Abstract
This article discusses standard and new disruptive strategies in the race to develop an anti-COVID-19 vaccine. We also included new bioinformatic data from our group mapping immunodominant epitopes and structural analysis of the spike protein. Another innovative approach reviewed here is the use of BCG vaccine as priming strategy and/or delivery system expressing SARS-CoV-2 antigens.
Keywords: COVID-19, SARS-CoV-2, RBD domain, Vaccines, BCG
The pandemic Coronavirus Disease 2019 (COVID-19) is the third global threat mediated by betacoronaviruses within this century. These enveloped viruses were thought to be restricted to animals until the first outbreak in 2002, where Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) infected around 8000 people reaching a fatality ratio of 9.6% [1]. Again in 2012, Middle East Respiratory Coronavirus (MERS-CoV) was the cause of an endemic in Middle Eastern countries, affecting almost 2500 people with approximately 35% of fatality ratio and outbreaks outside the region [1]. Less than a decade later, SARS-CoV-2 spread COVID-19 worldwide, so far with more than 18, 847, 261 confirmed cases and 708,469 deaths (data updated on 08/06/2020, available at Johns Hopkins Coronavirus Resource Center, https://coronavirus.jhu.edu/map.html). Currently, the only way to deal with this disease is through supportive care to treat the symptoms and mandatory isolation to slow down the transmission, that includes even those not affected. The course of COVID-19 and other coronaviruses diseases outbreaks can overload worldwide health systems and have severe implications in lives lost and economics, underscoring the need for effective vaccines.
SARS-CoV-2, together with SARS-CoV and MERS-CoV, belong to the cluster of betacoronoviruses, presenting large positive-sense RNA genomes. Cellular infection initiates when the spike glycoprotein binds to its cellular receptor, angiotensin-converting enzyme 2 (ACE2) for SARS-CoV and SARS-CoV-2 [2], or the dipeptidyl peptidase 4 (DPP4) for MERS-CoV [3]. After membrane fusion, the viral RNA genome is released into the cytoplasm. The replication cycle begins with the translation of non-structural proteins (nsps), whose main function is to form the replication-transcription polyprotein complex. Afterwards, a nested set of subgenomic mRNAs is transcribed into structural and accessory proteins [4,5]. A unique feature of coronaviruses is the exoribonuclease function of nsp14 that provides proofreading function to the polyprotein complex and contributes to the maintenance of the large RNA genome [6,7]. The subgenomic mRNAs encode the four structural proteins spike (S), envelope (E), membrane (M) and nucleocapsid (N), as well as proteins that interfere with the host innate immune response [[8], [9], [10]]. At the 5’ prime of SARS-Cov-2 genome we can find ORF1a (polyprotein pp1a) and ORF1b (Fig. 1 ). The ORF1a encodes 2 important proteases: (i) the papain-like and (ii) the 3C-like protease, also called Mpro [[11], [12], [13]].
Fig. 1.
Schematic organization of SARS-Cov-2 genome. The ORF1a and ORF1b comprise about 67% of the genome encoding 16 nonstructural proteins (nsps). In addition to nsps, the genome encodes the four major structural proteins: spike (S), membrane (M), nucleocapsid (N) and envelope (E).
Newly assembled structural glycoproteins are inserted in the intracellular traffic membranes located between endoplasmic reticulum and Golgi apparatus, and combine with the genomic RNA to form virions. Vesicles containing these particles are released through fusion with the plasma membrane [14]. As yet, there is no available data concerning the specifics of the SARS-CoV-2 replication cycle.
1. Lessons from the past: promising host immune components for protection against SARS-CoV-2
Currently, there are seven coronaviruses (CoVs) known to infect humans and cause respiratory diseases. Four of them, alphacoronaviruses HCoV-229E and HCoV-NL63, and betacoronaviruses HCoV-OC43 and HCoV-HKU1, cause mild upper respiratory system disease, progressing to severe infection in rare cases [15]. By contrast, SARS-CoV, MERS-CoV and SARS-CoV-2 can infect the lower respiratory tract causing acute lung injury and acute respiratory distress syndrome (ARDS). The pathogenesis of these three highly pernicious CoVs is driven by a combination of viral replication and an excessive host immune response, that leads to respiratory failure and death [[16], [17], [18]]. The progress of the disease towards a critical outcome is associated with extreme rise in proinflammatory cytokines and chemokines [18,16,19]. Genomic comparison places SARS-CoV-2 in closer proximity to SARS-CoV than to MERS-CoV [20]. It is reported that SARS-CoV-2 has around 80% genome similarity and about 95% protein homology to SARS-CoV, except for the orf10 protein, that have no biological function known so far in SARS-CoV-2 [21,20]. However, SARS-CoV-2 has an estimated 10- to 20-fold higher affinity to ACE2 and replicates more robustly, generating around 3 fold more infectious virus particles over a 48 h period than SARS-CoV. These observations can be related to the increased person-to-person transmissibility compared to the other coronaviruses [20,12].
The previous outbreaks of CoVs (SARS-CoV and MERS-CoV) gave us insights into the immune response induced during disease. These lessons can be applied to drive the efforts against the new COVID-19 pathology. During SARS (Severe Acute Respiratory Syndrome), the interaction between S protein and ACE2 leads to the downregulation of the host receptor, which contributes to lung injury [22]. This process results in the excessive production of angiotensin II, ultimately resulting in increased pulmonary vascular permeability and lung pathology [23].
The detection of RNA viruses by host innate immune system depends on recognition of pathogen associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) [24]. For CoVs in general, the most relevant PAMP is the viral double stranded (ds)-RNA, that can be sensed mainly by Toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) [25]. During SARS and MERS, the activation of the RLRs RIG-I and MDA5 (Melanoma differentiation-associated protein 5) is important to induce type I (α/β) and type III (λ) interferons (IFNs) expression and other innate responses, such as viral RNA degradation by activation of the 2′,5′-oligoadenilate synthetase (OAS)/RNAse L pathway or reduced translation in infected cell due to activation of protein kinase R (PKR) [26]. TLRs also contribute to the recognition and activation of innate responses against CoVs; both TRIF (TIR domain-containing adaptor-inducing interferon-β) and MyD88 (Myeloid differentiation factor 88) -dependent pathways are related to protection against severe disease in mice [27,28]. The engagement of cytosolic RLRs or membrane associated TLRs results in expression of interferon stimulated genes (ISGs) and NF-κB-dependent proinflammatory cytokines production [25]. Among all immune cells only macrophages have detectable, but low levels of ACE2 expression [29]. In human lung explant tissue, SARS-CoV-2 can infect pneumocytes (both type I and II cells) and alveolar macrophages [20]. However, in contrast to SARS-CoV, there is no significant trigger of IFN-β or λ expression. In this investigation of 13 proinflammatory mediators analyzed, IL-6, CCL2/MCP1, CXCL1, CXCL5 and CXCL10/IP-10 were upregulated compared to uninfected control. Among these, CXCL10/IP-10 presented higher levels compared to SARS-CoV infected tissue [20]. The lack of antiviral cytokines in the presence of intense chemokine upregulation is similarly observed in dendritic cells (DCs) exposed to SARS-CoV infection and may be related to viral immune evasion [30]. Indeed, several SARS-CoV strategies may hamper innate activation. This virus avoids detection of dsRNA by replicating in viral-induced double membrane vesicles that lack PRRs [31,4] and by capping the viral mRNAs due to methylation activities of some nsps [32]. The N protein and some accessory proteins antagonize IFN production or signaling [33,34]. Additionally, the nsp15 is an inhibitor of MAVS-induced apoptosis [35]. On the other hand, orf8 protein of SARS-CoV triggers intracellular stress pathway and activate NLRP3 inflammasome [36]. While SARS-CoV-2 may also share these activation and evasion mechanisms, their exact relevance remains undetermined.
