With a great diversity in gene composition, including multiple putative antibiotic resistance genes, AbaR islands are potential contributors to multidrug resistance in Acinetobacter baumannii. However, the effective contribution of AbaR to antibiotic resistance and bacterial physiology remains elusive. To address this, we sought to accurately remove AbaR islands and restore the integrity of their insertion site. To this end, we devised a versatile scarless genome editing strategy.
KEYWORDS: AbaR, Acinetobacter baumannii, natural transformation, resistance island
ABSTRACT
With a great diversity in gene composition, including multiple putative antibiotic resistance genes, AbaR islands are potential contributors to multidrug resistance in Acinetobacter baumannii. However, the effective contribution of AbaR to antibiotic resistance and bacterial physiology remains elusive. To address this, we sought to accurately remove AbaR islands and restore the integrity of their insertion site. To this end, we devised a versatile scarless genome editing strategy. We performed this genetic modification in two recent A. baumannii clinical strains: the strain AB5075 and the nosocomial strain AYE, which carry AbaR11 and AbaR1 islands of 19.7 kbp and 86.2 kbp, respectively. Antibiotic susceptibilities were then compared between the parental strains and their AbaR-cured derivatives. As anticipated by the predicted function of the open reading frame (ORF) of this island, the antibiotic resistance profiles were identical between the wild type and the AbaR11-cured AB5075 strains. In contrast, AbaR1 carries 25 ORFs, with predicted resistance to several classes of antibiotics, and the AYE AbaR1-cured derivative showed restored susceptibility to multiple classes of antibiotics. Moreover, curing of AbaRs restored high levels of natural transformability. Indeed, most AbaR islands are inserted into the comM gene involved in natural transformation. Our data indicate that AbaR insertion effectively inactivates comM and that the restored comM is functional. Curing of AbaR consistently resulted in highly transformable and therefore easily genetically tractable strains. Emendation of AbaR provides insight into the functional consequences of AbaR acquisition.
INTRODUCTION
Acinetobacter baumannii is responsible for health care-associated infections and concerns because of its intrinsic resistance to many antimicrobials and its ability to acquire resistance genes (1, 2). A potential contributor to multidrug resistance in A. baumannii is a genomic island, named AbaR, with a great diversity in gene content and carrying multiple putative antibiotic resistance genes (3, 4). The first report of AbaR described a large genomic island of 86 kbp in the epidemic A. baumannii strain AYE in 2006 (5). With a transposon backbone, AbaR islands are considered mobile genetic elements and present a modular structure (3, 4, 6). Although AbaR may contain up to 25 putative antibiotic resistance genes, most AbaR carry genes predicted to confer resistance mainly to aminoglycosides, tetracycline, and sulfonamide (4, 6, 7). Despite increasing genomic data, the actual contribution of AbaR-type islands to antibiotic resistance has not been fully evaluated. Deletion mutants of the AbaR27 island (originally published as TnAbaR23) were previously obtained and revealed a restored susceptibility to sulfamethoxazole and a partial increased susceptibility to tetracycline (4, 8). The authors also suggested a paradoxical role of AbaR27 in increasing susceptibility to ciprofloxacin. However, deletions of AbaR27 were obtained by allelic replacement of this AbaR with the aacC1 cassette, conferring resistance to gentamicin and confounded susceptibility testing against aminoglycosides. In addition, the original insertion site of AbaR was not restored, which prevented analysis of the physiological consequence of the insertion of AbaR in the targeted gene. AbaR islands are found inserted in the comM gene (4). This gene encodes a putative ATPase involved in natural transformation in a few transformable species, including nonpathogenic Acinetobacter baylyi (9, 10). Natural transformation allows bacteria to actively import exogenous DNA, and if the sequence of the imported DNA presents sufficient identity, it integrates into the recipient cell’s chromosome by homologous recombination (11, 12). Interestingly, the comM gene is not strictly essential for transformation, but its mutation decreases the transformation efficiency by 2 orders of magnitude (9, 10). A. baumannii and related species A. nosocomialis were described as transformable in 2013 (13, 14). In A. baumannii, natural transformation appears as a conserved trait among clinical and nonclinical isolates of human and animal origin (13, 15, 16), providing the bacteria with a major route for acquisition of antibiotic resistance genes.
