Electrically conductive protein nanowires appear to be widespread in the microbial world and are a revolutionary “green” material for the fabrication of electronic devices. Electrically conductive pili (e-pili) assembled from type IV pilin monomers have independently evolved multiple times in microbial history as have electrically conductive archaella (e-archaella) assembled from homologous archaellin monomers. A role for e-pili in long-range (micrometer) extracellular electron transport has been demonstrated in some microbes.
KEYWORDS: Geobacter, Syntrophus, cable bacteria, e-biologics, electromicrobiology, nanowire, syntrophy
ABSTRACT
Electrically conductive protein nanowires appear to be widespread in the microbial world and are a revolutionary “green” material for the fabrication of electronic devices. Electrically conductive pili (e-pili) assembled from type IV pilin monomers have independently evolved multiple times in microbial history as have electrically conductive archaella (e-archaella) assembled from homologous archaellin monomers. A role for e-pili in long-range (micrometer) extracellular electron transport has been demonstrated in some microbes. The surprising finding of e-pili in syntrophic bacteria and the role of e-pili as conduits for direct interspecies electron transfer have necessitated a reassessment of routes for electron flux in important methanogenic environments, such as anaerobic digesters and terrestrial wetlands. Pilin monomers similar to those found in e-pili may also be a major building block of the conductive “cables” that transport electrons over centimeter distances through continuous filaments of cable bacteria consisting of a thousand cells or more. Protein nanowires harvested from microbes have many functional and sustainability advantages over traditional nanowire materials and have already yielded novel electronic devices for sustainable electricity production, neuromorphic memory, and sensing. e-pili can be mass produced with an Escherichia coli chassis, providing a ready source of material for electronics as well as for studies on the basic mechanisms for long-range electron transport along protein nanowires. Continued exploration is required to better understand the electrification of microbial communities with microbial nanowires and to expand the “green toolbox” of sustainable materials for wiring and powering the emerging “Internet of things.”
INTRODUCTION
Electrification (the supply of electricity through a distribution system of conductive wires) was one of the most important advances in human civilization (1). It is becoming increasingly apparent that a broad diversity of bacteria and archaea function as “micro-Edisons” running electrically conductive “wires” and “cables” throughout their environment to electrify their communities. Although these microbes do not use the current to power their own lights (at least as far as we know), the electricity conducted through their electric grid can efficiently provide energy to support microbial metabolism and growth. Electric signals transmitted along the wires may also deliver important environmental information to cells (2). The most intensively studied of these “microbial nanowires” are the electrically conductive pili (e-pili) of the Geobacter species Geobacter sulfurreducens and Geobacter metallireducens (2, 3). The conductivity of Geobacter e-pili can be quite high (277 S/cm at pH 7 [4]), which is surprising considering that they are comprised solely of protein. Proteins are typically regarded as an insulating material. Yet, e-pili are conductive even though they lack the metal cofactors that are common electron transfer moieties of proteins involved in short-range electron transfer within a single molecule or between two proteins.
Geobacter e-pili are type IV pili (3) that have only recently evolved the characteristics that yield high electrical conductivity (5). However, type IV pili are among the most ancient and widespread outer-surface microbial structures (6), and it is now becoming apparent that over the course of microbial history, type IV pili and homologous protein structures have independently evolved into conductive filaments multiple times (7–9). In one remarkable instance, e-pili may have been modified to produce superstructures, forming electrically conductive cables (10). Although physiological function has been confirmed for only a few microbial nanowires, it seems likely that, in general, the evolved conductivity confers a selective advantage. High conductivity requires an increased abundance of aromatic amino acids, and synthesizing aromatic amino acids is metabolically costly (9). In this minireview, we discuss the rapidly expanding catalog of electrically conductive protein nanowires in the microbial world, their microbiological and environmental importance, and emerging practical applications.
GEOBACTER e-PILI
The e-pili of G. sulfurreducens are comprised of the type IV pilin monomer protein, PilA. This is evident from the following observations. (i) Deleting pilA prevents e-pilus expression, and complementation by expressing pilA in trans restores nanowire expression (11). (ii) PilA was the only protein in highly purified e-pili (12, 13). (iii) Introducing a short peptide “tag” to the carboxyl end of PilA yields e-pili with the peptide tag (14). (iv) Heterologous expression of PilA in Pseudomonas aeruginosa (15) or Escherichia coli (16) yields e-pili with the same physical and conductive properties as the e-pili recovered from G. sulfurreducens. (v) Replacing the native PilA gene with a synthetic PilA gene with an increased or decreased aromatic amino acid abundance, respectively, increases or decreases e-pili conductivity (4, 13, 17–21).
The G. sulfurreducens PilA is homologous to the pilin monomers that are assembled into pili in many bacteria, but it contains only 61 amino acids (11). This is considerably fewer amino acids than most type IV pilins, which have a longer C-terminal portion that forms a large globular head group (5, 11). Correspondingly, G. sulfurreducens e-pili are thinner (3-nm diameter) than most type IV pili. The truncated type IV pilin found in G. sulfurreducens appears to be a relatively recent evolutionary event, with strong selection for this feature in Geobacter species and other members of the order Desulfuromonadales (5). The few microbes outside the Desulfuromonadales that have a similar PilA appear to have acquired the gene for a homologous PilA via horizontal gene transfer (5).
A key feature of G. sulfurreducens e-pili is a higher density of aromatic amino acids than that found in most type IV pili that exhibit poor conductivity (2). The conductivity of individual e-pili at pH 7 has been tuned over 6 million-fold (40 μS/cm to 277 S/cm) simply by modifying the density of aromatic amino acids in the PilA pilin monomer (4, 19, 21). As previously reviewed in detail (2), it is generally agreed that pilins with a higher density of aromatic amino acids assemble into pili in which the aromatic amino acids are more tightly packed. Theoretical structural models have suggested that the aromatic amino acids of multiple pilins align in the assembled e-pili to form a central conduit of aromatic amino acids that could account for electron transport along the length of G. sulfurreducens e-pili (22, 23). This is consistent with experimental evidence for close association of aromatic amino acids and the finding that the increase in e-pili conductivity following exposure to low pH is associated with closer packing of the aromatic amino acids (22). A similar core of closely packed aromatic amino acids is present in the conductive protein nanowires of Methanospirillum hungatei (discussed in a subsequent section) for which a structure has been experimentally determined (8).
There is debate about whether aromatic amino acids pack sufficiently close to enable π-π stacking of the aromatic rings to confer a metallic-like conductivity or that closer packing simply enhances electron hopping between aromatics. Although there are multiple lines of evidence consistent with the metallic-like conductivity hypothesis (22, 24, 25), further research is required (2). Regardless of the fine-scale conduction mechanisms, the understanding that high aromatic amino acid density can yield highly conductive e-pili has served as a good empirical tool for identifying pilin gene sequences likely to yield e-pili as discussed in a subsequent section.
