Abstract
BipA is a conserved translational GTPase of bacteria recently implicated in ribosome biogenesis. Here we show that E. coli ΔbipA cells grown at suboptimal temperature accumulate immature large subunit particles missing several proteins. These include L17 and L17-dependent binders, suggesting that structural block 3 of the subunit folds late in the assembly process. Parallel analysis of the control strain revealed accumulation of nearly identical intermediates, albeit at lower levels, suggesting qualitatively similar routes of assembly. This came as a surprise, because earlier analogous studies of wild-type E. coli showed early binding of L17. Further investigation showed that the main path of 50S assembly differs depending on conditions of growth. Either supplementation of the media with lysine and arginine or suboptimal temperature appears to delay block 3 folding, demonstrating the flexible nature of the assembly process. We also show that the variant BipA-H78A fails to rescue phenotypes of the ΔbipA strain, indicating a critical role for GTP hydrolysis in BipA function. In fact, BipA-H78A confers a dominant negative phenotype in wild-type cells. Controlled production of BipA-H78A causes accumulation of 70S monosomes at the expense of polysomes, suggesting that the growth defect stems from a shutdown of translation.
Keywords: ribosome biogenesis, protein synthesis, GTPase, TypA, YchF, HflX
INTRODUCTION
The ribosome is a large two-subunit enzyme, which is composed of three ribosomal RNAs (rRNAs) and ~50 ribosomal (r) proteins in bacteria. Assembly of the ribosome begins during rRNA transcription and entails concurrent r protein binding, rRNA modification, and rRNA processing events. Early studies showed that r proteins bind to rRNA in a hierarchical manner, where the binding of the primary proteins nucleates the folding of rRNA to allow subsequent binding of secondary and tertiary proteins [1–3]. More recent kinetic studies have shown that, while generally hierarchical, many r-protein binding events occur in random order, resulting in multiple parallel pathways of assembly [3–5]. Although the ribosome is intrinsically capable of self-assembly [1,2,6], the process is facilitated in the cell by many trans-acting factors including ribonucleoprotein-binding proteins, helicases, chaperones, RNases, modification enzymes, and GTPases.
The small subunit (SSU, 30S) contains three main structural domains—the body, platform, and head—which correspond to the 5’, central, and 3’ major regions of 16S rRNA secondary structure. Each of these structural domains can be independently reconstituted, using the appropriate fragment of 16S rRNA, yielding a compact ribonucleoprotein particle with the correct r proteins [7–9]. Accordingly, analysis of the kinetics of 30S assembly by cryo-EM showed that the subunit forms in a modular way, with the body and platform domains folding prior to the head domain [5,10].
The structure of the large subunit (LSU, 50S), on the other hand, is monolithic, with the various secondary structural regions of 23S rRNA (regions I-VI) interlaced with one another [11]. Yet, remarkably, LSU subunit assembly is also modular [12]. Using cryo-EM and quantitative mass spectrometry (qMS), Williamson and coworkers have shown that particular groups of rRNA helices and associated r proteins, termed blocks, assemble as separate units [12,13]. Block 1 corresponds to a large portion of the “back” (solvent) side of the subunit, block 2 corresponds to the central protuberance region, block 3 corresponds to the “lower” portion of the interface side, and block 4 includes the peptidyl transferase center (PTC) and L7/12 stalk region (Figure 1(a), Figure S1). While the kinetics of 50S assembly involves several parallel paths, block 1 generally folds first, followed by block 2 or 3. Block 4 tends to fold last, even though the PTC is thought to be the oldest portion of the ribosome [14].
Figure 1. Modular assembly of the 50S subunit.
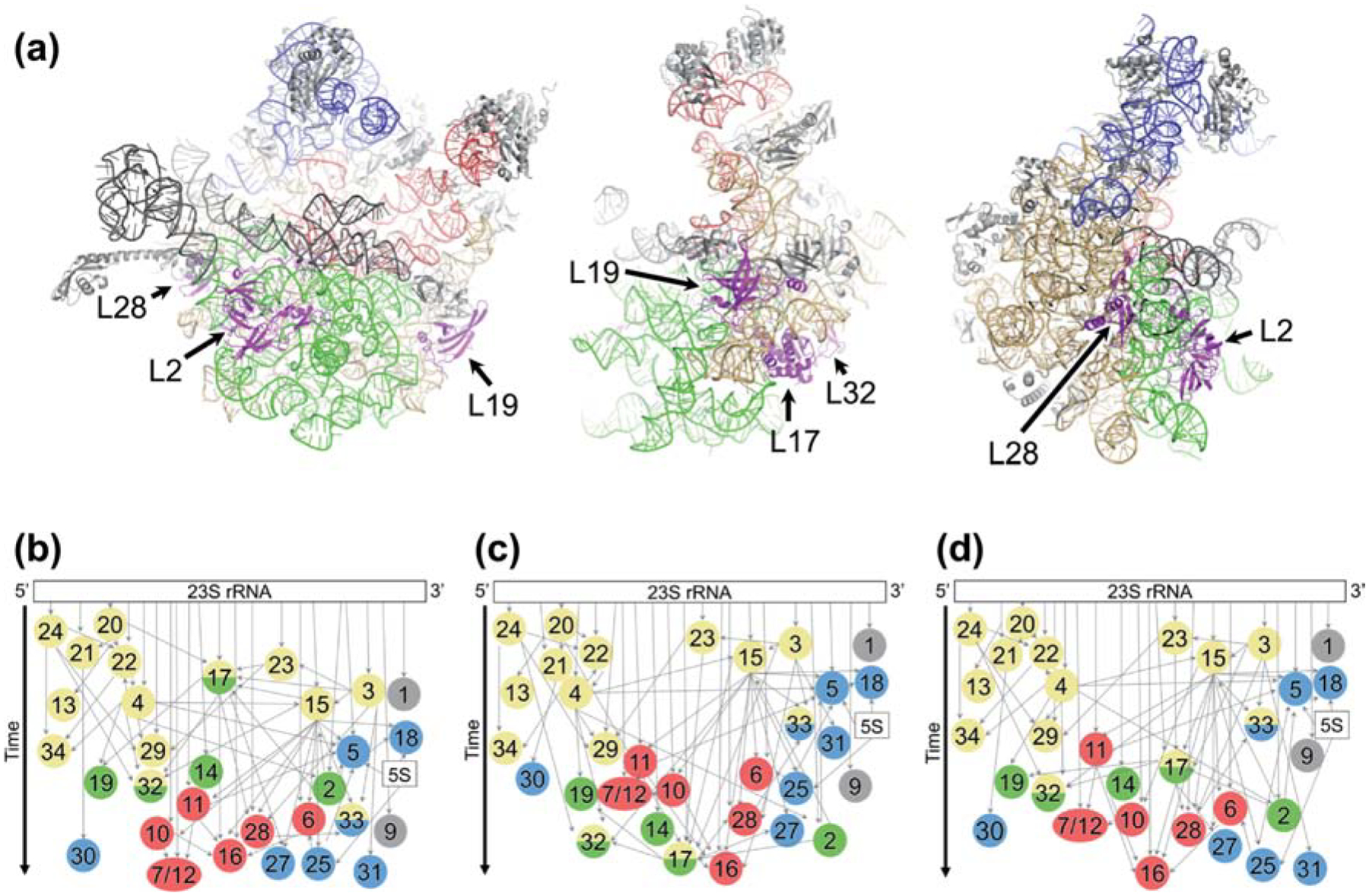
(a) Three views of the structure of the 50S subunit (PDB 4YBB) showing rRNA assembly block 1 (tan), block 2 (blue), block 3 (green), and block 4 (red). Other regions of rRNA, shown in gray, include flexible elements rarely resolved in intermediates. L17 and L17-dependent proteins are highlighted in magenta and labeled. (b-d) LSU assembly maps depicting the relative rates of r protein incorporation by vertical position. Colors in circles represent the block(s) with which each protein is associated, based on experimental evidence (Davis et al. 2016) or inferred from the structure of the mature LSU. Block 1, yellow; block 2, blue; block 3, green; block 4, red; unassigned, gray. Narrow gray arrows, thermodynamic binding dependencies; wide black arrow, time. (b) Map based on data from WT cells grown at 37° C in the absence of amino acid supplementation (Chen and Williamson, 2013). (c) Map based on data from ΔbipA cells grown at 20° C. (d) Map based on data from WT cells grown at 37° C with Lys and Arg supplementation.
BipA (BPI-inducible protein A) was initially identified in Salmonella enterica serovar Typhimurium, where its expression is induced in response to treatment with bactericidal/permeability increasing protein (BPI), an antimicrobial secreted by human neutrophils [15]. Independent work on pathogenic Escherichia coli identified BipA as tyrosine phosphoprotein A (TypA) [16,17]. Phosphorylation of BipA/TypA appears to be strain specific, as neither E. coli K-12 nor S. enterica produce phospho-BipA/TypA [16,17]. A number of phenotypes have been attributed to ΔbipA in pathogenic E. coli, including reduced cytoskeletal rearrangements in host cells, hypersensitivity to host defense peptides, and hypermotility [17]. It was proposed that BipA acts as a regulator of virulence in pathogenic E. coli because its absence alters expression of capsule gene clusters (K antigens) and pathogenicity islands [18,19].
