Abstract
Background
Based on MLST analyses the global population of C. difficile is distributed in eight clades, of which Clade 2 includes the “hypervirulent” NAP1/RT027/ST01 strain along with various unexplored sequence types (STs).
Methods
To clarify whether this clinically relevant phenotype is a widespread feature of C. difficile Clade 2, we used the murine ileal loop model to compare the in vivo pro-inflammatory (TNF-α, IL-1β, IL-6) and oxidative stress activities (MPO) of five Clade 2 clinical C. difficile isolates from sequence types (STs) 01, 41, 67, and 252. Besides, we infected Golden Syrian hamsters with spores from these strains to determine their lethality, and obtain a histological evaluation of tissue damage, WBC counts, and serum injury biomarkers (LDH, ALT, AST, albumin, BUN, creatinine, Na+, and Cl−). Genomic distances were calculated using Mash and FastANI to explore whether the responses were dictated by phylogeny.
Results
The ST01 isolate tested ranked first in all assays, as it induced the highest overall levels of pro-inflammatory cytokines, MPO activity, epithelial damage, biochemical markers, and mortality measured in both animal models. Statistically indistinguishable or rather similar outputs were obtained for a ST67 isolate in tests such as tissue damage, neutrophils count, and lethal activity. The results recorded for the two ST41 isolates tested were of intermediate magnitude and the ST252 isolate displayed the lowest pathogenic potential in all animal experiments. This ordering matched the genomic distance of the ST01 isolate to the non-ST01 isolates.
Conclusions
Despite their close phylogenic relatedness, our results demonstrate differences in pathogenicity and virulence levels in Clade 2 C. difficile strains, confirm the high severity of infections caused by the NAP1/RT027/ST01 strain, and highlight the importance of C. difficile typing.
Keywords: Pathogenicity, Virulence, Clostridium difficile, MLST Clade 2, NAP1/RT027/ST01
Introduction
The main causative agent of nosocomial diarrhea associated with the use and abuse of antibiotics is Clostridium difficile; an anaerobic, Gram-positive, spore-forming bacterium. C. difficile infection (CDI) symptoms include watery diarrhea, anorexia and leukocytosis and in severe disease presentations these features could be accompanied by dehydration, hypoalbuminemia, acute kidney injury, and hypotension or death [1]. Different genotypes of this bacterium have been isolated worldwide and the prevalence rates varied between the countries [2].
Most C. difficile strains produce toxin A (TcdA) and/or toxin B (TcdB), which display cytotoxic activity through glycosylation of small, monomeric GTPases. In addition, a subset of strains synthesize an actin-ADP-ribosylating toxin known as binary toxin (CDT) that targets the actin cytoskeleton [3]. Genes encoding TcdA and TcdB are located on a so-called Pathogenicity Locus (PaLoc). The genes for CDT, by contrast, are typically located elsewhere on the chromosome [4] or on extrachromosomal molecules [5, 6].
TcdA and TcdB induce the secretion of several proinflammatory cytokines (e.g. IL-1β, IL-12, and TNF-α) and lead to neutrophil and macrophage infiltration to intestinal tissues that can result in host damage, as seen in intestinal histopathology specimens [7, 8].
The global population of C. difficile is distributed in eight multilocus sequence typing (MLST) clades, of which five contain strains of common detection in human hosts [9]. In the last decade, many countries have reported outbreaks of CDI, partly due to the emergence and rapid spread of hypervirulent or epidemic strains, such as one classified as NAP1 by PFGE, RT027 by ribotyping, and ST01 by MLST [10–13].
Besides its epidemic potential, the NAP1/RT027/ST01 strain has received particular attention due to its link to CDI cases of increased severity and mortality [14]. This strain overproduces TcdA and TcdB, secretes binary toxin (CDT), harbors mutations in genes encoding S-layer proteins (SLPs) that increase its adherence to the gut epithelium and overproduce spores [15]. Some reports have highlighted these characteristics as likely contributors to its increased virulence [10]. However, the magnitude of these phenotypes is not equal in all isolates from this genotype [16], and their relative contribution to its epidemicity remains to be elucidated.
The NAP1/RT027/ST01 strain is classified into the so-called hypervirulent MLST Clade 2 [17, 18] along with 66 additional STs [19, 20]. It is yet unknown whether “hypervirulence” is a universal trait of the members of this clade. To answer this open question and to widen our current knowledge on the pathogenesis of C. difficile Clade 2 strains, we performed a comprehensive in vivo analysis of the pathogenicity and lethality of clinical C. difficile strains typed as ST01, ST41, ST67, and ST252 in two animal models and interpreted the results on the basis of their phylogenomic relatedness.
