Abstract
Bacteria have a remarkable ability to sense environmental changes, swiftly regulating their transcriptional and posttranscriptional machinery as a response. Under conditions that cause growth to slow or stop, bacteria typically stabilize their transcriptomes in what has been shown to be a conserved stress response. In recent years, diverse studies have elucidated many of the mechanisms underlying mRNA degradation, yet an understanding of the regulation of mRNA degradation under stress conditions remains elusive. In this review we discuss the diverse mechanisms that have been shown to affect mRNA stability in bacteria. While many of these mechanisms are transcript-specific, they provide insight into possible mechanisms of global mRNA stabilization. To that end, we have compiled information on how mRNA fate is affected by RNA secondary structures; interaction with ribosomes, RNA binding proteins, and small RNAs; RNA base modifications; the chemical nature of 5′ ends; activity and concentration of RNases and other degradation proteins; mRNA and RNase localization; and the stringent response. We also provide an analysis of reported relationships between mRNA abundance and mRNA stability, and discuss the importance of stress-associated mRNA stabilization as a potential target for therapeutic development.
Keywords: ribonucleic acid, stress response, carbon starvation, nutrient starvation, hypoxia, mRNA degradation, mRNA stability, bacteria
Introduction
Bacterial adaptation to stress is orchestrated by complex responses to specific environmental stimuli, capable of rapidly regulating transcription, transcript degradation, and translation, which increases the organism’s survival opportunities. Historically, regulation mechanisms for transcriptional and translational pathways have been the most studied, providing insight into the genes and protein products needed for bacterial adaptation to unfavorable growth environments. These findings have been key for our understanding of bacterial biology, allowing us, for example, to develop tools to tune bacterial machinery for biotechnology processes (such as Tao et al., 2011; Courbet et al., 2015; Daeffler et al., 2017; Martinez et al., 2017; Riglar et al., 2017), and to discover and develop new antibacterial drugs (for example, Yarmolinsky and Haba, 1959; Wolfe and Hahn, 1965; Maggi et al., 1966; Olson et al., 2011). However, the role of RNA degradation in stress responses is not well understood.
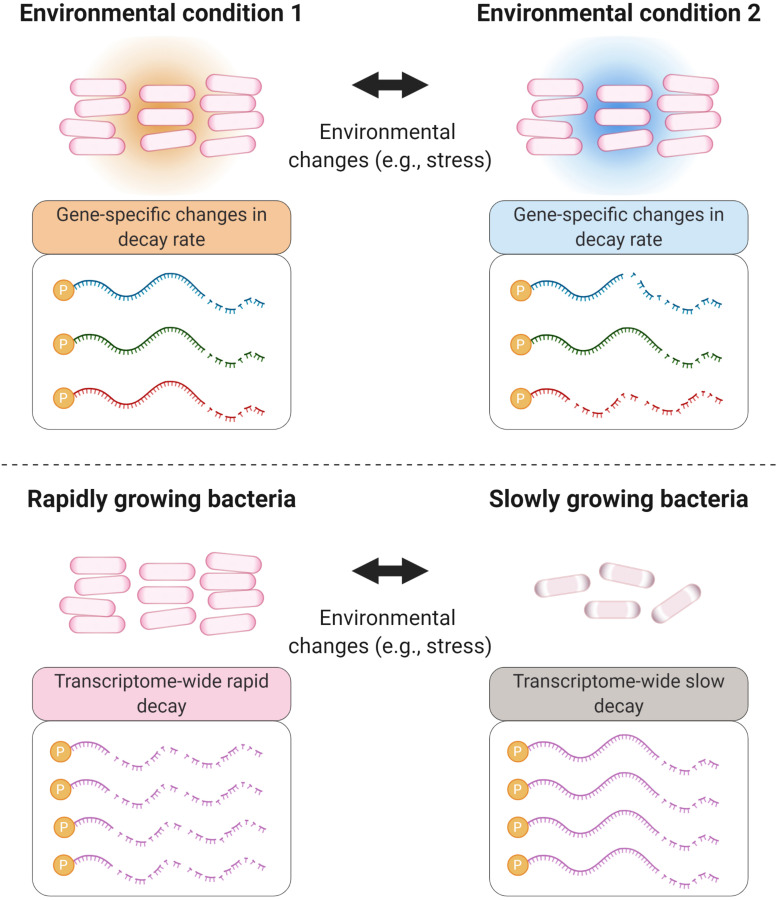
Modulation of mRNA degradation has been associated with various stress conditions in bacteria, such as temperature changes, growth rate, nutrient starvation, and oxygen limitation (see Table 1). Transcript stability – also referred as mRNA or transcript half-life – was shown to be globally altered in response to some stressors, while in other cases, gene-specific modulation of transcript stability contributes to specific expression changes that bacteria need to adapt to and survive in new environments (Figure 1).
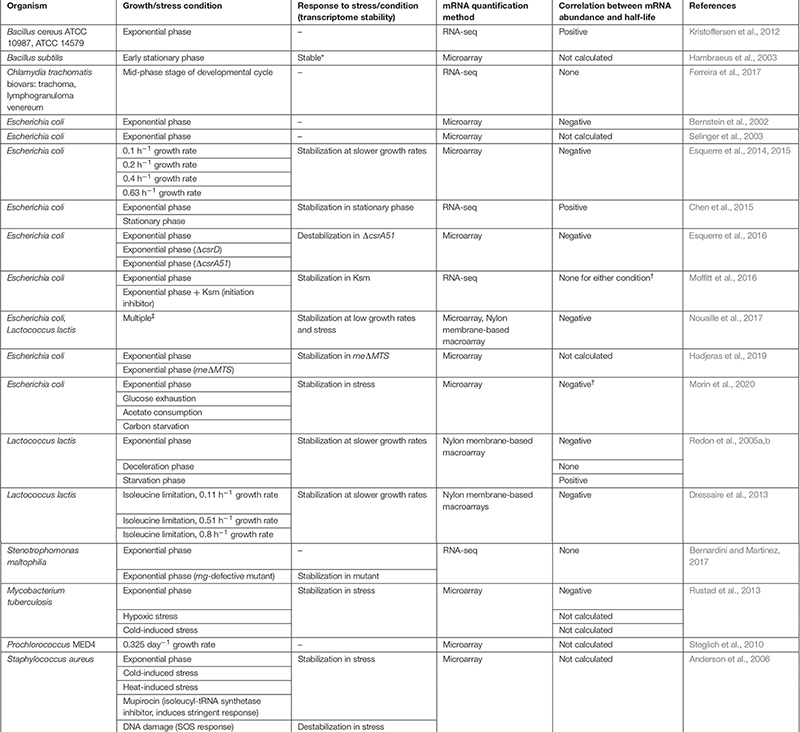
TABLE 1.
Transcriptome-wide studies on mRNA half-life in bacteria.
|
*Not compared to an exponential phase culture within the same study. Stabilization report based on previously reported studies. †Our analysis of the source data. ‡Includes data for L. lactis at growth rates of 0.09, 0.24, 0.35, and 0.47 h–1 (Dressaire et al., 2013); and unpublished and previously published data for E. coli at growth rates of 0.04, 0.11, 0.38, 0.51, and 0.80 h–1 and stationary phase (Esquerre et al., 2014).
FIGURE 1.
Environmental changes cause mRNA degradation rates to change in both global and gene-specific ways. Bacterial adaptation to many stressors, and other changes in environment, involve modulation of degradation rates of specific transcripts encoding proteins relevant to the changing conditions (top panel). Some stressors, particularly those causing severe energy stress, trigger global stabilization of the mRNA pool (bottom panel). These scenarios are not mutually exclusive; stressors that cause global transcriptome stabilization typically also cause gene-specific changes in relative degradation rates.
In this review, we will discuss a range of reported situations in which bacterial mRNA stability is modulated in response to various stress conditions, with a focus on known and suspected mechanisms underlying such regulation. We will also discuss the ways in which known gene-specific mechanisms shape our thinking on the unanswered question of how mRNA pools are globally stabilized in response to energy stress. Furthermore, we will discuss the ways in which regulation of mRNA stability in clinically relevant bacteria, such as Mycobacterium tuberculosis, shape their responses to the host environment.
RNases and Other Degradation Proteins
The Degradosome
RNA degradation is carried out by a wide range of RNases, enzymes with strong activities and relatively low specificities toward their targets (reviewed in Carpousis, 2007). There are two main types of RNases: endonucleases and exonucleases. The former cleave RNA sequences at internal points, while the latter carry out nucleolytic attacks from either end of the RNA chain (deemed 5′ or 3′ exonucleases based on their enzymatic directionality). Some bacteria possess both 5′ and 3′ exonucleases – M. tuberculosis and Mycobacterium smegmatis, for example – while others such as E. coli have only 3′ exonucleases.
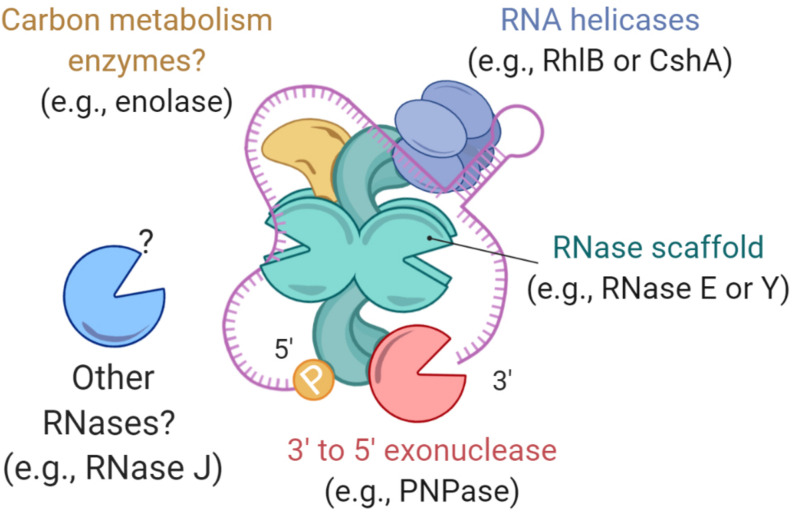
With respect to RNA degradation systems, E. coli is perhaps the most studied organism. In fact, it was in E. coli that a multiprotein complex, deemed the degradosome (Figure 2), was first reported (Carpousis et al., 1994; Py et al., 1994). In E. coli, the main degradosome components are two RNases (RNase E and PNPase), a DEAD-box RNA helicase (RhlB), and a glycolytic enzyme (enolase) (Carpousis et al., 1994; Py et al., 1994; Marcaida et al., 2006; Carpousis, 2007). RhlB facilitates RNase activity by unwinding stem-loops within RNA targets (Py et al., 1996). Both RNases carry out RNA degradation (Mohanty and Kushner, 2000; Deutscher, 2006; Unciuleac and Shuman, 2013). Moreover, in this bacterium the C-terminal region of RNase E acts as a scaffold for other degradosome components (Kido et al., 1996; Vanzo et al., 1998; Lopez et al., 1999; Morita et al., 2004). However, not all of the degradosome components are well defined or have known roles. For example, enolase is suspected to have a regulatory role in mRNA degradation under low phosphosugar levels (Morita et al., 2004; Chandran and Luisi, 2006) and anaerobic conditions (Murashko and Lin-Chao, 2017).
FIGURE 2.
Bacterial degradosomes. The bacterial degradosome is scaffolded by an RNase such as RNase E in E. coli and RNase Y in B. subtilis. The RNase scaffolds have catalytic domains and natively disordered scaffold domains that bind other degradosome proteins. Typical degradosome components in both gram-positive and gram-negative bacteria are RNA helicases, carbon metabolism enzymes, and other RNases.
While RNases can degrade RNA substrates on their own, it has been suggested that degradosomes increase the efficiency of RNA degradation, for example by facilitating processing of structures such as stem-loops and repeated extragenic palindromic sequences (Newbury et al., 1987; McLaren et al., 1991; Py et al., 1996). Alteration of the degradosome components leads to changes in transcriptome stability; for example, deletion of RhlB in E. coli results in longer mRNA half-lives (Bernstein et al., 2004). Similarly, mRNA stability is dramatically increased when the arginine−rich RNA binding region or the scaffolding region of RNase E are deleted (Kido et al., 1996; Ow et al., 2000). While the RNA degradosome of E. coli has been extensively studied, the composition and function of degradosomes in other gram-negatives and in gram-positives may differ, and new studies are still uncovering this information. In the Firmicute Bacillus subtilis, there is no RNase E homolog. Instead, RNase Y serves as a degradosome scaffold for PNPase, the helicase CshA (Lehnik-Habrink et al., 2010), phosphofructokinase (Commichau et al., 2009), and RNase J1 and RNase J2 – two bifunctional enzymes with both endonucleolytic and 5′–3′ exoribonuclease activity (Even et al., 2005; Shahbabian et al., 2009; Mathy et al., 2010; Durand et al., 2012). Interestingly, the B. subtilis degradosome interactions have been shown mainly by bacterial 2-hybrid assays and immunoprecipitation of complexes stabilized by formaldehyde crosslinking (Commichau et al., 2009; Lehnik-Habrink et al., 2010), in contrast to the E. coli degradosome which can be immunoprecipitated without a crosslinking agent (Carpousis et al., 1994; Py et al., 1994, 1996). This suggests that B. subtilis degradosomes could be more transient in nature. A recent report on the Actinomycete M. tuberculosis provided insight into its elusive degradosome structure, which appears to be composed of RhlE (an RNA helicase), PNPase, RNase E, and RNase J (Plocinski et al., 2019). Overall, the degradosome is considered to be the ultimate effector of bulk mRNA degradation in bacterial cells, but it has also been implicated in regulating the stability of specific mRNAs and sRNAs, as will be discussed in later sections. For further details on the degradosome, we encourage reading the following reviews (Carpousis, 2007; Bandyra et al., 2013; Ait-Bara and Carpousis, 2015; Cho, 2017; Tejada-Arranz et al., 2020).
An Overview of RNase Regulation
There are multiple ways in which transcript levels can be regulated. Alteration of mRNA steady-state abundance is ultimately a consequence of changes in transcription, changes in mRNA half-life, or both. In the process of mRNA degradation, the roles of different RNases may be defined in part by their preferred cleavage sequences. In Staphylococcus aureus, RNase Y cleavage is usually in the R↓W sequence, near AU rich regions (Khemici et al., 2015). This pattern seems to be conserved in B. subtilis (Shahbabian et al., 2009). Furthermore, in these two gram-positive organisms, RNase Y cleavage appears to be influenced by proximity to a secondary structure. In E. coli, RNase E cleaves single-stranded RNA with a strong preference for the +2 sites in RN↓AU (Mackie, 1992; McDowall et al., 1994), or in RN↓WUU in Salmonella enterica (Chao et al., 2017). In M. smegmatis, a strong preference for cleavage 5′ of cytidines was detected in a transcriptome-wide RNA cleavage analysis (Martini et al., 2019). RNase E could be responsible for these cleavage events, given its major role in mycobacteria, however, we cannot yet exclude the possibility that they are produced by another endonuclease. In contrast, RNase III in E. coli has optimal activity on double-stranded RNA, where the cleavage site is specified by both positive and negative sequence and secondary structure determinants (Pertzev and Nicholson, 2006). While the preferred cleavage sites of various RNases seem highly represented in the mRNA pool, some transcripts are more resistant to cleavage than others, indicating the presence of mechanisms that regulate not only bulk RNA stability, but also differential stabilities among transcripts.
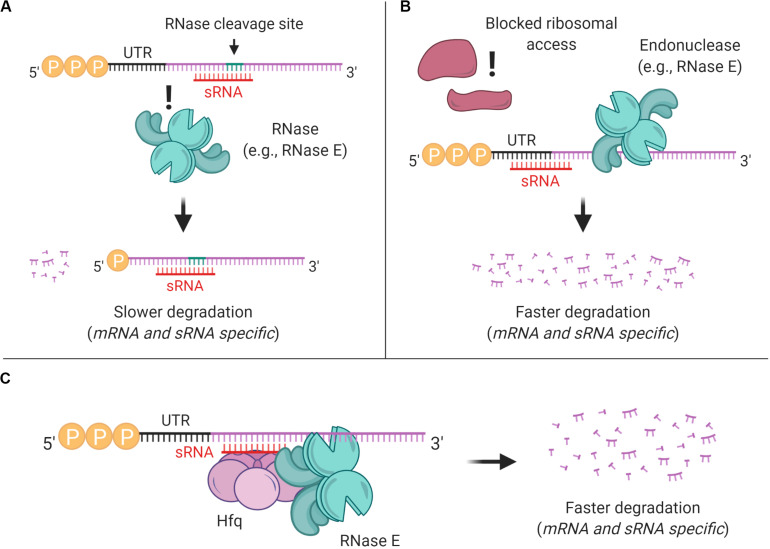
Studies of various mRNAs have identified multiple features that confer protection against RNase cleavage (Figures 3, 4A). These include stem-loops (Emory et al., 1992; McDowall et al., 1995; Arnold et al., 1998; Hambraeus et al., 2002), 5′ UTRs and leader/leaderless status (Chen et al., 1991; Arnold et al., 1998; Unniraman et al., 2002; Nguyen et al., 2020), subcellular compartmentalization (Khemici et al., 2008; Montero Llopis et al., 2010; Murashko et al., 2012; Khemici et al., 2015; Moffitt et al., 2016), 5′ triphosphate groups (Bouvet and Belasco, 1992; Emory et al., 1992; Arnold et al., 1998; Mackie, 1998), 5′ NAD+/NADH/dephospho-coenzyme A caps (Chen et al., 2009; Kowtoniuk et al., 2009; Bird et al., 2016; Frindert et al., 2018), NpnN caps (Luciano et al., 2019; Hudecek et al., 2020), and association with regulatory proteins and sRNAs (Braun et al., 1998; Gualerzi et al., 2003; Moll et al., 2003; Afonyushkin et al., 2005; Daou-Chabo et al., 2009; Nielsen et al., 2010; Morita and Aiba, 2011; Faner and Feig, 2013; Liang and Deutscher, 2013; Deng et al., 2014; Sinha et al., 2018; Zhao et al., 2018; Cameron et al., 2019; Chen H. et al., 2019; Richards and Belasco, 2019). For example, in Streptococcus pyogenes the sRNA FasX binds to the 5′ end of ska – a transcript coding for streptokinase – increasing its mRNA half-life, thus allowing an extended period of time in which translation of streptokinase can occur (Ramirez-Pena et al., 2010). In other cases, the product of an mRNA can regulate its own transcript stability. In E. coli, the fate of the lysC transcript is regulated by a dual-acting riboswitch that, under low levels of lysine, promotes translation initiation while simultaneously sequestering RNase E cleavage sites. In the presence of lysine, the riboswitch folds into an alternative conformation that exposes RNase E cleavage motifs, in addition to blocking translation (Caron et al., 2012). In these examples, it is ultimately the conformational structure of the mRNA that allows regulation of its half-life, independently from the stability of the bulk mRNA pool.
