Abstract
Noncanonical amino acids form a highly diverse pool of building blocks that can render unique physicochemical properties to peptides and proteins. Here, four methionine analogues with unsaturated and varying side chain lengths were successfully incorporated at four different positions in nisin in Lactococcus lactis through force feeding. This approach allows for residue-specific incorporation of methionine analogues into nisin to expand their structural diversity and alter their activity profiles. Moreover, the insertion of methionine analogues with biorthogonal chemical reactivity, e.g., azidohomoalanine and homopropargylglycine, provides the opportunity for chemical coupling to functional moieties and fluorescent probes as well as for intermolecular coupling of nisin variants. All resulting nisin conjugates retained antimicrobial activity, which substantiates the potential of this method as a tool to further study its localization and mode of action.
Keywords: nisin, methionine analogues, click chemistry, dimers, fluorescence
Lantibiotics are antimicrobial peptides harboring unusual post-translationally modified amino acid residues such as dehydroalanine (Dha) and dehydrobutyrine (Dhb), lanthionine (Lan) and methyllanthionine (MeLan), that are introduced by a promiscuous post-translational modification (PTM) machinery.1,2 The unique biosynthetic pathways and relatively low genetic complexity of biosynthesis make lantibiotics good candidates for synthetic biology and bioengineering, to expand the antimicrobial arsenal.2 Various synthetic and biosynthetic strategies have been developed to increase the diversity of lantibiotics.3−5 The uncommon amino acids (Dha, Dhb, Lan, MeLan) in lantibiotics play an important role in their biological activity and structural stability. Other noncanonical amino acids (ncAAs) offer a further highly diverse pool of building blocks that can introduce unique physicochemical properties.6 By incorporating non-natural functional groups with unique features, we can dramatically expand the chemical and functional space of lantibiotic structures and enable the design of novel lantibiotics with enhanced properties (e.g., stability, specificity, bioavailability, and half-life).7−9 The use of this approach allows for the in vivo production of new lantibiotics with an expanded amino acid repertoire.8 Among ncAAs, the analogues of methionine are of particular interest, as some of them (e.g., azidohomoalanine and homopropargylglycine) possess unique reactive groups which can serve as chemical handles to conjugate with fluorophores, glycans, PEGs, lipids, peptide moieties, and other antimicrobial moieties through click chemistry.10
Copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC), referred to as “click chemistry”, was first reported by Sharpless and co-workers in 2001.11 It is a region-selective copper(I) catalytic cycloaddition reaction between an azide and an alkyne that gives rise to a triazole. Peptide modification using click chemistry has been the subject of several studies for the development of target-specific bacterial probes and for expanding their bioactivity and application.12−17 Prompted by these recent reports, we used nisin as a model to explore the potential of this approach for lantibiotic engineering. Nisin is the best studied lanthipeptide to date.18 It is produced by Lactococcus lactis and has potent activity against a broad spectrum of Gram-positive bacteria, including many antibiotic-resistant organisms, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE). Mature nisin, encoded by nisA as a linear precursor peptide (57 aa) that consists of a leader peptide (23 aa) and a propeptide to be modified (34 aa), is released after modification and cleavage of the leader.19 Gratifyingly, the modification machinery of nisin has a broad substrate specificity, which allows for the divergence from the original core-peptide to produce variant peptides.
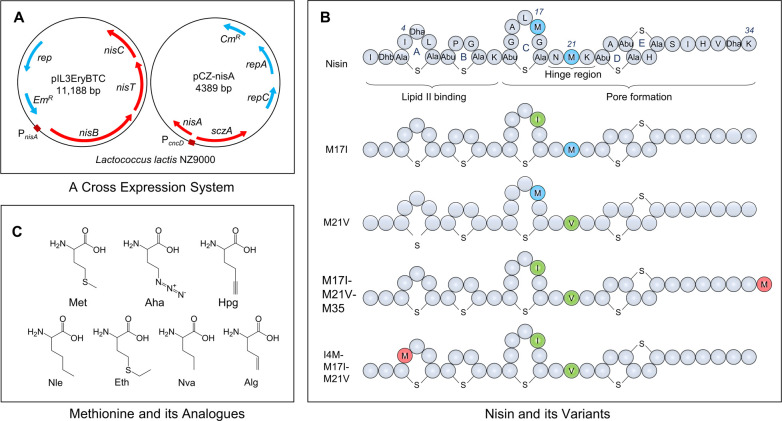
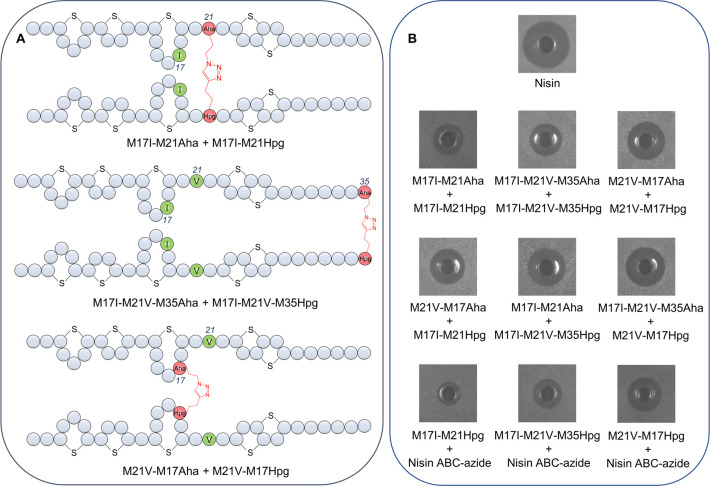
Here, we describe the incorporation of six different methionine analogues with unsaturated, unique chemical handles and varying side chain length, i.e., Aha (azidohomoalanine), Hpg (homopropargylglycine), Nle (norleucine), Eth (ethionine), Nva (norvaline), and Alg (allyglycine), at four different positions in the lantibiotic nisin by using a methionine auxotrophic strain of Lactococcus lactis. Previous studies have shown that with mutations at sites I4, M17, and M21, nisin can retain or even have enhanced bioactivity.20 To broaden the structural diversity and test the effects of single replacements of methionine with respective analogues, four single methionine nisin mutants, i.e., M17I, M21V, M17I-M21V-M35, and I4M-M17I-M21V, were constructed. As methionine is essential for the synthesis of post-translational modification enzymes, a cross-expression system was developed utilizing separate promoters, allowing for the separate induction of expression of target genes and biosynthetic enzymes. The amino acid replacement and incorporation efficiency of ncAAs into nisin derivatives were determined by matrix assisted laser desorption/ionization time-of-flight analyzer (MALDI-TOF) and liquid chromatography–mass spectrometry (LC-MS). Twelve nisin derivatives were purified in large scale by HPLC and their antimicrobial activities were determined. In addition, six Aha- or Hpg-containing nisin derivatives were coupled either mutually or with nisin ABC-azide (A, B, and C denoting the first three lanthionine rings of nisin; Figure 1), Cy5-azide and 6-FAM-alkyne through click chemistry to obtain six dimeric nisin constructs, three nisin hybrids, and six fluorescently labeled nisin variants.
Figure 1.
