ABSTRACT
Chronic wasting disease (CWD) is caused by prions, infectious proteinaceous particles, PrPCWD. We sequenced the PRNP gene of 2,899 white-tailed deer (WTD) from Illinois and southern Wisconsin, finding 38 haplotypes. Haplotypes A, B, D, E, G and 10 others encoded Q95G96S100N103A123Q226, designated ‘PrP variant A.’ Haplotype C and five other haplotypes encoded PrP ‘variant C’ (Q95S96S100N103A123Q226). Haplotype F and three other haplotypes encoded PrP ‘variant F’ (H95G96S100N103A123Q226). The association of CWD with encoded PrP variants was examined in 2,537 tested WTD from counties with CWD. Relative to PrP variant A, CWD susceptibility was lower in deer with PrP variant C (OR = 0.26, p < 0.001), and even lower in deer with PrP variant F (OR = 0.10, p < 0.0001). Susceptibility to CWD was highest in deer with both chromosomes encoding PrP variant A, lower with one copy encoding PrP variant A (OR = 0.25, p < 0.0001) and lowest in deer without PrP variant A (OR = 0.07, p < 0.0001). There appeared to be incomplete dominance for haplotypes encoding PrP variant C in reducing CWD susceptibility. Deer with both chromosomes encoding PrP variant F (FF) or one encoding PrP variant C and the other F (CF) were all CWD negative. Our results suggest that an increased population frequency of PrP variants C or F and a reduced frequency of PrP variant A may reduce the risk of CWD infection. Understanding the population and geographic distribution of PRNP polymorphisms may be a useful tool in CWD management.
KEYWORDS: CWD, haplotype, Illinois, incomplete dominance, non-synonymous SNPs, p.(Gln95His), p.(Gly96Ser), PRNP, synonymous SNPs
Introduction
Chronic Wasting Disease (CWD) that affects cervids appears to be the only prion disease that has emerged, persisted, and spread in populations of free-ranging or wild animals [1]. Even scrapie, the prototypical prion disease described for hundreds of years in domestic animals, has not been reported to persist or spread in free-ranging ovines or caprines [2]. Free-ranging cervids such as deer, elk, and moose are important species for recreational hunting and wildlife tourism [3], although these animals can become expensive nuisance animals when insufficient predation or hunting results in overpopulation. These characteristics of cervids make epidemiology of CWD particularly unique both from a biological standpoint and from an ecological or wildlife management standpoint. Variations in sequences of the prion protein gene (PRNP), or PRNP genotypes, are essential for understanding CWD susceptibility and transmission; after all, variation in protein sequence and conformation can impact the auto-catalytic conversion of host cellular prion protein (PrPC; PRNP) to disease-causing, and infectious, PrPCWD [4].
CWD was first identified in mule deer (O. hemionus hemionus) and black-tailed deer (O. h.columbianus) in captivity in Colorado and Wyoming in 1967 [5-7]. Since then, CWD has spread to additional species and affects free-ranging Cervidae in 24 US states, two Canadian provinces, Norway, Finland, and Sweden [8]. Including captive populations, CWD has been reported in an additional two US states, a Canadian province, and South Korea [8,9]. In Illinois, CWD was first detected in free-ranging white-tailed deer (WTD) in 2002 [10] and has since expanded to 17 counties in northern Illinois. As of 30 June 2018, the CWD prevalence rate for hunter-harvested deer across the affected counties is 1.07% for adult males and 0.54% for adult females, while the prevalence rate among adult deer taken by the Illinois Department of Natural Resources (IDNR) during targeted culling operations was 2.11% in 2018 [11]. Disease management strategies in Illinois have kept CWD prevalence rates at low levels since discovery [9-12].
Likely routes of disease transmission are via direct contact with a CWD infected animal, which would tend to be reflected by disease spread across the landscape, or by ingesting or inhaling contaminated soil, water, or food containing infectious PrPCWD [13,14]. Disease-associated PrPCWD results in rapid prion accumulation in lymphoid tissues [15,16] and infected deer may shed PrPCWD in saliva, urine, and faeces [17-21].
Variation among PRNP sequences may impact disease susceptibility by various mechanisms. Structural differences in prion proteins resulting from non-synonymous single nucleotide polymorphisms (SNPs) may alter the efficiency of prion-prion binding or conversion [22-24]. PRNP variants show different susceptibility to different strains in mouse model studies [24,25]. In many prion diseases, including kuru and Creutzfeldt-Jakob disease that affect humans, the polymorphisms in PRNP influence susceptibility and disease progression [26-33]. A scrapie eradication programme based on genetic susceptibility has been implemented in the United States [34]. While such a programme would be impractical for free-ranging animals, a better understanding of disease susceptibility would inform potential management intervention [6,7,35,36].
The PRNP gene in cervids consists of three exons, with the third exon encoding an open reading frame (771 bp) that encodes 257 amino acids. We and others have reported two SNPs, c.285A>C, and c.286G>A, that cause non-synonymous substitutions, respectively, from glutamine to histidine at codon 95, p.(Gln95His) (encoded by haplotype F, reported by Brandt et al. [37,38]), and from glycine to serine at codon 96, p.(Gly96Ser) (encoded by haplotype C, reported by Brandt et al. [37,38]), associated with a reduced incidence of CWD [37-42]. We have also reported that the frequency of protective PRNP haplotypes may have contributed to the way CWD has spread through Illinois [37]. While deer with protective variants may still be infected with CWD, albeit at a lower frequency, Otero et al. [43] reported that peripheral accumulation of infectious proteins is reduced in the deer that carry p.(Gln95His) reducing potential transmission. Henderson et al. [20] found little difference in prion shedding between deer that carry p.(Gly96Ser) and deer without it after inoculation, while Mathiason et al. [21] detected PrPCWD in deer with p.[(Gly96=)];[(Gly96=)] but failed to detect this in deer with p.[(Gly96=)];[(Gly96Ser)] after 18 months of inoculation.
