Summary
In a published case-control study (GSE152075) from SARS-CoV-2-positive (n = 403) and -negative patients (n = 50), we analyzed the response to infection assessing gene expression of host cell receptors and antiviral proteins. The expression analysis associated with reported risk factors for COVID-19 was also assessed. SARS-CoV-2 cases had higher ACE2, but lower TMPRSS2, BSG/CD147, and CTSB expression compared with negative cases. COVID-19 patients' age negatively affected ACE2 expression. MX1 and MX2 were higher in COVID-19 patients. A negative trend for MX1 and MX2 was observed as patients' age increased. Principal-component analysis determined that ACE2, MX1, MX2, and BSG/CD147 expression was able to cluster non-COVID-19 and COVID-19 individuals. Multivariable regression showed that MX1 expression significantly increased for each unit of viral load increment. Altogether, these findings support differences in ACE2, MX1, MX2, and BSG/CD147 expression between COVID-19 and non-COVID-19 patients and point out to MX1 as a critical responder in SARS-CoV-2 infection.
Subject Areas: Health Informatics, Virology
Graphical Abstract

Highlights
-
•
COVID-19 patients present a distinct expression pattern for antiviral genes
-
•
MX1 antiviral gene expression is triggered by SARS-CoV-2
-
•
MX1 can be induced by hemin, an FDA-approved drug
-
•
MX1 rises as a potential druggable target
Health Informatics; Virology
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel virus that emerged in late 2019 in Wuhan, China (World Health Organization, 2020). The World Health Organization (WHO) declared the SARS-CoV-2 infection a pandemic health emergency as of January 31, 2020.
As of July 28, 2020, there are 16,812,755 confirmed cases of COVID-19 (coronavirus disease 2019), including 662,905 deaths reported to WHO from 216 countries and territories. The current worldwide COVID-19 fatality rate (4%) is lower than that of SARS-CoV-1 (10%) and Middle East respiratory syndrome (MERS)-CoV (32%) (World Health Organization, 2020). However, the total number of infected and dead people is much higher than that due to the latter viruses.
Analyses for viral stability in aerosols and surfaces showed high similarity among SARS-CoV-2 and SARS-CoV-1 (van Doremalen et al., 2020). This might imply that the differences in the epidemiologic viral features probably arise from viral and host factors such as sex, comorbidities (Zheng et al., 2020), higher viral loads, and/or the potential for asymptomatic individuals to spread the virus (Bai et al., 2020; Zou et al., 2020).
SARS-CoV-2 invades host cells mainly via two receptors: angiotensin-converting enzyme 2 (ACE2) (Letko et al., 2020; Yan et al., 2020) and Basigin or EMMPRIN (BSG/CD147) (Wang et al., 2020). This mechanism results from the interaction between the host receptors and the viral spike glycoproteins (S), after priming of this viral protein by the serine protease TMPRSS2 (Hoffmann et al., 2020). TMPRSS2 inhibition has been shown to block SARS-CoV-2 cell entry (Hoffmann et al., 2020). In addition to ACE2, Wang et al. (2020) demonstrated that SARS-CoV-2 S protein also binds to BSG/CD147 (Ulrich and Pillat, 2020).
Sequential protease cleavage models have been proposed for activation of S protein of SARS-CoV and MERS-CoV (Belouzard et al., 2009; Millet and Whittaker, 2014). Depending on the virus strain and the cell types infected, S protein may be cleaved by one or several host proteases, including cathepsins, TMPRSS2, and BSG/CD147 (Park et al., 2016; Ulrich and Pillat, 2020). Thus, coronavirus cell entry, either through plasma membrane or endocytosis, appears to be largely determined by the availability of these proteases (Ou et al., 2020).
Of note, ADAM metallopeptidase domain 17 (ADAM17) may exert a protective effect on SARS-CoV-2 infection. ADAM17 can release the ectodomains of a plethora of membrane-anchored factors, such as ACE2 (Lambert et al., 2005; Palau et al., 2020).
However, the lead roles of these factors in promoting viral infection in humans still remain elusive.
Although much attention has been placed in virus host cell receptors, little has been commented on antiviral effector proteins. It is well accepted that type I interferon (IFN) is essential in fighting viral infection by induction of IFN-stimulated genes (ISGs), which work in synergy to inhibit viral replication via multiple mechanisms (Li et al., 2018; Teijaro, 2016). The human myxovirus resistance genes (MX) encode GTPases that are part of the antiviral response induced by type I/III IFNs (Verhelst et al., 2012). Humans encode two different MX proteins, MX1 and MX2, which differ considerably in viral specificities and mechanisms of action. MX1 has been shown to have wide antiviral activity against RNA and DNA viruses (Haller et al., 2015), whereas MX2 action is limited to certain viruses such as HIV (Bhargava et al., 2018). Specifically, MX1 has a direct effect on the viral ribonucleoprotein complex and its GTPase activity is essential for its antiviral function. Greater MX1 expression correlated with better response to influenza A H1N1 pandemic in 2009 (Verhelst et al., 2012).