Patients who recovered from SARS had early expression of IFNs and chemokines followed by gene expression profiles indicative of development of an adaptive immune response [37,38]. By contrast, those patients who died maintained high levels of ISG-encoded proteins, CXCL10/IP-10 and CCL2/MCP1, associated with low or absent production of spike-specific antibodies, which suggests that severe disease is related to the lack of a switch from an innate to an acquired immune response [19]. The infection with SARS-CoV induces an adaptive immune response against predominantly the structural antigens of the virus rather than nsps [39,40]. The S protein acts as a major antigen for both humoral and cellular acquired immunity [41]. Consistent with this, most peptides identified as CD4+ or CD8+ T cell epitopes are derived from the S protein, although there are also peptides from N and M proteins [42]. This observation may be related to the distribution and physiology of the structural proteins. The S protein is more exposed to the host immune system and influences virus entry; the M protein is the most abundant protein in the virion, and N protein is conserved among different coronaviruses [42]. T cell responses often target highly conserved internal proteins and promote long-term protection. Memory CD4+ T cells are more numerous at sites of infection than CD8+ T cells and have multiple roles in initiation and maintenance of efficient immune response, while cytotoxic CD8+ T cells (CTLs) eliminate infected cells [[43], [44], [45]]. CD4+ T cells protect against lethal disease through rapid local IFN-γ production and induction of neutralizing antibodies. These cells can also facilitate CD8+ T cell responses by stimulating migration of innate cells and CD8+ T cell mobilization [46]. CTL mediated protection through increased production of IFN-γ and cytotoxicity [40]. The antibody response is crucial for preventing viral infection and to reduce viral titers during ongoing infection. A subset of these antibodies is defined as neutralizing antibodies due to their capacity of blocking the entry of the virus into a healthy cell by binding to surface viral epitopes. Besides that, enveloped viruses can be eliminated through complement activation or antibody-mediated opsonization [47]. During SARS-CoV infection it seems that neutralization of viral invasion and elimination by phagocytic macrophages are the main mechanisms of protection, with neutralizing IgGs playing a major role [48]. Memory B cells provide long-lasting protection in SARS in association with cellular immune responses [49]. However, the protective ability of T cells is desirable since SARS-CoV antibodies levels decline rapidly after recovery. SARS-CoV-specific memory T cells persisted for at least 6 years in patients who have recovered from the disease while memory B cell response was not detected by this time [50].
The early research on SARS-CoV-2 infection identified some peculiarities that should be considered for medical interventions, such as vaccine development. Some COVID-19 patients secreted excessive IL-4, IL-13 and IL-10, which may be an indicator of inflammatory suppression via T-helper 2 or regulatory immune responses [16]. In most cases lymphopenia occurs, with significant reduction in T cells and NK cells numbers in severe cases. This clinical outcome is associated with functional exhaustion of cytotoxic lymphocytes [16,51]. Moreover, the high CXCL10/IP-10 production during SARS-CoV-2 infection is a sign of virulence since high and persistent levels of this chemokine are associated with death of patients in all recent epidemic CoVs infections. Some patients with COVID-19 develop a defective immune response that may lead to accumulation of immune cells in the lungs and overproduction of pro-inflammatory cytokines/chemokines, causing severe damage to the lung and a systemic pathogenesis [52]. A simple and direct approach that is been investigated to combat rapidly the COVID-19 is passive immunity through the use of plasma from convalescent patients [53]. Even though promising, the outcomes of this therapy are unpredictable due to variability of sera from different patients. Antibody-based therapies and vaccines should dedicate great effort to safety evaluation given the reports of antibody-dependent enhanced SARS-CoV entry and antibody-mediated lung injury during SARS in experimental models [[54], [55], [56]]. However, the viral peptides that induce protection can be identified and distinguished from the detrimental ones through epitope design [55]. Vaccine strategies include live attenuated strains, inactivated whole virions, nucleic acids (DNA and RNA) and recombinant proteins, most of them focusing on the highly immunogenic aspect of the spike protein.
2. The spike protein: the main target of host immune response
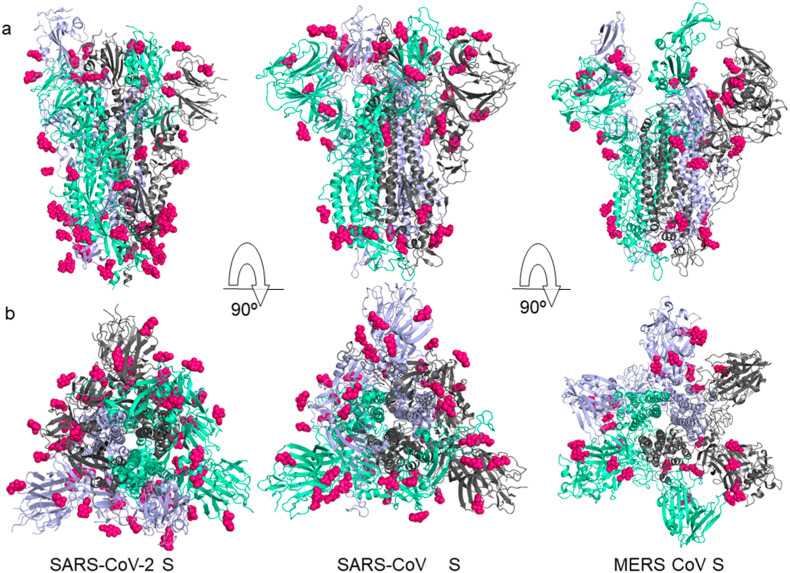
It is known that SARS-CoV-2 enters the host cells by the fusion of viral and cellular membranes [[57], [58], [59], [60], [61]] with the densely glycosylated spike protein. The S protein is a class I viral fusion protein and an important target for antibody neutralization and vaccine development. The role of glycosylation in camouflaging immunogenic protein epitopes has been studied for other coronaviruses. High viral glycan density and local protein architecture can influence the trafficking of recombinant immunogen to germinal centers [62]. Fusion S proteins of SARS-CoV-2, SARS-CoV and MERS-CoV are examples of viral proteins that are widely glycosylated [63]. The spike protein is comprised of three protein domains (Fig. 2 ) [12]. They participate in the cell recognition process by binding to host cell receptors and facilitating membrane fusion [58]. Analysis shows around 69-87 N-linked glycosylation sites identified on the surface of each trimeric peak in SARS-CoV-2 S protein, and approximately 22 N-linked glycosylated amino acids per domain (PDB ID 6VSB) [62]. The receptor binding sites formed by amino acids and glycan residues are a common feature of viral glycoproteins, as observed on SARS-CoV S and MERS-CoV S (respectively, PDB ID 5X58 and PDB ID 5X59) [64]. This structural glycosylation has functions in viral pathology such as mediating protein folding and stability, besides shaping viral tropism [65]. Thus, these glycosylation sites show selective pressure, as they facilitate evasion of the host immune system by protecting specific epitopes that induce antibodies or T cell responses [66,67].