FIGURE 3.
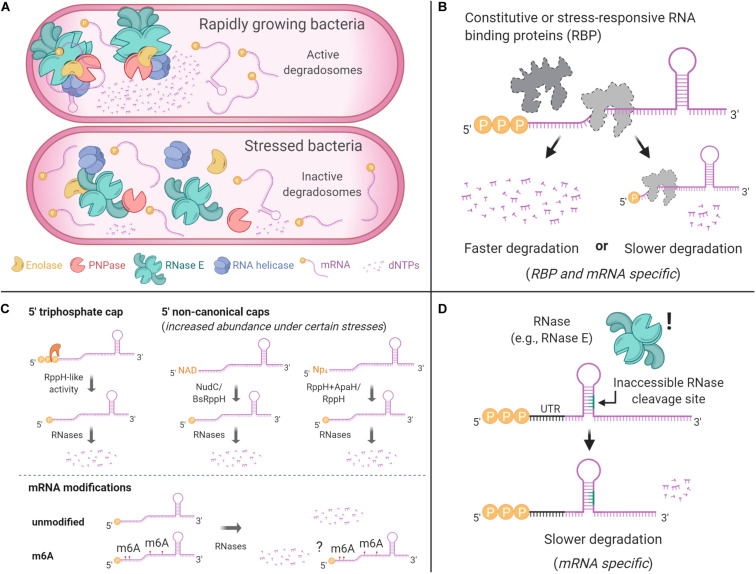
Common mechanisms that can protect mRNAs from degradation. (A) Degradosome localization can influence its RNA degradation activity. In E. coli, the degradosome is anchored to the cytoplasmic membrane via RNase E’s N-terminal domain, where it displays higher RNA processing activity in degradation foci. A cytoplasmic RNase E is less efficient in degradosome assembly and RNA processing. In B. subtilis, RNase Y is associated with the membrane and is more active when in smaller foci and less active when in larger foci. (B) RNA binding proteins can modulate mRNA degradation. Some of them, such as CsrA in γ-Proteobacteria, have regulatory roles as a response to environmental changes. (C) The chemical nature of mRNA 5′ ends can protect transcripts from degradation. These caps may vary depending on stress conditions. Nucleotide modifications in the bodies of transcripts have also been reported, but they have not been shown to alter mRNA stability. (D) RNA degradation depends on RNase accessibility to cleavage sites. Secondary structures that block cleavage sites can result in slower RNA degradation.
FIGURE 4.
sRNAs can affect mRNA stability through multiple mechanisms. (A) sRNA binding can mask preferred RNase cleavage sites, thereby stabilizing transcripts. (B) sRNA binding can block ribosome access to Shine-Dalgarno sites, reducing translation and typically destabilizing transcripts. (C) In E. coli and some other gram-negative bacteria, sRNA-mRNA pairing is often mediated by Hfq, which typically leads to mRNA degradation.
The activity of RNases does not always result in RNA decay. Some mRNA precursors can be processed by RNases to create mature, functional forms of the transcript (Condon et al., 1996). In a similar manner, polycistronic transcripts can be cleaved by endonucleases to produce transcripts with varying degrees of stability; some examples include (Belasco et al., 1985; Baga et al., 1988; Nilsson and Uhlin, 1991; Nilsson et al., 1996; Ludwig et al., 2001; Esquerre et al., 2014; Xu et al., 2015). While this is a fascinating mechanism of gene-specific regulation, it is beyond the scope of this review.
mRNA Stabilization as a Response to Stress
When bacteria are forced to slow or stop growth in response to stress, they must reduce their rates of protein synthesis. This can be done by direct modulation of translation or by regulation of transcription and transcript degradation rates. In recent decades, there have been many reports of mRNA stabilization as a response to different stressors, usually conditions that alter growth rate (see Table 1). In E. coli, the outer membrane protein A precursor transcript, ompA, is very stable in rapidly growing cells (Nilsson et al., 1984), but its half-life is significantly decreased in conditions of slow growth rate (Nilsson et al., 1984; Emory et al., 1992; Vytvytska et al., 2000). An inverse phenomenon was observed in stationary phase E. coli cells for
rpoS and rmf, transcripts coding for the transcription factor σ38 and the ribosome modulation factor, respectively (Zgurskaya et al., 1997; Aiso et al., 2005). Research conducted in other organisms also showed regulation of degradation rates of specific mRNAs according to growth rate: sdh, coding for succinate dehydrogenase in B. subtilis, and rpoS in Salmonella dublin had mRNA half-lives negatively correlated with growth rate (Melin et al., 1989; Paesold and Krause, 1999). Furthermore, cell growth studies using chemostats revealed that most transcripts in E. coli stabilize at low growth rates (Esquerre et al., 2014), with those belonging to the COGs “Coenzyme transport and metabolism” and “Intracellular trafficking, secretion and vesicular transport” being enriched among the most highly stabilized transcripts. On the other hand, genes in “Cell motility” and “Secondary metabolites biosynthesis, transport and catabolism” had shorter half-lives than the transcript population mean (Esquerre et al., 2015). This reinforces the ideas that transcript half-lives may be linked to gene function and can be regulated as conditions require. For example, in E. coli, genes from the COGs “Carbohydrate transport and metabolism” and “Nucleotide transport and metabolism” are amongst the most stable at normal growth rates (Esquerre et al., 2014, 2015, 2016). Although these findings propose a link between growth rate and mRNA stability, it is possible that metabolic status rather than growth rate per se is the key determinant of global mRNA stability. In M. smegmatis, a drug-induced increase in metabolic activity resulted in accelerated mRNA decay and vice versa, even though growth was halted in both conditions (Vargas-Blanco et al., 2019). Another study supported these findings, showing that mRNA stabilization upon changes in nutrient availability could be dissociated from changes in growth rate (Morin et al., 2020).
Growth rate is altered as a consequence of metabolic changes as bacteria adapt to different environments. Because the ultimate goal of an organism is to survive and multiply, we can assume that in stress conditions – such as low-nutrient environments – bacteria trigger mechanisms that regulate energy usage and preserve energetically expensive macromolecules, such as mRNA. Thus, transcript stabilization is a logical response to various forms of energy stress. Indeed, E. coli stabilizes most of its transcriptome in anaerobic conditions (Georgellis et al., 1993) as well as in carbon starvation and stationary phase (Esquerre et al., 2014; Chen et al., 2015; Morin et al., 2020). Studies on Rhizobium leguminosarum, Vibrio sp. S14, and Lactococcus lactis also showed increased transcriptome half-lives when the bacteria are subjected to nutrient starvation (Albertson et al., 1990; Thorne and Williams, 1997; Redon et al., 2005a,b). S. aureus induces global mRNA stabilization in response to low and high temperatures, as well as during the stringent response (Anderson et al., 2006). Under hypoxic conditions, the median mRNA half-life in M. tuberculosis increases from ∼9.5 min to more than 30 min, and cells shifted from 37°C to room temperature stabilized their transcriptomes so dramatically that half-lives could not be measured (Rustad et al., 2013). Similarly, transcript stabilization occurs in M. smegmatis in response to carbon starvation and hypoxia (Smeulders et al., 1999; Vargas-Blanco et al., 2019). Intriguingly, transcript destabilization can be resumed within seconds upon re-oxygenation of hypoxic M. smegmatis cultures, suggesting a highly sensitive mechanism regulating mRNA degradation in response to stress and energy status (Vargas-Blanco et al., 2019).
This response seems to be conserved even in some eukaryotes such as Saccharomyces cerevisiae, where the mRNA turnover rate is slower under stress than in log phase (Jona et al., 2000), and in plants as part of their immune response (Yu et al., 2019). However, the adaptive mechanism(s) underlying global mRNA stabilization as a stress response remain unknown. In the following sections we will discuss in more detail diverse bacterial strategies that contribute to global and gene-specific regulation of RNA stability. Our intent is to highlight recent findings on regulation of RNA degradation, to serve as a base for development of experiments to uncover how mRNA stabilization occurs as a response to stress.
Regulation of RNA Degradation Proteins
In this section we will discuss factors that have been shown to regulate the abundance and activity of endo- and exonucleases. We invite the reader to consult some excellent reviews (Condon, 2003; Arraiano et al., 2010; Bechhofer and Deutscher, 2019) for additional information on the roles and activities of RNases.
As we described in a previous section, RNases have preferred cleavage sequences. These patterns can be either masked or exposed by alternative RNA folding configurations as a result of intracellular changes, allowing modulation of specific cleavage events, e.g., the lysC riboswitch which is sensitive to lysine concentration (Caron et al., 2012). However, this regulatory paradigm tends to be used to control specific messages rather than the overall transcriptome stability. Hence, a major open question is: Are there elements that control RNase abundance or RNase activity that regulate transcriptome stability globally?
Abundance of key RNases that catalyze rate-limiting steps in mRNA degradation can affect bulk mRNA decay. For example, depletion or mutation of RNase E caused bulk mRNA stabilization in E. coli (Lopez et al., 1999; Sousa et al., 2001); depletion or mutation of RNase Y caused bulk mRNA stabilization in B. subtilis and S. pyogenes (Shahbabian et al., 2009; Chen et al., 2013); depletion of RNase J caused bulk mRNA stabilization in Helicobacter pylori (Redko et al., 2016); and deletion of RNases J1 and J2 caused mRNA stabilization in B. subtilis (Even et al., 2005). Mechanisms for regulation of RNase abundance have been reported in some bacteria. In E. coli, RNase III autoregulates its abundance by cleaving its own operon to induce its degradation when RNase III protein levels are high (Bardwell et al., 1989; Matsunaga et al., 1996, 1997; Xu et al., 2008). Similarly, in E. coli a stem-loop located in the 5′ UTR of rne responds to changes in RNase E levels, allowing this enzyme to autoregulate its own production (Diwa et al., 2000; Diwa and Belasco, 2002). There is evidence that in some cases, stability of other mRNAs can be regulated by changes in RNase abundance. In E. coli, the betT and proP transcripts, encoding osmoregulators, showed increased abundance and stability when cells were subject to osmotic stress, apparently as a consequence of lower RNase III concentrations (Sim et al., 2014). However, there is not yet evidence that global stress-induced mRNA stabilization can be attributed to reduced RNase abundance. In M. tuberculosis, a quantitative proteomics study comparing exponentially growing and hypoxic cultures showed no alteration in levels of RNase E, RNase J, RNase III, PNPase, or the helicase HelY even after 20 days under hypoxia (Schubert et al., 2015). Only one RNA helicase, RhlE, had reduced levels in hypoxia (Schubert et al., 2015). Similarly, a study of M. smegmatis showed no variation in levels of RNase E, PNPase, or the predicted RNA helicase msmeg_1930 under hypoxia, re-aeration, or exponential growth (Vargas-Blanco et al., 2019). Because mycobacterial transcriptomes are rapidly stabilized upon encountering hypoxia and other stress conditions (Rustad et al., 2013; Vargas-Blanco et al., 2019), it is unlikely that alteration of RNase abundance is part of the early RNA stabilization responses in these organisms.
It is possible that the activity of existing RNA degradation enzymes is regulated. RNA helicases are ATP-dependent, and ATP levels decrease in some bacteria in severe energy stress (Rao et al., 2008; Vargas-Blanco et al., 2019). This raises the possibility that RNA degradation could be directly modulated by ATP levels. However, when this hypothesis was tested in M. smegmatis, mRNA stabilization was found to occur prior to a decrease in intracellular ATP levels upon exposure to hypoxic conditions (Vargas-Blanco et al., 2019). While these findings suggest that nucleotide sensing – particularly changes in ATP concentrations – does not influence the initial global stabilization response in mycobacteria, it is possible that ATP concentrations or ATP/ADP ratios could be responsible for further stabilization in later stages of dormancy, and/or that ATP levels contribute to global mRNA stabilization in other bacteria. The roles of nucleotides associated with the stringent response are discussed separately below.
In E. coli, inhibition of RNase E activity by RraA and RraB (Regulator of ribonuclease activity A and B) result in increased bulk mRNA half-life (Lee et al., 2003). However, in the case of RraA, the effect was observed after a significant overexpression of the inhibitor (Lee et al., 2003), something not observed under stress. Alternatively, inhibition of RNase activity by other factors may regulate transcript degradation. RNase E was recently shown to have a 5′ linear scanning function, and its cleavage activity is impaired upon encountering obstacles, such as sRNAs or ribosomes (Richards and Belasco, 2019). Furthermore, in E. coli, the activity of RNase E has been shown to depend on its anchorage to the inner membrane (Figure 3A). YFP-tagged RNase E forms small foci localized at the inner membrane (Strahl et al., 2015) which are dependent on metabolic activity; in anaerobic conditions RNase E rapidly dissociates from the membrane and diffuses in the cytoplasm, a response apparently dependent on enolase (Murashko and Lin-Chao, 2017). A cytoplasmic version of RNase E was unstable, and led to increased mRNA half-lives (Hadjeras et al., 2019). Interestingly, the cytoplasmic RNase E was able to assemble a degradosome and had a comparable in vitro activity to wild type RNase E, supporting the role of membrane attachment and cellular localization in RNase E activity (Moffitt et al., 2016; Hadjeras et al., 2019). Conversely, in Caulobacter crescentus, RNase E is cytoplasmic and forms bacterial ribonucleoprotein (BR) bodies, which dynamically assemble and disassemble in the presence of mRNA (Al-Husini et al., 2018). BR body formation was dependent on the RNase E scaffold domains and the presence of mRNA, while disassembly of the bodies required mRNA cleavage (Al-Husini et al., 2018). Intriguingly, the formation of BR-bodies increased under some stress conditions but was unaffected by others, suggesting they play an as-yet undefined role in stress response (Al-Husini et al., 2018). Further work is needed to understand the extent to which RNase localization contributes to regulation of mRNA degradation rates in various species.
In B. subtilis, the activity of RNase Y appears to be regulated by both subcellular localization and association with proteins termed the Y-complex (YaaT, YlbF, and YmcA). The Y-complex affects expression of genes involved in biofilm formation, sporulation, and competence, and in some cases, this was shown to be a direct consequence of altered mRNA degradation rates for the relevant genes (Tortosa et al., 2000; Carabetta et al., 2013; DeLoughery et al., 2016; Dubnau et al., 2016). The Y complex has been viewed as a specificity factor for RNase Y, required in particular for processing of polycistronic transcripts (DeLoughery et al., 2018). RNase Y also localizes in the cell membrane, where it can form RNase Y foci (Hunt et al., 2006; Lehnik-Habrink et al., 2011; Hamouche et al., 2020). These foci seem to represent a less active form of the enzyme, as they increased in size in absence of RNA or in Y-complex mutants (Hamouche et al., 2020).
The Stringent Response and mRNA Degradation
The stringent response is perhaps one of the most well-studied mechanisms of prokaryotic stress adaptation. This response is modulated by guanosine-3′,5′-bisphosphate (ppGpp) and/or guanosine-3′-diphosphate-5′-triphosphate (pppGpp), alarmones collectively referred to as (p)ppGpp. In gram-negative bacteria, (p)ppGpp is synthesized by RelA in response to uncharged-tRNAs binding ribosomes, or by SpoT, a (p)ppGpp synthase/hydrolase, during fatty acid starvation (Seyfzadeh et al., 1993; Battesti and Bouveret, 2009). In some gram-positive bacteria, (p)ppGpp is synthesized by a dual RelA/SpoT homolog (Atkinson et al., 2011; Frederix and Downie, 2011; Corrigan et al., 2016). Once produced, (p)ppGpp halts the synthesis of stable RNA (tRNAs and ribosomes) while upregulating stress-associated genes and downregulating those associated with cell growth (Gentry et al., 1993; Chakraburtty and Bibb, 1997; Martinez-Costa et al., 1998; Avarbock et al., 2000; Artsimovitch et al., 2004; Corrigan et al., 2016). Intriguingly, (p)ppGpp was reported to inhibit PNPase in the actinomycetes Nonomuraea sp. and Streptomyces coelicolor but not in E. coli (Gatewood and Jones, 2010; Siculella et al., 2010), suggesting the stringent response may have a previously overlooked role in directly regulating mRNA degradation in some groups of bacteria. However, a recent study on the stringent response in M. smegmatis showed that (p)ppGpp was not required for mRNA stabilization in response to carbon starvation or hypoxia (Vargas-Blanco et al., 2019).
In the pathogen Borrelia burgdorferi, a connection between the stringent response and the expression of 241 sRNAs was recently stablished, 187 of which were upregulated during nutrient stress (Drecktrah et al., 2018). The authors of the aforementioned study described potential mechanisms of regulation by RelBbu on transcription and fate of some transcripts, such as destabilization of the glycerol uptake facilitator transcript, glpF. The SR0546 sRNA is among the sRNAs induced by nutrient starvation; the upregulation of its target, bosR, encoding a transcriptional regulator, may suggest a regulatory role of (p)ppGpp on specific mRNA stabilization. However, the effects of these stringent response-induced sRNAs on mRNA stability have not yet been directly tested.
