Enterococcus faecalis is a Gram-positive commensal bacterium native to the gastrointestinal tract and an opportunistic pathogen of increasing clinical concern. E. faecalis also colonizes the female reproductive tract, and reports suggest vaginal colonization increases following antibiotic treatment or in patients with aerobic vaginitis. Currently, little is known about specific factors that promote E. faecalis vaginal colonization and subsequent infection.
KEYWORDS: E. faecalis, Enterococcus, T7SS, TnSeq, colonization, ethanolamine catabolism, vaginal tract
ABSTRACT
Enterococcus faecalis is a Gram-positive commensal bacterium native to the gastrointestinal tract and an opportunistic pathogen of increasing clinical concern. E. faecalis also colonizes the female reproductive tract, and reports suggest vaginal colonization increases following antibiotic treatment or in patients with aerobic vaginitis. Currently, little is known about specific factors that promote E. faecalis vaginal colonization and subsequent infection. We modified an established mouse vaginal colonization model to explore E. faecalis vaginal carriage and demonstrate that both vancomycin-resistant and -sensitive strains colonize the murine vaginal tract. Following vaginal colonization, we observed E. faecalis in vaginal, cervical, and uterine tissue. A mutant lacking endocarditis- and biofilm-associated pili (Ebp) exhibited a decreased ability to associate with human vaginal and cervical cells in vitro but did not contribute to colonization in vivo. Thus, we screened a low-complexity transposon (Tn) mutant library to identify novel genes important for E. faecalis colonization and persistence in the vaginal tract. This screen revealed 383 mutants that were underrepresented during vaginal colonization at 1, 5, and 8 days postinoculation compared to growth in culture medium. We confirmed that mutants deficient in ethanolamine catabolism or in the type VII secretion system were attenuated in persisting during vaginal colonization. These results reveal the complex nature of vaginal colonization and suggest that multiple factors contribute to E. faecalis persistence in the reproductive tract.
INTRODUCTION
Enterococcus faecalis is an opportunistic pathogen that resides in the human gastrointestinal and urogenital tracts (1, 2). While E. faecalis colonization is normally asymptomatic, certain populations are at risk for severe disease, including urinary tract infections (3), wound infections, pelvic inflammatory disease (PID), infective endocarditis, and adverse birth effects during pregnancy (reviewed in references 4 and 5). Enterococcal infections are often associated with the production of biofilms, assemblages of microbes enclosed in an extracellular polymeric matrix that exhibit cell-to-cell interactions (reviewed in reference 6). These biofilms have been observed on catheters, diabetic ulcers, and wounds, resulting in severe infection. Treatment of enterococcal infections is becoming increasingly problematic due to their augmented ability to acquire mobile genetic elements, resulting in increased resistance to antibiotics, including last-line-of-defense antibiotics such as vancomycin (reviewed in references 7 and 8). Recently, there has been an increase in the emergence of vancomycin-resistant enterococci (9), putting immunocompromised individuals at risk for developing severe chronic enterococcal infections. The emergence of vancomycin-resistant enterococci (10) and its prevalence in both community and nosocomial settings is concerning and necessitates the development of alternative therapeutics to treat enterococcal infections.
E. faecalis encodes a multitude of virulence factors that allow the bacterium to colonize and persist in different sites of the human body. Surface proteins such as the adhesin to collagen (Ace), enterococcal fibronectin binding protein A (EfbA), aggregation substance (AS), and the endocarditis- and biofilm-associated pilin (Ebp) previously have been shown to play important roles in infective endocarditis and urinary tract infections (UTIs) (reviewed in reference 11). Secreted factors, such as gelatinase, autolysin A, and serine protease (SprE), are biofilm-associated factors that are involved in the degradation of host substrates, including collagen, fibrin, and certain complement components (14). Many of these virulence factors are regulated via quorum sensing, which may be responsible for the switch from a commensal to pathogenic state (15–17).
Certain risk factors are associated with the transition of E. faecalis from commensalism to pathogenicity, such as immune status, prolonged hospital stay, and the use of antibiotics (18). E. faecalis colonization and infection is often polymicrobial, and these interactions have been observed in the intestine, bloodstream, and wounds (reviewed in reference 19). Furthermore, E. faecalis is frequently found in the vaginal tract of healthy women (20, 21), and its prevalence is increased in women diagnosed with aerobic vaginitis (AV), an inflammatory response accompanied by depletion of commensal Lactobacillus sp. and increased presence of opportunistic pathogens, such as E. faecalis, group B Streptococcus (GBS), Staphylococcus aureus, and Escherichia coli (22, 23). Symptoms of AV include malodor and discomfort, but AV can transition to more serious complications, such as PID, severe UTIs, and complications during pregnancy. While it is evident that E. faecalis colonizes the human vaginal tract, the molecular determinants that allow enterococci to colonize and persist in the vaginal tract remain to be identified.
In this study, we modified our previously established GBS vaginal colonization model to analyze E. faecalis vaginal colonization and persistence. We determined that E. faecalis OG1RF (a rifampin and fusidic acid derivative of strain OG1) and vancomycin-resistant E. faecalis V583 can colonize and persist in the vaginal tract of CD1 and C57BL/6 mouse strains. We detected fluorescent E. faecalis in the vaginal lumen as well as the cervical and uterine tissues of colonized mice. Further, we demonstrated that an E. faecalis strain lacking Ebp pili is less adherent to vaginal cervical epithelium in vitro but not attenuated in vivo. Thus, we screened an E. faecalis OG1RF transposon (Tn) mutant library for mutants that are underrepresented in the vaginal tract compared to the culture input, revealing multiple factors for E. faecalis persistence within the vagina. These factors include sortase-dependent proteins (SDPs), ethanolamine (EA) utilization genes, and genes involved in type VII secretion system (T7SS) machinery. We confirmed that a strain mutated in ethanolamine catabolism was significantly attenuated in the ability to colonize the vaginal epithelium, and T7SS was required to ascend in the female reproductive tract. This work is an important first step in identifying factors required for enterococcal vaginal colonization and will provide insight into potential therapeutic targets aimed at mitigating E. faecalis vaginal colonization in at-risk individuals.
