Abstract
RGD sequence is a tripeptide composed of three amino acids: arginine (R), glycine (G), and aspartic acid (D). The RGD peptide has a high affinity to the integrin alpha v beta 3, which is overexpressed on the membrane of many cancer cells and is attracted to areas of angiogenesis. Proteinoids are biodegradable polymers based on amino acids which are formed by bulk thermal step-growth polymerization mechanism. Hollow proteinoid nanoparticles (NPs) may be formed via self-assembly process of the proteinoid polymers. We propose using novel RGD-based proteinoid polymers to manufacture NPs in which the RGD motif is self-incorporated in the proteinoid backbone. Such P(RGD) NPs can act both as a drug carrier (by encapsulation of a desired drug) and as a targeting delivery system. This article presents the synthesis of four RGD proteinoids with different RGD optical configurations, (d) or (l) arginine, glycine, and (d) or (l) aspartic acid, in order to determine which configuration is optimal as a drug-targeting carrier. These new RGD proteinoid polymers possess high molecular weights and molecular weight monodispersity. Homonuclear nuclear magnetic resonance methods were employed to predict the expected concentration of RGD tripeptide sequence in the polymer. Near infrared fluorescent NPs have been prepared by the encapsulation of indocyanine green (ICG) dye within the different P(RGD) NPs. The dry diameters of the hollow P(RdGDd), P(RdGD), P(RGD), and P(RGDd) NPs are 55 ± 13, 48 ± 9, 45 ± 11, and 42 ± 9 nm, respectively, whereas those of the ICG-encapsulated NPs were significantly higher, 141 ± 24, 95 ± 13, 86 ± 11, and 87 ± 12 nm, respectively. The ICG-encapsulated P(RdGD) NPs exhibited higher selectivity toward epithelial injury, as demonstrated using an in vitro scratch assay, because the P(RdGD) NPs accumulated in the injured area at higher concentrations when compared to other P(RGD) NPs with different chiralities. Therefore, the P(RdGD) polymer configuration is the polymer of choice for use as a targeted drug carrier to areas of angiogenesis, such as in tumors, wounds, or cuts.
Introduction
RGD sequence is a tripeptide composed of three amino acids: arginine (Arg, R), glycine (Gly, G), and aspartic acid (Asp, D). The sequence was initially discovered in 1985 by Pierschbacher, Hayman, and Ruoslahti to be the active component in the fibronectin protein.1 It has been reported that many cancer cells overexpress the alpha v beta 3 (αvβ3) integrin. This integrin is also highly upregulated on the surfaces of growing tumor blood vessels. The RGD peptide has a high affinity to the integrin αvβ3 and is attracted to areas of angiogenesis.2−4 These properties of the RGD motif has led to the development of integrin-targeted nanodrugs for imaging and treatment of tumors. Today, there are two accepted ways for conjugating RGD peptides to nanoparticles (NPs) for targeted drug-delivery applications, linear RGD or cyclic RGD peptides.5
Amino acids are categorized by their optical activity, where l and d configurations correspond to the ability to rotate polarized light to the left (l) or right (d) directions. It has been reported that the configuration of the amino acids directly influences the RGD peptide activity, which can be reflected in the cell attachment. For example, it has been demonstrated that when l-aspartic acid is replaced with the d-isomer, the RGD peptide is inactive; however, when l-arginine is replaced by the d-isomer, the binding efficiency increases 10-fold.6,7
Proteinoids are polymers which consist of natural or synthetic amino acids and are synthesized by bulk thermal step-growth polymerization mechanism. This special procedure was discovered by Fox and Harada.8−13 The formation of proteinoids is carried out in the absence of a catalyst or solvent by heating in an inert atmosphere of certain amino acids, such as aspartic acid, glutamic acid, or lysine, to their melting point, leading to the initiation of the polymerization of the amino acids.14−17 The suggested explanation for the polymerization process is that aspartic acid, for example, serves as a solvent for the other amino acid monomers as it is condensed through cyclization upon heating into 2,2′-(3,6-dioxopiperazine-2,5-diyl)diacetic acid, which initiates the polymerization with the rest of the amino acids, thus forming the proteinoid polymer, as shown in Figure 1.18
Figure 1.
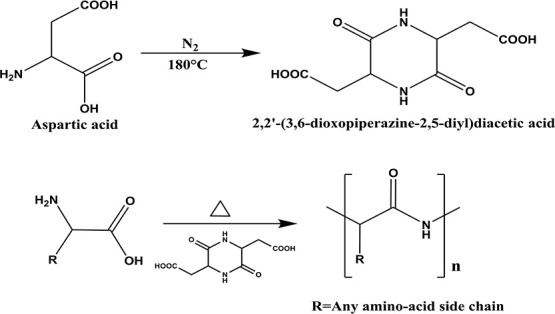
Thermal step-growth polymerization of amino acids through aspartic cyclization catalysis.
The proteinoids are considered to be nonimmunogenic (as illustrated in our previous work by cytokines induction assay), biodegradable, and nontoxic.18,19 Hence, they serve as a delivery carrier in the human body. Hollow proteinoid NPs can be produced by a self-assembly process of the proteinoids in an aqueous solution. The self-assembly process of the proteinoid occurs because of the many functional groups, which are part of the random polymer backbone.20 When proteinoids precipitate to form particles in an aqueous solution, the hydrophobic residues (e.g., phenyl groups) form an internal hydrophobic core to minimize their contact with water, whereas the hydrophilic groups (e.g., carboxylic or lysine ε-amino groups) will form hydrogen bonds with the aqueous continuous phase and reside on the particle surface.
To date, most of the recent studies using the RGD peptide for biomedical applications have not emphasized the effect of configuration on targeting.21−23 Finding the right RGD configuration can significantly improve the results of this study.
In the present research, new proteinoid polymers composed of arginine (R), glycine (G), and aspartic acid (D) were synthesized by thermal step-growth polymerization of the amino acid monomers. Different optical configurations were used with the intention of randomly achieving the RGD sequence as part of the proteinoid backbone.
In this fashion, novel RGD-based proteinoid NPs, where the RGD motif is self-incorporated in the proteinoid backbone, were prepared by a self-assembly process in an aqueous continuous phase. These P(RGD) may act both as a drug carrier (by encapsulation of a desired drug) and as a targeting delivery system because of the RGD sequence displayed on the proteinoid shell. This work offers a fast and economical method for the synthesis of RGD-based proteinoid NPs that can be suitable for therapeutic uses as well as biomedical imaging and diagnostics.
