Abstract
Molecular and functional characterization of a series of three bacteriophages, vB_SenM-1, vB_SenM-2, and vB_SenS-3, infecting various Salmonella enterica serovars and strains is presented. All these phages were able to develop lytically while not forming prophages. Moreover, they were able to survive at pH 3. The phages revealed different host ranges within serovars and strains of S. enterica, different adsorption rates on host cells, and different lytic growth kinetics at various temperatures (in the range of 25 to 42 °C). They efficiently reduced the number of cells in the bacterial biofilm and decreased the biofilm mass. Whole genome sequences of these phages have been determined and analyzed, including their phylogenetic relationships. In conclusion, we have demonstrated detailed characterization of a series of three bacteriophages, vB_SenM-1, vB_SenM-2, and vB_SenS-3, which reveal favorable features in light of their potential use in phage therapy of humans and animals, as well as for food protection purposes.
Keywords: bacteriophages, Salmonella, lytic development, genomic analysis
1. Introduction
The problem of infections caused by antibiotic-resistant bacteria is global [1]. The appearance of bacterial strains resistant to most or even all known antibiotics has become a real difficulty in medicine [2,3], and the need to find alternative methods of treatment of human and animal diseases caused by such bacteria is obvious [4,5]. The use of bacteriophages (or phages), viruses infecting bacterial cells, as alternative therapeutic agents is currently a hot topic; however, apart from many advantages of the use of phages in the treatment of humans and animals, including specificity of phages to selected bacteria without affecting the natural microbiota, their propagation restricted only to the presence of hosts, possibility to kill antibiotic-resistant cells, and a lack of documented considerable adverse effects related to phage administration, there are also controversies about both the efficacy and safety of such a method [6].
When considering phage therapy as an alternative treatment of human and animal infectious diseases, some conditions and requirements appear crucial [6,7,8]. Firstly, a large collection of phages infecting various bacterial species and strains must be established to offer an effective means of treatment of patients or animals suffering from diseases caused by different bacteria. Secondly, lytic rather than temperate bacteriophages should be used to achieve effective killing of bacterial pathogens and to avoid formation of lysogens which become resistant to phages. Thirdly, phages used in therapeutic approaches should not bear genes coding for toxins or other agents which are deleterious for humans and animals. These requirements indicate that isolation and detailed characterization of many bacteriophages is necessary to develop potentially effective phage therapy procedures [6,7,8].
Salmonella enterica is a foodborne pathogen, with poultry derived products being the main transmission factor to humans [9]. S. enterica can live in the chicken gastrointestinal tract and in most cases it does not cause any illness symptoms. However, depending on the type of serovar and the composition of the chicken’s gut microflora, it can quickly spread, resulting in later contamination in the abattoir during the process of meat preparation [10]. Serotypes most commonly responsible for the disease in humans are S. Typhimurium and S. Enteritidis. Other serotypes may vary depending on such factors as economy and the geography of a specific region [11]. As detection of S. enterica serotypes Typhimurium and Enteritidis results in many countries in immediate liquidation of the flock and legal restrictions are pushing towards prohibiting the use of antibiotics in farming animals, alternative methods of treatment—phages, in this case—are being tested for animal and food protection [9,12].
In fact, many papers have been published recently which described bacteriophages that might be used in phage therapy against Salmonella. They include reports on characterization of previously unknown bacteriophages having the potential for use in phage therapy [13,14,15,16,17,18,19,20], experimental studies with bacteriophages applied to poultry [21,22,23,24,25,26], and the use of bacteriophages in experimental phage therapy in mouse [27,28,29,30] or pig [31] models. Although promising results were obtained in these studies, and economic analyses have been performed to assess costs and benefits of the use of phage therapy for the control of Salmonella in poultry [9,32], it is evident that the host range of the vast majority of Salmonella phages is restricted to specific strains or serovars. Therefore, characterization of more bacteriophages infecting various Salmonella serovars and creation of large collections of different bacteriophages capable of killing such differential hosts appears to be necessary for the introduction of effective anti-Salmonella phage therapy.
The aim of this work was to characterize three phages infecting strains of S. enterica, a pathogen of humans and animals. These phages were isolated previously from urban sewage and preliminarily characterized, including classification, sizes of virions, and plaque morphology and size [33]. It appeared that, although infecting the same bacterial species, they can differ in many aspects of the optimal conditions for lytic development; therefore, each of them might be used for different purposes, as food protection or treatment of humans or animals. Hence, these phages might be potential tools in the development of anti-Salmonella therapies, and their detailed characterization is presented in this report.
2. Results
2.1. Morphology of Phage Virions and Plaque Morphology
Virion dimensions of vB_SenM-1, vB_SenM-2, and vB_SenS-3 were reported previously, as were morphologies of plaques [33]. Here, we present electron micrographs of vB_SenM-1, vB_SenM-2, and vB_SenS-3 virions which were not published earlier (Supplementary Figure S1).
2.2. Lysogenization Ability Assessment
To assess whether vB_SenM-1, vB_SenM-2, and vB_SenS-3 are able to form prophages, lysogenization tests were performed as described in Section 4.10. However, among bacteria which survived infection by tested phages, no lysogens could be identified as no plaques were observed after the induction of phage-treated bacteria with mitomycin C. Therefore, these results indicate that vB_SenM-1, vB_SenM-2, and vB_SenS-3 are virulent phages, unable to lysogenize host cells. This was additionally confirmed by analysis of phage genomes, as described in Section 2.8, where no lysogenization-specific genes could be detected in these genomes.
2.3. Host Range and Adsorption on Host Cells
To determine the host range of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3, we have employed a collection of S. enetrica strains representing various serovars. All these strains are clinical isolates; thus, performed tests might indicate the potential usefulness of investigated bacteriophages in phage therapy. As demonstrated in Table 1, there are various host ranges for each tested bacteriophage. However, most of the investigated S. enetrica strains were sensitive to at least one phage, with only one strain, S. Virchow, resistant to vB_SenM-1, vB_SenM-2, and vB_SenS-3. Such profiles of host sensitivities make the tested phages promising in light of their potential use in phage therapy.
Table 1.
Host range of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3.
| Bacterial Strain | Sensitivity to Phage c | |||
|---|---|---|---|---|
| Salmonella enterica Serotype a | Source and Identification Number b | vB_SenM-1 | vB_SenM-2 | vB_SenS-3 |
| Heidelberg | DMB UG collection; no. UGSA2 | 100% | 100% | 100% |
| Panama | DMB UG collection; no. UGSA3 | 97.82% ± 3.28% | 93.23% ± 4.21% | 95.12% ± 6.16% |
| Reading | DMB UG collection; no. UGSA4 | <0.01% | <0.01% | 15.21% ± 1.11% |
| London | DMB UG collection; no. UGSA5 | <0.01% | <0.01% | 1.25% ± 0.09% |
| Anatum | NSC, no. 78 | 28.11% ± 3.16% | 95.12% ± 4.11% | <0.01% |
| Typhimurium * | NSC, no. 12 | 67.13% ± 7.28% | 98.20% ± 4.77% | 95.13% ± 2.22% |
| Typhimurium | NSC, no. 13 | 23.28% ± 1.70% | 22.40% ± 3.41% | 98.61% ± 1.17% |
| Stanley * | NSC, no. 15 | 0.80% ± 0.06% | 97.28% ± 2.16% | 0.63% ± 0.11% |
| Heidelberg | NSC, no. 16 | 27.91% ± 8.09% | 98.66% ± 1.06% | 100% ± 1.03% |
| Cholerasuis | NSC, no. 34 | 17.22% ± 0.47% | 97.81% ± 3.22% | 99.82% ± 7.57% |
| Cholerasuis | NSC, no. 39 | 4.80% ± 0.88% | 89.16% ± 7.22% | 93.27% ± 4.21% |
| Cholerasuis var kunznedorf | NSC, no. 37 | 88.46% ± 2.61% | 80.52% ± 7.36% | 93.17% ± 4.16% |
| Cholerasuis | NSC, no. 1439 | <0.01% | <0.01% | 9.11% ± 0.76% |
| Virchow | NSC, no. 41 | <0.01% | <0.01% | <0.01% |
| Newport | NSC, no. 50 | 17.26% ± 2.16% | 10.08% ± 3.25% | <0.01% |
| Newport | NSC, no. 51 | <0.01% | <0.01% | 98.26% ± 3.89% |
| Enteritidis * | NSC, no. 64 | 88.87% ± 6.12% | 95.18% ± 2.16% | 98.26% ± 3.15% |
| Enteritidis * | NSC, no. 1392 | 66.21% ± 4.15% | 81.22% ± 4.33% | 86.36% ± 2.53% |
| Dublin | NSC, no. 65 | 55.21% ± 1.17% | 88.25% ± 3.21% | 97.16% ± 5.21% |
| Gallinarum | NSC, no. 74 | 63.55% ± 2.11% | 91.66% ± 5.11% | 89.21% ± 6.10% |
| Seftenberg | NSC, no. 87 | <0.01% | <0.01% | 94.66% ± 2.22% |
| Infantis * | NSC, no. 155 | 10.71% ± 1.06% | <0.01% | 6.15% ± 0.91% |
| Bovismorbificans * | NSC, no. 300 | <0.01% | <0.01% | 94.27% ± 2.31% |
| Saintpaul | NSC, no. 435 | 63.28% ± 4.95% | 96.21% ± 5.13% | 89.92% ± 2.89% |
| Kentucky * | NSC, no. 1368 | 51.22% ± 7.33% | 49.97% ± 3.47% | 24.25% ± 2.97% |
| Agona * | NSC, no. 1408 | 29.18% ± 3.15% | 98.36% ± 3.67% | 95.22% ± 5.11% |
| Hadar * | NSC, no. 1784 | <0.01% | <0.01% | 7.66% |
a Resistance to the following antibiotics was tested using minimal inhibitory concentration (MIC) and disc diffusion tests: tetracycline, rifampicin, ampicillin, chloramphenicol, colistin, enrofloxacin, and kanamycin. The resistance/susceptibility was determined on the basis of EUCAST guidelines. Investigated strains were susceptible to all tested antibiotics, with the exception of colistin for strains indicated by asterisks. b Abbreviations: NSC, National Salmonella Centre, Poland; DMB UG, Department of Molecular Biology of University of Gdansk, Poland. c Phage titer on S. Heidelberg UGSA2 was used as a reference value (100%). Mean values from three independent experiments are shown with SD.
