Abstract
The EAG (ether-à-go-go) family of voltage-gated K+ channels are important regulators of neuronal and cardiac action potential firing (excitability) and have major roles in human diseases such as epilepsy, schizophrenia, cancer, and sudden cardiac death. A defining feature of EAG (Kv10–12) channels is a highly conserved domain on the N terminus, known as the eag domain, consisting of a Per–ARNT–Sim (PAS) domain capped by a short sequence containing an amphipathic helix (Cap domain). The PAS and Cap domains are both vital for the normal function of EAG channels. Using heme-affinity pulldown assays and proteomics of lysates from primary cortical neurons, we identified that an EAG channel, hERG3 (Kv11.3), binds to heme. In whole-cell electrophysiology experiments, we identified that heme inhibits hERG3 channel activity. In addition, we expressed the Cap and PAS domain of hERG3 in Escherichia coli and, using spectroscopy and kinetics, identified the PAS domain as the location for heme binding. The results identify heme as a regulator of hERG3 channel activity. These observations are discussed in the context of the emerging role for heme as a regulator of ion channel activity in cells.
Keywords: heme, hERG3, ion channel, PAS domain, Cap domain, heme regulation, hERG, X-ray crystallography, protein crystallization, heme regulation
Per–ARNT–Sim (PAS) domains, first identified by sequence homology in the Drosophila proteins period and single-minded, and the vertebrate aryl hydrocarbon receptor nuclear transporter (ARNT), are well-known sensory modules present in a variety of signaling proteins in organisms ranging from bacteria to humans (1–3). PAS domains demonstrate considerable plasticity in binding different biologically relevant ligands. Relevant in the context of this work is their ability to bind heme as a ligand. The ligand-binding event is significant because it can act as a primary trigger to initiate a cellular signaling/regulatory response, or it can provide the PAS domain with a capacity to respond to secondary physical or chemical signals (such as binding of gas molecules, changes in redox potential, or light activation). In the case of heme as a ligand, there are several examples of heme-binding PAS domains that act as biological regulators in cells, for example the O2 sensor proteins (e.g. FixL, Escherichia coli DOS) and the transcriptional regulators involved in circadian control (e.g. nPAS2, CLOCK, Rev-ERB, Per) (4, 5).
Several layers of regulatory control can be envisaged in the process of heme binding to a PAS domain protein. Heme binding might, for example, induce a conformational change in the PAS domain, which affects the interactions with partner proteins, and there is evidence for this in the heme-binding circadian proteins (6). Because known cell-signaling gases such as CO, NO, and O2 all bind to heme (and with different affinities), the formation of a heme-bound PAS domain opens opportunities for the molecule to also respond to changes in concentrations of any or all of these cell-signaling gases. Thinking in these terms, it may not be a coincidence that CO and NO production in cells is catalyzed by two heme-containing proteins, heme oxygenase and nitric oxide synthase, respectively, both of which are themselves O2-dependent. This interplay of heme- and gas-responsive PAS domains could provide the cell with highly versatile mechanisms for regulatory control that are at the same time responsive to changing concentrations of heme, O2, CO, or NO. How these mechanisms play out has yet to be fully established.
In this paper, we present evidence from electrophysiological experiments for heme-dependent modulation of hERG3 (human ether-à-go-go–related gene 3, alternatively known as Kv11.3). We identify an interaction of heme with a PAS domain contained within the channel. Our results provide insight on the role of heme in channel regulation; we use this to discuss how heme-induced structural changes might be used to control channel function.
Results
Proteomics analyses
We sought to identify potential heme-binding proteins in neuronal lysates from primary cortical neurons, using heme-affinity chromatography and MS. Using this approach, we identified a number of proteins known to bind heme (e.g. mitochondrial enzymes), as well as proteins with unknown heme affinity; this included hERG3 (Fig. S1). In the structure of the overall hERG3 channel, there are four subunits, each containing a transmembrane region, as well as cytoplasmic N- and C-terminal regions (Fig. 1) (7). The N-terminal cytoplasmic region (∼400 residues) contains a highly conserved eag domain (residues 1–135), which itself comprises a Cap domain (residues 1–25) and a Per–ARNT–Sim (PAS) domain (residues 26–135).
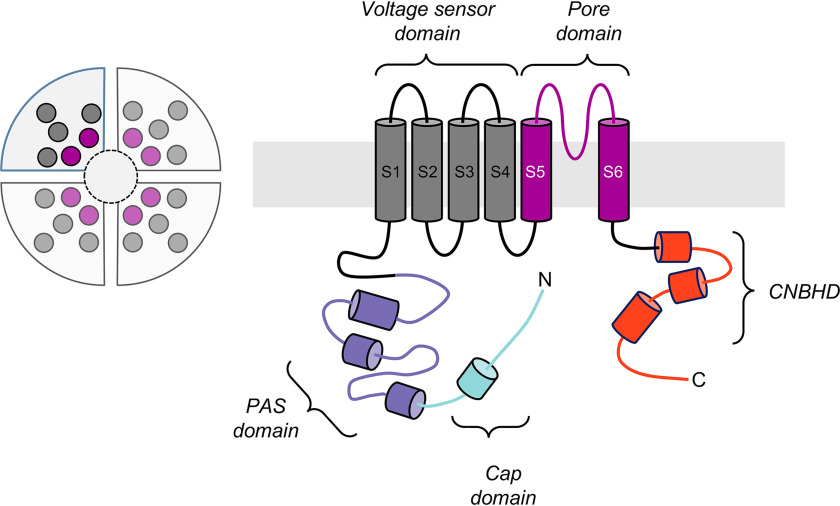
Figure 1.
Schematic of the hERG3 channel subunit. Left side, hERG, which is part of the EAG family of ion channels, has four subunits, each shown as a quadrant, that assemble to form a tetrameric structure as shown in the circular schematic on the left. Right side, each subunit contains a transmembrane region, as well as cytoplasmic N- and C-terminal regions. The transmembrane region contains voltage sensor (helices S1–S4, dark gray) and pore-forming (helices S5 and S6, magenta) domains inside the membrane bilayer. The C-terminal cytoplasmic region contains a cyclic nucleotide-binding homology domain (CNBHD, red), which is connected to the transmembrane region via a structured domain usually referred to as the C-linker. The N-terminal cytoplasmic region (∼400 residues) contains a region that is known as the eag domain (residues 1–135), which itself comprises a Cap domain (residues 1–26, light blue) and a PAS domain (residues 27–135, purple). This eag domain fragment (residues 1–135) has been expressed in this work and is referred to as hERG3-eag in this paper.
Heme binding to the hERG3-eag domain
We expressed the Cap domain (residues 1–26) and the PAS (residues 27–135) domain of the hERG3 protein as an N-terminal His6/S-tagged fusion protein in E. coli (Fig. 1 and Fig. S2); we will refer to this protein as hERG3-eag. Upon addition of heme to hERG3-eag, Fig. 2A, a species with Soret bands at 420 and 370 nm is observed, which we assign as arising from six- and five-coordinated heme, respectively. There is also a broad absorption envelope at 500–600 nm and a ligand-to-metal charge transfer band at 650 nm (Fig. 2A). These bands are similar to those reported for heme bound to proteins through Cys, Cys/His, or Cys/Xaa coordination (Table S1) (8). The presence of two bands in the Soret region is indicative of heterogeneity, which may arise either from changes in axial ligands or from different orientations of the bound heme (8–10). The ligands to the heme in hERG3 were not identified. The affinity of ferric heme for hERG3-eag was determined by titration (Kd = 7.02 ± 0.35 μm; Fig. 2A, inset, top panel). Heme affinity can also be extracted from the first-order rate constant for transfer of heme to apo-myoglobin (which has a very high affinity for heme (11)); we measured this rate constant as koff = 0.03 s−1 (Fig. 2A, inset, bottom panel), which is several orders of magnitude higher than for the globins and more in the range observed for other regulatory heme proteins (12).
Figure 2.