A surprising role of RelZ (initially called MS_RHII-RSD), a dual (p)ppGpp synthase and RNase HII, was reported for M. smegmatis (Murdeshwar and Chatterji, 2012). R-loops (RNA/DNA hybrids) are harmful structures that cause replication stress and can be removed by the RNase H domain of RelZ, while stalled ribosome removal is attributed to their alarmone synthase domain. RelZ was shown to be upregulated under short UV exposure in M. smegmatis (Krishnan et al., 2016), and while its role is suspected to increase cell viability under stress conditions (Petchiappan et al., 2020), the stringent response seems to not intervene in transcriptome stability regulation. This pathway leads to degradation of transcripts involved in R-loops, but given the low frequency of R-loop formation, the effects on mRNA pools are likely to be minimal.
Overall, there is much evidence that the stringent response regulates expression of specific transcripts in various bacteria. However, the extent to which control of mRNA stability contributes to these effects is mostly untested. The stringent response also plays important roles in mediating global responses to starvation and other forms of energy stress, but there is not yet evidence that it contributes to global mRNA stabilization, which is a consistent component of these stress responses. This suggests that the stringent response may not be the mediator of global mRNA stabilization in response to stress, or that its involvement in this process is species-specific.
Transcript Modifications as Regulators of mRNA Decay
Bacterial mRNA is primarily transcribed using nucleoside triphosphates as initiating nucleotides, making mRNAs triphosphorylated at their 5′ ends. In S. aureus, RNase J1 exhibits strong in vitro exo- and endonucleolytic activities on 5′ triphosphorylated transcripts (Hausmann et al., 2017). However, in most other organisms studied to date, RNases E, J, and Y more efficiently cleave mRNAs with 5′ monophosphates (Figure 3C). RNase E is an endoribonuclease, but has a binding pocket for monophosphorylated 5′ ends (Callaghan et al., 2005) that strongly stimulates its activity in organisms including E. coli and M. tuberculosis (Mackie, 1998; Zeller et al., 2007). Similarly, in B. subtilis, RNase J1, and to a lesser extent J2, show a strong preference toward 5′ monophosphorylated substrates (Even et al., 2005). RNase Y also shows preference toward monophosphorylated 5′ substrates, but to a lesser extent (Shahbabian et al., 2009). These findings contributed to the discovery of RppH, an RNA pyrophosphohydrolase. Similar enzymes were later found in other bacteria, such as Bdellovibrio bacteriovorus (Messing et al., 2009) and B. subtilis (Richards et al., 2011). However, while the role of 5′ triphosphate pyrophosphohydrolysis was initially attributed to RppH (Celesnik et al., 2007; Deana et al., 2008), recent findings have shown that the primary substrate of RppH in E. coli is 5′ diphosphorylated RNAs, and that 5′ diphosphorylated RNAs are abundant in the transcriptome (Luciano et al., 2017). As RppH cannot convert 5′ triphosphates to diphosphates, this suggests the existence of an unknown 5′ triphosphate to diphosphate phosphorylase. Given that 5′ monophosphates make transcripts more susceptible to degradation in multiple organisms, one could envision regulation of 5′ triphosphate pyrophosphohydrolysis as a potential mechanism for regulation of mRNA stability. However, to our knowledge there are not yet reports of if and how pyrophosphohydrolysis or γ-phosphate removal are regulated.
The presence of non-canonical mRNA 5′ ends has recently been reported for subsets of mRNAs in several bacterial species, suggesting another possible mechanism for regulation of mRNA stability (Figure 3C). Examples include NADH and NAD+ (Chen et al., 2009; Cahova et al., 2015), and less commonly, dephospho-CoA, succinyl-CoA, acetyl-CoA, and methylmalonyl-CoA (Kowtoniuk et al., 2009). We will refer to these as 5′ caps, with the understanding that they are structurally and functionally distinct from eukaryotic mRNA caps. Other studies have shown additional types of 5′ capping, as well as potential mechanisms behind it (Bird et al., 2016; Zhang et al., 2016; Julius and Yuzenkova, 2017). In most cases, bacterial caps are incorporated directly into mRNAs during transcription initiation. RNA polymerase can initiate transcription with non-canonical nucleotides such as NAD in E. coli (Bird et al., 2016; Vvedenskaya et al., 2018) and B. subtilis (Frindert et al., 2018). Furthermore, E. coli RNA polymerase seems to initiate with dinucleoside tetraphosphates (Np4N), Np4A in particular, with an efficiency almost 60 times higher than for NAD (Luciano and Belasco, 2020). Alternative, posttranscriptional mechanisms may also contribute to Np4 capping formation, as in vitro experiments using LysU (lysyl-tRNA synthetase) from E. coli suggest (Luciano et al., 2019).
The intracellular concentration of Np4As were shown to be affected by overproduction of aminoacyl-tRNA synthetases (Brevet et al., 1989). Interestingly, some stress conditions also induce higher levels of Np4Ns, for example heat shock (Lee et al., 1983), oxidative stress (Bochner et al., 1984), cadmium stress (Coste et al., 1987; Luciano et al., 2019) and disulfide stress (Bochner et al., 1984; Luciano et al., 2019). 5′ mRNA decapping was shown to require Nudix enzymes, such as NudC and BsRppH, to hydrolyze NAD-RNA substrates (Hofer et al., 2016; Frindert et al., 2018). On the other hand, hydrolysis of Np4As requires RppH and ApaH, the latter carrying out the hydrolysis of Np4As into two NDPs (Farr et al., 1989); in this context ApaH generates a diphosphorylated 5′ end that can be readily converted to monophosphate 5′ end by RppH (Figure 3C). Non-canonical mRNA 5′ ends also occur when transcription initiates with short RNA degradation products, resulting in mRNAs with 5′ hydroxyls (Druzhinin et al., 2015). Such transcripts have been found in E. coli and Vibrio cholerae and are present at increased abundance in stationary phase (Vvedenskaya et al., 2012; Druzhinin et al., 2015). However, the effects of these alternate 5′ ends on transcript stability have not been reported.
Some mRNA caps have been shown to stabilize mRNAs in E. coli (Bird et al., 2016; Luciano et al., 2019) and in B. subtilis (Frindert et al., 2018). For example, after increasing the cellular concentration of Np4Ns in cadmium-stressed cells and in ΔapaH mutants, RNA stability was increased, suggesting that Np4 caps have a stabilizing role (Luciano et al., 2019). Additionally, in this study Np4 caps were suggested to be more abundant than NAD caps. Similarly, in the E. coli ΔnudC mutant strain there is an increase of up to fourfold in RNA stability for transcripts with non-canonical 5′ caps (Bird et al., 2016). Furthermore, NAD 5′ caps were almost twofold more abundant for cells in stationary phase when compared to exponential phase (Bird et al., 2016). Together, these findings present a potential mechanism for stabilization of mRNA under stress conditions. An interesting regulatory mechanism behind Np4 decapping in E. coli was recently linked to methylation in m7Gp4Gm and m6Ap3A 5′ caps, which protects them from RppH cleavage but not from AppH (Hudecek et al., 2020). Methylated NpnN caps were shown to be more abundant in stationary phase than exponential phase (Hudecek et al., 2020), consistent with the idea that these caps protect mRNA from degradation. Interestingly, the NpnN caps found in that study did not include Ap4N (Hudecek et al., 2020), presumably due to different stress conditions and detection techniques than those in Luciano et al. (2019). Since capped mRNAs appear to be generally more stable than canonical mRNAs, it is logical to infer that when stress conditions cause growth to slow or stop and transcription to slow or stop concomitantly, the proportion of capped mRNAs will increase as a result of their inherently longer half-lives. One could therefore speculate that the global mRNA stabilization observed in non-growing bacteria is due in part to an mRNA pool that is largely protected by 5′ caps. This is plausible assuming capping frequency remains constant or increases under stress. But, a recent study argues against this idea. Rapid transcript destabilization occurred in hypoxic M. smegmatis cultures after re-exposure to oxygen, even when transcription was blocked prior to re-aeration (Vargas-Blanco et al., 2019). Thus, mRNA capping does not explain the transcript stabilization observed in these conditions (early-stage hypoxia) – at least in M. smegmatis – but could be involved in mRNA stabilization in other conditions and/or other bacteria.
Another possible mechanism of mRNA stabilization involves posttranscriptional nucleotide modifications (Figure 3C). N6-methyladenosine (m6A) is a common base modification in mice and humans (Meyer et al., 2012; Linder et al., 2015). This methylation is enriched near stop codons and in 3′ UTRs (Yue et al., 2018), and is dependent on the consensus motif DRACH (Linder et al., 2015). Recent studies revealed m6A to be an important part of a transcript stability regulatory mechanism, as it facilitates mRNA degradation in association with RBP in mice, zebra fish, and human cells (Schwartz et al., 2014; Wang et al., 2014; Zhao et al., 2017). Moreover, the levels of m6A methylation are responsive to stress conditions, as shown for human cancer cells under hypoxic conditions (Panneerdoss et al., 2018), suggesting a posttranscriptional regulatory role. In E. coli and Pseudomonas aeruginosa, m6A is present at similar levels, ∼0.2–0.3% of adenines (Deng W. et al., 2015), to those reported for yeast and other eukaryotes (Wei et al., 1975; Bodi et al., 2010). However, in contrast to mammals, m6A appears distributed throughout the gene, with modest enrichments near the 5′ ends and centers of transcripts, and with a similar m6A motif for E. coli and P. aeruginosa (UGCCAG and GGYCAG, respectively) (Deng X. et al., 2015). Contrary to eukaryotes, m6A methylation has not been shown to have a global role in mRNA degradation in bacterial stress responses. A deep analysis in E. coli and P. aeruginosa revealed no difference in the m6A levels for cells growing in LB when compared to other (unspecified) growth media, or oxidative stress; interestingly, increasing the temperature from 37 to 45°C lowered m6A methylation levels, but only for P. aeruginosa (Deng X. et al., 2015). Furthermore, the m6A levels were lower in other bacteria (∼0.02–0.08%, for S. aureus, B. subtilis, Anabaena sp., and Synechocystis sp.) (Deng X. et al., 2015), suggesting that this particular base modification may not be conserved across bacteria. In E. coli, codon modifications of the ermCL mRNA with m6A blocked translation, though it had no impact on mRNA degradation rates (Hoernes et al., 2016). While it is conceivable that m6A has a role in the regulation of bacterial translation, current evidence does not suggest it regulates mRNA fate.
5-methylcytosine (m5C) has also been found in mRNA. In eukaryotes, m5C has been shown to increase transcript stability (Arango et al., 2018; Chen X. et al., 2019; Yang et al., 2019; Schumann et al., 2020), while reports on translation regulation are controversial (Huang et al., 2019; Yang et al., 2019; Schumann et al., 2020). m5C modifications have been found in mRNA and 23S rRNA in the archaeon Solfolobus solfataricus (Edelheit et al., 2013). However, there is no defined role of m5C in S. solfataricus, and evidence of m5C in bacteria or regulatory roles in RNA degradation have not been reported.
Another modification, and perhaps the most abundant in RNA, is pseudouridine (Ψ) (Rozenski et al., 1999). Ψ is present at the position U55 in all E. coli tRNAs (Gutgsell et al., 2000), and is widespread across kingdoms (Nishikura and De Robertis, 1981; Becker et al., 1997; Ishida et al., 2011). In E. coli, deletion of truB, encoding a tRNA Ψ 55 synthase (Nurse et al., 1995), was shown reduce viability after a temperature shock (37–50°C); however, no viability changes were observed during exponential growth at 37°C (Kinghorn et al., 2002). In Thermus thermophilus, a ΔtruB mutant showed a growth defect when cultured at 50°C (Ishida et al., 2011). Thus, it is possible that the presence of tRNA modifications under stress conditions contributes to survival in other bacteria. Other tRNA modifications have been also reported in bacteria and yeast during stress, contributing to a translational bias with implications for translation regulation (Chan et al., 2010, 2012; Laxman et al., 2013; Deng W. et al., 2015; Chionh et al., 2016). However, while stress may alter tRNA modifications, ultimately these changes lead to translational regulation without clear evidence, at least in bacteria, of effects on mRNAs. On the other hand, Ψ modifications on mRNA have been shown to increase mRNA stability in yeast and human cells (Carlile et al., 2014) and in Toxoplasma gondii (Nakamoto et al., 2017). A broad study involving E. coli and human cells found that even a single replacement of U with Ψ in mRNA can interfere with translation (Eyler et al., 2019). Whether these modifications ultimately regulate mRNA stability in bacteria as a response to stress is an open question. Based on evidence aforementioned for M. smegmatis regarding the rapidity of transcript destabilization after stress alleviation (Vargas-Blanco et al., 2019), we speculate that base modifications are unlikely to be the primary mechanism of mRNA stabilization in hypoxic mycobacteria, although it could play roles in other organisms or conditions.
Roles of Ribosomes, Translation, sRNAs, and RNA-Binding Proteins in Regulation of mRNA Decay
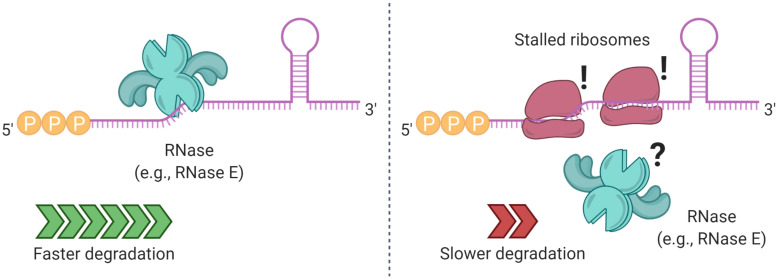
Experiments conducted by Bechhofer and others in B. subtilis showed that ribosome stalling can increase ermC half-life. In this scenario, ribosomes acted as obstacles at the 5′ ends of transcripts, resulting in protection from endonucleolytic cleavage downstream (Shivakumar et al., 1980; Bechhofer and Dubnau, 1987; Bechhofer and Zen, 1989). These findings would become early evidence of a 5′–3′ polarity for endonucleolytic activity, dependent upon or enhanced by (1) interaction with a 5′ monophosphate, and (2) RNase linear scanning mechanisms, as it would be later reported by others (Bouvet and Belasco, 1992; Jourdan and McDowall, 2008; Kime et al., 2010; Richards and Belasco, 2016, 2019). In E. coli, the use of puromycin or kasugamycin – translation inhibitors that cause ribosomes to dissociate from transcripts – caused faster mRNA decay in the absence of new transcription (Varmus et al., 1971; Pato et al., 1973; Schneider et al., 1978). On the other hand, the use of chloramphenicol, fusidic acid or tetracycline – elongation inhibitors that cause ribosomes to stall on transcripts – resulted in transcript stabilization (Varmus et al., 1971; Fry et al., 1972; Pato et al., 1973; Schneider et al., 1978), findings also later shown in M. smegmatis (Vargas-Blanco et al., 2019). These results are consistent with ribosome binding having a protective effect on mRNAs (Figure 5). In experiments where transcription was not blocked, it is possible that the mRNA stabilization seen in response to elongation inhibitors may also be conferred in part by the sudden increase in rRNA synthesis that these drugs cause, which increases the abundance of potential RNase substrates and could therefore titrate the activity of RNases such as PNPase and RNase E (Lopez et al., 1998). However, the increase in rRNA synthesis cannot fully explain these effects.
FIGURE 5.
Ribosome binding and stalling can alter mRNA degradation. In some cases, ribosome stalling can mask RNase cleavage sites, increasing the half-life of a transcript. Elements that prevent ribosome binding, such as translation initiation inhibitors, lead to shorter mRNA half-lives.
In B. subtilis, the stability of gsiB, encoding general stress protein, and ermC, encoding erythromycin resistance leader peptide, are associated with ribosome binding (Sandler and Weisblum, 1989; Hambraeus et al., 2000). Mutations to the RBS sites of gsiB, aprE (coding for subtilisin), and SP82 phage mRNA resulted in reductions of their mRNA half-lives (Hue et al., 1995; Jurgen et al., 1998; Hambraeus et al., 2002). Transcript stability conferred by ribosomes does not always require productive translation, at least for ermC (Hambraeus et al., 2002) and ompA (Emory and Belasco, 1990), where transcripts were stable in the absence of start codons as long as strong Shine-Dalgarno (SD) sequences were present (Arnold et al., 1998). A later study also in E. coli reported that ribosome protection is independent of translation for another transcript (Wagner et al., 1994). Transcript stabilization in a translation-independent manner was also shown for B. subtilis, with the insertion of an alternative SD (not involved in translation) to the gene reporter cryIII (Agaisse and Lereclus, 1996). These findings suggest that binding of a 30S subunit to a transcript, regardless of translation, may suffice to impair RNase degradation.
However, other studies did find a correlation between translation itself and stability. In E. coli, codon composition can influence translation rate and mRNA stability; codon-optimized transcripts were more stable than their corresponding non-modified, inefficiently-translated versions (Boel et al., 2016). Similar results were shown for S. cerevisiae (Presnyak et al., 2015). A transcriptome-wide analysis in E. coli also identified a positive correlation between mRNA stability and codon content optimality, for bacteria growing at different rates (Esquerre et al., 2015). This directly contradicted a previous report that codon optimality and half-life were inversely correlated (Lenz et al., 2011), possibly due to use of different codon optimality metrics. In B. subtilis, translation initiation is necessary to prevent swift degradation of the hbs transcript, which encodes the DNA binding protein HBsu (Daou-Chabo et al., 2009; Braun et al., 2017). In M. smegmatis and M. tuberculosis, RNase E cleaves the furA-katG operon, producing an unstable furA message that is rapidly degraded while the katG transcript is stabilized as it becomes readily accessible for translation (Sala et al., 2008). Overall, regulation of mRNA stability by translation initiation and SD strength seems to be gene-specific.
