Abstract
The concept that type I interferons (IFN-I) are essential to antiviral immunity derives from studies on animal models and cell lines. Virtually all pathogenic viruses have evolved countermeasures to IFN-I restriction, and genetic loss of viral IFN-I antagonists leads to virus attenuation. But just how important is IFN-I to antiviral defence in humans? The recent discovery of genetic defects of IFN-I signalling illuminates this and other questions of IFN biology, including the role of the mucosa-restricted type III IFNs (IFN-III), informing our understanding of the place of the IFN system within the concerted antiviral response. Here we review monogenic lesions of IFN-I signalling pathways and summarise the organising principles which emerge.
Keywords: type I interferons, interferon-stimulated genes, JAK–STAT signalling, IFNAR, inborn errors of immunity, antiviral immunity
Is IFN-I Essential to Human Antiviral Immunity?
Interferon (IFN, see Glossary) was discovered more than 60 years ago by Isaacs and Lindemann [1]. This soluble factor, produced by virally infected cells in culture, conferred an antiviral state when applied to uninfected naïve cells prior to infection. Its discovery led to the clinical application of IFN-I (IFNα) as the first host-directed antiviral therapy [2]. Over the intervening six decades, the molecular basis of IFN-I activity has been carefully dissected (Box 1 ) and its essential function in antiviral defence has been confirmed in model organisms [3]. In parallel with these discoveries has come the realisation that virtually all human viral pathogens encode strategies to evade and/or subvert the antiviral activity of IFN-I [4,5]. Furthermore, loss of viral IFN-I antagonists leads to virus attenuation and is being exploited for the development of novel live viral vaccines [6]. The ‘arms race’ between host and virus plays a decisive role in viral pathogenesis, driving viral evolution and restricting interspecies transmission [4,5].
Box 1. Canonical IFN-I Signalling.
In the current paradigm, the type I IFNs (IFN-Is) – comprising 13 subtypes of IFNα and one of IFNβ, IFNε, IFNκ, and IFNω – all signal at the single heterodimeric IFN alpha/beta receptor (IFNAR) expressed by all nucleated cells. By contrast, responses to the type III IFNs (IFNλ1–4, IFN-III), identified first in 2003 [8., 9., 10.], are constrained by the restricted expression of the IFN lambda receptor (IFNLR) on epithelial cells, (human) hepatocytes, and some immune cell subsets [11,16,60,61,71]. In the canonical IFN-I pathway, IFN-I binding initiates an intracellular signalling cascade in which reciprocal transphosphorylation of the receptor-associated kinases JAK1 and TYK2 is accompanied by phosphorylation of the signal transducers and activators of transcription STAT1 and STAT2 [59,83,95]. The majority of the transcriptional response to IFN-I is attributable to the heterotrimer comprising phosphorylated STAT1 and 2 together with IRF9 [56,95,96]. This complex, known as ISGF3, translocates to the nucleus where it interacts with an ISRE [97] in the promoter of multiple ISGs [97]. A fraction of the phosphorylated STAT1 also homodimerises to form the GAF [98], agonising a distinct but overlapping set of genes bearing GAS elements, typically associated with type II IFN (IFNγ) signalling [98]. A broadly similar pathway is activated in response to IFN-III [8., 9., 10.]. This simplified model omits multiple STAT-dependent and STAT-independent signalling pathways activated downstream of IFNAR, some of which involve unphosphorylated ISGF3 components assembled in distinct transcriptional complexes (e.g., STAT2:IRF9, U-ISGF3) [99].
Alt-text: Box 1
The IFN System
IFN-I does not operate in isolation. Additional IFNs have been identified; namely, IFNγ (type II IFN) [7] and the mucosa-restricted IFNs (IFNλ1–4, or type III IFNs) [8., 9., 10.]. These IFN types differ in their range of activity in tissues and their tendency to cause immunopathology and can be considered part of an integrated ‘IFN system’. IFN-III is produced by most cells but acts mainly at epithelial surfaces, due to constrained receptor expression [11]. IFN-III restricts virus replication at the point of initial encounter without inducing systemic immune activation or immunopathology [10., 11., 12., 13., 14., 15., 16.] – a relatively high-yield, low-cost outcome. By contrast, most tissues respond to IFN-I, which assumes greater importance when viruses breach epithelial barriers (e.g., invading into the lymphatics, bloodstream, or brain), but with an inherently greater risk of toxicity, particularly for the brain [17]. Finally, type II IFN (IFN-II) (IFNγ) is an extremely potent immunostimulatory cytokine, with the greatest potential for immunopathology. While expression of the IFNγ receptor (IFNGR) is, like that of the IFN-I receptor (IFNAR), widespread, the production of IFNγ is tightly controlled and restricted to specialised immune cells [18]. Since all IFNs share the ability to induce an antiviral state in responding cells, there is scope for compensation if one or more IFN type is disabled. The brain parenchyma is a notable exception since it cannot respond to IFN-III, and cells capable of making IFNγ are generally absent from the brain.
Given the ability of viruses to evade IFN-I in their natural hosts, how important is this response for antiviral immunity in humans? This question has added relevance given that dysregulation of IFN-I immunity may promote viral pathogenesis [17,19]. In humans, inborn errors of immunity that compromise the expression and/or function of genes such as TLR3, IFIH1, IRF3, and IRF7 involved in synthesis of IFN-I, IFN-III, and other proinflammatory mediators in response viral infection, are associated with heightened clinical susceptibility to viral disease [20., 21., 22., 23., 24., 25., 26., 27., 28., 29., 30.]. These disorders underline a central principle: that innate immunity makes an essential contribution to viral defence in humans. However, the fact that these pathways engage a range of innate immune effector mechanisms beyond the IFN-I response means that the precise involvement of IFN-I in this process remains uncertain. To dissect this, we turn to defects of the cellular response to IFN-I itself.
Within this group of autosomal recessive (AR) diseases are loss-of-expression variants negating the response to IFN-I alone (IFNAR1 or IFNAR2), responses to both IFN-I and IFN-III (i.e., STAT2, IRF9), and STAT1 variants, which negate responses to all IFNs (Figure 1 ). These inborn errors of immunity produce a phenotype of vulnerability to severe and/or recurrent viral disease (Table 1 ), the clinical significance of which depends on the extent of compromise to the IFN system as a whole. However, variable expressivity is also recognised; for example, ranging from death in infancy to survival into adulthood with no apparent phenotype in some signal transducer and activator of transcription (STAT)2-deficient patients [31]. This may, among other factors, be due to environmental differences in the range and dose of viruses encountered by individual patients and/or the effectiveness of compensatory immune pathways against specific pathogens [32].