The main therapeutic avenues to halt respiratory virus infection are targeting the virus directly or targeting the host system. Although the first strategy is highly efficient, it is limited by the sensitivity/resistance of the virus strain (McKimm-Breschkin, 2005; Monto, 2003). On the other hand, the second strategy is impaired by having shown relatively low efficacy (Darwish et al., 2011; Ramos and Fernandez-Sesma, 2015). As the clinical picture has evidenced that the “cytokine storm” triggered by these viruses causes severe lung injury and lethality (Jose and Manuel, 2020), and that antiviral drugs are not effective in preventing such lung injury, it is imperative to spawn new drugs that have the capacity to boost the host immune response while controlling tissue damage as a consequence of these infections (Zheng et al., 2008).
There are no conclusive data evidencing the shifts in expression of host cell receptors and viral effectors produced by SARS-CoV-2 infection in humans. Thus, in this work we analyzed gene expression profiles in response to coronavirus infection in COVID-19 versus non-COVID-19 patients. We evaluated the expression of the main host cell receptors described for SARS-CoV-2 and potential antiviral genes. Our results evidence high discrepancies regarding the expression of the reported main viral host cell receptors associated with COVID-19, and point out to the relevance of MX1 to fight this disease.
Results
Expression of Host Cell Receptors in SARS-CoV-2-Positive and -Negative Patients
We used the publicly available RNA sequencing (RNA-seq) dataset GSE152075 to evaluate the expression of known host SARS-CoV-2 receptor genes: ACE2, TMPRSS2, BSG/CD147, CTSB, CTSL, and ADAM17 (Heurich et al., 2014; Hoffmann et al., 2020; Simmons et al., 2005; Ulrich and Pillat, 2020; Zhou et al., 2015). This set consists of transcriptome data from nasopharyngeal swabs from 430 SARS-CoV-2-positive and 54 SARS-CoV-2-negative patients. We excluded 27 SARS-CoV-2-positive and 4 SARS-CoV-2-negative samples from the downstream analyses because the RNA-seq data from these individuals was of low quality (>70% of genes resulting in 0 sequencing reads). Patient demographics are available in Table S1.
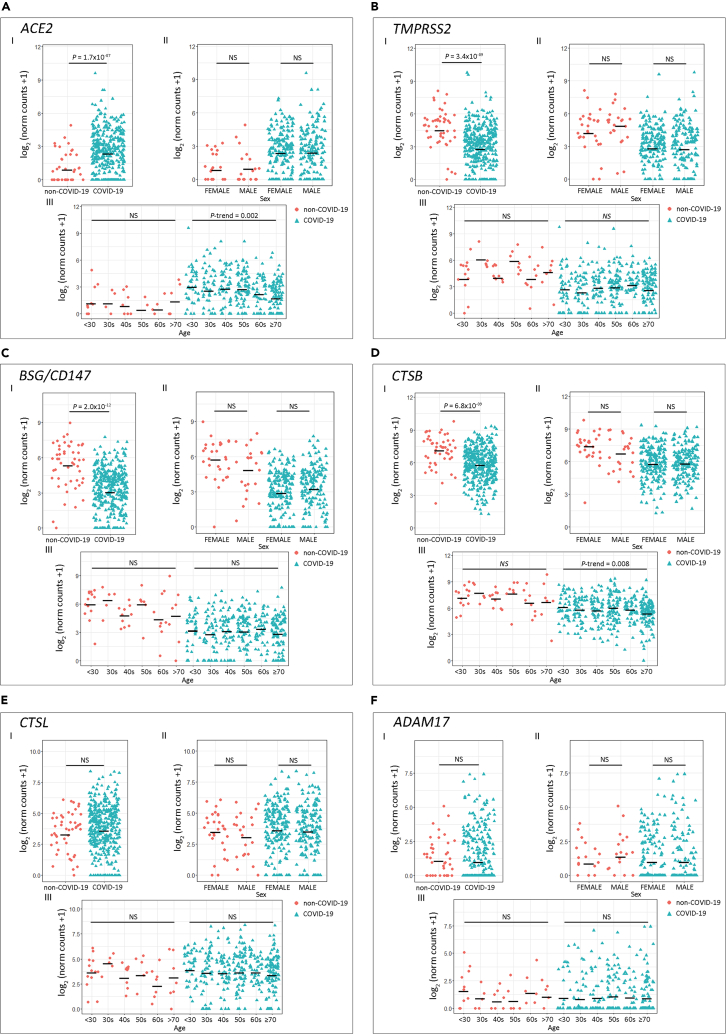
Results evidenced that COVID-19 patients had higher ACE2 expression compared with non-COVID-19 patients (Figure 1A.I; p = 1.7 × 10−7). On the contrary, TMPRSS2 (Figure 1B.I), BSG/CD147 (Figure 1C.I), and CTSB (Figure 1D.I) expression was lower in the COVID-19 group (p = 3.4 × 10−9, p = 2.0 × 10−12, and p = 6.8 × 10−9, respectively). Non-statistical differences for CTSL and ADAM17 expression were observed between SARS-CoV-2-positive and -negative patients (Figures 1E.I and 1F.I, respectively).
