Abstract
Chronic inflammation is a highly prevalent consequence of changes in environmental and lifestyle factors that contribute to the development of cancer. The basis for this critical association has largely remained unclear. The MUC1 gene evolved in mammals to protect epithelia from the external environment. The MUC1-C subunit promotes responses found in wound healing and cancer. MUC1-C induces EMT, epigenetic reprogramming, dedifferentiation and pluripotency factor expression, which when prolonged in chronic inflammation promote cancer progression. As discussed in this review, MUC1-C also drives drug resistance and immune evasion, and is an important target for cancer therapeutics now under development.
The oncogenic MUC1-C protein activates responses associated with wound healing, which if prolonged as in chronic inflammation, promote cancer progression, drug resistance and immune evasion, establishing MUC1-C as a highly important target for cancer treatment.
Introduction
The evolution of metazoans required the formation of simple epithelia for interacting with the external environment (1). In complex multicellular organisms, such as mammals, epithelia further evolved to define the architecture of tissues with an apical cell membrane facing luminal spaces and with basolateral membranes contacting adjacent cells and the extracellular matrix. Simple epithelia were thus the first organized tissues found in evolution and, over time with diversification of organs, epithelial layers required a protective barrier from external insults to maintain homeostasis (1).
Transmembrane mucins appeared in vertebrates to protect epithelia by forming a physical mucous barrier at the apical cell surface (2–5). The MUC1 transmembrane mucin is unique among the others in that it is the only one with expression restricted to mammalian species. MUC1 is also notable for having evolved with a capacity to respond to inflammation. In this way, MUC1 activates wound healing associated responses with proliferation and remodeling. As a consequence of this protective role and the highly prevalent emergence of chronic inflammation, prolonged MUC1 activation drives multiple hallmarks of the cancer cell, such as EMT, epigenetic reprogramming, chromatin remodeling, stemness and pluripotency factor expression.
Section summary
MUC1 represents an evolutionary adaptation of mammals to environmental challenges. Evolutionary adaptations occurring from natural selection, if successful, are beneficial for survival. This review addresses how the MUC1 ‘protectogene’ became an adverse adaptation that, as a result of changes in environmental factors, emerged as an oncogene.
MUC1 acts as a sensor of the microbiome and epithelial cell homeostasis
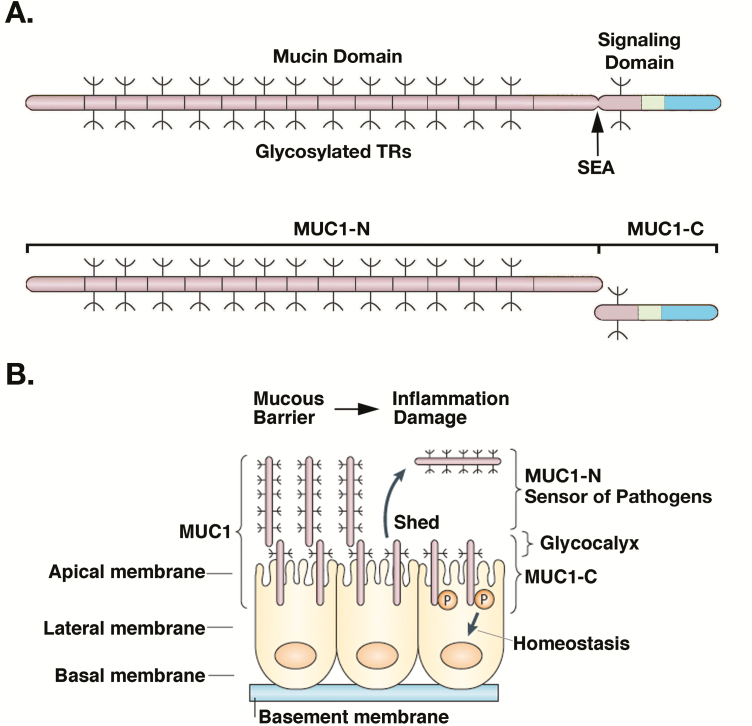
The MUC1 gene, located at 1q22 in a region that is frequently amplified in human cancers, encodes a single polypeptide containing an ectodomain with variable numbers of tandem repeats (TRs), a transmembrane domain and a cytoplasmic domain (CD; Figure 1A) (6). The MUC1 ectodomain includes a sea urchin sperm protein, enterokinase and agrin sequence that is subject to a unique process of autoproteolytic cleavage in the endoplasmic reticulum (ER), resulting in the generation of MUC1 N-terminal (MUC1-N) and C-terminal (MUC1-C) subunits (Figure 1A) (6,7). In turn, MUC1-N and MUC1-C form a non-covalent heterodimeric complex (Figure 1A) that is transported from the ER to the Golgi, where it is modified by glycosylation, and then for positioning at the epithelial cell membrane (Figure 1B).
Figure 1.
The MUC1 protein is cleaved into MUC1-N and MUC1-C subunits that form a complex at the epithelial cell apical membrane to respond to the microbiome and loss of homeostasis. (A) MUC1 is translated as a single polypeptide that includes (i) a characteristic mucin-like domain of glycosylated proline, threonine and serine (PTS) rich TRs and (ii) a signaling domain that evolved in mammals as an adaptation to environmental stress. MUC1 undergoes auto-cleavage at a SEA domain, resulting in MUC1 N-terminal (MUC1-N) and C-terminal (MUC1-C) subunits that, in turn, form a non-covalent heterodimer (8). The MUC1-N and MUC1-C nomenclature defines positioning of the subunits after cleavage and distinguishes them from genetic isoforms designated by Greek characters, such as ERα and ERβ, among others. Figure modified from Kufe (15). (B) MUC1-N extends as a rod-like structure into and beyond the glycocalyx as a component of the protective mucous barrier. MUC1-N is tethered to the cell membrane in a complex with the transmembrane MUC1-C subunit. Mechanical disruption of the complex in the response to loss of homeostasis results in shedding of MUC1-N into the mucous barrier and activation of MUC1-C for the intracellular transduction of signals to reestablish homeostasis. Figure modified from Kufe (6).
Under non-stressed conditions, the MUC1-N/MUC1-C complex is positioned in an inactive state at the apical borders of polarized epithelial cells where it contributes to the composition, organization and function of the glycocalyx. The MUC1-N subunit, consisting of highly glycosylated TRs ranging from 20 to >100 in number, forms a rigid structure that extends over 100 nm from the cell surface and beyond the ~10 nm glycocalyx into the mucous gel barrier (Figure 1B) (6).
The MUC1-N/MUC1-C complex functions in communication between the glycocalyx and apical cell membrane, and acts as a sensor of entropic forces within the extracellular matrix (9). As a result, epithelial cells are protected in part against mechanical forces and loss of homeostasis by disruption of the non-covalent association between MUC1-N and MUC1-C (Figure 1B) (10). In this way, the MUC1-N/MUC-C complex is poised to respond to infections, as well as toxins, physical damage and other forms of stress, that threaten integrity of the epithelial layer (Figure 1B).
MUC1 also evolved to play a role in protecting the epithelium from viral and bacterial infections (11). In responding to threats from the microbiome, MUC1-N acts as an adhesion ligand for the flagellin of Pseudomonas aeruginosa (11). In addition, MUC1-N functions as an adhesion receptor for the enteric pathogens Campylobacter jejuni, enteroaggregative Escherichia coli, and Helicobacter pylori, the latter of which has been linked to chronic gastritis and gastric cancer (12). Binding of MUC1-N to these microbes limits their proximity to the epithelial cell surface and, with shedding of MUC1-N (Figure 1B), the bound pathogens are released into the respiratory and gastrointestinal tracts, where they are excreted by mucociliary transport (13). The MUC1-N ectodomain thus acts as a barrier against microbial pathogens and shedding of this subunit affords protection against physical damage to epithelia.