There is substantial evidence that the e-pili of G. sulfurreducens and the closely related G. metallireducens confer the capacity for extracellular electron transfer at distance (up to ca. 20 μm) from the cells to extracellular electron acceptors such as Fe(III) oxides (11, 26) or electrodes (27, 28) and serve as electrical connections with other species for direct interspecies electron transfer (DIET) (29–32). Although these initial conclusions were based on the phenotypes of pilA-deficient mutants, more convincing is the finding that strains expressing pili with reduced aromatic amino acid density, and thus lower conductivity, are also defective in these forms of extracellular electron transfer (17, 18, 33).
PROTEIN NANOWIRES COMPRISED OF THE c-TYPE CYTOCHROME OmcS
Another type of G. sulfurreducens protein nanowire, filaments comprised of the mutiheme c-type cytochrome OmcS, has been detected with cryo-electron microscopy of outer surface protein preparations (34, 35). However, the available evidence suggests that, under physiological conditions, OmcS filaments are rare compared to e-pili and that OmcS filaments are not important conduits for long-range electron transport at a distance beyond the cell surface. For example, in studies in which the gene for the G. sulfurreducens PilA pilin monomer was modified to encode short peptide tags (14), all of the filaments observed emanating from the cells contained the peptide tag. This demonstrated that e-pili, not OmcS filaments, were the primary nanowire extension. In accordance with these observations, deleting the gene for OmcS had no impact on the recovery of conductive filaments from G. sulfurreducens (36, 37). In contrast, the nanowires of strains of G. sulfurreducens that produce abundant OmcS, but express pilins expected to yield poorly conductive pili, exhibit low conductivity (7, 17–19). Studies on G. sulfurreducens conductive nanowires have routinely reported that their diameter was 3 nm, consistent with the expected diameter of e-pili but too thin to be OmcS filaments, which have a diameter of 4 nm (3). The finding that nanowire conductivity can be tuned by modifying the aromatic amino acid content of PilA (discussed in the previous section) clearly demonstrates that the nanowires were e-pili, not OmcS filaments.
Unlike pilins for e-pili, which appear to be widely distributed throughout the microbial world (see subsequent sections), OmcS homologs are found in few microbes, including Geobacter species. For example, G. metallireducens, a close relative of G. sulfurreducens and highly effective in extracellular electron transfer, lacks OmcS (38). The role of OmcS in extracellular electron transfer is even limited within the species G. sulfurreducens. For example, e-pili are essential for long-range electron transfer and high-density current production by G. sulfurreducens biofilms, but deleting the gene for OmcS does not inhibit current production and actually increases biofilm conductivity (17, 18, 20, 28, 39, 40). Deleting the gene for OmcS in the DL-1 strain of G. sulfurreducens did inhibit Fe(III) oxide reduction and DIET (29, 41), but the same OmcS gene deletion had no impact on any form of extracellular electron transfer in G. sulfurreducens strain KN400 (37). As previously reviewed in detail (3, 34), OmcS is found attached to the outer surface of strain DL-1, possibly as filaments running along the surface, and may also be associated with e-pili. Therefore, additional studies on OmcS localization and function are of interest for better understanding extracellular electron transfer in one strain of G. sulfurreducens (strain DL-1), but the available evidence suggests that an important role for OmcS filaments as extensions for long-range electron transport at a distance from the cells is unlikely.
WHEN ARE PILI e-PILI?
There are multiple methods for assessing pili conductivity. The most informative measurements have been conducted with nanoelectrode arrays (Fig. 1A) that provide information on conductivity along the length of individual pili (4, 19, 21, 40). This approach requires advanced skills in nanofabrication and electronics and is very labor-intensive. The few nanoelectrode array measurements that are available have yielded rough guidelines on the conductivity required for pili to facilitate long-range electron transport in G. sulfurreducens. A strain of G. sulfurreducens expressing a synthetic pilin, known as Aro-5, was unable to reduce Fe(III) oxide and produced current densities ca. 10-fold lower than G. sulfurreducens expressing wild-type pilin (17). Five of the aromatic amino acids in the G. sulfurreducens PilA have been replaced with alanine in the Aro-5 pilin (17). The conductivity of individual pili comprised of the Aro-5 pilin (40 μS/cm, pH 7) is more than 1,000-fold less than the conductivity of e-pili (51 mS/cm, pH 7) comprised of G. sulfurreducens PilA (19).
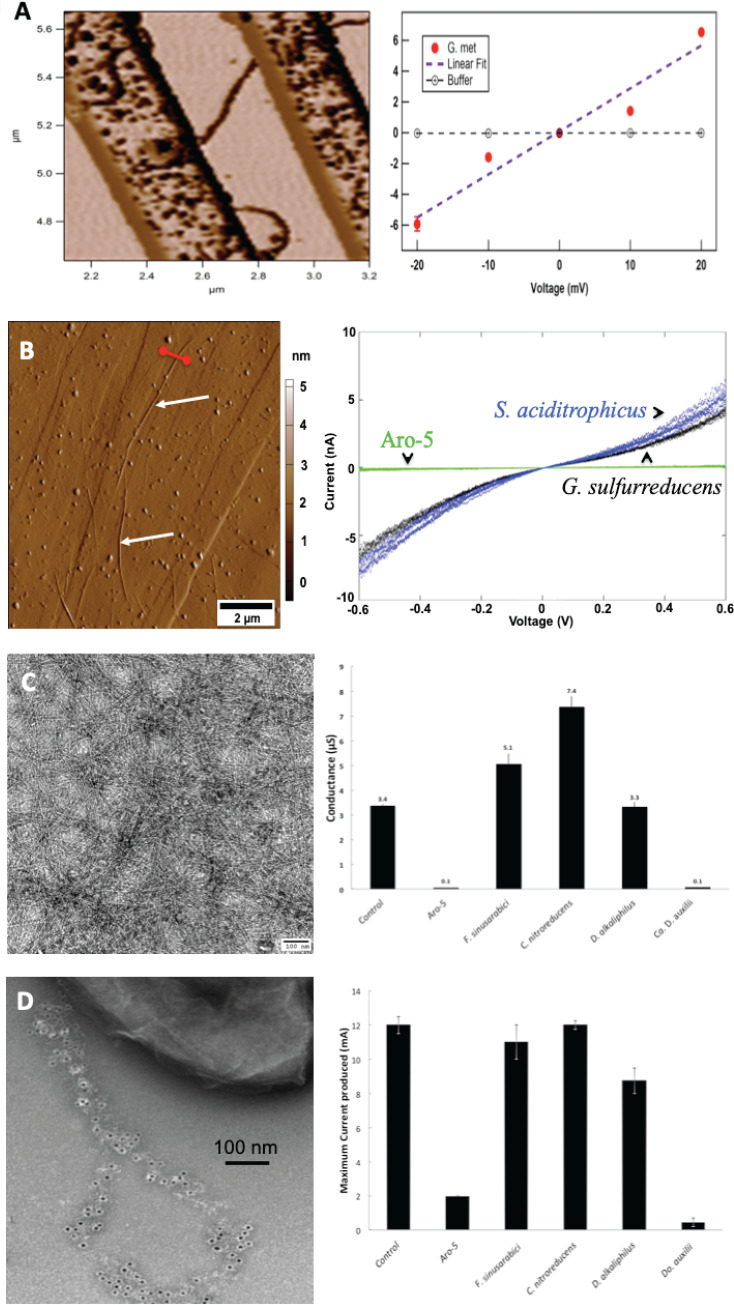
FIG 1.