(A) A cross expression system with two plasmids. SczA, encoding the repressor of PczcD; PczcD, a zinc inducible promoter; nisA, encoding NisA; repA and repC, encoding plasmid replication proteins; CmR, chloramphenicol resistance gene; PnisA, a nisin inducible promoter; nisB, encoding NisB; nisT, encoding NisT; nisC, encoding NisC; EmR, erythromycin resistance gene. (B) Peptide sequence of nisin and nisin derivatives. Lipid II binding site (rings AB), pore formation domain (rings CDE), and hinge region (NMK) are indicated; Positions 17, 21, and 35, which served to incorporate methionine analogues of nisin are indicated; Dha, dehydroalanine; Dhb, dehydrobutyrine; A-S-A, lanthionine; Abu-S-A, methyllanthionine; In blue, wildtype Met positions; In green, Met residues replaced by Ile or Val; In red, Met residues at novel positions. (C) Structures of methionine and its analogues. Met, l-methionine; Aha, l-azidohomoalanine; Hpg, l-homopropargylglycine; Nle, l-norleucine; Eth, l-ethionine; Nva, l-norvaline; Alg, l-allyglycine.
Results and Discussion
A Cross Expression System to Incorporate Methionine Analogues into Nisin by Use of L. lactis as a Host
Two in vivo approaches have been developed for incorporating ncAAs into peptides.8 The first approach is “site-specific incorporation”.21 For this method, the coexpression of orthogonal amber suppressor aminoacyl-tRNA synthetase (AARS/tRNA) pairs is necessary. Specific mutations can be introduced into the peptide sequence by reassigning the amber nonsense stop codon during translation. However, the screening and development of orthogonal AARS/tRNA pairs is time-consuming and the production yield of this method is extremely low. Conversely, “residue-specific incorporation”, the second approach, that generally does not suffer from such drawback, is a more promising strategy.22 This method typically involves replacing natural amino acids with the ncAAs of interest by using auxotrophic strains. It is able to generate broad and efficient structural diversity by directly incorporating ncAAs via translation into bioactive peptides.
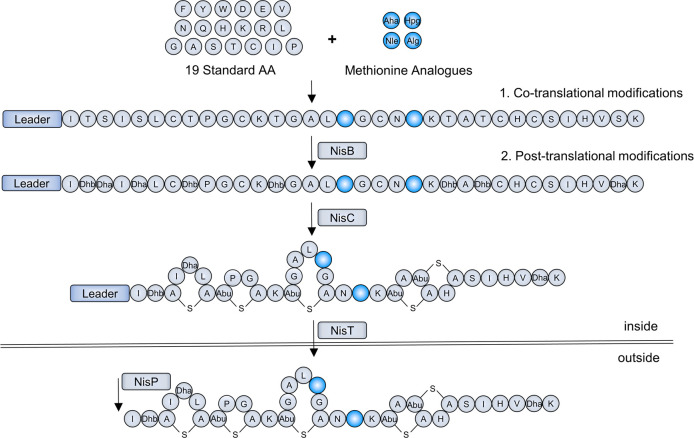
Various expression hosts have been developed for the incorporation of ncAAs.7 Until now, the Gram-negative Escherichia coli is the only prokaryotic expression host used for the incorporation of methionine analogues.23−25 Here, the Gram-positive expression host L. lactis, a methionine-auxotrophic strain, is used for the incorporation of methionine analogues into the lantibiotic nisin. After ribosomal synthesis of the precursor peptide with 19 standard amino acids and with variable methionine analogues, the unmodified prenisin is processed by its dedicated modification machinery (Scheme 1). First, the serine and threonine residues in the core peptide are Dha and Dhb by the dehydratase NisB. The dehydrated residues are then coupled to specific cysteine by the cyclase NisC to form lanthionine rings. Subsequently, the modified prenisin is transported out of the cell by the ABC-type transporter NisT, and then the leader is cleaved off by the extracellular protease NisP to liberate the active peptide.
Scheme 1. Incorporation of Methionine Analogues into Nisin.
(1) Cotranslational modifications, insertion of methionine analogues into the precursor peptide. (2) Post-translational modifications, converting the linear precursor peptide into an active polycyclic peptide.
As methionine is essential for the translation of post-translational modification (PTM) enzymes and transporter, a cross expression system, which allows for the expression of prenisin derivatives and PTM enzymes and transporter at different time was used for this study. L. lactis NZ9000 was transformed with a plasmid encoding the expression of NisBTC under the control of the PnisA promoter and the other plasmid encoding the expression of prenisin derivatives was controlled by the PczcD promoter. The expression of NisBTC was first conducted by the supplementation of methionine, after which the medium was replaced by new medium lacking methionine, but containing methionine analogues to express prenisin derivatives (Figure 1A). Although the expression of the modification machinery NisBTC was induced in advance, no effect on the modification efficiency was observed.
Production of Nisin and Its Derivatives
There are two methionine residues in the core peptide of nisin, which are located at sites 17 and 21. Previous studies showed that nisin with the mutation at sites I4, M17, and M21 could retain or even have increased antimicrobial activity.20 To test the effects of single methionine replacement with analogues in bioactive nisin, four single methionine mutants, i.e., M17I, M21V, M17I-M21V-M35, and I4M-M17I-M21V were constructed (Figure 1B). The choice for Ile or Val as substituents was to retain good antimicrobial activity, since both residues share the hydrophobic character of Met and are sterically not very different, though branched. Six methionine analogues, i.e., Aha, Hpg, Nle, Eth, Nva, and Alg were selected for the incorporation (Figure 1C), and each combination was tested.
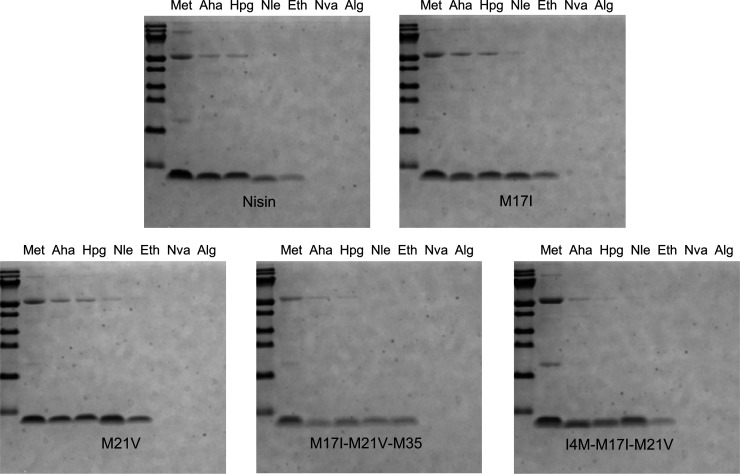
The expression level of nisin and its derivatives is shown in Figure 2. The protein quantities in the first five lanes showed that Aha, Hpg, Nle, and Eth can be incorporated into nisin and its derivatives at varying levels. However, incorporation of Nva and Alg was not observed at any moment, and addition of these analogues to a culture lacking methionine led to arrested cell growth. These results strongly indicate that Nva and Alg cannot be incorporated by L. lactis. The highest production yield was observed when normal methionine was supplemented. Effectively, a lower production yield was observed in the presence of methionine analogues, in particular with Eth, regardless of the position within the molecule. Methionine analogue incorporation in nisin, M17I, and M21V gave much higher yields of fully modified peptides than M17I-M21V-M35 and I4M-M17I-M21V, which may be due to the intolerance of the modification machinery and/or the transporter to a change of chemical structures at sites I4 and M35. Surprisingly, the production yield of fully modified M21V is even higher than that of WT nisin. To assess the presence of post-translational modifications and incorporation of ncAAs, all samples were further analyzed by HPLC and MALDI-TOF. The resulting spectra showed that the production yield of nisin and its derivatives with Aha and Hpg are much higher than the ones with Nle and Eth. In addition, we found that Nle and Eth had a negative influence on the dehydration rate, as large fractions of 7 times dehydrated peptides were observed.
Figure 2.