Flower and coworkers have provided compelling evidence that BipA is important for 50S subunit assembly [20–22]. At suboptimal temperature, loss of BipA causes accumulation of pre-50S particles and 23S rRNA precursors in E. coli K-12 [22]. These phenotypes are exacerbated by deletion of deaD, a DEAD-box helicase that acts on 23S rRNA, and suppressed by ΔrluC [22]. RluC is a pseudouridine synthase that modifies nucleotides U955, U2504, and U2580 in 23S rRNA. Mutation of these nucleotides also suppresses the ΔbipA phenotypes, indicating that the assembly defect seen in the absence of BipA depends on these pseudouridines [22].
BipA is a translational GTPase (trGTPase), structurally similar to elongation factor EF-G and to LepA, a protein that functions in SSU biogenesis [23,24]. These proteins share homologous domains G, II, III, and V. Both BipA and LepA lack domain IV, and each protein contains a unique C-terminal domain. All of these trGTPases bind the ribosome similarly, with domains G and II contacting the 50S and 30S subunits, respectively. Domain IV of EF-G protrudes into the 30S A site, whereas the C-terminal domains of BipA and LepA are oriented toward the PTC. Similar to other trGTPases, the GTPase activity of BipA is activated to a larger degree by the 70S ribosome than by either subunit alone [25]. In the presence of ppGpp, BipA binds specifically to 30S subunits, raising the hypothesis that BipA acts as a stress factor [25]. The 3’ phosphate of ppGpp is predicted from structural studies to present a steric clash with the 50S in the context of the 70S ribosome [26], which likely explains the earlier binding data, but the biological role of BipA•ppGpp remains unclear.
Here we use quantitative mass spectrometry (qMS) to characterize the pre-50S intermediates that accumulate in the absence of BipA. We find that these particles lack L17 and L17-dependent binders (L2, L19, L28, and L32), suggesting that block 3 assembles late in the process. Qualitatively similar data are seen with control cells, an unexpected result, given analogous earlier studies. This apparent discrepancy was resolved by further analysis, which revealed that the main path of 50S assembly changes depending on the growth conditions of the cell. These results illustrate the robust and flexible nature of the assembly process.
RESULTS
Pre-50S particles lacking several r proteins accumulate in ΔbipA cells at 20 °C
We confirmed that ΔbipA confers a growth defect at 20 °C, as reported previously [20]. Sucrose gradient sedimentation analysis showed a large peak at ~40S and almost complete disappearance of the 50S peak (Figure 2(a)). A plasmid encoding BipA (pBIPA) fully complemented these phenotypes, showing that they are due to the absence of BipA. At 37 °C, growth rates and relative LSU protein levels across the various ribosomal fractions were indistinguishable in ΔbipA and WT cells (Figure S2). Therefore, the assembly defect conferred by ΔbipA is only seen at suboptimal temperature.
Figure 2. Composition of ribosomal particles in control and ΔbipA cells.
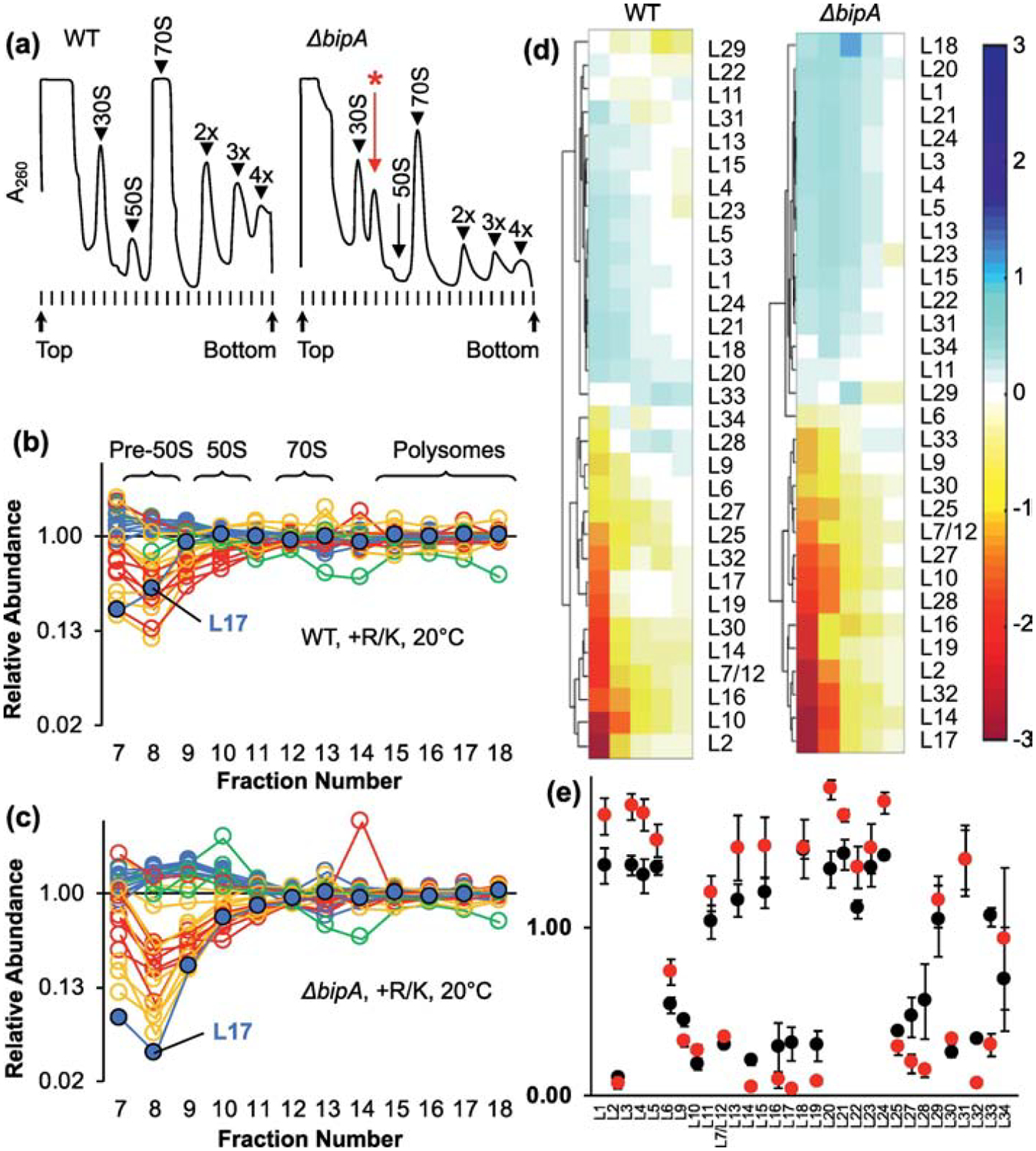
(a) Representative A260 traces of sucrose gradients from WT (left) and ΔbipA (right) cells grown at 20 °C. Peaks corresponding to subunits (30S, 50S), monosomes (70S), and polysomes (2x, 3x, 4x) are indicated. A prominent pre-50S peak seen in the ΔbipA case is indicated with a red asterisk. (b, c) Plotted are the normalized isotope ratios indicating the relative abundance of each LSU protein in each fraction (7–18) for WT (b) and ΔbipA cells (c) across the gradient. Color coding corresponds to temporal stages of assembly as defined by Chen and Williamson 2013 (blue, early; green, middle; yellow, middle-late; red, late). L17 is highlighted with a black outline. Data represent the mean of 3 independent experiments. Full datasets are included in Table S1. (d) Hierarchical clustering analysis of LSU abundance data using fractions 8–12. Color bar on right indicates degree of representation from blue (overrepresented) to red (underrepresented). (e) Protein composition of pre-50S fraction 8 from WT (black circles) and ΔbipA (red circles) cells. Data represents mean ± SEM from three independent replicates.
Williamson and coworkers have shown that the relative timing of r protein incorporation during subunit assembly can be inferred from qMS analysis of sucrose gradient fractions [4,27]. LSU proteins that bind early are overrepresented (relative to the median) in the pre-50S region, whereas those that bind late are underrepresented. To better understand the assembly defect of the ΔbipA strain, we followed the approach of Williamson [27], except that SILAC (stable isotope labeling of amino acids in culture [28]) was used rather than all-nitrogen (15NH4Cl) labeling. Proteins in auxotrophic (Arg− Lys−) control (WT) and mutant (ΔbipA) cells were isotopically labeled, lysates were run on sucrose gradients, unlabeled mature ribosomes (70S) were spiked into each ribosomal fraction, and samples were analyzed by mass spectrometry. This yielded isotope ratios reflecting the relative abundance of each r protein in each gradient fraction. In this experimental setup, it was important to use auxotrophic (Arg−Lys−) strains to ensure complete labeling of the proteome by imported (medium- or heavy-weight) amino acids.
Plots of these data are shown in Figure 2(b–c), color-coded according to the temporal stages assigned previously by Williamson. As expected, isotope ratios for the LSU proteins converged to 1.0 as a function of fraction number, reflecting fully-assembled subunits in the heavier fractions. In the pre-50S region there was considerable spread in the data, indicative of incompletely assembled particles. The degree of spread was larger in the ΔbipA case, presumably because a higher abundance of intermediates enhances the signal-to-noise ratio. Hierarchical clustering of data from the key region of the gradient (fractions 8–12 in this experiment) revealed that the r-protein profiles fell into two major groups, with highly similar representation in both WT and ΔbipA cases (Figure 2(d)). Most primary proteins (L1, L3, L4, L15, L20, L22, L23, L24) were overrepresented in the pre-50S region, indicating early incorporation, in line with Williamson’s assignments (blue data points in Figure 2(b–c)). One clear exception was L17, which was underrepresented in the pre-50S fractions (Figure 2(b–e)), indicative of late-stage incorporation. Proteins L2, L19, and L32—which are thermodynamically-linked to L17 [2,29]—also appeared to assemble later than expected from earlier analogous experiments. Collectively, these data suggest that LSU assembly follows similar paths in the WT and ΔbipA cells, although some kinetic bottleneck increases accumulation of the intermediates in the latter case. Remarkably, in both strains, LSU assembly differs from that observed previously [27], with block 3 folding late (Figure 1).