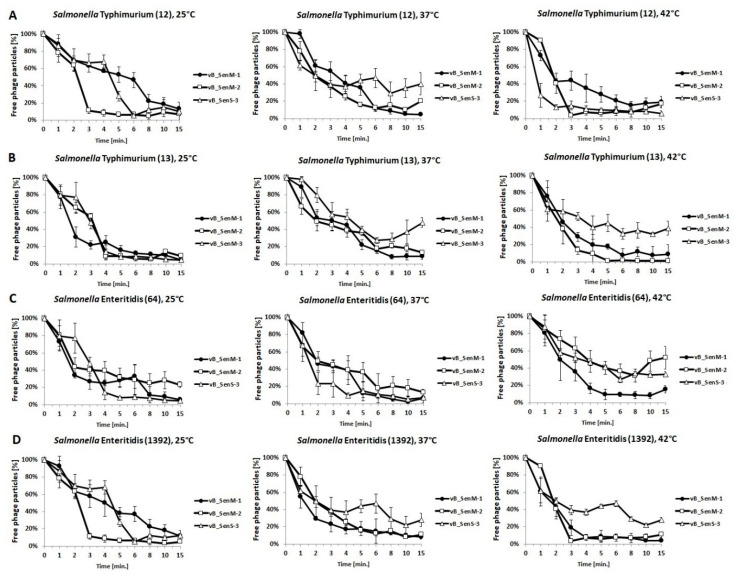
Efficiency of adsorption of tested phages on host cells was assessed using four selected S. enterica strains, two belonging to the Typhimurium serotype and two to Enteritidis serotype. Experiments were performed at three temperatures: 25, 37, and 42 °C. We have shown that titers of all phages were stable after incubation of virions for 60 min at all tested temperatures (Supplementary Table S1), indicating that any observed decrease in the number of free phages in experiments with host strains resulted from the adsorption of virions on bacterial cells rather than from virion inactivation. The most efficient adsorption was found for vB_SenM-1 on S. Typhimurium 13 at 25 °C and on S. Enteritidis 1392 at 42 °C, vB_SenM-2 on S. Typhimurium 12 at 25 °C and on S. Enteritidis 1392 at 25 and 42 °C, and vB_SenS-3 on Typhimurium 12 at 43 °C and on S. Enteritidis 64 at 37 °C (Figure 1). Nevertheless, all investigated phages adsorbed efficiently on bacterial cells within 15 min (Figure 1). These results indicated that the first stage of infection by vB_SenM-1, vB_SenM-2, and vB_SenS-3 was effective.
Figure 1.
Adsorption rate of phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares), and vB_SenS-3 (open triangles) on S. Typhimurium (A,B) and S. Enteritidis (C,D) at 25, 37, and 42 °C. Number of free phage particles at time 0 was used as reference value (100%). Mean values from three independent experiments are shown, with error bars representing SD.
2.4. One-Step Growth Experiments
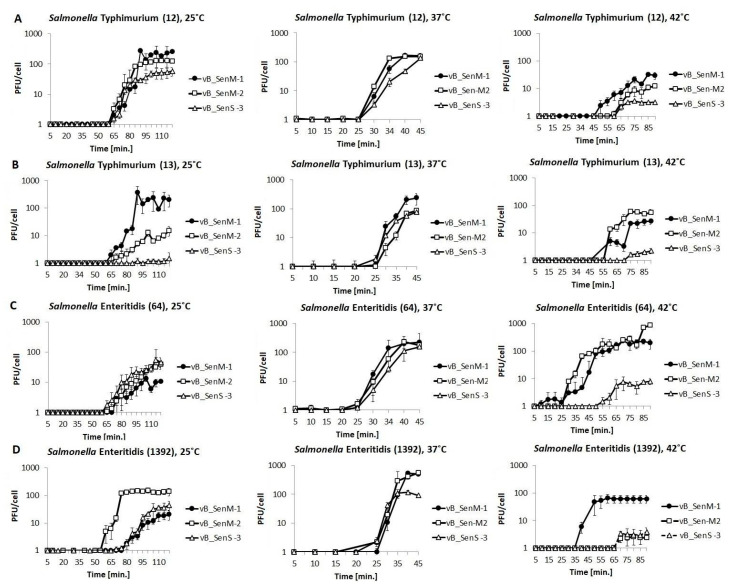
To assess the kinetics of intracellular phage development, one-step growth experiments were performed. As in phage adsorption experiments, four S. eneterica strains were used as hosts, and experiments were performed at 25, 37, and 42 °C. A temperature of 37 °C appeared to be optimal for all tested phages (Figure 2), which allowed the most efficient propagation of phages on all investigated hosts, giving phage yield between 58 and 499 per infected cell, on average (Table 2). The tested phages could also develop at either 25 or 42 °C, but under these conditions, the development was less effective in various host strains. For example, vB_SenS-3 developed poorly at 25 and 42 °C in S. Typhimurium 13 and S. Enteritidis 64, and lytic growth of both vB_SenM-2 and vB_SenS-3 was impaired in S. Enteritidis 1392 (Figure 2, Table 2). Nevertheless, each investigated S. enterica strain could be efficiently infected with at least one assessed phage at various temperatures, with the optimal temperature being 37 °C. These results were confirmed in independent experiments where PFU/mL of each phage was determined at 25, 37, and 42 °C and at multiplicity of infection (m.o.i.) 1, 0.5, and 0.1 (Supplementary Figures S2–S4).
Figure 2.
One-step growth experiments with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares), and vB_SenS-3 (open triangles) on S. Typhimurium (A,B) and S. Enteritidis (C,D) at 25, 37, and 42 °C, at m.o.i = 1. Mean values from three independent experiments are shown, with error bars representing SD.
Table 2.
Yield of progeny of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 per infected cell at different temperatures and on different S. enterica strains. Mean values from three independent experiments are shown with SD.
| Phage | S. enterica Strain | Yield of Phage Progeny (PFU/Cell) | |||
|---|---|---|---|---|---|
| Serotype | No. | Temperature | |||
| 25 °C | 37 °C | 42 °C | |||
| vB_SenM-1 | Typhimurium | 12 | 275.34 ± 30.27 | 58.31 ± 0.06 | 21.31 ± 5.56 |
| 13 | 375.34 ± 75.85 | 206.20 ± 68.51 | 22.20 ± 3.06 | ||
| Enteritidis | 64 | 13.35 ± 3.05 | 139.40 ± 3.55 | 105.48 ± 20.06 | |
| 1392 | 18.08 ± 6.98 | 499.22 ± 7.16 | 60.78 ± 18.37 | ||
| vB_SenM-2 | Typhimurium | 12 | 109.70 ± 15.98 | 131.24 ± 17.88 | 12.29 ± 2.40 |
| 13 | 12.76 ± 3.01 | 85.07 ± 4.91 | 59.06 ± 7.80 | ||
| Enteritidis | 64 | 23.75 ± 5.34 | 229.79 ± 54.27 | 174.59 ± 10.06 | |
| 1392 | 153.94 ± 34.07 | 283.73 ± 71.35 | 2.41 ± 0.35 | ||
| vB_SenS-3 | Typhimurium | 12 | 50.34 ± 18.98 | 136.21 ± 16.63 | 3.12 ± 0.91 |
| 13 | 1.18 ± 0.21 | 77.15 ± 12.80 | 2.23 ± 0.49 | ||
| Enteritidis | 64 | 28.75 ± 7.35 | 156.45 ± 31.15 | 7.40 ± 1.01 | |
| 1392 | 31.44 ± 19.17 | 117.38 ± 21.38 | 2.91 ± 1.57 | ||
2.5. Lysis Profile
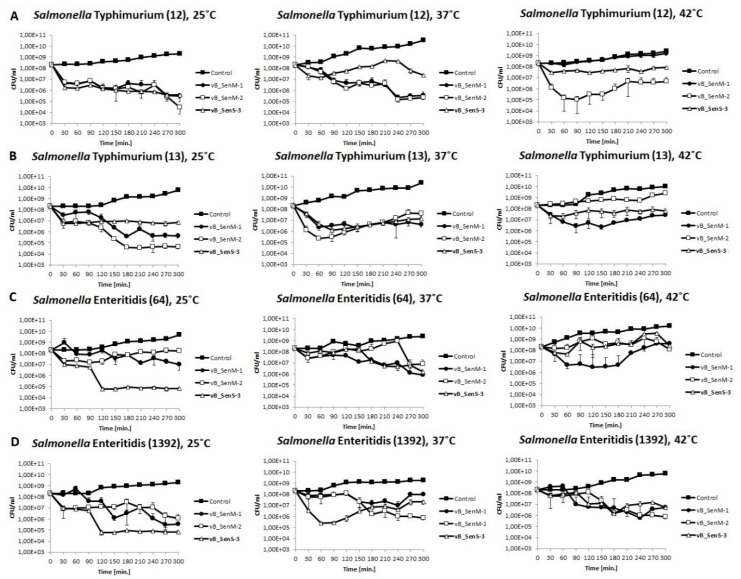
We have tested lysis profiles of the four bacterial strains (two S. Typhimurium strains and two S. Enteritidis strains) by all investigated phages at 25, 37, and 42 °C and at different m.o.i. Bacterial growth was monitored by determination of CFU/mL. Results of experiments performed at m.o.i. = 1 are presented in Figure 3, and results of those conducted at m.o.i. 0.5 or 0.1 are shown in Supplementary Figures S5 and S6. In addition, measurement of OD600 of bacterial cultures was monitored under the same conditions (Supplementary Figures S7–S9). These experiments generally corroborated the conclusions made on the basis of one-step growth experiments and indicated that the number of living bacterial cells of each of the host strains could be efficiently reduced by at least one of the tested phages at every assessed temperature. There were some ostensible discrepancies between results of one-step growth experiments and bacterial culture growth as assessed by measurement of OD600 (phages vB_SenM-1 and vB_SenM-2 gave high bursts size on S. Enteriditis 1392 at 37 °C and on S. Enteriditis 64 at 42 °C, while under these conditions, OD600 values resemble those of uninfected cultures). They might arise because, in one-step growth, we used chloroform to liberate phages from cells; thus, OD600 of bacterial cultures might in some cases remain relatively high if natural cell lysis is delayed.
Figure 3.
Changes in CFU/mL values of bacterial cultures of S. Typhimurium (A,B) and S. Enteritidis (C,D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares), and vB_SenS-3 (closed triangles) at m.o.i = 1, compared with uninfected control (closed squares). Mean values from three independent experiments are shown, with error bars representing SD.