Fig. 2.
Structural comparison between the SARS-CoV-2 S (PDB ID: 6VSB), SARS-CoV S (PDB ID: 5X58) and MERS-CoV S (PDB ID: 5X59) proteins. The individual protomer of S protein trimer domain is represented in a different color: violet, clear green and gray, respectively. a) Side and b) top conformation views of the prefusion structure of the S glycoproteins with N-linked glycans rendered as hot pink spheres.
S protein allows the virus to attach on the target cell and fuse membranes, and it is presented as a trimeric structure on the viral surface [68,69]. During the infection phase, S proteins are cleaved into two subunits: (a) receptor binding subunit, S1, and (b) membrane fusion subunit, S2. It uses the N-terminal region as a signal sequence to access the endoplasmic reticulum of cells and this region is highly N-glycosylated. The S1 subunit of S protein has a receptor binding domain (RBD) composed of a central subdomain (core) and a connection motif for the receptor. The central subdomain has 5 beta antiparallel sheets, connected by alpha-helices, and stabilized by 3 disulfide bridges. The protein’s S2 subunit is also similar to that of SARS-CoV and is responsible for the fusion of membranes from severe conformational changes [69,59,70]. S2 has two regions of heptad repeats (HR). During the fusion process, S2 dissociates from S1 and the HR1 and HR2 regions and form a 6-helix bundle (6-HB) structure, exposing a hydrophobic fusion peptide inserted in the host membrane and allowing the membrane to approach the virus for fusion.
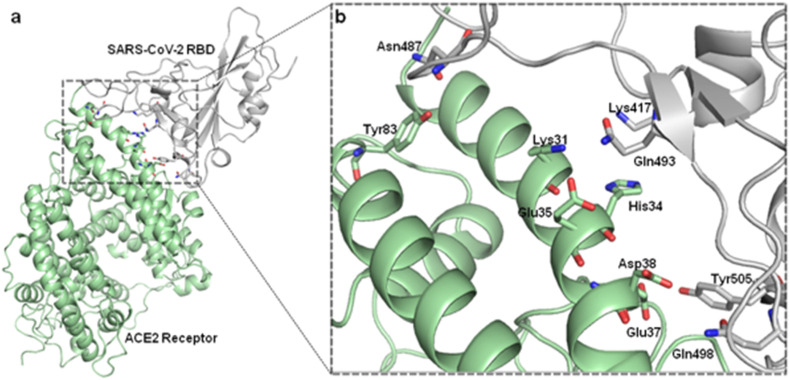
Recent experimental data support biophysical and structural evidence that the glycosylated S protein of SARS-CoV-2 binds to the ACE2 receptor with greater affinity than that of SARS-CoV [12]. Hence, the atomic level understanding of these interactions is important for the structural and biophysical elucidation of the initial virus infection process in human cells. By screening the experimentally-determined SARS-CoV-derived B cell and T cell epitopes in the immunogenic structural proteins of SARS-CoV, many authors identified a set of B cell and T cell epitopes derived from the spike and the nucleocapsid proteins that map identically to SARS-CoV-2 proteins. As no mutation has been observed in these identified epitopes among the 120 available SARS-CoV-2 sequences (as of February 2020), immune targeting of these epitopes may potentially offer protection against this novel virus. These results provide a screened set of epitopes that can help guide experimental efforts towards the development of vaccines against SARS-CoV-2. The identification of critical residues involved in receptor binding is important for the development of vaccines and inhibitor targets [63]. These studies provide regions to be modeled and investigated either experimentally or theoretically by computer simulations [42]. For this purpose, Briell et al. compared the interaction between ACE2 and peak SARS-CoV-2 S protein with that of other pathogenic coronaviruses using classical molecular dynamics (MD) simulations [71]. The authors observed that SARS-CoV and SARS-CoV-2 have comparable binding affinities. However, the complex formed by the interaction and fusion between SARS-CoV-2 and ACE2 contains a greater number of contacts between the amino acid residues of the side chains of the proteins. These data imply an evolution due to mutations to increase cell recognition and has implications for therapeutic strategies [72]. Veeramachaneni et al. evaluated through computational analysis the structural changes caused by specific mutations, hot spots binding residues and their interactions between the SARS-CoV-2 S RBD protein receptor and ACE2 [73]. We performed molecular dynamic simulations of the complexes. The major amino acids involved in the binding identified by interaction analysis after simulations, include the Glu 35, Tyr 83, Asp 38, Lys 31, Glu 37, His 34 amino acid residues of the ACE2 receptor and Gln 493, Gln 498, Asn 487, Tyr 505 and Lys 417 residues in the SARS-CoV-2 S protein RBD. By locating these amino acid residues, the authors propose that blockers can be designed to inhibit binding and interrupt the entry of the SARS-CoV-2 virus into host cells (Fig. 3 ).
Fig. 3.
Structure of SARS-CoV-2 RBD in fusion conformation and interaction with the angiotensin-converting enzyme 2 (ACE2) (PDB ID: 6M0J). Proteins are individually colored in gray (SARS-CoV-2 RBD) and green (ACE2 Receptor). Sticks representation with colors by elements show the amino acid residues Tyr83, Lys31, Glu35, His34, Asp38, Glu37 in ACE2 receptor and Asn487, Lys417, Gln493, Tyr505 and Gln498, residues in SARS-CoV-2 RBD.
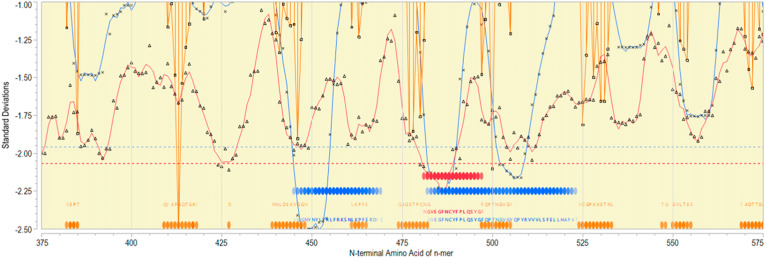
By using a computational approach, the RBD region of the SARS-CoV-2 S protein was explored to identify various immunodominant epitopes for the development of diagnostics and vaccines. B cell linear epitope probability and MHC binding affinity were determined for all sequential peptides with a single amino acid displacement by our group, using an updated version of methods previously described [74]. The results obtained here could also help us to understand the SARS-CoV-2 surface protein response towards T- and B-cells. Mapping of predicted B and T cell epitopes indicates that the most probable B cell epitopes are located throughout the RBD region of Spike protein (Fig. 4 ). In the region of amino acids 450–525 there are also two regions where sequential 15-mers are predicted to have high affinity binding for many human MHC II DRB alleles. Within the region of amino acids 475–500 there is also a region of sequential 9-mers predicted to bind to multiple MHC class I alleles. This indicates that the RBD region of Spike from SARS-CoV-2 is most likely to elicit a strong antibody response due to the number of B cell epitopes with associated T cell help predicted and may also elicit CTLs.
