Summary
Spatial maps in the brain are most accurate when they are linked to external sensory cues. Here we show that the compass in the Drosophila brain is linked to the direction of the wind. Shifting the wind rightward rotates the compass as if the fly were turning leftward, and vice versa. We describe the mechanisms of several computations that integrate wind information into the compass. First, an intensity-invariant representation of wind direction is computed by comparing left-right mechanosensory signals. Then, signals are reformatted to reduce the coding biases inherent in peripheral mechanics, and wind cues are brought into the same circular coordinate system that represents visual cues and self-motion signals. Because the compass incorporates both mechanosensory and visual cues, it should enable navigation under conditions where no single cue is consistently reliable. These results show how local sensory signals can be transformed into a global, multimodal, abstract representation of space.
Keywords: mechanosensation, sensorimotor integration, central complex, ellipsoid body, Ring neuron, Johnston’s organ, AMMC, wedge, lateral accessory lobe
eTOC Blurb
Okubo et al. show that wind influences the compass in the Drosophila brain. They describe a pathway that connects mechanoreceptors to compass neurons, and they show how mechanosensory signals are transformed within this pathway. Their results demonstrate that the compass is a multimodal map, like the mammalian head direction map. (50 words)
Graphical Abstract
Introduction
Traveling in a straight path to a destination saves time and energy. It is difficult to maintain a straight path without using external sensory cues for feedback control (Cheung et al., 2007; Dacke and el Jundi, 2018; Souman et al., 2009). Thus, many animals use external compass cues, for example, adopting a particular angle relative to the sun (Figure 1A; el Jundi et al., 2019; Guilford and Taylor, 2014; Warren et al., 2019). Some animals also use the wind as a compass cue, by maintaining a fixed angle to the wind during walking (Bell et al., 1989; Bell and Kramer, 1979; Birukow, 1958; Böhm, 1995; Böhm et al., 1991; Dacke et al., 2019; Heinzel and Böhm, 1984, 1989; Linsenmair, 1969, 1972, 1973; Müller and Wehner, 2007). When an experimenter shifts the wind direction, the animal makes a corrective turn to regain its initial angle to the wind. Different individuals adopt different angles to the wind, implying that the absolute wind direction is incidental; the key is that a steady wind provides a feedback signal that indicates a drift from a straight path. Wind can be the primary compass cue when solar compass cues are unreliable (Dacke et al., 2019; Müller and Wehner, 2007).
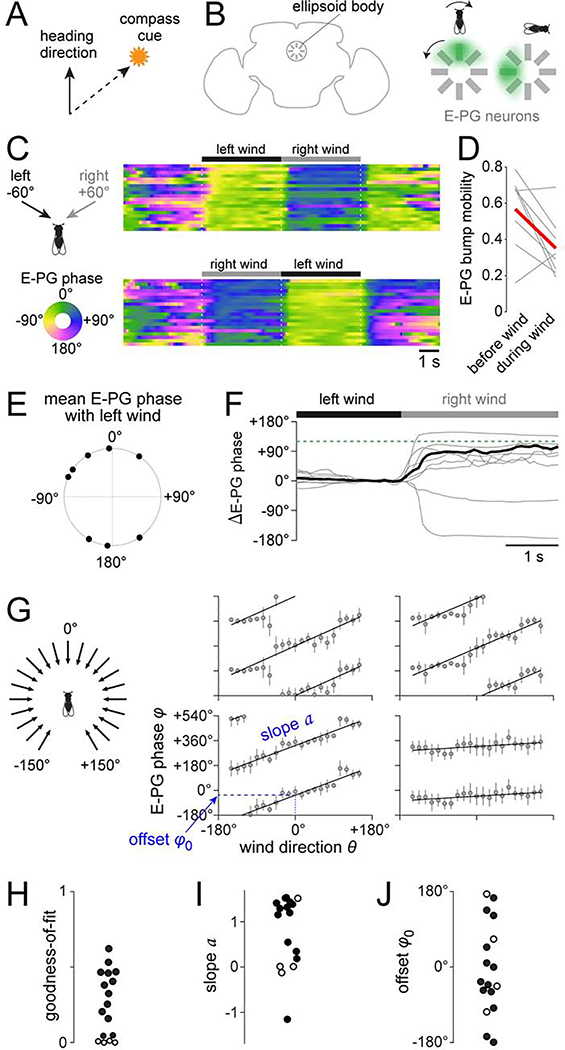
Figure 1. Wind influences the brain’s heading compass.
(A) Schematic of compass behavior. Animals can maintain a straight course by keeping a fixed angle between their heading and a compass cue.
(B) Schematic of E-PG neuron dendrites, which form a circular array in the ellipsoid body. A “bump” of activity (green shading) rotates as the fly turns.
(C) Heat map of E-PG phase (i.e. bump position) as we delivered wind from −60° or +60°. Each row is a different trial.
(D) E-PG bump mobility quantifies how much the bump moves over time. We computed mobility within each trial during the baseline period (“before wind”). We computed mobility separately for each wind direction, and then averaged these values (“during wind”). Each gray line indicates one fly and red is the mean across all the flies. Wind significantly decreased bump mobility in 6 of 8 flies; it had the opposite effect in one fly, and no significant effect in one fly; significance assessed at criterion of 0.05, Wilcoxon rank-sum tests).
(E) Mean E-PG phase during wind from −60°. Each symbol is a fly (n=8). The distribution of phases was not significantly different from uniform (p = 0.61, Rayleigh test).
(F) Change in E-PG phase during the wind shift from left to right, expressed as a change from baseline (0–1 s before the wind shift). Each gray line is trial-averaged data for one fly, and the black line is mean across flies (n=8). Dashed line indicates 120°, which was the separation between left and right wind.
(G) Mean E-PG phase verus wind direction for four flies, with fits to φ = [a ·θ + φ0] (mod 360°). Error bars show angular deviation. See Figure S1F for all flies.
(H) Goodness-of-fit of the linear model. Each symbol is a fly. Fit was statistically significant in 13 flies (solid symbols) and not significant in 4 flies (open symbols). The same 4 flies are marked with open symbols in (I) and (J).
(I) Value of the slope a.
(J) Value of the offset φ0 between the wind and the bump.
In insects, the brain area that mediates compass navigation is the central complex (Heinze et al., 2018; Pfeiffer and Homberg, 2014), which contains a topographic map of heading direction – i.e., an internal compass. In Drosophila, the neurons that form this map are called E-PG neurons (Figure 1B; Seelig and Jayaraman, 2015). Genetically silencing E-PG neurons eliminates compass navigation (Giraldo et al., 2018; Green et al., 2019). E-PG dendrites are arranged in a circle, and at any given moment, there is a single “bump” of activity which rotates around the circle as the fly turns. This occurs even in the absence of external compass cues, because E-PG neurons receive updates about the fly’s angular velocity, and they integrate this signal over time to generate a running estimate of the fly’s angular position (Green et al., 2017; Turner-Evans et al., 2017).
External sensory cues provide an important input to the compass, because the compass accumulates errors when these cues are absent. In particular, a prominent visual cue in a closed-loop virtual reality environment can make the E-PG compass more accurate (Green et al., 2017; Seelig and Jayaraman, 2015). The influence of a visual cue can be shown by a cue shift: if the cue is shifted rightward, the bump rotates as if the fly were turning leftward (Green et al., 2019; Seelig and Jayaraman, 2015). Whether wind cues provide input to the compass has not been investigated.
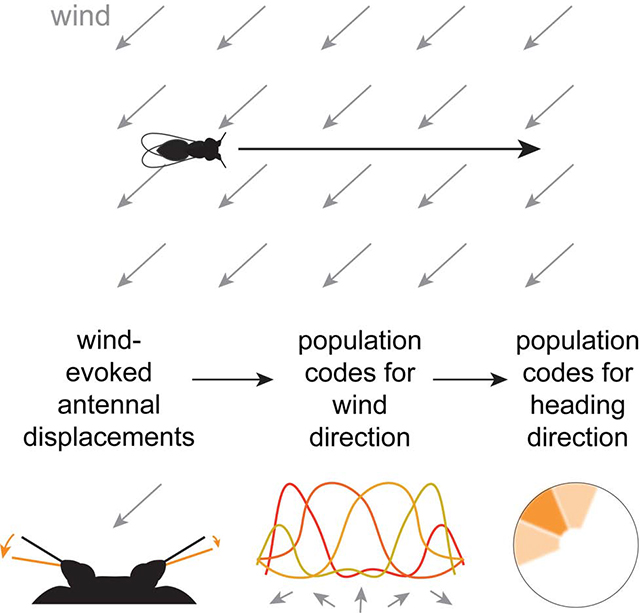
Here we show that the Drosophila heading compass is indeed influenced by wind. Moreover, we identify a neural pathway that connects mechanosensory centers with the compass. Finally, we show how this pathway transforms wind-induced antennal displacements into a global, abstract representation of space.
Results
Influence of wind on the E-PG compass
We began by asking whether wind can influence the compass in the brain. We performed two-photon calcium imaging using GCaMP6f to monitor the activity of the E-PG ensemble while delivering wind alternately from two horizontal angles, −60° and +60° (Figures 1C, S1A and S1B). We focused initially on flies whose legs were suspended in the air to minimize the contribution of feedback signals associated with walking. All experiments were performed in the dark, in the absence of visual cues.
Before wind onset, the E-PG bump was typically moving spontaneously. After wind onset, the bump moved to a fixed location and stayed there (Figures 1C, 1D, and S1D). Typically, the bump moved consistently to the same location in response to wind from a given direction. However, that same wind direction drove the bump to different locations in different individuals (Figure 1E). When we switched the wind direction, the bump of activity often moved to a new location, and this was consistent over multiple trials (Figures 1C, 1F, and S1D). That said, we occasionally saw abrupt spontaneous changes in the “offset” between wind direction and bump position (Figure S1E), reminiscent of the changes that can occur in the offset between visual cues and bump position (Seelig and Jayaraman, 2015). Wind offset also elicited bump movement (Figure 1C), which may be explained by adaptation in the mechanosensory inputs to E-PG neurons (see below).
The effect of wind was most consistent when the fly’s legs were suspended in the air. It was less consistent when the fly was walking, suggesting that wind cues compete with walking-related signals for control of the compass. Nonetheless, wind could influence the compass even in walking flies (Figures S1H and S1I).
A systematic relationship between wind direction and the E-PG compass
If wind is acting as a compass cue, it should be able to move the bump through its full dynamic range, with a one-to-one mapping between wind direction and bump position. We measured the bump’s position while delivering wind from 21 directions in a random sequence (Figure 1G). As a control, we verified that the bump only responded to the movement of the wind-delivery tube when air was flowing through the tube (Figure S1G). In most flies (13 of 17), the relationship between wind direction (θ) and E-PG phase (φ) was well-fit by a circular-linear model (Figure 1H):
| Equation 1 |
The sign of the slope (a) indicates the direction of the bump’s circular movement. In most cases, a was positive (Figure 1I), meaning that clockwise wind shifts produce clockwise bump rotations, as viewed from the posterior side of the head. Clockwise visual cue shifts produce clockwise bump rotations (Seelig and Jayaraman, 2015), and so wind has the directional effect we would predict for a compass cue. It is also known that the bump rotates clockwise when flies make counter-clockwise behavioral turns in darkness (Green et al., 2017; Seelig and Jayaraman, 2015; Turner-Evans et al., 2017). Thus, the compass rotates in the same direction in response to (1) counter-clockwise behavioral turns, (2) clockwise shifts of the wind, and (3) clockwise shifts of the visual scene.
The magnitude of the slope (a) indicates how far the bump moves when the wind shifts. In most individuals, a was close to 1 (Figure 1I). This implies that, on average, any given wind shift produces an equal angular displacement of the E-PG bump, which is what we predict if wind is acting as a compass cue. In four individuals, the model fit was good but the slope (a) was close to 0 or negative. In these individuals, the effects of wind may be opposed by motor commands.
Finally, φ0 indicates the offset of the bump relative to the wind (Figure 1J). Different individuals had different values of φ0. This is reminiscent of the observation that different individuals have different bump offsets relative to a visual cue (Fisher et al., 2019; Giraldo et al., 2018; Green et al., 2017; Seelig and Jayaraman, 2015). It suggests that the mapping of wind cues onto the compass is flexible, not fixed.
R neurons that carry wind signals to the heading compass
Next, we looked for cells that send wind direction information to the compass. The known sensory inputs to E-PG neurons originate with R neurons, also called Ring neurons because each axon forms a ring around the circular E-PG array. Most R neurons have dendritic arbors in the bulb (BU; Figure S2A), a visual brain region (Ito et al., 2014; Omoto et al., 2017; Seelig and Jayaraman, 2013). However, there are also “atypical” R neurons whose putative dendritic arbors reside in the lateral accessory lobe (LAL; Franconville et al., 2018; Omoto et al., 2018; Renn et al., 1999). The LAL has received little study in Drosophila (Namiki and Kanzaki, 2016), and nothing is known about the physiology of these neurons. We hypothesized that these R neurons might carry wind direction information, given that the LAL receives wind input in other insects (Homberg, 1994).
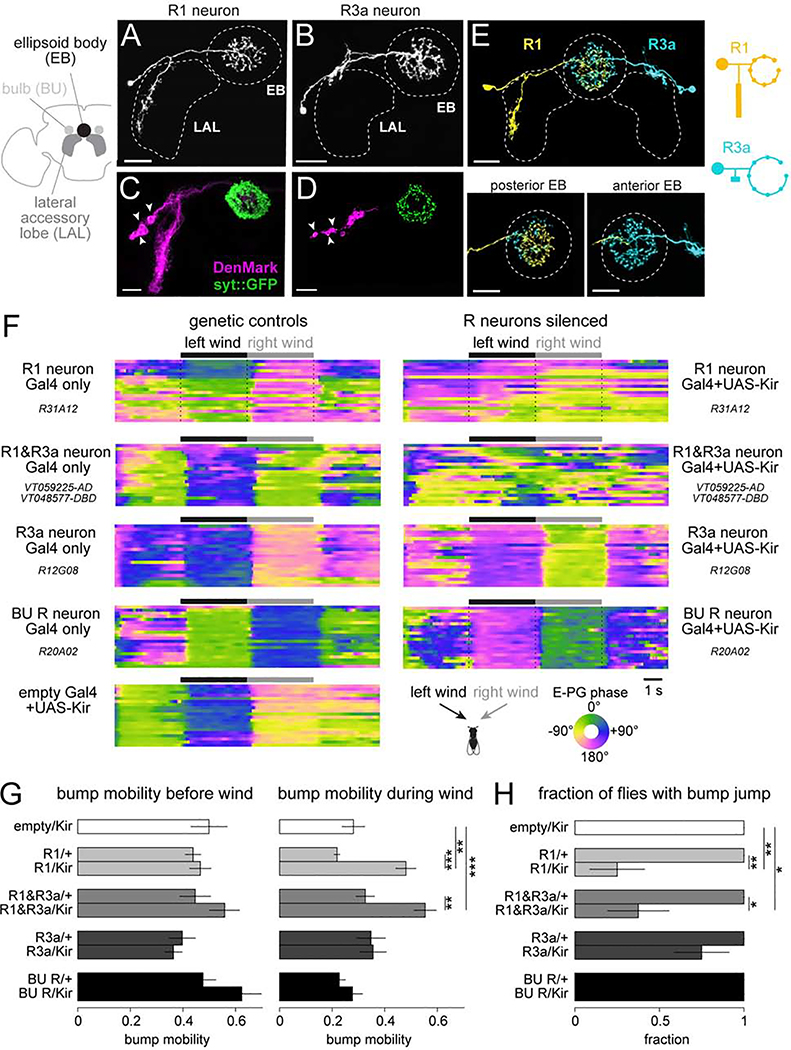
There are two R neuron types in the LAL, R1 in the lateral LAL (R1 neurons; Figures 2A and S2B) and R3a in the dorsal LAL (Figures 2B and S2C). R1 and R3a neurons innervate the posterior and anterior EB, respectively (Figure 2E). We used polarity markers to confirm that the LAL arbors of these neurons are dendrites, and their EB arbors are axons (Figures 2C and 2D). Both R1 and R3a neurons have been described by Omoto et al. (2018); we follow the naming conventions in that study, but it should be noted that the name “R1” has been used to refer to other R neuron types in the literature (see Table S1).
Figure 2. The influence of wind on E-PG neurons requires R1 neurons.
(A) Single-cell labeling of an R1 neuron using MultiColor FlpOut (MCFO).
(B) MCFO labeling of an R3a neuron. Both R1 and R3a have smooth neurites in the LAL and boutons in the EB.
(C) In R1 neurons, DenMark localizes to the LAL, whereas synaptotagmin::GFP localizes to the EB. LAL labeling is bilaterally symmetric, but only one LAL is shown. Arrowheads indicate somata.
(D) Same but for R3a neurons.
(E) Simultaneous two-color labeling of an R1 and R3a neuron using MCFO. R1 innervates the posterior EB; R3a innervates the anterior EB. Schematic at right compares R1 and R3a neuron morphologies.
(F) Heat map of the E-PG phase (i.e. bump location) as we delivered wind from −60° and +60°. Shown are 9 example flies, one per genotype. Within a block, each row is a trial.
(G) E-PG bump mobility before wind (left) and during wind (right), for each genotype, mean ± SEM across flies (n = 8 flies per genotype). Before wind, there is no significant difference between the Gal4/+ and Gal4/Kir conditions (two-way ANOVA with Gal4 line as one factor and the presence of Kir as another factor, p=0.09). During wind, there is a significant difference between the Gal4/+ and Gal4/Kir conditions (two-way ANOVA with Gal4 line as one factor and Kir as another factor, p=9.2×10–6) and the interaction term is also significant (p=0.004). Asterisks show results from post-hoc two-sample t-tests. See Figure S2J for individual flies.
(H) Fraction of flies where a wind direction switch evoked a significant E-PG phase change; mean ± SEM across flies, n = 8 flies per genotype. Fisher’s exact test. *p<0.05, **p<0.01, ***p<0.001. Scale bars in (A)-(E) are 20 μm.
To silence these neurons, we expressed the inwardly rectifying potassium channel Kir2.1 under the control of selective Gal4 lines for R1 or R3a (Figure S2D), as well as a split-Gal4 that we constructed to target both cell types (Figure S2E). We confirmed that Kir2.1 hyperpolarized R neurons and decreased their input resistance (Figures S2F and S2G). We then imaged the E-PG neuron ensemble while delivering wind alternately from −60° and +60°. In flies where R1 neurons were silenced, steady wind no longer stabilized the E-PG bump (Figures 2F, 2G, and S2H-J), although bump stability during the baseline period was not affected (Figure 2G). Moreover, a smaller proportion of these flies exhibited a bump jump after a wind direction shift (Figures 2H, S2H, and S2I). Silencing R1 and R3a neurons together produced the same phenotype. In both genotypes, there was a small residual effect of wind; this may reflect incomplete silencing (Figures S2F and S2G), incomplete coverage of R1/R3a neurons in these Gal4 lines (Figure S2D), and/or a small contribution from other cell types. We did not find a significant effect of silencing R3a neurons only (Figures 2G and 2H). We also did not find a significant effect of silencing R neurons that have dendrites in the BU (Figures 2G and 2H), which is a manipulation that severely attenuates visual responses in E-PG neurons (Fisher et al., 2019).
Direction-selective wind responses in R neurons
Our silencing experiments implied that R1 neurons are the primary pathway that relays wind signals to E-PG neurons. This motivated us to investigate wind responses in R1 neurons. We also examined wind responses in R3a neurons, because pilot imaging experiments revealed that both cell types respond to wind (Figures S3A-D).