While it is generally accepted in E. coli that occlusion of RNase cleavage sites by ribosome occupancy may protect a transcript from degradation (Joyce and Dreyfus, 1998), ribosome association with mRNA has not been shown to regulate mRNA stability globally in response to stress. However, data from B. subtilis suggest an interesting mechanism by which RNase activity could affect translation and therefore mRNA degradation on a transcriptome-wide scale (Bruscella et al., 2011). The infC-rpmI-rplT operon, which encodes translation initiation factor 3 (IF-3) along with two ribosomal proteins, is expressed from two promoters. The resulting transcripts have different sensitivities to RNase Y, and the RNase Y-sensitive transcript is not competent for translation of IF-3. As a result, inhibition of RNase Y expression alters the relative abundance of the two transcript and causes reduced translation of IF-3. If this were to cause globally reduced translation due to IF-3 deficiency, mRNA decay could be globally increased as a result, although this effect would presumably be counteracted by the globally reduced RNase Y activity. Complex interplays between RNase levels and translation may therefore have the potential to globally impact mRNA decay in B. subtilis.
RNA-binding proteins (RBPs), stalled ribosomes, and SD-like sequences in close proximity to transcript 5′ ends can also alter mRNA fate (Sharp and Bechhofer, 2005). In B. subtilis, interaction of the RBP Glp with the 5′ UTR of glpD, encoding glycerol-3-phosphate dehydrogenase, increases the transcript’s stability (Glatz et al., 1996). Other RBPs can modulate the stability of target genes during stress conditions (Figure 3B). For example, H-NS, a histone-like protein, regulates the RNA stability of rpoS in E. coli and V. cholerae in stressful environments (Brescia et al., 2004; Silva et al., 2008; Wang et al., 2012). The carbon storage regulator CsrA is an RBP that regulates gene expression posttranscriptionally in E. coli and other γ-Proteobacteria in response to environmental changes, described in Timmermans and Van Melderen (2010) and Romeo and Babitzke (2018). CsrA regulatory roles are best studied in E. coli. The glgCAP transcript, encoding genes implicated in the biosynthesis of glycogen, is destabilized when bound by CsrA (Liu et al., 1995). This response is halted when E. coli enters stationary phase, where CsrA is sequestered by the sRNA CsrB in a ribonucleoprotein complex (Liu et al., 1997). Conversely, CsrA was shown to stabilize some transcripts. CsrA directly binds the pgaA transcript, increasing its half-life along with the rest of the pgaABC polycistron, encoding genes associated to biofilm formation (Wang et al., 2005). Similarly, CsrA stabilizes the flhDC transcript, encoding the flagellar activation genes FlhD2C2 (Wei et al., 2001). More recently, a transcriptome-wide study together with bioinformatics predictions showed a major role for CsrA as an mRNA stabilization factor in E. coli (M9 minimal media, doubling time of 6.9 h) for more than a thousand transcripts, of which many were predicted to have at least one putative CsrA binding site (Esquerre et al., 2016). CsrA could directly bind transcripts and protect them from RNases, or could affect mRNA stability indirectly by modulating expression or activity of other post-transcriptional regulators, e.g., the RNA chaperone Hfq, encoded by hfq. In E. coli, CsrA can bind the hfq mRNA at a single binding site that overlaps its SD region, preventing ribosome access and decreasing its half-life, however, in stationary phase CsrA is sequestered, allowing higher expression of Hfq (Baker et al., 2007). Regulatory roles for CsrA in gram-positive bacteria have only recently been reported. In B. subtilis, CsrA mediates the interaction of the sRNA SR1 and the ahrC mRNA, encoding a transcription regulator of arginine metabolism, to regulate the expression of the arginine catabolic operons (Muller et al., 2019). However, CsrA-SR1 only mildly increased ahrC half-life, and it had no impact on SR1 degradation, indicating that the regulation was primarily at the level of protein synthesis (Muller et al., 2019).
The homohexameric Hfq, highly studied in E. coli and present in a large number of bacteria (Sun et al., 2002), is an important regulator of mRNA-sRNA pairing. The multiple roles of Hfq include modulation of sRNA-mediated translation blockage or promotion, and regulation of transcript degradation as a direct consequence of altered translation or through translation-independent mechanisms. For example, guiding a cognate sRNA to the 5′ region of mRNAs can result either in translation disruption by preventing the 30S subunit from binding (Figure 4B), or the opposite outcome by disruption of stem-loops that inhibit its binding (Wassarman et al., 2001; Arluison et al., 2002; Moller et al., 2002; Schumacher et al., 2002; Zhang et al., 2003; Afonyushkin et al., 2005; Sittka et al., 2008). Hfq can also allow RNase E access to specific mRNAs, or modulate the synthesis of Poly(A) tails, assisting PNPase in 3′–5′ degradation, as it will be discussed shortly. The physical properties, sequence specificity, protein interaction partners, sRNAs/mRNAs binding kinetics, and other important aspects of Hfq function will not be described here, as they are well described elsewhere; we refer the reader to the following detailed reviews (Vogel and Luisi, 2011; Updegrove et al., 2016; Kavita et al., 2018; Santiago-Frangos and Woodson, 2018).
A common outcome of Hfq sRNA/mRNA interactions is specific regulation of mRNA half-life (Figure 4C). For example, the destabilization of ptsG, encoding a glucose permease, in E. coli is mediated by the sRNA SgrS as a response to phosphosugar accumulation (Vanderpool and Gottesman, 2004). Similarly, degradation of ompA was also shown to be impacted by the specific binding of the sRNA MicA to its translational start site, blocking binding of the 30S ribosomal subunit and recruiting Hfq to promote RNase E cleavage (Lundberg et al., 1990; Vytvytska et al., 2000; Udekwu et al., 2005). While the regulatory roles of Hfq are widely accepted for other gram-negative bacteria as well (Sonnleitner et al., 2006; Cui et al., 2013), in gram-positive bacteria Hfq is less well characterized. Hfq rescue experiments in E. coli and S. enterica serovar Typhimurium using Hfq from B. subtilis and S. aureus, respectively, failed at rescuing the phenotypes (Vecerek et al., 2008; Rochat et al., 2012). These findings suggest important structural and/or functional differences in Hfq across evolutionarily divergent groups of bacteria. A study in B. subtilis found that the absence of Hfq does not impair growth under almost 2000 conditions including different carbon, nitrogen, phosphorus and sulfur sources, osmolarity or pH changes in a large phenotypic analysis (Rochat et al., 2015). Similar findings were shown for S. aureus (Bohn et al., 2007). However, Hfq became necessary for survival in stationary phase (Hammerle et al., 2014; Rochat et al., 2015). Surprisingly, the absence of Hfq in rich media conditions did not alter the transcriptome of B. subtilis (Rochat et al., 2015), while in minimal media, 68 mRNAs and a single sRNA were affected (Hammerle et al., 2014). Both of these studies reported transcriptome changes in the absence of Hfq for B. subtilis in stationary phase, particularly for sporulation and TA systems. Nevertheless, these changes do not necessarily confer fitness or increased survival (Rochat et al., 2015). Overall, while Hfq was shown to impact the B. subtilis transcriptome under certain stress conditions, its role as a regulator of transcript stability seems to greatly vary across species. In another gram-positive, the pathogen Listeria monocytogenes, Hfq interacts with the sRNA LhrA, increasing its stability and controlling the fate of its target mRNAs. But, ∼50 other sRNA seem to function in an Hfq-independent manner (Christiansen et al., 2006; Nielsen et al., 2010; Nielsen et al., 2011). Unexpectedly, hypoxia, stationary phase and low temperature (30°C) did not affect sRNA levels in a Δhfq strain (Toledo-Arana et al., 2009). Hence, it seems that Hfq may have a smaller role in control of mRNA stability, and an overall restricted role in sRNA/mRNA regulation in gram-positive bacteria; and it appears to not be required at all in some bacteria, such as mycobacteria, that lack identified Hfq orthologs (Sun et al., 2002).
mRNA Folding Alters mRNA Decay
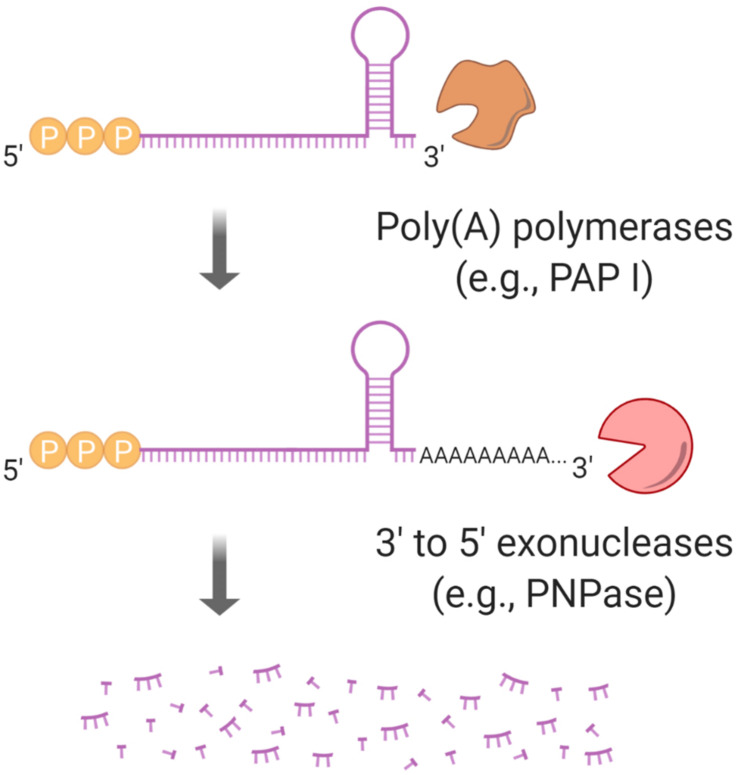
mRNA secondary structures can modulate translation and transcript stability (Figure 3D). Previously, we have discussed how specific 5′ UTR folding prevents RNase and ribosome accessibility to the lysC transcript (Caron et al., 2012). In other transcripts, secondary structures can also prevent RNase E from carrying out the first endonucleolytic cleavage, delaying subsequent steps in the decay pathways. In Rhodobacter capsulatus, formation of multiple hairpins can prevent endonucleolytic cleavage of the puf operon (Klug and Cohen, 1990). A stem-loop at the 5′ UTR confers stability to recA, coding for the nucleoprotein filament RecA in Acinetobacter baumannii (Ching et al., 2017), as well as vacA, coding for vacuolating cytotoxin A in Helicobacter pylori (Amilon et al., 2015). In the case of vacA, the stem-loop is also essential for transcript stabilization in acidic and osmotic stress (Amilon et al., 2015). The distance between the start codon and secondary structures can also affect mRNA half-life, as was shown for the ΔermC mRNA in B. subtilis, where placing a stem-loop too close to the SD decreased transcript stability (Sharp and Bechhofer, 2005). Secondary structure at transcript 3′ ends also affects stability. The mRNA 3′ end hairpins formed by Rho-independent transcriptional terminators typically stabilize transcripts, as 3′–5′ RNases have difficulty initiating decay without a single-stranded substrate (Adhya et al., 1979; Farnham and Platt, 1981; Abe and Aiba, 1996). In E. coli, the poly(A) polymerase (PAP I) is an enzyme responsible for synthesizing poly(A) tails in mRNA (Li et al., 1998). The addition of poly(A) tails to bacterial mRNAs facilitates degradation of transcripts with 3′ hairpins, allowing PNPase – an enzyme that also has a minor polyadenylation role – and other enzymes to carry out exonucleolytic activity (Donovan and Kushner, 1986; Blum et al., 1999; Figure 6).
FIGURE 6.
Polyadenylation regulates mRNA half-life. Stem-loops at mRNA 3′ ends block 3′–5′ exoribonucleases such as PNPase. PAP I, a poly(A) polymerase, can facilitate an exoribonuclease “grip” by synthesizing a poly(A) tail.
Thus, it is possible for poly(A) tails to act as regulators of mRNA stability, making PAP I a promising candidate for posttranscriptional regulation. However, while this enzyme has been characterized in E. coli, PAP I homologs in B. subtilis have not yet been identified (Campos-Guillen et al., 2005). An interesting role of Hfq in E. coli was reported for transcripts carrying long poly(A) tails, as binding to the tail prevents the access of PNPase, thereby increasing mRNA stability (Hajnsdorf and Regnier, 2000; Folichon et al., 2005). However, on shorter poly(A) tails (<10 nt), Hfq has poor accessibility, making the transcripts susceptible to the activity of PNPase and RNase II (Regnier and Hajnsdorf, 2013). Interestingly, in E. coli, the absence of PAP I disrupts the regulatory role of some sRNAs, leading to an unexpected destabilization of some sRNAs and transcripts, e.g., RyhB and MicA (Sinha et al., 2018). This appears to result from accumulation of transcripts that are normally degraded in a PAP I-dependent fashion. The accumulated transcripts participate in non-specific interactions with sRNAs, leading to degradation of the sRNA-mRNA pairs. Thus, it is suggested that many PAP I targets are transcripts that do not normally interact with sRNAs (Cameron et al., 2019).
Regulation of PNPase abundance has been shown for E. coli, as its transcript pnp is post-transcriptionally regulated by its own product and RNase III. This mechanism can be disrupted by transcript association with the ribosomal protein S1 (Briani et al., 2008; Carzaniga et al., 2015). Moreover, an increase of the pool of polyadenylated transcripts increases pnp half-life, an effect attributed to PNPase titration (Mohanty and Kushner, 2000, 2002). Regardless of this autoregulatory characteristic, changes in PNPase abundance were not detected as a response to hypoxic stress in M. smegmatis (Vargas-Blanco et al., 2019), despite increased mRNA stability. While these findings suggest that regulation by mRNA polyadenylation via PNPase abundance is not a mechanism of transcriptome stabilization in mycobacteria, it is possible that polyadenylation activity by other enzymes, such as PcnA and PcnB, Adilakshmi et al. (2000) might have a role in regulation of mRNA turnover in stress. Further research is needed to investigate this possibility.
The Relationship Between mRNA Abundance and mRNA Decay Rates
In bacteria, the steady-state mRNA concentration is a function of transcription rates and transcript degradation rates, and to a lesser extent, of mRNA dilution. The contribution of mRNA dilution occurring during cell growth is usually ignored, given that doubling times are significantly longer than the median mRNA half-life. For example, in L. lactis mRNA half-lives complied with this assumption for 85% of the measured transcripts, at multiple growth rates (Dressaire et al., 2013). In stress conditions, bacterial growth is generally impaired, making the impact of mRNA dilution even smaller and reinforcing the roles of transcription and RNA turnover as the major determinants of mRNA abundance. Also under stress conditions, transcript abundance per cell is typically lower than in conditions of rapid growth. For example, low transcript abundance was observed for S. aureus in cold shock, heat shock, and stringent response when compared to unstressed exponential phase (Anderson et al., 2006). The per-cell mRNA concentration decreased in L. lactis during progressive adaptation to carbon starvation (Redon et al., 2005a) or isoleucine starvation (Dressaire et al., 2013). The mRNA concentration was three times higher for E. coli growing in LB when compared to growth in in minimal media (Bartholomaus et al., 2016). For M. smegmatis in early hypoxic stress, the levels of atpB, atpE, rnj, rraA, and sigA ranged between ∼5 and 75% of those in cells growing in aerobic conditions, and after extended periods of hypoxic or carbon starvation stress, mRNA levels dropped to under 5% of those in log phase (Vargas-Blanco et al., 2019). Given the generally longer half-lives of mRNAs in stressed bacteria, the observation of reduced mRNA concentrations in these conditions may seem counter-intuitive. However, these observations can be reconciled if transcription is also greatly reduced. It is possible that maintaining lower overall mRNA abundance in stress conditions is an adaptive mechanism to favor translation of genes needed for survival of that particular stressor. For example, in a transcriptome-wide study in E. coli, mRNA abundance decreased in response to osmotic stress (from ∼2,400 to 1,600 transcripts per cell), a change that may allow specific transcripts – associated with stress response – to be more accessible to ribosomes and translated (Bartholomaus et al., 2016). Interestingly, transcripts with higher copy numbers per cell in normal conditions (>2 copies/cell) were downregulated the most in osmotic stress (Bartholomaus et al., 2016).
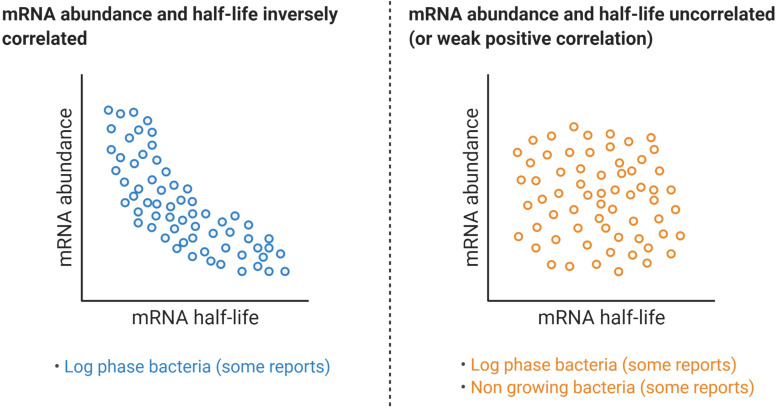
The question has arisen if lower mRNA concentrations can actually cause their degradation to be slowed. This idea is suggested by an observation made by several groups, in several species, that in log phase growth, mRNA half-lives are inversely correlated with steady-state abundance (Figure 7). For example, a weak negative correlation was shown between mRNA concentration and mRNA half-life for E. coli cells in exponential phase (Bernstein et al., 2002). Stronger negative correlations were reported in L. lactis (Redon et al., 2005b), and in M. tuberculosis (Rustad et al., 2013), both in exponentially growing bacteria. Moreover, in the latter study the overexpression of genes in the DosR regulon resulted in transcripts with shorter half-lives. Other reports in E. coli and L. lactis showed that cells growing at different growth rates also show a negative correlation between these parameters (Dressaire et al., 2013; Esquerre et al., 2015). For example, changes in growth rate from 0.1 to 0.63 h–1 – using chemostats – resulted in increased mRNA levels and a decreased median mRNA half-life from 4.2 to 2.8 min, respectively (Esquerre et al., 2014, 2015). Transcription modulation using five constructs with distinct 5′ UTRs in lacLM mRNA also depicted a similar trend in L. lactis in exponential phase, and a similar outcome was obtained for lacZ in E. coli, using PBAD-mediated transcription regulation (Nouaille et al., 2017). Two of the studies described here (Rustad et al., 2013; Nouaille et al., 2017) reported inverse relationships between mRNA abundance and half-life in defined systems where expression was modulated by inducible promotors and growth rate was not affected. This strongly suggested that transcription rate can directly influence degradation rate. However, contradictory findings have been reported.
