Ehrlichia chaffeensis, a tick-transmitted obligate intracellular rickettsial agent, causes human monocytic ehrlichiosis. In recent reports, we described substantial advances in developing random and targeted gene disruption methods to investigate the functions of E. chaffeensis genes. We reported earlier that the Himar1 transposon-based random mutagenesis is a valuable tool in defining E. chaffeensis genes critical for its persistent growth in vivo in reservoir and incidental hosts.
KEYWORDS: Ehrlichia chaffeensis, mutagenesis, transposon, in vivo screening, tick-borne diseases, rickettsial, tick-borne pathogens
ABSTRACT
Ehrlichia chaffeensis, a tick-transmitted obligate intracellular rickettsial agent, causes human monocytic ehrlichiosis. In recent reports, we described substantial advances in developing random and targeted gene disruption methods to investigate the functions of E. chaffeensis genes. We reported earlier that the Himar1 transposon-based random mutagenesis is a valuable tool in defining E. chaffeensis genes critical for its persistent growth in vivo in reservoir and incidental hosts. The method also aided in extending studies focused on vaccine development and immunity. Here, we describe the generation and mapping of 55 new mutations. To define the critical nature of the bacterial genes, infection experiments were carried out in the canine host with pools of mutant organisms. Infection evaluation in the physiologically relevant host by molecular assays and by xenodiagnoses allowed the identification of many proteins critical for the pathogen’s persistent in vivo growth. Genes encoding proteins involved in biotin biosynthesis, protein synthesis and fatty acid biosynthesis, DNA repair, electron transfer, and a component of a multidrug resistance (MDR) efflux pump were concluded to be essential for the pathogen’s in vivo growth. Three known immunodominant membrane proteins, i.e., two 28-kDa outer membrane proteins (P28/OMP) and a 120-kDa surface protein, were also recognized as necessary for the pathogen’s obligate intracellular life cycle. The discovery of many E. chaffeensis proteins crucial for its continuous in vivo growth will serve as a major resource for investigations aimed at defining pathogenesis and developing novel therapeutics for this and related pathogens of the rickettsial family Anaplasmataceae.
INTRODUCTION
During the past 3 decades, rickettsial diseases caused by Anaplasmataceae family pathogens in the genera Ehrlichia and Anaplasma have emerged as a growing public health concern (1–8), and they are now considered the second leading human tick-borne diseases in the United States and many parts of the world. These diseases include human monocytic and granulocytic ehrlichiosis caused by Ehrlichia chaffeensis and Ehrlichia ewingii, respectively, which are transmitted by Amblyomma americanum, and human granulocytic anaplasmosis resulting from infection with Anaplasma phagocytophilum, which is transmitted by Ixodes species ticks. Recently, another Ixodes scapularis-borne pathogen, Ehrlichia muris subsp. eauclairensis, has been recognized as the causative agent for another ehrlichial disease in people (7, 8). Despite the complex life cycle involving tick vectors and vertebrate hosts, the rickettsial pathogens evolved strategies to evade clearance by both vertebrate and acarine hosts.
Like human beings, dogs acquire E. chaffeensis from infected A. americanum ticks (9). The limited availability of genetic tools to study obligate intracellular rickettsiae of the genera Ehrlichia, Anaplasma, Rickettsia, and Orientia is a major constraint for investigations focused on defining the functions of genes contributing to bacterial pathogenesis (10). In this context, genetic factors associated with persistent infections are of particular interest (11–13). Lack of well-established mutagenesis methods for rickettsial agents has remained a major impediment for advancing research to understand the functions of many uncharacterized bacterial genes. Recently, we reported the development of random and targeted mutagenesis methods for E. chaffeensis (14). We described targeted mutagenesis resulting in disruption of gene function, as well as in restoration of the function of a mutated gene (15). We further reported that Himar1 transposase-based random mutagenesis is efficient in creating mutations in both protein-coding and noncoding regions of the pathogen (14). We demonstrated that Himar1 mutagenesis is a valuable tool in elucidating host-pathogen interactions and in developing attenuated mutant vaccines (16–18). Similarly, random mutagenesis is described for other members of the alphaproteobacterial order Rickettsiales (19–24).
In the current study, we generated a random mutagenesis library for E. chaffeensis and mapped 55 insertion mutations. The transposon insertion mutants were utilized for in vivo screening experiments in a physiologically relevant canine host infection model. The study aided in identifying many essential genes and genomic regions of E. chaffeensis.
RESULTS
Transposon mutagenesis library of E. chaffeensis.
We previously reported the application of Himar1 transposon mutagenesis in creating mutations spanning 9 genomic regions of E. chaffeensis (14). The mutants served as a resource in mapping genes critical for the pathogen and in studies focused on developing a live attenuated vaccine (16, 17). In the current study, we extended the transposon mutagenesis by performing several independent mutational experiments using the ISE6 tick cell line and with three different mutagenesis constructs; all three constructs had the aadA gene to confer resistance to spectinomycin/streptomycin, while two constructs contained an mCherry expression cassette and the third construct had a green fluorescent protein (GFP) expression cassette (25–28). One of the two mCherry-expressing constructs (pHimar1 A7 loxP plasmid) also included Cre-loxP flanking sequences; it was generated by insertion into mismatched loxP sites flanking the transposon segment in pCis mCherry-SS Himar A7 (Fig. 1). The mutagenesis experiments with all three plasmids aided in the identification of many insertion mutations, as judged from Southern blot analysis of genomic DNAs recovered from the mutant organisms (Fig. 2). The DNA blot analysis was performed using genomic DNAs of the mutants digested with BglII, as the insertion sequence lacked the recognition sequence for this restriction enzyme. All three mutagenesis constructs performed similarly in generating mutants, with few mutations identified after each experiment. The majority of the mutants were clonally pure, with the exception of a few having a mix of two or more mutants (Fig. 2).
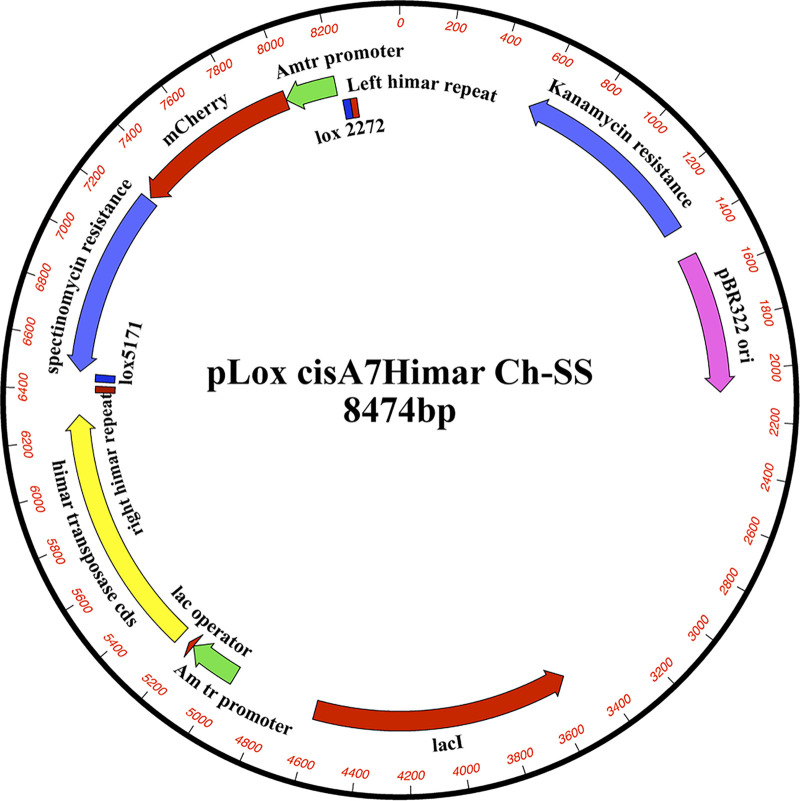
FIG 1.
Plasmid map of pHimar1 A7 loxP used for the electroporation to generate a subset of mutants in E. chaffeensis.
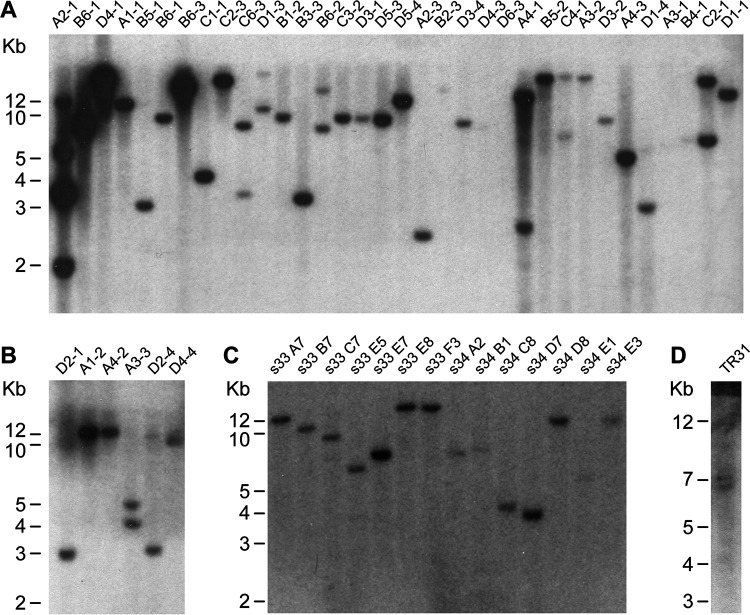
FIG 2.