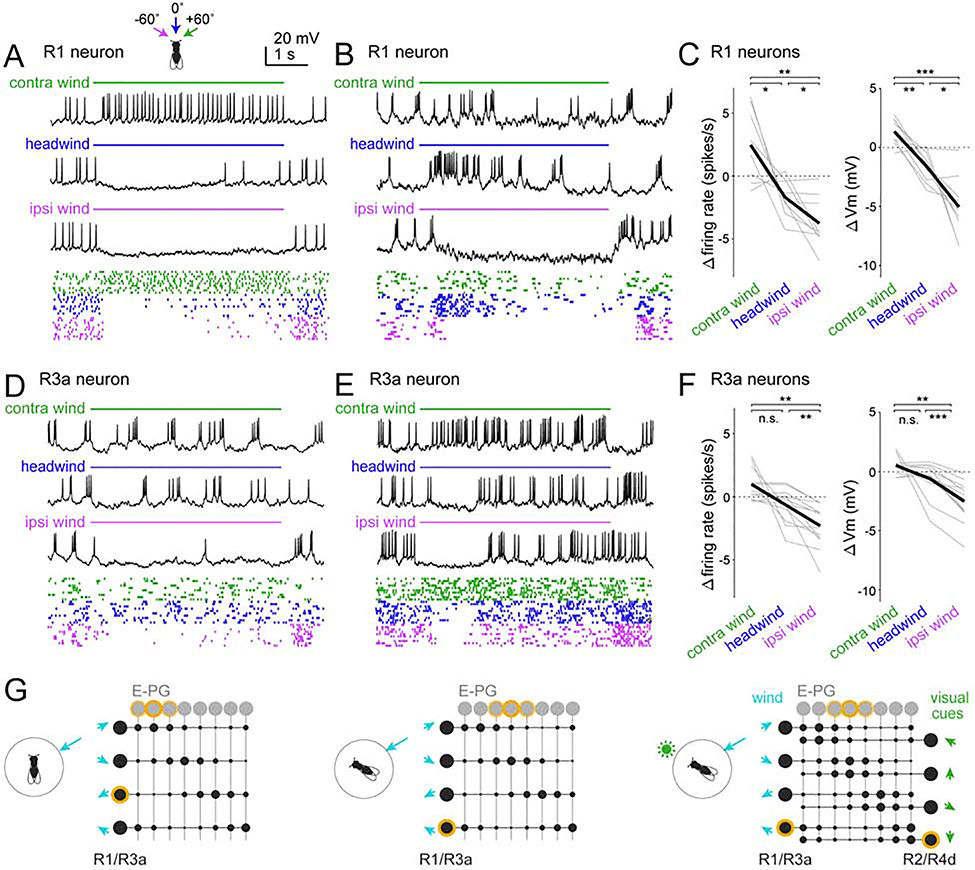
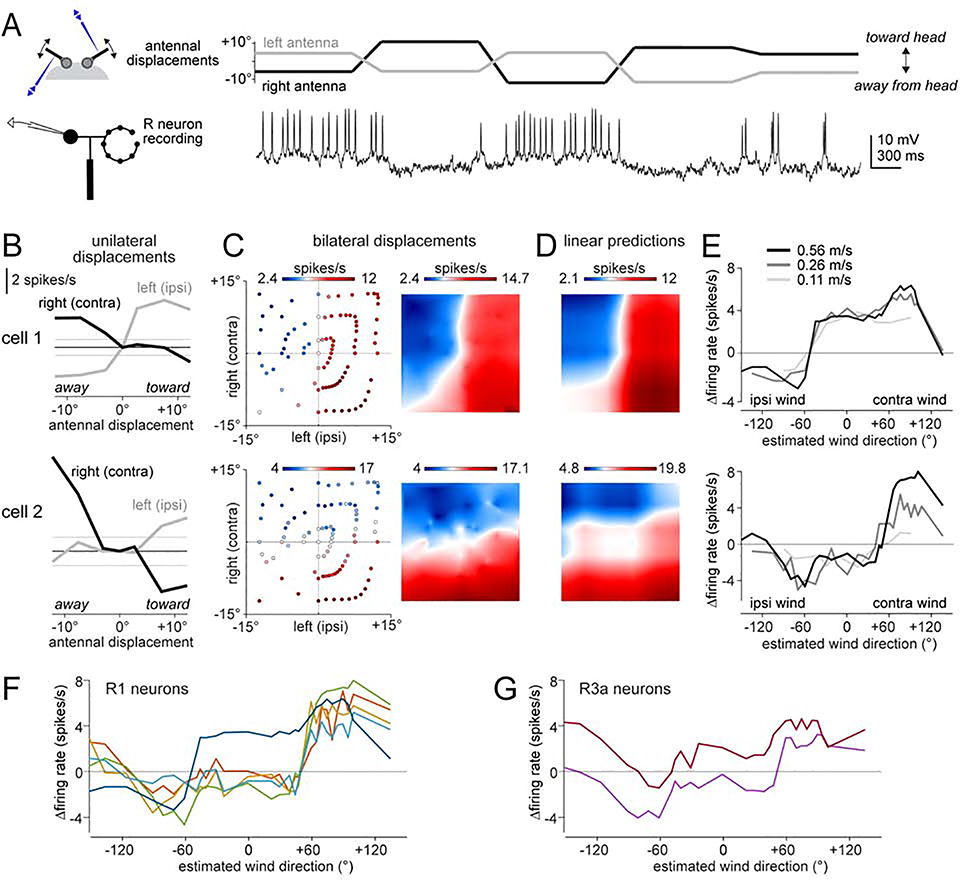
We performed in vivo GFP-targeted whole-cell recordings from R1 and R3a neurons while delivering wind from −60°, 0°, and +60° (Figure 3A-D). We found that wind from the ipsilateral side generally suppressed baseline activity in these cells (baseline spike rates were 4.5 ± 1.7 spikes/s for R1, n=9 neurons; 5.2 ± 2.7 spikes/s for R3a, n=12 neurons; mean ± S.D. ). Conversely, contralateral wind generally increased spike rates. Headwind could have either effect. Wind generally evoked steady responses, although we also observed some transient responses (Figure 3E and S3H). Responses were generally smaller in R3a neurons than R1 neurons (Figures 3C and 3F), supporting the notion that R1 neurons are the primary driver of E-PG wind responses. Within both the R1 population and the R3a population, we found varied wind direction preferences.
Figure 3: R1 and R3a neurons show direction-selective wind responses.
(A) R1 neuron responses to three wind directions. Raster plots show 10 randomly chosen trials.
(B) Same but for another R1 neuron with different tuning.
(C) Responses of all R1 neurons. Within each panel, each gray line is a neuron (1 per fly), black line is the mean across flies. Responses are averaged over the entire stimulus period and all trials and are expressed as changes from baseline. Responses depended on wind direction for both firing rate (p=2.4×10−4) and membrane potential (p=9.7×10−6); both tests one-way repeated measures ANOVA followed by paired t-tests with Bonferroni corrections; n = 9 neurons.
(D-F) Same as (A-C) but for R3a. Responses depended on wind direction for both firing rate (p=1.3×10−4) and membrane potential (p=3.5×10−5); both tests one-way repeated measures ANOVA followed by paired t-tests with Bonferroni corrections; n = 12 neurons.
R1 and R3a are significantly different (p=0.006, two-way repeated measures ANOVA with R1/R3a as one factor and wind direction as the within-subject factor, interaction between neuron type and wind direction). *p<0.05, **p<0.01, ***p<0.001
(G) Model of R→E-PG connectivity. Each R neuron inhibits a subset of E-PG neurons, disinhibiting other E-PG neurons. R neurons with adjacent preferred wind directions target adjacent E-PG subsets. Co-activated mechanosensory R neurons (R1/R3a neurons) and visual R neurons (R2/R4d neurons) connect to the same E-PG neurons. Active neurons are highlighted in yellow. Synaptic weights are represented as circles, with larger circles denoting larger weights (stronger inhibition).
At wind offset, an excitatory wind response in these neurons was often followed by an inhibitory off-response, and vice versa (Figures 3A-E and S3A, C, H). These off-responses are suggestive of adaptation. R neuron off-responses may account for E-PG off-responses (Figures 1C and 2F), because anything that causes differential firing rates in different R neurons should tend to drive the E-PG bump to a new location.
In summary, both R1 neurons and R3a neurons encode wind direction. Both cell types exhibit bidirectional responses: wind can elevate or suppress their firing rates, depending on the direction of the wind. Different neurons have varied wind direction preferences. We propose that there is a systematic relationship between R neuron wind direction preferences and R→E-PG synaptic weights, so that wind direction is mapped topographically onto the E-PG array (Figure 3G). If this topographical map were properly aligned with the visual map in E-PG neurons, then mechanosensory and visual cues could work together to drive the bump of E-PG activity to the appropriate location.
Sensitivity to bilateral antennal displacements in R1 and R3a neurons
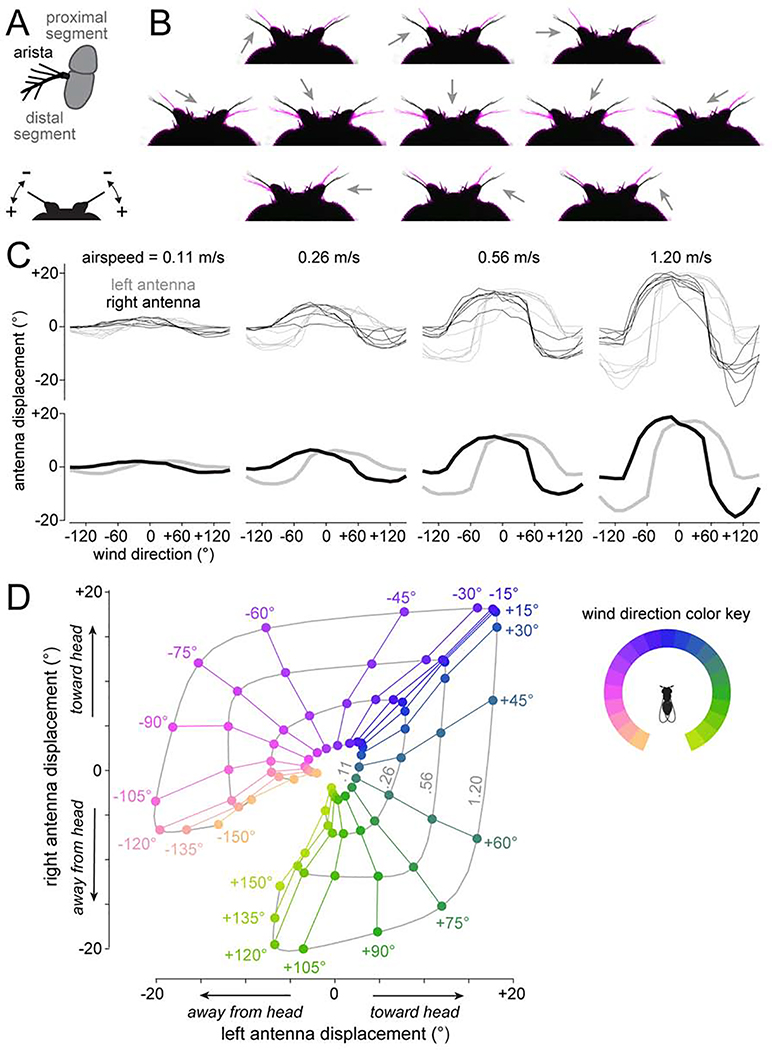
To understand the origins of wind signals in R neurons, we next considered peripheral wind responses. Walking Drosophila sense wind direction via displacements of their antennae (Yorozu et al., 2009). Wind displaces each antenna by exerting force on the arista, a sail-like structure rigidly coupled to the distal antennal segment (Figure 4A). This causes the distal antennal segment to rotate, and these rotations are encoded by peripheral mechanosensory neurons in the antennae (Matsuo et al., 2014; Patella and Wilson, 2018; Suver et al., 2019; Yorozu et al., 2009).
Figure 4: Antennal mechanics are nonlinearly sensitive to lateral wind directions.
(A) Top: schematic of the arista and antenna in frontal-medial view. Wind exerts force on the arista and rotates the distal antennal segment relative to the proximal segment. Bottom: schematic of the head in dorsal view. Wind can push the antenna toward (+) or away from the head (−).
(B) Dorsal view of the antennae with resting position in black, and wind-induced positions in magenta. Arrows are wind direction (wind speed: 1.20 m/s).
(C) Antennal displacements as a function of wind direction and wind speed. Top: individual flies. Bottom: mean across flies.
(D) Data from one fly in (C), displayed as trajectories in 2-D displacement space. In this plot, we pooled mirror-reflected data from the left and right to generate a symmetric map. Data points are color-coded by wind direction. Displacements measured at the same wind speed are connected, with the corresponding wind speeds (m/s) in gray type.
We used video tracking to measure bilateral antennal displacements evoked by 21 different wind directions and four wind speeds (Figures 4B-D, and Video S1). We found that the antennae are especially sensitive to lateral wind directions, meaning that there is a large change in antennal displacement for a small change in wind direction. In contrast, the antennae are less sensitive for frontal directions near 0°.
Note that wind direction can only be deduced by combining information from both antennae (Figure 4D; Patella and Wilson, 2018; Suver et al., 2019; Yorozu et al., 2009). For a given windspeed, there are typically two distinct wind directions that can produce any given antennal displacement. Moreover, increasing the windspeed increases the antennal displacements produced by any given wind direction. Bilateral integration would allow these ambiguities to be resolved.
To determine whether R neurons perform bilateral integration, we attached a piezoelectric actuator to each antenna (Figure 5A). We found that every R1 and R3a neuron responded to displacement of either antenna alone. Moreover, every neuron in the same brain hemisphere had essentially the same direction preferences: excitation when the ipsilateral antenna was moved toward the head, and inhibition when it was moved away from the head (Figures 5B and S4A). This direction preference was reversed for the contralateral antenna. Accordingly, the best bilateral stimulus was virtually always “ipsi-toward, contra-away” (Figures 5C and S4B).
Figure 5: R1 and R3a neurons combine displacement signals from both antennae.
(A) Example recording of an R1 neuron in the left hemisphere. Antennal displacements were ordered pseudo-randomly, and were maintained for 1 -s periods, with ramps in between.
(B) Top: responses of an R1 neuron in the left hemisphere to unilateral stimuli (i.e., stimuli where one antenna was displaced while holding the other antenna was held in its resting position). The horizontal line is this cell’s mean firing rate when both antennae are at rest (±95% confidence interval). Bottom: same for another R1 neuron in the left hemisphere. Whereas cell 1 is more sensitive to the ipsilateral antenna, cell 2 is more sensitive to the contralateral antenna.
(C) Left: scatterplots show responses of the same two R1 neurons, for all tested combinations of left and right displacements. Right: continuous maps obtained by 2D-interpolation of these scatterplots.
(D) Predicted bilateral responses of the same two R1 neurons, obtained by linearly combining each cell’s responses to unilateral stimuli alone, and then interpolating. See Figure S4 for other R1 neurons and R3a neurons.
(E) Estimated wind direction tuning curves of the same two example R1 neurons, at three different wind speeds. The transformation from antennal coordinates to wind coordinates was calculated from the measurements in Figure 4D.
(F) Estimated wind direction tuning curves for five recorded R1 neurons (wind speed 0.56 m/s).
(G) Estimated wind direction tuning curves for two recorded R3a neurons.
That said, we found variability across neurons in their relative sensitivity to each antenna. Some neurons in the left hemisphere that were slightly more sensitive to the left antenna (e.g., cell 1, Figures 5B-D) and other neurons in the left hemisphere that were more sensitive to the right antenna (e.g., cell 2, Figures 5B-D). Differential sensitivity to the two antennae can explain why these neurons have diverse tuning curves in antennal displacement space (Figures 5C and S4B).
Next, we asked whether signals from the two antennae are combined linearly. We used each neuron’s responses to unilateral stimuli to generate a linear prediction of the same neuron’s responses to bilateral stimuli. This linear prediction was a good fit to the data (Figures 5D and S4C).
Finally, we took the wind direction and wind intensity corresponding to each pattern of bilateral antennal displacements (Figure 4D), and we transformed the results from each piezoelectric actuator experiment into a wind direction tuning curve for each wind intensity. We found that different R neurons recorded on the same side of the brain had distinct wind direction tuning curves. Notably, these tuning curves were relatively invariant to wind intensity (Figure 5E). This invariance is attributable, in part, to bilateral integration. For example, if we increase windspeed while holding wind direction at 0°, both antennae are displaced more toward the head, which increases excitatory input (from the ipsilateral antenna) as well as inhibitory input (from the contralateral antenna), resulting in little net change in the summed bilateral input.
Modeling the transformation from peripheral mechanics to R neurons to E-PG neurons
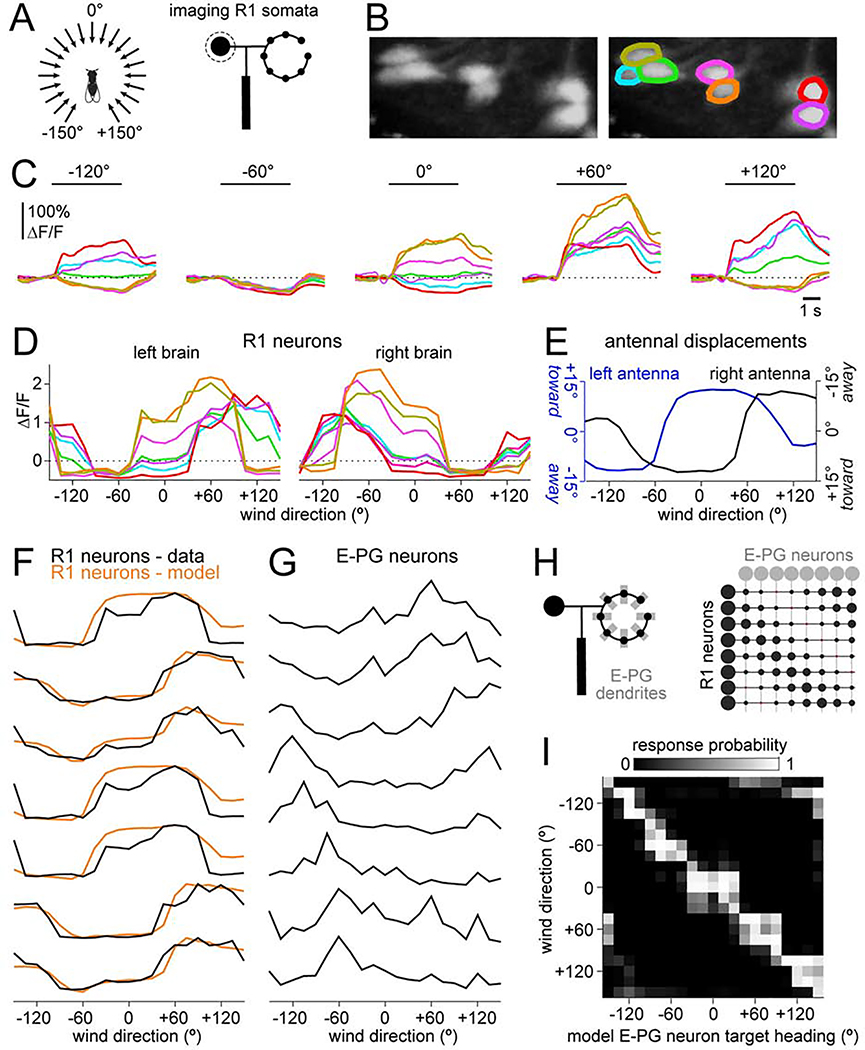
Thus far, we have been assessing R neuron diversity by comparing R neuron recordings in different individuals. A better approach is to compare R neuron responses within the same brain. We did this by simultaneously measuring GCaMP6f signals in many R neuron somata using volumetric two-photon imaging, while delivering wind from 21 directions in a random sequence (Figures 6A and 6B). Here we focused on R1 neurons because their wind responses are stronger than those of R3a neurons (Figure 3), and because R1 neurons make the primary contribution to E-PG wind responses (Figure 2).
Figure 6: R neuron tuning reflects antennal mechanics, whereas E-PG neuron tuning does not.
(A) Schematic: we used calcium imaging to monitor R1 somata while delivering wind from 21 angles in pseudo-random order.
(B) Maximum z-projection showing R1 somata in the left brain and corresponding ROIs.
(C) Time course of ΔF/F responses to several wind directions for these same ROIs.
(D) ΔF/F versus wind direction for R1 neurons in the left brain, and also for the right brain in the same individual. See Figure S5A for all R1 soma imaging experiments.