Results and Discussion
Synthesis and Characterization of RdGD RdGDd, RGD, and RGDd Proteinoids
Kieffer et al. reported that the optical configuration of the amino acids (d, l) has a direct influence on the biological activity of the RGD peptide, which can be reflected in the cell attachment.7 In order to determine the optimal optical configuration of RGD NPs as a targeted delivery vehicle, four types of RGD proteinoids were synthesized with different configurations: P(RdGDd), P(RdGD), P(RGD), and P(RGDd). The first letter corresponds to the configuration of the arginine in all cases. The aspartic acid serves as part of the RGD building blocks as well as a solvent and a linker, which helps to reduce the polymerization energy. A glassy mass was formed following the proteinoid polymerization synthesis, and the water-soluble proteinoid polymer was separated from the insoluble cross-linked proteinoid polymer mass. In comparison to previously published proteinoids, no cross-linked proteinoids were obtained, and all four RGD proteinoid configurations were water-soluble with 100% yield. Table 1 exhibits the molecular weights and polydispersity indices (PDIs) of the RdGD, RdGDd, RGD, and RGDd proteinoid polymers. The relative high molecular weights (67,660–69,066 Da) and the very narrow molecular weight monodispersity are not expected but fit to our previous publications18,24,25 because common step-growth polymerization processes result in low-molecular-weight polymers with broad molecular weight polydispersity.26
Table 1. Mw, Mn, Mp, and PDIs of the Proteinoids and the NP Diametere.
| polymer | Mw (Da)a | Mn (Da)b | Mp (Da)c | PDId | hollow NP diameters (nm) | ICG-encapsulated NP diameters (nm) |
|---|---|---|---|---|---|---|
| P(RdGDd) | 69,066 | 69,049 | 68,118 | 1.0002 | 55 ± 13 | 96 ± 12 |
| P(RdGD) | 67,660 | 67,639 | 66,464 | 1.0003 | 48 ± 9 | 95 ± 13 |
| P(RGD) | 68,722 | 68,714 | 67,707 | 1.0001 | 45 ± 11 | 86 ± 11 |
| P(RGDd) | 68,378 | 68,353 | 68,091 | 1.0004 | 42 ± 9 | 87 ± 12 |
Molecular weights were measured by GPC: weight average molecular weight (Mw).
Molecular weights were measured by GPC: number average molecular weight (Mn).
Molecular weights were measured by GPC: molecular weight at the peak (Mp).
Molecular weights were measured by GPC: PDI is the polydispersity index.
Dry particle diameters were analyzed by using ImageJ software, an open source Java image processing program, as described in the Materials and Methods section.
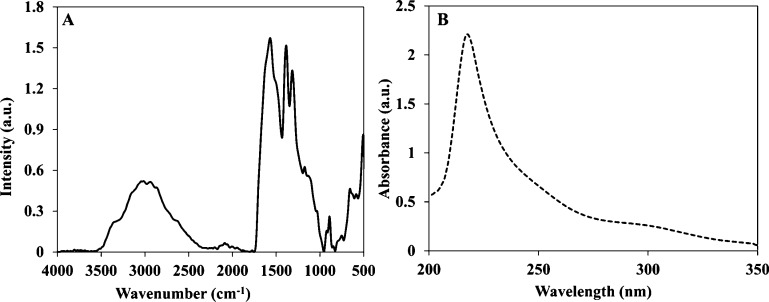
The RdGD, RdGDd, RGD, and RGDd proteinoids were characterized by Fourier transform infrared (FTIR) spectroscopy and ultraviolet–visible (UV–vis) spectroscopy. All of the RGD proteinoids showed similar peaks of NH stretching at 3356 and 2951 cm–1, amide CO stretching at 1570 cm–1, an amide NH bending band at 1490 cm–1, and CO bending at 500–660 cm–1, as shown in Figure 2A. A characteristic absorbance peak was observed for all RGD proteinoids at 218 nm, reflecting the absorbance of the peptide bonds, as shown in Figure 2B.27 Similar results of the RdGDd, RGD, and RGDd proteinoids by FTIR and UV–Vis spectroscopies are shown in Figure S1.
Figure 2.
FTIR spectrum (A) and UV–vis absorption spectra (B) of P(RdGD).
Determination of P(RdGD, RdGDd, RGD, and RGDd) Amino Acid Content by NMR
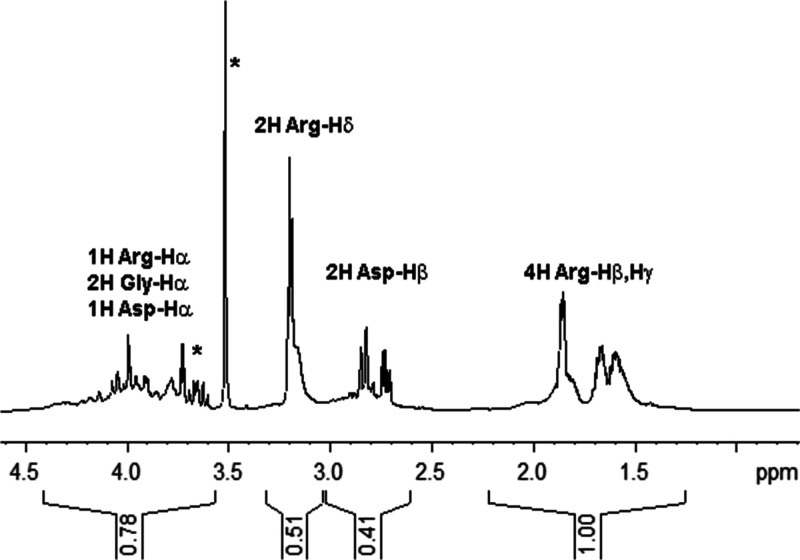
Further characterization of the P(RdGD) proteinoid in terms of amino acid incorporation levels was performed using 1H nuclear magnetic resonance (NMR) spectroscopy. In doing so, we utilized the strictly quantitative nature of the spectrum (i.e., the signal is proportional to the number of spins of a certain type) and exploited the sufficiently unique NMR footprints of the three amino acids in question. P(RdGD) was dissolved in neat 2H2O at a concentration of 10 mg/mL for acquisition of 1D 1H spectra (Figure 3). To ensure integration of proteinoid peaks only while avoiding solvent impurities, 2D-1H,1H-COSY and 2D-1H,13C-HMQC spectra were recorded for verification of typical chemical shifts of Arg/Gly/Asp amino acid protons (see Figure S2) (COSY: correlation spectroscopy; HMQC: heteronuclear multiple quantum coherence). Similar impurities were observed for all proteinoids throughout our study, facilitating their identification and exclusion from the integration analysis. As shown in Table 2, the integration of peaks in the aliphatic region of the spectrum is consistent with a molar Argd/Gly/Asp ratio of 0.25:0.145:0.205 (or 1.72:1:1:41) in the proteinoid. For P(RdGD) prepared at 1:1:1 weight ratio or 0.43:1.00:0.56 molar ratio, this indicates a strong preference for incorporation of Argd (4-fold over Gly) and Asp (2.5-fold over Gly).
Figure 3.
NMR analysis of P(RdGD) proteinoid amino acid composition. 1D 1H NMR spectrum of 10 mg/mL proteinoid in 2H2O acquired at 300 K and 16.4 T. Signals emanating from arginine, glycine, and aspartate protons, as well as integrated signal areas are shown. Asterisks denote nonproteinoid peaks excluded from the integration analysis.
Table 2. NMR Analysis of Amino Acid Composition in P(RdGD).
| PPM-1 | PPM-2 | 1H nuclei | relative intensity | normalized intensitya |
|---|---|---|---|---|
| 1.25 | 2.21 | 4H Argd(Hβ,Hγ) | 1.00 | 0.25 |
| 2.60 | 3.02 | 2H Asp(Hβ) | 0.41 | 0.205 |
| 3.04 | 3.32 | 2H Argd(Hδ) | 0.51 | 0.255 |
| 3.56 | 4.41 | 1H Argd(Hα), 2H | 0.74b | Argd—0.25c |
| Gly(Hα), 1H | Gly—0.145 | |||
| Asp(Hα) | Glu—0.205c |
Signal intensity per proton nucleus.
Excluding the signal from a solvent impurity at 3.65 ppm.
Relative contributions of Arg/Gly/Asp were based on appropriate stoichiometric ratios.