Southern blot analysis of E. chaffeensis Himar1 mutants. Genomic DNA from Himar1 transposon E. chaffeensis mutants was assessed by DNA blot analysis using a spectinomycin resistance gene (aadA) probe. Panels A to D represent four DNA blot analysis experimental data generated four independent times to locate all insertion mutations within E. chaffeensis genome. Genomic DNAs from the cultured mutants were digested with BglII restriction enzyme (mutants’ codes are identified at the top). Genomic locations for the DNA fragments (identified in Fig. S1) were established by sequence analysis.
Mapping the genomic insertion sites.
To establish the identity of the mutant insertion sites, genome-walking PCRs and sequencing analyses were performed using the genomic DNAs recovered from the mutant organisms as templates. We mapped 55 transposon insertion sites to the E. chaffeensis genome from the mutant library (Fig. 3 and Tables 1 and 2), while the identity of a few insertion mutations remains to be defined (see Fig. S1 in the supplemental material). Including the previously reported 9 transposon mutations (14), the total number of insertion mutations in E. chaffeensis genome is 64 (Fig. 3). The mutation sites were distributed randomly throughout the E. chaffeensis genome, although we did not identify mutations in some major genomic segments spanning regions of about 25 to 50 kbp (Fig. 3). Furthermore, there were about equal numbers of insertions found within the open reading frames (ORFs) of genes (31 mutants) (Table 1) and in the intergenic spaces (24 mutants) (Table 2).
FIG 3.
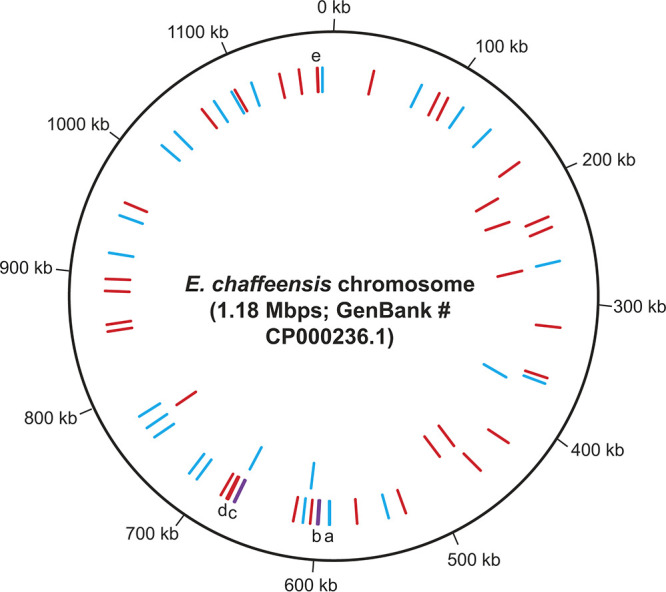
E. chaffeensis chromosomal map showing all 55 transposons insertion sites identified in the current study. The circular representation of E. chaffeensis chromosomal map was created using DNAplotter (79). The red lines indicate mutations within the coding regions of genes, and the blue lines refer to mutations within intergenic regions of genes. (The letters a, b, c, d, and e refer to the mutant lines where more than one insertion mutation is present at close proximities and they could not be separated in the image. Purple lines b and c are to represent the presence of intergenic and intragenic mutations at close vicinity.) The previously mapped 9 mutations (14) are also included (inner circle lines).
TABLE 1.
E. chaffeensis Himar1 transposon insertion mutants in ORFs (total, 31)
| Insertion cassette | Mutant no. | Mutant code | Gene no.a | Gene product | Genomic insertion locationa | Insertion orientationb | Insertion location in ORF/ORF length (bp) |
|---|---|---|---|---|---|---|---|
| mCherry loxP | 1 | A1-1 | ECH_0113 | Hypothetical protein | 99608 | + | 924/2,382 |
| 2 | A3-2 | ECH_0187 | Hypothetical protein | 176793 | − | 1154/1,692 | |
| 3 | D4-1 | ECH_0242 | Hypothetical protein | 226758 | + | 115/162 | |
| 4 | s33 E5 | ECH_0251 | Hypothetical protein | 236424 | + | 137/618 | |
| 5 | D3-2 | ECH_0368 | Dioxygenase family protein | 360362 | + | 243/675 | |
| 6 | s34 C8 | ECH_0445 | Queuine tRNA-ribosyltransferase | 423364 | + | 352/1,191 | |
| 7 | C1-1 | ECH_0475 | Signal recognition particle protein | 454669 | − | 1302/1,347 | |
| 8 | s33 C7 | ECH_0525 | Hypothetical protein | 525880 | − | 1037/2,001 | |
| 9 | s34 E3 | ECH_0592 | Coproporphyrinogen III oxidase, aerobic, truncation | 598900 | − | 83/147 | |
| 10 | B5-1 | ECH_0600 | Hypothetical protein | 606371 | + | 70/144 | |
| 11 | D3-1 | ECH_0614 | Hypothetical protein | 619640 | − | 334/696 | |
| 12 | B1-2 | ECH_0655 | RNA polymerase σ32 factor | 671099 | + | 869/894 | |
| 13 | B4-1 | ECH_0665 | Phage uncharacterized protein | 676091 | − | 907/1,410 | |
| 14 | C2-1 | ECH_0666 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | 678007 | − | 1182/1,281 | |
| 15 | A4-1 | ECH_0669 | 3-Oxoacyl-(acyl-carrier-protein) reductase | 683268 | − | 740/744 | |
| 16 | s34 A2 | ECH_0843 | Recombination protein RecR | 861495 | + | 581/588 | |
| 17 | s33 E8 | ECH_0878 | Hypothetical protein | 898815 | − | 456/1,230 | |
| 18 | C4-2 | ECH_1038 | Hypothetical protein | 1065224 | − | 5217/5,892 | |
| 19 | A1-2 | ECH_1067 | d-Alanyl-d-alanine carboxypeptidase family protein | 1095813 | + | 695/1,146 | |
| 20 | s34 D8 | ECH_1110 | Dethiobiotin synthetase | 1134587 | − | 98/693 | |
| 21 | C3-2 | ECH_1127 | Major outer membrane protein OMP-1V | 1150113 | + | 117/840 | |
| 22 | B6-1 | ECH_1144 | Major outer membrane protein P28-1/OMP-20 | 1165318 | − | 712/816 | |
| mCherry | 23 | C2-3 | ECH_0039 | 120-kDa immunodominant surface protein | 34759 | + | 1305/1,647 |
| 24 | A4-3 | ECH_0561 | AcrB/AcrD/AcrF family protein | 566002 | + | 1781/3,099 | |
| 25 | D4-3 | ECH_0837 | tRNA-i(6)A37 modification enzyme MiaB | 854867 | − | 56/1,329 | |
| 26 | C6-3 | ECH_0945 | Hypothetical protein | 964068 | + | 3741/4,050 | |
| 27 | TR31 | ECH_1144 | Major outer membrane protein P28-1/OMP-20 | 1165587 | + | 444/816 | |
| GFPuv | 28 | D1-4 | ECH_0104 | Hypothetical protein | 90734 | + | 89/126 |
| 29 | D5-4 | ECH_0329 | Hypothetical protein | 317141 | + | 649/684 | |
| 30 | D4-4 | ECH_0666 | Adenosylmethionine -8-amino-7-oxononanoate aminotransferase | 676872 | + | 47/1,281 | |
| 31 | D3-4 | ECH_0866 | Hypothetical protein | 888668 | + | 651/993 |
As per GenBank accession no. CP000236.
The “+” and “−” refer to an insertion mutation in the same orientation of the ORF and in the opposite orientation, respectively.
TABLE 2.
E. chaffeensis Himar1 transposon insertion mutants in intergenic region (total, 24)
|
Insertion cassette |
Mutant no. | Mutant code | Gene no.a | Gene product name | Genomic insertion location | Flanking gene orientationb | Mutation distances to flanking up-/downstream ORFs (bp) |
|---|---|---|---|---|---|---|---|
| mCherry loxP | 32 | B6-2 | ECH_0124/0125 | Citrate synthase I/glutamate-cysteine ligase | 113250 | → + → | 294/10 |
| 33 | A3-1 | ECH_0282/0283 | Hypothetical protein/hypothetical protein | 264190 | → + ← | 492/737 | |
| 34 | A2-1 | ECH_0372/0373 | Hypothetical protein/dihydroorotase | 364550 | → + ← | 356/159 | |
| 35 | s34 D7 | ECH_0537/0538 | Arginyl-tRNA synthetase/isoleucyl-tRNA synthetase | 539098 | → + ← | 78/34 | |
| 36 | s33 F3 | ECH_0579/0580 | Type IV secretion system protein VirB8/hypothetical protein | 589238 | → − ← | 46/424 | |
| 37 | s33 B7 | ECH_0593/0594 | Hypothetical protein/acetylglutamate kinase | 600196 | ← + ← | 21/248 | |
| 38 | D1-1 | ECH_0605/0606 | Glutamyl-tRNA synthetase/hypothetical protein | 611987 | → + → | 193/293 | |
| 39 | s34 B1 | ECH_0657/0658 | tRNA-Ser/hypothetical protein | 671821 | ← + ← | 120/26 | |
| 40 | s33 E7 | ECH_0750/0751 | DNA topoisomerase I/YjeF family protein | 756968 | → − → | 253/143 | |
| 41 | D2-1 | ECH_0769/0770 | Exopolysaccharide synthesis protein/hypothetical protein | 779062 | ← − ← | 414/59 | |
| 42 | B5-2 | ECH_0930/0931 | Putative BolA protein/pyridoxamine 5′-phosphate oxidase | 953314 | → + ← | 243/158 | |
| 43 | s34 E1 | ECH_1008/1009 | Preprotein translocase, YajC subunit/DNA polymerase III, β subunit | 1034973 | → + ← | 119/10 | |
| 44 | B6-2 | ECH_1044/1045 | Hypothetical protein/hypothetical protein | 1076550 | ← + ← | 1020/108 | |
| 45 | A4-2 | ECH_1065/1066 | 2-Oxoglutarate dehydrogenase/hexapeptide transferase family protein | 1094096 | ← + ← | 246/187 | |
| 46 | s33 A7 | ECH_1081/1082 | SURF1 family protein/hypothetical protein | 1109518 | ← + → | 103/10 | |
| 47 | D1-3 | ECH_0083/0084 | Hypothetical protein/hypothetical protein | 74911 | → − → | 83/386 | |
| 48 | B2-3 | ECH_0149/0150 | Pyruvate dehydrogenase subunit beta/hypothetical protein | 141187 | ← + ← | 476/15 | |
| 49 | B6-3 | ECH_0579/0580 | Type IV secretion system protein VirB8/hypothetical protein | 589575 | → − → | 383/87 | |
| mCherry | 50 | A3-3 | ECH_0699/0670 | Hypothetical protein/hypothetical protein | 708076 | → + → | 75/170 |
| 51 | A2-3 | ECH_0705/0706 | Peptide chain release factor 2/hypothetical protein | 716170 | → − ← | 63/49 | |
| 52 | D5-3 | ECH_0760/0761 | RNA polymerase sigma factor RpoD/DNA primase | 768120 | → + ← | 61/387 | |
| 53 | C6-3 | ECH_0894/0895 | Conserved domain protein/conserved hypothetical protein | 920356 | ← + ← | 133/133 | |
| 54 | D6-3 | ECH_0995/0996 | Hypothetical protein/ATP-dependent protease peptidase subunit | 1020175 | → − → | 26/667 | |
| 55 | B3-3 | ECH_1148/1149 | Hypothetical protein/preprotein translocase subunit SecA | 1169030 | → − → | 248/157 |
As per GenBank accession no. CP000236.