Methods for assessing the conductivity of microbial nanowires. (A) e-pilus assembled from G. metallireducens pilin bridging the 500-nm nonconducting gap between two electrodes in a nanoelectrode array (left) and a current-voltage plot (right) demonstrating ohmic-like conductivity of the pilus (G. met). (B) Atomic force microscope (AFM) image of pili from Syntrophus aciditrophicus (designated with white arrows) on a highly oriented pyrolytic graphite (HOPG) surface (left) and conductance measured between a conductive AFM tip contacting the upper surface of an individual pilus and the HOPG for individual pili from S. aciditrophicus, wild-type G. sulfurreducens, and the Aro-5 strain of S. aciditrophicus (right). (C) Transmission electron micrograph of a thin-film of G. sulfurreducens e-pili (left) and conductance across a 15-μm electrode-to-electrode gap of thin films of pili harvested from strains of G. sulfurreducens heterologously expressing the pilin monomer gene from the designated microbes (right). (D) Transmission electron micrograph demonstrating heterologous expression of a synthetic pilin gene in G. sulfurreducens (left). The wild-type pilin was modified with six histidines (His tag) at the carboxyl end, and the His tag on the pili was visualized with immunogold labeling. Current production of various strains of G. sulfurreducens heterologously expressing the pilin monomer gene from the designated microbes (right). The left image in panel C is unpublished data provided by our laboratory colleague Joy Ward. The remaining images were reproduced from previously published images with permission as follows: panel A, reference 4; panel B, reference 9; panel C, right, reference 7; panel D, left, reference 14; panel D, right, reference 7.
A more common approach to assessing the capacity of pili for long-range electron transport (8, 9, 11, 15) is to deposit pili-containing cells, or pili sheared from cells, onto highly oriented pyrolytic graphite (HOPG). The conductive HOPG serves as one electrode for the analysis (Fig. 1B). The pili are located with atomic force microscopy (AFM) and then the upper surface of an individual pilus is contacted with a conductive AFM tip that serves as the other electrode. Conductance between the two electrodes provides a qualitative indication of pili conductivity, which is suitable for comparing different pili types. For example, with the AFM-HOPG approach, wild-type G. sulfurreducens e-pili at pH 7 had a conductance (4.5 nS) that was 1,125-fold higher than the conductance (0.004 nS) of pili comprised of the Aro-5 pilin (8). This compares well with the 1,276-fold higher conductivity of the wild-type pili when the conductivity along the length of wild-type and Aro-5 pili was measured with the more technically difficult nanoelectrode array approach (19).
An even simpler approach (Fig. 1C) is to purify pili and then deposit an aqueous suspension of pili on microelectrode arrays. The spacing between electrodes is typically on the order of 10 to 50 μm, and thus, microelectrode arrays are much easier to fabricate than nanoelectrode arrays (50- to 500-nm spacing between electrodes). Air drying yields a thin (micrometer) conductive film of e-pili (24). The microelectrode array approach is more qualitative than measurements on individual pili because the thin-film measurements include the electrical resistance of electron transfer between pili within the film. Accordingly, the conductance difference between wild-type and Aro-5 pilin films (34-fold) was much less than the difference found with measurements of individual pilus conductance (1,125-fold); however, the obvious qualitative difference between the two types of pili remained clear (7).
A microbiologist-friendly yet empirical approach to assess whether a pilin of interest can assemble into functional e-pili (Fig. 1D) is to heterologously express the gene for the pilin of interest in G. sulfurreducens (4, 7, 9, 18, 40). Pilins that yield pili with conductivities similar to G. sulfurreducens e-pili enable G. sulfurreducens strains to produce high current densities in bioelectrochemical systems that have a potentiostat-poised anode and a continuous electron-donor feed (28). As described above for the G. sulfurreducens strain expressing the Aro-5 pilin gene, strains expressing poorly conductive pili generate currents that are much lower than those of strains expressing e-pili (7, 9, 18, 40).
e-PILI ASSEMBLED FROM TRUNCATED PilAs IN OTHER BACTERIA
To date, the pili of only a few bacteria have been examined in detail for their potential to function as e-pili. Notably, a number of Geobacter species and other microbes within the order Desulfuromonadales have short (ca. 60-amino-acid) pilin monomers that are homologous to the G. sulfurreducens PilA, with a high abundance of aromatic amino acids, and thus are expected to be highly conductive (5, 42). Of these, only the pilin monomer of G. metallireducens has been studied in detail. Expression of the G. metallireducens pilA in G. sulfurreducens yielded e-pili 5,000-fold more conductive than G. sulfurreducens wild-type e-pili (4) in accordance with the higher abundance of aromatic amino acids in the G. metallireducens PilA (15.3%) versus the G. sulfurreducens PilA (9.8%).
Flexistipes sinusarabici is one of a small number of bacteria outside the Desulfuromonadales that possess a truncated pilin monomer homologous to the G. sulfurreducens PilA (5). The pili of F. sinusarabici could not be directly studied because this microbe grows poorly in pure culture. However, heterologous expression of the F. sinusarabici pilA in G. sulfurreducens demonstrated that the F. sinusarabici pilin monomer assembles into e-pili (7). Examination of pili from a broader diversity of microbes with PilAs homologous to G. sulfurreducens is desirable, but from the limited data set available, it seems likely that microbes with truncated G. sulfurreducens PilA homologs are likely to have the capacity to produce e-pili.
e-PILI ASSEMBLED FROM LONGER PILIN MONOMERS
It was initially considered that type IV pilin monomers longer than the G. sulfurreducens PilA homologs could not assemble into e-pili because the pili assembled from longer pilin monomers initially examined (from Shewanella oneidensis and P. aeruginosa) were poorly conductive (11). The aromatic amino acid abundance in S. oneidensis and P. aeruginosa pilins are less than 6%, which may be too low to establish the hypothesized requirement for close packing of aromatic amino acids in the pilus structure (22).
