Graphical abstract
Keywords: SARS CoV, MERS, SARS CoV-2, Pandemic, Heterocyclic scaffolds, Coronaviruses
Abbreviations: 2019-nCoV-2, 2019-novel coronavirus; 3CLpro, 3chymotrypsin-like protease; 9-O-SIA, 9-O-acetyl-N-acetylneuraminic acid; ACE2, Angiotensin converting enzyme 2; COMFA, Comparative molecular field analysis; COMSIA, Comparative molecular similarity indices analysis; COVID 19, Corona virus disease 2019; CPE, Cytopathic effect inhibition assay; CRFK cells, Confluent crandel feline kidney cells; CVB3 3Cpro, Coxsackievirus B 3 cysteine protease; Dabcyl-KTSAVLQSGFRKME-Edans, Dabcyl-Lys-Thr-Ser-Ala-Val-Leu-Gln-Ser-Gly-Phe-Arg-Lys-Met-GluEdans, [4-(4-dimethylaminophenylazo) benzoic acid]-KTSAVLQSGFRKME-[5-[2′-(aminoethyl)amino]-naphthalenesulfonic acid]; DTT, 1,4-Dithio-d,l-threitol; E protein, Envelope protein; FDA, Food and Drug Administration; FIPV, Feline infectious peritonitis virus; FRET analysis, Fluorescence resonance energy transfer analysis; HBTU, Hexafluorophosphate benzotriazole tetramethyluronium; M protein, Membrane protein; MD simulation, Molecular dynamics simulation; MERS CoV, Middle east respiratory syndrome corona virus; MM-GBSA, Molecular Mechanics/Generalized Born Surface Area; MM-PBSA, Molecular mechanics Poisson–Boltzmann surface area; Mpro, Main protease; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; N protein, Nucleocapsid protein; NFkB, Nuclear factor kappa-light-chain-enhancer of activated B cells; NIH (MLPCN) screening, National Institutes of Health (Molecular libraries probe production centers network) screening; nsp10, non-structural protein 10; nsp12, non-structural protein 12, RNA dependent RNA polymerase; nsp13, non-structural protein 13, helicase; nsp14, non-structural protein 14, N-terminal exoribonuclease and C-terminal guanine-N7 methyl transferase; nsp15, non-structural protein 15, uridylate-specific endoribonuclease; nsp16, non-structural protein 16, 2′-O-methyl transferase; NTD, N-terminal domain; ORFs, open reading frames; PC 3Cpro, Picornavirus 3 cysteine protease; PDB, Protein data bank; PHEIC, Public Health Emergency of International Concern; PLpro, papain-like protease; PP1a, Polyprotein1a; PP1ab, Polyprotein1b; qRT-PCR, Quantitative reverse transcription polymerase chain reaction; QSAR, Quantitative structure-activity relationship; RASPD, Rapid Screening with Physicochemical Descriptors; RdRp, RNA dependent RNA polymerase; S protein, Spike protein; SARS CoV, Severe acute respiratory syndrome corona virus; WHO, World Health Organisation; Z-RLRGG-AMC, Z-Arg-Leu-Arg-Gly-Gly-7-amido-4-methylcoumarin
Abstract
Coronaviruses have led to severe emergencies in the world since the outbreak of SARS CoV in 2002, followed by MERS CoV in 2012. SARS CoV-2, the novel pandemic caused by coronaviruses that began in December 2019 in China has led to a total of 24,066,076 confirmed cases and a death toll of 823,572 as reported by World Health Organisation on 26 August 2020, spreading to 213 countries and territories. However, there are still no vaccines or medications available till date against SARS coronaviruses which is an urgent requirement to control the current pandemic like situations. Since many decades, heterocyclic scaffolds have been explored exhaustively for their anticancer, antimalarial, anti-inflammatory, antitubercular, antimicrobial, antidiabetic, antiviral and many more treatment capabilities. Therefore, through this review, we have tried to emphasize on the anticipated role of heterocyclic scaffolds in the design and discovery of the much-awaited anti-SARS CoV-2 therapy, by exploring the research articles depicting different heterocyclic moieties as targeting SARS, MERS and SARS CoV-2 coronaviruses. The heterocyclic motifs mentioned in the review can serve as crucial resources for the development of SARS coronaviruses treatment strategies.
1. Introduction
Heterocyclic scaffolds play a pivotal role in drug discovery and development as they constitute the key structural component of a majority of biologically active moieties. Their ability to interact with almost every cellular mechanism in living organism has been responsible for their versatile nature. Their interaction with different mechanistic pathways in viruses has continuously been exploited by researches for the designing of heterocycle-based antiviral agents. Several FDA approved drugs currently in the market comprise of different heterocyclic scaffolds [1].
Coronaviruses (CoV) are a family of viruses capable of causing mild to severe symptoms of respiratory distress. In the last two decades, the outbreak of two of the coronaviruses, Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS), have emerged as epidemics with severe mortality. Both epidemics were of zoonotic origin, with SARS CoV transmission from civet cats to humans in 2002 in China and MERS CoV transmission from dromedary camels to humans in 2012 in Saudi Arabia [2]. There was emergence of cluster of pneumonia cases of unknown etiology in Wuhan city, Hubei province, China on 31 December 2019 and later declared by China that the outbreak is associated with a seafood market in Wuhan. China shared the genetic sequence of novel coronavirus responsible for the outbreak for diagnostic purposes on 12 January 2020 [3].
On 30 January 2020, World Health Organisation (WHO) declared this 2019-nCoV outbreak as a PHEIC (Public Health Emergency of International Concern) which was declared pandemic on 11 March 2020 [4]. On 11 February 2020, WHO named this novel coronavirus as COVID-19 (corona virus disease 2019) and later International Committee on Taxonomy of viruses renamed it as SARS CoV-2 [5]. There were 24,066,076 confirmed cases of COVID-19 and 823,572 deaths, globally as on 26 August 2020 [6].
Coronaviruses are single stranded positive sense RNA viruses. COVID-19 is caused by seventh of known coronaviruses which have infected humans, in the sequence: 229E, NL63, OC43, KKU1, MERS-CoV, SARS-CoV, and 2019-nCoV-2. The latter is a betacoronavirus of subgenus sarbecovirus, and is a severe acute respiratory syndrome coronavirus 2 with 96% genome similarity to other bat coronaviruses [7], [8]. Among other coronaviruses, virus causing COVID-19 has an advantage of the presence of a unique polybasic cleavage site leading to its increased pathogenicity [9]. It has the largest RNA genome (30 kb) among all other RNA viruses with six to ten open reading frames (ORFs). It consists of some structural and some non-structural proteins. The structural proteins include: spike (S protein), envelope (E protein), membrane (M protein), nucleocapsid (N protein) while the non-structural proteins include: main protease (Mpro), papain-like protease (PLpro), non-structural protein 13 (nsp13, helicase), non-structural protein 12 (nsp12, RNA dependent RNA polymerase), N-terminal exoribonuclease and C-terminal guanine-N7 methyl transferase (nsp14), uridylate-specific endoribonuclease (nsp15), 2′-O-methyl transferase (nsp16) and nsp10 [10]. The N protein consists of viral genome while the other three proteins, S, E and M, make the viral envelope. The processing of two viral replicase polyproteins produced by ORF1a/b of COVID-19, PP1a and PP1ab, leads to the production of sixteen non-structural proteins while the mRNA encodes for the formation of structural proteins [11]. The spike protein is responsible for attachment to the host cell membrane using the host cell’s angiotensin converting enzyme-2 receptor thus initiating the infection process. Upon entering the host cells, the viral genome undergoes translation into viral polyproteins. The viral 3CLpro and PLpro then cleave these translated proteins into effector proteins. PLpro has the capability to deubiquinate host’s NFkB and interferon factor 3, resulting in suppression of host cell immunity. The binding of SARS-CoV-2 and ACE-2 has been found to be 10–20 times greater than that of SARS-CoV and ACE-2, favouring the higher transmissibility of SARS-CoV-2 [12].
Like other betacoronaviruses, SARS-CoV and MERS-CoV, SARS-CoV-2 also attacks the lower respiratory system of the patient, release the nucleocapsid in host cellular machinery and further undergoes replication in host cytoplasm leading to viral pneumonia. It can also lead to multiple organ damage affecting heart, kidney, gastrointestinal tract, liver and central nervous system of the patient [4], [12].
The RNA genome sequence of SARS CoV-2 (GenBank ID: MN908947.3) has shown to exhibit 82% similarity with SARS CoV (GenBank ID: NC_004718.3) and also it is often seen that the key target enzymes in coronaviruses reveal some sequence similarities, like RdRp of SARS CoV-2 shows 96% identity with SARS CoVRdRp, 3CLpro of SARS CoV-2 revealed 96.08% and 87.00% similarity with that of SARS CoV and MERS CoV, respectively, and though only 83% sequence similarity is seen between PLpro of SARS CoV-2 and SARS CoV but the active site of both the proteins do not show much variation, therefore the heterocyclic scaffolds showing effectiveness against these targets in SARS CoV and MERS CoV might also be repurposed and modified for healthcare emergencies like SARS CoV-2 and other such outbreaks of coronaviruses in future [13], [14], [15].
As the heterocyclic compounds have been rigorously involved in the ailments including viral infections, AIDS, cancer, there exists a profound scope of exploring these multiple nuclei to curb coronaviruses. Therefore, through this review we have tried to summarise some of the treatment options based on the heterocyclic nuclei researched and developed against SARS CoV, MERS-CoV and SARS-CoV-2 epidemics using in vitro, in vivo and in silico approaches, which may be of immense value at this hour of global emergency and in future.