Coomassie-blue stained Tricine-SDS-PAGE gel. Each well contained TCA-precipitated prepeptides from 1 mL supernatant.
LC-MS Analysis of Nisin Derivatives
In order to estimate the incorporation efficiency of methionine analogues at different positions, the precipitated precursor peptides were subjected to LC-MS. The LC-MS data showed that the incorporation efficiency of Aha and Hpg into mutants M17I, M21V, and I4M-M17I-M21V was more than 91%, while the incorporation efficiency of Nle and Eth was 88% and 71–73%, respectively. Remarkably, the incorporation efficiency of Aha and Hpg into M17I-M21V-M35 was at least 99.5%, and the peaks of peptides containing methionine were undetectable. However, the incorporation efficiency of Nle and Eth was only 51 and 71%, respectively. In the case of nisin, the incorporation efficiency was 88% for Aha, 87% for Hpg, 77% for Nle, and 56% for Eth. Generally, the incorporation efficiency of ncAAs declined in the order Aha > Hpg > Nle > Eth (Table 1).
Table 1. Incorporation Efficiency of Nisin and Its Derivatives Analyzed by LC-MSa.
| peptide | incorporation efficiency (%) | |
|---|---|---|
| Nisin | Aha | 88 |
| Hpg | 87 | |
| Nle | 77 | |
| Eth | 56 | |
| M17I | Aha | 96 |
| Hpg | 92 | |
| Nle | 88 | |
| Eth | 71 | |
| M21V | Aha | 99 |
| Hpg | 91 | |
| Nle | 88 | |
| Eth | 73 | |
| M17I-M21V-M35 | Aha | >99.5 |
| Hpg | >99.5 | |
| Nle | 51 | |
| Eth | 71 | |
| I4M-M17I-M21V | Aha | 95 |
| Hpg | 93 | |
| Nle | 88 | |
| Eth | 71 | |
>99.5% means the peak of peptides containing methionine is undetectable. The incorporation efficiency indicates the percentage of the produced peptide with methionine analogues incorporated completely.
Incorporation of Aha and Hpg into nisin, M17I, and M21V did not affect the dehydration efficiency, as peptides with 7 times dehydrated residues were nearly undetectable, suggesting they are excellent methionine surrogates. It may be due to the rate of activation of Aha and Hpg by methionyl-tRNA synthetase (MetRS) during translation, which finally results in the higher yield and efficient modification. However, introducing Nle and Eth resulted in a large fraction of peptides with 7 times dehydration. It may be that the integration speed of Nle and Eth during translation is relatively slow which leads to a lower yield and insufficient modification. The dehydration of M17I-M21V-M35 and I4M-M17I-M21V was dramatically affected by the mutation. Additionally, methionine and ethionine can be oxidized, and peaks corresponding to oxidized products were indeed observed. Furthermore, the first methionine of prenisin is usually cleaved by the enzyme methionine aminopeptidase (MAP). However, a large portion of precursor peptide produced by this system contained the N-terminal Met. The molecular weight of both peaks is shown in Table 2.
Table 2. MS Analysis of Prenisin and Its Derivativesa.
| measured
mass (Da) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| predicted
mass (Da) |
Met |
Aha |
Hpg |
Nle |
Eth |
|||||||||
| peptide | methionine analogue | modification | +Met1 | –Met1 | +Met1 | –Met1 | +Met1 | –Met1 | +Met1 | –Met1 | +Met1 | –Met1 | +Met1 | –Met1 |
| Nisin | Met | –8H2O | 5818.85 | 5687.66 | 5818.80 | 5687.76 | ||||||||
| –8H2O + Oxi | 5834.85 | 5703.66 | 5835.80 | 5704.76 | ||||||||||
| –8H2O + 2Oxi | 5850.85 | 5719.66 | 5850.79 | 5720.75 | ||||||||||
| Aha | –8H2O | 5803.63 | 5677.51 | 5803.84 | 5677.79 | |||||||||
| Hpg | –8H2O | 5752.66 | 5643.53 | 5752.84 | 5643.79 | |||||||||
| Nle | –8H2O | 5764.75 | 5651.59 | 5764.93 | 5651.85 | |||||||||
| –7H2O | 5782.77 | 5669.61 | 5782.90 | 5669.81 | ||||||||||
| Eth | –8H2O | 5860.93 | 5715.71 | 5861.84 | 5716.79 | |||||||||
| –8H2O + Oxi | 5876.93 | 5731.71 | 5877.84 | 5732.79 | ||||||||||
| –8H2O + 2Oxi | 5892.93 | 5747.71 | 5893.83 | 5748.78 | ||||||||||
| M17I | Met | –8H2O | 5800.81 | 5669.62 | 5800.85 | 5669.81 | ||||||||
| –8H2O + Oxi | 5816.81 | 5685.62 | 5816.84 | 5685.80 | ||||||||||
| Aha | –8H2O | 5790.66 | 5664.54 | 5790.87 | 5664.82 | |||||||||
| Hpg | –8H2O | 5756.68 | 5647.55 | 5756.87 | 5647.82 | |||||||||
| Nle | –8H2O | 5764.74 | 5651.58 | 5764.93 | 5651.85 | |||||||||
| –7H2O | 5782.76 | 5669.60 | 5782.90 | 5669.82 | ||||||||||
| Eth | –8H2O | 5828.86 | 5683.64 | 5829.87 | 5683.82 | |||||||||
| –8H2O + Oxi | 5844.86 | 5699.64 | 5845.87 | 5700.82 | ||||||||||
| –8H2O + 2Oxi | 5860.86 | 5861.87 | ||||||||||||
| M21V | Met | –8H2O | 5786.79 | 5655.60 | 5786.83 | 5655.79 | ||||||||
| –8H2O + Oxi | 5802.79 | 5671.60 | 5802.82 | 5672.78 | ||||||||||
| Aha | –8H2O | 5776.64 | 5650.52 | 5776.85 | 5650.80 | |||||||||
| Hpg | –8H2O | 5742.66 | 5633.53 | 5742.85 | 5633.50 | |||||||||
| Nle | –8H2O | 5750.72 | 5637.56 | 5750.91 | 5637.83 | |||||||||
| –7H2O | 5768.74 | 5655.58 | 5768.89 | 5655.81 | ||||||||||
| Eth | –8H2O | 5814.84 | 5669.62 | 5815.85 | 5670.80 | |||||||||
| –8H2O + Oxi | 5830.84 | 5685.62 | 5831.85 | 5685.80 | ||||||||||
| –8H2O + 2Oxi | 5846.84 | 5846.85 | ||||||||||||
| M17I-M21V-M35 | Met | –8H2O | 5899.95 | 5768.76 | 5899.91 | 5768.87 | ||||||||
| –7H2O | 5917.97 | 5786.77 | 5917.81 | 5786.88 | ||||||||||
| –7H2O + Oxi | 5933.97 | 5802.77 | 5933.91 | 5802.87 | ||||||||||
| Aha | –8H2O | 5889.80 | 5763.68 | 5889.94 | 5763.89 | |||||||||
| –7H2O | 5907.82 | 5781.70 | 5906.94 | 5781.89 | ||||||||||
| Hpg | –8H2O | 5855.82 | 5746.69 | 5855.97 | 5746.88 | |||||||||
| –7H2O | 5873.84 | 5764.71 | 5873.94 | 5764.89 | ||||||||||
| Nle | –8H2O | 5863.88 | 5750.72 | 5864.00 | 5750.91 | |||||||||
| –7H2O | 5881.90 | 5768.74 | 5882.01 | 5768.93 | ||||||||||
| Eth | –8H2O | 5928.00 | 5782.78 | 5928.94 | 5782.89 | |||||||||
| –7H2O | 5946.02 | 5800.80 | 5945.94 | 5800.89 | ||||||||||
| –7H2O + Oxi | 5962.02 | 5816.80 | 5961.94 | 5816.89 | ||||||||||
| I4M-M17I-M21V | Met | –8H2O | 5786.79 | 5655.60 | 5787.83 | 5655.79 | ||||||||
| –7H2O | 5804.81 | 5673.61 | 5804.82 | 5672.78 | ||||||||||
| –7H2O + Oxi | 5820.81 | 5689.61 | 5819.82 | 5689.78 | ||||||||||
| Aha | –8H2O | 5776.64 | 5650.52 | 5776.85 | 5650.80 | |||||||||
| –7H2O | 5794.66 | 5792.85 | ||||||||||||
| Hpg | –8H2O | 5742.66 | 5633.53 | 5742.85 | 5633.80 | |||||||||
| –7H2O | 5760.68 | 5758.85 | ||||||||||||
| Nle | –8H2O | 5750.72 | 5637.56 | 5750.91 | 5637.83 | |||||||||
| –7H2O | 5768.74 | 5655.58 | 5767.90 | 5655.81 | ||||||||||
| Eth | –8H2O | 5814.84 | 5669.62 | 5815.85 | 5670.80 | |||||||||
| –8H2O + Oxi | 5830.84 | 5685.62 | 5830.85 | 5686.80 | ||||||||||
| –8H2O + 2Oxi | 5846.84 | 5846.84 | ||||||||||||
+Met1, with N-terminal Met; −Met1, without N-terminal Met; −8H2O, eight times dehydrated; −8H2O + Oxi, eight times dehydrated and one time oxidized; −8H2O + 2Oxi, eight times dehydrated and two times oxidized; −7H2O, seven times dehydrated; −7H2O + Oxi, seven times dehydrated and one time oxidized.