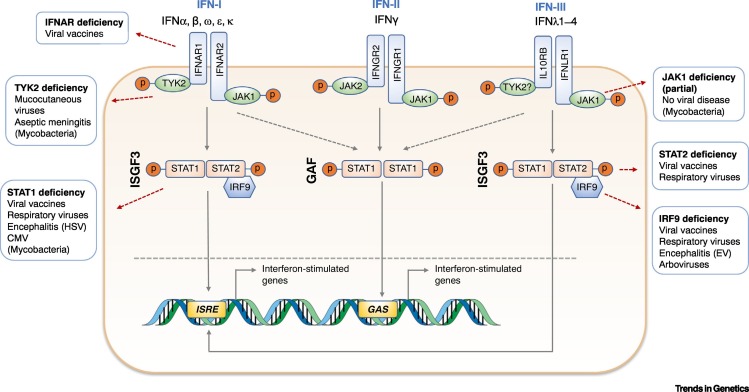
Figure 1.
Canonical Interferon (IFN) Signalling Pathways and Their Genetic Lesions.
Displayed are the three IFN pathways and a brief summary of the viral susceptibility phenotypes that accompany molecular defects of these pathways. IFN-I receptor (IFNAR) deficiency selectively impairs the IFN-I response, predisposing to disease secondary to inoculation with live-attenuated viral vaccines. Since IFN-I responses play a part in preventing the systemic dissemination of viruses, IFNAR-deficient individuals would hypothetically be vulnerable to arboviruses transmitted via the bloodstream. STAT2 and IRF9 are central to the response to both IFN-I and IFN-III, the latter mediating antiviral immunity at mucosal surfaces. Thus, these molecular defects are accompanied by problems in handling both live-attenuated viral vaccines and mucosally transmitted viruses such as influenza. In the case of STAT2 deficiency, variable expressivity of the phenotype is recognised. STAT1 deficiency is the most clinically serious defect since it compromises the response to all IFNs simultaneously. This is associated with susceptibility to a broad range of viruses, including herpesviruses. Because IFN-II is also critical to responses to mycobacteria, STAT1 deficiency is accompanied by life-threatening complications of mycobacterial infection. Tyrosine kinase 2 (TYK2) and (partial) Janus kinase 1 (JAK1) deficiency are also associated with mycobacterial susceptibility, alongside a much milder phenotype of viral disease. In the case of TYK2 deficiency, this may be due to the ability of IFN-III, and to a lesser extent IFN-I, to signal independently of TYK2. Complete JAK1 deficiency has not been reported. Abbreviations: CMV, cytomegalovirus; EV, enterovirus; GAF, IFNγ-activated factor; HSV, herpes simplex virus; ISGF3, interferon-stimulated gene factor 3.
Table 1.
| Gene | Protein expression | Severe/recurrent viral disease | Uncomplicated infection | Asymptomatic exposure | Other clinical manifestation | Refs |
|---|---|---|---|---|---|---|
| IFNAR1 | Absent | P1: MMR encephalitis P2: disseminated 17D YFV (MMR without incident) |
P1: none P2: none |
P1: CMV P2: CMV, HSV1, HSV2 |
No | [35] |
| IFNAR2 | Absent | P1: MMR encephalitis (fatal) P2: none (MMR withheld) |
P1: HHV6 P2: none |
P1: CMV, EBV P2: unknown (SCIG) |
No | [33] |
| STAT1 | Absent | P1: recurrent HSV encephalitis (fatal) P2: unknown viral pathogen (fatal) |
Unknown | Unknown | Both: disseminated BCG | [43] |
| STAT1 | Absent | Vaccine-strain polio virus shedding EBV-driven LPD post-HSCT |
HRV, PIV2, polio vaccine | None | Disseminated BCG Severe hepatitis Multiorgan failure post-HSCT (fatal) |
[45] |
| STAT1 | Reduced expression of truncated nonfunctional protein (∆Ex3) | Recurrent CMV pneumonitis Encephalitis (cause not identified) |
Viral GI infections and RTIs Cutaneous HSV1 |
Unknown | Pulmonary NTM infection Sepsis Recurrent pneumonia (BCG naïve) |
[44] |
| STAT1 | Absent | HLH, possibly related to MMR and/or HHV6 | Nil stated | Unknown | (BCG naïve) | [41] |
| STAT1 | Splicing defect (∆Ex23) with low-level expression of WT protein | P1: None noted P2: HSV1 gingivostomatitis, hospitalisation for CMV and VZV infection |
Unknown | Unknown | P1: recurrent NTS infection (BCG naïve) P2: Salmonella meningitis (BCG naïve) |
[46] |
| STAT1 | Splicing defect (∆Ex8) with low-level expression of WT protein | P1: severe varicella P2: none noted |
Unknown | P1: CMV, EBV P2: unknown |
P1: disseminated NTM infection (BCG naïve), Candida line infection P2: disseminated BCG Septic shock (fatal) |
[47] |
| STAT2 | Absent (multiple splicing defects) | P1: MMR pneumonitis/hepatitis HSV1 gingivostomatitis IAV pneumonia P2: fatal viral illness (10 weeks) P3: none noted (childhood history unknown) P4: bronchiolitis MMR encephalitis (SNHL) P5: hospitalisation with viral illness |
P1: none P2: N/A P3: unknown P4: varicella P5: varicella |
P1: EBV P2: N/A P3: MMR, VZV, CMV P4: Unknown P5: Unknown |
None | [31] |
| STAT2 | Absent | P1: opsoclonus–myoclonus syndrome post-MMR with late recurrence P2: MMR encephalitis (MuV) |
P1: unknown P2: unknown |
P1: Unknown P2: Unknown |
Mild renal tubulopathy Defect of mitochondrial fission |
[48] |
| STAT2 | Absent (premature stop/splicing defect) | P1: MMR hepatitis Severe RSV, EV, AdV Fatal febrile illness (7 years) P2: severe varicella MMR pneumonitis/hepatitis EV meningitis Prolonged primary EBV |
P1: unknown P2: RSV, IAV, EV, AdV |
P1: Unknown P2: Unknown |
‘Inflammatory’ responses to viral infection | [49] |
| STAT2 | Absent | HLH post-MMR | Unknown | HHV6, CMV, VZV, RSV, AdV, PIV1–3 | No | [50] |
| IRF9 | Reduced expression of truncated product (∆Ex7) | Severe IAV pneumonitis Biliary perforation post-MMR Recurrent bronchiolitis |
HMPV, RSV, AdV, PIV1-4 | HSV1, CMV, HRV, EV | (i) Periodic fever (ii) C1q autoantibody |
[52] |
| IRF9 | Absent | P1: disseminated VZV post vaccination Severe dengue fever and Zika virus disease EV encephalitis IAV pneumonia RSV bronchiolitis P2: none (on SCIG) |
P1: dengue, IBV pneumonia P2: none |
P1: Unknown P2: Unknown |
P1: recurrent pneumonia and bronchitis, septic shock with purpura fulminans P2: none |
[53] |
| JAK1 | Unaffected | None | VZV (shingles), HPV | Unknown | Recurrent mycobacterial infection Metastatic bladder carcinoma (fatal) |
[57] |
| TYK2 | Absent | P1: PIV3 pneumonia, recurrent oropharyngeal HSV1 P2: none P3: none P4: none P5: none P6: unspecified viral infection of skin P7: none P8: recurrent herpes gingivostomatitis and aseptic meningitis |
P1: molluscum contagiosum P2: shingles |
Unknown | P1: eosinophilia, elevated IgE Disseminated BCG NTS infection Candidiasis Ocular sarcoidosis P2: disseminated BCG Recurrent Brucella meningitis P3: TB, bacterial meningitis (fatal) P4: meningitis, otitis media, UTI, asthma P5: disseminated BCG P6: disseminated BCG P7: TB P8: none |
[60] |
| TYK2 | Absent | None | Unknown | HHV6, IAV, parvo-B19 MMR vaccinated |
Recurrent bronchopneumonia Duodenal perforation Anal/skin abscesses |
[61] |
| TYK2 | Absent | Recurrent herpes gingivostomatitis and aseptic meningitis | VZV | Unknown | Disseminated BCG | [62] |
All of the variants included above are pathogenic in homozygosity or compound heterozygosity.