RESULTS
E. faecalis colonization of the female reproductive tract.
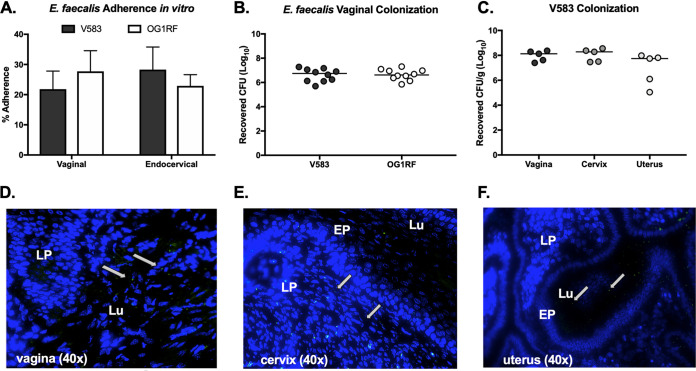
To characterize the ability of E. faecalis to interact with the epithelial cells of the lower female reproductive tract, we performed in vitro quantitative adherence assays using E. faecalis strains V583 (24) and OG1RF (25). An inoculum of 105 CFU/well (multiplicity of infection [MOI], 1) was added to a confluent monolayer of immortalized human vaginal and endocervical epithelial cells. Following 30 min of incubation, the cells were washed to remove nonadherent bacteria, the epithelial cells were detached from the plates, and adherent bacteria were plated on agar. Both strains exhibited substantial adherence to both cell lines (Fig. 1A). Next, we assessed the ability of E. faecalis to establish colonization of the murine vaginal tract. The vaginal lumen of C57BL/6 were swabbed and swabs were plated on CHROM agar to determine the presence of native enterococci. While native enterococci are detected on CHROM agar, no mice were colonized with strains that resemble E. faecalis V583 or OG1RF, as no colonies appeared on agar supplemented with antibiotics that select for V583 and OG1RF. Next, C57BL/6 mice were treated with β-estradiol 1 day prior to inoculation with 107 CFU of E. faecalis V583 or OG1RF. After 1 day postinoculation, the vaginal lumen was swabbed and bacteria were plated to enumerate E. faecalis V583 and OG1RF vaginal colonization levels (Fig. 1B). Swabs were plated on selective agar to ensure quantification of only the enterococcal strains of interest, restricting growth of native enterococcus. To determine whether E. faecalis ascends into reproductive tissues during colonization, murine vaginal, cervical, and uterine tissues were collected and homogenized to enumerate E. faecalis V583 abundance. E. faecalis was recovered from all mice 1 day postinoculation in all tissues tested (Fig. 1C), and the numbers of CFU recovered from the vaginal swabs were similar to the total CFU counts from the vaginal tissue homogenates (Fig. 1B and C). This level and range of recovered numbers of E. faecalis CFU is similar to what we have observed using this mouse model for GBS and S. aureus vaginal colonization (26, 27). To visualize E. faecalis within the murine reproductive tract, mice were inoculated with either wild-type (WT) E. faecalis V583 or a V583 strain expressing green fluorescent protein (GFP) (28). These strains both colonize the vaginal tract (see Fig. S1A in the supplemental material). We harvested the female reproductive tract 1 day postinoculation to avoid the loss of the GFP plasmid, made serial sections of these tissues, and performed fluorescence microscopy to visualize E. faecalis. We observed numerous fluorescent bacteria in the vaginal and uterine lumen (Fig. 1D and F) and embedded in the cervical lamina propria (Fig. 1E). We did not observe background green fluorescence in naive mice (Fig. S1B, C, and D), which coincides with previous experiments performed with GBS and S. aureus (26, 27). The presence of fluorescent E. faecalis in the cervix and uterus shows that E. faecalis can move from the vaginal lumen to the superior organs of the female reproductive tract.
FIG 1.
E. faecalis colonizes the murine female reproductive tract. (A) Adherence of E. faecalis V583 and OG1RF to human vaginal and endocervical cells. Data are expressed as percent recovered cell-associated E. faecalis relative to the initial inoculum. Experiments were performed in triplicate, and error bars represent standard deviations (SDs); the results of a representative experiment are shown. (B) CFU counts of V583 and OG1RF recovered from vaginal swabs 1 day postinoculation. (C) CFU counts of V583 from vaginal, cervical, and uterine tissue 1 day postinoculation. (D, E, and F) Mice were inoculated with V583 expressing gfp, and 7-μm sections of vaginal (D), cervical (E), and uterine (F) tissue were collected 1 day postinoculation and stained with DAPI for fluorescence microscopy. White arrows indicate green fluorescent bacteria present in tissue sections. Images were all taken at ×40 magnification. LP, lamina propria; EP, epithelial layer; Lu, lumen.
E. faecalis persists in the vaginal tract.
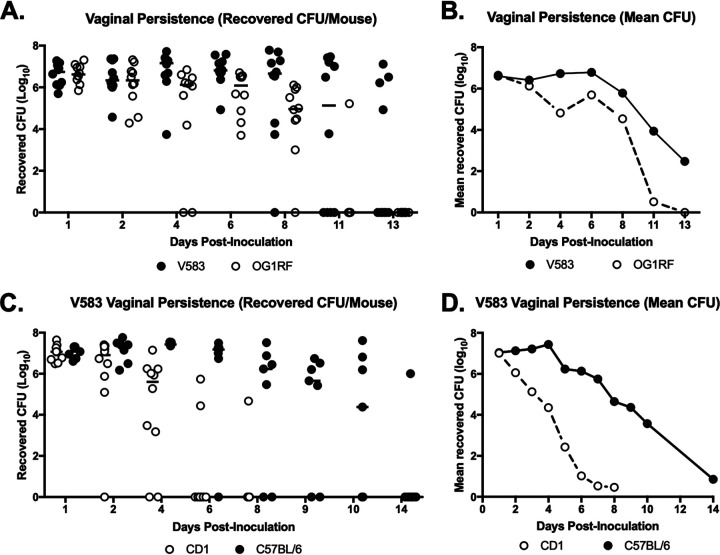
To assess vaginal persistence, C57BL/6 mice were colonized with E. faecalis V583 or OG1RF and swabbed to determine bacterial load over time. Mice were swabbed daily until no colonies appeared on agar selective for V583 or OG1RF, indicating bacterial clearance from the vaginal lumen. While V583 persisted longer in the mouse vaginal tract, the mean number of CFU recovered for both V583 and OG1RF remained constant for the first week and then declined as mice eventually cleared both strains by 11 to 13 days (Fig. 2A and B). To determine if enterococcal vaginal persistence differs across mouse strains, C57BL/6 and CD1 mice were inoculated with V583 and swabbed over time. Both mouse strains were initially colonized with V583, but bacteria in C57BL/6 mice persisted longer (Fig. 2C and D). By day six, only 20% of CD1 mice remained colonized compared to 85% of C57BL/6 mice. Differences in vaginal persistence may be due to differences in the native vaginal microbiota or immune status between mouse strains. It is also possible that bacteria occupy different niches within the reproductive tract of different mouse strains, which warrants further investigation. Overall, these results show that mouse strain background influences E. faecalis vaginal colonization and that C57BL/6 mice are a sufficient model to assess prolonged E. faecalis vaginal colonization and persistence.
FIG 2.
E. faecalis persists in the murine vaginal tract. (A and B) E. faecalis V583 and OG1RF in the murine vaginal tract. C57BL/6 mice (n = 10) were inoculated with 107 V583 or OG1RF CFU, the vaginal lumen of each mouse was swabbed daily, and swabs were serially diluted and plated on selective media to quantify CFU numbers. Data are presented as number of recovered CFU per mouse (A) and mean recovered number of CFU (B). Data were analyzed using a two-way ANOVA; **, P < 0.001. (C and D) CD1 (n = 10) and C57BL/6 (n = 7) mice were inoculated with V583, and the vaginal lumen of each mouse was swabbed daily, serially diluted, and plated on selective media to quantify CFU. Data are presented as number of recovered CFU per mouse (C) and mean number of recovered CFU (D). Black lines indicate the median CFU values.
Enterococcal pili contribute to interaction with reproductive tract tissues.
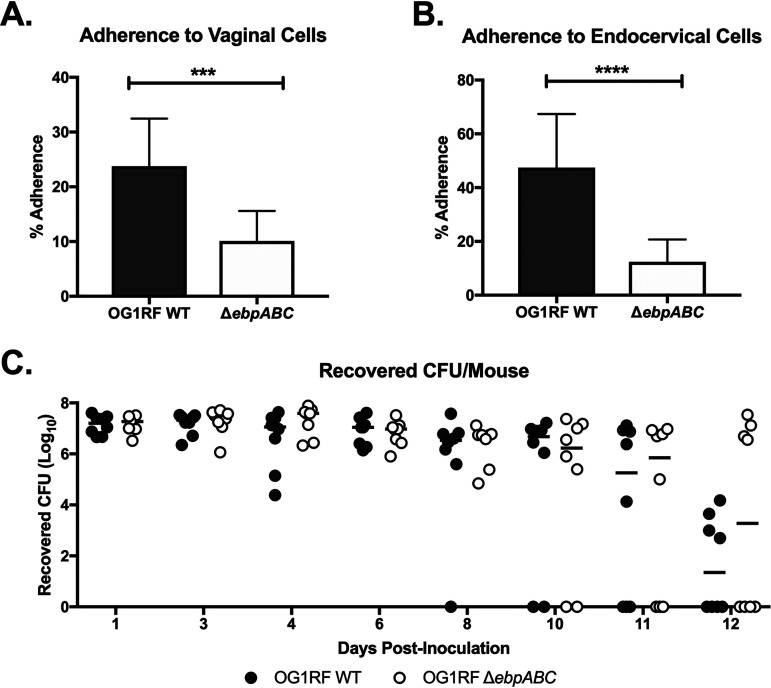
The endocarditis- and biofilm-associated pilin (Ebp) of E. faecalis mediates infective endocarditis and UTIs (29–32); thus, we hypothesized that Ebp similarly contributes to vaginal colonization. To determine whether Ebp is important for facilitating interaction with the vaginal epithelium, we used a deletion mutant of E. faecalis OG1RF lacking all pilin structural components (ΔebpABC) (33). We observed that the pilus mutant exhibited significantly reduced adherence to human vaginal and endocervical cells in vitro (Fig. 3A and B). To determine if Ebp is important for in vivo vaginal colonization and persistence, mice were inoculated with either the WT OG1RF or OG1RF ΔebpABC strain, and colonization was quantified over the course of 12 days. We observed no differences in the number of CFU recovered from the vaginal lumen between WT OG1RF and OG1RF ΔebpABC strains (Fig. 3C). Taken together, these data suggest that Ebp contributes to E. faecalis attachment to reproductive tract tissues, but additional factors are likely required for persistence in the vaginal lumen in vivo.