Incorporation of L17 and L17-dependent proteins is slower under conditions of amino acid supplementation
There were three differences between our experiments and those of Williamson—temperature, genotype, and growth media. To investigate temperature, we grew the control strain at 37 °C and repeated the analysis. Again, L17 behaved like a late-stage binder, being underrepresented in the pre-50S fractions, and thus change in growth temperature alone (under these conditions) does not appear to shunt assembly (Figure S3). To test genotype and growth media, we compared r-protein abundance in the auxotroph (Lys− Arg−) to that in the parental prototroph (Lys+ Arg+) grown with or without amino acid supplementation (Figure 3). For these experiments, we used isotopically-labeled mature ribosomes for the spike-in standard, allowing any growth condition to be tested. When either the auxotroph or the prototroph was grown with lysine and arginine, L17 again behaved as a later binder (Figure 3(a–b)). However, when the prototroph was grown without amino acid supplementation, the profile for L17 resembled that of early binders, with relative abundance >1 in fraction 18 (Figure 3(c)). In fact, the collective data were largely consistent with those of Williamson and coworkers (note color-coding, Figure 3(c)). This indicates that the kinetics of LSU assembly shifts in response to conditions of growth.
Figure 3. Amino acid supplementation influences the kinetics of LSU assembly.
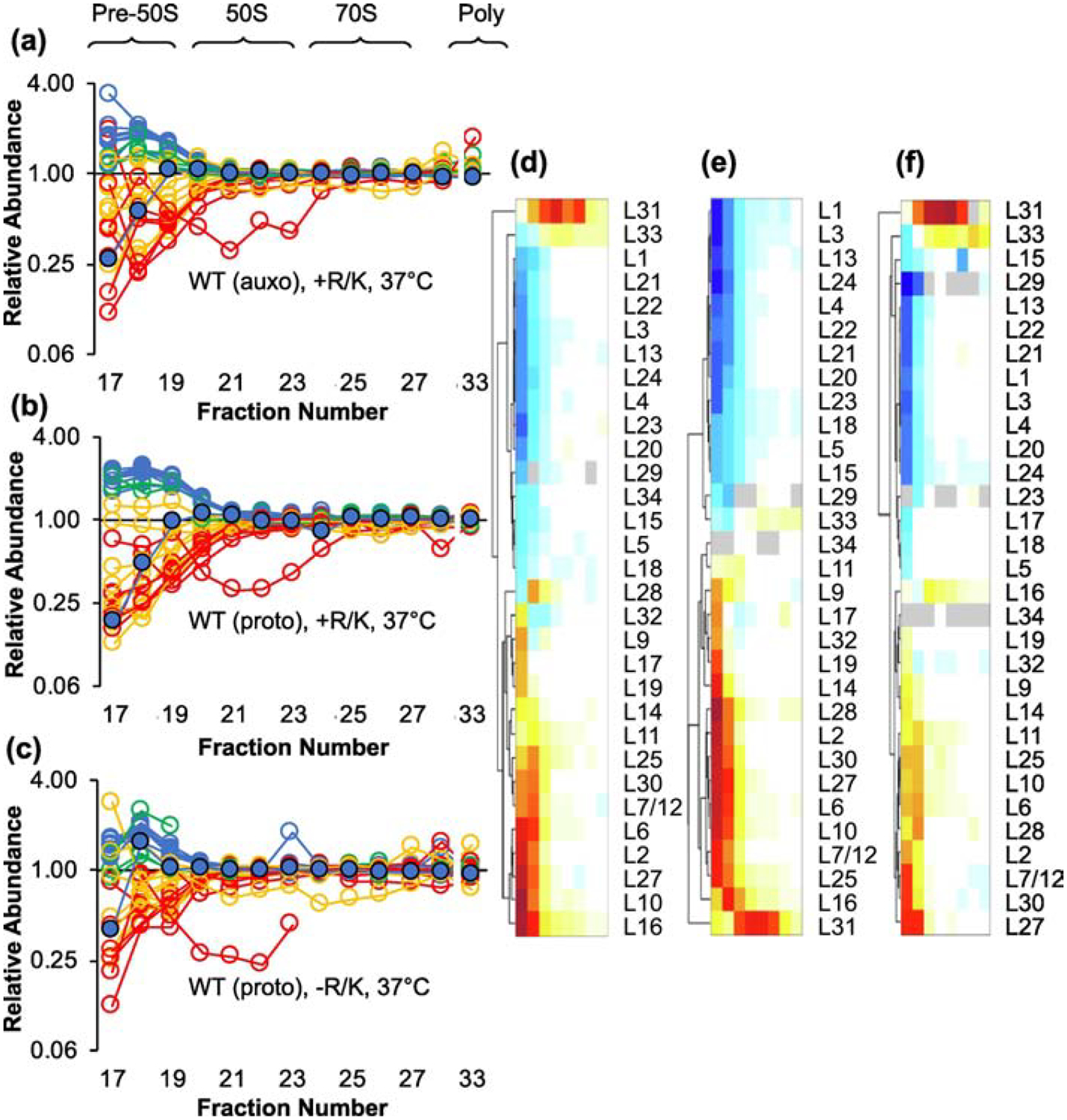
Shown are the relative abundance of LSU proteins in pre-50S and 50S particles from the control (WT) auxotroph grown with Arg and Lys supplementation (a), the control (WT) prototroph grown with Arg and Lys supplementation (b), and the control (WT) prototroph grown without supplementation (c). Color coding is based on temporal stages defined by Chen and Williamson, 2013 (blue, early; green, middle; yellow, middle-late; red, late). L17 is highlighted with a black outline. Data represent the mean of 3 independent experiments. Full datasets are included in Table S1. Hierarchical clustering of the data in (a), (b), and (c) is shown in (d), (e), and (f), respectively. Color scale as in Figure 2(d).
We then tested amino acid supplementation in the prototrophic ΔbipA strain grown at 20 °C, a condition in which 50S assembly is compromised. In this case, the protein abundance profiles were virtually identical in cells grown with or without amino acids, with L17 and L17-dependent r proteins showing delayed incorporation (Figure 4). These data suggest that without BipA at 20 °C, block 3 folds slowly regardless of amino acid supplementation.
Figure 4. Block 3 folds slowly in the ΔbipA strain regardless of media supplementation.
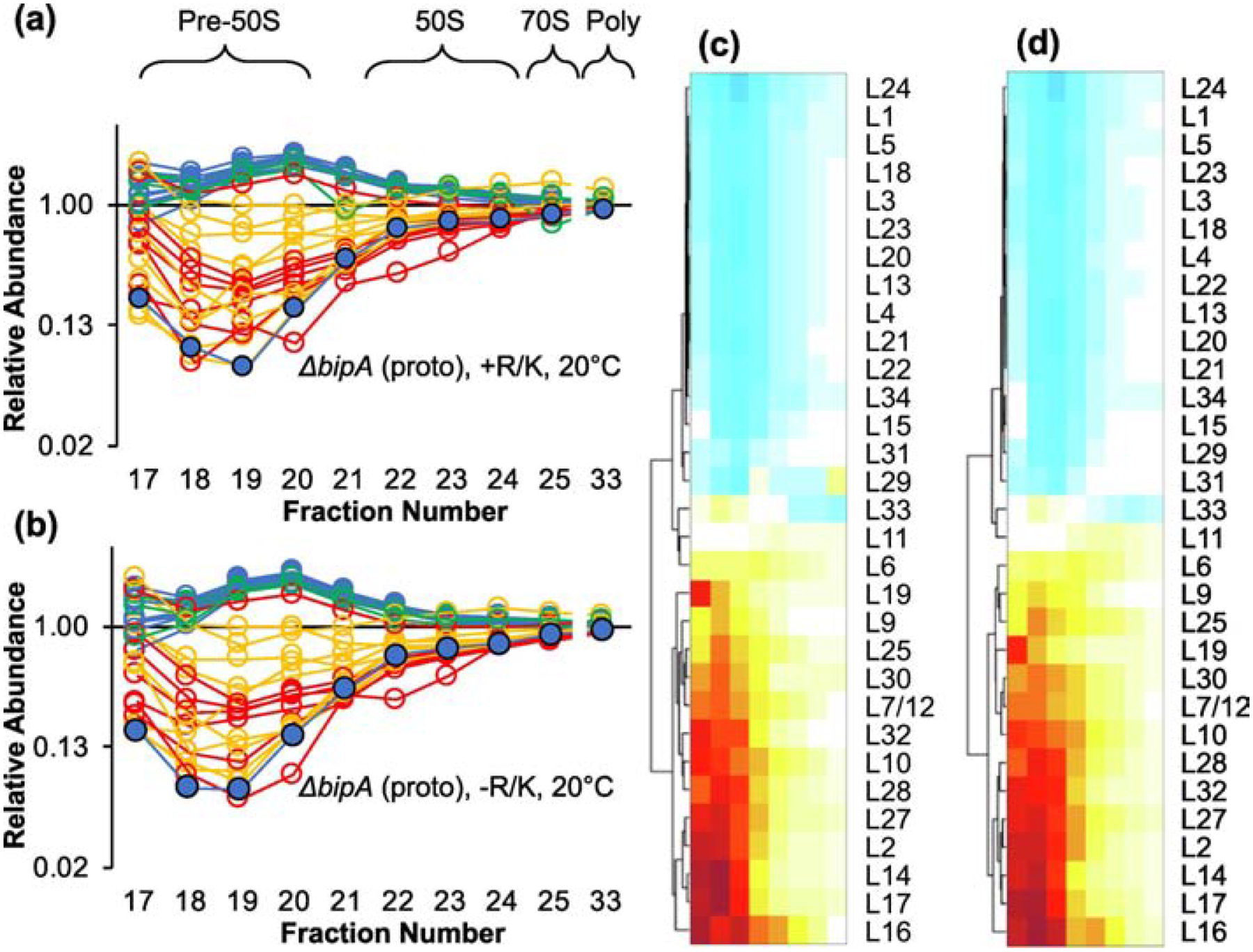
Shown are the relative abundance of LSU proteins in various ribosomal particles from the ΔbipA prototroph grown with (a) or without (b) arginine and lysine. Color coding is based on temporal stages defined by Chen and Williamson, 2013 (blue, early; green, middle; yellow, middle-late; red, late). L17 is highlighted with a black outline. Data represent the mean of 3 independent experiments. Full datasets are included in Table S1. Hierarchical clustering of the data in (a) and (b) is shown in (c) and (d), respectively. Color scale as in Figure 2(d).