(E) Antennal displacement versus wind direction for a typical experiment (Figure 4). The right axis is inverted; this follows the preferences of left brain R1 neurons, which are excited when the left antenna is moved toward the head or the right antenna is moved away from the head.
(F) R1 tuning curves from (D), normalized so all curves have the same range. Overlaid are model fits. The only free parameters in the model were the weights of the curves in (E).
(G) E-PG tuning curves from an example experiment (from Figure 1).
(H) Schematics showing an individual R1 neuron forming potential synapses with every E-PG dendrite (left) and a matrix of patterned R1→E-PG connection weights (right).
(I) Response probabilities of model E-PG neurons, modeled as binary units. R1→E-PG weights are adjusted to maximize the probability that each E-PG neuron responds to its target heading while minimizing its responses to other headings.
These imaging experiments provided more evidence that R1 neuron responses are functionally diverse. Indeed, some wind stimuli excited one R1 neuron while inhibiting an adjacent neuron in the same hemisphere (Figure 6C). We were able to capture the shape of each tuning curve f (Figures 6D and S5A) using the measured mechanical displacements of the ipsi- and contralateral antennae (di(θ) and dc(θ), Figure 6E)
| Equation 2 |
where wl and wr are non-negative. These fits captured the diverse shapes of these tuning curves, as well as the general tendency for sharp transitions around +60° and −60° (Figure 6F).
In contrast to R1 tuning curves, E-PG tuning curves did not show a tendency for sharp transitions around any stimulus angle (Figure 6G). This implies that the R→E-PG transformation can mitigate the constraints inherent in peripheral mechanics, so that the compass is equally sensitive to wind direction changes in all compass directions. How could this occur? Each R axon overlaps with every E-PG dendrite, implying an all-to-all matrix of “potential” R→E-PG synapses (Figure 6H). Sensorimotor experience can change the sensory inputs to E-PG neurons in a manner that depends on E-PG activity, suggesting that there is associative plasticity at R→E-PG synapses (Fisher et al., 2019; Kim et al., 2019). We propose that a consistent association between wind direction and heading direction creates a pattern of R→E-PG functional connections that renders the compass equally sensitive to all wind directions.
We devised a simple model to make this hypothesis concrete. In the model, R neuron wind responses were taken from data. Each E-PG neuron was a binary unit, with output=0 if its total R neuron input was below threshold, and 1 otherwise. Thresholds and R→E-PG weights were adjusted so that each E-PG neuron responded as accurately as possible to its target heading, with different E-PG neurons for different headings. R→E-PG weights were constrained to be negative (or zero) because most R neurons are GABAergic (Xie et al., 2017; Zhang et al., 2013). We found that it was possible to find synaptic weights that allowed each model E-PG neuron to respond fairly accurately to its target heading (Figure 6I and S5C). Overall, E-PG neuron accuracy was similar for all compass angles (Figure S5D). Thus, even though R1 tuning curves are relatively flat in the region around 0°, there is still enough information in the R1 ensemble to allow reasonably accurate compass performance in this region. Whether R→E-PG weights are actually patterned in this manner is of course unknown.
To summarize, much of the diversity in R1 neuron tuning can arise from diversity in antennal input weights. On the other hand, antennal mechanics also constrain the diversity of the R1 population: these neurons inherit the mechanical sensitivity profile of the antennae. The effects of antennal mechanics appear to be mitigated in E-PG neurons, which respond to all wind direction with approximately equal accuracy. Our model shows that this transformation could be accomplished – in theory – via an appropriate pattern of R→E-PG synaptic weights.
Synaptic inputs to R1 neurons
How do antennal displacement signals reach R neurons? Peripheral antennal mechanoreceptors project mainly to a region called the antennal mechanosensory and motor center (AMMC; Kamikouchi et al., 2006). There they synapse onto second-order mechanosensory neurons (Matsuo et al., 2016). Several types of second-order mechanosensory neurons in the AMMC are wind-responsive, and project to the wedge (WED) (Chang et al., 2016; Suver et al., 2019).
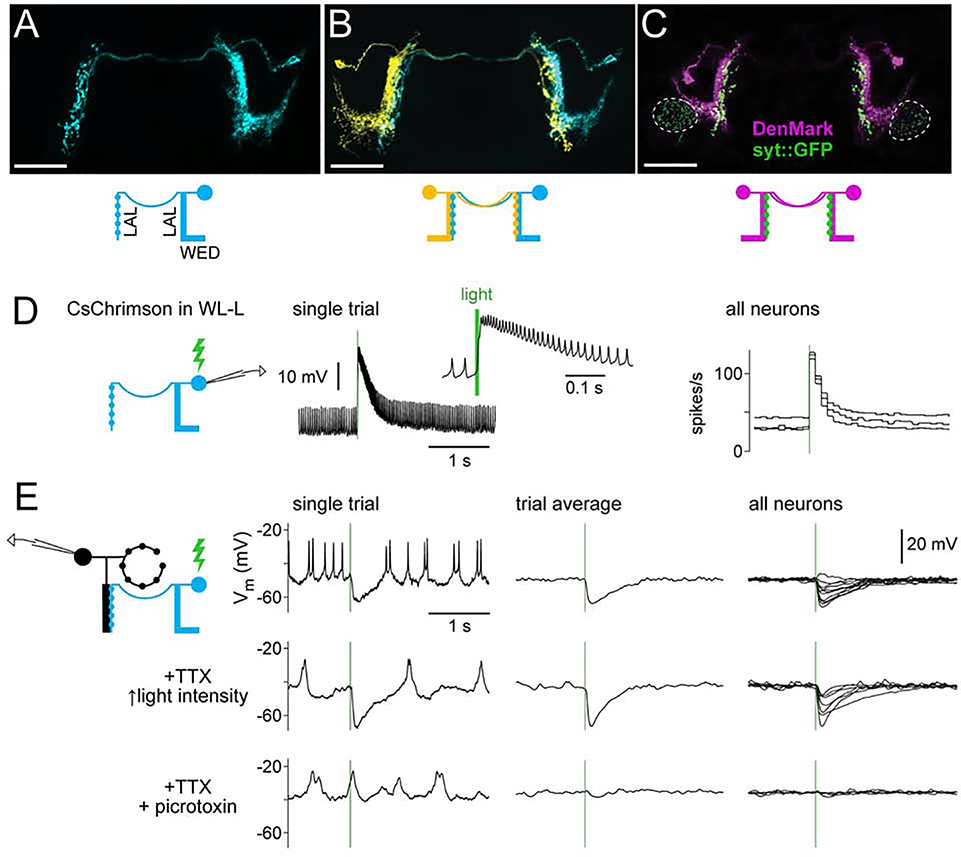
Notably, there is a WED cell type (called WL-L; Franconville et al., 2018) that projects to the LAL and overlaps with R1 neuron dendrites (Figures 7A and S6A). WL-L neurons are GABAergic cells (Figure S6B) with dendrites in the WED and the ipsilateral LAL, and axons in the contralateral LAL (Figures 7A–7C). We found one WL-L neuron per brain hemisphere.
Figure 7: R1 neurons receive direct GABAergic inhibition from WL-L neurons.
(A) MCFO labeling of an WL-L neuron, showing smooth neurites in the ipsilateral WED and ipsilateral LAL, and boutons in the contralateral LAL. Scale bars in (A-C) are 50 μm.
(B) Two-color MCFO labeling of two WL-L neurons in the same brain. Note that each cell’s bouton-rich arbor is just medial to the smooth arbor of the contralateral cell.
(C) Polarity markers expressed in WL-L neurons. The dendritic marker DenMark localizes to smooth arbors, whereas synaptotagmin::GFP localizes to bouton-rich arbors. Dashed lines indicate synaptotagmin::GFP in another cell type in the same Gal4 line (not WL-L).
(D) Recording from a CsChrimson+ WL-L neuron. Left: light-evoked response in an example trial (enlarged in inset). Right: firing rates in 3 neurons (in 3 flies). Green bars indicate 10-ms light pulses.
(E) Recording from R1 neurons while activating WL-L neurons. Responses were recorded first without antagonists, then in 1 μM TTX, and then after adding 5 μM picrotoxin (retaining TTX). Stimulus intensity was increased after adding TTX, to compensate for decreased excitability of presynaptic terminals. Left: single-trial responses from one neuron. Middle: trial-averaged responses for the same neuron. Right: trial-averaged responses for all neurons (n=11, 7, 4 neurons for the top, middle, and bottom rows).
When we expressed CsChrimson in WL-L neurons and depolarized them with light, R1 neurons were inhibited (Figures 7E and S6C). This inhibition persisted in TTX, indicating it is likely monosynaptic. Inhibition was essentially abolished by the GABAA antagonist picrotoxin (5 pM; Figures 7E, and S6C-D). Together, these results indicate that WL-L neurons inhibit contralateral R1 neurons via GABAA receptors.
We also observed that WL-L neurons are dye-coupled to ipsilateral R1 neurons (Figure S6E-H), indicating the presence of gap junctions. Indeed, optogenetically activating WL-L neurons with higher intensities could elicit both inhibition and excitation in the same R1 neuron (Figure S6I). These observations suggest that individual R1 neurons receive a sign-inverted input from the contralateral WL-L neuron and a sign-conserved input from the ipsilateral WL-L neuron (Figure S6J).
Wind responses in WL-L neurons
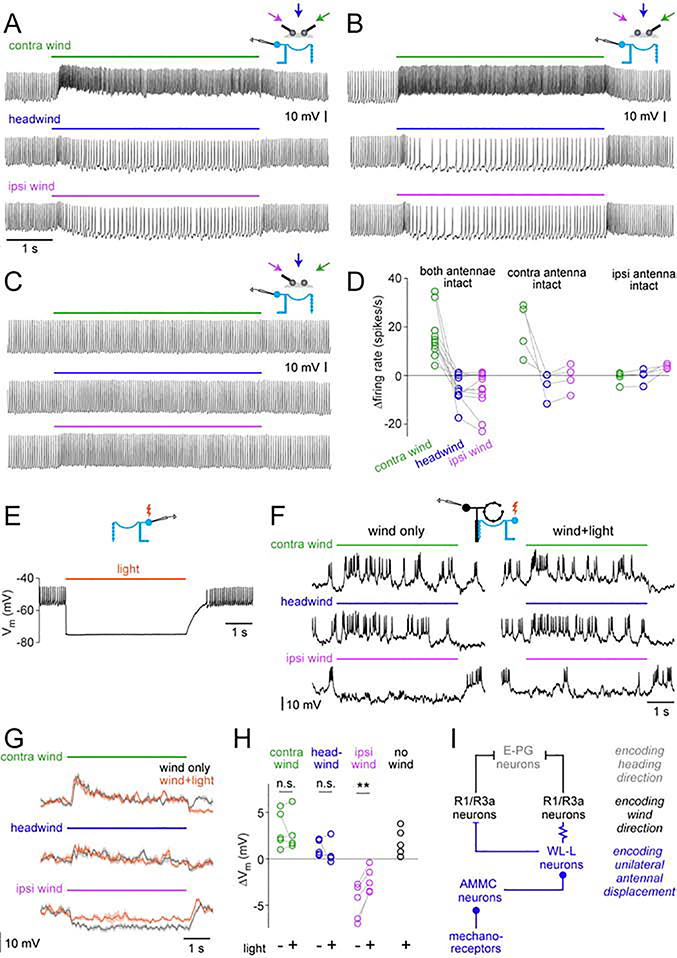
Using in vivo whole-cell recordings, we found that WL-L neurons respond to wind in a direction-dependent manner. In general, wind from 60° contralateral increased spike rates, whereas wind from 60° ipsilateral decreased spike rates below baseline (baseline = 31 ± 12.6 spikes/s, n=11 neurons; mean ± S.D; Figures 8A and S7A-B). Headwind produced essentially the same response as wind from 60° ipsilateral (Figures 8A and 8D). A few WL-L neurons fired a brief nonspecific burst of spikes at wind onset, regardless of wind direction (Figure 8A), but their sustained wind responses were direction-dependent.
Figure 8: WL-L neurons encode unilateral antennal displacement and contribute to R1 wind responses.
(A) An example whole-cell recording from an WL-L neuron in response to wind from −60°, 0°, and +60°.
(B) Same as (A) but with only the contralateral antenna intact.
(C) Same as (A) but with only the ipsilateral antenna intact. Here, excitation in response to contralateral wind is abolished. Inhibition in response to ipsilateral wind has also been abolished and is replaced by weak excitation.
(D) WL-L summary data. Responses are averaged over the stimulus period and expressed as changes from baseline. Within each condition, a gray line is a fly. The interaction between the stimulus (−60°, 0°, +60°) and the antennal condition (both, contra, ipsi) was not significant comparing “both-antennae-intact” with “contra-antenna-intact” (p=0.87), but it was significant comparing “both-antenna-intact” with “ipsi-antenna-intact” (p = 0.0006; two-way repeated measures ANOVA).
(E) Recording from a GtACR1+ WL-L neuron. Light (620 nm, 4.5 s) hyperpolarizes the neuron and suppresses spiking.
(F) R1 recording to test the effect of silencing WL-L neurons using GtACR1. We measured responses to wind from +60°, 0°, and 60°, with and without light.
(G) Mean membrane potential of the same R1 neuron (mean ± SEM; 8 trials per condition).
(H) Summary of wind responses in R1 neurons with and without WL-L silencing. Responses are averaged over the stimulus period and expressed as changes from baseline. Each line is a different neuron (n = 5). The effect of light is significant for ipsilateral wind (p=0.003; paired t-test). Also plotted is the mean response to light alone (no wind); note that light alone is depolarizing, implying that WL-L neurons are tonically inhibiting R1 neurons. Figure S7G-I shows data for genetic controls (lacking GtACR expression).
(I) Schematic showing the cell types involved in the transformation from wind-induced antennal displacements to compass signals. Filled circle, bar, and resistor symbols indicate excitatory connections, inhibitory connections, and gap junctions, respectively.
Next, we tested the effect of removing one antenna (Figures 8B-D, and S7C). When only the contralateral antennal was intact, WL-L wind responses were essentially normal. However, when only the ipsilateral antenna was intact, responses were nearly eliminated. Thus, WL-L neurons are driven mainly by the contralateral antenna (Figure S7D). Contralateral wind pulls the contralateral antenna away from the head (Figure S7E), and this excites WL-L neurons. Ipsilateral wind pushes the contralateral antenna toward the head (Figure S7E), and this inhibits WL-L neurons.
Recall that individual R1 neurons receive antagonistic input from the two antennae (Figure S7D and S7E). This is consistent with our finding that R1 neurons receive antagonistic input from left and right WL-L neurons (Figure S6J). If different R1 neurons received different ipsi/contra WL-L input weights, this could contribute to the diverse wind direction preferences of R1 neurons.
Contribution of WL-L to R1 wind responses
When we expressed GtACRl in WL-L neurons and hyperpolarized them with light (Figures 8E and S7F), R1 neurons were depolarized. This result implies that WL-L neurons tonically inhibit R1 neurons (Figure 8H), consistent with the high baseline firing rates of WL-L neurons. In the control genotype (empty-Gal4), light had no effect on R1 neurons (Figures S7G-I), indicating that the effect of light was not a visual artifact.
In these experiments, we presented wind from three directions (−60°, 0°, and +60°), with or without light. Light pulses began before wind presentation, and wind responses were measured relative to the cell’s new baseline with light. Notably, hyperpolarizing WL-L neurons significantly attenuated responses to ipsilateral wind (Figures 8F-H). However, we found no systematic effect on responses to contralateral wind or headwind. Thus, WL-L neurons clearly contribute to R1 wind responses, but there must be other wind inputs to R1 neurons.
When this manuscript was in revision, the FlyEM Project released a connectome of the dorsal part of the right central brain (Xu et al., 2020). In this dataset, we found an axon which matches the WL-L morphology (Figure S8A). This axon connects directly to many R1 neurons, and for these neurons, it is often their largest source of input in the LAL (Figures S8B-E). That said, every R1 neuron also receives other inputs in the LAL, consistent with our GtACR experiments. Interestingly, some R3a neurons also receive direct input from the putative WL-L axon (Figures S8C-E). To summarize, EM data support the conclusion that WL-L neurons are an important pathway from the antennae to R1 and R3a neurons, but they are not the only pathway. EM data help explain the diversity of R1/R3a responses, by revealing diverse inputs to individual R1/R3a neurons.
Discussion
In this study, we demonstrated that the heading compass in the Drosophila brain is influenced by wind direction cues. We showed how wind cues can reach the heading compass, via a WED→LAL→EB pathway. First, wind signals are integrated across the two antennae to extract wind direction information. Next, signals are reformatted to reduce the biases inherent in antennal mechanics. Ultimately, wind space is mapped uniformly onto the same coordinate system used to represent visual space and self-motion. These results provide a potential neural substrate for wind compass navigation, and they open the door to studying the mechanisms of mechanosensory-visual integration during navigation and spatial learning.
R neurons connect the compass to the sensory environment
R neurons are key inputs to the central complex (Honkanen et al., 2019; Pfeiffer and Homberg, 2014). Their primary role is to bring external sensory signals to the heading compass. A recent study in Drosophila identified 11 morphological types of R neurons (Omoto et al., 2018), most of which are functionally uncharacterized.
Our results show that both R1 and R3a neurons encode wind direction. When R1 neurons are silenced, this severely disrupts the effect of wind on the heading compass. R3a neuron responses are weaker than R1 responses, which might account for why silencing R3a neurons alone has no effect.
The R neurons that have been characterized have a key property in common: they encode sensory cues related to the fly’s angular position. Meanwhile, angular velocity information is relayed to E-PG neurons by a distinct cell type (P-EN neurons; Green et al., 2017; Turner-Evans et al., 2017). E-PG neurons integrate this angular velocity over time to generate a continuous estimate of angular position. This integral is prone to drift due to accumulation of errors. The function of R neurons is to improve this estimate by using external sensory signposts indicative of angular position whenever available.
Extracting wind direction information via bilateral integration in R neurons
Bilateral integration is a key step in extracting wind direction information. If the displacements of both antennae are known, then both wind direction and wind intensity can be inferred (Figure 4D). It is therefore notable that R1/R3a neurons integrate mechanosensory signals from both antennae. Bilateral integration helps explain why the shapes of R1/R3a tuning curves are relatively invariant to windspeed (Figure 5E).
By contrast, WL-L neurons are almost exclusively responsive to just one antenna (Figure 8D). This means that WL-L neurons are primarily encoders of unilateral antennal displacements, rather than encoders of wind direction. The same is true of more peripheral neurons, which are also sensitive to one antenna alone (antennal mechanoreceptors and AMMC neurons; Figure 9A; Chang et al., 2016; Patella and Wilson, 2018; Suver et al., 2019).
R1/R3a neurons are not the only cell types in the Drosophila brain that encode wind direction. A recent study identified wind-direction-sensitive WED projection neurons (“WPNs”) that integrate inputs from both antennae and project to higher brain regions, but not the central complex (Suver et al., 2019). It will be interesting to understand how different wind-direction pathways might be specialized for different functions.
From wind direction to heading direction
At the next step, wind direction representations (in R1/R3a neurons) are transformed to heading direction representations (in E-PG neurons; Figure 8I). E-PG neurons combine wind direction cues and visual object position cues (Figure 3G; Fisher et al., 2019; Green et al., 2017; Kim et al., 2019; Seelig and Jayaraman, 2015; Sun et al., 2017) as well as angular velocity cues (Green et al., 2017; Turner-Evans et al., 2017). It is the combination of different cues that makes E-PG neurons encoders of heading direction in a generalized sense, and not simply encoders of wind direction.
We also show that the R→E-PG transformation reduces the impact of constraints imposed by peripheral mechanics. Specifically, found that small changes in wind direction produce large changes in antennal displacement (Figure 4). These features were reflected in R1 wind direction tuning curves, but not in E-PG tuning curves. This makes sense, because an ideal compass should be equally sensitive to all directions. We were able to model the transformation from R1 neurons to E-PG neurons by adjusting the R1→E-PG synaptic weights. In the real network, recurrent connections also play a role in shaping E-PG selectivity and compass dynamics (Kim et al., 2017).