Fig. 4.
Epitope mapping of RBD region of Spike protein. X axis indicates sequential peptides with single amino acid displacement. Y axis indicates predicted binding affinity in SD units for the protein. The red line shows the permuted average predicted MHC-IA and B (37 alleles) binding affinity by index position of sequential 9-mer peptides with single amino acid displacement. The blue line shows the permuted average predicted MHC-II DRB allele (16 most common human alleles) binding affinity by index position of sequential 15-mer peptides. Both are plotted in standard deviation units, shown on the Y axis. Orange lines show the predicted probability of B-cell receptor binding for an amino acid centered in each sequential 9-mer peptide. Low numbers for MHC data represent high binding affinity, whereas low numbers equate to high B cell receptor contact probability. Ribbons (red: MHC-I, blue: MHC-II) indicate the 10% highest predicted MHC affinity binding. Orange ribbons indicate the top 25% predicted probability B-cell binding. Horizontal dotted lines demarcate the top 5% of binding affinity for the protein (red MHC I, blue MHC II).
3. Vaccine strategies against SARS-CoV-2
The spread of COVID-19 challenged the world to accelerate research in companies and universities in the search for a safe and effective vaccine against SARS-CoV-2 [75]. At the time of writing (August, 2020), at least 165 teams are working on different projects towards the same goal of developing a COVID-19 vaccine. The development process has been optimized with at least 26 groups already starting to test their vaccine candidates in clinical safety trials and many others are testing their candidates in cells and animals. Researchers are investing in different strategies and technologies, but most of these novel approaches have not been extensively tested for safety and do not have large-scale manufacturing capacity to produce the high number of doses needed. For this reason, the many candidates still in the race are creating real possibilities and new knowledge in vaccine design [76,77].
Twenty-six vaccine candidates are in clinical trials, six of which are in final clinical tests (phase 3) using different designs and vaccine strategies (Table 1 ). Among the SARS-CoV-2 vaccines under development are whole virus vaccines, nucleic acids vaccines, viral vector vaccines and protein-based vaccines. Virus vaccine are the most common platform used for other diseases. SARS-Cov-2 could be used as an inactivated or live attenuated virus that conserves most of the virus antigens. There are currently three vaccines based on inactivated virus already in phase 3 clinical trials, including one vaccine from SINOVAC (NCT04456595) and two from Sinopharm (ChiCTR2000034780). Instead of using the whole virus, many studies use protein subunit vaccines including antigens with strong immunogenicity, most focusing on the spike protein or only the receptor binding domain. Another protein-based vaccine strategy consists of virus-like particles (VLP) that mimic the SARS-CoV-2 structure on the surface of a non-replicative empty virus shell lacking genetic material [78,76,79]. Several virus (e.g.: vesicular stomatitis virus (VSV), influenza, measles and adenovirus) could also be engineered as replicative or non-replicative recombinant vector expressing coronavirus S protein. A live-attenuated recombinant vesicular stomatitis virus (VSV) expressing the Ebola glycoprotein (VSV-EBOV) successfully completed a phase III clinical trial and was shown to be safe and immunogenic against the Ebola virus [80]. The same strategy was used to make an attenuated VSV recombinant expressing the SARS-CoV spike protein (VSV–S) able to control a challenge with SARS-CoV and stimulating neutralizing antibody in mice [81,82]. The ChAdOx1 nCov19 vaccine developed at the University of Oxford is a very promising candidate that uses a replication-deficient chimpanzee adenovirus to deliver SARS-CoV-2 antigen protein. A single dose of ChAdOx1 nCov19 has protected six rhesus macaques from pneumonia caused by SARS-CoV-2, pushing the vaccine to a phase 3 (ISRCTN89951424) clinical trial [83,77]. Another phase 2 candidate from CanSino Biological Inc./Beijing Institute of Biotechnology uses non replicating Adenovirus type 5 as a vector based on the same platform strategy used for Ebola (Phase 2 - ChiCTR2000031781) [77,84].
Table 1.
Major COVID-19 candidate vaccines in clinical evaluation.
| Platform vaccinea | Consortium | Candidate vaccine | Clinical Stage | Clinical trial register |
|---|---|---|---|---|
| Inactivated | Sinovac | Adsorbed COVID-19 (inactivated) | Phase 3 | NCT04456595 |
| Wuhan Institute of Biological Products/Sinopharm | Inactivated novel coronavirus pneumonia (COVID-19) | Phase 3 | ChiCTR2000034780 | |
| Beijing Institute of Biological Products/Sinopharm | Inactivated novel coronavirus (2019-CoV) | Phase 3 | ChiCTR2000034780 | |
| Institute of Medical Biology, Chinese Academy of Medical Sciences | Inactivated SARS-CoV-2 | Phase 1/2 | NCT04470609 | |
| Bharat Biotech | Whole-Virion Inactivated SARS-Cov-2 Vaccine (BBV152) | Phase 1/2 | NCT04471519 | |
| Non-Replicating Viral Vector | University of Oxford/AstraZeneca | ChAdOx1 nCoV-19 | Phase 3 | ISRCTN89951424 |
| CanSino Biological Inc./Beijing Institute of Biotechnology | Recombinant Novel Coronavirus (2019-nCOV) Vaccine (Adenovirus Vector) | Phase 2 | ChiCTR2000031781 | |
| Janssen Pharmaceutical Companies | Ad26COVS1 | Phase 1/2 | NCT04436276 | |
| Gamaleya Research Institute | Gam-COVID-Vac Lyo | Phase 1 | NCT04437875 | |
| RNA | Moderna/NIAID | mRNA-1273 | Phase 3 | NCT04470427 |
| BioNTech/Fosun Pharma/Pfizer | BNT162b1 | Phase 3 | NCT04368728 | |
| Arcturus/Duke-NUS | ARCT-021 | Phase 1/2 | NCT04480957 | |
| Imperial College London | LNP-nCoVsaRNA | Phase 1 | SRCTN17072692 | |
| Curevac | CVnCoV | Phase 1 | NCT04449276 | |
| People’s Liberation Army (PLA) Academy of Military Sciences/Walvax Biotech. | SARS-CoV-2 mRNA | Phase 1 | ChiCTR2000034112 | |
| DNA | Inovio Pharmaceuticals/International Vaccine Institute | INO-4800 | Phase 1/2 | NCT04336410 |
| Osaka University/AnGes/Takara Bio | AG0301-COVID19 | Phase 1/2 | NCT04463472 | |
| Cadila Healthcare Limited | Novel Corona Virus-2019-nCov | Phase 1/2 | CTRI/2020/07/026352 | |
| Genexine Consortium | GX-19 | Phase 1/2 | NCT04445389 | |
| Protein Subunit | Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | Recombinant New Coronavirus | Phase 2 | NCT04466085 |
| Novavax | SARS-CoV-2 rS (COVID-19) Nanoparticle | Phase 1/2 | NCT04368988 | |
| Kentucky Bioprocessing, Inc | KBP-COVID-19 | Phase 1/2 | NCT04473690 | |
| Clover Biopharmaceuticals Inc./GSK/Dynavax | SCB-2019 | Phase 1 | NCT04405908 | |
| Vaxine Pty Ltd/Medytox | Monovalent Recombinant COVID19 Vaccine (COVAX19) | Phase 1 | NCT04453852 | |
| University of Queensland/CSL/Seqirus | SARS-Cov-2 Sclamp Protein Subunit Vaccine | Phase 1 | ACTRN12620000674932p | |
| Medicago Inc. | Coronavirus-Like Particle COVID-19 | Phase 1 | NCT04450004 | |
| Medigen Vaccine Biologics Corporation/NIAID/Dynavax | MVC-COV1901 | Phase 1 | NCT04487210 |
Source: WHO - DRAFT landscape of COVID-19 candidate vaccines (07/31/2020).