However, aromatic amino acid density cannot be the only consideration, as evidenced from studies on the pilin of Geobacter uraniireducens (40). Unlike the pilins of most Geobacter species, the length the G. uraniireducens pilin is more typical of canonical type IV pilins (193 amino acids), yet the aromatic amino acid density in the pilin (9.1%) is close to that of G. sulfurreducens (9.8%) (40). Heterologous expression of the G. uraniireducens pilin monomer gene in G. sulfurreducens yielded pili that were much less conductive (300 μS/cm at pH 7) than G. sulfurreducens pili (50 mS/cm). This lower conductivity is associated with multiple lines of evidence that suggest that G. uraniireducens has adopted strategies other than e-pili for long-range electron transport (40).
Further consideration suggested that not only aromatic amino acid abundance but also the position and spacing of aromatic amino acids in the pilin could be important (7). For example, there is a stretch of 53 amino acids without an aromatic amino acid within the G. uraniireducens pilin. This aromatic-free gap might prevent close packing of aromatic amino acids within the assembled pili. Heterologous expression in G. sulfurreducens of other type IV pilins with high aromatic amino acid abundance and smaller aromatic-free gaps yielded e-pili that appeared to be as conductive as wild-type G. sulfurreducens e-pili and functioned well in extracellular electron transfer (7). These included pilins from Calditerrivibrio nitroreducens (119 amino acids; 13.4% aromatic amino acid abundance; largest aromatic-free gap, 22 amino acids) and Desulfurivibrio alkaliphilus (182 aromatic amino acids; 11% aromatic amino acid abundance; largest aromatic-free gap, 27 amino acids). Other characteristics of these two microbes suggest that they may be capable of extracellular electron transfer (7).
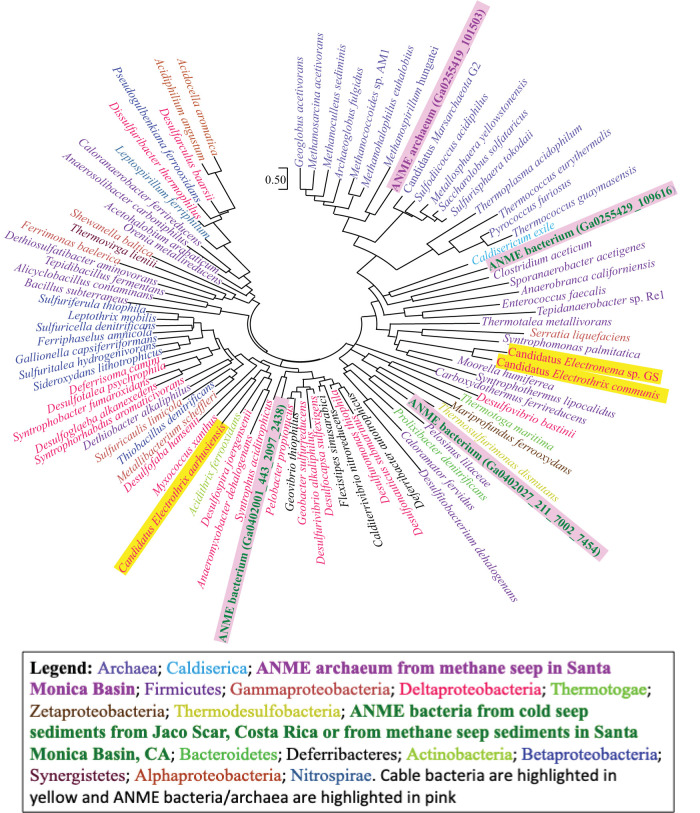
Prospecting through the microbial world for additional e-pili will be laborious with the methods currently available. A potential strategy for narrowing the search is to extrapolate from previous results on the abundance and placement of aromatic amino acids in previously described e-pilins (i.e., pilins that yield e-pili). For example, such criteria revealed a broad phylogenetic diversity of bacteria with genes for e-pilins in environmental samples and enrichment cultures in which reduction of Fe(III) or Mn(III) was expected (42). This included marine zetaproteobacteria, Nitrospinae, betaproteobacteria, and Firmicutes. Further analysis of available genomes of putative electroactive microorganisms (see Table S1 in the supplemental material) identified e-pili in an even wider diversity of bacteria (Fig. 2), including additional genera in the Proteobacteria as well as Actinobacteria, Bacteroidetes, Synergistetes, Aquificae, Caldiserica, Thermodesulfobacteria, and Thermotogae. Additional investigation into the possibility of e-pili in these microbes may lead to newly discovered capabilities.
FIG 2.
Phylogenetic tree comparing predicted e-pilins and e-archaellins from various bacteria and archaea inferred using the maximum likelihood method and JTT matrix-based model (75). An initial tree(s) for the heuristic search was obtained by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 92 amino acid sequences. Evolutionary analyses were conducted in MEGA X (76).
An example of the potential fruits of such labors is the study of the pili of Syntrophus aciditrophicus (9). S. aciditrophicus has been intensively studied as a model microbe for syntrophic bacteria that function as the electron-donating partner in H2/formate interspecies transfer (HFIT) to methanogens (43, 44). Although the original descriptions of S. aciditrophicus suggested that it did not produce pili and lacked genes for pilus expression, further examination revealed genes likely to be responsible for pilus formation as well as pili emanating from cells (9). Furthermore, the PilA sequence of S. aciditrophicus fit the empirical criteria for e-pili (7), with aromatic amino acids in the expected key positions as well as a high abundance of aromatic amino acids (10.9%) and a relatively short maximum aromatic-free gap (22 amino acids). S. aciditrophicus pili have a somewhat greater diameter (4 nm) than G. sulfurreducens pili (3 nm) but a similar conductance (9). Heterologous expression of the S. aciditrophicus pilA in G. sulfurreducens yielded e-pili with a diameter and conductance comparable to those of S. aciditrophicus e-pili and enabled effective long-range electron transport in G. sulfurreducens. The presence of e-pili suggested that DIET might be an alternative possibility for syntrophic growth of S. aciditrophicus. DIET was experimentally confirmed (9). Many other bacteria known to grow via HFIT also have pilin sequences that fit the criteria for assembly into e-pili (9). This is significant because previous studies on H2 fluxes in methanogenic environments have routinely been interpreted with the HFIT paradigm but actually are more readily interpretable if DIET predominates (9). Thus, a complete reevaluation of electron flux in diverse methanogenic environments seems warranted.