2.6. pH Sensitivity
Sensitivity of the tested phages to various pH conditions was preliminarily assessed previously [13]. However, here, we investigated the stability of the phages under conditions of extremely low pH in more detail. Therefore, we have tested the effects of incubation of virions of vB_SenM-1, vB_SenM-2, and vB_SenS-3 in Lysogeny Broth (LB) medium with pH between 1 and 3. This can be important for the potential use of studied bacteriophages in phage therapy, as oral administration of phages leads to their exposition to acidic environment in the stomach. Although the use of pH below 2 resulted in effective inactivation of all tested phages, vB-SenM-1, vB_SenM-2, and vB_SenS-3 could survive 1-h incubation at pH 3 at the levels of 3.2%–5.7%, 6.3%–18.3%, and 6.2%–13.3%, respectively, depending on temperature (Table 3).
Table 3.
Sensitivity of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 to different pH levels and temperatures during 1-h incubation. pH = 7.0 was used as control value (100%).
| Phage | Temperature (°C) | Phage Survival (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| pH | ||||||||
| 7.0 | 1.8 | 2.0 | 2.2 | 2.5 | 2.8 | 3.0 | ||
| vB_SenM-1 | 25 °C | 100% ± 0.59% | <0.01% | <0.01% | <0.01% | 3.23% ± 0.03% | 4.83% ± 0.01% | 4.51% ± 0.09% |
| 37 °C | 100% | <0.01% | <0.01% | <0.01% | <0.01% | 2.22% ± 0.01% | 5.71% ± 0.12% | |
| 42 °C | 95.12% ± 2.12% | <0.01% | <0.01% | <0.01% | <0.01% | <0.01% | 3.26% ± 0.01% | |
| vB_SenM-2 | 25 °C | 98.66% ± 1.1% | <0.01% | <0.01% | 3.14% ± 0.02% | 2.23% ± 0.06% | 3.62% ± 0.11% | 12.83% ± 1.13% |
| 37 °C | 100% | <0.01% | <0.01% | 2.13% ± 0.12% | 2.7% ± 0.03% | 2.54% ± 0.06% | 18.27% ± 3.16% | |
| 42 °C | 86.93% ± 6.27% | <0.01% | <0.01% | <0.01% | 1.7% ± 0.01% | 2.13% ± 0.12% | 6.32% ± 0.42% | |
| vB_SenS-3 | 25 °C | 100% ± 2.19% | <0.01% | <0.01% | 1.92% ± 0.24% | 2.43% ± 0.33% | 2.32% ± 0.07% | 7.93% ± 0.04% |
| 37 °C | 100% | <0.01% | <0.01% | <0.01% | 1.71% ± 0.04% | 4.31% ± 0.24% | 6.15% ± 1.13% | |
| 42 °C | 81.26% ± 3.44% | <0.01% | <0.01% | <0.01% | <0.01% | 12.14% ± 3.16% | 13.33% ± 0.72% | |
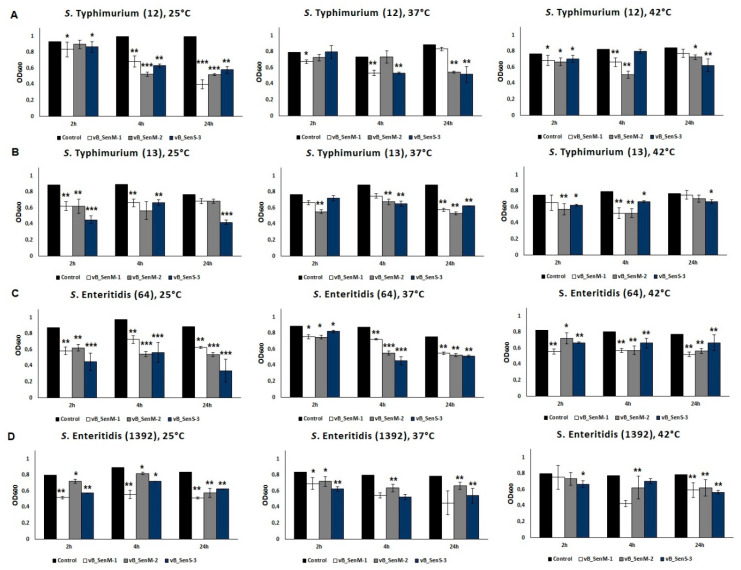
2.7. Effects of Phages on Bacterial Biofilm
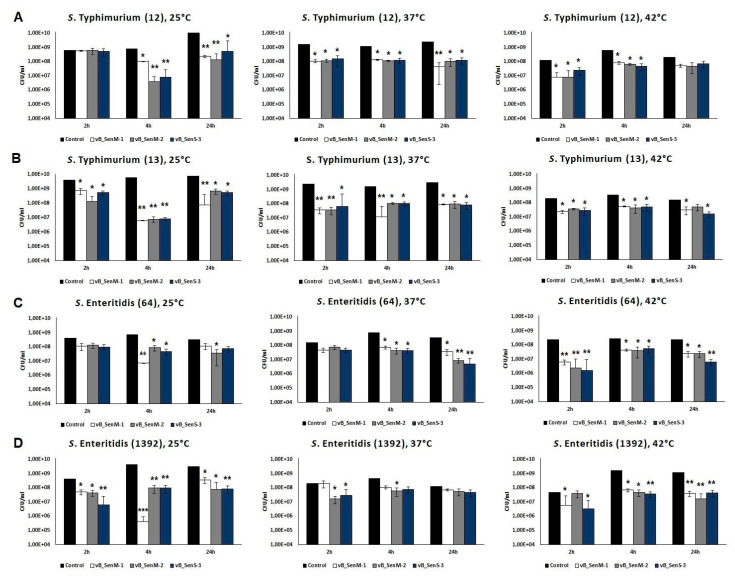
The efficiency of reducing the number of bacterial cells included in a biofilm by phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 was tested. We found that each tested phage was able to eliminate 47%–99% of bacterial cells (depending on the host and temperature) from the biofilm formed by any of the four tested S. enterica strains (Figure 4). This phage activity occurred at all tested temperatures. The optical density of biofilms after phage treatment is presented in Figure 5. A decrease in the biofilm mass was evident after treatment by phages vB_SenM-1, vB_SenM-2, and vB_SenS-3.
Figure 4.
Reduction in the number of bacteria (S. enterica serotypes Typhimurium (A,B) and Enteritidis (C,D)) from biofilm by phages vB_SenM-1 (white bars), vB_SenM-2 (grey bars), and vB_SenS-3 (navy blue bars) at 25, 37, and 42 °C. CFU/mL of bacterial biofilm untreated with bacteriophages was used as control and reference value (100%). Mean values from three independent experiments are shown, with error bars representing SD. Statistical analysis was performed using t-test; p < 0.05 (*), p < 0.01 (**).
Figure 5.
Changes in biofilm mass of different S. enterica serotypes Typhimurium 12 (A), 13 (B) and Enteritidis 64 (C), 1392 (D) caused by phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 after 4 h of incubation at 25, 37, or 42 °C. Mean values from three independent experiments are shown, with error bars representing SD. Statistical analysis was performed using t-test; p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
2.8. Analysis of Phage Genomes
The whole genomes of phages vB_SenM-1 and vB_SenS-3 have been sequenced. The genome of phage vb_SenM-2 has been described previously [33], and it is deposited in GenBank (accession number: KX171211).
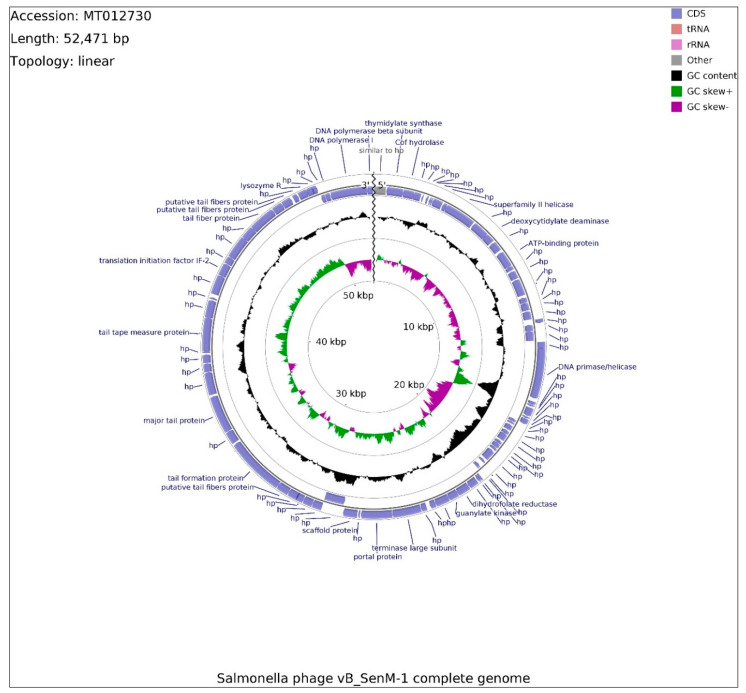
The genome of phage vB_SenM-1 contains 52,471 bp, with overall GC content of 45.9%, and is presented in the linear topology in Figure 6. Determination of open reading frames (ORFs) distinguished 87 putative protein-coding genes, among which 22 might be classified as known proteins (similar to previously reported ones) and 65 as putative. In total, 47 ORFs are located on the leading strand and 40 on the complementary strand.
Figure 6.
Schematic linear map of phage vB_SenM-1 genome (ends of the genome are indicated by the wavy line). The inner rings show genome location, GC skew + (green) and — (purple) and GC content (black). Two the most external rings show identified open reading frames (blue arrows) and results of genome annotation process.
Out of 87 ORFs, 76 initiate with the ATG start codon, whereas GTG and TTG occur less frequently, seven and four times, respectively. Stop codons were found to be more variable, from the most frequently observed codon TAA (46), throughout TGA (32), to the least frequent TAG (9). The total number of functionally assigned ORFs (18) was enriched by domain analysis, revealing four hypothetical proteins bearing domains, three of which are putatively engaged in bacteriophage morphogenesis (vB_SenM-1_68, vB_SenM-1_76, vB_SenM-1_79). Furthermore, ORFs were divided into four functional groups due to their assigned functions: morphogenesis (8), lysis (1), DNA packaging (3), and DNA replication (4), assembled in a tile-like manner. The lack of genes unambiguously related to lysogeny suggests that the phage is virulent; however, it was impossible to classify the life cycle of vB_SenM-1 phage based solely on the genome analysis. Besides hypothetical proteins with domains of tail chaperone function (vB_SenM-1_68) and baseplate J-like protein (vB_SenM-1_76), the rest of the morphogenesis ORFs are putatively engaged in tail assembly (vB_SenM-1_62, vB_SenM-1_63, vB_SenM-1_65, vB_SenM-1_78, and vB_SenM-1_79) and with tail tape-measure protein (vBSenM-1_70). No capsid proteins were identified. This section of the genome encompasses OFR74, potentially coding for translation initiation factor IF-2, which also bears a domain putatively involved in the process of “opening” the host cell membrane (vB_SenM-1_74). Coding DNA sequences (CDSs) involved in DNA packaging consist of genes for the terminase large subunit, portal and scaffold proteins located within 23869-27772 bp span. DNA replication encompasses four CDSs, two polymerase subunits (vB_SenM-1_87 and 88) within close proximity, and two CDSs scattered around the genome putatively coding for helicase (vB_SenM-1_11) and primase/helicase (vB_SenM-1_27). This section is intersected by a sequence similar to the gene of hypothetical protein (vBSenM1_01) missing a stop codon, which is typically located downstream of DNA polymerase CDSs (MT074444.1, MK770411.1). Using the PhageTerm software and Li’s method, the COS packaging strategy was identified for this linear genome. The sequences of the genome of phage vB_SenM-1 have been deposited in GenBank (accession number: MT012730.1).