In this study, we aim at assessing the contribution of AbaR islands to antibiotic resistance of two human clinical strains of the main global clone 1 bearing small and large AbaRs. We also analyzed the consequences of AbaR-mediated comM gene disruption on the transformation efficiency of these strains. We bring evidence that interruption of the comM gene by AbaR results in a decrease in natural transformation. Using genetic approaches, we further confirm a role for the comM gene in natural transformation in A. baumannii.
RESULTS AND DISCUSSION
Curing AbaR in A. baumannii using a scarless genome editing strategy.
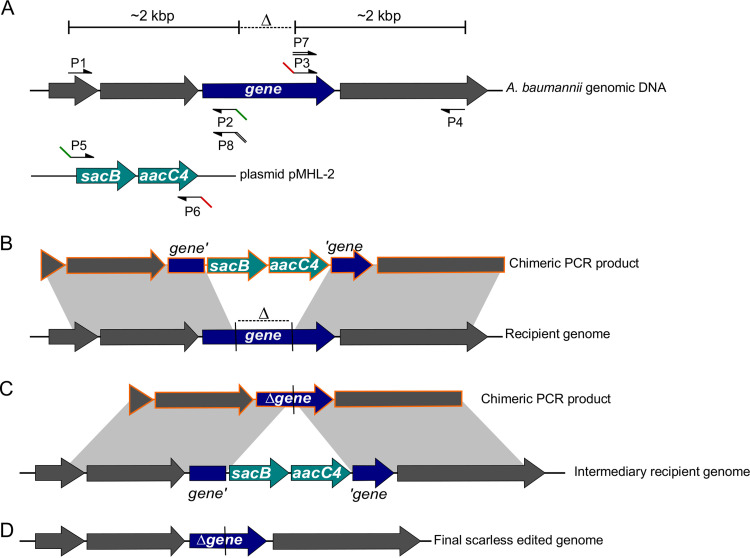
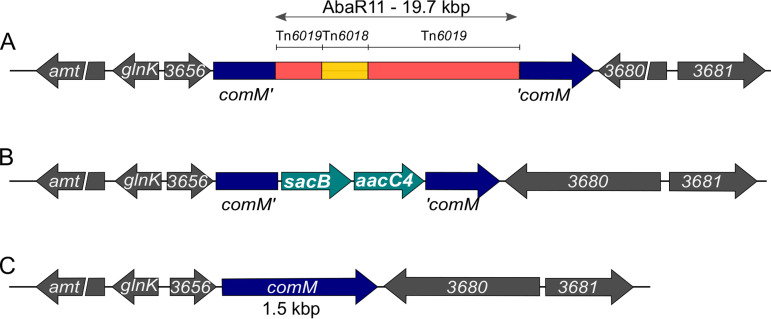
To cure A. baumannii of the AbaR island while restoring the original insertion site, we devised a scarless genome editing strategy. This method, depicted in Fig. 1, allows deletion, insertion, or single-base mutation. In contrast to other methods, it does not require cloning or prior genetic engineering of the target strain (17–19). Rather, it relies on the use of chimeric PCR products and takes advantage of natural transformation, a phenotypic trait exhibited by most isolates of A. baumannii. The method is based on a two-step selection (Fig. 1): first the introduction of a selectable/counterselectable sacB_aacC4 cassette at the loci of interest (Fig. 1B), and then the replacement of the cassette by the desired sequence (Fig. 1C). First, a chimeric PCR product is assembled, consisting of the sacB_aacC4 cassette bearing a selection marker (aacC4) conferring resistance to apramycin, flanked by 2-kbp-long sequences identical to the target region (Fig. 1B). This chimeric PCR product is introduced by natural transformation, and A. baumannii transformants are selected using apramycin, an antibiotic to which most clinical isolates are sensitive. A second PCR product is assembled, still carrying the homologous flanking regions but whose sequence is designed to produce deletion, insertion, or single-base mutation (deletion in Fig. 1C). Genetic replacement and the loss of the previously inserted sacB gene is then selected using sucrose resistance selection. We applied this method to delete AbaR11 in strain AB5075. The strain AB5075 is an A. baumannii ST1 strain isolated from a bone infection in 2008 with multidrug resistance (aminoglycosides, fluoroquinolones, and β-lactams, including carbapenems) (20). This strain bears a 19.7-kbp-long AbaR11 inserted in the comM gene with 19 predicted genes corresponding to five modules found in other AbaRs (Fig. 2A) (4, 8, 21). In a previous study, we identified the strain AB5075 as naturally transformable (15). Using natural transformation of PCR chimeric products, the AbaR island was replaced upon genetic recombination by the sacB_aacC4 cassette (Fig. 2B). Then, the strain bearing the sacB_aacC4 cassette in the comM gene was subjected to transformation with an assembly PCR encompassing the insertion region cured of AbaR, thereby carrying an intact comM gene. Then we performed genetic analysis of sucrose-resistant and apramycin-susceptible clones to select an AbaR-cured strain (named AB5075-R) with a restored comM gene presumably encoding a full-length ComM protein (Fig. 2C).