Results
5758-ST01 and 5757-ST67 bacteria-free supernatants induced the strongest levels of pro-inflammatory response and tissue damage in the murine ileal loop model
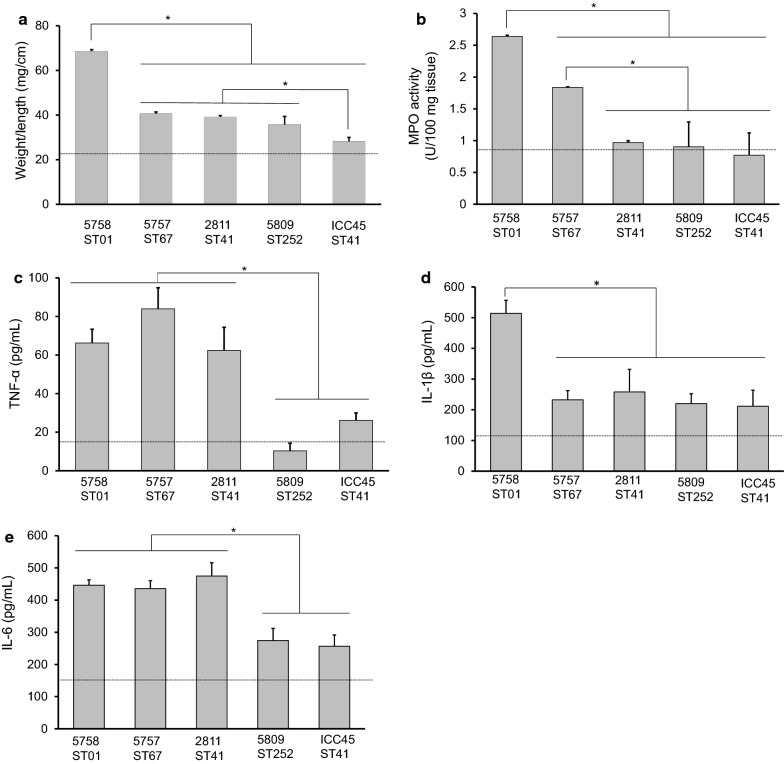
The pathogenic potential of the strains was assessed through inoculation of bacterial cell-free supernatants in murine ileal loops and subsequent measurement of the normalized weight of ileal sections as a proxy for edema, MPO (myeloperoxidase) activity as an indicator of neutrophilic degranulation, and several cytokines as markers of acute inflammation. A normalization of supernatant volumes was not required because all the supernatants were taken at equivalent time points of the growth curve. Supernatants from strain 5758-ST01 induced the highest level of edema (69 mg/cm, p < 0.05, Fig. 1a). Strains 5757-ST67, 2811-ST41, and 5809-ST252 induced intermediate responses (39–41 mg/cm), which in all cases were above the levels determined for ICC45-ST41 (28 mg/cm) and the controls (PBS, TYT broth, or non-toxigenic C. difficile ATCC® 700057 supernatant, 24 mg/cm) (Fig. 1a).
Fig. 1.
Effect of bacterial cell-free supernatants of Clade 2 C. difficile strains in the ligated murine ileal loop. Bacteria-free supernatants from the indicated genotypes were prepared in TYT-broth. 7–10 mice per group were inoculated with 0.3 ml of supernatant in ligated ileal loops. Four hours after inoculation, mice were euthanized and data was recorded. a ratio weight/length, bars represent means of the normalized weight of ileal sections. b myeloperoxidase activity (MPO) for each group. Cytokines levels were measured on intestinal tissue. c TNF-α, d IL-1β, and e IL-6. *p <0.05 One-way ANOVA followed by Bonferroni’s correction. The mean of measurements obtained for control groups (PBS, TYT-broth and non-toxigenic supernatant) is indicated by a dashed line
As to MPO activity, 5758-ST01 and 5757-ST67 supernatants caused a significant increase (2.6 and 1.8 U/100 mg of tissue) in its activity compared to supernatants from the other isolates with the former strain causing the highest MPO release overall (p < 0.05) (Fig. 1b).
Regarding the detection of cytokines in ileal tissue, 5758-ST01, 5757-ST67, and 2811-ST41 supernatants induced an increase in TNF-α (62–83 pg/mL) and IL-6 (446–475 pg/mL) levels when compared with supernatants from the other strains (256–274 pg/mL). Additionally, 5758–ST01 induced a higher level of IL-1β (514 pg/mL, p < 0.05) (Fig. 1c, d, e). Finally, and in line with the results described above, the histopathologic analyses revealed a greater level of cellular infiltration and epithelial damage in tissue exposed to supernatants from 5758-ST01 or 5757-ST67 (Fig. 2a and Additional file 1: Fig. S1).
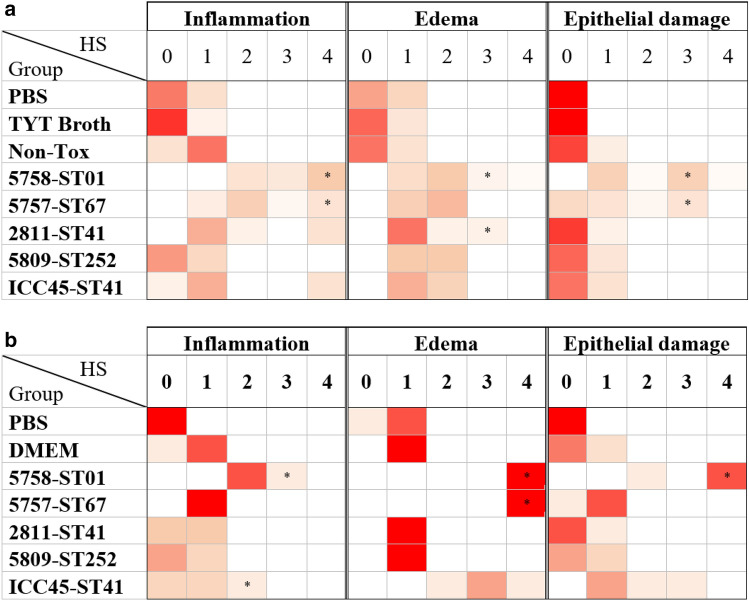
Fig. 2.