Section summary
MUC1 emerged in mammals as an evolutionary adaptation to protect epithelia from the external environment. The MUC1-N subunit contains glycosylated TRs that contribute to a protective physical barrier. MUC1-N interacts with pathogens to limit their access to the epithelial cell surface. Importantly, the MUC1 gene also acquired sequences encoding the MUC1-C transmembrane subunit which, acting in concert with MUC1-N, functions as a sensor of forces within the glycocalyx associated with disruption of homeostasis.
MUC1-C integrates inflammatory and proliferative responses
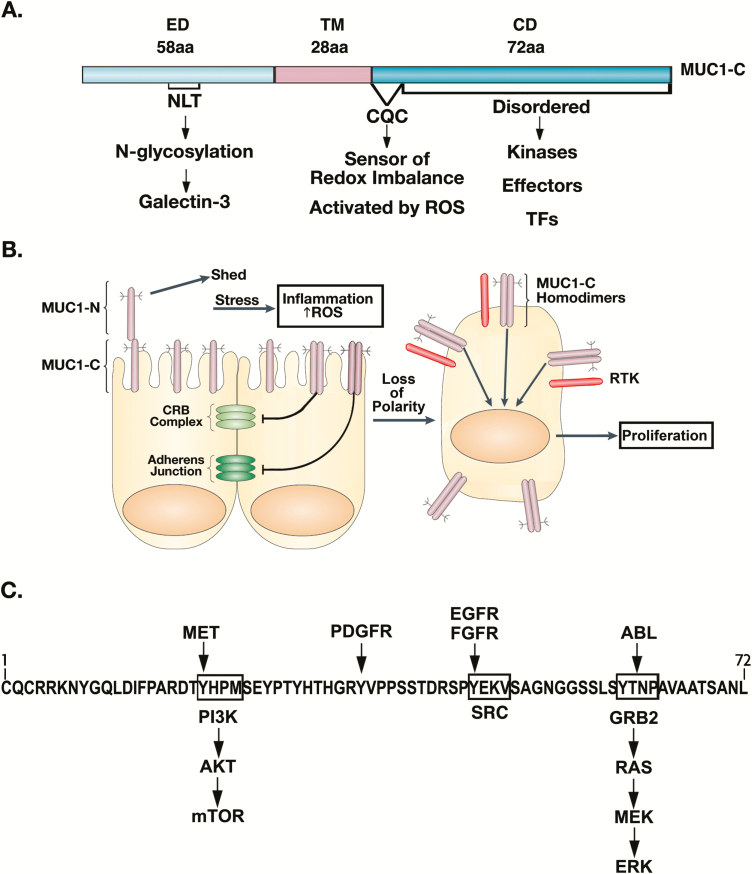
Release of MUC1-N from the apical cell membrane leaves the transmembrane MUC1-C subunit poised for activation in response to inflammation. MUC1-C consists of a 58 aa extracellular domain which, as noted above, forms a complex with MUC1-N that is disrupted by entropic forces at the apical cell membrane. The MUC1-C extracellular domain extends into a 28 aa transmembrane domain and a 72 aa cytoplasmic tail (Figure 2A). The MUC1-C CD is an intrinsically disordered protein (14), which given this lack of structure has the capacity for modifications by diverse kinases and thereby interactions with effectors that have been linked to cancer progression (Figure 2A) (15,16). In this respect, the activation of MUC1-C in response to loss of homeostasis holds potentially important implications regarding the subversion of these pathways for the initiation and progression of cancer cells.
Figure 2.
MUC1-C promotes loss of polarity in the integration of inflammatory and proliferative responses. (A) The MUC1-C subunit consists of the 58 aa extracellular domain (ED), 28 aa transmembrane region and 72 CD. The CD includes a CQC motif, which is activated by increases in ROS in responses to infection, damage and other forms of stress. The CQC motif is necessary for MUC1-C homodimerization, nuclear import and function as an oncoprotein. The CD is otherwise an intrinsically disordered protein that has the capacity for modifications by diverse kinases and thereby structural alterations for direct interactions with signaling molecules and transcription factors (TFs). (B) Inflammation is associated with disruption of redox balance and increases in ROS. MUC1-C is activated by ROS-induced homodimerization and, in turn, disrupts the (i) CRB complex by suppressing CRB3, HUGL2 and PATJ and (ii) adherens junction by downregulating E-cadherin. As a result, MUC1-C contributes to loss of polarity and is repositioned from the apical surface to over the entire cell membrane. There, it interacts with effectors, such as RTKs, that are otherwise restricted to the basolateral borders of polarized epithelial cells. Figure modified from Kufe (6). (C) MUC1-C contributes to RTK activation and their downstream proliferative signaling pathways. MUC1-C CD is phosphorylated by RTKs, resulting in SH2 domain binding motifs for (i) PI3K and activation of AKT→mTOR signaling, (ii) SRC, which can promote transformation and (iii) GRB2 with induction of the RAS→MEK→ERK pathway, linking inflammation and loss of polarity with proliferative responses.
Inflammation is associated with the production of reactive oxygen species (ROS), which activate MUC1-C by inducing the formation of MUC1-C homodimers (17). The MUC1-C CD contains a CQC motif that is adjacent to the cell membrane and acts as a sensor of redox imbalance (Figure 2A). ROS-induced activation of the Cys residues confers the formation of homodimeric MUC1-C complexes (17) and heterodimeric complexes with other proteins, such as MYC (18). MUC1-C has the capacity to regulate intracellular ROS levels, at least in part, through induction of NADPH and GSH production (19). As a result, MUC1-C can control its own activation in the reestablishment of homeostasis.
Epithelial wound healing includes phases of (i) inflammation, (ii) proliferation and (iii) remodeling (20–23). The extracellular role of MUC1-N/MUC1-C in protecting epithelia from stress is integrated with intracellular activation of innate inflammatory responses (6,11). MUC1-C coordinates this initial inflammatory phase with loss of polarity and induction of proliferative responses. Apical-basal polarity and integrity of the epithelial layer are maintained in part by (i) the Crumbs (CRB) complex including the CRB3 tumor suppressor and (ii) the adherens junction, which includes E-cadherin and β-catenin (Figure 2B). Activation of MUC1-C represses CRB3 expression and thereby disrupts function of the CRB complex in maintaining polarity (24). CRB3 activates the core complex of the HIPPO tumor suppressor pathway and, as a result of repressing CRB3, MUC1-C induces YAP (24), a transducer of mechanical signals critical for driving stem cells and tissue regeneration (25). MUC1-C also represses the E-cadherin tumor suppressor with disruption of the adherens junction, thereby further contributing to loss of polarity (Figure 2B).
As a consequence of the loss of polarity, MUC1-C forms complexes with RTKs, such as EGFR, HER2 and FGFR3, and contributes to their activation (Figure 2B) (26,27). In turn, the MUC1-C CD is modified by RTK-mediated phosphorylation and thereby acquires the capacity to interact with downstream effectors of proliferation (Figure 2C) (6,15). In this context, the phosphorylated MUC1-C CD binds directly to the PI3K SH2 domain and activates the AKT→mTOR pathway (Figure 2C) (6,15). MUC1-C also plays a role in activation of MEK/ERK signaling and repression of the RASSF1A tumor suppressor, which inhibits the RAF/MEK/ERK pathway and is one of the most frequently inactivated genes in human cancers (28). This capacity for MUC1-C to drive proliferation, which is necessary for wound healing, holds potentially important implications, if unchecked, for involvement of MUC1-C in promoting sustained progression to carcinoma cells.
