit amputation wounds in mice. Nat. Commun 10, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