The fact that supplementation had no effect on 50S assembly in this case (ΔbipA, 20 °C) came as a surprise and reopened the question of which variable(s) were important for shunting the pathway. Hence, we next tested the control strain grown at 20 °C in supplemented and unsupplemented media. Although the data were noisier in this experiment, L17 remained underrepresented across the pre-50S fractions, regardless of amino acid supplementation (Figure S4). Thus, the slow relative folding of block 3 was observed in nearly all tested conditions. The only exceptional case entailed WT cells grown at 37 °C in the absence of supplementation, conditions that Williamson used to deduce the temporal assembly map [27].
Mature 30S subunits accumulate in ΔbipA cells
Protein composition of SSU particles was also measured for supplemented cells grown at 20 °C. In WT cells, pre-30S particles were enriched with primary binders (S4, S6, S8, S15, S16, S17, S18, S20) and depleted of proteins S2, S3, S10, S14, and S21 (Figure S5(a), fraction 6), generally consistent with previous results of Williamson. The levels of S3, S10, S14, and S21 remained low through the 30S fractions, increasing gradually to stoichiometric levels in heavier fractions (10 and 11). In ΔbipA cells, these proteins (aside from S21) were present at near stoichiometric amounts by fraction 7 (Figure S5(b)) suggesting a smaller proportion of assembly intermediates migrating at 30S.
To further investigate this, we looked at processing of precursor 17S rRNA, one of the last steps of 30S subunit biogenesis [30]. In WT cells, 17S rRNA accounted for at least half of the SSU rRNA in the pre-30S and 30S fractions and decreased to ~6% in the 70S fractions (Figure 5). The ΔbipA strain exhibited substantially lower levels of precursor 17S rRNA, particularly in the 30S region. Introduction of pBIPA restored the levels of 17S rRNA and r proteins across the gradient fractions to WT values. These collective data indicate that the proportion of 30S particles that are mature increases in the absence of BipA. We suspect that mature 30S subunits accumulate because they are unable to enter the translationally-active pool without sufficient 50S partners.
Figure 5. Loss of BipA leads to lower levels of 17S precursor rRNA.
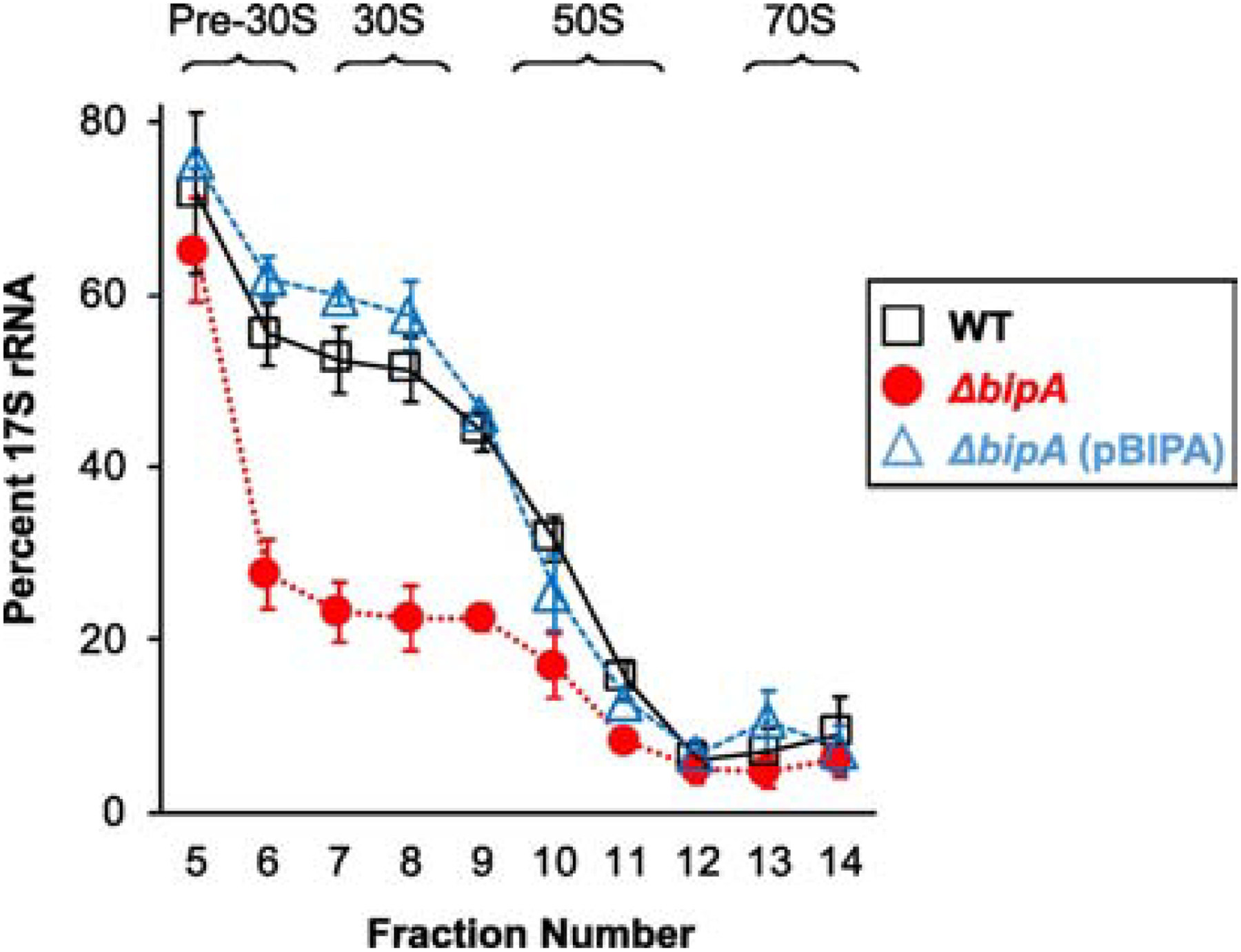
Total RNA was extracted from sucrose gradients fractions 5–14 (as indicated), SSU rRNA was resolved by PAGE, and 17S rRNA and 16S rRNA bands were quantified. The amount of 17S rRNA (percent of SSU rRNA) is plotted as a function of fraction number. Data represent mean ± SEM for three independent replicates. Black squares, WT; red circles, ΔbipA; blue triangles, ΔbipA (pBIPA).
BipA function depends on its GTPase activity
Translational GTPases contain a critical histidine residue that is required for GTPase activity [31–34]. We mutated the corresponding His codon in pBIPA to encode the variant BipA-H78A. Production of BipA-H78A in ΔbipA cells failed to complement the cold sensitive growth phenotype, indicating that the function of BipA depends on its GTPase activity (Figure 6(a)). In fact, production of BipA-H78A in ΔbipA cells slowed growth at both 20 °C and 37 °C (Figure 6(a–b)). In WT cells, production of BipA-H78A conferred a dominant growth defect (Figure 6(b)). These data indicate that a GTPase-inactive variant of BipA is deleterious to the cell regardless of strain background or growth temperature.
Figure 6. Production of BipA-H78A confers a dominant negative phenotype.