Figure 1.
Expression of Host Cell Receptor Genes in COVID-19 and Non-COVID-19 Patients
(A–F) Gene expression analysis for host cell receptor genes (A) ACE2, (B) TMPRSS2, (C) BSG/CD147, (D) CTSB, (E) CTSL, and (F) ADAM17.
(I) COVID-19 versus non-COVID-19 patients (p values correspond to Wilcoxon rank-sum test); (II) COVID-19 and non-COVID-19 patients by sex (p values correspond to Wilcoxon rank-sum test); (III) COVID-19 and non-COVID-19 patients categorized by age groups (p values correspond to decreasing Jonckheere-Terpstra trend test). Statistical significance was set at p < 0.05. NS: not significant.
As the reported risk factors for SARS-CoV-2 infection include age and sex, we next assessed the association between these risk factors and the host receptors' gene expression. We did not find gene expression differences between sex (Figures 1A.II–1F.II). When analyzing the association between gene expression and age, we found that the older the COVID-19 patient, the lower the expression for ACE2 (Figure 1A.III; P-trenddecreasing = 0.002) and CTSB (Figure 1D.III; P-trenddecreasing = 0.008). We did not observe an association between age and gene expression for the other receptors in the COVID-19 group (Figures 1B.III, 1C.III, 1E.III, and 1F.III). For SARS-CoV-2-negative donors, no variation in gene expression associated with age was observed (Figures 1A.III–1F.III).
Expression of Antiviral-Associated Genes in SARS-CoV-2-Positive and -Negative Patients
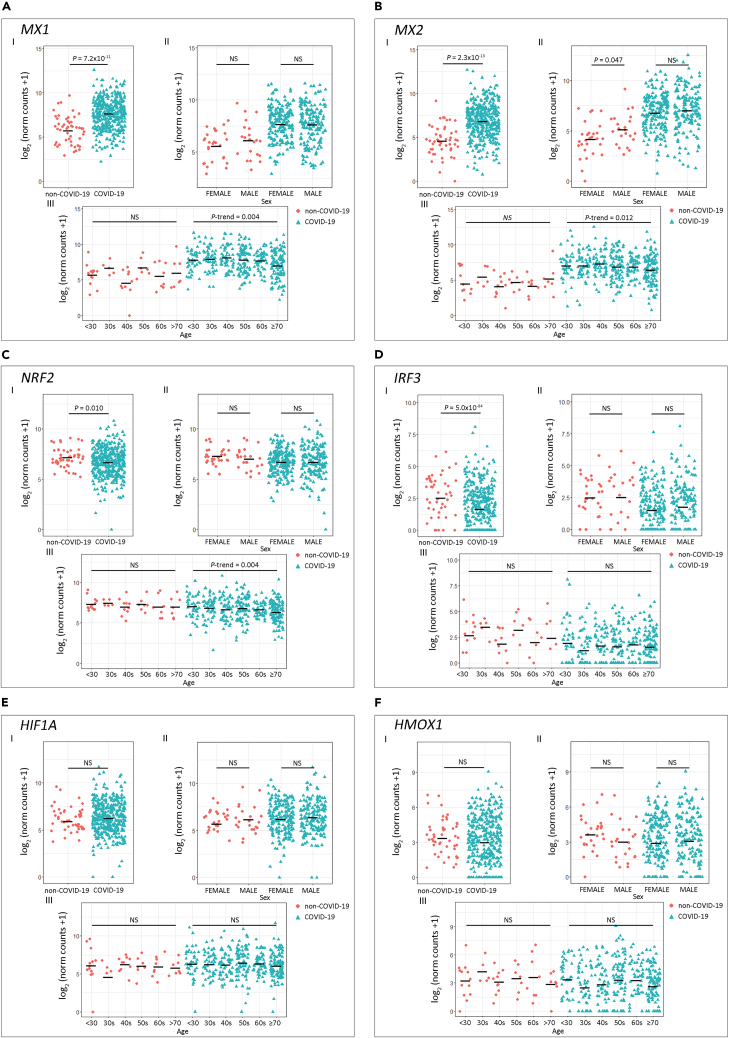
Next, we assessed antiviral-associated genes that play a crucial role in halting viral replication via multiple mechanisms: MX1, MX2, NRF2, IRF3, HIF1A, and HMOX1 (Chiang and Liu, 2018; Espinoza et al., 2017; Haller et al., 2015; Hassan et al., 2020; Hong et al., 2009) (Figure 2). MX1 (Figure 2A.I) and MX2 (Figure 2B.I) expression was significantly higher in COVID-19 patients (p = 7.2 × 10−11 and p = 2.3 × 10−13, respectively) compared with the non-COVID-19 group. No significant differences were observed between patients of different sex for any of the genes assessed (Figures 2A.II–2F.II). Interestingly, we observed a negative trend for MX1 (Figure 2A.III; P-trenddecreasing = 0.004) and MX2 (Figure 2B.III; P-trenddecreasing = 0.012) expression as patients' age increased. In addition, NRF2 and IRF3 expression was significantly lower in COVID-19 patients (Figures 2C.I and 2D.I; p = 0.010 and p = 5.0 × 10−4, respectively), and NRF2 also showed a significant P-trend in COVID-19 patients grouped by age (Figure 2C.III; P-trenddecreasing = 0.004). No statistical differences were observed when comparing HMOX1 and HIF1A expressions (Figures 2E and 2F). For SARS-CoV-2-negative donors no variation in gene expression associated with age was observed (Figures 2A.III–2F.III).