Another strategy is focused on nucleic acid manipulation to build DNA or RNA vaccines. A genetic construct coding for specific antigen(s) can be easily synthetized in DNA or RNA and inserted into human cells to generate many copies of the immunogenic virus protein. DNA vaccines are typically generated by a plasmid DNA containing eukaryotic expression elements that encode one or more antigens. The plasmid contains components that allows growth and selection of the vector in bacteria, such as Escherichia coli, followed by a purification process. The eukaryotic expression cassette contains a 5′ promotor, the gene of interest and a 3′ polyadenylation (poly A) signal, important for nuclear export, translation and stability of the transcript mRNA [[85], [86], [87]]. DNA vaccines are stable but have the challenge of needing to cross two cellular membranes before entering the nucleus, and may also bring the risk of vector integration in the human genome. The most common route of DNA vaccine administration is intramuscular or intradermal injection. However, DNA vectors alone generally lead to relatively low immunogenicity. Therefore, different delivery methods have been developed to improve DNA uptake, including the gene gun, needle free injection (jet injection) and in vivo electroporation [88,86].
The mRNA vaccines manufacturing process is essentially chemical, comprising an in vitro transcription from a linearized DNA template, then removing the template by digestion with DNAses to get a purified mRNA. mRNA comprises a 5′ cap, a 5′ untranslated region (UTR) (leader RNA), the coding sequence with a stop signal, a 3’ UTR, and a poly(A) tail. mRNA enters into the cytoplasm as a template to be translated making multiple copies of the antigen(s) protein(s) [87]. Various mRNA vaccine platforms have been developed to render the synthetic RNA sequence more translatable, stable and non-toxic. The use of “naked” mRNA is not recommended because RNA is highly unstable under physiological conditions, due to the extracellular ribonucleases which catalytically hydrolyze RNA, and due to the hydrophilicity and strong negative charge of RNA it is not taken up efficiently by the cells. Strategies for optimizing mRNA vaccines includes: synthetic cap analogues and capping enzymes to stabilize mRNA and increase protein translation, poly(A) tail to stabilizes mRNA, modified nucleosides that decrease innate immune activation, modulation of target cells that alters translation and immunogenicity, etc. mRNA vaccine delivery systems often encase RNA in a lipid coat so it can enter cells or alternatively involves ex vivo loading of mRNA to target cells (e.g. dendritic cells) followed by re-infusing the cells into the autologous vaccine recipient to initiate the immune response [89]. Furthermore, vesicles containing mRNA could display some specific proteins on their surface which direct target the vaccine to certain tissues or cells.
Nucleic acid vaccines (DNA and RNA) have a number of immunostimulatory mechanisms, which may be useful or detrimental for vaccine purposes. DNA vaccines activate both humoral and cellular immune responses, respectively through CD4+ and CD8+ T cells activation. DNA vaccine inside the cells have the potential to activate the innate immune pathways, mainly related to TLR9/MyD88, STING/TBK1/IRF3 and the AIM2 inflammasome [[90], [91], [92], [93], [94], [95],86]. Exogenous mRNA is also recognized by endosomal and cytosolic innate immune receptors with a high immunostimulatory capacity. The innate immune pathway sensed by mRNA includes TLR3, TLR7, TLR8 located in the endosomes and RIG-I, MDA-5, NLRP3 and NOD2 located in the cytoplasm [86,[96], [97], [98]]. Immunization with nucleic acids vaccines upregulates the expression of important cytokines such as type I interferons and chemokines (CXCL9, CXCL10, and CXCL11) that recruit innate immune cells such as dendritic cells and macrophages, thereby enhancing adaptive immune responses against the expressed antigens.
DNA vaccines were first tested in 1993, showing protective immunity against influenza in mice [99]. In the same year, a liposome-entrapped mRNA vaccine in mice was shown to induce virus-specific cytotoxic T lymphocytes response [100]. After decades, new techniques and formulations have improved, so SARS-CoV-2 candidates could possibly be the first licensed human nucleic acid vaccine. Previous studies of SARS-CoV and MERS-CoV paved the way for several groups to start working on SAR-CoV-2 vaccine right after recognition of the outbreak [101]. The target antigen (full-length S protein or S domain subunits) selection and vaccine platform are based on this previous knowledge [11,66,102]. In silico studies showed that SARS-CoV-2 structural proteins are genetically similar to SARS-CoV, but more distant from MERS. The previous knowledge and comparison among well-known virus (SARS-CoV and MERS) and SAR-CoV-2 provided a screened set of epitopes candidates that can help guide experimental efforts and accelerate the development of specific SARS-CoV-2 vaccine [67,103,104].
The first full genome sequence of SARS-CoV-2 was made public in January 2020, and within a few days the U.S. National Institutes of Health (NIH) and Moderna’s infectious disease research team finalized the sequence for mRNA-1273. Two months later they started a safety phase I clinical trial to test a lipid nanoparticle (LNP) dispersion containing an mRNA that encodes for the prefusion stabilized spike protein 2019-nCoV and they are now conducting the final clinical testes (Phase 3 - NCT04470427). Inovio Pharmaceuticals also focused on the S protein to build a DNA plasmid vaccine administered intradermally followed by electroporation that is being tested on healthy volunteers (Phase 1/2 - NCT04336410). So far, there are 15 projects involving DNA vaccines and 19 involving RNA vaccines (Table 1) [78,77]. Nucleic acid vaccines are promising, but there is still no licensed manufacturing platform. However, recent improvement in nucleic acid vaccine stability and protein translation efficiency combined with large financial investments may bring a new disruptive vaccine technology.
4. BCG as a priming strategy or delivery system for SARS-CoV-2 vaccine
Mycobacterium bovis BCG (Bacillus Calmette-Guerin) vaccine against tuberculosis (TB) is the most widely used vaccine in the world, has an excellent safety standard and has been shown to be also an effective adjuvant inducing cellular immunity in animals and humans. BCG is a live attenuated vaccine produced in more than 40 sites around the world using different substrains (Connaught, Danish, Glaxo, Moreau, Moscow, Pasteur and Tokyo) which are not identical in efficacy, safety and immunogenicity. The World Health Organization (WHO) adopted requirements for BCG vaccine in 1965 and still maintains lyophilized seed of the vaccine strains to prevent deviation from the original BCG [[105], [106], [107], [108]]. BCG is a slow-growing organism and provides low-level and persistent antigenic exposure, favoring the induction of a long-lasting cellular and/or humoral T-cell immune response with just one dose [109,110]. In addition, BCG vaccine induces protection in neonates, has high stability, well-established large-scale production and low cost.