Histopathological scores obtained from intestinal tissue inoculated with bacteria-free supernatants in the murine ligated ileal loop model and of cecal tissue from infected hamsters. a Ligated-ileal loop in 7–10 mice per group were inoculated with bacteria-free supernatants of the indicated genotypes. Four hours after treatment the animals were euthanized and intestinal tissues were collected and processed as described in the Methods section. Tissue alterations were scored on coded slides, using a histopathological score (HS) scale from 0 (absence of alterations) to 4 (severe) for three different effects: inflammation (neutrophil infiltration), edema, and epithelial damage. b Syrian Golden hamsters previously treated with clindamycin were orally inoculated with spores from the indicated strains. A chirurgical resection of the cecum of the deceased individuals was performed to record histopathological damage. These alterations were scored on coded slides, using a histopathological score (HS) scale that ranges from 0 (absence of alterations) to 4 (severe) for three different effects: inflammation (neutrophil infiltration), edema, and epithelial damage. Results are presented using a heatmap with a scale that ranges from red (100% of samples) to white (0% of samples). *p < 0.05 Kruskall-Wallis and Dunn’s multiple comparison test with Benjamini–Hochberg correction
5758-ST01 strain was more lethal, caused more epithelial destruction, and a stronger pro-inflammatory response in the hamster model
Clindamycin-treated hamsters were administered with C. difficile spores and their survival rate was measured daily for 15 days. Whereas 5758-ST01 strains colonized the animals within 1 to 3 days, the remaining strains required 3 to 5 days to achieve the same output. Hamsters infected with 5758-ST01 spores showed wet-tail 1.5 days post-inoculation. A similar result was recorded for animals that received spores from strains 5757-ST67 or 2811-ST41, as they developed wet-tail after 2 to 3 days. This sign was not observed after 5 days for the other strains and the controls.
The survival rates of hamsters inoculated with 5758-ST01 decreased rapidly and none of them survived by day 4 (Fig. 3). By contrast, hamsters infected with 5757- ST67 or 2811-ST41 spores reached 30% survival at day 7. Hamsters inoculated with spores from strains ICC45-ST41 or 5809-ST252 presented survival rates of 90% and 100% by day 12, respectively (Fig. 3).
Fig. 3.
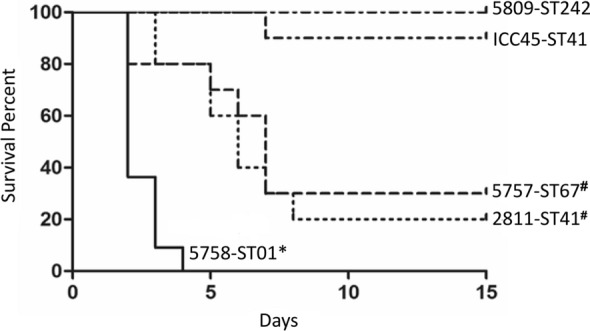
Kaplan-Meier survival curves of hamsters infected with spores of strains from different STs within the C. difficile Clade 2. Syrian Golden hamsters previously treated with clindamycin were orally inoculated with spores from the indicated strains. Hamsters were monitored at 12 h intervals for signs of C. difficile infection, and the numbers of deceased animals were recorded. C. difficile isolates obtained from fecal pellets were typed to confirm the inoculated strain. *,#p < 0.05 Mantel-Cox test
In agreement with these results, the histopathologic evaluation showed an increased cell infiltration and epithelial damage in the cecum of hamsters infected with 5758-ST01 when compared to the other strains (p < 0.5, Fig. 2b and Additional file 1: Fig. S2). Furthermore, the edema produced by 5758-ST01 was similar to the one measured in 5757-ST67 infected animals (HS = 4).
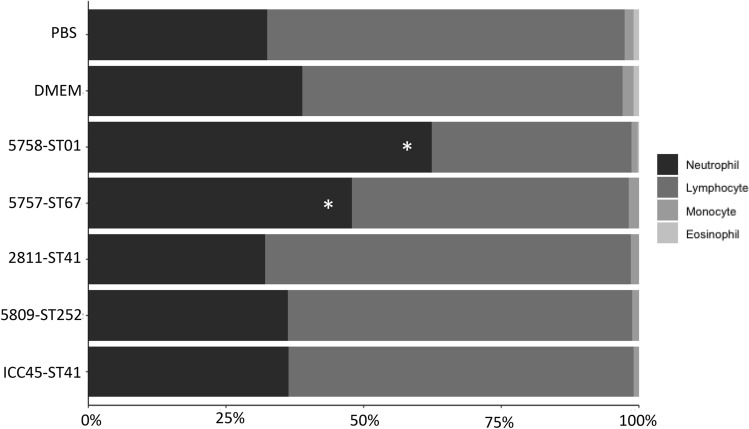
Strains 5758-ST01 and 5757-ST67 triggered an increase in neutrophil counts and biochemical alterations in peripheric blood in hamsters
After 1 or 2 days of a C. difficile positive feces culture of all animals from each group the euthanasia was performed, then white blood cells (WBC) differential and blood biochemical tests were obtained. Compared to the other groups, hamsters inoculated with strains 5758-ST01 or 5757-ST67 showed signs of infection, such as an increase in peripheral neutrophils (50–60%, p < 0.5) (Fig. 4).
Fig. 4.