The “+” and “−” refer to insertion mutation in the forward orientation and reverse orientation, respectively.
Impact of mutations on the RNA expression for gene disruption mutations and for the genes flanking insertion sites.
To assess the impact of the transposon insertion mutations on gene expression, reverse transcription-PCR (RT-PCR) analysis was performed targeting genes with open reading frame disruptions and the flanking genes for the intergenic sequence mutations. Transcriptional inactivation was observed for all gene disruption mutations downstream from the mutation insertion sites. For the intergenic sequence mutations, transcripts were detected for all genes located upstream and downstream of insertion sites when tested by RT-PCR in the subset of mutations assessed.
In vitro growth defects assessed for the mutants.
As the mutagenesis experiments were executed in the ISE6 tick cell line, all identified mutants are considered to have no detrimental effect on E. chaffeensis growth in these cells. When we attempted to adapt the growth of the mutants to a macrophage-like cell line (DH82), four mutants displayed delayed growth compared to the wild-type E. chaffeensis, while the remaining mutants grew similarly to the wild type. The mutants with disruption of gene activities for ECH_0837, encoding the tRNA-i(6)A37 modification enzyme MiaB (a metal ion binding protein), and ECH_1144, encoding a member of the 28-kDa outer membrane protein (P28/OMP) gene family, P28-1/OMP-20, had significantly slower growth in the macrophage cell line (DH82) as assessed by comparing the growth of wild-type E. chaffeensis (Fig. 4). The mutants with insertions into ECH_1127 (the gene encoding another P28/OMP protein [OMP-1V/OMP-6]) and ECH_0039 (the gene encoding the 120-kDa immunodominant surface protein) had an initial lag phase in DH82 cell culture but recovered thereafter.
FIG 4.
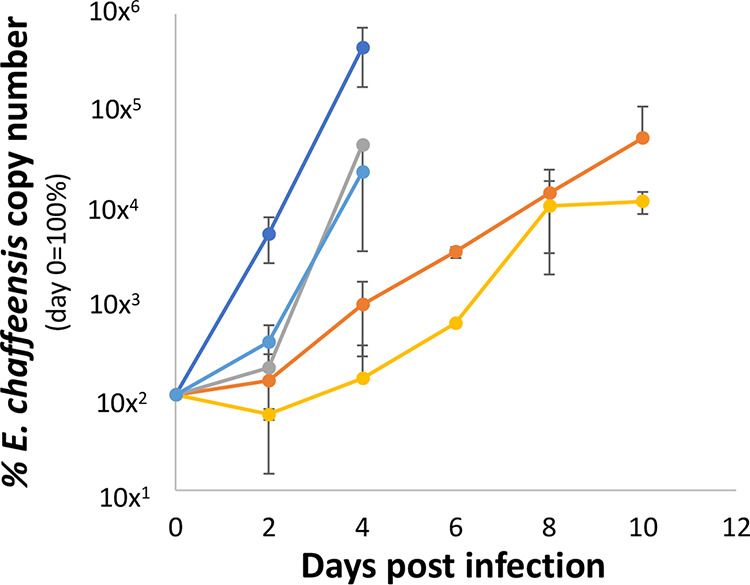
Culture assessment of four mutants having retarded growth in the canine macrophage cell line DH82. The growth of the mutants having insertion mutations in ECH_0039 (light blue line), ECH_0837 (yellow line), ECH_1127 (gray line), and ECH_1144 (orange line) is compared with that of wild-type E. chaffeensis (dark blue line). The number of bacteria at each time point postinoculation in DH82 was estimated by determining the copy numbers of the 16S rRNA gene. Each data point represents the average percentage of bacterial number relative to day 0 of inoculation from triplicate samples. The vertical bar represents standard deviation. P values were estimated by comparing the growth of the wild type with ECH_1127 (P = 0.06), ECH_0039 (P = 0.1), ECH_1144 (P = 0.04), and ECH_0837 (P = 0.04) on days 2 and 4.
Infection study in the canine host to define the importance of E. chaffeensis genes critical for the pathogen’s persistent growth.
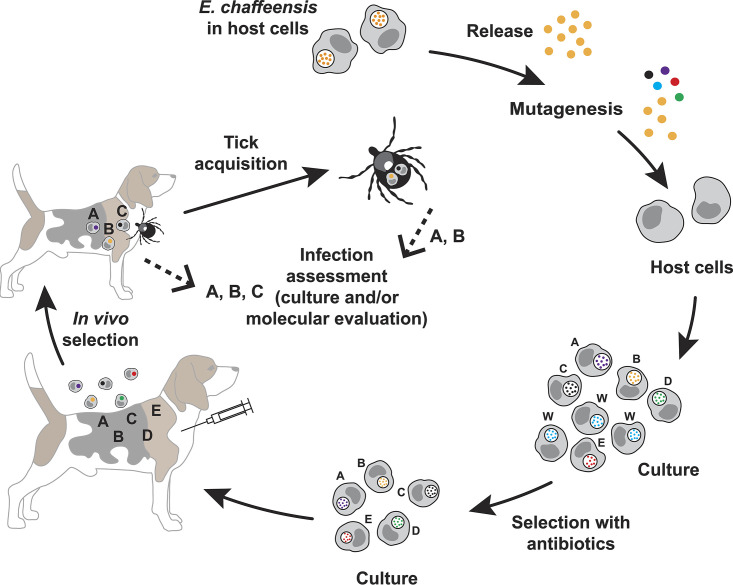
We investigated how mutations within both coding and noncoding regions impacted E. chaffeensis growth and persistence in a mammalian host and their acquisition from the host by A. americanum. A physiologically relevant canine host model was infected with pools of mutants and assessed for the presence and absence of mutants circulating in the blood of infected animals (Fig. 5). Xenodiagnosis was also performed using A. americanum ticks. This approach was similar to that described in our prior studies, which aided in mapping genes required for E. chaffeensis persistent growth (14). Similar methods have been employed in defining virulence-associated proteins for several other pathogenic bacteria (29–31). In the current study, we followed the same strategy as in our prior investigations of infection assessment of E. chaffeensis mutants (14). We used three beagle dogs per pool of randomly selected mutants; about equal numbers of mutants with gene disruption mutations and those with insertions into intergenic spaces were used in each pool. A total of 51 mutants were tested in 6 infection pools; 2 pools each containing 8 and 9 mutants and 1 pool each with 7 and 10, respectively. (Mutant numbers 2, 9, 36, and 44 [listed in Tables 1 and 2] were not part of the infection experiment.) In vitro cultures of mutants were mixed in each pool with approximately equal numbers of mutant organisms and used as infection inocula. Blood was sampled twice a week for 8 weeks from all dogs and used to assess infection by aadA-specific PCR and by performing insertion region-specific PCR analysis to detect mutants. Similarly, several tissue samples were assessed at the terminal point of the study (after 8 weeks). To assess if the mutants persisting in the canine host were acquired by a tick host (A. americanum), flat nymphal ticks were allowed to acquisition feed to repletion on dogs starting from day 5 postinfection, which lasted until about day 12. Following molting, adult ticks of both sexes were randomly selected and evaluated individually for the presence of mutants by performing insertion-specific and aadA gene-specific PCR assays to detect the mutants acquired by A. americanum (Table 3).
FIG 5.
Schematic representation of E. chaffeensis transposon mutants assessed to identify genes important for the pathogen’s in vivo growth. The method involves recovering ISE6 culture-derived E. chaffeensis organisms, subjecting them to transposon mutagenesis to identify mutants in cultures resistant to antibiotic clearance, infecting the canine host, and acquisition feeding assessment of A. americanum ticks.
TABLE 3.