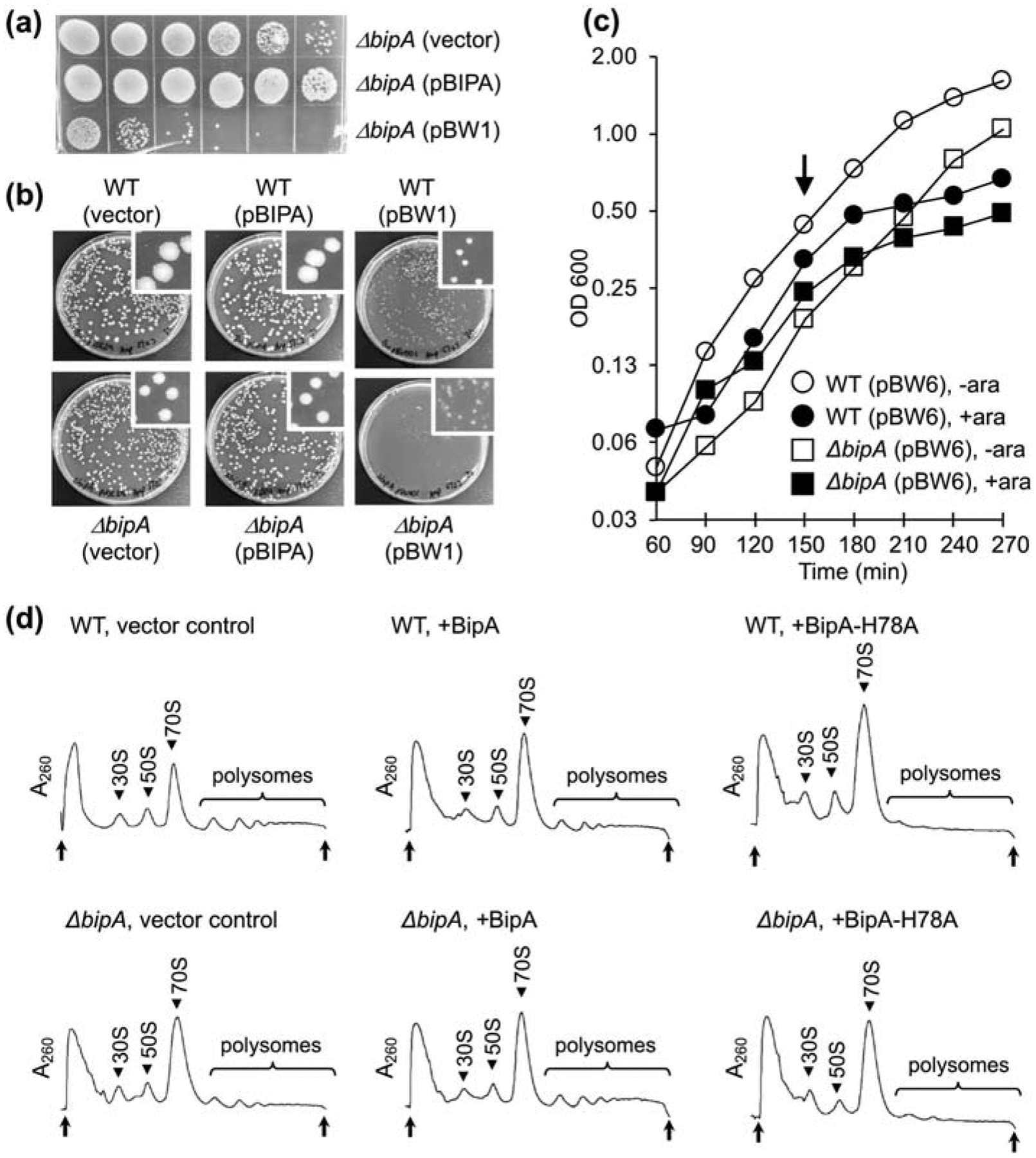
(a) A spot assay comparing growth at 20 °C of ΔbipA cells harboring the empty vector, pBIPA (encoding BipA), or pBW1 (encoding BipA-H78A), as indicated. Cultures of equivalent OD600 were serially diluted, and corresponding diluted cells (20 μL) were spotted onto plates and incubated for 3 days. (b) Transformation plates showing the deleterious effects of BipA-H78A. Cells (WT or ΔbipA) were transformed with the empty vector, pBIPA (encoding BipA), or pBW1 (encoding BipA-H78A), as indicated, and plated on selective media and grown at 37 °C. (c) Growth at 37 °C of WT and ΔbipA cells containing pBW6, encoding BipA-H78A, with or without arabinose induction (as indicated). Arrow indicates time of induction. (d) Inhibition of translation by BipA-H78A. Cells (WT or ΔbipA, as indicated) containing pBAD24 (empty vector), pBW2 (encoding BipA), or pBW6 (encoding BipA-H78A) (as indicated) were grown at 37 °C to mid-log phase, induced with arabinose (0.2% W/V) to express protein for 70 min, and subjected to sucrose gradient sedimentation analysis. Peaks corresponding to subunits (30S, 50S), monosomes (70S), and polysomes are indicated. Top and bottom of each gradient are marked with arrows.
When propagating strains containing BipA-H78A on plates, we frequently observed the appearance of large colonies, presumably the result of suppressor mutations. Consistent with this interpretation, liquid cultures inoculated from fresh transformants exhibited slow growth for some (variable) time, followed by rapid growth. Sequencing of plasmids from fast-growing suppressors showed that true reversions (GCC to CAC, codon 78) were frequent, presumably due to gene conversion. Hence, we moved in Δ(recA-srl)306 to prevent homologous recombination but still obtained suppressors, albeit at a lower frequency. To further characterize BipA-H78A, we cloned the bipA and bipA-H78A coding regions downstream of an arabinoseinducible promoter in pBAD24. Upon induction with arabinose, production of BipA-H78A halts growth of WT and ΔbipA cells (Figure 6(c)). To look at effects on translation, we grew cell cultures at 37 °C until they reached an OD600 of ~0.1, induced with arabinose (0.2%), grew cultures an additional 70 min, and then analyzed cell lysates by sucrose gradient sedimentation. Production of BipA-H78A in both WT and ΔbipA cells led to accumulation of 70S monosomes and loss of polysomes (Figure 6(d)). We suspect that BipA-H78A•GTP binds 70S ribosomes but cannot hydrolyze GTP to allow for factor release, which leads to inhibition of translation in the cell.
YchF and HflX have no apparent role in ribosome assembly at 37 °C
YchF and HflX are universally-conserved GTPases that bind the ribosome but whose functions in the cell remain poorly understood. YchF is a member of the P-loop GTPase family but specifically binds ATP, and its ATPase activity is stimulated by 70S ribosomes [35]. HflX interacts with 50S and 70S ribosomes [36] and is proposed to be a ribosome splitting factor [37,38]. We tested the role of YchF and HflX in ribosome assembly using qMS as described previously [23]. Differentially-labeled control and mutant cells were grown to mid-log phase at 37 °C, and corresponding lysates were independently fractionated via sucrose gradient sedimentation. Equivalent fractions were then combined, and the corresponding proteins analyzed by qMS. Deletion of either ychF or hflX caused no obvious change in the relative abundance of any LSU or SSU protein across the fractions (Figures S6 and S7). These data suggest that neither YchF nor HflX are important for ribosome assembly under these conditions.
DISCUSSION
Ribosome assembly is a complex and highly coordinated process. Recent studies have shown that, for both subunits, assembly occurs in a modular way. Each of several rRNA blocks folds cooperatively, with particle-to-particle differences in the order of block folding, resulting in multiple parallel pathways of subunit assembly [3–5,12]. Loss or depletion of certain assembly factors or r proteins has been shown to alter the assembly landscape [10,39–42]. But whether changes in environmental conditions influence ribosome assembly has remained an open question. In this study, we evaluated 50S assembly in control (WT) and ΔbipA cells grown at suboptimal or optimal temperature in the presence or absence of amino acid supplementation. In most cases, we found L17 and L17-dependent proteins to be underrepresented in the pre-50S fractions, consistent with late folding of block 3 (Figure 1(c–d)). The only exception entailed WT cells grown without supplementation at 37° C, equivalent to the standard conditions of Williamson, when L17 behaved as an early-binding protein (Figure 1(b)). Thus, the kinetics of 50S assembly differs appreciably in WT cells, depending on nutrient availability and growth temperature.
How supplementation of cells with arginine and lysine can shunt the 50S assembly pathway remains unclear. One possibility is that uptake of these amino acids alters the levels of polyamines in the cell and thereby influences assembly. E. coli produces putrescine, spermidine, and cadaverine—polyamines known to interact with rRNA and play important roles in ribosome function and biogenesis [43–45]. Arginine is a precursor to putrescine and spermidine, lysine is the precursor to cadaverine, and both Arg and Lys act as regulatory effectors at the gene and/or protein level [46]. While reasonable, this hypothesis should be considered speculative. Whether the assembly landscape is altered by Arg and/or Lys specifically remains unclear, as other amino acids have not yet been tested.
While exogenous Arg and/or Lys changes the 50S assembly landscape in cells grown at 37° C, supplementation has no apparent effect during growth at 20° C. In either the WT or mutant cells, incorporation of L17 and L17-dependent proteins occurs relatively late, whether or not the medium contains Arg and Lys. Thus, temperature also influences 50S assembly kinetics. For WT cells growing prototrophically, a shift to suboptimal temperature is sufficient to slow block 3. These effects of temperature may stem from altered rRNA dynamics and/or changes in gene expression.