Figure 2.
Expression of Host Antiviral Effector Genes in COVID-19 and Non-COVID-19 Patients
(A–F) Gene expression analysis for the selected host cell receptor genes (A) MX1, (B) MX2, (C) NRF2, (D) IRF3, (E) HIF1A, and (F) HMOX1.
(I) COVID-19 versus non-COVID-19 patients (p values correspond to Wilcoxon rank-sum test), (II) COVID-19 and non-COVID-19 patients by sex (p values correspond to Wilcoxon rank-sum test), and (III) COVID-19 and non-COVID-19 patients categorized by age groups (p values correspond to decreasing Jonckheere-Terpstra trend test). Statistical significance was set at p < 0.05. NS: not significant.
Gene Correlation and Principal-Component Analysis
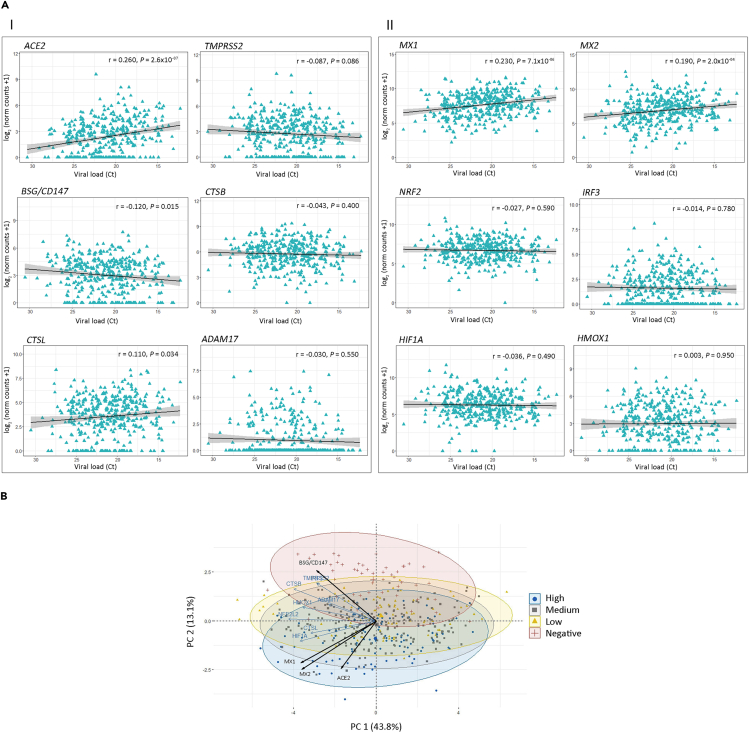
Next, we performed pairwise Spearman correlation analyses (Figure 3A and Table S2) for host receptors and antiviral gene expression levels of the host receptors and antiviral genes. We considered all patients (Corr.), only non-COVID-19 patients (Neg.), and only COVID-19 patients (Pos.). Black boxes highlight statistically significant correlation among genes for COVID-19 patients, whereas correlations for the same pairs in non-COVID-19 patients appeared non-significant (Figure 3A). Of note, ACE2 showed an r = 0.416 (p < 1 × 10−15) with MX1, whereas MX2 showed an r = 0.423 (p < 1 × 10−15) with HMOX1, an r = 0.595 (p < 1 × 10−15) with CTSB, and an r = 0.450 (p < 1 × 10−15) with CTSL (Figure 3A and Table S2).
Figure 3.
Gene Correlation and Principal-Component Analysis (PCA)
(A) Pairwise Spearman correlation matrix analysis between all genes of interest. The upper half displays the Spearman coefficients (r) considering all patients (Corr), non-COVID-19 patients (Neg.), or COVID-19 patients (Pos.). Black boxes highlight genes that have significant correlation only in COVID-19 patients. Statistical significance ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. The lower half displays the scatterplots.
(B) PCA biplot of gene expression data showing a rough segregation of non-COVID-19 and COVID-19 samples. Each point represents one individual, and the arrows depict the gene expression profile; black arrows show the 4 genes that have the greatest weight in driving the difference between the groups.
(C) 3D scatter plots for (I) ACE2, MX1, and BSG/CD147 and (II) ACE2, MX2, and BSG/CD147.
Interestingly, although MX1 and MX2 respond to SARS-CoV-2 infection (Figures 2A and 2B), the correlation analysis highlights MX2 positive correlation for both non-COVID-19 and COVID-19 patients (Figure 3A and Table S2; r = 0.642, p = 4.97 × 10−7 and r = 0.899, p < 1 × 10−15, respectively), whereas MX1 only displays positive correlation for COVID-19 patients. These data suggest that MX1 might be more specific in the response to SARS-CoV-2 than MX2.