Our study of CWD in white-tailed deer is novel in several respects. First, we did not analyse each of the DNA haplotype sequences separately, but instead grouped together all of the haplotypes encoding for the same amino acid sequence (i.e., which differed due to synonymous but not non-synonymous differences). Second, some previous studies have examined the presence or absence of a single nucleotide polymorphism (SNP). The current study examines the effects of different encoded polypeptides, i.e., PrP proteins that may differ at one or more than one amino acid site. We report that the DNA haplotypes among deer encode many PrP variants, of which three were common: PrP variant A, encoded by haplotypes A, B, D, E, G as well as other (rare) haplotypes; PrP variant C, encoded by haplotype C and other haplotypes; and PrP variant F, encoded by haplotype F and other haplotypes. Third, previous studies have examined the effects of diplotypes (combinations of the two DNA sequences encoded by a diploid individual). Our study examines the association of CWD with the combination of the two protein variants that the two haplotypes encode, i.e., considers the pair of encoded proteins rather than the pair of DNA haplotypes.
Our objectives in the current work were to examine (i) whether PrP variant C or PrP variant F are associated with lower CWD susceptibility compared to PrP variant A in free-ranging WTD; (ii) whether CWD susceptibility differs between PrP variant C and PrP variant F; (iii) whether protective haplotypes show dominance effects, by determining how the combination of proteins encoded by PRNP in an individual impacts susceptibility to CWD; and (iv) whether the frequency of PrP variants changed over time after CWD entered the Illinois white-tailed deer population.
Results
The complete coding region of PRNP (771 bp encoding 257 amino acids) was sequenced in 466 white-tailed deer samples collected in Illinois from 2015 to 2017 (calendar years) from regions known to be infected with CWD. Of the 466 samples, 157 were CWD positive, 308 were CWD negative, and 1 sample was not tested for CWD. In our previous studies, part of the PRNP coding region (621 bp encoding 207 amino acids) had been sequenced in 2433 samples collected between 2002 and 2014 [37,38,41]. Within the region for which sequences went beyond the coding region previously reported, we did not detect any SNPs in the newly sequenced coding regions in either the 5ʹ side (58 bp) or the 3ʹ side (89 bp). However, SNPs were detected in untranslated regions (UTRs), in both 5ʹ and 3ʹ UTRs.
For the analysis in this paper, the 466 samples sequenced from deer collected between 2015 and 2017 were combined with the 2433 samples from deer collected between 2002 and 2014 for a total of 2899 samples from Illinois and southern Wisconsin. Of these, 407 samples were CWD positive, 2347 samples were CWD negative, and 145 samples were not tested for CWD.
A total of 38 haplotypes were detected across the 2899 deer samples (Table 1). Many of these haplotypes had been previously reported (26 haplotypes, designated with letters A through Z) and had been deposited in GenBank (MG856905-MG856930) [37,38]. The newly identified novel haplotypes were rare and designated as ‘PRNP-Odvi27, PRNP-Odvi28,’ and so on. ‘Odvi’ is derived from Odocoileus virginianus. These were entered in GenBank (accession number: MN577934-MN577945). Nucleotide diversity was low (π = 0.00225). However, because of the large number of haplotypes present, haplotype diversity was high (Hd = 0.798).
Table 1.
Polymorphic sites within the coding region of PRNP in 2899 WTD collected from 2002 to 2017 in Illinois and Wisconsin, and haplotype frequencies for the 2754 CWD tested deer.