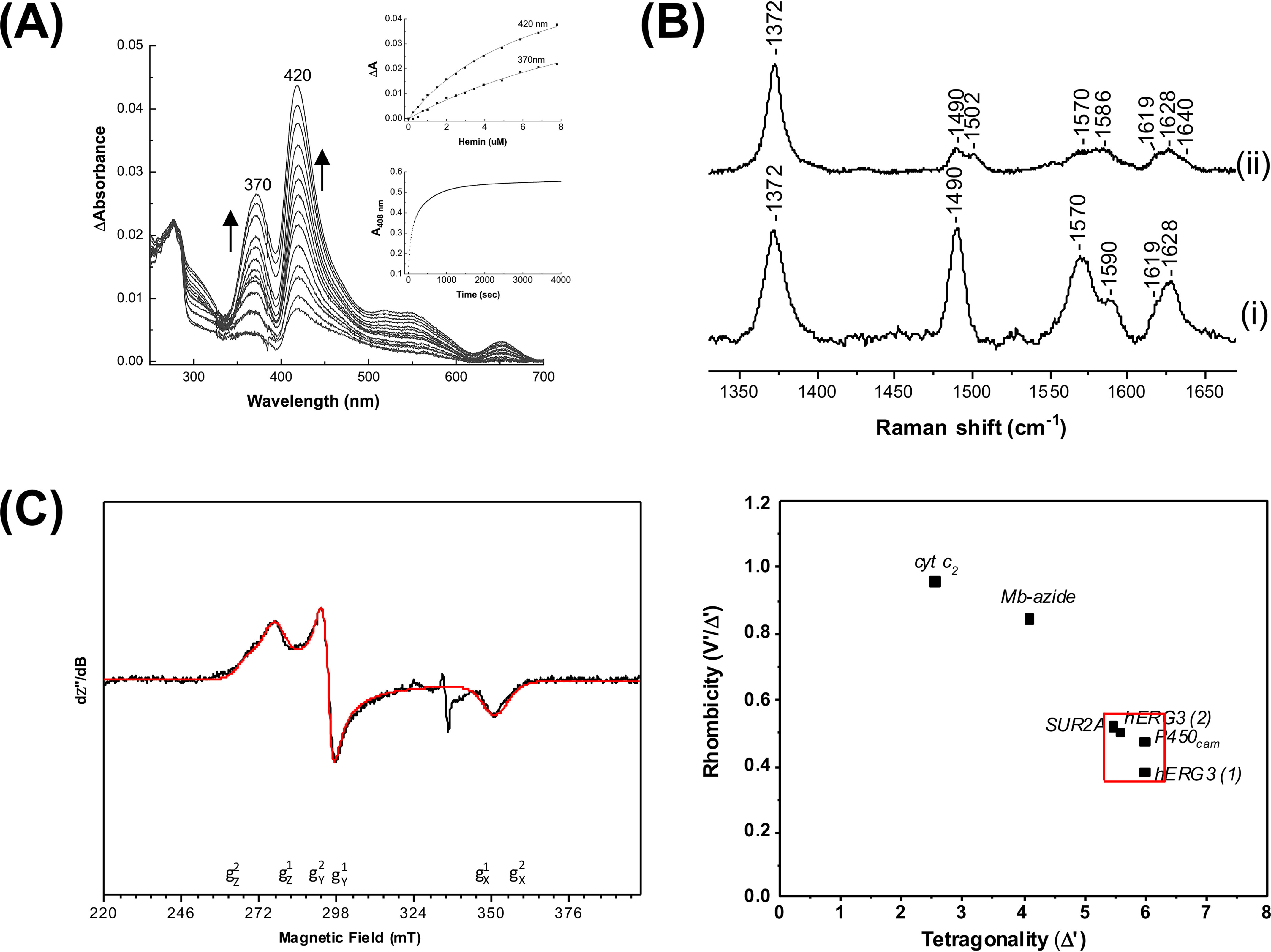
Analysis of heme binding to the hERG3-eag domain. A, difference spectra obtained on titration of the hERG3-eag domain with ferric heme; the arrows represent the directions of the absorbance changed with increasing heme concentration. Inset, top panel, hyperbolic fitting of the heme-binding data at both wavelength maxima to a 1:1 model for heme binding. Inset, bottom panel; absorbance changes at 408 nm on binding of the ferric hERG3-eag complex to apo-myoglobin. The data were fitted to the first order decay process, yielding kobs = 0.03 s−1. B, room temperature high-frequency resonance Raman spectra of free heme (panel (i)) and the ferric hERG3-eag–heme complex (panel (ii)). All spectra were collected with 413.1-nm laser excitation. C, left panel, X-band EPR spectrum (black trace) of the ferric hERG3-eag–heme complex in the low-spin region along with the simulated spectrum (red trace). The experimental conditions were as follows: microwave frequency, 9.38 GHz; microwave power, 0.064 mW; field modulation amplitude, 2 mT; field modulation frequency, 100 kHz; temperature, 15 K; [heme] = 100 μm, 5-fold excess of protein in 50 mm HEPES buffer, pH 7.5, 50 mm NaCl. The simulation parameters are as follows: species 1 (75%) g values (g strain) are gz1 = 2.42 (0.06), gy1 = 2.27 (0.00), and gx1 = 1.91 (0.04); species 2 (25%) g values (g strain) are gz2 = 2.50 (0,07), gy2 = 2.28 (0.00), and gx2 = 1.90 (0.06); Lorentzian linewidth full-width at half-maximum 4 mT. Right panel, Blumberg–Peisach correlation diagram showing EPR parameters plotted for various heme proteins.
Spectroscopic analysis of heme binding
Resonance Raman spectroscopy was used to provide more quantitative insight into the interaction of hERG3-eag with heme. In the high-frequency region, the spectrum of free heme displays the distinctive features of a ferric five-coordinate high-spin complex (5c-HS), with characteristic bands at 1372 (v4), 1490 (v3), and 1570 cm−1 (v2) (Fig. 2B, panel (i)). The v10 mode of the 5c-HS species is hidden by the vinyl stretching bands expected in the 1610–1640-cm−1 region (1618 cm−1 for the in-plane and 1628 cm−1 for the out-of-plane vinyl group (13)). The ferric hERG3-eag–heme complex shows significant differences from free heme (Fig. 2B, panel (ii)). All the modes described for 5c-HS are present for the hERG3-eag–heme complex. Additionally, we observe bands at 1502 (v3), 1553 and 1585 cm−1 (v2), and 1640 cm−1 (v10). These modes indicate the presence of a six-coordinate low-spin complex (Table S1) (14). These resonance Raman data are in agreement with the absorption spectra above and demonstrate the formation of a six-coordinated heme species with a specific binding environment.
EPR analyses support these conclusions, because the spectrum of the hERG3-eag–heme complex shows the formation of a low-spin heme complex (Fig. 2C). The spectrum is indicative of a mixture of species and was best simulated with two components (Fig. 2C). The g values for both are in good agreement with those of other ferric heme proteins bearing cysteine coordinated opposite a neutral sixth ligand (8). A Cys/His ligation is favored for both components as suggested by the placement in the Blumberg–Peisach diagram (Fig. 2C).
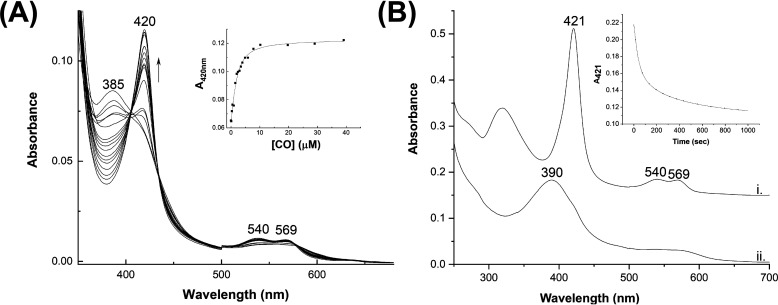
CO binds tightly (Kd = 1.03 ± 0.37 μm; Fig. 3A) to the ferrous hERG3-eag–heme complex and gives a species (Soret band at 420 nm; Q-bands at 540 and 569 nm) that is characteristic of a normal CO-bound heme species. An identical species (Soret band at 420 nm; Q-bands at 540 and 569 nm) is formed upon reaction of apo-hERG3-eag domain with a preformed heme–CO complex and with an affinity (Kd = 10.55 ± 1.34 μm, data not shown) that is within reasonable range of the Kd for binding of CO to the ferrous hERG3-eag complex (Kd = 1.03 ± 0.37 μm) as above. These spectra for the hERG3–heme–CO complex formed by two different routes are significantly different from that of a free heme–CO complex (λmax = 407, 537, and 567 nm (12)), which indicates formation of a specific protein–heme–CO complex. The CO ligand is easily dissociated (koff = 0.03 s−1) in the presence of nitric oxide (Soret band at 390 nm; Fig. 3B), which is characteristic of a five-coordinated NO-bound heme protein complex (15, 16). These data indicate that the hERG3-eag–heme complex is competent for binding of biologically relevant gaseous ligands.