Our work shows that BipA plays an important role in 50S biogenesis at suboptimal temperature, in line with earlier evidence from the Flower laboratory. At 20 °C, ΔbipA cells exhibit an obvious growth defect and accumulate pre-50S particles which lack L17 and other proteins associated with block 3. Under identical conditions, WT cells contain pre-50S particles of similar composition. The relative levels of L17 and L17-dependent proteins are slightly higher, indicating that block 3 folding is somewhat faster (Figure 1(c–d)). But this difference, while genetically attributable to bipA, does not mean that BipA acts specifically to facilitate block 3 folding. Loss of BipA causes accumulation of pre-50S intermediates that closely resemble those of WT cells, consistent with a kinetic bottleneck that slows the overall process without appreciable qualitative perturbations. BipA production is increased at low temperature [47] and ΔbipA only confers an assembly defect at suboptimal temperature, indicating that either BipA or thermal energy can alleviate the bottleneck. While the molecular basis of this bottleneck remains unclear, it depends on pseudouridines Ψ955, Ψ2504, and Ψ2580 [21]. Hence it likely entails misfolding of the PTC of the 23S rRNA, part of block 4 of the subunit.
No structures of pre-50S particles bound by BipA have been reported. However, a cryo-EM structure of BipA·GDPCP bound to the mature 70S ribosome provides clues to how the factor might function [26]. BipA·GDPCP binds the intersubunit cleft on the A-site side of the ribosome, much like other trGTPases, with the G domain contacting the sarcin-ricin-loop (SRL) of H95 of 23S rRNA (Figure S8). Helix H95 is one of the most conserved portions of the ribosome and is part of block 1 of the LSU. BipA makes numerous additional contacts with the LSU, most of which involve components of block 4 of the subunit. We envisage that BipA·GTP may be able to engage the SRL once block 1 is folded and then facilitate the folding of block 4, perhaps by inhibiting formation of an alternative (and Ψ-dependent) rRNA structure. BipA may act at a late step, after the parallel paths converge. Hence, loss of the factor at suboptimal temperature would back up flow along all paths, causing a largely unbiased accumulation of intermediates, as we observe.
Recently, Hwang and coworkers reported that overexpression of L20 suppresses the assembly defects of the ΔbipA strain [48]. L20 is a primary binder and one of the first proteins to be incorporated during LSU assembly. Part of block 1, L20 binds near L21 and L13 on the “back” side of subunit, away from the BipA-binding site (Figure S1). L20 is composed of an extended N-terminal region that penetrates into the rRNA and a more globular C-terminal domain (CTD) that lies at the solvent-exposed surface. Aside from its critical role in LSU assembly, L20 regulates translation of its own operon, an activity dependent on the CTD [49,50]. How increased levels of L20 can functionally compensate for the absence of BipA remains unclear. Suppression does not require the CTD and hence seems unrelated to L20’s regulatory function [48]. It is possible that high concentration of L20 favors a particular route of block 1 assembly, which influences the kinetics of subsequent folding events (in blocks 2–4) in a way that bypasses the need for BipA.
We also find reduced levels of premature (relative to mature) 30S particles in the ΔbipA strain at suboptimal temperature. This is indicated by the qMS data and by the decreased proportion of 17S rRNA in the pre-30S and 30S fractions (Figure 5, Figure S5). We propose that slow 50S biogenesis results in accumulation of mature 30S subunits, unable to enter the actively-translating pool. The other formal possibility—that the efficiency of 30S biogenesis is somehow increased under these conditions—is highly improbable in our view. Oddly, Flower and coworkers also looked 16S rRNA processing but saw no significant effect of ΔbipA [22]. However, they grew cells in rich media, which appears (based on sucrose gradient sedimentation analysis) to attenuate the 50S assembly defect; and they analyzed total RNA, which may mask differences in the 30S subpopulation.
BipA is a trGTPase, which probably hydrolyzes GTP late in the assembly process, in the context of the 70S ribosome [51]. We show here that the GTPase-deficient variant BipA-H78A is unable to rescue phenotypes of the ΔbipA strain. These data are in line with a recent report [47] that the GTPase-deficient BipA-N128D also fails to restore normal growth to the null mutant. Interestingly, we additionally find that production of BipA-H78A confers a dominant negative phenotype. Sucrose gradient sedimentation analysis reveals that BipA-H78A causes the loss of polysomes and accumulation of 70S monosomes. We hypothesize that BipA-H78A•GTP binds the ribosome but fails to hydrolyze GTP and dissociate, effectively trapping ribosomes on mRNA and shutting down translation overall. Whether BipA-H78A acts on mature ribosomes as well as newly-formed ribosomes remains an open question, one worth pursuing in future work.
MATERIALS AND METHODS
Plasmids
To make pBIPA, a ~2.0 kb DNA fragment containing the bipA gene and its native promoter was amplified from E. coli strain BW25113 using primers #1 (5’-GGATCTCGAGCCCTAAAAGGCGTTATCATGC-3’) and #2 (5’-GTCTAGACGGTGGATTGATTCTCCGCC-3’) and cloned into the low-copy vector pWSK29[52], via the Xho I and Xba I restriction sites. To make pBW1, a mutation was introduced into pBIPA using Phusion mutagenesis (New England Biolabs) and primers #3 (5’-GCCGCCGACTTCGGTGGTGAAGTTGAAC-3’) and #4 (5’-CCCCGGGGTATCAACGATGTTGATAC-3’), changing codon 78 of bipA from CAC (His) to GCC (Ala). From pBIPA and pBW1, the wild-type and mutant bipA coding regions were amplified, using primers #5 (5’-GAGAATTCAGTTGTGATCGAAAAATTGC-3’) and #6 (5’-GGGGCTCTAGATTTATGACTAAAAAACGAAATT-3’), and cloned into the Eco RI and Xba I sites of pBAD24 [53], generating pBW2 and pBW6, respectively.
Strains
All bacterial strains used in this study are listed in Table 1. For SILAC analysis, the ΔbipA::kan [54] mutation was moved by P1 transduction into MRG01-KanS [23], an Arg− and Lys− auxotroph. Plasmids pWSK29 and pBIPA were moved into this strain to create MC0030 and MRG71, respectively. Empty vector pWSK29 was moved into the MRG01-KanS to generate MRG06 [23]. MRG01 containing the kanamycin resistance marker was transformed with pWSK29 to generate MRG03 [23].
Table 1.
Bacterial strains used in this study
| Strain | Description of genotype | Plasmid | Reference | Experiments |
|---|---|---|---|---|
| BW25113 | F-, λ−, Δ(amD-araB)567, ΔlacZ4787::rrnB-3, Δ(rhaD-rhaB)568, rph-1, hsdR514 | none | [54] | |
| MRG46 | BW25113 | pWSK29 | This study | Figures 3(b–c), S4 |
| MRG01 | BW25113 argE::Tn10 ΔlysA::kan | none | [23] | |
| MRG01-KanS | BW25113 argE::Tn10 ΔlysA::frt | none | [23] | |
| MRG06 | BW25113 argE::Tn10 ΔlysA::frt | pWSK29 | [23] | Figures 2, 5, S2, S5(a), S6, S7 |
| MC0030 | BW25113 argE::Tn10 ΔlysA::frt ΔbipA::kan | pWSK29 | This study | Figures 2, 5, S2, S5(b) |
| MRG71 | BW25113 argE::Tn10 ΔlysA::frt ΔbipA::kan | pBIPA | This study | Figure 5 |
| MRG03 | BW25113 argE::Tn10 ΔlysA::kan | pWSK29 | [23] | Figures 3(a), S3 |
| MRG36 | BW25113 argE::Tn10 ΔlysA::frt ΔychF::kan | pWSK29 | This study | Figure S6 |
| MRG39 | BW25113 argE::Tn10 ΔlysA::frt ΔhflX::kan | pWSK29 | This study | Figure S7 |
| MRG61 | BW25113 ΔbipA::kan | none | This study | |
| MRG103 | BW25113 ΔbipA::kan | pWSK29 | This study | Figure 4 |
| BR1 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | none | This study | |
| MRG137 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | none | This study | |
| MRG141 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pWSK29 | This study | Figure 6(b) |
| MRG142 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pBIPA | This study | Figure 6(b) |
| MRG143 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pBW1 | This study | Figure 6(b) |
| MRG144 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pWSK29 | This study | Figure 6(a–b) |
| MRG145 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pBIPA | This study | Figure 6(a–b) |
| MRG146 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pBW1 | This study | Figure 6(a–b) |
| MRG147 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pBAD24 | This study | Figure 6(c–d) |
| MRG148 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pBW2 | This study | Figure 6(c–d) |
| MRG149 | BW25113 Δ(recA-srl)306 srlR301::Tn10 | pBW6 | This study | Figure 6(c–d) |
| MRG150 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pBAD24 | This study | Figure 6(c–d) |
| MRG151 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pBW2 | This study | Figure 6(c–d) |
| MRG152 | BW25113 Δ(recA-srl)306 srlR301::Tn10 ΔbipA::kan | pBW6 | This study | Figure 6(c–d) |
Mutations ΔychF::kan and ΔhflX::kan [54] were individually moved by P1 transduction into the MRG01-KanS. Transductants were transformed with empty vector pWSK29 to generate MRG36 and MRG39, respectively.
For experiments involving the prototroph [Figures 3(b–c), 4, and S4], E. coli strain BW25113 was transformed with pWSK29, generating MRG46. The ΔbipA::kan mutation was moved into BW25113 and the resistance marker was removed by FLP-mediated recombination, generating MRG61. This strain was transformed with pWSK29 to create MRG103. For experiments involving BipA-H78A, recombination-deficient strains were generated. Mutation Δ(recA-srl)306 srlR301::Tn10 was moved into BW25113 and MRG61 via P1 transduction, resulting in BR1 and MRG137, respectively. BR1 and MRG137 were each transformed with pWSK29, pBIPA, and pBW1 to obtain the strains used in Figure 6(a–b). BR1 and MRG137 were each transformed with pBAD24, pBW2, and pBW6 to obtain the strains used in Figure 6(c–d).