| Nucleotide position within the PRNP coding region |
Number of chromosomes |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | GenBank | 60 | 153 | 243 | 285 | 286 | 299 | 308 | 324 | 367 | 372 | 378 | 438 | 441 | 555 | 676 | CWD (–) | CWD (+) | Total | Freq. |
| A | MG856905 | C | C | T | A | G | G | A | A | G | G | G | C | C | C | C | 1368 | 322 | 1690 | 0.307 |
| B | MG856906 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | 1135 | 258 | 1393 | 0.253 |
| C | MG856907 | . | . | . | . | A | . | . | . | . | . | . | . | . | T | . | 878 | 48 | 926 | 0.168 |
| D | MG856908 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 477 | 101 | 578 | 0.105 |
| E | MG856909 | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | 177 | 34 | 211 | 0.038 |
| F | MG856910 | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | 277 | 6 | 283 | 0.051 |
| G | MG856911 | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 179 | 22 | 201 | 0.036 |
| H | MG856912 | . | . | . | . | . | . | . | . | . | A | . | . | . | T | . | 46 | 3 | 49 | <0.01 |
| I | MG856913 | . | . | A | . | A | . | . | . | . | . | . | . | . | T | . | 22 | 4 | 26 | <0.005 |
| J | MG856914 | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | 30 | 8 | 38 | <0.01 |
| K | MG856915 | T | . | . | . | . | . | . | . | . | . | . | . | . | . | A | 15 | 0 | 15 | <0.005 |
| L | MG856916 | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | 20 | 1 | 21 | <0.005 |
| M | MG856917 | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | 3 | 0 | 3 | <0.001 |
| N | MG856918 | T | . | . | C | A | . | . | . | . | . | . | . | . | . | . | 5 | 0 | 5 | <0.001 |
| O | MG856919 | T | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 15 | 4 | 19 | <0.005 |
| P | MG856920 | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | 9 | 1 | 10 | <0.005 |
| Q | MG856921 | . | . | . | . | A | . | . | . | A | . | . | . | . | T | . | 2 | 0 | 2 | <0.0005 |
| R | MG856922 | . | T | . | . | . | . | . | . | . | . | . | . | . | T | . | 5 | 1 | 6 | <0.005 |
| S | MG856923 | . | . | . | . | A | A | . | . | . | . | . | . | . | T | . | 1 | 0 | 1 | <0.0005 |
| T | MG856924 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | 3 | 0 | 3 | <0.001 |
| U | MG856925 | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | 8 | 0 | 8 | <0.005 |
| V | MG856926 | . | T | . | . | A | . | . | . | . | . | . | . | . | . | . | 3 | 0 | 3 | <0.001 |
| Y | MG856929 | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | 2 | 0 | 2 | <0.0005 |
| PRNP-Odvi28 | MN577935 | . | . | . | . | A | . | . | . | . | . | . | T | . | T | . | 3 | 0 | 3 | <0.001 |
| PRNP-Odvi29 | MN577936 | . | . | . | . | C | . | . | . | . | . | . | . | . | T | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi30 | MN577937 | . | . | . | . | C | . | . | G | . | . | . | . | . | . | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi31 | MN577938 | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi32 | MN577939 | . | T | . | . | . | . | . | . | . | . | . | . | T | . | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi33 | MN577940 | . | T | . | . | . | . | . | . | . | . | . | T | . | . | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi34 | MN577941 | T | . | . | . | A | . | . | . | . | . | . | . | . | T | . | 3 | 0 | 3 | <0.001 |
| PRNP-Odvi35 | MN577942 | T | . | . | C | . | . | . | . | . | . | . | . | T | . | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi36 | MN577943 | T | . | . | G | . | . | . | . | . | . | . | . | . | . | . | 0 | 1 | 1 | <0.0005 |
| PRNP-Odvi37 | MN577944 | T | T | . | C | . | . | . | . | . | . | . | . | . | . | . | 1 | 0 | 1 | <0.0005 |
| PRNP-Odvi38 | MN577945 | T | . | . | . | . | . | . | . | . | . | . | . | . | T | . | 1 | 0 | 1 | <0.0005 |
| W* | MG856927 | T | . | . | . | A | . | . | . | . | . | . | . | . | . | . | ||||
| X* | MG856928 | T | . | A | . | A | . | . | . | . | . | . | . | . | . | . | ||||
| Z* | MG856930 | . | . | . | . | A | . | . | . | . | . | . | T | . | . | . | ||||
| PRNP-Odvi27* | MN577934 | . | . | A | . | . | . | . | . | . | . | . | . | . | . | |||||
| Total | 4694 | 814 | 5508 | |||||||||||||||||
Only the polymorphic sites within the coding region of PRNP are shown. Numbers across the row refer to position within the coding region. The numbers under CWD columns refer to number of chromosomes carrying each haplotype, but the counts of the haplotypes that were detected in samples that were not tested for CWD are not shown. Frequency shows the haplotype frequencies in 2754 WTD tested for CWD; the samples that were not tested for CWD were not used in the calculations. Nucleotide positions matching those in haplotype A are shown as dots, while nucleotides that differ from those of haplotype A are shown. Dark boxes indicate non-synonymous substitutions; the other substitutions are synonymous. Haplotypes A to Z were reported by Brandt et al. [37]. PRNP-Odvi27 to PRNP-Odvi38 are novel haplotypes; (*) Haplotypes W, X, Z, and PRNP-Odvi27 were detected only among deer were not tested for CWD.
Alignment of the deer sequences revealed nucleotide variation at 15 positions (Table 1). Three different nucleotides were detected at position 285 (including 1 non-synonymous substitution), and at position 286 (all 3 non-synonymous). Only two different nucleotides were detected at other variable positions: non-synonymous substitutions at nucleotide positions: 299, 308, 367, 676; and synonymous substitutions at nucleotide positions 60, 153, 243, 324, 372, 378, 438, 441, and 555 (Table 1). Among the 2754 deer that had been tested for CWD, we detected 34 haplotypes (there were 4 other, very rare haplotypes detected only among untested deer). Of the 34 haplotypes detected among tested deer, only 7 had a frequency greater than 0.01 (Table 1); the rest were rare haplotypes.