FIG 1.
PCR-based genome editing of transformable A. baumannii. Schematic representations of steps for gene inactivation (dark blue gene) using natural transformation. (A) Annealing sites for primers of PCR assemblies on their respective templates: either A. baumannii genomic DNA for amplifications of 2 kbp upstream and downstream of the deleted region (Δ) or the laboratory plasmid pMHL-2 that bears the counterselectable cassette (light blue) with a sacB gene conferring sucrose sensitivity and the aacC4 gene encoding resistance to apramycin. Note that primers P2, P3, and P7 share annealing sequences with primers P5, P6, and P8, respectively. (B) Chimeric PCR product (orange) obtained after two-step PCR assembly of products P1 + P2 with P5 + P6 and P3 + P4. This chimeric product bears the sacB_aacC4 cassette flanked by two 2-kbp-long regions of homology with the recipient cell’s genome (gray areas). These homologous regions allow the recombination of the chimeric PCR product into the recipient cell’s genome, resulting, upon apramycin selection of the transformants, in the intermediary genotype represented in panel C. (C) Chimeric PCR product obtained after two-step assembly PCR of products P1 + P8 with P7 + P4 bearing the deleted gene flanked by homologous regions (gray areas), allowing recombination and replacement of the counterselectable cassette in the intermediary recipient genome. (D) The transformants are selected based on their sucrose resistance resulting in the final scarless edited genome.
FIG 2.
Genetic engineering of AbaR curing in A. baumannii. (A) Schematic representation of AbaR11 inserted in the comM gene in A. baumannii AB5075 genome. All the sequences and genes in gray color belong to the wild-type AB5075 chromosome, the interrupted comM gene is, however, highlighted in dark blue. Transposons Tn6019 and Tn6018 constituting AbaR11 are indicated as in reference 4. For illustration purposes, the scale of the AbaR island is not respected. (B) Intermediary genetic construct with the counterselectable cassette replacing the AbaR11 island. (C) Final genetic structure of the locus after restoring the comM gene integrity.
Resistance profiles associated with AbaR curing in two clinically relevant multidrug-resistant strains.
We then sought to investigate the role of AbaR island in the multidrug resistance profile of strain AB5075. This AbaR contains 19 open reading frames (ORFs) with, besides transposon-related proteins, seven ORFs predicted to confer resistance to metals, three of unknown function, and the sup gene encoding a putative sulfate permease present in most AbaRs (Fig. 2A). Antibiotic susceptibilities were thus compared between the AB5075 parental strain and its AbaR-cured derivative, denoted AB5075-T (Table 1). As anticipated, the resistance profiles to 14 antibiotics belonging to five antibiotic classes were identical between the wild type and its AbaR-cured derivative. We then investigated the contribution to antibiotic resistance of a larger AbaR found only in another A. baumannii ST1 strain, the clinical strain AYE resistant to aminoglycosides, β-lactams (excluding imipenem, piperacillin-tazobactam, and ticarcillin-clavulanate), fluoroquinolones, and tetracycline and was of intermediate susceptibility to rifampicin (5, 22). This strain carries the largest AbaR described so far, AbaR1, at 86 kbp long, containing 25 genes putatively involved in resistance to antibiotics of several classes. By exploiting natural transformation of the strain AYE (15), we cured this strain of its AbaR1 island and compared the resistance profiles to 21 antibiotics of the parental strain to that of its AbaR-cured AYE-T derivative (Table 2). In contrast to strain AB5075, curing the AbaR island from the strain AYE resulted in restored susceptibility to aminoglycosides, tetracycline, and sulfonamide. The resistance armamentarium conferred by the 86-kbp-long AbaR1 may be attributed to specific genes that are listed in Table 2 according to previous analyses (4, 5, 23). Restored resistances to sulfonamides and tetracycline are consistent with the previous report of AbaR27 deletion in strain A424 leading to the loss of two copies of sul1 genes and one tetA gene (8).