Recombinant BCG (rBCG) maintains all BCG’s characteristics. rBCG has also been investigated as a vehicle for antigen delivery strategy against different pathogens. It is possible to construct rBCG strains expressing different levels of viral, bacterial or parasitic pathogens antigens, resulting in the activation of cellular and/or humoral immune response depending on the vector and antigen [111]. BCG vaccine expressing HIV immunogens demonstrated its efficiency in activating the production of cytokines and T cell responses in mice, showing a strong potential as an integrative vaccine against HIV-1/TB or as a priming associated with other virus vector boost vaccines [[112], [113], [114], [115]]. Different rBCG strains expressing specific antigens have induced protection against the challenge with the respective pathogen, including Borrelia burgdorferi, Streptococcus pneumoniae, Leishmania major and Plasmodium falciparum [[116], [117], [118], [119], [120]]. Currently, rBCG studies are also focused on improving the BCG vaccine efficacy against tuberculosis [121,122].
In addition to inducing a specific anti-TB immune response, BCG vaccination appears to have other effects that have been associated with decreased infant mortality and a lower prevalence of other infections [[123], [124], [125]]. The revaccination with BCG in adults or the elderly can reduce respiratory tract infections [126,127]. This may be due to the trained innate immunity mechanism caused by metabolic changes, epigenetic reprogramming, cytokines releases, monocyte activation and improved host immune response after BCG vaccination [[128], [129], [130], [131]]. The COVID-19 pandemic raised the question whether BCG vaccine could offer protection or be used as a tool in the fight against SAR-Cov-2. The phenomenon has recently been correlated with the possibility that countries where vaccination for TB is mandatory would (so far) have a lower incidence of COVID-19 cases. However, as we are comparing very different cultures and people, it is difficult to exclude other variables that could confuse the analysis and some results are contradictory. Another aspect that needs to be taken into account is that the persistence and immunostimulatory properties of BCG strains differ and it could lead to a variable trained immunity response [[132], [133], [134], [135], [136], [137], [138]]. A recent study compared infection rates and proportions of severe COVID-19 in two similar populations in Israel, comprising one group with individuals born during the 3 years before and other group born 3 years after cessation of the Universal BCG vaccine program, resulting in no statistically significant difference between BCG-vaccinated vs unvaccinated group [139]. To better investigate the correlation between BCG vaccine and SARS-Cov-2 infection, large clinical trials, including one already started in Australia (https://clinicaltrials.gov/ct2/show/NCT04327206) and others in different countries, propose to test BCG vaccination in health professionals naturally exposed to COVID-19 infection to determine whether heterologous protection exists or not. The heterologous effect of BCG vaccination remains a vast field for research.
Our group is working in collaboration with national and international institutes to develop a new vaccine strategy using rBCG to express the immunodominant epitopes in the RBD region of the S protein from SARS-CoV-2 (Fig. 4) that can lead to immune system activation synergistically with BCG recognition. In addition, another interesting strategy would be to use a BCG vector to express other agonists that activate important innate immunity pathways involving type I IFN, which play an important role in the anti-viral response and in the recruitment of lymphocytes [[140], [141], [142]]. The concern that people already vaccinated with BCG could mount an immune response against the vector, preventing it from delivering the spike protein antigen into human, does not seem to be relevant given the satisfactory results with other studies involving rBCG persistence in the host. Another concern involves the release of IL-6 and other inflammatory cytokines that could aggravate COVID-19 pathology. However, previous studies related to other diseases suggests that BCG could lead to a decrease in viremia at the beginning of the SARS-Cov-2 infection, before a possible cytokine storm happens, thus preventing a systemic inflammation and severe disease [131,143]. Netea and collaborators suggest that BCG vaccine could be used at the beginning of a pandemic as a booster of the host defense, even if effective for a limited period of time, it might contribute reducing SARS-CoV-2 spread and help fight the pandemic until a specific vaccine against COVID-19 can be developed [135,134]. rBCG vaccine can be explored as a priming strategy together with other virus specific vaccines, as a potent adjuvant and/or as a delivery system for SARS-CoV-2 proteins in host cells. The exploration of rBCG’s potential for the development of new products can help to solve this and other epidemic outbreaks that may arise.
5. Conclusions
During SARS-CoV-2 infection, immune responses are believed to be essential for viral infection clearance and immunological memory. However, they also cause collateral damage to the lung tissue that can be detrimental and even fatal in some cases. This is a comprehensive review that has focused on host immune responses to SARS- CoV-2 infection, potential epitope targets for vaccine development and different vaccine strategies from live viral vectors, protein-based vaccines, nucleic acid vaccines and the use of BCG as potential delivery system to boost antiviral response via trained immunity. Additionally, we added original data on prediction of B cell and T cell epitopes on RBD region of spike protein, structural comparison analysis between the SARS-CoV-2, SARS-CoV and MERS CoV S proteins and interaction of SARS-CoV-2 RBD with ACE2 receptor. Here, we discussed the hypothesis that BCG vaccination might be a potent preventive measure against SARS-CoV-2 infection and/or may reduce COVID-19 disease severity. One critical vaccine target is to raise antibodies directed to the SARS-CoV-2 spike protein and its receptor-binding domain, the component required for virus binding to its host cell entry receptor ACE2. Given the immediate threat of the SARS-CoV-2 pandemic, vaccine trials should be designed and started as pragmatic studies with feasible primary end points that can be performed rapidly and that could provide results in a short period. Due to limitations in vaccine development, randomized controlled trials are needed to provide the highest quality proof that these vaccines can protect against COVID-19. Additionally, we must also recognize that there are potential safety issues that could slow the clinical development path and testing. Since, there is a desperately urgent need to develop strategies to restrain SARS-CoV-2 and limit the pandemic, worldwide efforts are gathered to move forward with all these vaccine candidates already in clinical testing and development.
Declaration of competing interest
EJH is an employee and equity holder in ioGenetics LLC. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgements
This study was carried out with the financial support of CNPq - Brazil (grants# 302660/2015-1, 401209/2020-2 and 465229/2014-0), CAPES - Brazil (grants# 88887.506611/2020-00, #88887.504420/2020-00 and #88887-364940/2019-00), National Institutes of Health – United States (grant# R01 AI116453), FAPESP - Brazil (grant# 2017/24832-6) and FAPEMIG Rede Mineira de Imunobiologicos - Brazil (grant# REDE-00140-16).