At one time, S. oneidensis, a microbe well-known for extracellular electron transfer, was thought to produce conductive protein nanowires (45). However, subsequent studies demonstrated that the proposed S. oneidensis wires were, in fact, extensions of the outer membrane and periplasm (46). This complex mixture of lipids, cytochromes, and other proteins forms conductive filaments when dried (47). There is no evidence that the membrane extensions confer long-range conduction under physiological, hydrated conditions. Furthermore, the membrane extensions cannot possibly extend electron transport beyond the outer surface of the cell because the membrane extensions are the outer cell surface. The available evidence suggests that S. oneidensis excels at extracellular electron transfer through the release of soluble flavin electron shuttles (48). This appears to be a common strategy for microbes that lack e-pili, including some Geobacter species (40, 49). It is still a common mistake in the literature to suggest that S. oneidensis produces nanowires that have functions similar to Geobacter protein nanowires, such as mediating long-range electron transport to other microbial cells. There is no evidence that S. oneidensis is capable of interspecies electron transfer.
e-ARCHAELLA
The archaella of archaea are homologous to the type IV pili of bacteria (6, 50), and many archaea are capable of electron exchange with their extracellular environment. For example, the capacity for Fe(III) oxide reduction is one of the most highly conserved physiological characteristics of hyperthermophilic archaea (51–54), many of which display abundant filaments. Diverse methanogens are known to reduce extracellular electron acceptors (see references 55 and 56 for recent examples and a literature review) or establish electrical connections with bacteria for DIET (30, 31, 57, 58). It has been proposed that the archaella of anaerobic methanotrophic archaea (ANME) may be electrical conduits for electron transfer to sulfate-reducing partners (59). Therefore, it may not be surprising if highly conductive archaella (e-archaella) have evolved one or more times.
The archaellum of Methanospirillum hungatei is the only archaellum for which conductivity has been examined in detail (8). It was chosen for study because it is one of the few archaella for which a structure is known (60). The conductance of individual M. hungatei archaella emanating from cells is more than 3-fold higher than the conductance of G. sulfurreducens e-pili (8). A tightly packed core of phenylalanines is a likely route for conductance (8).
Analysis of the aromatic amino acids in archaellin sequences (see Table S1 in the supplemental material) suggests that e-archaella may be found in a broad diversity of electroactive archaea (Fig. 2). Putative e-archaellins are present in the euryarchaotal orders Methanosarcinales, Thermococcales, Thermoplasmatales, Methanomicrobiales, Methanococcales, Haloferacales, and Archaeoglobales as well as the crenarchaotal orders Thermoproteales, Sulfolobales, and Desulfurococcales. The study of e-archaella could be accelerated with the identification of a robust, easily grown host for heterologous expression of candidate e-archaellin genes to provide a ready supply of archaella for conductivity measurements and to enable archaellin gene modifications to study factors conferring conductivity.
e-CABLES
The micrometer distances that electrons can be transported over e-pili and e-archaella are remarkable compared to typical electron transport proteins, but cable bacteria have taken long-range electron transport to a completely new dimension. Thousands of cable bacteria can connect end-to-end to form filaments that transport electrons over distances up to 7 cm (61–63). Cells at one end of the filament accept electrons from sulfide found in the anaerobic zones of sediments, and the electrons are transported through the multicellular filaments to cells in the overlying aerobic zone that dump the electrons onto oxygen (61). The conduit for the electron transport is an electrically conductive cable (e-cable) comprised of ca. 60 radially arranged parallel periplasmic fibers, each with a diameter of ca. 50 nm (64). At the cell-to-cell junctions, cartwheel-shaped structures electrically connect the individual fibers (65) to form the e-cable.
The key conductive material in e-cables may be protein similar to the pilin monomers that assemble into e-pili (10). The possibility that e-cable fibers are comprised of cytochromes has been eliminated (64). Cable bacteria appear to lack pili on the outer cell surface, yet the most abundant protein expressed by cable bacteria was an aromatic-rich PilA (10). e-cable fibers may be bundles of e-pili or composites of e-pili and carbohydrate (10). Cellular assembly of relatively thin e-pili into 60-nm-diameter conductive fibers is conceivable based on in vitro studies that demonstrated the potential for bundling G. sulfurreducens e-pili (3-nm diameter) into fibers with diameters of ca. 100 nm and the fabrication of conductive composite materials from a combination of e-pili and a nonconducting polymer (66; Y.-L. Sun, B. Montz, R. Selhorst, H.-Y. Tang, J.-X. Zhu, K. P. Nevin, T. P. Russell, S. Nonnenmann, T. Emrick, and D. R. Lovley, submitted for publication).
Another notable connection between e-pili and cable bacteria is the microbe Desulfurivibrio alkaliphilus. Studies on the physiology of cable bacteria are challenging because they are not available in pure culture. D. alkaliphilus has been proposed as a model microbe to infer cable bacteria physiology because of its close phylogenetic relationship and metabolic similarities with cable bacteria (67). As noted in a previous section, the pilin monomer of D. alkaliphilus assembles into e-pili when heterologously expressed in G. sulfurreducens (7). It is likely that D. alkaliphilus employs e-pili for extracellular electron transfer (7). As more information becomes available, it will be interesting to determine whether there is evidence for relatives common to D. alkaliphilus and cable bacteria evolving a strategy for bundling e-pili first into fibers and then eventually into the sophisticated structure of intracellular e-cables.
The conductivity of individual fibers, estimated from measurements on e-cables, was most commonly 1 to 10 S/cm, with estimates as high as 20 S/cm (64). These conductivity values fall between the conductivities of individual G. sulfurreducens (50 mS/cm) and G. metallireducens (277 S/cm) e-pili at pH 7. Unlike that of e-pili, the conductivity of e-cable fibers rapidly deteriorates in air (64), limiting their usefulness as the conductive component for most electronic devices. However, in vivo e-cables are an extraordinarily effective conduit for rapid microbial electron transport over centimeter distances (64).
ELECTRIFYING THE INTERNET OF THINGS WITH PROTEIN NANOWIRES
The emerging concept of ubiquitous interconnected monitoring of human health, the environment, and mechanical/electronic devices, known as the “Internet of things,” will be unsustainable with current electronic materials (68). Fabrication of the necessary nanoelectronic materials typically requires energy-intensive mining and/or processing and/or harsh chemicals. Electronic devices and the batteries that power them are often considered disposable but are not recyclable and are often filled with toxic materials, contributing to the mounting problem of electronic waste (68).