TABLE 1.
Antimicrobial resistance profiles of A. baumannii strain AB5075 deleted of AbaR11 (AB5075-T) in comparison to the wild-type strain
| Antibiotica | Inhibition zone diam (mm)b |
|
|---|---|---|
| AB5075 WT | AB5075-T | |
| AMK | 20 | 20 |
| GEN | 11 | 12 |
| TOB | 10 | 10 |
| CIP | 6 | 6 |
| PIP | 6 | 6 |
| TZP | 6 | 6 |
| TIC | 6 | 6 |
| TIM | 6 | 6 |
| CAZ | 6 | 6 |
| FEP | 6 | 6 |
| IMP | 6 | 6 |
| ATM | 6 | 6 |
| TET | 18 | 18 |
| SUF | 6 | 6 |
AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; PIP, piperacillin; TZP, piperacillin plus tazobactam; TIC, ticarcillin; TIM, ticarcillin plus clavulanic acid; CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; ATM, aztreonam; TET, tetracycline; SUF, sulfonamide.
WT, wild type; AB5075-T, strain AB5075 deleted of AbaR11.
TABLE 2.
Antimicrobial resistance profiles of A. baumannii strain AYE deleted of AbaR1 in comparison to the wild-type strain and genes carried by AbaR1 potentially involved in resistance
| Antibiotica | Inhibition zone diam (mm) |
MIC (μg/ml) |
MIC/Cd (μg/ml) |
Predicted AbaR1 resistance gene involvede | |||
|---|---|---|---|---|---|---|---|
| AYE WTb | AYE-Tc | AYE WT | AYE-T | AYE WT | AYE-T | ||
| AMK | 8 | 21 | 64 | 8 | —f | — | aadB, aacC1, aphA1b, aacA |
| GEN | 6 | 19 | 128 | 6 | — | — | |
| TOB | 6 | 18 | 24 | 1.5 | — | — | |
| KAN | — | — | >256 | 2 | — | — | |
| STR | — | — | 128 | 0.64 | — | — | Two aadA1 copies, strA, strB |
| CIP | 6 | 6 | — | — | — | — | |
| PIP | 6 | 6 | — | — | — | — | |
| TZP | 14 | 15 | — | — | — | — | |
| TIC | 6 | 19 | — | — | — | — | blaVEB-1, blaoxa-10g |
| TIM | 15 | 18 | — | — | — | — | blaoxa-10g |
| CAZ | 6 | 11 | >256 | 12 | >256 | 3 | blaVEB-1 |
| FEP | 6 | 16 | 256 | 12 | >256 | 16 | blaVEB-1 |
| CTX | — | — | >32 | >32 | >32 | >32 | |
| ATM | 6 | 11 | — | — | — | — | blaVEB-1 |
| IMP | 28 | 28 | — | — | — | — | |
| MEM | 20 | 22 | — | — | — | — | |
| SUF | 6 | 23 | — | — | — | — | Five sul1 copies |
| SXT | — | — | >32 | 0.25 | — | — | Five sul1 copies, dhfrI, dhfrX |
| TET | — | — | 128 | 8 | — | — | tetA(A), tetA(G) |
| CHL | — | — | >256 | >256 | — | — | cmlA1, cmlA5, cmlA9, catA1 |
| RIF | — | — | 12 | 8 | — | — | arr-2 |
AMK, amikacin; GEN, gentamicin; TOB, tobramycin; STR, streptomycin; KAN, kanamycin; CIP, ciprofloxacin; PIP, piperacillin; TZP, piperacillin plus tazobactam; TIC, ticarcillin; TIM, ticarcillin plus clavulanic acid; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; IMP, imipenem; MEM, meropenem; ATM, aztreonam; SUF, sulfonamide; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; CHL, chloramphenicol; RIF, rifampicin.
WT, wild type.
AYE-T, strain AYE deleted of AbaR1.
C, cloxacillin at 250 μg/ml.
—, not done.