Figure 3.
A, spectrophotometric titration of the ferrous hERG3-eag–heme complex with CO. Inset, quadratic (Morrison) fitting to the binding curve to give Kd = 1.03 ± 0.37 μm. B, formation of a heme–NO–hERG3 complex (λmax = 390 nm) on dissociation of CO from the ferrous heme–hERG3 complex (3 μm, λmax = 421 nm) in the presence of NO. NO was formed from the NO releasing molecule, S-nitroso-N-acetyl penicillamine (see “Experimental procedures”). Inset, the 421-nm time course, fitted to a three-exponential process; using the dominant (50%) phase a rate constant for dissociation of CO, koff(CO), was determined (koff(CO) = 0.03 s−1).
Structure of the hERG3-eag domain
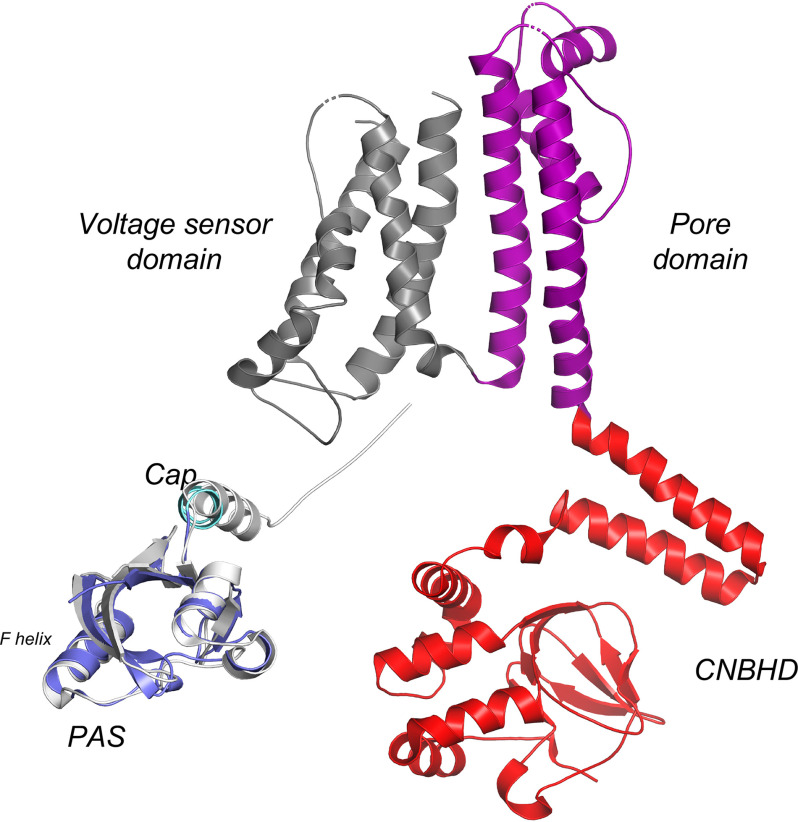
The purified hERG3-eag protein was crystallized in the apo-form, in a monomeric state. The structure of the PAS domain (residues 18–135) is shown in Fig. 4A The hERG3-eag domain has a canonical PAS fold that comprises of five antiparallel β-sheets flanked by three α-helices, forming a hydrophobic cleft. This cleft is formed between the inner face of the β−sheet and the conformationally mobile F-helix. It is conserved in PAS domains known to bind ligands, including heme (3). The hERG3-eag structure contains a Cys39–Cys64 disulfide bond (Fig. 4A) situated 12 residues from the F-helix and 13 residues from the Cap domain. The role of the disulfide bond is currently unknown and was not tested in this work, but it may impart a redox-sensing functionality to the PAS domain (as previously suggested (17–20)).
Figure 4.
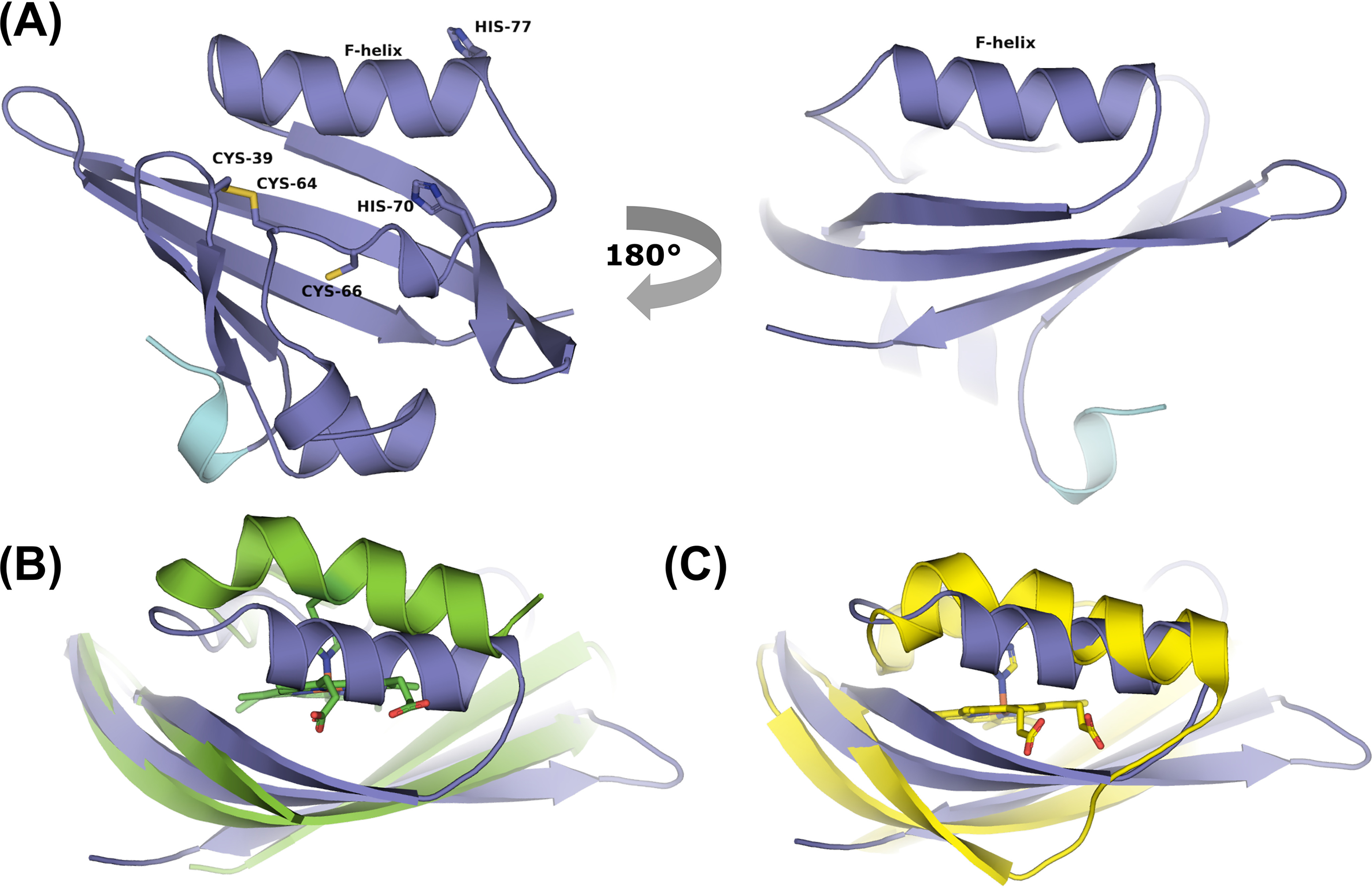
A, cartoon representation of the hERG3-eag crystal structure showing the PAS (purple) and Cap (light blue) domains; the color scheme used is the same as that used in Fig. 1 for the PAS and Cap domains. Potential heme-binding residues are labeled. B and C, structural alignment of hERG3-eag with structures of heme-bound PAS domains in E. coli DOS (Protein Data Bank code 1V9Y, green in B) and FixL (Protein Data Bank code 1DP6, yellow in C). Movement of the F-helix in E. coli DOS and FixL allows binding of the heme.