AdV, adenovirus; CNS, central nervous system; ECMO, extracorporeal membrane oxygenation; EV, enterovirus; HHV6, human herpesvirus 6; HPV, human papillomavirus; HRV, human rhinovirus; IAV/IBV, influenza A/B virus; LPD, lymphoproliferative disease; MuV, mumps virus; NTM, nontuberculous mycobacteria; NTS, non-typhoidal Salmonella; PIV, parainfluenza virus; RTI, respiratory tract infection; SCIG, subcutaneous immunoglobulin; SNHL, sensorineural hearing loss; TB, tuberculosis.
IFNAR Deficiency
Deficiency of IFNAR acts as the most specific readout of the involvement of IFN-I in human antiviral defence, revealing an essential but surprisingly narrow function attributable to IFN-I. IFNAR comprises two subunits, IFNAR1 and IFNAR2. Homozygosity for a nonsense IFNAR2 variant was first reported in two siblings, coming to light when the proband developed fatal encephalitis, complicated by haemophagocytic lymphohistiocytosis (HLH), following receipt of the live-attenuated measles, mumps, and rubella (MMR) vaccine [33]. Analysis of patient dermal fibroblasts revealed undetectable IFNAR2 protein expression accompanied by absent transcriptional responses to IFNα/IFNβ. This resulted in failure to mount an antiviral state in response to recombinant IFNα and an inability to control the replication of genetically modified IFN-sensitive viruses in vitro. These defects were rescued by complementing patient cells with wild-type (WT) IFNAR2. Notably, and surprisingly, prior to receipt of MMR the proband had been healthy with no apparent phenotype of heightened sensitivity to common childhood viral diseases. This was true despite serological testing revealing evidence of prior infection with the herpesviruses Epstein–Barr virus (EBV) and cytomegalovirus (CMV). Supporting this unexpectedly narrow vulnerability to viral disease, the patient’s IFNAR2-deficient sister remained well without a clinically apparent defect of viral resistance (MMR was withheld). It was suggested that this narrow phenotype might reflect residual IFNAR1-dependent signalling, as reported in Ifnar2 −/- mice [34]; however, a similar clinical phenotype was recognised in two unrelated children bearing homozygous nonsense mutations in IFNAR1 [35]. Their clinical phenotype – healthy until exposure to live-attenuated viruses – recapitulated IFNAR2 deficiency. Both suffered life-threatening dissemination of live viral vaccines, either MMR or yellow fever vaccine [35]. In cells (dermal fibroblasts and EBV-transformed B cells) from these cases, the IFNAR1 protein and transcriptional response to IFNα/IFNβ was absent, conferring a defect of antiviral state induction to recombinant IFNα or IFNβ, rescued by complementation with IFNAR1. As in IFNAR2-deficient cases, IFNAR1 deficiency was not associated with clinically evident vulnerability to viruses encountered in the natural environment such as influenza, other common respiratory viruses, or herpesviruses including CMV, despite serological evidence of past infection [35]. Thus, unlike in mice, the clinical and molecular consequences of human IFNAR1 and IFNAR2 deficiency appeared to be indistinguishable. Responses to IFNγ were intact in both IFNAR1- and IFNAR-deficient fibroblasts in vitro [33,35], again contrasting with findings in murine cells [36,37]. Although not systematically assessed, none of the defects of development or homeostasis reported in Ifnar1 −/- mice [38] have been observed to date in IFNAR-deficient humans.
IFN-III Compensation?
The apparent lack of vulnerability to naturally occurring viral pathogens initially encountered at mucosal surfaces in IFNAR-deficient patients is striking and at first glance appears in stark contrast to IFNAR-deficient mice, which are profoundly susceptible to multiple viruses. However, it is important to consider the route of viral exposure in judging these apparent phenotypic differences. Ifnar1 −/- mice are indeed highly susceptible to viruses when challenged by intravenous or intraperitoneal inoculation [3]. However, when challenged with influenza or respiratory syncytial virus (RSV) via the respiratory mucosa they displayed no increase in viral susceptibility compared with WT controls [12]. This has been attributed to the capacity of IFN-III to compensate entirely for the loss of IFN-I antiviral activity at the respiratory mucosa [12,14,39]. Deletion of Ifnar and Ifnlr together renders mice extremely vulnerable to infection of the respiratory tract [12,14,39]. An obvious hypothesis for why IFNAR-deficient patients become severely ill following immunization with some live-attenuated vaccines is that intramuscular injection bypasses the IFN-III response, which would normally be encountered during natural infections, leading to widespread dissemination of the attenuated virus and disease. This has been (indirectly) tested in mice, where Ifnar1 −/- mice are vulnerable to the dissemination of vaccine-strain YFV (YFV-17D) when it is injected by the intramuscular or intraperitoneal routes [40]. An interesting corollary is that patients with defective IFN-I immunity may be susceptible to viruses that naturally bypass the mucosa; for example, arthropod-borne pathogens such as dengue virus. As with any susceptibility state, pathogen exposure is a key factor governing disease expressivity [32], and IFNAR-deficient patients have not been knowingly exposed to arboviruses [33,35]. An open question is whether similar compensation occurs in exclusive defects of IFN-III (Box 2 ).