FIGURE 7.
Relationships between mRNA abundance and mRNA decay rates. While some reports have shown a clear negative correlation between a transcript half-life and its abundance, a similar number of reports have found no correlation at all or a modest positive correlation, even for the same organism. Table 1 compiles transcriptome-wide analyses of mRNA decay in different organisms, techniques used, and information on the reported relationships between mRNA abundance and mRNA half-life.
An E. coli transcriptome-wide mRNA half-life study by a different group reported that the rate of mRNA degradation had a very weak positive correlation with mRNA abundance for both exponential phase (R2 = 0.07) and stationary phase (R2 = 0.19) (Chen et al., 2015), in contrast to other E. coli studies (Bernstein et al., 2002; Esquerre et al., 2014, 2015). In Bacillus cereus, mRNA half-life had a positive correlation with expression level (Kristoffersen et al., 2012), while in Stenotrophomonas maltophilia and Chlamydia trachomatis trachoma and lymphogranuloma venereum biovars no correlations were found (Bernardini and Martinez, 2017; Ferreira et al., 2017). In M. smegmatis, induced overexpression of dCas9 (in the absence of a gene-targeting sgRNA) did not alter its half-life in log phase (Vargas-Blanco et al., 2019). Surprisingly, overexpressing dCas9 under hypoxic stress increased its mRNA stability by approximately twofold (Vargas-Blanco et al., 2019). Moreover, re-exposure of hypoxic M. smegmatis cultures to oxygen caused half-lives of several tested genes to immediately return to log-phase like levels, despite transcription being blocked by rifampicin and transcript levels therefore remaining low (Vargas-Blanco et al., 2019). Other reports have indicated that the relationship between mRNA abundance and half-life differs in various stress conditions. In carbon-starved L. lactis there was a positive correlation between mRNA degradation and abundance (Redon et al., 2005b), while the opposite was observed during isoleucine starvation (Dressaire et al., 2013). Work in eukaryotes suggests complexities that could conceivably occur in bacteria as well. In S. cerevisiae, under DNA damaging conditions, upregulated genes are usually stabilized and repressed genes are prone to degradation (Shalem et al., 2008). Conversely, under oxidative stress upregulated genes are destabilized, with the opposite scenario for repressed genes (Shalem et al., 2008). Furthermore, an in-depth analysis in that work revealed a trend between these two stress conditions: Genes with a rapid transcriptional regulation show a negative correlation between mRNA abundance and mRNA degradation. On the other hand, genes subject to a slow transcriptional response follow a positive correlation between mRNA abundance and degradation (Shalem et al., 2008).
Clearly, further work is needed to reconcile contradictory findings in bacteria with respect to the relationships between mRNA abundance and stability. Some reported differences may be attributable to differences between species, while others may result from differences in methodology for measuring half-life. Most studies measure half-life by measuring decreases in mRNA abundance following transcription blockage by rifampicin. Variability may arise from the time-points chosen to assay abundance following transcriptional block, given that we and others have reported multiphasic decay kinetics (Hambraeus et al., 2003; Selinger et al., 2003; Chen et al., 2015; Nguyen et al., 2020). Methodology for normalization and for calculating half-lives also vary (see Table 1).
The Importance of RNA Decay in Clinically Important Species
Pathogenic bacteria have developed mechanisms that allow them to survive often-hostile host environments by sensing cues and mounting specific responses at both transcriptional and posttranscriptional levels. These pathogens exhibit highly specific responses to some stressors, as well as broader responses to conditions such as energy stress, where resources are preserved by global modulation of processes including translation, protein degradation, transcription, and RNA stabilization (Bohne et al., 1994; Sherman et al., 2001; Park et al., 2003; Christiansen et al., 2004; Wood et al., 2005; Papenfort et al., 2006; Liu et al., 2010; Fritsch et al., 2011; Galagan et al., 2013; Guo et al., 2014; Sievers et al., 2015; Quereda et al., 2018; Ignatov et al., 2020).
In L. monocytogenes, PrfA serves as a transcriptional regulator of multiple virulence factors, such as phospholipases PlcA and PlcB, and the toxin listeriolysin O (Leimeister-Wachter et al., 1990, 1991; Quereda et al., 2018). Expression of PrfA itself is regulated by several mechanisms at the translational and transcriptional level. For example, PrfA translation is temperature-regulated by a stem-loop in its transcript, prfA, that prevents ribosome access to the SD sequence at 30°C but not at 37°C (Johansson et al., 2002). prfA is also regulated by an S-adenosylmethionine riboswitch and its product, the sRNA SreA, that blocks translation after binding the 5′ UTR (Loh et al., 2009). Additionally, while the stem-loop increases prfA stability (Loh et al., 2012), the binding of SreA to prfA triggers transcript degradation (Loh et al., 2009). Also in L. monocytogenes, posttranscriptional regulation of Tcsa, the T cell-stimulating antigen encoded by tcsA, was recently reported to be under the control of the sRNA LhrC in a translation-independent manner, by recruiting an undefined RNase (Ross et al., 2019). In S. aureus SarA, a histone-like protein, influences mRNA turnover of virulence factors, such as protein A (spa) and the collagen adhesion protein (cna) during exponential growth (Roberts et al., 2006; Morrison et al., 2012). Also in S. aureus, the multifunctional RNAIII binds other RNAs, recruiting RNase III to initiate transcript degradation. Some of RNAIII’s targets are spa, coa (encoding coagulase), sbi (encoding the IgG-binding protein Sbi), and SA1000 (encoding the fibrinogen-binding protein SA1000) (Huntzinger et al., 2005; Boisset et al., 2007; Chevalier et al., 2010), playing an important role in S. aureus virulence and response to stress. In S. enterica, under low Mg2+ conditions synthesis of the antisense AmgR RNA leads to interaction and destabilization of the mgtC transcript (encoding the virulence protein MgtC), in an RNase E-dependent manner (Lee and Groisman, 2010). Hence, regulation of the stabilities of specific mRNAs has a major role in the survival and virulence responses of pathogens.
Recent reports have suggested unexpected relationships between RNases and drug resistance. Nonsense and INDEL mutations in Rv2752c, encoding RNase J, were associated with drug resistance in a GWAS study that identified resistance-associated mutations in whole-genome sequences of hundreds of M. tuberculosis clinical isolates (Hicks et al., 2018), as well as an earlier study performing similar analyses on a smaller set of clinical isolates (Zhang et al., 2013). Another study, reporting whole-genome sequences of 154 M. leprae clinical isolates from 25 countries, found a disproportionately high number of polymorphisms in ML1040c, encoding RNase D, and ML1512c, encoding RNase J (Benjak et al., 2018). These mutations were not directly associated with drug resistance, but appeared to be under positive selection (Benjak et al., 2018).
Global mRNA stabilization is another feature associated with bacterial stress response and non-growing conditions (see Table 1). Cells in quiescent states contain relatively low levels of mRNA, with greatly reduced transcriptional and translational activity (Betts et al., 2002; Wood et al., 2005; Kumar et al., 2012; Rittershaus et al., 2013). In some cases, these states share similarities with B. subtilis spores, in which the bacteria have dramatically reduced mRNA turnover (Segev et al., 2012). This can be interpreted as a concerted cellular effort to downregulate global gene expression and preserve cellular resources, until encountering a suitable environment to resume growth. At the same time, having paused translational machinery may permit allocation of resources toward specific responses needed to survive a given condition, such as those described in the previous paragraph. Importantly, stress responses that establish and maintain non-growing states not only allow pathogens to survive these stressors, but also induce broad antibiotic tolerance, since most antibiotics are relatively ineffective at killing non-growing cells (for example, Rao et al., 2008). This relationship between growth arrest and antibiotic tolerance may be one of the reasons why months of multidrug therapy are required to prevent relapse in tuberculosis patients, where large numbers of bacteria are likely semi-dormant in hypoxic granulomas (Garton et al., 2008). The apparent universality of mRNA stabilization as a response to energy stress and other stressors that inhibit growth, compared to gene-specific mRNA regulation, brings up fascinating possibilities as a prospective target for therapeutic development. There has been a surge in antimicrobial resistance in recent decades, prompting collaborative efforts between academia and industry to develop new antimicrobials (Ventola, 2015a,b; World Health Organization [WHO], 2019). As we approach an understanding of the mechanisms behind mRNA turnover – and strive to unveil how transcript fate is regulated under stress conditions – we would like to emphasize the essentiality of mRNA degradation in bacteria, and the roles of RNases in the virulence and survival responses of pathogens. Many clinically important antibiotics target transcription and translation, highlighting the potential of targeting these central dogma processes from the opposite angle. In early steps in this direction, a protein degradation inhibitor was found to have strong activity against mycobacteria (Gavrish et al., 2014) and inhibitors of RNase E have been reported (Kime et al., 2015).
Conclusion
Transcriptome stabilization as a stress response is widespread across the bacterial domain. This globally concerted response is implicated in gene regulation and survival, as well as pathogenesis in bacteria. We have described and discussed various mechanisms of mRNA degradation and stabilization, many of which have established roles in regulation of specific genes, but have not yet been able to explain transcriptome-wide half-life alterations. We hope that the information presented here helps to inspire further study that will uncover the mechanism(s) behind global transcriptome stabilization in stress, which so far remains elusive. Finally, we hope to inspire the reader to find these mysteries as scientifically stimulating as we do.
Author Contributions
DV-B and SS wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Shell lab for technical assistance and helpful discussions. The figures were created with BioRender.com.
Footnotes
Funding. This work was supported by the NSF CAREER award 1652756 to SS. DV-B was partially supported by the Fulbright Foreign Student Program.
References
- Abe H., Aiba H. (1996). Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie 78 1035–1042. 10.1016/s0300-9084(97)86727-2 [DOI] [PubMed] [Google Scholar]
- Adhya S., Sarkar P., Valenzuela D., Maitra U. (1979). Termination of transcription by Escherichia coli RNA polymerase: influence of secondary structure of RNA transcripts on rho-independent and rho-dependent termination. Proc. Natl. Acad. Sci. U.S.A. 76 1613–1617. 10.1073/pnas.76.4.1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adilakshmi T., Ayling P. D., Ratledge C. (2000). Polyadenylylation in mycobacteria: evidence for oligo(dT)-primed cDNA synthesis. Microbiology 146(Pt 3), 633–638. 10.1099/00221287-146-3-633 [DOI] [PubMed] [Google Scholar]
- Afonyushkin T., Vecerek B., Moll I., Blasi U., Kaberdin V. R. (2005). Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33 1678–1689. 10.1093/nar/gki313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H., Lereclus D. (1996). STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20 633–643. 10.1046/j.1365-2958.1996.5401046.x [DOI] [PubMed] [Google Scholar]
- Aiso T., Yoshida H., Wada A., Ohki R. (2005). Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J. Bacteriol. 187 1951–1958. 10.1128/JB.187.6.1951-1958.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Bara S., Carpousis A. J. (2015). RNA degradosomes in bacteria and chloroplasts: classification, distribution and evolution of RNase E homologs. Mol. Microbiol. 97 1021–1135. 10.1111/mmi.13095 [DOI] [PubMed] [Google Scholar]
- Albertson M. H., Nyström T., Kjelleberg S. (1990). Functional mRNA half-lives in the marine Vibrio sp. S14 during starvation and recovery. Microbiology 136 2195–2199. 10.1099/00221287-136-11-2195 [DOI] [Google Scholar]
- Al-Husini N., Tomares D. T., Bitar O., Childers W. S., Schrader J. M. (2018). alpha-proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71 1027–1039.e14. 10.1016/j.molcel.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amilon K. R., Letley D. P., Winter J. A., Robinson K., Atherton J. C. (2015). Expression of the Helicobacter pylori virulence factor vacuolating cytotoxin A (vacA) is influenced by a potential stem-loop structure in the 5′ untranslated region of the transcript. Mol. Microbiol. 98 831–846. 10.1111/mmi.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. L., Roberts C., Disz T., Vonstein V., Hwang K., Overbeek R., et al. (2006). Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188 6739–6756. 10.1128/JB.00609-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D., Sturgill D., Alhusaini N., Dillman A. A., Sweet T. J., Hanson G., et al. (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175 1872–1886.e24. 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arluison V., Derreumaux P., Allemand F., Folichon M., Hajnsdorf E., Regnier P. (2002). Structural modelling of the m-like protein Hfq from Escherichia coli. J. Mol. Biol. 320 705–712. 10.1016/s0022-2836(02)00548-x [DOI] [PubMed] [Google Scholar]
- Arnold T. E., Yu J., Belasco J. G. (1998). mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA 4 319–330. [PMC free article] [PubMed] [Google Scholar]
- Arraiano C. M., Andrade J. M., Domingues S., Guinote I. B., Malecki M., Matos R. G., et al. (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34 883–923. 10.1111/j.1574-6976.2010.00242.x [DOI] [PubMed] [Google Scholar]
- Artsimovitch I., Patlan V., Sekine S., Vassylyeva M. N., Hosaka T., Ochi K., et al. (2004). Structural basis for transcription regulation by alarmone ppGpp. Cell 117 299–310. 10.1016/s0092-8674(04)00401-5 [DOI] [PubMed] [Google Scholar]
- Atkinson G. C., Tenson T., Hauryliuk V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock D., Avarbock A., Rubin H. (2000). Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry 39 11640–11648. 10.1021/bi001256k [DOI] [PubMed] [Google Scholar]
- Baga M., Goransson M., Normark S., Uhlin B. E. (1988). Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell 52 197–206. 10.1016/0092-8674(88)90508-9 [DOI] [PubMed] [Google Scholar]
- Baker C. S., Eory L. A., Yakhnin H., Mercante J., Romeo T., Babitzke P. (2007). CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189 5472–5481. 10.1128/JB.00529-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyra K. J., Bouvier M., Carpousis A. J., Luisi B. F. (2013). The social fabric of the RNA degradosome. Biochim. Biophys. Acta 1829 514–522. 10.1016/j.bbagrm.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Regnier P., Chen S. M., Nakamura Y., Grunberg-Manago M., Court D. L. (1989). Autoregulation of RNase III operon by mRNA processing. EMBO J. 8 3401–3407. 10.1002/j.1460-2075.1989.tb08504.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomaus A., Fedyunin I., Feist P., Sin C., Zhang G., Valleriani A., et al. (2016). Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. A Math. Phys. Eng. Sci. 374:20150069. 10.1098/rsta.2015.0069 [DOI] [PubMed] [Google Scholar]
- Battesti A., Bouveret E. (2009). Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191 616–624. 10.1128/JB.01195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Deutscher M. P. (2019). Bacterial ribonucleases and their roles in RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 54 242–300. 10.1080/10409238.2019.1651816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. (1987). Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 84 498–502. 10.1073/pnas.84.2.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Zen K. H. (1989). Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171 5803–5811. 10.1128/jb.171.11.5803-5811.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H. F., Motorin Y., Planta R. J., Grosjean H. (1997). The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 25 4493–4499. 10.1093/nar/25.22.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. (1985). Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell 40 171–181. 10.1016/0092-8674(85)90320-4 [DOI] [PubMed] [Google Scholar]
- Benjak A., Avanzi C., Singh P., Loiseau C., Girma S., Busso P., et al. (2018). Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat. Commun. 9:352. 10.1038/s41467-017-02576-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini A., Martinez J. L. (2017). Genome-wide analysis shows that RNase G plays a global role in the stability of mRNAs in Stenotrophomonas maltophilia. Sci. Rep. 7:16016. 10.1038/s41598-017-16091-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Khodursky A. B., Lin P. H., Lin-Chao S., Cohen S. N. (2002). Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 99 9697–9702. 10.1073/pnas.112318199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Lin P. H., Cohen S. N., Lin-Chao S. (2004). Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 101 2758–2763. 10.1073/pnas.0308747101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43 717–731. 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- Bird J. G., Zhang Y., Tian Y., Panova N., Barvik I., Greene L., et al. (2016). The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535 444–447. 10.1038/nature18622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum E., Carpousis A. J., Higgins C. F. (1999). Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274 4009–4016. 10.1074/jbc.274.7.4009 [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Lee P. C., Wilson S. W., Cutler C. W., Ames B. N. (1984). AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37 225–232. 10.1016/0092-8674(84)90318-0 [DOI] [PubMed] [Google Scholar]
- Bodi Z., Button J. D., Grierson D., Fray R. G. (2010). Yeast targets for mRNA methylation. Nucleic Acids Res. 38 5327–5335. 10.1093/nar/gkq266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel G., Letso R., Neely H., Price W. N., Wong K. H., Su M., et al. (2016). Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529 358–363. 10.1038/nature16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C., Rigoulay C., Bouloc P. (2007). No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 7:10. 10.1186/1471-2180-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne J., Sokolovic Z., Goebel W. (1994). Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol. Microbiol. 11 1141–1150. 10.1111/j.1365-2958.1994.tb00390.x [DOI] [PubMed] [Google Scholar]
- Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., et al. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21 1353–1366. 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Belasco J. G. (1992). Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360 488–491. 10.1038/360488a0 [DOI] [PubMed] [Google Scholar]
- Braun F., Durand S., Condon C. (2017). Initiating ribosomes and a 5′/3′-UTR interaction control ribonuclease action to tightly couple B. subtilis hbs mRNA stability with translation. Nucleic Acids Res. 45 11386–11400. 10.1093/nar/gkx793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F., Le Derout J., Regnier P. (1998). Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J. 17 4790–4797. 10.1093/emboj/17.16.4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brescia C. C., Kaw M. K., Sledjeski D. D. (2004). The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 339 505–514. 10.1016/j.jmb.2004.03.067 [DOI] [PubMed] [Google Scholar]