Interestingly, the visual inputs to the compass are also biased, disproportionately emphasizing lateralized visual cue positions (Fisher et al., 2019; Seelig and Jayaraman, 2013; Sun et al., 2017) and specific angles of polarized light (Labhart et al., 2001). These biases are not detectable in the patterns of visually-evoked excitation in the E-PG ensemble (Fisher et al., 2019; Sakura et al., 2008; Seelig and Jayaraman, 2015). Thus, it seems that “de-biasing” occurs in the transformation from visual R neurons to E-PG neurons, as well as the transformation from mechanosensory R neurons to E-PG neurons. We propose that de-biasing is a basic step in the integration of local sensory cues into global maps of space.
Because E-PG neurons integrate multiple heading cues (Green and Maimon, 2018; Hulse and Jayaraman, 2019), we would expect the effects of wind to be less consistent when multiple cues are present. Most of our experiments were designed to maximize the effect of wind – specifically, we removed visual cues, and we lifted the fly’s legs off the ground to eliminate feedback signals associated with walking. In walking flies, the effects of wind were more variable, probably because wind cues were conflicting with angular velocity cues (i.e., the wind stimulus was not locked to the fly’s angular velocity). In the future, it will be important to investigate how the compass system resolves conflicts between angular velocity cues, wind direction cues, and visual object position cues.
Flexibility in the wind-heading transformation
Our results imply that there is flexibility in how E-PG compass responds to wind. First, wind had a strong effect on E-PG neurons in some individuals, but almost no effect in other individuals (Figures 1G and S1F). Second, the mapping from wind direction to E-PG neuron location was smoothly circular in some individuals, and disordered in others (Figures 1G and S1F). Third, the offset between wind direction and E-PG bump location was variable across individuals (Figure 1J), similar to the variable offset between visual cue location and E-PG bump location (Fisher et al., 2019; Giraldo et al., 2018; Green et al., 2017; Seelig and Jayaraman, 2015). Finally, the offset between wind direction and E-PG bump location can change spontaneously and abruptly (Figure S1E), similar to what has been reported for visual cues (Seelig and Jayaraman, 2015).
Notably, recent studies have shown that after a few minutes of visuomotor training, visual inputs to E-PG neurons can be modified (Fisher et al., 2019; Kim et al., 2019). It was proposed that the locus of plasticity is the synapse between visual R neurons and E-PG neurons. Our results raise the possibility that mechanosensory neurons also form plastic synapses onto E-PG neurons. This type of plasticity would enable the compass system to learn new environmental scenes, including new wind/sun configurations (Figure 3G). Indeed, there is behavioral evidence that insects can learn arbitrary configurations of compass cues, including the direction of a steady wind, the position of the sun, and the orientation of e-vectors in the sky (Dacke et al., 2019; el Jundi et al., 2016; Müller and Wehner, 2007). Once a multi-cue configuration has been learned, it can be used for continuous navigation even when individual cues falter.
Wind compass behavior
Many arthropods perform wind compass behavior (Bell et al., 1989; Bell and Kramer, 1979; Birukow, 1958; Böhm, 1995; Böhm et al., 1991; Dacke et al., 2019; Heinzel and Böhm, 1984, 1989; Linsenmair, 1969, 1972, 1973; Müller and Wehner, 2007; Wehner and Duelli, 1971). In this behavior, the direction of the wind is used as a feedback signal to achieve a straight walking path. When the animal drifts off course, the wind appears to shift relative to the body, causing the organism to execute a corrective turn to regain its original heading relative to the wind.
Wind compass behavior implies that there are neurons in the brain that encode the animal’s heading relative to the wind. We show here that E-PG neurons perform this function. When the wind shifts rightward, the E-PG bump rotates the compass as if the fly were turning leftward, and vice versa. In the future, it will be interesting to investigate whether Drosophila exhibit wind compass behavior, as other arthropods do. If so, we would predict that silencing E-PG neurons disrupts wind compass behavior, just as it disrupts visual compass behavior (Giraldo et al., 2018; Green et al., 2019).
How are wind shifts translated into corrective turns? It seems likely that a “goal heading” is stored downstream from E-PG neurons (Green and Maimon, 2018; Green et al., 2019). If the brain registers an apparent mismatch between the goal heading and the current heading (as represented by E-PG neurons), this should trigger an error, which then drives a corrective behavioral turn to eliminate the error.
It important to note that some wind-guided behaviors may not involve the compass system. Although Drosophila typically walk straight upwind in response to an attractive odor (Alvarez-Salvado et al., 2018; Bell and Wilson, 2016; Handler et al., 2019; Steck et al., 2012), we would predict that this behavior does not require E-PG neurons. This is because upwind locomotion is conceptually analogous to locomotion straight toward a visual object, which does not require E-PG neurons (Giraldo et al., 2018; Green et al., 2019).
The wind input to the compass system is also probably not used during flight behavior. A flying animal cannot sense a steady wind, except indirectly, by observing drift in its flight path relative to visual landmarks (Schöne, 2014). Therefore, extracting wind direction during free-flight involves the visual system, not the mechanosensory system.
Our understanding of the navigational systems in the insect brain is expanding rapidly (Collett, 2019; el Jundi et al., 2019; Heinze et al., 2018; Hulse and Jayaraman, 2019; Warren et al., 2019). Behavioral studies are uncovering the rules of compass-based navigation, while physiological studies are revealing the cellular mechanisms of the compass. This comparative approach will be essential for obtain a satisfying neural explanation of complex and flexible spatial navigation algorithms in natural environments.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rachel I. Wilson (rachel_wilson@hms.harvard.edu).
Materials Availability
Fly lines generated in this study are available from the Lead Contact.
Data and Code Availability
Data and software may be obtained upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flies (Drosophila melanogaster) were raised on cornmeal agar medium or molasses food (Archon Scientific) under a 12 h light, 12 h dark cycle at 25 °C. Female flies were used for all experiments.
Genotypes
Genotypes of flies used in each figure are as follows:
Figure 1: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R60D05-Gal4}attP2/+
Figure 2A: (R31A12-Gal4 > MCFO-4)
P{R57C10-FLPG5}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA} VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R31A12-Gal4}attP2
Figure 2B: (VT017183-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT017183-Gal4}attP2
Figure 2C: w; repo-Gal80 / +; P{UAS-DenMark}, P{UAS-syt.eGFP} / P{R31A12-Gal4}attP2
Figure 2D: w; repo-Gal80 / +; P{UAS-DenMark}, P{UAS-syt.eGFP} / P{R12G08-Gal4}attP2
Figure 2E: (LE001 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; + / P{VT059225-p65.AD} attP40; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT048577-Gal4.DBD}attP2
For Gal4-only control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; + / X-Gal4 where X-Gal4 was either P{R31A12-Gal4}attP2, P{R12G08-Gal4}attP2, or P{R20A02-Gal4}attP2.
For R31A12-Gal4, the following genotype (no recombination on the second chromosome) was also used: w / +; P{R60D05-LexA}attP40 / P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5; + / P{R31A12-Gal4}attP2
For LE001-only control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / P{VT059225-p65.AD}attP40; + / P{VT048577-Gal4.DBD}attP2
For empty-Gal4 control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-Hsap \ KCNJ2.EGFP} / P{GAL4.1 Uw}attP2
For the experimental flies with Kir2.1 were:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-Hsap \ KCNJ2.EGFP} / X-Gal4
where X-Gal4 was either P{R31A12-Gal4}attP2, P{R12G08-Gal4}attP2, or P{R20A02-Gal4}attP2.
For the experimental flies using the LE001 line:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / P{VT059225-p65.AD}attP40;
P{UAS-HsapKCNJ2.EGFP} / P{VT048577-Gal4.DBD}attP2
For R1 recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For R3a recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R12G08-Gal4}attP2
Figure 4: Dickinson wild type
For R1 recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For R3a recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
Figure 6B-D, F: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R31A12-Gal4}attP2 / +
Figure 6E: Dickinson wild type
Figure 6G: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R60D05-Gal4}attP2 / +
Figure 7A, B: (R26B07-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R26B07-Gal4}attP2
Figure 7C: w; repo-Gal80 / +; P{UAS-DenMark}, P{UAS-syt.eGFP} / P{R26B07-Gal4}attP2
Figure 7D, E: w; P{R53B02-LexA}attP40 / +; P{R26B07-Gal4}attP2 / P{13XLexAop2-IVS-myr::GFP}VK00005,
P{20XUAS-CsChrimson-mCherry}su(Hw)attP1
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2
or
P{13XLexAop2-IVS-CsChrimson.mVenus}attP18 / w; P{20XUAS-IVS-mCD8::GFP}attP40; P{Orco-LexA} / P{VT040354-Gal4}attP2
w; P{R53B02-LexA}attP40 / P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2 / P{20XUAS-IVS-GtACR1-EYFP}attP2, P{13XLexAop2-IVS-myr::GFP}su(Hw)attP1
Figure S1A: w; +; P{R60D05-Gal4}attP2 / P{pJFRC2–10XUAS-IVS-mCD8::GFP}attP2
Figures S1B, D w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R60D05-Gal4}attP2 / +
Figure S1E (top)
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; + / P{R31A12-Gal4}attP2
Figure S1E (bottom)
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-Hsap \ KCNJ2.EGFP} / P{GAL4.1 Uw}attP2
Figures S1F, G: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R60D05-Gal4}attP2 / +
Figures S1H, I: +; +; P{R60D05-Gal4}attP2 / PBac{20XUAS-IVS-jGCaMP7f} VK00005
Figure S2A: (R54E12-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R54E12-Gal4}attP2
Figure S2B: (VT017183-Gal4>MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT017183-Gal4}attP2
Figure S2C: (VT017183-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA} VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT017183-Gal4}attP2
For R31A12-Gal4: (R31A12-Gal4 > MCFO-4)
P{R57C10-FLPG5}su(Hw)attP8 / w; +; PBac{10×UAS(FRT. stop)myr: :sm GdP-HA} VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R31A12-Gal4}attP2
or (R31A12-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R31A12-Gal4}attP2
For VT017183-Gal4: (VT017183-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT017183-Gal4}attP2
For R12G08-Gal4: (R12G08-Gal4 > MCFO-4)
P{R57C10-FLPG5}su(Hw)attP8 / w; +; PBac{10×UAS(FRT. stop)myr: :sm GdP-HA} VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R12G08-Gal4}attP2
or (R12G08-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA} VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R12G08-Gal4}attP2
For LE001-Gal4: (LE001 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; + / P{VT059225-p65.AD}attP40; PBac{10×UAS(FRT. stop)myr: :sm GdP-HA} VK00005 P{10×UAS(FRT. stop)myr: :smGdP- V5-THS- 10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{VT048577-Gal4.DBD}attP2
For R20A02-Gal4: (R20A02-Gal4 > MCFO-5)
P{R57C10-FLPG5.PEST}su(Hw)attP8 / w; +; PBac{10×UAS(FRT.stop)myr::smGdP-HA}VK00005 P{10×UAS(FRT.stop)myr::smGdP-V5-THS-10×UAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 / P{R20A02-Gal4}attP2
P{10XUAS-IVS-mCD8::GFP}su(Hw)attP8 / w; P{VT059225-p65.AD}attP40/CyO; P{VT048577-Gal4.DBD}attP2/TM6B
Figure S2F, G:
For controls:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For Kir2.1:
w / +; P{20XUAS-IVS-mCD8::GFP}attP40 / P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5; P{UAS-Hsap \ KCNJ2.EGFP} / P{R31A12-Gal4}attP2
Figure S2H: w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-Hsap \ KCNJ2.EGFP} /R31A12-Gal4
Figure S2I: w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / P{VT059225-p65.AD}attP40; P{UAS-HsapKCNJ2.EGFP} / P{VT048577-Gal4.DBD}attP2
For Gal4-only control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; + / X-Gal4 where X-Gal4 was either P{R31A12-Gal4}attP2, P{R12G08-Gal4}attP2, or P{R20A02-Gal4}attP2.
For R31A12-Gal4, the following genotype (no recombination on the second chromosome) was also used: w / +; P{R60D05-LexA}attP40 / P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5; + / P{R31A12-Gal4}attP2
For LE001-only control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / P{VT059225-p65.AD}attP40; + / P{VT048577-Gal4.DBD}attP2
For empty-Gal4 control:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-HsapKCNJ2.EGFP} / P{GAL4.1 Uw}attP2
For the experimental flies with Kir2.1 were:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / +; P{UAS-HsapKCNJ2.EGFP} / X-Gal4
where X-Gal4 was either P{R31A12-Gal4}attP2, P{R12G08-Gal4}attP2, or P{R20A02-Gal4}attP2.
For the experimental flies using the LE001 line:
w / +; P{R60D05-LexA}attP40, P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 / P{VT059225-p65.AD}attP40; P{UAS-Hsap \ KCNJ2.EGFP} / P{VT048577-Gal4.DBD}attP2
Figure S3A, B: w / +: P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R31A12-Gal4}attP2 / +
Figure S3C, D: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R12G08-Gal4}attP2 / +
For R1 recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For R3a recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R12G08-Gal4}attP2
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For R1 recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
or
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R31A12-Gal4}attP2
For R3a recordings:
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{VT017183-Gal4}attP2
Figure S5: w / +; P{20XUAS-IVS-GCaMP6f}attP40 / +; P{R31A12-Gal4}attP2 / +
Figure S6A, C, D, and I: w; P{R53B02-LexA}attP40 / +; P{R26B07-Gal4}attP2 / P{13XLexAop2-IVS-myr::GFP} VK00005, P{20XUAS-CsChrimson-mCherry}su(Hw)attP1
Figure S6B: P{10XUAS-IVS-mCD8::GFP}su(Hw)attP8 / w; + / CyO; P{R26B07-Gal4}attP2 / TM6B
Figure S6E, F, and H: w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2
Figure S6G: w; +; P{R31A12-Gal4}attP2 / P{pJFRC2–10XUAS-IVS-mCD8::GFP}attP2
Figure S7A, B: w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2
w; P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2
or
P{13XLexAop2-IVS-CsChrimson.mVenus}attP18 / w; P{20XUAS-IVS-mCD8::GFP}attP40; P{Orco-LexA} / P{VT040354-Gal4}attP2
w; P{R53B02-LexA}attP40 / P{20XUAS-IVS-mCD8::GFP}attP40; P{R26B07-Gal4}attP2 / P{20XUAS-IVS-GtACR1-EYFP}attP2, P{13XLexAop2-IVS-myr::GFP}su(Hw)attP1
w; P{R53B02-LexA}attP40 / P{20XUAS-IVS-mCD8::GFP}attP40; P{empty-Gal4}attP2 / P{20XUAS-IVS-GtACR1-EYFP}attP2, P{13XLexAop2-IVS-myr::GFP}su(Hw)attP1
or
w; P{R53B02-LexA}attP40 / +; P{empty-Gal4}attP2 / P{20XUAS-IVS-GtACR1-EYFP}attP2, P{13XLexAop2-IVS-myr::GFP}su(Hw)attP1
Origins of stocks: driver lines
Gal4 lines with an “R” prefix (GMR collection, Janelia Research Campus) were obtained from Bloomington Drosophila Stock Center (BDSC) and are described in Jenett et al. (2012). P{GAL4.1Uw}attP2 (also known as empty-Gal4) was used as a control for Kir2.1 experiments. This stock carries control construct carrying Gal4 in the vector used in the GMR collection (Pfeiffer et al., 2008) but with no regulatory fragment present. Gal4 lines with a “VT” prefix (Vienna Tiles collection) were obtained from Vienna Drosophila Resource Center (VDRC) and are described in Kvon et al. (2014) and Tirian et al. (2017).
Driver lines that target E-PG neurons
The name of this cell type refers to the fact that it has dendrites in the ellipsoid body, and axonal projections to the protocerebral bridge and gall (Seelig and Jayaraman, 2015; Wolff et al., 2015). P{R60D05-GAL4}attP2 targets E-PG neurons (Seelig and Jayaraman, 2015; Wolff et al., 2015). P{R60D05-LexA}attP40 also targets E-PG neurons (Green et al., 2017; Turner-Evans et al., 2017).
Driver lines that target R neurons
R neurons are also known as Ring neurons (Hanesch et al., 1989). Note that the term “R1 neurons” has been used previously to refer to different cell types. In this study, we use the new nomenclature of Omoto et al. (2018). Table S1 provides a lookup table for previously used Gal4 lines and the corresponding R neuron types that these Gal4 lines target (Baker et al., 2007; Chen et al., 2012; Dus et al., 2013; Franconville et al., 2018; Kottler et al., 2019; Kuntz et al., 2012; Kuntz et al., 2017; Liang et al., 2019; Lin et al., 2013; Martin-Pena et al., 2014; Neuser et al., 2008; Ofstad et al., 2011; Omoto et al., 2018; Renn et al., 1999; Shiozaki and Kazama, 2017; Xie et al., 2017; Young and Armstrong, 2010). P{R31A12-GAL4}attP2 targets R1 neurons, and P{R12G08-GAL4}attP2 targets R3a neurons (Omoto et al., 2018) (Figure S2D). P{R20A02-GAL4}attP2 targets many types of R neurons that arborize in the bulb (Fisher et al., 2019; Kunst et al., 2014). P{R54E12-GAL4}attP2 labels R neurons that arborize in the bulb (Fisher et al., 2019). P{R53B02-LexA}attP40 was identified through a visual screen of GMR LexA line collection (Pfeiffer et al., 2010). This line targets R1 neurons and was used for functional connectivity experiments. P{VT017183-Gal4}attP2 was identified through a visual screen of the VT collection and targets both R1 and R3a neurons (Figure S2D). This line was used for patch-clamp experiments but was not used for Kir2.1 experiments due to lethality when crossed with P{UAS-Hsap\KCNJ2.EGFP}7. We made a split-Gal4 line, which we call “LE001” since it innervates the LAL and EB, by combining two hemidrivers, P{VT059225-p65.AD}attP40 and P{VT048577-Gal4.DBD}attP2. These hemidrivers were obtained from BDSC, and are described in Tirian et al. (2017). The expression pattern of this split-Gal4 line is shown in Figure S2E. MCFO analysis showed that R neurons labeled in this line are exclusively R1 and R3a. However, the cell body count in this LE001 line (16 ± 0.4, mean ± SEM., neurons per hemisphere) is less than the sum of R1 (13 ± 2 neurons) and R3a (11 ± 0 neurons) (Omoto et al., 2018), suggesting that LE001 might not label all the R1 and R3a neurons.
Driver lines that target WL-L neurons
The name of this cell type refers to the fact that it has dendrites in the wedge and lateral accessory lobe, and an axonal projection to the contralateral lateral accessory lobe (Franconville et al., 2018). P{R26B07-GAL4}attP2 targets WL-L neurons (Franconville et al., 2018). While Franconville et al. (2018) mention that the line P{R26B07-GAL4}attP2 drives expression in two WL-L neurons per hemisphere, we only saw one WL-L neuron per hemisphere, potentially due to a difference in the UAS reporter that was used. Another Gal4 line that also targets WL-L neurons, P{VT040354-Gal4}attP2, was identified through a visual screen, and was used for initial patch-clamp experiments. We confirmed that P{R26B07-GAL4}attP2 and P{VT040354-Gal4}attP2 target the same WL-L neurons (as opposed to morphologically similar yet distinct neurons) by expressing UAS-mCD8::GFP using two drivers P{R26B07-GAL4}attP2 and P{VT040354-Gal4}attP2 in the same fly, and observing that only one neuron per hemisphere was labeled (data not shown).
Origins of stocks: effector lines and miscellaneous lines
For calcium imaging experiments, we used P{20XUAS-IVS-GCaMP6f}attP40 (Chen et al., 2013), which we obtained from BDSC via Thomas Clandinin. We also used P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 obtained from BDSC. For imaging E-PG neurons with flies on the ball (Figures S1H and S1I), we used PBac{20XUAS-IVS-jGCaMP7f}VK00005 (Dana et al., 2019), which we obtained from BDSC.
For polarity marker staining, we used P{UAS-DenMark} and P{UAS-syt.eGFP} (Nicolai et al., 2010). To suppress polarity marker expression in glial cells, we used repo-Gal80 (Awasaki et al., 2008). This stock was a gift from Tzumin Lee via Michael Crickmore.