The first 17 residues at the N terminus are not seen in interpretable electron density in the crystal; furthermore, residues 18–23 are of weak density, indicating heterogenetity in this region (the N-terminal Cap). MS data (Fig. S2C) show there is a mixture of molecular weights with varying degrees of truncation at the N terminus (Fig. S2D), and the observed density most likely reflects this mixture. Because a significant proportion of the protein is unaccounted for in the model, this probably explains the relatively high R values for a structure at this resolution (see Table S2). Despite attempts to obtain a co-crystal structure of heme-bound hERG3-eag, extensive co-crystallization and soaking experiments were unsuccessful. As we have noted previously (6), this does not mean that heme cannot bind to the protein, because the spectroscopic data clearly demonstrate that it does. It is more likely that the particular conformation selected for in the crystal structure is incompatible with heme binding, so that heme cannot bind to the protein during the crystallization process. Structural flexibility of the protein that is linked to the heme-binding event (see “Discussion”) might also affect whether heme co-crystallizes with the protein.
Effect of heme on hERG3 channels
To measure the functional response of hERG3 channels to heme, hERG3 was heterologously expressed in Chinese hamster ovary cells. In whole-cell recordings, K+ currents were elicited with the characteristic features of the hERG family (Fig. 5A), including slow voltage-dependent activation and deactivation, rapid onset, and recovery from inactivation and inhibition by astemizole (data not shown). Application of heme (Fig. 5, B–E) caused a substantial inhibition of hERG3 current at all potentials (shown here after >3 min). Mean (± S.E.) heme inhibition of peak tail currents with a test potential to +40 mV was 63 ± 12% (p < 0.05, n = 3). Heme caused a small but significant negative shift in the voltage for half-maximal activation of 6.7 mV (p < 0.05, n = 3) (Fig. 5E). The lipophilic properties of heme and its ability to interact with membrane phospholipids allows it to be transported across the plasma membrane via passive diffusion (21). To test whether addition of intracellular heme also inhibited hERG3 channels, currents were recorded using the inside-out configuration of the patch clamp technique, and heme was applied directly to the cytoplasmic side of excised membrane patches. As reported with many other ionic currents, upon excision, a rapid decrease of hERG3 currents was observed in inside-out macropatches, consistent with loss of channel activity (rundown) upon removing the channels from the cellular environment. The addition of phosphatidylinositol 4,5-bisphosphate (10 μm) to the perfusate ameliorated rundown and resulted in stable recording conditions (Fig. 5, G and H). Subsequent application of heme (Fig. 5, F–H) to the intracellular side of the membrane caused inhibition of hERG3 currents in a similar manner as extracellularly applied heme in whole cell recordings. The time courses of heme inhibition were comparable, suggesting factors other than membrane diffusion of heme determine the rates of hERG3 current inhibition. These data demonstrate that heme inhibits hERG3 channel activity, thus connecting the heme-binding event to modulation of channel function.
Figure 5.
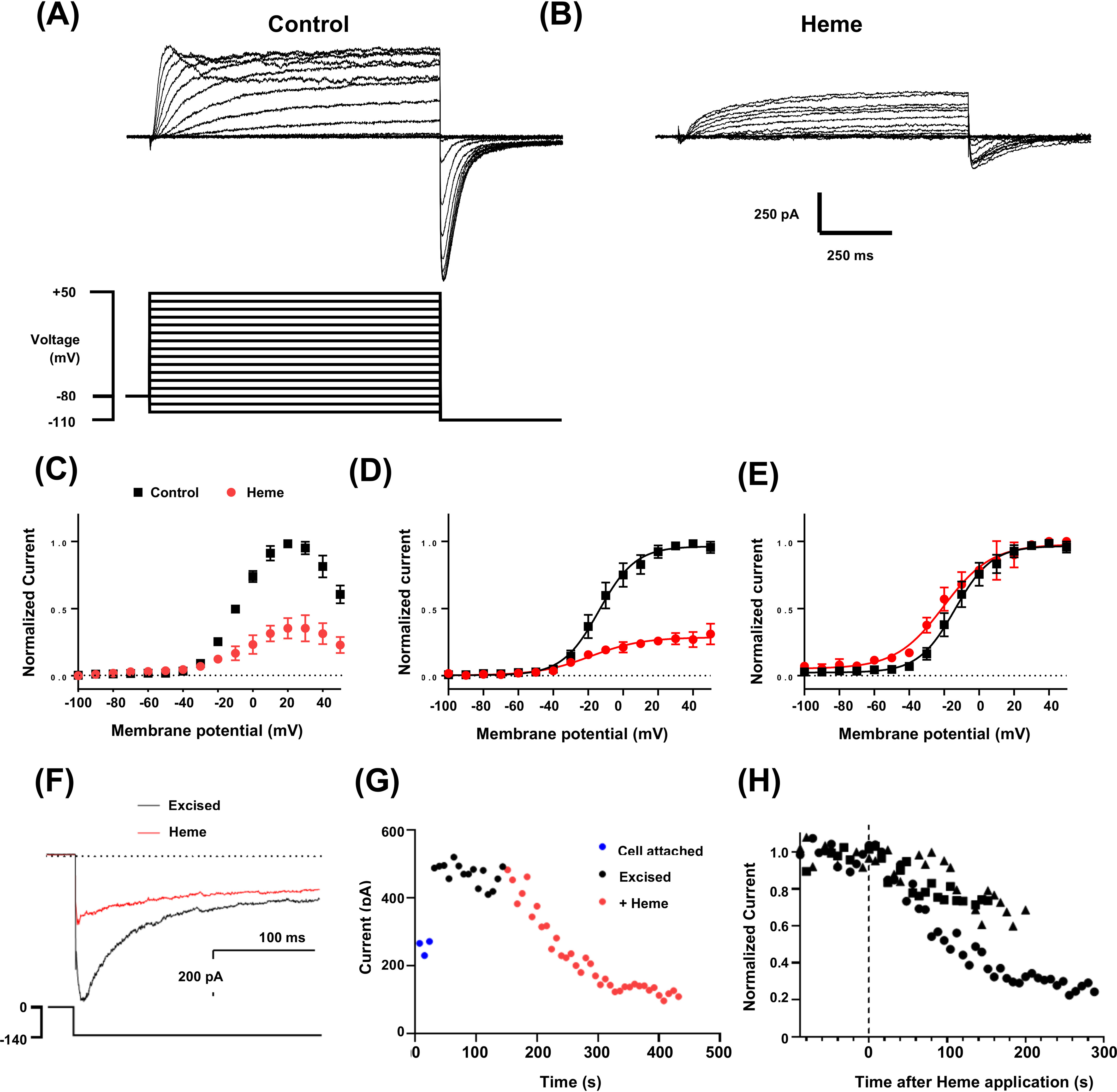
hERG3 currents are inhibited by heme. A, representative whole-cell hERG3 currents (upper panel) elicited with voltage protocol (lower panel) consisting of 1-s test pulses from −100 to +50 mV in 10-mV increments and from a holding potential of −80 mV. Inward tail currents were measured at a potential of −110 mV. The start-start interval for the voltage protocol was 5 s. B, a family of hERG3 current traces from the same cell shown in A after superfusion of heme (500 nm). C, mean end-pulse current-voltage relationship with and without heme (500 nm) (n = 3). D and E, mean tail current (normalized to maximum control current in individual cells (D) or normalized to maximum current (E) in each recording solution) and plotted against test-pulse potential, with and without heme (500 nm) (n = 3). The data are fitted with Boltzmann functions (solid lines). Half-maximal activation (V0.5) and slope factor were −12.3 ± 4.7 mV and 10.7 ± 1.0 mV, respectively, before heme and −19.0 ± 4.4 mV and 14.1 ± 2.6 mV with heme (n = 3). F, representative traces of excised inside-out macro-patch recordings of hERG3 tail currents before and during application of heme (1 μm). Patches were excised into solutions containing 10 μm phosphoinositol 4,5-bisphosphate to attenuate current rundown. Intracellular and extracellular solutions contained equimolar K+ concentrations. Tail currents were measured at a potential of −140 mV and following 2-s test pulses to 0 mV (see voltage protocol in lower panel). Horizontal dotted lines in D–F indicate zero current. G, representative plot of changes in amplitude of the deactivating component of the tail current plotted against time. The time between traces was 8 s. H, scatter plot of hERG3 tail current amplitudes in three separate excised patches plotted against time after heme application. The time of heme application is indicated by the vertical dashed line.