A similar approach was used to characterize proteinoids with different chirality combinations, P(RdGDd), P(RGDd), and P(RGD). As shown in Figure S3, similar 1H spectra were obtained for these proteinoids and analyzed to determine the relative incorporation rates of the three amino acids. Table S1a–c summarizes these results, showing the three proteinoids to exhibit molar incorporation ratios of (Arg/Gly/Asp) 1.39:1:1.11, 1.92:1:1.69, and 2.94:1:2.7, respectively.
While Arg/Asp ratios are relatively constant (ranging 1.09–1.25) and both exhibit increased incorporation relative to Gly, significant differences were apparent in molar incorporation ratios for the various proteinoids, particularly, Arg/Gly and Asp/Gly ratios. To determine whether this spread of molar ratios is significant, we compared the amino acid content of several proteinoids synthesized from the same 1:1:1 amino acid mixture (by weight) in different P(RdGD) batches. A comparison of four different preparations indicated that even with standard initial conditions, a range of obtained proteinoids is possible; specifically, Arg/Gly and Asp/Gly ratios spanned value ranges of 1.35–2.88 and 1.06–2.94, respectively. Thus, we attribute the incorporation ratio spread to small and difficult-to-control changes in reaction conditions between batches. This has an obvious effect on the frequency of occurrence of RGD tripeptides, and as a result, it becomes important to determine this frequency for a given proteinoid prior to an assay of its biological activity.
Although the broad nature of the 1H spectral dispersion does reflect certain differences in the local amino acid environment in the random proteinoid sequence, these are not a priori expected to afford an exact assignment of neighboring residues. As a result, the NMR spectrum is insufficiently resolved for detecting specific RGD occurrences in the proteinoids. Therefore, in order to estimate the occurrence of RGD tripeptides within a given P(RdGD, RdGDd, RGD, RGDd), we simulated such occurrences using the NMR-derived relative populations of the three amino acid types under the assumption that their incorporation is to a first approximation, independent of the sequence. Results of the simulation at various molar incorporation ratios are shown in Figure S4. Using the 1.72:1.00:1:41 ratio determined above for the P(RdGD) polymer, our simulation predicted 343 ± 16 (mean ± SD) occurrences of RdGD for every 10,000 amino acids in the proteinoid or 10.3 ± 0.5% of residues involved in these tripeptides. We also note that even a 10% error in the estimation of the limiting amino acid glycine (an over-estimated upper-bound for integration errors) would result in a small change in expected RGD occurrence (±0.3% of total residues), whereas the same change in estimation of either unlimiting amino acid would result in a negligible change (<0.05% of total residues). Thus, simulations that are based on the NMR-derived amino acid ratios are a reliable measure of RGD content in our proteinoids. A similar procedure yielded the RGD contents of RdGDd, RGD, and RGDd proteinoids taken for biological assays and were found to be 10.7 ± 0.5, 8.1 ± 0.4, and 9.8 ± 0.5%, respectively. These values were used to normalize the biological assay results (vide infra).
Preparation and Characterization of RdGD, RdGDd, RGD, and RGDd NPs
Diameter and Size Distribution of Hollow P(RdGD, RdGDd, RGD, RGDd) NPs
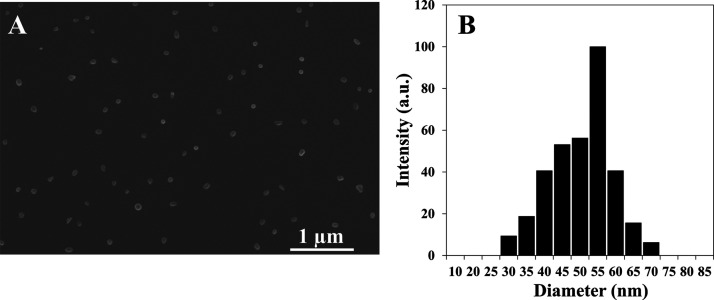
Proteinoids NPs can be formed by a simple self-assembly of the crude proteinoid polymer by heating the aqueous proteinoid polymer solution and then slow cooling to room temperature. Figure 4 shows the scanning electron microscopy (SEM) images (A) and diameter and size distribution histogram calculated by ImageJ 1.52a software (B) of the hollow P(RdGD) NPs. The measurements obtained indicate that the diameters of the hollow P(RdGDd), P(RdGD), P(RGD), and P(RGDd) NPs are similar and were found to be 55 ± 13, 48 ± 9, 45 ± 11, and 42 ± 9 nm, respectively (see Table 2).
Figure 4.
HR-SEM image (A) and dry diameter and size distribution histogram (B) of hollow P(RdGD)NPs.
Encapsulation of ICG within P(RdGD, RdGDd, RGD, RGDd) NPs
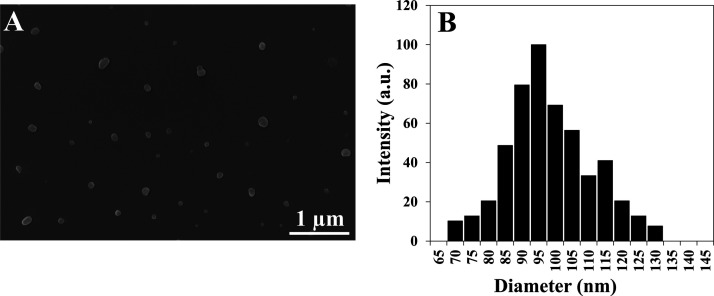
In this research, indocyanine green (ICG) near infrared (NIR) dye (700–900 nm) was used as it exhibits low auto-fluorescence and higher penetration in the biological tissue, compared to UV and visible light, because of lower light scattering, hence enabling the NPs to be used for diagnosis purposes.28 ICG was successfully encapsulated in all four NPs P(RdGDd), P(RdGD), P(RGD), and P(RGDd) giving a dry diameter of 141 ± 24, 95 ± 13, 86 ± 11, 87 ± 12 nm, respectively (see Table 2). Figure 5 shows for illustration the SEM image (A) and diameter distribution histogram (B) of the ICG-encapsulated P(RdGD) NPs. It is interesting that the ICG-encapsulated NPs own higher diameters than the nonencapsulated NPs, for example, 87 ± 12 nm compared to 42 ± 9 nm for the P(RGDd) NPs. It seems that the ICG encapsulation process swells the capsules to a relatively higher diameter.29
Figure 5.
HR-SEM image (A) and diameter distribution histogram (B) of ICG-encapsulated P(RdGD)NPs.
The concentration of the encapsulated ICG was 0.1 mg/mL per 10 mg NPs (1% w/w ICG relative to the proteinoid NPs).