Protein composition of ribosomal particles
For the experiments of Figure 2 and Figure S5, MRG06 and MC0030 were grown in M9 enriched minimal media plus glucose [M9E+G; 50 mM Na2HPO4, 200 mM KH2PO4, 8.6 mM NaCl, 7.5 mM (NH4)2SO4, 1 mM MgCl2, 1 mM MgSO4, 80 μM Na2EDTA, 50 μM CaCl2, 150 μM FeCl3, 0.94 μM ZnSO4, 0.96 μM CuSO4, 1.0 μM MnSO4, 1.1 μM CoCl2, 0.08 μM biotin, 0.04 μM folic acid, 0.15 μM thiamine, 0.41 μM nicotinamide, 0.13 μM riboflavin, 0.21 μM calcium pantothenate, 0.74 nM vitamin B12, 11 mM glucose] supplemented with medium (Arg 13C6 and Lys 2H4; MC0030), or heavy (Arg 13C6 15N4 and Lys 13C6 15N2; MRG06) amino acids, respectively. Each strain was grown overnight at 37 °C in the appropriate M9E+G media then diluted 1:100 in 50 mL of the same media. Cells were grown at 20 °C to OD600 ~0.5, and rapidly chilled, pelleted, and lysed as previously described [55]. Clarified lysate (0.3 mL) was loaded onto an 11 mL 10–35% sucrose gradient and subjected to ultracentrifugation at 35,000 rpm for 4 hours to separate ribosome complexes. Gradients were pumped using a syringe-pump system (Brandel) with in-line UV absorbance detector (UA-6, ISCO) and equivalent volume (0.5 mL) fractions were collected. In parallel, unlabeled 70S ribosomes were prepared and collected as previously described [56] from MRG06 grown in M9E+G supplemented with unlabeled Arg and Lys. Corresponding fractions from WT and mutant strains were mixed and 1.5 μM of 70S ribosomes were added to each mixed fraction. Proteins were precipitated using 13% (W/V) trichloroacetic acid (TCA).
For experiments in which corresponding gradient fraction were compared without any spike-in standard (Figures S2, S6, S7), cells were grown in M9E+G supplemented with light (unlabeled Arg and Lys; MRG06), medium (Arg 13C6 and Lys 2H4; MC0030, MRG36, MRG39), or heavy (Arg 13C6 15N4 and Lys 13C6 15N2; MRG71) amino acids. Sucrose gradient fractions were collected as described above. Corresponding fractions from WT, mutant, and complemented strains were mixed, and proteins were precipitated and subjected to qMS analysis.
For experiments in Figure S3, MRG03 was grown in M9E+G supplemented with medium-weight (Arg 13C6 and Lys 2H4 ; 37 °C) or heavy (Arg 13C6 15N4 and Lys 13C6 15N2; 20 °C) amino acids. Sucrose gradient fractions were prepared as described above, except that gradients were centrifuged for 6 hours and 0.3 mL fractions were collected (33 total fractions). In parallel, polysomes were prepared for use as the spike-in standard. These were collected as described [55] from cells grown in M9E+G supplemented with light amino acids (unlabeled Arg and Lys). Corresponding gradient fractions from cells grown at 37 °C and 20 °C were mixed and an equal volume (0.3 mL) of spike-in polysomes, and proteins were precipitated with TCA.
For experiments of Figures 3, 4, and S4, isotopically-labeled mature ribosomes (polysomes) were used as the spike-in standard, enabling sedimentation analysis of cells grown in any media. These polysomes were prepared from MRG06 grown in M9E+G with heavy (Arg 13C6 15N4 and Lys 13C6 15N2) amino acids. For experiments of Figure 3, cells were grown in M9E+G supplemented with light amino acids (unlabeled Arg and Lys; MRG03 and MRG46), or without supplementation (MRG46), at 37 °C. For the experiments of Figures 4 and S4, cells were grown with light amino acids (unlabeled Arg and Lys; MRG103 and MRG46), or without supplementation (MRG103 and MRG46) at 20 °C. At mid-log phase, cell lysates were prepared and subjected to sucrose gradient sedimentation, as described above. Each gradient fraction was mixed and an equal volume (0.3 mL) of spike-in polysomes, and proteins were precipitated.
Mass spectrometry
Protein precipitates were boiled in SDS sample buffer and run on a short 10% SDS-PAGE gel. Proteins were visualized by colloidal Coomassie [57] and digested out of the gel as described [58]. Peptide samples were purified by solid phase extraction on C-18 STop And Go Extraction (STAGE) Tips [59], and analyzed by a quadrupole – time of flight mass spectrometer (Impact II; Bruker Daltonics) coupled to an Easy nano LC 1000 HPLC (ThermoFisher Scientific) using an analytical column that was 40–50cm long, 75-μm-inner diameter fused silica with an integrated spray tip pulled with P-2000 laser puller (Sutter Instruments) and packed with 1.9 μm-diameter Reprosil-Pur C-18-AQ beads (Dr. Maisch, www.Dr-Maisch.com). Buffer A consisted of 0.1% aqueous formic acid, and buffer B consisted of 0.1% formic acid and 80% acetonitrile in water. A standard 90 min peptide separation was done and the column was washed with 100% Buffer B before re-equilibration with Buffer A. The Impact II was set to acquire in a data-dependent auto-MS/MS mode with inactive focus fragmenting the 20 most abundant ions (one at the time at 18 Hz rate) after each full-range scan from m/z 200 Th to m/z 2000 Th at 5 Hz rate. The isolation window for MS/MS was 2 to 3 Th depending on parent ion mass to charge ratio and the collision energy ranged from 23 to 65 eV depending on ion mass and charge. Parent ions were then excluded from MS/MS for the next 0.4 min and reconsidered if their intensity increased more than 5 times. Singly charged ions were excluded from fragmentation.
Data analysis
Analysis of Mass Spectrometry Data was performed using MaxQuant 1.5.1.0. The search was performed against a database comprised of the protein sequences from Uniprot’s E. coli K12 entries plus common contaminants with variable modifications of methionine oxidation, and N-acetylation of the proteins, in addition to the heavy amino acids used for quantitation. Only those peptides exceeding the individually calculated 99% confidence limit (as opposed to the average limit for the whole experiment) were considered as accurately identified. Data were deposited at the ProteomeXchange (accession number PXD017917).
Raw isotope ratios for proteins S2-S21 and L1-L36 were normalized to the median of all LSU or SSU proteins, respectively, in each fraction. In initial experiments (Figure 2), L10 and L7/12 appeared to be overrepresented across the entire polysome region, presumably due to partial stripping of these proteins from the (high-salt-washed) 70S ribosomes used as the spike-in standard. Hence, in experiments employing the 70S spike-in (Figure 2), we corrected for this by normalizing isotope ratios of L10 and L7/12 to their median values in the polysome region. In subsequent experiments, we used polysomes as the spike-in standard to avoid this complication. Significance testing was performed on Microsoft Excel using Student’s t test [=T.TEST(array1, array2, tails, type] where array 1 is the sample data, array 2 is 1.0, tails is 2 (two-tailed distribution) and type is 3 (two sample unequal variance). Isotope ratios of r-protein abundance were hierarchically clustered with MATLAB and the Bioinformatics toolbox [60], using a Euclidean distance metric and average linkage method.
Analysis of SSU rRNA processing
Relative levels of 17S rRNA were quantified as previously described [23] for strains MRG06, MC0030, and MRG71, except that RNA was extracted from each fraction 5–14, spanning from pre-30S to 70S.
Analysis of cells producing BipA-H78A
Cultures inoculated with a single colony were grown in 2 mL LB media plus 100 μg/mL ampicillin with shaking at 37 °C until visibly turbid (OD600 ~0.2). Cells from the starting culture were diluted 100-fold into identical fresh media (2 mL), and further grown for 150 min. Then, cultures were induced with 0.2% (W/V) arabinose and allowed to grow for another 120 min. Growth was monitored by measuring OD600.
For sucrose gradient sedimentation analysis, cells were grown in 50 mL LB media with 100 μg/ml ampicillin at 37 °C. When cultures reached an OD600 ~0.1, arabinose was added to 0.2% (W/V) and growth was continued for another 70 min. Then, cells were rapidly chilled, pelleted, and lysed as previously described [55]. Sucrose gradient sedimentation was performed as described above except that lysate was loaded onto a 10–40% sucrose gradient and subjected to ultracentrifugation at 35,000 rpm for 3 hours.
Supplementary Material
Highlights.
In E. coli, the landscape of 50S assembly changes in response to growth conditions.
BipA facilitates assembly of the 50S subunit at suboptimal growth temperature.
A GTPase-deficient variant of BipA shuts down translation and cell growth.