White blood cell (CBC) counts in infected hamsters measured 1 or 2 days after C. difficile detection in their feces. Animals were euthanized on day 6 after infection (those with 5758-ST01 were euthanized on day 3 due to their rapid mortality and detection of C. difficile in the feces at this time point). Blood samples were collected and WBC counts were performed on stained smears. Counts are expressed in percentages of cells observed. *p < 0.05 Kruskall-Wallis and Dunn’s multiple comparison test with Benjamini–Hochberg correction
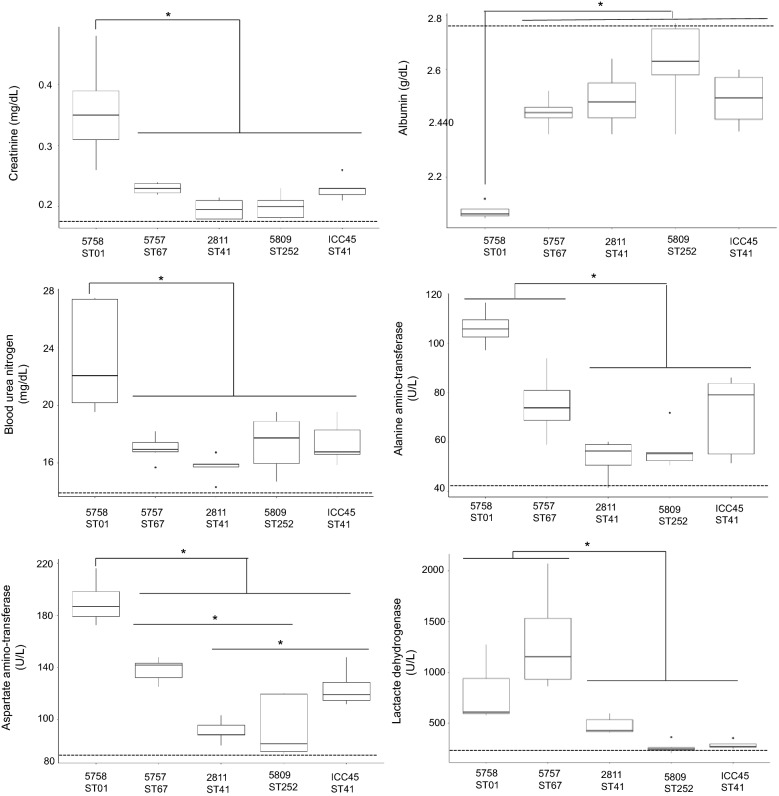
The biochemical testing suggested a general multiorgan alteration in hamsters infected with 5758-ST01. Unlike other groups, animals that received 5758-ST01 spores presented alterations in liver enzymes (ALT and AST), serum proteins synthesized in the liver (albumin), renal function impairment (creatinine and BUN), tissue damage (LDH), and serum electrolytes (sodium and chloride) (p < 0.5) (Fig. 5). Animals infected with 5757-ST067 showed similar alterations in ALT and LDH as hamsters that received 5758-ST01 spores (p < 0.5). The biochemical analytes measured in all other animals were indistinguishable from those recorded in the control groups that received PBS or DMEM (p > 0.5) (Fig. 5).
Fig. 5.
Serological parameters of infected hamsters measured 1 or 2 days after C. difficile detection in their feces. Animals were euthanized 1 or 2 days after C. difficile detection on feces (day 3 for 5758-ST01 and day 6 for all other groups). The results are presented as the average ± standard error. The following biomarkers were measured in hamster serum: a creatinine, b blood urea nitrogen (BUN), c alanine amino-transferase (ALT), d aspartate amino-transferase (AST), e lactate dehydrogenase (LDH), and f albumin. * p < 0.05 One-way ANOVA with Bonferroni’s correction. The mean of measurements for control groups (DMEM and PBS) is indicated by a dashed line
5757-ST67 showed the smallest genomic distance of all tested strains to 5758-ST01
The magnitude of the in vivo phenotypes matched their genomic distance of all non-ST01 isolates to 5758-ST01. The smallest Mash distance (0.0048) and highest ANI value (99.34) was determined for strain 5757-ST67, which was followed by the two ST41 strains tested (2811-ST41 and ICC45-ST41, Mash = 0.0061–0.0062, ANI = 99.18-99.30) and finally by strain 5809-ST252 (Mash = 0.0096, ANI = 98.86). (Table 1).
Table 1.
Genomic relatedness of Clade 2 C. difficile strains compared to strain 5758-ST01
| Strain | ANIa | Mashb |
|---|---|---|
| 5757-ST67 | 99.34 (1296/1354) | 0.0048 (826/1000) |
| ICC45-ST41 | 99.30 (1313/1427) | 0.0061 (786/1000) |
| 2811-ST41 | 99.18 (1296/1402) | 0.0062 (781/1000) |
| 5809-ST252 | 98.86 (1262/1381) | 0.0096 (690/1000) |
a Average nucleotide identity (ANI) values (bidirectional fragment mapping/total query fragments)
b Mash distance (shared-hashes)
Discussion
The use of MLST technique has facilitated the classification of isolates into several clades. Clade 2 includes ST01 strains [17, 18], which have caused outbreaks worldwide [21, 22]. This clade has been divided into subgroups 2i (ST01, ST67, ST41, ST62 or ST140) and 2ii (ST32, ST47, ST61, ST95, ST97, ST123 or ST252) [23]. This work revealed a broad spectrum of virulence and pathogenic potential among five clinical isolates from different STs within the C. difficile “hypervirulent” Clade 2.
Strain 5758-ST01 from subgroup 2i was the most virulent strain in all in vivo experiments, as it induced higher cytokine secretion in intestinal tissue, tissue damage, multiorgan alterations, and mortality. This could be due a combination of already reported characteristics of the NAP1/RT027/ST01 strain, such as toxin overproduction [24], binary toxin secretion [25], and increased sporulation [26]. However, other unexplored mechanisms such as polymorphisms and/or regulation patterns of colonization determinants like S-layer protein, flagellin, β-glucosidases and other adhesins [27–30] could also have played a role in the pathogenesis. A detailed studied of those mechanisms is beyond the scope of this study.
An even wider pathogenicity spectrum could have been observed if more isolates from other STs had been studied. Nonetheless, our results suggest that the relative virulence of the non-ST01 strains goes in line with their phylogenomic distance to the well-characterized NAP1/RT027/ST01 genotype.
Strain 5757-ST67 was second in terms of the damage inflicted in animal experiments. This is somewhat expectable, as ST-67 strains also belong to subgroup 2i and have been reported to cause severe CDI [31]. Despite having a variant TcdB, 5757-ST67 produced an elevated inflammatory response and alteration of biochemical markers that could be explained by toxin overproduction [32]. Moreover, mutations in treR (trehalose repressor), CpG depleted genomes [31] and a PaLoc rather similar to that of strain R202091-ST01 [33] have been described in this ST, all of which are characteristics of hypervirulent C. difficile strains.