Mutants that persisted in the canine host
| Mutant no. | Mutant code | Gene no. | Protein identifier | Dogs | No. of ticks positive/no. tested |
|---|---|---|---|---|---|
| 12 | B1-2a | ECH_0655 | RNA polymerase σ32 factor | + − − | 0/10 |
| 13 | B4-1 | ECH_0665 | Phage uncharacterized protein | + + + | 2/10 |
| 17 | S33 E8 | ECH_0878 | Hypothetical protein | + + − | 7/17 |
| 18 | C4-1a | ECH_1038 | Hypothetical protein | + − − | 0/10 |
| 19 | A1-2 | ECH_1067 | d-Alanyl-d-alanine carboxypeptidase family protein | + + + | 3/10 |
| 26 | C6-3 | ECH_0945 | Hypothetical protein | + + + | 0/17 |
| 34 | A2-1 | ECH_0372/0373 | Hypothetical protein/dihydroorotase | + + + | 4/17 |
| 38 | D1-1a | ECH_0605/0606 | Glutamyl-tRNA synthetase/hypothetical protein | + + + | 0/10 |
| 40 | s33 E7 | ECH_0750/0751 | DNA topoisomerase I/YjeF family protein | + − − | 2/17 |
| 45 | A4-2 | ECH_1065/1066 | 2-Oxoglutarate dehydrogenase/hexapeptide transferase family protein | + + + | 5/10 |
| 48 | B2-3a | ECH_0149/0150 | Pyruvate dehydrogenase subunit beta/hypothetical protein | + + − | 0/10 |
| 53 | C6-3a | ECH_0894/0895 | Conserved domain protein/conserved hypothetical protein | + + + | 0/10 |
| 55 | B3-3 | ECH_1148/1149 | Hypothetical protein/preprotein translocase subunit SecA | + + + | 1/10 |
Tested negative by xenodiagnosis.
All dogs tested positive for the aadA gene several times throughout the study period in blood and also in several tissue samples at the study’s terminal time point, suggesting the persistence of one or more mutant organisms used in each pool. PCR analysis targeting specific mutants using the respective mutant insertion-specific PCRs resulted in the identification of only a subset of mutants in each pool. If a mutant was detected in blood after a week for at least one of the three dogs and/or in one or more tissue samples, then the respective insertion mutation was considered to have minimal impact on the persistence of E. chaffeensis growth (Table 3). If a mutant was not detected in dogs any time during the 8-week study period and was also negative in tissue samples, then the mutation was regarded as detrimental for the pathogen’s persistence in the canine host. Thirteen mutants persisted in the canine host, which included 6 gene disruption mutations and 7 mutations within the intergenic spaces (Table 3). The persisting mutants included three each with gene disruptions in hypothetical protein genes and in genes with predicted annotated protein names. The only notable gene disruption mutant organism that persisted was with a mutation in the gene for RNA polymerase sigma factor σ32 (RpoH) (ECH_0655). Insertion in this gene was located near the 3′ end of the open reading frame, 26 nucleotides upstream from the stop codon (Table 1). Eight of the 13 persistent mutants were also detected in ticks allowed to feed on the dogs (Table 3). Of the five persistent mutants which tested negative by xenodiagnosis, two were gene disruptions; one each in a hypothetical protein gene (ECH_1038) and in the RpoH gene and the remaining three are located within the intergenic spacers (mutant numbers 38, 48, and 53) (Table 3).
There were significantly more rapidly cleared mutant organisms than those that persisted (Table 3). The rapidly cleared mutants included 23 with gene disruption mutations (Table 4) and 15 with insertion mutations in noncoding regions (Table 5). The gene disruption mutations included mutations in genes encoding 9 hypothetical proteins, an oxidoreductase family protein, two proteins each involved in the protein synthesis machinery and biosynthesis of biotin, one protein each with a role in fatty acid metabolism, a multidrug resistance (MDR) efflux pump protein, a DNA repair protein, three immunodominant outer membrane proteins (two belonging to the P28/OMP protein family and one representing the 120-kDa immunodominant surface protein), and a metal ion binding protein (tRNA-i[6]A37 modification enzyme protein [MiaB]). Several intergenic spacer mutations were also identified with insertion mutations near genes likely engaged as virulence determinants. They included genes encoding various synthases and proteins involved in transcription and translation. Three mutants tested positive by xenodiagnosis, while they were undetected in the canine host. They included one gene disruption mutation in a hypothetical protein gene (ECH_0113) and two within the noncoding regions (mutants 46 and 47) (Table 4). It is likely that these mutants may have circulated in the canine host at a very low level.
TABLE 4.
E. chaffeensis gene disruption mutants cleared from the canine host
| Mutant no. | Mutant code | Gene no. | Protein identifier |
|---|---|---|---|
| 1 | A1-1a | ECH_0113 | Hypothetical protein |
| 3 | D4-1 | ECH_0242 | Hypothetical protein |
| 4 | s33 E5 | ECH_0251 | Hypothetical protein |
| 5 | D3-2 | ECH_0368 | Dioxygenase family protein (oxidoreductase) |
| 6 | s34 C8 | ECH_0445 | Queuine tRNA-ribosyltransferase (protein synthesis) |
| 7 | C1-1 | ECH_0475 | Signal recognition particle protein (protein synthesis) |
| 8 | s33 C7 | ECH_0525 | Hypothetical protein |
| 10 | B5-1 | ECH_0600 | Hypothetical protein |
| 11 | D3-1 | ECH_0614 | Hypothetical protein |
| 14 | C2-1 | ECH_0666 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase (biotin biosynthesis) |
| 15 | A4-1 | ECH_0669 | 3-Oxoacyl-(acyl-carrier-protein) reductase (fatty acid biosynthesis) |
| 16 | s34 A2 | ECH_0843 | Recombination protein RecR (DNA repair) |
| 20 | s34 D8 | ECH_1110 | Dethiobiotin synthetase (biotin biosynthesis) |
| 21 | C3-2 | ECH_1127 | Major outer membrane protein OMP-1V/OMP-6 (immunogenic outer membrane protein) |
| 22 | B6-1 | ECH_1144 | Major outer membrane protein P28-1/OMP-20 (immunogenic outer membrane protein) |
| 23 | C2-3 | ECH_0039 | 120-kDa immunodominant surface protein (immunogenic outer membrane protein) |
| 24 | A4-3 | ECH_0561 | AcrB/AcrD/AcrF family protein (MDR efflux protein) |
| 25 | D4-3 | ECH_0837 | tRNA-i(6)A37 modification enzyme MiaB (metal ion binding/tRNA modification) |
| 27 | TR31 | ECH_1144 | Major outer membrane protein P28-1/OMP-20 (immunogenic outer membrane protein) |
| 28 | D1-4 | ECH_0104 | Hypothetical protein |
| 29 | D5-4 | ECH_0329 | Hypothetical protein |
| 30 | D4-4b | ECH_0666 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase (biotin biosynthesis) |
| 31 | D3-4 | ECH_0866 | Hypothetical protein |
Tested positive by xenodiagnosis.
D4-4 tested positive only in tissue samples of two animals on day 56.
TABLE 5.
E. chaffeensis intergenic mutants cleared from the canine host
| Mutant no. | Mutant code | Gene no. | Protein identifier |
|---|---|---|---|
| 32 | B6-2 | ECH_0124/0125 | Citrate synthase I/glutamate-cysteine ligase |
| 33 | A3-1 | ECH_0282/0283 | Hypothetical protein/hypothetical protein |
| 35 | s34 D7 | ECH_0537/0538 | Arginyl-tRNA synthetase/isoleucyl-tRNA synthetase |
| 37 | s33 B7 | ECH_0593/0594 | Hypothetical protein/acetylglutamate kinase |
| 39 | s34 B1 | ECH_0657/0658 | tRNA-Ser/hypothetical protein |
| 41 | D2-1 | ECH_0769/0770 | Exopolysaccharide synthesis protein/hypothetical protein |
| 42 | B5-2 | ECH_0930/0931 | Putative BolA protein/pyridoxamine 5′-phosphate oxidase |
| 43 | s34 E1 | ECH_1008/1009 | Preprotein translocase, YajC subunit/DNA polymerase III, β subunit |
| 46 | s33 A7a | ECH_1081/1082 | SURF1 family protein/hypothetical protein |
| 47 | D1-3 | ECH_0083/0084 | Hypothetical protein/hypothetical protein |
| 49 | B6-3a | ECH_0579/0580 | Type IV secretion system protein VirB8/hypothetical protein |
| 50 | A3-3 | ECH_0699/0670 | Hypothetical protein/hypothetical protein |
| 51 | A2-3 | ECH_0705/0706 | Peptide chain release factor 2/hypothetical protein |
| 52 | D5-3 | ECH_0760/0761 | RNA polymerase sigma factor RpoD/DNA primase |
| 54 | D6-3 | ECH_0995/0996 | Hypothetical protein/ATP-dependent protease peptidase subunit |
Tested positive by xenodiagnosis.
DISCUSSION
E. chaffeensis is among several tick-transmitted rickettsial bacteria responsible for causing zoonosis in people. Despite several recent advances in performing molecular genetic studies, only limited progress is documented in understanding pathogenesis and in identifying rickettsial proteins posited to contribute to bacterial virulence and host-pathogen interactions and those involved in supporting the immune evasion mechanisms (14, 16, 20, 23, 24, 32–34). One of the challenges is that the pathogenic organisms are difficult to grow in axenic culture media (35). Furthermore, the mutational success rate remains extremely limited even with Himar1 mutagenesis plasmids, possibly because Ehrlichia and Anaplasma species are not known to naturally harbor extrachromosomal plasmids. For example, several attempts are required to generate a moderately sized mutant library for Ehrlichia and Anaplasma species, and often none to only a few mutant organisms are recovered following a typical mutational experiment (14, 19, 23, 24). We previously reported 9 random mutations within the E. chaffeensis genome in our prior study (14). Subsequent research has focused on detailed characterization of three mutant E. chaffeensis organisms (16, 17, 36, 37). These studies aided in the identification of one mutation in the ECH_0660 gene causing the rapid clearance of the pathogen and inducing a sufficient host immune response to serve as a live attenuated vaccine to protect against wild-type infection challenge by intravenous (i.v.) injection, as well as by tick transmission (16, 17). Mutagenized E. chaffeensis organisms are also valuable in studies focused on understanding the host response against the pathogen (17, 18).
