Abstract
Background & Aims:
In patients with inflammatory bowel diseases (IBD), symptoms do not always associate with the severity of endoscopic inflammation and can persist after mucosal healing. We investigated whether symptoms in patients with successfully treated IBD are related to composition of the intestinal microbiome.
Methods:
We analyzed 590 tissue biopsies from 215 patients with IBD and 48 healthy individuals (controls). We obtained mucosal biopsies from 2 colon sites (ascending and rectosigmoid) and from the terminal ileum along with clinical data. Bacterial DNA was extracted from the biopsies and the V4 region of 16s rRNA sequenced by Miseq and processed using the QIIME v1.9 pipeline.
Results:
Mucosal biopsies from patients with Crohn’s disease (CD) who achieved mucosal healing (Mayo scores of 0–1 or SES-CD scores of 0–5) had lower Chaol diversity than biopsies from patients with ulcerative colitis (UC) or unclassified IBD (IBD-U), or controls. After endoscopic evidence of improvement in patients with UC or IBD-U, diversity of the tissue-associated microbiota did not differ significantly from that of controls. Colon biopsies from patients with CD had lower microbial diversity, before and after healing (segmental endoscopic severity-CD scores, 0–2), than colon biopsies from controls (P<.002). In patients with CD who achieved mucosal healing, residual clinical activity (CD activity index scores above 150; P=.03) and persistent diarrhea was associated with reduced microbial diversity (P=.01). Continued diarrhea was associated with a trend towards dysbiosis, based on the microbial dysbiosis index (P=.059). In patients with UC or IBD-U with moderate to severe inflammation, increasing severity of diarrhea was associated with reduced microbial diversity (P=.03).
Conclusions:
In an analysis of biopsies from patients with IBD and controls, we found that despite endoscopic evidence of improvement or remission, alpha diversity of the tissue-associated intestinal microbiome remained lower in patients with CD than in controls. This observation, along with the reduced Chao1 diversity and greater dysbiosis in intestinal microbiota of patients with residual symptoms of IBD, indicates that microbiome composition could be associated with persistent diarrhea.
Keywords: outcome, response to therapy, microbiome, prognostic factor
INTRODUCTION
External environmental factors, the intestinal microbiome, inherited genetic risk and dysregulation of the innate and adaptive immune responses participate in a complex interplay, influencing disease development and prognosis in inflammatory bowel disease (IBD), and dysbiosis has been described in both single time-point and dynamic longitudinal studies of IBD 1–3. It is unclear if increased abundance of pathobionts in the presence of active inflammation impacts severity of patient symptoms and plays a role in the conundrum of unexplained residual clinical activity in patients with treated IBD. Several hypotheses have been proposed to explain the persistence of clinical symptoms in patients who have had objectively successful treatment. Given the link between dysbiosis and diarrhea-predominant irritable bowel syndrome (IBS-D) 4, we sought evidence of an association between alterations in the tissue-associated microbiome and specific symptoms in IBD patients.
We analyzed 590 tissue biopsies taken from 215 recruited patients with colonic and ileal CD, ulcerative colitis or IBD-Unclassified (UC/IBD-U) and 48 healthy controls (HC) and have characterized the tissue-associated microbiome at two colon sites (ascending and rectosigmoid) and at the terminal ileum. In biopsy samples, analysis of the microbiome by disease location is valuable since the detected bacteria are in direct contact with the host tissue. Analyzing features of tissue-associated microbiome diversity and composition may differentiate between IBD phenotypes both in active and endoscopically quiescent disease, determining the relationship between patient-reported symptoms and the intestinal microbiome.
MATERIALS AND METHODS
Patient selection and recruitment
Patients with confirmed IBD and asymptomatic HC who were attending for colorectal cancer screening were recruited at colonoscopy. Study activities were conducted with ethical approval. Biopsies were taken from sigmoid, ascending colon and terminal ileum when technically possible and snap frozen in liquid nitrogen. Endoscopic scores were determined by the endoscopist (IBD specialists and advanced IBD fellows) using Mayo scores (UC/IBD-U) and in CD, the segmental endoscopic severity score (SES-CD) 5, 6. Severity of endoscopic disease was graded on a 4-point scale (Table 1). The ordinal score reflects the segment of maximal inflammation (i.e. severe terminal ileal disease with normal colon). Patients with a maximum Mayo Score 1 or SES-CD score 0-5 were considered to have mucosal healing/endoscopic improvement. Endoscopic remission was defined as Mayo 0 or SES-CD 0-2.