Microbially produced protein nanowires, sheared from cells, offer a potential “green” solution as electronic conductors, sensing materials, and a sustainable power source. For example, protein nanowires have many advantages over traditional nanowire materials, such as silicon nanowires, carbon nanotubes, and conducting polymers. Microbial fabrication of protein nanowires with inexpensive, renewable organic feedstocks requires 100-fold less energy than producing silicon nanowires or carbon nanotubes (68). No toxic chemicals are required for wire production, and the final product is biocompatible, environmentally benign, and recyclable (68). The flexibility in modifying protein nanowire properties is unrivaled among nanowire materials. With the simple design of new synthetic pilin genes, it has been possible to tune the conductivity of protein nanowires by over 1 million-fold (3) and to modify protein nanowire binding properties by decorating the outer wire surface with peptide ligands (14). Yet, protein nanowires are remarkably robust, maintaining function even under harsh complementary metal oxide semiconductor (CMOS)-compatible fabrication conditions (66), and they are stable in bodily fluids, unlike silicon nanowires. They can be processed into thin-film electronics (69–72), nanocables (Sun et al., submitted), and flexible conductive composites (66). The construction of a strain of E. coli that can mass produce protein nanowires from pilin monomers has overcome limitations associated with the growth and genetic manipulation of G. sulfurreducens and other native strains (16). The unique properties of microbially produced nanowires have already led to the development of novel electronic devices with unrivaled capabilities in electronic memory (69) and sensing (70, 72) as well as a strategy for sustainably powering electronics (71).
FUTURE MICROBIOLOGICAL DIRECTIONS
Technical challenges in growing anaerobes that express protein nanowires, coupled with laborious nanowire purification procedures, have greatly limited progress in developing a basic understanding of the properties of microbially produced protein nanowires as well as the innovation of applications. However, the ease of producing protein nanowires with an E. coli chassis that can be aerobically mass cultured and a simplified nanowire purification procedure have overcome these limitations (16). Thus, it can be expected that, in the near future, we will see an enhanced understanding of electron transport mechanisms in protein nanowires as well as the design of an array of novel synthetic pilins tailored for unique nanowire functionalization and new and improved applications.
The study of the diversity of conductive protein filaments and their function in the microbial world is clearly in its infancy. There are a substantial number of microbial filaments that appear to be electrically conductive but whose composition is unknown (73, 74). Not only is there still a poor understanding of the portion of the microbial world that is wired together for sharing energy, but other potential functions of conductive wires, such as signaling environmental information to microbes about contacted surfaces, have yet to be explored (2).
Supplementary Material
ACKNOWLEDGMENTS
Our research on microbial nanowires is supported with exploratory research funds from the College of Natural Sciences, UMass-Amherst, a UMASS Manning/Institute for Applied Life Sciences Award, and a National Science Foundation grant through the DMREF program (NSF CMMI-1921839).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Morris E. 2019. Edison. Random House, New York. [Google Scholar]
- 2.Lovley DR. 2017. Electrically conductive pili: biological function and potential applications in electronics. Curr Opin Electrochem 4:190–198. doi: 10.1016/j.coelec.2017.08.015. [DOI] [Google Scholar]
- 3.Lovley DR, Walker DJF. 2019. Geobacter protein nanowires. Front Microbiol 10:2078. doi: 10.3389/fmicb.2019.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan Y, Adhikari RY, Malvankar NS, Ward JE, Woodard TL, Nevin KP, Lovley DR. 2017. Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens yields pili with exceptional conductivity. mBio 8:e02203-16. doi: 10.1128/mBio.02203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes DE, Dang Y, Walker DJF, Lovley DR. 2016. The electrically conductive pili of Geobacter species are a recently evolved feature for extracellular electron transfer. Microb Genom 2:e000072. doi: 10.1099/mgen.0.000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, Koonin EV, Albers S-V. 2016. Diversity and evolution of type IV pili systems in Archaea. Front Microbiol 7:667. doi: 10.3389/fmicb.2016.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker DJF, Adhikari RY, Holmes DE, Ward JE, Woodard TL, Nevin KP, Lovley DR. 2018. Electrically conductive pili from pilin genes of phylogenetically diverse microorganisms. ISME J 12:48–58. doi: 10.1038/ismej.2017.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker DJF, Martz E, Holmes DE, Zhou Z, Nonnenmann SS, Lovley DR. 2019. The archaellum of Methanospirillum hungatei is electrically conductive. mBio 10:e00579-19. doi: 10.1128/mBio.00579-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker DJF, Nevin KP, Holmes DE, Rotaru A-E, Ward JE, Woodard TL, Zhu J, Ueki T, Nonnenmann SS, McInerney MJ, Lovley DR. 2020. Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. ISME J 14:837–846. doi: 10.1038/s41396-019-0575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kjeldsen KU, Schreiber L, Thorup CA, Boesen T, Bjerg JT, Yang T, Dueholm MS, Larsen S, Risgaard-Petersen N, Nierychlo M, Schmid M, Bøggild A, van de Vossenberg J, Geelhoed JS, Meysman FJR, Wagner M, Nielsen PH, Nielsen LP, Schramm A. 2019. On the evolution and physiology of cable bacteria. Proc Natl Acad Sci U S A 116:19116–19125. doi: 10.1073/pnas.1903514116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 12.Cologgi DL, Lampa-Pastirk S, Speers AM, Kelly SD, Reguera G. 2011. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc Natl Acad Sci U S A 108:15248–15252. doi: 10.1073/pnas.1108616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lampa-Pastirk S, Veazey JP, Walsh KA, Feliciano GT, Steidl RJ, Tessmer S, Reguera G. 2016. Thermally activated charge transport in microbial protein nanowires. Sci Rep 6:23517. doi: 10.1038/srep23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueki T, Walker DJF, Tremblay P-L, Nevin KP, Ward JE, Woodard TL, Nonnenmann SS, Lovley DR. 2019. Decorating the outer surface of microbially produced protein nanowires with peptides. ACS Synth Biol 8:1809–1817. doi: 10.1021/acssynbio.9b00131. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Wang S, Xu A, Zhang L, Liu H, Ma LZ. 2019. Biological synthesis of high-conductive pili in aerobic bacterium Pseudomonas aeruginosa. Appl Microbiol Biotechnol 103:1535–1544. doi: 10.1007/s00253-018-9484-5. [DOI] [PubMed] [Google Scholar]