Optical Spectra
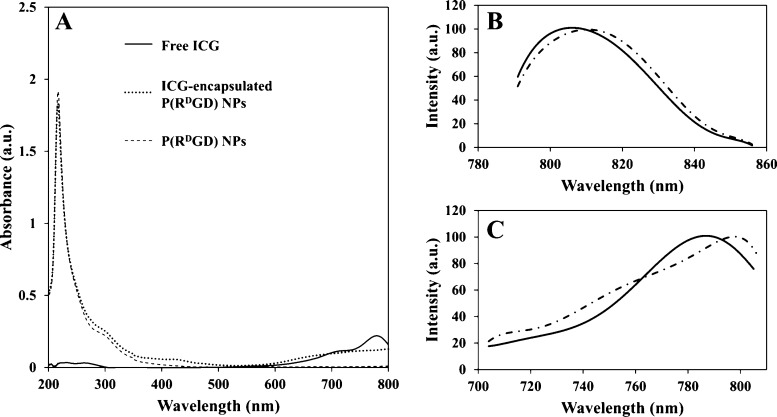
The ICG loading in the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs was determined using a calibration curve of free ICG in water, as mentioned in the experimental section. The encapsulation was found to be successful, with a loading capacity of 100%, meaning the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs contain 1% ICG w/w (relative to the NPs). Figure 6A exhibits the UV–vis absorption spectra of the ICG-encapsulated P(RdGD) NPs compared to the hollow NPs and free ICG. Hollow NPs and ICG-encapsulated NPs both show the characteristic peak of proteinoids at 218 nm. The absorbance at the region of 600–800 nm demonstrates that ICG was successfully encapsulated within the NPs. It should be noted that ICG is a NIR dye, which absorbs between 600 and 900 nm. Figure 6B demonstrates the shift in the emission peak from 812 nm for the free ICG dye to 806 nm for ICG-encapsulated P(RdGD) NPs, and the shift in excitation peak from 798 to 786 nm is exhibited in Figure 6C. The blue shift in the fluorescence is probably due to the encapsulation within the small interior space of the NPs, causing aggregation of the dye molecules.30,31
Figure 6.
Absorbance spectra of free ICG (solid line), ICG-encapsulated P(RdGD) NPs (dotted line), and hollow P(RdGD) NPs (dashed line), (A). Emission (B) and excitation (C) peaks for free ICG (dashed line) and ICG-encapsulated P(RdGD) NPs (solid line).
Photostability
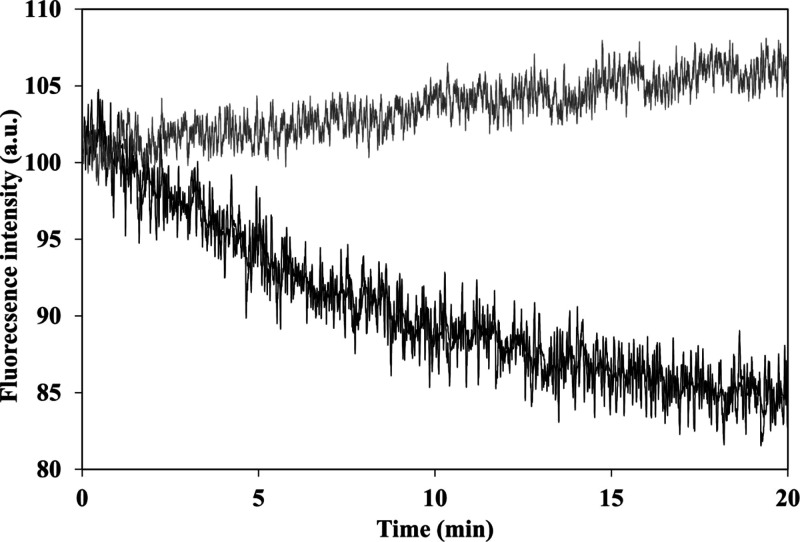
It has been reported that encapsulation of a fluorophore can increase its photostability.32 Illumination was performed continuously at 780 nm for a period of 20 min for all four different configurations of the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs and was compared to the photostability of the free ICG. While the fluorescence intensity of all four ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs was not affected by the continuous illumination, the free ICG fluorescence decreased by approximately 15% (see Figure S5). This indicates that ICG is encapsulated within the NPs and thus is protected against oxidation, reducing agents, heat, illumination levels, or exposure time which may decrease the fluorescence intensity, as shown in Figure 7 for P(RdGD).33,34 The fluorescence intensities of the free ICG and the four ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs were normalized to 100% for visual comparison.
Figure 7.
Photostability of the ICG-encapsulated P(RdGD) NPs (gray line) and free ICG (black line) as a function of time.
Cytotoxicity of P(RdGD, RdGDd, RGD, RGDd) NPs
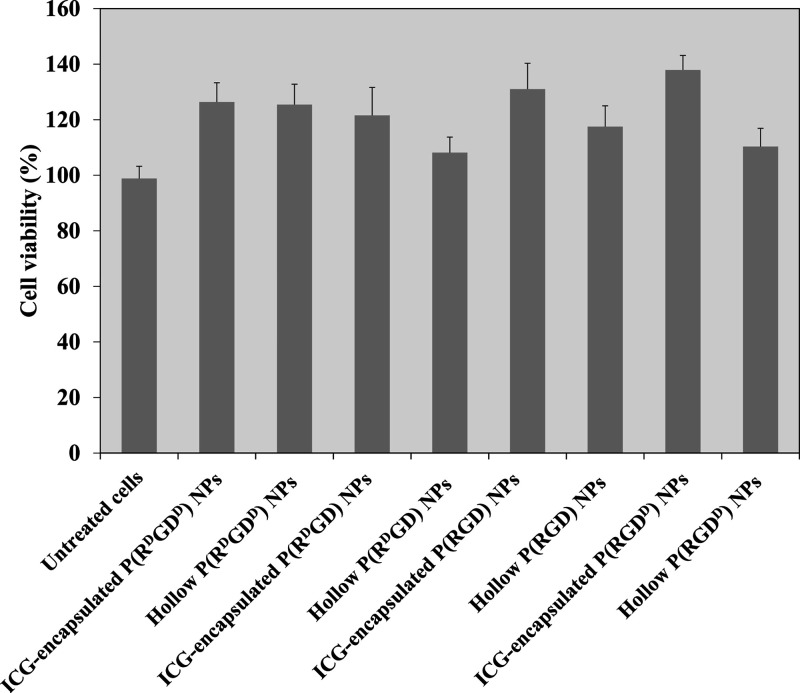
Cell cytotoxicity toward all four different optical configurations of the hollow and ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs on human umbilical vein endothelial cells (HUVECs) was evaluated using the XTT cell viability test.35 XTT is a colorimetric method which utilizes the ability of metabolic active cells to reduce the tetrazolium salt XTT to formazan. The intensity of formazan is proportional to the number of metabolic active cells. HUVECs were treated for 48 h with hollow and ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs of various optical configurations (1 mg/mL). Figure 8 shows that the cell viability levels post treatment of HUVECs with 1 mg/mL of either hollow P(RdGD, RdGDd, RGD, RGDd) or ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs did not decrease the cell viability after 48 h of treatment, compared to untreated cells. In addition, the HUVEC viability levels increased (above 100%) after 48 h of treatment, probably due to the uptake of the P(RdGD, RdGDd, RGD, RGDd) NPs as a nutrient (e.g., glucose, amino acids, etc.).36
Figure 8.
Cell viability levels of HUVECs after exposure to hollow and ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs (1 mg/mL) measured by the XTT assay. Cells (104) were incubated for 48 h with NPs dispersed in PBS in accordance with the experimental section. Untreated cells (positive control) were similarly incubated. Each bar represents the mean ± standard deviations of six separate samples.