The proinflammatory response and mortality levels recorded for strain 2811-ST41, also belonging to subgroup 2i, did not reach 5758-ST01 and 5757-ST67 levels. Furthermore, strain ICC45-ST41 produced weaker proinflammatory responses than 2811-ST41. Among multiple explanations, this difference could be tracked to the synthesis of a variant TcdB in the former [34]. However, information on the amount of secreted TcdA and CDT by strains from this ST is still lacking, though it could clarify the observed differences in proinflammatory potential.
Strain 5809-ST252, from subgroup 2ii, presented the weakest proinflammatory response and lethal potential of the studied strains. We did not find reports in the literature on the pathogenesis of this type of C. difficile strains and therefore cannot provide explanations for its behavior.
Our results could be explained by undescribed differences in the strains´ exoproteomes and attributed to structural particularities on the bacterial surface that alter human intestinal homeostasis [29]. Other factors, unexplored in this work, such as germination potential, colonization efficiency, the number of metabolically active C. difficile cells in the intestine, and bile acids metabolism could explain the differences in the damage observed in the infection models [35–37].
Contradictory observations on in vitro phenotypes expressed by isolates from the same ribotype, such as RT027, have been reported, particularly with regard to sporulation and toxin production [16, 38, 39]. Therefore, in vivo studies, such as the work herein presented, have the potential to clarify pending issues and should be carried out with two or more animal models and including others STs from the “hypervirulent” Clade 2.
We observed indications of extraintestinal organic damage, mainly in renal and hepatic functions, as well as an increase in leukocytes, these alterations were recorded after 1 or 2 days of C. difficile detection on feces. The animals infected with 5758-ST01 showed altered biochemical parameters sooner than those infected with non-ST01 strains (exposed for 3 additional days). As such, in an early infection, the general health condition is altered by 5758-ST01 infection but not by non-ST01 strains. These findings have been described in the severe outcome of CDI in humans [40, 41], which suggest a severe pathology in animals infected by the 5758-ST01 strain.
We conclude that “hypervirulence” is not a widespread feature in C. difficile Clade 2 strains. This result lay the foundations for interpretation of future in vitro and genomic comparisons of these and other Clade 2 strains. Moreover, it highlights the importance of C. difficile typing and of targeted diagnosis of NAP1/027/ST01 strains.
Materials and methods
C. difficile strains
This study was done with five C. difficile MLST Clade 2 strains that were isolated from stool samples of symptomatic patients. Briefly, samples were treated with 96% ethanol and inoculated onto Cefoxitin-Cycloserine-Fructose agar plates (Oxoid) that were later incubated in anaerobic chamber (90% N2, 5% CO2, 5% H2). Colonies were identified phenotypically with the RapID 32A system (bioMerieux) and by PCR-amplification of the tpi gene. These bacteria were previously analyzed by PFGE, an end-point PCR targeting different PaLoc fragments, and toxigenic culture following the previously reported protocols [42]. All strains were cryopreserved at − 80° C and recovered through culturing on Brucella agar plates supplemented with vitamin K and laked horse blood under anaerobic conditions (Table 2).
Table 2.
C. difficile MLST Clade 2 strains used in this study
| Strain | ST | PFGE-typea | PCR PaLoc profile | CPEb |
|---|---|---|---|---|
| 5758 | 01 | NAP1 | tcdA+/tcdB+/cdtB+/tcdC+, del | COv |
| 2811 | 41 | New type | tcdA+/tcdB+/cdtB+/tcdC+ | C |
| ICC45 | 41 | New type | tcdA+/tcdB+/cdtB+/tcdC+ | V |
| 5757 | 67 | NAP1 | tcdA+/tcdB+/cdtB+/tcdC+, del | VOv |
| 5809 | 252 | New type | tcdA+/tcdB+/cdtB+/tcdC+ | C |
Preparation of bacterial cell-free supernatants
All strains were grown in TYT-broth (3% tryptose, 2% yeast extract, 0.1% thioglycolate, and pH 6.8) for 24 h under anaerobic conditions. At this time point, viable bacterial cells counts for all strains were in the 107 CFU/mL order (Additional file 1: Fig. S3). Bacterial cells were then pelleted by centrifugation for 30 min at 20,000 g and the resulting supernatants were filtered using 0.2 µm pore membranes [32]. These bacterial cell-free supernatants were used in the ileal loop assay in mice (see below).
Murine ileal loop model
Four to 5 weeks old, male, Hsd:ICR mice with a body weight of 18–25 g were used. These animals were grouped in polycarbonate cages and maintained under controlled conditions of temperature (19.9 ± 0.7 °C and 23.5 ± 1.0 °C), relative moisture (75 ± 5% and 89 ± 3%), noise (81.4 ± 2.2 dB); and photoperiod (12 h). Water and feed were available ad libitum. All animal proceedings were in compliance with local legislation (Ley de Bienestar de los Animales N° 7451) and approved by the Comité Institucional de Cuido y Uso de Animales (CICUA) from the Universidad de Costa Rica (CICUA 52-15). We used the maximum amount of animals authorized by CICUA based on the 3R’s principles.
Mice were fasted overnight and then anesthetized with ketamine (60 mg/kg) and xylazine (5 mg/kg). Through a midline laparotomy, a 4 cm ileal loop was ligated and injected with 0.3 ml of bacterial cell-free supernatants or control solutions (PBS, TYT-broth or non-toxigenic C. difficile ATCC® 700057 supernatant). Four hours after inoculation mice were euthanized and the length and weight of the intestinal loops were recorded [43, 44].