In parallel, we performed a principal-component analysis (PCA), which is a dimensionality reduction technique, used to increase the interpretability of the dataset, minimizing at the same time information loss and maximizing variance. In this dataset, the principal component (PC) 1 explained 43.8% of the variance among samples (Figure 3B). Only the infection status is shown because none of the other assessed variables (age, sex, and RNA-seq batch) clustered the samples on PC1. The PC2 is responsible for 13.1% of the variability, and the expression of ACE2, MX1, MX2, and BSG/CD147 seemed to drive the difference between non-COVID-19 and COVID-19 individuals (Figure 3B). Furthermore, we plotted the combined expression of ACE2, BSG/CD147, and MX1 (Figure 3C.I) and ACE2, BSG/CD147, and MX2 (Figure 3C.II). These combinations were selected because MX1 and MX2 gene expression were highly correlated for both non-COVID-19 and COVID-19 patients (Figure 3A and Table S2). The graphs show distinct 95% confidence ellipsoids, generated by 3-gene expression patterns, able to cluster non-COVID-19 and COVID-19 individuals (Figure 3C).
SARS-CoV-2 Viral Load Association with Gene Expression
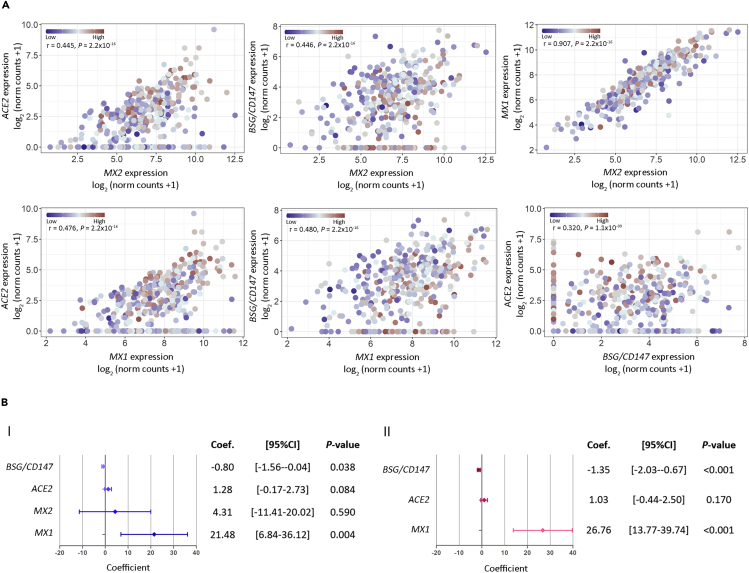
To assess the correlation between gene expression and SARS-CoV-2 viral load, we used the PCR cycle threshold (Ct) of the N1 viral gene amplification as a surrogate variable for viral load. We found a positive correlation between expression of ACE2 (Figure 4A.I; r = 0.260, p = 2.6 × 10−7), CTSL (Figure 4A.I; r = 0.110, p = 0.034), MX1 (Figure 4A.II; r = 0.230, p = 7.1 × 10−6), and MX2 (Figure 4A.II; r = 0.190, p = 2.0 × 10−4), with SARS-CoV-2 viral load. Of note, MX1 correlation with viral load appears to be stronger than the correlation observed for MX2, further supporting MX1 enhanced response to SARS-CoV-2, compared with MX2. On the other hand, we observed a negative correlation between BSG/CD147 expression (Figure 4A.I; r = −0.120, p = 0.015) and viral load. These results were further supported by PCA that showed a rough clustering of COVID-19 samples according to patients' viral load (high, medium, low) mainly by ACE2, BSG/CD147, MX1, and MX2 (Figure 4B).
Figure 4.
Correlation Analysis between Gene Expression and SARS-CoV-2 Viral Load in COVID-19 Patients
(A) Viral load and gene expression correlation for (I) ACE2, TMPRSS2, BSG/CD147, CTSB, CTSL, and ADAM17 and for (II) MX1, MX2, NRF2, IRF3, HIF1A, and HMOX1. Viral load was defined by the cycle threshold (Ct) of the N1 viral gene amplification during diagnostic PCR. Spearman coefficient (r) is shown for each correlation. Statistical significance was set at p < 0.05. NS: not significant.
(B) Principal-component analysis biplot of gene expression data showing a rough segregation of non-COVID-19 and COVID-19 patients stratified by low (Ct > 24), medium (Ct = 24–19), and high (Ct < 19; blue) viral load. Each point represents one individual, and the arrows depict the gene expression profile; black arrows show the 4 genes that have the greatest weight in driving the difference between the groups.
Next, we analyzed the pairwise correlations for BSG/CD147, ACE2, MX1, and MX2 considering viral load in COVID-19 patients. As expected, results showed positive and significant correlations for all genes in all comparisons (Figure 5A). Although a high correlation was observed for MX1 versus MX2 + viral load (Figure 5A; r = 0.907, p = 2.2 × 10−16), this is probably due to the collinearity between MX1 and MX2.