In this study, we carried out investigations in generating a mutant library consisting of 55 mapped insertion mutations within the E. chaffeensis genome. We used three different Himar1 mutagenesis constructs for generating the mutational library. Further, several independent mutagenesis experiments were performed to generate more mutations in the E. chaffeensis genome. While all constructs worked similarly in producing a low mutation rate, there was no bias observed toward generating gene open reading frame disruption mutations or intergenic mutations, as nearly equal numbers of intergenic and intragenic mutations were observed. Our previously well-defined physiologically relevant canine infection model, molecular assessment, and xenodiagnosis methods (14–17, 38, 39) were valuable in the current study to identify many E. chaffeensis gene sequences associated with persistent infection of dogs. Of the 29 gene open reading frame disruption mutations tested, only 6 mutants (21%) persisted in the host, while mutations in the remaining 23 mutants (79%) were rapidly cleared. These data suggest that disruptions in the majority of the pathogen genes can be detrimental to the pathogen’s persistence in vivo and that the disrupted genes causing the rapid clearance are among the many essential genes of E. chaffeensis. Nine of the 23 genes identified as critical for E. chaffeensis persistent growth encode hypothetical proteins. The list of essential genes included two coding for proteins known to be involved in protein synthesis machinery, three coding for outer membrane-expressed immunogenic proteins, two genes belonging to biotin biosynthesis, and one each representing the DNA repair machinery, fatty acid biosynthesis, MDR efflux pump, and an oxidoreductase (dioxygenase family protein; ECH_0368). Rapid clearance of E. chaffeensis from the canine host resulting from the gene function disruption mutations suggests that the pathogen has retained many important pathway genes to support its obligate parasitic lifestyle.
Outer membranes of Gram-negative bacteria contain many proteins that perform essential functions, such as for the nutrient uptake mediated by porin activities, cell adhesion, cell signaling, and waste export, as well as to support the evasion of host defense mechanisms by pathogenic bacteria (40). Several P28 outer membrane proteins (P28/OMP) encoded from a multigene locus are identified as immunodominant proteins in E. chaffeensis (41–47). Similarly, the 120-kDa surface protein (also known as TRP 120) is recognized as an immunodominant protein of E. chaffeensis (41–47). Mutations reported in the current study included two P28/OMP genes (p28-1/OMP-20 and OMP-1v/OMP-6) and TRP 120. Disruption mutations in these three immunogenic outer membrane protein genes also caused in vitro growth defects in DH82 cultures, suggesting that the membrane proteins are essential for the pathogen’s replication in macrophages both in vitro and in vivo. The discovery of the two P28/OMP proteins and TRP 120 as essential for E. chaffeensis persistent growth is consistent with the prior studies demonstrating the importance of these immunogenic outer membrane-associated proteins for the pathogen’s replication in macrophages in vitro and in vivo. Indeed, two E. chaffeensis P28/OMPs (P28/OMP-19 and OMP-1F/OMP-18) have been reported to possess porin-like structures and porin activities (48). Further to this, two independent studies demonstrated that antibodies targeting P28/OMP-19 can block the infection progression in vivo and in vitro (41, 49). TRP 120 has been extensively investigated by the McBride group for its multiple roles, with recent evidence pointing to the protein being likely essential for the continued replication of E. chaffeensis in phagosomes (47). E. chaffeensis TRP 120 is expressed on the cell surface as well as a type 1 secretory system-mediated translocated effector having several defined functions, such as being engaged in the pathogen’s host cell entry (46), as a host cell nuclear translocation to serve as a nucleomodulin in regulating gene expression associated with signal transduction and apoptosis (50), and in interacting within the host cytoplasm as a moonlighting effector, a ubiquitin ligase targeting host nuclear proteins (47).
We expected that that the disruption mutation in the RpoH gene encoding the RNA polymerase sigma factor (σ32) would be detrimental for E. chaffeensis, but this mutant persisted, although it was detected less frequently and tested negative by xenodiagnosis. Two possibilities for this outcome are that (i) the E. chaffeensis σ70 protein may have complemented the function of σ32 in the mutant and (ii) a modified but functional version of the protein lacking the last 8 amino acids is formed. The first hypothesis is supported by our previous study demonstrating that E. chaffeensis gene promoters can be recognized by RNA polymerase holoenzyme containing either σ32 or σ70 in initiating transcription from a gene (51). The mutant tested positive for the RpoH transcript when assessed by RT-PCR targeting the region upstream of the mutation insertion site (data not shown). Thus, assuming that the truncated version of the transcript is translated, it is also highly likely that the mutated version of the protein is functionally active.
The current study is the first to demonstrate that mutations in two different biotin pathway enzyme genes have similar impacts on rapidly clearing a rickettsial pathogen from the vertebrate host. The E. chaffeensis genome includes the biotin biosynthesis pathway protein genes (52–54). Similarly, biotin pathway genes are conserved in other related rickettsiae (53, 55). A recent study suggested that E. chaffeensis biotin pathway genes are functionally active, as judged from experiments performed using the Escherichia coli complementation system (52). Biotin is an essential cofactor for several key metabolic pathways in bacteria, such as fatty acid biosynthesis and amino acid metabolism (56, 57). Indeed, many microorganisms synthesize biotin de novo (56, 58–60). Considering the absence of biotin synthesis machinery in mammals (56, 61), E. chaffeensis and other related Rickettsiales may have evolved to maintain their own functional biotin synthesis pathway. The biotin pathway enzymes are also attractive targets in generating antibacterial inhibitors (62, 63). Considering the availability of only one class of drugs (tetracycline derivatives) to treat rickettsial infections (64, 65) and that doxycycline-treated patients may remain persistently infected with Ehrlichia species (66, 67), studies may be extended in developing novel drugs targeting the biotin synthesis pathway.
We also discovered that the mutation in a fatty acid biosynthesis gene, 3-oxoacyl-(acyl-carrier-protein) reductase gene, results in E. chaffeensis rapid clearance from the vertebrate host. Similarly, MDR efflux pump, DNA repair, and protein synthesis pathway proteins are among the proteins essential for E. chaffeensis persistent growth in vivo. The efflux pump is known to play a critical role in conferring resistance to antibiotics in several bacteria (68–70). Efflux pump proteins are, therefore, commonly known as a requirement for bacterial virulence and also serve as an attractive target for designing novel therapeutics (68–70). It is not yet defined if E. chaffeensis has an active efflux pump to eliminate antibiotic accumulation from its cytoplasm. Reflecting on the essential nature of the identified efflux pump protein of the pathogen, it is highly likely that the human monocytic ehrlichiosis agent uses this protein for the benefit of clearing host defense proteins from its cytoplasmic space.
As with gene disruption mutants, only about one-third of intergenic spacer mutations (7 of 22) were identified as nonessential, while the majority (15 of 22) were among the rapidly cleared mutants. This is a surprising outcome, as the mutations did not appear to impact the transcription of the genes upstream and downstream from the insertion sites. However, in our prior studies, we reported that intergenic mutations can cause polar effects in altering the gene expression from genes located proximal to the mutation insertion sites (38). Interestingly, mutations in genomic regions found to be essential for the pathogen’s in vivo growth included several genes upstream and downstream of the insertion sites coding for proteins likely involved in the protein synthesis machinery and DNA replication and transcription and type IV secretion system-associated proteins. While it is unclear how the intergenic mutations impact the expression of genes proximal to insertion mutations, it is conceivable that the mutations can alter gene expression and potentially render the transcripts less stable, thus interfering with protein synthesis. Also, we cannot rule out the possibility that mutations within intergenic spacers may have impacted gene expression by disrupting the normal function of regulatory elements, including those involving the contributions of microRNAs. Several recent studies described the existence of microRNAs in Rickettsiales having a functional role in bacterial gene regulation (71, 72).
Five mutant organisms persisted in the canine host, while they were undetectable by xenodiagnoses in ticks. It is likely that the mutations caused defective growth of E. chaffeensis in its tick vector. Similarly, several genes/genomic regions found to be critical for the pathogen persistence in the canine host may also be critical for its replication in the tick host. This hypothesis remains to be tested. We recently described a needle inoculation method of infecting ticks, which bypasses the need for tick infection acquisition from a vertebrate host (39). This method will be valuable in determining which genomic regions of the pathogen are critical for A. americanum to harbor E. chaffeensis infection.
The current study demonstrates that Himar1 mutagenesis and in vivo screening methods using a physiologically relevant incidental host are ideally suited for mapping many essential bacterial proteins associated with E. chaffeensis virulence and persistent growth. The discovery of many genes essential for the continuous in vivo growth of E. chaffeensis opens the path for studies to define pathogenesis and develop novel therapeutics targeting critical pathways of the organism and also to extend such studies to other important Ehrlichia and Anaplasma pathogens impacting human and animal health.
MATERIALS AND METHODS
E. chaffeensis in vitro cultivation.
Wild-type E. chaffeensis isolate Arkansas and the mutated organisms were continuously cultivated in an Ixodes scapularis cell line (ISE6) at 34°C in the absence of CO2 (73). Where applicable, the organisms were also cultivated in a canine macrophage cell line (DH82) at 37°C with 5% CO2 as described earlier (74, 75).
Generation of E. chaffeensis transposon mutant library.