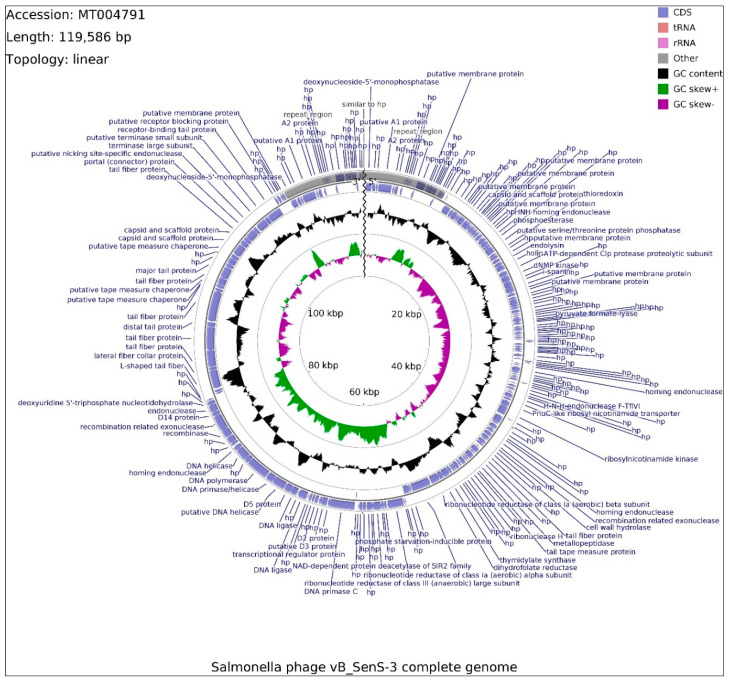
The genome of phage vB_SenS-3 consists of 119,586 bp, with overall GC content of 39.97% and a linear topology with repeat regions (9455 bp) located at sequence ends. The genome was found to be circularly permutated with redundant ends. The linear map of this genome is presented in Figure 7.
Figure 7.
Schematic linear map of genome of the vB_SenS-3 phage (ends of the genome are indicated by the wavy line). The inner rings show genome location, GC skew + (green) and—(purple) and GC content (black). The two most external rings show identified open reading frames (blue arrows) and results of genome annotation process. Repeated sequence at the beginning and the end of phage genome is marked in gray.
Determination of ORFs distinguished 229 putative protein-coding genes, of which 23 are embedded in the repeat region. Within the total ORFs, 78 are located on the leading strand and 151 are located on the complementary strand. Functions of 83 proteins could be determined on the basis of similarity to previously published sequences, and 146 remain putative. Out of 229 ORFs, 185 initiate with the ATG start codon, whereas GTG and TTG occur less frequently, 18 and 26 times, respectively. Frequency of observed stop codons is set out as follows: TAA (158), TGA (46), TAG (25). ORFs with assigned functions mainly use ATG start codons, with the exception of genes coding for holin (vB_SenS3_51) and DNA polymerase (vB_SenS3_167), utilizing GTG and TTG codons, respectively. ORFs with assigned functions were divided into four functional groups: morphogenesis (16), lysis (2), DNA packaging (9), and DNA replication (9). Two CDSs putatively coding for endolysin and holin (vB_SenS3_50 and 51) represent genes involved in host cell lysis. Morphogenesis CDSs are scattered around the genome, mainly gathered within positions 86,460-103,386 bp (8), where three CDSs code for head, capsid, and scaffold proteins (vB_SenS3_194, vB_SenS3_195, vB_SenS3_196). CDS with similar function was found in position vB_SenS3_32, with a significantly shorter sequence. The rest of the morphogenesis-related CDSs are involved in tail assembly, while the vB_SenS3_186 sequence was found to encompass Rho-independent transcription terminator. The DNA packaging group contains endonucleases (vB_SenS3_116, vB_SenS3_170, vB_SenS3_176), ribonuclease H (vB_SenS3_129), with portal protein (vBSenS3_198) and terminase large and small subunits (vB_SenS3_200, vB_SenS3_201), located upstream of the morphogenesis-related CDS conglomerate. Within positions 66,561-76,182 bp, genes encoding two ligases (vB_SenS3_161, vB_SenS3_163), primase (vB_SenS3_166), DNA polymerase (vB_SenS3_167), and DNA helicase (vB_SenS3_165, vB_SenS3__69) were found, and all these genes which are putatively responsible for phage DNA replication were predicted to be under the control of the same promoter, 66,350-66,395 bp, with the exception of vB_SenS3_169. The primase gene was found to possess Rho-independent transcription terminator. Additionally, DNA primase C gene (vB_SenS3_153) was found downstream of genes engaged in DNA replication. The vB_SenS3_173 sequence classified as putative recombinase gene was subjected to domain analysis at the amino acid level of the gene product, and the sequence was found to be partially similar to DNA repair exonuclease SbcCD nuclease subunit, responsible for cleaving DNA hairpin structures. The genome of phage vB_SenS-3 has been deposited in GenBank (accession number: MT004791.1).
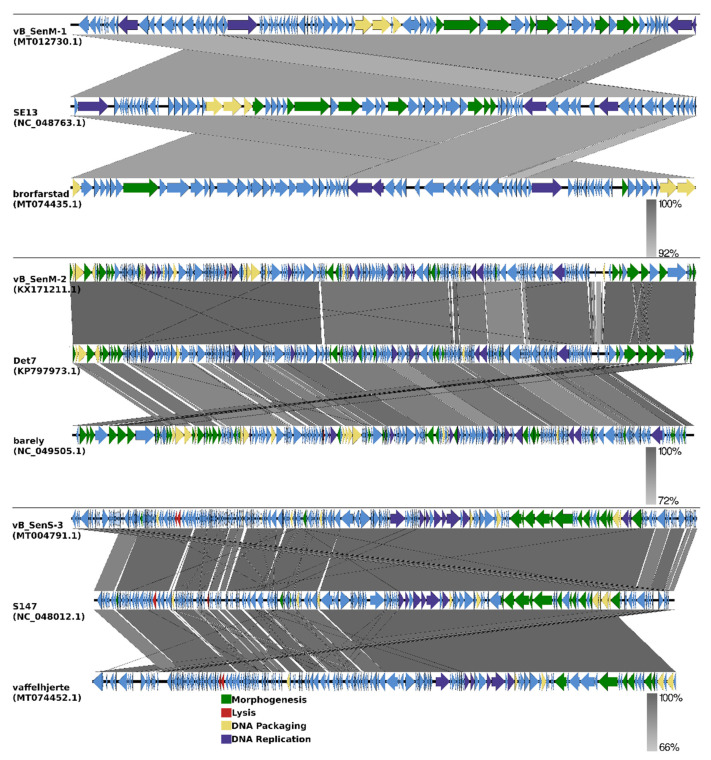
Annotations of genomes of phages vB_SenM-1 and vB_SenS-3 are presented in Supplementary Tables S2 and S3, respectively. In addition, genomic comparisons between phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 and the most closely related phages are shown in Figure 8 (see also Section 2.9).
Figure 8.
EasyFig output image of the genomic comparison between phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 and the most closely related phages. Phage genomes are presented by linear visualization, with coding regions shown as arrows. Selected open reading frames are colored in relation to their functions. The percentage of sequence similarity is indicated by the intensity of the gray color.
2.9. Phylogenetic Analyses of the Phages
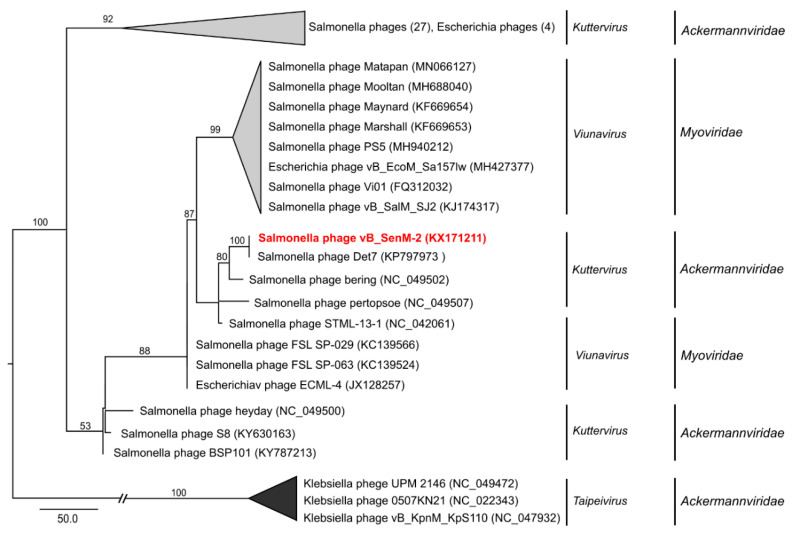
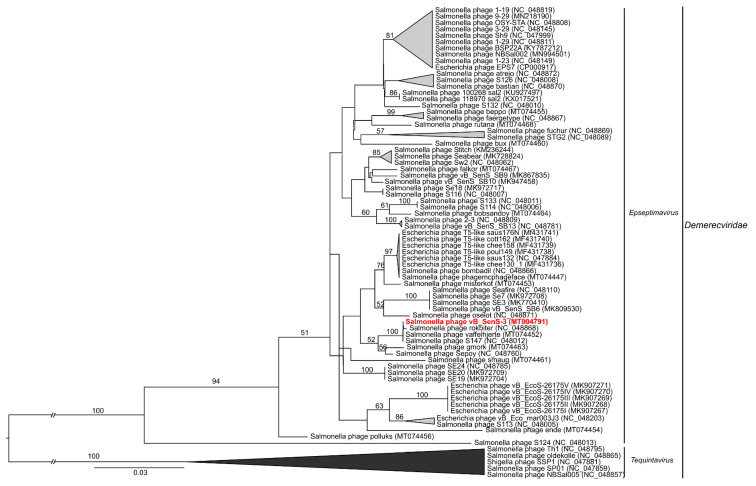
To analyze phylogenetic relationships between investigated phages and other viruses, we have compared the nucleotide sequences of genes coding for large terminase subunits of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 with the sequences of corresponding genes of other phages (Figure 9, Figure 10, and Figure 11, respectively).