2. Isatin and indole-based derivatives
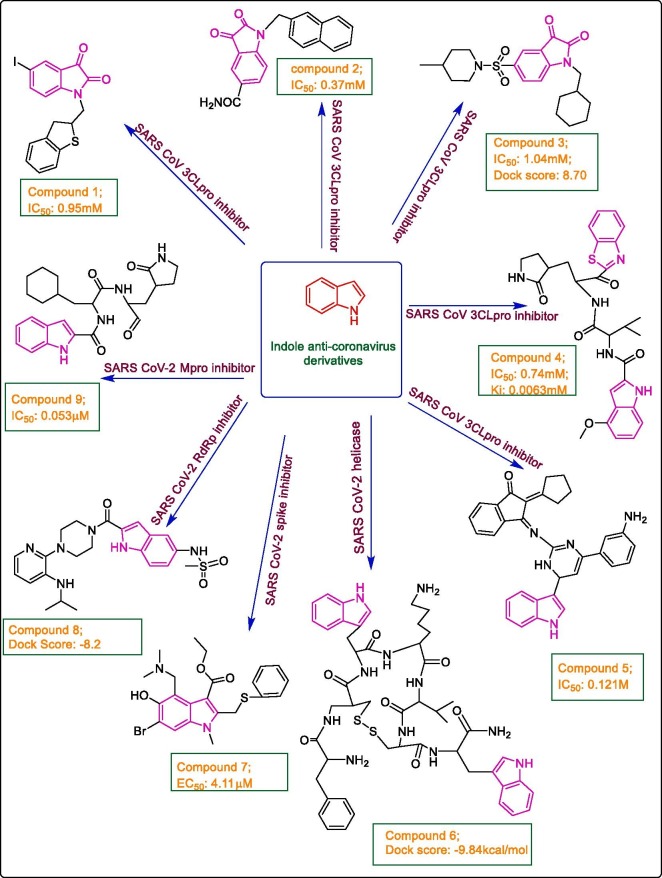
As some isatin based compounds have shown potent activity against 3C protease of rhinoviruses and the cysteine proteases of both rhinovirus and SARS CoV possessed structural similarity at the active site, some isatin derivatives were designed and assayed for inhibitory activity against SARS CoV 3CLpro by fluorescence resonance energy transfer FRET analysis. The assay revealed the higher potency of 5-iodo or 7-bromo isatin derivatives with benzothiophenemethyl side chain rather than with benzyl or alkyl side chains. The IC50 value of the most potent inhibitor 1 came to be 0.95 μM. The compound exhibited efficient binding within the active site of the enzyme. The carbonyl and amine groups of isatin scaffold were involved in forming hydrogen bond with Gly-143, Ser-144 and Cys-145 as shown by the docking analysis. The authors reported that the bulky nature of side chain on isatin derivatives decreased the inhibitory activity due to steric hinderance of the side chain with His-164 and Met-165 of 3CLpro [16]. After a year, N-substituted-5-carboxamide derivatives of isatin scaffold were designed and compared with 5-iodo substituted analogues using in vitro and computational approaches. The colorimetric inhibition assay results reported 2 as the potent inhibitor with lowest IC50value of 0.37 μM. The hydrophobic naphthyl group at N-1 of isatin moiety was found to be well fitted in hydrophobic pocket, thus increasing the activity. Also the C-5 carboxamide substitution imparted 3 to 4 times higher potential compared to the 5-iodo derivatives while the C-5 ester and carboxylic analogues did not display any inhibitory activity which was explained to be due to their poor binding at P1 site of 3CLpro as a result of strong charge repulsion from Glu-166. In addition, the carboxamide group successfully formed two hydrogen bonds with His 163 and Phe 140. The synthesized derivatives possessed noncovalent reversible binding with 3CLpro of SARS CoV [17]. Other group of scientists synthesized 5-sulfonamide derivatives of isatin and docked within SARS CoV 3CLpro active pocket with an improvement in inhibitory potential. The results of FRET inhibition assay suggested a better activity of 5-substituted analogues compared to substitutions at other positions of isatin scaffold. The 5-(piperazin-1-ylsulfonyl)isatin analogues displayed the higher potential than the 5-halogen substituted derivatives. Replacing the 5-piperazinyl moiety with 5-piperidinyl substitution further enhanced the antiviral potency with an IC50 of <5 μM. The effect of substitutions at N1 of isatin moiety revealed the most promising activity of compound 3 (IC50 = 1.04 μM) having an N-benzyl group which was further confirmed by docking analysis using Glide 5.5 software. Compound 3 extended hydrogen bonds with residues Gly143 and Cys145 of 3CLpro (PDB ID: 1UK4), with a dock score of 8.70. The docking results unleashed the crucial role of N-1 and C O at position 2 of isatin nucleus in hydrogen bond formation. The 5-sulfonyl and N1-benzyl substituents fitted well into the S2 and S1 hydrophobic pockets of 3CLpro, respectively, thus improving the inhibitory potential of the derivatives [18]. Reports are also available with tripeptidic inhibitors of SARS CoV 3CL protease. The dipeptide-type analogues with position-3 N-arylglycyl moiety were developed resulting in enhanced activity due to the hydrogen bond formation between amine of glycyl and Glu166 residue of 3CL pro. The fluorometric inhibition assay showed that the presence of dl-pyroglutamyl or pyrrole-2-carbonyl in place of N-(3-methoxyphenyl) glycyl moiety as rigid position-3 analogue, decreased the inhibitory potential dramatically while the indole-2-carbonyl group as rigid position-3 moiety remarkably improved the potential. Compound 4 with substitutions on position-3 indole nucleus with a 4-methoxy substituent was reported as the most potent derivative (IC50 value = 0.74 μM) when compared to 5- or 6-methoxy or 4-hydroxyl/4-isopropoxyl/4-isobutyloxyl derivatives. Replacing the rigid position-3 indole scaffold with other heterocycles like benzothiazole or benzofuran greatly reduced the activity suggesting the crucial role of amine of indole nucleus in hydrogen bond formation. The docking analysis of compound 4 (Ki = 0.0063 μM) with SARS CoVMpro (PDB ID: 1WOF) highlighted a favourable conformation of the rigid P3 4-methoxyindole moiety in the active site, compared to the flexible N-(3-methoxyphenyl)glycyl unit providing a 65-fold greater potential to the indole derivative [19]. Two series of pyrazole and pyrimidine fused indole derivatives were designed and analysed for their antiviral activity against SARS CoV 3CLpro, biosterically replacing isatin with indole. The FRET inhibition assay results revealed compound 5 as the most potent derivative with an IC50 value of 0.12lM. It was concluded that 2,3-dihydroinden-1-one was a required for anti-SARS CoV activity whereas it’s clubbing with pyrimidine nucleus provided greater enhancement in potency compared to that with pyrazole moiety and at the same time, isoxazole fusion lead to compounds with lesser potency [20]. The SARS CoV-2 helicase was homology modelled using 2019-nCoV/USA-WA1-F6/2020 (Gen Bank: QHU79203.1) helicase amino acid sequence against SARS CoV helicase (PDB: 6jyt.2.A) as template due to a 99.78% sequence similarity between the two and the modelled protein was analysed for antiviral properties of 23 clinically approved antiviral drug candidates using MOE software. The docking study revealed the highest binding affinity of compound 6 with a dock score of −9.84 kcal/mol involving a hydrogen bond formation with Gly79 of SARS CoV-2 helicase thus enabling the compound to be an efficient inhibitor of SARS CoV-2 helicase, thus interfering with the viral replication potential of helicase [21]. The disruption of the trimerization of SARS CoV-2 spike protein which is reported to be involved in host cell membrane fusion, can lead to antiviral activity. A portion of spike protein trimer S2 domain (947–1027 residues) was found to be structurally similar to influenza virus H3N2 haemagglutinin by comparative analysis, to which Arbidol7 binds for its anti-influenza action. The binding mode and mechanism of action of compound7, against SARS CoV-2 spike protein trimer (PDB: 6VSB), was analysed using HADDOCK2.2 and SWISS-DOCK molecular docking servers. The results of docking analysis revealed the potential interaction of compound 7 with the SARS CoV-2 spike protein S2 domain residues (K776, K780, K947, E1017, R1019, S1021, N1023, L1024, T1027) in the same way as with H3N2 HA protein (PDB: 5T6N), thus interfering with SARS CoV-2 trimerization and suggesting a potential role of compound 7 in the treatment of SARS CoV- 2 infection [22]. The homology modelling of SARS CoV-2 RdRp structure using SWISS-MODEL server revealed its 97.05% sequence similarity with the template, SARS CoVRdRp (PDB: 6NUR). The modelled structure was then docked against 74 antiviral drugs using Autodock Vina and the binding interactions were analysed by PyMol and Chimera. Delavirdine 8, an anti-HIV drug, exhibited pronounced inhibitory activity against SARS CoV-2 RdRp forming hydrogen bonds with Ala576 and Asn 582, with a dock score of −8.5, thus emphasizing the imperative role of 8 in SARS CoV-2 treatment therapy by interfering with RdRp dependant viral replication [23]. Novel inhibitors of SARS CoV-2 Mpro (PDB: 2H2Z) were designed by analyzing the active site, which basically comprise of S1′, S1, S2 and S4 subunits. In the newly synthesized compounds, aldehyde was taken as a warhead required to form covalent bond with the thiol of cysteine which keeps the inhibitor anchored at S1′ site of the protein. The FRET inhibition assay displayed a high potential of compound 9 with an IC50 value of 0.053 μM with no cytotoxicity. The electron density map examination of the crystal structure of SARS CoV-2 Mpro (PDB: 6LZE) in complex with compound 9 revealed a favourable conformation of compound 9 in the active site, involving a hydrogen bond of its indole moiety with Glu166 and hydrophobic interactions with Pro168 and Gln189. In vivo pharmacokinetic study in mice with 5 mg/kg i.p and 5 mg/kg i.v administration, demonstrated 87.8% bioavailability value while in vivo toxicity analysis in SD rats and beagle dogs indicated no toxicity and mortality at a dose of 40 mg/kg. Therefore, the results of analysis suggested a promising role of compound 9 for SARS CoV-2 clinical trial studies [24]. Six anti-influenza drugs namely arbidol, oseltamivir, baloxavir, zanamivir, peramivir and laninamivir were evaluated for their antiviral potential against SARS CoV-2. The cell counting kit-8 (CCK-8) assay demonstratedarbidol7 as a potent inhibitor with an EC50 value of 4.11 μM while a CC50 value of 31.79 μM. A sharp decline in the SARS CoV-2 induced cytopathic effect and viral NP expression by compound 7 was also observed using immunofluorescence staining assay. The qRT-PCR examination displayed an efficient inhibition of viral replication stages with more pronounced effect at entry (41%) compared to post-entry stage (61%) using viral multiplicity of infection (MOI) of 0.05. Arbidol 7 lowered the binding efficiency of virions, in virus infected Vero E6 cells (MOI of 0.05) upto 67%. The result of viral intracellular trafficking analysis by immunofluorescence microscopy revealed a decrease in release of SARS CoV-2 from endo lysosomes. Since no neuraminidase analogue was found in SARS CoV-2, therefore the neuraminidase inhibitors tested in this study failed to show any result. Also, since the cap snatching effect of endonuclease of viral polymerases is absent in coronaviruses, as a result of this, baloxavir which acts by inhibiting this particular mechanism of endonuclease, also failed in SARS CoV-2 infection [25]. Fig. 1 outlines various indole and isatin derivatives.
Fig. 1.
Isatin and indole-based derivatives.