FIG 3.
Role of enterococcal pili during vaginal colonization. (A and B) E. faecalis OG1RF WT and OG1RF ΔebpABC strain adherence to human vaginal (A) and endocervical (B) cells. Data are expressed as percent recovered cell-associated E. faecalis relative to the initial inoculum. Experiments were performed with four technical replicates, and error bars represent SD; the results of three combined biological replicates are shown and analyzed using an unpaired t test; ***, P < 0.0001; ****, P < 0.00001. (C) C57BL/6 mice were inoculated with either the OG1RF WT or OG1RF ΔebpABC strain, and the vaginal lumen was swabbed daily. Data were analyzed using a two-way ANOVA with Sidak’s multiple comparisons (P > 0.05). Black lines indicate the median CFU values.
Identification of additional vaginal colonization factors by transposon mutagenesis analysis.
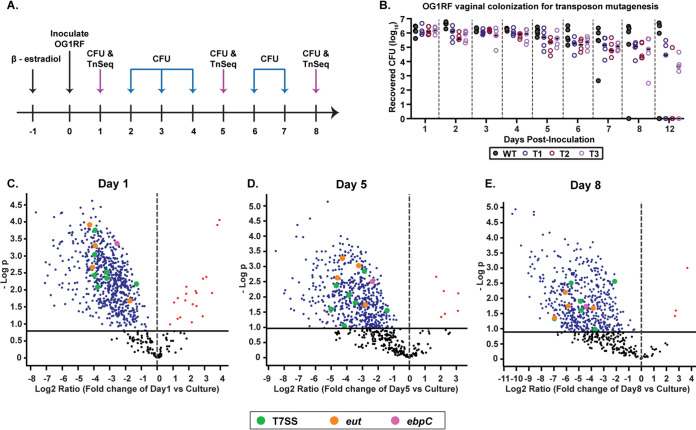
To identify genetic determinants that confer enterococccal vaginal persistence, we used sequence-defined mariner technology transposon sequencing (SMarT TnSeq) to screen an E. faecalis OG1RF Tn library consisting of 6,829 unique mutants (34). The library was grown to mid-log phase in triplicate, and 107 CFU of each replicate was vaginally inoculated into a group of 5 C57BL/6 mice (Fig. 4A). Vaginal swabs were plated on selective media daily for 8 days, and numbers of CFU were quantified to assess colonization of the OG1RF Tn library compared to that of WT OG1RF (Fig. 4A and B). Genomic DNA was isolated from pooled Tn mutants recovered on days 1, 5, and 8 postinoculation, and Tn insertion junctions in E. faecalis genomic DNA were sequenced as described by Dale et al. (34). Sequenced reads were mapped to the E. faecalis OG1RF genome to identify genes that are necessary for E. faecalis vaginal colonization.
FIG 4.
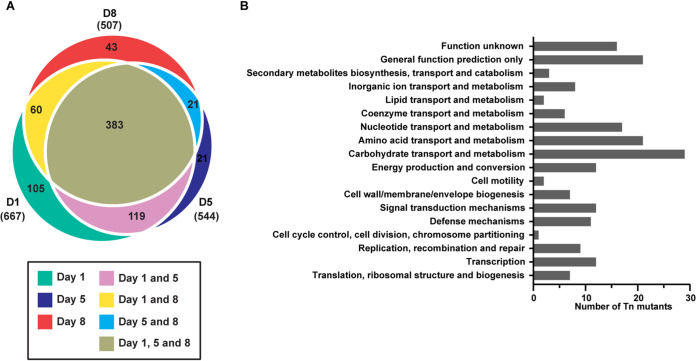
Identification of additional factors required for vaginal colonization and persistence by transposon mutant library screen. (A) Schematic representing experimental approach. C57BL/6 mice were treated with 17β-estradiol prior to inoculation with the OG1RF Tn library in triplicate groups or WT OG1RF. The vaginal lumen was swabbed, and the number of CFU was enumerated daily. DNA from recovered OG1RF Tn mutants was sequenced on days 1, 5, and 8. (B) CFU recovered from vaginal swabs of triplicate groups of mice colonized with OG1RF Tn mutagenesis library (T1, T2, and T3) and OG1RF WT (n = 5 mice per group). (C, D, and E) Volcano plots depicting underrepresented and overrepresented mutants in vivo compared to culture on day 1 (C), day 5 (D), and day 8 (E) postinoculation. (C) A total of 667 underrepresented genes and 21 overrepresented genes in vivo compared to culture. (D) A total of 404 underrepresented genes and 6 overrepresented genes in vivo compared to culture. (E) A total of 507 underrepresented genes and 3 overrepresented genes in vivo compared to culture. Colored dots represent mutants of interest. Pink, ebpC Tn mutant; orange, eut Tn mutants; green, T7SS gene Tn mutants. The black solid line represents the cutoff for statistical significance.
We observed that the in vivo vaginal environment altered the abundance of select mutants from the Tn library pool compared to the original culture input (Fig. 4C to E) (Tables S1 to S5). At day 1, a total of 667 depleted mutants were identified (Table S1), along with 544 (Table S2) and 507 (Table S3) at days 5 and 8, respectively; 383 of these mutants were identified at all three time points (Fig. 5A and Table S4A). Classification by clusters of orthologous groups of proteins (COGs) could be identified in 196 mutants from all 3 time points, the majority of which are involved in carbohydrate, amino acid, lipid, and nucleotide transport/metabolism, as well as those involved in transcription and defense mechanisms (Fig. 5B). Of the remaining 187 mutants, 85 had insertions in intergenic regions and 102 were not assigned a COG domain. Interestingly, the ebpC transposon mutant (OG1RF_10871, which encodes the shaft component of Ebp) was underrepresented at all time points (Fig. 4C to E and Table S4A). Additional mutants of interest included those involved in ethanolamine catabolism and T7SS components, in which various components of these systems were underrepresented at all three time points (Table 1 and Table S4A). Furthermore, mutants for multiple sortase-dependent proteins (SDPs), including ebpC, were underrepresented at all time points (Table 1 and Table S4A), suggesting that these factors play important roles in vaginal colonization and persistence.
FIG 5.
Classification of transposon insertion mutants by COGs. (A) Euler plot representing number of underrepresented mutants at all time points. (B) COGs from underrepresented mutants common to all three time points categorized into functional categories. The total of 196 represents all common mutants that were assigned a COG domain.
TABLE 1.