Three different plasmid constructs encoding the Himar1 transposase, antibiotic resistance conferred by aadA, and a fluorescent protein (mCherry or GFP) driven by the Anaplasma marginale Am-tr promoter were used for mutagenesis of E. chaffeensis: (i) pCis mCherry-SS Himar A7 containing the mCherry and aadA genes (14, 25), (ii) pHimar1 A7 loxP plasmid containing the mCherry and aadA genes flanked by loxP sites, and (iii) pCis GFPuv-SS Himar A7 containing the gfpuv and aadA genes (14, 27). The pHimar1 A7 loxP plasmid was generated by inserting mismatched loxP sites (76, 77) flanking the transposon segment into pCis mCherry-SS Himar A7. Host cell-free E. chaffeensis suspensions recovered from ISE6 tick cell cultures were subjected to mutagenesis by following the protocol we described earlier (14, 15). Briefly, a 5-ml culture of E. chaffeensis-infected ISE6 cells was transferred to microcentrifuge tubes containing 0.2 ml of silicon carbide (no. 1 coarse rock tumbling grit; Loretone Inc., Mukilteo, WA) and vortexed for 30 s at high speed. The supernatant was passed through a 2-μm-pore-size filter (Whatman Ltd., Piscataway, NJ), and bacteria were collected by centrifugation at 11,000 × g at 4°C for 5 min. Bacteria were washed twice in 0.3 M sucrose and kept on ice between washes. Aliquots of purified E. chaffeensis (∼5 × 108) were resuspended in 50 μl of cold 0.3 M sucrose containing 1 μg of plasmid DNA, transferred to a 1-mm-gap electroporation cuvette, and incubated on ice for 15 min (19). (Plasmid DNAs were prepared using a Maxiprep plasmid DNA isolation kit by following the manufacturer’s instructions [Qiagen, Valencia, CA.]) E. chaffeensis organisms were electroporated at 2,000 V, 25 μF, and 400 Ω. The mixture was then combined with 0.5 ml of fetal bovine serum and 1 ml of ISE6 cell suspension containing about 1 × 106 cells. The sample was centrifuged at 5,000 × g for 5 min, incubated at room temperature for 15 min, and then mixed with ISE6 cells from a confluent culture (∼1 × 107 cells). Cells were then seeded into all wells of a 48-well plate, incubated at 30°C overnight, and then transferred to a 34°C incubator for ISE6 cells for the continuous growth of the organisms. After 48 h, 100 μg/ml each of spectinomycin and streptomycin was added to the culture medium to select mutants. The culture medium containing antibiotics was replaced once a week. When infectivity reached 80% or higher, cell-free Ehrlichia was prepared for inoculating a new flask of uninfected host cells with medium containing antibiotics. This procedure was repeated until all wild-type bacteria were eliminated. The presence of insertion mutations was monitored for 60 days or longer.
Southern blot analysis to identify mutations in E. chaffeensis.
Genomic DNA from the transformant cultures was isolated using a genomic DNA isolation kit as per manufacturer instructions (Qiagen, Valencia, CA). About 100 ng of genomic DNA recovered from cultured organisms was digested with BglII restriction enzyme for 2 to 3 h at 37°C. The digested DNA samples were resolved on a 0.9% agarose gel for about 6 h at 60 V and transferred to a nylon membrane. Blots were then hybridized with a 32P-labeled aadA gene probe at 68°C overnight, followed by washing steps to identify specific DNA-probe interactions as per our previously described protocol (14). The hybridized membranes were exposed to X-ray film to observe radioactive signals emitting from hybridized blots.
Mapping and verification of transposon insertions.
The genomic locations of the insertions within the mutated bacteria were mapped with the help of a Universal Genome Walker 2.0 kit (Clontech Laboratories). SspI restriction enzyme was used for DNA fragmentation and for genomic library construction. Inserted fragment-specific primers for PCR amplifications were GSP1 and GSP2. A third primer, GSP3, was used to sequence the PCR products (primers are listed in Table S1). Sequence data were then subjected to BLAST search analysis to localize the insertion sites within the E. chaffeensis genome (GenBank accession no. CP000236.1). Subsequently, inserted fragment-specific PCRs were performed using primers targeting genomic regions either 5′ or 3′ to insertion sites and to an inserted fragment-specific sequence. The primers targeting the genomic region of each mutant are listed in Table S1; the inserted fragment-specific primers are Amtr R1 (RG92), mCherry R1 (RG97), and aadA F1 (RG1202). The expected PCR product sizes are indicated in Table S1.
Transcriptional analysis to assess the impact of mutations.
Total RNAs from E. chaffeensis mutants grown in ISE6 cell cultures were isolated using the Tri-reagent RNA isolation method as per the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO). Total RNA was treated with RQ1 DNase at 37°C for 60 min to remove any genomic DNA contamination. Primers targeting each insertion-specific mutation were designed for use in RT-PCR analysis. For each set of RT-PCRs, controls included reactions without reverse transcriptase, reactions with genomic DNA as a template, or reactions with no DNA or RNA added. For verifying RNA expression, the presence or absence of specific products in the assays containing RNA with reverse transcriptase was assessed and compared with the products generated from genomic DNA-positive controls. Similarly, RT-PCR assays were performed to assess transcriptional changes from genes upstream and downstream from insertion sites for the insertion mutations located in the intergenic spaces. (All RT-PCR primers are listed in Table S1.)
In vitro growth analysis of mutants in the canine macrophage cell line DH82.
To assess the effect of different gene mutations on E. chaffeensis growth in macrophage cells, we attempted to regrow mutants in the canine macrophage cell line DH82, as described earlier (14). Nearly all mutants could be cultured in DH82 cells and exhibited growth patterns similar to that of wild-type E. chaffeensis, except for a few mutant organisms. The growth of mutants having insertions in certain genes was considerably retarded in DH82 cells. To further assess the growth variations of the slow-growing mutants compared to the wild type, cell-free bacteria recovered from about 90% infected 25-cm2 flasks from slow-growing mutants and wild-type E. chaffeensis were recovered from ISE6 cell cultures and used to assess their growth in DH82 cells. Briefly, bacteria from infected ISE6 cells were recovered by repeated passing the cultures through a bent 27-gauge needle and then filtered using a 2-μm filter. Cell-free bacteria from the filtrate were then recovered following centrifugation at 10,000 × g for 5 min. The pellets were resuspended in 3 ml each of culture medium. About 100 μl of the cell-free bacteria was then used to infect confluent DH82 monolayers in 12-well plates in triplicates for each mutant. After 24 h, the monolayers were washed to remove any cell-free bacteria. The cultures were monitored for up to 12 days. At 2-day intervals, the cultures were recovered from the individual triplicate wells representing each mutant or the wild type, and DNAs were purified and then assessed for bacterial growth by quantitative PCR. Bacterial numbers were estimated by real-time quantitative PCR targeting the16S rRNA gene segment, as we described previously (78). This experiment was performed three independent times and using data collected from triplicate well samples each time.
Dog infections with pools of E. chaffeensis mutants.
Animal experiments with dogs were performed in compliance with the Public Health Service (PHS) Policy on the Humane Care and Use of Laboratory Animals (https://olaw.nih.gov/policies-laws/phs-policy.htm), the U.S. Department of Agriculture’s (USDA) Animal Welfare Act & Regulations, and with the prior approval of the university Institutional Animal Care and Use Committee (IACUC). At the end of each experiment, all animals were euthanized in accordance with the IACUC recommendations, which are consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
About 6-month-old female beagle dogs were obtained from a USDA-certified commercial breeder. Dogs were housed indoors at a climate-controlled animal facility at Kansas State University and ad libitum feed and water were provided. All dogs were placed in individual housing pens with adequate space to allow regular exercise/activity. In addition, all dogs were permitted to socialize in groups several times each day. The animals were also monitored daily for health and behavioral changes and twice weekly for body temperature and hematological changes during the study period. Veterinary care for the animals was overseen by a university veterinarian.
E. chaffeensis mutants grown in ISE6 cultures to about 80 to 90% infection in T75 flasks were harvested by centrifugation at 15,000 × g for 10 min at 4°C, supernatants were discarded, and the cultures were resuspended in 15 ml of 1× phosphate-buffered saline (PBS). The washing steps were repeated twice, and the final cell pellet was suspended to concentrate the infected ISE6 cells to about 2 × 106 per ml, yielding an estimated concentration of Ehrlichia organisms of ∼2 × 108 per ml. Equal volumes of the culture suspensions of randomly selected mutants were mixed for preparing mutant pools having equal ratios of the mutants in each pool. One milliliter of each mutant pool per dog was inoculated by i.v. injection.
Evaluation of canine blood samples over time for the presence of mutants.
About 2 ml of blood was recovered from all dogs into sterile EDTA tubes on day 0 (prior to infection) and twice a week starting from the day 3 postinfection and until the end of 8 weeks. The blood samples were used immediately or stored at 4°C until use (maximum of 1 day). The samples were centrifuged at 3,000 rpm in a Clay Adams Sero-fuge (Becton, Dickinson, Sparks, MD) for 5 min, and buffy coats were transferred to a 15-ml sterile Falcon centrifuge tube containing 10 ml of erythrocyte (RBC) lysis buffer (155 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA) and mixed several times until complete lysis of erythrocytes. The samples were then centrifuged at 5,000 × g for 5 min. The buffy coat pellet from each sample was mixed in 300 μl of 1× PBS. One-hundred-microliter volumes of the buffy coats recovered from blood samples were used to recover total genomic DNA using the DNeasy blood and tissue kit (Qiagen, Germantown, MD). Purified DNA from each sample was dissolved in 200 μl of elution buffer. The DNAs were used to assess E. chaffeensis infection status by performing nested PCR targeting the inserted fragment-specific spectinomycin resistance gene (aadA) (primers for this experiment are listed in Table S1) as we described previously (14). Samples testing positive for the aadA gene were subsequently evaluated by nested PCRs targeting the transposon insertion fragment and the respective flanking genomic regions for the mutants using the insertion-specific primer sets (primers listed in Table S1).