Figure 5.
Association between Viral Load and Gene Expression
(A) Dot plots representing pairwise correlations for BSG/CD147, ACE2, MX1, and MX2 considering viral load in COVID-19 patients. For each comparison, the independent variable is plotted in the x axis and the dependent variable is plotted in the y axis. For all cases, viral load was considered as an independent variable. Viral load is represented as a color scale.
(B) Forest plots representing multivariable regression analysis. Model (I) considering as covariates: individual gene expression, viral load, and age; and model (II) considering as covariates: viral load, age, and all genes. Statistical significance was set at p < 0.05. NS: not significant.
Finally, we assessed the association between each gene with viral load using a linear multivariable regression analysis adjusted by age. Results indicate that viral load can explain variations in MX1, BSG/CD147, and ACE2 expression (Figure 5B.I; p = 0.004, p = 0.038, and p = 0.084, respectively). However, MX2 does not appear to behave dependently from viral load when considering patient's age (Figure 5B.I; p = 0.590). When further adjusting the model to consider the viral load-dependent genes, we found that for each unit of increase in viral load, MX1 expression increased 26.76 units, ACE2 expression increased 1.03 units, and BSG/CD147 expression decreased 1.35 units (Figure 5B.II; p < 0.001, p = 0.170 and p < 0.001, respectively). Altogether these results point out to MX1 as the critical responder in SARS-CoV-2 infection.
Discussion
Respiratory epithelium is the primary barrier that defends the host against respiratory pathogens. When respiratory viruses overcome the first barrier, attachment and internalization into epithelial cells is the next step to start its replication.
ACE2 mediates SARS-CoV-2 entry to the cells by binding to the S protein (Hoffmann et al., 2020). Its primary physiological function is the conversion of angiotensin II to angiotensin-(1–7). There are controversial data concerning therapies that could modulate ACE2 expression, like angiotensin receptor blockers (ARBs), as they may cause an initial up-regulation with further down-regulation of ACE2 expression during SARS-CoV-2 infection, still not ascertaining whether this may lead to harmful or beneficial effects (Vaduganathan et al., 2020). Our analysis evidenced an increase of ACE2 expression in COVID-19 versus non-COVID-19 patients correlating with viral load.
TMPRSS2 is currently also in the main scene regarding key SARS-CoV-2 host cell receptors (Hoffmann et al., 2020). TMPRSS2-mediated cleavage of S protein has been described to be necessary for SARS-CoV and MERS-CoV productive infections (Glowacka et al., 2011; Matsuyama et al., 2010; Shulla et al., 2011). Recently, this same process has been reported for SARS-CoV-2 (Hoffmann et al., 2020). However, our analysis reflects a down-regulation of TMPRSS2 in COVID-19 patients. Similarly, BSG/CD147, another key receptor that binds to the S protein (Wang et al., 2020), appears to be down-regulated in COVID-19 patients with lower levels associated with higher viral load. These results may suggest that SARS-CoV-2 infection may prompt a selective mechanism of host cell receptors, and thus, targeting TMPRSS2 and BSG/CD147 might not necessarily be the most effective strategy to halt infection.
Despite host cell receptors being critical targets to prevent viral infection, it is also important to consider the second line of defense in response to the pathogen. One of the antiviral effectors is MX1 (Haller et al., 2015). MX genes exist in nearly all vertebrate genomes, and they are active mainly against RNA viruses. MX1 is an effector protein of the IFN system (Verhelst et al., 2012; Zav'yalov et al., 2019). Studies report that MX1 generates a protective antiviral response controlling the expression of key modulator molecules associated with influenza A virus lethality; effects increased under IFN-ɑ stimulation (Cilloniz et al., 2012; Haller et al., 2015). MX2 is also a key contributor to the type I IFN-induced post-entry inhibition of HIV-1 infection (Dicks et al., 2018). Currently, there are several ongoing clinical trials for COVID-19 prevention and/or treatment using type I or III IFNs (NCT04343976 [Phase II], NCT04385095 [Phase II], NCT04354259 [Phase II], NCT04293887 [Early Phase I], NCT04344600 [Phase II], NCT04320238 [Phase III], NCT04388709 [Phase II]). However, IFN administration could enhance a “cytokine-storm” causing a hyper-inflammatory response and contributing to multiple organ failure. Of note, recent reports showed that COVID-19 patients present a defective type I IFN response characterized by decreased production and activity, with consequent deregulation of ISGs. The authors suggest type I IFN deficiency in the blood as a hallmark of severe COVID-19 and also discuss the risk of using drugs that interfere with the IFN pathway (Hadjadj et al., 2020).
As shown in this work, MX1 and MX2 expression levels are higher in COVID-19 patients. Interestingly, both MX1 and MX2 expression increases significantly with viral load and on the contrary, their expression is reduced with age. Epidemiologic reports by the Centers for Disease Control and Prevention (CDC) have shown increased severity in older patients (Centers for Disease Control and Prevention, 2020). Although an important limitation of this study is the lack of clinical data accounting for disease severity, there is no correlation between age and viral load in the COVID-19 patients assessed (r = 0.031, p = 0.58). Thus, we might speculate that the loss of MX1 and MX2 with age may be associated with higher risk for severe illness.