According to reference 24, oxa-10 gene expression is probably weak.
In the AYE-T strain, partially decreased resistance to ticarcillin, ceftazidime, cefepime, and aztreonam was also observed. Only a partial loss of resistance to β-lactams upon AbaR1 curing is likely explained by the additional resistance mechanisms the AYE strain possesses, such as the overexpression of the blaADC-11 gene encoding a narrow-spectrum chromosomal cephalosporinase (22, 24). In AYE-T strain ceftazidime, Etest values decreased from 12 μg/ml to 3 μg/ml in medium supplemented with cloxacillin (250 μg/ml), the latter known to inhibit the cephalosporinase, whereas no significant differences were observed for cefepime, which is not a substrate for ADC-11 (25). Other chromosomal resistance determinants most likely contribute to the reduced susceptibility to cefotaxime, for instance, the predicted new class A β-lactamase on AYE chromosome (24). Despite the four putative chloramphenicol resistance genes on AbaR1, their deletion did not modify the high intrinsic resistance of A. baumannii to this antibiotic mediated by efflux (26). Deletion of AbaR1 resulted in a minor change of rifampicin susceptibility.
Contrarily to the previous report on AbaR27 deletion, we did not observe for both strains, AB5075 and AYE, an increased resistance to fluoroquinolone of the AbaR-cured strains (8).
Beyond the role of AbaR in GC1 resistance, this method should allow investigation of other genomic islands found in A. baumannii as long as strains are transformable. For instance, as strains of GC2 are seemingly transformable, scarless removal of the AbGRI1 resistance island found in this clone is anticipated to be feasible (27, 28).
Curing of AbaR and repair of the comM gene result in increased transformation ability.
AbaR islands are inserted in the comM gene (4). In the nonpathogenic species A. baylyi, a mutation of the latter gene is associated with reduced transformability (9). We therefore pursued this by investigating the role of comM in A. baumannii as well as the physiological consequence of AbaR-island disruption of this gene. To do so, we performed transformation assays comparing the levels of transformation frequency between the wild-type and AbaR-cured strains using an rpoB mutation (I581F) conferring resistance to rifampicin as a genetic marker (Fig. 3). For both clinical strains, curing AbaR and thus restoring the comM gene increased significantly the ability to acquire the genetic marker, with a mean increase of approximately 1,000-fold in strain AB5075-T (Fig. 3A) and of approximately 300-fold in strain AYE-T (Fig. 3B). With one bacterium of 100 that performed natural transformation, strain AB5075-T presented the highest transformation ability (Fig. 3A). Using the same genome editing strategy as depicted in Fig. 1, we generated markerless deletions of the comM gene (ΔcomM) in both A. baumannii clinical strains. Consistently, deletion of the comM gene in both strains resulted in transformation levels comparable to levels observed for the wild-type strains bearing a genuine insertion in the comM gene by the AbaR island (Fig. 3A and B). Thus, prior to acquisition of AbaR, both strains were transformable at higher levels, and the subsequent disruption generated transformation defects that are consistent with previous observations made in other species or genera (9, 10). The data show that the AbaR-inactivated comM gene, once cured of AbaR, is still functional. This suggests that it has not been affected by mutational drifts, confirming that acquisition of AbaR occurred relatively recently (4).
FIG 3.
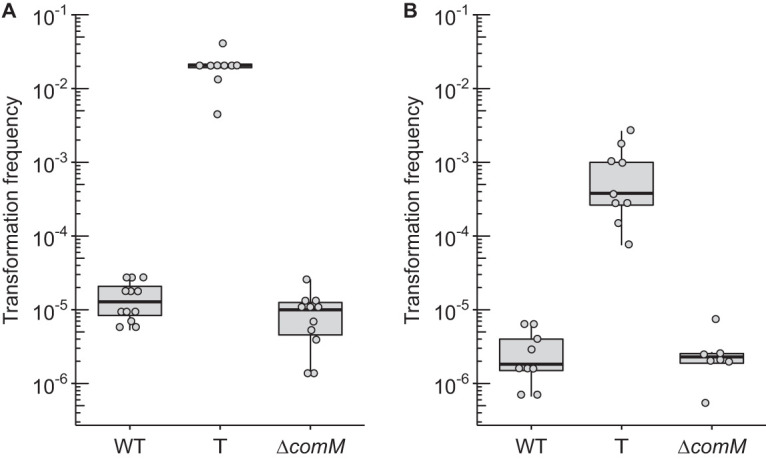
Transformation frequencies of A. baumannii strains AB5075 (A) and AYE (B) of a 3.8-kbp-long PCR product carrying a mutation in the rpoB gene conferring resistance to rifampicin (final concentration of 50 ng/μl, 125 ng per sample). For both strains, either the wild type or their “T” (ΔAbaR/repaired comM) or ΔcomM derivatives were tested. At least seven independent transformation assays were performed on at least two separate occasions represented by separate dots and corresponding boxplots. All results were above the limit of detection (10−8). Pairwise comparisons using the nonparametric Mann-Whitney-Wilcoxon test (two-tailed) gave P values of at least <0.005 between transformation frequencies obtained for the wild-type/ΔcomM strains and the T derivatives.