- Brevet A., Chen J., Leveque F., Plateau P., Blanquet S. (1989). In vivo synthesis of adenylylated bis(5′-nucleosidyl) tetraphosphates (Ap4N) by Escherichia coli aminoacyl-tRNA synthetases. Proc. Natl. Acad. Sci. U.S.A. 86 8275–8279. 10.1073/pnas.86.21.8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briani F., Curti S., Rossi F., Carzaniga T., Mauri P., Deho G. (2008). Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 14 2417–2429. 10.1261/rna.1123908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruscella P., Shahbabian K., Laalami S., Putzer H. (2011). RNase Y is responsible for uncoupling the expression of translation factor IF3 from that of the ribosomal proteins L35 and L20 in Bacillus subtilis. Mol. Microbiol. 81 1526–1541. 10.1111/j.1365-2958.2011.07793.x [DOI] [PubMed] [Google Scholar]
- Cahova H., Winz M. L., Hofer K., Nubel G., Jaschke A. (2015). NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519 374–377. 10.1038/nature14020 [DOI] [PubMed] [Google Scholar]
- Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., Luisi B. F. (2005). Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437 1187–1191. 10.1038/nature04084 [DOI] [PubMed] [Google Scholar]
- Cameron T. A., Matz L. M., Sinha D., De Lay N. R. (2019). Polynucleotide phosphorylase promotes the stability and function of Hfq-binding sRNAs by degrading target mRNA-derived fragments. Nucleic Acids Res. 47 8821–8837. 10.1093/nar/gkz616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Guillen J., Bralley P., Jones G. H., Bechhofer D. H., Olmedo-Alvarez G. (2005). Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 187 4698–4706. 10.1128/JB.187.14.4698-4706.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Tanner A. W., Greco T. M., Defrancesco M., Cristea I. M., Dubnau D. (2013). A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol. Microbiol. 88 283–300. 10.1111/mmi.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T. M., Rojas-Duran M. F., Zinshteyn B., Shin H., Bartoli K. M., Gilbert W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron M. P., Bastet L., Lussier A., Simoneau-Roy M., Masse E., Lafontaine D. A. (2012). Dual-acting riboswitch control of translation initiation and mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 109 E3444–E3453. 10.1073/pnas.1214024109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A. J. (2007). The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61 71–87. 10.1146/annurev.micro.61.080706.093440 [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. (1994). Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76 889–900. 10.1016/0092-8674(94)90363-8 [DOI] [PubMed] [Google Scholar]
- Carzaniga T., Deho G., Briani F. (2015). RNase III-independent autogenous regulation of Escherichia coli polynucleotide phosphorylase via translational repression. J. Bacteriol. 197 1931–1938. 10.1128/JB.00105-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H., Deana A., Belasco J. G. (2007). Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27 79–90. 10.1016/j.molcel.2007.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty R., Bibb M. (1997). The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179 5854–5861. 10.1128/jb.179.18.5854-5861.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. T., Dyavaiah M., DeMott M. S., Taghizadeh K., Dedon P. C., Begley T. J. (2010). A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6:e1001247. 10.1371/journal.pgen.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. T., Pang Y. L., Deng W., Babu I. R., Dyavaiah M., Begley T. J., et al. (2012). Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3:937. 10.1038/ncomms1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V., Luisi B. F. (2006). Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 358 8–15. 10.1016/j.jmb.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Chao Y., Li L., Girodat D., Forstner K. U., Said N., Corcoran C., et al. (2017). In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65 39–51. 10.1016/j.molcel.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Previero A., Deutscher M. P. (2019). A novel mechanism of ribonuclease regulation: GcvB and Hfq stabilize the mRNA that encodes RNase BN/Z during exponential phase. J. Biol. Chem. 294 19997–20008. 10.1074/jbc.RA119.011367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shiroguchi K., Ge H., Xie X. S. (2015). Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11:781. 10.15252/msb.20145794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. H., Emory S. A., Bricker A. L., Bouvet P., Belasco J. G. (1991). Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J. Bacteriol. 173 4578–4586. 10.1128/jb.173.15.4578-4586.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li A., Sun B. F., Yang Y., Han Y. N., Yuan X., et al. (2019). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21 978–990. 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- Chen Y. G., Kowtoniuk W. E., Agarwal I., Shen Y., Liu D. R. (2009). LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5 879–881. 10.1038/nchembio.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Itzek A., Malke H., Ferretti J. J., Kreth J. (2013). Multiple roles of RNase Y in Streptococcus pyogenes mRNA processing and degradation. J. Bacteriol. 195 2585–2594. 10.1128/jb.00097-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Boisset S., Romilly C., Masquida B., Fechter P., Geissmann T., et al. (2010). Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 6:e1000809. 10.1371/journal.ppat.1000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching C., Gozzi K., Heinemann B., Chai Y., Godoy V. G. (2017). RNA-Mediated cis Regulation in Acinetobacter baumannii Modulates Stress-Induced Phenotypic Variation. J. Bacteriol. 199:e00799-16. 10.1128/JB.00799-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chionh Y. H., McBee M., Babu I. R., Hia F., Lin W., Zhao W., et al. (2016). tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7:13302. 10.1038/ncomms13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. H. (2017). The structure and function of the gram-positive bacterial RNA degradosome. Front. Microbiol. 8:154. 10.3389/fmicb.2017.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. K., Larsen M. H., Ingmer H., Sogaard-Andersen L., Kallipolitis B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186 3355–3362. 10.1128/JB.186.11.3355-3362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. K., Nielsen J. S., Ebersbach T., Valentin-Hansen P., Sogaard-Andersen L., Kallipolitis B. H. (2006). Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12 1383–1396. 10.1261/rna.49706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau F. M., Rothe F. M., Herzberg C., Wagner E., Hellwig D., Lehnik-Habrink M., et al. (2009). Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics 8 1350–1360. 10.1074/mcp.M800546-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. (2003). RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67 157–174. 10.1128/mmbr.67.2.157-174.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C., Putzer H., Grunberg-Manago M. (1996). Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 93 6992–6997. 10.1073/pnas.93.14.6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Bellows L. E., Wood A., Grundling A. (2016). ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 113 E1710–E1719. 10.1073/pnas.1522179113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste H., Brevet A., Plateau P., Blanquet S. (1987). Non-adenylylated bis(5′-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 262 12096–12103. [PubMed] [Google Scholar]
- Courbet A., Endy D., Renard E., Molina F., Bonnet J. (2015). Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 7:289ra283. 10.1126/scitranslmed.aaa3601 [DOI] [PubMed] [Google Scholar]
- Cui M., Wang T., Xu J., Ke Y., Du X., Yuan X., et al. (2013). Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PLoS One 8:e71933. 10.1371/journal.pone.0071933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daeffler K. N., Galley J. D., Sheth R. U., Ortiz-Velez L. C., Bibb C. O., Shroyer N. F., et al. (2017). Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13:923. 10.15252/msb.20167416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou-Chabo R., Mathy N., Benard L., Condon C. (2009). Ribosomes initiating translation of the hbs mRNA protect it from 5′-to-3′ exoribonucleolytic degradation by RNase J1. Mol. Microbiol. 71 1538–1550. 10.1111/j.1365-2958.2009.06620.x [DOI] [PubMed] [Google Scholar]
- Deana A., Celesnik H., Belasco J. G. (2008). The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451 355–358. 10.1038/nature06475 [DOI] [PubMed] [Google Scholar]
- DeLoughery A., Dengler V., Chai Y., Losick R. (2016). Biofilm formation by Bacillus subtilis requires an endoribonuclease-containing multisubunit complex that controls mRNA levels for the matrix gene repressor SinR. Mol. Microbiol. 99 425–437. 10.1111/mmi.13240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoughery A., Jean-Benoît L., Losick R., Gene-Wei L. (2018). Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 115 E5585–E5594. 10.1073/pnas.1803283115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Babu I. R., Su D., Yin S., Begley T. J., Dedon P. C. (2015). Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet. 11:e1005706. 10.1371/journal.pgen.1005706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Chen K., Luo G. Z., Weng X., Ji Q., Zhou T., et al. (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43 6557–6567. 10.1093/nar/gkv596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Liu Z., Bi Y., Wang X., Zhou D., Yang R., et al. (2014). Rapid degradation of Hfq-free RyhB in Yersinia pestis by PNPase independent of putative ribonucleolytic complexes. Biomed Res. Int. 2014:798918. 10.1155/2014/798918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. (2006). Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34 659–666. 10.1093/nar/gkj472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwa A., Bricker A. L., Jain C., Belasco J. G. (2000). An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 14 1249–1260. [PMC free article] [PubMed] [Google Scholar]
- Diwa A. A., Belasco J. G. (2002). Critical features of a conserved RNA stem-loop important for feedback regulation of RNase E synthesis. J. Biol. Chem. 277 20415–20422. 10.1074/jbc.M202313200 [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. (1986). Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 83 120–124. 10.1073/pnas.83.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D., Hall L. S., Rescheneder P., Lybecker M., Samuels D. S. (2018). The stringent response-regulated sRNA transcriptome of Borrelia burgdorferi. Front. Cell. Infect. Microbiol. 8:231. 10.3389/fcimb.2018.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressaire C., Picard F., Redon E., Loubiere P., Queinnec I., Girbal L., et al. (2013). Role of mRNA stability during bacterial adaptation. PLoS One 8:e59059. 10.1371/journal.pone.0059059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinin S. Y., Tran N. T., Skalenko K. S., Goldman S. R., Knoblauch J. G., Dove S. L., et al. (2015). A conserved pattern of primer-dependent transcription initiation in Escherichia coli and Vibrio cholerae revealed by 5′, RNA-seq PLoS Genet. 11:e1005348. 10.1371/journal.pgen.1005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E. J., Carabetta V. J., Tanner A. W., Miras M., Diethmaier C., Dubnau D. (2016). A protein complex supports the production of Spo0A-P and plays additional roles for biofilms and the K-state in Bacillus subtilis. Mol. Microbiol. 101 606–624. 10.1111/mmi.13411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S., Gilet L., Bessieres P., Nicolas P., Condon C. (2012). Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 8:e1002520. 10.1371/journal.pgen.1002520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S., Schwartz S., Mumbach M. R., Wurtzel O., Sorek R. (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9:e1003602. 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Belasco J. G. (1990). The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 172 4472–4481. 10.1128/jb.172.8.4472-4481.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. (1992). A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6 135–148. 10.1101/gad.6.1.135 [DOI] [PubMed] [Google Scholar]
- Esquerre T., Bouvier M., Turlan C., Carpousis A. J., Girbal L., Cocaign-Bousquet M. (2016). The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli. Sci. Rep. 6:25057. 10.1038/srep25057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerre T., Laguerre S., Turlan C., Carpousis A. J., Girbal L., Cocaign-Bousquet M. (2014). Dual role of transcription and transcript stability in the regulation of gene expression in Escherichia coli cells cultured on glucose at different growth rates. Nucleic Acids Res. 42 2460–2472. 10.1093/nar/gkt1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerre T., Moisan A., Chiapello H., Arike L., Vilu R., Gaspin C., et al. (2015). Genome-wide investigation of mRNA lifetime determinants in Escherichia coli cells cultured at different growth rates. BMC Genomics 16:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even S., Pellegrini O., Zig L., Labas V., Vinh J., Brechemmier-Baey D., et al. (2005). Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 33 2141–2152. 10.1093/nar/gki505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler D. E., Franco M. K., Batool Z., Wu M. Z., Dubuke M. L., Dobosz-Bartoszek M., et al. (2019). Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. U.S.A. 116 23068–23074. 10.1073/pnas.1821754116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faner M. A., Feig A. L. (2013). Identifying and characterizing Hfq-RNA interactions. Methods 63 144–159. 10.1016/j.ymeth.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. (1981). Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 9 563–577. 10.1093/nar/9.3.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr S. B., Arnosti D. N., Chamberlin M. J., Ames B. N. (1989). An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86 5010–5014. 10.1073/pnas.86.13.5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Borges V., Borrego M. J., Gomes J. P. (2017). Global survey of mRNA levels and decay rates of Chlamydia trachomatis trachoma and lymphogranuloma venereum biovars. Heliyon 3:e00364. 10.1016/j.heliyon.2017.e00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M., Allemand F., Regnier P., Hajnsdorf E. (2005). Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 272 454–463. 10.1111/j.1742-4658.2004.04485.x [DOI] [PubMed] [Google Scholar]
- Frederix M., Downie A. J. (2011). Quorum sensing: regulating the regulators. Adv. Microb. Physiol. 58 23–80. 10.1016/B978-0-12-381043-4.00002-7 [DOI] [PubMed] [Google Scholar]
- Frindert J., Zhang Y., Nubel G., Kahloon M., Kolmar L., Hotz-Wagenblatt A., et al. (2018). Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep. 24 1890–1901.e8. 10.1016/j.celrep.2018.07.047 [DOI] [PubMed] [Google Scholar]
- Fritsch F., Mauder N., Williams T., Weiser J., Oberle M., Beier D. (2011). The cell envelope stress response mediated by the LiaFSRLm three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaSLm. Microbiology 157(Pt 2), 373–386. 10.1099/mic.0.044776-0 [DOI] [PubMed] [Google Scholar]
- Fry M., Israeli-Reches M., Artman M. (1972). Stabilization and breakdown of Escherichia coli messenger ribonucleic acid in the presence of chloramphenicol. Biochemistry 11 3054–3059. 10.1021/bi00766a017 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., et al. (2013). The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499 178–183. 10.1038/nature12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton N. J., Waddell S. J., Sherratt A. L., Lee S. M., Smith R. J., Senner C., et al. (2008). Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:e75. 10.1371/journal.pmed.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood M. L., Jones G. H. (2010). (p)ppGpp inhibits polynucleotide phosphorylase from streptomyces but not from Escherichia coli and increases the stability of bulk mRNA in Streptomyces coelicolor. J. Bacteriol. 192 4275–4280. 10.1128/JB.00367-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrish E., Sit C. S., Cao S., Kandror O., Spoering A., Peoples A., et al. (2014). Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21 509–518. 10.1016/j.chembiol.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry D. R., Hernandez V. J., Nguyen L. H., Jensen D. B., Cashel M. (1993). Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 175 7982–7989. 10.1128/jb.175.24.7982-7989.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D., Barlow T., Arvidson S., von Gabain A. (1993). Retarded RNA turnover in Escherichia coli: a means of maintaining gene expression during anaerobiosis. Mol. Microbiol. 9 375–381. 10.1111/j.1365-2958.1993.tb01698.x [DOI] [PubMed] [Google Scholar]
- Glatz E., Nilsson R. P., Rutberg L., Rutberg B. (1996). A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol. Microbiol. 19 319–328. 10.1046/j.1365-2958.1996.376903.x [DOI] [PubMed] [Google Scholar]
- Gualerzi C. O., Giuliodori A. M., Pon C. L. (2003). Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331 527–539. 10.1016/s0022-2836(03)00732-0 [DOI] [PubMed] [Google Scholar]
- Guo M. S., Updegrove T. B., Gogol E. B., Shabalina S. A., Gross C. A., Storz G. (2014). MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28 1620–1634. 10.1101/gad.243485.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgsell N., Englund N., Niu L., Kaya Y., Lane B. G., Ofengand J. (2000). Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6 1870–1881. 10.1017/s1355838200001588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjeras L., Poljak L., Bouvier M., Morin-Ogier Q., Canal I., Cocaign-Bousquet M., et al. (2019). Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RNase E and increased turnover of ribosome-free transcripts. Mol. Microbiol. 111 1715–1731. 10.1111/mmi.14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E., Regnier P. (2000). Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U.S.A. 97 1501–1505. 10.1073/pnas.040549897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambraeus G., Karhumaa K., Rutberg B. (2002). A 5′ stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology 148(Pt 6), 1795–1803. 10.1099/00221287-148-6-1795 [DOI] [PubMed] [Google Scholar]
- Hambraeus G., Persson M., Rutberg B. (2000). The aprE leader is a determinant of extreme mRNA stability in Bacillus subtilis. Microbiology 146(Pt 12), 3051–3059. 10.1099/00221287-146-12-3051 [DOI] [PubMed] [Google Scholar]
- Hambraeus G., von Wachenfeldt C., Hederstedt L. (2003). Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269 706–714. 10.1007/s00438-003-0883-6 [DOI] [PubMed] [Google Scholar]
- Hammerle H., Amman F., Vecerek B., Stulke J., Hofacker I., Blasi U. (2014). Impact of Hfq on the Bacillus subtilis transcriptome. PLoS One 9:e98661. 10.1371/journal.pone.0098661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouche L., Billaudeau C., Rocca A., Chastanet A., Ngo S., Laalami S., et al. (2020). Dynamic membrane localization of RNase Y in Bacillus subtilis. mBio 11:e03337-19. 10.1128/mBio.03337-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Guimaraes V. A., Garcin D., Baumann N., Linder P., Redder P. (2017). Both exo- and endo-nucleolytic activities of RNase J1 from Staphylococcus aureus are manganese dependent and active on triphosphorylated 5′-ends. RNA Biol. 14 1431–1443. 10.1080/15476286.2017.1300223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks N. D., Yang J., Zhang X., Zhao B., Grad Y. H., Liu L., et al. (2018). Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3 1032–1042. 10.1038/s41564-018-0218-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes T. P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., et al. (2016). Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44 852–862. 10.1093/nar/gkv1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer K., Li S., Abele F., Frindert J., Schlotthauer J., Grawenhoff J., et al. (2016). Structure and function of the bacterial decapping enzyme NudC. Nat. Chem. Biol. 12 730–734. 10.1038/nchembio.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Chen W., Liu J., Gu N., Zhang R. (2019). Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26 380–388. 10.1038/s41594-019-0218-x [DOI] [PubMed] [Google Scholar]