Box 2. IFN-I Compensation?
Compensatory interactions between IFN-I and IFN-III go both ways. The IFNLR is formed from IL10RB, the ubiquitously expressed subunit of the IL10 receptor, and IFNLR1, which displays tissue-restricted expression mainly in epithelial cells and hepatocytes. AR loss-of-expression mutations of IL10RB confer susceptibility to severe, early-onset inflammatory bowel disease, a phenotype indistinguishable from null mutations of IL10RA or IL10 itself [100], presumably reflecting the dominant role of loss of IL10 signalling in this phenotype. Notably, there have been no reports of clinical susceptibility to mucosal viral disease [either respiratory or gastrointestinal (GI)] in over 20 patients reported to date with deficiency of IL10RB (reviewed in [101]). This implies, but does not prove, that defects in IFN-III signalling are compensated by IFN-I, in apparent contrast to the situation in mice, where IFN-III plays an nonredundant role in defence against rotavirus and norovirus in the intestine [13,15] and in restricting influenza virus to the upper airway [16]. An open question is whether IL10RB deficiency might confer a subtler defect of mucosal antiviral immunity in specific circumstances.
Alt-text: Box 2
The implication of these experiments of nature is that there is redundancy between IFN-I and IFN-III in mediating the antiviral state at mucosal surfaces. One would predict a more significant phenotype to arise in defects that compromise both IFN-I and IFN-III responses simultaneously. For such an example, we turn to defects of the shared signal transduction pathway downstream of these receptors, beginning with defects of the interferon-stimulated gene factor 3 (ISGF3) complex [STAT1, STAT2, and interferon regulatory factor 9 (IRF9)].
Homozygous STAT1 Deficiency
AR complete STAT1 deficiency leads to a profound defect of immunity with broad susceptibility to infectious pathogens, extending to herpesviruses as well as intracellular bacteria (including mycobacteria). This pattern reflects the participation of STAT1 in signalling by all IFNs [i.e., IFN-I, IFN-II (IFNγ), and IFN-III], thereby disrupting critical functions of IFNγ among specialised cells of the immune system. It is the only disorder described in this review where haematopoietic stem cell transplantation (HSCT) is indicated and has documented life-saving potential in patients [41,42]. Heterozygosity for hypomorphic variants in human STAT1 produces clinical vulnerability to mycobacteria but not viruses due to the selective impact on IFNγ signalling and thus will not be considered further.
Complete AR STAT1 deficiency was first identified in unrelated infants presenting with disseminated bacille Calmette–Guérin (BCG) infection who succumbed to severe viral illness [caused by recurrent herpes simplex virus 1 (HSV1) encephalitis in one and an unknown viral pathogen in the other] [43]. Sequencing of the candidate disease gene STAT1 revealed in one child a homozygous frameshift deletion (c.1757-1758delAG) while the other bore a homozygous missense variant (p.L600P) and preserved mRNA expression. Consistent with the complete absence of STAT1 protein in each child, resulting in loss of ISGF3 and IFNγ-activated factor (GAF) DNA-binding activity, patient cells were completely refractory to IFN in the induction of an antiviral state in vitro. Subsequent cases of complete STAT1 deficiency broadly recapitulated this phenotype [44], although sometimes with additional inflammatory features such as severe hepatitis [45] or HLH [41] provoked, at least in the latter case, by viral infection. Homozygosity for a hypomorphic STAT1 allele was shown to produce a less severe phenotype both in vitro and in vivo [46,47]. The profound clinical impact of STAT1 deficiency and the therapeutic benefit of HSCT reflect the broad importance of this transcription factor in immunity.
Homozygous STAT2 Deficiency
Biallelic loss-of-expression variants of the transcription factor STAT2 produce a milder clinical phenotype than STAT1 deficiency comprising viral susceptibility without associated problems in handling bacteria, owing to STAT2’s more restricted role in IFN-I and IFN-III signalling. Compared with defects of IFNAR, STAT2 deficiency has a broader clinical phenotype, encompassing viral disease caused by vaccine-strain viruses as well as common viral pathogens encountered at mucosal surfaces [31,48., 49., 50.]. Presumably, this is due to the parallel disruption of IFN-I and IFN-III responses. As previously mentioned, the phenotype of viral susceptibility in STAT2 deficiency also has variable expressivity, for reasons that remain unclear.
STAT2 deficiency was originally described in a kindred of five individuals with a spectrum of disease ranging from death in infancy to survival into middle age with no apparent phenotype [31]. Homozygosity for a variant (c.381+5G>C) causing aberrant STAT2 mRNA splicing and undetectable STAT2 protein expression was accompanied by a profound in vitro defect of the expression of IFN-stimulated response element (ISRE)-containing genes and failure to mount an IFN-I-mediated antiviral state [31], defects that were rescued by STAT2 complementation. Patient cells expressed a small subset (10%) of interferon-stimulated genes (ISGs) induced in WT cells [31], most of which contained gamma activation site (GAS) elements within their promoters, presumably reflecting a residual amount of IFNAR-dependent signalling via GAF. Whilst STAT2 deficiency almost certainly compromises ISGF3 function in response to IFN-III, this prediction has yet to be formally tested owing to the lack of available patient cell types that reliably express a functional IFN lambda receptor (IFNLR). Treatment with IFNγ protected STAT2-deficient cells against cytopathic effects of vesicular stomatitis virus [49], suggesting the compensatory induction of an antiviral state, as described in IFNAR-deficient cells.
Complete deficiency of STAT2 was subsequently reported in a further five children in three kindreds [48., 49., 50.], totalling ten individuals (additional patients have been identified but their details remain unpublished). Some of these individuals had more obvious problems with common childhood viruses, such as viral pneumonia and enteroviral meningitis, than in the original kindred [49]. In total, viral disease had a fatal outcome in two of ten cases. All six children known to have been exposed to MMR have experienced dissemination of vaccine-strain viruses to the lungs and/or brain (Table 1). The outcome of herpesvirus infections in these patients was seemingly unremarkable, except for one child with prolonged EBV replication following primary infection [49] and another with herpes gingivostomatitis [31]. There were no reports of CMV disease despite serological evidence of exposure. Measures of adaptive immune cell number and immunoglobulin production were normal, with one or two minor exceptions [49]. This suggests that adaptive immunity, and/or IFN-II signalling, provides compensatory immunity to CMV and possibly other herpesviruses in STAT2-deficient patients. In some cases, viral disease has also been associated with significant systemic inflammation, reaching the criteria for HLH [50], apparently responsive to immunoglobulin therapy [49,50]. This and related inflammatory phenotypes are observed in other disorders (IFNAR2, STAT1, IRF9), implying immunopathology caused by aberrant activity of compensatory immune pathways.