There were a total of 11 different PrP variants encoded (i.e., different amino acid sequences, Table 2). Of the 34 haplotypes, 15 encoded p.[(Gln95=);(Gly96=);(Ser100=);(Asn103=);(Ala123=);(Gln226=)], including the haplotypes A, B, D, E, and G, along with 10 rare haplotypes. We designated the protein encoded by these 15 haplotypes as PrP variant A (Table 2, Figure 1). Six haplotypes encoded amino acids p.[(Gln95=);(Gly96Ser);(Ser100=);(Asn103=);(Ala123=);(Gln226=)], including haplotype C and five rare haplotypes. We designated the protein encoded by these six haplotypes as PrP variant C (Table 2, Figure 1). Four haplotypes encoded amino acids p.[(Gln95His);(Gly96=);(Ser100=);(Asn103=);(Ala123=);(Gln226=)], including haplotype F and three rare haplotypes. The protein encoded by this set of haplotypes was designated PrP variant F (Table 2, Figure 1). For some rare haplotypes, the translated amino acid sequences differed from those of PrP variants A, C and F; these uncommon PrP variants were categorized as ‘others.’
Table 2.
Amino acid variation in WTD prion protein.
| Nucleotide position | 285 | 286 | 299 | 308 | 367 | 676 | CWD | ||
|---|---|---|---|---|---|---|---|---|---|
| Amino acid position |
95 |
96 |
100 |
103 |
123 |
226 |
Negative |
Positive |
Total |
| PrP variant A | Q | G | S | N | A | Q | 3439 | 754 | 4193 |
| PrP variant C | . | S | . | . | . | . | 918 | 53 | 971 |
| PrP variant F | H | . | . | . | . | . | 281 | 6 | 287 |
| Other protein variants | |||||||||
| K | . | . | . | . | . | K | 15 | 0 | 15 |
| L | . | . | . | . | T | . | 20 | 1 | 21 |
| M | . | . | N | . | . | . | 3 | 0 | 3 |
| N | H | S | . | . | . | . | 5 | 0 | 5 |
| Q | . | S | . | . | T | . | 2 | 0 | 2 |
| S | . | S | N | . | . | . | 1 | 0 | 1 |
| U | . | . | . | I | . | . | 8 | 0 | 8 |
| PrP-Odvi29 | . | R | . | . | . | . | 2 | 0 | 2 |
| Total | 4694 | 814 | 5508 | ||||||
Polymorphic sites are shown for each protein variant. Amino acids that match those of protein variant A are indicated by dots, and the amino acids that differ from protein variant A are shown. Amino acid polymorphism associated with lower incidence of CWD are shaded [39-42]. Numbers in CWD positive, CWD negative, and total columns refer to number of chromosomes encoding each protein variant.
Figure 1.
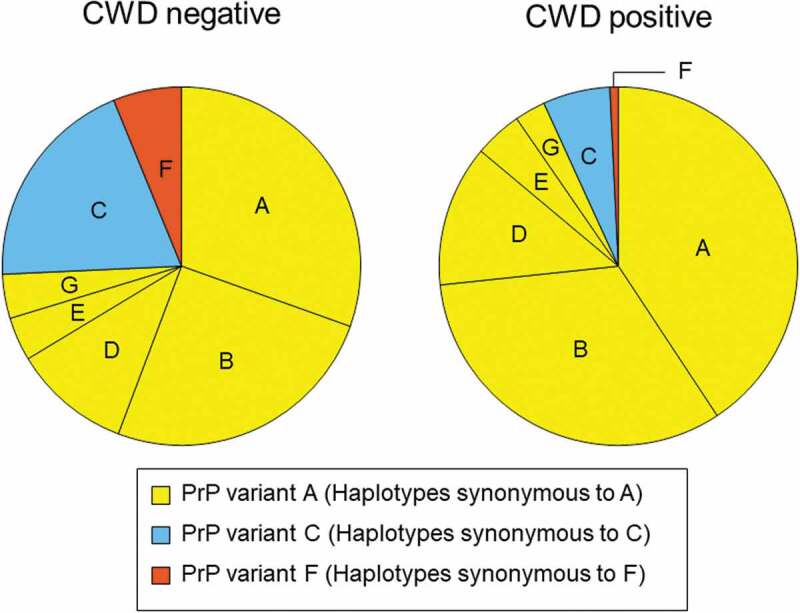
PRNP haplotype frequencies for deer collected between 2002 and 2017 and tested for CWD. Pie charts show frequencies for CWD negative (left) and positive (right) deer that carried PRNP haplotypes A through G. Haplotypes are coloured and arranged based on the encoded protein variants. Haplotypes B, D, E, and G were synonymous to haplotype A and shown in yellow (PrP variant A), haplotype C is in blue (PrP variant C), haplotype F is in orange (PrP variant F). The reduced frequency of PrP variants C and F in positive deer is evident. Rare haplotypes with frequencies <0.01 are not shown.
The associations of PrP variants and CWD susceptibility were tested using the data only from samples collected after CWD was found in a county, and only from deer samples that did not carry uncommon protein variants (n = 2376 after these criteria were applied). PrP variant A was detected significantly more frequently than PrP variant C in CWD positive WTD (odds ratio [OR] = 0.26, 95% confidence interval [CI]: 0.187–0.341, p < 0.0001, Fisher’s exact test, two-tailed) or PrP variant F (OR = 0.10, 95% CI: 0.035–0.213, p < 0.0001, Fisher’s exact test, two-tailed) (Figure 2). In addition, PrP variant F was less frequent in CWD positive deer than PrP variant C (OR = 0.37, 95% CI: 0.128–0.872, p = 0.016, Fisher’s exact test, two-tailed) (Figure 2).
Figure 2.