Myeloperoxidase (MPO) assay
Neutrophils degranulation in homogenized ileal tissue was evaluated using a colorimetric MPO activity assay [45]. Briefly, ileal tissue (100 mg) was homogenized in hexadecyltrimethylammonium bromide (HTAB) (Sigma) buffer (PBS, HTAB 50% w/v, and H2O2 0.1% v/v) and cleared by centrifugation at 4500 g for 7 min at 4 °C. The resulting supernatants were incubated with a 0.017% o-dianisidine solution (Sigma) and after 5 min the absorbance was determined at 450 nm. Results were reported as MPO/100 mg of ileal tissue.
Detection of TNF-α, IL-1β, IL-6, and IL-10
The concentration of proinflammatory interleukins (IL-1β, IL-6, IL-10) and tumor necrosis factor alpha (TNF-α) in ileal tissue homogenates was determined with commercial enzyme-linked immunosorbent assays (ELISA) following the instructions of the manufacturer (R&D Systems).
Hamster infection model
Six to eight weeks old, male, Golden Syrian hamsters (Mesocricetus auratus) with a body weight of 85–120 g were used. Similarly to previous studies [46, 47], the hamsters were separated into groups of 5 animals.
The animals were maintained in polycarbonate cages under the conditions mentioned above for mice. Two clindamycin doses were administered to each individual through the oral route 10 (30 mg/kg) and 5 days (50 mg/kg) prior to the beginning of the experiment. On day 0, each animal was inoculated orally with 104 spores that were prepared as previously described [48]. Seven groups of animals were used, including five treatments (five strains) and two negative controls (PBS or DMEM).
Animals were monitored for 15 days for infection signs, such as weight loss and wet tail (diarrhea), and to record their mortality. Stool samples were collected every 2 days throughout the experiment for isolation of C. difficile following published protocols [49]. When an experimental subject died, or at day 15 (when euthanasia was performed), a chirurgical resection of the ascendant colon and the cecum was performed to monitor colonization by C. difficile.
Analysis of blood biomarkers
Additional groups of 5 Golden Syrian hamsters were inoculated with spores of each bacterial strain and negative controls (PBS or DMEM) as described above. All animals infected with 5758-ST01 strain were euthanized on day 3 (due to its rapid lethality rate associated with this strain) and all other groups on day 6 post-spore inoculation. The criteria in this matter was that euthanasia (and subsequent blood collection) should be performed 1 or 2 days after successful colonization or infection by C. difficile (as demonstrated by a positive fecal C. difficile culture).
Then, a portion of whole blood was collected after decapitation to measure serum levels of albumin, creatinine, blood urea nitrogen (BUN), sodium, chloride, lactate dehydrogenase (LDH), alanine amino-transferase (ALT), and aspartate amino-transferase (AST) levels with an automated analyzer (Roche Cobas® 3c11). Another portion of total blood was collected in microtubes with EDTA to obtain white blood cells (WBC) differential counts through visualization of Wright-stained blood smears.
Histopathologic assessment of tissue sections
Murine ileal and hamsters’ cecum samples were fixed in 10% buffered formalin, processed, and stained with hematoxylin and eosin (H&E) for histopathological evaluation. These preparations were evaluated for epithelial damage, edema, and neutrophil infiltration using a histopathological scoring (HS) system that ranged from 0 (absence of alterations) to 4 (severe) [50]. All histopathological assays were performed by an expert DVM pathologist in a single blind setting.
Phylogenomic analyses
Whole genomes were reconstructed from 2 × 250 bp paired-end Illumina reads from the NCBI BioProject PRJNA293889. The genomic relatedness of the non-ST01 isolates to 5757-ST01 strain was determined using Mash [51], which is based on a MinHash dimensionality-reduction technique, and through calculation of average nucleotide identity (ANI) values with an alignment-free method [52].
Statistical analyses
Data from the animal models are presented as mean ± standard error (SEM) or as medians. Means and median were compared using one-way ANOVA tests with Bonferroni correction or Kruskall-Wallis tests followed by Dunn’s multiple comparison tests with Benjamini–Hochberg adjustment, respectively. Mortality was evaluated using Kaplan–Meier curves. P values < 0.05 were considered statistically significant.
Supplementary information
Additional file 1: Figure S1. Histopathological analysis of the effects induced by bacteria-free supernatants in the murine model of ileal ligated loop. Tissue was fixed in 10% buffered formalin and stained with H&E for histological evaluation of the following groups: A) PBS, B) TYT-broth, C) Non-toxigenic C. difficile ATCC® 700057, D) 5758-ST01, E) 5757-ST67, F) 2811-ST41, G) 5809-ST252 and H) ICC45-ST41. Figure S2. Histopathological analysis of cecum from infected hamsters. Cecum sections were fixed in 10% buffered formalin and stained with H&E for histological evaluation of the following groups: A) PBS, B) DMEM, C) 5758-ST01, D) 5757-ST67, E) 2811-ST41, F) 5809-ST252, and G) ICC45-ST41. Figure S3. Counts of viable C. difficile cells by strain. Growth curves were performed in TYT-broth under the described conditions. At 0, 8 and 24h, an aliquot was taken, serially diluted and inoculated onto Brucella agar plates supplemented with vitamin K agar plates. The number of colonies that appeared after 48 h of incubation was recorded to obtain CFU/mL values.
Acknowledgements
We thank Pablo Vargas and Robin Cárdenas for their technical assistance. We further thank Dr. Sara González-Camacho and the LEBi-UCR staff for their support with the animal experiments.