Table 1:
Endoscopic score ranges used to generate a continuous 4-point scale of endoscopic inflammation for microbiome analysis
| Ulcerative colitis/IBD unclassified | Mayo score |
|---|---|
| Endoscopic Score 0 | 0 |
| Endoscopic Score 1 | 1 |
| Endoscopic Score 2 | 2 |
| Endoscopic Score 3 | 3 |
| Crohn’s disease | SES-CD score |
| Endoscopic Score 0 | 0-2 |
| Endoscopic Score 1 | 3-5 |
| Endoscopic Score 2 | 6-15 |
| Endoscopic Score 3 | >15 |
Clinical and demographic data were recorded and use of antibiotics at endoscopy was a criterion for exclusion. Clinical activity was defined as partial Mayo score ≥2 in UC/IBD-U or CD Activity Index >150 7–9. Stool frequency, abdominal pain, use of anti-diarrheal medications, nocturnal diarrhea and rectal bleeding were recorded at endoscopy. Daily diarrhea was classified as 2 loose stools more than normal or >3 loose stools daily and residual diarrhea reflects daily diarrhea despite mucosal healing.
Microbiome analysis
Total microbial DNA was extracted from biopsies in two batches using the DNeasy blood and tissue kit (Qiagen), as per the manufacturer’s protocol, with an additional bead beating step to ensure adequate cell lysis. Bead beating was performed using both 5 mm stainless steel beads to disrupt tissue (Qiagen 69989) and glass beads (Mo-Bio, Mississauga, ON, Canada) to disrupt bacterial cells, in conjunction with the FastPrep tissue homogenizer (MP Biomedicals, Santa Ana, CA, USA). Additional enzymatic lysis was conducted through the addition of proteinase K (as per the Qiagen protocol) and incubation of samples at 65°C.
Amplicon sequencing of the V4 hypervariable region of 16s rRNA bacterial DNA was completed using primers 515F/806R 10 and on an Illumina MiSeq platform (Illumina, San Diego). Paired-end sequences were processed using the QIIME 1.9 pipeline using an algorithm of closed-reference operational taxonomic unit (OTU) -picking which included reference-based chimera searching 11. OTUs were assigned using the Greengenes database 13_8 11, 12. Alpha diversity was calculated using Chao1 and Shannon index after rarefaction depth of 8500 reads per sample. Associations between outcomes and taxa were performed for each taxonomic level from phylum to genus. Association testing corrected for covariates including smoking status, sex, disease location and activity. The Microbial Dysbiosis Index (MD-Index) is a unique framework for assessment of ileal dysbiosis in CD based on log abundance of specific taxa and was applied to quantify dysbiosis2.
Statistical analysis
Shannon and Chao1 alpha diversity indices were compared using a t-test, and Bray-Curtis beta diversity using Adonis within QIIME software. The Deseq2 model 13 was used while accounting for sex, age, and inflammation. A logistic regression was applied for binary traits adjusting for age, sex, inflammation and total number of reads.
Correlation between MD-index scores and endoscopic inflammation used non-parametric Spearman’s correlation coefficient2. Taxa associations were considered significant when corrected q-value was <5% corresponding to a raw p-value <2.1 x 10−4 (considering 235 taxa from Phylum down to genus). Paired analysis of sigmoid and ascending colon tissue was performed in UC patients using corrected paired t-test.
RESULTS
Description of the cohort
We enrolled 215 patients with IBD (n=114 UC/IBD-U, n=101 CD) and 48 HC (Table 2). Patients with UC (n=99) and IBD-U (n=15) were considered together given phenotypic similarities.
Table 2:
Data represent patients included in this study with relevant clinical and demographic data as well as samples available from each of the three ileal and colon biopsy sites.
| VARIABLE | UC/IBDU (%), N=114 | CD (%), N=101 | HC (%), N=48 |
|---|---|---|---|
| MALE | 64 (56%) | 52 (51%) | 30 (62.5%) |
| MEDIAN AGE (YRS, RANGE) | 36 (18-72) | 28 (17-56) | 56 (31-71) |
| MEDIAN AGE AT DIAGNOSIS (YRS, RANGE) | 25 (11-59) | 20 (5-57) | |
| SMOKER (CURRENT) | 10 (9%) | 9(9%) | 3 (6%) |
| WHITE (SELF-DESCRIBED ETHINICITY) | 90 (79%) | 91 (90%) | 47 (98%) |
| MONTREAL CLASS | |||
| A1 | 24 (21%) | 35 (35%) | - |
| A2 | 69 (61%) | 57 (58%) | - |
| A3 | 16 (14%) | 9 (7%) | - |
| UNKNOWN | 5 | 2 | - |
| B1 | - | 21 (21%) | - |
| B2 | - | 20 (20%) | - |
| B3 | - | 60 (60%) | - |
| PERIANAL | - | 28 (28%) | - |
| L/E1 | 13 (11%) | 15 (15%) | - |
| L/E2 | 32 (28%) | 22 (22%) | - |
| L/E3 | 69 (61%) | 64 (64%) | |
| MEDICATIONS | |||
| METHOTREXATE | 0 | 11 (11%) | - |
| AZATHIOPRINE/6-MP | 13 (11%) | 20 (20%) | - |
| ANTI-TNF BIOLOGIC | 20 (17.5%) | 33 (33%) | - |
| ANTI-INTEGRIN BIOLOGIC | 1 (1%) | 1 (1%) | - |
| STEROIDS (ORAL) | 11 (10%) | 2 (2%) | - |
| SURGERY (INTRAABDOMINAL) | - | 20 (20%) | - |
| EXTRA-INTESTINAL MANIFESTATIONS | 19 (16.5%) | 20 (19.8%) | - |
Seven patients on antibiotics at endoscopy were excluded, leaving 590 samples with complete metadata. Sixty-four percent of the CD cohort and 68% of UC/IBD-U had endoscopic improvement or remission (Mayo 0-1 or SES-CD 0-5, figure 1).