To hyperpolarize R neurons, we overexpressed the inward-rectifying potassium channel Kir 2.1 using P{UAS-Hsap\KCNJ2.EGFP}7 (Hardie et al., 2001), which we obtained from BDSC.
To express GFP for visual targeting of patch-clamp recordings or for anatomy, we used P{20XUAS-IVS-mCD8::GFP}attP40 or P{13XLexAop2-IVS-myr::GFP}VK00005 (Pfeiffer et al., 2010), which were gifts from Barret Pfeiffer and Gerry Rubin. We also used P{10XUAS-IVS-mCD8::GFP}su(Hw)attP8 obtained from BDSC.
For functional connectivity experiments, we expressed CsChrimson (Klapoetke et al., 2014) in WL-L neurons. P{20XUAS-CsChrimson-mCherry}su(Hw)attP1 (Franconville et al., 2018) was a gift from Vivek Jayaraman’s lab.
For optogenetic silencing experiments, we expressed GtACR1 (Govorunova et al., 2015) in WL-L neurons. P{20XUAS-IVS-GtACR1-EYFP}attP2 (Mohammad et al., 2017) was a gift from Adam Claridge-Chang via Michael Crickmore.
For MultiColor FlpOut (MCFO), we used fly stocks in which Flp variants are expressed pan-neuronally (see genotype details above). These were obtained from BDSC and are described in Nern et al. (2015). To achieve sparse labeling, we used the version of Flp (FLPL in MCFO-3; FLPG5 in MCFO-4; FLPG5.PEST in MCFO-5) that is appropriate for a given Gal4 line and adjusted the age of dissection.
For initial pilot experiments, we used P{Orco-LexA} and P{13XLexAop2-IVS-CsChrimson.mVenus}attP18 to optogenetically activate the majority of olfactory receptor neurons to test for responses to fictive odor in WL-L neurons. P{Orco-LexA} (also known as P{Or83b-LexA}) was a gift from Tzumin Lee. However, with the light intensity we used, we did not observe any fictive odor responses (data not shown), and these data were combined with other WL-L recordings performed in flies that did not express these transgenes.
For measurements of the antennal movements, we used wild type flies originating from the lab of Michael Dickinson (strain ‘Dickinson wild type’). This stock was established from 200 wild-caught females (Reiser and Dickinson, 2010) and was a gift from Michael Dickinson.
METHOD DETAILS
Stimulus delivery and antennal displacement measurements
Air tubes for wind delivery
To deliver wind from three directions, three stainless steel dispensing needles (45° bend, 20 gauge, ½” long; McMaster-Carr, 75165A35) were attached to the underside of the horizontal titanium foil platform used for imaging and electrophysiology. The luer lock adaptor attached to the needle was cut off and the remaining needle was used as a wind tube. One tube was placed directly in front of the fly, and two tubes were placed ± 60 deg from the midline (Figure 1C). The tips of the wind tubes were 2 mm away from the head of the fly.
A custom flow-control device was used to control the timing of air flow through the air tubes. Charcoal-filtered medical-grade air was introduced into the device via a three-way solenoid valve (Numatics MPG, LO03L7H00E). The air flow rate was set using flow meters (32003–00 or 03227–06, Cole-Parmer). A set of tubes connected the three-way-solenoid valve to the wind tubes (silicone tube, I.D. 1/16”, O.D. 1/8”, 06411–62, Cole-Parmer; Tygon tube, I.D. 1/32”, O.D. 3/32”, 07407–70, Cole-Parmer). The direction of the wind was specified by closing one of the three pinch valves (Bio-Chem Valve, 075P2NC24–02SQ). The timing of the valves was controlled through the Data Acquisition Toolbox in MATLAB (MathWorks) and a data acquisition card (PCIe-6351, National Instruments). The TTL output of the data acquisition card was converted to a 24V control signal using a custom electronic circuit.
Three different wind directions (−60°, 0°, +60°) were presented, 20 trials each, in a pseudo-random order. For two-photon imaging experiments, each trial consisted of 4 s of baseline, 4 s of wind delivery, 4 s of baseline, followed by a 6 s inter-trial interval. For patch clamp experiments, each trial consisted of 2 s of baseline, 4.5 s of wind delivery, 5 s of baseline followed by a 3 s inter-trial interval. Some recordings were lost before the entire set of 60 trials were completed. We included these recordings as long as there were two trials for 3 wind directions (see Data exclusion criteria below).
Wind speed at the location of the fly was measured with a hot-wire anemometer (A004, Kanomax). The wind speed used in the experiment was 0.12 m/s, except for R neuron recordings where it was 0.18 m/s. Air speed was slightly increased for R neuron recordings because the head angle was different in these experiments, and we needed to increase the wind speed to 0.18 m/s in order to achieve similar antennal deflections we would otherwise obtain at 0.12 m/s. Note that our simulated wind data shows that R1 neurons are relatively insensitive to changes in wind speed (Figure 5E). Antennal movements were monitored with two cameras, one on each side (FMVU-03MTM, Point Grey; InfiniStix 94 mm/1.5× lens, Infinity Photo-Optical). When the wind was delivered either +60° or −60°, the left and the right antennae typically produced a coherent movement (i.e. both antennae rotating clockwise for left wind and counterclockwise for right wind), consistent with our antennal movement measurements in head-fixed flies without the horizontal recording platform (Figure 4, Video S1). We discarded a minority of flies where only one antenna moved in response to wind, or where the left and right antennae produced incoherent movements.
Rotating wind delivery device
To deliver wind from multiple horizontal angles (Figure 1G), we constructed a device that allows an air tube to rotate on a motorized platform. The rotating platform was an acrylic disk (outer diameter 23 cm, inner diameter 6 cm) placed on top of a round turn table (1544T12, McMaster-Carr), housed on a bearing layer and capable of rotating with little friction about a vertical axis passing through the center of the disk. To rotate the platform, we attached a laser-cut acrylic gear. In Figure 4, the surface was rotated manually to a set of directions between −150° and 150° in 15° intervals, either in monotonically increasing or decreasing order. In Figures 1 and 6, it was rotated by means of a stepper motor (#1205, Polulu) controlled by an Arduino Uno board through custom software, with all directions between −150° and 150° (in 15° intervals, 21 different positions) interleaved pseudo-randomly. The zero position was reset at the beginning of each block of 21 trials by means of a reflective optical encoder (AEDR-8300, Agilent) or a mechanical limit switch (CH166-ND, ZF electronics). The zero position was also checked at the end of each block to ensure that no error had accumulated due to transmission slips. Each trial consisted of 4 s of baseline, 4 s of wind delivery, 4 s of baseline, followed by a 13 s inter-trial interval. A block of 21 trials was repeated 4–8 times.
To deliver the wind stimulus using the rotating device, we attached a wind tube made from a 16G stainless steel dispensing needle with a 90°-bend (75165A97, McMaster-Carr). The tip of the wind tube pointed toward the center of the disk and was 5 mm away from the fly. A custom-flow control device, almost identical to the one mentioned above, was used to deliver air through the tube; the only difference was the model of the flow meter we used (32003–08, Cole-Parmer). For two-photon calcium imaging of E-PG and R1 somata, the wind speed was set to 0.56 m/s.
Measurements of the antennal movements in response to wind
To measure the movements of the antennae as a function of wind blowing from different horizontal angles (Figure 4), flies were tethered to a tungsten rod, and precisely positioned above the center of the rotating platform by means of a set of micromanipulators with 6 degrees of freedom (x, y and z translations; pitch, roll and yaw rotations; PT3 and GNL20, Thorlabs). An acrylic platform was placed underneath the legs to mimic the air-flow boundary conditions that would be normally experienced by a walking fly. To reduce the movement artifact due to the fly kicking the platform, all the legs were gently removed with forceps. To prevent head movements, the posterior part of each eye was glued to the anterior part of the thorax. To prevent active movement of the antennae, the first and second segments of the antennae (a1 and a2) were glued to the head with a small drop of UV-cured glue (KOA 300, Kemxert).
Antennal movements were recorded using a camera placed underneath the fly (Blackfly BFLY-PGE-20E4M, FLIR; InfiniStix 90-degree 44 mm/3.00× lens with a DL Doubler Tube, Infinity Photo-Optical). Two more cameras (Blackfly BFLY-PGE-13E4M, FLIR; InfiniStix 96 mm/1× lens, Infinity Photo-Optical) were used to guide the precise alignment of the fly to the center of the rotating stage and with respect to the air dispensing needle. One camera pointed toward the front of the fly was used to correct for yaw and roll imbalances. The other camera was placed at a horizontal angle of 90°, pointing toward one side of the fly; this camera was used to standardize the fly’s pitch angle.
The wind tube was rotated from −150° to +150° with respect to the fly in 15° intervals and at four different wind speeds (0.11, 0.26, 0.56, and 1.20 m/s). Stimuli were delivered in sequential order of horizontal angle for each level of intensity. At any given direction, the stimulus was turned on for 1 s and off for 1 s, and this was repeated 5 times. The platform was then rotated to the next position (inter-trial interval: 3 s).
Videos of antennal movements were acquired at 10 Hz. This frame rate allowed to capture the steady component of the antennal displacement produced by the wind stimuli.
Stimulation of the antennae using two piezoelectric actuators
In order to displace both antennae independently and in a controlled manner (Figure 5A), we used two preloaded piezoelectric actuators (90 pm travel, servo-controlled; P-840.6, Physik Instrumente). A custom-milled attachment was used to linearly couple each piezoelectric actuator to a sharpened tungsten filament (diameter 0.150 mm), which was then attached to the respective arista perpendicular to its plane of rotation. To increase the stability of the tungsten filament, it was inserted into a glass pipette filled with epoxy and rigidly coupled to the custom-milled attachment, with the tungsten extending 8 mm past the end of the epoxy. The tip of the tungsten filament was coated with a thin layer of flange sealant (Loctite 515, Henkel Adhesive Technologies) and brought into contact with the arista as described below.
Our goal was to orient each filament perpendicular to the arista it contacts, so that linear displacement of the filament would produce rotation of the antenna about its long axis (Gopfert and Robert, 2001) or, more precisely, rotation about the antenna’s funicular stalk (Pezier and Blagburn, 2013). Given this goal, it was not possible to approach both aristae from the front, because the two filaments would interfere with each other. Therefore, we approached the right antenna from the posterior side (and lateral to the eye), and we approached the left antenna from the anterior-medial side. The vertical and horizontal angles of each linear actuator were adjusted in order to reach each arista from the correct direction. Specifically, each piezoelectric actuator was mounted on a 3-axis micromanipulator with manual control for vertical angle (MP-225, Sutter Instruments). The manipulator was mounted on a custom-built arm arranged to rotate about the fly. We mounted a camera on the same arm (FLIR Blackfly BFLY-PGE-13E4M, equipped with an InfiniStix 44 mm/3× lens with a DL Doubler Tube, Infinity Photo-Optical) pointed toward the fly along a line perpendicular to the piezoelectric actuator. This side camera was used to precisely attach the actuator to the antenna, along with another camera (FLIR Blackfly BFLY-PGE-20E4M, equipped with an InfiniStix 44 mm/3× lens with a DL Doubler Tube and a 90° Mirror module, Infinity Photo-Optical) that was pointed toward both antennae from below.
We delivered mechanical waveforms consisting of sustained backward and forward displacements of different amplitudes. We used the camera under the fly to visualize each arista in the horizontal plane; using this projection, we computed the estimated angular displacement of each arista in response to each stimulus waveform. In flies 1–7 (Figures 5 and S4: neurons 1–7), we independently calibrated the relative rotational amplitude of each antenna at the beginning of each experiment, in order to produce stimuli of intended rotational amplitudes. In two other flies (Figure S4: neurons 8–10), we delivered fixed linear displacements and used video recordings to estimate the corresponding angular amplitudes post hoc. The antennal positions corresponding to each stimulus were maintained for 1 s. Both antennae were linearly ramped to the next stimulus position over a time window of 0.3 s. Stimuli were repeated and randomly permuted within blocks. The stimulus that brought both antennae to their resting positions was overrepresented 2 to 7 times with respect to other stimuli, depending on the experiment, in order to improve our estimate of the cell’s firing rate when both antennae are at rest.
Physiology
Fly selection and housing
Unless otherwise noted, patch-clamp or imaging experiments were performed on adult female flies 12–48 hours after eclosion. Virgin female flies were anesthetized on CO2 at least 12 hours before the experiment and allowed to recover from anesthesia in a new culture vial containing standard food. We found that cells were easier to patch in flies <48 hrs old (as compared to older flies) because there was less fat inside the head capsule and the peri-neural sheath was thinner, but we found that flies that eclosed <12 hrs before the experiment) had cuticles that were too soft to allow reliable antennal stimulation.
Functional connectivity experiments using CsChrimson (Klapoetke et al., 2014) were performed on adult female flies 5–72 hours after eclosion. These flies were raised on a standard-size vial of molasses food supplemented with rehydrated potato flakes (Formula 4–24, Carolina Biological Supply) and 100 μL of all-trans-retinal (35 mM in ethanol; R2500, Sigma-Aldrich). These vials were covered with aluminum foil to prevent photoconversion of all-trans-retinal by ambient light.
Fly preparation for two-photon imaging
At the beginning of the experiment, a fly was briefly anesthetized in a glass vial (V7005–500EA, Sigma-Aldrich) on ice, and was placed inside a fly-sized hole in a horizontal titanium foil platform, which was itself secured in a hole within an acrylic platform. In this platform, the body is positioned dorsal-side up with its legs suspended in the air; because the wings were not free to move, the fly did not engage in flight behavior. To image the protocerebral bridge, the head was tilted forward so that the posterior surface of the head was roughly parallel to the foil. The fly was secured to the platform by gluing the lateral edge of the eye, the anterior edge of the scutum, and the posterior-lateral edges of the scutellum to the foil using a UV-curable acrylic-based adhesive (Loctite AA 3972, Henkel Adhesive Technologies; UV light delivered with LED-100 or LED-200, Electro-Lite). To prevent antennal grooming, the forelegs were gently removed with fine forceps. To reduce brain movement, the proboscis was fixed with wax (Electra Waxer, Almore) in an extended position. Then, the external saline was applied to the upper side of the platform, and the cuticle on the posterior surface of the head was gently removed with fine forceps. Fat cells obstructing the access to the brain were removed. To prevent brain movement, muscle 16 was removed using a hook made from an electrolytically sharpened tungsten wire (diameter: 0.125mm; W559516, Advent Research Materials). The external saline contained (in mM): 103 NaCl, 3 KCl, 5 TES, 8 trehalose dihydrate, 10 glucose, 26 NaHCO3, 1 NaH2PO4, 1.5 CaCl2, 4 MgCl2 with osmolarity adjusted to 270–275 mOsm and pH adjusted to ~7.3. The saline was bubbled with 95% O2 and 5% CO2 and was superfused during dissection. To make sure that the antennae were not damaged during dissection, we visually confirmed their movement by blowing air. After the dissection, the acrylic platform was mounted on a 3-axis motorized translational stage (MT3-Z8, Thorlabs) to finely adjust its position with respect to the objective and the wind-delivery device. Flies moved their mid legs and hind legs freely under the foil.
Two-photon calcium imaging
In vivo calcium imaging was performed with a two-photon laser scanning microscope equipped with a resonant scanner (Bergamo II, Thorlabs). Two-photon excitation was achieved with a femtosecond Ti:Sapphire Laser with dispersion precompensation (Vision-S, Coherent) tuned to 940 nm. Imaging was done using a 20× objective (N.A. 1.0; XLUMPLFLN, Olympus) mounted on a fast objective scanner (P-725, Physik Instrumente). Images (256×128 pixels) were acquired using ScanImage 2015 or 2018 (Vidrio Technologies) (Pologruto et al., 2003). We chose to image the axon terminals of E-PG in the protocerebral bridge rather than the dendrites of these cells in the ellipsoid body, because the Kir2.1 construct used for R neuron silencing experiments was tagged with EGFP, and EGFP in the axon terminals from R neurons in the ellipsoid body prevented us from measuring GCaMP fluorescence changes accurately in E-PG dendrites. We scanned through 6 z-planes (4–5 μm intervals for E-PG axons in the protocerebral bridge; 5–7 μm intervals for R1/R3a dendrites in the LAL; 2–3 μm intervals for R1 cell bodies) at a volumetric scanning rate of 12 Hz.
Measurement of locomotion
To image the activity of E-PG neurons during locomotion, we placed a fly on a 9-mm ball made of white foam (FR-4615, General Plastics) supported on a column of air. The ball was painted with black shapes and its rotational movement was tracked with a camera (CM3-U3–13Y3M-CS, FLIR) with a zoom lens (0.5×, 94 mm WD, InfiniStix). We used FicTrac (Moore et al., 2014) (v.2 running at 50 Hz on a Windows machine) to convert the image of the ball to an estimate of ball position in all three axes of rotation. Note that for these experiments, the front legs of the fly were removed just prior to the experiment, as in all other experiments. This was to prevent the fly from grooming the antennae and interfering with the detection of the wind stimuli. Removing the front legs reduced locomotion, but we nonetheless observed systematic wind-evoked turning maneuvers in most flies (Figures S1H and S1I). These turning maneuvers were more prominent in the sideways axis (ball roll), rather than the rotational axis (ball yaw), probably because the fly’s front legs were removed; this is why we show the sideways axis in Figure S1H, I.
Fly preparation for patch-clamp recordings
The preparations for patch-clamp recordings were performed as described above for two-photon imaging, with several differences. First, the head angle was adjusted for each cell type of interest. For example, to access the cell bodies of R neurons, the head was oriented so that the dorsal surface of the head (above the first antennal segment) was positioned on the upper side of the titanium foil. To access the cell bodies of R neurons while stimulating the antennae with two piezoelectric actuators, the fly was placed in a fly-sized hole at the base of a conical titanium foil, secured in a hole within an acrylic platform. To access the cell bodies of WL-L neurons, the head was tilted forward so that the posterior surface of the head was roughly parallel to the foil. For experiments involving the removal of one antenna (Figures 8B-D and S7C), second and third segments (a2 and a3) were gently removed with forceps. Rather than securing the proboscis, we instead gently removed the proboscis by cutting the soft tissues surrounding the palatal plate with sharp forceps (11252–20 or Dumont #5CO, Fine Science Tools); this improved optical resolution of cell bodies during patching while also reducing brain movement. Air sacs around the brain were also gently removed with forceps. Then, the external solution was applied, and a window was cut in the cuticle. Specifically, for R neuron recordings, we removed the dorsal head cuticle and the dorsal part of the eye that is ipsilateral to the recording site; whereas for WL-L neuron recordings, we moved the cuticle on the posterior side of the head. The perineural sheath near the location of the cell bodies was gently removed with fine forceps. For R neuron recordings, muscle 16 was removed by inserting a hook or with fine forceps in between antennal nerves, whereas for WL-L neuron recordings, muscle 16 was removed by clipping the posterior attachment site. In other respects, the dissection was the same as for imaging experiments.
For R neuron recordings with wind stimulation (Figure 3), flies were placed on a spherical treadmill which allowed them to walk, similar to the setup described previously (Seelig et al., 2010). However, the dissection was optimized for obtaining good optical access to the R neuron somata for patch-clamp recordings, meaning it was not optimized for obtaining a good walking. Therefore, most flies made only small movements on the ball and did not exhibit much walking. For all the other patch-clamp recordings, we removed the forelegs to prevent the fly from grooming its antennae; in these experiments, flies typically made intermittent spontaneous movements of their middle legs and back legs.
Whole-cell patch-clamp recordings
Pipettes were pulled from thick-wall borosilicate glass (O.D. 1.5 mm, I.D. 0.86 mm; BF150–86-7.5HP, Sutter) using a micropipette puller (P-97, Sutter). Pipettes used for cleaning the brain prior to patch clamping were pulled from thin-wall borosilicate glass (O.D. 1.5 mm, I.D. 1.12 mm; TW150F-3, World Precision Instruments).