Selected list of differentially abundant transposon mutants during vaginal colonization compared to in vitro cultures
| Old locus tag | NCBI description | Difference by day of culture |
||
|---|---|---|---|---|
| 1 | 5 | 8 | ||
| SDPs | ||||
| OG1RF_10811 | Collagen adhesion protein | −4.33 | −4.14 | −5.72 |
| OG1RF_10871 | Cell wall surface anchor family protein, ebpC | −2.54 | −2.37 | −4.44 |
| OG1RF_11531 | Glycosyl hydrolase | −3.04 | −3.15 | −4.25 |
| OG1RF_11764 | Cell wall surface anchor family protein | −2.80 | −3.48 | −5.24 |
| OG1RF_12054 | Cell wall surface anchor family protein | −2.62 | −1.50 | −6.01 |
| EA utilization | ||||
| OG1RF_11342 | Ethanolamine utilization protein EutL | −3.92 | −3.25 | −6.10 |
| OG1RF_11343 | Ethanolamine ammonia-lyase small subunit | −4.09 | −4.60 | −5.85 |
| OG1RF_11344 | Ethanolamine ammonia-lyase large subunit | −4.27 | −4.28 | −6.94 |
| OG1RF_11347 | Response regulator EutV | −1.72 | −2.82 | −3.84 |
| T7SS | ||||
| OG1RF_11103 | YukD superfamily protein, esaB | −3.19 | −5.02 | −6.95 |
| OG1RF_11109 | Putative LXG-containing toxin | −3.93 | −3.47 | −4.87 |
| OG1RF_11111 | Putative T7SS toxin | −3.76 | −4.15 | −4.80 |
| OG1RF_11113 | Putative T7SS toxin | −3.19 | −2.87 | −3.70 |
| OG1RF_11114 | Putative immunity protein | −3.96 | −3.88 | −5.62 |
| OG1RF_11122 | Immunity protein | −1.31 | −1.43 | −2.14 |
Potential gain-of-function mutations have also been discovered during genome-wide library screens of fitness determinants in other bacteria (35–39). In addition to mutations that adversely impact vaginal colonization, our data show that Tn insertions in 11 protein-coding genes and 11 intergenic regions potentially enhance bacterial fitness in vivo. Nineteen of the 22 enriched mutants were common between days 1 and 5, whereas the other 3 were unique to day 8 (Table S5A and B). Since the majority of the mutants with increased fitness encode hypothetical proteins, the relationship between these genes and vaginal colonization is currently unclear and requires further investigation.
EA utilization and T7SS genes contribute to E. faecalis persistence in the reproductive tract.
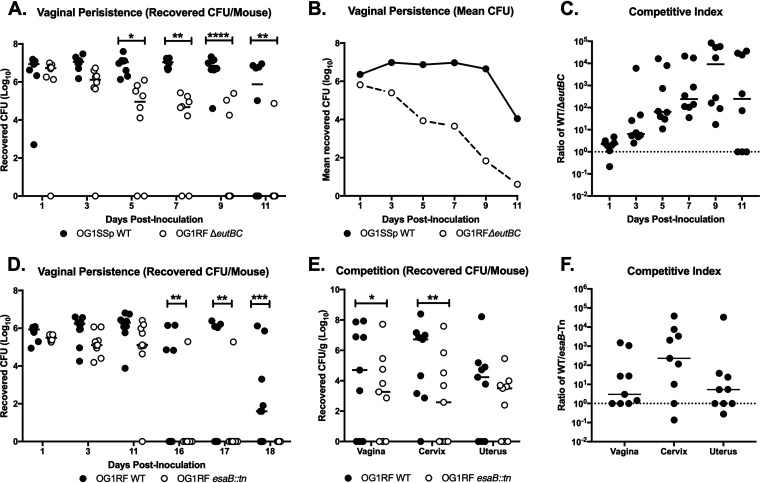
Our TnSeq analysis revealed many potential mutants that exhibited decreased colonization in the murine vaginal tract. We sought to confirm these results by analyzing mutants from systems with multiple affected genes. One significantly affected operon was EA utilization (eut), which consists of 19 genes in E. faecalis (40). Mutants in 4 eut genes were significantly underrepresented in vivo compared to the culture input at all time points. These included transposon mutants of the genes encoding both subunits for ethanolamine ammonia lyase, eutB (OG1RF_11344) and eutC (OG1RF_11343) (41), a carboxysome structural protein, eutL (OG1RF_11342) (42), and the response regulator, eutV (OG1RF_11347), of the two-component system involved in the regulation of EA utilization (43) (Table S4A). To assess the importance of EA utilization on E. faecalis vaginal colonization, we cocolonized mice with E. faecalis OG1SSp (a derivative of OG1 that is resistant to streptomycin and spectinomycin) and an OG1RF ΔeutBC mutant (44). We note that both WT strains, OG1RF and OG1SSp, were able to colonize the vaginal tract at similar levels (Fig. S2). Further chromosomal DNA sequence comparison of OG1RF and OG1SSp revealed multiple single-nucleotide polymorphisms (SNPs) in OG1SSp but no SNPs in genes in the eut locus (Table S6). Compared to the WT OG1SSp strain, the ΔeutBC mutant was cleared significantly faster from the mouse vagina, as seen by the number of CFU from individual mouse swabs, the mean number of CFU recovered, and the competitive index (CI) over time (Fig. 6A to C). These results suggest that the utilization of ethanolamine is important for enterococcal persistence in the vaginal tract.
FIG 6.
Ethanolamine utilization and type VII secretion system genes contribute to enterococcal persistence in the vaginal tract. (A, B, and C) C57BL/6 mice were coinoculated with OG1SSp WT and OG1RF ΔeutBC strains, and vaginal lumen was swabbed to quantify CFU. Data are presented as recovered number of CFU per swab (A), mean number of CFU recovered (B), and CI between WT and mutant strains (C). Data were analyzed using a two-way ANOVA with Sidak’s multiple comparisons; *, P < 0.05; **, P < 0.005; ****, P < 0.00005. (D) C57BL/6 mice were coinoculated with OG1RF WT and OG1RF esaB::tn strains, and vaginal lumen was swabbed to quantify CFU. Data were analyzed using a two-way ANOVA with Sidak’s multiple comparisons; **, P < 0.005; ***, P < 0.0005. (E and F) C57BL/6 mice were coinoculated with OG1RF WT and OG1RF esaB::tn strains, and reproductive tissue was collected at 11 days postinoculation. Data are presented as recovered log10 CFU/gram (E) and CI between WT and Tn mutant strain (F). CI is enumerated by calculating the ratio of WT to mutant E. faecalis recovered from the mouse reproductive tract. A CI of >1 indicates an advantage to WT E. faecalis. Values below the limit of detection were enumerated as one-half the limit of detection. Data were analyzed using a paired t test; *, P < 0.05; **, P < 0.005. Black lines indicate the median of CFU values.
We also observed that Tn insertion mutants in the T7SS locus were significantly underrepresented at all time points in vivo compared to the culture input. The T7SS has been shown to play an important role in virulence in multiple bacterial species, such as Staphylococcus, Listeria, and Bacillus (45). In E. faecalis, genes in the T7SS locus have been shown to be induced during phage infection (46). We observed that transposon mutants for esaB (OG1RF_11103), a putative cytoplasmic accessory protein; OG1RF_11109 and OG1RF_11111, putative toxin effector proteins; OG1RF_11113, a putative toxin; OG1RF_11114, a transmembrane protein; and OG1RF_11122, a potential antitoxin protein, were all significantly underrepresented (Table S4A). To confirm the role of the T7SS in vaginal colonization, we utilized an esaB (esaB::tn) mutant in which esaB is disrupted by a transposon; thus, Tn-mediated polar effects may exist for this strain. Following cocolonization with E. faecalis OG1RF and the OG1RF esaB::tn mutant strain, we observed that while there were no differences in initial colonization, fewer esaB::tn mutant bacteria were recovered from the vaginal lumen at later time points (Fig. 6D). Since we only observed differences in colonization between WT OG1RF and OG1RF esaB::tn at later time points, we performed subsequent experiments to determine whether there were differences in ascension between the two strains. Mice were cocolonized with the two strains and we harvested tissues at day 11, before any colonization differences were observed between the two strains. Here, we observed that WT OG1RF outcompeted the esaB::tn mutant strain and was better able to access reproductive tract tissues (Fig. 6E and F), indicating that the T7SS is involved in vaginal persistence and ascension in the female reproductive tract.