Figure 9.
Neighbor-joining phylogenetic tree of the terminase large subunit (TerL) gene nucleotide sequences data showing the phylogenetic position of phage vB_SenM-1 (in red color) within Rosemountvirus. The reference sequences were collected from NCBI database. The tree was constructed using PAUP* with GTR + G+ I model of DNA substitution. Bootstrap values >50%, calculated based on 1000 resamplings, are shown above branches.
Figure 10.
Neighbor-joining phylogenetic tree of the terminase large subunit (TerL) gene nucleotide sequences data showing the phylogenetic position of phage vB_SenM-2 (in red color) within Kuttervirus. The reference sequences were collected from NCBI database. The tree was constructed using PAUP* with GTR + G+ I model of DNA substitution. Bootstrap values >50%, calculated based on 1000 resamplings, are shown above branches.
Figure 11.
Neighbor-joining phylogenetic tree of the terminase large subunit (TerL) gene nucleotide sequences data showing the phylogenetic position of phage vB_SenS-3 (in red color) within Epseptimavirus. The reference sequences were collected from NCBI database. The tree was constructed using PAUP* with GTR + G+ I model of DNA substitution. Bootstrap values >50%, calculated based on 1000 resamplings, are shown above branches.
As shown in Figure 9, the sequence of the gene encoding the large terminase subunit of vB_SenM-1 bacteriophage presents the highest identity with that of Salmonella phage brorfarstad (MT074435). Sequence similarity searches revealed that the aforementioned phages show a very high level of genome sequence identity (~96%). vB_SenM-1 also shows a high level of identity with other Salmonella phages such as SE13 (~96%) (NC_048763), yarpen (~95% identity) (NC_048863), brik (~95% identity) (NC_048864), and LSE7621 (~95% identity) (MK568062) (see also Figure 8). All of the phages are classified as belonging to the family Myoviridae, genus Rosemountvirus [34,35].
The sequence of the gene encoding the large terminase subunit of the phage vB_SenM-2 shows the highest identity with Salmonella phage Det7 (KP797973) [36], belonging to the family Ackermannviridae, of the genus Kuttervirus, according to the newest update of ICTV (International Committee on Taxonomy of Viruses) [37]. The phages also show 100% identity at the terminase gene level (Figure 10). This is an update of previous data according to which vB_SenM-2 had presented the highest identity with ViO1 (FQ312032) belonging to Myoviridae [33,38]. Thus, in this report, we classify the phage vB_SenM-2 as belonging to Ackermannviridae.
The nucleotide sequence of the gene encoding the large terminase subunit of vB_SenS-3 shows 100% identity with the sequences of two other phages, Salmonella phage vaffelhjerte (MT074452) and Salmonella phage S147 (NC_048012), both belonging to the family Demerecviridae, of the genus Epseptimavirus [39,40,41] (Figure 11; see also Figure 8). Sequence similarity searches revealed that the aforementioned phages also show a very high level of genome sequence identity (~99%). vB_SenS-3 shows high identity (~99%) with other phages such as S113 (MH370366) and rokbiter (NC_048868) [41].
3. Discussion
Phage therapy is one of the most promising alternatives to treat patients suffering from infectious diseases caused by antibiotic-resistant bacteria [42]. However, apart from legal difficulties which currently impair formal possibilities of the common use of phage therapy in clinical practice [43], there are several microbiological issues which must be considered when applying such therapeutic procedures [44]. Particularly, the crucial requirements are as follows: (i) the presence of a large collection of bacteriophages, (ii) the use of virulent, rather than temperate, phages, (iii) the use of phages devoid of genes coding for toxins and other potential deleterious agents in their genomes. Moreover, it is advantageous to use phages which adsorb efficiently on host cells, propagate rapidly, and give relatively high burst size. In addition, phages whose virions are stable under various environmental conditions are valuable.
S. enterica is a pathogenic bacterium, often revealing antibiotic resistance, infecting human and animal gastrointestinal tracts; thus, development of phage therapy against this bacterium is desirable. Phages infecting Salmonella strains can also be used in controlling this bacterium in poultry [9], as well as in food protection and dishwashing procedures [2,12,45]. However, each of the abovementioned applications require different properties of phages, like various optimal growth temperatures or resistance to different pH conditions.
In this report, we present characterization of a series of three bacteriophages infecting S. enterica. These phages have been isolated from urban sewage, but they were described only preliminarily, in light of biodiversity rather than phage therapy [33]. Nevertheless, we found that they have various properties which make them potentially useful agents in therapeutic and food protection purposes. These phages, named vB_SenM-1, vB_SenM-2, and vB_SenS-3, have virion morphologies characteristic for the Myoviridae (vB_SenM-1, vB_SenM-2) and Siphoviridae (vB_SenS-3) families (Supplementary Figure S1). Importantly, they are virulent phages, as indicated experimentally by their inability to lysogenize host cells. This is corroborated by genomic analyses which demonstrated a lack of genes potentially involved in lysogenization as well as toxin genes or virulence factors that could pose risk to other prokaryotic or eukaryotic cells (Figure 6 and Figure 7; Supplementary Tables S2 and S3). These features make them appropriate candidates for phage therapy and food protection purposes. The vB_SenM-1, vB_SenM-2, and vB_SenS-3 phages revealed different but relatively broad host ranges within the species S. enterica, being able to efficiently infect many different serovars and strains (Table 1), which again is an advantage in light of their practical use.
Developmental parameters of vB_SenM-1, vB_SenM-2, and vB_SenS-3 were characterized. All tested phages adsorbed effectively on various host strains within 15 min at different temperatures (Figure 1), demonstrating desirable features for phage therapy and food protection. One of the crucial properties of phages in light of these applications is the ability to perform effective lytic development at various temperatures and at different m.o.i. values. We found flexibility of the tested phages for these features, as demonstrated by one-step growth experiments and lysis profile determination (Figure 2 and Figure 3; Supplementary Figures S2–S9). Although some preferences of particular phages were evident, the crucial message is that effective reduction in the number of cells of different S. enterica strains was possible under every tested condition by at least one of the investigated bacteriophages. One should note that our one-step growth experiments did not estimate the precise latent period and classical burst size, defined as the number of phages liberated by spontaneous lysis of an infected bacterium. Regarding the latent period, we have conducted phage adsorption in an ice-bath and then centrifugation at 4 °C; however, we cannot exclude that phage development might proceed to some extent in cells during these 15 min (5 min phage adsorption, followed by 10 min centrifugation). Classical inhibition of phage development at the initial stages of one-step growth experiment by the use of a metabolic toxin like NaN3 could not be achieved as we found that S. enterica strains did not restart growth after washing such a compound out. Regarding burst size, we have used chloroform to lyse the cells and to estimate number of mature virions at each experimental time point. This allowed us to determine actual yield of phage progeny (both intra and extracellular) but did not fulfill the classical definition of the burst size as the lysis of hosts cells was not spontaneous.
In light of the potential use of the investigated bacteriophages in phage therapy, some level of resistance to pH levels of 3 (Table 3) and higher [33] may also be important, indicating that the use of compound(s) neutralizing acid conditions in human and animal stomach should be sufficient for the successful delivery of phages into the intestine. Moreover, effective reduction of number of cells of S. enterica from a biofilm, revealed by all tested phages at all tested temperatures (Figure 4), as well as reduction in the biofilm mass (Figure 5), are additional advantages when considering applications of vB_SenM-1, vB_SenM-2, and vB_SenS-3 in medicine, animal care, and food protection.
Phylogenetic analysis and sequence similarity searches revealed the phage vB_SenM-1 genome to be similar to previously described phage SE13. This phage has been characterized as being of broad host range among various S. enterica serotypes as well as having been effectively used as part of an experimental phage cocktail against Salmonella infection in food, especially cantaloupe and lettuce [34,46]. Moreover, phage LSE7621, to which phage vB_SenM-1 shows ~95% identity, has also been effectively tested in S. Enteritidis biocontrol on lettuce. However, this phage shows narrower host range than phage vB_SenM-1 [35]. Based on available data and experiments performed in this work, it can be assumed that phage vB_SenM-1 has the potential to be effectively used in food protection.
Phylogenetic and genomic analysis allowed the revision of classification based on the morphology of investigated phages. As a result, SenM-2 is now classified as Ackermannviridae instead of Myoviridae and SenS-3 is classified as Demerecviridae instead of Siphoviridae. The classification of SenM-1 remains unchanged—Myoviridae. In fact, viral classification is very dynamic due to the growing amount of viral genome data being deposited; therefore, it requires regular updates.
In conclusion, we have demonstrated detailed characterization of a series of three bacteriophages, vB_SenM-1, vB_SenM-2, and vB_SenS-3, which reveal favorable features in light of their potential use in phage therapy of humans and animals, as well as for food protection purposes. They are lytic phages, able to adsorb and propagate on various S. enterica strains under different conditions of temperature and m.o.i. Virions of these phages can partially (in the range of 3.2%–13.3%) survive 1-h incubation at pH 3.0. These phages can efficiently decrease both the number of S. enterica cells in the biofilm and the biofilm mass. Genomic and bioinformatic analyses allowed us to determine the principles of the organizations of the genomes of the investigated phages and to identify the potential functions of ORFs, revealing similarity to previously described and characterized genes coding for proteins involved in various processes during the development of different bacteriophages. Finally, the construction of phylogenetic trees based on DNA sequences led us to classify SenM-1, SenM-2, and SenS-3 as Myoviridae, Ackermannviridae and Demerecviridae, respectively.
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
Salmonella enterica strains used in this study come from the National Salmonella Centre at Medical University of Gdansk (Gdansk, Poland) and Bacterial Strain and Plasmid Collection at the University of Gdansk. More detailed description of strains used in this study is presented in Table 1.
4.2. Bacterial Culture Conditions
Bacteria were cultivated in LB medium at 25, 37, or 42 °C in a water bath at 150 rpm. LB with 1% agar was used for solid media and 0.7% agar was used in top agar for double-layer agar plates. Overnight incubation was performed at 37 °C.