For R1/R3a neuron recordings, pipettes were typically pressure polished (Goodman and Lockery, 2000) using a microforge (CPM-2, ALA Scientific Instruments). The pipette resistance was in the range of 5–15 MΩ. The internal patch pipette solution contained (in mM): 140 potassium aspartate, 10 HEPES, 1 EGTA, 1 KCl, 4 MgATP, 0.5 Na3GTP, 13 biocytin hydrazide. To improve post hoc morphological reconstruction, we also used internal solution with Neurobiotin (Vector Laboratories) (Kita and Armstrong, 1991) which contained (in mM): 140 potassium aspartate, 10 HEPES, 1 EGTA, 1 KCl, 4 MgATP, 0.5 Na3GTP, 15 Neurobiotin citrate. We obtained Neurobiotin citrate from Vector Laboratories as a custom synthesis (rather than using the more standard Neurobiotin chloride) in order to avoid changing the cell’s chloride reversal potential. Since it was difficult to obtain a good morphological reconstruction of R1/R3a neurons, in a subset of recordings, we also used internal solution where the concentration of Neurobiotin citrate was increased two-fold. This internal solution contained (in mM): 110 potassium aspartate, 10 HEPES, 1 EGTA, 1 KCl, 4 MgATP, 0.5 Na3GTP, 30 Neurobiotin citrate. Note that potassium aspartate was lowered in this solution in order to avoid changing the osmolarity of the solution.
Somata were visualized on an upright compound microscope (BX51F, Olympus) equipped with a 40× water-immersion objective (LMUPlanFL, Olympus), a 100W mercury arc lamp for fluorescence excitation (BH2-RFL-T3, Olympus), and a camera (BFLY-PGE-13E4M, FLIR). For a subset of WL-L recordings, the brain was illuminated with IR light from the microscope substage condenser; for all other recordings, the brain was illuminated with IR light from below-oblique using an LED coupled to a fiber adapter (850 nm; SXF30–850, Smart Vision Lights).
Recordings were obtained using a patch-clamp amplifier (Axopatch 200B, Molecular Devices) with a matched headstage (CV 203BU, Molecular Devices). Whole-cell recordings were performed in current-clamp mode. Signals were low-pass filtered at 5 kHz (4-pole Bessel filter) and were acquired at 10 kHz using a data acquisition card (PCIe-6351, National Instruments). A small amount of hyperpolarizing current was applied to compensate for the depolarizing leak conductance and thereby match the spontaneous firing rate measured in cell-attached mode prior to break-in (Gouwens and Wilson, 2009) (see below); this current was held constant throughout a given recording. In cases where the cell-attached spikes were not resolvable, or the break-in occurred spontaneously immediately after seal formation, the amount of hyperpolarizing current was adjusted to match the current used in other cell-attached recordings from that cell type. The liquid junction potential was corrected post hoc by subtracting 13 mV from recorded voltages (Gouwens and Wilson, 2009).
R1/R3a neurons and WL-L neurons exhibited spontaneous spiking in the absence of wind stimulation. To estimate the spontaneous firing rate unaffected by the seal conductance, we attempted to obtain cell-attached recordings (Gouwens and Wilson, 2009) prior to break-in. Cell-attached recordings were performed in voltage-clamp mode and the holding potential was adjusted such that the net current across the electrode was close to zero. Signals obtained with cell-attached recordings were low-pass filtered at 1 kHz (4-pole Bessel filter) and were acquired at 10 kHz using a data acquisition card (PCIe-6351, National Instruments). We did not analyze cell-attached data if there was a sign of a partial break-in (i.e., if spike waveforms looked monophasic and the current signal ramped in between spikes).
Optogenetic activation
To activate CsChrimson (Klapoetke et al., 2014) in functional connectivity experiments, we delivered light from the microscope’s 100 W mercury arc lamp through the 40× water-immersion objective. This light was band-pass filtered between 545 ± 12.5 nm (ET545/25×, Chroma). The power density of the light was varied using a series of neutral density filters and was measured at the level of the sample using a power meter (PM100D with the sensor S120C, Thorlabs). Unless otherwise notes, a single 10 ms pulse of light was delivered using a mechanical shutter (VCM-D1, Uniblitz). The physiology rig was covered with blackout fabric (BK5, Thorlabs) or black posterboards (TB5, Thorlabs) to shield it from ambient light.
We confirmed that the CsChrimson can activate WL-L neurons by performing whole-cell current-clamp recordings in WL-L expressing CsChrimson tagged with mCherry. WL-L neurons have a high rate of baseline spiking, and the firing rate increased upon delivery of 10 ms light pulse (8 μW/mm; Figure 7D). We observed that a much stronger light intensity (>250 μW/mm2) led to a depolarization block in WL-L neurons.
When we recorded from R1 neurons while activating WL-L neurons, we were able to observe responses using the same light intensity (8 μW/mm2; Figure 7E). However, after 1 μM tetrodotoxin (TTX) was applied to the bath solution, the light intensity often had to be increased to 32 μW/mm2 to be able to observe responses in R1 neurons. This is likely due to decreased excitability of presynaptic terminals after TTX application.
Optogenetic silencing
To recruit GtACR1 (Govorunova et al., 2015) in optogenetic silencing experiments, we delivered light from the microscope’s 100 W mercury arc lamp through the 40× water-immersion objective (LUMPlanFL N, Olympus). This light was band-pass filtered between 620 ± 30 nm (ET620/60×, Chroma). Although GtACR1 is maximally activated by 515 nm light (Govorunova et al., 2015), we chose 620 nm to reduce the effect of the light on the visual system (Mauss et al., 2017). The power density of the light was measured at the level of the sample using a power meter (PM100D with the sensor S120VC, Thorlabs). Light was turned on either 1 or 2 s before wind onset, kept on during the 4.5-s wind stimulus, and then turned off 1 or 2 s after wind offset. Light delivery was turned on and off using a mechanical shutter (VCM-D1, Uniblitz).
We confirmed that the GtACR1 can silence WL-L neurons by performing whole-cell current-clamp recordings. In these recordings, we targeted WL-L neurons for recording by taking advantage of the fact that the GtACR1 construct we used was tagged with EYFP (Mohammad et al., 2017). At light onset, WL-L neurons immediately hyperpolarized, and their spikes were immediately and completely suppressed (Figures 8E and S7D). We used a power density of 5 μW/mm2 for all our R neuron recordings (Figures 8F-H and S7G-I), except in one experiment in Fig. 8H where the light level was 0.3 μW/mm2. We confirmed that WL-L spiking was completely suppressed at both 5 and 0.3 μW/mm2.
Pharmacology
To block spiking, we added 1 μM TTX in the bath. A 1 mM stock solution of TTX was prepared either by dissolving TTX (BML-NA120–0001, Enzo) in sodium citrate or dissolving TTX citrate (ab120055, Abcam) in distilled water. In TTX, R1 neurons stopped spiking (Figure S6C), but they often still exhibited slow depolarization events (Figure 7E).
To block inhibition mediated by GABAa receptors, we used 5 μM picrotoxin. While picrotoxin is a broad-spectrum chloride channel pore blocker, at this low concentration, it preferentially blocks GABA-gated chloride conductances (Wilson and Laurent, 2005) rather than glutamate-gated chloride conductances (Liu and Wilson, 2013). Picrotoxin (P1675, Sigma-Aldrich) was dissolved in aqueous 140 mM NaCl to make a 5 mM stock solution that was stored in the dark at room temperature.
Stock solutions were diluted to the final concentration in external saline solution immediately before use. Drugs were bath-applied via the saline perfusate.
Data exclusion criteria
For calcium imaging of E-PG neurons, flies were discarded at the beginning of the experiment if the fluorescence in the protocerebral bridge was too dim to identify glomeruli. In a minority of flies (n=4 flies), the E-PG bump started to move in one direction at a relatively constant speed in the middle of the experiment. We discarded all the trials that contained such a movement.
For patch clamp recordings, neurons that had resting membrane potential above −33 mV at the time of break-in were excluded (one R3a neuron, one WL-L neuron). In addition, neurons that did not have at least two trials per wind direction were excluded (two WL-L neurons). One recording from the driver line that targets both R1 and R3a cells (VT017183-Gal4) was not included because we were not able to recover the morphology of the recorded neuron. We did include two R neuron recordings from VT017183-Gal4 in Figure 5 where the morphology was not recovered, because these recordings were extremely difficult to obtain, and because we were able to obtain a reasonably confident estimate of the cell’s identity based on its inter-spike interval (ISI) distribution (see Figure S3E-G and Quantification of burstiness in spiking below).
Immunohistochemistry and anatomy
3–8 day old female flies were used for all immunohistochemistry experiments. Brains were dissected and fixed for 15 min at room temperature in 4% paraformaldehyde (15714, Electron Microscopy Sciences) in phosphate buffered saline (PBS; 46–013-CM, Thermo Fisher Scientific), then rinsed in PBS and incubated in blocking solution, which was 5% normal goat serum (G9023, Sigma-Aldrich) in PBST (PBS + 0.44% Triton-X, T8787, Sigma-Aldrich) for at least 20 min. Brains were then incubated for ~24 hours at room temperature in a solution of primary antibodies dissolved in blocking solution. After rinsing with PBST, brains were incubated for ~24 hours at room temperature in a solution of secondary antibodies dissolved in blocking solution. Brains were rinsed with PBST and were mounted on a microscope slide in antifade mounting medium (Vectashield, H-1000, Vector Laboratories).
Examining expression pattern of Gal4 lines
The expression patterns of Gal4 lines were examined by crossing these lines with a UAS-mCD::GFP reporter line, and were visualized using standard immunostaining protocols. Primary antibody solution contained anti-Bruchpilot antibody (1:30, nc82, Developmental Studies Hybridoma Bank), chicken anti-GFP (1:1000, ab13970, Abcam), and 5% normal goat serum (NGS) in PBST. Secondary antibody solution contained Alexa Fluor 488-conjugated goat anti-chicken (1:250, A-11039, Thermo Fisher Scientific), Alexa Fluor 633-conjugated goat anti-mouse (1:250, A-21050, Thermo Fisher Scientific), and 5% NGS in PBST.
MultiColor FlpOut (MCFO)
MCFO was used to identify the morphologies of single neurons and was performed essentially as described previously (Nern et al., 2015). Primary incubation solution contained mouse anti-Bruchpilot antibody (1:30, nc82, Developmental Studies Hybridoma Bank), rat anti-FLAG (1:200, NBP1–06712B, Novus Biologicals), rabbit anti-HA (1:300, Cell Signaling Technologies), and 5% normal goat serum (NGS) in PBST. Secondary incubation solution contained Alexa Fluor 488-conjugated goat anti-rabbit (1:250, 3724, Thermo Fisher Scientific), ATTO-647-conjugated goat anti-rat (1:400, 612–156-120, Rockland), and Alexa Fluor 405-conjugated goat anti-mouse (1:500, 3724, Thermo Fisher Scientific) and 5% NGS in PBST. Tertiary incubation solution contained DyLight 500-conjugated mouse anti-V5 (1:500, MCA1360D550GA, Bio-Rad), and 5% normal mouse serum in PBST.
Polarity marker staining
DenMark (Nicolai et al., 2010) was used to determine the polarity of neurons. Primary antibody solution contained anti-Bruchpilot antibody (1:30, nc82, Developmental Studies Hybridoma Bank), chicken anti-GFP (1:1000, ab13970, Abcam), rabbit-anti DsRed (1:500, 632496, Living Colors), and 5% normal goat serum (NGS) in PBST. Secondary antibody solution contained Alexa Fluor 488-conjugated goat anti-chicken (1:250, A-11039, Thermo Fisher Scientific), Alexa Fluor 568-conjugated goat anti-rabbit (1:250, A-11011, Thermo Fisher Scientific), Alexa Fluor 633-conjugated goat anti-mouse (1:250, A-21050, Thermo Fisher Scientific), and 5% NGS in PBST.
GABA immunostaining
Immunostaining against GABA was performed essentially as described previously (Wilson and Laurent, 2005). Primary antibody solution contained rabbit anti-GABA (1:100, A2052, Sigma-Aldrich), anti-Bruchpilot antibody (1:30, nc82, Developmental Studies Hybridoma Bank), chicken anti-GFP (1:1000, ab13970, Abcam), and 5% normal goat serum (NGS) in PBST. Secondary antibody solution contained Alexa Fluor 568-conjugated goat anti-rabbit (1:250, Thermo Fisher Scientific), Alexa Fluor 633-conjugated goat anti-mouse (1:250, A-11011, Thermo Fisher Scientific), Alexa Fluor 488-conjugated goat anti-chicken (1:250, A-11039, Thermo Fisher Scientific), and 5% NGS in PBST.
Morphological reconstruction following whole-cell recordings
To visualize the morphology of the recorded neurons, we processed the brains in a primary incubation solution containing anti-Bruchpilot antibody (nc82, 1:30, Developmental Studies Hybridoma Bank), chicken anti-GFP (1:1000, ab13970, Abcam) and 5% normal goat serum (NGS) in PBST. Secondary antibody solution contained Alexa Fluor 488-conjugated goat anti-chicken (1:250, A-11039, Thermo Fisher Scientific), Alexa Fluor 633-conjugated goat anti-mouse (1:250, A-11011, Thermo Fisher Scientific), Alexa Fluor 568-conjugated streptavidin (1:1000, S11226, Thermo Fisher Scientific), and 5% NGS in PBST.
In subset of the functional connectivity experiments (Figure 7 and S6), we used four fluorophores to visualize GFP, CsChrimson-tdTomato, Neurobiotin fill, and Bruchpilot. Primary incubation solution contained anti-Bruchpilot antibody (nc82, 1:30, Developmental Studies Hybridoma Bank), chicken anti-GFP (1:1000, ab13970, Abcam), rabbit anti-DsRed (1:500; 632496, Takara Bio) and 5% normal goat serum (NGS) in PBST. Secondary antibody solution contained Alexa Fluor 488-conjugated goat anti-chicken (1:250, A-11039, Thermo Fisher Scientific), Alexa Fluor 405-conjugated goat anti-mouse (1:500; A-31553, Thermo Fisher Scientific), Alexa Fluor 594-conjugated donkey anti-rabbit (1:500; 711–585-152, Jackson ImmunoResearch Laboratories), Alexa Fluor 633-conjugated streptavidin (1:1000, S21375, Thermo Fisher Scientific), and 5% NGS in PBST.
Confocal microscopy and classification of R neuron morphologies
Whole-mounted brains were imaged using a confocal microscope (FV1000, Olympus; SPE or SP8, Leica). Series of 50 – 100 optical sections (1–2 pm spacing) were imaged using either a UPLFLN 40× oil-immersion lens (N.A. 1.3) or a PLAPON 60× oil-immersion lens (N.A. 1.42). The images were analyzed in Fiji (Schindelin et al., 2012).
R neuron MCFO clones were classified as one of 11 subtypes following (Omoto et al., 2018) based on the consensus of two experts. For morphological reconstruction of the recorded neurons, the dendrites of R1/R3a neurons were often difficult to fill, but even in these cases, we were often able to observe the fills of axon terminals in the ellipsoid body. Since MCFO analysis revealed that R1 neurons innervate the posterior EB and R3a neurons innervate the anterior EB (Figure 2E), we were able to determine whether the recorded neuron was R1 or R3a from these partial axonal fills in the EB.
QUANTIFICATION AND STATISTICAL ANALYSIS
General policies for statistical tests
We used parametric tests unless Lilliefors test (Lilliefors, 1967) indicated the data was not normally distributed at the significance value of α = 0.05. One exception was the use of ANOVA for Figure 3F for the membrane potential: the null hypothesis that the distribution of membrane potential during headwind came from normal distribution was rejected, but we chose to use ANOVA to keep it consistent with all the other tests we used for comparing wind direction. All the tests were two-tailed. We corrected for multiple comparisons using Bonferroni correction. If the post-hoc test was a planned comparison, we did not correct for multiple comparisons. To analyze circular variables (e.g. E-PG phase), we used the CircStat toolbox in MATLAB (Berens, 2009).
Analysis of antennal movements
The positions of the base and tip of both aristae over time were extracted from the video recordings using LEAP, a deep learning-based method for pose estimation (Pereira et al., 2019). Movies were first preprocessed with spatial reduction and downsampling. Subsequently, a training set was selected by concatenating representative blocks of frames from movies of all five experimental flies. This set was composed of 6674 frames (out of 135,080 frames total). This set was then further reduced to 240 representative frames by means of PCA and cluster sampling. From this point, two models were independently trained.
For the first model, we manually annotated the position of each antenna in 90 of 240 frames of the smallest training set. We then trained the network with these 90 annotated frames and applied the model predictions to the rest of the smaller training set. We visually inspected the model predictions and manually corrected and validated the estimates in all 240 frames of this set. We then trained the model again with all the 240 annotated frames, and we applied it to the entire dataset to obtain the first set of final estimates of antennal positions. This model performed well, except for rare moments where video quality was low (e.g. smeared, or under-exposed images), and so the model generated large errors. Additionally, we found that consecutive and virtually identical frames would often produce slightly different estimates. These errors were smaller but also more common. These errors are due to the probabilistic nature of the model and the fact that even experts had difficulty measuring antennal poses precisely from the videos.
To ameliorate these problems, we trained a second model. We first initialized the 6674 frames of our larger training set with the estimates from model 1. We then visually inspected the frames and manually corrected and validated 225 frames out of the 6674 total. These frames and the corresponding annotations were largely distinct from the ones that were used to train model 1. In particular, these frames were selected to be both generally representative of the dataset, but also to over-represent in this training set frames in which model 1 exhibited a poorer performance than average. We used this new set of 225 annotated frames to train model 2, then we applied this model to the entire dataset to obtain the second set of final estimates of antennal positions. Model 2 also performed generally very well, tending to outperform model 1 in the rare but large-error condition.
For each frame f, antenna f, and model m, we used the base and tip positions of the arista to calculate its angular orientation α(f, a,m) with respect to a line in the horizontal plane which runs tangent to the anterior edge of the fly head. For each frame and antenna, we then quantified the model error (e) by looking at the deviation between the two model estimates. Because we only had two estimates, we simply considered the absolute value of the difference between the two:
We excluded from further analysis all the time points with e(f, a) > 3° (1.6% for the right antenna, 1.8% for the left antenna). For each frame and antenna, we then averaged the two model estimates to obtain
This allowed us to reduce the effect of the small noisy variations in pose estimates.
For each fly, we concatenated all the baseline periods (i.e., periods when no stimulus was present) over the entire course of the experiment, to check that the resting position of each antenna was stable. (Recall that active movements were absent because the first and second segments of the antennae were glued to the head.) We then averaged these values to estimate of the resting position of each antenna in our dataset.
Next, for each fly and each stimulus presentation, we time-averaged α(f, a) across all the frames when the wind was presented, and we did the same for each immediately preceding baseline period. Finally, we subtracted the antenna’s baseline angle from the antenna’s wind-evoked angle to produce tuning curves of antennal displacement as a function of wind direction and intensity.
Modeling antennal movements in response to wind
For each fly and for each wind intensity level, we generated tuning curves of antennal displacement as a function of wind direction. The shape of the tuning curves was generally similar across flies and mirror-symmetric across the left and right antennae. Variations were likely due to imprecision in stimulus delivery and recording noise, or to slight variations in the angular position of each antenna at rest. This resting position was fixed for each antenna (between 20° and 40°) and imposed on the fly by gluing the proximal part of each antenna to the fly’s head. Changes in the resting position of the antenna produced small but systematic translations of the tuning curve along the wind azimuth axis, and so imprecision in our gluing procedure is another source of measurement error.
To build a model of bilateral antennal displacements as a function of wind direction and intensity, we selected a fly whose antennae were both fixed at 30°. To model symmetric displacements of the two antennae, we averaged the tuning curve of the right antenna with the flipped tuning curve of the left antenna. We then fit a smoothing spline model (smoothing parameter 0.0027) to the data. Finally, we mirrored the resulting tuning curve to represent symmetric left and right antennal displacements.