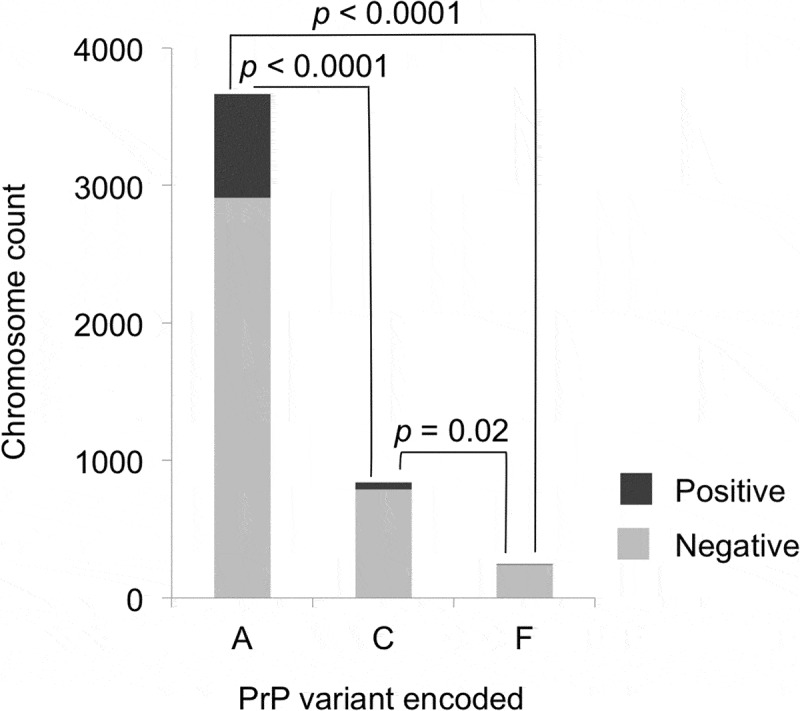
PrP variants and CWD susceptibility, for PrP variants A, C, and F. Only the samples collected after CWD spread to each county and that did not encode uncommon protein variants were used (n = 2376). Light shading indicates CWD negative and darker shading indicates CWD positive cases. PrP variant A was detected significantly more frequently than PrP variant C (OR = 0.26) or PrP variant F (OR = 0.10) in CWD positive deer than in CWD negative deer. PrP variant C showed a significantly smaller relative reduction than PrP variant F in CWD positive samples (OR = 0.37). For each protein variant, deer with one chromosome encoding the variant added one to the total shown, while deer with both chromosomes encoding a variant added two to the total; p-values are based on Fisher’s exact tests (two-tailed) and adjusted using the Benjamini-Hochberg procedure.
We also examined the association between CWD susceptibility and the combinations of PrP variants encoded by the two chromosomes of individual deer to test whether the effects of PRNP alleles are dominant or incompletely dominant [44] (Figures 3 and 4). To examine this, we grouped the deer into seven different categories: (1) deer with both chromosomes encoding PrP variant A, labelled AA; (2) deer in which one of the two chromosomes encoded PrP variant A while the other encoded protein variant C, labelled AC; (3) deer that encoded PrP variants A and F, labelled AF; (4) deer with both chromosomes encoding PrP variant C, labelled CC; (5) deer with both chromosomes encoding variant F, labelled FF; (6) deer carrying haplotypes encoding PrP variants C and F, labelled CF; and (7) deer that encoded a different PrP variant (from A, C or F) in at least one of the chromosomes, labelled ‘others.’ These categories are listed in Figure 4(b) except for ‘others.’ Fisher’s exact tests were conducted only using the samples collected after CWD spread to each county. We found significant associations between CWD susceptibility and the PrP variant(s) encoded by the two chromosomes in a deer (Figure 4). Deer in which PrP variant A was encoded by both chromosomes (AA) showed significantly higher susceptibility to CWD compared to deer that carried only 1 chromosome encoding PrP variant A (AC or AF) (OR = 0.25, 95% CI: 0.178–0.339, p < 0.0001, Fisher’s exact test, two-tailed), or to deer that did not carry any chromosomes encoding PrP variant A (CC, FF, or CF) (OR = 0.07, 95% CI: 0.019–0.182, p < 0.0001, Fisher’s exact test, two-tailed) (Figure 4). Deer with no chromosomes encoding PrP variant A (CC, FF, or CF) showed significantly reduced susceptibility to CWD compared to deer that encoded PrP variant A in just one chromosome (AC or AF) (OR = 0.28, 95% CI: 0.073–0.777, p = 0.009, Fisher’s exact test, two-tailed) (Figure 4).
Figure 3.
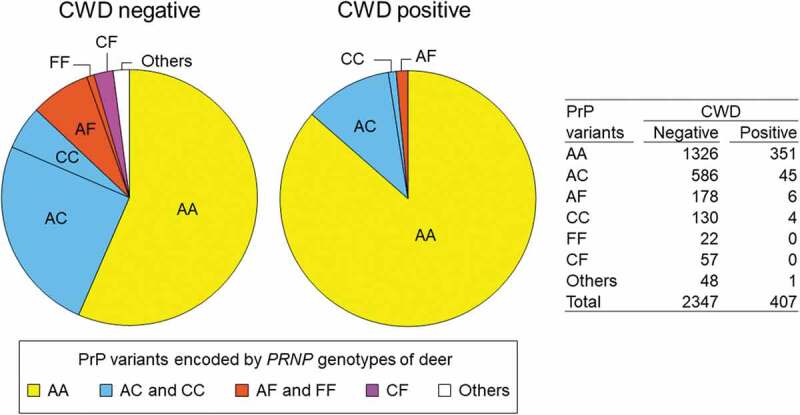
PrP variant combinations in CWD positive and negative deer. Combinations of protein variants are colour-coded; deer with both chromosomes encoding PrP variant A (AA) are shown in yellow, deer with PrP variant C encoded by at least one chromosome are shown in blue (AC or CC), deer with PrP variant F encoded by at least one chromosome are shown in orange (AF or FF), deer with one chromosome encoding PrP variant C and the other encoding F are shown in purple (CF), and deer with other protein variants encoded by at least one chromosome are in white (others). The much higher proportion of AA relative to other combinations is evident among CWD positive relative to CWD negative deer. No FF or CF deer were detected among CWD positive deer.