DISCUSSION
E. faecalis is associated with a wide spectrum of infections, particularly under immunocompromised states and during compositional shifts in the host microbiota (47, 48). Although an increasing body of evidence links enterococci with bacterial vaginosis (BV) and aerobic vaginitis (AV) (22, 23, 49–51), the molecular determinants that facilitate E. faecalis colonization and persistence in the vaginal tract are largely unknown. Here, we employed in vitro and in vivo systems to acquire genome-scale interactions that confer E. faecalis fitness within the female reproductive tract. We show that both vancomycin-sensitive enterococci (VSE) and vancomycin-resistant enterococci (VRE) adhere to cell types of vaginal and cervical origin, a signature of bacterial colonization that precedes tissue invasion and systemic spread. Further, genetic features involved in biofilm formation, ethanolamine utilization, and polymicrobial interactions influence E. faecalis vaginal carriage.
Previous studies have demonstrated the importance of Ebp pili in enterococcal virulence and biofilm formation (19). We found that deletion of ebpABC attenuated binding to human vaginal and endocervical cells but did not influence bacterial burden in the vaginal lumen, similar to the observed function of Ebp pili in the intestine (52). Enhanced E. faecalis adherence in tissue culture compared to in vivo colonization may reflect the lack of liquid and mucus flow that bacteria encounter within the vaginal tract, emphasizing the significance of our animal model for investigating E. faecalis-vaginal interactions. Consistent with this observation, an in vivo Tn library screen revealed only two underrepresented biofilm-associated mutants, ebpC-Tn and OG1RF_10506-Tn, encoding a putative polysaccharide deacetylase homolog implicated in low biofilm formation in vitro (53, 54). Together, these results show that individual mutations in ebp or other well-characterized biofilm genes are not sufficient to impair vaginal niche establishment and/or persistence of enterococci, which likely depends on the concerted effort of multiple factors. Furthermore, similar to ebpC-Tn, we observed that genes for multiple sortase-dependent proteins (SDPs) were underrepresented at all time points during vaginal colonization. The genome of OG1RF contains 21 sortase-dependent proteins, including Ebp (52). Other than ebpC (OG1RF_10871), we observed that 4 other SDPs are underrepresented during vaginal colonization, including OG1RF_10811, OG1RF_11531, OG1RF_11764, and OG1RF_12054 (Table 1). Previous reports indicate the importance of SDPs during gastrointestinal colonization by enterococci (52), implying that multiple SDPs also play a role during vaginal colonization.
Transition from nutrient-rich laboratory media to the vaginal tract likely imparts dramatic alterations in E. faecalis metabolism. In support of this hypothesis, our high-throughput Tn mutant screen showed that mutations in carbohydrate, amino acid, and nucleotide metabolic pathways were indispensable in the vaginal tract. Specifically, we showed that WT bacteria outcompete a eut locus mutant during vaginal colonization. In contrast, Kaval and colleagues demonstrated that mutations in eut genes leads to a slight increase in fitness within the intestine (55). This observation likely reflects various metabolic requirements of enterococci in different host environments. While a number of reports exist on the contributions of EA catabolism in host-bacterium interactions within the intestine (56), studies are lacking for the relevance of this EA metabolism in other host-associated environments. Our results raise important questions regarding EA utilization in the female reproductive tract. Although commensal microbes and the epithelium are rich sources of EA, the composition and source of EA in the vaginal tract remain unknown. A recent report showed that E. faecalis EA utilization attenuates intestinal colitis in interleukin-10 knockout mice in the presence of a defined microbiota (57). Whether EA utilization promotes virulence or commensalism for enterococci in the context of vaginal tissue remains to be determined. Considering that the by-product of EA metabolism, acetate, is anti-inflammatory and promotes IgA production in the intestine (58, 59), it is intriguing to consider that enterococcal EA catabolism might modulate immune responses within the female reproductive tract.
T7SSs have been implicated in the maintenance of bacterial membrane integrity, virulence, and interbacterial antagonism (45, 60–66). In S. aureus, T7SS-encoded proteins confer protection from membrane damage caused by host fatty acids (65, 66). Although E. faecalis T7SS genes were shown to be induced in response to phage-driven membrane damage, direct contributions of these genes in cell envelop barrier function and/or virulence in the context of animal models are poorly defined. Our TnSeq analysis revealed that insertional mutations in six T7SS genes diminished early and late vaginal colonization by E. faecalis OG1RF. In vaginal cocolonization competition experiments, an esaB::tn strain reached WT colonization levels early and showed a defect in long-term persistence. The incongruence in the colonization kinetics of T7SS mutant strains compared to T7SS-Tn library mutants, which were observed early after inoculation, presumably stem from the inherent differences in the vaginal milieu in these two experiments. The TnSeq library employed in this screen is a complex population of approximately 7,000 unique mutants, and it is very likely that direct or indirect interactions between mutants influence fitness. LXG-domain toxins, which are part of the T7SS, have been shown to antagonize neighboring non-kin bacteria (63). The fact that two mutants with LXG-domain-encoding gene mutations, OG1RF_11109 and OG1RF_11111, were underrepresented across all time points during vaginal colonization suggests that these putative antibacterial proteins influence enterococcal interactions with the resident microbes of the vaginal tract.
In addition to genes encoding SDPs, ethanolamine utilization, and T7SS, other Tn mutants that were underrepresented at all time points are worth discussing. For example, OG1RF_12241, a homolog of the oxidative stress regulator hypR, was underrepresented at all time points (Table S4A). We have recently shown that this gene is involved in phage VPE25 infection of E. faecalis OG1RF (46). Furthermore, enterococcal mutants in the CRISPR/cas9 locus (OG1RF_10404 and OG1RF_10407) were underrepresented at all three time points (Table S4A). While the role of CRISPR-Cas systems in providing prokaryotic immunity to mobile genetic elements has been extensively investigated, some evidence suggests that this system is involved in prokaryotic processes besides adaptive immunity. Cas9 has been shown to have various functions in regulation of virulence in a number of bacteria, including Francisella novicida, Campylobacter jejuni, and Streptococcus agalactiae (67–69). Our TnSeq analysis further reveals the potential importance for Cas9 during vaginal colonization, which warrants follow-up studies.
While a majority of underrepresented mutants were common to all time points, we identified certain mutants unique to either early or late colonization. For example, the ethanolamine utilization protein EutQ (OG1RF_11333), a classified acetate kinase in Salmonella enterica (71), was significantly underrepresented at day 1 but not the later time points. We also observed that Tn mutants for the transmembrane signaling protein kinase IreK (OG1RF_12384) were underrepresented only at day 1. In E. faecalis, IreK is involved in regulation of cell wall homeostasis (72) and long-term persistence in the gut, and it also has been shown to be essential for enterococcal T7SS expression and subsequent activity (73). Therefore, these proteins may be important contributors to enterococcal vaginal colonization, although further investigation is required. Our TnSeq analysis also identified Tn mutants that were unique to day 8 postinoculation, indicating that these factors are important for later-stage colonization and persistence. One observed mutant was the fsrB gene (OG1RF_11528) of the Fsr quorum-sensing system, which directly regulates virulence factors such as serine protease and gelatinase while also indirectly regulating other virulence factors involved in surface adhesion and biofilm development (74–76). Although it is not well understood whether biofilms are being formed during vaginal colonization, certain hits in our TnSeq (i.e., ebpC and fsrB) analysis suggests that biofilm-associated factors play a role in enterococcal persistence in the vaginal tract. Bacterial mutants for the response regulator croR (OG1RF_12535) were also underrepresented only at the later time point. CroR has shown to be involved in virulence regulation, cell wall homeostasis and stress response, and antibiotic resistance (77–79). Finally, underrepresentation of the sortase-associated gene (OG1RF_10872) was also unique to late colonization. The underrepresentation of enterococcal mutants late in vaginal colonization suggests these factors are important for long-term persistence of E. faecalis in the vaginal tract. While the majority of underrepresented mutants were common to all time points, mutants unique to certain time points indicate that some factors are important for either initial colonization or enterococcal survival in the vaginal tract.