Analysis of calcium imaging data
Alignment and selection of regions of interest (ROIs)
Images were aligned using an efficient subpixel motion registration (Guizar-Sicairos et al., 2008), as described previously (Patella and Wilson, 2018). ROIs for dendrites and somata of R1/R3a neurons were drawn manually. ROIs for E-PG axons were drawn in a semi-automated manner. Briefly, our goal in this analysis was to identify an ROI for each of the 16 glomeruli in the protocerebral bridge (PB) that receive E-PG input (Wolff et al., 2015). We first manually drew a mask around the entire PB. We computed candidate ROIs using the constrained non-negative matrix factorization to decompose the imaging data into spatial and temporal components for each plane (greedy initialization with number of spatial components = 30) (Pnevmatikakis et al., 2016). We then inspected spatial components that lay within the PB mask in 6 z-planes. We selected 16 spatial components that likely correspond to each of the 16 glomeruli (Figure S1B). Each of these 16 spatial components was defined in one z-plane, meaning that it did not necessarily encompass the entire z-depth of the glomerulus in question.
Quantification of ΔF/F
For E-PG neuron data, the baseline F for calculating ΔF/F was computed by taking the mean of the lowest 10% of the intensity values in a given ROI within each trial. For R neuron data, we defined the baseline F as the mean of the intensity values over 1 s prior to the wind onset in a given ROI. ΔF/F traces were temporally smoothed using a 3rd order Savitzky-Golay filter over 11 frames (0.9 s). To calculate the wind responses of R1 and R3a dendrites (Figure S3A-D), we averaged ΔF/F during the wind presentation excluding the first 1 sec. To calculate the wind responses of R1 somata, we calculated the mean of ΔF/F during the last 1 sec of wind presentation to capture the peak immediately before wind offset (Figure 6C).
Quantification of bump location and E-PG phase
Following previous work (Fisher et al., 2019; Green et al., 2017), we estimated the location of the E-PG bump by analyzing the spatial frequency of the activity patterns in the PB. Specifically, we took the spatial Fourier transform (Hanning window; FFT with N = 1,024 points after zero-padding) of ΔF/F values across all 16 PB glomeruli. We defined the phase of the Fourier component that has a spatial period of 8 glomeruli as the E-PG phase. The sign convention of the E-PG phase is depicted in Figure S1C.
To quantify how much the E-PG bump moves over time, we defined “bump mobility” as the angular deviation of the E-PG phase during an epoch when the stimulus condition is not changing. Bump mobility takes on a large value when the E-PG bump moves around over time and is bounded by . Angular deviation was computed using circ_std function in Circular Statistics Toolbox in MATLAB (Berens, 2009). We computed bump mobility within each trial during the baseline period (“before wind”). We computed mobility separately during each of the two wind presentations, and then averaged these values (“during wind”) (Figures 2G and S2J).
Quantification of bump jump (Figure 2H)
We used the following procedure to calculate whether the E-PG bump location changed significantly after shifting the wind direction. For each trial, we calculated the mean of the E-PG phase and the mean resultant vector during the left wind epoch and the right wind epoch. Trials where the wind direction changed from left to right and trials where the wind trials changed from right to left were pooled. For this calculation, we excluded the initial 1 sec after the wind shift to avoid the period when the bump is typically moving or jumping. We then tested the null hypothesis that the mean of mean E-PG phase during the left wind epoch is the same as the mean of mean E-PG phase during the right wind epoch. Note that because we are dealing with mean of means, we used a second-order analysis (Zar, 2010). Therefore, we used a Hotelling test on paired second-order data (Zar, 2010) to test this null hypothesis at α = 0.05.
Analysis of electrophysiology data
Spike detection
To detect spikes, we first removed the slow trend in the membrane potential. This was done by median-filtering the membrane potential (50 ms window) and then subtracting the median-filtered from the original membrane potential Vm. We then took a time-derivative of , and set a threshold to detect spikes. In a few dorsal LAL neuron recordings, neurons exhibited wide bursts (~200 ms) with multiple fast spikes riding on top, which made it difficult to detect individual spikes. In those cases, we may be underestimating the firing rates during those epochs.
Quantification of wind responses
To quantify spiking responses to wind, we calculated the spike rate during the entire wind presentation (4.5 s). We then calculated Δ spike rate by subtracting baseline spike rate during 2 s prior to wind onset. To quantify changes in membrane potential in response to wind, we calculated the mean median-filtered membrane potential during the entire wind presentation. We then calculated Δ membrane potential by subtracting the baseline median-filtered membrane potential averaged over 2 s prior to wind onset. To test whether stimulus- evoked responses (either Δ spike rate or Δ membrane potential) depended on wind direction, we used one-way repeated measures ANOVA with wind direction as the within-subject factor. To test whether the wind responses differed between R1 and R3a neurons, we used two-way ANOVA with wind direction and neuron type (R1/R3a) as two factors.
Quantification of burstiness in spiking
We quantified burstiness by examining the inter-spike interval (ISI) distribution on a logarithmic x-axis. Neurons that exhibited bursting tended to have two peaks, one corresponding to ISIs within bursts, and the other corresponding to ISIs between bursts (Figures S3E and S3F). Neurons that did not burst tended to have a broad ISI distribution with a single peak. To characterize the bimodality of the log-transformed ISI distribution (base 10), we used a bimodality coefficient (BC). BC is calculated based on skewness and excess kurtosis, and it ranges from 0 to 1, with higher values indicating bimodality (Freeman and Dale, 2013; Pfister et al., 2013).
We used the BC of the ISI to classify R1 or R3a when the morphological reconstruction of the recorded neuron was not available (a total of five neurons in Figure S4, two of which also appear in Figure 5). If the recording had BC less than 0.5, it was classified as R1, otherwise it was classified as R3a. This threshold value of 0.5 was chosen to minimize the classification error (19%) on a dataset where ground-truth was available (Figure S3G).
Fitting the relationship between E-PG phase and wind direction (Figure 1 and S1)
Visual inspection of the data suggested that, in some flies, the relationship between wind direction and E-PG phase was relatively linear (Figure S1F). To quantify this relationship, we fitted the following model:
where θ is wind direction in degrees, and φ is E-PG phase in degrees. Slope a and the y-intercept φ0 are the regression coefficients that we estimated from sets of observations (θk, φk), where k is the index for individual trials.
Note that we treat wind direction as a linear variable rather than a circular variable, to avoid building a complex model based on circular-circular regression. This is reasonable because in our setup, wind direction is limited to −150° to +150° and does not wrap around (Figure 1G).
The regression coefficients a and φ0 were found by minimizing the mean circular distance between the observed data and the model prediction. Circular distance d between two angles φ1 and φ2 was defined as
following (Kempter et al., 2012). Note that this distance is non-negative. Using this notation, the mean circular distance between the observed data φk and the model prediction a · θk + φ0 can be written as:
Since the estimate of slope is not unique for a given dataset (Kempter et al., 2012), we restricted the range of possible slopes to the interval [amin, amax] = [−2,2]. Finding a slope a that minimizes D(a) under this constraint was found numerically using the fmincon function in Optimization Toolbox in MATLAB. To search for the global minimum, the search was performed 20 times with different initial values of a, drawn from a standard normal distribution N(0,1), and the value of a that minimized D(a) was chosen as an estimate of the slope . Using this , we calculated based on methods described previously (Kempter et al., 2012).
To quantify how well this model explains the data, we devised a goodness-of-fit metric (GOF), which is similar to coefficient of determination r2 in linear regression. If the model is a good fit to the data, the sum of circular distances between the actual data φk and the predicted value should be small (analogous to the residual sum of squares). We normalized this distance by dividing it by the sum of circular distances between the actual data φk and the circular mean of the data (analogous to the total sum of squares).
If the model provides a perfect fit, GOF = 1. On the other hand, if the model is a poor fit, GOF will be close to zero. Note that because we are using circular distance instead of Euclidian distance, the total sum of squares cannot be decomposed into the explained sum of squares and the residual sum of squares.
To assess the significance of the model fit, we compared the GOF values obtained from the actual data and those obtained from shuffled data. Shuffled data was generated by pairing E-PG phase with wind directions that were randomly permuted independently on different trials. GOF for this shuffled data was calculated as described above, and this shuffling was repeated 1,000 times to obtain the distribution of GOF values. Finally, based on this distribution, probability p for obtaining GOF larger than that observed from actual data was calculated. If p<0.05, we considered GOF calculated from actual data to be significantly different from the shuffled data.
Analysis of locomotion (Figures S1H, I)
We processed the output of the FicTrac signal (3-DOF orientation of the ball) as follows: we took the derivative of the unwrapped angle to calculate the angular velocity, removed outliers (defined as three median absolute deviations away from the median) and replaced them with linearly interpolated values, and low-pass filtered the output at 10 Hz.
Analysis of R1/R3a neuron responses to piezoelectric displacements of the antennae (Figures 5 and S4)
Representation of neural responses in 2-D antennal displacement space
For all tested combinations of independent left and right antennal displacements, firing rates were averaged across all presentations of the same displacement. The mean spike rate measured when the antennae were at their resting position (0°,0°) was used as a baseline. This baseline was subtracted from all measured firing rates to generate Δ firing rate in Figures 5E-G. In Figures 5C, D and S4B, C, scatterplots of 2-D neural responses were interpolated using a Delaunay triangulation with natural neighbor interpolation and nearest neighbor extrapolation, through the scatteredInterpolant function of MATLAB (MathWorks). To plot responses of R1/R3a neuron as a function of wind direction and speed (Figures 5E-G) given their responses to piezoelectric displacements, we used a subset of displacements directly drawn from the three lowest-intensity model tuning curves in Figure 4D. For each of those stimuli, we mapped back the corresponding wind parameters in the model tuning curve.
Single antenna contribution to wind responses in R1/R3a neurons
Our stimulus set for piezoelectric actuator experiments included stimuli where one antenna was displaced away from its resting position while the other antenna was held at its resting position. To compare R1/R3a neuron responses to unilateral stimuli against “resting” activity in R1/R3a neurons (i.e., baseline firing when both antennae are held at their resting positions), we computed the 95% confidence interval of the mean of an equal-sized data set (N repetitions) of firing rates recorded in the same neuron when both antennae were held at rest (0°, 0°). Specifically, the confidence interval was calculated from an empirical bootstrap distribution of the mean of 100,000 samples of N repetitions of (0°, 0°). Presentations of (0°, 0°) were randomly drawn from within the stimulus block where unilateral stimuli were being analyzed, to reduce the effect of any drift in neural activity across blocks. Note that we deliberately over-represented the (0°, 0°) condition in our stimulus set, in order to improve our estimates of firing rates in this condition within each stimulus block.
Predicted bilateral responses were obtained by linearly combining each cell’s responses to unilateral stimuli alone, assuming that each antenna contributes equally to the neuron’s firing rate. The resulting scatterplots were then interpolated as described above.
Binary classifiers for detecting wind direction (Figure 6 and S5)
To test whether an ensemble of E-PG neurons can respond to all heading directions with roughly equal accuracy, given only R1 neurons as inputs, we created a simple feedforward network model. We modeled 21 E-PG neurons, one for each wind direction in the R1 imaging experiments (from −150° to +150° in 15° increments). E-PG neurons were modeled as as binary units, with each designed to respond specifically to one wind direction. Each received input from all R1 neurons in both hemispheres measured in a single fly (Figure S5B). One model E-PG network was created for each fly. For flies where only the left hemisphere R1 neurons were imaged (Figure S5A), we simulated right hemisphere R1 neurons by mirroring the left hemisphere data. R→E-PG synaptic weights (w) were initialized with random numbers drawn from a uniform distribution [−1 0].
The activity of each model R1 neuron xi was drawn from a Gaussian distribution whose mean and standard deviation were matched to the observed data for one R1 neuron in the corresponding experiment. R1 data were peak ΔF/F values obtained from calcium imaging (Figure S5A). To generate a training set for each model, we generated 5,000 R1 ensemble activity patterns corresponding to the target wind direction, and 5,000 R1 ensemble activity patterns corresponding to non-target directions. Each activity pattern was designated as the model input (x) on one training trial.
The binary output of the network y was determined by taking the dot product between w and x, and comparing it against a threshold γ.
After each training trial, weights were modified based on the following perceptron learning rule with normalization.
where ϵ is the learning rate, which was set to ϵ = 0.01. Weights were constrained to be non-positive: if the update rule made a weight positive, the weight was set to 0 (Amit et al., 1989). The threshold γ was also updated by treating it as a weight that always received an input of −1, noting that w · x > γ is equivalent to w · x + γ · (−1)>0. Once the weights and the threshold were determined from the training set, the performance of the network was tested on a separate test set that was generated in the same way as the training data set (n=1,000 trials for each wind direction). We quantified the performance of the model E-PG neurons by computing the true positive rate and the false positive rate for each model E-PG neuron in each network (Figure S5D).
Analysis of EM dataset (Figure S8)
To analyze the connectivity between WL-L and R1/R3a neurons, we analyzed a partial connectome of the dorsal part of the right central brain of an adult female fly obtained by the FlyEM Project at Janelia Research Campus (https://www.janelia.org/project-team/flyem/hemibrain) (Xu et al., 2020). We performed the analysis in R using neuprintr (Bates et al., 2019a; Bates et al., 2019b). R1 neurons and R3a neurons were already annotated in this dataset (hemibrain v1.0.1). sA partial skeleton of a WL-L neuron was identified by comparing the single-cell morphology of WL-L neurons at the light microscopy level and the reconstructed skeleton in the EM data (Figure S8A). Using this dataset, we focused on presynaptic inputs to R1/R3a neuron dendrites in the LAL, excluding inputs to R1/R3a axons in the EB. Neurons that make three or fewer synapses onto R1 or R3a neurons were omitted from our analysis, based on the recommendation of (Xu et al., 2020).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Bruchpilot (nc82) | Developmental Studies Hybridoma Bank | Cat# nc82; RRID: AB_2314866 |
| chicken anti-GFP | Abcam | Cat# ab13970; RRID:AB_300798 |
| rabbit-anti DsRed (Living Colors) | Takara Bio | Cat# 632496; RRID: AB_10013483 |
| rat mAb anti-FLAG | Novus Biologicals | Cat# NBP1-06712B; RRID: AB_10006034 |
| Rabbit anti-HA | Cell Signaling Technologies | Cat# 3724; RRID:AB_1549585 |
| rabbit anti-GABA | Sigma-Aldrich | Cat# A2052; RRID: AB_477652 |
| DyLight 500-conjugated mouse anti-V5 | Bio-Rad | Cat# MCA1360D550GA; RRID: AB_2687576 |
| Alexa Fluor 405-conjugated goat anti-mouse | Thermo Fisher Scientific | Cat# A-31553, RRID:AB_221604 |
| Alexa Fluor 488-conjugated goat anti-chicken | Thermo Fisher Scientific | Cat# A-11039; RRID: AB_2534096 |
| Alexa Fluor 488-conjugated goat anti-rabbit | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_143165 |
| Alexa Fluor 568-conjugated goat anti-rabbit | Thermo Fisher Scientific | Cat# A-11011; RRID: AB_143157 |
| Alexa Fluor 594-conjugated donkey anti-rabbit | Jackson ImmunoResearch Laboratories | Cat# 711-585-152; RRID:AB_2340621 |
| Alexa Fluor 633-conjugated goat anti-mouse | Thermo Fisher Scientific | Cat# A-21050; RRID: AB_2535718 |
| ATTO-647-conjugated goat anti-rat | Rockland | Cat# 612-156-120; RRID: AB_10893386 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin, Alexa Fluor 568 conjugate | Thermo Fisher Scientific | Cat# S11226; RRID: AB_2315774 |
| Streptavidin, Alexa Fluor 633 conjugate | Thermo Fisher Scientific | Cat# S21375; RRID: AB_2313500 |
| Tetrodotoxin (TTX) | Enzo | BML-NA120-0001 |
| Tetrodotoxin citrate | Abcam | ab120055 |
| Picrotoxin | Sigma-Aldrich | P1675 |
| Vectashield mounting medium | Vector Laboratories | Cat# H-1000, RRID:AB_2336789 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: Dickinson wild type | Michael Dickinson lab | N/A |
| D. melanogaster: P{pBDPGAL4.1Uw}attP2 | Bloomington Drosophila Stock Center (BDSC) | RRID:BDSC_68384 |
| D. melanogaster: P{R12G08-Gal4}attP2 | BDSC | RRID:BDSC_47855 |
| D. melanogaster: P{R20A02-Gal4}attP2 | BDSC | RRID:BDSC_48870 |
| D. melanogaster: P{R26B07-Gal4}attP2 | BDSC | RRID:BDSC_49160 |
| D. melanogaster: P{R31A12-Gal4}attP2 | BDSC | RRID:BDSC_49661 |
| D. melanogaster: P{R54E12-Gal4}attP2 | BDSC | RRID:BDSC_39077 |
| D. melanogaster: P{R60D05-Gal4}attP2 | BDSC | RRID:BDSC_39247 |
| D. melanogaster: P{VT017183-Gal4}attP2 | Vienna Drosophila Resource Center (VDRC) | RRID:FlyBase_FBst0484665 |
| D. melanogaster: P{VT040354-Gal4}attP2 | VDRC | RRID:FlyBase_FBst0487578 |
| D. melanogaster: P{VT059225-p65.AD}attP40 | BDSC | RRID:BDSC_71395 |
| D. melanogaster: P{VT048577-GAL4.DBD}attP2 | BDSC | RRID:BDSC_72828 |
| D. melanogaster: P{repo-Gal80} | Tzumin Lee lab | N/A |
| D. melanogaster: P{R53B02-LexA}attP40 | BDSC | RRID:BDSC_53615 |
| D. melanogaster: P{R60D05-LexA}attP40 | BDSC | RRID:BDSC_52867 |
| D. melanogaster: P{Or83b-LexA} | Tzumin Lee lab | N/A |
| D. melanogaster: P{20XUAS-GCaMP6f}attP40 | BDSC | RRID:BDSC_42747 |
| D. melanogaster: Pbac{20XUAS-IVS-jGCaMP7f}VK00005 | BDSC | RRID:BDSC_79031 |
| D. melanogaster: P{UAS-DenMark}3, P{UAS-syt.eGFP}3 | BDSC | RRID:BDSC_33065 |
| D. melanogaster: P{UAS-Hsap\KCNJ2.EGFP}7 | BDSC | RRID:BDSC_6595 |
| D. melanogaster: P{20×UAS-CsChrimson-mCherry-trafficked}su(Hw)attP1 | Vivek Jayaraman lab | N/A |
| D. melanogaster: P{20XUAS-GtACR1-EYFP}attP2 | Adam Claridge-Chang lab | N/A |
| D. melanogaster: P{20XUAS-IVS-mCD8::GFP}attP40 | Gerry Rubin lab | RRID:BDSC_32194 |
| D. melanogaster: P{10XUAS-IVS-mCD8::GFP}su(Hw)attP8 | BDSC | RRID:BDSC_32189 |
| D. melanogaster: P{13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 | BDSC | RRID:BDSC_44277 |
| D. melanogaster: P{13XLexAop2-IVS-myr::GFP}VK00005 | Gerry Rubin lab | RRID:BDSC_32209 |
| D. melanogaster: P{13XLexAop2-IVS-myr::GFP}su(Hw)attP1 | BDSC | RRID:BDSC_32212 |
| D. melanogaster: P{13XLexAop2-IVS-CsChrimson.mVenus}attP18 | BDSC | RRID:BDSC_55137 |
| D. melanogaster: MCFO-3 | BDSC | RRID:BDSC_64087 |
| D. melanogaster: MCFO-4 | BDSC | RRID:BDSC_64088 |
| D. melanogaster: MCFO-5 | BDSC | RRID:BDSC_64089 |
| Software and Algorithms | ||
| MATLAB | MathWorks | RRID:SCR_001622 |
| ScanImage | Pologruto et al., 2003 | RRID:SCR_014307 |
| LEAP (LEAP Estimates Animal Pose) | https://github.com/talmo/leap | Pereira et al., 2018 |
| Constrained nonnegative matrix factorization for calcium imaging data | https://github.com/flatironinstitute/CaImAn-MATLAB | Pnevmatikakis et al., 2016 |
| FicTrac v2 | http://rjdmoore.net/fictrac/ | Moore et al., 2014 |
| Fiji | http://fiji.sc | RRID:SCR_002285 |
Highlights.