Figure 1:
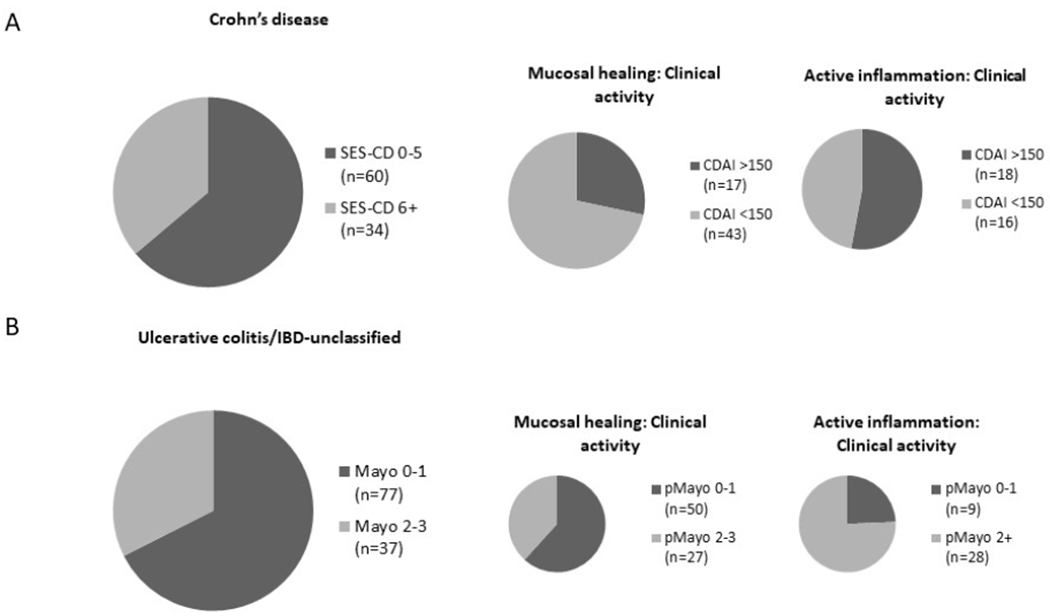
Proportion of IBD patients with active disease. 1A: Mucosal healing (MH) in 64% patients with CD (n=60/94). 28% of patients with MH and 53% with moderate-severe inflammation had CDAI >150. 1B: A similar proportion (68%) of patients with UC or UC/IBD-U(n=77/114) had MH. Approximately one third of patients with mucosal healing had pMayo score 2 or greater. Despite moderate-severe inflammation in 37 patients, 28% had pMayo scores of 0-1.
In the absence of endoscopic inflammation, the tissue-derived microbiota is less diverse in CD patients compared with healthy controls
Lower alpha diversity metrics in patients with CD has previously been described 3, 14, 15. Here, when compared with UC/IBD-U or HC, patients with CD had reduced alpha diversity in both sigmoid (p= .01 and p= 9.4 x 10−6 for CD vs. UC/IBD-U and CD vs. HC respectively) and terminal ileum mucosa (p= .03; p= .001 for CD vs. UC/IBD-U and CD vs. HC respectively; supplementary figure 1). Similarly, Bray-Curtis beta diversity in sigmoid mucosal samples was lower in CD patients when compared with either UC/IBD-U or HC (p<0.001 CD vs UC/IBD-U, p<0.001 CD vs HC, supplementary figure 2A). The MD-index has been validated in terminal ileum samples, but in our cohort, both terminal ileal and sigmoid colon inflamed samples demonstrated increased dysbiosis in CD compared with UC/IBD-U or HC (.01≤p≤ .03, supplementary figure 3).