Abbreviations
- ALT
Alanine amino-transferase
- AST
Aspartate amino-transferase
- ANI
Average nucleotide identity
- BUN
Blood urea nitrogen
- CDI
Clostridium difficile infection
- CDT
Binary toxin
- DMEM
Dulbecco’s Modified Eagle’s medium
- H&E
Hematoxylin and eosin
- HS
Histopathological score
- HTAB
Hexadecyltrimethylammonium bromide
- IL
Interleukin
- LDH
lactate dehydrogenase
- MLST
Multilocus sequence typing
- MPO
Myeloperoxidase
- NAP
North American PFGE type
- PaLoc
Pathogenecity locus
- PBS
Phosphate buffered saline
- RT
Ribotype
- SLP
S-layer
- ST
Sequence type
- TcdA
Toxin A
- TcdB
Toxin B
- TNF
Tumor necrosis factor
- TYT
Tryptose-yeast extract-thioglycolate
- WBC
White blood cells
Authors’ contributions
JOA: performed experiments, analyzed data. AAA: performed histopathological analyses. LAA: analyzed data, wrote manuscript. ECO: Provided reagents, analyzed data, edited manuscript. CR: provided advices in experimental design, critically discussed results, performed genomic analyses, wrote manuscript. CQG: designed, supervised and performed experiments, analyzed data, wrote final paper. All authors read and approved the final manuscript.
Funding
This work was funded by the Vicerrectory of Research of the University of Costa Rica.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file 1.
Ethics approval and consent to participate
All animal experiments were conducted in accordance with the local legislation (Ley de Bienestar de los Animales N° 7451) following approval of the protocol by the Comité Institucional de Cuido y Uso de Animales (CICUA) from the Universidad de Costa Rica (CICUA 52-15).
Consent for publication
Not applicable.
Competing interests
The authors declare that they do not have competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13099-020-00383-4.
References
- 1.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 2.Seugendo M, Janssen I, Lang V, Hasibuan I, Bohne W, Cooper P, et al. Prevalence and strain characterization of clostridioides (Clostridium) difficile in representative regions of germany, ghana, tanzania and indonesia—a comparative multi-center cross-sectional study. Front Microbiol. 2018;9:1843. doi: 10.3389/fmicb.2018.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012;3:121–134. doi: 10.4161/gmic.19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jank T, Giesemann T, Aktories K. Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J Biol Chem. 2007;282:35222–35231. doi: 10.1074/jbc.M703138200. [DOI] [PubMed] [Google Scholar]
- 5.Riedel T, Wittmann J, Bunk B, Schober I, Spröer C, Gronow S, et al. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J Biotechnol. 2017;250:23–28. doi: 10.1016/j.jbiotec.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Ramírez-Vargas G, López-Ureña D, Badilla A, Orozco-Aguilar J, Murillo T, Rojas P, et al. Novel Clade C-I Clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-32390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genth H, Dreger SC, Huelsenbeck J, Just I. Clostridium difficile toxins: more than mere inhibitors of Rho proteins. Int J Biochem Cell Biol. 2008;40:592–597. doi: 10.1016/j.biocel.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Madan R, Jr, Petri WA. Immune responses to Clostridium difficile infection. Trends Mol. Med. 2012;18(11):658–666. doi: 10.1016/j.molmed.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol. 2014;6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–1924. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 11.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goorhuis A, Van der Kooi T, Vaessen N, Dekker FW, Van den Berg R, Harmanus C, et al. Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin Infect Dis. 2007;45:695–703. doi: 10.1086/520984. [DOI] [PubMed] [Google Scholar]
- 13.Kuijper EJ, Coignard B, Tüll P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;6:2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 14.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 15.Akerlund T, Persson I, Unemo M, Norén T, Svenungsson B, Wullt M, et al. Increased sporulation rate of epidemic Clostridium difficile Type 027/NAP1. J Clin Microbiol. 2008;46:1530–1533. doi: 10.1128/JCM.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson PE, Walk ST, Bourgis AET, Liu MW, Kopliku F, Lo E, et al. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe. 2013;24:109–116. doi: 10.1016/j.anaerobe.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingle KE, Griffiths D, Didelot X, Evans J, Vaughan A, Kachrimanidou M, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS ONE. 2011;6:e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janezic S, Rupnik M. Genomic diversity of Clostridium difficile strains. Res Microbiol. 2015;166:353–360. doi: 10.1016/j.resmic.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Valiente E, Cairns MD, Wren BW. The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move. Clin Microbiol Infect. 2014;20:396–404. doi: 10.1111/1469-0691.12619. [DOI] [PubMed] [Google Scholar]
- 21.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson LF, Valiente E, Donahue EH, Birchenough G, Wren BW. Hypervirulent Clostridium difficile PCR-ribotypes exhibit resistance to widely used disinfectants. PLoS ONE. 2011;6:e25754. doi: 10.1371/journal.pone.0025754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stabler RA, Dawson LF, Valiente E, Cairns MD, Martin MJ, Donahue EH, et al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS ONE. 2012;7:e31559. doi: 10.1371/journal.pone.0031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanis JM, Heinlen LD, James JA, Ballard JD. Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog. 2013;9:e1003523. doi: 10.1371/journal.ppat.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aktories K, Papatheodorou P, Schwan C. Binary Clostridium difficile toxin (CDT)—a virulence factor disturbing the cytoskeleton. Anaerobe. 2018;53:21–29. doi: 10.1016/j.anaerobe.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batah J, Kobeissy H, Pham PTB, Denève-Larrazet C, Kuehne S, Collignon A, et al. Clostridium difficile flagella induce a pro-inflammatory response in intestinal epithelium of mice in cooperation with toxins. Sci Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-03621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BAP, et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS ONE. 2013;8:e78404. doi: 10.1371/journal.pone.0078404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janoir C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe. 2016;37:13–24. doi: 10.1016/j.anaerobe.