Controlled Release of ICG-Encapsulated P(P(RdGD, RdGDd, RGD, RGDd) NPs in Human Serum or PBS
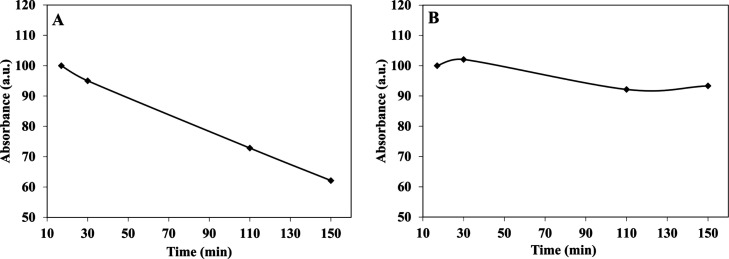
A common method for studying the in vitro drug release is by incubation in human serum or phosphate-buffered saline (PBS).37 In order to evaluate the ICG (a drug model) release of ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs in different physiological conditions, the NPs were incubated in PBS or human serum for 2.5 h at 37 °C. Figure 9 illustrates the controlled release kinetics of the UV absorbance intensities of ICG from P(RdGD) NPs following treatment in human serum (Figure 9A) or PBS (Figure 9B). This figure shows, as expected, that ICG hardly releases from the particles to PBS while a significant release during time is illustrated in the serum, probably due to the enzymatic cleavage of peptide bonds. The absorbance of ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs was normalized to 100% for visual comparison.
Figure 9.
Relative absorbance of ICG-encapsulated P(RdGD) NPs in various physiological solutions: human serum (A) and PBS (B) over 2.5 h incubated at 37 °C.
Scratch Test Assay
The development of new blood vessels is known as angiogenesis and is essential in the pathogenesis of over 50 different medical conditions including cancer, psoriasis, and rheumatoid arthritis. Angiogenesis is a well-organized cellular cascade of events including vascular initiation, formation, maturation, remodeling, and regression.38,39 Integrins are important adhesion receptors used by endothelial cells to interact with their extracellular microenvironment and regulate the growth or repair of blood vessels.39 The RGD peptide has a high affinity to the integrin αvβ3 and is attracted to areas of angiogenesis.2−4 Scratch assay is a commonly used method for studying angiogenesis in vitro.40,41
Scratch assay was used in order to determine the optimal optical configuration of P(RdGD, RdGDd, RGD, RGDd) NPs toward areas of angiogenesis. Figure 10 exhibits the confocal images of live cells following 15 min of incubation of HUVECs with ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs (1 mg/mL); each column demonstrates a different optical configuration [Figure 10(I–IV)]. Line of HUVEC nuclei stained with Hoechst dye (blue line Figure 10A), line of ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs (red line Figure 10B), and overlay images of lines in Figure 10A,B are presented in Figure 10C. The dashed line represents the scratch border between the area cleared of cells (left side) and the HUVEC cells (right side). From the overlay images, it can be clearly noticed that the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs accumulate specifically in the injured scratch area (marked in red). ICG-encapsulated P(RdGD) NPs have presented the strongest fluorescent signal in the scratch region compared to the other ICG-encapsulated P(RdGDd, RGD, RGDd) NPs. Furthermore, following the analysis of the fluorescence intensity in the area of the scratch normalized for proteinoid RGD content (Figure 10D), it is clearly demonstrated that cells treated with P(RdGD) NPs exhibited a significantly higher fluorescence intensity compared to P(RdGDd, RGD) NPs and P(RGDd) NPs. This effect can be explained by the high affinity of d-arginine in the ICG-encapsulated P(RdGD) NPs to the expressed integrins on the cell membrane of HUVECs. However, when comparing the effect of l and d-aspartic acid in ICG-encapsulated P(RdGD) and ICG-encapsulated P(RdGDd) NPs, the images exhibited less fluorescence in the scratch area when treated with ICG-encapsulated P(RdGDd), which contains d-aspartic acid. Hence, it can be deduced that d-aspartic acid interferes with the cell attachment. These results correlate with the results reported by Kieffer et al. and Ruoslahti, which suggested that the optical configuration of the amino acids (d, l) has a direct influence on the biological activity of the RGD peptide.6,7 Since P(RdGD) NPs exhibited the highest fluorescence in the scratch region, it can be concluded that they are the best candidates to be used as a targeting drug carrier to areas of angiogenesis, such as in tumors, wounds, or other angiogenesis-based medical conditions.
Figure 10.
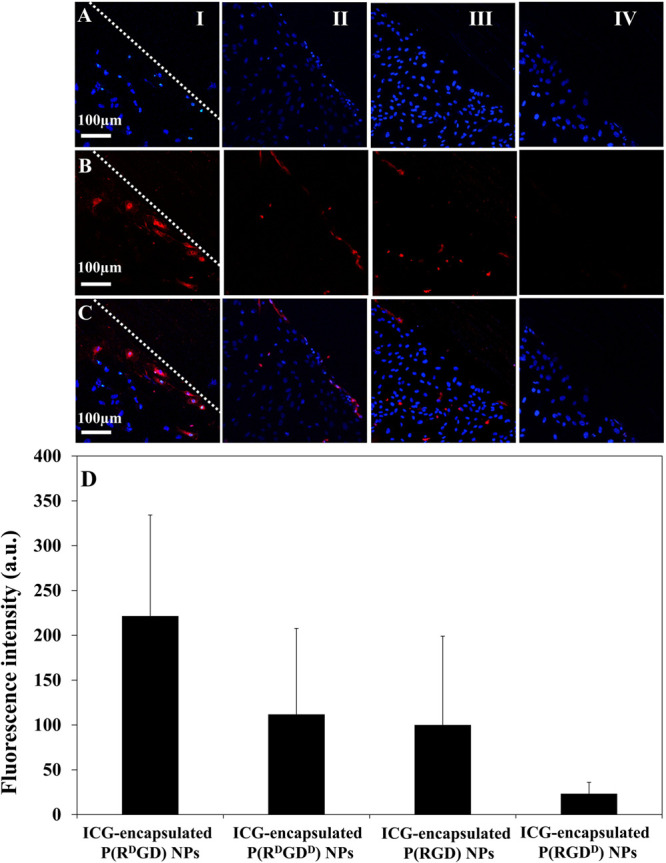
Scratch test assay, fluorescent confocal microscopy images of HUVEC cultures treated with ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs; line of HUVEC nuclei stained with Hoechst dye in blue (A), line of fluorescent ICG-encapsulated P(RdDGD, RdGDd, RGD, RGDd) NPs in red (B), and overlay images of (A) and (B) lines (C). Scale bar 100 μm. ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs; each column demonstrates a different optical configuration [(I–IV) respectively]. Fluorescence intensity of scratch region images with the four optical configurations of ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs was analyzed by ImageJ 1.52a (D) and normalized for proteinoid RGD content.
Conclusions
In this study, four new proteinoids with different optical configurations were prepared using (d) or (l) arginine, glycine, and (d) or (l) aspartic acid. The polymerization was performed by simple step-growth polymerization under a nitrogen atmosphere. The self-assembly of the proteinoids to hollow P(RdGD, RdGDd, RGD, RGDd) NPs yielded NPs with an average diameter of 47 nm with a narrow size distribution. The NPs were found to be nontoxic and stable over a period of 4 months. The NIR dye ICG was encapsulated within the P(RdGD, RdGDd, RGD, RGDd) NPs. Photostability tests demonstrated that following encapsulation of ICG within the NPs, the fluorescence of the encapsulated ICG was stable in comparison to the free ICG, indicating that the ICG was successfully encapsulated within the NPs. All four optical configurations of the P(RdGD, RdGDd, RGD, RGDd) NPs exhibited similar chemical characterization, that is, similar diameter, concentration of encapsulated ICG, stability, photostability, and cytotoxicity. However, when evaluating the targeting ability toward areas of angiogenesis in a scratch assay, the ICG-encapsulated P(RdGD) NPs exhibited selectivity toward the scratch region and a high concentration of the NPs accumulated in the injured area, compared to the ICG-encapsulated P(RdGDd, RGD, RGDd) NPs. Therefore, P(RdGD) NPs are the best candidates to be used as a targeting drug carrier to areas of angiogenesis, such as in tumors, wounds, cuts, and so forth.