Acknowledgements
We thank members of the Ibba laboratory for use of their incubator shaker, L. Ying for help with the clustering analysis, and B. Roy for providing P1 carrying Δ(recA-srl)306 srlR301::Tn10. This work was supported by grants from the National Institutes of Health (R01 GM072528 to K.F. and T32 GM086252 to M.R.G.) and Genome Canada and Genome British Columbia (214PRO to L.J.F.). Mass spectrometry infrastructure was supported by the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Traub P, Nomura M, Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins, Proc. Natl. Acad. Sci. U. S. A 59 (1968) 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nierhaus KH, Dohme F, Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli, Proc. Natl. Acad. Sci. U. S. A 71 (1974) 4713–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Adilakshmi T, Bellur DL, Woodson SA, Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly, Nature 455 (2008) 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Talkington MW, Siuzdak G, Williamson JR, An assembly landscape for the 30S ribosomal subunit, Nature 438 (2005) 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mulder AM, Yoshioka C, Beck AH, et al. , Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit, Science 330 (2010) 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Held WA, Mizushima S, Nomura M, Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components, J. Biol. Chem 248 (1973) 5720–5730. [PubMed] [Google Scholar]
- [7].Samaha RR, O’Brien B, O’Brien TW, Noller HF, Independent in vitro assembly of a ribonucleoprotein particle containing the 3’ domain of 16S rRNA, Proc. Natl. Acad. Sci. U. S. A 91 (1994) 7884–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J, Chemical evidence for domain assembly of the Escherichia coli 30S ribosome, FASEB J 7 (1993) 177–180. [DOI] [PubMed] [Google Scholar]
- [9].Agalarov SC, Zheleznyakova EN, Selivanova OM, et al. , In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit, Proc. Natl. Acad. Sci. U. S. A 95 (1998) 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR, A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli, Elife 3 (2014) 10.7554/eLife.04491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ban N, Nissen P, Hansen J, Moore PB, Steitz TA, The complete atomic structure of the large ribosomal subunit at 2.4 A resolution, Science 289 (2000) 905–920. [DOI] [PubMed] [Google Scholar]
- [12].Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR, Modular Assembly of the Bacterial Large Ribosomal Subunit, Cell 167 (2016) 1610–1622.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davis JH, Williamson JR, Structure and dynamics of bacterial ribosome biogenesis, Philos. Trans. R. Soc. Lond. B. Biol. Sci 372 (2017) 10.1098/rstb.2016.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bokov K, Steinberg SV, A hierarchical model for evolution of 23S ribosomal RNA, Nature 457 (2009) 977–980. [DOI] [PubMed] [Google Scholar]
- [15].Qi SY, Li Y, Szyroki A, Giles IG, Moir A, O’Connor CD, Salmonella typhimurium responses to a bactericidal protein from human neutrophils, Mol. Microbiol 17 (1995) 523–531. [DOI] [PubMed] [Google Scholar]
- [16].Freestone P, Trinei M, Clarke SC, Nystrom T, Norris V, Tyrosine phosphorylation in Escherichia coli, J. Mol. Biol 279 (1998) 1045–1051. [DOI] [PubMed] [Google Scholar]
- [17].Farris M, Grant A, Richardson TB, O’Connor CD, BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells, Mol. Microbiol 28 (1998) 265–279. [DOI] [PubMed] [Google Scholar]
- [18].Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O’Connor CD, Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC), Mol. Microbiol 48 (2003) 507–521. [DOI] [PubMed] [Google Scholar]
- [19].Rowe S, Hodson N, Griffiths G, Roberts IS, Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli, J. Bacteriol 182 (2000) 2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pfennig PL, Flower AM, BipA is required for growth of Escherichia coi K12 at low temperature, Mol. Genet. Genomics 266 (2001) 313–317. [DOI] [PubMed] [Google Scholar]
- [21].Krishnan K, Flower AM, Suppression of DeltabipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA, J. Bacteriol 190 (2008) 7675–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Choudhury P, Flower AM, Efficient assembly of ribosomes is inhibited by deletion of bipA in Escherichia coli, J. Bacteriol 197 (2015) 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gibbs MR, Moon KM, Chen M, Balakrishnan R, Foster LJ, Fredrick K, Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K, The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli, Nucleic Acids Res 42 (2014) 13370–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].deLivron MA, Robinson VL, Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes, J. Bacteriol 190 (2008) 5944–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kumar V, Chen Y, Ero R, et al. , Structure of BipA in GTP form bound to the ratcheted ribosome, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 10944–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen SS, Williamson JR, Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry, J. Mol. Biol 425 (2013) 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ong SE, Blagoev B, Kratchmarova I, et al. , Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics, Mol. Cell. Proteomics 1 (2002) 376–386. [DOI] [PubMed] [Google Scholar]
- [29].Rohl R, Nierhaus KH, Assembly map of the large subunit (50S) of Escherichia coli ribosomes, Proc. Natl. Acad. Sci. U. S. A 79 (1982) 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shetty S, Varshney U, An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation, Proc. Natl. Acad. Sci. U. S. A (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV, Dual use of GTP hydrolysis by elongation factor G on the ribosome, Translation (Austin) 1 (2013) e24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].De Laurentiis EI, Wieden HJ, Identification of two structural elements important for ribosome-dependent GTPase activity of elongation factor 4 (EF4/LepA), Sci. Rep 5 (2015) 8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Daviter T, Wieden HJ, Rodnina MV, Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome, J. Mol. Biol 332 (2003) 689–699. [DOI] [PubMed] [Google Scholar]
- [34].Simonetti A, Marzi S, Fabbretti A, et al. , Structure of the protein core of translation initiation factor 2 in apo, GTP-bound and GDP-bound forms, Acta Crystallogr. D Biol. Crystallogr 69 (2013) 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Becker M, Gzyl KE, Altamirano AM, Vuong A, Urban K, Wieden HJ, The 70S ribosome modulates the ATPase activity of Escherichia coli YchF, RNA Biol 9 (2012) 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shields MJ, Fischer JJ, Wieden HJ, Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli: a kinetic approach, Biochemistry 48 (2009) 10793–10802. [DOI] [PubMed] [Google Scholar]
- [37].Coatham ML, Brandon HE, Fischer JJ, Schummer T, Wieden HJ, The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome, Nucleic Acids Res 44 (2016) 1952–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Basu A, Yap MN, Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E8165–E8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Guo Q, Goto S, Chen Y, et al. , Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process, Nucleic Acids Res 41 (2013) 2609–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Leong V, Kent M, Jomaa A, Ortega J, Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement, RNA 19 (2013) 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thurlow B, Davis JH, Leong V, Moraes TF, Williamson JR, Ortega J, Binding properties of YjeQ (RsgA), RbfA, RimM and Era to assembly intermediates of the 30S subunit, Nucleic Acids Res 44 (2016) 9918–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA, In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates, Mol. Cell 52 (2013) 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dever TE, Ivanov IP, Roles of polyamines in translation, J. Biol. Chem 293 (2018) 18719–18729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Igarashi K, Kashiwagi K, Effects of polyamines on protein synthesis and growth of Escherichia coli, J. Biol. Chem 293 (2018) 18702–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Michael AJ, Biosynthesis of polyamines and polyamine-containing molecules, Biochem. J 473 (2016) 2315–2329. [DOI] [PubMed] [Google Scholar]
- [46].Charlier D, Glansdorff N, Biosynthesis of Arginine and Polyamines, EcoSal Plus 1 (2004) 10.1128/ecosalplus.3.6.1.10. [DOI] [PubMed] [Google Scholar]
- [47].Choi E, Hwang J, The GTPase BipA expressed at low temperature in Escherichia coli assists ribosome assembly and has chaperone-like activity, J. Biol. Chem 293 (2018) 18404–18419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Choi E, Jeon H, Oh JI, Hwang J, Overexpressed L20 Rescues 50S Ribosomal Subunit Assembly Defects of bipA-Deletion in Escherichia coli, Front. Microbiol 10 (2020) 2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guillier M, Allemand F, Graffe M, et al. , The N-terminal extension of Escherichia coli ribosomal protein L20 is important for ribosome assembly, but dispensable for translational feedback control, RNA 11 (2005) 728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guillier M, Allemand F, Raibaud S, Dardel F, Springer M, Chiaruttini C, Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry, RNA 8 (2002) 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gibbs MR, Fredrick K, Roles of elusive translational GTPases come to light and inform on the process of ribosome biogenesis in bacteria, Mol. Microbiol 107 (2018) 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang RF, Kushner SR, Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli, Gene 100 (1991) 195–199. [PubMed] [Google Scholar]
- [53].Guzman LM, Belin D, Carson MJ, Beckwith J, Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter, J. Bacteriol 177 (1995) 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Baba T, Ara T, Hasegawa M, et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection, Mol. Syst. Biol 2 (2006) 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Qin D, Fredrick K, Analysis of polysomes from bacteria, Methods Enzymol 530 (2013) 159–172. [DOI] [PubMed] [Google Scholar]
- [56].Lancaster L, Kiel MC, Kaji A, Noller HF, Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing, Cell 111 (2002) 129–140. [DOI] [PubMed] [Google Scholar]
- [57].Candiano G, Bruschi M, Musante L, et al. , Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis, Electrophoresis 25 (2004) 1327–1333. [DOI] [PubMed] [Google Scholar]
- [58].Chan QW, Howes CG, Foster LJ, Quantitative comparison of caste differences in honeybee hemolymph, Mol. Cell. Proteomics 5 (2006) 2252–2262. [DOI] [PubMed] [Google Scholar]
- [59].Ishihama Y, Rappsilber J, Andersen JS, Mann M, Microcolumns with self-assembled particle frits for proteomics, J. Chromatogr. A 979 (2002) 233–239. [DOI] [PubMed] [Google Scholar]
- [60].The MathWorks Inc., MATLAB, (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