The fact that acquisition of the AbaR element in the comM gene alters the natural transformability of A. baumannii and its capacity for further acquisitions by natural transformation is intriguing. Interestingly, acquisition of AbaR and consequent comM interruption did not hinder GC1 subsequent acquisition of different capsule gene clusters or gyrA or parC alleles leading to fluoroquinolone resistance (29). The significance of lower transformability is not known, but theoretical work suggests that intermediate transformation rates allow bacteria to maximize fitness in fluctuating stressful environments such as repeated antibiotic stress (30).
Conclusion.
By exploiting natural transformation, we set up a genetic system to easily edit the genome of A. baumannii. We used this strategy to perform small-gene deletion but also to cure large genomic islands in two multidrug-resistant clinical A. baumannii strains. Moving beyond predicted resistance, this system allowed experimental determination of the antibiotic resistance conferred by the islands. We conclude that acquisition of AbaR islands may increase A. baumannii strain resistance to antibiotics, but it comes at the cost of decreasing the cell’s ability to perform natural transformation by disrupting the comM gene. Thus, acquisition of AbaR may impact genome evolution of A. baumannii. From an applied perspective, the simultaneous deletion of AbaR and restoration of the comM function in two clinically relevant strains made them even more genetically amenable, blazing the trail for mutational analysis of these model A. baumannii strains.
MATERIALS AND METHODS
Bacterial strains, typing, and growth conditions.
The bacterial strains used in this study are listed in Table 3. Unless specified, A. baumannii isolates and strains were grown in Lennox broth (LB). All experiments were performed at 37°C. Antibiotic concentrations in selective media were 30 μg/ml for apramycin and 100 μg/ml for rifampicin.
TABLE 3.
Acinetobacter baumannii strains used in this study
| Strain name | Genotype | Description/phenotype | Reference |
|---|---|---|---|
| AB5075 WT | A. baumannii AB5075 wild type | 31 | |
| AB5075 comM::sacB_aacC4 | A. baumannii AB5075 comM::sacB_aacC4 | Replacement of the AbaR island with the sacB_aacC4 cassette, resistant to apramycin and sensitive to sucrose | This study |
| AB5075-T | A. baumannii AB5075 ΔAbaR11 | Deletion of the AbaR11 island and a repaired comM gene, highly transformable strain | This study |
| AB5075 ΔcomM | A. baumannii AB5075 ΔcomM | Markerless 231-bp deletion of the comM gene | This study |
| AB5075 Rifr | A. baumannii AB5075 Rifr | Spontaneous rpoB mutant I581F; rifampicin resistant | This study |
| AYE WT | A. baumannii AYE wild type | Wild type | 22 |
| AYE comM::sacB_aacC4 | A. baumannii AYE comM::sacB_aacC4 | Strain with a replacement of the AbaR island with the sacB_aacC4 cassette, resistant to apramycin and sensitive to sucrose | This study |
| AYE-T | A. baumannii AYE ΔAbaR1 | Strain with a deletion of the AbaR1 island and a repaired comM gene, highly transformable and antibiotic susceptible strain | This study |
| AYE ΔcomM | A. baumannii AYE ΔcomM | Markerless 231-bp deletion of the comM gene | This study |
Construction of bacterial strains.