4.3. Preparation of Phage Lysates
Phage lysates were prepared as follows: overnight culture of S. Heidelberg was diluted in fresh LB medium at a ratio of 1:100 and incubated with shaking at 37 °C until reaching OD600 = 0.15. Phages were then added to final m.o.i = 0.25. The culture was further incubated until lysis of bacterial cells was observed (OD600 = 0.05). Lysates were centrifugated at 5 min, 3500 g, 4 °C. The pellet was discarded. Lysates were stored at 4 °C with addition of 1% chloroform.
4.4. Electron Microscopy
Purification of bacteriophage vB_SenM-1, vB_SenM-2, and vB_SenS-3 particles was carried out by using the cesium chloride density gradient centrifugation method, according to the procedure described previously [47]. Transmission electron microscopy analysis of the phages was performed in the Laboratory of Electron Microscopy, Faculty of Biology, University of Gdansk, Gdansk, Poland. Virions were negatively stained with uranyl acetate; then, micrographs were taken under a Philips CM 100 electron microscope.
4.5. Phage Host Range
Host range of bacteriophages was determined using spot-test method described previously, with some modifications [33,48]. Phages were serially diluted in TM buffer (10 mM Tris-HCl pH 7.2., 10 mM MgSO4). The dilutions were then overlaid on top of double-layer agar plates with top agar containing 200 µL of bacterial culture, 10 mM CaCl2, and 1% casamino acids. Plates were incubated overnight at 37 °C and then scanned for the presence of single phage plaques. Phage titer was then counted, and results are presented relative to values obtained with the S. Heidelberg strain.
4.6. Influence of Low pH on Phage Viability
Susceptibility of phages to low pH levels was performed as described previously [33], with some modifications. In order to determine how low pH affects the viability of bacteriophages, 100 µL of phage lysate was added to 900 µL of LB at pH: 1.8, 2.0, 2.2, 2.5, 2.8, and 3.0. The samples were mixed and incubated for 1 h at 25, 37, or 42 °C. Afterwards, samples were serially diluted and overlaid on top of LB-agar plates with top agar containing 200 µL of bacterial culture, 10 mM CaCl2, and 1% casamino acids. Plates were incubated overnight at 37 °C and then scanned for plaques.
4.7. Phage Adsorption to Bacterial Host Cells
The adsorption assay was performed according to the protocol described before [48,49], with some modifications. Adsorption rate of bacteriophages vB_SenM-1, vB_SenM-2, and vB_SenS-3 was analyzed at three different temperatures (25, 37, and 42 °C) and on four S. enterica strains: S. Typhimurium 12, S. Typhimurium 13, S. Enteritidis 64, and S. Enteritidis 1392 (Table 1). Next, 109 cells were infected with a phage lysate to final m.o.i. = 0.1 and then incubated at given temperatures. At indicated time points, three individual samples per phage were collected and centrifuged at 6000 g for 30 s, to sediment the bacterial cells. The supernatant was diluted in TM buffer and assayed for free, unadsorbed phage particles. The number of viruses mixed with bacterial host cells at time 0 was considered 100% non-adsorbed phages. Other values were compared to this sample.
4.8. One-Step Growth Experiment
One-step growth experiments were performed as described previously, with some modifications [50]. In brief, S. enterica strains S. Typhimurium 12, S. Typhimurium 13, S. Enteritidis 64, and S. Enteritidis 1392 were grown at given temperatures (25, 37, or 42 °C) until reaching OD600 = 0.2. Then, 10 mL of bacterial culture was centrifuged (4000× g, 10 min, 4 °C), and the pellet was suspended in 1 mL of LB medium. Phage particles were added to the host culture at an m.o.i. = 0.5 and allowed to adsorb for 5 min in an ice-bath. The mixture was centrifuged (4000× g, 10 min, 4 °C) to remove unadsorbed viruses. Then, 50 µL of the bacterial mixture was added to 25 mL of LB medium (time 0) and cultivated at 25, 37, or 42 °C. The number of infective centers was estimated from samples taken 1, 2.5, and 5 min after infection by plating under permissive conditions. Then, 100 µL sample was collected at given temperatures, mixed with equal volume of chloroform, cleared by centrifugation (6000× g, 30 s) and titrated to determine the number of PFU per ml. The plates were incubated at 37 °C overnight. Yield of progeny phages was calculated as the ratio of phage titer to the number of infection centers. This value should not be interpreted as burst size, since, according to the classical definition, burst size indicates the number of phages liberated by spontaneous lysis of an infected bacterium.
4.9. Lysis Profile of Host Bacteria after Phage Infection
The experiments were performed in accordance with previously described procedures using four strains of S. enterica [51]. In brief, S. Typhimurium 12, S. Typhimurium 13, S. Enteritidis 64, and S. Enteritidis 1392 were cultivated to OD600 = 0.2 at 25, 37, or 42 °C. Then, phage stock solution was added to the host at an m.o.i. = 0.1, 0.5, or 1. In negative control experiments, bacteria were inoculated with LB medium instead of phage lysate. Bacterial growth was monitored by measuring the OD600 in 15 min intervals over period of 300 min. During this experiment, the number of bacterial cells per ml (CFU/mL) and phage titer (PFU/mL) were also determined. To calculate the number of surviving cells after virus infection, 100 µL of bacterial culture was collected at every 30 min, and serial dilutions were made in 0.89% sodium chloride and overlaid onto LB agar plates. The CFU/mL was calculated based on the counted colonies after overnight incubation at 37 °C. To estimate the PFU/mL, samples were taken every 30 min and diluted in TM buffer. The samples were overlaid onto a double-layer agar plate. The phage titer was determined by counting single plaques.
4.10. Efficiency of Lysogenization
The experiments were performed in accordance with previously described protocols [52,53], with some modifications. In brief, S. Typhimurium 12, S. Typhimurium 13, S. Enteritidis 64, and S. Enteritidis 1392 were cultivated to OD600 = 0.2 at 37 °C. Then, 1 mL of bacterial culture was centrifugated (2000× g, 10 min, room temperature) and the pellet was resuspended in 1 mL of LB medium containing 10 mM CaCl2. After incubation for 5 min at 30 °C, phage lysate was added to m.o.i = 1. In control experiments, LB was added to the culture instead of phage lysate. Bacteria were incubated for 2 h at 30 °C. Serial dilutions were prepared in TM buffer and 40 µL of each dilution was poured onto LB plates. After overnight incubation at 37 °C, 96 colonies were passaged separately each in a well of a 96-well plate with 200 µL of LB medium (if less colonies were visible on the plates all of them were passaged). The plates were incubated with shaking at 37 °C until bacteria reached OD600 = 0.2.
In order to test the resistance to superinfection, 50 µL of bacterial culture was mixed with a solution composed of 10 mM CaCl2 and 10% casamino acids and spread onto LB agar. Then, 2.5 µL of phage lysate was spotted on top of it. The plates were then incubated overnight at 37 °C and then scanned for plaques. If plaques were not visible, the clone was recognized as resistant to phage. The resistance to infection was determined as a percent of bacteria uninfected by the phage. Each experiment was performed in triplicate.
For estimation of efficiency of lysogenization, 1 µg/mL mitomycin C was added to 150 µL of remaining bacterial culture. The plates were then incubated for another 3 h. Afterwards, 10 µL of chloroform was added, the plates were centrifugated (2000× g, 10 min, 4 °C) and 5 µL of top layer was spotted onto double-layer LB agar plates. The plates were incubated overnight at 37 °C. The presence of lysogen was determined by formation of plaque on double-layer agar plate. The efficiency of lysogenization was determined as percent of lysogens among all tested 96 bacterial colonies. The experiment was performed in triplicate.
4.11. Effects of Phages on Bacterial Cells in Biofilm
Biofilm cell biomass after phage infection was determined according to the protocols described previously [48,54], with some modifications. The experiment was performed at three different temperatures (25, 37, and 42 °C) and on four S. enterica strains: S. Typhimurium 12, S. Typhimurium 13, S. Enteritidis 64, and S. Enteritidis 1392. For biofilm cell culture, S. enterica was grown at 37 °C or 42 °C in 12-well polystyrene plate with 0.5 × LB medium for 24 h or at 25 °C for 48 h. After the incubation period, the liquid medium containing planktonic cells was removed. The biofilm was washed with 1 mL of distilled water. In the next step, phage lysate was added to each well, except for controls, to the final titer of 109 PFU/well, and then plates were incubated at 25, 37, or 42 °C for 2, 4, and 24 h. In the case of control wells, the medium was added instead of phage lysate. Following incubation, phage lysate was removed, and biofilms were washed with 1 mL of distilled water and then resuspended in 1 mL of distilled water. Serial dilutions were made and overlaid on top of LB-agar medium and left to dry down. The plates were incubated at 37 °C overnight and then scanned for bacterial colonies.
4.12. Assessment of Biofilm Biomass by Crystal Violet Staining after Phage Infection
Biofilms were prepared according to the procedure described above. After incubation with phage lysate (added to final titer 109 PFU/well), the liquid medium was removed and surface-attached cells were treated with 1 mL of 0.1% crystal violet solution (Sigma-Aldrich). Plates were incubated in the dark for 20 min at 42 °C. Crystal violet was then carefully removed and biofilms were washed 2 times with 1 mL of distilled H2O. Then, biofilms were fixed by additional incubation at 42 °C for 20 min. Afterwards, crystal violet was dissolved by the addition of 0.5 mL of 96% ethanol. The absorbance was measured in a plate reader at 600 nm (VICTOR, PerkinElmer, Waltham, MA, USA) in order to assess biofilm biomass. Biofilms were also photographed following the fixation in order to visualize the differences in biofilm biomass after phage treatment.
4.13. Isolation and Sequencing of Phages vB_SenM-1 and vB_SenS-3 Genomes
The DNA isolation was performed as described previously [33]. Briefly, phage lysates were purified using cesium chloride density gradient. Then, 300 µL of purified lysate was treated with DNase and RNase for 30 min at 37 °C to digest the exogenous bacterial RNA and DNA. After thermal inactivation, the suspension was treated with proteinase K for 60 min at 37 °C. The phage genome DNA was isolated by using the MasterPure™ Complete DNA and RNA Purification Kit (Epicentre) in accordance with the manufacturer’s protocol. DNA concentration was determined spectrophotometrically by measuring the absorbance at a wavelength of 260 nm. The genomic DNA of phages vB_SenM-1, vB_SenM-2, and vB_SenS-3 were sequenced by the Genomed company with next generation sequencing (NGS) and MiSeq (Illumina) genome sequencing. Sequences of genomes were deposited in the GenBank database, and accession numbers for genomes of vB_SenM-1, vB_SenM-2, and vB_SenS-3 are MT012730, KX171211, and MT004791, respectively.