Further investigations should be carried out in order to evaluate the targeting ability of the P(RdGD) NPs in vivo. Our future plans include investigating the ability of P(RdGD) NPs as a targeted drug carrier by encapsulating various chemotherapeutics such as doxorubicin or paclitaxel within the proteinoid NPs.
Materials and Methods
Materials
The following chemicals were purchased from Sigma and used without further purification: NaCl, d-arginine, l-arginine, glycine, d-aspartic acid, l-aspartic acid, Hoechst dye (bisBenzimide H33342 trihydrochloride), and ICG. HUVECs and their EBM-2 medium were purchased from Lonza (Switzerland). PBS and XTT kit were ordered form Biological Industries (Israel). Water was purified by passing deionized water through an Elgastat Spectrum reverse osmosis system (Elga Ltd., High Wycombe, UK).
Synthesis of RdGD, RdGDd, RGD, and RGDd Proteinoids by Step-Growth Polymerization Mechanism
Mixtures of amino acids (total weight of 5 g) under a nitrogen atmosphere were heated in a three-neck flask by a heating mantle to 180 °C to full dissolution of the solids. The mixture was stirred by a mechanical stirrer at 250 rpm for 20 min, producing a highly viscous yellowish to brownish paste. The paste was allowed to cool to room temperature and harden until a glassy mass was formed. After cooling, the residual material was extracted by 30 mL of distilled water and lyophilized to yield the crude proteinoid. The different configurations of RGD proteinoid were synthesized as described in Table 3. The superscript letters correspond to the configuration of arginine and aspartic acid in all cases.
Table 3. Amino Acid Composition of the Different Proteinoids.
| amino acid content (g) |
|||||
|---|---|---|---|---|---|
| polymer | (Arg)d | (Arg) | Gly | (Asp)d | (Asp) |
| P(RdGDd) | 1.666 | − | 1.66 | 1.66 | − |
| P(RdGD) | 1.66 | − | 1.66 | − | –1.66 |
| P(RGD) | − | 1.66 | 1.66 | − | 1.66 |
| P(RGDd) | − | 1.66 | 1.66 | 1.66 | − |
Proteinoid Analysis and Characterization
The molecular weights and PDIs of the proteinoid polymers were determined by gel permeation chromatography (GPC) consisting of a Waters Spectra Series P100 isocratic high-pressure liquid chromatography (HPLC) pump with an ERMA ERC-7510 refractive index detector and a Rheodyne injection valve (Coatati, CA) with a 20 μL loop (Waters, MA). The samples were dissolved with superpure HPLC water (Sigma) through a linear BioSep SEC-s3000 column (Phenomenex) at a flow rate of 1 mL/min. The molecular weights of the proteinoids were determined relative to poly(ethylene glycol) standards (Polymer Standards Service-USA, Silver Spring, MD, USA) with a molecular weight range of 100–450,000 Da, bovine plasma fibrinogen (340 kDa), and human serum albumin (67 kDa) using Clarity chromatography software.
The absorption spectra of the proteinoids were obtained by using a Cary 100 UV–vis spectrophotometer (Agilent Technologies Inc.). All of the measurements were performed in water at 25 °C. Excitation and emission spectra were recorded using a Cary Eclipse spectrophotometer (Agilent Technologies Inc.). FTIR measurements of the proteinoids were performed by the attenuated total reflectance (ATR) technique using a Bruker ALPHA-FTIR QuickSnapTM sampling module equipped with a Platinum ATR diamond module (Bruker, Germany).
Preparation and Characterization of Hollow and NIR Fluorescent Proteinoid RdGD, RdGDd, RGD, RGDd Nanoparticles
RGD Proteinoid NPs were prepared by a self-assembly process. Briefly, 50 mg of dried proteinoids were added to 5 mL of 0.01 mM NaCl aqueous solution. We refer to the RGD proteinoid as “P(RdGD, RdGDd, RGD, RGDd)” dependent on the configuration throughout this manuscript. The mixture was then stirred at 250 rpm and heated to 80 °C until the proteinoid dissolved completely. After 30 min, heating was stopped, and the proteinoid solution was allowed to cool slowly to room temperature, producing thereby the proteinoid NPs.
NIR fluorescent NPs were prepared in a similar manner substituting the 5 mL, 0.01 mM NaCl aqueous solution by a similar solution containing 0.5 mg of ICG (1% relative to the proteinoid weight). After preparation, the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NP aqueous dispersions went through extensive dialysis to remove undesired reagents using a cellulose membrane (1000 Da molecular weight cutoff) against distilled water.
Size and Size Distribution Determination
The dry diameter and size distribution of the P(RdGD, RdGDd, RGD, RGDd) NPs were measured with high-resolution SEM (HR-SEM) and analyzed by ImageJ software, an open source Java image processing program, according to our previous publication.16
NMR Characterization of Proteinoids
For preparation of proteinoid samples for NMR measurements, proteinoids were dissolved in neat 2H2O at a concentration of 10 mg/mL. All spectra were acquired on a DRX 700 MHz spectrometer system at 16.4 T and 298 K. One-dimensional 1H spectra were acquired with 8k complex points and an acquisition time of 973 ms with presaturation of the residual HDO peak. After processing, phasing, and baseline correction, regions-of-interest were integrated. Two-dimensional NMR spectra, 2D-1H,1H-COSY and 2D-1H,13C-HMQC spectra, were acquired using standard acquisition parameters, typically 512–2048 complex points and 50–200 ms in the direct 1H dimension, 180–200 complex points and 26–28 ms acquisition time in the indirect 1H dimension, and 128 complex points and 13.7 ms in the 13C indirect dimension. All spectra were acquired, processed, and visualized using the Bruker TopSpin 3.6 suite of programs.
Determination of the ICG Loading Capacity
Calibration curve of free ICG was obtained by measuring the integrals of absorbance peaks of standard solutions (0.5–10 μg/mL) in water at a wavelength of 700–900 nm. The concentration of the encapsulated ICG was determined by measuring the integral of the corresponding absorbance of a 10 mg/mL dispersion of the ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs in water. An estimation of the encapsulated material per milligram of NPs was determined according to the calibration curve. All of the ICG encapsulated P(RdGD) NPs were prepared to give a similar fluorescence intensity.
In Vitro XTT Cell Viability Assay
The XTT assay was performed to determine the viability of the HUVECs after treatment with hollow and ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs. Cells were seeded in a 96-well plate at a density of 104 cells/well in 100 μL culture medium and grown in a humidified 5% CO2 atmosphere at 37 °C. After 48 h at 37 °C, the proteinoid NPs dispersed in double-distilled water were diluted in cell medium and added to the cells, giving final concentrations of 1 mg/mL per well. After incubation for 48 h at 37 °C, 50 μL of XTT solution was added to each well according to the kit manufacturer’s instructions. The absorbance was read at 490 nm. Cell viability was determined using the procedure recommended in the manufacturer’s protocol.42
Controlled Release of ICG-Encapsulated P(RdGD, RdGDd, RGD, RGDd) NP Models
ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs (500 μL, 10 mg/mL) were incubated with PBS buffer (4.5 mL) or human serum (500 μL) and PBS buffer (4 mL), at 37 °C, giving a final concentration of 1 mg/mL NPs in a total volume of 5 mL. Samples were collected at several periods of time, and the released ICG, as a model, was measured by a UV spectrophotometer.