Discussion
The EAG (Kv10–12) family of voltage-gated K+ channels are regulators of neuronal and cardiac cell action potential firing (excitability). In humans, the family comprises hEAG, hERG, and hELK channels (22, 23). In this work, we have identified heme-dependent regulation of hERG3 channel activity. Formation of a six-coordinated heme species is supported from resonance Raman data, as well as EPR and UV-visible spectra, and a heme-binding PAS domain within the eag domain of the channel has been identified.
Recent cryo-EM structural models of rat EAG1 and hERG1 confirm that the four eag domains are arranged around the periphery of the tetrameric intracellular assembly formed by the CNBHD and C-linkers (24, 25). The function of this large intracellular complex is to modulate voltage-dependent gating; profoundly influencing the time course and amplitudes of potassium currents during physiological processes such as action potentials. It sits underneath the transmembrane domains, with the C-linker connecting directly with the S6 activation gate. The PAS domain makes contacts predominantly with the CNBHD, whereas the Cap domain extends toward the interface between the voltage sensor, C-linker, and channel pore and thus is perfectly positioned to influence voltage- and time-dependent gating properties. Interestingly, functional studies reveal that deletion of the PAS and Cap domains has differential effects on the gating properties of hERG1 and hEAG1, suggesting that specific contacts with gating machinery of the two channels are different (26, 27). In hEAG1 channels, the eag domain is also required for calcium-calmodulin–dependent inhibition of current (26). hERG3 gating is similar to hERG1, and it is likely that the eag domain has a similar role in both channels, which is to slow deactivation gating and enhance inactivation gating.
PAS domains, which are known to bind heme (4, 5), have well-established roles in regulating the function of the related hERG1 channel (7, 28, 29), but heme binding to this channel has not been established. Heme has been shown to potently modulate the activity of a range of other potassium ion channels such as Kv1.4, large conductance Ca2+-activated K channels, and ATP-activated K channels (12, 30–36). Our observation that heme binding reduces hERG3 activity furthers the notion that heme plays a deliberate modulatory role in these ion channel systems. As we noted in the introduction, the capacity for the heme group to also bind cell-signaling gases (CO, NO, O2, or even H2S) vastly broadens the range of stimuli that these ion channels could potentially be modulated by.
It is worth noting that heme is important for neuronal survival (37), and so a role for heme in the regulation of the “neuronal” channels hERG2 and hERG3 might reasonably be anticipated. At present, the biological basis for an interaction of heme with hERG3 is unknown, but a significant portion of brain damage caused during hemorrhagic stroke is believed to be due to the longer-lasting, cytotoxic effects of heme (38). More generally, an abundance of heme after traumatic events such as stroke and ischemia could conceivably disrupt the regulatory balance of ion channels in the brain and has cytotoxic implications for the cells involved (39). Heme-dependent inhibition of hERG3 would reduce repolarizing K+ currents, potentially increasing neuronal excitability and susceptibility to cell death. An abundance of heme after traumatic events thus has the capacity to disrupt the dynamic processes surrounding the regulation of ion channel function if they are sensitive to heme.
We did not identify the heme-binding location in hERG3-eag. However, we have compared the structure of apo-hERG3-eag with those of other heme-binding PAS domain proteins (EcDOS (40) and FixL (41)). Overlay of hERG3 with those of EcDOS and FixL shows that, compared with the apo-hERG3 structure, there is a large movement of the F-helix in the heme-bound EcDOS and FixL structures to accommodate binding of the heme group (Fig. 4, B and C). It is feasible, therefore, that a similar mode of heme binding might occur in hERG3 if its F-helix is similarly flexible. Relevant in this context is the disulfide bond between Cys39 and Cys64, which is located 12 residues from the F-helix via a flexible loop. It has been proposed that disulfide bridges might serve as redox sensors in cells, by switching between thiol and disulfide structures (17–20). Loss of the Cys39–Cys64 disulfide could facilitate movement of the F-helix, allowing heme to bind to the protein. The ligands to the bound heme are not identified as yet but candidate residues are His70, His77, and Cys66 (Fig. 4A), which are in the vicinity of the proposed heme-binding site based on the comparison with EcDOS and FixL. Cys/His axial ligations would be consistent with our spectroscopic data and with known ligations in other regulatory heme proteins (4, 5).
Based on our structure, we identify one further possible mechanism of heme-dependent regulation. The N-terminal Cap domain (residues 1–26) in hERG3-eag is mostly unresolved in our structure. In hERG1 the Cap domain, which contains a flexibly linked but stable amphipathic helix, is already known to affect channel activity (27, 42, 43). So in addition to affecting the conformation of the F-helix (as above), it is possible that heme binding to the PAS domain in hERG3 could also affect the orientation of the adjacent and potentially highly mobile Cap domain (Fig. 1). This N terminus is oriented toward the pore in the structure of the channel (25), and movements of the Cap domain might affect the conformations of the nearby cyclic nucleotide-binding homology domain, C-linker, voltage-sensing domain, or pore-forming domains as shown Fig. 6. Structural adjustments of this kind, in response to heme binding, would provide a mechanism for closure of the channel, because of the proximity of the N-terminal Cap domain to vital regulatory domains of channel gating (25). Relevant in this context is the observation that free heme affects channel function in the A-type potassium channels and, crucially, that the heme binds to the inactivation domain on the N-terminal region (30). We have no evidence for heme binding directly to the Cap domain in hERG3-eag, but changes in conformation of the N-terminal regions—induced either directly (by heme binding to N termini) or indirectly (by heme binding to adjacent domains, such as PAS)—might plausibly provide a mechanism for ion channel regulation.
Figure 6.
Cryo-EM structure of the hERG1 (Kv11.1) channel (Protein Data Bank 5VA1), showing the CNBHD (red), the pore domain (magenta), and the voltage-sensing domain (dark gray). For direct comparison, the color schemes used for the CNBHD, pore, and voltage-sensing domains are the same as those shown schematically in Fig. 1. In addition, the PAS and Cap (in light gray) domains are also shown for hERG1, aligned with hERG3-eag crystal structure (PAS domain, in purple; Cap domain in light blue, color scheme as Fig. 1). Movements of the F-helix (labeled) or the Cap domain, induced by heme binding, might conceivably affect the conformations of the nearby CNBDH, pore, and voltage-sensing domains, which could provide a mechanism for channel regulation. Refer also to Fig. 1, which shows a schematic of the locations of each domain in relation to the overall channel structure. We have drawn cartoon schematics of heme-dependent regulation of several different ion channels previously (KATP, Slo1 (BK) and Kv1.4 channels (36)), and we envisage a similar mechanism of control here.
Experimental procedures
Chemicals and reagents
All reagents were from Sigma–Aldrich unless otherwise stated. CO solutions were prepared by bubbling bath solution with CO gas. Iron protoporphyrin IX (hemin; Fig. S1A) was purchased from Sigma–Aldrich. Aqueous stock solutions of heme for all spectroscopic and electrophysiological experiments were prepared by dissolving solid heme in 0.1 m NaOH; final concentrations were calculated spectrophotometrically (ε385 = 58.4 mm−1 cm−1 (44)) and diluted accordingly to 500 nm prior to use.
Heme–agarose affinity purification and MS of neuronal lysates
Neuronal lysates were screened for possible heme-binding proteins using a modification of a previously published heme–agarose affinity (45). Binding of target proteins to heme–agarose was performed as previously (46), with modifications (Fig. S1B) that minimize pulling down proteins that bind to the heme–agarose resin nonspecifically. Full details are given in the supporting information. Using this approach, only proteins that were displaced from the agarose beads to bind specifically to the (heme) competitor (i.e. more likely to be bona fide heme-binding proteins) were identified and subjected to proteomics analysis (Fig. S1C). The data are available via ProteomeXchange with identifier PXD019887.