To determine whether microbiome composition is associated with clinical symptoms, we identified patients in endoscopic remission (Mayo 0 or SES-CD 0-2). In sigmoid colon samples, patients with CD (n=32) exhibited reduced Chao1 alpha diversity when compared with UC/IBD-U (n=38; p= .03) or HC (n=48; p= 4×10−4, figure 2A). Biopsy-specific scoring of inflammation diversity was not mirrored by lower Chao1 alpha diversity or increased dysbiosis using MD-index (p= .24, rho= .12 in CD; p= .06, rho= .2 in UC/IBD-U, Supplementary Figure 4).
Figure 2:
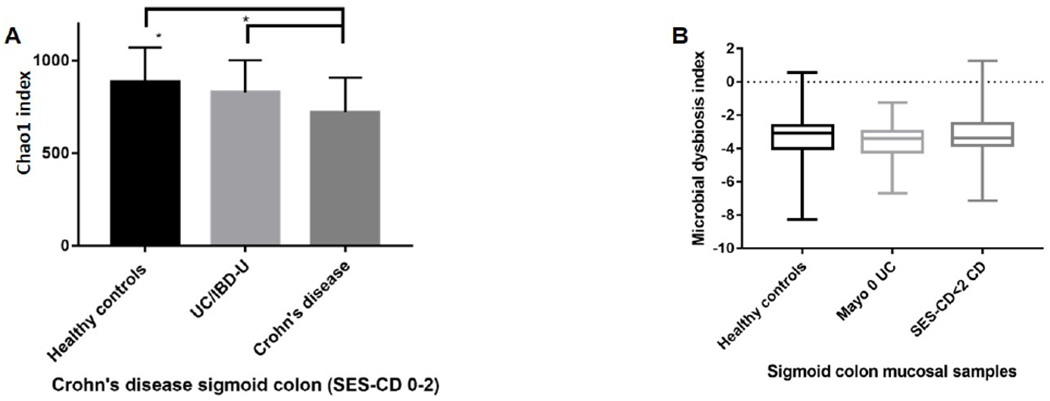
Alpha diversity analysis and MD-index scores of sigmoid mucosa from patients with endoscopic remission (Mayo 0 UC/IBD-U, n=27 and SES-CD 0-2 in CD, n=26). 2A: Chao1 alpha diversity of sigmoid mucosa, *CD vs HC, p=0.001 and #CD vs UC/IBD-U p=0.002. Data show mean observations after rarefaction at 8500 sequences. 2B: Comparison of MD-index of sigmoid colon between IBD (CD, n=26, UC n=38) and controls (n=48, CD vs HC, p=0.4; CD vs UC, p=0.48 ; UC vs HC, p=0.75).
Increased relative abundance of Fusobacteria and Proteobacteria and other taxa variations are associated with CD patients with mucosal healing
We analyzed individual taxa at three sites in remission. Patients with CD had increased relative abundance of Fusobacteria at the phylum and lower taxonomic ranks in the terminal ileum and sigmoid colon (p< 7×10−5) and increased Proteobacteria genera in ileal samples (p< 1 x 10−4) compared with UC/IBD-U and HC. Two Prevotella species were more abundant in UC/IBD-U than in CD patients at the terminal ileum (p≤ 1 x 10−4, supplementary data 2). In terminal ileum, Fusobacteriaceae cetobacterium abundance (p= 2.7 x 10−5) was increased in CD vs HC, while a lower abundance of Bacteroidetes genera including Akkermansia muciniphila (p= 2 x 10−4, supplementary data 2) was observed. Comparing CD sigmoid samples with HC, genera from Fusobacteria and Actinobacteria phyla have increased relative abundance in CD, as did Dorea from the Firmicutes phylum (6.9 x 10−10 ≤p≤ 2 x 10−5). In Mayo 0 UC/IBD-U there was increased prevalence of Lactobacillus at the terminal ileum compared with HC (p= 9.7 x 10−4, supplementary data 2).
UC/IBD-U patients with Mayo 0-1 disease had increased relative abundance of mucin-residing, Akkermansia muciniphila among UC/IBD-U (1×10−4≤p≤ 2×10−4, supplementary data 3). Paired analysis of sigmoid and ascending colon samples in patients with quiescent left-sided disease in endoscopic remission identified no differentially abundant taxa in previously diseased mucosa when compared with historically unaffected right colon (supplementary data 4).