3. Quinoline and isoquinoline derivatives
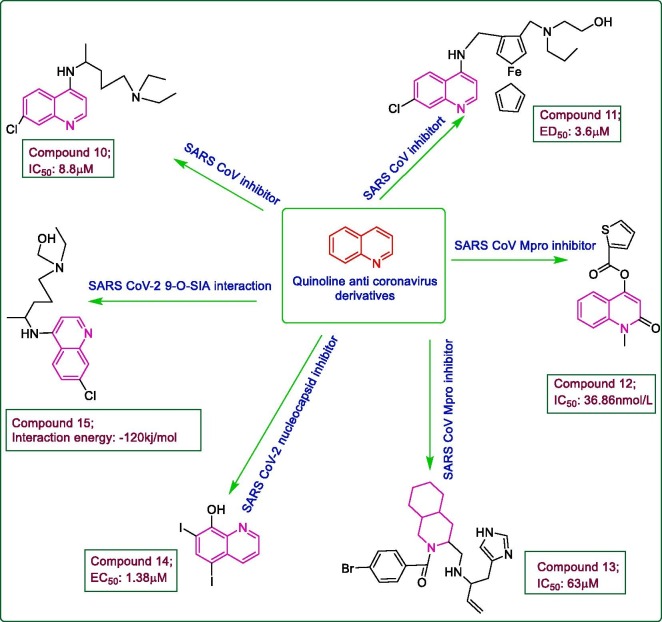
In 2003, with the outbreak of SARS CoV epidemic, several drugs were repurposed against it. One such anti-malarial drug, chloroquine 10, was also evaluated for its antiviral potential against SARS CoV infection. The results of MTT assay in Vero E6 cells revealed a good potency with an IC50 value of 8.8 μM while a CC50 value of 261.3 μM, suggesting the use of chloroquine in SARS CoV like infections. The drug proved to be more effective in later stages of viral replication rather than effecting viral attachment or penetration [26]. Almost a year later, chloroquine 10 was further reported for its prophylactic and therapeutic use against SARS CoV activity by treating Vero E6 cells at a concentration of 0.1–10 μM, both 20–24 h prior and 3–5 h after the viral infection. Immunofluorescence assay results indicated a 100% viral inhibition chloroquine pre-treated cells at 10 μM concentration while a 90–94% decrease in virus antigen-positive cells at 33–100 μM in post-treated cells with an ED50 of 4.4 μM. The mechanism of action for antiviral action was depicted as the abrogation of terminal glycosylation of ACE-2 receptor thus interfering with SARS CoV spike-ACE-2 receptor binding necessary for viral entry in host cells by flow cytometry and immunoprecipitation analysis. As chloroquine is basic in nature, the inhibition of virus-endosome fusion due to increase in endosomal pH was presumed to be responsible for post-treatment anti-SARS CoV effects [27]. Keeping in view the antimalarial potential of ferroquine, even on chloroquine resistant strains and the antiviral efficacy of chloroquine was well established against SARS CoV, some ferroquine derivatives have been reported for their antimalarial and antiviral potential. The analysis results revealed a high anti-malarial potency of compound 11, a hydroxyferroquine analogue, with high lipophilicity and lesser toxicity than ferroquine. From the in vitro study results of ferroquine showing the good potency of its metabolites against chloroquine resistant Plasmodium falciparum strain, it was suggested that the de-alkylated metabolite of the metallocene 11 namely mono-N-desethyl-ferroquine, facilitated the activity of parent molecule. The compound 11 exhibited good anti-HIV and anti-SARS CoV activity with an EC50 value of 3.6 μM in SARS CoV infected Vero cells, compared to the standard chloroquine showing an EC50 value of 6.5 μM though it exhibited poor potential against other viruses like reovirus-1 and HSV-2, herpes simplex virus-1, sindbis virus, influenza A virus, vesicular stomatitis virus and vaccinia virus, thus revealing its high selectivity towards HIV and SARS CoV infections [28]. From the virtual screening of over six lakh compounds in a database, a quinolinone derivative was selected for its promising anti-SARS CoV 3CLpro activity, for which the in vitro SARS CoV inhibition analysis revealed an IC50 value of 0.44 μM/L. Therefore, taking it as lead, based on its structure and activity, 23 novel 4-quinolinone ester derivatives were synthesized and in vitro and in silico studies were conducted to evaluate their SARS CoV 3CLpro inhibition potential. Compound 12 with a methyl substitution at NH moiety of the quinolinone lead compound, demonstrated nearly 12-fold greater anti-SARS CoV 3CLpro activity using FRET analysis, with an IC50 of 36.86 nM/L in comparison to lead. The docking analysis with SARS CoV 3CLpro (PDB: 3SN8) via Discovery Studio 3.0, indicated a more favourable binding of 12 in the active site of SARS CoV 3CLpro compared to the lead molecule which is proposed to be due to a hydrogen bond of amide oxygen with His 163 which remains conserved in both molecules and hydrogen bonds of ester carbonyl oxygen of 12 with Cys 145 and His 41 (the catalytic enzymes in SARS CoV 3CLpro active site necessary for proteolytic function). The structure activity relationship study of synthesized derivatives displayed a decrease in anti SARS CoV potential with the replacement of thiophene moiety of lead molecule with benzene, substituted benzene or alkyl carbon chains which is considered to be due to large size of these substituents which failed to fit within small S1′ pocket of the protease [29]. With the outbreak of MERS CoV infection in 2012, some FDA approved drugs which have shown activity against other coronaviruses, were repurposed against MERS CoV infection also. One such drug namely chloroquine 10 from a database of 348 FDA approved compounds, was also investigated for its in vitro potential against MERS CoV infected Vero and Huh7 cells using a colorimetric cell viability assay. The results unleashed the inhibition of MERS CoV mediated cytopathic effect by chloroquine with an EC50 value of 3 μM while it exhibited an EC50 value of 4.1 μM in SARS CoV infected Vero E6 cells. To determine the pre- and post- treatment effects of chloroquine, the time-of-addition experiment using plaque assay was conducted by adding the drug both 1 h prior and 1 h post viral infection (multiplicity of infection, 1) which revealed an approximate decrease of 2-log in viral production in chloroquine pre-treated Vero cells with no effect in post-treated cells, thus suggesting an early stage inhibition of MERS CoV infection via inhibition of clathrin mediated endocytosis [30]. The catalytic site of SARS CoV at Arg188/Gln189, was reported to be degradation sensitive while its mutant strain with isoleucine residue at 188 in place of arginine renders the protease highly stable and catalytically more efficient. At the same time, a peptide-mimetic substrate-based inhibitor with an aldehyde group at C-terminal demonstrated a potent competitive inhibition of the mutant R188I SARS CoV 3CLpro strain.
Novel decahydroisoquinoline based non-peptide derivatives were designed taking the above peptide mimetic inhibitor as lead and evaluated for their anti-R188I SARS CoV 3CLpro activity. The compound 13, a (4aR,8aS) isomer of trans-decahydroquinoline diastereomers with N-p-bromo-benzoyl substitution, displayed a high potential with an IC50 value of 63 μM. The X-ray crystallographic study of compound 13 in complex with 3CLpro (PDB: 4TWW), revealed a compact fitting of compound 13 within the active site of SARS CoV 3CLpro comparable to the active lead molecule. The S2 pocket was largely covered with decahydroisoquinoline fused ring while the hydrogen bond between the N atom of imidazole ring of compound 13 and His163 of 3CLpro provided it a favourable conformation at the S1 site of protease. Also, the N-p-bromobenzoyl moiety lies outward from 3CLpro where hydrophobic interactions might be possible [31]. From a dataset of FDA approved drugs, a set of 1528 compounds were screened and evaluated for their anti-SARS CoV-2 activity by combining cell viability assay and SARS CoV-2 ELISA. The Cell Titer Glo assay and cytopathic effect (CPE) inhibition assay (MOI: 0.01) in Vero E6 cells revealed promising effects of compound diiodohydroxyquinoline 14, (a quinoline derivative) used as a luminal amebicide, with a CC50 value of >100 μM and approximately 70% CPE inhibition, respectively. Compound 14 also showed an EC90 value of 4.50 μM in viral load assay using MOI of 0.01 in Vero E6 cells while an EC50 value of 1.38 μM through plaque reduction assay. It was also observed that treatment with compound 14 greatly reduced the SARS CoV-2 N antigen expression at a non-toxic concentration of <10 μM just like remdesivir which was taken as reference. The replication of SARS CoV-2 was also inhibited effectively at post entry. The in vivo studies revealed that compound 14 could be used as a potent luminal antiviral agent [32]. As it has been already reported that apart from using ACE-2 receptor, coronaviruses use sialic acid-containing glycoproteins and glycosides also as a source for entry into the host cell membrane and also that MERS CoV entry can be inhibited by depleting these sialic acids content. The efficiency of chloroquine 10 and hydroxychloroquine 15 against SARS CoV-2 was studied in silico (Hyperchem and Molegrow molecular viewer) using the mechanism of depletion of cell surface sialic acids containing gangliosides. Merging chloroquine with Neu5Ac depicted a good fit between the two, through interaction between negatively charged carboxylate of Neu5Ac and a cationic charge of chloroquine. The molecular modelling of 9-O-SIA (9-O-acetyl-N-acetylneuraminic acid), the preferable sialic acid for interaction with coronaviruses, also demonstrated a favourable fit among the two via interaction of nitrogen containing cationic ring of chloroquine with carboxylate of 9-O-SIA (interaction energy: −47 kJ/mol). Compound 15 showed a similar interaction with 9-O-SIA with enhanced binding owing to a hydrogen bond formation (interaction energy: −46 kJ/mol). The interaction affinities of the two drugs with ganglioside sialic acids, as sialic acid mostly forms a part of human respiratory tract gangliosides and glycoproteins was studied through molecular modelling using ganglioside GM1. The results revealed good interaction of both the drugs within the two drug-binding sites in GM1, showing a high interaction energy of −108 kJ/mol and −120 kJ/mol, respectively. Finally to determine the ability of both the drugs to prevent the binding of SARS CoV-2 spike NTD with cell surface ganglioside, the NTD-GM1 complex was superposed with drug-GM1 complex, which indicated that both NTD and the drug binds at the same position in GM1 and with the same mechanism involving a hydrogen bond and a CHπ interaction, thus unleashing the anti-SARS CoV-2 potential of the drugs [33]. Some active quinoline and isoquinoline derivatives are shown Fig. 2 .
Fig. 2.
Some of the promising Quinoline and Isoquinoline derivatives.
4. Pyridine derivatives
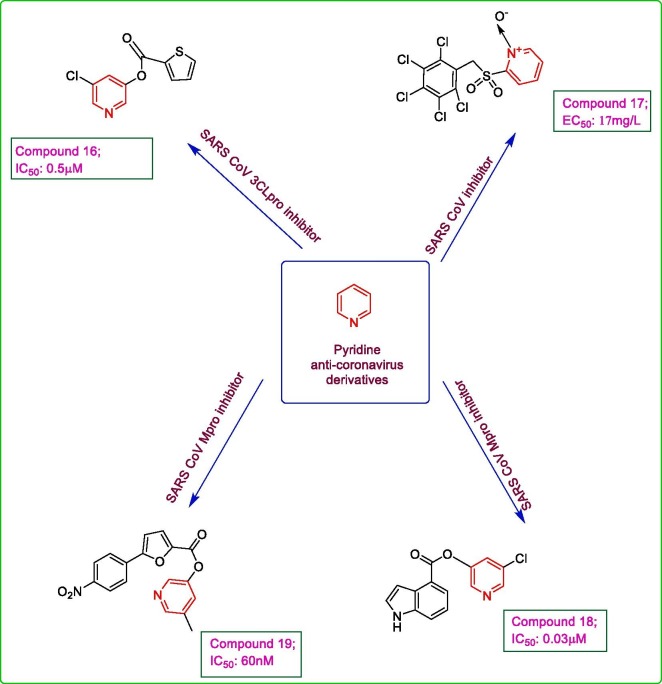
A library of 50,000 compounds was screened for their activity against SARS CoV 3CLpro using FRET analysis. The non-specific compounds were further screened by examining their activity against SARS CoV 3CLpro in the presence of BSA (bovine serum albumin) and out of the screened compounds, 69 compounds show specificity against SARS CoV 3CLpro. All of these screened compounds showed potential electrophilic centres like amides, nitriles which may be capable of forming covalent bond with the nucleophilic thiol of Cys 145 at the active site of 3CLpro. Therefore, they were further analysed in the presence of DTT (1,4-dithio-d,l-threitol) for their specificity against SARS CoV 3CLpro which depicted 5 inhibitors whose activities were least affected by DTT. Finally, the selected compounds were evaluated against other coronaviruses also namely hepatitis A virus 3Cpro, chymotrypsin, hepatitis C NS3pro and papain, to test their selectivity for SARS CoV 3CLpro. Compound 16 displayed the highest selectivity and potency against SARS CoV 3CLpro with an IC50 value of 0.5 μM, thus emphasizing on the role of 5-chloropyridine derivatives as anti-SARS CoV agents [34]. The derivatives were evaluated for their activities against SARS CoV (Frankfurt-1 strain) and FIPV (feline infectious peritonitis virus) infections by MTT assay in Vero E6 cells and CRFK (confluent crandel feline kidney) cells infected with 100 CCID50 (50% cell culture infective dose) SARS CoV and FIPV, respectively. The highest potency was revealed by compound 17, with an EC50 of 17 mg/L against SARS CoV and with almost no cytotoxic effect in Vero cells (MIC > 100 mg/L). The compound showed post entry inhibition of viral infection in time of addition experiment in FIPV infected CRFK cells. The structure activity relationship study revealed that the sulphide and sulphoxide analogues displayed poor activity against both viruses. The reduced pyridine-N-oxide analogues exhibited a complete loss of antiviral potential against both SARS CoV and FIPV, at their subtoxic concentrations, thus emphasizing on the imperative role of N-oxide in pyridine nucleus [35]. The 5-chloropyridinyl indolecarboxylate analogues were synthesized as anti-SARS CoV evaluated for their in vitro and in silico anti-SARS CoV 3CLpro inhibitory activity [36]. The FRET inhibition assay demonstrated compound 18, as the most active analogue having a 4-carboxylate indole substitution, with an IC50 value 0.03 μM. The analysis results indicated a decrease in potential with the shift of carboxylate moiety on positions other than 4 in indole nucleus. The acylation of indole N also resulted in loss of anti-viral potency. In silico study was also conducted by docking of compound 18 within the binding pocket of SARS CoV 3CLpro (PDB: 2HOB) using GOLD 3.2 program which further confirmed its promising role as an anti-SARS CoV agent. The results of docking analysis revealed a good fit of compound 18 within the active site of protease due to the formation of three hydrogen bonds by carbonyl oxygen with Cys 145, Ser 144 and Gly 143 while the indole moiety fits into the hydrophobic S2 site involving the interaction of imidazole N with His 41 [37]. Two series of derivatives of 5 chloropyridine were synthesized using MAC-5576 16 (a potent anti-SARS CoV agent) as lead. In vitro SARS CoVMpro inhibition assay showed the derivatives of series 1 (derivatized benzene substituted furan fused to chloropyridyl ester) to be possessing higher inhibitory potential than those of series 2 (derivatized, six membered cyclic aromatic moiety clubbed with chloropyridyl ester) with compound 19 as the most promising inhibitor exhibiting an IC50 value of 60 nM. The analysis results revealed a decrease in anti-SARS CoV potential with a change in the position of nitro group in compound 19 from para position to ortho or meta position of benzene ring. The molecular docking study using AutoDock depicted the binding of chloropyridine group within the S1 pocket of SARS CoV 3CLpro, with N of pyridine forming a hydrogen bond with N of His 163 while the furan moiety fits into the space between S1 and S2 subsites involving van der Waals interaction with Met 165. The p-substituted benzene moiety (ΔG: −9.34 kcal/mol) fitted well into the S2 pocket while the o- or m-substituted benzene rings in other derivatives were forced to move outside from S2 pocket due to steric hinderance with SARS CoVMpro residues, His 41 and Met 49 [38]. Fig. 3 . shows various pyridine derivatives.
Fig. 3.
Potential Pyridine derivatives.
5. Purine and pyrimidine derivatives
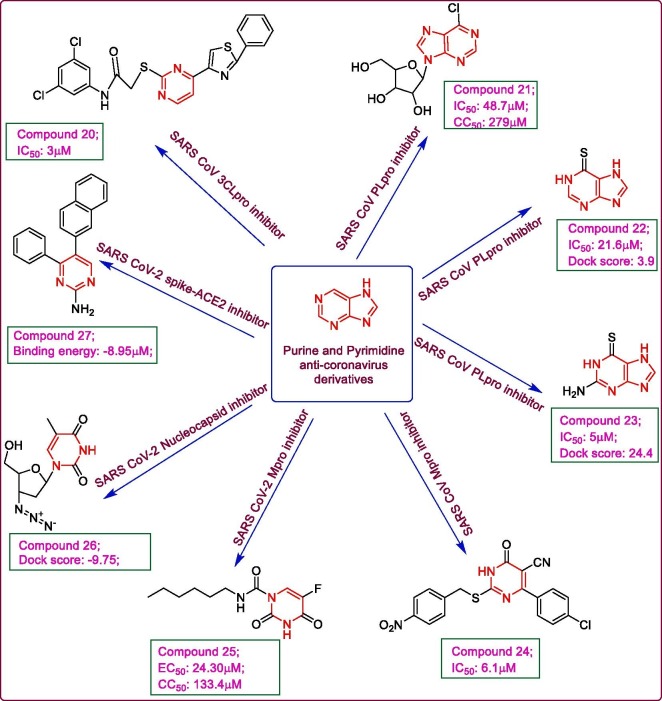
The Maybridge compound database, was virtually screened for a set of anti-SARSCoV 3CLpro (PDB: 1UK4) inhibitors within the binding site containing the catalytic dyad Cys 145 and His 41 which forms a part of subsites S1, S2 and S3, using DOCK 4.0.2 server. The top 93 compounds involving more than two hydrogen bonds with Mpro, were further analysed by SARS CoVMpro inhibition assay, among which 21 derivatives were found active exhibiting an IC50 value of <30 μM. Three of these 21 compounds shared a similar core structure, N-phenyl-2-(2-pyrimidinylthio)acetamide, using which 25 more structural analogues were screened from Maybridge, SPECS_SC and ChemBridge databases and a total of 28 analogues were analysed for anti-SARS CoV inhibitory potential which displayed compound 20 as the most promising analogue with an IC50 value of 3 μM. The results furnished from 3D-QSAR modelling using COMFA and COMSIA analysis completely matched with those from experimental analysis. The MM/GBSA analysis results also revealed the best fit of compound 20 within the binding pocket of SARS CoV 3CLpro with a binding free energy of –23.17 kcal/mol, due to the strong interactions formed by its thiazole and benzene moieties with protease residues namely, Gln 192, Leu 167, Glu 166 and Pro 168 [39]. 6-Chloropurine derivatives have been reported to exhibit potent antiviral activity against many viruses. Nucleoside analogues based on 6-chloropurine moiety were designed and analysed for their activity against SARS CoV using plaque reduction assay in SARS CoV Frankfurt-1 strain infected Vero E6 cells. Compound 21 exhibited promising activity with an IC50 value of 48.7 μM and a decrease in virus yield to less than one-hundredth of the control, at 20 μM concentration. The structure activity relationship study showed the importance of chlorine atom at position-6 of purine nucleus and a loss of anti-SARS CoV activity with the 2-amino group substitution in 6-chloropurine moiety. The replacement of chlorine group with weaker leaving groups like —SMe or —OMe, led to a decrease in anti-SARS CoV potential which is attributed to be due to lack of formation of an irreversible covalent bond which 6-chloropurine can form with the active site of SARS CoV owing to its electrophilic nature. The unprotected 5′-hydroxyl substitution in ribofuranosyl moiety is also imperative as it gets converted to the active triphosphate form leading to the antiviral activity. Also, the replacement of ribofuranosyl moiety with 2′-deoxy- or 3′-deoxyribonucleoside analogues led to a detrimental effect on the anti-SARS CoV potential of compound 21 [40]. From the compound library, Genesis plus collection, 960 compounds were screened against SARS CoVPLpro using in vitro and in silico approaches. The results of deubiquitination assay demonstrated that out of the screened compounds, only two derivatives, 6-Mercaptopurine 22, (IC50 value: 21.60 μM) and 6-Thioguanine 23 (IC50 value 5 μM) exhibited pronounced SARS CoVPLpro inhibition. The thiocarbonyl group of 22 and 23 was found to be crucial for SARS CoVPLpro inhibition using structure function relationship study by ISIS, as replacing it with hydroxyl or methylthio moiety led to loss of inhibition. The docking analysis confirmed the inhibition inhibition mechanism within the active sites of SARS CoVPLpro (PDB:2FE8) and 3CLpro (PDB: 1UK2) using DS modelling 1.7 program. The docking result suggested a good fit of both the compounds with a dock score for 22 (23.9) and 23 (24.4) within the active site of PLpro with the probability of formation of a hydrogen bond between sulphur atom of the compounds and Cys 1651 residue of SARS CoVPLpro resulting in reversible competitive inhibition. While the dock scores with SARS CoV 3CLpro were found to be 17.8 for 22 and 18.4 for 23, much lower than with SARS COV PLpro, thus emphasizing on the selectivity of both the compounds towards PLpro [41]. Another series of pyrimidine derivatives was synthesized and evaluated for their SARS CoV 3CLpro inhibitory potential. The FRET analysis using Dabcyl-KTSAVLQSGFRKME-Edans as fluoregenic substrate demonstrated compound 24 to be endowed with marked SARS CoV 3CLpro inhibitory potential exhibiting an IC50 value of 6.1 μM, with no cytotoxicity as depicted by MTT assay. The molecular docking analysis of compound 24 with SARS CoV 3CLpro (PDB: 1UK4) using Discovery studio modelling 1.2 SBD program unleashed a good fit of phenylnitro group within the S1 pocket with O atom of nitro group forming a hydrogen bond with Cys 145 and Gly 143 at the binding site of 3CLpro. The lack of nitro group led to a loss of inhibitory potential, emphasizing on the imperative role of nitro substituent. Moreover, electron withdrawing groups like chloro in phenyl ring favoured the inhibitory potential more than the electron releasing groups like methoxy or methyl [42]. The antineoplastic drug carmofur 25, a pyrimidine analogue was analysed for SARS CoV-2 Mpro inhibitory potential using the X-ray crystallographic study. The electron density map of the 25-SARS CoV-2 Mpro complex indicated a favourable conformation of the drug within the active site of protease with its fatty acid moiety forming a covalent bond with Cys 145 of the catalytic dyad leading to the release of 5-fluorouracil. The carbonyl O of 25 was observed to be involved in hydrogen bond formation with Cys 145 and Gly 143 while the fatty acid tail showed hydrophobic interaction with His 41, Met 165 and Met 49 at the S2 subsite of the protease. The in vitro inhibition assay in SARS CoV-2 infected Vero E6 cells revealed an EC50 value of 24.30 μM while the cytotoxicity study indicated the selectivity of compound 25 with a CC50 value of 133.4 μM [43]. A set of 6799 compounds from Pubchem and Asinex library were virtually screened and analysed by molecular docking against SARS CoV-2 nucleocapsid RNA binding domain (PDB: 6VYO). The high throughput virtual screening with docking was done using Schrodinger’s molecular docking module at the active sites 1, 2 and 3 of the protein. From the screened ligands, compound zidovudine 26, a thymidine analogue, demonstrated the highest potential with a dock score of −9.75 while a binding free energy of −59.43 kcal/mol at site 3, as depicted by MM-GBSA approach. The molecular dynamic simulation study also revealed a stable interaction of 26-SARS CoV-2 N protein complex, thus suggesting the exploration of this potential nucleus as anti-SARS CoV-2 agent [44]. A series of diaryl pyrimidine derivatives were evaluated for their SARS CoV-2 inhibitory potential using in silico approaches. The molecular docking analysis of the derivatives with the SARS CoV-2 spike glycoprotein-human ACE-2 complex (PDB: 6VW1) was carried out using AutoDock Vina while the results were analysed by MGL tools and PyMol. The pyrimidine derivative 27 showed best binding affinity at the interface of the complex with a binding energy of −8.95 kcal/mol which was attributed to be due to formation of a hydrogen bond with ASP 350, Arg 393 and Asn 394 residues and hydrophobic interactions of its naphthyl and phenyl moieties with Phe 40, Trp 69, Leu 73, Phe 390 and Leu 391 amino acid residues. The molecular dynamic simulation study with GROMACS v5.1.4 biomolecular simulation package demonstrated a stable conformation of 27 within the spike glucoprotein-ACE2 complex throughout the simulation. Its high binding affinity of with the complex was further confirmed by binding free energy calculation using MM-PBSA, the results of which came out to be lowest (ΔG: −30.89 kcal/mol), thus suggesting further evaluation of diaryl pyrimidines as effective anti-SARS CoV-2 agents [45]. Fig. 4 highlights some of the purine and pyrimidine derivatives active against coronaviruses.
Fig. 4.
Purine and Pyrimidine derivatives acting on coronaviruses.
6. Pyrazole derivatives
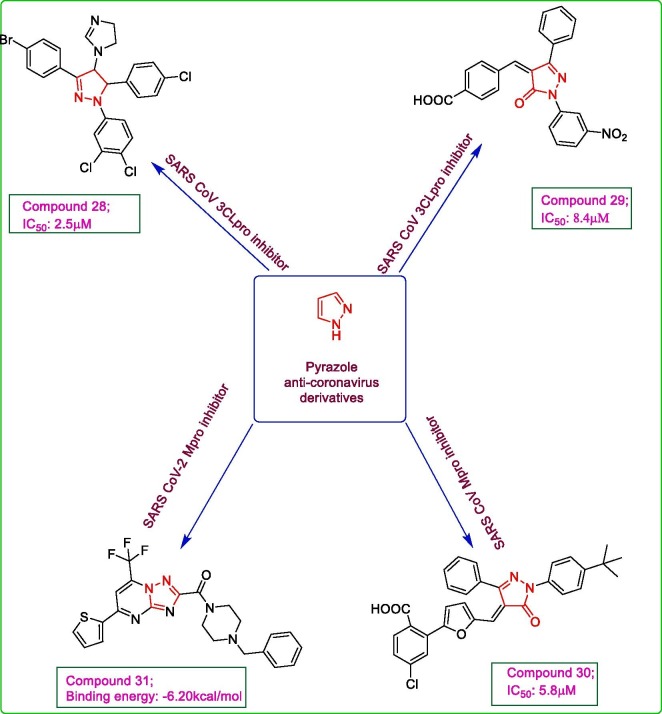
Pyrazolones have been exploited for activity against viruses. A set of 6800 compounds from Korea chemical bank database underwent high throughput screening to obtain the most potent compound against SARS CoV 3CLpro. After primary screening using Dabcyl-KTSAVLQSGFRKME-Edans as fluoregenic substrate, the compounds that showed > 50% inhibition of protease activity at 50 μM further undergo secondary screening at 10 μM. The inhibition assay revealed compound 28 as the most potent inhibitor of SARS CoV 3CLpro with an IC50 value of 2.5 μM. The docking analysis of compound 28 within the active site of SARS CoV 3CLpro (PDB: 1UK4) using Discovery studio modelling 1.2 SBD program, depicted a good fit of the molecule with its 4,5-dihydro-1H-pyrazole moiety occupying S1′ and S2 sites and remaining part resting at S3 site of the protease [46]. Another series of pyrazolone analogues were reported for the antiviral potential against SARS CoV 3CLpro and CVB3 3Cpro. As a result of in vitro FRET analysis, compound 29, with a p-benzylidene aryl ring substitution at C4 of pyrazolone moiety, was observed to be endowed with marked activity against both 3CLpro and 3Cpro with IC50 values of 8.4 μM and 9.6 μM, respectively. The MTT cytotoxicity assay results revealed no cytotoxicity of the target compounds at 200 μM. To further confirm the inhibitory potential of the target compounds, molecular docking analysis via Discovery studio modelling 1.2 SBD program of Accelrys, of the derivatives within the binding site of SARS CoV 3CLpro (PDB: 1UK4) was performed which unleashed a favourable conformation of compound 29 with its N1 phenyl substituent fitted well in S1′ pocket while the O of nitro group formed a hydrogen bond with Gly 143. The carbonyl oxygen of pyrazolone moiety was involves in H-bond formation with Glu 166. The phenyl ring at C3 of pyrazolone fitted well in hydrophobic S2 site while the p-carboxybenzylidene substituent at C4 of pyrazolone nucleus fitted easily in S3 pocket with carboxyl oxygen forming a hydrogen bond to Gln 192. The in vitro and in silico study results emphasized on the imperative role of carboxyl group in benzylidene ring as pyrazolone derivatives lacking this substituent lost their inhibitory potential. It was found that the presence of electron withdrawing substituents like nitro, cyano or fluoro at N1 phenyl ring led to an increase in inhibitory potential [47]. Some 5-pyrazolone derivatives were designed and evaluated for their inhibitory potential against SARS CoV and MERS CoV 3CLpro. The in vitro fluorometric analysis using Dabcyl-KTSAVLQSGFRKME-Edans as peptide substrate for 50 nM SARS CoV 3CLpro or 300 nM MERS CoV 3CLpro, unleashed the highest potential of compound 30 with IC50 values of 5.8 μM and 7.4 μM, respectively. The structure activity relationship study proved that the replacement of bulky phenyl substituent from position 3 of pyrazolone moiety with smaller groups like methyl or CF3 will led to loss of inhibitory potential. Also, the removal of carboxylate group from compound 30 resulted in derivatives with no inhibitory activity. The substitution with lipophilic group at p-position of phenyl ring at N1 of pyrazolone also led to a marked increase in activity. The in vitro assay results were further confirmed by docking analysis of the ligand within the active site of SARS CoV 3CLpro (PDB: 2ALV) using iGemdock v2.1 program. The docking results revealed a good fit of compound 30 within the active site with carboxylate moiety resting in S1 site forming hydrogen bonds to Gly 143, Ser 144 and Cys 145 while furan moiety interacting with hydrophobic S1′ site. The lipophilic tert-butyl group of compound 30 showed good interaction with hydrophobic S2 site, thus enhancing its inhibitory potential. Also, the carbonyl group of pyrazolone moiety is involved in hydrogen bond formation with His 41, thus destabilizing the catalytic dyad at the active site of SARS CoV 3CLpro [48]. The ZINC database was screened using RASPD web server with the aim of searching for potent SARS CoV-2 main protease inhibitors. The best two hit molecules selected as a result of RASPD score, were screened for their drug likeness using SwissADME and Molinspiration servers which revealed a good bioactive score and pharmacokinetics of both the ligands. Therefore, these two ligands were analysed for their binding affinities with SARS CoV-2 main protease (PDB: 6LU7) using ParDOCK server. The docking study results showed a good fit of both the ligands within the active site of protease with a higher binding affinity of compound 31 (Binding energy: −6.20 kcal/mol) comparable to ligand N3 (Binding energy: −6.43 kcal/mol), involving π-alkyl interactions with histidine residue of main protease [49]. Fig. 5 depicts pyrazole derivatives.
Fig. 5.
Pyrazole derivatives acting as inhibitors of SARS CoV or SARS-CoV-2.
7. Thiazole derivatives
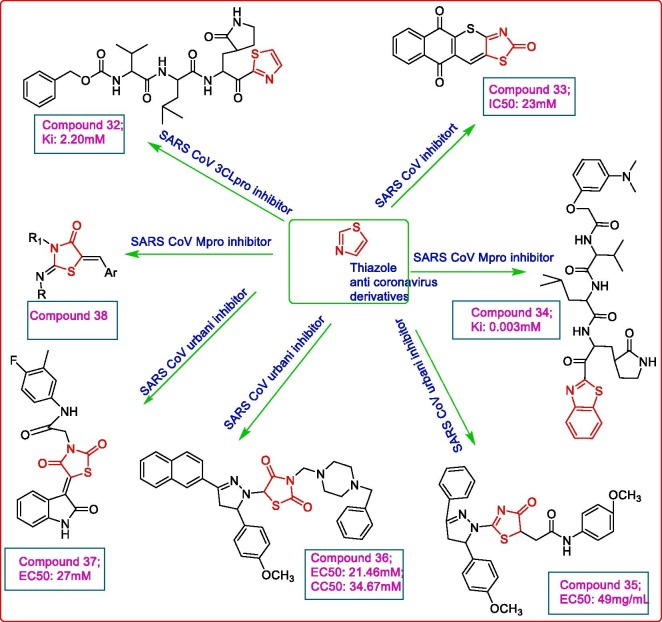
Thiazoles and their derivatives have been explored for activity against corona viruses. A series of trifluoromethyl ketone peptide derivatives were designed and evaluated for their activity against SARS CoV 3CLpro. The inhibition assay results revealed low inhibition potential of these derivatives with a minimum Ki value of 21 μM. Regnier et al. designed another series of derivatives by replacing the trifluoromethyl ketone group with electron withdrawing thiazolyl or benzothiazolyl ketone moieties, with the objective of improving the covalent bond formation with Cys 145 of catalytic dyad at the active site of 3CLpro. The inhibition assay results proved the highest potency of compound 32, a thiazolyl ketone peptide derivative, possessing a Ki value of 2.2 μM. This inhibitory potential was further confirmed by computational docking study of 32 with SARS CoV 3CLpro (PDB: 1WOF) using MOE 2007.09 modelling package which demonstrated a good fit of ligand within the active site of protease with N of thiazole forming a hydrogen bond to His 41 while the benzyloxycarbonyl and thiazolyl ketone groups involved in interaction with Cys 145 of the catalytic dyad [50]. The 5-arylidene-4-thiazolidinones were considered less selective towards biological targets because of the high reactivity of exocyclic double bond with the nucleophilic protein residues favouring the Michael addition reaction. Therefore, isosteric thiopyrano[2,3-d][1,3]thiazole derivatives were synthesized taking 5-substituted-4-thioxo-2-thiazolidinone as precursors. Compound 33 displayed the highest anti-cancer potential with GI50 value of 0.309 μM against MCF-7 breast cancer cell lines while it demonstrated a moderate potency against SARS CoV with IC50 value of 23 µM (visual assessment) and IC50 value of 14 µM (neutral red dye assessment) [51]. Konno et al. took compound 32 as lead and carried out the molecular modelling with 3CLpro (PDB: 1WOF) and found a vacant space in S1′ pocket carrying the thiazolyl group of compound 32 and a protruding benzyloxycarbonyl (P4) moiety from the active site while cyclic amide (P1) and electron withdrawing thiazolyl (P1′) moieties were observed to be crucial for inhibitory activity. The authors carried out optimization of P1′ and P4 moieties to get more efficient anti-SARS CoV 3CLpro inhibitors. The fluorescence-based peptide cleavage assay results of the newly optimized derivatives using Dabcyl-KTSAVLQSGFRKME-Edans as fluorogenic substrate, indicated the pronounced inhibition effect of compound 34 (against SARS CoV 3CLpro possessing Ki value of 0.003 μM. The authors reported that a benzothiazolyl moiety led to enhancement of activity owing to its good fit in the large S1′ pocket. Also the presence of 4-N,N'-dimethylaminophenoxy acetyl moiety resulted in favourable hydrophobic interactions with Ala 191 at S4 pocket, with a unique folding conformation for improved anti-SARS CoV potential [52]. As 4-thiazolidinone and pyrazoline analogues exhibit potent antiviral activity, 2-pyrazoline-4-thiazolidinone hybrid derivatives were designed and evaluated in vitro for their anticancer and antiviral potential. Compound 35 emerged as the most potent anticancer and antiviral agent based on AACF (antimicrobial acquisition coordinating facility) programme, against Tacaribe TRVL 11,753 strain (EC50: 0.71 μg/mL) but showed mild activity towards SARS CoV urbani strain (EC50: 49 μg/mL) [53]. The same group (Havrylyuk et al., 2013) synthesized, another series of 5-pyrazoline conjugated 4-thiazolidinones analogues to get more active anticancer and antiviral agents. The evaluation of antiviral potency of compound 36 exhibited an EC50 value of 21.46 μM and a CC50 value of 34.67 μM against SARS CoV urbani strain [54]. A set of 5-ylidene-4-thiazolidinone-3-carboxylic acid derivatives were synthesized and found to have low or moderate inhibitory potential of the target compounds where compound 37 displayed marked anti-SARS CoV activity with an EC50 value of 27 μM and a selectivity index of >3.7 against SARS CoV urbani strain in Vero 76 cells [55]. A commercial database was screened to identify the novel inhibitors of SARS CoVMpro. Using the 3D structure of transmissible gastroenteritis Mpro (PDB: 1LVO) as template, the structure of SARS CoVMpro was simulated and its active site predicted owing to the sequence similarity between the two proteases. The molecular docking analysis result revealed the analogues bearing the core nucleus as compound 38 as inhibitors of SARS CoVMpro had the potential to inhibit Mpro infected Vero-E6 cells [56]. Some of such derivatives are depicted in Fig. 6 .
Fig. 6.
Thiazole derivatives active against coronaviruses.
8. Triazole derivatives
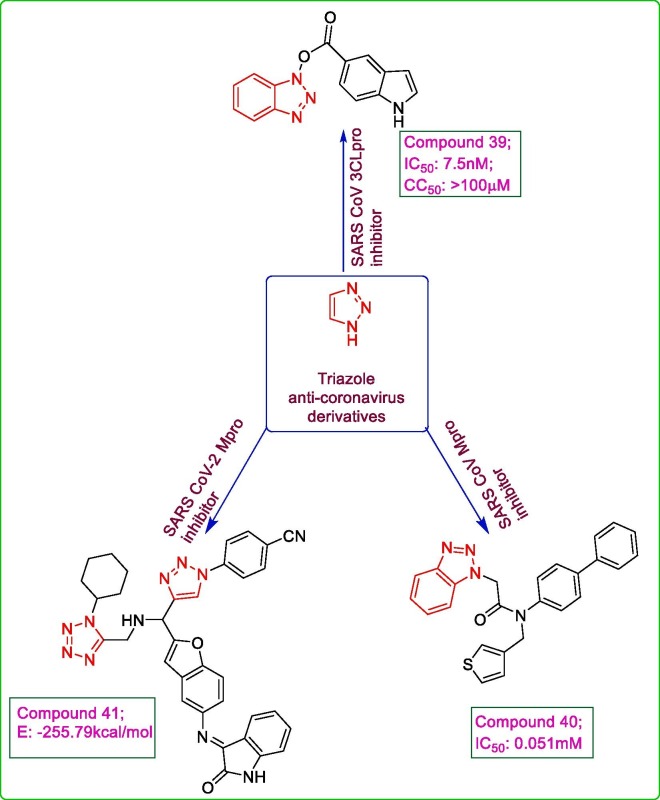
Triazoles have been reported in literature to possess remarkable antimicrobial activity. A series of benzotriazole esters was prepared by condensing (HBTU Hexafluorophosphate benzotriazole tetramethyluronium) with carboxylic acids and the target compounds were evaluated for their potential against SARS CoV 3CLpro using in vitro and in silico studies. The in vitro fluorometric assay revealed compound 39 to be highly active anti-viral agent (Ki value: 7.5 nM; CC50: >100 μM) with an irreversible inhibition of SARS CoV 3CLpro. The in silico molecular docking exposed a favourable conformation of compound 39 within the active site of SARS CoV3CLpro (PDB: 1uk4) with benzotriazole scaffold resting in pocket containing Gly 143, Ser 144 and Cys 145, facilitating a nucleophilic attack by the Cys 145 to the carbonyl group in benzotriazole ester. Also, the hydrogen bond formation between indole of 39 and —OH group of Thr 25 further stabilized the complex formation with SARS CoV 3CLpro. The benzotriazole ester derivatives exhibited a good inhibition of SARS CoV 3CLpro via acylation of Cys 145 in the catalytic dyad at active site [36]. Triazole based non-covalent inhibitors of SARS CoV 3CLpro were developed and analysed for their anti-SARS CoV 3CLpro inhibitory activity. Compound 40, a biaryl substituted triazole derivative possessed a ligand efficiency of >0.3 which on analysis demonstrated the highest potency with an IC50 value of 0.051 μM [57]. Some 1,5-disubstituted tetrazole-1,2,3-triazole conjugates were screened via docking analysis against SARS CoV-2 main protease (PDB:6LU7) possessing the favourable interactions similar to the co-crystalized ligand with the active site catalytic triad, Gly 143, Ser 144 and Cys 145 residues involving the hydrogen bonds and hydrophobic interactions. Ten 1,5-disubstituted tetrazole-1,2,3-triazole hybrids as SARS CoV-2 Mpro inhibitors were generated. Among the proposed hybrids, compound 41 having an isatin moiety exhibited the highest interaction energy (E: −255.79 kcal/mol) within the active site of 6LU7 involving hydrogen bond formation of 1,2,3 triazole moiety with Ser 144 and Cys 145 and that of tetrazole moiety with Ser 1 and Asn 142 and also electrostatic interactions with His 41, His 172, Glu 166 and His 163 while the higher number of rings in its structure enhance the hydrophobic interactions [58]. Fig. 7 depicts some triazole derivatives active against corona viruses.
Fig. 7.
Some active triazole anti-coronavirus derivatives.
9. Miscellaneous heterocycles
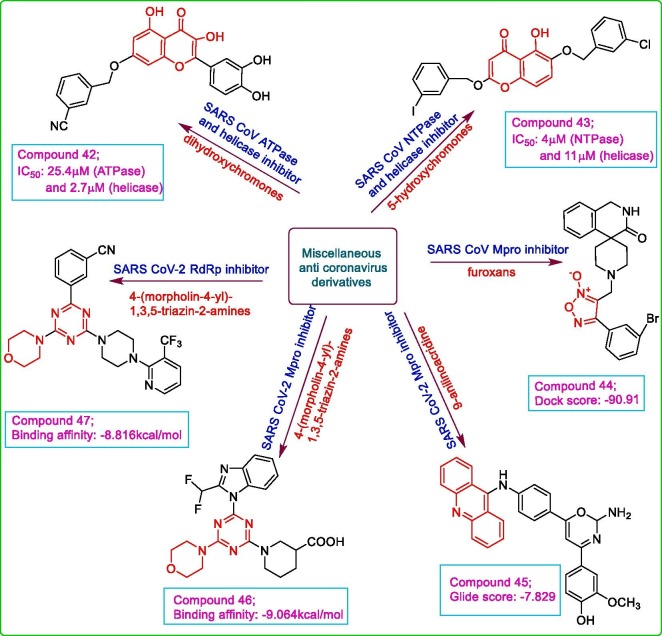
Keeping in view the anti-SARS CoV activity of aryl diketo acids, a series of bioisosteric dihydroxy chromones were designed and examined for their SARS COV inhibitory potential against ATPase and helicase using phosphate release assay method and FRET-based analysis, respectively. The in vitro assay results revealed a good inhibitory potential of compound 42, a flavonol analogue with free catechol group, with IC50 values of 25.4 μM and 2.7 μM against ATPase and helicase, respectively which proposed the involvement of two binding sites in Mpro i.e. one for hydrophobic interaction with arylmethyl moiety while other for hydrogen bond interaction with free catechol moiety [59]. Kim et al. developed a series of 2,6-bis-arylmethyloxy-5-hydroxychromones as anti-SARS CoV agents. The compounds displayed dual inhibitory activities against both nucleoside triphosphatases and helicases of SARS CoV. The compound 43 was found to be endowed with highest potential against both HCV (EC50: 4 μM) and SARS CoV (IC50: 4 μM for ATPase and 11 μM for helicase) with no cytotoxicity in HS27 (human normal fibroblast) cells (CC50 > 50 μM). According to structure activity relationship study, 3-iodo- or chloro-substituted benzyloxy moiety on 5-hydroxy chromone scaffold played a major role whereas derivatives with substitution at position 4 of benzyloxy ring displayed lower inhibition potential [60]. As nitric oxide (NO) derivatives have shown good antiviral effect against SARS CoV, hence phenyl furoxan derivatives, as NO donors, were evaluated by in silico approaches to test their potency against SARS CoV. The molecular docking analysis of some compounds with SARS CoVMpro (PDB: 6 W63) disclosed the best binding pose of compound 44 with binding affinity of −9.8 kcal/mol and dock score of −90.91. The docked conformation revealed hydrogen bond formation of 44 with Cys 145 and Ser 144 residues of the catalytic triad and also aromatic interactions involving His 41 and His 163. It was also concluded from docking results that spiroisoquinolino-piperidine substituted furoxan analogues exhibited better binding interactions compared to benzhydrylpiperazine substituted furoxan analogues. The molecular dynamic simulation demonstrated the stable complex formation between 44 and Mpro comparable to the co-crystalized ligand with ΔG value of −171.972 kj/mol [61]. A series of oxazine conjugate 9-anilinoacridine derivatives was designed using molecular docking approach of Schrodinger suite 2019. All the docked ligands represented favourable conformation within the active site of SARS CoV-2 Mpro (PDB: 5R82) while compound 45 exhibited the highest binding affinity with a Glide score of −7.829 greater than the standard hydroxychloroquine (Glide score: −5.47) which could be attributed to enhanced lipophilicity and hydrogen bonding of the ligand. The —OH group of phenyl ring was found to be involved in H-bond formation with Asn 119 residue of Mpro. The determination of ADMET properties of all the designed ligands were within the acceptable values. The molecular dynamic simulation study depicted a stabilized complex formation of A38-5R82, with —OCH3 group of 45 forming a hydrogen bond with Asn 119 while N of acridine hydrogen bonded with Arg 188 and Ser 144 and acridine moiety also showed π-π interaction with His 41 [62]. With the help of computational methods, the CAS COVID-19 antiviral compound database was screened for nearly 50,000 compounds to examine their potencies against SARS CoV-2 Mpro and RdRp. The compounds were virtually screened by docking analysis using the crystal structures of main protease (PDB: 6LU7) and RdRp (PDB: 6LM7) proteins, by SMINA software. The selected hits were further analysed for their pharmacokinetic and pharmacodynamic parameters using pkCSM model. The best possible compounds were subjected to molecular dynamic simulation for stabilization of the complexes with the proteins via GROMACS 2018 program and compounds 46 (Binding affinity: −9.064 kcal/mol) and 47 (Binding affinity 8.816 kcal/mol) showed best binding poses and good ADMET against Mpro and RdRp, respectively. The compound 46 was stabilized well in the active site of Mpro forming hydrogen bonds with Gly 143, Ser 144 and Cys 145 while the morpholine moiety and 1,3,5-triazine group were involved in hydrophobic interactions with His 41 and Met 49 of catalytic dyad. The compound 47 also showed stable conformation within RdRp active site involving hydrogen bonds with Arg 553, Tyr 619 and Ser 682. There was formation of stable complexes 46 with 6LU7 and 47 with 6MU7 as per MD simulation studies [63]. Miscellaneous compounds are shown in Fig. 8 .
Fig. 8.
Miscellaneous heterocyclic compounds with anti-coronavirus activity.
10. Natural products
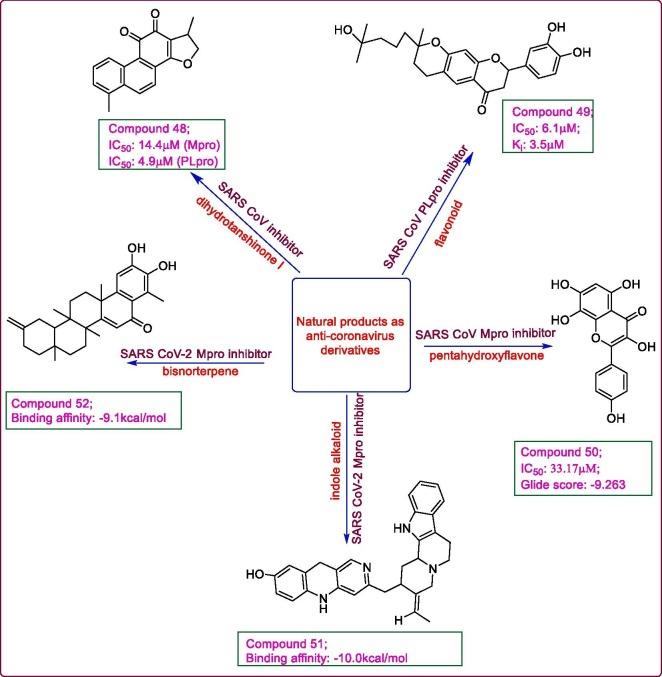
Natural products are being used as natural remedies for various ailments since time immemorial. Scientists have been actively involved in derivatization of these natural products to yield active compounds. A class of quinoid derivatives, tanshinones from Salvia miltiorrhiza, were evaluated for their antiviral potential against SARS CoV cystein proteases, 3CLpro and PLpro. The FRET based peptide cleavage assay results showed that compound 48, a dihydrotanshinone I, was found to be endowed with highest competitive inhibition potential against both SARS CoVMpro (IC50: 14.4 μM) and PLpro (IC50: 4.9 μM) and a good inhibition of deubiquitination (IC50: 1.2 μM). The structure activity relationship study revealed the presence of a naphthalene ring and a dihydrofuran moiety in tanshinone 1 analogues to be crucial for anti-SARS CoV activity [64]. Twelve geranylated flavonoids were isolated from Paulownia tomentosa fruit extract with the view to examine their anti-SARS CoV potential against PLpro. The FRET inhibition assay results demonstrated compound 49 possessing a 3,4-dihydro-2H-pyran moiety, to be the most potent mixed type reversible inhibitor of SARS CoVPLpro exhibiting an IC50 value of 6.1 μM and Ki value of 3.5 μM [65]. Another library of 64 flavonoids was examined using FRET based inhibition assay and induced fit docking analysis, to determine the inhibitory potential of the target compounds against SARS CoV 3CLpro. Herbacetin 50, a pentahydroxyflavone, showed good inhibitory activity with an IC50 value of 33.17 μM. It showed good inhibition even in the presence of 0.01% Triton X-100, which is used to avoid the false bioassay results owing to the aggregating tendency of flavonoids. The induced-fit docking analysis of compound 50 within the active site of SARS CoV 3CLpro (PDB: 4WY3) revealed its good binding interaction with a glide score of −9.263. The 8 —OH group of 50 formed hydrogen bonds with Glu 166 (S1 pocket) and Gln 189 (S2 pocket) which imparted additional binding affinity to 50 with the 4WY3 compared to other analogues, kaempferol and morin which lack the hydroxyl group at position 8 [66]. Some plant alkaloids and terpenoids of African origin were analysed by molecular docking studies, for their antiviral potential against SARS CoV-2 main protease (PDB: 6LU7). Among the alkaloids, 10-hydroxyusambarensine (51) an indole alkaloid from Strychnos usambarensis (Binding affinity: −10.0 kcal/mol) and among the terpenoids, compound 6-oxoisoiguesterin (52), a bisnorterpene from Bisnorterpenes (Binding affinity: −9.1 kcal/mol) exhibited the highest binding potentials against SARS CoV-2 3CLpro even higher than the references taken, lopinavir (-8.3 kcal/mol) and ritonavir (−6.8 kcal/mol). There was hydrogen bond formation with Cys 145, Gln 166 and Gln 189 in 51 and in 52 with Thr 111 and Thr 292 leading to a favourable conformational fit within the active site of SARS CoV-2 Mpro. The ADMET study revealed good pharmacokinetics of both the ligands with a high gastrointestinal absorption index and no toxicity [67]. Fig. 9 highlights some natural products.
Fig. 9.
Natural products as SARS CoV/CoV2 inhibitors.
10.1. Authors’ perspective
Based on the literature available, it was observed that some of the key enzymes of coronaviruses show high sequence similarity among themselves which can be exploited to target these coronaviruses by compounds with similar scaffolds and their bioisosteric analogues over a wide range. Therefore, keeping this in mind, this review gives an insight into the crucial role of heterocyclic moieties as anti-SARS CoV and anti-SARS Cov-2 agents. Isatin nucleus has shown tremendous effects against SARS CoV Mpro involving hydrogen bond formation by carbonyl and amine groups of isatin scaffold within the active site of protease as demonstrated by docking analysis [16]. 5-carboxamide and 5-sulfonamide analogues of isatin also revealed better inhibitory potential compared to 5-iodo analogues [17], [18]. As hybrids of different heterocycles have often been used to increase potency, similarly, the pyrimidine fused indole derivatives demonstrated great potential against SARS CoVMpro [20]. Drugs like vapreotide, arbidol and delavirdine have also shown good inhibition activities against SARS CoV-2 helicase, spike and RdRp proteins, respectively [21], [22], [23]. Quinoline based heterocycles are in great demand with the researches on chloroquine and hydroxychloroquine as anti-SARS CoV-2 agents [33]. The quinolinone derivatives have also shown promise against SARS CoV Mpro, thus emphasizing the need to think of more such derivatives as anti-SARS CoV-2 agents [29]. The 5-chloropyridines like MAC-5576 and pyridine N-oxides also exhibited good inhibition of SARS CoVMpro [34], [35]. Further, the docking analysis revealed a good fit of 4-indolecarboxylate fused 5-chloropyrine derivatives within the binding pocket of SARS CoV Mpro [37]. Therefore, 5-chloropyridine moiety proved to be effective against SARS CoVMpro and can be further exploited against SARS CoV-2 infections. Purine analogues, 6-mercaptopurine and 6-thioguanine demonstrated reversible competitive inhibition of SARS CoV PLpro through the formation of strong hydrogen bonds within the active site of protease [41]. The in silico analysis of pyrimidine based drugs, carmofur, zidovudine and AP-NP unleashed a potent inhibition of SARS CoV-2 Mpro, SARS CoV-2 nucleocapsid RNA binding domain and SARS CoV-2 spike-ACE-2 complex, respectively, thus providing an insight into the need for greater research on purine and pyrimidine analogues as SARS CoV-2 inhibitors [43], [44], [45]. The pyrazole and 5-pyrazolone derivatives underwent a favourable conformation within the active site of SARS CoV 3CLpro through hydrogen bonds and hydrophobic interactions with S1, S1′, S2 and S3 pockets of the protease, emphasizing on a crucial role of this moiety as SARS CoV inhibitory agent [46], [47], [48]. This review discloses a mild to moderate SARS CoV inhibitory potential of pyrazoline conjugated 4-thiazolidinones while 5-benzylidene-4-oxo-1,3-thiazolidine derivatives demonstrated good binding affinity with SARS CoV Mpro [havrylyuk; shen], thus asserting on a need for developing better 4-thiazolidinone derivatives as inhibitors of SARs coronaviruses. Some triazole derivatives also revealed potent inhibition of SARS CoV as indicated by docking analysis with SARS CoV Mpro active site [36]. Heterocyclic compounds of natural origin also play a crucial role in inhibiting the SARS CoV and SARS CoV-2 infections as depicted in this review [64], [65], [66], [67].
11. Conclusion
With the emergence of SARS CoV-2 pandemic there is an urgent need for designing and developing safe, low-cost and potent anti-SARS CoV-2 agents and therapies with the aim to put an end to this global health crisis as early as possible. At present, a number of pharmaceutical industries and research centres throughout the world are working persistently to find a solution to the current pandemic situation. Some drugs are being repurposed based on their current therapeutic application while some are undergoing clinical trials to investigate their safety, efficacy and toxicity against SARs CoV-2 infections. Much focus has been given on pharmacotherapy, immunotherapy and plasma therapy as a means to combat SARS CoV-2 and other such life-threatening infections at present and in future. However, there is no vaccine or medication approved by FDA till date. Therefore, keeping in view the immense role of heterocyclic compounds as antiviral agents specially against coronaviruses, the better knowledge and understanding of the mechanism of heterocyclic scaffolds as anti-SARS CoV, anti-MERS CoV and anti-SARS CoV-2 agents may led to the discovery of an effective antiviral treatment thus minimizing the morbidity and mortality. As SARS CoV-2 show 82% sequence similarity with SARS CoV genome and also many target enzymes in SARS CoV, MERS CoV and SARS CoV-2 also show similarity, therefore, many existing approved drugs and compounds revealing inhibitory potential against SARs CoV or MERS CoV can be exploited to analyse their potential against SARS CoV-2 pandemic. This review focuses on FDA approved or unapproved heterocyclic compounds involved in inhibiting SARS coronaviruses through in vitro or in silico approaches which may act as lead structures for the design and development of potent SARS CoV-2 inhibitors and other such pathogenic infections in future.
Consent for publication
Not applicable.
Funding
None.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
None.
References
- 1.The importance of heterocyclic compounds in anti-cancer drug design. https://www.ddw-online.com/therapeutics/p320375-the-importance-of-heterocyclic-compounds-in-anti-cancer-drug-design.html (accessed July 10, 2020).
- 2.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) Outbreak. J. Autoimmun. 2020:1–4. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novel coronavirus (2019-nCoV) situation report – 1, 21 JANUARY 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 (accessed on July 10, 2020).
- 4.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:1–12. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A distinct name is needed for the new coronavirus. https://www.thelancet.com/pdfs/journals/lancet/PIIS0140-6736(20)30557-2.pdf (accessed on August 24, 2020).
- 6.Coronavirus disease (COVID-19) pandemic. World Health Organization (WHO). https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on August 24, 2020).
- 7.Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). World Health Organization (WHO). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf (accessed on August 24, 2020).
- 8.9 novel coronavirus (9-nCov) Update: uncoating the virus. https://asm.org/Articles/2020/January/9-Novel-Coronavirus-9-nCoV-Update-Uncoating (accessed on August 24, 2020).
- 9.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. Correspondence: the proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong R., Yang G., Xue R., Liu M., Wang F., Hu J., Guo X., Chang S. COVID-19 docking server: an interactive server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa645. https://arxiv.org/ct?url=https%3A%2F%2Fdx.doi.org%2F10.1093%2Fbioinformatics%2Fbtaa645&v=22737329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prajapa M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Li Z., Liu S., Sun J., Chen Z., Jiang M., Zhang Q., Wei Y., Wang X., Huang Y., Shi Y., Xu Y., Xian H., Bai F., Ou C., Xiong B., Lew A.M., Cui J., Fang R., Huang H., Zhao J., Hong X., Zhang Y., Zhou F., Luo H. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B. 2020;811:1–11. doi: 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko W., Rolain J., Lee N., Chen P., Huang C., Lee P., Hsueh P. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents. 2020;55:1–3. doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020:1–7. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Wang Y., Lin Y.W., Chou S., Chen S., Liu L.T., Wu Y., Kuo C., Chen T.S., Juang S. Synthesis and analysis of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L., Liu Y., Zhang W., Wei P., Huang C., Pei J., Yuan Y., Lai L.J. Isatin compounds as noncovalent SARS coronavirus 3C-like protease inhibitors. J. Med. Chem. 2006;49:3440–3443. doi: 10.1021/jm0602357. [DOI] [PubMed] [Google Scholar]
- 18.Liu W., Zhu H., Niu G., Shi E., Chen J., Sun B., Chen W., Zhou H., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014;22:292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K., Yakushiji F., Akaji K., Chen S., Naser-Tavakolian A., Schön A., Freire E., Hayashi Y. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, synthesis, biological evaluation, and docking studies. Eur. J. Med. Chem. 2013;68:372–384. doi: 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed S.F., Ibrahiem A.A., Amr A.E., Abdalla M.M. SARS-CoV 3C-like protease inhibitors of some newly synthesized substituted pyrazoles and substituted pyrimidines based on 1-(3 Aminophenyl)-3-(1Hindol-3-yl)prop-2-en-1-one. Int. J. Pharmacol. 2015;11:749–756. doi: 10.3923/ijp.2015.749.756. [DOI] [Google Scholar]
- 21.Borgio J.F., Alsuwat H.S., Otaibi W.M.A., Ibrahim A.M., Almandil N.B., Asoom L.I.A., Salahuddin M., Kamaraj B., AbdulAzeez S. State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Arch Med Sci. 2020;16:508–518. doi: 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;4:1–3. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beg M.A., Athar F. Anti-HIV and Anti-HCV drugs are the putative inhibitors of RNA-dependent-RNA polymerase activity of NSP12 of the SARS CoV-2 (COVID-19) Pharm. Pharmacol. Int. J. 2020;8:163–172. doi: 10.15406/ppij.2020.08.00292. [DOI] [Google Scholar]
- 24.Dai W., Zhang B., Jiang X., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X., Yang X., Shi Z., Deng F., Hu Z., Zhong W., Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:1–5. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keyaerts E., Vijgen L., Maes P., Neyts J., Ranst M.V. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Bioph. Res. Co. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and Spread. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biot C., Daher W., Chavain N., Fandeur T., Khalife J., Dive D., de Clercq E. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J. Med. Chem. 2006;49:2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y., Zhang N., Wang J., Guo Y., Sun B., Liu W., Zhou H., Yang C. Synthesis and biological evaluation of quinolinone compounds as SARS CoV 3CLpro inhibitors. Chin. J. Chem. 2013;31:1199–1206. doi: 10.1002/cjoc.201300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinoline as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23:876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan S., Chan J.F.W., Chik K.K.H., Chan C.C.Y., Tsang J.O.L., Liang R., Cao J., Tang K., Chen L., Wen K., Cai J., Ye Z., Lu G., Chu H., Jin D., Yuen K. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol. Res. 2020;159:1–9. doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55:1–8. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balzarini J., Keyaerts E., Vijgen L., Vandermeer F., Stevens M., de Clercq E., Egberink H., Ranst M.V. Pyridine N-oxide derivatives are inhibitory to the human SARS and feline infectious peritonitis coronavirus in cell culture. J. Antimicrob. Chemother. 2006;57:472–481. doi: 10.1093/jac/dki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C., King K., Kuo C., Fang J., Wu Y., Ho M., Liao C., Shie J., Liang P., Wong C. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006;13:261–268. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh A.K., Gong G., Grum-Tokars V., Mulhearn D.C., Baker S.C., Coughlin M., Prabhakar B.S., Sleeman K., Johnson M.E., Mesecar A.D. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008;18:5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu C., Yin J., Zhang J., Vederas J.C., James M.N.G. Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1pocket of SARS-CoV Mpro. Bioorrg. Med. Chem. 2008;16:293–302. doi: 10.1016/j.bmc.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai K., Chen S., Liang P., Lu I., Mahindroo N., Hsieh H., Chao Y., Liu L., Liu D., Lien W., Lin T., Wu S. Discovery of a novel family of SARS-CoV protease inhibitors by virtual screening and 3D-QSAR studies. J. Med. Chem. 2006;49:3485–3495. doi: 10.1021/jm050852f. [DOI] [PubMed] [Google Scholar]
- 40.Ikejiri M., Saijo M., Morikawa S., Fukushi S., Mizutani T., Kurane I., Maruyama T. Synthesis and biological evaluation of nucleoside analogues having 6-chloropurine as anti-SARS-CoV agents. Bioorg. Med. Chem. Lett. 2007;17:2470–2473. doi: 10.1016/j.bmcl.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou C., Chien C., Han Y., Prebanda M.T., Hsieh H., Turk B., Chang G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramajayam R., Tan K., Liu H., Liang P. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010;20:3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 44.Yadav R., Imran M., Dhamija P., Suchal K., Handu S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1778536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rane J.S., Pandey P., Chatterjee A., Khan R., Kumar A., Prakash A., Ray S. Targeting virus–host interaction by novel pyrimidine derivative: an in silico approach towards discovery of potential drug against COVID-19. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1794969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo C., Liu H., Lo Y., Seong C., Lee K., Jung Y., Liang P. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583:549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramajayam R., Tan K., Liu H., Liang P. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorg. Med. Chem. 2010;18:7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar V., Tan K., Wang Y., Lin S., Liang P. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar D., Ramappa V.K., Patel R., Kumar V., Dass S.K., Chandra R., Singh P. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regnier T., Sarma D., Hidaka K., Bacha U., Freire E., Hayashi Y., Kiso Y. New developments for the design, synthesis and biological evaluation of potent SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2009;19:2722–2727. doi: 10.1016/j.bmcl.2009.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atamanyuk D., Zimenkovsky B., Atamanyuk V., Lesyk R. 5-Ethoxymethylidene-4-thioxo-2-thiazolidinone as versatile building block for novel biorelevant small molecules with thiopyrano[2,3-d] [1,3]thiazole core. Synth. Commun. 2014;44:237–244. doi: 10.1080/00397911.2013.800552. [DOI] [Google Scholar]
- 52.Konno S., Thanigaimalai P., Yamamoto T., Nakada K., Kakiuchi R., Takayama K., Yamazaki Y., Yakushiji F., Akaji K., Kiso Y., Kawasaki Y., Chen S., Freire E., Hayashi Y. Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorg. Med. Chem. 2013;21:412–424. doi: 10.1016/j.bmc.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Havrylyuk D., Zimenkovsky B., Vasylenko O., Lesyk R. Synthesis and anticancer and antiviral activities of new 2-pyrazoline-substituted 4-thiazolidinones. J. Heterocycl. Chem. 2013;50:E55–E62. doi: 10.1002/jhet.1056. [DOI] [Google Scholar]
- 54.Havrylyuk D., Zimenkovsky B., Vasylenko O., Day C.W., Smee D.F., Grellier P., Lesyk R. Synthesis and biological activity evaluation of 5-pyrazoline substituted 4-thiazolidinones. Eur. J. Med. Chem. 2013;66:228–237. doi: 10.1016/j.ejmech.2013.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaminskyy D.V. Screening of the antiviral activity in the range of C5 and N3 substituted 4-thiazolidinone derivatives. J. Org. Pharm. Chem. 2015;13:64–69. doi: 10.24959/ophcj.15.819. [DOI] [Google Scholar]
- 56.J. Shen, H. Jiang, X. Shen, S. Li, W. Huang, C. Gui, J. Chen, T. Sun, F. Ye, D. Bai, H. Liu, X. Luo, K. Chen, Patent CN200410018418, 2005.
- 57.Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P., Baez-Santos Y.M., Lindsley C.W., Hodder P., Mesecar A.D., Stauffer S.R. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido) phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cortés-García C.J., Chacón-García L., Mejía-Benavides J.E., Díaz-Cervantes E. Tackling the SARS-CoV-2 main protease using hybrid derivatives of 1,5-disubstituted tetrazole-1,2,3-triazoles: an in silico assay. PeerJ Phys. Chem. 2020;2(e10):1–16. doi: 10.7717/peerj-pchem.10. [DOI] [Google Scholar]
- 59.Lee C., Lee J.M., Lee N., Kim D., Jeong Y., Chong Y. Investigation of the pharmacophore space of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone Derivatives. Bioorg. Med. Chem. Lett. 2009;19:4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim M.K., Y M., Parka H.R., Kim K.B., Lee C., Cho S.Y., Kang J., Yoon H., Kim D., Choo H., Jeong Y., Chong Y. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV) Eur. J. Med. Chem. 2011;46:5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Sehemi A.G., Pannipara M., Parulekar R.S., Patil O., Choudhari P.B., Bhatia M.S., Zubaidh P.K., Tamboli Y. Potential of NO donor furoxan as SARS-CoV-2 main protease (Mpro) inhibitors: in silico analysis. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1790038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajagopal K., Varakumar P., Aparna B., Byran G., Jupudi S. Identification of some novel oxazine substituted 9-anilinoacridines as SARS-CoV-2 inhibitors for COVID-19 by molecular docking, free energy calculation and molecular dynamics studies. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1798285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aouidate A., Ghaleb A., Chtita S., Aarjane M., Ousaa A., Maghat H., Sbai A., Choukrad M., Bouachrine M., Lakhlifi T. Identification of a novel dual-target scaffold for 3CLpro and RdRp proteins of SARS-CoV-2 using 3Dsimilarity search, molecular docking, molecular dynamics and ADMET evaluation. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1779130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H., Park S., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1007/s12272-012-0108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keun Cho J., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park Ki. H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioor. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jo S., Kim S., Shin D.H., Kim M. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]