Figure 4.
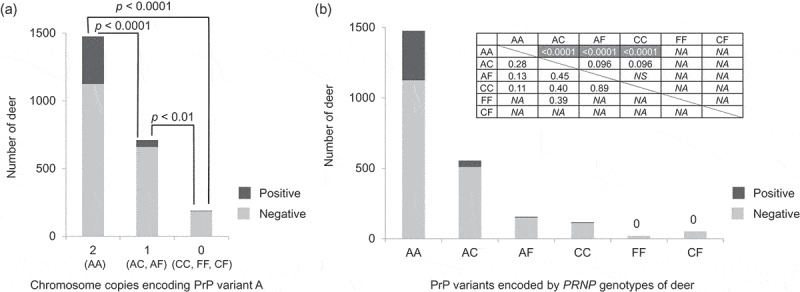
PrP variant combinations and CWD susceptibility. Only the samples collected after CWD spread to each county and that did not carry uncommon protein variants were used (n = 2376). (a) Samples were grouped into deer with two chromosomes encoding PrP variant A (AA), deer with one chromosome encoding PrP variant A (AC and AF), and deer without PrP variant A (CC, FF, CF). Deer with both chromosomes encoding PrP variant A showed a significantly higher susceptibility to CWD compared to deer that carried only 1 chromosome encoding PrP variant A (AC or AF) (OR = 0.25), or to deer that did not carry any chromosomes encoding PrP variant A (CC, FF, or CF) (OR = 0.07). Deer with no chromosomes encoding PrP variant A (CC, FF, or CF) showed significantly reduced susceptibility to CWD compared to deer that encoded PrP variant A in just one chromosome (AC or AF) (OR = 0.28). The p-values are based on Fisher’s exact tests (two-tailed) and adjusted using the Benjamini-Hochberg procedure. (b) CWD positive and negative deer numbers are shown in a histogram for each protein variant combination: AA, AC, AF, CC, FF, and CF. The association between each PrP variant combination and CWD susceptibility was tested with Fisher’s exact test (two-tailed). The upper right table shows the p-value (above diagonal) and odds ratio (below diagonal) for each comparison. NS indicates that the p-value that was more than 0.10 and NA indicates not applicable, because CWD was not detected for deer encoding FF or CF. Shading indicates significant results after the Benjamini-Hochberg procedure was applied.
The presence of PrP variant F showed a stronger effect in lowering CWD susceptibility compared to PrP variant C. Only six CWD positive deer carried a haplotype encoding PrP variant F, and all six also carried a haplotype encoding PrP variant A in the other chromosomes (AF) (Figure 3). When deer with AC and CC were compared, there was a marginally significant association to CWD susceptibility and CC deer showed a reduced CWD susceptibility compared to AC deer (OR = 0.40, 95% CI: 0.103–1.135, p = 0.096, Fisher’s exact test, two-tailed) (Figure 4(b)). There was also a marginally significant difference between AC and AF (OR = 0.45, 95% CI: 0.156–1.095, p = 0.096, Fisher’s exact test, two-tailed) (Figure 4(b)), with AF being relatively lower in CWD positive deer than AC. The sample sizes were small for deer with one chromosome encoding protein variant C and the other encoding F (CF); and for deer in which both chromosomes encoded protein variant F (FF), and any conclusions may therefore be tentative. Yet none of the CF (n = 52) or FF (n = 20) deer were CWD positive in the counties where CWD had spread. Haplotypes that encode PrP variant F may have complete or almost complete dominance in lowering the CWD susceptibility. Haplotypes that encode PrP variant C have an incomplete dominance effect over haplotypes that encode PrP variant A.
In the ten counties in IL that experienced CWD for more than five years, we examined whether the frequency of PrP variant A changed in the years after CWD spread in each county. There was no significant correlation between the frequency of PrP variant A and the years after CWD spread into each county (p > 0.05, generalized linear mixed-effects model) (Figure S1(a)). We also compared the frequency of PrP variant A in the first five years of CWD infection, to the frequency after more than five years of CWD, finding no significant difference (p > 0.05, df = 1, common OR = 0.92, Cochran-Mantel-Haenszel test) (Figure S1(b)). Before conducting a Cochran-Mantel-Haenszel test, we made sure that there were no differences of odds ratios across counties (p > 0.05, X2 = 14.824, df = 9, Woolf test).