4.14. Annotation and Bioinformatic Analysis of Phages vB_SenM-1 and vB_SenS-3
Putative open reading frames (ORFs) that may encode gene products were predicted by using RASTtk [55], backed up by prodigal incorporated in Prokka annotation suite [56]. To confirm the selection of correct start codon, the ribosome binding sites were analyzed 4–12 nucleotides upstream of the start codon. Putative functions of translated products were then verified by BLASTn analysis of the Nucleotide Collection (nr/nt) database, additionally utilizing the CDD database pinpointing conserved domains [57], pVOGs database where (97) Salmonella phage genomes were obtained (http://dmk-brain.ecn.uiowa.edu/pVOGs/, last accessed on 7 August, 2020), and HMMer Reference Proteomes database [58]. The CGView Comparison Tool [59] was used to create a map of virus genome with GC skew and GC content analyses. To determine phage termini, packaging mode and topology of genomes PhageTerm was employed [60].
Comparison of ORFs from relative phages was performed by using the EasyFig program (http://mjsull.github.io/Easyfig/files.html, last accessed on 16 August, 2020). Classification of phages’ lifestyle was conducted in PHACTS [61]. Phage-specific promoters and Rho-independent transcriptional terminators in viral DNA sequences were predicted by the Neural Network Promoter Prediction NNPP method (http://www.fruitfly.org/seq_tools/promoter.html, last accessed on 7 August, 2020) and the Arnold tool [62], respectively.
Salmonella phages’ (vB_SenM-1, vB_SenS-3) genomes were sequenced on Illumina platform MiSeq, with Whole Genome Shotgun (WGS) strategy. The library was prepared using NEBNext DNA Library Prep Master Mix Set for Illumina; selection was random, with read length ranging from 36 to 251 bp. Raw reads were deposited in SRA databases under BioProject ID: PRJNA655386.
4.15. Phylogenetic Analysis
To construct phylogenetic trees, the nucleotide sequences of genes coding for terminase large subunits (genetic marker for the order Caudovirales) of vB_SenM-1, vB_SenM-2, and vB_SenS-3 viruses were compared with the sequences of other reference bacteriophages within the order Caudovirales that were deposited in the NCBI database. DNA sequences were aligned and adjusted by eye using Seaview [63]. The jModelTest v. 2.1.1 (Phylogenomics Lab, University of Vigo, Vigo, Spain) [64] was used in order to choose the best-fitting evolutionary model by the Akaike information criterion. All matrices were analyzed using PAUP (Phylogenetic Analysis Using Parsimony and Other Methods) version 4.0a [65]. The optimality criterion was set to distance using the neighbor-joining algorithm (NJ). The robustness of the tree topology was assessed by bootstrap analyses based on 1000 replicates.
Acknowledgments
The authors would like to thank Malwina Richert for making electron microscopic images and Jakub Kaźmierczak for providing software enabling phage capsid measurements and help with its use.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/17/6152/s1. Table S1. Genome annotations of phage vB_SenM-1. Table S2. Genome annotations of phage vB_SenS-3. Figure S1. Electron micrographs of phages vB_SenM-1 (A), vB_SenM-2 (B) and vB_SenS-3 (C). Scale bars represent 50 nM. Figure S2. Changes in PFU/ml values of phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) after infection of bacterial cultures of S. Typhimurium (A, B) and S. Enteritidis (C, D) at m.o.i=1. Mean values from three independent experiments are shown, with error bars representing SD. Figure S3. Changes in PFU/ml values of phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) after infection of bacterial cultures of S. Typhimurium (A, B) and S. Enteritidis (C, D) at m.o.i=0.5. Mean values from three independent experiments are shown, with error bars representing SD. Figure S4. Changes in PFU/ml values of phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) after infection of bacterial cultures of S. Typhimurium (A, B) and S. Enteritidis (C, D) at m.o.i=0.1. Mean values from three independent experiments are shown, with error bars representing SD. Figure S5. Changes in CFU/ml values of cultures of S. Typhimurium (A, B) and S. Enteritidis (C, D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (closed triangles) at m.o.i=0.5, compared with uninfected control (closed squares). Mean values from three independent experiments shown, with error bars representing SD. Figure S6. Changes in CFU/ml values of bacterial cultures of S. Typhimurium (A, B) and S. Enteritidis (C, D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (closed triangles) at m.o.i=0.1, compared with uninfected control (closed squares). Mean values from three independent experiments are shown, with error bars representing SD. Figure S7. Lysis profile of S. Typhimurium (A, B) and S. Enteritidis (C, D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) at m.o.i=1 compared with uninfected control (closed squares). Mean values from three independent experiments are shown, with error bars representing SD. Figure S8. Lysis profile of S. Typhimurium (A, B) and S. Enteritidis (C, D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) at m.o.i=0.5 compared with uninfected control (closed squares). Mean values from three independent experiments are shown, with error bars representing SD. Figure S9. Lysis profile of S. Typhimurium (A, B) and S. Enteritidis (C, D) infected with phages vB_SenM-1 (closed circles), vB_SenM-2 (open squares) and vB_SenS-3 (open triangles) at m.o.i=0.1 compared with uninfected control (closed squares). Mean values from three independent experiments are shown, with error bars representing SD.
Author Contributions
Conceptualization, K.K.-K., G.W. and A.W.; methodology, K.K.-K., M.G. (Michał Grabski), M.G. (Marcin Górniak), and A.J.-K.; validation, K.K.-K. and M.G. (Michał Grabski); experimental investigation, K.K.-K., K.C., Ł.G.; data analysis, K.K.-K., M.G. (Michał Grabski), M.G. (Marcin Górniak), and A.W.; data interpretation, K.K.-K., M.G. (Michał Grabski), M.G. (Marcin Górniak), G.W. and A.W.; writing—original draft preparation, K.K.-K. and A.W.; writing—review and editing, K.K.-K., M.G. (Michał Grabski), G.W. and A.W.; visualization, K.K.-K. and M.G. (Michał Grabski); supervision, A.W.; funding acquisition, A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Science Center (Poland), grants No. 2015/17/B/NZ9/01724 and 2017/27/B/NZ9/00393.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Davies J., Davies D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okocha R.C., Olatoye I.O., Adedeji O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018;39:21. doi: 10.1186/s40985-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchings M.I., Truman A.W., Wilkinson B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019;51:72–80. doi: 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Bengtsson B., Greko C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Upsala J. Med Sci. 2014;119:96–102. doi: 10.3109/03009734.2014.901445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventola C.L. The Antibiotic Resistance Crisis part 1: Causes and threats. Pharm. Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 6.Górski A., Międzybrodzki R., Jończyk-Matysiak E., Borysowski J., Letkiewicz S., Weber-Dąbrowska B. The fall and rise of phage therapy in modern medicine. Expert Opin. Biol. Ther. 2019;19:1115–1117. doi: 10.1080/14712598.2019.1651287. [DOI] [PubMed] [Google Scholar]
- 7.Górski A., Międzybrodzki R., Węgrzyn G., Jończyk-Matysiak E., Borysowski J., Weber-Dąbrowska B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2019;40:459–463. doi: 10.1002/med.21593. [DOI] [PubMed] [Google Scholar]
- 8.Rohde C., Resch G., Pirnay J.-P., Blasdel B.G., Debarbieux L., Gelman D., Górski A., Hazan R., Huys I., Kakabadze E., et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses. 2018;10:178. doi: 10.3390/v10040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Acosta M., Clavijo V., Vaglio C., González-Barrios A.F., Vives-Flórez M.J., Rito-Palomares M. Economic evaluation of the development of a phage therapy product for the control of Salmonella in poultry. Biotechnol. Prog. 2019;35:e2852. doi: 10.1002/btpr.2852. [DOI] [PubMed] [Google Scholar]
- 10.Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gal-Mor O., Boyle E.C., Grassl G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014;5:319. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan T., Liang L., Yin P., Zhou Y., Sharoba A.M., Lu Q., Dong X.X., Liu K., Connerton I., Li J. Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms. 2020;8:400. doi: 10.3390/microorganisms8030400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakabadze E., Makalatia K., Grdzelishvili N., Bakuradze N., Goderdzishvili M., Kusradze I., Phoba M.-F., Lunguya O., Lood C., Lavigne R., et al. Selection of Potential Therapeutic Bacteriophages that Lyse a CTX-M-15 Extended Spectrum β-Lactamase Producing Salmonella enterica Serovar Typhi Strain from the Democratic Republic of the Congo. Viruses. 2018;10:172. doi: 10.3390/v10040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Virk S.M., Shi J., Zhou Y., Willias S.P., Morsy M.K., Abdelnabby H.E., Liu J., Wang X., Li J. Isolation, Characterization, and Application of Bacteriophage LPSE1 Against Salmonella enterica in Ready to Eat (RTE) Foods. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sritha K.S., Bhat S.G. Genomics of Salmonella phage ΦStp1: Candidate bacteriophage for biocontrol. Virus Genes. 2018;54:311–318. doi: 10.1007/s11262-018-1538-3. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Sun E., Song J., Tong Y., Wu B. Three Salmonella enterica serovar Enteritidis bacteriophages from the Siphoviridae family are promising candidates for phage therapy. Can. J. Microbiol. 2018;64:865–875. doi: 10.1139/cjm-2017-0740. [DOI] [PubMed] [Google Scholar]
- 17.Patil K., Zeng C., O’Leary C., Lessor L., Kongari R., Gill J.J., Liu M. Complete Genome Sequence of Salmonella enterica Serovar Typhimurium Siphophage Seabear. Microbiol. Resour. Announc. 2019;8:e01160-19. doi: 10.1128/MRA.01160-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holguín A.V., Cárdenas P., Prada-Peñaranda C., Leite L.R., Buitrago C., Clavijo V., Oliveira G.C., Leekitcharoenphon P., Aarestrup F.M., Vives M. Host Resistance, Genomics and Population Dynamics in a Salmonella Enteritidis and Phage System. Viruses. 2019;11:188. doi: 10.3390/v11020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao H., Shahin K., Zhang Q., Zhang H., Wang Z., Zhou Y., Zhang X., Zhu S., Stefan S., Wang R. Morphologic and genomic characterization of a broad host range Salmonella enterica serovar Pullorum lytic phage vB_SPuM_SP116. Microb. Pathog. 2019;136:103659. doi: 10.1016/j.micpath.2019.103659. [DOI] [PubMed] [Google Scholar]