All oligonucleotides used in this study for genetic modification are listed in Table S1 in the supplemental material. Gene disruptions were performed using overlap extension PCR to synthesize large chimeric DNA fragments carrying the selection marker flanked by 2-kbp-long fragments that are homologous to sequences flanking the insertion site. The PCRs were performed with a high-fidelity DNA polymerase (PrimeStarMax; TaKaRa) (see “Detailed protocol of chimeric PCR for mutation in A. baumannii” in the supplemental material). When used as the template for the generation of PCR products, genomic DNA was extracted and purified using the Wizard Genomic DNA purification kit (Promega) according to the manufacturer’s guidelines. The AB5075 wild-type strain was naturally transformed with an assembly PCR product to generate the AB5075 comM::sacB_aacC4 strain. The AYE comM::sacB_aacC4 was then obtained by transforming the AYE wild-type strain with genomic DNA extracted from the AB5075 comM::sacB_aacC4 strain. For both strains, transformants were selected on LB plates supplemented with apramycin, and susceptibility to sucrose was verified on M63 plates supplemented with 10% sucrose. The AB5075 and AYE comM::sacB_aacC4 strains were naturally transformed with an assembly PCR product (obtained from assembly PCR on wild-type genomic DNA) to generate their T and ΔcomM derivatives. Transformants were selected on M63 plates supplemented with 10% sucrose, and susceptibility to apramycin was verified on LB plates supplemented with apramycin. All chromosomal modifications were verified in transformants using colony PCR (cf. Table S1). Removal of AbaR and restoration of the comM gene were verified by Sanger sequencing.
The AB5075 rifampicin-resistant (Rifr) strain was generated by selecting spontaneous mutants after plating an overnight culture of the AB5075 wild-type strain on plates supplemented with rifampicin.
Antibiotic susceptibility testing.
Susceptibility of the strains to a panel of 15 antibiotics (ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, meropenem, imipenem, gentamicin, tobramycin, amikacin, ciprofloxacin, tetracycline, aztreonam, and sulfonamide) was evaluated by disc diffusion on Muller-Hinton agar (Bio-Rad, France) according the CA-SFM 2013 recommendations (https://resapath.anses.fr/resapath_uploadfiles/files/Documents/2013_CASFM.pdf). Inhibition values were interpreted according to CA-SFM 2013 breakpoints for all antibiotics but aztreonam, for which Pseudomonas spp. breakpoints are given. Susceptibilities of wild-type AYE and deleted AbaR1 mutant were also compared by Etest strips (bioMérieux, France) for tetracycline, streptomycin, trimethoprim-sulfamethoxazole, chloramphenicol, rifampicin, and β-lactams, including cefotaxime, ceftazidime, and cefepime. The strain Pseudomonas aeruginosa CIP 7110 was used as control for all antibiotic susceptibility testing.
Transformation assay.
The method used in this study was described in reference 15 with the following modifications. After overnight incubation at 37°C on Lennox broth agar, the strains were cultured for a few hours in 2 ml of Lennox broth (LB) until an optical density at 600 nm (OD600) of at least 1 was reached. The bacterial broths were then diluted to an OD600 of 0.01 in phosphate-buffered saline (PBS). Then, small aliquots of bacterial suspensions (5 to 10 μl) were mixed with DNA substrate. To measure transformation frequency, a final PCR concentration of 50 ng/μl was used. The mixtures (2.5 μl) were deposited on the surface of 1 ml of transformation medium (2% agarose D3 [Euromexdex], 5 g/liter of tryptone, 2.5 g/liter NaCl), poured in 2-ml microtubes, and incubated overnight at 37°C. The next day, bacteria were recovered by resuspension in 300 μl of PBS. For genome edition experiments, the suspensions were spread on selective media (see “Construction of bacterial strains” for details). To measure transformation frequency, the bacterial suspensions were serial diluted and spread on LB agar plates without antibiotic or supplemented with rifampicin. All the transformation assays were performed on at least two separate experiments. On each occasion, at least three independent transformation reactions were conducted (three different transformation tubes). All the independent data points are plotted. As normality of the distribution of transformation frequency does not apply for transformation frequency analysis, nonparametric tests were performed (Mann-Whitney-Wilcoxon).
Supplementary Material
ACKNOWLEDGMENTS
We thank the LABEX ECOFECT for building an “ecofectian” community.