References
- 1.WHO Summary of MERS cases with onset of illness from 1 September 2012. https://www.who.int/emergencies/mers-cov/en/ Available at:
- 2.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi P., Su Y., Li R., Liang Z., Dong S., Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 14.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020:12. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto M., Tsukamoto H., Kouwaki T., Seya T., Oshiumi H. Recognition of viral RNA by pattern recognition receptors in the induction of innate immunity and excessive inflammation during respiratory viral infections. Viral Immunol. 2017;30:408–420. doi: 10.1089/vim.2016.0178. [DOI] [PubMed] [Google Scholar]
- 25.Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.tenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Totura A.L., Whitmore A., Agnihothram S., Schafer A., Katze M.G., Heise M.T. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6 doi: 10.1128/mBio.00638-15. e00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law H.K., Cheung C.Y., Sia S.F., Chan Y.O., Peiris J.S., Lau Y.L. Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol. 2009;10:35. doi: 10.1186/1471-2172-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Gene. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J. MAVS-mediated apoptosis and its inhibition by viral proteins. PloS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Dis. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H.S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.S. Immune responses to middle east respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis. 2019;68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antivir Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner D.L., Farber D.L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front Immunol. 2014;5:331. doi: 10.3389/fimmu.2014.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Peng H., Zhu Z., Li G., Huang Z., Zhao Z. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J Gen Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coughlin M.M., Prabhakar B.S. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev Med Virol. 2012;22:2–17. doi: 10.1002/rmv.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454–455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 51.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruse R.L. Vol. 9. 2020. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res; p. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci U S A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letko M., Munster V. Functional assessment of cell entry and receptor usage for lineage B beta-coronaviruses, including 2019-nCoV. bioRxiv. 2020 doi: 10.1101/2020.01.22.915660. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang Y., Wang M.L., Chien C.S., Yarmishyn A.A., Yang Y.P., Lai W.Y. Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 infection. Front Immunol. 2020;11:1022. doi: 10.3389/fimmu.2020.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ou X., Guan H., Qin B., Mu Z., Wojdyla J.A., Wang M. Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1. Nat Commun. 2017;8:15216. doi: 10.1038/ncomms15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kandeel M., Ibrahim A., Fayez M., Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92:660–666. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020 doi: 10.1126/science.abd4251. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of Coronaviruses. J Biol Chem. 2020 doi: 10.1074/jbc.REV120.013930. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brielle E.S., Schneidman-Duhovny D., Linial M. The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses. 2020;12:497. doi: 10.3390/v12050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Veeramachaneni G.K., Thunuguntla V., Bobbillapati J., Bondili J.S. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bremel R.D., Homan E.J. An integrated approach to epitope analysis II: a system for proteomic-scale prediction of immunological characteristics. ImmunomeRes. 2010;6:8. doi: 10.1186/1745-7580-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. N Engl J Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 76.Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 77.WHO Draft landscape of Covid-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines Date last accessed: June 18, 2020. Date last updated: September 2020.
- 78.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines (Basel) 2020;8:153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henao-Restrepo A.M., Camacho A., Longini I.M., Watson C.H., Edmunds W.J., Egger M. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kapadia S.U., Rose J.K., Lamirande E., Vogel L., Subbarao K., Roberts A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology. 2005;340:174–182. doi: 10.1016/j.virol.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kapadia S.U., Simon I.D., Rose J.K. SARS vaccine based on a replication-defective recombinant vesicular stomatitis virus is more potent than one based on a replication-competent vector. Virology. 2008;376:165–172. doi: 10.1016/j.virol.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.05.13.093195. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu F.C., Wurie A.H., Hou L.H., Liang Q., Li Y.H., Russell J.B. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 85.Williams J.A. Vector design for improved DNA vaccine efficacy, safety and production. Vaccines (Basel) 2013;1:225–249. doi: 10.3390/vaccines1030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines (Basel) 2019;7:37. doi: 10.3390/vaccines7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lambricht L., Lopes A., Kos S., Sersa G., Preat V., Vandermeulen G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expet Opin Drug Deliv. 2016;13:295–310. doi: 10.1517/17425247.2016.1121990. [DOI] [PubMed] [Google Scholar]
- 89.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tudor D., Dubuquoy C., Gaboriau V., Lefevre F., Charley B., Riffault S. TLR9 pathway is involved in adjuvant effects of plasmid DNA-based vaccines. Vaccine. 2005;23:1258–1264. doi: 10.1016/j.vaccine.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Ishii K.J., Kawagoe T., Koyama S., Matsui K., Kumar H., Kawai T. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 92.Rottembourg D., Filippi C.M., Bresson D., Ehrhardt K., Estes E.A., Oldham J.E. Essential role for TLR9 in prime but not prime-boost plasmid DNA vaccination to activate dendritic cells and protect from lethal viral infection. J Immunol. 2010;184:7100–7107. doi: 10.4049/jimmunol.0803935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suschak J.J., Wang S., Fitzgerald K.A., Lu S. A cGAS-independent STING/IRF7 pathway mediates the immunogenicity of DNA vaccines. J Immunol. 2016;196:310–316. doi: 10.4049/jimmunol.1501836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev Vaccines. 2016;15:313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen N., Xia P., Li S., Zhang T., Wang T.T., Zhu J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. 2017;69:297–304. doi: 10.1002/iub.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edwards D.K., Jasny E., Yoon H., Horscroft N., Schanen B., Geter T. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J Transl Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lazzaro S., Giovani C., Mangiavacchi S., Magini D., Maione D., Baudner B. CD8 T-cell priming upon mRNA vaccination is restricted to bone-marrow-derived antigen-presenting cells and may involve antigen transfer from myocytes. Immunology. 2015;146:312–326. doi: 10.1111/imm.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ulmer J.B., Donnelly J.J., Parker S.E., Rhodes G.H., Felgner P.L., Dwarki V.J. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 100.Martinon F., Krishnan S., Lenzen G., Magne R., Gomard E., Guillet J.G. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 101.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 102.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lucchese G. Epitopes for a 2019-nCoV vaccine. Cell Mol Immunol. 2020;17:539–540. doi: 10.1038/s41423-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee C.H., Koohy H. Vol. 9. 2020. In silico identification of vaccine targets for 2019-nCoV. F1000Res; p. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bloom B.R., Jacobs W.R., Jr. New strategies for leprosy and tuberculosis and for development of bacillus Calmette-Guerin into a multivaccine vehicle. Ann N Y Acad Sci. 1989;569:155–173. doi: 10.1111/j.1749-6632.1989.tb27366.x. [DOI] [PubMed] [Google Scholar]
- 106.Milstien J.B., Gibson J.J. Quality control of BCG vaccine by WHO: a review of factors that may influence vaccine effectiveness and safety. Bull World Health Organ. 1990;68:93–108. [PMC free article] [PubMed] [Google Scholar]
- 107.Oettinger T., Jorgensen M., Ladefoged A., Haslov K., Andersen P. Development of the Mycobacterium bovis BCG vaccine: review of the historical and biochemical evidence for a genealogical tree. Tuber Lung Dis. 1999;79:243–250. doi: 10.1054/tuld.1999.0206. [DOI] [PubMed] [Google Scholar]
- 108.WHO BCG (tuberculosis) 2018. https://www.who.int/biologicals/areas/vaccines/bcg/Tuberculosis/en/ Date last accessed: May 20, 2020. Date last updated: July 5, 2018.