The compass in the Drosophilia brain is influenced by wind direction
Wind direction is conveyed to the compass via a specialized Ring neuron population
Ring neurons extract wind direction based on the displacements of both antennae
Compass neurons integrate wind and visual cues to produce a multimodal map of space
Acknowledgments
We are grateful to T. Clandinin, A. Claridge-Chang, M. Crickmore, M. Dickinson, V. Jayaraman, T. Lee, B. Pfeiffer, and G. Rubin for providing fly stocks. We thank S. Rayshubskiy, H. Haberkern, J. Drugowitsch, A. Kutschireiter, Y. Fisher, J. Lu, and M. Basnak, for discussions, and A. Bates for help with analysis of EM data. We thank the Neurobiology Machine Shop (NEI Core grant for Vision Research EY012196), and O. Mazor and P. Gorelik at the HMS Research Instrumentation Core (NEI Core grant for Vision Research EY012196) for advice on constructing the wind stimulus device, and the HMS Neurobiology Imaging Facility (NINDS P30 Core Center grant NS072030) for help in confocal imaging. We thank the Wilson lab for comments on the manuscript. T.S.O. was supported by Harvard Mahoney Neuroscience Institute Fund and an HHMI fellowship from the Helen Hay Whitney Foundation. This work is supported by NIH grants R01NS101157 and U19NS104655, and an HMS Dean’s Innovation Award. R.I.W. is an HHMI Investigator.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Salvado E, Licata AM, Connor EG, McHugh MK, King BM, Stavropoulos N, Victor JD, Crimaldi JP, and Nagel KI (2018). Elementary sensory-motor transformations underlying olfactory navigation in walking fruit-flies. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit DJ, Wong KYM, and Campbell C (1989). Perceptron learning with sign-constrained weights. J Phys A-Math Gen 22, 2039–2045. [Google Scholar]
- Awasaki T, Lai SL, Ito K, and Lee T (2008). Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci 28, 13742–13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Beckingham KM, and Armstrong JD (2007). Functional dissection of the neural substrates for gravitaxic maze behavior in Drosophila melanogaster. J Comp Neurol 501, 756–764. [DOI] [PubMed] [Google Scholar]
- Bates AS, Franconville R, and Jefferis GSXE (2019a). neuprintr: R client utilities for interacting with the neuPrint connectome analysis service. R package version 0.4.0. [Google Scholar]
- Bates AS, Manton JD, Jagannathan SR, Costa M, Schlegel P, Rohlfing T, and Jefferis GSXE (2019b). The natverse: a versatile computational toolbox to combine and analyse neuroanatomical data. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JS, Tobin TR, and Sorensen KA (1989). Orientation responses of individual larder beetles, Dermestes ater (Coleoptera, Dermestidae), to directional shifts in wind stimuli. J Insect Behav 2, 787–801. [Google Scholar]
- Bell JS, and Wilson RI (2016). Behavior reveals selective summation and max pooling among ofactory processing channels. Neuron 91, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell WJ, and Kramer E (1979). Search and anemotactic orientaton of cockroaches. J Insect Physiol 25, 631–640. [Google Scholar]
- Berens P (2009). CircStat: A MATLAB Toolbox for Circular Statistics. J Stat Softw 31. [Google Scholar]
- Birukow G (1958). Zur Funktion der Antennen beim Mistkäfer (Geotrupes silvaticus Panz.). Ethology 15, 265–276. [Google Scholar]
- Böhm H (1995). Dynamic properties of orientation to turbulent air current by walking carrion beetles. J Exp Biol 198, 1995–2005. [DOI] [PubMed] [Google Scholar]
- Böhm H, Heinzel H-G, Scharstein H, and Wendler G (1991). The course control system of beetles walking in an air-current field. J Comp Physiol [A] 169, 671–683. [Google Scholar]
- Chang AEB, Vaughan AG, and Wilson RI (2016). A mechanosensory circuit that mixes opponent channels to produce selectivity for complex stimulus features. Neuron 92, 888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Chen WY, and Chien CT (2012). The POU-domain protein Pdm3 regulates axonal targeting of R neurons in the Drosophila ellipsoid body. Dev Neurobiol 72, 1422–1432. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A, Zhang S, Stricker C, and Srinivasan MV (2007). Animal navigation: the difficulty of moving in a straight line. Biol Cybern 97, 47–61. [DOI] [PubMed] [Google Scholar]
- Collett TS (2019). Path integration: how details of the honeybee waggle dance and the foraging strategies of desert ants might help in understanding its mechanisms. J Exp Biol 222. [DOI] [PubMed] [Google Scholar]
- Dacke M, Bell ATA, Foster JJ, Baird EJ, Strube-Bloss MF, Byrne MJ, and el Jundi B (2019). Multimodal cue integration in the dung beetle compass. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacke M, and el Jundi B (2018). The dung beetle compass. Curr Biol 28, R993–R997. [DOI] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16, 649–657. [DOI] [PubMed] [Google Scholar]
- Dus M, Ai M, and Suh GS (2013). Taste-independent nutrient selection is mediated by a brain-specific Na+ /solute co-transporter in Drosophila. Nat Neurosci 16, 526–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Jundi B, Baird E, Byrne MJ, and Dacke M (2019). The brain behind straight-line orientation in dung beetles. J Exp Biol 222. [DOI] [PubMed] [Google Scholar]
- el Jundi B, Foster JJ, Khaldy L, Byrne MJ, Dacke M, and Baird E (2016). A snapshot-based mechanism for celestial orientation. Curr Biol 26, 1456–1462. [DOI] [PubMed] [Google Scholar]
- Fisher YE, Lu J, D’Alessandro I, and Wilson RI (2019). Sensorimotor experience remaps visual input to a heading-direction network. Nature 576, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franconville R, Beron C, and Jayaraman V (2018). Building a functional connectome of the Drosophila central complex. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JB, and Dale R (2013). Assessing bimodality to detect the presence of a dual cognitive process. Behav Res Methods 45, 83–97. [DOI] [PubMed] [Google Scholar]
- Giraldo YM, Leitch KJ, Ros IG, Warren TL, Weir PT, and Dickinson MH (2018). Sun navigation requires compass neurons in Drosophila. Curr Biol 28, 2845–2852 e2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, and Lockery SR (2000). Pressure polishing: a method for re-shaping patch pipettes during fire polishing. J Neurosci Methods 100, 13–15. [DOI] [PubMed] [Google Scholar]
- Gopfert MC, and Robert D (2001). Biomechanics. Turning the key on Drosophila audition. Nature 411, 908. [DOI] [PubMed] [Google Scholar]
- Gouwens NW, and Wilson RI (2009). Signal propagation in Drosophila central neurons. J Neurosci 29, 6239–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Janz R, Liu X, and Spudich JL (2015). Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Adachi A, Shah KK, Hirokawa JD, Magani PS, and Maimon G (2017). A neural circuit architecture for angular integration in Drosophila. Nature 546, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, and Maimon G (2018). Building a heading signal from anatomically defined neuron types in the Drosophila central complex. Curr Opin Neurobiol 52, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Vijayan V, Mussells Pires P, Adachi A, and Maimon G (2019). A neural heading estimate is compared with an internal goal to guide oriented navigation. Nat Neurosci 22, 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford T, and Taylor GK (2014). The sun compass revisited. Anim Behav 97, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizar-Sicairos M, Thurman ST, and Fienup JR (2008). Efficient subpixel image registration algorithms. Opt Lett 33, 156–158. [DOI] [PubMed] [Google Scholar]
- Handler A, Graham TGW, Cohn R, Morantte I, Siliciano AF, Zeng J, Li Y, and Ruta V (2019). Distinct dopamine receptor pathways underlie the temporal sensitivity of associative learning. Cell 178, 60–75 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, and Heisenberg M (1989). Neuronal architecture of the central complex nin Drosophila melanogaster. Cell Tissue Res 257, 343–366. [Google Scholar]
- Hardie RC, Raghu P, Moore S, Juusola M, Baines RA, and Sweeney ST (2001). Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron 30, 149–159. [DOI] [PubMed] [Google Scholar]
- Heinze S, Narendra A, and Cheung A (2018). Principles of insect path integration. Curr Biol 28, R1043–R1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel H-G, and Bohm H (1984). Wind orientation in walking carrion beetles (Necrophorus humator F.) under closed and open loop conditions In Localization and Orientation in Biology and Engineering, Varju D, and Schnitzler HU, eds. (Berlin Heidelberg: Springer Verlag; ), pp. 173–176. [Google Scholar]
- Heinzel H-G, and Bohm H (1989). The wind-orientation of walking carrion beetles. J Comp Physiol [A] 164, 775–786. [Google Scholar]
- Homberg U (1994). Flight-correlated activity changes in neurons of the lateral accessory lobes in the brain of the locust Schistocerca gregaria. J Comp Physiol [A] 175, 597–610. [Google Scholar]
- Honkanen A, Adden A, da Silva Freitas J, and Heinze S (2019). The insect central complex and the neural basis of navigational strategies. J Exp Biol 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse BK, and Jayaraman V (2019). Mechanisms underlying the neural computation of head direction. Annu Rev Neurosci. [DOI] [PubMed] [Google Scholar]
- Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. (2014). A systematic nomenclature for the insect brain. Neuron 81, 755–765. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikouchi A, Shimada T, and Ito K (2006). Comprehensive classification of the auditory sensory projections in the brain of the fruit fly Drosophila melanogaster. J Comp Neurol 499, 317–356. [DOI] [PubMed] [Google Scholar]
- Kempter R, Leibold C, Buzsaki G, Diba K, and Schmidt R (2012). Quantifying circular-linear associations: hippocampal phase precession. J Neurosci Methods 207, 113–124. [DOI] [PubMed] [Google Scholar]
- Kim SS, Hermundstad AM, Romani S, Abbott LF, and Jayaraman V (2019). Generation of stable heading representations in diverse visual scenes. Nature 576, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Rouault H, Druckmann S, and Jayaraman V (2017). Ring attractor dynamics in the Drosophila central brain. Science 356, 849–853. [DOI] [PubMed] [Google Scholar]
- Kita H, and Armstrong W (1991). A biotin-containing compound N-(2-aminoethyl)biotinamide for intracellular labeling and neuronal tracing studies: comparison with biocytin. J Neurosci Methods 37, 141–150. [DOI] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nat Methods 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler B, Faville R, Bridi JC, and Hirth F (2019). Inverse control of turning behavior by dopamine D1 Receptor signaling in columnar and ring neurons of the central complex in Drosophila. Curr Biol 29, 567–577 e566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, and Nitabach MN (2014). Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 24, 2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz S, Poeck B, Sokolowski MB, and Strauss R (2012). The visual orientation memory of Drosophila requires Foraging (PKG) upstream of Ignorant (RSK2) in ring neurons of the central complex. Learn Mem 19, 337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz S, Poeck B, and Strauss R (2017). Visual working memory requires permissive and instructive NO/cGMP signaling at presynapses in the Drosophila central brain. Curr Biol 27, 613–623. [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yanez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, and Stark A (2014). Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512, 91–95. [DOI] [PubMed] [Google Scholar]
- Labhart T, Petzold J, and Helbling H (2001). Spatial integration in polarization-sensitive interneurones of crickets: a survey of evidence, mechanisms and benefits. J Exp Biol 204, 2423–2430. [DOI] [PubMed] [Google Scholar]
- Liang X, Ho MCW, Zhang Y, Li Y, Wu MN, Holy TE, and Taghert PH (2019). Morning and evening circadian pacemakers independently drive premotor centers via a specific dopamine relay. Neuron 102, 843–857 e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilliefors HW (1967). On the Kolmogorov-Smirnov test for normality with mean and variance unknown. J Am Stat Assoc 62, 399–402. [Google Scholar]
- Lin CY, Chuang CC, Hua TE, Chen CC, Dickson BJ, Greenspan RJ, and Chiang AS (2013). A comprehensive wiring diagram of the protocerebral bridge for visual information processing in the Drosophila brain. Cell Rep 3, 1739–1753. [DOI] [PubMed] [Google Scholar]
- Linsenmair KE (1969). Anemomenotaktische Orientierung bei Skorpionen (Chelicerata, Scorpiones). Z Vgl Physiol 60, 445–449. [Google Scholar]
- Linsenmair KE (1972). Anemomenotatic orientation in beetles and scorpions In Animal Orientation and Navigation (Wallops Station, VA, NASA Scientific and Technical Information Office; ), pp. 501–510. [Google Scholar]
- Linsenmair KE (1973). Die Windorientierung laufender Insekten. Fortschr Zool 21, 59–79. [PubMed] [Google Scholar]
- Liu WW, and Wilson RI (2013). Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc Natl Acad Sci U S A 110, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Pena A, Acebes A, Rodriguez JR, Chevalier V, Casas-Tinto S, Triphan T, Strauss R, and Ferrus A (2014). Cell types and coincident synapses in the ellipsoid body of Drosophila. Eur J Neurosci 39, 1586–1601. [DOI] [PubMed] [Google Scholar]
- Matsuo E, Seki H, Asai T, Morimoto T, Miyakawa H, Ito K, and Kamikouchi A (2016). Organization of projection neurons and local neurons of the primary auditory center in the fruit fly Drosophila melanogaster. J Comp Neurol 524, 1099–1164. [DOI] [PubMed] [Google Scholar]
- Matsuo E, Yamada D, Ishikawa Y, Asai T, Ishimoto H, and Kamikouchi A (2014). Identification of novel vibration- and deflection-sensitive neuronal subgroups in Johnston’s organ of the fruit fly. Front Physiol 5, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss AS, Busch C, and Borst A (2017). Optogenetic Neuronal Silencing in Drosophila during Visual Processing. Sci Rep 7, 13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Stewart JC, Ott S, Chlebikova K, Chua JY, Koh TW, Ho J, and Claridge-Chang A (2017). Optogenetic inhibition of behavior with anion channelrhodopsins. Nat Methods 14, 271–274. [DOI] [PubMed] [Google Scholar]
- Moore RJ, Taylor GJ, Paulk AC, Pearson T, van Swinderen B, and Srinivasan MV (2014). FicTrac: a visual method for tracking spherical motion and generating fictive animal paths. J Neurosci Methods 225, 106–119. [DOI] [PubMed] [Google Scholar]
- Müller M, and Wehner R (2007). Wind and sky as compass cues in desert ant navigation. Naturwissenschaften 94, 589–594. [DOI] [PubMed] [Google Scholar]
- Namiki S, and Kanzaki R (2016). Comparative neuroanatomy of the lateral accessory lobe in the Insect brain. Front Physiol 7, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A 112, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuser K, Triphan T, Mronz M, Poeck B, and Strauss R (2008). Analysis of a spatial orientation memory in Drosophila. Nature 453, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, and Hassan BA (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A 107, 20553–20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofstad TA, Zuker CS, and Reiser MB (2011). Visual place learning in Drosophila melanogaster. Nature 474, 204–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Keles MF, Nguyen BM, Bolanos C, Lovick JK, Frye MA, and Hartenstein V (2017). Visual input to the Drosophila central complex by developmentally and functionally distinct neuronal populations. Curr Biol 27, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Nguyen B-CM, Kandimalla P, Lovick JK, Donlea JM, and Hartenstein V (2018). Neuronal constituents and putative interactions within the Drosophila ellipsoid body neuropil. Front Neural Circuits 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patella P, and Wilson RI (2018). Functional maps of mechanosensory features in the Drosophila brain. Curr Biol 28, 1189–1203 e1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira TD, Aldarondo DE, Willmore L, Kislin M, Wang SS, Murthy M, and Shaevitz JW (2019). Fast animal pose estimation using deep neural networks. Nat Methods 16, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezier A, and Blagburn JM (2013). Auditory responses of engrailed and invected-expressing Johnston’s Organ neurons in Drosophila melanogaster. PLoS One 8, e71419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, and Homberg U (2014). Organization and functional roles of the central complex in the insect brain. Annu Rev Entomol 59, 165–184. [DOI] [PubMed] [Google Scholar]
- Pfister R, Schwarz KA, Janczyk M, Dale R, and Freeman JB (2013). Good things peak in pairs: a note on the bimodality coefficient. Front Psychol 4, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, and Svoboda K (2003). ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser MB, and Dickinson MH (2010). Drosophila fly straight by fixating objects in the face of expanding optic flow. J Exp Biol 213, 1771–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, and Taghert PH (1999). Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol 41, 189–207. [PubMed] [Google Scholar]
- Sakura M, Lambrinos D, and Labhart T (2008). Polarized skylight navigation in insects: model and electrophysiology of e-vector coding by neurons in the central complex. J Neurophysiol 99, 667–682. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöne H (2014). Spatial Orientation: The Spatial Control of Behavior in Animals and Man (Princeton, NJ: Princeton University Press; ). [Google Scholar]
- Seelig JD, Chiappe ME, Lott GK, Dutta A, Osborne JE, Reiser MB, and Jayaraman V (2010). Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat Methods 7, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, and Jayaraman V (2013). Feature detection and orientation tuning in the Drosophila central complex. Nature 503, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, and Jayaraman V (2015). Neural dynamics for landmark orientation and angular path integration. Nature 521, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki HM, and Kazama H (2017). Parallel encoding of recent visual experience and self-motion during navigation in Drosophila. Nat Neurosci 20, 1395–1403. [DOI] [PubMed] [Google Scholar]
- Souman JL, Frissen I, Sreenivasa MN, and Ernst MO (2009). Walking straight into circles. Curr Biol 19, 1538–1542. [DOI] [PubMed] [Google Scholar]
- Steck K, Veit D, Grandy R, Badia SB, Mathews Z, Verschure P, Hansson BS, and Knaden M (2012). A high-throughput behavioral paradigm for Drosophila olfaction - The Flywalk. Sci Rep 2, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nern A, Franconville R, Dana H, Schreiter ER, Looger LL, Svoboda K, Kim DS, Hermundstad AM, and Jayaraman V (2017). Neural signatures of dynamic stimulus selection in Drosophila. Nat Neurosci 20, 1104–1113. [DOI] [PubMed] [Google Scholar]
- Suver MP, Matheson AMM, Sarkar S, Damiata M, Schoppik D, and Nagel KI (2019). Encoding of wind direction by central neurons in Drosophila. Neuron 102, 828–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirian L, and Dickson BJ (2017). The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. In bioRxiv http://dxdoiorg/101101/198648. [Google Scholar]
- Turner-Evans D, Wegener S, Rouault H, Franconville R, Wolff T, Seelig JD, Druckmann S, and Jayaraman V (2017). Angular velocity integration in a fly heading circuit. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TL, Giraldo YM, and Dickinson MH (2019). Celestial navigation in Drosophila. J Exp Biol 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner R, and Duelli P (1971). The spatial orientation of desert ants, Cataglyphis bicolor, before sunrise and after sunset. Experientia 27, 1364–1366. [Google Scholar]
- Wilson RI, and Laurent G (2005). Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe. J Neurosci 25, 9069–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Iyer NA, and Rubin GM (2015). Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J Comp Neurol 523, 997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Tabuchi M, Brown MP, Mitchell SP, Wu MN, and Kolodkin AL (2017). The laminar organization of the Drosophila ellipsoid body is semaphorin-dependent and prevents the formation of ectopic synaptic connections. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang G, Shinomiya K, Maitin-Shepard J, Ackerman D, Berg S, et al. (2020). A connectome of the adult Drosophila central brain. In bioRxiv https://doiorg/101101/20200121911859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorozu S, Wong A, Fischer BJ, Dankert H, Kernan MJ, Kamikouchi A, Ito K, and Anderson DJ (2009). Distinct sensory representations of wind and near-field sound in the Drosophila brain. Nature 458, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, and Armstrong JD (2010). Structure of the adult central complex in Drosophila: organization of distinct neuronal subsets. J Comp Neurol 518, 1500–1524. [DOI] [PubMed] [Google Scholar]
- Zar JH (2010). Biostatistical analysis, 5th edn (Upper Saddle River, N.J: Prentice-Hall/Pearson; ). [Google Scholar]
- Zhang Z, Li X, Guo J, Li Y, and Guo A (2013). Two clusters of GABAergic ellipsoid body neurons modulate olfactory labile memory in Drosophila. J Neurosci 33, 5175–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and software may be obtained upon reasonable request.