Persistent clinical activity and daily diarrhea in the absence of endoscopic activity in CD is associated with reduced diversity
After mucosal healing, 28% CD (n=17/60) and 35% UC/IBD-U patients (n=28/77) met criteria indicating active clinical disease (CDAI>150 or pMayo >1). There were no associations between individual taxa abundance and specific symptoms or clinical activity in CD (supplementary data 5) or UC/IBD-U (supplementary data 6). In CD patients with endoscopic improvement, we noted Bray-Curtis beta diversity dissimilarity in patients with CDAI >150 compared with patients in clinical remission (p=0.01, r=0.24, supplementary figure 2B). Furthermore, alpha diversity was lower in patients with CDAI >150 (n= 17/43, p< .03, figure 3A). When alpha diversity was analyzed according to presence of rectal bleeding, nocturnal diarrhea, daily diarrhea or pain, only daily diarrhea in endoscopically improved CD was associated with a trend towards lower alpha diversity (p= .06, figure 3B). In endoscopic remission (SES-CD 0-2), the association between reduced diversity and daily diarrhea (n=8/26) was significant (p= .01, figure 3C), with no relationship between histological activity and residual diarrhea (p= .70, figure 3D). We noted a trend towards dysbiosis of sigmoid samples in patients with residual diarrhea (p= .059, figure 3E). Daily diarrhea was associated with the presence of Lactobacillus (q= .04, figure 3F).
Figure 3:
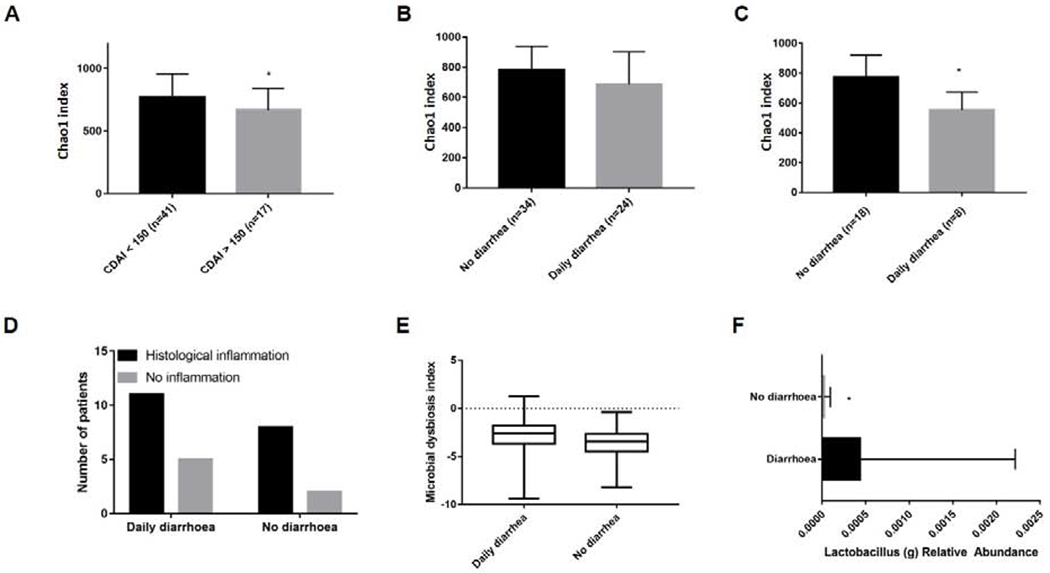
Alpha diversity analysis and MD-index scores of sigmoid samples in CD after mucosal healing 3A: Chao1 alpha diversity +/−SEM in CD patients with mucosal healing (SES-CD 0-5), comparing CDAI >150 with clinical remission, p=0.03. Data show mean observations after rarefaction at 8500 sequences. 3B: Comparison of Chao1 diversity according to presence or absence of residual diarrhea in patients with SES-CD 0-5 (p=0.06) and 3C: SES-CD 0-2 (p=0.01). 3D: Proportion of patients with histological inflammation according to presence of diarrhea. 3E: MD-index in CD patients with mucosal healing and daily diarrhea, p=0.059. 3F: Increased Lactobacillus abundance in presence/absence taxa analysis in daily diarrhea (q=0.04).
In moderate to severe UC/IBD-U, increasing clinical activity is associated with reduced diversity
Despite active disease, 24% with Mayo 2-3 and 47% of CD patients with SES-CD score > 6 were in clinical remission (pMayo 0-1 or CDAI <150, figure 1). Neither diversity nor dysbiosis in moderate-severe endoscopic inflammation were associated with clinical activity or specific symptoms in CD (supplementary data 7). Alpha diversity was lower in patients with daily diarrhea in moderate-severe UC/IBD-U in sigmoid colon samples (p= .03, figure 4A), accompanied by a higher MD-index (p= .03, Figure 4B). Daily stool frequency negatively correlated with abundance of Firmicutes Anaerostipes (q= .037, rho= − .61) and two unidentified members of the Christensenellaceae family, one in ileum (q= .04, rho=− .6) and the other in sigmoid colon (q=.046, rho=− .40, supplementary data 8). Relative abundance of a genus from the phylum Proteobacteria, Luteimonas also, correlated with increasing diarrhea in ileal samples in UC/IBD-U (q= .047, rho= .6 supplementary data 8).