- 109.Ravn P., Boesen H., Pedersen B.K., Andersen P. Human T cell responses induced by vaccination with Mycobacterium bovis bacillus Calmette-Guerin. J Immunol. 1997;158:1949–1955. [PubMed] [Google Scholar]
- 110.van Faassen H., Dudani R., Krishnan L., Sad S. Prolonged antigen presentation, APC-, and CD8+ T cell turnover during mycobacterial infection: comparison with Listeria monocytogenes. J Immunol. 2004;172:3491–3500. doi: 10.4049/jimmunol.172.6.3491. [DOI] [PubMed] [Google Scholar]
- 111.Ohara N., Yamada T. Recombinant BCG vaccines. Vaccine. 2001;19:4089–4098. doi: 10.1016/s0264-410x(01)00155-4. [DOI] [PubMed] [Google Scholar]
- 112.Aldovini A., Young R.A. Humoral and cell-mediated immune responses to live recombinant BCG-HIV vaccines. Nature. 1991;351:479–482. doi: 10.1038/351479a0. [DOI] [PubMed] [Google Scholar]
- 113.Ami Y., Izumi Y., Matsuo K., Someya K., Kanekiyo M., Horibata S. Priming-boosting vaccination with recombinant Mycobacterium bovis bacillus Calmette-Guerin and a nonreplicating vaccinia virus recombinant leads to long-lasting and effective immunity. J Virol. 2005;79:12871–12879. doi: 10.1128/JVI.79.20.12871-12879.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Matsuo K., Yasutomi Y. Mycobacterium bovis Bacille Calmette-Guerin as a vaccine vector for global infectious disease control. Tuberc Res Treat. 2011;2011:574591. doi: 10.1155/2011/574591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kilpelainen A., Saubi N., Guitart N., Olvera A., Hanke T., Brander C. Recombinant BCG expressing HTI prime and recombinant ChAdOx1 boost is safe and elicits HIV-1-specific T-cell responses in BALB/c Mice. Vaccines (Basel) 2019;7:78. doi: 10.3390/vaccines7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stover C.K., de la Cruz V.F., Fuerst T.R., Burlein J.E., Benson L.A., Bennett L.T. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 117.Langermann S., Palaszynski S.R., Burlein J.E., Koenig S., Hanson M.S., Briles D.E. Protective humoral response against pneumococcal infection in mice elicited by recombinant bacille Calmette-Guerin vaccines expressing pneumococcal surface protein A. J Exp Med. 1994;180:2277–2286. doi: 10.1084/jem.180.6.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanson M.S., Bansal G.P., Langermann S., Stover C.K., Orme I. Efficacy and safety of live recombinant BCG vaccines. Dev Biol Stand. 1995;84:229–236. [PubMed] [Google Scholar]
- 119.Nascimento I.P., Dias W.O., Mazzantini R.P., Miyaji E.N., Gamberini M., Quintilio W. Recombinant Mycobacterium bovis BCG expressing pertussis toxin subunit S1 induces protection against an intracerebral challenge with live Bordetella pertussis in mice. Infect Immun. 2000;68:4877–4883. doi: 10.1128/iai.68.9.4877-4883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Varaldo P.B., Leite L.C., Dias W.O., Miyaji E.N., Torres F.I., Gebara V.C. Recombinant Mycobacterium bovis BCG expressing the Sm14 antigen of Schistosoma mansoni protects mice from cercarial challenge. Infect Immun. 2004;72:3336–3343. doi: 10.1128/IAI.72.6.3336-3343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grode L., Seiler P., Baumann S., Hess J., Brinkmann V., Nasser Eddine A. Increased vaccine efficacy against tuberculosis of recombinant Mycobacterium bovis bacille Calmette-Guerin mutants that secrete listeriolysin. J Clin Invest. 2005;115:2472–2479. doi: 10.1172/JCI24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kanno A.I., Goulart C., Rofatto H.K., Oliveira S.C., Leite L.C.C., McFadden J. New recombinant Mycobacterium bovis BCG expression vectors: improving genetic control over mycobacterial promoters. Appl Environ Microbiol. 2016;82:2240–2246. doi: 10.1128/AEM.03677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roth A., Gustafson P., Nhaga A., Djana Q., Poulsen A., Garly M.L. BCG vaccination scar associated with better childhood survival in Guinea-Bissau. Int J Epidemiol. 2005;34:540–547. doi: 10.1093/ije/dyh392. [DOI] [PubMed] [Google Scholar]
- 124.Higgins J.P., Soares-Weiser K., Lopez-Lopez J.A., Kakourou A., Chaplin K., Christensen H. Association of BCG, DTP, and measles containing vaccines with childhood mortality: systematic review. BMJ. 2016;355:i5170. doi: 10.1136/bmj.i5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Butkeviciute E., Jones C.E., Smith S.G. Heterologous effects of infant BCG vaccination: potential mechanisms of immunity. Future Microbiol. 2018;13:1193–1208. doi: 10.2217/fmb-2018-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nemes E., Geldenhuys H., Rozot V., Rutkowski K.T., Ratangee F., Bilek N. Prevention of M. tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N Engl J Med. 2018;379:138–149. doi: 10.1056/NEJMoa1714021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wardhana, Datau E.A., Sultana A., Mandang V.V., Jim E. The efficacy of Bacillus Calmette-Guerin vaccinations for the prevention of acute upper respiratory tract infection in the elderly. Acta Med Indones. 2011;43:185–190. [PubMed] [Google Scholar]
- 128.Netea M.G., Quintin J., van der Meer J.W. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 129.Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A., Ifrim D.C., Saeed S. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Arts R.J.W., Moorlag S., Novakovic B., Li Y., Wang S.Y., Oosting M. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100 e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 132.Gursel M., Gursel I. Is global BCG vaccination-induced trained immunity relevant to the progression of SARS-CoV-2 pandemic? Allergy. 2020;75:1815–1819. doi: 10.1111/all.14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miyasaka M. Is BCG vaccination causally related to reduced COVID-19 mortality? EMBO Mol Med. 2020;12 doi: 10.15252/emmm.202012661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O’Neill L.A.J., Netea M.G. BCG-induced trained immunity: can it offer protection against COVID-19? Nat Rev Immunol. 2020;20:335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Netea M.G., Giamarellos-Bourboulis E.J., Dominguez-Andres J., Curtis N., van Crevel R., van de Veerdonk F.L. Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181:969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miller A., Reandelar M.J., Fasciglione K., Roumenova V., Li Y., Otazu G.H. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020 doi: 10.1101/2020.03.24.2004293. [Preprint] [DOI] [Google Scholar]
- 137.Green C.M., Fanucchi S., Fok E.T., Moorlag S.J.C.F.M., Dominguez-Andres J., Negishi Y. COVID-19: a model correlating BCG vaccination to protection from mortality implicates trained immunity. medRxiv. 2020 doi: 10.1101/2020.04.10.20060905. [Preprint] [DOI] [Google Scholar]
- 138.Berg M.K., Yu Q., Salvador C.E., Melani I., Kitayama S. Mandated Bacillus Calmette-Guérin (BCG) vaccination predicts flattened curves for the spread of COVID-19. Sci Adv. 2020 doi: 10.1126/sciadv.abc1463. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hamiel U., Kozer E., Youngster I. SARS-CoV-2 rates in BCG-vaccinated and unvaccinated young adults. J Am Med Assoc. 2020;323:2340–2341. doi: 10.1001/jama.2020.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maringer K., Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014;25:669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ceron S., North B.J., Taylor S.A., Leib D.A. The STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) stimulates an antiviral state and protects mice against herpes simplex virus-induced neurological disease. Virology. 2019;529:23–28. doi: 10.1016/j.virol.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]