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Shaw HA, Preston MD, Vendrik KEW, Cairns MD, Browne HP, Stabler RA, et al. The recent emergence of a highly related virulent Clostridium difficile clade with unique characteristics. Clin Microbiol Infect. 2020;26:492–498. doi: 10.1016/j.cmi.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao H, Wong SCY, Yam WC, Liu MCJ, Chow KH, Wu AKL, et al. Genomic investigation of a sequence type 67 Clostridium difficile causing community-acquired fulminant colitis in Hong Kong. Int J Med Microbiol. 2019;309:270–273. doi: 10.1016/j.ijmm.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Quesada-Gómez C, López-Ureña D, Chumbler N, Kroh HK, Castro-Peña C, Rodríguez C, et al. Analysis of TcdBs within the hypervirulent clade 2 reveals an impact of RhoA glucosylation in Clostridium difficile pro inflammatory activities. Infect Immun. 2016;84:856–865. doi: 10.1128/IAI.01291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito R, Usui Y, Ayibieke A, Nakajima J, Prah I, Sonobe K, et al. Hypervirulent clade 2, ribotype 019/sequence type 67 Clostridioides difficile strain from Japan. Gut Pathog. 2019;11:54. doi: 10.1186/s13099-019-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa CL, López-Ureña D, de Oliveira Assis T, Ribeiro RA, Silva ROS, Rupnik M, et al. A MLST Clade 2 Clostridium difficile strain with a variant TcdB induces severe inflammatory and oxidative response associated with mucosal disruption. Anaerobe. 2016;40:76–84. doi: 10.1016/j.anaerobe.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of Bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE. 2010;5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant Clostridium difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burns DA, Heeg D, Cartman ST, Minton NP. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS ONE. 2011;6:e24894. doi: 10.1371/journal.pone.0024894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns DA, Heap JT, Minton NP. The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe. 2010;16:618–622. doi: 10.1016/j.anaerobe.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Bauer MP, Hensgens MPM, Miller MA, Gerding DN, Wilcox MH, Dale AP, et al. Renal failure and leukocytosis are predictors of a complicated course of Clostridium difficile infection if measured on day of diagnosis. Clin Infect Dis. 2012;55:149–153. doi: 10.1093/cid/cis340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pant C, Madonia P, Minocha A, Manas K, Jordan P, Bass P. Laboratory markers as predictors of mortality in patients with Clostridium difficile infection. J Investig Med. 2010;58:43–45. doi: 10.2310/JIM.0b013e3181bca525. [DOI] [PubMed] [Google Scholar]
- 42.Quesada-Gómez C, López-Ureña D, Acuña-Amador L, Villalobos-Zúñiga M, Du T, Freire R, et al. Emergence of an outbreak-associated Clostridium difficile variant with increased virulence. J Clin Microbiol. 2015;53:1216–1226. doi: 10.1128/JCM.03058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlowski SW, Calabrese G, Kolling GL, Platts-Mills J, Freire R, Warren C, et al. Murine model of Clostridium difficile infection with aged gnotobiotic C57BL/6 mice and a BI/NAP1 strain. J Infect Dis. 2010;202:1708–1712. doi: 10.1086/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavalcante IC, Castro MV, Barreto ARF, Sullivan GW, Vale M, Almeida PRC, et al. Effect of novel A2A adenosine receptor agonist ATL 313 on Clostridium difficile toxin A-induced murine ileal enteritis. Infect Immun. 2006;74:2606–2612. doi: 10.1128/IAI.74.5.2606-2612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Araújo Junqueira AFT, Dias AAM, Vale ML, Spilborghs GMGT, Bossa AS, Lima BB, et al. Adenosine deaminase inhibition prevents Clostridium difficile toxin A-induced enteritis in mice. Infect Immun. 2011;79:653–662. doi: 10.1128/IAI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckley AM, Spencer J, Maclellan LM, Candlish D, Irvine JJ, Douce GR. Susceptibility of hamsters to Clostridium difficile isolates of differing toxinotype. PLoS ONE. 2013;8:e64121. doi: 10.1371/journal.pone.0064121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goulding D, Thompson H, Emerson J, Fairweather NF, Dougan G, Douce GR. Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1. Infect Immun. 2009;77:5478–5485. doi: 10.1128/IAI.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambol SP, Tang JK, Merrigan MM, Johnson S, Gerding DN. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis. 2001;183:1760–1766. doi: 10.1086/320736. [DOI] [PubMed] [Google Scholar]
- 49.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. Colonization for the prevention of Clostridium difficile disease in Hamsters. J Infect Dis. 2002;186:1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 50.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile Infections. MBio. 2015;6:e00551-15. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Histopathological analysis of the effects induced by bacteria-free supernatants in the murine model of ileal ligated loop. Tissue was fixed in 10% buffered formalin and stained with H&E for histological evaluation of the following groups: A) PBS, B) TYT-broth, C) Non-toxigenic C. difficile ATCC® 700057, D) 5758-ST01, E) 5757-ST67, F) 2811-ST41, G) 5809-ST252 and H) ICC45-ST41. Figure S2. Histopathological analysis of cecum from infected hamsters. Cecum sections were fixed in 10% buffered formalin and stained with H&E for histological evaluation of the following groups: A) PBS, B) DMEM, C) 5758-ST01, D) 5757-ST67, E) 2811-ST41, F) 5809-ST252, and G) ICC45-ST41. Figure S3. Counts of viable C. difficile cells by strain. Growth curves were performed in TYT-broth under the described conditions. At 0, 8 and 24h, an aliquot was taken, serially diluted and inoculated onto Brucella agar plates supplemented with vitamin K agar plates. The number of colonies that appeared after 48 h of incubation was recorded to obtain CFU/mL values.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Additional file 1.