Figure 4:
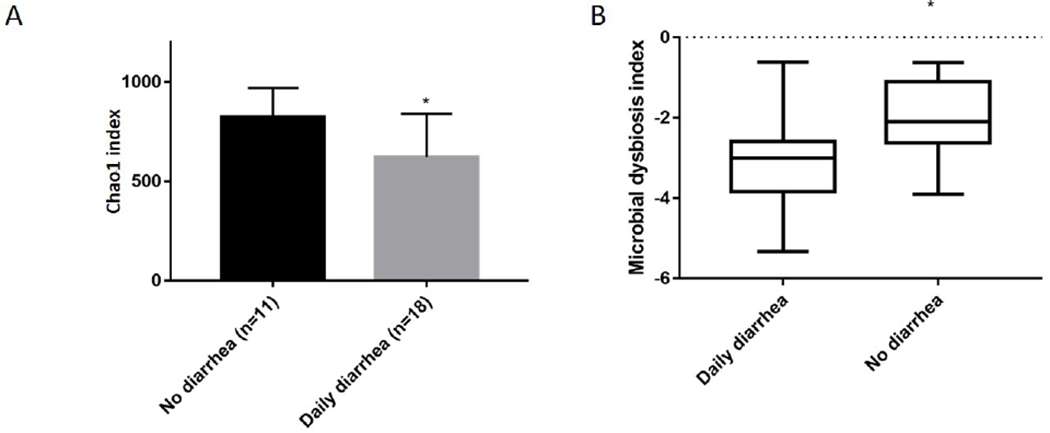
Alpha diversity and MD-index from sigmoid mucosa in moderate-severe UC/IBD-U. 4A: Chao1 diversity in Mayo 2-3 disease. Data represent comparison of Chao1 diversity according to daily diarrhea, (diarrhea [n=18], no diarrhea [n=11], p=0.03). Mean observations after rarefaction at 8500 sequences. 4B: MD-index in moderate-severe UC/IBD-U reporting daily diarrhea, (diarrhea [n=18], no diarrhea [n=11], p=0.03).
DISCUSSION
In CD patients in endoscopic remission, the mucosa-associated microbiome has less species richness and greater dysbiosis than UC/IBD-U or controls. Furthermore, in CD with endoscopic improvement, patients with persistent clinical symptoms have dissimilar beta diversity and reduced Chao1 alpha diversity than those in remission. Echoing a previous study1, we found no relationship between persistent symptoms and histological activity, previous treatments or flares over a 12-month follow-up. The detailed reporting of clinical activity, endoscopic inflammation and extensive metadata leaves this cohort well placed to compare the microbiome between IBD phenotypes in the presence and absence of inflammation.
Despite variable sites of disease activity, differences in alpha diversity and MD-Index scores were evident in ileum and rectosigmoid samples (supplementary figures 1 & 3). Fusobacterium abundance was increased in both colon and TI samples in CD relative to UC/IBD-U, but other differential taxa at one location were not replicated at the other sites. This suggests, even in active inflammation, abundance of tissue-associated taxa may differ depending on sampling site. We noted increased relative abundance of Akkermansia muciniphila in ileal CD and in UC/IBD-U patients with endoscopic improvement relative to those with moderate-severe inflammation. This bacterium effects epithelial response and goblet cell function influencing gut protective mucus layer, which is compromised in UC 16, 17 and was predictive of endoscopic remission in IBD 18.
There was no relationship between either alpha diversity or specific taxa abundance and the severity of inflammation at the biopsy site (supplementary figure 4). In our analysis, the lack of correlation between either diversity or taxa abundance and endoscopic inflammation supports the hypothesis that the microbiome may predict IBD phenotype through analysis of community structure 19 or specific OTU combinations, rather than individual species abundance. However, dysbiosis and diversity indices may have potential to differentiate between phenotypes. The MD-index was lower in CD compared with UC/IBD-U and control patients in this population. This dysbiosis score has not been validated for use in IBD phenotypes other than CD, but we propose that this may have potential to differentiate between IBD phenotypes in active inflammation. Chao1 alpha diversity in sigmoid samples was persistently lower in CD relative to UC/IBD-U and HC, even in patients with healed colons (Mayo 0, SES-CD 0-2, figure 2). In addition, paired analysis of ascending and sigmoid colon samples in patients with quiescent left sided UC/IBD-U showed no differentially abundant taxa. These findings suggest that the microbial community is different long after successful treatment in CD whereas healed UC/IBD-U patients cannot be distinguished from controls. This may reflect effects of pervasive bowel damage in CD.
Non-invasive clinical activity indices do not reliably identify patients who have achieved endoscopic remission, with 19-39% of these patients reporting persistent symptoms 20–22. We hypothesized that there may be a relationship between microbiome composition and persistent symptoms. In CD patients with endoscopic improvement, both residual clinical activity (CDAI > 150), and patients who reported daily diarrhea had significantly reduced alpha diversity and greater dysbiosis (MD-index) as compared to patients in clinical remission (Figure 3). In those reporting diarrhea, we found an association with Lactobacillus, an aero-tolerant Firmicute, often reported in greater abundance in active CD, with over 180 species having variable strain-specific effects on gut health 23. This genus is generally bile salt resistant due in part to bile salt hydrolase capacity, and CD is associated with bile-salt mediated diarrhea 24. Furthermore, the microbiota plays a role in bile acid metabolism through deconjugation and dehydroxylation of primary bile acids 25. Select Lactobacillus species can thrive in a bile-salt rich environment 26 due to resistance mechanisms preventing protein misfolding and facilitating bile salt efflux 27 It is possible that greater dysbiosis in our CD patients with residual diarrhea may be related to altered bile salt metabolism.
Another explanation for residual symptoms is an irritable bowel syndrome (IBS) overlap. The IBD in South Eastern Norway (IBSEN) study group found 27% patients in remission met criteria for co-existing IBS 28. In diarrhea-predominant IBS (IBS-D), community-level dysbiosis and reduced alpha-diversity are recognized 29, 30. The presence of loose stool itself has been associated with altered fecal species richness and community structure 31–33. The extent to which microbiome community composition in our CD cohort is a cause or consequence of residual diarrhea is unclear. Depletion of genera from the Christensenellaceae family, as in our cohort (supplementary data 8), has been reported in IBS-D 34. Although microbiome composition and dynamic stability differs in IBS and IBD, the association between altered diversity and daily diarrhea in patients without IBD suggests that the microbiome could play a role in residual symptoms in CD.
While there was no association between residual symptoms and dysbiosis in quiescent UC/IBD-U, in moderate-severe endoscopic disease (Mayo score 2-3), lower alpha diversity associated with higher clinical activity, daily diarrhea and increasing stool frequency (Figure 4). This was associated with reduced relative abundance of butyrate-producing genera and increased relative abundance of Luteimonas, a Proteobacterium. Similar changes in the microbiome have been reported in intestinal inflammation from a variety of etiologies 35–37. The association between lower diversity, greater dysbiosis and increasing diarrhea in our inflamed UC/IBD-U cohort may point to a relationship between the microbiome and clinical symptoms given the lack of correlation between diversity and site-specific Mayo scores here. The extent to which this is merely a consequence of ongoing intestinal inflammation is unclear.
In healed colon and ileal samples, CD patients have lower alpha diversity than UC/IBD-U or HC, suggesting long-term microbiota changes in CD in remission (figure 2). Residual clinical activity and daily diarrhea were associated with lower alpha diversity of the tissue-associated microbiome (figure 3). This study is strengthened by detailed phenotyping facilitating analysis of the tissue-associated microbiome relative to both validated clinical activity scores and specific symptoms. Furthermore, site-specific microbiome variability was considered, with sampling of 3 distinct sites. Limitations include the small sample size in groups with endoscopic remission, the potential influence of colonoscopy preparation on the gut microbiome, and the lack of longitudinal data which would go some way to evaluate microbiome stability. Furthermore, visceral hypersensitivity data and depression as predictors for persistent symptoms or IBD/IBS overlap are lacking. Fecal calprotectin was not widely available for patients at recruitment and may be useful to confirm remission. Our findings associate persistent clinical symptoms after mucosal healing with the gut microbiome, particularly lower diversity and species richness. The potential to control symptoms with a microbiome-based strategy should be further explored in prospective studies and may represent a novel therapeutic approach for these patients.
Supplementary Material
Need to Know.
Background:
In patients with inflammatory bowel diseases (IBD), symptoms do not always associate with the severity of endoscopic inflammation and can persist after mucosal healing.
Findings:
In an analysis of biopsies from patients with IBD and controls, the authors found that despite patients’ endoscopic improvement and remission, alpha diversity of intestinal microbes remained lower in patients with CD than in controls.
Implications for patient care:
Continued alterations in intestinal microbiomes of patients with IBD after treatment might contribute to persistent diarrhea.