Homozygous IRF9 Deficiency
The clinical and molecular impact of IRF9 deficiency echoes STAT2 deficiency, reflecting the intimate cooperation of these transcription factors in IFN-I and IFN-III signalling. IRF9 is the main DNA-binding component of ISGF3. Alone, IRF9 has minimal ISRE-binding capacity or transcriptional activity [51]. AR IRF9 deficiency has been described in three children in two kindreds [52,53]. In the original report, the proband bore a homozygous IRF9 missense variant producing a truncated IRF9 protein lacking the IRF association domain (IAD), a critical domain for STAT interaction. Full-length IRF9 lacks a nuclear export signal and is predominantly localised to the cytosol through its constitutive interaction with STAT2 [51,54]. Loss of the IRF9 IAD led to an inability to bind STAT2 and was associated with constitutive nuclear localisation of IRF9∆Ex7 [52]. In the second kindred, homozygosity of an IRF9 variant allele (c.577+1G>T) in two siblings was accompanied by undetectable IRF9 protein expression [53]. An elder sibling who died with pathological dissemination of yellow fever vaccine was probably similarly affected. In cells from IRF9-mutated patients, STAT1 and STAT2 were appropriately phosphorylated on IFNα treatment; however, binding to an ISRE probe was completely impaired, consistent with the failure to form a functional ISGF3 complex. Consistent with this defect of ISGF3 assembly in IRF9-deficient cells, the transcriptional induction of classical ISRE-containing ISGs was abolished [52,53]. RNA-seq studies of IRF9∆Ex7 cells revealed the expression of a subset of ISGs in patient cells, some to a higher magnitude than controls, implying the activity of ISGF3-independent complexes such as GAF [52]. Responses to IFNγ were preserved in cells bearing the truncated IRF9 protein (IFNγ response was not reported in IRF9-null cells), in contrast to reports in Irf9 −/- mouse embryonic fibroblasts [55] and in U2A cells, an IRF9-deficient fibrosarcoma cell line [56].
As in STAT2 deficiency, IRF9-deficient children experienced life-threatening problems in controlling vaccine-strain viruses and evidence of increased susceptibility to mucosal viral pathogens but not herpesviruses [52,53] (Table 1). Adaptive immune parameters were normal when monitored during health in the first child; the proband in the second kindred had mildly reduced IgG and was treated with intravenous immunoglobulin (IVIG) [53]. In another similarity with some STAT2-deficient patients, periodic fevers and severe systemic inflammatory response syndrome (SIRS) suggestive of autoinflammation were noted, the trigger for which was not identified [52,53].
Receptor-Associated Kinases
The receptor associated kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2) play key roles in linking innate IFN receptor ligation to transcriptional activation. However, unlike most of the signalling molecules considered in this review, which have specific functions in IFN signalling, JAK1 and TYK2 participate in a wide range of additional cytokine and growth factor signalling pathways. Thus, from our IFN-centric perspective, these disorders are nonspecific and less informative. Nevertheless, they provide some interesting insights.
Partial JAK1 Deficiency
Biallelic hypomorphic mutations in JAK1 have been reported in a single individual to date [57]. The impact of these variants on IFN-I signalling was modest, with minimal compromise of the transcriptional response. Consistent with this partial defect of IFN-I signal transduction, no evidence of a clinically relevant defect of antiviral immunity was observed in this patient, beyond a single episode of uncomplicated varicella zoster virus (VSV) reactivation (shingles). JAK1 participates in signalling through a large range of cytokines, including IFNγ, and compromise of these cytokine pathways appears to dictate the clinical phenotype, which was dominated by susceptibility to mycobacterial infection and early-onset bladder carcinoma (Table 1). Complete JAK1 deficiency has not been described in humans. Based on observations in Jak1 −/- mice, this might be expected to be incompatible with postnatal life [58].
AR TYK2 Deficiency
A different picture emerges for patients deficient in TYK2. In the canonical model of IFN-I signalling, based on studies in TYK2-deficient U1A fibrosarcoma cells [59], TYK2 is essential. However, complete deficiency of TYK2 in humans has a relatively mild clinical impact on antiviral immunity (Table 1). Reported in 11 individuals, in all cases accompanied by absent TYK2 protein expression, the clinical phenotype is dominated by susceptibility to mycobacterial and/or fungal infections [60., 61., 62., 63.], again presumably due to the involvement of TYK2 in other cytokine pathways. Initially considered a cause of hyper-IgE syndrome [63], this was refuted by the subsequent identification of multiple TYK2-deficient cases lacking this phenotype [60]. No patients experienced problems handling live-viral vaccines. Naturally encountered viral infections reported in TYK2-deficient patients were in most cases mild, with herpes gingivostomatitis in three patients accompanied in one case by viral pneumonia; two individuals developed aseptic meningitis.
Studies in virally transformed fibroblasts, T cells, and B cells from TYK2-deficient patients demonstrated reductions in IFNAR1 and IL10RB expression [60,61], consistent with the loss of a scaffolding function of TYK2 towards these receptor subunits [64]. Interestingly, in response to IFN-I there was residual – albeit substantially reduced – STAT1 phosphorylation, ISGF3 and GAF DNA binding, and ISG expression [60,61]. Cell-type variation was apparent, since responses to IFNα in transformed T cells (as well as monocytes from a single case) were absent [60,61]. Unexpectedly, IFN-III signalling (assayed in patient EBV-transformed B cells and TYK2 knockout HAP1 cells) was preserved, suggesting that it may be TYK2 independent [60,61]. While these findings are inconclusive, they imply that residual IFN-III (and possibly also IFN-I) responses in TYK2-deficient patients are of a sufficient magnitude to sustain antiviral immunity. These results are broadly compatible with Tyk2 −/- mice, which are less susceptible than Ifnar1 −/- mice to experimental VSV infection [65], and where IFN-I signal transduction is only partially impaired [65,66]. Thus, IFN-III and IFN-I responses appear to be at least partially TYK2 independent.
Interspecies Differences in IFN-I Biology?
Since much of our current understanding of IFN-I biology is derived from mouse models, we briefly consider how the phenotypes of deficient mice and humans compare. Human and mouse genomes contain 17 IFN-I and one IFNG genes; humans have four IFN-III genes (IFNL1–4), whereas mice have two (Ifnl2/3) [8., 9., 10.]. IFN-I signalling molecules show variable interspecies diversity. Human and mouse IFNAR1 and IFNAR2 proteins share only 50% homology [67]. While STAT1 is highly conserved, STAT2 has 70% homology, the lowest of all STATs [68]. Consequently, certain viral IFN-I antagonists (e.g., flaviviral NS5 proteins) do not function in the mouse because they fail to bind murine STAT2 [69,70]. Another difference is that human but not murine hepatocytes respond to IFN-III [39,71,72]. Accordingly, recombinant human IFNλ1 is being explored as a therapy for viral hepatitis [73].
Despite the inherent limitation of comparing results from experimental and natural infection, and the fact that most viral pathogens are human adapted, viral susceptibility phenotypes of deficient humans and mice broadly correspond, particularly in the case of TYK2, STAT1, and STAT2 [65,66,74., 75., 76.]. Viral susceptibility has not been systematically analysed in Irf9 −/- mice [55]. The apparent exception is IFNAR-deficient mice, which demonstrate broader susceptibility to herpesviruses, such as HSV [77] and (murine) CMV [78,79], than humans. In addition, a range of defects in haematopoietic stem cell (HSC) maintenance [80,81], immune cell number [e.g., reduced natural killer (NK) cells [82], expanded CD11c+ myeloid cells [83]], increased bone turnover [84], and altered responses to alternative cytokines [colony stimulating factor 1(CSF-1), interleukin (IL)6, and IFNγ [36,37,85]], attributed to the loss of constitutive IFN-I signalling in IFNAR-deficient mice [38], were not observed in human patients [33,35]. Despite several caveats (low patient numbers, the absence of deep immunophenotyping data), an obvious question is why human immunity is seemingly not impacted to the same degree by defective IFN-I signalling.
Concluding Remarks
Human genetic disorders reveal that IFN-I is an important, and in certain circumstances essential, component of the concerted antiviral response, at least in early life. However, IFNAR-deficient patients appear significantly less susceptible to viruses that are restricted in their replication to mucosal surfaces than patients deficient in STAT2 or IRF9. This is likely to be because IFNAR-deficient patients will have preserved IFN-III responses in mucosal sites, whereas STAT2- and IRF9-deficient patients will not, although this has yet to be proved in patient material (see Outstanding Questions).
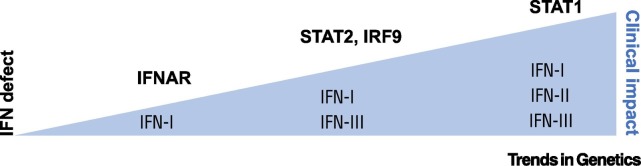
A spectrum of disease severity is also apparent, whereby the impact of STAT1 deficiency is more serious than loss of STAT2 or IRF9, seeming to correlate with the extent of compromise to the IFN system as a whole (Figure 2 ). Another notable difference between complete STAT1 deficiency and other disorders is the vulnerability of patients with the former to herpesviruses, and in particular CMV – presumably due to defective IFNγ responses in STAT1 deficiency. The apparent lack of CMV disease in patients with defects in IFN-I responses is striking (Table 1) and implies that CMV has evolved such sophisticated immune evasion strategies so as to render IFN-I effectively redundant, in contrast to the situation in vitro, where CMV has an early growth advantage in IFN-I/IFN-III-incompetent fibroblasts [86]. CMV susceptibility is a clinical feature of defects of IFNγ signalling such as IFNGR- or STAT1- deficiency [87,88]. The only monogenic defect of IFN-I signalling in which CMV susceptibility was reported – involving a hypomorphic IFNAR1 variant [89] – was probably explained by digenic inheritance of a null mutation in IFNGR2.
Figure 2.
The Extent of the Defect in the Interferon (IFN) System Correlates with Its Clinical Impact.
IFN-I receptor (IFNAR) deficiency, selectively impairing IFN-I, results in susceptibility to a more limited range of viruses than signal transducer and activator of transcription (STAT)1 deficiency, which disables responses to all IFNs. STAT2 and interferon regulatory factor 9 (IRF9) deficiencies, impeding both IFN-I and IFN-III responses, are intermediate.
An intriguing phenotype of immune dysregulation accompanies monogenic defects in IFNAR2, STAT2, STAT1, and IRF9, which warrants further investigation. Hyperinflammatory manifestations range from periodic fever [52] and hepatitis [45] to HLH [33,41,50] and severe SIRS [53]. While temporally associated with viral infection, there is not always sustained viral replication, and in some instances no obvious infectious precipitant could be identified [53]. We speculate that a combination of distinct aetiological factors may contribute, including increased pathogen load and overactivity of compensatory immune pathways, such as IFNγ and/or inflammasome-dependent IL1 signalling (which is negatively regulated by IFN-I [90,91]).
While providing insight, these disorders also raise several interesting avenues for further enquiry (see Outstanding Questions). Data from TYK2-deficient humans are intriguing, implying that responses to IFN-III and possibly also to IFN-I proceed independently of TYK2 in certain cell types and that the residual IFN-I/IFN-III signalling in TYK2 deficiency may be sufficient for antiviral defence. This would be consistent with the capacity of very low concentrations of IFN-I to induce an antiviral state [92]. From the clinical perspective, IFNγ might offer a potential therapeutic option in some of the molecular disorders described. IFNγ provides a degree of antiviral compensation in IFNAR-, STAT2-, and IRF9-deficient cells in vitro, although in vivo its use may come at a cost of immunopathology. Some of the findings in patient cells – for example, the preserved responses to IFNγ – contrast with what has been observed in IFNAR- or IRF9-deficient murine or fibrosarcoma cell systems [36,37,55,56], calling into question the clinical relevance of a large body of literature derived from these models.
Viruses face a race between the speed of virus replication and the establishment of an antiviral state mediated by any of the three IFN types. Any viral defect has the potential to tip the balance in this race against the virus. Consequently, in a normal acute infection, the IFN system is likely to constitute a constant selective pressure that keeps viruses maximally fit in terms of replication speed and competence, as well as in the maintenance of mechanisms to circumvent the IFN response [93,94]. The outcome of this race is also likely to be extremely important in governing viral pathogenesis and host range. Collectively, the insights from human genetics summarised here reveal the essential nature of the human IFN system as a whole and how certain viral pathogens (e.g., CMV) subvert restriction by some, but not all, IFN types. These insights are helping to transform our understanding of the function and interconnectedness of the IFN system and its place in the concerted antiviral response in humans, which should inform the development of next-generation vaccines and antiviral therapies.
Outstanding Questions.
Why do disease outcomes vary significantly between patients with the same genetic defect? Is this explained by differences in exposure or do these disorders highlight variation in other immune compensatory mechanisms?
How relevant are IFN-I/IFN-III defects to antiviral immunity across an individual’s lifespan? Does susceptibility diminish with age, as seen in other monogenic innate immune disorders (often attributed to the immunological education of the adaptive immune response)?
To what extent, if any, do defects in IFN-I signalling impact subtler aspects of adaptive immune maintenance and/or function in humans?
Does IFN-III protect IFNAR-deficient human epithelial surfaces from viral infection? Epithelial cells respond to both IFN-I and IFN-III, and IFNAR-deficient patients appear no more vulnerable to mucosally acquired viruses than the general population.
Is the route of live-viral vaccine delivery a determinant of pathogenicity? Does the process of attenuation facilitate mutations that confer enhanced replication capacity, pathogenicity, or tropism in IFN-I-incompetent hosts?
What pathomechanism underlies hyperinflammatory phenotypes in some patients deficient in STAT2 or IRF9? Inflammatory disease in this scenario may reflect failure to control viral replication, the exaggerated immunopathogenic activity of compensatory immune pathways, a defect of negative regulation, or a combination of these.
Can knowledge of the organisation of the IFN system be harnessed in antiviral therapy or human vaccine design? For example, IFN-III may provide an effective agent for prophylaxis or therapy of mucosal viral disease, free from the systemic toxicity associated with IFN-I, and is being explored in three Phase II clinical trials currently recruiting for novel coronavirus disease (COVID-19) (NCT04343976i, NCT04354259ii, NCT04344600iii). Rationally designed replication-competent viruses deleted for key IFN-evasion proteins are showing promise as viral oncotherapy and as vectors in animal vaccines and may play an important role in next-generation vaccines or gene-therapy vectors.
Alt-text: Outstanding Questions
Acknowledgments
Funding was provided by the Wellcome Trust [211153/Z/18/Z (C.J.A.D.), 207556/Z/17/Z (S.H.), 101788/Z/13/Z (R.E.R.)], Sir Jules Thorn Trust [12/JTA (S.H.)], and the British Medical Association (C.J.A.D).
Glossary
- Antiviral state
a cellular state of resistance to virus entry, replication, assembly and/or release, governed by the expression of multiple antiviral effector gene products (ISGs), many of them induced by interferons.
- Bacille Calmette–Guérin (BCG)
a live-attenuated form of Mycobacterium bovis developed in 1921 as a vaccine against Mycobacterium tuberculosis.
- Haemophagocytic lymphohistiocytosis (HLH)
a life-threatening systemic immune disorder characterised by widespread immune cell activation (predominantly lymphocytes, NK cells, and macrophages) and dysregulated cytokine signalling, leading to end-organ damage.
- Inborn errors of immunity
rare genetic disorders of human immune function, due to germline pathogenic variants, typically monogenic. These are usually inherited, although they can also arise de novo or result from somatic mutation.
- Interferons (IFNs)
the term applied to secreted cytokines mediating ‘viral interference’, defined as the ability of virus-infected cells to produce a substance that, when added to uninfected cells, ‘interferes with’ viral infection.
- Interferon-stimulated gene factor 3 (ISGF3)
a heterotrimeric complex of STAT1, STAT2, and IRF9 and the principal transcription factor responsible for ISG induction in response to IFN-I and IFN-III.
- Interferon-stimulated genes (ISGs)
a group of genes induced in response to IFNs, responsible for the functional activity of IFNs.
- Interferon-stimulated response element (ISRE)
ISRE in the promoters of ISGs is bound by ISGF3 to activate transcription in response to IFN-I and IFN-III. This is distinct from the IFNγ activation site (GAS), which is bound by homodimers of tyrosine phosphorylated STAT1, also known also GAF.
- Systemic inflammatory response syndrome (SIRS)
a syndromic description for systemic inflammation arising from a broad range of noxious stimuli (including trauma, infection, malignancy, immune-mediated disease, etc) that, if unchecked, can lead to multiple organ dysfunction, cardiovascular collapse, and death.
- Type I interferons (IFN-I)
a group of cytokines comprising 13 subtypes of IFNα, IFNβ, IFNε, IFNκ, and IFNω, with a range of antiviral, growth inhibitory, and immunoregulatory properties in vertebrate immune systems; increasingly recognised to have immunopathological potential, subject to stringent negative feedback control.
- Type II interferon (IFN-II)
a single cytokine, IFNγ, produced mainly by lymphocytes, which in addition to its primary role in immunomodulation has antiviral properties through the induction of a transcriptional programme distinct from but overlapping that of IFN-I and IFN-III.
- Type III interferons (IFN-III)
comprises four cytokines – IFNλ1, IFNλ2, IFNλ3, and IFNλ4. IFN-III activates an intracellular signalling cascade similar to that of IFN-I. It is produced by most cells in response to pattern recognition receptor signalling but acts on a more restricted range of cell types.