In Vitro Scratch Assay43,44
Epithelial injury was emulated using an in vitro scratch assay. The NIR fluorescent NPs were added to a scratched monolayer of endothelial cells to investigate the specific targeting of the P(RGD) NPs with different optical configurations to the scratched region.43,44 HUVECs (30 × 103 cells) were seeded in an eight-well glass bottom plate. The scratch was performed using a 100 μL plastic tip. Free ICG and ICG-encapsulated P(RdGD, RdGDd, RGD, RGDd) NPs were suspended in 3 mL of cell medium to a final concentration of 1 mg/mL and then added to the cell cultures. The cell nuclei were stained with Hoechst. The presence of P(RdGD, RdGDd, RGD, RGDd) NPs in cells was examined using confocal microscopy. Cells were observed 15 min post NP addition with a Leica confocal live SP8 microscope equipped with an NIR filter (excitation/emission: 638 nm/721–774 nm).
Acknowledgments
The authors would like to thank Dr. Avi Jacob for his help and guidance with confocal microscopy.
Glossary
Abbreviations
- αvβ3
alpha v beta 3
- ICG
indocyanine green
- HUVEC
human umbilical vein endothelial cells
- NIR
near infra-red
- HPLC
high pressure liquid chromatography
- GPC
gel permeation chromatography
- FTIR
Fourier transform infrared
- ATR
attenuated total reflectance
- PBS
phosphate buffered saline
- MWCO
molecular weight cut-off
- HR-SEM
high resolution scanning electron microscopy
- mL
milliliter
- DDW
double distilled water
- NMR
nuclear magnetic resonance
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01916.
FTIR spectrum and UV–Vis absorption spectra of P(RdGDd, RGD, RGDd) proteinoids; two-dimensional NMR analysis of P(RdGD) proteinoids; NMR analysis of P(RdGDd, RGD, RGDd) composition; simulations for prediction of RGD content in P(RdGD, RdGDd, RGD, RGDd) NPs; NMR analysis of amino acid incorporation into P(RGD)s; and photostability of the encapsulated ICG P(RdGD, RdGDd, RGD, RGDd) NPs and free ICG (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Pierschbacher M. D.; Ruoslahti E. Cell Attachment Activity of Fibronectin Can Be Duplicated by Small Synthetic Fragments of the Molecule. Nature 1984, 309, 30–33. 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- Dubey P. K.; Mishra V.; Jain S.; Mahor S.; Vyas S. P. Liposomes Modified with Cyclic RGD Peptide for Tumor Targeting. J. Drug Target. 2004, 12, 257–264. 10.1080/10611860410001728040. [DOI] [PubMed] [Google Scholar]
- Weis S. M.; Cheresh D. A. Tumor Angiogenesis: Molecular Pathways and Therapeutic Targets. Nat. Med. 2011, 17, 1359–1370. 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- Fu X.; Yang Y.; Li X.; Lai H.; Huang Y.; He L.; Zheng W.; Chen T. RGD Peptide-Conjugated Selenium Nanoparticles : Antiangiogenesis by Suppressing VEGF-VEGFR2-ERK / AKT Pathway. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1627–1639. 10.1016/j.nano.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Yin H.-Q.; Mai D.-S.; Gan F.; Chen X.-J. One-Step Synthesis of Linear and Cyclic RGD Conjugated Gold Nanoparticles for Tumour Targeting and Imaging. RSC Adv. 2014, 4, 9078–9085. 10.1039/c3ra47729k. [DOI] [Google Scholar]
- Ruoslahti E. RGD and Other Recognition Sequences For Integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Kieffer B.; Mer G.; Mann A.; Lefèvre J.-F. Structural Studies of Two Antiaggregant RGDW Peptides by LH and 13C NMR. Int. J. Pept. Protein Res. 2009, 44, 70–79. 10.1111/j.1399-3011.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- Fox S. W. How Did Life Begin?. Science 1960, 132, 200–208. 10.1126/science.132.3421.200. [DOI] [PubMed] [Google Scholar]
- Fox S. W. The Proteinoid Theory of the Origin of Life and Competing Ideas. Am. Biol. Teach. 1974, 36, 161–181. 10.2307/4444705. [DOI] [Google Scholar]
- Fox S. W. Thermal Synthesis of Amino Acids and the Origin of Life. Geochim. Cosmochim. Acta 1995, 59, 1213–1214. 10.1016/0016-7037(95)00037-z. [DOI] [PubMed] [Google Scholar]
- Fox S. W.; Jungck J. R.; Nakashima T. From Proteinoid Microsphere to Contemporary Cell: Formation of Internucleotide and Peptide Bonds by Proteinoid Particles. Orig. Life 1974, 5, 227–237. 10.1007/bf00927027. [DOI] [PubMed] [Google Scholar]
- Fox S. W., Mccauley R. J., Fukushim T., Windsor C. R., Montgome P.. Selective Action in Boundaries of Particles of Thermal Proteinoid. Federation Proceedings, 1967; Vol. 26, p 749.
- Fox S. W.; Nakashima T.; Przybylski A.; Syren R. M. The Updated Experimental Proteinoid Model. Int. J. Nanomed. 1982, 22, 195–204. 10.1002/qua.560220719. [DOI] [Google Scholar]
- Harada K.Thermal Polymerization of Amino Acid Mixtures Containing Aspartic Acid or a Thermal Precursor of Aspartic Acid. U.S. Patent 3,052,655 A, 1962.