Other ways of increasing and sustaining MX1 expression to fight SARS-CoV-2 infection may be the use of a previously US Food and Drug Administration-approved drug known as hemin, a well-known inducer of HMOX1, and a modulator of MX1 expression independent from IFN activation. Although our results show no significant alteration in HMOX1 expression, there is sound literature referring to its interference with viral infections (Espinoza et al., 2017). Some examples include the use of CoPP, which induces HMOX1, promoting IRF3 phosphorylation and nuclear translocation, and subsequently expression of MX1 (Ma et al., 2019). The protective role of HMOX1 against viral infection has also been demonstrated for dengue, hepatitis C, hepatitis B, Ebola, Zika, and HIV-1 infections, revealing that HMOX1 signaling pathway was a promising strategy for treating these infections (Devadas and Dhawan, 2006; Hill-Batorski et al., 2013; Huang et al., 2017; Lee et al., 2014; Protzer et al., 2007; Tseng et al., 2016). Furthermore, andrographolide, an anti-inflammatory and anti-oxidative compound that induces HMOX1, was suggested as a potential inhibitor of the main protease of SARS-CoV-2 (Enmozhi et al., 2020).
To increase the interpretability of the data we performed pairwise Spearman correlation and PCA. Results obtained were able to cluster non-COVID-19 and COVID-19 individuals based on ACE2, MX1, and BSG/CD147 expression. Furthermore, when performing a multivariable regression analysis including viral load, age, and selected genes, MX1 and BSG/CD147 appear to significantly correlate with viral load independently from the other gene expressions included in the model and age. However, the increase in MX1 compared with BSG/CD147, per unit of viral infection, is strikingly higher (>15-fold).
In conclusion, these results highlight MX1 as a solid responder to SARS-CoV-2 pointing out to the relevance of evaluating approved drugs able to boost MX1 expression and halt infection. Further experimental research will be needed to ascertain the antiviral function of MX1 against SARS-CoV-2.
Limitations of the Study
Our analysis in this article has limitations worth mentioning. Although we performed an extensive analysis with a public RNA-seq dataset containing 484 human samples, we were not able to access complete clinical records because they are not publicly available (e.g., illness severity). In addition, RNA-seq raw data were unavailable; therefore, we were not able to perform the pre-processing and alignment of sequences. However, we were able to curate samples excluding those that did not have reads for >70% of genes. Finally, some individuals suffered from other bacterial or viral infections that might confound the findings.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Geraldine Gueron (ggueron@iquibicen.fcen.uba.ar).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate new datasets.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by grants from AGENCIA-PICT-2016-0056 (Argentina) and AGENCIA-PICT-RAICES-2018-02639 (Argentina).
Author Contributions
Conceptualization, J.C., E.V., and G.G.; Formal Analysis (Figures 1 and 2: J.B., S.L.-V., and S.O.; Figure 3: J.B., S.L.-V., R.L., M.A., and S.O.; Figure 4: J.B., S.L.-V., S.O., and R.L.; Figure 5: J.B., P.S., S.L.-V., and R.L.); Investigation, J.B., P.S., S.L.-V., R.L., A.S., A.T., M.A., F.C., S.O., E.V., J.C., and G.G; Resources, E.V., J.C., and G.G.; Data Curation, J.B., P.S., S.L.-V., R.L., A.S., A.T., M.A., F.C., S.O., J.C., E.V., and G.G.; Writing – Original Draft preparation, J.C., G.G., and E.V.; Writing – Review and Editing, J.B., P.S., S.L.-V., R.L., A.S., A.T., M.A., F.C., S.O., E.V., J.C., and G.G.; Funding, E.V., J.C., and G.G.; All authors have read and agreed to the published version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101585.
Contributor Information
Elba Vazquez, Email: elba@qb.fcen.uba.ar.
Javier Cotignola, Email: jcotignola@qb.fcen.uba.ar.
Geraldine Gueron, Email: ggueron@iquibicen.fcen.uba.ar.