Antimicrobial Activity of Nisin and Its Derivatives
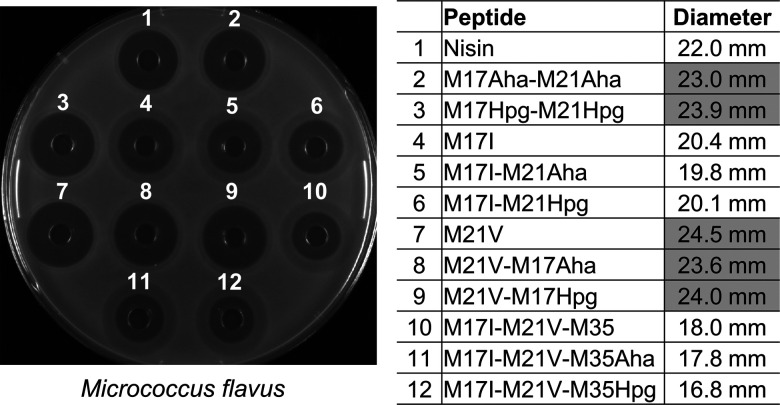
In consideration of a sufficient production yield of the fully modified peptides, 12 peptides containing methionine, Aha or Hpg were purified in large scale for antimicrobial activity tests (Figure 3). M. flavus was used as a first indicator strain in an agar-well diffusion assay to assess the antimicrobial activity. The results showed that M17Aha-M21Aha, M17Hpg-M21Hpg, M21V, M21V-M17Aha, and M21V-M17Hpg have higher antimicrobial activity compared to WT nisin, and mutant M21V showed the best activity with any of the three amino acids. However, in all the cases, the activity of mutants M17I and M17I-M21V-M35 decreased dramatically.
Figure 3.
Antimicrobial activity of nisin and its derivatives against M. flavus. In gray: values that are improved in comparison to nisin.
The MIC values were determined for L. lactis and six Gram-positive pathogenic strains. The tested strains included two Staphylococci, two Enterococci, Bacillus cereus and Listeria monocytogenes (Table 3). The results showed that the replacement of Met with Met analogues can alter the antimicrobial activity and spectrum. Certain peptides retained or even displayed higher activity against a specific strain, while showing reduced activities against others, suggesting a possible increase in selectivity. For example, M17Aha-M21Aha showed improved activity against L. monocytogenes, but the activity against the other strains was reduced when compared to nisin. M21V has been reported to have enhanced bioactivity and specific activity against all tested Gram-positive pathogens including four VRE strains compared to WT nisin.26,27 In our study, M21V showed reduced activities against two Enterococci strains, but retained a high activity against others. Improving the antimicrobial activity of nisin turned out to be difficult. However, engineering nisin can generate new nisin derivatives that have different properties and can be used for specific targets. In addition, some nisin derivatives showed different inhibition activity in solid media tests compared to the broth MIC test. This phenomenon can be related to the difference in diffusion ability.
Table 3. MIC Values (μM) of Nisin and Its Derivatives.
| peptide | CAL-MRSA | MW2-MRSA | B. cereus | E. faecalis | E. faecium | L. monocytogenes | L. lactis |
|---|---|---|---|---|---|---|---|
| Nisin | 10.39 | 5.19 | 5.19 | 2.60 | 0.32 | 2.60 | 0.020 |
| M17Aha-M21Aha | 19.99 | 13.33 | 6.66 | 3.33 | 0.42 | 1.67a | 0.026 |
| M17Hpg-M21Hpg | 19.41 | 19.41 | 9.70 | 4.85 | 0.61 | 4.85 | 0.019a |
| M17I | >19.92 | >19.92 | >19.92 | 19.92 | 2.49 | 4.98 | 0.622 |
| M17I-M21Aha | >17.42 | >17.42 | 8.71 | 17.42 | 2.18 | 4.36 | 0.544 |
| M17I-M21Hpg | >19.08 | >19.08 | 19.08 | 19.08 | 2.38 | 4.77 | 0.596 |
| M21V | 9.81a | 2.45a | 4.90a | 4.90 | 0.61 | 2.45a | 0.019a |
| M21V-M17Aha | >19.72 | 19.72 | 19.72 | 9.86 | 0.62 | 4.93 | 0.039 |
| M21V-M17Hpg | >19.74 | 19.74 | 19.74 | 9.87 | 0.62 | 4.93 | 0.019a |
| M17I-M21V-M35 | >18.33 | >18.33 | >18.33 | >18.33 | 2.29 | >18.33 | 0.573 |
| M17I-M21V-M35Aha | >19.96 | >19.96 | >19.96 | >19.96 | 2.49 | >19.96 | 0.624 |
| M17I-M21V-M35Hpg | >16.82 | >16.82 | >16.82 | 16.82 | 2.10 | >16.82 | >0.526 |
MIC values that are retained or improved in comparison to nisin.