- Harada K.; Matsuyama M. Polycondensation of Thermal Precursors of Amino Acids and Characterization of Constituent Amino Acids. BioSystems 1979, 11, 47–53. 10.1016/0303-2647(79)90019-4. [DOI] [PubMed] [Google Scholar]
- Kolitz-Domb M.; Margel S. Recent Advances of Novel Proteinoids and Proteinoid Nanoparticles and Their Applications in Biomedicine and Industrial Uses. Isr. J. Chem. 2018, 58, 1277–1285. 10.1002/ijch.201800021. [DOI] [Google Scholar]
- Sasson E.; Pinhasi R. V. O.; Margel S.; Klipcan L. Engineering and Use of Proteinoid Polymers and Nanocapsules Containing Agrochemicals. Sci. Rep. 2020, 10, 1–13. 10.1038/s41598-020-66172-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile S.; Kolitz-Domb M.; Corem-Salkmon E.; Margel S. Engineered Doxorubicin Delivery System Using Proteinoid-Poly ( L-Lactic Acid ) Polymeric Nanoparticles of Narrow Size Distribution and High Molecular Weight for Cancer Treatment. Int. J. Nanotechnol. Nanomed. 2017, 2, 1–11. 10.33140/ijnn.02.01.00008. [DOI] [Google Scholar]
- Madhan Kumar A. B.; Panduranga Rao K. Preparation and Characterization of PH-Sensitive Proteinoid Microspheres for the Oral Delivery of Methotrexate. Biomaterials 1998, 19, 725–732. 10.1016/s0142-9612(97)00188-9. [DOI] [PubMed] [Google Scholar]
- Yang D.-P.; Oo M. N. N. L.; Deen G. R.; Li Z.; Loh X. J. Nano-Star-Shaped Polymers for Drug Delivery Applications. Macromol. Rapid Commun. 2017, 38, 1700410. 10.1002/marc.201700410. [DOI] [PubMed] [Google Scholar]
- Zeng C.; Shang W.; Wang K.; Chi C.; Jia X.; Fang C.; Yang D.; Ye J.; Fang C.; Tian J. Intraoperative Identification of Liver Cancer Microfoci Using a Targeted Near-Infrared Fluorescent Probe for Imaging-Guided Surgery. Sci. Rep. 2016, 6, 1–10. 10.1038/srep21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo L.; Paradossi G.; Cerroni B.; Ben-Harush C.; Ariel E.; Di Meco F.; Ram Z.; Grossman R. In Vivo Biodistribution of Engineered Lipid Microbubbles in Rodents. ACS Omega 2019, 4, 13371–13381. 10.1021/acsomega.9b01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J.; Yan C.; Yan Y.; Chen L.; Song L.; Zang F.; An Y.; Teng G.; Gu N.; Zhang Y. Multi-Modal Mn-Zn Ferrite Nanocrystals for Magnetically-Induced Cancer Targeted Hyperthermia: A Comparison of Passive and Active Targeting Effects. Nanoscale 2016, 8, 16902–16915. 10.1039/c6nr03916b. [DOI] [PubMed] [Google Scholar]
- Sason E.; Kolitz-Domb M.; Chill J. H.; Margel S. Engineering of Durable Antifog Thin Coatings on Plastic Films by UV-Curing of Proteinoid Prepolymers with PEG-Diacrylate Monomers. ACS Omega 2019, 4, 9352–9360. 10.1021/acsomega.9b00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz-Domb M.; Margel S. Recent Advances of Novel Proteinoids and Proteinoid Nanoparticles and Their Applications in Biomedicine and Industrial Uses. Isr. J. Chem. 2018, 58, 1277–1285. 10.1002/ijch.201800021. [DOI] [Google Scholar]
- Yokoyama A.; Yokozawa T. Converting Step-Growth to Chain-Growth Condensation Polymerization. Macromolecules 2007, 40, 4093–4101. 10.1021/ma061357b. [DOI] [Google Scholar]
- Walker J. M.The Protein Protocols Handbook; Springer, 1996. [Google Scholar]
- Rudnick-Glick S.; Corem-Salkmon E.; Grinberg I.; Margel S. Targeted Drug Delivery of near IR Fluorescent Doxorubicin-Conjugated Poly(Ethylene Glycol) Bisphosphonate Nanoparticles for Diagnosis and Therapy of Primary and Metastatic Bone Cancer in a Mouse Model. J. Nanobiotechnol. 2016, 14, 80. 10.1186/s12951-016-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-J.; Bae P. K.; Chung B. H. Self-Assembled Levan Nanoparticles for Targeted Breast Cancer Imaging. Chem. Commun. 2015, 51, 107–110. 10.1039/c4cc07679f. [DOI] [PubMed] [Google Scholar]
- Haritoglou C.; Freyer W.; Priglinger S. G.; Kampik A. Light Absorbing Properties of Indocyanine Green (ICG) in Solution and after Adsorption to the Retinal Surface — an Ex-Vivo Approach. Graefe’s Arch. Clin. Exp. Ophthalmol. 2006, 244, 1196–1202. 10.1007/s00417-005-0216-z. [DOI] [PubMed] [Google Scholar]
- Zweck J.; Penzkofer A. Microstructure of Indocyanine Green J-Aggregates in Aqueous Solution. Chem. Phys. 2001, 269, 399–409. 10.1016/S0301-0104(01)00368-8. [DOI] [Google Scholar]
- Cohen S.; Pellach M.; Kam Y.; Grinberg I.; Corem-Salkmon E.; Rubinstein A.; Margel S. Synthesis and Characterization of near IR Fluorescent Albumin Nanoparticles for Optical Detection of Colon Cancer. Mater. Sci. Eng., C 2013, 33, 923–931. 10.1016/j.msec.2012.11.022. [DOI] [PubMed] [Google Scholar]
- Sharma P.; Brown S.; Walter G.; Santra S.; Moudgil B. Nanoparticles for Bioimaging. Adv. Colloid Interface Sci. 2006, 123–126, 471–485. 10.1016/j.cis.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Altinoǧlu E. I.; Adair J. H. Near Infrared Imaging with Nanoparticles. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2010, 2, 461–477. 10.1002/wnan.77. [DOI] [PubMed] [Google Scholar]
- Scudiero D. a.; Shoemaker R. H.; Paull K. D.; Monks A.; Tierney S.; Nofziger T. H.; Currens M. J.; Seniff D.; Boyd M. R. Evaluation of a Soluble Tetrazolium/Formazan Assay for Cell Growth and Drug Sensitivity in Culture Using Human and Other Tumor Cell Lines. Cancer Res. 1988, 48, 4827–4833. [PubMed] [Google Scholar]
- van der Heiden M. G.; Cantley L. C.; Thompson C. B.; Mammalian P.; Exhibit C.; Metabolism A. Understanding the Warburg Effect : Cell Proliferation. Science 2009, 324, 1029. 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H.; Chen Y.; Mount C. W.; Gombotz W. R.; Li X.; Pun S. H. Evaluation of Temperature-Sensitive, Indocyanine Green-Encapsulating Micelles for Noninvasive near-Infrared Tumor Imaging. Pharm. Res. 2010, 27, 1900–1913. 10.1007/s11095-010-0190-y. [DOI] [PubMed] [Google Scholar]
- Staton C. A.; Reed M. W. R.; Brown N. J. A Critical Analysis of Current in Vitro and in Vivo Angiogenesis Assays. Int. J. Exp. Pathol. 2009, 90, 195–221. 10.1111/j.1365-2613.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G.Current Topics in Developmental Biology; Elsevier Academic, 2004; Vol. 64. [Google Scholar]
- Nowak-Sliwinska P.; Alitalo K.; Allen E.; Anisimov A.; Aplin A. C.; Auerbach R.; Augustin H. G.; Bates D. O.; van Beijnum J. R.; Bender R. H. F.; et al. Consensus Guidelines for the Use and Interpretation of Angiogenesis Assays. Angiogenesis 2018, 21, 425–532. 10.1007/s10456-018-9613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Wu Q.; Qin L.; Cai J.; Du B. Gold Nanoparticles Inhibit VEGF165-Induced Migration and Tube Formation of Endothelial Cells via the Akt Pathway. BioMed Res. Int. 2014, 2014, 418624. 10.1155/2014/418624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehm N. W.; Rodgers G. H.; Hatfield S. M.; Glasebrook A. L. An Improved Colorimetric Assay for Cell Proliferation and Viability Utilizing the Tetrazolium Salt XTT. J. Immunol. Methods 1991, 142, 257–265. 10.1016/0022-1759(91)90114-U. [DOI] [PubMed] [Google Scholar]
- Liang C.-C.; Park A. Y.; Guan J.-L. In Vitro Scratch Assay: A Convenient and Inexpensive Method for Analysis of Cell Migration in Vitro. Nat. Protoc. 2007, 2, 329–333. 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Salierno M. J.; García A. J.; Del Campo A. Photo-Activatable Surfaces for Cell Migration Assays. Adv. Funct. Mater. 2013, 23, 5974–5980. 10.1002/adfm.201300902. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.