Resources
iwww.clinicaltrials.gov/ct2/show/NCT04343976iiwww.clinicaltrials.gov/ct2/show/NCT04354259iiiwww.clinicaltrials.gov/ct2/show/NCT04344600References
- 1.Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Friedman R.M. Clinical uses of interferons. Br. J. Clin. Pharmacol. 2008;65:158–162. doi: 10.1111/j.1365-2125.2007.03055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNab F., et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann H.H., et al. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleming S.B. Viral inhibition of the IFN-induced JAK/STAT signalling pathway: development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines (Basel) 2016;4:23. doi: 10.3390/vaccines4030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheelock E.F. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science. 1965;149:310–311. [PubMed] [Google Scholar]
- 8.Kotenko S.V., et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 9.Sheppard P., et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 10.Prokunina-Olsson L., et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommereyns C., et al. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mordstein M., et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pott J., et al. IFN-λ determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotta S., et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldridge M.T., et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J. Virol. 2017;91 doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klinkhammer J., et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018;7 doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan C.J.A., et al. Severe type I interferonopathy and unrestrained interferon signaling due to a homozygous germline mutation in STAT2. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aav7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder K., et al. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 19.Yockey L.J., et al. Type I interferons instigate fetal demise after Zika virus infection. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aao1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casrouge A., et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S.Y., et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 22.Perez de Diego R., et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audry M., et al. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J. Allergy Clin. Immunol. 2011;128:610–617. doi: 10.1016/j.jaci.2011.04.059. e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman M., et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen L.L., et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciancanelli M.J., et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asgari S., et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamborn I.T., et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 2017;214:1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogunjimi B., et al. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J. Clin. Invest. 2017;127:3543–3556. doi: 10.1172/JCI92280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaki M., et al. Recurrent and prolonged infections in a child with a homozygous IFIH1 nonsense mutation. Front. Genet. 2017;8:130. doi: 10.3389/fgene.2017.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hambleton S., et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3053–3058. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nish S., Medzhitov R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34:629–636. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan C.J., et al. Human IFNAR2 deficiency: lessons for antiviral immunity. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Weerd N.A., et al. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 2013;14:901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez N., et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J. Exp. Med. 2019;216:2057–2070. doi: 10.1084/jem.20182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaoka A., et al. Cross talk between interferon-γ and -α/β signaling components in caveolar membrane domains. Science. 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 37.Gough D.J., et al. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. 2010;e1000361:8. doi: 10.1371/journal.pbio.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gough D.J., et al. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mordstein M., et al. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson A.K., Pfeiffer J.K. Spectrum of disease outcomes in mice infected with YFV-17D. J. Gen. Virol. 2015;96:1328–1339. doi: 10.1099/vir.0.000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns C., et al. A novel presentation of homozygous loss-of-function STAT-1 mutation in an infant with hyperinflammation – a case report and review of the literature. J. Allergy Clin. Immunol. Pract. 2016;4:777–779. doi: 10.1016/j.jaip.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 42.Naviglio S., et al. Long-term survival after hematopoietic stem cell transplantation for complete STAT1 deficiency. J. Clin. Immunol. 2017;37:701–706. doi: 10.1007/s10875-017-0430-6. [DOI] [PubMed] [Google Scholar]
- 43.Dupuis S., et al. Impaired response to interferon-α/β and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 44.Vairo D., et al. Severe impairment of IFN-γ and IFN-α responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118:1806–1817. doi: 10.1182/blood-2011-01-330571. [DOI] [PubMed] [Google Scholar]
- 45.Chapgier A., et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 46.Chapgier A., et al. A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong X.F., et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116:5895–5906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shahni R., et al. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 2015;138:2834–2846. doi: 10.1093/brain/awv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moens L., et al. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 2017;139:1995–1997.e9. doi: 10.1016/j.jaci.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 50.Alosaimi M.F., et al. A novel variant in STAT2 presenting with hemophagocytic lymphohistiocytosis. J. Allergy Clin. Immunol. 2019;144:611–613.e3. doi: 10.1016/j.jaci.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau J.F., et al. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7278–7283. doi: 10.1073/pnas.97.13.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez N., et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 2018;215:2567–2585. doi: 10.1084/jem.20180628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bravo Garcia-Morato M., et al. Impaired control of multiple viral infections in a family with complete IRF9 deficiency. J. Allergy Clin. Immunol. 2019;144:309–312.e10. doi: 10.1016/j.jaci.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 54.Banninger G., Reich N.C. STAT2 nuclear trafficking. J. Biol. Chem. 2004;279:39199–39206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 55.Kimura T., et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 56.John J., et al. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol. Cell. Biol. 1991;11:4189–4195. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eletto D., et al. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 2016;7 doi: 10.1038/ncomms13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodig S.J., et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 59.Velazquez L., et al. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 60.Kreins A.Y., et al. Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 2015;212:1641–1662. doi: 10.1084/jem.20140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuchs S., et al. Tyrosine kinase 2 is not limiting human antiviral type III interferon responses. Eur. J. Immunol. 2016;46:2639–2649. doi: 10.1002/eji.201646519. [DOI] [PubMed] [Google Scholar]
- 62.Sarrafzadeh S.A., et al. A new patient with inherited TYK2 deficiency. J. Clin. Immunol. 2020;40:232–235. doi: 10.1007/s10875-019-00713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minegishi Y., et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 64.Ragimbeau J., et al. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karaghiosoff M., et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 66.Shimoda K., et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 67.Harari D., et al. Bridging the species divide: transgenic mice humanized for type-I interferon response. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chowdhury F.Z., Farrar J.D. STAT2: a shape-shifting anti-viral super STAT. JAKSTAT. 2013;2 doi: 10.4161/jkst.23633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashour J., et al. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grant A., et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyle S.E., et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 72.Hermant P., et al. Human but not mouse hepatocytes respond to interferon-λ in vivo. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muir A.J., et al. A randomized Phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 2014;61:1238–1246. doi: 10.1016/j.jhep.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 74.Durbin J.E., et al. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 75.Meraz M.A., et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK–STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 76.Park C., et al. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 77.Leib D.A., et al. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 1999;189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gil M.P., et al. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tegtmeyer P.K., et al. STING induces early IFN-β in the liver and constrains myeloid cell-mediated dissemination of murine cytomegalovirus. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Essers M.A., et al. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 81.Sato T., et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 82.Swann J.B., et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J. Immunol. 2007;178:7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- 83.Hwang S.Y., et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takayanagi H., et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-β. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 85.Hamilton J.A., et al. Endogenous IFN-alpha beta suppresses colony-stimulating factor (CSF)-1-stimulated macrophage DNA synthesis and mediates inhibitory effects of lipopolysaccharide and TNF-alpha. J. Immunol. 1996;156:2553–2557. [PubMed] [Google Scholar]
- 86.McSharry B.P., et al. Abrogation of the interferon response promotes more efficient human cytomegalovirus replication. J. Virol. 2015;89:1479–1483. doi: 10.1128/JVI.02988-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bustamante J., et al. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 2014;26:454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dorman S.E., et al. Viral infections in interferon-γ receptor deficiency. J. Pediatr. 1999;135:640–643. doi: 10.1016/S0022-3476(99)70064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoyos-Bachiloglu R., et al. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 2017;127:4415–4420. doi: 10.1172/JCI93486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guarda G., et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Reboldi A., et al. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levin D., et al. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perez-Cidoncha M., et al. An unbiased genetic screen reveals the polygenic nature of the influenza virus anti-interferon response. J. Virol. 2014;88:4632–4646. doi: 10.1128/JVI.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young D.F., et al. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 2003;77:2174–2181. doi: 10.1128/JVI.77.3.2174-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muller M., et al. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung S., et al. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy D.E., et al. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 98.Decker T., et al. Cytoplasmic activation of GAF, an IFN-γ-regulated DNA-binding factor. EMBO J. 1991;10:927–932. doi: 10.1002/j.1460-2075.1991.tb08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheon H., et al. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glocker E.O., et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu L., et al. IL-10 and IL-10 receptor mutations in very early onset inflammatory bowel disease. Gastroenterol. Res. 2017;10:65–69. doi: 10.14740/gr740w. [DOI] [PMC free article] [PubMed] [Google Scholar]