Supplemental Information
References
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U S A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A., Lahaye X., Manel N. Let me in: control of HIV nuclear entry at the nuclear envelope. Cytokine Growth Factor Rev. 2018;40:59–67. doi: 10.1016/j.cytogfr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2020. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/older-adults.html
- Chiang H.-S., Liu H.M. The molecular basis of viral inhibition of IRF- and STAT-dependent immune responses. Front. Immunol. 2018;9:3086. doi: 10.3389/fimmu.2018.03086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloniz C., Pantin-Jackwood M.J., Ni C., Carter V.S., Korth M.J., Swayne D.E., Tumpey T.M., Katze M.G. Molecular signatures associated with Mx1-mediated resistance to highly pathogenic influenza virus infection: mechanisms of survival. J. Virol. 2012;86:2437–2446. doi: 10.1128/JVI.06156-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish I., Mubareka S., Liles W.C. Immunomodulatory therapy for severe influenza. Expert Rev. Anti. Infect. Ther. 2011;9:807–822. doi: 10.1586/eri.11.56. [DOI] [PubMed] [Google Scholar]
- Devadas K., Dhawan S. Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J. Immunol. 2006;176:4252–4257. doi: 10.4049/jimmunol.176.7.4252. [DOI] [PubMed] [Google Scholar]
- Dicks M.D.J., Betancor G., Jimenez-Guardeño J.M., Pessel-Vivares L., Apolonia L., Goujon C., Malim M.H. Multiple components of the nuclear pore complex interact with the amino-terminus of MX2 to facilitate HIV-1 restriction. PLoS Pathog. 2018;14:e1007408. doi: 10.1371/journal.ppat.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J. Biomol. Struct. Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza J.A., González P.A., Kalergis A.M. Modulation of antiviral immunity by heme oxygenase-1. Am. J. Pathol. 2017;187:487–493. doi: 10.1016/j.ajpath.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Hassan S.M., Jawad M.J., Ahjel S.W., Singh R.B., Singh J., Awad S.M., Hadi N.R. The Nrf2 activator (DMF) and covid-19: is there a possible role? Med. Arch. 2020;74:134–138. doi: 10.5455/medarh.2020.74.134-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill-Batorski L., Halfmann P., Neumann G., Kawaoka Y. The cytoprotective enzyme heme oxygenase-1 suppresses Ebola virus replication. J. Virol. 2013;87:13795–13802. doi: 10.1128/JVI.02422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.W., Yoo J.W., Kang H.S., Kim S., Lee D.-K. HIF-1alpha-dependent gene expression program during the nucleic acid-triggered antiviral innate immune responses. Mol. Cells. 2009;27:243–250. doi: 10.1007/s10059-009-0030-2. [DOI] [PubMed] [Google Scholar]
- Huang H., Falgout B., Takeda K., Yamada K.M., Dhawan S. Nrf2-dependent induction of innate host defense via heme oxygenase-1 inhibits Zika virus replication. Virology. 2017;503:1–5. doi: 10.1016/j.virol.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-C., Tseng C.-K., Young K.-C., Sun H.-Y., Wang S.-W., Chen W.-C., Lin C.-K., Wu Y.-H. Andrographolide exerts anti-hepatitis C virus activity by up-regulating haeme oxygenase-1 via the p38 MAPK/Nrf2 pathway in human hepatoma cells. Br. J. Pharmacol. 2014;171:237–252. doi: 10.1111/bph.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-F., Gong M.-J., Zhao F.-R., Shao J.-J., Xie Y.-L., Zhang Y.-G., Chang H.-Y. Type I interferons: distinct biological activities and current applications for viral infection. Cell. Physiol. Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- Ma L.-L., Zhang P., Wang H.-Q., Li Y.-F., Hu J., Jiang J.-D., Li Y.-H. Heme oxygenase-1 agonist CoPP suppresses influenza virus replication through IRF3-mediated generation of IFN-α/β. Virology. 2019;528:80–88. doi: 10.1016/j.virol.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKimm-Breschkin J.L. Management of influenza virus infections with neuraminidase inhibitors: detection, incidence, and implications of drug resistance. Treat. Respir. Med. 2005;4:107–116. doi: 10.2165/00151829-200504020-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U S A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S. The role of antivirals in the control of influenza. Vaccine. 2003;21:1796–1800. doi: 10.1016/s0264-410x(03)00075-6. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palau V., Riera M., Soler M.J. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol. Dial. Transpl. 2020;35:1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B.J., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U S A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protzer U., Seyfried S., Quasdorff M., Sass G., Svorcova M., Webb D., Bohne F., Hösel M., Schirmacher P., Tiegs G. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology. 2007;133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Ramos I., Fernandez-Sesma A. Modulating the innate immune response to influenza A virus: potential therapeutic use of anti-inflammatory drugs. Front. Immunol. 2015;6:361. doi: 10.3389/fimmu.2015.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016;16:31–40. doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-K., Lin C.-K., Wu Y.-H., Chen Y.-H., Chen W.-C., Young K.-C., Lee J.-C. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci. Rep. 2016;6:32176. doi: 10.1038/srep32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020:1–7. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012;86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Novel coronavirus (2019-nCoV) situation report - 1. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zav’yalov V.P., Hämäläinen-Laanaya H., Korpela T.K., Wahlroos T. Interferon-Inducible myxovirus resistance proteins: potential biomarkers for differentiating viral from bacterial infections. Clin. Chem. 2019;65:739–750. doi: 10.1373/clinchem.2018.292391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.-J., Chan K.-W., Lin Y.-P., Zhao G.-Y., Chan C., Zhang H.-J., Chen H.-L., Wong S.S.Y., Lau S.K.P., Woo P.C.Y. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc. Natl. Acad. Sci. U S A. 2008;105:8091–8096. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., Li Q., Jiang C., Zhou Y., Liu S. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J. Infect. 2020;81:e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R.J., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets.