Bacterial infections are increasingly being recognized as risk factors for the development of adenocarcinomas. The strong epidemiological evidence linking Helicobacter pylori infection to stomach cancer has paved the way to the demonstration that bacterial infections cause DNA damage in the host cells, initiating transformation. In this regard, the role of bacterial genotoxins has become more relevant. Salmonella enterica serovars Typhi and Paratyphi A have been clinically associated with gallbladder cancer. By harnessing the stem cell potential of cells from healthy human gallbladder explant, we regenerated and propagated the epithelium of this organ in vitro and used these cultures to model S. Paratyphi A infection. This study demonstrates the importance of the typhoid toxin, encoded only by these specific serovars, in causing genomic instability in healthy gallbladder cells, posing intoxicated cells at risk of malignant transformation.
KEYWORDS: DNA damage, gallbladder, mucosoid cultures, organoid cultures, Salmonella, typhoid toxin, gallbladder cancer
ABSTRACT
Carcinoma of the gallbladder (GBC) is the most frequent tumor of the biliary tract. Despite epidemiological studies showing a correlation between chronic infection with Salmonella enterica Typhi/Paratyphi A and GBC, the underlying molecular mechanisms of this fatal connection are still uncertain. The murine serovar Salmonella Typhimurium has been shown to promote transformation of genetically predisposed cells by driving mitogenic signaling. However, insights from this strain remain limited as it lacks the typhoid toxin produced by the human serovars Typhi and Paratyphi A. In particular, the CdtB subunit of the typhoid toxin directly induces DNA breaks in host cells, likely promoting transformation. To assess the underlying principles of transformation, we used gallbladder organoids as an infection model for Salmonella Paratyphi A. In this model, bacteria can invade epithelial cells, and we observed host cell DNA damage. The induction of DNA double-strand breaks after infection depended on the typhoid toxin CdtB subunit and extended to neighboring, non-infected cells. By cultivating the organoid derived cells into polarized monolayers in air-liquid interphase, we could extend the duration of the infection, and we observed an initial arrest of the cell cycle that does not depend on the typhoid toxin. Non-infected intoxicated cells instead continued to proliferate despite the DNA damage. Our study highlights the importance of the typhoid toxin in causing genomic instability and corroborates the epidemiological link between Salmonella infection and GBC.
INTRODUCTION
Gallbladder cancer (GBC) is an adenocarcinoma with very poor prognosis because early stages are often asymptomatic and few patients can be cured with surgery at initial presentation (1). Although uncommon in Western countries, it has relatively high incidence in the western parts of South America and in the northern part of the Indian subcontinent (2). An intriguing aspect is its putative link to chronic carriage of Salmonella enterica serovar Typhi/Paratyphi A. In these patients, Salmonella resides in the gallbladder (GB) both intracellularly and extracellularly by forming biofilms on gallstones (3–5), which serve as a reservoir from where bacteria are intermittently shed into the duodenum (6). A higher incidence of GBC in chronic carriers was first observed after an outbreak of Salmonella enterica in Aberdeen, Scotland (7), an observation confirmed by subsequent epidemiological studies (8, 9).
Epidemiological associations with cancer have also been shown for several other bacterial pathogens. However, studies that illuminate the underlying mechanisms are only just emerging and suggest that infection can lead to genomic instability, which may contribute to the development of cancer (10). Helicobacter pylori, Escherichia coli, and Chlamydia trachomatis have been shown to induce DNA double-strand breaks (DSBs) in host cells (11–15). Evidence suggests that infection with some species not only causes the production of reactive oxygen species (ROS) that induce DNA damage in the host, but can also modify the DNA damage response and thereby induce error-prone mechanisms of repair (10).
Salmonella enterica provokes direct genotoxicity through the action of a crucial effector, the typhoid toxin (16), which is only expressed by the human-specific serovars Typhi (17) and Paratyphi A (18). It has been hypothesized that Salmonella enterica delivers the typhoid toxin through secreted outer membrane vesicles after internalization into the host cell (19, 20). More recently, it has been found that a specific interaction of a subunit of the typhoid toxin (PtlB) with luminal receptors allows the loading of the toxin from the Salmonella-containing vacuoles into vesicle carriers (21).
Typhoid toxin is able to induce direct DNA DSBs via its CdtB subunit, a DNase that is translocated into the nucleus of the intoxicated cell (19, 20, 22). CdtB also exists as part of another bacterial toxin: the cytolethal distending toxin (CDT), which is produced by multiple Gram-negative bacterial species, including Helicobacter hepaticus (23). Here, as well, it has been directly linked to tumor development in vivo and in vitro (24, 25).
Commonly used cell lines in infection biology are mostly derived from cancerous tissues, limiting their utility for studies of early carcinogenic events, since they are already transformed and have alterations in key cellular signaling pathways. Since the epithelium is the prime target of infections and toxins, the development of organoid-based human primary cell models is an invaluable means for illuminating the molecular mechanisms by which bacteria could promote cancer. While organoid or derivative models of human gastrointestinal epithelia from the small intestine (26), colon (27), stomach (28, 29), and intrahepatic duct (30) are available, such a system was developed for murine (31) and human (32) gallbladders only very recently and has not yet been utilized for infection studies (33, 34). A robust in vitro model that recapitulates the infection dynamics in healthy human gallbladder epithelium would be of immense value in this regard.
Developing from the foregut, the outer lining of the gallbladder consists of a simple columnar epithelium without any gland or crypt structures. The cells tend to moderately produce mucins (35) and transport bile and organic ions (36–38). They share many similarities with the cholangiocytes of the intrahepatic bile duct (39), and therefore the stem cells of the adult gallbladder might express similar markers, such as CD44, CD13, and LGR5 (40, 41) and also depend on activation of the Wnt/β-catenin pathway for their maintenance (30).
Here, we describe the establishment of human gallbladder organoids and their adaptation into more physiological polarized monolayers. We use these systems to study the human-restricted, GBC-associated Salmonella enterica serovar Paratyphi A and, specifically, the effect of the typhoid toxin on healthy cells. These new models will serve as a useful resource to investigate the interaction of Salmonella and its toxin with authentic human tissue.
RESULTS
Maintenance of adult gallbladder epithelial stem cells depends on activation of the Wnt/β-catenin pathway.
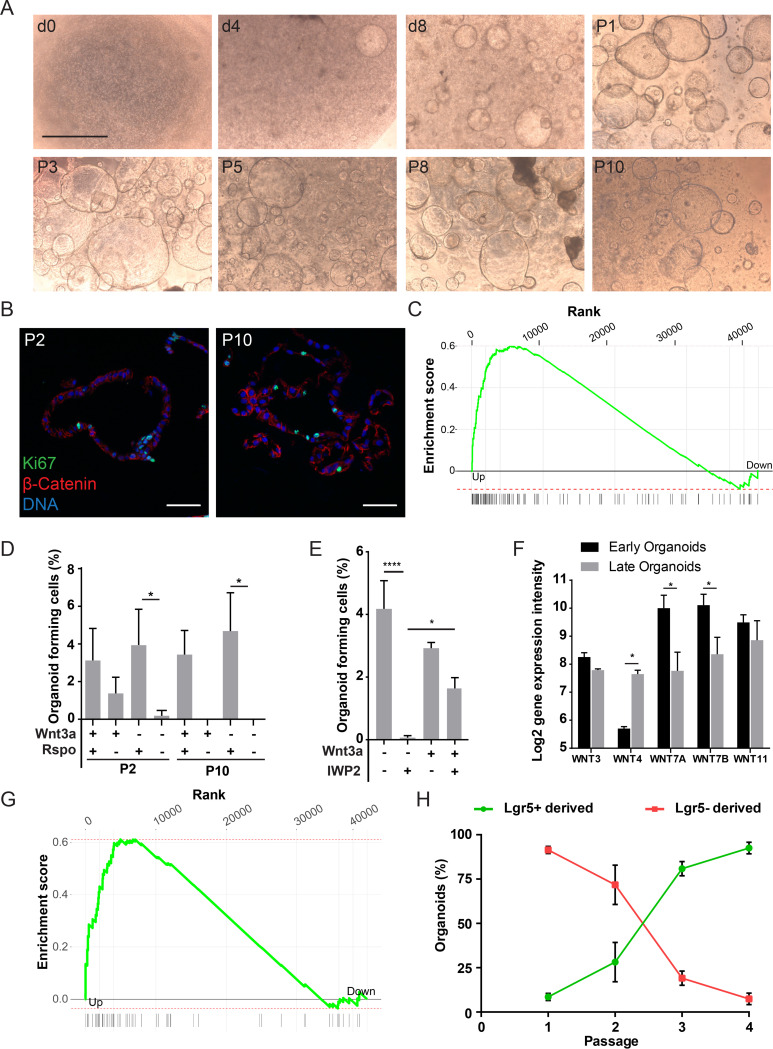
Primary epithelial cells from human and murine gallbladder (GB) were isolated and grown in Matrigel supplemented with defined medium (Table 1). After 3 to 5 days, the cells started to form hollow spheres, reaching up to 1 mm in diameter (Fig. 1A for humans; see Fig. S1A in the supplemental material for mice). Organoids were passaged every 7 to 10 days by enzymatic and mechanical shearing, and the resulting cells were seeded in fresh Matrigel for further expansion. Cultures expanded indefinitely for murine cells and for human cells. Fluorescence immunohistochemistry for the proliferation markers Ki67 and PCNA showed randomly distributed positive cells at early and late passages (Fig. 1B for humans; see Fig. S1B in the supplemental material for mice). Since only a small fraction of the cells has the ability to form organoids, we assumed that growing organoids accumulate mainly differentiated cells. We therefore analyzed the transcriptome profile of early (4-day-old) versus late (14-day-old) organoids, which confirmed that only the former were enriched in stem cell markers (42) (Fig. 1C and Table 2), indicating their undifferentiated state.
TABLE 1.
Cultivation medium composition
| Reagenta | Supplier | Catalog no. | Working concentration |
|---|---|---|---|
| Human medium | |||
| Advanced/DMEM/F-12 | Invitrogen | 12634-010 | |
| R-spondin 1 conditioned medium | In house | 25% | |
| B27 | Invitrogen | 17504-044 | 1× |
| N2 | Invitrogen | 17502-048 | 1× |
| Human epidermal growth factor (EGF) | Invitrogen | PHG0311 | 20 ng/ml |
| Human noggin | Peprotech | 120-10C | 150 ng/ml |
| Human fibroblast growth factor (FGF)-10 | Peprotech | 100-26 | 150 ng/ml |
| Nicotinamide (NIC) | Sigma | N0636 | 10 mM |
| A 83-01 (TGF-β type I receptor ALK-5 inhibitor) | Calbiochem | 2939 | 1 μM |
| Forskolin (FSK) | Tocris | 1099 | 10 μM |
| Human hepatocyte growth factor (HGF) | Peprotech | 100-39 | 25 ng/ml |
| Y-27632 (ROCK inhibitor)* | Sigma | Y0503 | 7.5 μM |
| Penicillin-streptomycin** | Invitrogen | 15140122 | 1 U/ml |
| IWP-2*** | Merck Millipore | 681671 | 10 μM |
| Wnt3a conditioned medium*** | In house | 25% | |
| Murine medium | |||
| Advanced/DMEM/F-12 | Invitrogen | 12634-010 | |
| R-spondin 1 conditioned medium | In house | 25% | |
| B27 | Invitrogen | 17504-044 | 1× |
| N2 | Invitrogen | 17502-048 | 1× |
| Murine epidermal growth factor (mEGF) | Invitrogen | PMG8044 | 50 ng/ml |
| Murine noggin | Peprotech | 250-38 | 100 ng/ml |
| Nicotinamide (NIC) | Sigma | N0636 | 10 mM |
| A 83-01 (TGF-β type I receptor ALK-5 inhibitor) | Calbiochem | 2939 | 1 μM |
| Y-27632 (ROCK inhibitor)* | Sigma | Y0503 | 7.5 μM |
| Penicillin-streptomycin** | Invitrogen | 15140122 | 1 U/ml |
*, Added only for the first 4 days after seeding; **, not added in infection experiments; ***, only if mentioned.
FIG 1.
Cultivation of human gallbladder organoids and dependence on the Wnt/β-catenin pathway activation. (A) Gallbladder epithelial cells were isolated and grown as described in Materials and Methods. Pictures were taken 0, 4, and 8 days after seeding and at passages 1, 3, 5, 8, and 10. Scale bar, 1 mm. (B) Gallbladder organoids were fixed 7 days after seeding. Organoids were paraffinized, sectioned, and immunostained for the proliferation marker Ki67 (green), β-catenin (red). DRAQ5 was used to stain the nuclei (blue). (C) Gene set enrichment analysis of human pluripotent stem cell genes published by Mallon et al. (42) among genes regulated in early versus late organoids, as identified by microarray. Adjusted P value = 0.00039, enrichment score = 0.6, normalized enrichment score = 1.9. (D) Organoids at passage 1 were split to single cells and seeded, and the number of resulting organoids was counted 5 to 7 days later (i.e., at passage 2), in media + or − Wnt3A and + or − Rspo1. The organoids were kept in culture and the procedure was repeated after 8 passages (i.e., at passage 10). *, P < 0.05 (t test). (E) Organoids were split to single cells which were seeded in Matrigel and provided with media + or − the Wnt inhibitor IWP-2 and + or − 25% of Wnt3a conditioned medium. The number of resulting organoids was counted 5 to 7 days later. *, P < 0.05; ****, P < 0.00005. (F) Change in expression levels of Wnt family members observed in a microarray comparing early versus late organoids. Only transcripts with an average log2 expression of >6 are shown. *, P < 0.05 (t test). (G) Gene set enrichment analysis of β-catenin targets published by Herbst et al. (44) among genes regulated in early versus late organoids as identified by microarray. Adjusted P value = 0.0015, enrichment score = 0.61, normalized enrichment score = 1.8. (H) Lineage tracing of murine organoids derived from the Lgr5 reporter mouse, Lgr5-EGFP-IRES-CreERT2, ROSA-mTmGfloxed after HT induction. The number of organoids derived from Lgr5+ cells (green) and Lgr5– cells (red) was counted at each passage 5 to 7 days after seeding. The plot shows the percentage of each population compared to the total number of organoids. Bars indicate the standard deviations (SD).
TABLE 2.
List of differentially regulated stem cell related genes in early organoids versus late organoids
| Probe | Gene symbol | RefSeq | Entrez ID | logFC | Avg expression | t score | P |
|---|---|---|---|---|---|---|---|
| A_23_P374844 | GAL | NM_015973 | 51083 | 5.55 | 9.38 | 23.45 | 0.00 |
| A_24_P225616 | RRM2 | NM_001034 | 6241 | 4.23 | 9.90 | 26.43 | 0.00 |
| A_24_P397107 | CDC25A | NM_001789 | 993 | 3.39 | 10.26 | 23.07 | 0.00 |
| A_32_P194264 | CHAC2 | NM_001008708 | 494143 | 2.75 | 10.21 | 9.17 | 0.01 |
| A_33_P3286208 | LRR1 | NM_203467 | 122769 | 2.61 | 8.83 | 20.53 | 0.00 |
| A_33_P3332126 | SCLY | ENST00000409736 | 51540 | 2.45 | 9.74 | 17.13 | 0.00 |
| A_33_P3387856 | CENPN | NM_001100625 | 55839 | 2.42 | 6.42 | 6.88 | 0.01 |
| A_23_P88740 | CENPN | NM_018455 | 55839 | 2.22 | 12.96 | 12.74 | 0.00 |
| A_24_P83678 | MMS22L | NM_198468 | 253714 | 2.14 | 8.86 | 8.09 | 0.01 |
| A_23_P325040 | TMPO | NM_003276 | 7112 | 2.14 | 8.95 | 13.02 | 0.00 |
| A_23_P56553 | METTL8 | NM_024770 | 79828 | 2.07 | 10.98 | 12.13 | 0.00 |
| A_33_P3253707 | LRR1 | NM_152329 | 122769 | 2.06 | 12.60 | 14.00 | 0.00 |
| A_33_P3379886 | FGF2 | NM_002006 | 2247 | 1.99 | 6.35 | 3.18 | 0.07 |
| A_23_P217637 | TIMM8A | NM_004085 | 1678 | 1.94 | 11.61 | 11.10 | 0.00 |
| A_33_P3419696 | FGF2 | NM_002006 | 2247 | 1.92 | 7.69 | 4.99 | 0.03 |
| A_24_P49747 | HMGB3P24 | ENST00000433260 | NAa | 1.89 | 6.61 | 15.70 | 0.00 |
| A_33_P3489737 | NLN | NM_020726 | 57486 | 1.86 | 10.59 | 5.83 | 0.02 |
| A_24_P244699 | NUDT15 | NM_018283 | 55270 | 1.81 | 9.55 | 6.06 | 0.02 |
| A_24_P178093 | TOMM40 | NM_006114 | 10452 | 1.77 | 8.62 | 10.80 | 0.00 |
| A_32_P95914 | MMS22L | NM_198468 | 253714 | 1.75 | 9.70 | 6.71 | 0.01 |
| A_23_P143958 | RPL22L1 | NM_001099645 | 200916 | 1.66 | 16.39 | 8.91 | 0.01 |
| A_24_P336853 | PNO1 | NM_020143 | 56902 | 1.61 | 10.31 | 12.46 | 0.00 |
| A_33_P3357082 | METTL8 | NM_024770 | 79828 | 1.59 | 9.10 | 13.84 | 0.00 |
| A_23_P82823 | PINX1 | NM_017884 | 54984 | 1.57 | 10.41 | 10.49 | 0.00 |
| A_23_P209337 | METTL21A | NM_145280 | 151194 | 1.56 | 10.87 | 6.70 | 0.01 |
| A_33_P3250861 | ZIC3 | ENST00000370606 | 7547 | 1.29 | 4.62 | 1.57 | 0.24 |
| A_23_P144337 | CCRN4L | NM_012118 | 25819 | 1.29 | 3.26 | 6.09 | 0.02 |
| A_32_P25273 | HSPD1 | NM_002156 | 3329 | 1.24 | 17.11 | 11.51 | 0.00 |
| A_23_P150092 | SEPHS1 | NM_012247 | 22929 | 1.21 | 12.66 | 7.32 | 0.01 |
| A_21_P0000006 | TOMM40 | NM_001128917 | 10452 | 1.21 | 13.86 | 9.57 | 0.01 |
| A_33_P3412613 | TMPO | NM_001032283 | 7112 | 1.18 | 5.82 | 6.27 | 0.02 |
| A_23_P202143 | NOLC1 | NM_004741 | 9221 | 1.13 | 12.02 | 4.96 | 0.03 |
| A_33_P3619171 | PMAIP1 | NM_021127 | 5366 | 1.09 | 10.98 | 4.95 | 0.03 |
| A_24_P253215 | EMG1 | NM_006331 | 10436 | 1.07 | 12.98 | 6.16 | 0.02 |
| A_32_P71788 | FKBP4 | NM_002014 | 2288 | 1.06 | 8.96 | 9.26 | 0.01 |
| A_24_P357266 | GRPR | NM_005314 | 2925 | 1.01 | 4.42 | 1.66 | 0.22 |
| A_23_P136504 | SLC25A21 | NM_030631 | 89874 | 1.01 | 5.54 | 3.14 | 0.07 |
| A_24_P297888 | MTAP | NM_002451 | 4507 | 1.00 | 9.97 | 7.12 | 0.01 |
| A_33_P3256425 | BICD1 | NM_001714 | 636 | 0.94 | 6.25 | 2.98 | 0.08 |
| A_23_P345065 | SCLY | NM_016510 | 51540 | 0.93 | 10.96 | 3.47 | 0.06 |
| A_23_P160881 | SMPDL3B | NM_001009568 | 27293 | 0.93 | 9.95 | 2.62 | 0.10 |
| A_23_P27167 | RNASEH1 | NM_002936 | 246243 | 0.93 | 11.31 | 5.67 | 0.02 |
| A_23_P365060 | MDN1 | NM_014611 | 23195 | 0.92 | 6.39 | 7.63 | 0.01 |
| A_33_P3388135 | MKKS | NM_170784 | 8195 | 0.92 | 13.58 | 3.97 | 0.04 |
| A_23_P164141 | PSME3 | NM_176863 | 10197 | 0.89 | 10.79 | 7.38 | 0.01 |
| A_23_P156842 | EEF1E1 | NM_004280 | 9521 | 0.89 | 13.42 | 7.96 | 0.01 |
| A_33_P3287815 | DDX21 | NM_004728 | 9188 | 0.87 | 13.48 | 8.11 | 0.01 |
| A_23_P43726 | NUP160 | NM_015231 | 23279 | 0.86 | 11.39 | 6.96 | 0.01 |
| A_21_P0011842 | EEF1E1 | NM_001135650 | 9521 | 0.86 | 13.35 | 5.47 | 0.02 |
| A_23_P131954 | SNX5 | NM_014426 | 27131 | 0.86 | 15.01 | 4.18 | 0.04 |
| A_23_P148484 | RLIM | NM_016120 | 51132 | 0.86 | 10.97 | 3.37 | 0.06 |
| A_23_P252362 | MRPS30 | NM_016640 | 10884 | 0.86 | 10.49 | 6.46 | 0.01 |
| A_24_P134727 | TFAM | NM_003201 | 7019 | 0.83 | 9.21 | 7.42 | 0.01 |
| A_23_P214907 | MTHFD1L | NM_015440 | 25902 | 0.82 | 9.75 | 2.11 | 0.15 |
| A_23_P256148 | AKIRIN1 | NM_024595 | 79647 | 0.81 | 11.04 | 3.58 | 0.05 |
| A_33_P3287502 | MSH2 | NM_000251 | 4436 | 0.77 | 11.49 | 5.50 | 0.02 |
| A_23_P128991 | SLIRP | NM_031210 | 81892 | 0.77 | 14.35 | 2.63 | 0.10 |
| A_33_P3285444 | TERF1 | NM_017489 | 7013 | 0.76 | 4.75 | 1.48 | 0.26 |
| A_24_P50458 | TERF1 | NM_017489 | 7013 | 0.76 | 12.20 | 3.75 | 0.05 |
| A_33_P3242659 | KIF13A | NM_022113 | 63971 | 0.70 | 5.56 | 3.56 | 0.05 |
| A_33_P3329108 | MTAP | NM_002451 | 4507 | 0.69 | 11.71 | 3.99 | 0.04 |
| A_23_P333951 | DNAH14 | NM_144989 | 127602 | 0.67 | 9.74 | 2.67 | 0.10 |
| A_23_P137484 | L1TD1 | NM_019079 | 54596 | 0.67 | 5.12 | 5.21 | 0.02 |
| A_23_P128372 | FKBP4 | NM_002014 | 2288 | 0.65 | 12.78 | 4.38 | 0.04 |
| A_33_P3294404 | AKIRIN1 | NM_024595 | 79647 | 0.64 | 10.32 | 3.56 | 0.05 |
| A_23_P216149 | TERF1 | NM_017489 | 7013 | 0.64 | 11.57 | 1.77 | 0.20 |
| A_23_P102471 | MSH2 | NM_000251 | 4436 | 0.59 | 12.22 | 5.40 | 0.02 |
| A_24_P854913 | METTL21A | NM_001127395 | 151194 | 0.56 | 11.08 | 1.39 | 0.28 |
| A_23_P54540 | EIF2AK4 | NM_001013703 | 440275 | 0.55 | 11.76 | 3.94 | 0.04 |
| A_23_P94636 | RC3H2 | NM_018835 | 54542 | 0.49 | 9.40 | 4.46 | 0.03 |
| A_23_P146997 | TXLNG | NM_018360 | 55787 | 0.48 | 9.65 | 1.61 | 0.23 |
| A_33_P3389188 | TFAM | NM_003201 | 7019 | 0.47 | 12.74 | 1.88 | 0.18 |
| A_33_P3354267 | AKIRIN1 | NM_024595 | 79647 | 0.47 | 11.81 | 3.21 | 0.07 |
| A_33_P3283906 | NIP7 | NM_016101 | 51388 | 0.46 | 12.12 | 3.33 | 0.06 |
| A_33_P3345504 | RC3H2 | NM_018835 | 54542 | 0.45 | 7.47 | 4.19 | 0.04 |
| A_33_P3299776 | NODAL | NM_018055 | 4838 | 0.45 | 3.81 | 1.54 | 0.25 |
| A_32_P220696 | TERF1 | NM_017489 | 7013 | 0.45 | 10.55 | 1.81 | 0.19 |
| A_23_P213908 | PHAX | NM_032177 | 51808 | 0.44 | 13.19 | 3.43 | 0.06 |
| A_24_P192434 | TERF1 | NM_017489 | 7013 | 0.44 | 10.44 | 1.56 | 0.24 |
| A_33_P3241786 | ADD2 | NM_017482 | 119 | 0.40 | 3.31 | 1.34 | 0.29 |
| A_32_P87531 | DNAH14 | NM_001145154 | 127602 | 0.35 | 8.81 | 1.17 | 0.35 |
| A_33_P3269453 | BPTF | ENST00000342579 | 2186 | 0.34 | 10.51 | 1.74 | 0.20 |
| A_21_P0000013 | TIMM8A | NM_001145951 | 1678 | 0.34 | 10.14 | 2.88 | 0.08 |
| A_33_P3278118 | CASP3 | NM_004346 | 836 | 0.32 | 8.43 | 0.86 | 0.47 |
| A_23_P134008 | USP45 | ENST00000472914 | 85015 | 0.31 | 9.59 | 1.34 | 0.29 |
| A_33_P3297978 | MYO1E | NM_004998 | 4643 | 0.30 | 14.27 | 2.24 | 0.13 |
| A_24_P127691 | DNAH14 | ENST00000495456 | 127602 | 0.29 | 4.94 | 1.91 | 0.18 |
| A_33_P3289996 | USP45 | NM_001080481 | 85015 | 0.27 | 7.74 | 2.52 | 0.11 |
| A_24_P281975 | GNPTAB | NM_024312 | 79158 | 0.25 | 11.60 | 1.16 | 0.35 |
| A_24_P215407 | DDX6 | NM_004397 | 1656 | 0.25 | 8.91 | 2.20 | 0.14 |
| A_33_P3289995 | USP45 | ENST00000369232 | 85015 | 0.24 | 4.91 | 2.23 | 0.14 |
| A_33_P3409506 | C9orf85 | NM_182505 | 138241 | 0.20 | 8.88 | 1.66 | 0.22 |
| A_32_P104478 | FGD6 | NM_018351 | 55785 | 0.17 | 11.00 | 0.86 | 0.47 |
| A_33_P3418294 | DNAH14 | NM_001373 | 127602 | 0.15 | 3.15 | 1.24 | 0.32 |
| A_24_P51118 | MTAP | NM_002451 | 4507 | 0.14 | 10.15 | 0.28 | 0.80 |
| A_23_P214354 | EXOC2 | NM_018303 | 55770 | 0.11 | 8.27 | 0.33 | 0.77 |
| A_33_P3235340 | DDX18 | NM_006773 | 8886 | 0.11 | 13.07 | 0.98 | 0.42 |
| A_33_P3269976 | GAL | ENST00000538401 | 51083 | 0.10 | 2.93 | 0.84 | 0.48 |
| A_23_P86504 | C10orf76 | NM_024541 | 79591 | 0.10 | 11.60 | 0.74 | 0.53 |
| A_33_P3291976 | TERF1 | ENST00000518695 | 7013 | 0.07 | 5.35 | 0.46 | 0.69 |
| A_23_P92410 | CASP3 | NM_004346 | 836 | 0.06 | 14.49 | 0.52 | 0.65 |
| A_32_P44775 | C9orf85 | NM_182505 | 138241 | 0.05 | 9.97 | 0.38 | 0.73 |
| A_33_P3414669 | RLIM | NM_183353 | 51132 | 0.04 | 6.26 | 0.40 | 0.72 |
| A_23_P351215 | SKIL | NM_005414 | 6498 | 0.04 | 7.47 | 0.34 | 0.76 |
| A_24_P152404 | C10orf76 | ENST00000311122 | 79591 | 0.03 | 10.23 | 0.16 | 0.89 |
| A_32_P135243 | MTHFD1L | NM_015440 | 25902 | 0.03 | 10.60 | 0.11 | 0.92 |
| A_32_P80255 | DDX6 | NM_004397 | 1656 | 0.03 | 10.53 | 0.15 | 0.89 |
| A_32_P528967 | RTP1 | NM_153708 | 132112 | 0.02 | 3.05 | 0.18 | 0.87 |
| A_21_P0013574 | MTHFD1L | NM_001242767 | 25902 | 0.02 | 11.20 | 0.10 | 0.93 |
| A_33_P3378972 | UNC5D | NM_080872 | 137970 | 0.01 | 3.02 | 0.12 | 0.92 |
| A_32_P741851 | GLB1L3 | NM_001080407 | 112937 | 0.01 | 2.96 | 0.11 | 0.92 |
| A_23_P140362 | VRTN | NM_018228 | 55237 | 0.01 | 2.86 | 0.11 | 0.92 |
| A_33_P3241782 | ADD2 | NM_001617 | 119 | 0.01 | 2.76 | 0.08 | 0.94 |
| A_23_P72817 | GDF3 | NM_020634 | 9573 | 0.01 | 2.91 | 0.05 | 0.96 |
| A_23_P329798 | CER1 | NM_005454 | 9350 | 0.00 | 2.67 | 0.04 | 0.97 |
| A_23_P5370 | RPRM | NM_019845 | 56475 | 0.00 | 2.84 | 0.03 | 0.98 |
| A_23_P327910 | ZIC3 | NM_003413 | 7547 | 0.00 | 2.83 | 0.03 | 0.98 |
| A_33_P3419632 | GLB1L3 | ENST00000389887 | 112937 | 0.00 | 3.57 | 0.01 | 1.00 |
| A_23_P216118 | UNC5D | NM_080872 | 137970 | 0.00 | 2.95 | 0.02 | 0.99 |
| A_21_P0014207 | LOC100506507 | XR_108853 | NA | 0.00 | 2.66 | 0.01 | 0.99 |
| A_23_P380526 | DPPA4 | NM_018189 | 55211 | 0.00 | 2.81 | −0.04 | 0.97 |
| A_23_P421436 | ADD2 | NM_017488 | 119 | −0.01 | 2.84 | −0.05 | 0.96 |
| A_19_P00318232 | SHISA9 | NM_001145205 | 729993 | −0.01 | 2.81 | −0.06 | 0.96 |
| A_33_P3280729 | SHISA9 | NM_001145204 | 729993 | −0.01 | 2.88 | −0.06 | 0.96 |
| A_23_P137573 | LEFTY2 | NM_003240 | 7044 | −0.01 | 2.87 | −0.06 | 0.96 |
| A_24_P235049 | MTHFD1L | NM_015440 | 25902 | −0.02 | 11.42 | −0.10 | 0.93 |
| A_32_P213091 | SHISA9 | NM_001145205 | 729993 | −0.03 | 4.56 | −0.26 | 0.82 |
| A_23_P375147 | RC3H2 | ENST00000373670 | 54542 | −0.05 | 11.16 | −0.18 | 0.87 |
| A_24_P380132 | G3BP2 | NM_203505 | 9908 | −0.06 | 14.44 | −0.28 | 0.80 |
| A_23_P70168 | TARS | NM_152295 | 6897 | −0.11 | 14.69 | −0.58 | 0.61 |
| A_23_P79962 | MKKS | NM_170784 | 8195 | −0.11 | 12.65 | −0.85 | 0.47 |
| A_23_P84070 | LARP7 | NM_016648 | 51574 | −0.11 | 12.56 | −0.98 | 0.42 |
| A_33_P3297245 | RRAS2 | NM_012250 | 22800 | −0.12 | 14.04 | −1.06 | 0.38 |
| A_24_P332230 | LARP7 | NM_016648 | 51574 | −0.12 | 13.09 | −0.72 | 0.54 |
| A_24_P943922 | CACHD1 | NM_020925 | 57685 | −0.14 | 4.65 | −0.18 | 0.87 |
| A_33_P3307775 | DENR | NM_003677 | 8562 | −0.14 | 7.10 | −0.44 | 0.69 |
| A_33_P3862375 | USP45 | NM_001080481 | 85015 | −0.14 | 9.20 | −0.32 | 0.78 |
| A_33_P3234317 | RRAS2 | NM_012250 | 22800 | −0.15 | 13.91 | −1.31 | 0.30 |
| A_33_P3378644 | PHC1 | NM_004426 | 1911 | −0.19 | 7.01 | −1.11 | 0.37 |
| A_23_P47058 | CUZD1 | NM_022034 | 50624 | −0.22 | 8.19 | −1.04 | 0.39 |
| A_23_P215484 | CCL26 | NM_006072 | 10344 | −0.26 | 4.06 | −1.35 | 0.29 |
| A_23_P427217 | JMJD1C | NM_032776 | 221037 | −0.46 | 9.97 | −3.89 | 0.05 |
| A_23_P346265 | GNPTAB | NM_024312 | 79158 | −0.47 | 9.02 | −1.41 | 0.28 |
| A_24_P940125 | CNOT6 | NM_015455 | 57472 | −0.50 | 11.75 | −4.61 | 0.03 |
| A_33_P3295523 | RAC3 | NM_005052 | 5881 | −0.50 | 12.37 | −3.43 | 0.06 |
| A_23_P25587 | LECT1 | NM_007015 | 11061 | −0.51 | 4.69 | −1.91 | 0.18 |
| A_24_P347624 | SNURF | NM_022804 | 8926 | −0.52 | 13.32 | −1.90 | 0.18 |
| A_23_P204246 | PHC1 | NM_004426 | 1911 | −0.55 | 4.48 | −0.98 | 0.42 |
| A_23_P259127 | ESRP1 | NM_017697 | 54845 | −0.60 | 11.38 | −1.91 | 0.18 |
| A_23_P366376 | TDGF1 | NM_003212 | 6997 | −0.65 | 7.90 | −2.86 | 0.08 |
| A_24_P144601 | POU5F1 | NM_002701 | 5460 | −0.66 | 7.98 | −2.07 | 0.15 |
| A_23_P156809 | METTL21A | NM_001127395 | 151194 | −0.66 | 11.71 | −5.71 | 0.02 |
| A_24_P104538 | BPTF | ENST00000342579 | 2186 | −0.67 | 9.15 | −2.77 | 0.09 |
| A_21_P0000084 | SLC25A21 | NM_030631 | 89874 | −0.68 | 3.26 | −1.42 | 0.27 |
| A_23_P72770 | USP44 | NM_032147 | 84101 | −0.79 | 7.71 | −7.30 | 0.01 |
| A_33_P3309206 | GABRB3 | ENST00000556166 | 2562 | −0.87 | 4.56 | −7.79 | 0.01 |
| A_23_P59138 | POU5F1 | NM_002701 | 5460 | −0.99 | 12.96 | −6.27 | 0.02 |
| A_33_P3227506 | BPTF | NM_182641 | 2186 | −1.01 | 9.64 | −6.30 | 0.02 |
| A_33_P3277075 | GABRB3 | NM_000814 | 2562 | −1.04 | 8.28 | −9.63 | 0.01 |
| A_24_P52921 | BCAT1 | NM_005504 | 586 | −1.06 | 3.48 | −4.10 | 0.04 |
| A_24_P314477 | TUBB2B | NM_178012 | 347733 | −1.14 | 7.23 | −10.24 | 0.01 |
| A_23_P323094 | PHC1 | NM_004426 | 1911 | −1.24 | 6.06 | −4.36 | 0.04 |
| A_33_P3242014 | PHC1 | NM_004426 | 1911 | −1.26 | 10.65 | −9.28 | 0.01 |
| A_23_P204640 | NANOG | NM_024865 | 79923 | −1.66 | 7.94 | −7.10 | 0.01 |
| A_24_P935986 | BCAT1 | NM_005504 | 586 | −1.77 | 9.14 | −9.18 | 0.01 |
| A_23_P160336 | LEFTY1 | NM_020997 | 10637 | −2.93 | 4.40 | −21.49 | 0.00 |
NA, not applicable.
We next tested whether the activation of the Wnt/β-catenin pathway is essential for maintenance of GB epithelial stem cells since they are phenotypically similar to adult cholangiocytes of the intrahepatic duct, which require activation of the LGR5 receptor by the Wnt agonist R-spondin for long-term culture (30). The fraction of cells able to give rise to new organoids remained at 3 to 4% for at least 10 passages for human cells only if R-spondin was added, irrespective of the presence of Wnt3A in the culture medium (Fig. 1D), and at 7 to 9% for 19 passages for murine organoids (see Fig. S1C) (30). Since R-spondins usually act synergistically with Wnt ligands, we next tested whether the epithelial cells themselves produce such ligands. Blocking Wnt ligand secretion through addition of the porcupine inhibitor IWP2 inhibited organoid formation from single cells (Fig. 1E). Organoid formation was partially rescued by the addition of exogenous Wnt3a, suggesting that GB epithelial cells or a subset of them might secrete Wnt agonists. Such a mechanism has been shown in mouse small intestinal organoids, where Paneth cells produce Wnt ligands, supporting organoid growth in the absence of exogenous Wnt agonists (43). Whether a similar subpopulation of cells is responsible for Wnt ligand production in the gallbladder is currently not known.
Cultivation of murine gallbladder organoids. (A) Murine gallbladder epithelial cells grown as organoids at 1, 2, and 4 days after seeding, and at passages 1, 5, 10, 16, and 19. Scale bar, 1mm. (B) Organoids on day 7 after seeding, fluorescently labeled with antibodies against the proliferation marker PCNA (green) and β-catenin (red); nuclei were stained with DRAQ5 (blue). Scale bar, 50 μm. (C) Organoids at passage 0 were split to single cells, seeded, and the number of resulting organoids was counted 5 to 7 days later (i.e., at passage 1). The organoids were kept in culture, and the procedure was repeated after 18 passages (i.e., at passage 19). Bars represent means ± the SD. (D) Lineage tracing of organoids derived from Lgr5-EGFP-IRES-CreERT2, ROSA-mTmGfloxed reporter mice after HT induction. Organoids derived from Lgr5+ cells express mGFP, while those derived from Lgr5− cells express mTomato. Scale bar, 200 μm. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next found that WNT3, -4, -7A, -7B, and -11 were expressed in GB organoids, but only WNT7A and WNT7B were significantly overexpressed in the stem cell-enriched early organoids, whereas late organoids were enriched in WNT4 (Fig. 1F). This indicates that different types of cells are secreting specific Wnt proteins and that WNT7A and B might play a specific role in stem cell maintenance, since they are abundantly expressed in early organoids (Fig. 1F).
Since the activation of the Wnt/β-catenin pathway is essential for stem cell maintenance we expected to find higher levels of target gene transcription in stem cells. We compared a published list of β-catenin target genes (44) with the results of our microarray (Table 3) and observed a dramatic enrichment of such genes in early organoids compared to older, more differentiated organoids (Fig. 1G). The most relevant differentially regulated genes were the secreted Wnt inhibitors Dickkopf-1 (DKK1) and DKK4, the transcription factor binding to nuclear β-catenin LEF1, and LGR5. In differentiated organoids, we observed upregulated expression of the intracellular Wnt inhibitor AXIN2, which may play a role in inhibiting the pathway in more differentiated cells (Table 3).
TABLE 3.
List of differentially regulated β-catenin target genes in early organoids versus late organoids
| Probe | Gene symbol | RefSeq | Entrez ID | logFC | Avg expression | t score | P |
|---|---|---|---|---|---|---|---|
| A_23_P118815 | BIRC5 | NM_001012271 | 332 | 4.68 | 13.18 | 42.25 | 0.00 |
| A_23_P94275 | DKK4 | NM_014420 | 27121 | 3.40 | 6.21 | 16.98 | 0.00 |
| A_23_P24129 | DKK1 | NM_012242 | 22943 | 3.17 | 14.38 | 19.03 | 0.00 |
| A_24_P20630 | LEF1 | NM_016269 | 51176 | 2.02 | 5.96 | 14.91 | 0.00 |
| A_33_P3329187 | DNMT1 | NM_001130823 | 1786 | 1.69 | 12.38 | 10.55 | 0.00 |
| A_23_P159191 | GAST | NM_000805 | 2520 | 1.58 | 7.69 | 5.45 | 0.02 |
| A_23_P98974 | LGR5 | NM_003667 | 8549 | 1.54 | 5.41 | 8.18 | 0.01 |
| A_33_P3258392 | EDN1 | NM_001955 | 1906 | 1.49 | 12.07 | 3.21 | 0.07 |
| A_23_P214821 | EDN1 | NM_001955 | 1906 | 1.48 | 14.90 | 7.19 | 0.01 |
| A_23_P202837 | CCND1 | NM_053056 | 595 | 1.44 | 11.40 | 3.97 | 0.04 |
| A_33_P3232828 | SRSF3 | NM_003017 | 6428 | 1.44 | 13.02 | 6.81 | 0.01 |
| A_23_P215956 | MYC | NM_002467 | 4609 | 1.37 | 14.06 | 6.25 | 0.02 |
| A_23_P24104 | PLAU | NM_002658 | 5328 | 1.16 | 14.30 | 6.33 | 0.02 |
| A_33_P3306146 | PLAU | NM_001145031 | 5328 | 1.10 | 9.48 | 2.68 | 0.10 |
| A_23_P160968 | LAMC2 | NM_018891 | 3918 | 1.08 | 10.17 | 3.70 | 0.05 |
| A_23_P413761 | SRSF3 | NM_003017 | 6428 | 1.03 | 15.83 | 8.66 | 0.01 |
| A_33_P3411075 | FSCN1 | NM_003088 | 6624 | 0.99 | 15.10 | 7.56 | 0.01 |
| A_23_P19673 | SGK1 | NM_005627 | 6446 | 0.94 | 12.66 | 4.51 | 0.03 |
| A_23_P135381 | SP5 | NM_001003845 | 389058 | 0.87 | 13.23 | 7.77 | 0.01 |
| A_33_P3381751 | TIAM1 | NM_003253 | 7074 | 0.86 | 11.85 | 6.00 | 0.02 |
| A_33_P3301514 | NRCAM | NM_001193582 | 4897 | 0.85 | 6.87 | 3.77 | 0.05 |
| A_23_P201636 | LAMC2 | NM_005562 | 3918 | 0.83 | 15.34 | 6.35 | 0.02 |
| A_23_P94800 | S100A4 | NM_002961 | 6275 | 0.81 | 12.16 | 7.43 | 0.01 |
| A_32_P69368 | ID2 | NM_002166 | 3398 | 0.72 | 12.89 | 2.62 | 0.10 |
| A_23_P54144 | BMP4 | NM_001202 | 652 | 0.71 | 11.68 | 2.37 | 0.12 |
| A_23_P201711 | S100A6 | NM_014624 | 6277 | 0.71 | 17.59 | 4.76 | 0.03 |
| A_23_P143143 | ID2 | NM_002166 | 3398 | 0.64 | 12.84 | 5.39 | 0.02 |
| A_23_P16469 | PLAUR | NM_001005377 | 5329 | 0.64 | 11.83 | 2.75 | 0.09 |
| A_33_P3294509 | CD44 | NM_000610 | 960 | 0.61 | 15.41 | 5.07 | 0.03 |
| A_23_P359245 | MET | NM_000245 | 4233 | 0.60 | 15.75 | 4.42 | 0.03 |
| A_23_P58788 | CDX1 | NM_001804 | 1044 | 0.58 | 3.62 | 4.50 | 0.03 |
| A_33_P3332414 | ABCB1 | NM_000927 | 5243 | 0.53 | 9.18 | 4.36 | 0.04 |
| A_23_P57784 | CLDN1 | NM_021101 | 9076 | 0.52 | 13.42 | 4.78 | 0.03 |
| A_24_P252364 | NRCAM | NM_001037132 | 4897 | 0.49 | 10.83 | 1.64 | 0.22 |
| A_24_P303989 | BMI1 | NM_005180 | 648 | 0.41 | 8.43 | 3.63 | 0.05 |
| A_23_P201655 | MYCBP | NM_012333 | 26292 | 0.39 | 13.57 | 2.96 | 0.08 |
| A_23_P412389 | FGF18 | NM_003862 | 8817 | 0.35 | 10.37 | 2.58 | 0.10 |
| A_23_P210763 | JAG1 | NM_000214 | 182 | 0.35 | 11.81 | 3.07 | 0.07 |
| A_23_P344555 | NEDD9 | NM_006403 | 4739 | 0.34 | 8.83 | 1.21 | 0.33 |
| A_23_P314115 | BMI1 | NM_005180 | 648 | 0.31 | 10.15 | 1.20 | 0.34 |
| A_23_P214681 | PPARD | NM_006238 | 5467 | 0.31 | 5.70 | 1.13 | 0.36 |
| A_33_P3374443 | L1CAM | NM_024003 | 3897 | 0.31 | 4.29 | 1.05 | 0.39 |
| A_23_P100883 | SUZ12 | NM_015355 | 23512 | 0.30 | 13.86 | 1.12 | 0.36 |
| A_33_P3323298 | JUN | NM_002228 | 3725 | 0.28 | 12.78 | 2.51 | 0.11 |
| A_23_P138631 | SMC3 | NM_005445 | 9126 | 0.27 | 12.56 | 1.97 | 0.17 |
| A_23_P82523 | ABCB1 | NM_000927 | 5243 | 0.27 | 12.31 | 2.01 | 0.16 |
| A_24_P207995 | L1CAM | NM_000425 | 3897 | 0.26 | 3.50 | 0.58 | 0.61 |
| A_32_P171061 | ASCL2 | NM_005170 | 430 | 0.23 | 9.18 | 1.38 | 0.28 |
| A_21_P0000152 | CD44 | NM_001202557 | 960 | 0.21 | 6.02 | 0.65 | 0.57 |
| A_33_P3243857 | ADAM10 | NM_001110 | 102 | 0.19 | 11.61 | 1.42 | 0.27 |
| A_23_P31073 | MYB | NM_005375 | 4602 | 0.19 | 12.36 | 1.03 | 0.40 |
| A_23_P26847 | SOX9 | NM_000346 | 6662 | 0.16 | 10.69 | 1.17 | 0.35 |
| A_24_P69095 | ENC1 | NM_003633 | 8507 | 0.13 | 13.73 | 0.20 | 0.86 |
| A_33_P3289848 | CDX1 | NM_001804 | 1044 | 0.11 | 8.23 | 0.72 | 0.54 |
| A_23_P402751 | COX2 | ENST00000361739 | 4513 | 0.08 | 15.40 | 0.28 | 0.80 |
| A_33_P3880302 | EPHB2 | NM_004442 | 2048 | 0.06 | 7.27 | 0.28 | 0.80 |
| A_24_P252130 | PPARD | NM_006238 | 5467 | 0.06 | 11.98 | 0.50 | 0.66 |
| A_33_P3245163 | MYC | M13930 | 4609 | 0.05 | 3.06 | 0.42 | 0.71 |
| A_33_P3311795 | MYB | ENST00000531845 | 4602 | 0.02 | 2.96 | 0.19 | 0.86 |
| A_24_P365807 | EFNB1 | NM_004429 | 1947 | −0.03 | 15.21 | −0.29 | 0.80 |
| A_24_P82106 | MMP14 | NM_004995 | 4323 | −0.05 | 10.03 | −0.36 | 0.75 |
| A_23_P48886 | ADAM10 | NM_001110 | 102 | −0.06 | 10.76 | −0.55 | 0.63 |
| A_33_P3370787 | EPHB2 | NM_004442 | 2048 | −0.18 | 8.02 | −1.69 | 0.21 |
| A_23_P6596 | HES1 | NM_005524 | 3280 | −0.19 | 7.93 | −1.74 | 0.20 |
| A_23_P95060 | EPHB3 | NM_004443 | 2049 | −0.19 | 11.52 | −0.99 | 0.41 |
| A_33_P3331376 | EPHB2 | NM_004442 | 2048 | −0.22 | 5.58 | −1.61 | 0.23 |
| A_33_P3411628 | CDKN2A | NM_000077 | 1029 | −0.33 | 10.69 | −3.01 | 0.08 |
| A_23_P52207 | BAMBI | NM_012342 | 25805 | −0.40 | 14.60 | −3.50 | 0.06 |
| A_21_P0014167 | NEDD9 | ENST00000379433 | 4739 | −0.41 | 4.24 | −2.81 | 0.09 |
| A_23_P27332 | TCF4 | NM_003199 | 6925 | −0.50 | 7.67 | −3.83 | 0.05 |
| A_33_P3258824 | NOTCH2 | NM_001200001 | 4853 | −0.54 | 12.30 | −2.72 | 0.09 |
| A_24_P298027 | AXIN2 | NM_004655 | 8313 | −0.56 | 7.02 | −2.16 | 0.14 |
| A_23_P43490 | CDKN2A | NM_058197 | 1029 | −0.60 | 12.54 | −4.48 | 0.03 |
| A_33_P3368358 | NEDD9 | NM_182966 | 4739 | −0.64 | 8.82 | −4.65 | 0.03 |
| A_23_P418373 | BCL2L2 | NM_004050 | 599 | −0.68 | 12.42 | −6.26 | 0.02 |
| A_23_P148015 | AXIN2 | NM_004655 | 8313 | −0.71 | 10.85 | −4.42 | 0.03 |
| A_23_P200792 | NOTCH2 | NM_024408 | 4853 | −1.06 | 13.84 | −8.94 | 0.01 |
| A_23_P52761 | MMP7 | NM_002423 | 4316 | −1.10 | 16.35 | −5.16 | 0.02 |
| A_23_P502464 | NOS2 | NM_000625 | 4843 | −2.40 | 4.30 | −9.83 | 0.01 |
Finally, to verify that expansion of GB organoids is driven by Lgr5+ cells, we took advantage of a Lgr5–EGFP-IRES-creERT2:ROSA-mTmG-floxed reporter mouse. In the gallbladder cells of this mouse, Cre-ERT2 is under the control of the Lgr5 promoter. After induction with 4-hydroxytamoxifen (4HT), Lgr5+ cells switch from red-Tomato to green-GFP expression. Induction with 4HT during culture of organoids derived from GBs of the reporter mice resulted in the generation of two distinct organoid populations. The majority derived from Lgr5− cells expressing mTomato, while 8.6% originated from Lgr5+ cells expressing mGFP (Fig. 1H; see also Fig. S1D in the supplemental material). The proportion of organoids derived from Lgr5+ cells steadily increased after the first passage, making up >90% by passage 4, confirming the crucial role of Wnt/β-catenin signaling through the Lgr5 receptor in the long-term maintenance of GB cells in vitro.
Gallbladder organoids are stable and resemble the cell structure and function of the organ in situ.
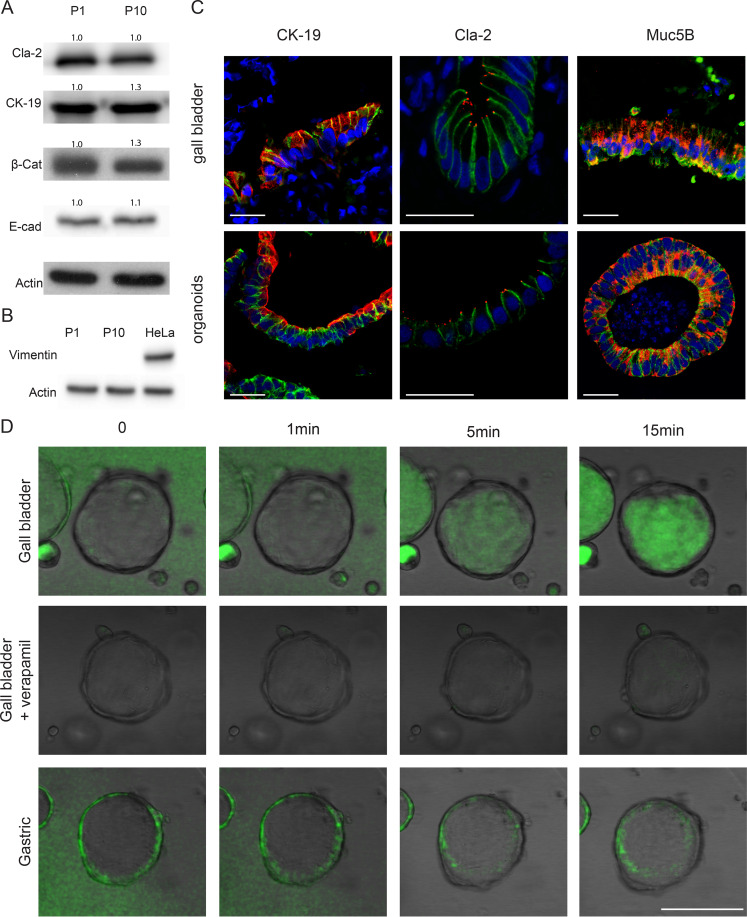
To confirm that GB organoids maintain their epithelial identity, we examined expression of the epithelial marker E-cadherin by Western blot (Fig. 2A). The levels of the GB markers claudin-2 and cytokeratin-19 did not change between early (passage 1) and late (passage 10 for human, 19 for mouse) passages (see Fig. 2A for humans and see Fig. S2A for mice). Previous attempts to cultivate epithelial primary cells were frustrated by fibroblast outgrowth (45, 46). In our system, we observed that fibroblasts do not grow in Matrigel, and at the end of passage 1 we could not detect the mesenchymal marker Vimentin (Fig. 2B and Fig. S2B). In order to assess the GB identity of organoids, we used fluorescence immunohistochemistry to examine a GB-specific combination of markers and compared their expression to that of GB tissue. The luminal mucosa of the GB consists of a simple columnar epithelium expressing cytokeratin-19 (47). Similarly, the GB organoids consist of an E-cadherin-positive cell monolayer, with apical cytokeratin-19 expression (Fig. 2C for humans and Fig. S2C for mice, left panel) and eccentric nuclei (Fig. 2C for humans and Fig. S2C for mice). These organoids also show luminal junctional expression of claudin-2 (Fig. 2C for humans and Fig. S2C for mice), a tight-junction protein expressed at higher levels in the gallbladder compared to other organs including the cholangiocytes of the bile duct (48). GB epithelial cells also produce mucins, with MUC5B being one of the most abundant (49, 50). As expected, we detected MUC5B expression in both the tissue sample and the organoids (Fig. 2C for humans and Fig. S2C for mice).
FIG 2.
Characterization of human organoids. (A) Western blot analysis of epithelial and gallbladder markers at early (P1) and late (P10) passages. Relative densitometry values, normalized to P1 (=1), are shown above the bands. (B) Western blot analysis as in panel A of the fibroblast marker Vimentin compared to HeLa cells. (C) Immunofluorescence analysis of human gallbladder tissue and organoids 7 days after seeding for the gallbladder markers cytokeratin-19, claudin-2, or mucin5B (red); the epithelial marker E-cadherin (green); and DRAQ5 (blue). Scale bar, 25 μm. (D) Transport assay of rhodamine-123 (green) in gallbladder organoids treated with the multidrug transporter inhibitor verapamil (middle row), and gastric organoids. Scale bar, 100 μm.
Characterization of murine gallbladder organoids. (A) Western blot analysis of murine epithelial and gallbladder markers at early (P1) and late (P19) passages. (B) Western blot analysis as in panel A of the fibroblast marker vimentin compared to HeLa cells. (C) Immunofluorescence analysis of murine gallbladder tissue and organoids at 7 days after seeding for the gallbladder markers cytokeratin-19, claudin-2, or mucin5B (red); the epithelial marker E-cadherin (green); and DRAQ5 (blue). Scale bar, 10 μm. Download FIG S2, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
One of the functions of the GB is to concentrate bile in the lumen (37, 38). The gallbladder expresses the ATP-dependent multidrug transporter MDR1, which transports organic cations back into the lumen (51–53), protecting the organ from high concentrations of potentially toxic organic ions. To test whether gallbladder organoids functionally recapitulate this physiological feature, we added rhodamine-123, a chemical dye substrate of MDR1 often used to monitor organoid function, to the medium (54). Gallbladder organoids actively transported the dye into the lumen, resulting in increased concentration of luminal fluorescence relative to the medium on the outside (Fig. 2D, top panel). Pretreating organoids with the MDR1 inhibitor verapamil prevented luminal dye accumulation (Fig. 2D, middle panel), confirming dependence on MDR1. In contrast, gastric organoids did not accumulate the dye (Fig. 2D, bottom panel).
Salmonella enterica serovar Paratyphi A induces paracrine CdtB-dependent DNA damage in GB organoids.
Since the gallbladder organoids accurately recapitulate the main molecular features of the epithelium of origin, we used them to model infection with S. enterica using the human restricted pathogenic serovar Paratyphi A, which has been epidemiologically linked to gallbladder cancer (7, 55). Previous observations of the genotoxic effects of S. Typhi/Paratyphi A were based on experiments in cell lines, using mostly ectopic expression of recombinant typhoid toxin (19, 20).
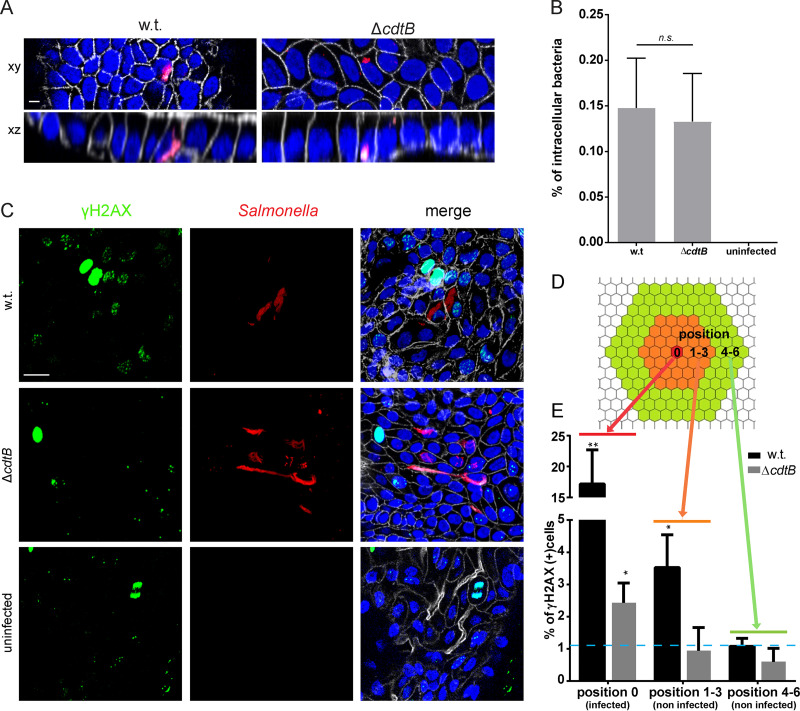
Since the genotoxicity of the bacterium resides in the CdtB subunit of the typhoid toxin, we generated a cdtB knockout. Organoids were mechanically sheared to expose the luminal side and cocultured with Salmonella enterica serovar Paratyphi A or with its isogenic cdtB knockout strain, before reseeding in Matrigel, with gentamicin-supplemented medium to eliminate extracellular bacteria. At 3 days post infection, organoids showed foci of infection with intracellular Salmonella (Fig. 3A). After verifying that the ΔcdtB mutant is capable of invading epithelial cells at a rate similar to the wild-type (w.t.) bacteria (Fig. 3B), we examined the induction of DNA damage.
FIG 3.
Infection and paracrine genotoxic effect of CdtB. (A) Reconstruction of whole-mount immunofluorescence labeling of organoids infected with Salmonella Paratyphi A carrying the mCherry-expressing plasmid pLS002 (red) at 3 days post infection, with phalloidin to detect F-actin (white) and Hoechst for DNA (blue). Scale bar, 10 μm. (B) Proportion of cells invaded after infection of organoids with wild-type Salmonella or a cdtB deletion mutant. (C) Whole-mount immunofluorescence labeling of organoids 3 days after infection with Salmonella Paratyphi A w.t. or ΔcdtB carrying the mCherry-expressing plasmid pLS002 using antibodies against γH2AX (green), phalloidin (white), and Hoechst (blue). Scale bar, 20 μm. (D) Model for categorization of uninfected cells according to the distance from the infected cell at position 0 (red). Orange represents the first three rings of non-infected cells (positions 1 to 3), and green represents the next three rings (positions 4 to 6). (E) Percentage of cells positive for the DNA damage marker γH2AX depending on their distance from the infected cell. The dashed blue line represents the average percentage of γH2AX-positive cells in uninfected organoids (SD = 0.97). *, P < 0.05; **, P < 0.01 (compared to uninfected cells). Infected cells are defined as cells with >5 bacteria, and γH2AX-positive cells are cells with >3 foci.
To this end, we tested organoids for phosphorylation of H2AX at serine 139 (γH2AX), a histone variant involved in detection of DSBs and recruitment of repair factors (56), and we quantified and mapped the number of γH2AX-positive cells after infection with the wild type and the ΔcdtB strain. The number of cells experiencing DNA damage was generally higher in the organoids infected with the wild-type strain compared to the ΔcdtB mutant (Fig. 3C). Quantification of the number of γH2AX-positive cells that are infected (defined in the map of Fig. 3D as position 0) revealed that both cells infected with the w.t. or ΔcdtB strain experience DNA damage (Fig. 3E, position 0). However, there is a significantly reduced number of γH2AX-positive cells among the ones infected with the mutant strain (Fig. 3E, position 0, ΔcdtB).
In addition, we noticed that in organoids infected with the wild-type strain, a number of uninfected neighboring cells also contained γH2AX foci (Fig. 3C to E). To quantify this paracrine genotoxic effect, uninfected cells were divided into two groups depending on the distance from the infected cell (Fig. 3D): Positions 1 to 3 include the first three rings of uninfected cells surrounding the infected focus, whereas positions 4 to 6 represent the rings 4 to 6 of the uninfected cells. The proportion of γH2AX-positive cells was higher in positions 1 to 3 than in positions 4 to 6 (Fig. 3E), but only for the organoids infected with wild-type bacteria. This confirms that the typhoid toxin is secreted from infected cells also in the primary polarized cells of the organoids (17) and that its genotoxic effects extend to the neighboring cells in a paracrine manner. In our system, this paracrine effect was limited to the first three rings of cells surrounding the infected one. Since γH2AX is also highly expressed during mitosis, cells that displayed chromosome condensation were excluded from the analysis. Our experiments suggest that infection with Salmonella Paratyphi A causes DNA damage and that a functional typhoid toxin increases the extent of damage in the infected cells and extends it to the neighboring uninfected cells.
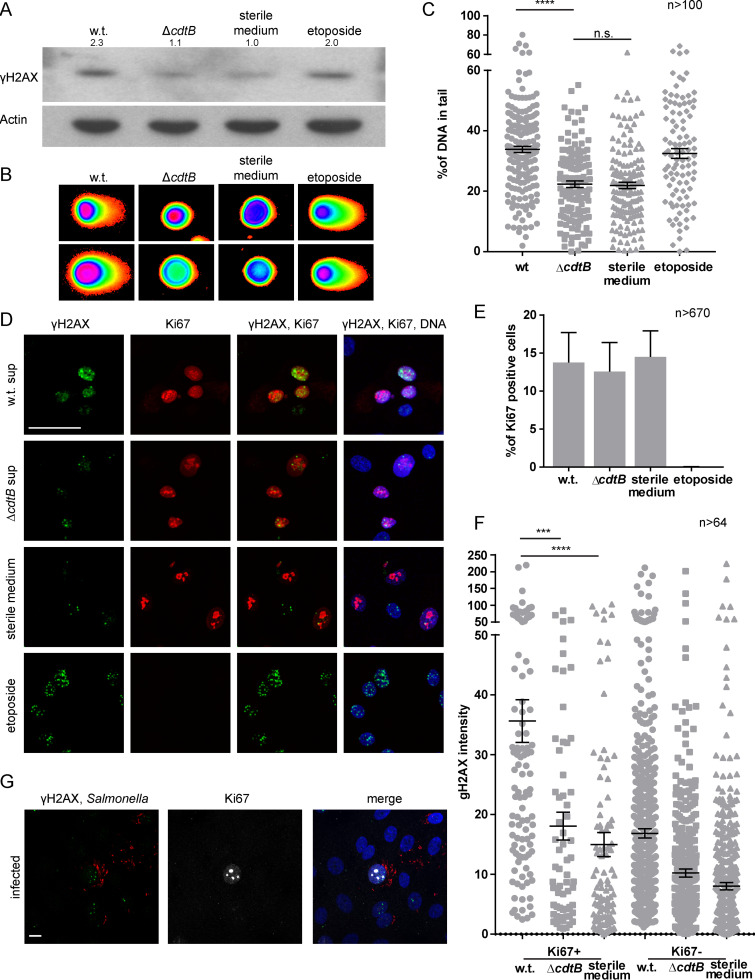
Infection with Salmonella Paratyphi A activates transcription programs associated with cell cycle arrest.
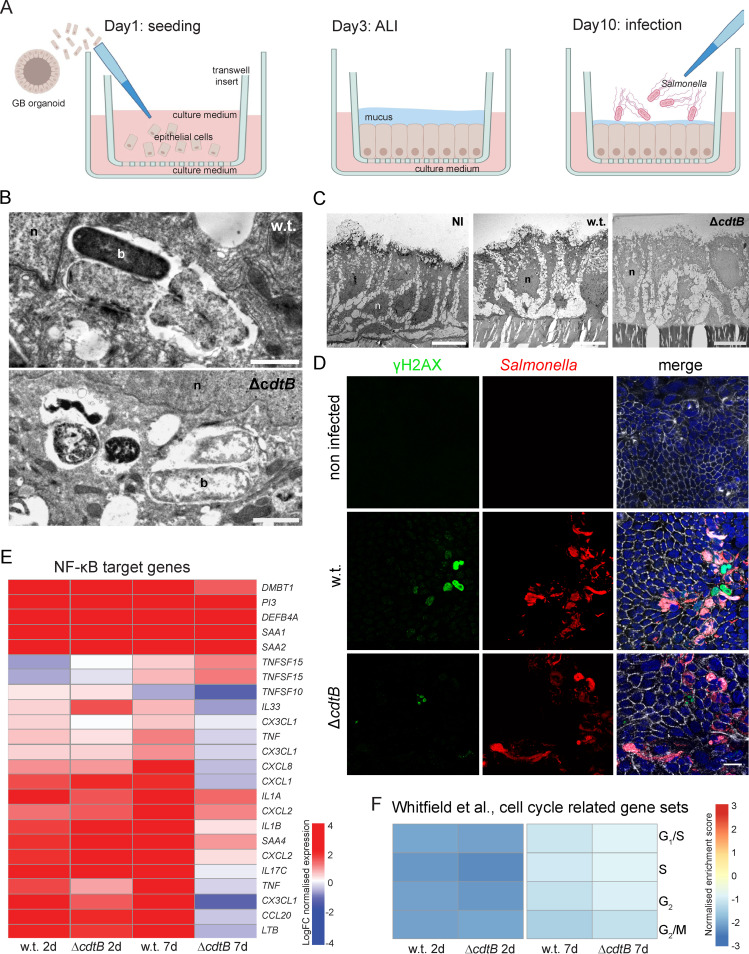
The risk of developing gallbladder cancer is higher in patients who are chronic carriers of typhoid Salmonella serovars. Therefore, to understand the fate of the infected cells, we sought to extend the duration of the infection using a more physiological model that mimics chronic infection in vitro. For the infection of the organoids, the cells must be disaggregated, and after 3 days we usually observed an overgrowth of bacteria or of cells, which impaired longer-term analysis. To understand the effect of the infection on a homeostatic gallbladder epithelial barrier and to allow longer term infection, we adapted the gallbladder organoids into mucosoid cultures, as previously done for the human stomach (34). Single cells derived from organoids were seeded on a collagen-coated polycarbonate filter in a standing cell culture insert (Fig. 4A). The cultivation cocktail was identical to that used for organoids and applied both below and above the filter. After 3 days, the apical medium was removed to start air-liquid interface cultivation (Fig. 4A). Primary gallbladder cells can be expanded on a monthly basis by deriving single cells from mucosoid cultures and restarting from the seeding procedure. Gallbladder mucosoids can be infected by applying a suspension of bacteria on top of the filter after removing excess mucus (Fig. 4A). The progress of the infection can be monitored using fluorescent transgenic Salmonella. Presence of intracellular Salmonella was detectable equally for both wild-type and ΔcdtB strains (Fig. 4B), and electron microscopy analysis of non-infected and infected mucosoid cultures revealed that the monolayer and the cell gross morphology remain intact during infection (Fig. 4C).
FIG 4.
Generation of gallbladder mucosoids and long-term infection experiments. (A) Schematic of gallbladder mucosoid cultivation and infection procedure. (From left to right) After seeding, a polarized cell layer of gallbladder cells begins to form on the collagen-coated polycarbonate filter in the transwell insert. Primary cell medium is provided around the cell culture insert and on top of the cells. At day 3, the upper medium is withdrawn, and cells start to produce mucus. From day 10 onward, the culture is stable, and infection experiments can be performed by administering Salmonella on the cell layer. (B) Detailed view of long-term infection of human gallbladder mucosoids with S. enterica Paratyphi A and transmission electron microscopy. Stable long-term infection can be reached with both the wild type and the cdtB deletion mutant by applying gentamicin for 24 h and then withdrawing it again from the medium. Internalization and perinuclear localization of the bacteria within lysosomal structures is visible. Two zoomed-in images of intracellular bacteria are shown. b, bacterium; n, nucleus. Scale bar, 1 μm. (C) Establishment of mucosoids. The development of a polarized monolayer of gallbladder cells in an air-liquid cultivation (“mucosoids”) and transmission electron microscopy images of non-infected control (NI) and infected with S. Paratyphi A w.t. and isogenic ΔcdtB KO strains for 2 days are shown. Scale bar, 10 μm. (D) Top view of infected and non-infected gallbladder mucosoids. Staining was performed for γH2AX (green), Salmonella (red), phalloidin (white), and nuclei (blue). Cultures infected for 6 days show DNA damage, whereas there is no damage visible in the non-infected control. Scale bar, 20 μm. (E) Heat map of manually selected NF-κB target genes. A comparison of w.t. and ΔcdtB infections at 2 and 7 days post infection is shown. The heatmap was plotted using the normalized expression values (log-normalized intensity) relative to the non-infected control at each time point (logFC). (F) Heatmap of normalized enrichment scores from GSEA for genes preferentially expressed in distinct cell cycle phases (58) for comparisons of mucosoid cultures with w.t. or ΔcdtB strain infections at 2 and 7 days post infection relative to non-infected controls.
Similar to what we observed with organoids, in the infected mucosoid cultures, we found that established colonies of w.t. Salmonella induce more DNA damage than the isogenic ΔcdtB strain, as measured using gH2AX staining (Fig. 4D). We performed a microarray analysis to compare the short versus the long-term effect of the infection on the gallbladder epithelial cells. We used gene set enrichment analysis (GSEA) to investigate any statistically significant consistent differences between gene set expression in the culture after infection with the w.t. strain versus infection with the ΔcdtB isogenic mutant. Infection with both strains induced similar expression of NF-κB target genes at 2 days post infection, indicating the expected initiation of an inflammatory response (Fig. 4E). Interestingly, in the cultures infected with the w.t. strain, NF-κB-controlled cytokines and chemokine genes continued to be highly expressed at 7 days, suggesting a role of the typhoid toxin in maintaining inflammation. It has previously been observed that the typhoid toxin reduces inflammation in mice infected with a transgenic Salmonella Typhimurium strain expressing the typhoid toxin (57). Inflammation is the result of a complex interaction between immune cells and the epithelium in the mucosa, and we observed here that typhoid toxin directly or indirectly maintains high transcription of NF-κB target genes in epithelial cells.
Analysis of the cell-cycle related gene sets (58) during infection (Table 4) revealed a strong underrepresentation of transcriptional programs related to each cell cycle phase (G1/S, S, G2, and G2/M) (Fig. 4F). As those genes are usually accumulated only in a specific phase of the cell cycle, the downregulation of all the G1/S, S, G2, and G2/M transcription programs implies that a proportion of cells in the infected mucosoids are not replicating (58, 59). This effect of the infection in stopping cell replication is particularly strong at 2 days after infection, but is attenuated after 1 week, indicating that an increasing number of cells are cycling again (Fig. 4F). The effect of the infection on the cell cycle was either independent from a functional typhoid toxin or any effect of the typhoid toxin on the infected culture was masked by other bacterial effectors.
TABLE 4.
List of differentially regulated genesa
| Probe | Gene symbol | RefSeq | Entrez ID | logFC at: |
Cell cycle phaseb | |||
|---|---|---|---|---|---|---|---|---|
| 2 days |
7 days |
|||||||
| w.t. vs NI | dCdtB vs NI | w.t. vs NI | dCdtB vs NI | |||||
| A_23_P39481 | ABCA7 | NM_019112 | 10347 | −0.22 | −0.02 | 0.20 | −0.66 | G1_S |
| A_23_P26375 | ACD | NM_022914 | 65057 | −0.11 | −0.01 | 0.10 | −0.48 | G1_S |
| A_23_P163143 | ACYP1 | NM_203488 | 97 | 0.05 | −0.13 | −0.18 | 0.71 | G1_S |
| A_23_P211039 | ADAMTS1 | NM_006988 | 9510 | 0.46 | 0.38 | −0.08 | 0.45 | G1_S |
| A_23_P342275 | ADAMTS1 | NM_006988 | 9510 | 0.37 | 0.16 | −0.22 | 0.48 | G1_S |
| A_24_P283395 | ADCK2 | NM_052853 | 90956 | −0.25 | −0.29 | −0.03 | −0.55 | G1_S |
| A_24_P291978 | ADCK2 | NM_052853 | 90956 | 0.23 | −0.02 | −0.26 | −0.10 | G1_S |
| A_23_P501547 | ADCY6 | NM_015270 | 112 | −0.21 | 0.10 | 0.31 | −0.73 | G1_S |
| A_24_P298077 | ANKRD10 | NM_017664 | 55608 | 0.75 | −0.03 | −0.78 | 0.94 | G1_S |
| A_23_P205046 | ANKRD10 | NM_017664 | 55608 | 0.03 | 0.56 | 0.53 | −0.36 | G1_S |
| A_23_P170331 | AP3M2 | NM_006803 | 10947 | 0.34 | 0.04 | −0.30 | 0.60 | G1_S |
| A_24_P64039 | AP3M2 | NM_006803 | 10947 | −0.02 | −0.32 | −0.30 | 0.47 | G1_S |
| A_23_P160729 | AP4B1 | NM_006594 | 10717 | 0.31 | 0.28 | −0.03 | 0.46 | G1_S |
| A_23_P256682 | APEX2 | NM_014481 | 27301 | −0.22 | 0.20 | 0.42 | −0.34 | G1_S |
| A_23_P162782 | ARGLU1 | NM_018011 | 55082 | 0.17 | −0.09 | −0.25 | 0.74 | G1_S |
| A_24_P159648 | BAIAP2 | NM_006340 | 10458 | −0.33 | −0.03 | 0.30 | −0.93 | G1_S |
| A_23_P315836 | BAIAP2 | NM_017451 | 10458 | 0.16 | 0.11 | −0.06 | −0.12 | G1_S |
| A_23_P61810 | BAIAP2 | NM_017450 | 10458 | −0.07 | 0.34 | 0.41 | −1.21 | G1_S |
| A_23_P67771 | BARD1 | NM_000465 | 580 | −1.44 | −1.07 | 0.37 | −0.47 | G1_S |
| A_32_P18824 | BRD7 | NM_013263 | 29117 | 0.04 | −0.13 | −0.17 | 0.07 | G1_S |
| A_23_P381378 | CAPN7 | NM_014296 | 23473 | 0.00 | −0.06 | −0.06 | 0.20 | G1_S |
| A_23_P58898 | CASP8AP2 | NM_012115 | 9994 | −0.42 | −0.45 | −0.03 | 0.12 | G1_S |
| A_32_P180315 | CCDC180 | NM_020893 | 1E+08 | −0.65 | −0.98 | −0.33 | −0.36 | G1_S |
| A_24_P280706 | CCDC180 | NM_020893 | 1E+08 | 0.27 | 0.28 | 0.01 | 0.50 | G1_S |
| A_23_P209200 | CCNE1 | NM_001238 | 898 | −0.40 | −0.33 | 0.08 | 0.17 | G1_S |
| A_23_P215976 | CCNE2 | NM_057749 | 9134 | −1.03 | −0.94 | 0.10 | −0.63 | G1_S |
| A_24_P397107 | CDC25A | NM_001789 | 993 | −1.34 | −1.13 | 0.21 | −0.75 | G1_S |
| A_23_P121423 | CDC25A | NM_001789 | 993 | −0.57 | −0.96 | −0.38 | 0.06 | G1_S |
| A_23_P49972 | CDC6 | NM_001254 | 990 | −1.14 | −1.40 | −0.26 | 0.39 | G1_S |
| A_23_P251421 | CDCA7 | NM_031942 | 83879 | −1.06 | −0.35 | 0.72 | −0.88 | G1_S |
| A_24_P171549 | CDCA7 | NM_031942 | 83879 | −1.00 | −0.16 | 0.84 | −1.13 | G1_S |
| A_24_P274795 | CDCA7L | NM_018719 | 55536 | −1.39 | −0.68 | 0.71 | −0.92 | G1_S |
| A_23_P20752 | CDK20 | NM_001039803 | 23552 | 0.20 | 0.17 | −0.02 | −0.03 | G1_S |
| A_24_P53519 | CHAF1A | NM_005483 | 10036 | −0.37 | −0.42 | −0.05 | −0.71 | G1_S |
| A_23_P57306 | CHAF1B | NM_005441 | 8208 | −0.81 | −0.45 | 0.37 | −0.54 | G1_S |
| A_23_P126212 | CLSPN | NM_022111 | 63967 | −0.82 | −1.06 | −0.24 | −0.08 | G1_S |
| A_23_P52556 | CTSD | NM_001909 | 1509 | −0.32 | −0.47 | −0.15 | −0.61 | G1_S |
| A_23_P139312 | DHFR2 | NM_176815 | 200895 | −0.54 | −0.71 | −0.17 | 0.09 | G1_S |
| A_24_P186065 | DHFR2 | NM_176815 | 200895 | −0.05 | 0.11 | 0.16 | −0.33 | G1_S |
| A_24_P219024 | DIS3 | NM_014953 | 22894 | 0.08 | 0.00 | −0.08 | 0.31 | G1_S |
| A_23_P48416 | DIS3 | NM_014953 | 22894 | 0.05 | 0.17 | 0.12 | −0.27 | G1_S |
| A_24_P395317 | DIS3 | NM_014953 | 22894 | 0.01 | −0.06 | −0.07 | −0.29 | G1_S |
| A_23_P36962 | DNAJC3 | NM_006260 | 5611 | 0.10 | 0.13 | 0.03 | 0.34 | G1_S |
| A_23_P10385 | DTL | NM_016448 | 51514 | −2.06 | −1.93 | 0.13 | −1.18 | G1_S |
| A_23_P80032 | E2F1 | NM_005225 | 1869 | −1.89 | −1.24 | 0.65 | −1.39 | G1_S |
| A_23_P408955 | E2F2 | NM_004091 | 1870 | −2.73 | −1.92 | 0.80 | −2.19 | G1_S |
| A_23_P125990 | E2F2 | NM_004091 | 1870 | −0.11 | −0.52 | −0.40 | 0.71 | G1_S |
| A_23_P44932 | EIF2A | NM_032025 | 83939 | 0.33 | 0.14 | −0.19 | 0.46 | G1_S |
| A_32_P197524 | EIF2A | NM_032025 | 83939 | 0.17 | −0.18 | −0.35 | 0.32 | G1_S |
| A_23_P87964 | ESD | NM_001984 | 2098 | −0.33 | −0.23 | 0.11 | 0.29 | G1_S |
| A_24_P841662 | ESD | AK093643 | 2098 | −0.38 | −0.21 | 0.17 | −0.17 | G1_S |
| A_24_P332314 | FAM111B | NM_198947 | 374393 | −2.21 | −2.52 | −0.31 | −0.18 | G1_S |
| A_23_P409516 | FAM122A | NM_138333 | 116224 | 0.03 | −0.28 | −0.31 | 0.34 | G1_S |
| A_23_P71644 | FANCG | NM_004629 | 2189 | −0.62 | −0.62 | −0.01 | −0.03 | G1_S |
| A_23_P141146 | FBXL20 | NM_032875 | 84961 | 0.56 | 0.06 | −0.49 | 0.23 | G1_S |
| A_32_P318086 | FLAD1 | NM_025207 | 80308 | −0.63 | −0.30 | 0.33 | −0.11 | G1_S |
| A_32_P6917 | FLAD1 | NM_025207 | 80308 | −0.68 | −0.18 | 0.50 | 0.02 | G1_S |
| A_23_P34527 | FLAD1 | NM_025207 | 80308 | −0.67 | −0.05 | 0.62 | −0.18 | G1_S |
| A_23_P118246 | GINS2 | NM_016095 | 51659 | −3.12 | −2.76 | 0.36 | −1.06 | G1_S |
| A_23_P152136 | GINS3 | NM_022770 | 64785 | −0.67 | −0.65 | 0.02 | 0.09 | G1_S |
| A_24_P159323 | GINS3 | NM_022770 | 64785 | 0.12 | 0.26 | 0.14 | −0.05 | G1_S |
| A_23_P19712 | GMNN | NM_015895 | 51053 | −0.77 | −0.85 | −0.08 | 0.45 | G1_S |
| A_23_P99579 | GON7 | NM_032490 | 84520 | −0.62 | −0.44 | 0.18 | 0.05 | G1_S |
| A_24_P567952 | HCG18 | NR_024052 | 414777 | −0.54 | −0.06 | 0.48 | −0.46 | G1_S |
| A_24_P934162 | HCG18 | A_24_P934162 | NA | 0.28 | −0.15 | −0.43 | 0.48 | G1_S |
| A_32_P181722 | HCG18 | NR_024052 | 414777 | −0.15 | 0.07 | 0.22 | −0.19 | G1_S |
| A_24_P567944 | HCG18 | NR_024052 | 414777 | −0.07 | 0.23 | 0.30 | −0.14 | G1_S |
| A_32_P199884 | HORMAD1 | NM_032132 | 84072 | 0.28 | 0.03 | −0.25 | 0.24 | G1_S |
| A_24_P416370 | HOXB4 | NM_024015 | 3214 | −0.84 | −0.78 | 0.06 | −0.48 | G1_S |
| A_24_P305067 | HOXB4 | NM_024015 | 3214 | −0.09 | 0.18 | 0.27 | −0.62 | G1_S |
| A_23_P98183 | HRAS | NM_005343 | 3265 | 0.32 | 0.49 | 0.17 | −0.12 | G1_S |
| A_32_P9963 | HSF2 | NM_004506 | 3298 | 0.28 | −0.09 | −0.37 | 0.80 | G1_S |
| A_23_P111360 | HSF2 | NM_004506 | 3298 | 0.21 | −0.23 | −0.44 | 0.62 | G1_S |
| A_23_P43079 | INTS8 | NM_017864 | 55656 | 0.04 | 0.25 | 0.21 | 0.64 | G1_S |
| A_23_P391506 | IVNS1ABP | NM_006469 | 10625 | −0.60 | −0.45 | 0.15 | −0.49 | G1_S |
| A_23_P137514 | IVNS1ABP | NM_006469 | 10625 | −0.36 | −0.18 | 0.18 | 0.63 | G1_S |
| A_24_P324787 | KANK2 | NM_015493 | 25959 | −0.33 | 0.05 | 0.38 | −0.90 | G1_S |
| A_23_P50426 | KANK2 | NM_015493 | 25959 | −0.35 | 0.16 | 0.51 | −1.00 | G1_S |
| A_23_P55897 | KANK2 | NM_015493 | 25959 | −0.05 | 0.24 | 0.29 | −0.50 | G1_S |
| A_23_P12079 | KCNC4 | NM_153763 | 3749 | −0.37 | −0.04 | 0.33 | −0.43 | G1_S |
| A_23_P404821 | KIAA1147 | NM_001080392 | 57189 | −0.25 | 0.09 | 0.34 | −0.52 | G1_S |
| A_24_P101047 | KIAA1586 | NM_020931 | 57691 | −0.24 | 0.03 | 0.27 | −0.25 | G1_S |
| A_24_P230965 | KIAA1586 | NM_020931 | 57691 | −0.20 | −0.22 | −0.02 | 0.24 | G1_S |
| A_24_P237559 | LNPEP | AK096804 | 4012 | 0.41 | 0.10 | −0.31 | 0.64 | G1_S |
| A_23_P144677 | LNPEP | ENST00000231368 | 4012 | −0.29 | −0.02 | 0.27 | −0.56 | G1_S |
| A_24_P132019 | LNPEP | ENST00000231368 | 4012 | 0.21 | 0.36 | 0.15 | −0.16 | G1_S |
| A_23_P156061 | LNPEP | NM_005575 | 4012 | −0.09 | −0.07 | 0.02 | 0.24 | G1_S |
| A_32_P69475 | LNPEP | ENST00000231368 | 4012 | 0.03 | 0.05 | 0.02 | −0.30 | G1_S |
| A_23_P207445 | MAP2K6 | NM_002758 | 5608 | −2.10 | −0.61 | 1.49 | −1.44 | G1_S |
| A_23_P408996 | MBOAT1 | NM_001080480 | 154141 | −0.45 | −0.03 | 0.42 | −0.57 | G1_S |
| A_32_P103633 | MCM2 | NM_004526 | 4171 | −1.61 | −1.56 | 0.06 | −1.32 | G1_S |
| A_23_P132277 | MCM5 | NM_006739 | 4174 | −1.85 | −1.65 | 0.20 | −1.36 | G1_S |
| A_23_P90612 | MCM6 | NM_005915 | 4175 | −1.27 | −1.79 | −0.52 | 0.06 | G1_S |
| A_23_P204782 | MDM1 | NM_020128 | 56890 | −0.85 | −0.55 | 0.29 | −0.15 | G1_S |
| A_23_P413180 | MDM1 | NM_017440 | 56890 | −0.66 | −0.11 | 0.55 | −0.69 | G1_S |
| A_23_P105730 | MDM1 | NM_020128 | 56890 | −0.14 | −0.02 | 0.12 | 0.24 | G1_S |
| A_24_P313678 | MED31 | NM_016060 | 51003 | 0.18 | 0.08 | −0.11 | 0.78 | G1_S |
| A_23_P341443 | MNT | NM_020310 | 4335 | 0.68 | 0.66 | −0.02 | −0.28 | G1_S |
| A_24_P350969 | MNT | AF318360 | 4335 | 0.08 | −0.20 | −0.28 | 0.56 | G1_S |
| A_32_P6015 | MNX1 | NM_005515 | 3110 | −0.33 | −0.42 | −0.10 | −0.36 | G1_S |
| A_23_P253331 | MNX1 | NM_005515 | 3110 | −0.07 | −0.33 | −0.26 | 0.05 | G1_S |
| A_24_P279797 | MRI1 | NM_001031727 | 84245 | 0.01 | 0.11 | 0.10 | 0.38 | G1_S |
| A_23_P102471 | MSH2 | NM_000251 | 4436 | −0.38 | −0.39 | −0.02 | −0.28 | G1_S |
| A_23_P34800 | NASP | NM_172164 | 4678 | −0.92 | −0.57 | 0.34 | −0.26 | G1_S |
| A_32_P28365 | NASP | NM_172164 | 4678 | −0.70 | −0.46 | 0.24 | −0.15 | G1_S |
| A_24_P926760 | NKTR | NM_005385 | 4820 | 0.53 | 0.36 | −0.17 | 0.78 | G1_S |
| A_23_P212002 | NKTR | NM_005385 | 4820 | 0.38 | 0.32 | −0.06 | 0.47 | G1_S |
| A_24_P171601 | NKTR | NM_005385 | 4820 | 0.37 | 0.20 | −0.16 | 0.83 | G1_S |
| A_23_P203013 | NPAT | NM_002519 | 4863 | −0.59 | −0.36 | 0.23 | −0.18 | G1_S |
| A_24_P273823 | NPAT | NM_002519 | 4863 | −0.22 | −0.31 | −0.09 | 0.27 | G1_S |
| A_24_P29641 | NSUN5P2 | NM_148936 | 260294 | 0.05 | 0.12 | 0.07 | 0.46 | G1_S |
| A_23_P161324 | NUDT13 | NM_015901 | 25961 | −0.39 | −0.32 | 0.07 | −0.01 | G1_S |
| A_32_P41471 | NUDT13 | NM_015901 | 25961 | −0.19 | −0.31 | −0.11 | 0.29 | G1_S |
| A_24_P200761 | NUP43 | NM_198887 | 348995 | −0.37 | −0.10 | 0.27 | −0.24 | G1_S |
| A_23_P31055 | NUP43 | NM_198887 | 348995 | −0.21 | 0.02 | 0.22 | 0.16 | G1_S |
| A_23_P45799 | ORC1 | NM_004153 | 4998 | −0.91 | −0.95 | −0.04 | −0.48 | G1_S |
| A_24_P371053 | ORMDL1 | NM_016467 | 94101 | −0.59 | −0.20 | 0.39 | −0.10 | G1_S |
| A_23_P120194 | ORMDL1 | NM_016467 | 94101 | −0.01 | 0.07 | 0.08 | 0.25 | G1_S |
| A_32_P220762 | OSBPL6 | ENST00000190611 | 114880 | 0.10 | −0.19 | −0.29 | 0.11 | G1_S |
| A_23_P108823 | OSBPL6 | NM_032523 | 114880 | 0.00 | −0.27 | −0.28 | 0.17 | G1_S |
| A_24_P414446 | OTULIN | NM_138348 | 90268 | −0.16 | −0.55 | −0.39 | 0.09 | G1_S |
| A_23_P353106 | OTULIN | NM_138348 | 90268 | 0.13 | −0.25 | −0.38 | 0.12 | G1_S |
| A_24_P142885 | PANK2 | ENST00000497424 | 80025 | −0.56 | −0.16 | 0.41 | −0.45 | G1_S |
| A_23_P79942 | PANK2 | NM_153638 | 80025 | −0.12 | 0.06 | 0.17 | −0.25 | G1_S |
| A_24_P299911 | PASK | NM_015148 | 23178 | −0.45 | −0.28 | 0.18 | −0.39 | G1_S |
| A_23_P28886 | PCNA | NM_002592 | 5111 | −1.10 | −0.88 | 0.22 | −0.11 | G1_S |
| A_24_P280029 | PDXP | NM_020315 | 57026 | 0.09 | −0.20 | −0.28 | 0.81 | G1_S |
| A_23_P61180 | PLCXD1 | NM_018390 | 55344 | 0.64 | 0.69 | 0.06 | 0.14 | G1_S |
| A_23_P99582 | PNN | NM_002687 | 5411 | 0.46 | 0.30 | −0.16 | 1.13 | G1_S |
| A_32_P182439 | POLD3 | NM_006591 | 10714 | −0.43 | −0.01 | 0.42 | −0.26 | G1_S |
| A_24_P75056 | POLD3 | NM_006591 | 10714 | −0.26 | 0.19 | 0.45 | −0.45 | G1_S |
| A_23_P70794 | RAB23 | NM_016277 | 51715 | −0.48 | −0.19 | 0.29 | −0.27 | G1_S |
| A_23_P71558 | RECQL4 | NM_004260 | 9401 | −1.56 | −0.91 | 0.65 | −0.90 | G1_S |
| A_23_P353717 | RMI2 | NM_152308 | 116028 | −1.49 | −1.51 | −0.02 | −1.39 | G1_S |
| A_23_P258071 | RNF113A | NM_006978 | 7737 | 0.21 | −0.05 | −0.26 | 0.49 | G1_S |
| A_23_P254970 | RNPC3 | AK057799 | 55599 | −0.24 | −0.35 | −0.11 | 0.31 | G1_S |
| A_24_P341504 | RNPC3 | NM_017619 | 55599 | 0.23 | 0.13 | −0.10 | 0.71 | G1_S |
| A_23_P34396 | RSRP1 | NM_020317 | 57035 | 0.66 | 0.09 | −0.57 | 0.97 | G1_S |
| A_24_P34155 | RUNX1 | NM_001122607 | 861 | 1.03 | 0.33 | −0.71 | 0.67 | G1_S |
| A_24_P96403 | RUNX1 | NM_001001890 | 861 | −0.68 | 0.18 | 0.87 | −1.35 | G1_S |
| A_23_P16944 | SDC1 | NM_001006946 | 6382 | −0.69 | −0.50 | 0.19 | −1.07 | G1_S |
| A_24_P97129 | SDC1 | NM_001006946 | 6382 | −0.35 | −0.44 | −0.08 | 0.50 | G1_S |
| A_23_P357856 | SEC62 | NM_003262 | 7095 | −0.56 | −0.28 | 0.28 | −0.71 | G1_S |
| A_23_P144224 | SEC62 | NM_003262 | 7095 | 0.54 | 0.07 | −0.47 | 0.48 | G1_S |
| A_24_P285880 | SEC62 | NM_003262 | 7095 | −0.35 | −0.26 | 0.10 | 0.05 | G1_S |
| A_23_P357860 | SEC62 | NM_003262 | 7095 | −0.52 | 0.04 | 0.56 | −1.67 | G1_S |
| A_24_P251704 | SEC62 | NM_003262 | 7095 | −0.05 | 0.04 | 0.09 | −0.96 | G1_S |
| A_23_P55632 | SERPINB3 | NM_006919 | 6317 | 1.00 | 1.78 | 0.78 | 1.52 | G1_S |
| A_23_P156310 | SKP2 | NM_032637 | 6502 | −0.38 | −0.75 | −0.37 | 0.37 | G1_S |
| A_23_P7101 | SLBP | NM_006527 | 7884 | 0.21 | 0.00 | −0.21 | 0.23 | G1_S |
| A_23_P40896 | SLC25A36 | NM_018155 | 55186 | 0.27 | −0.15 | −0.42 | 0.91 | G1_S |
| A_23_P408455 | SLC25A36 | NM_001104647 | 55186 | 0.24 | −0.37 | −0.61 | 0.85 | G1_S |
| A_24_P136725 | SPIN3 | NR_027139 | 169981 | −0.07 | 0.34 | 0.40 | 0.01 | G1_S |
| A_24_P494454 | SPIN3 | NM_001010862 | 169981 | 0.01 | 0.06 | 0.05 | 1.15 | G1_S |
| A_32_P222961 | SPIN4 | NM_001012968 | 139886 | 0.33 | 0.38 | 0.06 | 0.30 | G1_S |
| A_24_P467371 | SPIN4 | NM_001012968 | 139886 | 0.29 | 0.10 | −0.19 | −0.42 | G1_S |
| A_24_P222911 | SRSF7 | NM_001031684 | 6432 | −0.81 | −0.73 | 0.08 | −0.35 | G1_S |
| A_23_P39704 | SRSF7 | NM_001031684 | 6432 | −0.58 | −0.60 | −0.02 | −0.01 | G1_S |
| A_23_P155229 | SSR3 | NM_007107 | 6747 | 0.53 | −0.06 | −0.58 | 0.94 | G1_S |
| A_24_P319942 | SSR3 | NM_007107 | 6747 | −0.06 | −0.23 | −0.18 | −0.01 | G1_S |
| A_24_P928068 | TAF15 | DB509819 | NA | 0.46 | 0.45 | 0.00 | 0.02 | G1_S |
| A_23_P159305 | TAF15 | NM_139215 | 8148 | 0.11 | 0.26 | 0.15 | −0.66 | G1_S |
| A_32_P56525 | TCAF1 | NM_014719 | 9747 | 0.28 | 0.46 | 0.18 | −1.13 | G1_S |
| A_24_P380628 | TCAF1 | NM_014719 | 9747 | −0.08 | 0.10 | 0.18 | −0.69 | G1_S |
| A_24_P368023 | TCAF1 | ENST00000479870 | 9747 | −0.03 | 0.26 | 0.29 | −1.42 | G1_S |
| A_23_P99930 | TIPIN | NM_017858 | 54962 | 0.17 | −0.46 | −0.63 | 0.86 | G1_S |
| A_23_P157283 | TMEM243 | NM_024315 | 79161 | 0.17 | 0.30 | 0.14 | −0.37 | G1_S |
| A_23_P159390 | TOPBP1 | NM_007027 | 11073 | −0.46 | −0.26 | 0.21 | −0.01 | G1_S |
| A_23_P31389 | TRA2A | NM_013293 | 29896 | −0.67 | −0.32 | 0.34 | −0.04 | G1_S |
| A_23_P218879 | TREX1 | NM_016381 | 11277 | −0.44 | −0.10 | 0.34 | −0.38 | G1_S |
| A_24_P339858 | TSPEAR-AS2 | NR_026547 | 114043 | 0.36 | 0.72 | 0.36 | −0.13 | G1_S |
| A_24_P910854 | TTC14 | NM_001042601 | 151613 | 0.51 | 0.21 | −0.30 | −0.10 | G1_S |
| A_23_P212511 | TTC14 | NM_001042601 | 151613 | −0.10 | −0.25 | −0.14 | 0.52 | G1_S |
| A_24_P159094 | UBR7 | NM_175748 | 55148 | −0.97 | −0.65 | 0.33 | −0.45 | G1_S |
| A_23_P205393 | UBR7 | NM_175748 | 55148 | −0.43 | −0.63 | −0.21 | 0.41 | G1_S |
| A_23_P208880 | UHRF1 | NM_013282 | 29128 | −2.52 | −1.75 | 0.77 | −1.77 | G1_S |
| A_32_P101235 | UHRF1 | NM_013282 | 29128 | −0.47 | −0.43 | 0.04 | −0.15 | G1_S |
| A_24_P398585 | UNG | NM_003362 | 7374 | −0.28 | 0.01 | 0.29 | −0.27 | G1_S |
| A_24_P137522 | USP53 | NM_019050 | 54532 | 0.46 | 0.26 | −0.20 | 0.60 | G1_S |
| A_32_P128701 | USP53 | NM_019050 | 54532 | 0.40 | 0.15 | −0.25 | 0.38 | G1_S |
| A_23_P115215 | VPS72 | NM_005997 | 6944 | −0.35 | −0.10 | 0.25 | 0.01 | G1_S |
| A_23_P129075 | WDR76 | NM_024908 | 79968 | −0.19 | −0.48 | −0.29 | −0.09 | G1_S |
| A_24_P158385 | ZMYND19 | NM_138462 | 116225 | −0.29 | −0.28 | 0.01 | −0.38 | G1_S |
| A_32_P183218 | ZNF367 | NM_153695 | 195828 | −1.12 | −0.76 | 0.35 | −0.53 | G1_S |
| A_23_P410625 | ZNF367 | NM_153695 | 195828 | −0.41 | −0.50 | −0.09 | −0.08 | G1_S |
| A_23_P340922 | ZNF414 | NM_032370 | 84330 | −0.35 | 0.32 | 0.68 | −0.93 | G1_S |
| A_32_P85978 | ZNF414 | NM_001146175 | 84330 | 0.21 | 0.26 | 0.05 | −0.48 | G1_S |
| A_23_P85521 | ZRANB2 | NM_203350 | 9406 | 0.36 | 0.09 | −0.27 | 0.81 | G1_S |
| A_24_P242299 | ZRANB2 | NM_005455 | 9406 | 0.30 | 0.10 | −0.20 | 0.57 | G1_S |
| A_23_P120784 | TRMT2A | NM_022727 | 27037 | −0.61 | −0.27 | 0.34 | −0.58 | G1_S,G2 |
| A_24_P305662 | TRMT2A | NM_022727 | 27037 | −0.33 | −0.12 | 0.21 | −0.20 | G1_S,G2 |
| A_23_P370989 | MCM4 | NM_005914 | 4173 | −1.44 | −1.07 | 0.36 | −0.90 | G1_S,G2_M |
| A_24_P59596 | ATAD2 | NM_014109 | 29028 | −1.00 | −1.55 | −0.55 | 0.22 | G1_S,S |
| A_23_P216068 | ATAD2 | NM_014109 | 29028 | −0.82 | −0.97 | −0.15 | −0.21 | G1_S,S |
| A_23_P387943 | CASP2 | NM_032982 | 835 | −0.26 | 0.15 | 0.41 | −0.75 | G1_S,S |
| A_24_P269398 | CASP2 | NM_032982 | 835 | −0.17 | 0.04 | 0.21 | −0.43 | G1_S,S |
| A_23_P215701 | CASP2 | NM_032982 | 835 | −0.13 | 0.18 | 0.31 | −0.40 | G1_S,S |
| A_23_P203645 | CREBZF | NM_001039618 | 58487 | 0.69 | 0.06 | −0.63 | 0.90 | G1_S,S |
| A_23_P252740 | DSCC1 | NM_024094 | 79075 | −1.22 | −1.25 | −0.04 | −0.64 | G1_S,S |
| A_23_P162579 | HSPB8 | NM_014365 | 26353 | 1.38 | 0.02 | −1.35 | 1.06 | G1_S,S |
| A_23_P170110 | NEAT1 | AW806882 | NA | 0.73 | 0.39 | −0.34 | 1.00 | G1_S,S |
| A_24_P290999 | NEAT1 | NR_028272 | 283131 | 0.77 | −0.15 | −0.92 | 1.04 | G1_S,S |
| A_24_P566916 | NEAT1 | NR_028272 | 283131 | 0.38 | 0.03 | −0.34 | 0.03 | G1_S,S |
| A_23_P160518 | TRIM45 | NM_025188 | 80263 | −0.40 | −0.18 | 0.22 | −0.16 | G1_S,S |
| A_23_P160523 | TRIM45 | NM_025188 | 80263 | −0.22 | −0.20 | 0.02 | −0.47 | G1_S,S |
| A_23_P14543 | ALKBH1 | NM_006020 | 8846 | 0.21 | 0.05 | −0.16 | 0.40 | G2 |
| A_23_P108135 | AP3D1 | NM_003938 | 8943 | 0.58 | 0.63 | 0.05 | −0.50 | G2 |
| A_23_P119526 | AP3D1 | NM_003938 | 8943 | 0.46 | 0.51 | 0.05 | −0.18 | G2 |
| A_24_P30034 | ARHGEF39 | NM_032818 | 84904 | −0.48 | −0.49 | −0.01 | 0.19 | G2 |
| A_23_P216517 | ARHGEF39 | NM_032818 | 84904 | −0.48 | −0.91 | −0.43 | 0.45 | G2 |
| A_32_P234827 | ARMC1 | NM_018120 | 55156 | 0.00 | −0.04 | −0.05 | 0.23 | G2 |
| A_23_P415015 | ATL2 | NM_022374 | 64225 | 0.40 | 0.35 | −0.05 | 0.17 | G2 |
| A_23_P209619 | ATL2 | NM_022374 | 64225 | 0.07 | 0.40 | 0.33 | −0.17 | G2 |
| A_23_P130182 | AURKB | NM_004217 | 9212 | −1.55 | −1.32 | 0.24 | −0.60 | G2 |
| A_24_P89512 | BCLAF1 | NM_014739 | 9774 | −0.94 | −0.52 | 0.43 | −0.39 | G2 |
| A_24_P80915 | BCLAF1 | NM_014739 | 9774 | −0.85 | −0.89 | −0.04 | −0.03 | G2 |
| A_23_P111343 | BCLAF1 | NM_014739 | 9774 | −0.56 | −0.34 | 0.22 | 0.35 | G2 |
| A_23_P25626 | BORA | NM_024808 | 79866 | −1.02 | −0.89 | 0.12 | 0.28 | G2 |
| A_23_P145016 | BRD8 | NM_006696 | 10902 | −0.13 | 0.02 | 0.15 | −0.28 | G2 |
| A_23_P81280 | BTNL9 | NM_152547 | 153579 | 0.42 | 0.29 | −0.14 | 0.18 | G2 |
| A_32_P187951 | BTNL9 | NM_152547 | 153579 | −0.29 | −0.38 | −0.09 | −0.48 | G2 |
| A_23_P46924 | BUB3 | NM_001007793 | 9184 | −0.66 | −0.53 | 0.13 | −0.08 | G2 |
| A_23_P202316 | BUB3 | NM_001007793 | 9184 | −0.63 | −0.34 | 0.29 | −0.25 | G2 |
| A_23_P320658 | BUB3 | NM_004725 | 9184 | 0.20 | −0.10 | −0.29 | 0.52 | G2 |
| A_24_P413941 | C2orf69 | NM_153689 | 205327 | −0.24 | −0.07 | 0.17 | −0.37 | G2 |
| A_23_P142918 | C2orf69 | NM_153689 | 205327 | 0.10 | 0.06 | −0.04 | −0.04 | G2 |
| A_23_P92410 | CASP3 | NM_004346 | 836 | −0.51 | −0.13 | 0.38 | −0.08 | G2 |
| A_24_P664995 | CBX5 | NM_001127322 | 23468 | −0.34 | 0.24 | 0.57 | −1.23 | G2 |
| A_24_P620621 | CBX5 | NM_001127322 | 23468 | −0.15 | −0.02 | 0.13 | −0.14 | G2 |
| A_23_P2355 | CBX5 | NM_012117 | 23468 | −0.17 | −0.29 | −0.12 | 0.49 | G2 |
| A_24_P193592 | CCNF | NM_001761 | 899 | −0.89 | −0.41 | 0.48 | −0.46 | G2 |
| A_23_P37954 | CCNF | NM_001761 | 899 | −0.74 | −0.22 | 0.52 | −0.94 | G2 |
| A_23_P88083 | CDC16 | NM_003903 | 8881 | −0.36 | −0.13 | 0.23 | 0.02 | G2 |
| A_23_P70249 | CDC25C | NM_001790 | 995 | −2.28 | −1.87 | 0.41 | −0.78 | G2 |
| A_23_P385861 | CDCA2 | NM_152562 | 157313 | −1.95 | −1.77 | 0.18 | −0.24 | G2 |
| A_24_P323434 | CDCA2 | NM_152562 | 157313 | −1.36 | −1.32 | 0.04 | −0.07 | G2 |
| A_23_P375 | CDCA8 | NM_018101 | 55143 | −2.04 | −1.76 | 0.29 | −0.84 | G2 |
| A_23_P138507 | CDK1 | NM_001786 | 983 | −2.95 | −2.22 | 0.73 | −0.84 | G2 |
| A_24_P282343 | CDKL5 | NM_003159 | 6792 | 0.14 | −0.06 | −0.20 | −0.28 | G2 |
| A_24_P81841 | CDKN1B | NM_004064 | 1027 | −0.70 | −0.55 | 0.15 | −0.73 | G2 |
| A_23_P204696 | CDKN1B | NM_004064 | 1027 | −0.41 | −0.54 | −0.13 | 0.24 | G2 |
| A_23_P85460 | CDKN2C | NM_078626 | 1031 | −0.61 | −1.44 | −0.82 | −0.04 | G2 |
| A_23_P126120 | CENPL | NM_033319 | 91687 | −0.75 | −0.56 | 0.19 | −0.24 | G2 |
| A_24_P930100 | CENPL | AK056348 | 91687 | 0.01 | −0.68 | −0.69 | 0.61 | G2 |
| A_23_P201816 | CEP350 | NM_014810 | 9857 | −0.25 | −0.13 | 0.12 | −0.12 | G2 |
| A_23_P119562 | CFD | NM_001928 | 1675 | −0.36 | −0.22 | 0.13 | −0.26 | G2 |
| A_23_P109452 | CHEK2 | NM_001005735 | 11200 | −0.96 | −0.67 | 0.28 | 0.01 | G2 |
| A_23_P250313 | CIP2A | NM_020890 | 57650 | −0.85 | −0.77 | 0.08 | 0.01 | G2 |
| A_24_P351466 | CIP2A | NM_020890 | 57650 | 0.02 | −0.40 | −0.42 | 0.14 | G2 |
| A_23_P388812 | CKAP2L | NM_152515 | 150468 | −2.34 | −2.03 | 0.31 | −0.35 | G2 |
| A_23_P213745 | CXCL14 | NM_004887 | 9547 | −2.26 | −1.16 | 1.10 | −2.35 | G2 |
| A_23_P2181 | CYB5R2 | NM_016229 | 51700 | 0.31 | −0.19 | −0.49 | 1.01 | G2 |
| A_23_P119377 | CYTH2 | NM_004228 | 9266 | −0.42 | −0.11 | 0.31 | −0.64 | G2 |
| A_23_P422268 | DCAF7 | NM_005828 | 10238 | 0.32 | 0.42 | 0.10 | −0.21 | G2 |
| A_24_P916141 | DCAF7 | NM_005828 | 10238 | −0.20 | 0.23 | 0.44 | −1.47 | G2 |
| A_24_P91222 | DCAF7 | NM_005828 | 10238 | 0.13 | 0.10 | −0.02 | −0.15 | G2 |
| A_23_P26836 | DCAF7 | NM_005828 | 10238 | 0.10 | 0.41 | 0.31 | −1.19 | G2 |
| A_32_P430743 | DET1 | AK125793 | NA | 1.07 | 0.74 | −0.33 | 0.27 | G2 |
| A_23_P26184 | DET1 | NM_017996 | 55070 | 0.04 | 0.07 | 0.03 | 0.09 | G2 |
| A_23_P124224 | DHX8 | NM_004941 | 1659 | 0.03 | 0.04 | 0.02 | 0.16 | G2 |
| A_23_P119478 | EBI3 | NM_005755 | 10148 | 4.87 | 4.24 | −0.63 | 4.69 | G2 |
| A_24_P370201 | EBI3 | NM_005755 | 10148 | 1.35 | 0.70 | −0.65 | 1.40 | G2 |
| A_23_P117580 | ENTPD5 | NM_001249 | 957 | 0.12 | 0.09 | −0.03 | 0.16 | G2 |
| A_23_P32707 | ESPL1 | NM_012291 | 9700 | −0.40 | −1.11 | −0.70 | 0.46 | G2 |
| A_32_P119007 | ESPL1 | NM_012291 | 9700 | 0.48 | 1.80 | 1.33 | −0.18 | G2 |
| A_24_P278637 | FADD | NM_003824 | 8772 | −0.41 | −0.08 | 0.33 | −0.18 | G2 |
| A_23_P86917 | FADD | NM_003824 | 8772 | −0.33 | 0.05 | 0.37 | −0.32 | G2 |
| A_23_P386241 | FAM110A | NM_001042353 | 83541 | 0.01 | 0.38 | 0.37 | −0.66 | G2 |
| A_23_P323751 | FAM83D | NM_030919 | 81610 | −1.23 | −1.77 | −0.54 | 0.09 | G2 |
| A_23_P377888 | FAN1 | NM_014967 | 22909 | 0.24 | 0.15 | −0.09 | 0.33 | G2 |
| A_23_P345678 | FANCD2 | NM_033084 | 2177 | −0.61 | −1.11 | −0.50 | −0.17 | G2 |
| A_32_P24165 | FANCD2 | NM_001018115 | 2177 | −0.59 | −0.68 | −0.09 | −0.10 | G2 |
| A_23_P143994 | FANCD2 | NM_001018115 | 2177 | −0.59 | −0.56 | 0.03 | −0.90 | G2 |
| A_23_P142333 | FZR1 | NM_016263 | 51343 | 0.40 | 0.18 | −0.22 | 0.12 | G2 |
| A_23_P142325 | FZR1 | NM_001136198 | 51343 | 0.41 | 0.05 | −0.35 | 0.11 | G2 |
| A_24_P944291 | FZR1 | ENST00000395095 | 51343 | 0.04 | 0.25 | 0.21 | −0.49 | G2 |
| A_24_P318836 | FZR1 | NM_016263 | 51343 | −0.04 | 0.02 | 0.06 | −0.53 | G2 |
| A_23_P106280 | GABPB1 | NR_026891 | 55056 | −0.85 | −0.42 | 0.43 | −0.95 | G2 |
| A_23_P205789 | GABPB1 | NM_002041 | 2553 | 1.27 | 0.43 | −0.84 | 1.47 | G2 |
| A_24_P176255 | GABPB1 | NM_005254 | 2553 | −0.14 | −0.02 | 0.12 | 0.16 | G2 |
| A_23_P83134 | GAS1 | NM_002048 | 2619 | 0.50 | −0.71 | −1.21 | 0.26 | G2 |
| A_24_P38895 | H2AFX | NM_002105 | 3014 | −1.09 | −0.78 | 0.31 | −0.88 | G2 |
| A_23_P141965 | HAUS8 | NM_033417 | 93323 | −0.41 | −0.45 | −0.04 | 0.18 | G2 |
| A_32_P85500 | HCP5 | NM_006674 | 10866 | 0.72 | 0.39 | −0.32 | 0.76 | G2 |
| A_23_P111126 | HCP5 | L06175 | 10866 | 0.55 | 0.24 | −0.31 | 0.80 | G2 |
| A_24_P17870 | HCP5 | NM_006674 | 10866 | 0.52 | 0.14 | −0.37 | 0.70 | G2 |
| A_24_P238609 | HCP5 | NM_006674 | 10866 | 0.28 | 0.09 | −0.19 | 0.40 | G2 |
| A_23_P145574 | HINT3 | NM_138571 | 135114 | 0.38 | 0.18 | −0.20 | 0.80 | G2 |
| A_24_P681011 | HIPK2 | NM_022740 | 28996 | 0.20 | 0.72 | 0.52 | −2.31 | G2 |
| A_23_P169756 | HIPK2 | NM_022740 | 28996 | 0.20 | 0.39 | 0.19 | −1.41 | G2 |
| A_24_P500621 | HIPK2 | NM_022740 | 28996 | 0.10 | 0.39 | 0.29 | −1.71 | G2 |
| A_23_P169766 | HIPK2 | NM_022740 | 28996 | −0.02 | 0.20 | 0.22 | −1.50 | G2 |
| A_23_P149301 | HIST3H2A | NM_033445 | 92815 | 0.15 | 0.07 | −0.08 | 0.79 | G2 |
| A_24_P257099 | HJURP | NM_018410 | 55355 | −2.18 | −1.32 | 0.86 | −1.01 | G2 |
| A_23_P155765 | HMGB2 | NM_002129 | 3148 | −0.77 | −2.05 | −1.29 | 0.90 | G2 |
| A_23_P88303 | HSPA2 | NM_021979 | 3306 | 1.00 | 0.29 | −0.71 | 0.23 | G2 |
| A_23_P17633 | IFNAR1 | NM_000629 | 3454 | 0.54 | 0.11 | −0.43 | 0.40 | G2 |
| A_23_P113803 | KATNA1 | NM_007044 | 11104 | 0.17 | −0.33 | −0.50 | 0.62 | G2 |
| A_23_P77286 | KATNBL1 | NM_024713 | 79768 | −0.55 | −0.37 | 0.18 | −0.19 | G2 |
| A_32_P58163 | KATNBL1 | NM_024713 | 79768 | −0.34 | −0.15 | 0.20 | −0.11 | G2 |
| A_24_P12539 | KBTBD2 | NM_015483 | 25948 | 0.20 | −0.15 | −0.35 | 0.37 | G2 |
| A_23_P70951 | KBTBD2 | NM_015483 | 25948 | 0.06 | 0.00 | −0.07 | 0.06 | G2 |
| A_23_P74446 | KDM4A | NM_014663 | 9682 | −0.20 | 0.07 | 0.27 | −0.48 | G2 |
| A_24_P227091 | KIF11 | NM_004523 | 3832 | −2.63 | −1.71 | 0.93 | −1.86 | G2 |
| A_23_P52278 | KIF11 | NM_004523 | 3832 | −1.24 | −0.93 | 0.31 | −0.76 | G2 |
| A_23_P54622 | KIF22 | NM_007317 | 3835 | −1.46 | −1.13 | 0.33 | −0.79 | G2 |
| A_23_P133956 | KIFC1 | NM_002263 | 3833 | −2.20 | −1.79 | 0.41 | −1.11 | G2 |
| A_24_P252739 | KLF6 | NM_001300 | 1316 | −1.45 | −1.02 | 0.43 | −0.96 | G2 |
| A_24_P932981 | KLF6 | NM_001300 | 1316 | 0.59 | −0.18 | −0.77 | −0.30 | G2 |
| A_24_P69654 | KLF6 | NM_001300 | 1316 | 0.81 | −0.29 | −1.10 | 0.00 | G2 |
| A_23_P63798 | KLF6 | NM_001300 | 1316 | 0.57 | −0.29 | −0.86 | −0.26 | G2 |
| A_23_P125265 | KPNA2 | NM_002266 | 3838 | −1.25 | −0.63 | 0.62 | −0.25 | G2 |
| A_23_P342744 | LIX1L | NM_153713 | 128077 | 0.56 | 0.04 | −0.52 | 0.40 | G2 |
| A_24_P687594 | LIX1L | ENST00000369308 | 128077 | 0.30 | −0.02 | −0.31 | −0.45 | G2 |
| A_23_P258493 | LMNB1 | NM_005573 | 4001 | −1.90 | −1.59 | 0.31 | −1.26 | G2 |
| A_24_P264790 | LTBP3 | NM_021070 | 4054 | −0.85 | −0.57 | 0.28 | −0.36 | G2 |
| A_24_P298360 | LTBP3 | NM_021070 | 4054 | 0.36 | −0.16 | −0.51 | −0.64 | G2 |
| A_23_P92441 | MAD2L1 | NM_002358 | 4085 | −1.86 | −1.58 | 0.28 | −0.36 | G2 |
| A_24_P873659 | MALAT1 | NR_002819 | 378938 | 0.29 | −0.40 | −0.69 | 1.76 | G2 |
| A_23_P21143 | MALAT1 | NR_002819 | 378938 | −0.19 | −0.44 | −0.25 | 0.80 | G2 |
| A_24_P829261 | MALAT1 | NR_002819 | 378938 | 0.11 | 0.03 | −0.08 | 0.27 | G2 |
| A_24_P497244 | MALAT1 | NR_002819 | 378938 | 0.07 | −0.32 | −0.40 | 1.05 | G2 |
| A_23_P94422 | MELK | NM_014791 | 9833 | −3.11 | −2.07 | 1.04 | −1.29 | G2 |
| A_23_P42626 | MEPCE | NM_019606 | 56257 | −0.18 | 0.15 | 0.32 | −0.74 | G2 |
| A_24_P312189 | MEPCE | NM_019606 | 56257 | −0.16 | 0.24 | 0.40 | −0.71 | G2 |
| A_23_P145844 | MET | NM_000245 | 4233 | 0.51 | 0.41 | −0.10 | 0.86 | G2 |
| A_23_P145846 | MET | NM_000245 | 4233 | 0.27 | 0.27 | 0.00 | 0.71 | G2 |
| A_23_P359245 | MET | NM_000245 | 4233 | 0.22 | 0.30 | 0.08 | 0.11 | G2 |
| A_23_P65558 | MGAT2 | NM_002408 | 4247 | −0.19 | −0.39 | −0.20 | 1.27 | G2 |
| A_32_P128656 | MID1 | NM_000381 | 4281 | −0.31 | 0.14 | 0.46 | −0.59 | G2 |
| A_23_P170037 | MID1 | NM_033290 | 4281 | −0.01 | 0.50 | 0.50 | −0.87 | G2 |
| A_23_P133123 | MND1 | NM_032117 | 84057 | −2.67 | −2.06 | 0.61 | −1.08 | G2 |
| A_23_P360605 | MTCL1 | NM_015210 | 23255 | −1.12 | −0.38 | 0.74 | −1.41 | G2 |
| A_23_P137856 | MUC1 | NM_002456 | 4582 | −0.29 | 0.17 | 0.46 | −1.61 | G2 |
| A_32_P71447 | NCAPD3 | NM_015261 | 23310 | −0.92 | −0.59 | 0.32 | −0.53 | G2 |
| A_23_P415443 | NCAPH | NM_015341 | 23397 | −2.82 | −2.15 | 0.67 | −1.02 | G2 |
| A_23_P50108 | NDC80 | NM_006101 | 10403 | −3.12 | −2.49 | 0.64 | −0.69 | G2 |
| A_24_P14156 | NDC80 | NM_006101 | 10403 | −1.90 | −1.59 | 0.31 | −0.41 | G2 |
| A_23_P155711 | NEIL3 | NM_018248 | 55247 | −0.52 | −0.51 | 0.02 | 0.19 | G2 |
| A_24_P356830 | NFIC | AK129956 | 4782 | 0.20 | 0.15 | −0.06 | −0.83 | G2 |
| A_23_P131115 | NFIC | NM_005597 | 4782 | −0.18 | 0.31 | 0.49 | −0.92 | G2 |
| A_24_P180383 | NIPBL | NM_015384 | 25836 | −0.12 | 0.22 | 0.33 | −0.46 | G2 |
| A_23_P213883 | NIPBL | NM_133433 | 25836 | 0.09 | −0.04 | −0.13 | −0.15 | G2 |
| A_24_P357688 | NIPBL | NM_015384 | 25836 | 0.10 | −0.05 | −0.16 | 0.70 | G2 |
| A_24_P213161 | NLRP2 | NM_017852 | 55655 | −0.78 | −0.17 | 0.61 | −1.33 | G2 |
| A_23_P88522 | NMB | NM_021077 | 4828 | 1.35 | 0.91 | −0.44 | 2.12 | G2 |
| A_23_P127584 | NNMT | NM_006169 | 4837 | −0.01 | 0.25 | 0.27 | −0.30 | G2 |
| A_24_P787914 | NR3C1 | U25029 | 2908 | 1.13 | 0.58 | −0.55 | 1.02 | G2 |
| A_23_P214059 | NR3C1 | NM_001018077 | 2908 | 0.83 | 0.19 | −0.64 | 0.56 | G2 |
| A_24_P214754 | NR3C1 | NM_001018077 | 2908 | 0.80 | 0.04 | −0.76 | 1.23 | G2 |
| A_24_P216968 | NUCKS1 | NM_022731 | 64710 | −0.40 | −0.39 | 0.01 | −0.04 | G2 |
| A_24_P145122 | NUCKS1 | NM_022731 | 64710 | −0.35 | −0.36 | −0.01 | −0.07 | G2 |
| A_24_P216964 | NUCKS1 | NM_022731 | 64710 | −0.23 | −0.48 | −0.25 | −0.15 | G2 |
| A_24_P374652 | NUCKS1 | NM_022731 | 64710 | 0.09 | −0.04 | −0.14 | 0.21 | G2 |
| A_23_P149724 | NUCKS1 | NM_022731 | 64710 | −0.02 | −0.24 | −0.22 | 0.07 | G2 |
| A_23_P162120 | NUMA1 | NM_006185 | 4926 | 0.11 | 0.39 | 0.28 | −0.38 | G2 |
| A_23_P17471 | PCED1A | NM_022760 | 64773 | −0.13 | −0.27 | −0.14 | 0.08 | G2 |
| A_23_P416468 | PIF1 | NM_025049 | 80119 | −2.16 | −1.63 | 0.53 | −0.08 | G2 |
| A_23_P323749 | PIF1 | NM_025049 | 80119 | −0.37 | 0.19 | 0.56 | −0.14 | G2 |
| A_23_P323743 | PIF1 | NM_025049 | 80119 | −0.19 | −0.27 | −0.08 | 0.04 | G2 |
| A_24_P196534 | PKNOX1 | NM_004571 | 5316 | 0.32 | 0.19 | −0.14 | 0.10 | G2 |
| A_23_P211299 | PKNOX1 | NM_004571 | 5316 | 0.20 | 0.25 | 0.05 | 0.49 | G2 |
| A_24_P378907 | PKNOX1 | NM_004571 | 5316 | −0.23 | 0.25 | 0.48 | −0.15 | G2 |
| A_23_P333998 | POLQ | AF090919 | 10721 | −1.90 | −1.42 | 0.48 | −1.15 | G2 |
| A_23_P218827 | POLQ | NM_199420 | 10721 | −2.19 | −1.69 | 0.49 | −1.00 | G2 |
| A_24_P63109 | PPP1R2 | NM_006241 | 5504 | 0.30 | −0.18 | −0.48 | 0.89 | G2 |
| A_32_P17133 | PPP1R2 | NM_006241 | 5504 | 0.11 | −0.21 | −0.31 | 0.63 | G2 |
| A_24_P174367 | PPP1R2 | NM_006241 | 5504 | −0.06 | −0.17 | −0.11 | 0.34 | G2 |
| A_23_P46539 | PSRC1 | NM_032636 | 84722 | −0.92 | −0.66 | 0.26 | −0.48 | G2 |
| A_23_P106439 | RCCD1 | NM_033544 | 91433 | −0.52 | −0.10 | 0.41 | −0.66 | G2 |
| A_23_P106433 | RCCD1 | NM_033544 | 91433 | −0.23 | −0.05 | 0.17 | −0.62 | G2 |
| A_23_P25684 | RDH11 | NM_016026 | 51109 | −0.39 | −0.19 | 0.20 | −0.56 | G2 |
| A_24_P377775 | RGS3 | NM_017790 | 5998 | 0.85 | 0.37 | −0.48 | 0.24 | G2 |
| A_23_P219197 | RGS3 | NM_134427 | 5998 | −0.03 | −0.04 | −0.01 | −0.43 | G2 |
| A_23_P134714 | RIDA | NM_005836 | 10247 | −0.54 | −0.12 | 0.42 | −0.41 | G2 |
| A_23_P121602 | SAP30 | NM_003864 | 8819 | −0.39 | −0.22 | 0.16 | 0.11 | G2 |
| A_23_P147647 | SGCD | NM_000337 | 6444 | 0.23 | −0.19 | −0.41 | 0.51 | G2 |
| A_23_P136254 | SGCD | NM_172244 | 6444 | 0.17 | 0.37 | 0.20 | −0.63 | G2 |
| A_32_P4595 | SGCD | NM_000337 | 6444 | 0.05 | −0.38 | −0.43 | 0.33 | G2 |
| A_23_P340909 | SKA3 | BC013418 | 221150 | −3.76 | −2.71 | 1.05 | −1.64 | G2 |
| A_23_P327643 | SMC4 | A_23_P327643 | NA | 0.61 | 0.07 | −0.53 | 0.40 | G2 |
| A_23_P91900 | SMC4 | NM_005496 | 10051 | −0.46 | −0.37 | 0.09 | −0.62 | G2 |
| A_23_P87049 | SORL1 | NM_003105 | 6653 | 0.07 | 0.31 | 0.24 | −0.86 | G2 |
| A_23_P56630 | STAT1 | NM_007315 | 6772 | 0.35 | 0.58 | 0.23 | −0.36 | G2 |
| A_24_P274270 | STAT1 | NM_139266 | 6772 | 0.16 | 0.43 | 0.28 | −0.09 | G2 |
| A_24_P214231 | STIL | NM_001048166 | 6491 | −0.73 | 0.04 | 0.78 | −0.96 | G2 |
| A_23_P154367 | STK17B | NM_004226 | 9262 | −0.33 | −0.57 | −0.24 | 0.27 | G2 |
| A_24_P636882 | STK17B | NM_004226 | 9262 | −0.30 | 0.07 | 0.37 | −0.78 | G2 |
| A_23_P100022 | SV2B | NM_014848 | 9899 | −0.25 | 0.02 | 0.27 | 0.33 | G2 |
| A_23_P431381 | TEDC1 | NM_001134875 | 283643 | −0.52 | −0.16 | 0.37 | −0.24 | G2 |
| A_32_P14187 | TFAP2A | NM_001032280 | 7020 | 0.71 | 0.16 | −0.56 | 0.26 | G2 |
| A_23_P62115 | TIMP1 | NM_003254 | 7076 | 0.09 | 0.06 | −0.03 | 0.27 | G2 |
| A_23_P24716 | TMEM132A | NM_017870 | 54972 | 0.30 | 0.29 | 0.00 | −0.74 | G2 |
| A_24_P210244 | TMPO | NM_001032283 | 7112 | −0.85 | −0.65 | 0.20 | 0.42 | G2 |
| A_23_P325040 | TMPO | NM_003276 | 7112 | −0.33 | −0.59 | −0.26 | −0.17 | G2 |
| A_24_P44891 | TNPO2 | NM_013433 | 30000 | 0.63 | 0.13 | −0.50 | 1.02 | G2 |
| A_23_P354953 | TNPO2 | NM_013433 | 30000 | −0.43 | 0.05 | 0.48 | −0.11 | G2 |
| A_23_P170491 | TRAIP | NM_005879 | 10293 | −0.48 | −0.61 | −0.12 | 0.31 | G2 |
| A_23_P407718 | TRIM59 | NM_173084 | 286827 | −0.89 | −0.76 | 0.13 | −0.13 | G2 |
| A_32_P72341 | TRIM59 | NM_173084 | 286827 | −0.44 | −0.58 | −0.14 | 0.05 | G2 |
| A_23_P48826 | TRIM69 | NM_182985 | 140691 | 0.42 | 0.05 | −0.38 | 0.44 | G2 |
| A_24_P50543 | TRIM69 | A_24_P50543 | NA | −0.04 | −0.01 | 0.03 | 0.21 | G2 |
| A_23_P254193 | TTC38 | NM_017931 | 55020 | −0.45 | 0.19 | 0.64 | −0.76 | G2 |
| A_24_P156388 | TTC38 | NM_017931 | 55020 | −0.35 | 0.01 | 0.36 | −0.53 | G2 |
| A_32_P168388 | TTF2 | AK123765 | 8458 | −0.21 | −0.18 | 0.03 | −0.40 | G2 |
| A_23_P97161 | TTF2 | NM_003594 | 8458 | 0.23 | −0.05 | −0.28 | 0.25 | G2 |
| A_23_P139547 | TUBA1A | NM_006009 | 7846 | −0.34 | −0.17 | 0.17 | 0.19 | G2 |
| A_23_P128598 | TUBA3C | NM_006001 | 7278 | 0.22 | 0.66 | 0.44 | −0.22 | G2 |
| A_23_P154065 | TUBA4A | NM_006000 | 7277 | 0.66 | 0.48 | −0.18 | 0.76 | G2 |
| A_23_P102109 | TUBA4A | NM_006000 | 7277 | 0.56 | 0.48 | −0.09 | 0.81 | G2 |
| A_23_P154070 | TUBA4A | NM_006000 | 7277 | −0.45 | −0.15 | 0.30 | 0.06 | G2 |
| A_23_P84448 | TUBA4A | NM_006000 | 7277 | 0.22 | 0.14 | −0.08 | 0.52 | G2 |
| A_23_P387057 | TUBB | NM_178014 | 203068 | −0.46 | −0.41 | 0.05 | 0.07 | G2 |
| A_23_P81912 | TUBB | NM_178014 | 203068 | −0.37 | −0.37 | 0.00 | 0.25 | G2 |
| A_32_P78528 | TUBB | NM_178014 | 203068 | −0.28 | −0.16 | 0.12 | 0.07 | G2 |
| A_23_P19291 | TUBB2A | NM_001069 | 7280 | 1.18 | −0.07 | −1.25 | 0.79 | G2 |
| A_23_P501276 | TUBB2A | NM_001069 | 7280 | 0.42 | −0.32 | −0.74 | 0.72 | G2 |
| A_23_P26895 | TUBD1 | NM_016261 | 51174 | −0.02 | 0.13 | 0.15 | 0.14 | G2 |
| A_23_P423480 | TYSND1 | NM_173555 | 219743 | −0.21 | −0.16 | 0.05 | 0.11 | G2 |
| A_24_P290585 | UACA | NM_001008224 | 55075 | 0.28 | 0.01 | −0.27 | 1.38 | G2 |
| A_23_P360340 | UACA | NM_001008224 | 55075 | 0.10 | −0.23 | −0.33 | 0.55 | G2 |
| A_24_P90774 | UACA | NM_001008224 | 55075 | 0.03 | 0.11 | 0.07 | −0.41 | G2 |
| A_24_P297539 | UBE2C | NM_181803 | 11065 | −2.93 | −2.50 | 0.43 | −0.57 | G2 |
| A_23_P11936 | UBXN11 | NM_183008 | 91544 | −0.32 | −0.37 | −0.06 | −0.35 | G2 |
| A_24_P239811 | UBXN11 | NM_183008 | 91544 | −0.03 | 0.00 | 0.03 | 0.00 | G2 |
| A_23_P428298 | UNC5CL | NM_173561 | 222643 | 0.48 | 0.40 | −0.08 | −0.09 | G2 |
| A_23_P66599 | VPS25 | NM_032353 | 84313 | −0.33 | −0.20 | 0.13 | 0.02 | G2 |
| A_24_P294982 | VTA1 | NM_016485 | 51534 | −0.42 | −0.42 | 0.00 | 0.18 | G2 |
| A_23_P42368 | VTA1 | NM_016485 | 51534 | −0.36 | −0.20 | 0.16 | −0.24 | G2 |
| A_23_P393766 | WDR62 | NM_173636 | 284403 | 0.15 | −0.21 | −0.36 | 0.47 | G2 |
| A_23_P339705 | WDR62 | NM_173636 | 284403 | −0.02 | −0.17 | −0.15 | −0.03 | G2 |
| A_23_P101281 | ZNF587 | NM_032828 | 84914 | −0.17 | −0.06 | 0.11 | 0.19 | G2 |
| A_24_P313804 | ZNF587 | NM_032828 | 84914 | −0.19 | −0.14 | 0.05 | 0.74 | G2 |
| A_24_P373726 | ZNF587 | NM_032828 | 84914 | −0.01 | −0.30 | −0.28 | 0.42 | G2 |
| A_23_P329286 | ZNHIT2 | NM_014205 | 741 | −0.25 | −0.03 | 0.22 | −0.41 | G2 |
| A_23_P356684 | ANLN | NM_018685 | 54443 | −2.41 | −2.24 | 0.17 | −0.42 | G2,G2_M |
| A_23_P334845 | ARHGAP19 | NM_032900 | 84986 | 0.51 | 0.47 | −0.04 | 0.34 | G2,G2_M |
| A_23_P1387 | ARHGAP19 | NM_032900 | 84986 | 0.00 | −0.49 | −0.49 | 0.70 | G2,G2_M |
| A_24_P19337 | ASXL1 | NM_015338 | 171023 | 0.61 | 0.34 | −0.26 | 0.39 | G2,G2_M |
| A_32_P41021 | ASXL1 | NM_015338 | 171023 | −0.32 | 0.00 | 0.32 | −0.65 | G2,G2_M |
| A_23_P58321 | CCNA2 | NM_001237 | 890 | −2.44 | −1.76 | 0.69 | −0.71 | G2,G2_M |
| A_24_P218979 | CDCA3 | NM_031299 | 83461 | −2.19 | −2.02 | 0.17 | −0.56 | G2,G2_M |
| A_23_P162476 | CDCA3 | NM_031299 | 83461 | −1.69 | −1.43 | 0.27 | −0.36 | G2,G2_M |
| A_23_P209394 | CFLAR | NM_001127184 | 8837 | 1.55 | 0.95 | −0.60 | 2.06 | G2,G2_M |
| A_24_P120115 | CFLAR | NM_003879 | 8837 | 0.65 | 0.80 | 0.15 | 0.12 | G2,G2_M |
| A_23_P151405 | CKAP2 | NM_018204 | 26586 | −1.71 | −0.85 | 0.87 | −0.82 | G2,G2_M |
| A_24_P99090 | CKAP2 | NM_018204 | 26586 | −0.17 | −0.05 | 0.12 | 0.04 | G2,G2_M |
| A_23_P90062 | DNAJB1 | NM_006145 | 3337 | 1.18 | 0.26 | −0.92 | 0.32 | G2,G2_M |
| A_24_P941759 | G2E3 | NM_017769 | 55632 | −0.44 | −0.47 | −0.04 | −0.04 | G2,G2_M |
| A_23_P99604 | G2E3 | NM_017769 | 55632 | 0.22 | −0.14 | −0.36 | 0.64 | G2,G2_M |
| A_32_P189204 | GAS2L3 | NM_174942 | 283431 | −0.76 | −0.63 | 0.13 | 0.22 | G2,G2_M |
| A_32_P37143 | GAS2L3 | BX649059 | 283431 | −0.46 | −0.60 | −0.14 | 0.25 | G2,G2_M |
| A_24_P944616 | HP1BP3 | NM_016287 | 50809 | −0.33 | −0.08 | 0.25 | −0.95 | G2,G2_M |
| A_23_P137630 | HP1BP3 | NM_016287 | 50809 | −0.28 | −0.26 | 0.02 | 0.52 | G2,G2_M |
| A_24_P247660 | JPT1 | NM_001002033 | 51155 | −0.50 | −0.45 | 0.05 | −0.64 | G2,G2_M |
| A_23_P100632 | JPT1 | NM_001002033 | 51155 | −0.11 | 0.22 | 0.33 | −0.42 | G2,G2_M |
| A_23_P75071 | KIF20B | NM_016195 | 9585 | −1.72 | −1.70 | 0.02 | −0.31 | G2,G2_M |
| A_23_P86403 | KIF5B | NM_004521 | 3799 | 0.08 | −0.01 | −0.09 | −0.13 | G2,G2_M |
| A_23_P200493 | LBR | NM_002296 | 3930 | −0.26 | −0.23 | 0.03 | 0.20 | G2,G2_M |
| A_23_P106162 | MIS18BP1 | NM_018353 | 55320 | −0.89 | −0.34 | 0.55 | −0.14 | G2,G2_M |
| A_23_P364107 | MIS18BP1 | NM_018353 | 55320 | −1.10 | −0.59 | 0.52 | −0.69 | G2,G2_M |
| A_23_P315843 | NCOA5 | NM_020967 | 57727 | −0.63 | 0.19 | 0.82 | −1.05 | G2,G2_M |
| A_23_P210515 | NCOA5 | NM_020967 | 57727 | −0.66 | 0.45 | 1.12 | −1.46 | G2,G2_M |
| A_24_P416079 | NUSAP1 | NM_016359 | 51203 | −2.70 | −2.01 | 0.69 | −0.98 | G2,G2_M |
| A_23_P333420 | RANGAP1 | NM_002883 | 5905 | 0.07 | −0.20 | −0.27 | 0.29 | G2,G2_M |
| A_23_P139066 | RNF141 | NM_016422 | 50862 | 0.39 | −0.13 | −0.53 | 0.95 | G2,G2_M |
| A_24_P372625 | RNF141 | NM_016422 | 50862 | 0.06 | −0.02 | −0.08 | 0.14 | G2,G2_M |
| A_23_P100788 | STAT5B | NM_012448 | 6777 | 0.68 | 0.33 | −0.34 | 0.33 | G2,G2_M |
| A_24_P342178 | STAT5B | BC020868 | 6777 | 0.33 | 0.08 | −0.25 | 0.46 | G2,G2_M |
| A_32_P187327 | TUBB4B | NM_006088 | 10383 | −0.26 | 0.35 | 0.60 | −0.76 | G2,G2_M |
| A_23_P312863 | TUBB4B | NM_006088 | 10383 | −0.21 | 0.32 | 0.53 | −0.67 | G2,G2_M |
| A_23_P4353 | WSB1 | NM_015626 | 26118 | 1.02 | −0.01 | −1.03 | 1.36 | G2,G2_M |
| A_23_P214756 | ADGRG6 | NM_198569 | 57211 | 0.15 | 0.08 | −0.07 | 0.85 | G2,S |
| A_24_P942945 | ADGRG6 | NM_020455 | 57211 | −0.14 | −0.28 | −0.14 | −0.14 | G2,S |
| A_24_P411749 | ADGRG6 | NM_020455 | 57211 | −0.01 | −0.21 | −0.21 | 0.28 | G2,S |
| A_23_P118834 | TOP2A | NM_001067 | 7153 | −3.63 | −2.61 | 1.02 | −1.33 | G2,S |
| A_23_P502314 | ADGRE5 | NM_078481 | 976 | −0.13 | 0.18 | 0.32 | −0.67 | G2_M |
| A_23_P502312 | ADGRE5 | NM_078481 | 976 | 0.02 | 0.39 | 0.37 | −0.76 | G2_M |
| A_23_P30098 | ADH4 | NM_000670 | 127 | −0.24 | 0.51 | 0.75 | 0.10 | G2_M |
| A_23_P27688 | ADM5 | NM_001101340 | 199800 | −0.26 | −0.26 | 0.00 | 0.09 | G2_M |
| A_23_P436353 | AFDN | NM_001040001 | 4301 | 0.39 | 0.35 | −0.04 | −0.39 | G2_M |
| A_23_P256603 | AFDN | NM_005936 | 4301 | 0.11 | −0.83 | −0.93 | 0.75 | G2_M |
| A_24_P943484 | AHI1 | NM_017651 | 54806 | 0.27 | 0.29 | 0.01 | 0.72 | G2_M |
| A_24_P213710 | AHI1 | NM_017651 | 54806 | −0.24 | 0.10 | 0.34 | 0.06 | G2_M |
| A_24_P38143 | AHI1 | NM_017651 | 54806 | −0.19 | 0.11 | 0.30 | 0.14 | G2_M |
| A_23_P70746 | AHI1 | NM_017651 | 54806 | 0.07 | 0.27 | 0.19 | 0.16 | G2_M |
| A_23_P428819 | AKIRIN2 | NM_018064 | 55122 | 0.39 | 0.20 | −0.19 | −0.07 | G2_M |
| A_23_P428827 | AKIRIN2 | NM_018064 | 55122 | 0.11 | −0.07 | −0.18 | −0.37 | G2_M |
| A_23_P20615 | ANP32B | NM_006401 | 10541 | 0.43 | 0.07 | −0.36 | 0.24 | G2_M |
| A_24_P225468 | ANP32E | NM_030920 | 81611 | −0.32 | −0.68 | −0.37 | 0.30 | G2_M |
| A_23_P160934 | ANP32E | NM_030920 | 81611 | 0.01 | −0.39 | −0.41 | 0.90 | G2_M |
| A_23_P151075 | ARHGDIB | NM_001175 | 397 | −0.28 | 0.41 | 0.69 | 0.07 | G2_M |
| A_24_P356218 | ARL6IP1 | NM_015161 | 23204 | −0.14 | −0.27 | −0.13 | 0.08 | G2_M |
| A_23_P118150 | ARL6IP1 | NM_015161 | 23204 | −0.29 | −0.32 | −0.03 | 1.16 | G2_M |
| A_23_P91640 | ASPHD2 | NM_020437 | 57168 | −0.55 | 0.19 | 0.74 | −1.23 | G2_M |
| A_24_P245815 | ASPHD2 | NM_020437 | 57168 | −0.29 | 0.33 | 0.62 | −1.00 | G2_M |
| A_23_P112950 | ATF7IP | NM_018179 | 55729 | 0.17 | 0.54 | 0.37 | −0.51 | G2_M |
| A_24_P923757 | ATF7IP | NM_018179 | 55729 | 0.26 | 0.26 | 0.00 | 0.25 | G2_M |
| A_23_P48278 | ATF7IP | AK001001 | 55729 | −0.01 | 0.81 | 0.82 | −0.13 | G2_M |
| A_23_P163814 | ATXN1L | NM_001137675 | 342371 | 0.09 | −0.31 | −0.40 | 0.20 | G2_M |
| A_23_P131866 | AURKA | NM_198433 | 6790 | −1.56 | −1.01 | 0.56 | 0.03 | G2_M |
| A_24_P103803 | B4GALT1 | NM_001497 | 2683 | 0.90 | 0.68 | −0.22 | −0.45 | G2_M |
| A_23_P135271 | B4GALT1 | NM_001497 | 2683 | 0.49 | 0.22 | −0.27 | −0.06 | G2_M |
| A_24_P115774 | BIRC2 | NM_001166 | 329 | −0.12 | −0.28 | −0.16 | 0.32 | G2_M |
| A_23_P118815 | BIRC5 | NM_001012271 | 332 | −3.41 | −2.33 | 1.08 | −1.52 | G2_M |
| A_23_P143331 | BMP2 | NM_001200 | 650 | 1.44 | 0.57 | −0.87 | 0.93 | G2_M |
| A_23_P124417 | BUB1 | NM_004336 | 699 | −3.26 | −2.47 | 0.79 | −0.75 | G2_M |
| A_23_P163481 | BUB1B | NM_001211 | 701 | −3.08 | −2.32 | 0.76 | −1.02 | G2_M |
| A_23_P92928 | C6 | NM_000065 | 729 | −0.19 | −0.33 | −0.14 | −0.46 | G2_M |
| A_23_P28664 | CCDC88A | NM_018084 | 55704 | 0.71 | 0.22 | −0.49 | 0.70 | G2_M |
| A_24_P20120 | CCDC88A | NM_018084 | 55704 | 0.52 | 0.40 | −0.12 | 0.49 | G2_M |
| A_23_P17269 | CCDC88A | NM_018084 | 55704 | 0.36 | −0.01 | −0.37 | 0.82 | G2_M |
| A_24_P28739 | CCDC88A | NM_018084 | 55704 | 0.32 | −0.17 | −0.49 | 0.41 | G2_M |
| A_23_P122197 | CCNB1 | NM_031966 | 891 | −2.14 | −1.49 | 0.65 | −0.27 | G2_M |
| A_23_P65757 | CCNB2 | NM_004701 | 9133 | −3.05 | −2.42 | 0.64 | −0.70 | G2_M |
| A_32_P72822 | CCNB2 | AK023404 | 9133 | 0.52 | 0.74 | 0.22 | 0.59 | G2_M |
| A_23_P202837 | CCND1 | NM_053056 | 595 | −0.15 | −0.20 | −0.05 | −0.10 | G2_M |
| A_24_P124550 | CCND1 | NM_053056 | 595 | −0.03 | −0.15 | −0.12 | −0.75 | G2_M |
| A_24_P193011 | CCND1 | NM_053056 | 595 | −0.02 | 0.09 | 0.11 | −1.15 | G2_M |
| A_23_P366908 | CCSAP | NM_145257 | 126731 | 0.38 | −0.19 | −0.57 | 0.18 | G2_M |
| A_24_P930764 | CCSAP | BC039241 | 126731 | 0.40 | −0.35 | −0.75 | 0.71 | G2_M |
| A_32_P83776 | CCSAP | NM_145257 | 126731 | 0.42 | 0.13 | −0.29 | −1.01 | G2_M |
| A_23_P149200 | CDC20 | NM_001255 | 991 | −1.36 | −1.65 | −0.28 | 0.10 | G2_M |
| A_23_P210726 | CDC25B | NM_021873 | 994 | −0.75 | −0.03 | 0.72 | −0.74 | G2_M |
| A_23_P66777 | CDC27 | NM_001256 | 996 | 0.05 | 0.25 | 0.20 | 0.28 | G2_M |
| A_23_P166453 | CDC42EP1 | NM_152243 | 11135 | −0.06 | 0.25 | 0.31 | −0.98 | G2_M |
| A_24_P143032 | CDC42EP1 | NM_152243 | 11135 | 0.06 | 0.25 | 0.19 | −0.63 | G2_M |
| A_23_P89941 | CDKN2D | NM_001800 | 1032 | 0.50 | −0.51 | −1.01 | 0.79 | G2_M |
| A_24_P413884 | CENPA | NM_001809 | 1058 | −2.99 | −2.34 | 0.65 | −1.30 | G2_M |
| A_23_P253524 | CENPE | NM_001813 | 1062 | −2.90 | −1.84 | 1.06 | −1.03 | G2_M |
| A_23_P401 | CENPF | NM_016343 | 1063 | −3.83 | −2.46 | 1.37 | −1.17 | G2_M |
| A_24_P96780 | CENPF | NM_016343 | 1063 | −1.58 | −0.86 | 0.72 | −0.66 | G2_M |
| A_23_P115872 | CEP55 | NM_018131 | 55165 | −2.96 | −2.26 | 0.71 | −0.74 | G2_M |
| A_23_P420551 | CIT | NM_007174 | 11113 | −2.31 | −1.43 | 0.88 | −1.92 | G2_M |
| A_23_P135977 | CKAP5 | NM_001008938 | 9793 | −0.65 | −0.56 | 0.09 | −0.09 | G2_M |
| A_24_P300841 | CKAP5 | NM_001008938 | 9793 | −0.27 | −0.13 | 0.14 | −0.11 | G2_M |
| A_23_P45917 | CKS1B | NM_001826 | 1163 | −0.90 | −1.00 | −0.10 | 0.48 | G2_M |
| A_32_P206698 | CKS1B | NM_001826 | 1163 | −0.79 | −0.88 | −0.08 | 0.51 | G2_M |
| A_32_P192430 | CKS1B | NM_001826 | 1163 | −0.52 | −0.67 | −0.15 | 0.44 | G2_M |
| A_23_P71727 | CKS2 | NM_001827 | 1164 | −0.85 | −1.06 | −0.21 | 0.78 | G2_M |
| A_23_P16673 | CNN2 | NM_004368 | 1265 | 0.34 | 0.36 | 0.01 | −0.98 | G2_M |
| A_24_P142743 | CNN2 | NM_004368 | 1265 | 0.15 | 0.29 | 0.14 | 0.07 | G2_M |
| A_23_P9768 | CNTROB | NM_053051 | 116840 | −0.07 | −0.01 | 0.06 | 0.00 | G2_M |
| A_23_P9761 | CNTROB | NM_001037144 | 116840 | −0.02 | 0.10 | 0.12 | −0.45 | G2_M |
| A_23_P404606 | CREBRF | NM_153607 | 153222 | 0.59 | −0.30 | −0.88 | 1.08 | G2_M |
| A_23_P134835 | CSGALNACT1 | NM_018371 | 55790 | 1.04 | 0.75 | −0.29 | 1.04 | G2_M |
| A_24_P406525 | CSGALNACT1 | NM_018371 | 55790 | 0.10 | 0.31 | 0.21 | 0.25 | G2_M |
| A_23_P58647 | CTNNA1 | NM_001903 | 1495 | 0.06 | 0.36 | 0.30 | −0.44 | G2_M |
| A_24_P80633 | CTNNA1 | NM_001903 | 1495 | 0.03 | 0.05 | 0.02 | −0.27 | G2_M |
| A_24_P881527 | CTNND1 | NM_001085458 | 1500 | −0.14 | 0.09 | 0.23 | −0.46 | G2_M |
| A_23_P95080 | CTNND1 | NM_001085461 | 1500 | 0.13 | −0.15 | −0.28 | 0.76 | G2_M |
| A_23_P251316 | CTNND1 | NM_001331 | 1500 | −0.12 | −0.21 | −0.09 | −0.15 | G2_M |
| A_24_P38930 | CTNND1 | NM_001331 | 1500 | 0.07 | −0.03 | −0.10 | −0.22 | G2_M |
| A_23_P200310 | DEPDC1 | NM_017779 | 55635 | −2.48 | −2.08 | 0.40 | −0.16 | G2_M |
| A_24_P25872 | DEPDC1 | NM_017779 | 55635 | −1.13 | −1.41 | −0.28 | −0.32 | G2_M |
| A_23_P361419 | DEPDC1B | NM_018369 | 55789 | −1.89 | −1.25 | 0.63 | −0.44 | G2_M |
| A_23_P88331 | DLGAP5 | NM_014750 | 9787 | −3.42 | −2.36 | 1.06 | −1.16 | G2_M |
| A_24_P9671 | DNAJA1 | NM_001539 | 3301 | 0.95 | 0.69 | −0.26 | 1.02 | G2_M |
| A_24_P192586 | DNAJA1 | ENST00000330899 | 3301 | −0.49 | −0.02 | 0.47 | 0.14 | G2_M |
| A_23_P60479 | DNAJA1 | NM_001539 | 3301 | 0.31 | 0.26 | −0.05 | 0.90 | G2_M |
| A_23_P63205 | DR1 | NM_001938 | 1810 | 0.30 | −0.06 | −0.36 | 0.37 | G2_M |
| A_23_P391725 | DR1 | NM_001938 | 1810 | 0.13 | 0.16 | 0.03 | 0.01 | G2_M |
| A_23_P134935 | DUSP4 | NM_001394 | 1846 | −0.31 | −0.20 | 0.12 | −0.96 | G2_M |
| A_23_P144165 | DZIP3 | NM_014648 | 9666 | 0.64 | 0.09 | −0.55 | 0.75 | G2_M |
| A_24_P102504 | DZIP3 | NM_014648 | 9666 | −0.14 | −0.30 | −0.16 | 0.24 | G2_M |
| A_23_P435051 | DZIP3 | ENST00000463306 | 9666 | 0.06 | −0.22 | −0.28 | 0.25 | G2_M |
| A_23_P31721 | E2F5 | NM_001951 | 1875 | −0.48 | −0.02 | 0.46 | −0.36 | G2_M |
| A_23_P9574 | ECT2 | NM_018098 | 1894 | −0.96 | −0.37 | 0.59 | 0.16 | G2_M |
| A_23_P44684 | ECT2 | NM_018098 | 1894 | −1.09 | −0.50 | 0.59 | −0.44 | G2_M |
| A_24_P366033 | ECT2 | NM_018098 | 1894 | −0.29 | −0.31 | −0.02 | 0.38 | G2_M |
| A_24_P282251 | FGA | NM_021871 | 2243 | 0.26 | −0.16 | −0.42 | −0.04 | G2_M |
| A_23_P44274 | FGA | NM_000508 | 2243 | −0.08 | 0.01 | 0.09 | 0.24 | G2_M |
| A_23_P375372 | FGA | NM_021871 | 2243 | 0.07 | 0.62 | 0.55 | −0.54 | G2_M |
| A_23_P151150 | FOXM1 | NM_202002 | 2305 | −1.27 | −1.19 | 0.07 | −1.05 | G2_M |
| A_23_P363778 | FRZB | NM_001463 | 2487 | 1.24 | 0.54 | −0.70 | 1.02 | G2_M |
| A_23_P10902 | FRZB | NM_001463 | 2487 | 0.95 | 0.41 | −0.54 | 0.75 | G2_M |
| A_23_P502142 | FYN | NM_002037 | 2534 | −0.54 | −0.35 | 0.20 | −1.32 | G2_M |
| A_23_P23221 | GADD45A | NM_001924 | 1647 | 0.64 | 0.07 | −0.58 | −0.01 | G2_M |
| A_23_P146922 | GAS6 | NM_000820 | 2621 | 0.18 | 0.16 | −0.02 | −0.78 | G2_M |
| A_23_P105251 | GLI1 | NM_005269 | 2735 | 0.06 | 0.17 | 0.12 | −0.31 | G2_M |
| A_23_P63825 | GOT1 | NM_002079 | 2805 | 0.29 | 0.53 | 0.24 | 0.44 | G2_M |
| A_24_P81473 | GOT1 | NM_002079 | 2805 | 0.20 | 0.18 | −0.01 | 0.64 | G2_M |
| A_23_P63402 | GPSM2 | NM_013296 | 29899 | −1.24 | −0.96 | 0.28 | −0.28 | G2_M |
| A_24_P273132 | GPSM2 | NM_013296 | 29899 | −0.47 | −0.69 | −0.23 | −0.02 | G2_M |
| A_23_P257256 | GRK6 | NM_002082 | 2870 | −0.18 | 0.03 | 0.21 | −0.53 | G2_M |
| A_23_P152420 | GSE1 | NM_014615 | 23199 | 0.19 | 0.03 | −0.16 | −0.38 | G2_M |
| A_24_P943062 | GSE1 | NM_014615 | 23199 | −0.11 | 0.18 | 0.29 | −1.06 | G2_M |
| A_23_P57588 | GTSE1 | NM_016426 | 51512 | −2.01 | −1.57 | 0.43 | −0.89 | G2_M |
| A_23_P125771 | HCFC1 | NM_005334 | 3054 | −0.26 | 0.18 | 0.45 | −1.13 | G2_M |
| A_23_P168490 | HERPUD2 | NM_022373 | 64224 | 0.03 | 0.00 | −0.03 | −0.02 | G2_M |
| A_23_P119543 | HMG20B | NM_006339 | 10362 | −0.82 | −0.34 | 0.48 | −0.54 | G2_M |
| A_23_P217236 | HMGB3 | NM_005342 | 3149 | −0.75 | −0.79 | −0.04 | −0.06 | G2_M |
| A_23_P70007 | HMMR | NM_012484 | 3161 | −3.37 | −2.64 | 0.73 | −0.46 | G2_M |
| A_23_P109442 | HPS4 | NM_022081 | 89781 | 0.72 | 0.36 | −0.36 | −0.05 | G2_M |
| A_23_P109446 | HPS4 | NM_022081 | 89781 | 0.58 | 0.44 | −0.14 | −0.34 | G2_M |
| A_23_P17606 | HSPA13 | NM_006948 | 6782 | 1.40 | 0.56 | −0.85 | 2.40 | G2_M |
| A_24_P134392 | HSPA13 | NM_006948 | 6782 | 0.30 | −0.05 | −0.35 | 0.52 | G2_M |
| A_23_P70547 | HSPA1L | NM_005527 | 3305 | 1.11 | 0.14 | −0.96 | 0.90 | G2_M |
| A_32_P13728 | HSPA8 | NM_006597 | 3312 | 0.36 | 0.40 | 0.03 | 0.34 | G2_M |
| A_24_P295745 | HSPA8 | NM_153201 | 3312 | 0.13 | 0.12 | −0.01 | 0.78 | G2_M |
| A_24_P287129 | HSPA8 | NM_006597 | 3312 | 0.05 | 0.06 | 0.02 | 0.10 | G2_M |
| A_23_P24594 | HSPA8 | NM_006597 | 3312 | 0.03 | 0.12 | 0.09 | 0.17 | G2_M |
| A_23_P410600 | IDI2 | NM_033261 | 91734 | 0.14 | −0.01 | −0.16 | −0.21 | G2_M |
| A_23_P112026 | IDO1 | NM_002164 | 3620 | 0.69 | 0.85 | 0.16 | 0.41 | G2_M |
| A_23_P55076 | INPP5K | NM_130766 | 51763 | 0.47 | 0.28 | −0.19 | −0.09 | G2_M |
| A_24_P279328 | INPP5K | NM_130766 | 51763 | −0.09 | −0.12 | −0.02 | −0.42 | G2_M |
| A_23_P109184 | INSM1 | NM_002196 | 3642 | 1.19 | 0.87 | −0.32 | −0.25 | G2_M |
| A_24_P31676 | INSM1 | NM_002196 | 3642 | −0.05 | 0.13 | 0.17 | −0.08 | G2_M |
| A_23_P92042 | ITPR1 | NM_002222 | 3708 | 0.17 | 0.24 | 0.07 | −0.72 | G2_M |
| A_23_P156198 | JADE2 | NM_015288 | 23338 | 1.15 | 0.18 | −0.97 | 0.77 | G2_M |
| A_24_P226278 | JADE2 | NM_015288 | 23338 | 0.53 | −0.07 | −0.60 | 0.28 | G2_M |
| A_23_P416434 | JADE2 | NM_015288 | 23338 | −0.17 | 0.47 | 0.64 | −0.91 | G2_M |
| A_24_P927883 | JADE2 | A_24_P927883 | NA | −0.02 | 0.29 | 0.31 | −0.10 | G2_M |
| A_24_P324011 | KCTD2 | NM_015353 | 23510 | −0.20 | −0.03 | 0.17 | −0.55 | G2_M |
| A_32_P160693 | KCTD2 | NM_015353 | 23510 | −0.02 | −0.13 | −0.11 | −0.13 | G2_M |
| A_23_P149668 | KIF14 | NM_014875 | 9928 | −0.68 | −0.91 | −0.23 | −0.16 | G2_M |
| A_23_P34788 | KIF2C | NM_006845 | 11004 | −3.33 | −2.53 | 0.80 | −0.82 | G2_M |
| A_23_P415401 | KLF9 | NM_001206 | 687 | 0.04 | −0.15 | −0.20 | −0.41 | G2_M |
| A_23_P86100 | KLHDC9 | NM_001007255 | 126823 | −0.01 | −0.26 | −0.25 | 0.04 | G2_M |
| A_32_P82807 | KMT5A | NM_020382 | 387893 | 0.38 | 0.39 | 0.02 | 0.29 | G2_M |
| A_24_P238855 | KMT5A | NM_020382 | 387893 | −0.22 | 0.33 | 0.54 | −0.29 | G2_M |
| A_32_P191527 | KMT5A | NM_020382 | 387893 | −0.19 | 0.07 | 0.26 | −0.57 | G2_M |
| A_32_P191859 | KMT5A | NM_020382 | 387893 | −0.10 | 0.11 | 0.21 | −0.39 | G2_M |
| A_23_P217968 | KMT5B | NM_016028 | 51111 | 1.11 | 0.43 | −0.68 | 0.99 | G2_M |
| A_23_P326739 | KMT5B | NM_017635 | 51111 | −0.60 | −0.03 | 0.57 | −0.90 | G2_M |
| A_23_P96688 | KMT5B | NM_016028 | 51111 | −0.02 | −0.15 | −0.13 | 0.10 | G2_M |
| A_23_P140705 | KNSTRN | NM_001142761 | 90417 | −1.55 | −1.01 | 0.54 | 0.14 | G2_M |
| A_24_P162718 | LMNA | NM_005572 | 4000 | −0.08 | −0.12 | −0.04 | −0.35 | G2_M |
| A_23_P34835 | LMNA | NM_005572 | 4000 | −0.01 | −0.01 | 0.01 | −0.44 | G2_M |
| A_23_P251118 | LPP | NM_005578 | 4026 | 0.44 | 0.47 | 0.03 | 0.31 | G2_M |
| A_24_P916515 | LPP | A_24_P916515 | NA | 0.33 | 0.35 | 0.01 | −0.02 | G2_M |
| A_32_P70519 | LPP | ENST00000312675 | 4026 | 0.16 | 0.16 | 0.00 | −0.46 | G2_M |
| A_24_P114551 | LPP | NM_005578 | 4026 | −0.14 | 0.07 | 0.21 | −0.89 | G2_M |
| A_32_P39049 | LPP | ENST00000312675 | 4026 | −0.13 | 0.10 | 0.24 | −0.09 | G2_M |
| A_32_P193218 | LPP | ENST00000312675 | 4026 | −0.10 | 0.18 | 0.28 | −0.83 | G2_M |
| A_24_P778741 | LPP | ENST00000312675 | 4026 | 0.10 | 0.37 | 0.27 | −1.09 | G2_M |
| A_32_P56874 | LPP | ENST00000312675 | 4026 | 0.02 | 0.06 | 0.03 | −0.35 | G2_M |
| A_23_P200222 | LRP8 | NM_033300 | 7804 | 0.86 | 0.93 | 0.08 | 0.31 | G2_M |
| A_23_P34325 | LRP8 | NM_033300 | 7804 | 0.14 | 0.53 | 0.38 | −0.26 | G2_M |
| A_23_P253958 | LRRC17 | NM_005824 | 10234 | 0.43 | 1.01 | 0.58 | −0.15 | G2_M |
| A_23_P145376 | MAPK13 | NM_002754 | 5603 | 0.22 | −0.13 | −0.35 | 0.82 | G2_M |
| A_24_P406132 | MAPK13 | NM_002754 | 5603 | 0.08 | −0.03 | −0.12 | 0.20 | G2_M |
| A_23_P71328 | MATN2 | NM_030583 | 4147 | −0.50 | −0.30 | 0.20 | −1.34 | G2_M |
| A_24_P179225 | MATN2 | NM_030583 | 4147 | 0.24 | 0.06 | −0.17 | −0.24 | G2_M |
| A_23_P19455 | MDC1 | NM_014641 | 9656 | −0.14 | −0.15 | −0.01 | −0.40 | G2_M |
| A_23_P105227 | ME3 | NM_001014811 | 10873 | −0.40 | 0.15 | 0.55 | −0.26 | G2_M |
| A_23_P116614 | ME3 | NM_001014811 | 10873 | 0.04 | 0.39 | 0.35 | 0.08 | G2_M |
| A_24_P346855 | MKI67 | NM_002417 | 4288 | −1.66 | −1.70 | −0.04 | −0.93 | G2_M |
| A_32_P9382 | MZT1 | NM_001071775 | 440145 | −0.33 | −0.50 | −0.18 | 0.57 | G2_M |
| A_24_P532589 | MZT1 | NM_001071775 | 440145 | −0.12 | −0.28 | −0.16 | 0.18 | G2_M |
| A_24_P210675 | NDE1 | NM_017668 | 54820 | −0.60 | 0.01 | 0.61 | −1.05 | G2_M |
| A_23_P206901 | NDE1 | NM_017668 | 54820 | 0.07 | 0.63 | 0.56 | −0.46 | G2_M |
| A_23_P35219 | NEK2 | NM_002497 | 4751 | −1.67 | −1.54 | 0.13 | −0.25 | G2_M |
| A_24_P319613 | NEK2 | NM_002497 | 4751 | −0.86 | −0.50 | 0.36 | −0.09 | G2_M |
| A_23_P74349 | NUF2 | NM_145697 | 83540 | −3.08 | −2.39 | 0.69 | −1.08 | G2_M |
| A_23_P102320 | NUP35 | NM_138285 | 129401 | −0.43 | −0.18 | 0.25 | 0.55 | G2_M |
| A_23_P203586 | NUP98 | NM_016320 | 4928 | 0.32 | 0.34 | 0.02 | −0.05 | G2_M |
| A_24_P141522 | NUP98 | NM_016320 | 4928 | 0.29 | 0.24 | −0.06 | 0.12 | G2_M |
| A_23_P308032 | NUP98 | NM_005387 | 4928 | −0.12 | −0.28 | −0.16 | 0.14 | G2_M |
| A_23_P60488 | ODF2 | NM_002540 | 4957 | −0.72 | −0.22 | 0.50 | −0.90 | G2_M |
| A_23_P71889 | ODF2 | NM_153437 | 4957 | −0.21 | 0.30 | 0.51 | −0.15 | G2_M |
| A_23_P63777 | OIT3 | NM_152635 | 170392 | −0.28 | 0.06 | 0.34 | 0.06 | G2_M |
| A_24_P124624 | OLR1 | NM_002543 | 4973 | 0.54 | −0.05 | −0.59 | 0.51 | G2_M |
| A_24_P274072 | PATJ | NM_176877 | 10207 | 0.45 | 0.06 | −0.39 | 0.74 | G2_M |
| A_23_P126100 | PATJ | NM_176877 | 10207 | 0.39 | 0.15 | −0.24 | 0.60 | G2_M |
| A_32_P88958 | PATJ | NM_176877 | 10207 | 0.20 | −0.13 | −0.33 | 0.41 | G2_M |
| A_23_P321034 | PATJ | NM_176877 | 10207 | 0.17 | 0.33 | 0.17 | −0.25 | G2_M |
| A_24_P942454 | PATJ | NM_176877 | 10207 | −0.05 | 0.06 | 0.11 | 0.50 | G2_M |
| A_32_P62997 | PBK | NM_018492 | 55872 | −4.16 | −3.00 | 1.15 | −1.67 | G2_M |
| A_23_P116578 | PCF11 | NM_015885 | 51585 | −0.41 | −0.40 | 0.01 | 0.36 | G2_M |
| A_24_P835763 | PCF11 | A_24_P835763 | NA | 0.38 | −0.66 | −1.05 | 1.16 | G2_M |
| A_23_P33303 | PIK3CD | NM_005026 | 5293 | 0.56 | 0.53 | −0.03 | −0.27 | G2_M |
| A_24_P71244 | PIK3CD | NM_005026 | 5293 | 0.15 | −0.05 | −0.20 | −0.21 | G2_M |
| A_23_P259833 | PIK3CD | NM_005026 | 5293 | 0.02 | −0.13 | −0.15 | 0.13 | G2_M |
| A_23_P49878 | PIMREG | NM_019013 | 54478 | −2.67 | −2.40 | 0.27 | −0.54 | G2_M |
| A_23_P411723 | PLAG1 | NM_002655 | 5324 | 0.44 | −0.18 | −0.62 | 1.02 | G2_M |
| A_24_P313504 | PLK1 | NM_005030 | 5347 | −1.32 | −1.37 | −0.05 | −0.17 | G2_M |
| A_23_P118174 | PLK1 | NM_005030 | 5347 | −0.82 | −0.68 | 0.14 | −0.43 | G2_M |
| A_24_P354300 | POC1A | NM_015426 | 25886 | −1.21 | −0.81 | 0.40 | −0.63 | G2_M |
| A_23_P212284 | POC1A | NM_015426 | 25886 | −1.26 | −0.82 | 0.44 | −0.77 | G2_M |
| A_24_P349002 | POM121 | NM_172020 | 9883 | 0.32 | 0.50 | 0.19 | −0.52 | G2_M |
| A_32_P131940 | POM121 | NM_172020 | 9883 | 0.27 | 0.40 | 0.13 | −0.95 | G2_M |
| A_24_P417784 | POM121 | NM_172020 | 9883 | 0.35 | 0.57 | 0.22 | −0.86 | G2_M |
| A_24_P266273 | POM121 | NM_172020 | 9883 | 0.03 | −0.19 | −0.22 | 0.06 | G2_M |
| A_23_P156667 | PPP1R10 | NM_002714 | 5514 | −0.93 | 0.07 | 1.00 | −1.19 | G2_M |
| A_23_P15305 | PRPSAP1 | NM_002766 | 5635 | −0.51 | −0.31 | 0.20 | −0.68 | G2_M |
| A_23_P80382 | PRR5 | NM_015366 | 55615 | −0.21 | 0.00 | 0.21 | −0.48 | G2_M |
| A_24_P10890 | PRR5 | NM_015366 | 55615 | 0.20 | 0.06 | −0.14 | −0.15 | G2_M |
| A_24_P21044 | PSMG3 | NM_032302 | 84262 | −0.03 | 0.12 | 0.15 | −0.11 | G2_M |
| A_23_P103328 | PTGER3 | NM_198714 | 5733 | 1.30 | 1.41 | 0.11 | 0.82 | G2_M |
| A_24_P945365 | PTGER3 | NM_198715 | 5733 | 0.53 | 0.61 | 0.08 | −0.09 | G2_M |
| A_23_P81770 | PTP4A1 | NM_003463 | 7803 | 0.67 | 0.28 | −0.39 | 0.32 | G2_M |
| A_24_P294832 | PTP4A1 | NM_003463 | 7803 | −0.37 | −0.01 | 0.36 | −0.57 | G2_M |
| A_24_P252043 | PTP4A1 | NM_003463 | 7803 | 0.50 | −0.24 | −0.74 | 0.60 | G2_M |
| A_23_P124486 | PTPN9 | NM_002833 | 5780 | −0.51 | −0.22 | 0.29 | −1.07 | G2_M |
| A_23_P13632 | PYM1 | NM_032345 | 84305 | −0.42 | −0.12 | 0.30 | −0.64 | G2_M |
| A_23_P45106 | QRICH1 | NM_017730 | 54870 | 0.22 | 0.14 | −0.08 | −0.53 | G2_M |
| A_23_P45108 | QRICH1 | NM_017730 | 54870 | 0.17 | −0.10 | −0.27 | 0.53 | G2_M |
| A_23_P207014 | RAD51C | NM_002876 | 5889 | −0.96 | −0.75 | 0.21 | 0.02 | G2_M |
| A_23_P391344 | RASGEF1A | BC022548 | 221002 | 0.73 | 0.39 | −0.34 | 0.62 | G2_M |
| A_32_P223140 | RASGEF1A | NM_145313 | 221002 | 0.75 | 0.14 | −0.62 | 1.04 | G2_M |
| A_24_P79955 | RBM8A | NM_005105 | 9939 | −0.54 | −0.12 | 0.41 | −0.54 | G2_M |
| A_23_P305335 | RBM8A | BC017770 | 9939 | 1.29 | 0.20 | −1.09 | 1.74 | G2_M |
| A_32_P62571 | RBM8A | NM_005105 | 9939 | −0.41 | 0.02 | 0.44 | −0.81 | G2_M |
| A_23_P104116 | RBM8A | NM_005105 | 9939 | 0.09 | 0.16 | 0.07 | 0.49 | G2_M |
| A_23_P166248 | RCAN1 | NM_004414 | 1827 | −0.18 | −0.13 | 0.05 | −0.24 | G2_M |
| A_23_P14105 | RCBTB2 | NM_001268 | 1102 | −1.53 | −0.06 | 1.47 | −1.74 | G2_M |
| A_24_P342591 | RERE | NM_012102 | 473 | 0.16 | 0.39 | 0.23 | −1.12 | G2_M |
| A_23_P85414 | RERE | NM_012102 | 473 | 0.04 | 0.07 | 0.04 | −0.65 | G2_M |
| A_24_P725630 | RNPS1 | NM_006711 | 10921 | 0.85 | 0.54 | −0.31 | 0.39 | G2_M |
| A_24_P766577 | RNPS1 | NM_006711 | 10921 | 0.12 | 0.02 | −0.10 | −0.21 | G2_M |
| A_23_P152272 | RNPS1 | NM_006711 | 10921 | 0.02 | 0.08 | 0.06 | −0.38 | G2_M |
| A_23_P80129 | RRP1 | NM_003683 | 8568 | 0.63 | 0.52 | −0.11 | 0.04 | G2_M |
| A_23_P80136 | RRP1 | NM_003683 | 8568 | −0.09 | 0.26 | 0.34 | −0.03 | G2_M |
| A_24_P100517 | SAPCD2 | NM_178448 | 89958 | −0.91 | −0.60 | 0.31 | −0.42 | G2_M |
| A_23_P370625 | SELENON | NM_020451 | 57190 | −0.80 | −0.26 | 0.54 | −1.75 | G2_M |
| A_24_P231250 | SELENON | NM_020451 | 57190 | −0.37 | −0.07 | 0.30 | −1.06 | G2_M |
| A_24_P105283 | SFPQ | NM_005066 | 6421 | −0.18 | −0.09 | 0.09 | −0.28 | G2_M |
| A_23_P411335 | SGO2 | NM_152524 | 151246 | −1.95 | −1.54 | 0.41 | −0.59 | G2_M |
| A_32_P96719 | SHCBP1 | NM_024745 | 79801 | −1.68 | −1.83 | −0.14 | −0.60 | G2_M |
| A_23_P59051 | SLC17A2 | NM_005835 | 10246 | 0.13 | 0.43 | 0.29 | −0.36 | G2_M |
| A_24_P10657 | SLC44A2 | NM_020428 | 57153 | 0.24 | 0.31 | 0.07 | −0.65 | G2_M |
| A_23_P208340 | SLC44A2 | NM_020428 | 57153 | −0.08 | 0.15 | 0.23 | −0.96 | G2_M |
| A_23_P68824 | SMARCB1 | NM_003073 | 6598 | −0.33 | 0.02 | 0.35 | −0.82 | G2_M |
| A_24_P232696 | SMARCD1 | NM_139071 | 6602 | −0.23 | −0.01 | 0.22 | −0.68 | G2_M |
| A_23_P204745 | SMARCD1 | NM_139071 | 6602 | 0.16 | 0.65 | 0.49 | −0.24 | G2_M |
| A_23_P211428 | SMTN | NM_134269 | 6525 | −0.47 | −0.31 | 0.17 | −1.05 | G2_M |
| A_23_P5934 | SOGA1 | AB020696 | 140710 | −0.43 | −0.12 | 0.31 | −0.40 | G2_M |
| A_23_P5936 | SOGA1 | AB020696 | 140710 | −0.26 | −0.44 | −0.18 | −0.42 | G2_M |
| A_23_P5938 | SOGA1 | AB020696 | 140710 | 0.18 | 0.27 | 0.08 | −0.36 | G2_M |
| A_23_P89509 | SPAG5 | NM_006461 | 10615 | −2.17 | −1.39 | 0.78 | −0.41 | G2_M |
| A_23_P41948 | SPDL1 | NM_017785 | 54908 | −1.58 | −0.78 | 0.80 | −0.55 | G2_M |
| A_23_P102523 | SPTBN1 | NM_003128 | 6711 | −0.45 | −0.37 | 0.08 | −0.81 | G2_M |
| A_23_P339095 | SPTBN1 | NM_178313 | 6711 | −0.19 | −0.25 | −0.07 | −0.15 | G2_M |
| A_23_P30223 | SRD5A1 | NM_001047 | 6715 | −0.15 | −0.47 | −0.32 | −0.08 | G2_M |
| A_23_P413761 | SRSF3 | NM_003017 | 6428 | −0.31 | 0.03 | 0.33 | −0.18 | G2_M |
| A_23_P19702 | TAB2 | NM_015093 | 23118 | 0.32 | 0.20 | −0.12 | 0.03 | G2_M |
| A_24_P245778 | TFF3 | ENST00000291525 | 7033 | −0.63 | −0.48 | 0.15 | −0.45 | G2_M |
| A_23_P393099 | TFF3 | NM_003226 | 7033 | −0.11 | −0.09 | 0.02 | −0.05 | G2_M |
| A_24_P289208 | TFF3 | NM_003226 | 7033 | 0.14 | −0.41 | −0.54 | 0.60 | G2_M |
| A_23_P257296 | TFF3 | NM_003226 | 7033 | 0.03 | 0.03 | 0.00 | 0.06 | G2_M |
| A_23_P153197 | TGIF1 | NM_170695 | 7050 | −0.28 | −0.22 | 0.06 | −1.29 | G2_M |
| A_24_P926367 | THRAP3 | NM_005119 | 9967 | −0.61 | −0.46 | 0.15 | −0.71 | G2_M |
| A_24_P256863 | THRAP3 | NM_005119 | 9967 | −0.19 | −0.38 | −0.18 | 0.33 | G2_M |
| A_23_P160367 | THRAP3 | NM_005119 | 9967 | −0.18 | −0.42 | −0.24 | −0.01 | G2_M |
| A_23_P158277 | TMCO4 | NM_181719 | 255104 | −0.15 | 0.32 | 0.46 | −0.61 | G2_M |
| A_23_P24723 | TMEM138 | NM_016464 | 51524 | −0.43 | −0.25 | 0.17 | 0.17 | G2_M |
| A_23_P428875 | TNFAIP8L1 | NM_152362 | 126282 | −0.70 | −0.84 | −0.14 | −0.55 | G2_M |
| A_23_P156101 | TNPO1 | NM_002270 | 3842 | −0.17 | −0.20 | −0.04 | 0.49 | G2_M |
| A_32_P99097 | TNPO1 | NM_002270 | 3842 | −0.03 | 0.19 | 0.22 | 0.29 | G2_M |
| A_24_P260440 | TNPO1 | NM_002270 | 3842 | 0.00 | 0.10 | 0.10 | 0.32 | G2_M |
| A_24_P199097 | TOMM34 | NM_006809 | 10953 | 0.31 | 0.09 | −0.22 | 1.37 | G2_M |
| A_23_P57033 | TOMM34 | NM_006809 | 10953 | 0.08 | −0.19 | −0.27 | 0.78 | G2_M |
| A_23_P68610 | TPX2 | NM_012112 | 22974 | −2.37 | −1.66 | 0.71 | −1.18 | G2_M |
| A_24_P277576 | TRIP13 | NM_004237 | 9319 | −0.28 | −0.08 | 0.21 | −0.37 | G2_M |
| A_23_P75839 | TSG101 | NM_006292 | 7251 | 0.10 | 0.06 | −0.04 | 1.78 | G2_M |
| A_23_P162142 | TSKU | NM_015516 | 25987 | 0.51 | 0.27 | −0.24 | −0.24 | G2_M |
| A_23_P28105 | TSN | NM_004622 | 7247 | −0.28 | −0.10 | 0.18 | −0.10 | G2_M |
| A_24_P242820 | TSN | NM_004622 | 7247 | −0.19 | −0.19 | 0.00 | −0.29 | G2_M |
| A_23_P259586 | TTK | NM_003318 | 7272 | −2.35 | −2.45 | −0.10 | −0.31 | G2_M |
| A_24_P263524 | TXNDC9 | NM_005783 | 10190 | −0.65 | −0.41 | 0.24 | 0.04 | G2_M |
| A_23_P154330 | TXNDC9 | NM_005783 | 10190 | −0.51 | −0.20 | 0.32 | 0.23 | G2_M |
| A_24_P362646 | TXNDC9 | NM_005783 | 10190 | −0.32 | −0.08 | 0.24 | 0.18 | G2_M |
| A_23_P204581 | TXNRD1 | NM_003330 | 7296 | 0.59 | 0.55 | −0.04 | 0.70 | G2_M |
| A_23_P40989 | USP13 | NM_003940 | 8975 | −0.40 | −0.38 | 0.03 | −0.37 | G2_M |
| A_23_P257911 | USP16 | NM_001032410 | 10600 | −0.34 | −0.07 | 0.27 | 0.58 | G2_M |
| A_24_P199655 | VANGL1 | NM_138959 | 81839 | −0.63 | 0.04 | 0.66 | −0.93 | G2_M |
| A_23_P103795 | VANGL1 | NM_138959 | 81839 | −0.30 | 0.26 | 0.57 | −0.97 | G2_M |
| A_23_P369316 | VANGL1 | NM_138959 | 81839 | −0.17 | 0.00 | 0.18 | −0.46 | G2_M |
| A_23_P103070 | YWHAH | NM_003405 | 7533 | −0.19 | −0.35 | −0.16 | −0.35 | G2_M |
| A_23_P215088 | ZC3HC1 | NM_016478 | 51530 | 0.02 | 0.09 | 0.07 | 0.32 | G2_M |
| A_24_P290527 | ZFX | NM_003410 | 7543 | −0.28 | −0.08 | 0.21 | 0.18 | G2_M |
| A_23_P125639 | ZFX | NM_003410 | 7543 | 0.36 | 0.08 | −0.28 | 0.75 | G2_M |
| A_24_P940524 | ZFX | NM_003410 | 7543 | −0.16 | 0.13 | 0.29 | −0.31 | G2_M |
| A_23_P161091 | ZMYM1 | NM_024772 | 79830 | −0.12 | −0.30 | −0.18 | 0.27 | G2_M |
| A_24_P53985 | ZMYM1 | NM_024772 | 79830 | −0.10 | −0.25 | −0.15 | 0.38 | G2_M |
| A_23_P159027 | ZNF521 | NM_015461 | 25925 | −0.33 | 0.08 | 0.41 | −0.63 | G2_M |
| A_23_P78018 | ABCA5 | NM_018672 | 23461 | −0.15 | −0.02 | 0.13 | −0.05 | S |
| A_24_P67096 | ABCA5 | NM_018672 | 23461 | −0.15 | −0.01 | 0.14 | −0.10 | S |
| A_23_P158976 | ABCC2 | NM_000392 | 1244 | 0.45 | 0.07 | −0.38 | 0.36 | S |
| A_23_P44569 | ABCC2 | NM_000392 | 1244 | 0.12 | 0.14 | 0.02 | 0.26 | S |
| A_23_P212665 | ABCC5 | NM_005688 | 10057 | 0.29 | −0.21 | −0.50 | −0.14 | S |
| A_23_P258221 | ABCC5 | NM_005688 | 10057 | −0.08 | 0.07 | 0.15 | −0.03 | S |
| A_24_P268662 | ABHD10 | NM_018394 | 55347 | −0.37 | 0.38 | 0.75 | −0.08 | S |
| A_23_P92213 | ABHD10 | NM_018394 | 55347 | −0.18 | −0.37 | −0.19 | 0.56 | S |
| A_24_P308590 | ABHD10 | NM_018394 | 55347 | 0.11 | −0.07 | −0.19 | 0.04 | S |
| A_23_P23630 | ACAP3 | NM_030649 | 116983 | −0.69 | −0.34 | 0.35 | −0.14 | S |
| A_23_P23625 | ACAP3 | NM_030649 | 116983 | −0.61 | −0.11 | 0.50 | −0.52 | S |
| A_23_P12231 | ACAP3 | NM_030649 | 116983 | 0.26 | −0.45 | −0.71 | −0.20 | S |
| A_24_P281497 | ACAP3 | NM_030649 | 116983 | −0.15 | 0.41 | 0.55 | −0.17 | S |
| A_24_P355006 | ADAM22 | ENST00000398204 | 53616 | −0.54 | −0.45 | 0.09 | −0.53 | S |
| A_23_P215625 | ADAM22 | NM_021723 | 53616 | −0.37 | −0.61 | −0.24 | −0.10 | S |
| A_24_P243741 | ADAM22 | NM_021721 | 53616 | 0.22 | 0.27 | 0.06 | −0.17 | S |
| A_24_P203630 | ANKRD36 | NM_001164315 | 375248 | 0.93 | 0.78 | −0.15 | 0.57 | S |
| A_24_P6725 | ANKRD36 | NM_001164315 | 375248 | 0.78 | 0.66 | −0.12 | 0.32 | S |
| A_24_P686992 | ANKRD36 | NM_001164315 | 375248 | 0.67 | 0.50 | −0.17 | 0.58 | S |
| A_24_P336931 | ANKRD36 | NM_001164315 | 375248 | −0.02 | −0.39 | −0.36 | 0.53 | S |
| A_23_P119254 | ASF1B | NM_018154 | 55723 | −2.77 | −2.37 | 0.40 | −1.54 | S |
| A_23_P120629 | ASIP | NM_001672 | 434 | 0.04 | −0.21 | −0.25 | 0.38 | S |
| A_23_P106835 | BBS2 | NM_031885 | 583 | −0.11 | −0.25 | −0.14 | 0.10 | S |
| A_23_P105833 | BIVM | NM_017693 | 54841 | −0.28 | 0.14 | 0.43 | −0.75 | S |
| A_23_P88630 | BLM | NM_000057 | 641 | −2.28 | −1.89 | 0.39 | −0.83 | S |
| A_24_P303989 | BMI1 | NM_005180 | 648 | −0.26 | −0.03 | 0.23 | 0.05 | S |
| A_23_P314115 | BMI1 | NM_005180 | 648 | −0.12 | −0.01 | 0.10 | 0.02 | S |
| A_23_P207400 | BRCA1 | NM_007300 | 672 | −1.00 | −0.91 | 0.08 | −0.65 | S |
| A_23_P15844 | BRIP1 | NM_032043 | 83990 | −1.30 | −1.16 | 0.14 | 0.07 | S |
| A_24_P255524 | CALD1 | AF247820 | 800 | 0.54 | 0.69 | 0.15 | 0.28 | S |
| A_24_P921366 | CALD1 | NM_033138 | 800 | −0.51 | −0.17 | 0.34 | −0.79 | S |
| A_23_P42575 | CALD1 | NM_033138 | 800 | −0.33 | 0.02 | 0.35 | −0.35 | S |
| A_24_P313993 | CAPS | NM_004058 | 828 | −0.42 | 0.14 | 0.56 | −0.19 | S |
| A_23_P78958 | CAPS | NM_004058 | 828 | −0.22 | 0.44 | 0.66 | −0.43 | S |
| A_23_P384056 | CCDC14 | NM_022757 | 64770 | −0.59 | −0.46 | 0.13 | 0.41 | S |
| A_23_P39574 | CCDC150 | NM_001080539 | 284992 | −1.38 | −1.34 | 0.04 | 0.15 | S |
| A_24_P157156 | CCDC150 | NM_001080539 | 284992 | −0.47 | −0.52 | −0.05 | −0.24 | S |
| A_23_P320190 | CCDC150 | A_23_P320190 | NA | −0.34 | −0.24 | 0.10 | −0.53 | S |
| A_24_P636332 | CCDC84 | NM_198489 | 338657 | 0.17 | 0.21 | 0.04 | 0.76 | S |
| A_24_P693946 | CCDC84 | A_24_P693946 | NA | 0.05 | −0.01 | −0.06 | 0.62 | S |
| A_23_P57379 | CDC45 | NM_003504 | 8318 | −3.57 | −2.37 | 1.20 | −1.90 | S |
| A_23_P148807 | CDC7 | NM_003503 | 8317 | −0.70 | −0.50 | 0.20 | −0.07 | S |
| A_23_P104651 | CDCA5 | NM_080668 | 113130 | −2.58 | −2.15 | 0.42 | −1.16 | S |
| A_23_P405267 | CDH24 | AK057922 | 64403 | 1.03 | −0.05 | −1.08 | 1.04 | S |
| A_23_P25790 | CDH24 | NM_022478 | 64403 | 0.26 | −0.37 | −0.62 | 0.19 | S |
| A_23_P258002 | CDKN2AIP | NM_017632 | 55602 | 0.18 | −0.17 | −0.35 | 0.51 | S |
| A_24_P399888 | CENPM | NM_001002876 | 79019 | −2.65 | −2.27 | 0.38 | −0.97 | S |
| A_23_P70328 | CENPQ | NM_018132 | 55166 | −0.18 | −0.36 | −0.18 | 0.41 | S |
| A_23_P254733 | CENPU | NM_024629 | 79682 | −1.79 | −1.85 | −0.06 | −0.23 | S |
| A_24_P289366 | CERS6 | NM_203463 | 253782 | 0.10 | 0.18 | 0.08 | −0.46 | S |
| A_32_P5480 | CERS6 | NM_203463 | 253782 | −0.08 | 0.35 | 0.43 | −0.45 | S |
| A_23_P144071 | COL7A1 | NM_000094 | 1294 | −0.17 | 0.28 | 0.46 | −0.03 | S |
| A_24_P932308 | COQ9 | AK075438 | 57017 | 0.90 | 0.78 | −0.12 | 1.05 | S |
| A_23_P14928 | COQ9 | NM_020312 | 57017 | −0.08 | 0.22 | 0.31 | −0.15 | S |
| A_23_P87556 | CPNE8 | NM_153634 | 144402 | −0.23 | 0.33 | 0.56 | −0.20 | S |
| A_24_P56240 | CPNE8 | NM_153634 | 144402 | 0.05 | 0.58 | 0.53 | 0.06 | S |
| A_23_P144438 | DCAF16 | NM_017741 | 54876 | 0.31 | 0.11 | −0.20 | 0.31 | S |
| A_32_P104000 | DCUN1D3 | NM_173475 | 123879 | −0.18 | 0.11 | 0.30 | 0.02 | S |
| A_23_P429491 | DDIAS | NM_145018 | 220042 | −1.24 | −1.00 | 0.24 | −0.25 | S |
| A_24_P926543 | DDIAS | AK058145 | 220042 | 0.41 | 0.29 | −0.12 | −0.14 | S |
| A_23_P385126 | DEPDC7 | NM_139160 | 91614 | 0.99 | 0.06 | −0.93 | 1.16 | S |
| A_24_P320284 | DHFR | NM_000791 | 1719 | −1.51 | −1.20 | 0.32 | −0.38 | S |
| A_24_P942328 | DHFR | NM_000791 | 1719 | −1.91 | −1.52 | 0.39 | −1.10 | S |
| A_32_P211045 | DHFR | NM_000791 | 1719 | −2.16 | −1.40 | 0.76 | −1.27 | S |
| A_23_P167553 | DHFR | NM_000791 | 1719 | −1.58 | −1.13 | 0.45 | −0.59 | S |
| A_24_P343095 | DHFR | NM_000791 | 1719 | −1.26 | −0.87 | 0.39 | −0.36 | S |
| A_23_P327361 | DMXL2 | NM_015263 | 23312 | 0.07 | −0.14 | −0.21 | −0.02 | S |
| A_24_P366107 | DNA2 | NM_001080449 | 1763 | −0.86 | −0.69 | 0.17 | −0.56 | S |
| A_23_P51339 | DNAJB4 | NM_007034 | 11080 | 1.16 | −0.02 | −1.18 | 0.98 | S |
| A_24_P393958 | DNAJB4 | NM_007034 | 11080 | 1.11 | −0.10 | −1.21 | 1.23 | S |
| A_23_P95359 | DNAJC6 | NM_014787 | 9829 | −0.44 | −0.40 | 0.03 | −0.41 | S |
| A_23_P147479 | DNAJC6 | NM_014787 | 9829 | 0.35 | −0.02 | −0.38 | 0.58 | S |
| A_23_P500390 | DONSON | NM_017613 | 29980 | 0.52 | −0.11 | −0.64 | 1.07 | S |
| A_23_P425502 | DONSON | NM_017613 | 29980 | 0.30 | −0.34 | −0.64 | 0.96 | S |
| A_23_P35871 | E2F8 | NM_024680 | 79733 | −0.76 | −0.35 | 0.41 | −0.23 | S |
| A_23_P214291 | EFHC1 | NM_018100 | 114327 | −0.37 | −0.45 | −0.08 | 0.72 | S |
| A_32_P86245 | EFHC1 | NM_018100 | 114327 | −0.31 | −0.07 | 0.24 | −0.10 | S |
| A_24_P913374 | EIF4EBP2 | NM_004096 | 1979 | −0.66 | −0.29 | 0.36 | −1.22 | S |
| A_24_P4387 | EIF4EBP2 | NM_004096 | 1979 | −0.42 | −0.08 | 0.34 | −0.58 | S |
| A_24_P115621 | EIF4EBP2 | NM_004096 | 1979 | −0.25 | −0.04 | 0.21 | −0.90 | S |
| A_23_P115922 | EIF4EBP2 | NM_004096 | 1979 | −0.22 | 0.01 | 0.24 | −0.47 | S |
| A_24_P323598 | ESCO2 | NM_001017420 | 157570 | −2.00 | −1.95 | 0.05 | −0.79 | S |
| A_23_P23303 | EXO1 | NM_003686 | 9156 | −2.11 | −1.68 | 0.42 | −1.23 | S |
| A_23_P259641 | EZH2 | NM_004456 | 2146 | −0.74 | −0.91 | −0.16 | 0.13 | S |
| A_23_P99853 | FAM214A | NM_019600 | 56204 | 0.25 | 0.05 | −0.20 | −0.03 | S |
| A_24_P357576 | FAM214A | NM_019600 | 56204 | 0.17 | 0.09 | −0.08 | −0.27 | S |
| A_23_P206441 | FANCA | NM_000135 | 2175 | −0.49 | −0.42 | 0.07 | −0.58 | S |
| A_24_P73158 | FEN1 | NM_004111 | 2237 | −1.16 | −1.11 | 0.04 | −0.36 | S |
| A_24_P84898 | FEN1 | NM_004111 | 2237 | −1.27 | −1.29 | −0.02 | −0.33 | S |
| A_23_P103996 | GCLM | NM_002061 | 2730 | 0.55 | 1.42 | 0.87 | −0.32 | S |
| A_32_P177953 | GCLM | ENST00000370238 | 2730 | 0.05 | 1.10 | 1.05 | −0.67 | S |
| A_32_P2392 | GOLGA8A | NM_181077 | 23015 | 0.91 | 0.31 | −0.60 | 0.66 | S |
| A_23_P37623 | GOLGA8A | NM_181077 | 23015 | 0.54 | 0.52 | −0.02 | −0.12 | S |
| A_24_P910580 | GOLGA8A | NR_027409 | 23015 | 0.50 | 0.47 | −0.03 | 0.73 | S |
| A_23_P29257 | H1F0 | NM_005318 | 3005 | 0.40 | 0.02 | −0.39 | −0.58 | S |
| A_23_P349771 | HAUS5 | NM_015302 | 23354 | 0.41 | 0.89 | 0.48 | −0.01 | S |
| A_23_P12816 | HELLS | NM_018063 | 3070 | −0.95 | −1.12 | −0.17 | −0.02 | S |
| A_23_P372860 | HIST1H2AC | NM_003512 | 8334 | −0.38 | −0.57 | −0.19 | 0.76 | S |
| A_23_P167983 | HIST1H2AC | ENST00000314088 | 8334 | −0.22 | −0.24 | −0.02 | −0.03 | S |
| A_32_P221799 | HIST1H2AM | NM_003514 | 8336 | −1.05 | −1.41 | −0.36 | 0.21 | S |
| A_24_P86389 | HIST1H2AM | NM_003514 | 8336 | 0.10 | −0.07 | −0.17 | 0.83 | S |
| A_23_P93180 | HIST1H2BC | NM_003526 | 8347 | −0.04 | −0.26 | −0.22 | 0.90 | S |
| A_24_P166407 | HIST1H4B | NM_003544 | 8366 | −0.91 | −0.89 | 0.02 | −0.16 | S |
| A_23_P214487 | HIST1H4C | NM_003542 | 8364 | −1.27 | −1.00 | 0.27 | −0.66 | S |
| A_23_P323685 | HIST1H4H | NM_003543 | 8365 | −0.31 | −0.82 | −0.51 | 0.73 | S |
| A_23_P52266 | IFIT1 | NM_001548 | 3434 | 1.72 | 0.64 | −1.08 | 1.80 | S |
| A_23_P102454 | INSIG2 | NM_016133 | 51141 | 0.53 | 0.07 | −0.46 | 1.13 | S |
| A_24_P944458 | INSIG2 | NM_016133 | 51141 | −0.30 | −0.67 | −0.37 | 0.31 | S |
| A_23_P52082 | INTS7 | NM_015434 | 25896 | −0.49 | 0.16 | 0.64 | −0.44 | S |
| A_32_P159651 | KAT2B | NM_003884 | 8850 | −0.66 | −0.62 | 0.04 | −0.51 | S |
| A_23_P41128 | KAT2B | NM_003884 | 8850 | −0.38 | −0.33 | 0.05 | −0.36 | S |
| A_23_P358542 | KIFC2 | NM_145754 | 90990 | −0.21 | −0.01 | 0.20 | 0.33 | S |
| A_23_P165414 | KLHL23 | NM_144711 | 151230 | −0.63 | −0.12 | 0.51 | −1.29 | S |
| A_24_P923102 | KLHL23 | ENST00000392647 | 151230 | −0.62 | −0.02 | 0.60 | −0.81 | S |
| A_23_P165408 | KLHL23 | NM_144711 | 151230 | −0.70 | −0.36 | 0.34 | −1.03 | S |
| A_23_P74252 | LINC00339 | NR_023918 | 29092 | −0.41 | −0.62 | −0.20 | 0.04 | S |
| A_23_P84219 | LIPH | NM_139248 | 200879 | −0.15 | −0.11 | 0.04 | 0.38 | S |
| A_24_P799858 | LIPH | ENST00000296252 | 200879 | −0.03 | 0.14 | 0.17 | −0.11 | S |
| A_23_P380181 | LMO4 | NM_006769 | 8543 | −0.36 | −0.07 | 0.30 | −0.34 | S |
| A_32_P18159 | LYRM7 | NM_181705 | 90624 | 0.41 | 0.17 | −0.23 | −0.18 | S |
| A_32_P211141 | LYRM7 | NM_181705 | 90624 | 0.26 | 0.44 | 0.18 | 0.07 | S |
| A_24_P256603 | LYRM7 | NM_181705 | 90624 | 0.20 | 0.32 | 0.13 | 0.16 | S |
| A_23_P337464 | LYRM7 | NM_181705 | 90624 | −0.23 | 0.02 | 0.25 | −0.66 | S |
| A_24_P926195 | MAN1A2 | NM_006699 | 10905 | −0.52 | −0.01 | 0.51 | −0.55 | S |
| A_23_P103571 | MAN1A2 | NM_006699 | 10905 | −0.48 | 0.14 | 0.62 | −0.21 | S |
| A_32_P88603 | MAN1A2 | ENST00000356554 | 10905 | −0.49 | 0.09 | 0.59 | −0.78 | S |
| A_32_P88598 | MAN1A2 | ENST00000356554 | 10905 | −0.42 | 0.25 | 0.66 | −0.72 | S |
| A_24_P213548 | MAN1A2 | NM_006699 | 10905 | −0.11 | 0.41 | 0.52 | 0.02 | S |
| A_23_P313640 | MAP3K2 | NM_006609 | 10746 | 0.60 | 0.45 | −0.15 | 0.16 | S |
| A_32_P98887 | MAP3K2 | ENST00000409947 | 10746 | 0.42 | −0.11 | −0.53 | 0.32 | S |
| A_23_P313645 | MAP3K2 | NM_006609 | 10746 | 0.23 | 0.24 | 0.01 | 0.40 | S |
| A_23_P201988 | MASTL | NM_032844 | 84930 | −0.45 | −0.86 | −0.41 | 0.36 | S |
| A_24_P258051 | MASTL | NM_032844 | 84930 | −0.16 | −0.38 | −0.22 | 0.06 | S |
| A_23_P92154 | MBD4 | NM_003925 | 8930 | −0.37 | −0.14 | 0.24 | 0.14 | S |
| A_23_P68547 | MCM8 | NM_182802 | 84515 | −0.81 | −0.49 | 0.32 | −0.43 | S |
| A_24_P305556 | MCM8 | NM_182802 | 84515 | −0.50 | −0.48 | 0.03 | −0.09 | S |
| A_32_P129894 | MEGF9 | NM_001080497 | 1955 | −0.12 | 0.13 | 0.25 | −1.13 | S |
| A_23_P426663 | MITF | NM_198159 | 4286 | −0.17 | −0.15 | 0.02 | −0.04 | S |
| A_23_P73345 | MITF | NM_198159 | 4286 | 0.26 | 0.19 | −0.07 | 1.01 | S |
| A_24_P910310 | MITF | NM_198177 | 4286 | 0.14 | −0.39 | −0.53 | 0.72 | S |
| A_23_P61945 | MITF | NM_198159 | 4286 | −0.03 | 0.11 | 0.13 | −0.07 | S |
| A_24_P323815 | MYCBP2 | NM_015057 | 23077 | −0.48 | −0.28 | 0.20 | −0.08 | S |
| A_23_P151459 | MYCBP2 | NM_015057 | 23077 | 0.04 | 0.56 | 0.52 | −0.46 | S |
| A_23_P209805 | NAB1 | NM_005966 | 4664 | 0.67 | 0.61 | −0.06 | 1.30 | S |
| A_24_P191417 | NAB1 | NM_005966 | 4664 | 0.01 | 0.29 | 0.28 | 0.47 | S |
| A_23_P5761 | NFE2L2 | NM_006164 | 4780 | 0.04 | 0.26 | 0.22 | −0.14 | S |
| A_23_P23006 | NRDC | NM_002525 | 4898 | −0.42 | −0.16 | 0.27 | −0.07 | S |
| A_32_P213822 | NSUN3 | ENST00000314622 | 63899 | −0.66 | −0.31 | 0.36 | −0.19 | S |
| A_23_P21785 | NSUN3 | NM_022072 | 63899 | −0.08 | −0.10 | −0.01 | 0.55 | S |
| A_23_P382043 | NT5DC1 | NM_152729 | 221294 | −0.40 | −0.15 | 0.25 | −0.31 | S |
| A_23_P219004 | NT5DC1 | NM_152729 | 221294 | 0.14 | 0.15 | 0.01 | 0.37 | S |
| A_24_P922606 | NUP160 | NM_015231 | 23279 | 0.83 | 0.11 | −0.72 | 0.83 | S |
| A_23_P43726 | NUP160 | NM_015231 | 23279 | −0.06 | 0.06 | 0.12 | 0.15 | S |
| A_23_P381979 | OGT | NM_181672 | 8473 | −0.25 | −0.44 | −0.19 | 0.06 | S |
| A_23_P381976 | OGT | NM_181672 | 8473 | −0.17 | −0.08 | 0.09 | 0.12 | S |
| A_23_P42045 | ORC3 | NM_181837 | 23595 | −0.10 | −0.60 | −0.50 | 0.66 | S |
| A_23_P79818 | OSER1 | NM_016470 | 51526 | 0.27 | −0.36 | −0.63 | 0.95 | S |
| A_24_P261083 | OSGIN2 | NM_004337 | 734 | 0.89 | 0.41 | −0.47 | 0.84 | S |
| A_23_P82859 | OSGIN2 | NM_004337 | 734 | −0.01 | −0.06 | −0.05 | 0.18 | S |
| A_23_P117852 | PCLAF | NM_014736 | 9768 | −3.04 | −2.31 | 0.73 | −0.87 | S |
| A_32_P61339 | PHIP | BC036479 | 55023 | −0.57 | −1.00 | −0.43 | −0.08 | S |
| A_24_P196400 | PHIP | NM_017934 | 55023 | −0.36 | −0.34 | 0.01 | 0.15 | S |
| A_23_P145437 | PHIP | NM_017934 | 55023 | −0.18 | −0.09 | 0.09 | 0.20 | S |
| A_24_P630039 | PHIP | NM_017934 | 55023 | −0.22 | −0.03 | 0.20 | −0.13 | S |
| A_24_P931503 | PHIP | NM_017934 | 55023 | −0.21 | −0.12 | 0.09 | −0.61 | S |
| A_24_P175176 | PHTF2 | NM_020432 | 57157 | −0.76 | −0.05 | 0.70 | −0.58 | S |
| A_32_P409919 | PHTF2 | NM_020432 | 57157 | 0.57 | 0.05 | −0.52 | 0.50 | S |
| A_24_P323944 | PHTF2 | NM_020432 | 57157 | 0.08 | −0.05 | −0.13 | 0.21 | S |
| A_24_P403244 | PILRB | NM_013440 | 29990 | −0.31 | 0.08 | 0.38 | 0.15 | S |
| A_23_P19829 | PILRB | NM_013440 | 29990 | −0.32 | 0.06 | 0.38 | 0.08 | S |
| A_24_P105102 | PKMYT1 | NM_182687 | 9088 | −1.77 | −1.36 | 0.41 | −1.22 | S |
| A_23_P398515 | PKMYT1 | NM_004203 | 9088 | 0.12 | 0.19 | 0.07 | −0.19 | S |
| A_23_P25019 | PRIM1 | NM_000946 | 5557 | −1.04 | −1.21 | −0.17 | −0.32 | S |
| A_23_P44139 | PRIM2 | NM_000947 | 5558 | −0.53 | −0.21 | 0.32 | −0.16 | S |
| A_24_P282237 | PRIM2 | NM_000947 | 5558 | −0.33 | −0.25 | 0.08 | −0.17 | S |
| A_24_P75158 | PTAR1 | NM_001099666 | 375743 | −0.36 | −0.37 | 0.00 | 0.26 | S |
| A_23_P121222 | RAD18 | NM_020165 | 56852 | −0.58 | −0.47 | 0.11 | −0.13 | S |
| A_23_P88731 | RAD51 | NM_002875 | 5888 | −1.73 | −1.58 | 0.15 | −1.02 | S |
| A_23_P99292 | RAD51AP1 | NM_006479 | 10635 | −2.35 | −2.26 | 0.08 | −1.01 | S |
| A_23_P74115 | RAD54L | NM_003579 | 8438 | −2.14 | −1.79 | 0.34 | −0.94 | S |
| A_23_P252371 | RBBP8 | NM_002894 | 5932 | −0.40 | −0.41 | −0.01 | 0.30 | S |
| A_23_P96285 | REEP1 | NM_022912 | 65055 | −2.14 | −1.68 | 0.45 | −1.95 | S |
| A_23_P93823 | RFC2 | NM_181471 | 5982 | −0.45 | −0.33 | 0.12 | −0.47 | S |
| A_23_P18196 | RFC4 | NM_002916 | 5984 | −0.84 | −0.88 | −0.04 | −0.04 | S |
| A_23_P92710 | RHOBTB3 | NM_014899 | 22836 | 0.42 | 0.87 | 0.46 | −0.96 | S |
| A_23_P315386 | RHPN1 | NM_052924 | 114822 | −0.19 | −0.28 | −0.09 | 0.50 | S |
| A_23_P87432 | RHPN1 | NM_052924 | 114822 | 0.22 | −0.57 | −0.79 | 0.73 | S |
| A_23_P87435 | RHPN1 | NM_052924 | 114822 | −0.14 | −0.25 | −0.11 | −0.38 | S |
| A_23_P86133 | RPA2 | NM_002946 | 6118 | −0.52 | −0.77 | −0.25 | 0.22 | S |
| A_23_P87351 | RRM1 | NM_001033 | 6240 | −0.73 | −0.81 | −0.08 | 0.31 | S |
| A_24_P234196 | RRM2 | NM_001034 | 6241 | −3.49 | −2.56 | 0.93 | −1.85 | S |
| A_24_P225616 | RRM2 | NM_001034 | 6241 | −1.98 | −1.54 | 0.44 | −1.50 | S |
| A_24_P350160 | RSRC2 | NM_198261 | 65117 | 0.43 | 0.09 | −0.34 | 1.05 | S |
| A_23_P53267 | RSRC2 | NM_198261 | 65117 | −0.16 | −0.52 | −0.36 | 0.70 | S |
| A_24_P304987 | SAP30BP | NM_013260 | 29115 | −0.32 | −0.05 | 0.27 | −0.13 | S |
| A_23_P54953 | SAP30BP | NM_013260 | 29115 | −0.23 | −0.44 | −0.21 | 0.26 | S |
| A_23_P169351 | SH3GL2 | NM_003026 | 6456 | 0.07 | −0.50 | −0.57 | −0.13 | S |
| A_23_P200443 | SHC1 | NM_003029 | 6464 | 0.29 | 0.36 | 0.07 | −0.42 | S |
| A_24_P68585 | SHC1 | NM_183001 | 6464 | 0.06 | 0.14 | 0.08 | −0.06 | S |
| A_23_P202587 | SHTN1 | NM_018330 | 57698 | −0.27 | 0.15 | 0.42 | −0.46 | S |
| A_32_P309404 | SLC22A3 | NM_021977 | 6581 | −0.45 | 0.30 | 0.75 | −0.81 | S |
| A_23_P19733 | SLC22A3 | NM_021977 | 6581 | 0.20 | 0.72 | 0.52 | 0.12 | S |
| A_24_P246841 | SLC25A27 | NM_004277 | 9481 | 0.40 | 0.38 | −0.03 | 0.84 | S |
| A_23_P81721 | SLC25A27 | NM_004277 | 9481 | 0.26 | 0.25 | −0.01 | 0.57 | S |
| A_23_P218079 | SLC38A2 | NM_018976 | 54407 | 0.24 | 0.33 | 0.09 | 1.24 | S |
| A_24_P295963 | SLC38A2 | NM_018976 | 54407 | −0.09 | 0.13 | 0.22 | 0.23 | S |
| A_23_P104282 | SLF2 | NM_018121 | 55719 | −0.37 | −0.42 | −0.05 | −0.26 | S |
| A_32_P143516 | SLF2 | NM_018121 | 55719 | −0.14 | −0.16 | −0.02 | 0.17 | S |
| A_23_P356139 | SLF2 | NM_018121 | 55719 | −0.12 | 0.13 | 0.25 | −0.62 | S |
| A_23_P366312 | SP1 | NM_138473 | 6667 | 0.39 | 0.48 | 0.09 | −0.14 | S |
| A_23_P53397 | SP1 | NM_138473 | 6667 | 0.23 | 0.09 | −0.15 | −0.01 | S |
| A_32_P45493 | SRSF10 | NM_006625 | 10772 | −0.79 | −0.49 | 0.30 | 0.03 | S |
| A_23_P45737 | SRSF10 | NM_006625 | 10772 | −0.43 | −0.30 | 0.13 | 0.21 | S |
| A_23_P352291 | SRSF10 | NM_054016 | 10772 | −0.20 | −0.18 | 0.01 | −0.41 | S |
| A_24_P4795 | SRSF10 | NM_054016 | 10772 | −0.19 | −0.09 | 0.10 | 0.57 | S |
| A_23_P377819 | SRSF5 | NM_001039465 | 6430 | −0.29 | −0.21 | 0.08 | −0.40 | S |
| A_32_P45894 | STAG3L1 | NM_018991 | 54441 | 0.17 | 0.06 | −0.12 | 0.53 | S |
| A_24_P374962 | STAG3L1 | NM_018991 | 54441 | 0.05 | 0.20 | 0.15 | 0.21 | S |
| A_24_P111242 | SVIP | NM_148893 | 258010 | −0.53 | −0.55 | −0.03 | 0.11 | S |
| A_32_P41070 | TMCC1 | NM_015008 | 23023 | −0.10 | 0.09 | 0.20 | −0.29 | S |
| A_32_P41065 | TMCC1 | NM_001017395 | 23023 | −0.08 | −0.15 | −0.08 | −0.17 | S |
| A_24_P922288 | TMCC1 | NM_001017395 | 23023 | 0.08 | −0.29 | −0.37 | 0.60 | S |
| A_23_P170986 | TMCC1 | NM_001017395 | 23023 | −0.01 | −0.12 | −0.11 | 0.40 | S |
| A_23_P39813 | TTC31 | NM_022492 | 64427 | −0.34 | 0.04 | 0.38 | −0.46 | S |
| A_32_P204169 | TTLL7 | ENST00000260505 | 79739 | 0.29 | 0.86 | 0.57 | −0.42 | S |
| A_23_P97481 | TTLL7 | NM_024686 | 79739 | −0.18 | −0.38 | −0.21 | 0.08 | S |
| A_24_P165450 | TTLL7 | NM_024686 | 79739 | 0.02 | −0.02 | −0.03 | 0.15 | S |
| A_23_P50096 | TYMS | NM_001071 | 7298 | −1.55 | −1.70 | −0.15 | −0.34 | S |
| A_23_P115482 | UBE2T | NM_014176 | 29089 | −1.18 | −0.89 | 0.29 | 0.14 | S |
| A_24_P330234 | UBL3 | NM_007106 | 5412 | −0.30 | −0.27 | 0.03 | 0.04 | S |
| A_23_P140029 | UBL3 | NM_007106 | 5412 | 0.23 | 0.04 | −0.19 | 0.04 | S |
| A_23_P11652 | USP1 | NM_003368 | 7398 | −0.44 | −0.67 | −0.23 | 0.22 | S |
| A_23_P98483 | ZBED5 | NM_021211 | 58486 | −0.41 | −0.22 | 0.19 | 0.20 | S |
| A_23_P210608 | ZNF217 | NM_006526 | 7764 | 0.02 | −0.28 | −0.30 | 0.05 | S |
| A_23_P63789 | ZWINT | NM_032997 | 11130 | −2.88 | −2.07 | 0.82 | −1.15 | S |
From Whitfield et al. (58).
Phases: G1, G1; G2, G2; G2_M, G2/M; G2,G2_M, G2 or G2/M, etc.
Intoxication with Salmonella supernatant containing CdtB induces DNA damage not coupled with cell cycle arrest.
The majority of chronically infected carriers of typhoid Salmonella are usually diagnosed with gallstones and it has been found that Salmonella is able to grow on them forming biofilms. Salmonella covered gallstones might represent a reservoir for the bacteria but also a potential source of typhoid toxin (60). To understand the effect of the typhoid toxin on primary gallbladder cells, we sought to achieve a homogeneous typhoid toxin intoxication. To this aim, we seeded organoid-derived cells as 2D monolayers on collagen-coated plastic wells and supplemented them for 24 h with supernatant from Salmonella grown in MM5.8 medium, which is known to stimulate the production of typhoid toxin (19). Western blot analysis confirmed that only treatment with wild-type supernatant and etoposide, a chemical inducer of DSBs (61), resulted in an increased phosphorylation of H2AX, which is indicative of the presence of DSBs (Fig. 5A). The amount of DSBs was quantified using a neutral comet assay, which showed a significant increase in DNA in the tail of the comet analyzed from cells treated with supernatant from wild-type bacteria compared to supernatant from the ΔcdtB Salmonella or sterile medium (Fig. 5B, quantified in Fig. 5C). Cells treated with a genotoxic agent normally respond by arresting the cell cycle, and this was also reported for cells treated with recombinant CDT, a bacterial toxin that shares the CdtB subunit with the typhoid toxin (62). To examine whether cell cycle arrest was also induced in our intoxication model, we fluorescently labeled cells with antibodies against γH2AX and Ki67, a marker of proliferating cells. There was no difference in the percentage of Ki67+ cells in cultures treated with sterile, deletion mutant or wild-type supernatants (Fig. 5D, quantified in Fig. 5E). In stark contrast, cultures treated with etoposide contained little to no Ki67+ cells, although etoposide caused an amount of damage similar to that caused by the supernatant conditioned with wild-type typhoid toxin (Fig. 5A to C). Analysis of the γH2AX signal intensity in Ki67+ versus Ki67– cells showed that supernatant conditioned with typhoid toxin induced more DNA damage, especially in proliferating cells (Fig. 5D and F). Cells intoxicated with ΔcdtB supernatant showed non-significant differences in the distribution of γH2AX intensities compared to sterile medium, both in proliferating and non-proliferating cells. The presence of cells positive for both γH2AX and Ki67 was detected only after intoxication with the wild-type typhoid toxin, and this observation occurred up to 48 h after intoxication started (see Fig. S3A and B). Finally, not only intoxicated cells but also rare infected cells were positive for γH2AX but still in an active state of proliferation, as indicated by double labeling with a Ki67 antibody (Fig. 5G). Our data show that human primary gallbladder cells are subjected to a low but persistent level of DNA damage caused by the CdtB subunit of the S. Paratyphi A-encoded typhoid toxin. The DNA damage caused by the genotoxin does not induce cell cycle arrest but particularly affects proliferating cells.
FIG 5.
Intoxication of primary cell 2D monolayers with typhoid toxin-containing Salmonella supernatant. (A) Western blot analysis of γH2AX levels in primary cells after exposure to Salmonella supernatant or etoposide for 24 h. Relative densitometry values, normalized to the sterile medium condition (=1), are shown above the bands. (B) Comet assay showing that DNA damage, seen as a tail of DNA after electrophoresis, is higher after exposure to supernatant from w.t. Salmonella than from the cdtB deletion mutant. Etoposide served as a positive control. Pictures of two representative nuclei per condition are shown. (C) Quantification of the comet assay, shown as means ± the SEM. ****, P < 0.0001. (D) Immunofluorescence analysis of cells intoxicated or treated with etoposide for 24 h with antibodies against γH2AX (green) and Ki67 (red); nuclei were stained with Hoechst (blue). Scale bar, 25 μm. (E) Quantification of Ki67+ cells in intoxicated cells. Unlike cells treated with etoposide, cells intoxicated with w.t. Salmonella supernatant do not stop proliferation despite the presence of DNA DSBs. (F) Quantification of DNA damage in proliferating and non-proliferating cells. The intensity of the γH2AX signal was quantified for each Ki67 positive and negative nucleus, using ImageJ. Data shown as means ± the SEM. ***, P < 0.001; ****, P < 0.0001. (G) Primary cell monolayers infected for 3 days with Salmonella Paratyphi A transformed with the mCherry expressing vector pLS002 (red) and fluorescently labeled with antibodies against γH2AX (green) and Ki67 (white); nuclei were labeled with Hoechst (blue). Scale bar, 10 μm.
Long-term intoxication, 24 and 48 h. Human GB organoids were seeded in 2D and intoxicated for 24 or 48 h. For intoxication for 48 h, the bacterial supernatant was produced twice, and fresh supernatant was diluted in medium was added after 24 h. The cells seeded were less confluent than in normal 24-h intoxication experiments to avoid premature confluence of the culture. The figure shows double-positive cells for Ki67 and γH2AX at 24 h (A) and 48 h (B). N, the minimum number of counted cells per individual experiment. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Here, we present a long-lived organoid model for human and murine GB. We found that long-term maintenance of GB organoid cultures depends on the presence of R-spondin, which mediates the activation of the Wnt/β-catenin signaling pathway and the regeneration of the gallbladder epithelium from Lgr5+ cells. These results confirm a recent report from a murine organoid model, which showed that the addition of R-spondin and Noggin, but not of Wnt ligands, was necessary for the expansion of GB stem cells in vitro (31). Since Wnt ligands are crucial for the activation of the Wnt/β-catenin pathway, we have found that the epithelium is itself the source of secreted WNT7A/7B. Our data suggest, in addition, that the organoids are able to transport organic ions, emulating the concentration of bile typical of this organ, and that GB epithelial features are stable over time in culture.
Resembling the architecture of the organ in situ, this organoid model provides an advanced platform for investigating GB pathology in primary, non-transformed cells. We therefore used GB organoids to develop a novel infection model for the human-specific, cancer-associated bacterium S. Paratyphi A, focusing on the genotoxic effect of the typhoid toxin. Previous data have suggested that bacterial internalization is essential for the secretion of the typhoid toxin by the host cell (17, 19–21). Here, we confirm that by infecting healthy human GB cells with a wild-type strain that produces the typhoid toxin, the non-infected bystander cells experienced genomic instability. This paracrine DNA damage effect depended on the presence of the CdtB subunit, moving attention to non-infected but intoxicated cells as potential targets of cellular transformation.
It has been shown that Salmonella is able to promote cell division through activation of the AKT and mitogen-activated protein kinase (MAPK) pathways. S. Typhimurium, which lacks the typhoid toxin, is able to induce tumor growth in a genetically predisposed primary mouse fibroblast model (63). We previously suggested that chronic carriers, subjected to low levels of genotoxicity and DSBs for years, might develop a similar genetic predisposition (64). Together with the anti-apoptotic effects of Salmonella on host cells (65, 66) and persistent inflammation, the enhanced damage is likely to contribute to an increased risk of developing malignant mutations observed in chronic carriers.
By using the organoids a source of cells to develop mucosoid cultures (as previously done for the human stomach) (34), we could generate an advanced model for S. Paratyphi A chronic infection in vitro. The gene expression profile of long-term S. Paratyphi A-infected mucosoid revealed that infection induced an initial cell cycle arrest that did not depend on the DNA damage caused by the typhoid toxin. It has been reported that bacteria use particular cell cycle phases for their invasion or replication. For these reasons they are equipped with factors known as cyclomodulins. Salmonella is known to preferentially invade mitotic cells (67) and is equipped with diverse cyclomodulins, including SpvB and PheA, that induce cell cycle arrest at different phases of the cell cycle depending on the type of infected cell (68). The typhoid toxin and other CdtB containing toxins, such as the cytolethal distending toxins (CDT), are also cyclomodulins since the DNA damage that they induce is known to induce cell cycle arrest, typically at the G2/M checkpoint (69–72). In the physiological settings of the mucosoids and using S. Paratyphi A, a wild-type typhoid toxin-producing strain, we could again detect DNA damage. However, we could not observe any stronger or longer effect due to the typhoid toxin over other effectors in blocking the cell cycle.
To distinguish the effect of the typhoid toxin over other bacterial effectors, we intoxicated primary epithelial cells derived from the organoids with a functional typhoid toxin obtained from bacterial supernatants. Our data confirm that the DNA damage is due to the action of the CdtB subunit of the typhoid toxin, but we showed in addition that damaged cells failed to arrest their cell cycle, and cells with higher levels of damage actually maintained proliferation. Chronic exposure to sublethal levels of recombinant CDT was previously found to induce genomic instability and anchorage-independent growth in Big Blue rat fibroblasts (25), suggesting that the duration of the exposure rather than the dose of the toxin is the key element that increases the risk of cellular transformation. In chronic carriers, healthy GB cells might get intoxicated with the typhoid toxin from neighboring infected Salmonella Paratyphi A cells or from gallstones coated with the same bacterium. The secretion of the toxin is at such a level that does not impair the cell cycle but still provokes DNA damage.
Future investigations should seek to understand whether the typhoid toxin leaves a genetic mutational signature in the gallbladder, as has been observed for other cancer types that have a signature reflecting the original mutagenic insult (15, 73, 74). Such a signature would provide an important molecular link between Salmonella and associated GBC.
MATERIALS AND METHODS
Human organoid culture.
Human gallbladder epithelial cells were derived from patients that underwent cholecystectomy (for details, see Table 5). The samples were stored in ice-cold phosphate-buffered saline (PBS) for up to 2 h, and then epithelial cell isolation was performed as described previously (75). Briefly, the tissue was washed with PBS from the residual bile and mucus, and it was then incubated at 37°C with the mucosal side facing a solution of 0.2% collagenase type IV. The mucosa was abraded thoroughly with the end of a glass microscope slide held at an angle of 45° every 5 min four times. The isolated cells were passed through a cell strainer with 70-μm pores and spun down, and 1 to 3 million cells were resuspended in a drop of 50 μl of Matrigel. The polymerized Matrigel drop was then supplemented with a medium based on Advanced/DMEM/F-12 (Invitrogen; described in Table 1). The medium was replaced twice a week. Every 7 to 10 days, the organoids were split at a ratio 1 to 3 or 4 by treatment with trypsin and then passed 10 times through a heat-narrowed Pasteur pipette. In the experiments in which single cells were seeded, trypsin-treated organoids were also passed through a 40-μm-pore cell strainer before seeding them in Matrigel.
TABLE 5.
Patient information
| ID | Age (yr) | Gender | Comments |
|---|---|---|---|
| GB6 | 66 | F | Cholecystectomy due to presence of gallstones and inflammation. |
| GB10 | 61 | F | Cholecystectomy due to presence of gallstones and inflammation. |
| GB11 | 56 | M | Cholecystectomy due to presence of gallstones and inflammation. |
| GB12 | 37 | M | Cholecystectomy due to polyps in the gallbladder; we got a healthy piece |
| GB13 | 66 | M | Adenocarcinoma of the gastroesophageal junction, type III, otherwise healthy GB; patient received neoadjuvant chemotherapy before surgery and preventive cholecystectomy. |
| GB16 | 65 | F | Gastric carcinoma, otherwise healthy GB; patient underwent gastrectomy and preventive cholecystectomy. |
Murine organoid culture.
Murine gallbladders were derived from mice with C57BL/6J genetic background. After sacrificing the mouse, the gallbladder was resected, cut in four pieces, and incubated in a thermal mixer at 37°C in 2 ml of TrypLE (Thermo Scientific) for 45 min. The tissue pieces were pipetted up and down five times to release the cells, big pieces were removed, and isolated cells were centrifuged, washed with Dulbecco modified Eagle medium (DMEM), and then seeded in 50-μl Matrigel drops. The polymerized Matrigel drop was then supplemented with the medium described in Table 1. The medium was replaced twice a week. The splitting procedure was the same as that described for the human organoids.
Human gallbladder mucosoid culture.
The generation of the human gallbladder mucosoids follows a protocol that was previously published for the healthy human stomach (34). Briefly, single cells derived from organoid cultures were seeded on collagen-coated filters of Millicell standing cell culture inserts (Millipore, PIHP01250) at 150,000 cells/insert in primary cell medium (refer to Table 1 for more detail). Cells were incubated at 37°C, and the medium in the surrounding well was changed daily for the first 5 days, followed by twice a week. After 3 days, the medium on the filter was removed, and cells started to produce mucus that was withdrawn during medium change. Once a month, the mucosoids were split at a ratio of 1:3 by incubating the apical and basal sides of the mucosoids with trypsin-EDTA (0.5%). Single cells were reseeded again on new coated cell culture inserts.
Lineage tracing.
For lineage tracing experiments, we used murine gallbladder organoids derived from C57BL/6J, Lgr5-EGFP-IRES-CreERT2, ROSA-mTmGfloxed mice. At 5 days after seeding, 10 μM 4-hydroxytamoxifen (4HT; Sigma) was added to the medium, and the mixture was kept for 2 days. The induction was performed only once.
Microarray.
Organoids were harvested 4 or 14 days (small and big organoids, respectively) after seeding. The Matrigel drops containing the organoids were dissolved in 1 ml of TRIzol (Life Technologies), and RNA was isolated as described in the manufacturer’s protocol using glycogen as a coprecipitant. For mucosoids, filters were cut from the insert and dissolved thoroughly in 1 ml of TRIzol. Quality control and quantification of total RNA was assessed using a 2100 bioanalyzer (Agilent Technologies) and a NanoDrop 1000 UV-Vis spectrophotometer (Kisker).
(i) Organoids. Microarray experiments were performed as independent dual-color dye-reversal color-swap hybridizations using two biological replicates each. Total RNA was amplified and labeled with a dual-color Quick-Amp labeling kit (Agilent Technologies). In brief, mRNA was reverse transcribed and amplified using an oligo-dT-T7 promoter primer and labeled with cyanine 3-CTP or cyanine 5-CTP. After precipitation, purification, and quantification, 0.75 μg of each labeled cRNA was fragmented and hybridized to custom whole-genome human multipack microarrays (8 × 60k; Agilent, 048908) according to the supplier’s protocol (Agilent Technologies). Scanning of microarrays was performed at 3-μm resolution using a G2565CA high-resolution laser microarray scanner (Agilent Technologies). Microarray image data were processed with Image Analysis/Feature Extraction software G2567AA vA.11.5.1.1 (Agilent Technologies) using default settings and the GE2_1105_Oct12 extraction protocol. The extracted dual-color raw data .txt files were further analyzed using R and the associated BioConductor package limma (76). Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be assessed with the GEO accession number GSE100656.
For GSEA, a gene set of β-catenin target genes published previously (44) and human pluripotent stem cell genes published by Mallon et al. (42) (Tables 2 and 3) were used, and GSEA was performed on genes preranked by gene expression-based t score between early and differentiated organoids, using the fgsea R package (77) with 5,000 permutations. Wnt family member’s average intensities were calculated by global average in all the conditions. They were then filtered for an average intensity of >6. The differentially expressed ones were then identified as having a P value of <0.05.
(ii) Mucosoids. Single-color hybridizations using two technical replicates each were conducted. Microarrays used had design Agilent-014850 whole human genome microarray 4x44K G4112F (Agilent Technologies) and were read using the machines and software of the same manufacturer. The extracted raw data .txt files were further analyzed using R and the associated BioConductor package limma (76). Since MSigDB gene sets use human gene symbols to map genes to pathways, mouse symbols were translated to homologous human symbols using HomologeneDB from NCBI. GSEA was also performed on gene sets for cell cycle associated genes (58) from MSigDB v7.0 (PMID 21546393) (Table 4) between wild-type (w.t.) and ΔcdtB strain-infected mucosoid versus non-infected at 2 and 7 days post infection.
For human gene sets (i.e., MSigDB and those derived from human experiments), the full set of genes in the DGE results after collapsing t scores by gene and ranking was used. To analyze the mouse gene sets, the DGE data were restricted to probe sets that have a homologous gene in mice and humans. For these probe sets, the one with the highest t score and rank in the resulting list was selected and subsequently used for fGSEA analysis.
Expression data were analyzed as follows. For each of the selected comparisons, the replicates of the target condition were compared to the corresponding control using limma, producing differential expression statistics for all genes and comparisons. Analyses were performed as individual two-group unpaired comparisons: 2-day infection, w.t. versus NI; 2-day infection, ΔcdtB versus NI; 2-day infection, ΔcdtB versus w.t.; 7-day infection, w.t. versus NI; 7-day infection, ΔcdtB versus NI; and 7-day infection, ΔcdtB versus w.t.
The interpreting plotting of the results was done using Microsoft Excel, and the software R/R Studio was used to create the plots for the heatmaps. The heatmaps were plotted by using the normalized expression values (log-normalized intensity) again normalized on the non-infected control of each time point (logFC) when expression data from single genes were plotted and the calculated NES scores, respectively, for pathway analysis.
Immunofluorescence.
Organoids were removed from Matrigel at the indicated time point by washing with ice-cold PBS and then fixed with 3.7% paraformaldehyde. Tissue pieces were washed with PBS and fixed. After fixation, organoids and tissue pieces were embedded in paraffin and cut with a microtome to get 5-μm slices. Cells seeded in 2D (two dimensions) were washed with PBS and fixed. For whole-mount staining, the organoids were fixed directly in the Matrigel drop and then stained. The staining was performed with the antibodies and dyes listed in Table 6. Images were acquired with a Leica TCS SP-8 confocal microscope. For immunofluorescence of the mucosoids, the filters were cut form the insert, and pieces of the filters were blocked in a bovine serum albumin-containing blocking buffer for 3 h for whole-mount staining. Alternatively, the filters were fixed overnight in 4% paraformaldehyde (PFA) at 4°C, washed, embedded orthogonally in Histogel (HG-4000-144) inside a casting mold, and paraffinized overnight in a Leica TP1020 tissue processor. The paraffin blocks were generated inside a casting mold on a Paraffin console (Microm). Next, 5-μm sections were cut with a paraffin rotation microtome (Microm). For dewaxing and antigen retrieval, sample slides were washed twice with xylene (10 min), followed by a descending series of alcohols (20 s each), followed by two washes with water and 30 min in target retrieval solution (Dako) at 95°C and 20 min at room temperature and 5 min under running water. Primary antibodies were diluted in the blocking solution: in-house-made anti-γH2AX conjugated to ATTO488 (a green fluorescent dye; 1:500), phalloidin-Alexa 647 (lot 1731699; 1:100), and Hoechst (1:1,000; Sigma, B2261, lot 019K4029). Antibodies were incubated overnight at room temperature in the dark. Next, filter pieces were washed three times with blocking solution for 3 h at room temperature in the dark. The stained filters were mounted in Vectashield (Vector Laboratories, H-1500) on a glass slide, and the images were acquired using an SP-8 confocal microscope. The pictures are a result of a projection of multiple z-stacks analyzed with the software ImageJ.
TABLE 6.
Antibodies and dyes
| Antibody | Supplier | Catalog no. | Source | Application(s) (dilution)a |
|---|---|---|---|---|
| Anti-β-Actin (AC-15) | Sigma | A5441 | Mouse monoclonal | WB (1:10,000) |
| E-cadherin (clone CD324) | BD | 562869 | Mouse monoclonal | IF (1:300), WB (1:500) |
| Ki67 (D2H10) | Cell Signaling | 9027 | Rabbit monoclonal | IF (1:200) |
| PCNA (csPC10) | Cell Signaling | 2586 | Mouse monoclonal | IF (1:100) |
| Phospho-histone H2A.X (Ser139) (clone JBW301) | Millipore | 05-636-I | Mouse monoclonal | WB (1:200) |
| Phospho-histone H2A.X (Ser139)-conjugated FITCS | In house | Mouse monoclonal | IF (1:500) | |
| β-Catenin | Sigma | C2206 | Rabbit polyclonal | IF (1:300), WB (1:500) |
| Claudin-2 | Abcam | ab53032 | Rabbit polyclonal | IF (1:200), WB (1:500) |
| Cytokeratin 19 (EP1580Y) | Abcam | ab52625 | Rabbit monoclonal | IF: 1:500), WB (1:5,000) |
| Muc5B | Abcam | ab87376 | Rabbit polyclonal | IF (1:300) |
| Vimentin (D21H3) | Cell Signaling | 5741 | Rabbit monoclonal | WB (1:1,000) |
| Hoechst (bisbenzimide H 33258) | Sigma | H6024 | IF (1:1,000) | |
| Draq5 | Abcam | ab108410 | IF (1:1,000) | |
| Phalloidin 647 | Invitrogen | A22287 | IF (1:500) |
WB, Western blotting; IF, immunofluorescence.
Transmission electron microscopy.
Infected and non-infected gallbladder mucosoids were washed in-well with PBS, fixed with 4% PFA for 30 min, and washed twice with PBS. Filters were cut from the insert and cropped into pieces with bacterial patches under visual control. Cropped filter pieces were stored in PBS at 4°C until use. For fine structural analysis, cell layers on filters were fixed with 2.5% glutaraldehyde, postfixed with 0.5% osmium tetroxide, contrasted with uranyl acetate and tannic acid, dehydrated in a graded ethanol series, and infiltrated in Polybed (Polysciences). Cut-out pieces of the filters were stacked in flat embedding molds with Polybed. After polymerization, specimens were cut at 60 nm and contrasted with lead citrate. Specimens were analyzed in a Leo 906E transmission electron microscope (Zeiss, Oberkochen, Germany) equipped with a side-mounted digital camera (Morada, SIS-Olympus, Münster, Germany). Figures were assembled with the help of a FigureJ-Plugin (78).
Western blotting.
For the Western blots, organoids and cells seeded in 2D were harvested in Laemmli buffer, and 12% SDS-PAGE gels were run and transferred to a nitrocellulose membrane, which was then blotted with the antibodies listed in Table 6. Densitometry was calculated using ImageJ software.
Functional assay.
The functional assay is a modified version of a previously described assay (54). Briefly, 1 week after seeding, the organoids were incubated with DMEM/F-12 (Invitrogen) containing 100 μM rhodamine-123 (Sigma) for 5 min, washed with three times with PBS, and supplemented with the regular medium. Images were taken every minute with a Leica SP-E confocal microscope for 30 min. Temperature and CO2 concentration were kept at 37°C and 5%, respectively. To show that transport of rhodamine-123 depends on activity of multidrug-resistant (MDR) gene products, the organoids were incubated with 10 μM verapamil (Sigma), an MDR inhibitor, for 30 min before rhodamine-123 was added. As a negative control, gastric organoids were used, cultivated as previously described (28).
Bacterial strains.
Salmonella enterica serovar Paratyphi A (ATCC 9150) was used for the infection experiments. An isogenic mutant knockout of cdtB was generated by interrupting the gene with a kanamycin resistance cassette. Briefly, two sequences were amplified upstream and downstream cdtB using the primers TCTATAGTTGTCTCTTTGGTATTAAC and CGCGGATCCACCATAAGAATATCC for the region upstream and the primers CGCGGATCCATATAAGATATATCT and ACAGCTTCGTGCCAAAAAGG for the region downstream. After insertion in a pGEM-T Easy vector (Promega), a kanamycin resistance cassette was inserted in between by making use of the BamHI sites included in the primers. The resulting region was PCR amplified and electroporated in Salmonella, and the clones where homologous recombination occurred were selected, as described previously (79). If mentioned, to visualize the bacteria, the w.t. and ΔcdtB strains were additionally transformed with pLS002, a plasmid carrying the constitutively expressed mCherry gene and an ampicillin resistance cassette.
Infection experiments.
Organoids were removed from Matrigel by washing with ice-cold PBS, mechanically sheared by pipetting them three times through a heat-narrowed Pasteur pipette, and incubated at 37°C with 300 μl of primary medium containing log-phase Salmonella to a multiplicity of infection of 100 for 2 h. The cells were pelleted and washed twice with PBS before reseeding them in Matrigel. The gentamicin protection assay was performed by incubation for 1 h in primary medium supplemented with 100 μg/ml gentamicin. At this point, the invasion assay was performed. The organoids were removed from Matrigel and washed twice with PBS, the membrane was permeabilized by 2 min of incubation with 1% Triton X-100, and then sequential dilutions were plated on LB agar plates. The following day, colonies were counted as follows: invasion percentage = (CFU recovered from the infected organoids/bacteria used for infection) × 100. In the well with the remaining infected organoids, the concentration of gentamicin was decreased to 10 μg/ml for the duration of the experiment (80). Infection of mucosoids with Salmonella was done accordingly: log-phase mCherry-transformed Salmonella was administered on the filter to a multiplicity of infection of 100 for 24 h by using a penicillin-streptomycin-free 3D gallbladder medium (see Table 1). The infection medium was then removed, the filters and wells were washed with 37°C PBS, and the gentamicin protection assay was performed by incubation for 1 h in primary medium supplemented with 100 μg/ml gentamicin. The gentamicin concentration was then reduced to 10 μg/ml and withdrawn completely at 48 h post infection. The cells were washed, and the medium was refreshed every 2 days.
Intoxication experiments.
Organoids were split to single cells, seeded onto a type I collagen (Thermo Fisher, A10644-0)-coated plastic (10 μg/cm2) or glass (15 μg/cm2) surface, supplemented with the conventional 3D medium, and intoxicated when 50% confluence was reached. The typhoid toxin-containing Salmonella supernatant was prepared by using a modified version of a previously described protocol (19). Briefly, the bacteria were grown in Luria-Bertani overnight, diluted 1:50 in MM5.8 (19, 81), and then grown overnight until the optical density at 600 nm reached 0.4 to 0.5. The bacteria were then removed by centrifugation and subsequent filtration through 0.4-μm-pore filters. The supernatant was then concentrated 20-fold using an Amicon Ultra-15 column. It was then diluted 1 to 20 in primary medium and incubated for 24 h with the cells. As a positive control, the cells received 50 μM etoposide (Sigma) for 24 h.
Neutral comet assay.
The neutral comet assay was performed after intoxication using the kit from Trevigen according to the manufacturer´s protocol. Images were acquired using fluorescence microscope (Leica DMR). The percentage of DNA in the tail (which is a measure of DNA damage) was quantified using Comet Score software (TriTek).
Data availability.
Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be accessed under GEO accession number GSE100656.
ACKNOWLEDGMENTS
We thank Lothar Wieler and Anton Aebischer for critical revision of the project, Hans-Joachim Mollenkopf and Ina Wagner for support with the microarray experiments, and Rike Zietlow and Mary Muers for editing the manuscript.
Author contributions were as follows: L.P.S., study design, experiments design, generation, analysis and interpretation of data, drafting of the manuscript, statistical analysis, development of experimental procedures; K.H., development of experimental procedures, generation, analysis and interpretation of data, statistical analysis, drafting of the manuscript; A.I., experimental design and development of procedures, generation of data; H.B., statistical analysis of microarray data, bioinformatics support; N.K., generation and analysis of data; C.G., electron microscopy; S.C., preparation and provision of human specimens; S.C.S., preparation and provision of human specimens; R.K.G., conception, contribution to the study design and supervision; T.F.M., conception, design and supervision of the study, revision of manuscript, funding acquisition; and F.B., study conception and design, study supervision, analysis and interpretation of data, writing and revision of the manuscript.
We declare there are no conflicts of interest.
Footnotes
Citation Sepe LP, Hartl K, Iftekhar A, Berger H, Kumar N, Goosmann C, Chopra S, Schmidt SC, Gurumurthy RK, Meyer TF, Boccellato F. 2020. Genotoxic effect of Salmonella Paratyphi A infection on human primary gallbladder cells. mBio 11:e01911-20. https://doi.org/10.1128/mBio.01911-20.
REFERENCES
- 1.Kanthan R, Senger J-L, Ahmed S, Kanthan SC. 2015. Gallbladder cancer in the 21st century. J Oncol 2015:967472–967472. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randi G, Franceschi S, La Vecchia C. 2006. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer 118:1591–1602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 3.Crawford RW, Gibson DL, Kay WW, Gunn JS. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun 76:5341–5349. doi: 10.1128/IAI.00786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford RW, Rosales-Reyes R, Ramírez-Aguilar ML, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. 2010. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A 107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Escobedo G, Gunn JS. 2013. Gallbladder epithelium as a niche for chronic salmonella carriage. Infect Immun 81:2920–2930. doi: 10.1128/IAI.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Escobedo G, Marshall JM, Gunn JS. 2011. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9:9–14. doi: 10.1038/nrmicro2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caygill CPJ, Hill MJ, Braddick M, Sharp JCM. 1994. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 343:83–84. doi: 10.1016/S0140-6736(94)90816-8. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Kumar S, Kumar S. 2006. Infection as a risk factor for gallbladder cancer. J Surg Oncol 93:633–639. doi: 10.1002/jso.20530. [DOI] [PubMed] [Google Scholar]
- 9.Nagaraja V, Eslick GD. 2014. Systematic review with meta-analysis: the relationship between chronic Salmonella Typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther 39:745–750. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 10.Chumduri C, Gurumurthy RK, Zietlow R, Meyer TF. 2016. Subversion of host genome integrity by bacterial pathogens. Nat Rev Mol Cell Biol 17:659–673. doi: 10.1038/nrm.2016.100. [DOI] [PubMed] [Google Scholar]
- 11.Koeppel M, Garcia-Alcalde F, Glowinski F, Schlaermann P, Meyer TF. 2015. Helicobacter pylori infection causes characteristic DNA damage patterns in human cells. Cell Rep 11:1703–1713. doi: 10.1016/j.celrep.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 12.Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M, Muller A. 2011. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci U S A 108:14944–14949. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chumduri C, Gurumurthy RK, Zadora PK, Mi Y, Meyer TF. 2013. Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe 13:746–758. doi: 10.1016/j.chom.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziubańska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, Katainen R, Cajuso T, Crosetto N, Orozco M, Aaltonen LA, Meyer TF. 2020. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med 26:1063–1069. doi: 10.1038/s41591-020-0908-2. [DOI] [PubMed] [Google Scholar]
- 16.Song J, Gao X, Galán JE. 2013. Structure and function of the Salmonella Typhi chimeric A(2)B(5) typhoid toxin. Nature 499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spanò S, Ugalde JE, Galán JE. 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Rivera LD, Bowen BM, den Bakker HC, Duhamel GE, Wiedmann M. 2015. Characterization of the cytolethal distending toxin (typhoid toxin) in non-typhoidal Salmonella serovars. Gut Pathog 7:19–19. doi: 10.1186/s13099-015-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidi R, Levi L, Rouf SF, Puiac S, Rhen M, Frisan T. 2013. Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cell Microbiol 15:2034–2050. doi: 10.1111/cmi.12172. [DOI] [PubMed] [Google Scholar]
- 20.Haghjoo E, Galan JE. 2004. Salmonella Typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang SJ, Song J, Galán JE. 2016. Receptor-mediated sorting of typhoid toxin during its export from Salmonella Typhi-infected cells. Cell Host Microbe 20:682–689. doi: 10.1016/j.chom.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasso F, Frisan T. 2015. Bacterial genotoxins: merging the DNA damage response into infection biology. Biomolecules 5:1762–1782. doi: 10.3390/biom5031762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. 2011. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol 4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge Z, Feng Y, Ge L, Parry N, Muthupalani S, Fox JG. 2017. Helicobacter hepaticus cytolethal distending toxin promotes intestinal carcinogenesis in 129Rag2-deficient mice. Cell Microbiol 19. doi: 10.1111/cmi.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guidi R, Guerra L, Levi L, Stenerlöw B, Fox JG, Josenhans C, Masucci MG, Frisan T. 2013. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol 15:98–113. doi: 10.1111/cmi.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 27.Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco M, Sancho E, Clevers H, Batlle E. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat Med 17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 28.Schlaermann P, Toelle B, Berger H, Schmidt SC, Glanemann M, Ordemann J, Bartfeld S, Mollenkopf HJ, Meyer TF. 2016. A novel human gastric primary cell culture system for modeling Helicobacter pylori infection in vitro. Gut 65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148:126–136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huch M, Gehart H, Boxtel RV, Hamer K, Blokzijl F, Verstegen MMa, Ellis E, Wenum MV, Fuchs S, Ligt JD, Wetering MVD, Sasaki N, Boers SJ, Kemperman H, Jonge JD, Ijzermans JNM, Nieuwenhuis EES, Hoekstra R, Strom S, Vries RRG, Laan L, Cuppen E, Clevers H. 2015. Article long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lugli N, Kamileri I, Keogh A, Malinka T, Sarris ME, Talianidis I, Schaad O, Candinas D, Stroka D, Halazonetis TD. 2016. R-spondin 1 and noggin facilitate expansion of resident stem cells from non-damaged gallbladders. EMBO Rep 17:769–779. doi: 10.15252/embr.201642169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampaziotis F, Justin AW, Tysoe OC, Sawiak S, Godfrey EM, Upponi SS, Gieseck RL, 3rd, de Brito MC, Berntsen NL, Gómez-Vázquez MJ, Ortmann D, Yiangou L, Ross A, Bargehr J, Bertero A, Zonneveld MCF, Pedersen MT, Pawlowski M, Valestrand L, Madrigal P, Georgakopoulos N, Pirmadjid N, Skeldon GM, Casey J, Shu W, Materek PM, Snijders KE, Brown SE, Rimland CA, Simonic I, Davies SE, Jensen KB, Zilbauer M, Gelson WTH, Alexander GJ, Sinha S, Hannan NRF, Wynn TA, Karlsen TH, Melum E, Markaki AE, Saeb-Parsy K, Vallier L. 2017. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat Med 23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- 33.Bartfeld S. 2016. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev Biol 420:252–270. doi: 10.1016/j.ydbio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Boccellato F, Woelffling S, Imai-Matsushima A, Sanchez G, Goosmann C, Schmid M, Berger H, Morey P, Denecke C, Ordemann J, Meyer TF. 2019. Polarised epithelial monolayers of the gastric mucosa reveal insights into mucosal homeostasis and defence against infection. Gut 68:400–413. doi: 10.1136/gutjnl-2017-314540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo KS, Choi HS, Jun DW, Lee HL, Lee OY, Yoon BC, Lee KG, Paik SS, Kim YS, Lee J. 2016. MUC expression in gallbladder epithelial tissues in cholesterol-associated gallbladder disease. Gut Liver 10:851–858. doi: 10.5009/gnl15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi A, Lee SP. 1996. Bidirectional transport of cholesterol between gallbladder epithelial cells and model bile. Am J Physiol 271:G410–G414. doi: 10.1152/ajpgi.1996.271.3.G410. [DOI] [PubMed] [Google Scholar]
- 37.Nakanuma Y, Katayanagi K, Kawamura Y, Yoshida K. 1997. Monolayer and three-dimensional cell culture and living tissue culture of gallbladder epithelium. Microsc Res Tech 39:71–84. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Frizzell RA, Heintze K. 1980. Transport functions of the gallbladder. Int Rev Physiol 21:221–247. [PubMed] [Google Scholar]
- 39.Yoo KS, Lim WT, Choi HS. 2016. Biology of cholangiocytes: from bench to bedside. Gut Liver 10:687–698. doi: 10.5009/gnl16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manohar R, Li Y, Fohrer H, Guzik L, Stolz DB, Chandran UR, LaFramboise WA, Lagasse E. 2015. Identification of a candidate stem cell in human gallbladder. Stem Cell Res 14:258–269. doi: 10.1016/j.scr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpino G, Cardinale V, Gentile R, Onori P, Semeraro R, Franchitto A, Wang Y, Bosco D, Iossa A, Napoletano C, Cantafora A, D’Argenio G, Nuti M, Caporaso N, Berloco P, Venere R, Oikawa T, Reid L, Alvaro D, Gaudio E. 2014. Evidence for multipotent endodermal stem/progenitor cell populations in human gallbladder. J Hepatol 60:1194–1202. doi: 10.1016/j.jhep.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, Tyson LJ, Park K, Chen KG, Fann YC, McKay RD. 2013. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res 10:57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Goke B, Kolligs FT. 2014. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genomics 15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JC. 2008. Reduction of contamination of epithelial cultures by fibroblasts. CSH Protoc 2008:pdb.prot4478. doi: 10.1101/pdb.prot4478. [DOI] [PubMed] [Google Scholar]
- 46.Mitra A, Mishra L, Li S. 2013. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol 31:347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuver R, Savard CE, Lee SK, Haigh WG, Lee SP. 2007. Murine gallbladder epithelial cells can differentiate into hepatocyte-like cells in vitro. Am J Physiol Gastrointest Liver Physiol 293:G944–G955. doi: 10.1152/ajpgi.00263.2006. [DOI] [PubMed] [Google Scholar]
- 48.Németh Z, Szász AM, Tátrai P, Németh J, Gyorffy H, Somorácz A, Szíjártó A, Kupcsulik P, Kiss A, Schaff Z. 2009. Claudin-1, -2, -3, -4, -7, -8, and -10 protein expression in biliary tract cancers. J Histochem Cytochem 57:113–121. doi: 10.1369/jhc.2008.952291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kampf C, Mardinoglu A, Fagerberg L, Hallström BM, Danielsson A, Nielsen J, Pontén F, Uhlen M. 2014. Defining the human gallbladder proteome by transcriptomics and affinity proteomics. Proteomics 14:2498–2507. doi: 10.1002/pmic.201400201. [DOI] [PubMed] [Google Scholar]
- 50.van Klinken BJ, Dekker J, van Gool SA, van Marle J, Buller HA, Einerhand AW. 1998. MUC5B is the prominent mucin in human gallbladder and is also expressed in a subset of colonic goblet cells. Am J Physiol 274:G871–G878. doi: 10.1152/ajpgi.1998.274.5.G871. [DOI] [PubMed] [Google Scholar]
- 51.Gigliozzi A, Fraioli F, Sundaram P, Lee J, Mennone A, Alvaro D, Boyer JL. 2000. Molecular identification and functional characterization of Mdr1a in rat cholangiocytes. Gastroenterology 119:1113–1122. doi: 10.1053/gast.2000.18156. [DOI] [PubMed] [Google Scholar]
- 52.Pstrpw JD. 1967. Absorption of bile pigments by the gall bladder. J Clin Invest 46:2035–2052. doi: 10.1172/JCI105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scoazec JY, Bringuier AF, Medina JF, Martínez-Ansó E, Veissiere D, Feldmann G, Housset C. 1997. The plasma membrane polarity of human biliary epithelial cells: in situ immunohistochemical analysis and functional implications. J Hepatol 26:543–553. doi: 10.1016/s0168-8278(97)80419-9. [DOI] [PubMed] [Google Scholar]
- 54.Tanimizu N, Miyajima A, Mostov KE. 2007. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol Biol Cell 18:1472–1479. doi: 10.1091/mbc.e06-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nath G, Gulati AK, Shukla VK. 2010. Role of bacteria in carcinogenesis, with special reference to carcinoma of the gallbladder. World J Gastroenterol 16:5395–5404. doi: 10.3748/wjg.v16.i43.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 57.Del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, Wrande M, Candela M, Turroni S, Nastasi C, Consolandi C, Peano C, Tebaldi T, Viero G, Gorgoulis VG, Krejsgaard T, Rhen M, Frisan T. 2016. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog 12:e1005528. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13:1977–2000. doi: 10.1091/mbc.02-02-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Chen S, Wang S, Soares F, Fischer M, Meng F, Du Z, Lin C, Meyer C, DeCaprio JA, Brown M, Liu XS, He HH. 2017. Transcriptional landscape of the human cell cycle. Proc Natl Acad Sci U S A 114:3473–3478. doi: 10.1073/pnas.1617636114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. 2017. Biofilm-producing Salmonella Typhi: chronic colonization and development of gallbladder cancer. Int J Mol Sci 18:1887. doi: 10.3390/ijms18091887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wozniak AJ, Ross WE. 1983. DNA damage as a basis for 4′-demethylepipodophyllotoxin-9-(4,6-O-ethylidene-β-d-glucopyranoside) (etoposide) cytotoxicity. Cancer Res 43:120–124. [PubMed] [Google Scholar]
- 62.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. 2017. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol 15:109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 63.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song J-Y, Neefjes-Borst EA, Te Riele H, Holden DW, Nath G, Neefjes J. 2015. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Boccellato F, Meyer TF. 2015. Bacteria moving into focus of human cancer. Cell Host Microbe 17:728–730. doi: 10.1016/j.chom.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 65.Steele-Mortimer O, Knodler LA, Marcus SL, Scheid MP, Goh B, Pfeifer CG, Duronio V, Finlay BB. 2000. Activation of Akt/protein kinase B in epithelial cells by the Salmonella Typhimurium effector sigD. J Biol Chem 275:37718–37724. doi: 10.1074/jbc.M008187200. [DOI] [PubMed] [Google Scholar]
- 66.Roppenser B, Kwon H, Canadien V, Xu R, Devreotes PN, Grinstein S, Brumell JH. 2013. Multiple host kinases contribute to Akt Activation during Salmonella infection. PLoS One 8:e71015. doi: 10.1371/journal.pone.0071015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santos AJ, Meinecke M, Fessler MB, Holden DW, Boucrot E. 2013. Preferential invasion of mitotic cells by Salmonella reveals that cell surface cholesterol is maximal during metaphase. J Cell Sci 126:2990–2996. doi: 10.1242/jcs.115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buckner MM. 2016. Divide and conquer: Salmonella move into both daughter cells during mitosis. Virulence 7:616–619. doi: 10.1080/21505594.2016.1190063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pérès SY, Marchès O, Daigle F, Nougayrède JP, Herault F, Tasca C, De Rycke J, Oswald E. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol Microbiol 24:1095–1107. doi: 10.1046/j.1365-2958.1997.4181785.x. [DOI] [PubMed] [Google Scholar]
- 70.Comayras C, Tasca C, Pérès SY, Ducommun B, Oswald E, De Rycke J. 1997. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing cdc2 protein kinase dephosphorylation and activation. Infect Immun 65:5088–5095. doi: 10.1128/IAI.65.12.5088-5095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohguchi M, Ishisaki A, Okahashi N, Koide M, Koseki T, Yamato K, Noguchi T, Nishihara T. 1998. Actinobacillus actinomycetemcomitans toxin induces both cell cycle arrest in the G2/M phase and apoptosis. Infect Immun 66:5980–5987. doi: 10.1128/IAI.66.12.5980-5987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sert V, Cans C, Tasca C, Bret-Bennis L, Oswald E, Ducommun B, De Rycke J. 1999. The bacterial cytolethal distending toxin (CDT) triggers a G2 cell cycle checkpoint in mammalian cells without preliminary induction of DNA strand breaks. Oncogene 18:6296–6304. doi: 10.1038/sj.onc.1203007. [DOI] [PubMed] [Google Scholar]
- 73.Poon SL, McPherson JR, Tan P, Teh BT, Rozen SG. 2014. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Med 6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H, Genomics England Research Consortium. 2020. Mutational signature in colorectal cancer caused by genotoxic pks(+) Escherichia coli. Nature 580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Auth MK, Keitzer R, Scholz M, Blaheta R, Hottenrott EC, Herrmann G, Encke A, Markus BH. 1993. Establishment and immunological characterization of cultured human gallbladder epithelial cells. Hepatology 18:546–555. doi: 10.1002/hep.1840180311. [DOI] [PubMed] [Google Scholar]
- 76.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sergushichev AA, Loboda AA, Jha AK, Vincent EE, Driggers EM, Jones RG, Pearce EJ, Artyomov MN. 2016. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res 44:W194–W200. doi: 10.1093/nar/gkw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mutterer J, Zinck E. 2013. Quick-and-clean article figures with FigureJ. J Microsc 252:89–91. doi: 10.1111/jmi.12069. [DOI] [PubMed] [Google Scholar]
- 79.Datsenko K, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elhadad D, Desai P, Grassl GA, McClelland M, Rahav G, Gal-Mor O. 2016. Differences in host cell invasion and SPI-1 expression between Salmonella enterica serovar Paratyphi A and the non-typhoidal serovar Typhimurium. Infect Immun 84:1150–1165. doi: 10.1128/IAI.01461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nelson DL, Kennedy EP. 1971. Magnesium transport in Escherichia coli: inhibition by cobaltous ion. J Biol Chem 246:3042–3049. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cultivation of murine gallbladder organoids. (A) Murine gallbladder epithelial cells grown as organoids at 1, 2, and 4 days after seeding, and at passages 1, 5, 10, 16, and 19. Scale bar, 1mm. (B) Organoids on day 7 after seeding, fluorescently labeled with antibodies against the proliferation marker PCNA (green) and β-catenin (red); nuclei were stained with DRAQ5 (blue). Scale bar, 50 μm. (C) Organoids at passage 0 were split to single cells, seeded, and the number of resulting organoids was counted 5 to 7 days later (i.e., at passage 1). The organoids were kept in culture, and the procedure was repeated after 18 passages (i.e., at passage 19). Bars represent means ± the SD. (D) Lineage tracing of organoids derived from Lgr5-EGFP-IRES-CreERT2, ROSA-mTmGfloxed reporter mice after HT induction. Organoids derived from Lgr5+ cells express mGFP, while those derived from Lgr5− cells express mTomato. Scale bar, 200 μm. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of murine gallbladder organoids. (A) Western blot analysis of murine epithelial and gallbladder markers at early (P1) and late (P19) passages. (B) Western blot analysis as in panel A of the fibroblast marker vimentin compared to HeLa cells. (C) Immunofluorescence analysis of murine gallbladder tissue and organoids at 7 days after seeding for the gallbladder markers cytokeratin-19, claudin-2, or mucin5B (red); the epithelial marker E-cadherin (green); and DRAQ5 (blue). Scale bar, 10 μm. Download FIG S2, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Long-term intoxication, 24 and 48 h. Human GB organoids were seeded in 2D and intoxicated for 24 or 48 h. For intoxication for 48 h, the bacterial supernatant was produced twice, and fresh supernatant was diluted in medium was added after 24 h. The cells seeded were less confluent than in normal 24-h intoxication experiments to avoid premature confluence of the culture. The figure shows double-positive cells for Ki67 and γH2AX at 24 h (A) and 48 h (B). N, the minimum number of counted cells per individual experiment. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2020 Sepe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be accessed under GEO accession number GSE100656.