Production of Dimeric Nisin Constructs and Nisin Hybrids
The mode of action of nisin involves its binding to lipid II, followed by membrane insertion, which leads to pore formation. The pore-complex has a uniform and stable structure, consisting out of eight nisin and four lipid II molecules.28 In a previous study, a nisin dimer was prepared by connecting two nisin molecules at the C-terminus through a linker, which led to slightly increased pore-forming activity.13 As the nisin derivatives contain a clickable group (azide or alkyne) at positions 17, 21, and 35, a setup was devised to investigate how different orientations and multivalency patterns of nisin dimers affect antimicrobial activity.29 M17I-M21Aha, M17I-M21Hpg, M17I-M21V-M35Aha, M17I-M21V-M35Hpg, M21V-M17Aha, and M21V-M17Hpg were coupled either mutually or with nisin ABC-azide to generate six dimeric nisin constructs and three nisin hybrids which were characterized by MALDI-TOF (Supplementary Figure S1). The antimicrobial activity of these dimers was tested against M. flavus by agar diffusion assays. The resulting growth inhibition halos indicated the retainment of at least some degree of activity in all variants. We found that the activity of dimeric nisin constructs increased in order as reactions are performed at the hinge region (position 21), the C-terminus (position 35), and ring C (position 17). M17I-M21Aha + M17I-M21Hpg is the least active dimeric nisin construct (Figure 4A and 4B). Coupling at the hinge region may result in increased steric hindrance and decreased flexibility and therefore hindering its lipid II binding and pore formation features. It again proves that the flexibility of the hinge region is important for the activity, which is in accordance with previous studies.30,31 Also M17I-M21V-M35Aha + M17I-M21V-M35Hpg showed lower activity. Coupling at the C-terminus of nisin gave rise to a dimeric nisin construct containing two lipid II binding sites. However, the pore formation ability may be weakened or abolished as the C-terminus of nisin was involved in the connection since both C-termini must flip simultaneously and insert in the membrane. This involves the movement and reorientation of a bulky set of amino acids, including the intertwined rings DE of each monomer, through the membrane. M21V-M17Aha + M21V-M17Hpg is the most active dimeric nisin construct. Coupling at ring C gave the best activity, which may be due to the fact that rings AB are still able to bind lipid II, while the hinge region, rings DE and the linear C-terminus keeps their individual flexibility, allowing the C-terminus of nisin to form pores. Therefore, position 17 is the optimal site for coupling moieties out of the 3 chosen positions. This study shows the great potential of this strategy for linking active modules from different peptides. Moreover, nisin ABC was obtained by enzymatic digestion of nisin using chymotrypsin and it was subsequently C-terminally functionalized with azidopropylamine to generate nisin ABC-azide. Coupling M17I-M21Hpg, M17I-M21V-M35Hpg, and M21V-M17Hpg with nisin ABC-azide showed the same antimicrobial activity pattern as above; i.e., activity is altered in ascending order as reactions are performed to the hinge region, the C-terminus, and ring C, respectively (Figure 4B).
Figure 4.
(A) Structure of three representative dimeric nisin constructs with reactions performed at the hinge region (position 21), the C-terminus (position 35), and ring C (position 17). (B) Antimicrobial activity of six dimeric nisin and three nisin hybrids at equimolar concentrations against M. flavus with nisin as positive control. M17I-M21Aha + M17I-M21Hpg is the least active dimeric nisin construct whereas M21VM17Aha + M21V-M17Hpg is the most active. Similarly, M17I-M21Hpg + Nisin ABC-azide is the least active nisin hybrid, whereas M21V-M17Hpg + Nisin ABC-azide is the most active.
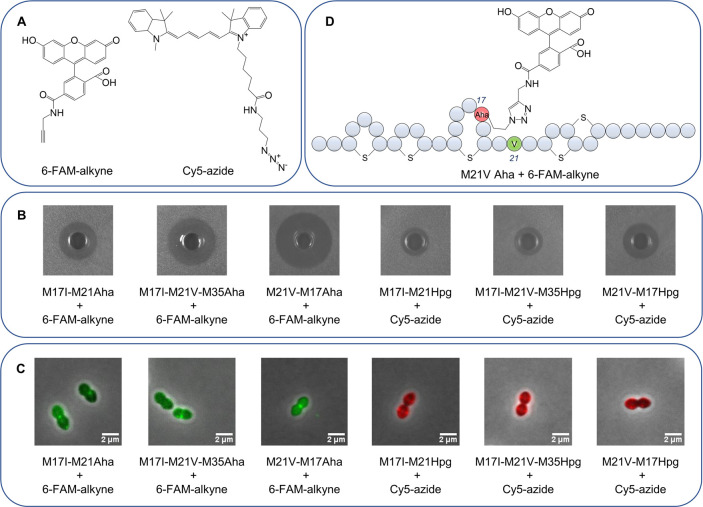
Fluorescently Labeled Nisin Variants
Labeling of nisin with fluorescent probes has greatly contributed to understanding its mechanism of action as shown in studies by Scherer et al.(32) and Descobry et al.(33) The C-terminus of nisin is the common site for labeling. However, introduction of a tag in this position poses a considerable perturbation in the structure and activity of nisin. Here, 6-FAM-alkyne and Cy5-azide (Figure 5A) were successfully coupled at three different positions (positions 17, 21, and 35) of nisin, and the resulting compounds were characterized by MALDI-TOF (Supplementary Figure S2). The antimicrobial activity of six fluorescently labeled nisin variants was retained (Figure 5B). Coupling 6-FAM-alkyne and Cy5-azide at different positions of nisin showed the same activity pattern as that obtained with dimeric nisin constructs. Thus, M21V-M17Aha was found to be the most suitable derivative for labeling with both 6-FAM-alkyne and Cy5-azide. The localization of six fluorescently labeled nisin variants interacting with E. faecium were studied by fluorescence microscopy (Figure 5C). Fluorescence intensity detection indicated that the labeled nisin conjugates were all located at the cell membrane. Cy5-azide labeled nisin variants showed lower activity than their 6-FAM-alkyne labeled counterparts, and no aggregation was observed in cell division sites. This may be due to the fact that Cy5-azide would affect the binding of nisin conjugates to lipid II. M21V-M17Aha + 6-FAM-alkyne (Figure 5D) was found to be the most potent fluorescently labeled nisin variant, as it showed similar activity to nisin. It was located at the septum of cell division sites where the membrane-bound cell wall precursor lipid II concentration is maximal. These results are in accordance with previous studies using fluorescently labeled nisin A and nisin Z, which indicated that both molecules were accumulating at the cell division sites of Bacillus subtilis and L. monocytogenes, respectively.33,34 M21V-M17Aha + 6-FAM-alkyne shows great potential as a tool to further study the antibacterial mechanism of action of nisin and for understanding the mechanism of synergy of nisin with other molecules on Gram-negative strain. Moreover, this strategy can be extended to modify other ribosomally synthesized and post-translationally modified peptides (RiPPs). While numerous novel RiPPs have been reported, little is known about the mechanism of action of these peptides. It would be highly appropriate to use this method to modify such RiPPs with biomarkers or fluorescence probes to investigate their mechanism of action.
Figure 5.
(A) Structure of fluorescent dyes 6-FAM-alkyne and Cy5-azide. (B) Antimicrobial activity of six fluorescently labeled nisin variants. (C) Localization of six fluorescently labeled nisin variants by fluorescence microscopy. (D) Structure of the most potent fluorescently labeled nisin variant M21V-M17Aha + 6-FAM-alkyne.
Conclusions
In summary, we have demonstrated for the first time the incorporation of methionine analogues into RiPPs in L. lactis. Four methionine analogues were successfully installed at four distinct positions of the lantibiotic nisin. The genetic code of L. lactis can be regarded to be expanded by incorporating methionine analogues. The structural diversity was enhanced, while retaining or even improving antimicrobial activity against specific pathogens or Gram-positive bacteria. In addition, this study underlines that the bio-orthogonal reactive groups of ncAAs can serve as a platform for post-biosynthetic modifications, such as conjugating with peptides, or functional labels (e.g., fluorescence). The insertion of ncAAs during translation along with the possibility for their subsequent modification (postsynthetic conjugation) could further expand the chemical and functional space of RiPPs. Overall, our experiments further exemplify one of the most important applications of ncAA incorporation, that is, the functional, structural, and chemical diversification of RiPPs. This study provides an efficient method for RiPPs engineering by incorporation of ncAAs and chemical coupling.
Methods
Bacterial Strains, Plasmids, and Growth Conditions
Strains and plasmids used in this study are listed in Supplementary Table S1. All L. lactis strains were grown in M17 broth supplemented with 0.5% (w/v) glucose at 30 °C for genetic manipulation. Five μg/mL erythromycin and/or chloramphenicol were added when it was necessary. Chemical defined medium lacking tryptone (CDM-P)5 was specially used for peptide expression and methionine analogues incorporation.
Construction of Expression Vectors
The primers used in this study are listed in Supplementary Table S2. The molecular cloning techniques were performed following standard protocols.35 The preparation of competent cells and transformation were performed according to Holo and Nes.36 Fast digest restriction enzymes and ligase were used as recommended by the manufacturer. Nisin derivatives with one mutation in the core peptide (pCZ-nisA-M17I and pCZ-nisA-M21V) were produced by spice overlap extension PCR. For the construction of pCZ-nisA-M17I-M21V-M35 and pCZ-nisA-I4M-M17I-M21V, nested PCR of pCZ-nisA was used to introduce the mutation. The amplification was performed using Phusion Polymerase (Thermo Scientific) following the provider’s instructions and the primers are listed in Supplementary Table S2. After amplification and digestion with NheI and PaeI, it was ligated in pCZ-nisA digested with the same enzymes. The ligation product was desalted and transformed into L. lactis NZ9000. The plasmid was isolated and sequenced to check the integrity of the sequence.
Methionine Analogues and Fluorescent Probes
The methionine analogue l-homopropargylglycine (Hpg) was purchased from Chiralix (Nijmegen, Netherlands). l-Azidohomoalanine (Aha), l-norleucine (Nle), l-norvaline (Nva), and l-allyglycine (Alg) were purchased from Iris Biotech GmbH (Marktredwitz, Germany). l-Ethionine (Eth) was purchased from Alfa Aesar (Karlsruhe, Germany). 6-FAM-alkyne and Cy5-azide were purchased from Jena Bioscience (Thuringia, Germany).
Precursor Peptide Precipitation
L. lactis strains harboring pIL3eryBTC and pCZ-nisA were grown overnight in CDM-P with 5 μg/mL erythromycin and 5 μg/mL chloramphenicol. Subsequently, the overnight culture was diluted in 20 mL fresh CDM-P back to OD600 = 0.1. When the OD600 reached 0.4–0.6, 10 ng/mL nisin was added to induce the expression of NisBTC. Three hours later, the cells were spun down at room temperature for 8 min at 5000 rpm and then washed three times with CDM-P lacking methionine and resuspended back in the initial volume of CDM-P lacking methionine. The medium was supplemented with either methionine (38 mg/L) or 50 mg/L methionine analogues, and 0.5 mM ZnSO4 was added to induce peptide expression. After overnight growth, the supernatant was harvested by centrifugation at 8500 rpm for 20 min at 4 °C. The precursor peptides were precipitated by Trichloroacetic acid (TCA) for further analysis according to Link et al.(37) Briefly, an ice-cold 100% TCA solution was added to the supernatant to reach a final concentration of 10% TCA, and the solution was stored overnight at 4 °C. The precipitated peptide was pelleted by centrifugation at 8000 rpm for 60 min at 4 °C. The supernatant was discarded and the pellet was washed with ice-cold acetone in half the original culture volume by a second centrifugation (8000 rpm, 45 min, 4 °C). The acetone was discarded, and the remaining acetone was evaporated off over several hours at room temperature. Dried pellets were suspended in 300 μL 0.05% aqueous acetic acid solution.
Tricine-SDS-PAGE Analysis
The precipitated precursor peptides were analyzed by Tricine-SDS-PAGE according to Schagger et al.(38) 15 μL of each sample mixed with 4 μL loading dye was loaded on the gel. Coomassie brilliant blue G-250 was used to stain the gel.
LC-MS Analysis of Nisin Derivatives
The precipitated precursor peptides were injected into the LC-MS system consisting of an Ultimate 3000 UHPLC system coupled via a HESI-II electrospray source with a Q-Exactive Orbitrap-based mass spectrometer (all Thermo Scientific, San Jose, CA, USA). Three μL of each sample was loaded onto a Kinetex EVO-C18 column (2.6 μm particles, 100 × 2.1 mm, Phenomenex). The eluents for the LC separation were (A) water and (B) Acetonitrile both containing 0.1% formic acid. The following gradient was delivered at a flow rate of 0.5 mL/min: 10% B until 1 min; then linear to 40% B in 9 min; linear to 80% B in 2 min; hold in 80% B for 2 min, after which a switch back to 10% B was performed in 0.1 min. After 5 min of equilibration the next injection was performed. The LC column was kept at 60 °C. The HESI-II electrospray source was operated with the parameters recommended by the MS software for the LC flow rate used (Spray voltage 3.5 kV (positive mode)); other parameters were sheath gas 50 AU, auxiliary gas 10 AU, cone gas 2 AU; capillary temperature 275 °C; heater temperature 400 °C. The samples were measured in positive mode from m/z 500–2000 at a Resolution of 140 000 @ m/z 200. The instrument was calibrated in positive mode using the Pierce LTQ Velos ESI positive-ion calibration solution (Thermo Fisher Scientific, Rockford, USA) (containing caffeine, the tetrapeptide MRFA and a mixture of fluorinated phosphazines ultramark 1621). The system was controlled using the software packages Xcalibur 4.1, SII for Xcalibur 1.3 and Q-Exactive Tune 2.9 (Thermo Fisher Scientific). The Xtract-algorithm within Xcalibur was used for deconvolution of the isotopically resolved data.
Purification of Nisin and Its Derivatives
To obtain pure peptides for activity test, the supernatant of 1 L culture was first incubated with purified NisP39 at 37 °C for 3 h to cleave off the nisin leader, and then the supernatant was loaded on a C18 open column (Spherical C18, particle size: 40–75 μm, Sigma-Aldrich). The column was washed and eluted with different concentrations of buffer B (buffer A, Milli-Q with 0.1% TFA; buffer B, acetonitrile with 0.1% TFA). The active fractions were lyophilized and further purified by HPLC using an Agilent 1200 series HPLC with a RP-C12 column (Jupiter 4 μm Proteo 90A, 250 × 4.6 mm, Phenomenex). The peak that is the fully modified peptide with the correct molecular weight was lyophilized and stored as powder until further use.
MALDI-TOF Mass Spectrometry Characterization
One μL of each sample was spotted, dried and washed with Milli-Q water on the target. Subsequently, 1 μL of 5 mg/mL a-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) was spotted on top of the sample. An ABI Voyager DE Pro (Applied Biosystems) matrix-assisted laser desorption/ionization time-of-flight analyzer (MALDI-TOF) operating in linear mode using external calibration was used to obtain mass spectra.40
Agar Well Diffusion Assay
Antimicrobial activity was tested against M. flavus according to protocols described previously.40 HPLC purified and lyophilized peptides were resuspended in 0.05% aqueous acetic acid solution and the peptide amount was quantified by HPLC according to Schmitt et al.(5) 0.15 nmol of sample was added to each well. The agar plate was incubated at 30 °C overnight, after which the zone of inhibition was measured.
Determination of the Minimal Inhibitory Concentration (MIC)
For the MIC assay, the indicator strains CAL-MRSA, MW2-MRSA, E. faecalis, E. faecium, B. cereus, L. monocytogenes, and L. lactis were first streaked on GM17 plate and cultured overnight. The peptide samples were diluted with 0.05% acetic acid to a concentration of 4–128 μg/mL (depending on the estimated activity of the peptide and the strain tested). GM17 broth was used for the activity test against E. faecium, L. monocytogenes, and L. lactis. MHB was used for the activity test against CAL-MRSA, MW2-MRSA, E. faecalis, and B. cereus. The MIC value test was performed according to Wiegand et al.(41)
Preparation of Nisin ABC-Azide
Nisin was digested using chymotrypsin to generate nisin ABC. The truncated nisin molecule can be readily purified in accordance with protocols reported previously.42 Nisin (180 mg) was dissolved in 150 mL Tris buffer (25 mmol Tris acetate, pH 7.5) and the solution was cooled on ice for 15 min. Then chymotrypsin (15 mg) was added and stirred at room temperature for 15 min. The reaction was performed at 30 °C for 16 h and an extra 15 mg chymotrypsin was added. After 24 h incubation, another 15 mg chymotrypsin was added and incubated for another 24 h. The reaction mixture was acidified with HCl (1 M) to pH 4.0 followed by adding 3 mL MeCN and concentrated in vacuo. The pure nisin ABC was purified from the mixture by RP-HPLC and lyophilized to obtain a white powder (20 mg). Nisin ABC (10 mg, 6.5 μmol) was dissolved in DMF (50 μL) and azidopropylamine (44 μL, 43.2 mg, 432 μmol), PyBOP (Benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate) (9 mg, 17.2 μmol), and DIPEA (N,N-diisopropylethylamine) (6 μL, 34.8 μmol) were added. The reaction was vortexed for 20 min and subsequently quenched with 5 mL buffer (H2O:MeCN, 95:5 + 0.1% TFA). The reaction mixture was purified by HPLC and pure nisin ABC-azide was lyophilized to obtain a white powder (8 mg).
Click Chemistry
A stock solution of CuSO4 (10 mg, 100 mM), BTTAA (2-(4-((bis((1-(tert-butyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)-acetic acid) (25 mg, 50 mM), and sodium ascorbate (200 mg, 1 M) in ddH2O were prepared, aliquoted and stored at −20 °C for further use. M17I-M21Aha (70 μg, 0.02 μmol) and M17I-M21Hpg (70 μg, 0.02 μmol) were dissolved in 100 mM phosphate buffer (pH 7.0, final reaction volume: 200 μL). Then, CuSO4 (4 μL, 0.4 μmol): BTTAA (40 μL, 2 μmol)-premix were added followed by the addition of sodium ascorbate (20 μL, 20 μmol). The reaction was performed at 37 °C for 1 h and purified directly by RP-HPLC. M17I-M21V-M35Aha, M17I-M21V-M35Hpg, M21V-M17Aha, and M21V-M17Hpg were reacted either mutually or with nisin ABC-azide (40 μg, 0.02 μmol), Cy5-azide (5 μL, 10 mg/mL), or 6-FAM-alkyne (4 μL, 10 mg/mL) at the above conditions. The reaction was further scaled up in ratio to obtain more products. The reaction products were purified directly by HPLC and the peak with the correct molecular weight was lyophilized and stored as powder until further use.
Fluorescence Microscopy
Cultures of overnight grown E. faecium were diluted 1:100 and incubated in GM17 at 37 °C for about 4 h to reach OD600 of 0.5. Then, 0.5 mL of culture were centrifuged at 7000 rpm for 3 min. Fluorescently labeled nisin variants were added into the Eppendorf tube with cells at desired concentration in 100 μL saline solution and cells were incubated at 37 °C for 30 min. After three other washes in saline buffer, 0.6 μL bacterial suspensions and 1% low-melting-point agar were added to a microscopy plate and the localization of nisin variants were inspected with a Delta Vision Elite inverted epifluorescence microscope (Applied Precision, GE Healthcare, Issaquah, WA, USA) equipped with a stage holder, a climate chamber, a seven-color combined set InsightSSI Solid-state Illumination module and an sCMOS camera (PCO AG, Kelheim, Germany). Excitation was set to 646 nm and emission to 662 nm to capture Cy5-azide fluorescence. For 6-FAM-alkyne fluorescence, we employed 490 nm for excitation and emission at 513 nm. Images were obtained by ImageJ software.
Acknowledgments
We thank Manuel Montalban-Lopez (Department of Microbiology, Faculty of Sciences, University of Granada, Spain) for his valuable suggestions during the project. JD and JC were supported by Chinese Scholarship Council (CSC). JHV was supported by The Netherlands Organization for Scientific Research (NWO, ALW OP.214).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00308.
Supporting figures that provide MALDI-TOF MS data of dimeric nisin constructs, nisin hybrids, and fluorescently labeled nisin variants; supporting tables that list strains, plasmids, and primers used in this study (PDF)
Author Contributions
JD designed and carried out the experiments, obtained and analyzed data, and wrote the manuscript. JHV contributed to the manuscript. JC contributed to the data interpretation. OPK conceived and supervised the project and corrected the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Willey J. M.; van der Donk W. A. (2007) Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501. 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Montalban-Lopez M.; van Heel A. J.; Kuipers O. P. (2017) Employing the promiscuity of lantibiotic biosynthetic machineries to produce novel antimicrobials. FEMS Microbiol. Rev. 41, 5–18. 10.1093/femsre/fuw034. [DOI] [PubMed] [Google Scholar]
- Escano J.; Smith L. (2015) Multipronged approach for engineering novel peptide analogues of existing lantibiotics. Expert Opin. Drug Discovery 10, 857–870. 10.1517/17460441.2015.1049527. [DOI] [PubMed] [Google Scholar]
- Ongey E. L.; Neubauer P. (2016) Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb. Cell Fact. 15, 97. 10.1186/s12934-016-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S.; Montalban-Lopez M.; Peterhoff D.; Deng J.; Wagner R.; Held M.; Kuipers O. P.; Panke S. (2019) Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat. Chem. Biol. 15, 437–443. 10.1038/s41589-019-0250-5. [DOI] [PubMed] [Google Scholar]
- Blaskovich M. A. (2016) Unusual amino acids in medicinal chemistry. J. Med. Chem. 59, 10807–10836. 10.1021/acs.jmedchem.6b00319. [DOI] [PubMed] [Google Scholar]
- Baumann T.; Nickling J. H.; Bartholomae M.; Buivydas A.; Kuipers O. P.; Budisa N. (2017) Prospects of in vivo incorporation of non-canonical amino acids for the chemical diversification of antimicrobial peptides. Front. Microbiol. 8, 124. 10.3389/fmicb.2017.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa N. (2013) Expanded genetic code for the engineering of ribosomally synthetized and post-translationally modified peptide natural products (RiPPs). Curr. Opin. Biotechnol. 24, 591–598. 10.1016/j.copbio.2013.02.026. [DOI] [PubMed] [Google Scholar]
- Sun S. B.; Schultz P. G.; Kim C. H. (2014) Therapeutic applications of an expanded genetic code. ChemBioChem 15, 1721–1729. 10.1002/cbic.201402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldach F.; Al Toma R.; Kuthning A.; Caetano T.; Mendo S.; Budisa N.; Sussmuth R. D. (2012) Congeneric lantibiotics from ribosomal in vivo peptide synthesis with noncanonical amino acids. Angew. Chem., Int. Ed. 51, 415–418. 10.1002/anie.201106154. [DOI] [PubMed] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem., Int. Ed. 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