Here, we report the utilization of a mouse model for investigating host-enterococcal interactions in the vaginal tract. This will be a useful model for analyzing the bacterial and host factors that govern enterococcal vaginal colonization as well as characterizing the polymicrobial interactions that may contribute to E. faecalis niche establishment and persistence. Transposon library screening of E. faecalis recovered from the mouse vagina has revealed new insights into our understanding of enterococcal vaginal carriage. Our results emphasize the importance of ethanolamine utilization and T7SS components for successful E. faecalis colonization of the female reproductive tract, highlighting the complex nature of this niche.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A detailed list of bacterial strains can be found in Table S7. E. faecalis strains V583 (80) and OG1 (OG1RF and OG1SSp) (70, 81) were used for these experiments. E. faecalis was grown in brain heart infusion (BHI [82]) broth at 37°C with aeration, and growth was monitored by measuring the optical density at 600 nm (OD600). For selection of E. faecalis V583, BHI agar was supplemented with gentamicin (100 μg/ml). For selection of E. faecalis OG1RF, OG1RF ΔebpABC (33), and OG1RF ΔeutBC (44) strains, BHI agar was supplemented with rifampin (50 μg/ml) and fusidic acid (25 μg/ml). For selection of E. faecalis OG1SSp, BHI agar was supplemented with streptomycin (150 μg/ml) and spectinomycin (100 μg/ml). E. faecalis OG1RF esaB::tn (34) was grown on BHI agar supplemented with rifampin (50 μg/ml), fusidic acid (25 μg/liter), and chloramphenicol (15 μg/ml).
In vitro adherence assays.
Immortalized VK2 human vaginal epithelial cells and End1 human endocervical epithelial cells were obtained from the American Type Culture Collection (VK2.E6E7, ATCC CRL-2616; End1/E6E7, ATCC CRL-2615) and were maintained in keratinocyte serum-free medium (KSFM; Gibco) with 0.1 ng/ml human recombinant epidermal growth factor (EGF; Gibco) and 0.05 mg/ml bovine pituitary extract (Gibco) at 37°C with 5% CO2. Assays to determine cell surface-adherent E. faecalis were performed as described previously when quantifying GBS adherence (83). Briefly, bacteria were grown to mid-log phase (OD600, 0.4 to 0.6) and added to cell monolayers (MOI of 1). After a 30-min incubation, cells were washed with phosphate-buffered saline (PBS) three times following detachment with 0.1 ml of 0.25% trypsin-EDTA solution and lysed with addition of 0.4 ml of 0.025% Triton X-100 in PBS by vigorous pipetting. The lysates were then serially diluted and plated on Todd Hewitt agar (THA) to enumerate the bacterial CFU levels. Experiments were performed at least three times with each condition in triplicate, and results from a representative experiment are shown.
Murine vaginal colonization model.
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Colorado-Anschutz Medical Campus, protocols 00316 and 00253, and performed using accepted veterinary standards. A mouse vaginal colonization model for GBS was adapted for our studies (26). Eight-week-old female CD1 (Charles River) or C57BL/6 (Jackson) mice were injected intraperitoneally with 0.5 mg 17β-estradiol (Sigma) 1 day prior to colonization with E. faecalis. Mice were vaginally inoculated by gently pipetting 107 CFU of E. faecalis in 10 μl PBS into the vaginal tract, avoiding contact with the cervix. On subsequent days, the vaginal lumen was swabbed with a sterile ultrafine swab (Puritan). For cocolonization, mice were inoculated with two of the following E. faecalis strains: OG1SSp, OG1RF, or deletion mutants in the OG1RF background. To assess numbers of tissue CFU, mice were euthanized according to approved veterinary protocols, and the female reproductive tract tissues were dissected, placed into 500 μl PBS, and bead beaten for 2 min to homogenize the tissues. The resulting homogenate was serially diluted and E. faecalis CFU enumerated on BHI agar supplemented with antibiotics to select for the strain of interest.
Histology.
Mice were inoculated with E. faecalis V583 containing a plasmid that expresses gfp (pMV158GFP) and contains resistance to tetracycline (15 μg/ml) (23). After 1 day postinoculation, the murine female reproductive tract was harvested, embedded into optimal cutting temperature (OCT) compound (Sakura), and sectioned at 7 μm with a CM1950 freezing cryostat (Leica). For fluorescence microscopy, coverslips were mounted with VECTASHIELD mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Labs). Images were taken with a BZ-X710 microscope (Keyence) (22).
Transposon mutant library growth and vaginal colonization.
The E. faecalis OG1RF transposon mutant library was generated previously (34). The E. faecalis OG1RF pooled transposon library was inoculated into 5 ml of BHI at a total of 108 CFU and grown with aeration to an OD600 of 0.5. The library was inoculated into the vaginal tracts of C57BL/6 mice at 107 CFU. The library was plated on BHI agar to confirm the inoculum for all groups of mice. Mice were swabbed daily, and swabs were plated on BHI supplemented with rifampin (50 μg/ml), fusidic acid (25 μg/ml), and chloramphenicol (20 μg/ml) to quantify the number of CFU. On days 1, 5, and 8, undiluted swabs were plated on BHI agar with antibiotics and grown to a bacterial lawn. Bacteria were scraped, resuspended in PBS, and pelleted. DNA from days 1, 5, and 8 and the input culture was extracted from pellets using a ZymoBIOMICS DNA miniprep kit (Zymo Research).
Transposon library sequencing and data analysis.
Transposon-junction DNA library preparation and Illumina NovaSeq 6000 DNA sequencing (150-bp paired-end mode) was performed by the Microarray and Genomics Core at the University of Colorado Anschutz Medical Campus as previously described (46). For downstream analysis of transposon junctions, we used only the R1 reads generated by paired-end sequencing. Illumina adapter trimmed raw reads were mapped to the E. faecalis OG1RF reference sequence (NC_017316.1), and differentially abundant transposon mutants were identified using statistical analysis scripts established by Dale et al. (34). The abundance of Tn mutants in culture was compared to that of the input library used for culture inoculation, and mutants that are not significantly different (P > 0.05) between these two samples were considered for the next steps of the analysis. For comparisons between in vivo and in vitro samples, mutants were considered significantly different if the adjusted P value was <0.05 and the log2 fold change was >1.
Genomic DNA sequencing and comparative analysis.
E. faecalis genomic DNA was purified using a ZymoBIOMICS DNA miniprep kit (Zymo Research), and 150-bp paired-end sequencing was performed on an Illumina NextSeq 550 by the Microbial Genome Sequencing Center, University of Pittsburgh. E. faecalis OG1SSp genomic DNA was purified using a Qiagen DNeasy kit and was sequenced on the MiSeq platform (2× 75 bp) at the University of Minnesota Genomics Center. All reads were mapped to the E. faecalis OG1RF reference sequence (NC_017316.1) using CLC Workbench (Qiagen). The basic variant caller tool in CLC Genomics Workbench was used to identify single-nucleotide polymorphisms using default settings (similarity fraction, 0.5; length fraction, 0.8).
Statistical analysis.
GraphPad Prism version 7.0 was used for statistical analysis, and statistical significance was accepted at P values of <0.05 (*, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005). Specific tests are indicated in the figure legends.
Data availability.