Discussion
As shown in other species where prion diseases occur naturally, the occurrence of CWD positive deer varies greatly depending on non-synonymous substitutions in the prion protein gene. Polymorphisms at codon 95 from glutamine to histidine and at codon 96 from glycine to serine in white-tailed deer have been suggested to lower the susceptibility to CWD [39-42]. Transgenic (Tg) mice with deer PRNP encoding serine at codon 96 have shown delayed or even no disease progression [45,46]. While Tg mice expressing p.[(Gly96=)];[(Gly96=)] deer PRNP were susceptible to CWD, Tg mice expressing p.[(Gly96=)];[(Gly96Ser)] showed delayed disease progression and p.[(Gly96Ser)];[(Gly96Ser)] mice even showed no transmission [46]. However, an orally infected deer [47] and a small number of cases of CWD in free-ranging deer [48,49] with p.[(Gly96Ser)];[(Gly96Ser)] have been reported. In studies that examined the DNA sequences of PRNP, two haplotypes, haplotype C and haplotype F, have been found to be associated with reduced susceptibility to CWD [37,38]. In the current study, we focused on protein variants encoded by these previously reported PRNP polymorphisms and haplotypes, grouping each of them with all other haplotypes that code for the same amino acid sequence, and increasing the power to detect the effect of the protein variants on CWD susceptibility.
Sheep expressing PrP relatively resistant to scrapie are susceptible to atypical scrapie [50,51]. In CWD, Duque Velásquez et al. [25] reported that transgenic mice expressing deer p.(Gly96Ser) developed disease only when inoculated intracerebrally with CWD agents derived from deer expressing p.(Gln95His) while the mice did not develop disease inoculated with CWD agents from deer expressing p.[(Gly96=)];[(Gly96=)] or p.[(Gly96=)];[(Gly96Ser)] at 700 days post inoculation. In our current dataset, there is no information available about differing pathologies to indicate possible strain differences; however, our data showed lower CWD frequency in deer expressing p.(Gly96Ser) or p.(Gln95His).
We demonstrated that deer that carry PrP variants C and F on at least one chromosome form a smaller proportion of the deer testing positive than of deer testing negative, consistent with what was previously reported for DNA haplotypes C and F [37,38], and for studies of non-synonymous SNPs in PRNP [39-42]. By grouping all haplotypes that encode PrP variants A, C and F, greater statistical power was possible for examining the effects of protein variants and of the number of chromosomes in a deer that encode a protective protein. PrP variants C and F significantly lowered CWD susceptibility compared to PrP variant A. Compared to PrP variant A, variant C has serine at amino acid position 96 (p.(Gly96Ser)) while PrP variant F has histidine at amino acid position 95 (p.(Gln95His)). PrP variant C was also less detected in CWD positive deer in prior studies (in which the designations were QSS or QSAS) [39,42]. When PrP variant C and PrP variant F were compared, the deer that carried PrP variant F were proportionately less common among positive than among negative deer than were deer carrying PrP variant C. The incubation period of CWD in naturally infected animals is unknown, but in captive elk most natural cases occur in animals 3 to 8 years old [52], and it has been estimated that the majority of Cervid species probably develop the disease within the first 3 years of infection [6]. However, under inoculation experiments in white-tailed deer, prion shedding as early as 3 months after CWD exposure was detected by a real-time quaking-induced conversion method [20,53]. Following oral inoculation, the average survival period of WTD that had glutamine at codon 95 and glycine at 96 was found to be 693 days while deer with p.[(Gly96=)];[(Gly96Ser)] survived 956 days, and the deer with p.[(Gln95=)];[(Gln95His)] started to show the disease symptoms much later and survived a much longer period (1508 days) after inoculation [54], suggesting that PrP variant F with p.(Gln95His) may slow disease progression more than PrP variant C, which has p.(Gly96Ser). A recent study reported that the deer with p.[(Gly96=)];[(Gly96Ser)] showed delayed disease progression but also showed the similar PrPCWD distribution in tissues at terminal stages of disease, while the deer with p.[(Gln95=)];[(Gln95His)] or p.(Gln95His)(;)p.(Gly96Ser) showed limited peripheral accumulation of PrPCWD [43].
The haplotypes that encode protective protein variants seem to have incomplete dominance. Deer that encode two copies of PrP variants C or F showed a reduced susceptibility to CWD compared to deer with only one chromosome encoding C or F (Figure 4). When the effects of PrP variants C and F were examined separately, the deer in which both chromosomes encoded PrP variant C were not completely resistant to CWD but tended to have lower susceptibility to CWD compared to deer in which one chromosome encoded PrP variant C (Figure 4(b)). Compared to PrP variant C, PrP variant F has a greater impact in lowering the CWD susceptibility (only 6 of 184 AF deer were CWD positive). We detected only four cases of CWD in deer in which both chromosomes encode protective PrP variant C. None of the CWD-positive deer encoded PrP variant combinations FF or CF. Although no CWD positive deer had PrP variant combinations FF or CF in our samples from Illinois and southern Wisconsin, additional data would be needed to determine the degree to which deer with PrP variant combinations FF or CF may be resistant to CWD.