Expression and purification of hERG3-eag domain
The human KCNH7 clone (imaGenes Gmbh, Berlin, Germany) was used to obtain a sequence encoding residues 1–135 of the N terminus of the human hERG3 channel, which comprises the Cap (residues 1–26) and PAS (residues 27–135) domains (Fig. 1 and Fig. S2A). We refer to the fragment encoding residues 1–135 as hERG3-eag. The hERG3-eag domain was expressed as an N-terminal His6/S-tagged fusion protein in E. coli BL21(DE3) cells using a pLEICS-93 (PROTEX facility, University of Leicester) expression vector. Cell cultures in LB media were grown at 37 °C containing ampicillin (0.1%) to an A600 of 0.6–0.7, at which point the temperature was reduced to 18 °C and isopropyl β-d-thiogalactoside (0.2 mm) was added. After overnight incubation, the cells were harvested, suspended in lysis buffer (50 mm phosphate, 100 mm NaCl, pH 7.00, 10% glycerol, 0.1% Triton, 0.1 m MgSO4, 1 complete ULTRA Tablet Mini EASY pack (Roche), 1 mg/ml lysozyme, and 1 mg/ml DNase), lysed by sonication, and centrifuged to remove cell debris. The cell lysates were loaded onto a 5-ml Ni2+–nitrilotriacetic acid (Qiagen) affinity column equilibrated with 50 mm phosphate, 100 mm NaCl, pH 7.00, 10% (v/v) glycerol, and 15 mm imidazole. After washing with equilibration buffer, the His6/S-tagged proteins were eluted with 50 mm phosphate, 100 mm NaCl, pH 7.0, 10% glycerol, and 100 mm EDTA. The His6/S tag was cleaved with a His-tagged TEV-protease during overnight dialysis. The His6/S tag and His-tagged TEV protease were removed by reverse Ni2+–nitrilotriacetic acid affinity column, and the proteins were purified to homogeneity with size-exclusion chromatography (Superdex 200, GE Healthcare) using 50 mm phosphate, 100 mm NaCl, pH 7.0 (Fig. S2B). Protein concentrations were determined using a Bradford assay.
Optical absorption spectroscopy
Absorption spectra (25.0 °C) were obtained using a double beam spectrophotometer (PerkinElmer Lambda 40) or a Kontron Uvikon UV-visible spectrometer. Ferric heme-bound hERG3-eag was obtained by the addition of heme to hERG3-eag in 50 mm HEPES, 50 mm NaCl, pH 7.5. The reduced protein was obtained by anaeorobic addition of dithionite, and the ferrous-CO complex by gentle bubbling of CO through the reduced protein. Values of Kd for binding of ligands (heme, CO) to hERG3 (3–5 mm) were obtained at 25.0 °C in 50 mm HEPES, 50 mm NaCl, pH 7.5, as described previously (12). Heme dissociation was measured by reacting the ferric heme–hERG3-eag complex (6.4 μm) with a 5.6-fold excess of apo-myoglobin (36 μm); the reaction was followed at 408 nm (50 mm HEPES, 50 mm NaCl, pH 7.5).
Resonance Raman spectroscopy
Samples (50 μl, in 50 mm HEPES, 50 mm NaCl, pH 7.5) of ferric heme and its complexes were prepared in quartz EPR tubes and placed in a homemade spinning cell, at room temperature, to avoid local heating and to prevent photodissociation and degradation. Protein concentrations were in the range 90–120 μm, and substoichiometric concentrations of heme (to 0.8 equivalents) were added to the protein to ensure that no excess of heme was present. Raman excitation at 413.1 nm was achieved with a laser power <10 mW from an Innova 90 Kr+ laser (Coherent, Palo Alto, CA, USA). Resonance Raman spectra were recorded using a two-stage monochromator (U1000, Jobin Yvon, Longjumeau, France) equipped with a front illuminated, deep-depleted CCD detector (Jobin Yvon, Longjumeau, France). Spectra correspond to an average of three different 1-h accumulations. The spectral accuracy was estimated to be ±1 cm−1. Baseline correction was performed using GRAMS 32 (Galactic Industries, Salem, NH) and Origin®. Sample integrity was verified by following resonance Raman spectral evolution during the experiment.
EPR spectroscopy
EPR spectra were recorded on an Elexsys 500 X-band spectrometer (Bruker) equipped with a continuous-flow ESR 900 cryostat and an ITC504 temperature controller (Oxford Instruments, Abingdon, UK). Simulations were performed by using the Easyspin software package (47) and routines written in the Dorlet laboratory. Samples (100 μl, 100 μm total heme concentration, 4–5-fold excess protein) of the ferric heme complexes of hERG3-eag were prepared in 50 mm HEPES and 50 mm NaCl, pH 7.5, and transferred to quartz EPR tubes.
Electrophysiology
hERG3 currents were expressed in Chinese hamster ovary cells and currents were recorded using either whole-cell or macropatch (inside-out or cell attached) configurations of the patch clamp technique. hERG3 currents were recorded with an Axopatch 200 voltage clamp amplifier, digitized using a Digidata 1440A interface, and acquired and analyzed using pClamp 10.3 software (Molecular Devices, Sunnyvale, CA, USA). For whole cell recordings the intracellular solution contained 30 mm KOH, 110 mm KCl, 1.2 mm MgCl2, 1 mm CaCl2, 10 mm EGTA, 5 mm HEPES, 1 mm ATP, titrated to pH 7.2 with KOH. The cells were superfused with normal bath solution containing 135 mm NaCl, 10 mm KCl, 5 mm glucose, 2 mm MgCl2, 2 mm CaCl2 and 10 mm HEPES, titrated to pH 7.3 with NaOH. Macropatch currents were recorded in equimolar K+ solutions. The bath solution contained 40mm KOH, 80 mm KCl, 10 mm K2SO4, 10 mm HEPES, 5 mm EGTA, 5 mm HEDTA, and 10 glucose, pH 7.2. Pipette solution contained 140 mm KCl, 1 mm MgCl2, 0.1 mm CaCl2, and 10 mm HEPES, pH 7.4. To prevent rapid and complete current rundown, 10 μm phosphatidylinositol 4,5-bisphosphate was added to the bath solution in all excised inside-out macropatch recordings.
Aqueous stock solutions of ferric heme for electrophysiology experiments were prepared by dissolving heme in 0.1 m aqueous NaOH and mixing for 10 min, followed by centrifugation at 4,000 × g for 10 min and dilution as required. Final solutions contained 1 mm reduced GSH. Aliquots of heme were made fresh daily. All experiments were conducted at room temperature (22 ± 1 °C). The data are presented as means ± S.E. of n experiments.
Crystallization and data collection
A crystal of hERG3-eag (amino acids 1–135) was grown using sitting-drop vapor diffusion at 295 K. The crystallization drop was set up by mixing hERG3-eag (15 mg/ml, in 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine) in a 1:1 ratio with reservoir solution (0.1 m NaCl, 0.1 m HEPES, pH 7.5, 1.6 m (NH4)2SO4). A transparent crystal appeared after 4 weeks. The crystal was prepared for X-ray diffraction by brief immersion in the reservoir solution containing 25% glycerol for cryo-protection followed by flash vitrification in liquid nitrogen. X-ray diffraction data were collected at Diamond Light Source on Beamline I03. 3600 images of 0.05 degree were collected with an exposure time of 0.039 s/image using 0.976 Å radiation. The crystal belongs to the space group P43212 with unit cell dimensions a = b = 32.78 Å, c = 200.68 Å, α = β = γ = 90° (Table S2). The data were processed and scaled with DIALS software from the CCP4 program suite (48). The structure was determined by molecular replacement in PHASER (49) using the hERG1 PAS domain (Protein Data Bank code 4HQA (50)) as the search model and refined with REFMAC5 (51). The model was refined to a resolution of 1.39 Å with Rwork and Rfree values of 18.2 and 23.4%, respectively. The final cycles of refinement included extrapolated hydrogen atoms with anisotropic atomic displacement parameters (B) for all atoms. Data collection and refinement statistics are in Table S2 (Protein Data Bank code 6Y7Q), and the weighting scheme is given in Table S3.