- Hudecek O., Benoni R., Reyes-Gutierrez P. E., Culka M., Sanderova H., Hubalek M., et al. (2020). Dinucleoside polyphosphates act as 5′-RNA caps in bacteria. Nat. Commun. 11:1052. 10.1038/s41467-020-14896-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue K. K., Cohen S. D., Bechhofer D. H. (1995). A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J. Bacteriol. 177 3465–3471. 10.1128/jb.177.12.3465-3471.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A., Rawlins J. P., Thomaides H. B., Errington J. (2006). Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152(Pt 10), 2895–2907. 10.1099/mic.0.29152-0 [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., et al. (2005). Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24 824–835. 10.1038/sj.emboj.7600572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov D., Vaitkevicius K., Durand S., Cahoon L., Sandberg S. S., Liu X., et al. (2020). An mRNA-mRNA interaction couples expression of a virulence factor and its chaperone in Listeria monocytogenes. Cell Rep. 30 4027–4040.e7. 10.1016/j.celrep.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., et al. (2011). Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39 2304–2318. 10.1093/nar/gkq1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110 551–561. 10.1016/s0092-8674(02)00905-4 [DOI] [PubMed] [Google Scholar]
- Jona G., Choder M., Gileadi O. (2000). Glucose starvation induces a drastic reduction in the rates of both transcription and degradation of mRNA in yeast. Biochim. Biophys. Acta 1491 37–48. 10.1016/s0167-4781(00)00016-6 [DOI] [PubMed] [Google Scholar]
- Jourdan S. S., McDowall K. J. (2008). Sensing of 5′ monophosphate by Escherichia coli RNase G can significantly enhance association with RNA and stimulate the decay of functional mRNA transcripts in vivo. Mol. Microbiol. 67 102–115. 10.1111/j.1365-2958.2007.06028.x [DOI] [PubMed] [Google Scholar]
- Joyce S. A., Dreyfus M. (1998). In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol. 282 241–254. 10.1006/jmbi.1998.2027 [DOI] [PubMed] [Google Scholar]
- Julius C., Yuzenkova Y. (2017). Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 45 8282–8290. 10.1093/nar/gkx452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgen B., Schweder T., Hecker M. (1998). The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258 538–545. 10.1007/s004380050765 [DOI] [PubMed] [Google Scholar]
- Kavita K., de Mets F., Gottesman S. (2018). New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42 53–61. 10.1016/j.mib.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V., Poljak L., Luisi B. F., Carpousis A. J. (2008). The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70 799–813. 10.1111/j.1365-2958.2008.06454.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V., Prados J., Linder P., Redder P. (2015). Decay-initiating endoribonucleolytic cleavage by RNase y is kept under tight control via sequence preference and sub-cellular localisation. PLoS Genet. 11:e1005577. 10.1371/journal.pgen.1005577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido M., Yamanaka K., Mitani T., Niki H., Ogura T., Hiraga S. (1996). RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 178 3917–3925. 10.1128/jb.178.13.3917-3925.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime L., Jourdan S. S., Stead J. A., Hidalgo-Sastre A., McDowall K. J. (2010). Rapid cleavage of RNA by RNase E in the absence of 5′ monophosphate stimulation. Mol. Microbiol. 76 590–604. 10.1111/j.1365-2958.2009.06935.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime L., Vincent H. A., Gendoo D. M., Jourdan S. S., Fishwick C. W., Callaghan A. J., et al. (2015). The first small-molecule inhibitors of members of the ribonuclease E family. Sci. Rep. 5:8028. 10.1038/srep08028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn S. M., O’Byrne C. P., Booth I. R., Stansfield I. (2002). Physiological analysis of the role of truB in Escherichia coli: a role for tRNA modification in extreme temperature resistance. Microbiology 148(Pt 11), 3511–3520. 10.1099/00221287-148-11-3511 [DOI] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. (1990). Combined actions of multiple hairpin loop structures and sites of rate-limiting endonucleolytic cleavage determine differential degradation rates of individual segments within polycistronic puf operon mRNA. J. Bacteriol. 172 5140–5146. 10.1128/jb.172.9.5140-5146.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowtoniuk W. E., Shen Y., Heemstra J. M., Agarwal I., Liu D. R. (2009). A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl. Acad. Sci. U.S.A. 106 7768–7773. 10.1073/pnas.0900528106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S., Petchiappan A., Singh A., Bhatt A., Chatterji D. (2016). R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol. Microbiol. 102 168–182. 10.1111/mmi.13453 [DOI] [PubMed] [Google Scholar]
- Kristoffersen S. M., Haase C., Weil M. R., Passalacqua K. D., Niazi F., Hutchison S. K., et al. (2012). Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol. 13:R30. 10.1186/gb-2012-13-4-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Majid M., Kunisch R., Rani P. S., Qureshi I. A., Lewin A., et al. (2012). Mycobacterium tuberculosis DosR regulon gene Rv0079 encodes a putative, ‘dormancy associated translation inhibitor (DATIN)’. PLoS One 7:e38709. 10.1371/journal.pone.0038709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S., Sutter B. M., Wu X., Kumar S., Guo X., Trudgian D. C., et al. (2013). Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154 416–429. 10.1016/j.cell.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Groisman E. A. (2010). An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 76 1020–1033. 10.1111/j.1365-2958.2010.07161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhan X., Gao J., Qiu J., Feng Y., Meganathan R., et al. (2003). RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114 623–634. [PubMed] [Google Scholar]
- Lee P. C., Bochner B. R., Ames B. N. (1983). AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. U.S.A. 80 7496–7500. 10.1073/pnas.80.24.7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Newman J., Rothe F. M., Solovyova A. S., Rodrigues C., Herzberg C., et al. (2011). RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J. Bacteriol. 193 5431–5441. 10.1128/JB.05500-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Pfortner H., Rempeters L., Pietack N., Herzberg C., Stulke J. (2010). The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 77 958–971. 10.1111/j.1365-2958.2010.07264.x [DOI] [PubMed] [Google Scholar]
- Leimeister-Wachter M., Domann E., Chakraborty T. (1991). Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is co-ordinately expressed with listeriolysin in Listeria monocytogenes. Mol. Microbiol. 5 361–366. 10.1111/j.1365-2958.1991.tb02117.x [DOI] [PubMed] [Google Scholar]
- Leimeister-Wachter M., Haffner C., Domann E., Goebel W., Chakraborty T. (1990). Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 87 8336–8340. 10.1073/pnas.87.21.8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G., Doron-Faigenboim A., Ron E. Z., Tuller T., Gophna U. (2011). Sequence features of E. coli mRNAs affect their degradation. PLoS One 6:e28544. 10.1371/journal.pone.0028544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit S., Deutscher M. P. (1998). Polyadenylation of stable RNA precursors in vivo. Proc. Natl. Acad. Sci. U.S.A. 95 12158–12162. 10.1073/pnas.95.21.12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Deutscher M. P. (2013). Ribosomes regulate the stability and action of the exoribonuclease RNase R. J. Biol. Chem. 288 34791–34798. 10.1074/jbc.M113.519553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., Jaffrey S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Y., Gui G., Wei B., Preston J. F., III, Oakford L., Yuksel U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272 17502–17510. 10.1074/jbc.272.28.17502 [DOI] [PubMed] [Google Scholar]
- Liu M. Y., Yang H., Romeo T. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177 2663–2672. 10.1128/jb.177.10.2663-2672.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wu N., Dong J., Gao Y., Zhang X., Mu C., et al. (2010). Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS One 5:e13069. 10.1371/journal.pone.0013069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E., Dussurget O., Gripenland J., Vaitkevicius K., Tiensuu T., Mandin P., et al. (2009). A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139 770–779. 10.1016/j.cell.2009.08.046 [DOI] [PubMed] [Google Scholar]
- Loh E., Memarpour F., Vaitkevicius K., Kallipolitis B. H., Johansson J., Sonden B. (2012). An unstructured 5′-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res. 40 1818–1827. 10.1093/nar/gkr850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P. J., Marchand I., Joyce S. A., Dreyfus M. (1999). The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33 188–199. 10.1046/j.1365-2958.1999.01465.x [DOI] [PubMed] [Google Scholar]
- Lopez P. J., Marchand I., Yarchuk O., Dreyfus M. (1998). Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc. Natl. Acad. Sci. U.S.A. 95 6067–6072. 10.1073/pnas.95.11.6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D. J., Belasco J. G. (2020). Np4A alarmones function in bacteria as precursors to RNA caps. Proc. Natl. Acad. Sci. U.S.A. 117 3560–3567. 10.1073/pnas.1914229117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D. J., Levenson-Palmer R., Belasco J. G. (2019). Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell 75 957–966.e8. 10.1016/j.molcel.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D. J., Vasilyev N., Richards J., Serganov A., Belasco J. G. (2017). A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell 67 44–54.e6. 10.1016/j.molcel.2017.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H., Homuth G., Schmalisch M., Dyka F. M., Hecker M., Stulke J. (2001). Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41 409–422. 10.1046/j.1365-2958.2001.02523.x [DOI] [PubMed] [Google Scholar]
- Lundberg U., von Gabain A., Melefors O. (1990). Cleavages in the 5′ region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 9 2731–2741. 10.1002/j.1460-2075.1990.tb07460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. (1992). Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J. Biol. Chem. 267 1054–1061. [PubMed] [Google Scholar]
- Mackie G. A. (1998). Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395 720–723. 10.1038/27246 [DOI] [PubMed] [Google Scholar]
- Maggi N., Pasqualucci C. R., Ballotta R., Sensi P. (1966). Rifampicin: a new orally active rifamycin. Chemotherapy 11 285–292. 10.1159/000220462 [DOI] [PubMed] [Google Scholar]
- Marcaida M. J., DePristo M. A., Chandran V., Carpousis A. J., Luisi B. F. (2006). The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 31 359–365. 10.1016/j.tibs.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Martinez I., El-Said Mohamed M., Santos V. E., Garcia J. L., Garcia-Ochoa F., Diaz E. (2017). Metabolic and process engineering for biodesulfurization in Gram-negative bacteria. J. Biotechnol. 262 47–55. 10.1016/j.jbiotec.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Martinez-Costa O. H., Fernandez-Moreno M. A., Malpartida F. (1998). The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J. Bacteriol. 180 4123–4132. 10.1128/jb.180.16.4123-4132.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini M. C., Zhou Y., Sun H., Shell S. S. (2019). Defining the transcriptional and post-transcriptional landscapes of Mycobacterium smegmatis in aerobic growth and hypoxia. Front. Microbiol. 10:591. 10.3389/fmicb.2019.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N., Hebert A., Mervelet P., Benard L., Dorleans A., Li de la Sierra-Gallay I., et al. (2010). Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 75 489–498. 10.1111/j.1365-2958.2009.07004.x [DOI] [PubMed] [Google Scholar]
- Matsunaga J., Simons E. L., Simons R. W. (1996). RNase III autoregulation: structure and function of rncO, the posttranscriptional “operator”. RNA 2 1228–1240. [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J., Simons E. L., Simons R. W. (1997). Escherichia coli RNase III (rnc) autoregulation occurs independently of rnc gene translation. Mol. Microbiol. 26 1125–1135. 10.1046/j.1365-2958.1997.6652007.x [DOI] [PubMed] [Google Scholar]
- McDowall K. J., Kaberdin V. R., Wu S. W., Cohen S. N., Lin-Chao S. (1995). Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374 287–290. 10.1038/374287a0 [DOI] [PubMed] [Google Scholar]
- McDowall K. J., Lin-Chao S., Cohen S. N. (1994). A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269 10790–10796. [PubMed] [Google Scholar]
- McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. (1991). mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 221 81–95. 10.1016/0022-2836(91)90806-h [DOI] [PubMed] [Google Scholar]
- Melin L., Rutberg L., von Gabain A. (1989). Transcriptional and posttranscriptional control of the Bacillus subtilis succinate dehydrogenase operon. J. Bacteriol. 171 2110–2115. 10.1128/jb.171.4.2110-2115.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing S. A., Gabelli S. B., Liu Q., Celesnik H., Belasco J. G., Pineiro S. A., et al. (2009). Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus. Structure 17 472–481. 10.1016/j.str.2008.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J. R., Pandey S., Boettiger A. N., Wang S., Zhuang X. (2016). Spatial organization shapes the turnover of a bacterial transcriptome. eLife 5:e13065. 10.7554/eLife.13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B. K., Kushner S. R. (2000). Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97 11966–11971. 10.1073/pnas.220295997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B. K., Kushner S. R. (2002). Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol. Microbiol. 45 1315–1324. 10.1046/j.1365-2958.2002.03097.x [DOI] [PubMed] [Google Scholar]
- Moll I., Afonyushkin T., Vytvytska O., Kaberdin V. R., Blasi U. (2003). Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9 1308–1314. 10.1261/rna.5850703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T., Franch T., Hojrup P., Keene D. R., Bachinger H. P., Brennan R. G., et al. (2002). Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9 23–30. 10.1016/s1097-2765(01)00436-1 [DOI] [PubMed] [Google Scholar]
- Montero Llopis P., Jackson A. F., Sliusarenko O., Surovtsev I., Heinritz J., Emonet T., et al. (2010). Spatial organization of the flow of genetic information in bacteria. Nature 466 77–81. 10.1038/nature09152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin M., Enjalbert B., Ropers D., Girbal L., Cocaign-Bousquet M. (2020). Genomewide Stabilization of mRNA during a “Feast-to-Famine” Growth Transition in Escherichia coli. mSphere 5:e00276-20. 10.1128/mSphere.00276-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Aiba H. (2011). RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 25 294–298. 10.1101/gad.2030311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Kawamoto H., Mizota T., Inada T., Aiba H. (2004). Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54 1063–1075. 10.1111/j.1365-2958.2004.04329.x [DOI] [PubMed] [Google Scholar]
- Morrison J. M., Anderson K. L., Beenken K. E., Smeltzer M. S., Dunman P. M. (2012). The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front. Cell. Infect. Microbiol. 2:26. 10.3389/fcimb.2012.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Gimpel M., Wildenhain T., Brantl S. (2019). A new role for CsrA: promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 16 972–987. 10.1080/15476286.2019.1605811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashko O. N., Kaberdin V. R., Lin-Chao S. (2012). Membrane binding of Escherichia coli RNase E catalytic domain stabilizes protein structure and increases RNA substrate affinity. Proc. Natl. Acad. Sci. U.S.A. 109 7019–7024. 10.1073/pnas.1120181109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashko O. N., Lin-Chao S. (2017). Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc. Natl. Acad. Sci. U.S.A. 114 E8025–E8034. 10.1073/pnas.1703731114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdeshwar M. S., Chatterji D. (2012). MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 194 4003–4014. 10.1128/JB.00258-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M. A., Lovejoy A. F., Cygan A. M., Boothroyd J. C. (2017). mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 23 1834–1849. 10.1261/rna.062794.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. (1987). Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 48 297–310. 10.1016/0092-8674(87)90433-8 [DOI] [PubMed] [Google Scholar]
- Nguyen T. G., Vargas-Blanco D. A., Roberts L. A., Shell S. S. (2020). The impact of leadered and leaderless gene structures on translation efficiency, transcript stability, and predicted transcription rates in Mycobacterium smegmatis. J. Bacteriol. 202:e00746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. S., Larsen M. H., Lillebaek E. M., Bergholz T. M., Christiansen M. H., Boor K. J., et al. (2011). A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 6:e19019. 10.1371/journal.pone.0019019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. S., Lei L. K., Ebersbach T., Olsen A. S., Klitgaard J. K., Valentin-Hansen P., et al. (2010). Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38 907–919. 10.1093/nar/gkp1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. (1984). Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature 312 75–77. 10.1038/312075a0 [DOI] [PubMed] [Google Scholar]
- Nilsson P., Naureckiene S., Uhlin B. E. (1996). Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J. Bacteriol. 178 683–690. 10.1128/jb.178.3.683-690.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P., Uhlin B. E. (1991). Differential decay of a polycistronic Escherichia coli transcript is initiated by RNaseE-dependent endonucleolytic processing. Mol. Microbiol. 5 1791–1799. 10.1111/j.1365-2958.1991.tb01928.x [DOI] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. (1981). RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J. Mol. Biol. 145 405–420. 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- Nouaille S., Mondeil S., Finoux A. L., Moulis C., Girbal L., Cocaign-Bousquet M. (2017). The stability of an mRNA is influenced by its concentration: a potential physical mechanism to regulate gene expression. Nucleic Acids Res. 45 11711–11724. 10.1093/nar/gkx781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K., Wrzesinski J., Bakin A., Lane B. G., Ofengand J. (1995). Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. RNA 1 102–112. [PMC free article] [PubMed] [Google Scholar]
- Olson P. D., Kuechenmeister L. J., Anderson K. L., Daily S., Beenken K. E., Roux C. M., et al. (2011). Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathog. 7:e1001287. 10.1371/journal.ppat.1001287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M. C., Liu Q., Kushner S. R. (2000). Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38 854–866. 10.1046/j.1365-2958.2000.02186.x [DOI] [PubMed] [Google Scholar]