Xenodiagnosis of E. chaffeensis mutants by A. americanum.
About 200 each of the laboratory-reared nymphal A. americanum ticks (Ecto Services, Inc., Henderson, NC) were placed per each dog starting day 5 postinoculation. The ticks were allowed to complete the blood acquisition (about 7 days) and the recovered fed ticks were kept at room temperature and 14 h of light in a 96% humidity chamber for molting to the adult stage (which took between 36 and 50 days). Genomic DNAs from about 10 to 20 ticks (from each group of dogs) were isolated individually using the DNeasy blood and tissue kit (Qiagen). Purified DNA from each tick was resuspended in 200 μl of elution buffer. Two microliters of DNA derived from each tick was used for nested PCR analysis targeting the aadA gene, and those testing positive for the aadA gene were then retested for transposon insertion regions specific for each mutant, as described above.
Statistics.
Statistical analysis was carried out to assess differences in average copy numbers of bacteria present in the wild type and mutants at each time point following in vitro growth in DH82 cells. The analysis was performed using the 2-tailed unpaired Student t test (GraphPad Software, La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the PHS grant AI070908 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA (https://www.niaid.nih.gov/), to R.R.G.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Mal Rocks Hoover for her help in formatting the figures.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med 316:853–856. doi: 10.1056/NEJM198704023161406. [DOI] [PubMed] [Google Scholar]
- 2.Anderson BE, Dawson JE, Jones DC, Wilson KH. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol 29:2838–2842. doi: 10.1128/JCM.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SM, Dumler JS, Bakken JS, Walker DH. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol 32:589–595. doi: 10.1128/JCM.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumler JS, Bakken JS. 1995. Ehrlichial diseases of humans: emerging tick-borne infections. Clin Infect Dis 20:1102–1110. doi: 10.1093/clinids/20.5.1102. [DOI] [PubMed] [Google Scholar]
- 5.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 6.Ismail N, Bloch KC, McBride JW. 2010. Human ehrlichiosis and anaplasmosis. Clin Lab Med 30:261–292. doi: 10.1016/j.cll.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritt BS, Sloan LM, Johnson DKH, Munderloh UG, Paskewitz SM, McElroy KM, McFadden JD, Binnicker MJ, Neitzel DF, Liu G, Nicholson WL, Nelson CM, Franson JJ, Martin SA, Cunningham SA, Steward CR, Bogumill K, Bjorgaard ME, Davis JP, McQuiston JH, Warshauer DM, Wilhelm MP, Patel R, Trivedi VA, Eremeeva ME. 2011. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N Engl J Med 365:422–429. doi: 10.1056/NEJMoa1010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritt BS, Allerdice MEJ, Sloan LM, Paddock CD, Munderloh UG, Rikihisa Y, Tajima T, Paskewitz SM, Neitzel DF, Hoang Johnson DK, Schiffman E, Davis JP, Goldsmith CS, Nelson CM, Karpathy SE. 2017. Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int J Syst Evol Microbiol 67:2121–2126. doi: 10.1099/ijsem.0.001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw SE, Day MJ, Birtles RJ, Breitschwerdt EB. 2001. Tick-borne infectious diseases of dogs. Trends Parasitol 17:74–80. doi: 10.1016/s1471-4922(00)01856-0. [DOI] [PubMed] [Google Scholar]
- 10.McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra JHF. 2017. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol 15:544–558. doi: 10.1038/nrmicro.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson WR, Lockhart JM, Stallknecht DE, Howerth EW, Dawson JE, Rechav Y. 2001. Persistent Ehrlichia chaffeensis infection in white-tailed deer. J Wildl Dis 37:538–546. doi: 10.7589/0090-3558-37.3.538. [DOI] [PubMed] [Google Scholar]
- 12.Unver A, Rikihisa Y, Stich RW, Ohashi N, Felek S. 2002. The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts. Infect Immun 70:4701–4704. doi: 10.1128/iai.70.8.4701-4704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumler JS, Sutker WL, Walker DH. 1993. Persistent infection with Ehrlichia chaffeensis. Clin Infect Dis 17:903–905. doi: 10.1093/clinids/17.5.903. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C, Nair ADS, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR. 2013. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog 9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Wei L, Liu H, Cheng C, Ganta RR. 2017. A genetic system for targeted mutations to disrupt and restore genes in the obligate bacterium, Ehrlichia chaffeensis. Sci Rep 7:15801. doi: 10.1038/s41598-017-16023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair ADS, Cheng C, Jaworski DC, Ganta S, Sanderson MW, Ganta RR. 2015. Attenuated mutants of Ehrlichia chaffeensis induce protection against wild-type infection challenge in the reservoir host and in an incidental host. Infect Immun 83:2827–2835. doi: 10.1128/IAI.00487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGill JL, Nair ADS, Cheng C, Rusk RA, Jaworski DC, Ganta RR. 2016. Vaccination with an attenuated mutant of Ehrlichia chaffeensis induces pathogen-specific CD4+ T cell immunity and protection from tick-transmitted wild-type challenge in the canine host. PLoS One 11:e0148229. doi: 10.1371/journal.pone.0148229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGill JL, Wang Y, Ganta CK, Boorgula GDY, Ganta RR. 2018. Antigen-specific CD4(+)CD8(+) double-positive T cells are increased in the blood and spleen during Ehrlichia chaffeensis infection in the canine host. Front Immunol 9:1585. doi: 10.3389/fimmu.2018.01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol 6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ZM, Tucker AM, Driskell LO, Wood DO. 2007. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol 73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin A, Tucker AM, Hines A, Wood DO. 2004. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl Environ Microbiol 70:2816–2822. doi: 10.1128/aem.70.5.2816-2822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark TR, Lackey AM, Kleba B, Driskell LO, Lutter EI, Martens C, Wood DO, Hackstadt T. 2011. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol 193:4993–4995. doi: 10.1128/JB.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosby FL, Wamsley HL, Pate MG, Lundgren AM, Noh SM, Munderloh UG, Barbet AF. 2014. Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics 15:278. doi: 10.1186/1471-2164-15-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bekebrede H, Lin M, Teymournejad O, Rikihisa Y. 2020. Discovery of in vivo virulence genes of obligatory intracellular bacteria by random mutagenesis. Front Cell Infect Microbiol 10:2. doi: 10.3389/fcimb.2020.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn GE, Burkhardt NY, Felsheim RF, Nelson CM, Oliver JD, Kurtti TJ, Cornax I, O’Sullivan MG, Munderloh UG. 2019. Ehrlichia isolate from a Minnesota tick: characterization and genetic transformation. Appl Environ Microbiol 85:e00866-19. doi: 10.1128/AEM.00866-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtti TJ, Burkhardt NY, Heu CC, Munderloh UG. 2016. Fluorescent protein expressing Rickettsia buchneri and Rickettsia peacockii for tracking symbiont-tick cell interactions. Vet Sci 3:34. doi: 10.3390/vetsci3040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oki AT, Seidman D, Lancina MG, Mishra MK, Kannan RM, Yang H, Carlyon JA. 2015. Dendrimer-enabled transformation of Anaplasma phagocytophilum. Microbes Infect 17:817–822. doi: 10.1016/j.micinf.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felsheim R, Oliva Chavez A, Palmer G, Crosby L, Barbet A, Kurtti T, Munderloh U. 2010. Transformation of Anaplasma marginale. Vet Parasitol 167:167–174. doi: 10.1016/j.vetpar.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones AL, Knoll KM, Rubens CE. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol Microbiol 37:1444–1455. doi: 10.1046/j.1365-2958.2000.02099.x. [DOI] [PubMed] [Google Scholar]
- 30.Hudson P, Gorton TS, Papazisi L, Cecchini K, Frasca S Jr, Geary SJ. 2006. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect Immun 74:931–939. doi: 10.1128/IAI.74.2.931-939.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HLT. 2008. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol 57:1068–1078. doi: 10.1099/jmm.0.2008/002071-0. [DOI] [PubMed] [Google Scholar]