MS analysis was carried out on a hERG3-eag crystal. The crystal was dissolved in 50 mm HEPES, 50 mm NaCl, pH 7.5, and LC–MS was carried out using an RSLCnano HPLC system (Thermo Scientific) and an LTQ-Orbitrap-Velos mass spectrometer (Thermo Scientific). Samples were loaded at 0.1 ml/min onto a Vydac C8 5 μm 250-mm × 1-mm inner-diameter reverse-phase column (Grace Davison). The protein was desalted for 10 min in the loading buffer (0.1% formic acid) before elution using a 10-min linear gradient from 3 to 96% B (80% acetonitrile, 0.1% formic acid). The output of the column was sprayed directly into the H-ESI2 electrospray ion source of the mass spectrometer maintained at 5 kV. The FT analyzer was set to acquire 10 microscans over the m/z range 800–2000 Da in positive ion mode. The maximum injection time for MS was 150 ms, and the AGC target setting was 3e6. Protein charge-state distributions were deconvoluted using the Xtract function of the Xcalibur program (version 2.1.0.1139, Thermo Scientific).
Data availability
The atomic coordinates and diffraction data have been deposited in the Protein Data Bank (accession code 6Y7Q). The proteomics data are available via ProteomeXchange with identifier PXD019887. All other data are included in the article or are available on request from the corresponding author.
Supplementary Material
Acknowledgments
We thank the late Dr. Xiaowen Yang for preparing the expression clones and Dr. Fred Muskett for helpful discussions on the purification of hERG3-eag. We acknowledge Dr. Chris Millard, Chitra Seewooruthun, Kirsty Ford and Prof Marten Vos for help with the acquisition of data, and the Diamond Light Source staff at Beamline I03 (BAG MX14692-18).
This article contains supporting information.
Author contributions—M. J. B., J. C.-B., M. T., N. P., J. B., S. L. F., H. K., A. R. B., M. J. L.-P., A. A. P., T. C., R. S., P. D., and E. L. R. investigation; M. J. B. and E. L. R. writing-original draft; M. J. B. and E. L. R. project administration; M. J. B., P. C. E. M., J. S. M., and E. L. R. writing-review and editing; J. C.-B., J. B., A. R. B., M. J. L.-P., and R. S. methodology; J. B., R. S., N. W. D., N. M. S., P. C. E. M., and J. S. M. formal analysis; J. B., A. A. P., N. W. D., N. M. S., P. D., P. C. E. M., E. L. R., and J. S. M. supervision; J. B. and R. J.-J. validation; J. B. and H. K. visualization; E. L. R. and J. S. M. conceptualization; E. L. R., J. S. M., N. W. D, N. S., R. S., and P. C. E. M. funding acquisition.
Funding and additional information—This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/K000128/1 and BB/M018598/1 (to E. L. R.). This work benefited from the Biophysics Platform of Institute for Integrative Biology of the Cell, supported by iBiSA and by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PAS
- Per–ARNT–Sim
- hERG3-eag
- residues 1–135 of the N terminus of the human hERG3 channel, comprising the Cap and PAS domains
- 5c-HS
- ferric five-coordinate high-spin complex
- CNBHD
- cyclic nucleotide-binding homology domain.
References
- 1. Taylor B. L., and Zhulin I. B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Rev. 63, 479–506 10.1128/MMBR.63.2.479-506.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henry J. T., and Crosson S. (2011) Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65, 261–286 10.1146/annurev-micro-121809-151631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Möglich A., Ayers R. A., and Moffat K. (2009) Structure and signaling mechanism of Per–ARNT–Sim domains. Structure 17, 1282–1294 10.1016/j.str.2009.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shimizu T., Lengalova A., Martínek V., and Martínková M. (2019) Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 48, 5624–5657 10.1039/c9cs00268e [DOI] [PubMed] [Google Scholar]
- 5. Shimizu T., Huang D., Yan F., Stranava M., Bartosova M., Fojtíková V., and Martínková M. (2015) Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 115, 6491–6533 10.1021/acs.chemrev.5b00018 [DOI] [PubMed] [Google Scholar]
- 6. Freeman S. L., Kwon H., Portolano N., Parkin G., Venkatraman Girija U., Basran J., Fielding A. J., Fairall L., Svistunenko D. A., Moody P. C. E., Schwabe J. W. R., Kyriacou C. P., and Raven E. L. (2019) Heme binding to human CLOCK affects interactions with the E-box. Proc. Natl. Acad. Sci. U.S.A. 116, 19911–19916 10.1073/pnas.1905216116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morais Cabral J. H., Lee A., Cohen S. L., Chait B. T., Li M., and Mackinnon R. (1998) Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95, 649–655 10.1016/S0092-8674(00)81635-9 [DOI] [PubMed] [Google Scholar]
- 8. Smith A. T., Pazicni S., Marvin K. A., Stevens D. J., Paulsen K. M., and Burstyn J. N. (2015) Functional divergence of heme-thiolate proteins: a classification based on spectroscopic attributes. Chem. Rev. 115, 2532–2558 10.1021/cr500056m [DOI] [PubMed] [Google Scholar]
- 9. Reynolds M. F., Shelver D., Kerby R. L., Parks R. B., Roberts G. P., and Burstyn J. N. (1998) EPR and electronic absorption spectroscopies of the CO-sensing CooA protein reveal a cysteine-ligated low-spin ferric heme. J. Am. Chem. Soc. 35, 9080–9081 10.1021/ja981146p [DOI] [Google Scholar]
- 10. Pazicni S., Lukat-Rodgers G. S., Oliveriusová J., Rees K. A., Parks R. B., Clark R. W., Rodgers K. R., Kraus J. P., and Burstyn J. N. (2004) The redox behavior of the heme in cystathionine β-synthase is sensitive to pH. Biochemistry 43, 14684–14695 10.1021/bi0488496 [DOI] [PubMed] [Google Scholar]
- 11. Hargrove M. S., Singleton E. W., Quillin M. L., Ortiz L. A., Phillips G. N. Jr., Olson J. S., and Mathews A. J. (1982) His64(E7)-Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J. Biol. Chem. 269, 4207–4214 [DOI] [PubMed] [Google Scholar]
- 12. Kapetanaki S. M., Burton M. J., Basran J., Uragami C., Moody P. C. E., Mitcheson J. S., Schmid R., Davies N. W., Dorlet P., Vos M. H., Storey N. M., and Raven E. (2018) A mechanism for CO regulation of ion channels. Nat. Commun. 9, 907 10.1038/s41467-018-03291-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalsbeck W. A., Robertson D. E., Pandey R. K., Smith K. M., Dutton P. L., and Bocian D. F. (1996) Structural and electronic properties of the heme cofactors in a multi-heme synthetic cytochrome. Biochemistry 35, 3429–3438 10.1021/bi952662k [DOI] [PubMed] [Google Scholar]
- 14. Kitanishi K., Igarashi J., Hayasaka K., Hikage N., Saiful I., Yamauchi S., Uchida T., Ishimori K., and Shimizu T. (2008) Heme-binding characteristics of the isolated PAS: a domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochemistry 47, 6157–6168 10.1021/bi7023892 [DOI] [PubMed] [Google Scholar]
- 15. Stone J. R., Sands R. H., Dunham W. R., and Marletta M. A. (1995) Electron paramagnetic resonance spectral evidence for the formation of a pentacoordinate nitrosyl–heme complex on soluble guanylate cyclase. Biochem. Biophys. Res. Commun. 207, 572–577 10.1006/bbrc.1995.1226 [DOI] [PubMed] [Google Scholar]
- 16. Stone J. R., and Marletta M. A. (1994) Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry 33, 5636–5640 10.1021/bi00184a036 [DOI] [PubMed] [Google Scholar]