The TnSeq and genomic DNA reads have been deposited at the European Nucleotide Archive under accession numbers PRJEB37929 and PRJEB39171, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kimberly Kline for providing E. faecalis OG1RF ΔebpABC, Danielle Garsin for providing E. faecalis OG1RF ΔeutBC, Jennifer Dale for DNA purification and sequencing of OG1SSp and the University of Minnesota Genomics Center for DNA sequencing, Katrina Diener and Monica Ransom for custom TnSeq library preparation, and the Microarray and Genomics Core at the University of Colorado Anschutz Medical Campus for DNA sequencing.
This study was supported by NIH 5T32AI007405-28 to B.L.S., NIH/NIAID R21 AI130857 to K.S.D., NIH/NIAID R01 AI141479 to B.A.D., NIH/NIAID R01 AI122742 to G.M.D., American Heart Association grant 19POST34450124/Julia Willett/2018 to J.L.E.W., and a University of Colorado School of Medicine IMPA to K.S.D. and B.A.D.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lebreton F, Willems RJL, Gilmore MS. 2014. Enterococcus diversity, origins in nature, and gut colonization In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 2.Coolen MJ, Post E, Davis CC, Forney LJ. 2005. Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Appl Environ Microbiol 71:8729–8737. doi: 10.1128/AEM.71.12.8729-8737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaturvedi SK, Manche Gowda S, Ahmed HU, Alosaimi FD, Andreone N, Bobrov A, Bulgari V, Carra G, Castelnuovo G, de Girolamo G, Gondek T, Jovanovic N, Kamala T, Kiejna A, Lalic N, Lecic-Tosevski D, Minhas F, Mutiso V, Ndetei D, Rabbani G, Somruk S, Srikanta S, Taj R, Valentini U, Vukovic O, Wolwer W, Cimino L, Nouwen A, Lloyd C, Sartorius N. 2019. More anxious than depressed: prevalence and correlates in a 15-nation study of anxiety disorders in people with type 2 diabetes mellitus. Gen Psychiatr 32:e100076. doi: 10.1136/gpsych-2019-100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huycke MM, Sahm DF, Gilmore MS. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis 4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindlbeck C, Dziura D, Mylonas I. 2014. Diagnosis of pelvic inflammatory disease (PID): intra-operative findings and comparison of vaginal and intra-abdominal cultures. Arch Gynecol Obstet 289:1263–1269. doi: 10.1007/s00404-014-3150-7. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed JA, Huang DB. 2007. Biofilm formation by enterococci. J Med Microbiol 56:1581–1588. doi: 10.1099/jmm.0.47331-0. [DOI] [PubMed] [Google Scholar]
- 7.Arias CA, Murray BE. 2008. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther 6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 8.Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet 1:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 9.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 10.Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J. 1999. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis 180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 11.Madsen KT, Skov MN, Gill S, Kemp M. 2017. Virulence factors associated with Enterococcus faecalis infective endocarditis: a mini review. Open Microbiol J 11:1–11. doi: 10.2174/1874285801711010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted. [Google Scholar]
- 13.Reference deleted. [Google Scholar]
- 14.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. 2010. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun 78:4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali L, Goraya MU, Arafat Y, Ajmal M, Chen JL, Yu D. 2017. Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int J Mol Sci 18:960. doi: 10.3390/ijms18050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters CM, Antiporta MH, Murray BE, Dunny GM. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol 185:3613–3623. doi: 10.1128/jb.185.12.3613-3623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin X, Singh KV, Xu Y, Weinstock GM, Murray BE. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob Agents Chemother 42:2883–2888. doi: 10.1128/AAC.42.11.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tacconelli E, Cataldo MA. 2008. Vancomycin-resistant enterococci (VRE): transmission and control. Int J Antimicrob Agents 31:99–106. doi: 10.1016/j.ijantimicag.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Ch'ng JH, Chong KKL, Lam LN, Wong JJ, Kline KA. 2019. Biofilm-associated infection by enterococci. Nat Rev Microbiol 17:82–94. doi: 10.1038/s41579-018-0107-z. [DOI] [PubMed] [Google Scholar]
- 20.Leyva-Gomez G, Prado-Audelo MLD, Ortega-Pena S, Mendoza-Munoz N, Urban-Morlan Z, Gonzalez-Torres M, Gonzalez-Del Carmen M, Figueroa-Gonzalez G, Reyes-Hernandez OD, Cortes H. 2019. Modifications in vaginal microbiota and their influence on drug release: challenges and opportunities. Pharmaceutics 11:217. doi: 10.3390/pharmaceutics11050217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donders GGG, Bellen G, Grinceviciene S, Ruban K, Vieira-Baptista P. 2017. Aerobic vaginitis: no longer a stranger. Res Microbiol 168:845–858. doi: 10.1016/j.resmic.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Kaambo E, Africa C, Chambuso R, Passmore JS. 2018. Vaginal microbiomes associated with aerobic vaginitis and bacterial vaginosis. Front Public Health 6:78. doi: 10.3389/fpubh.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gold OG, Jordan HV, van Houte J. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch Oral Biol 20:473–477. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- 26.Patras KA, Doran KS. 2016. A murine model of group B Streptococcus vaginal colonization. J Vis Exp 117:54708. doi: 10.3791/54708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng L, Schilcher K, Burcham LR, Kwiecinski JM, Johnson PM, Head SR, Heinrichs DE, Horswill AR, Doran KS. 2019. Identification of key determinants of Staphylococcus aureus vaginal colonization. mBio 10:e02321-19. doi: 10.1128/mBio.02321-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto C, Espinosa M. 2003. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 49:281–285. doi: 10.1016/s0147-619x(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 29.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nallapareddy SR, Sillanpaa J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, Sullam PM, Murray BE. 2011. Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun 79:2911–2920. doi: 10.1128/IAI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen HV, Guiton PS, Kline KA, Port GC, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. 2012. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. mBio 3:e00177-12. doi: 10.1128/mBio.00177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. 2011. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun 79:2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonina I, Lim XN, Tan R, Kline KA. 2018. Planktonic interference and biofilm alliance between aggregation substance and endocarditis- and biofilm-associated pili in Enterococcus faecalis. J Bacteriol 200:e00361-18. doi: 10.1128/JB.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dale JL, Beckman KB, Willett JLE, Nilson JL, Palani NP, Baller JA, Hauge A, Gohl DM, Erickson R, Manias DA, Sadowsky MJ, Dunny GM. 2018. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3:e00062-18. doi: 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HL. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775. doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung HJ, Littmann ER, Seok R, Leiner IM, Taur Y, Peled J, van den Brink M, Ling L, Chen L, Kreiswirth BN, Goodman AL, Pamer EG. 2019. Genome-wide screening for enteric colonization factors in carbapenem-resistant ST258 Klebsiella pneumoniae. mBio 10:e02663-18. doi: 10.1128/mBio.02663-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shames SR, Liu L, Havey JC, Schofield WB, Goodman AL, Roy CR. 2017. Multiple Legionella pneumophila effector virulence phenotypes revealed through high-throughput analysis of targeted mutant libraries. Proc Natl Acad Sci U S A 114:E10446–E10454. doi: 10.1073/pnas.1708553114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, Pier GB. 2013. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog 9:e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiles TJ, Norton JP, Russell CW, Dalley BK, Fischer KF, Mulvey MA. 2013. Combining quantitative genetic footprinting and trait enrichment analysis to identify fitness determinants of a bacterial pathogen. PLoS Genet 9:e1003716. doi: 10.1371/journal.pgen.1003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Papa MF, Perego M. 2008. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol 190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roof DM, Roth JR. 1988. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol 170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO. 2014. Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor. Acta Crystallogr F Struct Biol Commun 70:1584–1590. doi: 10.1107/S2053230X1402158X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A 106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaval KG, Gebbie M, Goodson JR, Cruz MR, Winkler WC, Garsin DA. 2019. Ethanolamine utilization and bacterial microcompartment formation are subject to carbon catabolite repression. J Bacteriol 201:e00703-18. doi: 10.1128/JB.00703-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. 2016. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee A, Willett JLE, Nguyen UT, Monogue B, Palmer KL, Dunny GM, Duerkop BA. 2020. Parallel genomics uncover novel enterococcal-bacteriophage interactions. mBio 11:e03120-19. doi: 10.1128/mBio.03120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dezwaan DC, Mequio MJ, Littell JS, Allen JP, Rossbach S, Pybus V. 2007. Purification and characterization of enterocin 62–6, a two-peptide bacteriocin produced by a vaginal strain of Enterococcus faecium: potential significance in bacterial vaginosis. Microb Ecol Health Dis 19:241–250. doi: 10.1080/08910600701538240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelly MC, Mequio MJ, Pybus V. 2003. Inhibition of vaginal lactobacilli by a bacteriocin-like inhibitor produced by Enterococcus faecium 62–6: potential significance for bacterial vaginosis. Infect Dis Obstet Gynecol 11:147–156. doi: 10.1080/10647440300025513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tempera G, Abbadessa G, Bonfiglio G, Cammarata E, Cianci A, Corsello S, Raimondi A, Ettore G, Nicolosi D, Furneri PM. 2006. Topical kanamycin: an effective therapeutic option in aerobic vaginitis. J Chemother 18:409–414. doi: 10.1179/joc.2006.18.4.409. [DOI] [PubMed] [Google Scholar]
- 52.Banla LI, Pickrum AM, Hayward M, Kristich CJ, Salzman NH. 2019. Sortase-dependent proteins promote gastrointestinal colonization by enterococci. Infect Immun 87:e00853-18. doi: 10.1128/IAI.00853-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willett JLE, Ji MM, Dunny GM. 2019. Exploiting biofilm phenotypes for functional characterization of hypothetical genes in Enterococcus faecalis. NPJ Biofilms Microbiomes 5:23. doi: 10.1038/s41522-019-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balomenou S, Fouet A, Tzanodaskalaki M, Couture-Tosi E, Bouriotis V, Boneca IG. 2013. Distinct functions of polysaccharide deacetylases in cell shape, neutral polysaccharide synthesis and virulence of Bacillus anthracis. Mol Microbiol 87:867–883. doi: 10.1111/mmi.12137. [DOI] [PubMed] [Google Scholar]
- 55.Kaval KG, Singh KV, Cruz MR, DebRoy S, Winkler WC, Murray BE, Garsin DA. 2018. Loss of ethanolamine utilization in Enterococcus faecalis increases gastrointestinal tract colonization. mBio 9:e00790-18. doi: 10.1128/mBio.00790-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaval KG, Garsin DA. 2018. Ethanolamine utilization in bacteria. mBio 9:e00066-18. doi: 10.1128/mBio.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lengfelder I, Sava IG, Hansen JJ, Kleigrewe K, Herzog J, Neuhaus K, Hofmann T, Sartor RB, Haller D. 2019. Complex bacterial consortia reprogram the colitogenic activity of Enterococcus faecalis in a gnotobiotic mouse model of chronic immune-mediated colitis. Front Immunol 10:1420. doi: 10.3389/fimmu.2019.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, Huang X, Xiao Y, Yao S, Zhao Q, Liu Z, Cong Y. 2017. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol 10:946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burts ML, Williams WA, DeBord K, Missiakas DM. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A 102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jager F, Cargill JS, Chalmers J, van der Kooi-Pol MM, van Dijl JM, Ryan RP, Hunter WN, Palmer T. 2014. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol 93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohr RJ, Anderson M, Shi M, Schneewind O, Missiakas D. 2017. EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. J Bacteriol 199:e00528-16. doi: 10.1128/JB.00528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, Goo YA, Surette MG, Borenstein E, Vollmer W, Mougous JD. 2017. A broadly distributed toxin family mediates contact-dependent antagonism between Gram-positive bacteria. Elife 6:e26938. doi: 10.7554/eLife.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishii K, Adachi T, Yasukawa J, Suzuki Y, Hamamoto H, Sekimizu K. 2014. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect Immun 82:1500–1510. doi: 10.1128/IAI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez MS, Tan IS, Yan D, Kang J, McCreary M, Modrusan Z, Austin CD, Xu M, Brown EJ. 2017. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc Natl Acad Sci U S A 114:11223–11228. doi: 10.1073/pnas.1700627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tchoupa AK, Watkins KE, Jones RA, Kuroki A, Alam MT, Perrier S, Chen Y, Unnikrishnan M. 2020. The type VII secretion system protects Staphylococcus aureus against antimicrobial host fatty acids. bioRxiv doi: 10.1101/572172. [DOI] [PMC free article] [PubMed]
- 67.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shabbir MAB, Tang Y, Xu Z, Lin M, Cheng G, Dai M, Wang X, Liu Z, Yuan Z, Hao H. 2018. The involvement of the Cas9 gene in virulence of Campylobacter jejuni. Front Cell Infect Microbiol 8:285. doi: 10.3389/fcimb.2018.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer BL, Deng L, Patras KA, Burcham ZM, Sanches GF, Nagao PE, Doran KS. 2019. Cas9 contributes to group B streptococcal colonization and disease. Front Microbiol 10:1930. doi: 10.3389/fmicb.2019.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moore TC, Escalante-Semerena JC. 2016. The EutQ and EutP proteins are novel acetate kinases involved in ethanolamine catabolism: physiological implications for the function of the ethanolamine metabolosome in Salmonella enterica. Mol Microbiol 99:497–511. doi: 10.1111/mmi.13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banla IL, Kommineni S, Hayward M, Rodrigues M, Palmer KL, Salzman NH, Kristich CJ. 2017. Modulators of Enterococcus faecalis cell envelope integrity and antimicrobial resistance influence stable colonization of the mammalian gastrointestinal tract. Infect Immun 86:e00381-17. doi: 10.1128/IAI.00381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chatterjee A, Willett JLE, Dunny GM, Duerkop BA. 2020. Phage infection mediates inhibition of bystander bacteria. bioRxiv doi: 10.1101/2020.05.11.077669. [DOI] [PMC free article] [PubMed]
- 74.Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol 188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teixeira N, Varahan S, Gorman MJ, Palmer KL, Zaidman-Remy A, Yokohata R, Nakayama J, Hancock LE, Jacinto A, Gilmore MS, de Fátima Silva Lopes M. 2013. Drosophila host model reveals new enterococcus faecalis quorum-sensing associated virulence factors. PLoS One 8:e64740. doi: 10.1371/journal.pone.0064740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kellogg SL, Kristich CJ. 2016. Functional dissection of the CroRS two-component system required for resistance to cell wall stressors in Enterococcus faecalis. J Bacteriol 198:1326–1336. doi: 10.1128/JB.00995-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muller C, Le Breton Y, Morin T, Benachour A, Auffray Y, Rincé A. 2006. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J Bacteriol 188:2636–2645. doi: 10.1128/JB.188.7.2636-2645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Comenge Y, Quintiliani R Jr, Li L, Dubost L, Brouard JP, Hugonnet JE, Arthur M. 2003. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J Bacteriol 185:7184–7192. doi: 10.1128/jb.185.24.7184-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 81.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morgan DJ, Aboud CJ, McCaffrey IM, Bhide SA, Lamont RF, Taylor-Robinson D. 1996. Comparison of Gram-stained smears prepared from blind vaginal swabs with those obtained at speculum examination for the assessment of vaginal flora. Br J Obstet Gynaecol 103:1105–1108. doi: 10.1111/j.1471-0528.1996.tb09591.x. [DOI] [PubMed] [Google Scholar]
- 83.Deng L, Mu R, Weston TA, Spencer BL, Liles RP, Doran KS. 2018. Characterization of a two-component system transcriptional regulator, LtdR, that impacts group B streptococcal colonization and disease. Infect Immun 86:e00822-17. doi: 10.1128/IAI.00822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TnSeq and genomic DNA reads have been deposited at the European Nucleotide Archive under accession numbers PRJEB37929 and PRJEB39171, respectively.