- Koopmans T.; Wood T. M.; Hart P.; Kleijn L. H.; Hendrickx A. P.; Willems R. J.; Breukink E.; Martin N. I. (2015) Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent Compound. J. Am. Chem. Soc. 137, 9382–9389. 10.1021/jacs.5b04501. [DOI] [PubMed] [Google Scholar]
- Slootweg J. C.; van der Wal S.; Quarles van Ufford H. C.; Breukink E.; Liskamp R. M.; Rijkers D. T. (2013) Synthesis, antimicrobial activity, and membrane permeabilizing properties of C-terminally modified nisin conjugates accessed by CuAAC. Bioconjugate Chem. 24, 2058–2066. 10.1021/bc400401k. [DOI] [PubMed] [Google Scholar]
- Slootweg J. C.; Peters N.; Quarles van Ufford H. L.; Breukink E.; Liskamp R. M.; Rijkers D. T. (2014) Semi-synthesis of biologically active nisin hybrids composed of the native lanthionine ABC-fragment and a cross-stapled synthetic DE-fragment. Bioorg. Med. Chem. 22, 5345–5353. 10.1016/j.bmc.2014.07.046. [DOI] [PubMed] [Google Scholar]
- Arnusch C. J.; Bonvin A. M. J. J.; Verel A. M.; Jansen W. T. M.; Liskamp R. M. J.; de Kruijff B.; Pieters R. J.; Breukink E. (2008) The vancomycin-nisin (1–12) hybrid restores activity against vancomycin resistant Enterococci. Biochemistry 47, 12661–12663. 10.1021/bi801597b. [DOI] [PubMed] [Google Scholar]
- Bolt H. L.; Kleijn L. H. J.; Martin N. I.; Cobb S. L. (2018) Synthesis of antibacterial nisin (−) peptoid hybrids using click methodology. Molecules 23, 1566. 10.3390/molecules23071566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoganathan S.; Sit C. S.; Vederas J. C. (2011) Chemical synthesis and biological evaluation of gallidermin-siderophore conjugates. Org. Biomol. Chem. 9, 2133–2141. 10.1039/c0ob00846j. [DOI] [PubMed] [Google Scholar]
- Lubelski J.; Rink R.; Khusainov R.; Moll G. N.; Kuipers O. P. (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65, 455–476. 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheigh C. I.; Pyun Y. R. (2005) Nisin biosynthesis and its properties. Biotechnol. Lett. 27, 1641–1648. 10.1007/s10529-005-2721-x. [DOI] [PubMed] [Google Scholar]
- Field D.; Cotter P. D.; Ross R. P.; Hill C. (2015) Bioengineering of the model lantibiotic nisin. Bioengineered 6, 187–192. 10.1080/21655979.2015.1049781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromgaard A.; Jensen A. A.; Stromgaard K. (2004) Site-specific incorporation of unnatural amino acids into proteins. ChemBioChem 5, 909–916. 10.1002/cbic.200400060. [DOI] [PubMed] [Google Scholar]
- Johnson J. A.; Lu Y. Y.; Van Deventer J. A.; Tirrell D. A. (2010) Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr. Opin. Chem. Biol. 14, 774–780. 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hest J. C. M.; Kiick K. L.; Tirrell D. A. (2000) Efficient incorporation of unsaturated methionine analogues into proteins in vivo. J. Am. Chem. Soc. 122, 1282–1288. 10.1021/ja992749j. [DOI] [Google Scholar]
- Abdeljabbar D. M.; Klein T. J.; Link A. J. (2011) An engineered methionyl-tRNA synthetase enables azidonorleucine incorporation in methionine prototrophic bacteria. ChemBioChem 12, 1699–1702. 10.1002/cbic.201100089. [DOI] [PubMed] [Google Scholar]
- Kiick K.L.; Weberskirch R.; Tirrell D.A. (2001) Identification of an expanded set of translationally active methionine analogues in Escherichia coli. FEBS Lett. 502, 25–30. 10.1016/S0014-5793(01)02657-6. [DOI] [PubMed] [Google Scholar]
- Field D.; Connor P. M.; Cotter P. D.; Hill C.; Ross R. P. (2008) The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol. Microbiol. 69, 218–230. 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- Field D.; Quigley L.; O’Connor P. M.; Rea M. C.; Daly K.; Cotter P. D.; Hill C.; Ross R. P. (2010) Studies with bioengineered nisin peptides highlight the broad-spectrum potency of nisin V. Microb. Biotechnol. 3, 473–486. 10.1111/j.1751-7915.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasper H. E.; de Kruijff B.; Breukink E. (2004) Assembly and stability of nisin-lipid II pores. Biochemistry 43, 11567–11575. 10.1021/bi049476b. [DOI] [PubMed] [Google Scholar]
- Pieters R. J. (2009) Maximising multivalency effects in protein-carbohydrate interactions. Org. Biomol. Chem. 7, 2013–2025. 10.1039/b901828j. [DOI] [PubMed] [Google Scholar]
- Kuipers O. P.; Bierbaum G.; Ottenwälder B.; Dodd H. M.; Horn N.; Metzger J.; Kupke T.; Gnau V.; Bongers R.; van den Bogaard P.; Kosters H.; Rollema H. S.; de Vos W. M.; Siezen R. J.; Jung G.; Götz F.; Sahl H. G.; Gasson M. J. (1996) Protein engineering of lantibiotics. Antonie van Leeuwenhoek 69, 161–170. 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- Zhou L.; van Heel A. J.; Kuipers O. P. (2015) The length of a lantibiotic hinge region has profound influence on antimicrobial activity and host specificity. Front. Microbiol. 6, 11. 10.3389/fmicb.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer K.; Wiedemann I.; Ciobanasu C.; Sahl H. G.; Kubitscheck U. (2013) Aggregates of nisin with various bactoprenol-containing cell wall precursors differ in size and membrane permeation capacity. Biochim. Biophys. Acta, Biomembr. 1828, 2628–2636. 10.1016/j.bbamem.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Imran M.; Revol-Junelles A. M.; de Bruin M.; Paris C.; Breukink E.; Desobry S. (2013) Fluorescent labeling of nisin Z and assessment of anti-listerial action. J. Microbiol. Methods 95, 107–113. 10.1016/j.mimet.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Hasper H. E.; Kramer N. E.; Smith J. L.; Hillman J. D.; Zachariah C.; Kuipers O. P.; de Kruijff B.; Breukink E. (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637. 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press. [Google Scholar]
- Holo H.; Nes I. F. (1995) Transformation of lactococcus by electroporation. Methods Mol. Biol. 47, 195–199. 10.1385/0-89603-310-4:195. [DOI] [PubMed] [Google Scholar]
- Faoro V., and Stanta G. (2011) Trichloroacetic acid (TCA) precipitation of proteins. In Guidelines for Molecular Analysis in Archive Tissues, pp 257–258, Springer. [Google Scholar]
- Schagger H. (2006) Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22. 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Montalban-Lopez M.; Deng J.; van Heel A. J.; Kuipers O. P. (2018) Specificity and application of the lantibiotic protease NisP. Front. Microbiol. 9, 160. 10.3389/fmicb.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel A. J.; Mu D.; Montalban-Lopez M.; Hendriks D.; Kuipers O. P. (2013) Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth. Biol. 2, 397–404. 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- Wiegand I.; Hilpert K.; Hancock R. E. (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Slootweg J. C.; Liskamp R. M.; Rijkers D. T. (2013) Scalable purification of the lantibiotic nisin and isolation of chemical/enzymatic cleavage fragments suitable for semi-synthesis. J. Pept. Sci. 19, 692–699. 10.1002/psc.2551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.