- 20.Mohamed A., Taha O., El-Sherif H., Connerton I., Hooton S.P., Bassim N., Connerton I., El-Shibiny A. Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy. Viruses. 2020;12:424. doi: 10.3390/v12040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevilla-Navarro S., Marín C., Cortés V., García C., Vega S., Catalá-Gregori P. Autophage as a control measure for Salmonella in laying hens. Poult. Sci. 2018;97:4367–4373. doi: 10.3382/ps/pey294. [DOI] [PubMed] [Google Scholar]
- 22.Tie K., Yuan Y., Yan S., Yu X., Zhang Q., Xu H., Zhang Y., Gu J., Sun C., Lei L., et al. Isolation and identification of Salmonella pullorum bacteriophage YSP2 and its use as a therapy for chicken diarrhea. Virus Genes. 2018;54:446–456. doi: 10.1007/s11262-018-1549-0. [DOI] [PubMed] [Google Scholar]
- 23.Clavijo V., Baquero D., Hernandez S., Farfan J., Arias J., Arévalo A., Donado-Godoy P., Vives M. Phage cocktail SalmoFREE® reduces Salmonella on a commercial broiler farm. Poult. Sci. 2019;98:5054–5063. doi: 10.3382/ps/pez251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaz C.S.L., Voss-Rech D., Alves L., Coldebella A., Brentano L., Trevisol I.M. Effect of time of therapy with wild-type lytic bacteriophages on the reduction of Salmonella Enteritidis in broiler chickens. Vet. Microbiol. 2020;240:108527. doi: 10.1016/j.vetmic.2019.108527. [DOI] [PubMed] [Google Scholar]
- 25.Li M., Lin H., Jing Y., Wang J. Broad-host-range Salmonella bacteriophage STP4-a and its potential application evaluation in poultry industry. Poult. Sci. 2020;99:3643–3654. doi: 10.1016/j.psj.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Żbikowska K., Michalczuk M., Dolka B. The Use of Bacteriophages in the Poultry Industry. Animals. 2020;10:872. doi: 10.3390/ani10050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikkhahi F., Dallal M.M.S., Alimohammadi M., Foroushani A.R., Rajabi Z., Fardsanei F., Imeni S.M., Bonab P.T. Phage therapy: Assessment of the efficacy of a bacteriophage isolated in the treatment of salmonellosis induced by Salmonella enteritidis in mice. Gastroenterol. Hepatol. Bed Bench. 2017;10:131–136. [PMC free article] [PubMed] [Google Scholar]
- 28.Tang F., Zhang P., Zhang Q., Xue F., Ren J., Sun J., Qu Z., Zhuge X., Li D., Wang J., et al. Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb. Pathog. 2019;126:193–198. doi: 10.1016/j.micpath.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 29.Dallal M.M.S., Nikkhahi F., Alimohammadi M., Douraghi M., Rajabi Z., Foroushani A.R., Azimi A., Fardsanei F. Phage Therapy as an Approach to Control Salmonella enterica serotype Enteritidis Infection in Mice. Rev. Soc. Bras. Med. Trop. 2019;52:20190290. doi: 10.1590/0037-8682-0290-2019. [DOI] [PubMed] [Google Scholar]
- 30.Bao H., Zhou Y., Shahin K., Zhang H., Cao F., Pang M., Zhu S., Olaniran A., Schmidt S., Wang R., et al. The complete genome of lytic Salmonella phage vB_SenM-PA13076 and therapeutic potency in the treatment of lethal Salmonella Enteritidis infections in mice. Microbiol. Res. 2020;237:126471. doi: 10.1016/j.micres.2020.126471. [DOI] [PubMed] [Google Scholar]
- 31.Seo B.-J., Song E.-T., Lee K., Kim J.-W., Jeong C.-G., Moon S.-H., Son J.S., Kang S.H., Cho H.-S., Jung B.Y., et al. Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella Typhimurium. J. Vet. Med Sci. 2018;80:851–860. doi: 10.1292/jvms.17-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamal M., Bukhari S.M.A.U.S., Andleeb S., Ali M., Raza S., Nawaz M.A., Hussain T., Rahman S.U., Shah S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2018;59:123–133. doi: 10.1002/jobm.201800412. [DOI] [PubMed] [Google Scholar]
- 33.Jurczak-Kurek A., Gąsior T., Nejman-Faleńczyk B., Bloch S., Dydecka A., Topka G., Necel A., Jakubowska-Deredas M., Narajczyk M., Richert M., et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016;6:34338. doi: 10.1038/srep34338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fong K., Tremblay D.M., Delaquis P., Goodridge L.D., Levesque R.C., Moineau S., Suttle C.A., Wang S. Wang Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica. Viruses. 2019;11:854. doi: 10.3390/v11090854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu A., Liu Y., Peng L., Cai X., Shen L., Duan M., Ning Y., Liu S., Li C., Liu Y., et al. Characterization of the narrow-spectrum bacteriophage LSE7621 towards Salmonella Enteritidis and its biocontrol potential on lettuce and tofu. LWT. 2020;118:108791. doi: 10.1016/j.lwt.2019.108791. [DOI] [Google Scholar]
- 36.Casjens S.R., Jacobs-Sera D., Hatfull G.F., Hendrix R.W. Genome Sequence of Salmonella enterica Phage Det7. Genome Announc. 2015;3:e00279-15. doi: 10.1128/genomeA.00279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefkowitz E., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acid. Res. 2017;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickard D., Toribio A., Petty N.K., Van Tonder A.J., Yu L., Goulding D., Barrell B., Rance R., Harris D., Wetter M., et al. A Conserved Acetyl Esterase Domain Targets Diverse Bacteriophages to the Vi Capsular Receptor of Salmonella enterica Serovar Typhi. J. Bacteriol. 2010;192:5746–5754. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradiso R., Lombardi S., Iodice M.G., Riccardi M.G., Orsini M., Censi S.B., Galiero G., Borriello G. Complete Genome Sequence of a Lytic Siphoviridae Bacteriophage Infecting Several Serovars of Salmonella enterica. Genome Announc. 2016;4:e00943-16. doi: 10.1128/genomeA.00943-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paradiso R., Lombardi S., Iodice M.G., Riccardi M.G., Orsini M., Censi S.B., Galiero G., Borriello G. Complete Genome Sequences of Three Siphoviridae Bacteriophages Infecting Salmonella enterica Serovar Enteritidis. Genome Announc. 2016;4:e00939-16. doi: 10.1128/genomeA.00939-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gencay Y.E., Gambino M., Prüssing T.F., Brøndsted L. The genera of bacteriophages and their receptors are the major determinants of host range. Environ. Microbiol. 2019;21:2095–2111. doi: 10.1111/1462-2920.14597. [DOI] [PubMed] [Google Scholar]
- 42.Górski A., Międzybrodzki R., Lobocka M., Głowacka-Rutkowska A., Bednarek A., Borysowski J., Jończyk-Matysiak E., Łusiak-Szelachowska M., Weber-Dąbrowska B., Bagińska N., et al. Phage Therapy: What Have We Learned. Viruses. 2018;10:288. doi: 10.3390/v10060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero-Calle D., Benevides R.G., Góes-Neto A., Billington C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics. 2019;8:138. doi: 10.3390/antibiotics8030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber-Dąbrowska B., Jończyk-Matysiak E., Żaczek M., Lobocka M., Łusiak-Szelachowska M., Górski A. Bacteriophage Procurement for Therapeutic Purposes. Front. Microbiol. 2016;7:1177. doi: 10.3389/fmicb.2016.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imklin N., Nasanit R. Characterization of Salmonella bacteriophages and their potential use in dishwashing materials. J. Appl. Microbiol. 2020;129:266–277. doi: 10.1111/jam.14617. [DOI] [PubMed] [Google Scholar]
- 46.Wong C.W., Delaquis P., Goodridge L., Lévesque R.C., Fong K., Wang S. Inactivation of Salmonella enterica on post-harvest cantaloupe and lettuce by a lytic bacteriophage cocktail. Curr. Res. Food Sci. 2020;2:25–32. doi: 10.1016/j.crfs.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J.F., Russell D.W. Molecular Cloning: A Laboratory Manual. Volume 1 Cold Spring Harbor Laboratory; New York, NY, USA: 2001. [Google Scholar]
- 48.Topka G., Bloch S., Nejman-Faleńczyk B., Gąsior T., Jurczak-Kurek A., Necel A., Dydecka A., Richert M., Węgrzyn G., Węgrzyn A. Characterization of Bacteriophage vB-EcoS-95, Isolated from Urban Sewage and Revealing Extremely Rapid Lytic Development. Front. Microbiol. 2019;9:3326. doi: 10.3389/fmicb.2018.03326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Necel A., Bloch S., Nejman-Faleńczyk B., Grabski M., Topka G., Dydecka A., Kosznik-Kwaśnicka K., Grabowski Ł., Jurczak-Kurek A., Wolkowicz T., et al. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020;10:3743. doi: 10.1038/s41598-020-60568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J., Zhang Z., Tian C., Chen X., Hu L., Wei X., Li H., Lin W., Jiang A., Feng R., et al. Characterizing the Biology of Lytic Bacteriophage vB_EaeM_φEap-3 Infecting Multidrug-Resistant Enterobacter aerogenes. Front. Microbiol. 2019;10:420. doi: 10.3389/fmicb.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abedon S.T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J. Bacteriol. 1992;174:8073–8080. doi: 10.1128/JB.174.24.8073-8080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.León M., Santander J., Curtiss R., Robeson J. Natural lysogenization and transduction in Salmonella enterica serovar Choleraesuis by bacteriophage P1. Res. Microbiol. 2013;164:1–5. doi: 10.1016/j.resmic.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dydecka A., Nejman-Faleńczyk B., Bloch S., Topka G., Necel A., Donaldson L., Węgrzyn G., Węgrzyn A. Roles of orf60a and orf61 in Development of Bacteriophages λ and Φ24B. Viruses. 2018;10:553. doi: 10.3390/v10100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramachandran G., Aheto K., Shirtliff M.E., Tennant S.M. Poor biofilm-forming ability and long-term survival of invasive Salmonella Typhimurium ST313. Pathog. Dis. 2016;74:49. doi: 10.1093/femspd/ftw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldeira J.C., Peabody D.S. Stability and assembly in vitro of bacteriophage PP7 virus-like particles. J. Nanobiotechnol. 2007;5:10. doi: 10.1186/1477-3155-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acid. Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acid. Res. 2011;39(Suppl. 2):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stothard P., Wishart D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2004;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 60.Garneau J.R., Depardieu F., Fortier L.-C., Bikard D., Monot M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-07910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McNair K., Bailey B.A., Edwards R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics. 2012;28:614–618. doi: 10.1093/bioinformatics/bts014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011;8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 63.Galtier N., Gouy M., Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 64.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swofford D.L. PAUP. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates; Sunderland, MA, USA: 2003. pp. 160–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.