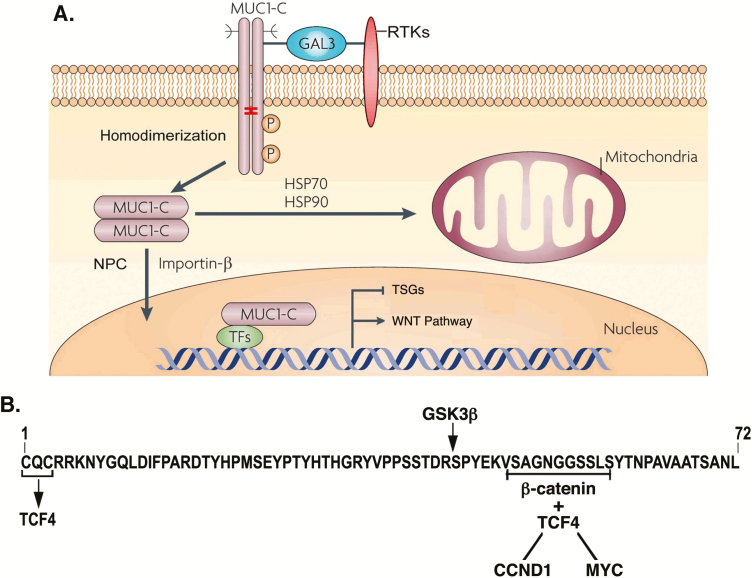
RTKs, such as EGFR and others, are released from the cell membrane by palmitoylation and are imported into the nucleus as uncleaved holoenzymes (29). Localization of cell membrane-bound EGFR to the nucleus is conferred by dysregulation of redox balance and oxidation of Cys residues (30). Endocytic homeostasis is dysregulated in human cancers and, like EGFR, cell membrane-bound MUC1-C is also subject to ROS-induced activation, palmitoylation and thereby similar aberrant intracellular trafficking (31–33). Intracellular MUC1-C homodimers are transported by HSP70/HSP90 complexes to the outer mitochondrial membrane, where they block the intrinsic apoptotic pathway and cell death (Figure 3A) (15). In addition, MUC1-C homodimers are imported into the nucleus by importin-β and the nuclear pore complex nucleoporin 62 (NUP62) (32), NUP358/RANBP2, NUP214 and NUP88 proteins, indicating a direct interaction with the nuclear pore complex cytoplasmic face (Figure 3A). These findings supported a model in which inflammation and the disruption of redox balance extend the role for MUC1-C at the cell membrane to mitochondria and the nucleus.
Figure 3.
MUC1-C transduces cell membrane-associated signaling to mitochondria and the nucleus. (A) Activated MUC1-C homodimers that form complexes with RTKs, such as EGFR, at the cell membrane are internalized by endocytic trafficking. Intracellular MUC1-C homodimers are transported by HSP70/HSP90 to the mitochondrial outer membrane, where they block BAX, activation of the intrinsic apoptotic pathway and cell death. MUC1-C homodimers are imported to the nucleus by interactions with importin-β and nucleoproteins at the cytoplasmic face of the nuclear pore complex. Figure modified from Kufe (6). (B) In the nucleus, the MUC1-C CQC motif interacts directly with the TCF4 E-tail and the SAGNGGSSLS region binds to β-catenin Arm repeats to form a MUC1-C/TCF4/β-catenin complex that activates CCND1 and MYC, and expression of their target genes (34,35). Nuclear MUC1-C also interacts with effectors that promote the repression of TSGs (36).
Early studies showed that the MUC1-C CD binds directly to β-catenin and stabilizes β-catenin by a GSK3β-mediated mechanism (37), establishing involvement of MUC1-C in regulation of the WNT signaling pathway. The significance of this interaction was extended by findings that MUC1-C forms a complex with β-catenin and TCF4 on the CCND1 promoter and activates cyclin D1 expression (Figure 3B) (34). MUC1-C/β-catenin/TCF4 complexes also occupy the MYC promoter and drive MYC expression (24,35,38), linking MUC1-C to aberrant induction of cyclin D1 and MYC in cancers and conferring the capacity for sustained proliferative responses (Figure 3B).
Chronic inflammation of epithelia has been well established as an important driver of cancer initiation and progression (39). As one example, prolonged inflammation and cycles of injury and repair in the intestinal mucosa are associated with the expansion of intestinal stem cells (ISCs) in colitis and the development of colorectal cancer (CRC). Findings that MUC1-C integrates inflammatory and proliferative responses underscored a potential role for MUC1-C in the link between chronic inflammation and cancer (6). In support of that notion, MUC1-C contributes to the progression of colitis to dysplasia and CRC (40,41). MUC1-C promotes intestinal carcinogenesis by inducing (i) the TGF-β-activated kinase 1 (TAK1), NF-κB→p65 and inflammation (40) and (ii) MYC in driving stemness in colitis and CRC (41). In parallel with these findings, MUC1-C-induced activation of NF-κB→p65 and MYC signaling is involved in linking inflammation and proliferation with the induction of EMT and epigenetic reprogramming (36).
Section summary
MUC1-C represents an evolutionary adaptation for protecting mammalian epithelial cells by acting as a sensor of stress and redox imbalance. MUC1-C contributes to loss of polarity and promotes the integration of inflammatory and proliferative responses associated with wound healing. In this way, MUC1-C interacts with diverse effectors and TFs, such as NF-κB, STAT3 and MYC, to activate those responses.
MUC1-C integrates EMT, epigenetic reprogramming and chromatin remodeling
In the remodeling phase of wound healing, migratory cells that have acquired characteristics of the EMT program contribute to re-epithelialization of the damaged area (20,21,23). EMT is driven in part by EMT-TFs, such as TWIST1, ZEB1 and SNAIL, and reprogramming of the epigenome (42). The findings that MUC1-C forms direct complexes with TFs driving inflammatory and proliferative pathways invoked the possibility that the unstructured CD might also interact with EMT-TFs and contribute to the EMT program. Along these lines, activation of MUC1-C→NF-κB→p65 pathway results in induction the ZEB1 gene (Figure 4) (36). In turn, MUC1-C binds directly to ZEB1 and represses miR-200c, which encodes a suppressor of EMT (36) and CRB3 (24), integrating EMT with disruption of the Crumbs complex. Other work has linked MUC1-C with induction of the TWIST1 and SNAIL EMT-TFs (Figure 4) (43,44). TWIST1 is a notable regulator of embryonic morphogenesis that induces EMT, loss of epithelial cell–cell adhesion and an invasive phenotype (45,46). MUC1-C activates the STAT3 pathway in inducing TWIST1 (Figure 4). As found for other TFs, MUC1-C binds directly to TWIST1 and, of significance, MUC1-C/TWIST1 signaling is sufficient for driving (i) ZEB1 and SNAIL expression and (ii) integration of the EMT program with stemness and drug resistance (44).
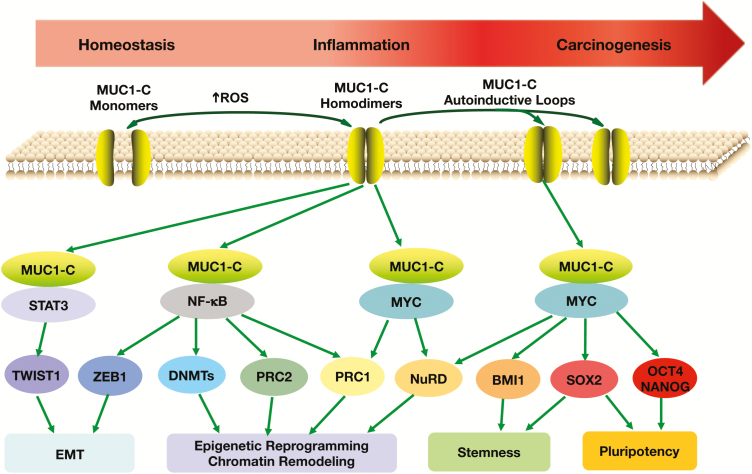
Figure 4.
MUC1-C promotes progression of inflammation and carcinogenesis by integrating EMT, epigenetic reprogramming and stemness. MUC1-C activates the IKK→NF-κB→p65 and JAK1→STAT3 pathways, which are linked to inflammatory responses and are aberrantly activated in cancer cells. The MUC1 gene is also activated by NF-κB→p65 and STAT3, resulting in the formation of auto-inductive circuits that can drive sustained signaling of these pathways in settings of chronic inflammatory responses. MUC1-C/STAT3 complexes activate TWIST1 and the EMT program (44). MUC1-C/NF-κB→p65 complexes induce (i) ZEB1 and EMT, (ii) DNMT1/3b and DNA methylation of TSGs and (iii) EZH2/PRC2 and epigenetic reprogramming (36). Binding of the MUC1-C CQC motif to the MYC HLH-LZ contributes to activation of MYC target genes, including those encoding components of the NuRD and BMI1/PRC1 complexes (18,36). MUC1-C/MYC complexes have also been linked to activation of SOX2, OCT4 and NANOG pluripotency factors (41,47). Prolonged auto-induction of MUC1-C in settings of chronic inflammation drives EMT, epigenetic reprogramming, chromatin remodeling, stemness and pluripotency that promote carcinogenesis (41).
TWIST1 and other EMT-TFs regulate EMT by integrating gene expression with effectors of epigenetic reprogramming (48,49). This integration holds important implications for directing reversible and irreversible regulation of genes that dictate EMT, stemness and plasticity (49). In this context, the MUC1-C→NF-κB pathway plays a role in activating the DNA methyltransferase 1 (DNMT1) and DNMT3b genes (Figure 4) (36), both of which are required for repressing TSGs in cancer cells (50). As a consequence, MUC1-C-induced DNMT1/3b expression results in hypermethylation of the (i) CHD1 promoter with downregulation of E-cadherin (36) and (ii) RASSF1A promoter with suppression of the KRAS→MEK→ERK pathway (28). The MUC1-C→NF-κB→p65 and MUC1-C→MYC pathways play roles in activating Polycomb Repressive Complex 1 (PRC1) and PRC2 (36) (Figure 4). In the regulation of PRC1, MUC1-C (i) induces BMI1, RING1 and RING2 transcription, (ii) binds to BMI1 and (iii) promotes H2A ubiquitylation (36). Regarding PRC2, MUC1-C (i) activates the EZH2, SUZ12 and EED genes, (ii) interacts directly with EZH2 and (iii) enhances EZH2-mediated H3K27 trimethylation on PRC2 target genes (36). In the hierarchical model, PRC1 is recruited to PRC2 sites and, as a result, MUC1-C contributes to the integration of PRC2- and PRC1-mediated repression of TSGs, such as CHD1, CDKN2A, PTEN and BRCA1 (36).
The highly conserved nucleosome remodeling and histone deacetylase (NuRD) complex regulates chromatin assembly and reorganization (51,52). NuRD is of importance for metazoan development and cancer progression (51,52). Components of the NuRD complex include (i) CHD4, which catalyzes ATP-dependent chromatin remodeling, (ii) the non-enzymatic methyl-CpG-binding domain 3 (MBD3) and (iii) metastasis-associated gene 1 (MTA1). Of these, MTA1 is widely overexpressed in human cancer cells with the induction of EMT (53,54) and promotion of invasion and metastases (55–57). An unanticipated finding was that MUC1-C/MYC complexes induce MTA1, MBD3 and CHD4 expression in cancer cells and that MUC1-C associates with the NuRD complex in driving a dedifferentiated phenotype (Figure 4) (18). MTA1 has also been linked to chromatin remodeling during DNA repair by interacting with poly(ADP ribose) polymerase 1 (PARP1) (58,59), in concert with the involvement of MUC1-C in the response to DNA damage and PARP1 activation (60).
Studies in mouse models of intestinal tumorigenesis have linked NF-κB to induction of dedifferentiation and the generation of cancer stem cells (CSCs) (61). In this regard, MUC1-C activates the TAK1→NF-κB pathway in mouse models of colitis-associated colon cancer (CACC) and in human CRC cells (40). Damage to the intestinal epithelium in mice is repaired by expansion of Lgr5+ ISCs, which also contribute to the development of colon cancer (62,63). Of importance, MUC1-C promotes the expansion of Lgr5+ ISCs in mouse models of CACC (41). Moreover, MUC1-C induces LGR5 expression in human CRC stem cells by a MYC-mediated mechanism (41). These findings established a role for MUC1-C in promoting progression of chronic inflammation to cancer by activation of the NF-κB and MYC pathways in the dedifferentiation of ISCs and their transformation to CSCs (Figure 4).
Section summary
Remodeling with re-epithelialization is an essential phase of wound healing. MUC1-C integrates inflammatory and proliferative stress responses with remodeling by activating EMT, epigenetic reprogramming and stemness. In this way, MUC1-C functions as a beneficial adaptation that contributes to repair of damaged epithelia and mammalian survival. In contrast, in settings of chronic inflammation with repetitive cycles of damage and repair, prolonged activation of MUC1-C and these responses promote cancer progression.
MUC1-C induces plasticity and pluripotency factor expression in cancer progression
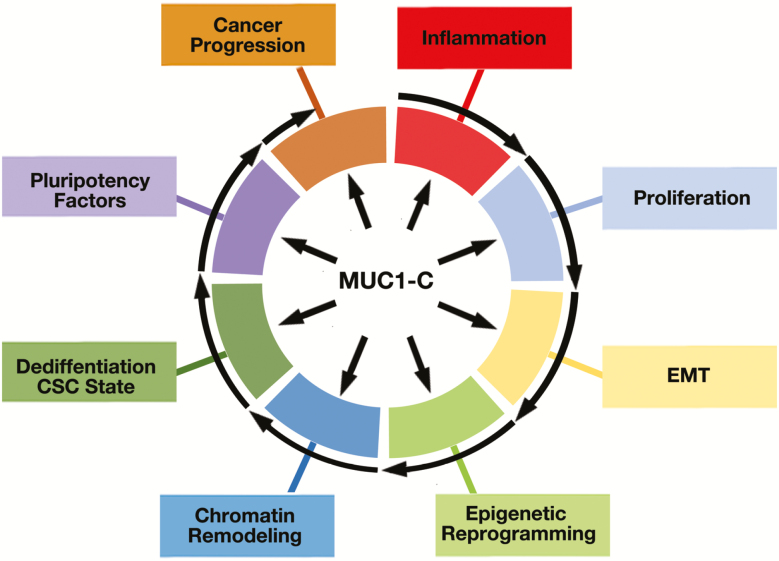
MUC1-C drives multiple hallmarks of the cancer cell, including the integration of inflammation and proliferation with EMT, epigenetic reprogramming and chromatin remodeling (Figure 5). MUC1-C also promotes plasticity as evidenced by involvement in dedifferentiation, transdifferentiation and the interconnectivity of these CSC states (Figure 5).
Figure 5.
MUC1-C drives hallmarks of the cancer cell and is an emerging target for cancer therapeutics. MUC1-C appeared in mammals to protect epithelia from inflammation and loss of homeostasis. MUC1-C activates a wound healing response that has been misappropriated by cancer cells which, in addition to loss of polarity, includes induction of proliferation, EMT and epigenetic reprogramming. MUC1-C also induces chromatin remodeling, dedifferentiation and pluripotency factor expression. Activation of these responses, if sustained as in chronic inflammation, can promote cancer progression and, in turn, drug resistance and immune evasion. Given this capacity, MUC1-C has emerged as an important target for the development of vaccines, CAR-T cells, ADCs, bispecific antibodies and small molecule inhibitors, among other therapeutic approaches.
Adult stem cells have the capacity for self-renewal and the generation of terminally differentiated cells (64,65). Phenotypic plasticity of somatic stem cells includes dedifferentiation that notably occurs during wound healing and cancer progression (66,67). MUC1-C drives luminal→basal dedifferentiation of TNBC cells in association with induction of CSC-associated factors, such as BMI1, ALDH1 and CD44, and the capacity for self-renewal and tumorigenicity (Figure 5) (44). MUC1-C driven plasticity in TNBC cells is linked to (i) NF-κB-mediated induction of EMT, and epigenetic reprogramming and (ii) MYC-mediated NuRD-mediated chromatin remodeling (18,44). As an additional model, castrate resistant prostate cancer often progresses to a more aggressive form of neuroendocrine prostate cancer (NEPC) as a consequence of resistance to treatment with androgen receptor pathway targeted agents (68). MUC1-C contributes to CRPC→NEPC plasticity by suppressing AR-mediated signaling, inducing BRN2 expression by a MYC-mediated mechanism and driving the capacity for self-renewal and tumorigenicity (47).
Another integral component linked to MUC1-C-induced plasticity is the activation of pluripotency networks (18,41,47). The seminal findings that somatic cells can be reprogrammed to pluripotency by expression of the Yamanaka factors, OCT4, SOX2, KLF4 and MYC (OSKM) has had a transformative impact on our understanding of cellular plasticity (69,70). Induced pluripotent stem cells and CSCs, defined by self-renewal and maintenance of tumors, have similar gene signatures and other properties (71–74). However, in contrast to induced pluripotent stem cells, relatively little is known about the regulation of pluripotent reprogramming factors in cancer cells.
Pluripotency factor expression is restricted in somatic cells to maintain lineage specification, but is reactivated in wound healing and in cancer progression (70,75–77). MUC1-C induces MYC by activating the canonical WNT/β-catenin pathway in cancer cells (24,35,38,78). In NSCLC cells, MUC1-C induces LIN28B and thereby downregulation of let-7 miRNA, which suppresses pluripotency factor expression (79,80). MUC1-C also induces SOX2, KLF4 and OCT4 in dedifferentiation of TNBC and NEPC cells (18,44,47) and in the progression of colitis to CRC (41), providing support for involvement of MUC1-C in conferring pluripotency in the progression of chronic inflammation to cancer (Figure 5).
Plasticity of somatic stem cells is essential for wound repair and tissue homeostasis, which is typically reversible as wounds heal (22,75). Importantly, in cancer, plasticity drives the capacity for sustained dedifferentiation and represents a major challenge that is responsible for poor clinical outcomes, heterogeneity, drug resistance and immune evasion (74,81–85).
Section summary
MUC1-C integrates multiple hallmarks of the cancer cell that include stemness, plasticity and pluripotency. MUC1-C has been linked to stemness, plasticity and pluripotency factor expression in the progression of recalcitrant cancers, such as TNBC and NEPC. In concert with these findings, MUC1-C is associated with unresponsiveness to cancer treatment.
MUC1-C confers drug resistance, immune evasion and poor clinical outcomes
MUC1 is overexpressed in multiple types of human cancers compared with their normal tissue counterparts. As initially found in breast carcinomas (86), MUC1 overexpression is associated with MUC1 gene alterations in multiple cancers. In contrast to other oncoproteins, such as RAS, which are frequently altered, mutations in MUC1-C are uncommon, consistent with the findings that MUC1-C mutants function as dominant-negatives for transformation (87). With regard to clinical outcomes, overexpression of MUC1-C in carcinomas is associated with induction of gene signatures that predict highly significant decreases in patient disease-free and overall survival (88–90). Multiple meta-analyses have further shown that MUC1 expression is linked to poor clinical outcomes in patients with diverse types of carcinomas (91–98).
Clearly, the upregulation of MUC1 expression per se is not a transforming event. Along these lines, MUC1 is highly expressed in the lactating mammary gland, which importantly occurs in association with suppression of the EMT program and stemness to preserve epithelial integrity and differentiation (99,100). In this setting, remodeling is a benign process during involution of the lactating mammary gland (101,102). In contrast, increases in MUC1-C during chronic inflammation and cancer progression are associated with EMT, dedifferentiation and pluripotency. Indeed, this distinction for involvement of MUC1-C in lactation and in promoting carcinogenesis is of importance as a focus for future investigation.
Early studies found that MUC1-C confers resistance of carcinoma cells to genotoxic anti-cancer agents, which was attributed in part to suppression of the DNA damage-induced apoptotic response (103). Consistent with those findings, targeting MUC1-C is synergistic with doxorubicin and taxol against cancer cells (104,105) and is effective in reversing resistance to these agents (44,60,106). Additional work identified involvement of MUC1-C in the repair of DNA damage and the integration of epigenetic reprogramming in that response (44,60). MUC1-C also promotes resistance to targeted agents, including tamoxifen, trastuzumab and afatinib, and inhibiting MUC1-C function is synergistic with these agents in treatment of drug resistant cells (26,27,107). The findings that MUC1-C drives resistance to genotoxic and targeted agents supported a pleotropic capacity to circumvent sensitivity of cancer cells to drugs with diverse structures and mechanisms of action.
Dedifferentiation also endows cancer cells with the capacity for resistance to immune recognition and destruction (83). In accord with those findings, the MUC1-C→NF-κB→p65 inflammatory pathway activates PD-L1/CD274 expression and represses the IFNG, TLR9, MCP-1 and GM-CSF genes in NSCLC cells (108). In an immunocompetent transgenic mouse model of NSCLC, targeting MUC1-C downregulated PD-L1 and induced IFN-γ in association with enhancing the effector function of anti-tumor CD8+ cells (109). Studies in human TNBCs and in a transgenic TNBC mouse model further showed that MUC1-C drives PD-L1 expression and suppression of the tumor immune microenvironment (38). Analysis of NSCLC and TNBC gene expression datasets has also demonstrated that upregulation of MUC1 expression correlates with immune cell depleted microenvironments and poor clinical outcomes (38,108,109). These findings have supported involvement of MUC1-C in immune evasion and have provided the basis for studying the association of MUC1-C with immune cell-depleted ‘cold’ TNBCs.
Section summary
MUC1-C promotes the progression of cancers with the capacity for stemness, plasticity and pluripotency, which have been increasingly linked to drug resistance, immune evasion and poor clinical outcomes. Targeting MUC1-C thereby represents a potential approach to inhibit plasticity of cancer cells and thereby circumvent the development of drug resistance and immune evasion in cancer treatment.
MUC1-C is a target for cancer treatment
Attempts at targeting MUC1 have been largely unsuccessful to date. In earlier work, the overexpression of MUC1 in diverse carcinomas established it as a highly attractive target, particularly for cancer immunotherapy (110). Despite this interest and increasing numbers of MUC1-directed clinical trials, there has been no immunotherapeutic agent with clinical activity sufficient for regulatory approval (111). In this respect, vaccines targeting MUC1 have been associated with inducing immune responses; however, they have not been effective in the treatment of solid tumors (111). A potential explanation for these findings is that, as noted above, MUC1 has the capacity for suppressing the tumor immune microenvironment.
One vaccine that has shown promise in inducing anti-MUC1 immunity and anti-cancer activity has been advanced to multi-center trials (NCT03059485 and CTN1401). This dendritic cell (DC)-based vaccine was initially developed to overcome MUC1 immune tolerance in human MUC1 transgenic mice and was found to induce anti-MUC1 CD8+ and CD4+ T cell responses (112,113). MUC1 was widely recognized as a carcinoma-associated antigen; however, largely unappreciated observations have been that MUC1 is expressed by acute myeloid leukemia (AML) stem cells and represents a potential target for AML treatment (114). Accordingly, a personalized AML/DC-based vaccine was developed to induce anti-MUC1 immunity. Vaccination of AML patients, who achieved remission after chemotherapy, was well-tolerated and associated with expansion and persistence of CD8+ T cells recognizing MUC1 and other shared AML antigens (115). Of 17 AML patients who received the vaccine, 12 remained alive without recurrence at a median follow-up of 57 months (115). Based on these provocative findings, a randomized multi-center trial is underway for AML patients (NCT03059485) and a national study is ongoing for multiple myeloma, another MUC1-expressing hematologic malignancy (116,117) (CTN1401).
Aberrant O-glycosylation of the MUC1-N subunit results in truncated Tn and sialyl-Tn glycoforms, which represent neoantigens for targeting with CAR-T cells (118,119). Anti-Tn-MUC1 CAR-T cells exhibited anti-tumor activity in mouse models of leukemia and pancreatic cancer (119), supporting the clinical development of this approach. A Phase 1 CART-TnMUC1-01 trial was thus initiated for TnMUC1 CAR-T therapy in combination with a lymphodepletion chemotherapy regimen for patients with TnMUC1-positive advanced cancers (Tmunity Therapeutics; NCT04025216). Treating solid tumors has been a major challenge in the field of CAR-T cells. Targeting MUC1-N, which is shed from the surface of cancer cells and circulates at increased levels in cancer patients as measured by the CA15-3 assay (120), could also pose an obstacle for directing anti-MUC1 CAR-T cells to tumors. Targeting MUC1-expressing carcinomas with MUC1-C-induced immune evasion may present an additional obstacle for the effectiveness of CAR-T therapeutics.
Targeting the oncogenic MUC1-C subunit, which is not shed from the cancer cell surface, has been faced with other challenges. The MUC1-C 58 amino acid extracellular domain includes an N-glycosylation site adjacent to two alpha-helices (8). Generating antibodies against this conformational structure posed an obstacle that was recently overcome by targeting alpha-helices (8). In this way, mouse MAb 3D1 reacts with the MUC1-C alpha-3 helix, binds to the surface of MUC1-C-expressing cancer cells and undergoes internalization (8). Conjugation of MAb 3D1 to MMAE generated an ADC that effectively kills MUC1-C-positive NSCLC, TNBC and other carcinoma cells (8). MAb 3D1-MMAE ADCs were also effective against human tumor xenografts in mice without associated overt toxicities (8). Of importance for the concern that MUC1 is expressed on the apical surface of normal epithelial cells, administration of MAb 3D1-MMAE ADCs to MUC1-transgenic mice further demonstrated a lack of toxicity (8). Based on these findings, MAb 3D1 was humanized without loss of reactivity for the clinical development of an anti-MUC1-C ADC. This first-in-class human MAb 3D1 targeting MUC1-C has provided new opportunities for the development of other immunotherapeutic approaches, such as an ADCC and CAR-T cells. CARs have been generated with the human MAb 3D1 sequences and, based on preclinical data, are being developed clinically for targeting MUC1-C-expressing malignancies.
The MUC1-C CD is an intrinsically disordered protein that is devoid of kinase activity (14). As a result, the identification of small molecules that selectively target this domain has been a challenge (121). The Achilles’ heel of MUC1-C is the cytoplasmic CQC motif that is adjacent to the transmembrane domain and is necessary for ROS-induced MUC1-C homodimerization, nuclear import and oncogenic function (6,15). Based on these observations, cell penetrating peptides containing the CQCRRK motif were evaluated in preclinical models, leading to the selection of GO-201 (L-amino acids: R9-CQCRRKN) as a candidate for targeting MUC1-C-expressing human prostate, breast, pancreatic and esophageal tumors (78,122–124). Investigations of the second generation GO-203 peptide [D-amino acids: (R9-CQCRRKN)] demonstrated dose-dependent activity in other human tumor xenograft models (26,35,125,126). Phase I clinical evaluation of daily intravenous GO-203 administration further showed that this agent has an acceptable safety profile for combination studies (127). GO-203 has thus been formulated in nanoparticles for more convenient dosing schedules of once or twice a week in the clinic (128).
Concluding remarks
MUC1 emerged an evolutionary adaptation of mammals to protect epithelia from inflammation and loss of homeostasis. As a first line of defense, the MUC1-N subunit physically contributes to a protective mucous barrier. As a second line of defense, the MUC1-C subunit promotes wound healing associated responses of inflammation, proliferation and remodeling. MUC1-C appeared in mammals to respond to intermittent inflammation for protection and survival. However, with chronic inflammation and prolonged cycles of damage and repair, MUC1-C drives stemness and pluripotency that promote carcinogenesis. Given this capacity for the MUC1-C ‘protectogen’ to also function as an oncogenic protein, an adverse consequence of mammalian evolution is that MUC1-C contributes to cancer in humans, particularly after reproductive age, by responding to the highly prevalent emergence of chronic inflammation promoted by changes in societal, environmental and lifestyle factors (129).
Glossary
Abbreviations
- CD
cytoplasmic domain
- CRC
colorectal cancer
- CRPC
castrate resistant prostate cancer
- CSCs
cancer stem cells
- iPSCs
induced pluripotent stem cells
- ISC
intestinal stem cells
- NEPC
neuroendocrine prostate cancer
- NPC
nuclear pore complex
- SEA
sperm protein, enterokinase and agrin
- TM
transmembrane domain
- TR
tandem repeats
Funding
This publication was supported by the National Cancer Institute of the National Institutes of Health under grant numbers CA97098, CA166480, CA229716, CA212649 and CA233084.
Conflict of Interest Statement: Author has equity interests in Genus Oncology, Reata Pharmaceuticals, Hillstream BioPharma and Victa Biotherapeutics, served as a member of the board of directors of Genus Oncology and is a paid consultant to Reata, Victa and CanBas.
References
- 1. Miller PW, et al. (2013) The evolutionary origin of epithelial cell-cell adhesion mechanisms. Curr. Top. Membr., 72, 267–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duraisamy S, et al. (2007) Evolution of the human MUC1 oncoprotein. Int. J. Oncol., 31, 671–677. [PubMed] [Google Scholar]
- 3. Duraisamy S, et al. (2006) Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 AND MUC16. Gene, 373, 28–34. [DOI] [PubMed] [Google Scholar]
- 4. Lang T, et al. (2007) Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. U. S. A., 104, 16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lang T, et al. (2016) Searching the evolutionary origin of epithelial mucus protein components-Mucins and FCGBP. Mol. Biol. Evol., 33, 1921–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kufe DW. (2009) Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer, 9, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Macao B, et al. (2006) Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol., 13, 71–76. [DOI] [PubMed] [Google Scholar]
- 8. Panchamoorthy G, et al. (2018) Targeting the human MUC1-C oncoprotein with an antibody-drug conjugate. JCI Insight, 3, e99880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shurer CR, et al. (2019) Physical principles of membrane shape regulation by the Glycocalyx. Cell, 177, 1757–1770.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pelaseyed T, et al. (2013) Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell-protective device. FEBS J., 280, 1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhar P, et al. (2019) The role of the cell surface Mucin MUC1 as a barrier to infection and regulator of inflammation. Front. Cell. Infect. Microbiol., 9, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butcher LD, et al. (2017) Oxidative stress resulting from helicobacter pylori infection contributes to gastric carcinogenesis. Cell. Mol. Gastroenterol. Hepatol., 3, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindén SK, et al. (2009) MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog., 5, e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raina D, et al. (2015) Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One, 10, e0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kufe DW. (2013) MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene, 32, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rajabi H, et al. (2017) MUC1-C Oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. Biochim. Biophys. Acta. Rev. Cancer, 1868, 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raina D, et al. (2012) Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int. J. Oncol., 40, 1643–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hata T, et al. (2019) MUC1-C activates the NuRD complex to drive dedifferentiation of triple-negative breast cancer cells. Cancer Res., 79, 5711–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahmad R, et al. (2017) Targeting MUC1-C inhibits the AKT-S6K1-elF4A pathway regulating TIGAR translation in colorectal cancer. Mol. Cancer, 16, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schäfer M, et al. (2008) Cancer as an overhealing wound: an old hypothesis revisited. Nat. Rev. Mol. Cell Biol., 9, 628–638. [DOI] [PubMed] [Google Scholar]
- 21. Arwert EN, et al. (2012) Epithelial stem cells, wound healing and cancer. Nat. Rev. Cancer, 12, 170–180. [DOI] [PubMed] [Google Scholar]
- 22. Ge Y, et al. (2018) Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet., 19, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brazil JC, et al. (2019) Innate immune cell-epithelial crosstalk during wound repair. J. Clin. Invest., 129, 2983–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alam M, et al. (2016) MUC1-C represses the crumbs complex polarity factor CRB3 and Downregulates the Hippo pathway. Mol. Cancer Res., 14, 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panciera T, et al. (2017) Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol., 18, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kharbanda A, et al. (2014) Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin. Cancer Res., 20, 5423–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raina D, et al. (2014) Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene, 33, 3422–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajabi H, et al. (2019) MUC1-C represses the RASSF1A tumor suppressor in human carcinoma cells. Oncogene, 38, 7266–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carpenter G, et al. (2013) Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol., 5, a008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Little AC, et al. (2019) Dysregulated Redox regulation contributes to nuclear EGFR localization and Pathogenicity in lung cancer. Sci. Rep., 9, 4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinlough CL, et al. (2006) Recycling of MUC1 is dependent on its palmitoylation. J. Biol. Chem., 281, 12112–12122. [DOI] [PubMed] [Google Scholar]
- 32. Leng Y, et al. (2007) Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J. Biol. Chem., 282, 19321–19330. [DOI] [PubMed] [Google Scholar]
- 33. Hanisch FG, et al. (2012) MUC1 membrane trafficking: protocols for assessing biosynthetic delivery, endocytosis, recycling, and release through exosomes. Methods Mol. Biol., 842, 123–140. [DOI] [PubMed] [Google Scholar]
- 34. Rajabi H, et al. (2012) MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J. Biol. Chem., 287, 10703–10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bouillez A, et al. (2016) Inhibition of MUC1-C Suppresses MYC Expression and Attenuates Malignant Growth in KRAS Mutant Lung Adenocarcinomas. Cancer Res., 76, 1538–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajabi H, et al. (2018) MUC1-C activates polycomb repressive complexes and downregulates tumor suppressor genes in human cancer cells. Oncogene, 37, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang L, et al. (2005) MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res., 65, 10413–10422. [DOI] [PubMed] [Google Scholar]
- 38. Maeda T, et al. (2018) MUC1-C induces PD-L1 and immune evasion in triple-negative breast cancer. Cancer Res., 78, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Greten FR, et al. (2019) Inflammation and cancer: triggers, mechanisms, and consequences. Immunity, 51, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takahashi H, et al. (2015) MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene, 34, 5187–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W, et al. (2020) MUC1-C drives stemness in progression of colitis to colorectal cancer. JCI Insight, 5, 137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nieto MA, et al. (2016) EMT: 2016. Cell, 166, 21–45. [DOI] [PubMed] [Google Scholar]
- 43. Gnemmi V, et al. (2014) MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/β-catenin pathway and interaction with SNAIL promoter. Cancer Lett., 346, 225–236. [DOI] [PubMed] [Google Scholar]
- 44. Hata T, et al. (2019) Targeting MUC1-C inhibits TWIST1 signaling in triple-negative breast cancer. Mol. Cancer Ther., 18, 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang J, et al. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell, 117, 927–939. [DOI] [PubMed] [Google Scholar]
- 46. Xu Y, et al. (2017) Breast tumor cell-specific knockout of Twist1 inhibits cancer cell plasticity, dissemination, and lung metastasis in mice. Proc. Natl. Acad. Sci. U. S. A., 114, 11494–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yasumizu Y, et al. (2020) MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat. Commun., 11, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tam WL, et al. (2013) The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med., 19, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skrypek N, et al. (2017) Epithelial-to-Mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet., 33, 943–959. [DOI] [PubMed] [Google Scholar]
- 50. Rhee I, et al. (2002) DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature, 416, 552–556. [DOI] [PubMed] [Google Scholar]
- 51. Clapier CR, et al. (2017) Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol., 18, 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bornelöv S, et al. (2018) The Nucleosome Remodeling and Deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell, 71, 56–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yan D, et al. (2012) Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires β-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. J. Biol. Chem., 287, 8598–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pakala SB, et al. (2011) TGF-β1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells. Oncogene, 30, 2230–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sen N, et al. (2014) Role of MTA1 in cancer progression and metastasis. Cancer Metastasis Rev., 33, 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Toh Y, et al. (2014) Identification and characterization of metastasis-associated gene/protein 1 (MTA1). Cancer Metastasis Rev., 33, 837–842. [DOI] [PubMed] [Google Scholar]
- 57. Malisetty VL, et al. (2017) MTA1 expression in human cancers—Clinical and pharmacological significance. Biomed. Pharmacother., 95, 956–964. [DOI] [PubMed] [Google Scholar]
- 58. Chou DM, et al. (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U. S. A., 107, 18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li DQ, et al. (2014) MTA family of proteins in DNA damage response: mechanistic insights and potential applications. Cancer Metastasis Rev., 33, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yamamoto M, et al. (2019) MUC1-C integrates chromatin remodeling and PARP1 activity in the DNA damage response of triple-negative breast cancer cells. Cancer Res., 79, 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwitalla S, et al. (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell, 152, 25–38. [DOI] [PubMed] [Google Scholar]
- 62. Barker N, et al. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature, 457, 608–611. [DOI] [PubMed] [Google Scholar]
- 63. de Sousa e Melo F, et al. (2017) A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature, 543, 676–680. [DOI] [PubMed] [Google Scholar]
- 64. Reya T, et al. (2005) Wnt signalling in stem cells and cancer. Nature, 434, 843–850. [DOI] [PubMed] [Google Scholar]
- 65. Post Y, et al. (2019) Defining adult stem cell function at its simplest: the ability to replace lost cells through mitosis. Cell Stem Cell, 25, 174–183. [DOI] [PubMed] [Google Scholar]
- 66. Gupta PB, et al. (2019) Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell, 24, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yuan S, et al. (2019) Cellular plasticity in cancer. Cancer Discov., 9, 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davies AH, et al. (2018) Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol., 15, 271–286. [DOI] [PubMed] [Google Scholar]
- 69. Takahashi K, et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. [DOI] [PubMed] [Google Scholar]
- 70. Takahashi K, et al. (2016) A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol., 17, 183–193. [DOI] [PubMed] [Google Scholar]
- 71. Ben-Porath I, et al. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet., 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Riggs JW, et al. (2013) Induced pluripotency and oncogenic transformation are related processes. Stem Cells Dev., 22, 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iglesias JM, et al. (2017) Linking pluripotency reprogramming and cancer. Stem Cells Transl. Med., 6, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hepburn AC, et al. (2019) The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene, 38, 4412–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wollenzien H, et al. (2018) Somatic pluripotent genes in tissue repair, developmental disease, and cancer. SPG Biomed, 1. doi: 10.32392/biomed.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Abad M, et al. (2013) Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature, 502, 340–345. [DOI] [PubMed] [Google Scholar]
- 77. Ohnishi K, et al. (2014) Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell, 156, 663–677. [DOI] [PubMed] [Google Scholar]
- 78. Xin Z, et al. (2018) Inhibition of MUC1-C entering nuclear suppresses MYC expression and attenuates malignant growth in esophageal squamous cell carcinoma. Onco. Targets. Ther., 11, 4125–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Melton C, et al. (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature, 463, 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Alam M, et al. (2015) MUC1-C induces the LIN28B→LET-7→HMGA2 Axis to regulate self-renewal in NSCLC. Mol. Cancer Res., 13, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith BA, et al. (2018) A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep., 24, 3353–3366.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. De Angelis ML, et al. (2019) Stem cell plasticity and dormancy in the development of cancer therapy resistance. Front. Oncol., 9, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miranda A, et al. (2019) Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. U. S. A., 116, 9020–9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Malta TM, et al. (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell, 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mohiuddin IS, et al. (2019) Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta. Mol. Basis Dis., 1866, 165432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Merlo GR, et al. (1989) Frequent alteration of the DF3 tumor-associated antigen gene in primary human breast carcinomas. Cancer Res., 49(24 Pt 1), 6966–6971. [PubMed] [Google Scholar]
- 87. Kufe DW. (2008) Targeting the human MUC1 oncoprotein: a tale of two proteins. Cancer Biol. Ther., 7, 81–84. [DOI] [PubMed] [Google Scholar]
- 88. Khodarev NN, et al. (2009) MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res., 69, 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pitroda SP, et al. (2009) MUC1-induced alterations in a lipid metabolic gene network predict response of human breast cancers to tamoxifen treatment. Proc. Natl. Acad. Sci. U. S. A., 106, 5837–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. MacDermed DM, et al. (2010) MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Med. Genomics, 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu F, et al. (2015) Prognostic significance of Mucin antigen MUC1 in various human epithelial cancers: a meta-analysis. Medicine (Baltimore)., 94, e2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zeng Y, et al. (2015) MUC1 predicts colorectal cancer metastasis: a systematic review and meta-analysis of case controlled studies. PLoS One, 10, e0138049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang XT, et al. (2016) MUC1 immunohistochemical expression as a prognostic factor in gastric cancer: meta-analysis. Dis. Markers, 2016, 9421571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Huang X, et al. (2017) MUC1 overexpression predicts worse survival in patients with non-small cell lung cancer: evidence from an updated meta-analysis. Oncotarget, 8, 90315–90326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Niv Y, et al. (2019) Mucin Expression in Colorectal Cancer (CRC): systematic review and meta-analysis. J. Clin. Gastroenterol., 53, 434–440. [DOI] [PubMed] [Google Scholar]
- 96. Li C, et al. (2019) Prognostic and clinicopathological value of MUC1 expression in colorectal cancer: A meta-analysis. Medicine (Baltimore)., 98, e14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jing X, et al. (2019) Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol. Rep., 41, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li C, et al. (2019) Prognostic and Clinicopathological significance of MUC family members in colorectal cancer: a systematic review and meta-analysis. Gastroenterol. Res. Pract., 2019, 2391670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang Y, et al. (2016) Numb and Numbl act to determine mammary myoepithelial cell fate, maintain epithelial identity, and support lactogenesis. FASEB J., 30, 3474–3488. [DOI] [PubMed] [Google Scholar]
- 100. Watanabe K, et al. (2014) Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev. Cell, 29, 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hughes K, et al. (2018) The multifaceted role of STAT3 in mammary gland involution and breast cancer. Int J Mol Sci, 19, E1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jena MK, et al. (2019) Molecular mechanism of mammary gland involution: An update. Dev. Biol., 445, 145–155. [DOI] [PubMed] [Google Scholar]
- 103. Ren J, et al. (2004) Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell, 5, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Uchida Y, et al. (2013) Inhibition of the MUC1-C oncoprotein is synergistic with cytotoxic agents in the treatment of breast cancer cells. Cancer Biol. Ther., 14, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jin W, et al. (2017) MUC1 induces acquired chemoresistance by upregulating ABCB1 in EGFR-dependent manner. Cell Death Dis., 8, e2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lv Y, et al. (2019) Erlotinib overcomes paclitaxel-resistant cancer stem cells by blocking the EGFR-CREB/GRβ-IL-6 axis in MUC1-positive cervical cancer. Oncogenesis, 8, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kharbanda A, et al. (2013) Oncogenic MUC1-C promotes tamoxifen resistance in human breast cancer. Mol. Cancer Res., 11, 714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bouillez A, et al. (2017) MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small-cell lung cancer. Oncogene, 36, 4037–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bouillez A, et al. (2017) MUC1-C promotes the suppressive immune microenvironment in non-small cell lung cancer. Oncoimmunology, 6, e1338998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cheever MA, et al. (2009) The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res., 15, 5323–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Taylor-Papadimitriou J, et al. (2018) Latest developments in MUC1 immunotherapy. Biochem. Soc. Trans., 46, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gong J, et al. (1997) Induction of antitumor activity by immunization with fusions of dendritic and carcinoma cells. Nat. Med., 3, 558–561. [DOI] [PubMed] [Google Scholar]
- 113. Gong J, et al. (1998) Reversal of tolerance to human MUC1 antigen in MUC1 transgenic mice immunized with fusions of dendritic and carcinoma cells. Proc. Natl. Acad. Sci. U. S. A., 95, 6279–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Stroopinsky D, et al. (2013) MUC1 is a potential target for the treatment of acute myeloid leukemia stem cells. Cancer Res., 73, 5569–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rosenblatt J, et al. (2016) Individualized vaccination of AML patients in remission is associated with induction of antileukemia immunity and prolonged remissions. Sci. Transl. Med., 8, 368ra171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rosenblatt J, et al. (2013) Vaccination with dendritic cell/tumor fusions following autologous stem cell transplant induces immunologic and clinical responses in multiple myeloma patients. Clin. Cancer Res., 19, 3640–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Stroopinsky D, et al. (2016) MUC1 in hematological malignancies. Leuk. Lymphoma, 57, 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Posey AD, Jr, et al. (2016) Distinguishing truncated and normal MUC1 Glycoform targeting from Tn-MUC1-specific CAR T cells: specificity is the key to safety. Immunity, 45, 947–948. [DOI] [PubMed] [Google Scholar]
- 119. Posey AD, Jr, et al. (2016) Engineered CAR T cells targeting the cancer-associated Tn-Glycoform of the membrane Mucin MUC1 control Adenocarcinoma. Immunity, 44, 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li X, et al. (2018) Clinicopathological and prognostic significance of cancer antigen 15-3 and Carcinoembryonic antigen in breast cancer: a meta-analysis including 12,993 patients. Dis. Markers, 2018, 9863092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhou J, et al. (2011) MUC1-C oncoprotein is a target for small molecule inhibitors. Mol. Pharm., 79, 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Joshi MD, et al. (2009) MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol. Cancer Ther., 8, 3056–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Raina D, et al. (2009) Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res., 69, 5133–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Banerjee S, et al. (2012) MUC1c regulates cell survival in pancreatic cancer by preventing lysosomal permeabilization. PLoS One, 7, e43020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Raina D, et al. (2011) Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol. Cancer Ther., 10, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. GongSun X, et al. (2019) Inhibition of MUC1-C regulates metabolism by AKT pathway in esophageal squamous cell carcinoma. J. Cell. Physiol., 234, 12019–12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Liegel J, et al. (2017) Phase I/Ib trial of MUC1-C inhibitor GO-203-2C with decitabine in acute myeloid leukemia. ASH 59th Annual Meeting, Abstract #616.
- 128. Hasegawa M, et al. (2015) Intracellular targeting of the Oncogenic MUC1-C protein with a novel GO-203 Nanoparticle formulation. Clin. Cancer Res., 21, 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Furman D, et al. (2019) Chronic inflammation in the etiology of disease across the life span. Nat. Med., 25, 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]