- 16.Ueki T, Walker DJF, Woodard TL, Nevin KP, Nonnenmann S, Lovley DR. 2020. An Escherichia coli chassis for production of electrically conductive protein nanowires. ACS Synth Biol 9:647–654. doi: 10.1021/acssynbio.9b00506. [DOI] [PubMed] [Google Scholar]
- 17.Vargas M, Malvankar NS, Tremblay P-L, Leang C, Smith JA, Patel P, Snoeyenbos-West O, Synoeyenbos-West O, Nevin KP, Lovley DR. 2013. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 4:e00105-13. doi: 10.1128/mBio.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Tremblay P-L, Malvankar NS, Nevin KP, Lovley DR, Vargas M. 2014. A Geobacter sulfurreducens strain expressing Pseudomonas aeruginosa type IV pili localizes OmcS on pili but Is deficient in Fe(III) oxide reduction and current production. Appl Environ Microbiol 80:1219–1224. doi: 10.1128/AEM.02938-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adhikari RY, Malvankar NS, Tuominen MT, Lovley DR. 2016. Conductivity of individual Geobacter pili. RSC Adv 6:8354–8357. doi: 10.1039/C5RA28092C. [DOI] [Google Scholar]
- 20.Steidl RJ, Lampa-Pastirk S, Reguera G. 2016. Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat Commun 7:12217. doi: 10.1038/ncomms12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y, Adhikari RY, Malvankar NS, Pi S, Ward JE, Woodard TL, Nevin KP, Xia Q, Tuominen MT, Lovley DR. 2016. Synthetic biological protein nanowires with high conductivity. Small 12:4481–4485. doi: 10.1002/smll.201601112. [DOI] [PubMed] [Google Scholar]
- 22.Malvankar NS, Vargas M, Nevin KP, Tremblay P-L, Evans-Lutterodt K, Nykypanchuk D, Martz E, Tuominen MT, Lovley DR. 2015. Structural basis for metallic-like conductivity in microbial nanowires. mBio 6:e00084-15. doi: 10.1128/mBio.00084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao K, Malvankar NS, Shu C, Martz E, Lovley DR, Sun X. 2016. Low energy atomic models suggesting a pilus structure that could account for electrical conductivity along the length of Geobacter sulfurreducens pili. Sci Rep 6:23385. doi: 10.1038/srep23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim B-C, Inoue K, Mester T, Covalla SF, Johnson JP, Rotello VM, Tuominen MT, Lovley DR. 2011. Tunable metallic-like conductivity in nanostructured biofilms comprised of microbial nanowires. Nat Nanotechnol 6:573–579. doi: 10.1038/nnano.2011.119. [DOI] [PubMed] [Google Scholar]
- 25.Malvankar NS, Yalcin SE, Tuominen MT, Lovley DR. 2014. Visualization of charge propagation along individual pili proteins using ambient electrostatic force microscopy. Nat Nanotechnol 9:1012–1017. doi: 10.1038/nnano.2014.236. [DOI] [PubMed] [Google Scholar]
- 26.Smith JA, Lovley DR, Tremblay PL. 2013. Outer cell surface components essential for Fe(III) oxide reduction by Geobacter metallireducens. Appl Environ Microbiol 79:901–907. doi: 10.1128/AEM.02954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevin KP, Kim B-C, Glaven RH, Johnson JP, Woodard TL, Methé BA, DiDonato RJ Jr, Covalla SF, Franks AE, Liu A, Lovley DR. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers ZM, Fogarty H, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 30.Rotaru A-E, Shrestha PM, Liu F, Markovaite B, Chen S, Nevin KP, Lovley DR. 2014. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl Environ Microbiol 80:4599–4605. doi: 10.1128/AEM.00895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotaru A-E, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, Zengler K, Wardman C, Nevin KP, Lovley DR. 2014. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7:408–415. doi: 10.1039/C3EE42189A. [DOI] [Google Scholar]
- 32.Shrestha PM, Rotaru A-E, Summers ZM, Shrestha M, Liu F, Lovley DR. 2013. Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl Environ Microbiol 79:2397–2404. doi: 10.1128/AEM.03837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueki T, Nevin KP, Rotaru A-E, Wang L-Y, Ward JE, Woodard TL, Lovley DR. 2018. Geobacter strains expressing poorly conductive pili reveal constraints on direct interspecies electron transfer mechanisms. mBio 9:e01273-18. doi: 10.1128/mBio.01273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filman DJ, Marino SF, Ward JE, Yang L, Mester Z, Bullitt E, Lovley DR, Strauss M. 2019. Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire. Commun Biol 2:219. doi: 10.1038/s42003-019-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Gu Y, O'Brien JP, Yi SM, Yalcin SE, Srikanth V, Shen C, Vu D, Ing NL, Hochbaum AI, Egelman EH, Malvankar NS. 2019. Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177:361–369. doi: 10.1016/j.cell.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Ye Y, Xiao K, Rensing C, Zhou S. 2020. Molecular evidence for the adaptive evolution of Geobacter sulfurreducens to perform dissimilatory iron reduction in natural environments. Mol Microbiol 113:783–793. doi: 10.1111/mmi.14443. [DOI] [PubMed] [Google Scholar]
- 37.Walker DJF, Li Y, Meier D, Pinches S, Holmes DE. 2020. Cytochrome OmcS is not essential for long-range electron transport in Geobacter sulfurreducens strain KN400. bioRxiv doi: 10.1101/2020.07.22.214791. [DOI] [PMC free article] [PubMed]
- 38.Butler JE, Young ND, Lovley DR. 2010. Evolution of electron transfer out of the cell: comparative genomics of six Geobacter genomes. BMC Genomics 11:40. doi: 10.1186/1471-2164-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malvankar NS, Tuominen MT, Lovley DR. 2012. Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy Environ Sci 5:8651–8659. doi: 10.1039/c2ee22330a. [DOI] [Google Scholar]
- 40.Tan Y, Adhikari RY, Malvankar NS, Ward JE, Nevin KP, Woodard TL, Smith JA, Snoeyenbos-West OL, Franks AE, Tuominen MT, Lovley DR. 2016. The low conductivity of Geobacter uraniireducens pili suggests a diversity of extracellular electron transfer mechanisms in the genus Geobacter. Front Microbiol 7:980. doi: 10.3389/fmicb.2016.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bray MS, Wu J, Padilla CC, Stewart FJ, Fowle DA, Henny C, Simister RL, Thompson KJ, Crowe SA, Glass JB. 2020. Phylogenetic and structural diversity of aromatically dense pili from environmental metagenomes. Environ Microbiol Rep 12:49–57. doi: 10.1111/1758-2229.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW, Gunsalus RP. 2007. The genome of Syntrophus aciditrohicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci U S A 104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 45.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci U S A 103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirbadian S, Barchinger SE, Leung KM, Byun HS, Jangir Y, Bouhenni RA, Reed SB, Romine MF, Saffarini DA, Shi L, Gorby YA, Golbeck JH, El-Naggar MY. 2014. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci U S A 111:12883–12888. doi: 10.1073/pnas.1410551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Naggar MY, Wanger G, Leung KM, Yuzvinsky TD, Southam G, Yang J, Lau WM, Nealson KH, Gorby YA. 2010. Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc Natl Acad Sci U S A 107:18127–18131. doi: 10.1073/pnas.1004880107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, Yu H-Q, Fredrickson JK. 2016. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol 14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 49.Thirumurthy MA, Hitchcock A, Cereda A, Liu J, Chavez MS, Doss BL, Ros R, El-Naggar MY, Heap JT, Bibby TS, Jones AK. 2020. Type IV pili-independent photocurrent production by the cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol 11:1344. doi: 10.3389/fmicb.2020.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albers S-V, Jarrell KF. 2018. The archaellum: an update on the unique archaeal motility structure. Trends Microbiol 26:351–362. doi: 10.1016/j.tim.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Vargas M, Kashefi K, Blunt-Harris EL, Lovley DR. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65–67. doi: 10.1038/25720. [DOI] [PubMed] [Google Scholar]
- 52.Lovley D, Kashefi K, Vargas M, Tor J, Blunt-Harris E. 2000. Reduction of humic substances and Fe(III) by hyperthermophilic microorganisms. Chem Geol 169:289–298. doi: 10.1016/S0009-2541(00)00209-6. [DOI] [Google Scholar]
- 53.Kashefi K, Tor JM, Holmes DE, Van Praagh CG, Reysenbach AL, Lovley DR. 2002. Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int J Syst Evol Bacteriol 52:719–728. doi: 10.1099/00207713-52-3-719. [DOI] [PubMed] [Google Scholar]
- 54.Kashefi K, Lovley DR. 2003. Extending the upper temperature limit for life. Science 301:934. doi: 10.1126/science.1086823. [DOI] [PubMed] [Google Scholar]
- 55.Holmes DE, Ueki T, Tang H-Y, Zhou J, Smith JA, Chaput G, Lovley DR. 2019. A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer. mBio 10:e00789-19. doi: 10.1128/mBio.00789-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prakash D, Chauhan SS, Ferry JG. 2019. Life on the thermodynamic edge: respiratory growth of an acetotrophic methanogen. Sci Adv 5:eaaw9059. doi: 10.1126/sciadv.aaw9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yee MO, Snoeyenbos-West O, Thamdrup B, Ottosen L, Rotaru A-E. 2019. Extracellular electron uptake by two Methanosarcina species. Front Energy Res 7:29. doi: 10.3389/fenrg.2019.00029. [DOI] [Google Scholar]
- 58.Yee MO, Rotaru A-E. 2020. Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes. Sci Rep 10:372. doi: 10.1038/s41598-019-57206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krukenberg V, Riedel D, Gruber-Vodicka HR, Buttigieg PL, Tegetmeyer HE, Boetius A, Wegener G. 2018. Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia. Environ Microbiol 20:1651–1666. doi: 10.1111/1462-2920.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poweleit N, Ge P, Nguyen HN, Loo RRO, Gunsalus RP, Zhou ZH. 2017. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat Microbiol 2:16222. doi: 10.1038/nmicrobiol.2016.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeffer C, Larsen S, Song J, Dong M, Besenbacher F, Meyer RL, Kjeldsen KU, Schreiber L, Gorby YA, El-Naggar MY, Leung KM, Schramm A, Risgaard-Petersen N, Nielsen LP. 2012. Filamentous bacteria transport electrons over centimetre distances. Nature 491:218–221. doi: 10.1038/nature11586. [DOI] [PubMed] [Google Scholar]
- 62.Bjerg JT, Boschker HTS, Larsen S, Berry D, Schmid M, Millo D, Tataru P, Meysman FJR, Wagner M, Nielsen LP, Schramm A. 2018. Long-distance electron transport in individual, living cable bacteria. Proc Natl Acad Sci U S A 115:5786–5791. doi: 10.1073/pnas.1800367115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meysman FJR. 2018. Cable bacteria take a new breath using long-distance electricity. Trends Microbiol 26:411–422. doi: 10.1016/j.tim.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 64.Meysman FJR, Cornelissen R, Trashin S, Bonné R, Martinez SH, van der Veen J, Blom CJ, Karman C, Hou J-L, Eachambadi RT, Geelhoed JS, Wael KD, Beaumont HJE, Cleuren B, Valcke R, van der Zant HSJ, Boschker HTS, Manca JV. 2019. A highly conductive fibre network enables centimetre-scale electron transport in multicellular cable bacteria. Nat Commun 10:4120. doi: 10.1038/s41467-019-12115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eachambadi RT, Bonné R, Cornelissen R, Hidalgo-Martinez S, Vangronsveld J, Meysman FJR, Valcke R, Cleuren B, Manca JV. 2020. An ordered and fail-safe electrical network in cable bacteria. Adv Biosys 4:2000006. doi: 10.1002/adbi.202000006. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y-L, Tang H-Y, Ribbe A, Duzhko V, Woodard TL, Ward JE, Bai Y, Nevin KP, Nonnenmann SS, Russell T, Emrick T, Lovley DR. 2018. Conductive composite materials fabricated with microbially produced protein nanowires. Small 14:1802624. doi: 10.1002/smll.201802624. [DOI] [PubMed] [Google Scholar]
- 67.Thorup CA, Schramm A, Findlay AJ, Finster KW, Schreiber L. 2017. Disguised as a sulfate reducer: growth of the deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. mBio 8:e00671-17. doi: 10.1128/mBio.00671-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovley DR. 2017. e-Biologics: fabrication of sustainable electronics with “green” biological materials. mBio 8:e00695-17. doi: 10.1128/mBio.00695-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu T, Liu X, Gao H, Ward JE, Liu X, Yin B, Wang Z, Zhuo Y, Walker DJF, Yang J, Chen J, Lovley DR, Yao J. 2020. Bioinspired bio-voltage memristors. Nat Commun 11:1861. doi: 10.1038/s41467-020-15759-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X, Fu T, Ward J, Gao H, Yin B, Woodard TL, Lovley DR, Yao J. 2020. Multifunctional protein nanowire humidity sensors. Adv Electron Mater 6:2000721. doi: 10.1002/aelm.202000721. [DOI] [Google Scholar]
- 71.Liu X, Gao H, Ward J, Liu X, Yin B, Fu T, Chen J, Lovley DR, Yao J. 2020. Power generation from ambient humidity using protein nanowires. Nature 578:550–554. doi: 10.1038/s41586-020-2010-9. [DOI] [PubMed] [Google Scholar]
- 72.Smith AF, Liu X, Woodard TL, Fu T, Emrick T, Jiménez JM, Lovley DR, Yao J. 2020. Bioelectronic protein nanowire sensors for ammonia detection. Nano Res 13:1479–1484. doi: 10.1007/s12274-020-2825-6. [DOI] [Google Scholar]
- 73.Sure S, Ackland ML, Torriero AJ, Adholeya A, Kochar M. 2016. Microbial nanowires: an electrifying tale. Microbiology 162:2017–2028. doi: 10.1099/mic.0.000382. [DOI] [PubMed] [Google Scholar]
- 74.Venkidusamy K, Megharaj M, Schröder U, Karouta F, Mohan SV, Naidu R. 2015. Electron transport through electrically conductive nanofilaments in Rhodopseudomonas palustris strain RP2. RSC Adv 5:100790–100798. doi: 10.1039/C5RA08742B. [DOI] [Google Scholar]
- 75.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.