- Paesold G., Krause M. (1999). Analysis of rpoS mRNA in Salmonella dublin: identification of multiple transcripts with growth-phase-dependent variation in transcript stability. J. Bacteriol. 181 1264–1268. 10.1128/jb.181.4.1264-1268.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss S., Eedunuri V. K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., et al. (2018). Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 4:eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J. C., Vogel J. (2006). SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62 1674–1688. 10.1111/j.1365-2958.2006.05524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. D., Guinn K. M., Harrell M. I., Liao R., Voskuil M. I., Tompa M., et al. (2003). Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48 833–843. 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato M. L., Bennett P. M., von Meyenburg K. (1973). Messenger ribonucleic acid synthesis and degradation in Escherichia coli during inhibition of translation. J. Bacteriol. 116 710–718. 10.1128/jb.116.2.710-718.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertzev A. V., Nicholson A. W. (2006). Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 34 3708–3721. 10.1093/nar/gkl459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petchiappan A., Naik S. Y., Chatterji D. (2020). RelZ-mediated stress response in Mycobacterium smegmatis: pGpp synthesis and its regulation. J. Bacteriol. 202:e00444-19. 10.1128/JB.00444-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plocinski P., Macios M., Houghton J., Niemiec E., Plocinska R., Brzostek A., et al. (2019). Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 47 5892–5905. 10.1093/nar/gkz251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V., Alhusaini N., Chen Y. H., Martin S., Morris N., Kline N., et al. (2015). Codon optimality is a major determinant of mRNA stability. Cell 160 1111–1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B., Causton H., Mudd E. A., Higgins C. F. (1994). A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14 717–729. 10.1111/j.1365-2958.1994.tb01309.x [DOI] [PubMed] [Google Scholar]
- Py B., Higgins C. F., Krisch H. M., Carpousis A. J. (1996). A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381 169–172. 10.1038/381169a0 [DOI] [PubMed] [Google Scholar]
- Quereda J. J., Andersson C., Cossart P., Johansson J., Pizarro-Cerda J. (2018). Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet. Res. 49:13. 10.1186/s13567-017-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E., Trevino J., Liu Z., Perez N., Sumby P. (2010). The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 78 1332–1347. 10.1111/j.1365-2958.2010.07427.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. P., Alonso S., Rand L., Dick T., Pethe K. (2008). The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 105 11945–11950. 10.1073/pnas.0711697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y., Galtier E., Arnion H., Darfeuille F., Sismeiro O., Coppee J. Y., et al. (2016). RNase J depletion leads to massive changes in mRNA abundance in Helicobacter pylori. RNA Biol. 13 243–253. 10.1080/15476286.2015.1132141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon E., Loubiere P., Cocaign-Bousquet M. (2005a). Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J. Bacteriol. 187 3589–3592. 10.1128/JB.187.10.3589-3592.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon E., Loubiere P., Cocaign-Bousquet M. (2005b). Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J. Biol. Chem. 280 36380–36385. 10.1074/jbc.M506006200 [DOI] [PubMed] [Google Scholar]
- Regnier P., Hajnsdorf E. (2013). The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3′ ends of RNAs resulting from Rho-independent termination: a tentative model. RNA Biol. 10 602–609. 10.4161/rna.23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Belasco J. G. (2016). Distinct requirements for 5′-monophosphate-assisted RNA cleavage by Escherichia coli RNase E and RNase G. J. Biol. Chem. 291 20825. 10.1074/jbc.A115.702555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Belasco J. G. (2019). Obstacles to scanning by RNase E govern bacterial mRNA lifetimes by hindering access to distal cleavage sites. Mol. Cell 74 284–295.e5. 10.1016/j.molcel.2019.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Liu Q., Pellegrini O., Celesnik H., Yao S., Bechhofer D. H., et al. (2011). An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell 43 940–949. 10.1016/j.molcel.2011.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar D. T., Giessen T. W., Baym M., Kerns S. J., Niederhuber M. J., Bronson R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35 653–658. 10.1038/nbt.3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus E. S., Baek S. H., Sassetti C. M. (2013). The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13 643–651. 10.1016/j.chom.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C., Anderson K. L., Murphy E., Projan S. J., Mounts W., Hurlburt B., et al. (2006). Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188 2593–2603. 10.1128/JB.188.7.2593-2603.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T., Bouloc P., Yang Q., Bossi L., Figueroa-Bossi N. (2012). Lack of interchangeability of Hfq-like proteins. Biochimie 94 1554–1559. 10.1016/j.biochi.2012.01.016 [DOI] [PubMed] [Google Scholar]
- Rochat T., Delumeau O., Figueroa-Bossi N., Noirot P., Bossi L., Dervyn E., et al. (2015). Tracking the elusive function of Bacillus subtilis Hfq. PLoS One 10:e0124977. 10.1371/journal.pone.0124977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T., Babitzke P. (2018). Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6:10.1128/microbiolspec.RWR-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Thorsing M., Lillebaek E. M. S., Teixeira Dos Santos P., Kallipolitis B. H. (2019). The LhrC sRNAs control expression of T cell-stimulating antigen TcsA in Listeria monocytogenes by decreasing tcsA mRNA stability. RNA Biol. 16 270–281. 10.1080/15476286.2019.1572423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski J., Crain P. F., McCloskey J. A. (1999). The RNA modification database: 1999 update. Nucleic Acids Res. 27 196–197. 10.1093/nar/27.1.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T. R., Minch K. J., Brabant W., Winkler J. K., Reiss D. J., Baliga N. S., et al. (2013). Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res. 41 509–517. 10.1093/nar/gks1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C., Forti F., Magnoni F., Ghisotti D. (2008). The katG mRNA of Mycobacterium tuberculosis and Mycobacterium smegmatis is processed at its 5′ end and is stabilized by both a polypurine sequence and translation initiation. BMC Mol. Biol. 9:33. 10.1186/1471-2199-9-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. (1989). Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171 6680–6688. 10.1128/jb.171.12.6680-6688.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Frangos A., Woodson S. A. (2018). Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9:e1475. 10.1002/wrna.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. (1978). Translation and mRNA decay. Mol. Gen. Genet. 160 121–129. [DOI] [PubMed] [Google Scholar]
- Schubert O. T., Ludwig C., Kogadeeva M., Zimmermann M., Rosenberger G., Gengenbacher M., et al. (2015). Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18 96–108. 10.1016/j.chom.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Schumacher M. A., Pearson R. F., Moller T., Valentin-Hansen P., Brennan R. G. (2002). Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21 3546–3556. 10.1093/emboj/cdf322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Zhang H. N., Sibbritt T., Pan A., Horvath A., Gross S., et al. (2020). Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 18:40. 10.1186/s12915-020-00769-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M. R., Jovanovic M., Wang T., Maciag K., Bushkin G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8 284–296. 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E., Smith Y., Ben-Yehuda S. (2012). RNA dynamics in aging bacterial spores. Cell 148 139–149. 10.1016/j.cell.2011.11.059 [DOI] [PubMed] [Google Scholar]
- Selinger D. W., Saxena R. M., Cheung K. J., Church G. M., Rosenow C. (2003). Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13 216–223. 10.1101/gr.912603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfzadeh M., Keener J., Nomura M. (1993). spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90 11004–11008. 10.1073/pnas.90.23.11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K., Jamalli A., Zig L., Putzer H. (2009). RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28 3523–3533. 10.1038/emboj.2009.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Dahan O., Levo M., Martinez M. R., Furman I., Segal E., et al. (2008). Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 4:223. 10.1038/msb.2008.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. S., Bechhofer D. H. (2005). Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol. Microbiol. 57 484–495. 10.1111/j.1365-2958.2005.04683.x [DOI] [PubMed] [Google Scholar]
- Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001). Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc. Natl. Acad. Sci. U.S.A. 98 7534–7539. 10.1073/pnas.121172498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. (1980). Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc. Natl. Acad. Sci. U.S.A. 77 3903–3907. 10.1073/pnas.77.7.3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siculella L., Damiano F., di Summa R., Tredici S. M., Alduina R., Gnoni G. V., et al. (2010). Guanosine 5′-diphosphate 3′-diphosphate (ppGpp) as a negative modulator of polynucleotide phosphorylase activity in a ‘rare’ actinomycete. Mol. Microbiol. 77 716–729. 10.1111/j.1365-2958.2010.07240.x [DOI] [PubMed] [Google Scholar]
- Sievers S., Lund A., Menendez-Gil P., Nielsen A., Storm Mollerup M., Lambert Nielsen S., et al. (2015). The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol. 12 985–997. 10.1080/15476286.2015.1071011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., Sultan S. Z., Liang W., Benitez J. A. (2008). Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae. J. Bacteriol. 190 7335–7345. 10.1128/JB.00360-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim M., Lim B., Sim S. H., Kim D., Jung E., Lee Y., et al. (2014). Two tandem RNase III cleavage sites determine betT mRNA stability in response to osmotic stress in Escherichia coli. PLoS One 9:e100520. 10.1371/journal.pone.0100520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D., Matz L. M., Cameron T. A., De Lay N. R. (2018). Poly(A) polymerase is required for RyhB sRNA stability and function in Escherichia coli. RNA 24 1496–1511. 10.1261/rna.067181.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A., Lucchini S., Papenfort K., Sharma C. M., Rolle K., Binnewies T. T., et al. (2008). Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeulders M. J., Keer J., Speight R. A., Williams H. D. (1999). Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181 270–283. 10.1128/jb.181.1.270-283.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E., Schuster M., Sorger-Domenigg T., Greenberg E. P., Blasi U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59 1542–1558. 10.1111/j.1365-2958.2006.05032.x [DOI] [PubMed] [Google Scholar]
- Sousa S., Marchand I., Dreyfus M. (2001). Autoregulation allows Escherichia coli RNase E to adjust continuously its synthesis to that of its substrates. Mol. Microbiol. 42 867–878. 10.1046/j.1365-2958.2001.02687.x [DOI] [PubMed] [Google Scholar]
- Steglich C., Lindell D., Futschik M., Rector T., Steen R., Chisholm S. W. (2010). Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol. 11:R54. 10.1186/gb-2010-11-5-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H., Turlan C., Khalid S., Bond P. J., Kebalo J. M., Peyron P., et al. (2015). Membrane recognition and dynamics of the RNA degradosome. PLoS Genet. 11:e1004961. 10.1371/journal.pgen.1004961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhulin I., Wartell R. M. (2002). Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30 3662–3671. 10.1093/nar/gkf508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F., Liu Y., Luo Q., Su F., Xu Y., Li F., et al. (2011). Novel organic solvent-responsive expression vectors for biocatalysis: application for development of an organic solvent-tolerant biodesulfurizing strain. Bioresour. Technol. 102 9380–9387. 10.1016/j.biortech.2011.08.015 [DOI] [PubMed] [Google Scholar]
- Tejada-Arranz A., de Crecy-Lagard V., de Reuse H. (2020). Bacterial RNA Degradosomes: molecular machines under tight control. Trends Biochem. Sci. 45 42–57. 10.1016/j.tibs.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne S. H., Williams H. D. (1997). Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 179 6894–6901. 10.1128/jb.179.22.6894-6901.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans J., Van Melderen L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67 2897–2908. 10.1007/s00018-010-0381-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- Tortosa P., Albano M., Dubnau D. (2000). Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol. Microbiol. 35 1110–1119. 10.1046/j.1365-2958.2000.01779.x [DOI] [PubMed] [Google Scholar]
- Udekwu K. I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E. G. (2005). Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19 2355–2366. 10.1101/gad.354405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unciuleac M. C., Shuman S. (2013). Distinctive effects of domain deletions on the manganese-dependent DNA polymerase and DNA phosphorylase activities of Mycobacterium smegmatis polynucleotide phosphorylase. Biochemistry 52 2967–2981. 10.1021/bi400281w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unniraman S., Chatterji M., Nagaraja V. (2002). A hairpin near the 5′ end stabilises the DNA gyrase mRNA in Mycobacterium smegmatis. Nucleic Acids Res. 30 5376–5381. 10.1093/nar/gkf697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove T. B., Zhang A., Storz G. (2016). Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30 133–138. 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool C. K., Gottesman S. (2004). Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54 1076–1089. 10.1111/j.1365-2958.2004.04348.x [DOI] [PubMed] [Google Scholar]
- Vanzo N. F., Li Y. S., Py B., Blum E., Higgins C. F., Raynal L. C., et al. (1998). Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12 2770–2781. 10.1101/gad.12.17.2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Blanco D. A., Zhou Y., Zamalloa L. G., Antonelli T., Shell S. S. (2019). mRNA degradation rates are coupled to metabolic status in Mycobacterium smegmatis. mBio 10:e00957-19. 10.1128/mBio.00957-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Perlman R. L., Pastan I. (1971). Regulation of lac transcription in antibiotic-treated E. coli. Nat. New Biol. 230 41–44. 10.1038/newbio230041a0 [DOI] [PubMed] [Google Scholar]
- Vecerek B., Rajkowitsch L., Sonnleitner E., Schroeder R., Blasi U. (2008). The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 36 133–143. 10.1093/nar/gkm985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. (2015a). The antibiotic resistance crisis: part 1: causes and threats. P T 40 277–283. [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. (2015b). The antibiotic resistance crisis: part 2: management strategies and new agents. P T 40 344–352. [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Luisi B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9 578–589. 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya I. O., Bird J. G., Zhang Y., Zhang Y., Jiao X., Barvik I., et al. (2018). CapZyme-seq comprehensively defines promoter-sequence determinants for rna 5′ capping with NAD. Mol. Cell 70 553–564.e9. 10.1016/j.molcel.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya I. O., Sharp J. S., Goldman S. R., Kanabar P. N., Livny J., Dove S. L., et al. (2012). Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 26 1498–1507. 10.1101/gad.192732.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O., Moll I., Kaberdin V. R., von Gabain A., Blasi U. (2000). Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14 1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Wagner L. A., Gesteland R. F., Dayhuff T. J., Weiss R. B. (1994). An efficient Shine-Dalgarno sequence but not translation is necessary for lacZ mRNA stability in Escherichia coli. J. Bacteriol. 176 1683–1688. 10.1128/jb.176.6.1683-1688.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ayala J. C., Benitez J. A., Silva A. J. (2012). Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J. Bacteriol. 194 1205–1215. 10.1128/JB.05900-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Dubey A. K., Suzuki K., Baker C. S., Babitzke P., Romeo T. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56 1648–1663. 10.1111/j.1365-2958.2005.04648.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman K. M., Repoila F., Rosenow C., Storz G., Gottesman S. (2001). Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15 1637–1651. 10.1101/gad.901001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B. L., Brun-Zinkernagel A. M., Simecka J. W., Pruss B. M., Babitzke P., Romeo T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40 245–256. 10.1046/j.1365-2958.2001.02380.x [DOI] [PubMed] [Google Scholar]
- Wei C. M., Gershowitz A., Moss B. (1975). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4 379–386. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- Wolfe A. D., Hahn F. E. (1965). Mode of action of chloramphenicol. Ix. Effects of chloramphenicol upon a ribosomal amino acid polymerization system and its binding to bacterial ribosome. Biochim. Biophys. Acta 95 146–155. 10.1016/0005-2787(65)90219-4 [DOI] [PubMed] [Google Scholar]
- Wood D. N., Chaussee M. A., Chaussee M. S., Buttaro B. A. (2005). Persistence of Streptococcus pyogenes in stationary-phase cultures. J. Bacteriol. 187 3319–3328. 10.1128/JB.187.10.3319-3328.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2019). No Time to Wait: Securing the Future from Drug-Resistant Infections. Available online at: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/ (accessed April 17, 2020). [Google Scholar]
- Xu C., Huang R., Teng L., Jing X., Hu J., Cui G., et al. (2015). Cellulosome stoichiometry in Clostridium cellulolyticum is regulated by selective RNA processing and stabilization. Nat. Commun. 6:6900. 10.1038/ncomms7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang J., Cohen S. N. (2008). Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 190 5526–5530. 10.1128/JB.00558-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang L., Han X., Yang W. L., Zhang M., Ma H. L., et al. (2019). RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA Decay. Mol. Cell 75 1188–1202.e11. 10.1016/j.molcel.2019.06.033 [DOI] [PubMed] [Google Scholar]
- Yarmolinsky M. B., Haba G. L. (1959). Inhibition by puromycin of amino acid incorporation into protein. Proc. Natl. Acad. Sci. U.S.A. 45 1721–1729. 10.1073/pnas.45.12.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li B., Jang G. J., Jiang S., Jiang D., Jang J. C., et al. (2019). Orchestration of Processing body dynamics and mrna decay in Arabidopsis immunity. Cell Rep. 28 2194–2205.e6. 10.1016/j.celrep.2019.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller M. E., Csanadi A., Miczak A., Rose T., Bizebard T., Kaberdin V. R. (2007). Quaternary structure and biochemical properties of mycobacterial RNase E/G. Biochem. J. 403 207–215. 10.1042/BJ20061530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurskaya H. I., Keyhan M., Matin A. (1997). The sigma S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24 643–651. 10.1046/j.1365-2958.1997.3961742.x [DOI] [PubMed] [Google Scholar]
- Zhang A., Wassarman K. M., Rosenow C., Tjaden B. C., Storz G., Gottesman S. (2003). Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50 1111–1124. 10.1046/j.1365-2958.2003.03734.x [DOI] [PubMed] [Google Scholar]
- Zhang D., Liu Y., Wang Q., Guan Z., Wang J., Liu J., et al. (2016). Structural basis of prokaryotic NAD-RNA decapping by NudC. Cell Res. 26 1062–1066. 10.1038/cr.2016.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li D., Zhao L., Fleming J., Lin N., Wang T., et al. (2013). Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45 1255–1260. 10.1038/ng.2735 [DOI] [PubMed] [Google Scholar]
- Zhao B. S., Wang X., Beadell A. V., Lu Z., Shi H., Kuuspalu A., et al. (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542 475–478. 10.1038/nature21355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. P., Zhu H., Guo X. P., Sun Y. C. (2018). AU-rich long 3′ untranslated region regulates gene expression in bacteria. Front. Microbiol. 9:3080. 10.3389/fmicb.2018.03080 [DOI] [PMC free article] [PubMed] [Google Scholar]