- 17. Gupta N., and Ragsdale S. W. (2011) Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erbβ. J. Biol. Chem. 286, 4392–4403 10.1074/jbc.M110.193466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yi L., Jenkins P. M., Leichert L. I., Jakob U., Martens J. R., and Ragsdale S. W. (2009) Heme regulatory motifs in heme oxygenase-2 form a thiol/disulfide redox switch that responds to the cellular redox state. J. Biol. Chem. 284, 20556–20561 10.1074/jbc.M109.015651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yi L., Morgan J. T., and Ragsdale S. W. (2010) Identification of a thiol/disulfide redox switch in the human BK channel that controls its affinity for heme and CO. J. Biol. Chem. 285, 20117–20127 10.1074/jbc.M110.116483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yi L., and Ragsdale S. W. (2007) Evidence that the heme regulatory motifs in heme oxygenase-2 serve as a thiol/disulfide redox switch regulating heme binding. J. Biol. Chem. 282, 21056–21067 10.1074/jbc.M700664200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Light W. R. 3rd., and Olson J. S. (1990) Transmembrane movement of heme. J. Biol. Chem. 265, 15623–15631 [PubMed] [Google Scholar]
- 22. Asher V., Sowter H., Shaw R., Bali A., and Khan R. (2010) Eag and HERG potassium channels as novel therapeutic targets in cancer. World J. Surg. Oncol. 8, 113 10.1186/1477-7819-8-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganetzky B., Robertson G. A., Wilson G. F., Trudeau M. C., and Titus S. A. (1999) The eag family of K+ channels in Drosophila and mammals. Ann. N.Y. Acad. Sci. 868, 356–369 10.1111/j.1749-6632.1999.tb11297.x [DOI] [PubMed] [Google Scholar]
- 24. Whicher J. R., and MacKinnon R. (2016) Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–669 10.1126/science.aaf8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W., and MacKinnon R. (2017) Cryo-EM structure of the open human ether-a-go-go–related K+ channel hERG. Cell 169, 422–430e410 10.1016/j.cell.2017.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lörinczi E., Helliwell M., Finch A., Stansfeld P. J., Davies N. W., Mahaut-Smith M., Muskett F. W., and Mitcheson J. S. (2016) Calmodulin regulates human ether a Go-Go 1 (hEAG1) potassium channels through interactions of the Eag domain with the cyclic nucleotide binding homology domain. J. Biol. Chem. 291, 17907–17918 10.1074/jbc.M116.733576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muskett F. W., Thouta S., Thomson S. J., Bowen A., Stansfeld P. J., and Mitcheson J. S. (2011) Mechanistic insight into human ether-a-go-go–related gene (hERG) K+ channel deactivation gating from the solution structure of the EAG domain. J. Biol. Chem. 286, 6184–6191 10.1074/jbc.M110.199364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Zou A., Splawski I., Keating M. T., and Sanguinetti M. C. (1999) Long QT syndrome-associated mutations in the Per–ARNT–Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J. Biol. Chem. 274, 10113–10118 10.1074/jbc.274.15.10113 [DOI] [PubMed] [Google Scholar]
- 29. Gianulis E. C., Liu Q., and Trudeau M. C. (2013) Direct interaction of eag domains and cyclic nucleotide-binding homology domains regulate deactivation gating in hERG channels. J. Gen. Physiol. 142, 351–366 10.1085/jgp.201310995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sahoo N., Goradia N., Ohlenschlager O., Schonherr R., Friedrich M., Plass W., Kappl R., Hoshi T., and Heinemann S. H. (2013) Heme impairs the ball-and-chain inactivation of potassium channels. Proc. Natl. Acad. Sci. U.S.A. 110, E4036–E4044 10.1073/pnas.1313247110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang X. D., Xu R., Reynolds M. F., Garcia M. L., Heinemann S. H., and Hoshi T. (2003) Haem can bind to and inhibit mammalian calcium-dependent Slo1 BK channels. Nature 425, 531–535 10.1038/nature02003 [DOI] [PubMed] [Google Scholar]
- 32. Jaggar J. H., Li A., Parfenova H., Liu J., Umstot E. S., Dopico A. M., and Leffler C. W. (2005) Heme is a carbon monoxide receptor for large-conductance Ca2+-activated K+ channels. Circ. Res. 97, 805–812 10.1161/01.RES.0000186180.47148.7b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burton M. J., Kapetanaki S. M., Chernova T., Jamieson A. G., Dorlet P., Santolini J., Moody P. C. E., Mitcheson J. S., Davies N. W., Schmid R., Raven E. L., and Storey N. M. (2016) A heme-binding domain controls regulation of ATP-dependent potassium channels. Proc. Natl. Acad. Sci. U.S.A. 113, 3785–3790 10.1073/pnas.1600211113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Augustynek B., Kudin A. P., Bednarczyk P., Szewczyk A., and Kunz W. S. (2014) Hemin inhibits the large conductance potassium channel in brain mitochondria: a putative novel mechanism of neurodegeneration. Exp. Neurol. 257, 70–75 10.1016/j.expneurol.2014.04.022 [DOI] [PubMed] [Google Scholar]
- 35. Wang S., Publicover S., and Gu Y. (2009) An oxygen-sensitive mechanism in regulation of epithelial sodium channel. Proc. Natl. Acad. Sci. U.S.A. 106, 2957–2962 10.1073/pnas.0809100106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu J. Y., Qu H. Y., Shang Z. L., Tao S. T., Xu G. H., Wu J., Wu H. Q., and Zhang S. L. (2011) Reciprocal regulation of Ca2+-activated outward K+ channels of Pyrus pyrifolia pollen by heme and carbon monoxide. New Phytol. 189, 1060–1068 10.1111/j.1469-8137.2010.03564.x [DOI] [PubMed] [Google Scholar]
- 37. Smith A. G., Raven E. L., and Chernova T. (2011) The regulatory role of heme in neurons. Metallomics 3, 955–962 10.1039/c1mt00085c [DOI] [PubMed] [Google Scholar]
- 38. Li Z. (2011) Heme Biology: The Secret Life of Heme in Regulating Diverse Biological Processes, World Scientific, Singapore [Google Scholar]
- 39. Robinson S. R., Dang T. N., Dringen R., and Bishop G. M. (2009) Hemin toxicity: a preventable source of brain damage following hemorrhagic stroke. Redox Rep. 14, 228–235 10.1179/135100009X12525712409931 [DOI] [PubMed] [Google Scholar]
- 40. Kurokawa H., Lee D. S., Watanabe M., Sagami I., Mikami B., Raman C. S., and Shimizu T. (2004) A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 279, 20186–20193 10.1074/jbc.M314199200 [DOI] [PubMed] [Google Scholar]
- 41. Gong W., Hao B., and Chan M. K. (2000) New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 39, 3955–3962 10.1021/bi992346w [DOI] [PubMed] [Google Scholar]
- 42. Ng C. A., Hunter M. J., Perry M. D., Mobli M., Ke Y., Kuchel P. W., King G. F., Stock D., and Vandenberg J. I. (2011) The N-terminal tail of hERG contains an amphipathic α-helix that regulates channel deactivation. PLoS One 6, e16191 10.1371/journal.pone.0016191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J., Myers C. D., and Robertson G. A. (2000) Dynamic control of deactivation gating by a soluble amino-terminal domain in HERG K+ channels. J. Gen. Physiol. 115, 749–758 10.1085/jgp.115.6.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dawson R. M. C., Elliot D. C., Elliot W. H., and Jones K. M. (1975) Data for Biochemical Research, Oxford University Press, Oxford, UK [Google Scholar]
- 45. Otto B. R., van Dooren S. J., Nuijens J. H., Luirink J., and Oudega B. (1998) Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188, 1091–1103 10.1084/jem.188.6.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee B. G. (1992) Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type B. Infect. Immun. 60, 810–816 10.1128/IAI.60.3.810-816.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stoll S., and Schweiger A. (2006) EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 10.1016/j.jmr.2005.08.013 [DOI] [PubMed] [Google Scholar]
- 48. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 49. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adaixo R., Harley C. A., Castro-Rodrigues A. F., and Morais-Cabral J. H. (2013) Structural properties of PAS domains from the KCNH potassium channels. PLoS One 8, e59265 10.1371/journal.pone.0059265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and diffraction data have been deposited in the Protein Data Bank (accession code 6Y7Q). The proteomics data are available via ProteomeXchange with identifier PXD019887. All other data are included in the article or are available on request from the corresponding author.