- 32.Crosby FL, Brayton KA, Magunda F, Munderloh UG, Kelley KL, Barbet AF. 2015. Reduced infectivity in cattle for an outer membrane protein mutant of Anaplasma marginale. Appl Environ Microbiol 81:2206–2214. doi: 10.1128/AEM.03241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Severo MS, Sakhon OS, Choy A, Herron MJ, Felsheim RF, Wiryawan H, Liao J, Johns JL, Munderloh UG, Sutterwala FS, Kotsyfakis M, Pedra JH. 2012. Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect Immun 80:3194–3205. doi: 10.1128/IAI.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamason RL, Kafai NM, Welch MD. 2018. A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS One 13:e0197012. doi: 10.1371/journal.pone.0197012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eedunuri VK, Zhang Y, Cheng C, Chen L, Liu H, Omsland A, Boyle D, Ganta RR. 2018. Protein and DNA synthesis demonstrated in cell-free Ehrlichia chaffeensis organisms in axenic medium. Sci Rep 8:9293. doi: 10.1038/s41598-018-27574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondethimmanahalli C, Ganta R. 2018. Impact of three different mutations in Ehrlichia chaffeensis in altering the global gene expression patterns. Sci Rep 8:6162. doi: 10.1038/s41598-018-24471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondethimmanahalli C, Liu H, Ganta RR. 2019. Proteome analysis revealed changes in protein expression patterns caused by mutations in Ehrlichia chaffeensis. Front Cell Infect Microbiol 9:58. doi: 10.3389/fcimb.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng C, Nair ADS, Jaworski DC, Ganta RR. 2015. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10:e0132657. doi: 10.1371/journal.pone.0132657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaworski DC, Cheng C, Nair ADS, Ganta RR. 2017. Amblyomma americanum ticks infected with in vitro cultured wild-type and mutants of Ehrlichia chaffeensis are competent to produce infection in naïve deer and dogs. Ticks Tick Borne Dis 8:60–64. doi: 10.1016/j.ttbdis.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rollauer SE, Sooreshjani MA, Noinaj N, Buchanan SK. 2015. Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci 370:20150023. doi: 10.1098/rstb.2015.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohashi N, Zhi N, Zhang Y, Rikihisa Y. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun 66:132–139. doi: 10.1128/IAI.66.1.132-139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge Y, Rikihisa Y. 2007. Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun 75:3833–3841. doi: 10.1128/IAI.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crocquet-Valdes PA, Thirumalapura NR, Ismail N, Yu X, Saito TB, Stevenson HL, Pietzsch CA, Thomas S, Walker DH. 2011. Immunization with Ehrlichia P28 outer membrane proteins confers protection in a mouse model of ehrlichiosis. Clin Vaccine Immunol 18:2018–2025. doi: 10.1128/CVI.05292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JZ, Guo H, Winslow GM, Yu XJ. 2004. Expression of members of the 28-kilodalton major outer membrane protein family of Ehrlichia chaffeensis during persistent infection. Infect Immun 72:4336–4343. doi: 10.1128/IAI.72.8.4336-4343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy GR, Sulsona CR, Barbet AF, Mahan SM, Burridge MJ, Alleman AR. 1998. Molecular characterization of a 28 kDa surface antigen gene family of the tribe Ehrlichiae. Biochem Biophys Res Commun 247:636–643. doi: 10.1006/bbrc.1998.8844. [DOI] [PubMed] [Google Scholar]
- 46.Popov VL, Yu X, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 47.Wang JY, Zhu B, Patterson LL, Rogan MR, Kibler CE, McBride JW. 2020. Ehrlichia chaffeensis TRP120-mediated ubiquitination and proteasomal degradation of tumor suppressor FBW7 increases oncoprotein stability and promotes infection. PLoS Pathog 16:e1008541. doi: 10.1371/journal.ppat.1008541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumagai Y, Huang H, Rikihisa Y. 2008. Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J Bacteriol 190:3597–3605. doi: 10.1128/JB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velayutham TS, Kumar S, Zhang X, Kose N, Walker DH, Winslow G, Crowe JE Jr, McBride JW. 2019. Ehrlichia chaffeensis outer membrane protein 1-specific human antibody-mediated immunity is defined by intracellular TRIM21-dependent innate immune activation and extracellular neutralization. Infect Immun 87:e00383-19. doi: 10.1128/IAI.00383-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitra S, Dunphy PS, Das S, Zhu B, Luo T, McBride JW. 2018. Ehrlichia chaffeensis TRP120 effector targets and recruits host polycomb group proteins for degradation to promote intracellular infection. Infect Immun 86:e00845-17. doi: 10.1128/IAI.00845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Von Ohlen T, Cheng C, Faburay B, Ganta RR. 2013. Transcription of Ehrlichia chaffeensis genes is accomplished by RNA polymerase holoenzyme containing either sigma 32 or sigma 70. PLoS One 8:e81780. doi: 10.1371/journal.pone.0081780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hang X, Zeng Q, Zeng L, Jia J, Bi H. 2019. Functional replacement of the BioC and BioH proteins of Escherichia coli biotin precursor biosynthesis by Ehrlichia chaffeensis novel proteins. Curr Microbiol 76:626–636. doi: 10.1007/s00284-019-01669-w. [DOI] [PubMed] [Google Scholar]
- 53.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rikihisa Y. 2015. Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu Rev Microbiol 69:283–304. doi: 10.1146/annurev-micro-091014-104411. [DOI] [PubMed] [Google Scholar]
- 55.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, Azad AF, Sobral BW, Caler E. 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol 194:376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salaemae W, Booker GW, Polyak SW. 2016. The role of biotin in bacterial physiology and virulence: a novel antibiotic target for Mycobacterium tuberculosis. Microbiol Spectr 4:VMBF-0008-2015. doi: 10.1128/microbiolspec.VMBF-0008-2015. [DOI] [Google Scholar]
- 57.Polyak SW, Abell AD, Wilce MCJ, Zhang L, Booker GW. 2012. Structure, function and selective inhibition of bacterial acetyl-CoA carboxylase. Appl Microbiol Biotechnol 93:983–992. doi: 10.1007/s00253-011-3796-z. [DOI] [PubMed] [Google Scholar]
- 58.Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I. 2015. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Y, Napier BA, Manandhar M, Henke SK, Weiss DS, Cronan JE. 2014. A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol Microbiol 91:300–314. doi: 10.1111/mmi.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guillén-Navarro K, Encarnación S, Dunn MF. 2005. Biotin biosynthesis, transport and utilization in rhizobia. FEMS Microbiol Lett 246:159–165. doi: 10.1016/j.femsle.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 61.Said HM. 2009. Cell and molecular aspects of human intestinal biotin absorption. J Nutr 139:158–162. doi: 10.3945/jn.108.092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soares da Costa TP, Tieu W, Yap MY, Zvarec O, Bell JM, Turnidge JD, Wallace JC, Booker GW, Wilce MC, Abell AD, Polyak SW. 2012. Biotin analogues with antibacterial activity are potent inhibitors of biotin protein ligase. ACS Med Chem Lett 3:509–514. doi: 10.1021/ml300106p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zlitni S, Ferruccio LF, Brown ED. 2013. Metabolic suppression identifies new antibacterial inhibitors under nutrient limitation. Nat Chem Biol 9:796–804. doi: 10.1038/nchembio.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cross R, Ling C, Day NPJ, McGready R, Paris DH. 2016. Revisiting doxycycline in pregnancy and early childhood—time to rebuild its reputation? Expert Opin Drug Saf 15:367–382. doi: 10.1517/14740338.2016.1133584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Purvis J, Edwards M. 2000. Doxycycline use for rickettsial disease in pediatric patients. Pediatr Infect Dis J 19:871–874. doi: 10.1097/00006454-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 66.Schaefer JJ, Needham GR, Bremer WG, Rikihisa Y, Ewing SA, Stich RW. 2007. Tick acquisition of Ehrlichia canis from dogs treated with doxycycline hyclate. Antimicrob Agents Chemother 51:3394–3396. doi: 10.1128/AAC.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qurollo BA, Buch J, Chandrashekar R, Beall MJ, Breitschwerdt EB, Yancey CB, Caudill AH, Comyn A. 2019. Clinicopathological findings in 41 dogs (2008–2018) naturally infected with Ehrlichia ewingii. J Vet Intern Med 33:618–629. doi: 10.1111/jvim.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blair JMA, Smith HE, Ricci V, Lawler AJ, Thompson LJ, Piddock LJV. 2015. Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J Antimicrob Chemother 70:424–431. doi: 10.1093/jac/dku380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cattoir V. 2004. Efflux-mediated antibiotics resistance in bacteria. Pathol Biol (Paris) 52:607–616. doi: 10.1016/j.patbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 70.Schweizer HP. 2012. Understanding efflux in Gram-negative bacteria: opportunities for drug discovery. Expert Opin Drug Discov 7:633–642. doi: 10.1517/17460441.2012.688949. [DOI] [PubMed] [Google Scholar]
- 71.Narra HP, Schroeder CLC, Sahni A, Rojas M, Khanipov K, Fofanov Y, Sahni SK. 2016. Small regulatory RNAs of Rickettsia conorii. Sci Rep 6:36728. doi: 10.1038/srep36728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schroeder CLC, Narra HP, Rojas M, Sahni A, Patel J, Khanipov K, Wood TG, Fofanov Y, Sahni SK. 2015. Bacterial small RNAs in the Genus Rickettsia. BMC Genomics 16:1075. doi: 10.1186/s12864-015-2293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singu V, Liu H, Cheng C, Ganta RR. 2005. Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect Immun 73:79–87. doi: 10.1128/IAI.73.1.79-87.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen SM, Popov VL, Feng HM, Wen J, Walker DH. 1995. Cultivation of Ehrlichia chaffeensis in mouse embryo, Vero, BGM, and L929 cells and study of Ehrlichia-induced cytopathic effect and plaque formation. Infect Immun 63:647–655. doi: 10.1128/IAI.63.2.647-655.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng C, Ganta RR. 2008. Laboratory maintenance of Ehrlichia chaffeensis and Ehrlichia canis and recovery of organisms for molecular biology and proteomics studies. Curr Protoc Microbiol Chapter 3:Unit 3A.1. doi: 10.1002/9780471729259.mc03a01s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee G, Saito I. 1998. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 77.Shmerling D, Danzer CP, Mao X, Boisclair J, Haffner M, Lemaistre M, Schuler V, Kaeslin E, Korn R, Burki K, Ledermann B, Kinzel B, Muller M. 2005. Strong and ubiquitous expression of transgenes targeted into the beta-actin locus by Cre/lox cassette replacement. Genesis 42:229–235. doi: 10.1002/gene.20135. [DOI] [PubMed] [Google Scholar]
- 78.Sirigireddy KR, Ganta RR. 2005. Multiplex detection of Ehrlichia and Anaplasma species pathogens in peripheral blood by real-time reverse transcriptase-polymerase chain reaction. J Mol Diagn 7:308–316. doi: 10.1016/S1525-1578(10)60559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.