Acknowledgments
Funding declaration: Research reported in this publication was partially supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number U01DK062423. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest
REFERENCES
- 1.Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nature microbiology. 2017;2:17004 Epub 2017/02/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell host & microbe. 2014;15(3):382–92. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut. 2017;66(5):813–22. Epub 2017/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62(1):159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. The New England journal of medicine. 1987;317(26):1625–9. Epub 1987/12/24. [DOI] [PubMed] [Google Scholar]
- 6.Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointestinal endoscopy. 2004;60(4):505–12. Epub 2004/10/09. [DOI] [PubMed] [Google Scholar]
- 7.Best WR, Becktel JM, Singleton JW, et al. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology. 1976;70(3):439–44. Epub 1976/03/11. [PubMed] [Google Scholar]
- 8.Best WR, Becktel JM, Singleton JW. Rederived values of the eight coefficients of the Crohn’s Disease Activity Index (CDAI). Gastroenterology. 1979;77(4 Pt 2):843–6. Epub 1979/10/01. [PubMed] [Google Scholar]
- 9.Lewis JD, Chuai S, Nessel L, et al. Use of the noninvasive components of the Mayo score to assess clinical response in ulcerative colitis. Inflammatory bowel diseases. 2008;14(12):1660–6. Epub 2008/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–4. Epub 2012/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–6. Epub 2010/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72(7):5069–72. Epub 2006/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550 Epub 2014/12/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manichanh C, Borruel N, Casellas F, et al. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9(10):599–608. Epub 2012/08/22. [DOI] [PubMed] [Google Scholar]
- 15.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. The ISME journal. 2008;2(7):716–27. Epub 2008/04/11. [DOI] [PubMed] [Google Scholar]
- 16.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell metabolism. 2014;20(5):779–86. Epub 2014/08/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes JM. Colonic mucus and ulcerative colitis. Gut. 1997;40(6):807–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedjo DI, Smolinska A, Savelkoul PH, et al. The fecal microbiota as a biomarker for disease activity in Crohn’s disease. Scientific reports. 2016;6:35216 Epub 2016/10/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7(6):e39242 Epub 2012/07/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peyrin-Biroulet L, Reinisch W, Colombel JF, et al. Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn’s disease in the SONIC trial. Gut. 2014;63(1):88–95. Epub 2013/08/27. [DOI] [PubMed] [Google Scholar]
- 21.Colombel JF, Keir ME, Scherl A, et al. Discrepancies between patient-reported outcomes, and endoscopic and histological appearance in UC. Gut. 2017;66(12):2063–8. Epub 2016/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang J, Leong RW, Wasinger VC, et al. Impaired Intestinal Permeability Contributes to Ongoing Bowel Symptoms in Patients With Inflammatory Bowel Disease and Mucosal Healing. Gastroenterology. 2017;153(3):723–31.e1. Epub 2017/06/12. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Chen L, Zhou R, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. Journal of clinical microbiology. 2014;52(2):398–406. Epub 2014/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyhlin H, Merrick MV, Eastwood MA. Bile acid malabsorption in Crohn’s disease and indications for its assessment using SeHCAT. Gut. 1994;35(1):90–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of lipid research. 2006;47(2):241–59. Epub 2005/11/22. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka H, Doesburg K, Iwasaki T, et al. Screening of lactic acid bacteria for bile salt hydrolase activity. Journal of dairy science. 1999;82(12):2530–5. Epub 2000/01/12. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz L, Margolles A, Sanchez B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Frontiers in Microbiology. 2013;4:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriksen M, Hoivik ML, Jelsness-Jorgensen LP, et al. Irritable bowel-like symptoms in ulcerative colitis are as common in patients in deep remission as with inflammation; results from a population-based study (the IBSEN study). Journal of Crohn’s & colitis. 2017. Epub 2017/12/01. [DOI] [PubMed] [Google Scholar]
- 29.Codling C, O’Mahony L, Shanahan F, et al. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Digestive diseases and sciences. 2010;55(2):392–7. Epub 2009/08/21. [DOI] [PubMed] [Google Scholar]
- 30.Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1792–801. Epub 2011/08/09. [DOI] [PubMed] [Google Scholar]
- 31.Gorkiewicz G, Thallinger GG, Trajanoski S, et al. Alterations in the colonic microbiota in response to osmotic diarrhea. PloS one. 2013;8(2):e55817 Epub 2013/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouhani S, Griffin NW, Yori PP, et al. Diarrhea as a Potential Cause and Consequence of Reduced Gut Microbial Diversity Among Undernourished Children in Peru. Clin Infect Dis. 2019. Epub 2019/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugerth LW, Andreasson A, Talley NJ, et al. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2019:gutjnl-2019-318717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Scientific reports. 2015;5:12693 Epub 2015/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2(3):204 Epub 2007/11/23. [DOI] [PubMed] [Google Scholar]
- 36.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in biotechnology. 2015;33(9):496–503. Epub 2015/07/27. [DOI] [PubMed] [Google Scholar]
- 37.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Current opinion in microbiology. 2011;14(1):82–91. Epub 2010/11/03. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