This work was supported by the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program Investissements d'Avenir (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR). A.-S.G. and M.-H.L. were also supported by Programme Jeune Chercheur from VetAgro Sup. A.L. and M.H. were supported by the French Agency for Food, Environmental and Occupational Health & Safety (ANSES).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed. WHO, Atlanta, GA. [Google Scholar]
- 3.Bonnin RA, Poirel L, Nordmann P. 2012. AbaR-type transposon structures in Acinetobacter baumannii. J Antimicrob Chemother 67:234–236. doi: 10.1093/jac/dkr413. [DOI] [PubMed] [Google Scholar]
- 4.Hamidian M, Hall RM. 2018. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 – structure, origin and evolution. Drug Resist Updat 41:26–39. doi: 10.1016/j.drup.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 7.Post V, Hamidian M, Hall RM. 2012. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J Antimicrob Chemother 67:1039–1040. doi: 10.1093/jac/dkr586. [DOI] [PubMed] [Google Scholar]
- 8.Kochar M, Crosatti M, Harrison EM, Rieck B, Chan J, Constantinidou C, Pallen M, Ou H-Y, Rajakumar K. 2012. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother 56:1845–1853. doi: 10.1128/AAC.05334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nero TM, Dalia TN, Wang JC-Y, Kysela DT, Bochman ML, Dalia AB. 2018. ComM is a hexameric helicase that promotes branch migration during natural transformation in diverse Gram-negative species. Nucleic Acids Res 46:6099–6111. doi: 10.1093/nar/gky343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gwinn ML, Ramanathan R, Smith HO, Tomb JF. 1998. A new transformation-deficient mutant of Haemophilus influenzae Rd with normal DNA uptake. J Bacteriol 180:746–748. doi: 10.1128/JB.180.3.746-748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau D, Blokesch M. 2019. Mechanisms of DNA uptake by naturally competent bacteria. Annu Rev Genet 53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 13.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godeux A-S, Lupo A, Haenni M, Guette-Marquet S, Wilharm G, Laaberki M-H, Charpentier X. 2018. Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J Bacteriol 200:e00181-18. doi: 10.1128/JB.00181-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domingues S, Rosário N, Cândido Â, Neto D, Nielsen KM, Da Silva GJ. 2019. Competence for natural transformation is common among clinical strains of resistant Acinetobacter spp. Microorganisms 7:30. doi: 10.3390/microorganisms7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trebosc V, Gartenmann S, Royet K, Manfredi P, Tötzl M, Schellhorn B, Pieren M, Tigges M, Lociuro S, Sennhenn PC, Gitzinger M, Bumann D, Kemmer C. 2016. A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob Agents Chemother 60:7263–7271. doi: 10.1128/AAC.01275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Wang Z, Chen Y, Hua X, Yu Y, Ji Q. 2019. A highly efficient CRISPR-Cas9-based genome engineering platform in Acinetobacter baumannii to understand the H2O2-sensing mechanism of OxyR. Cell Chem Biol 26:1732.e5–1742.e5. doi: 10.1016/j.chembiol.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Biswas I. 2015. Genetic tools for manipulating Acinetobacter baumannii genome: an overview. J Med Microbiol 64:657–669. doi: 10.1099/jmm.0.000081. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol 41:3542–3547. doi: 10.1128/jcm.41.8.3542-3547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coyne S, Guigon G, Courvalin P, Périchon B. 2010. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother 54:333–340. doi: 10.1128/AAC.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Martínez J-M, Nordmann P, Ronco E, Poirel L. 2010. Extended-spectrum cephalosporinase in Acinetobacter baumannii. Antimicrob Agents Chemother 54:3484–3488. doi: 10.1128/AAC.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamidian M, Hall RM. 2017. Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J Antimicrob Chemother 72:2944–2947. doi: 10.1093/jac/dkx206. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, He L, Tao X, Meng F, Zhang J. 2019. High DNA uptake capacity of international clone II Acinetobacter baumannii detected by a novel planktonic natural transformation assay. Front Microbiol 10:2165. doi: 10.3389/fmicb.2019.02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, Dougan G, Hall R. 2016. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2:e000052. doi: 10.1099/mgen.0.000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho G, Fouchet D, Danesh G, Godeux A-S, Laaberki M-H, Pontier D, Charpentier X, Venner S. 2020. Bacterial transformation buffers environmental fluctuations through the reversible integration of mobile genetic elements. mBio 11:e02443-19. doi: 10.1128/mBio.02443-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.