In a study of the oral inoculation of brain homogenate from CWD positive WTD, deer with two protective non-synonymous SNPs p.(Gln95His)(;)p.(Gly96Ser), i.e., the same as found, respectively, in PrP variants F and C, did develop CWD and survived 1596 days after inoculation [54]. However, under natural conditions, it is unlikely that deer would become infected due to direct contact with brain tissue from CWD infected deer. The CWD infected brain tissue carries a high infectious dose especially in advanced cases of disease, and extreme inoculation conditions may overcome the protective nature of PrP variants F and C. Furthermore, it is likely given our results that deer that have p.(Gln95His) on both chromosomes or deer that have two non-synonymous mutations may be highly resistant to CWD. Interestingly, haplotype N encodes both p.(Gln95His) and p.(Gly96Ser) and none of the deer carrying this haplotype was CWD positive (because this was a rare haplotype, each of the deer that carried it was heterozygous at the PRNP gene). The sample size was five, too small to analyse for effects of having two protective SNPs on the same chromosome. Overall, decreasing the dose of infectious CWD in the environment by reducing the number of infected animals may benefit populations in which deer carry haplotypes that encode protective protein variants.
We detected several rare haplotypes with frequencies lower than 0.01 that did not encode PrP variants A, C, or F (Tables 1 and 2). The conversion of PrPC to the infectious and abnormal PrPCWD is less efficient between different PrP protein variants due to binding interference [22]. Due to the small sample sizes of these rare haplotypes, we were not able to examine the association between the uncommon PrP variants and CWD susceptibility, but it is possible that they may also have a protective effect against CWD.
Robinson et al. [55] simulated the changes of PRNP allele frequency under selective pressure from CWD and predicted that alleles encoding glycine at codon 96 would decrease over time, while those encoding serine would increase. We did not find significant changes over time (Figure S1), although it is possible that changes may occur over longer time scales.
Controlling CWD prevalence rates at low levels and preventing the spread of CWD into new areas has been a significant challenge in managing deer populations [10,12]. Due to its high level of transmissibility and the persistence and accumulation of protease-resistant prion proteins in the soil and water [56-58], it will be difficult to eliminate PrPCWD from infected areas. However, it may be possible to decrease the number of newly infected animals, by reducing the risk of infection and the number of infected animals shedding PrPCWD. In addition to adopting regulations that promote ‘social distancing’ in deer (prohibitions on baiting, feeding, or artificial mineral licks that cause deer to congregate under conditions that increase the risk of environmental transmission [14,59]) and deer management approaches that reduce deer densities in CWD-affected areas, managers may find that altering the genetic composition of deer populations to be also critical for the management of CWD.
One study reported that among deer inoculated with PrPCWD, those infected with p.(Gln95His) or p.(Gly96Ser) (amino acid substitutions corresponding to PrP variants F and C, respectively) may demonstrate longer incubation periods for CWD and survive longer [54]. If so, there may be a risk that deer with protective protein variants may shed PrPCWD for a longer period than deer with PrP variant A. However, the much lower susceptibility to CWD of deer with PrP variants C or F may outweigh the risk of a prolonged shedding period, particularly for deer in which both chromosomes encode PrP variant F, or in which PrP variants F and C are both encoded. Furthermore, there are no data from free-ranging deer indicating whether there are any great differences in when and how prion proteins are shed by naturally infected deer that carry different protein variants. In inoculation experiments, Henderson et al. [20] found little difference in prion shedding between deer with p.[(Gly96=)];[(Gly96=)] and p.[(Gly96=)];[(Gly96Ser)] while Mathiason et al. [21] failed to detect PrPCWD in the deer with p.[(Gly96=)];[(Gly96Ser)] after 18 months of inoculation. Our results suggest that the PRNP haplotypes encoding PrP variants C or F are much less common in CWD positive deer than those encoding PrP variant A. Thus increased frequency of haplotypes encoding PrP variants C or F and reduced frequency of haplotypes encoding A may benefit the control of CWD in deer populations. Assessing the PRNP polymorphisms in deer populations may foster greater understanding of the role of protein differences encoded by the prion protein gene in controlling the spread of CWD, and may provide additional information to assess the risks of CWD infection in the population, offering clues to adopt management strategies that account for PRNP polymorphisms in deer populations on the landscape.
Materials and methods
Samples
We analysed tissue samples from 2899 free-ranging wild white-tailed deer from Illinois and southern Wisconsin from 2002 to 2017 from an archived collection at the University of Illinois. Samples were obtained through the CWD surveillance and government control programmes in Illinois and Wisconsin. Of 2899 samples, 2754 were tested for CWD. In Illinois, the obex and retropharyngeal lymph nodes were tested for CWD using immunohistochemistry (IHC) to detect PrPCWD at the Illinois Department of Agriculture Diagnostic laboratories in Galesburg or Centralia [10,12] and at the University of Illinois Veterinary Diagnostic Laboratory (VDL). Samples collected in Wisconsin were tested for CWD by the Wisconsin Veterinary Diagnostic Laboratory using IHC or an enzyme-linked immunosorbent assay. Detailed information including location, sex, and age was recorded at the time of sampling. We analysed all CWD positive samples and chose CWD negative control samples to match positive samples based on age, sex, and location to minimize confounding factors [37,38,41]. The laboratory work was conducted under the University of Illinois Institutional Biosafety Committee approved protocol.
PCR and sequencing of PRNP
Genomic DNA was extracted from muscle samples using the Wizard Genomic DNA Purification Kit (Promega Corporation, A1120) following the manufacturer’s protocol with some modifications. Part of the PRNP coding region (621 bp encoding 207 amino acids) had been sequenced previously in 2433 white-tailed deer samples collected between 2002 and 2014 [37,38,41]. We also amplified the complete coding region of PRNP (771 bp encoding 257 amino acids) in an additional 466 deer samples collected in Illinois from 2015 to 2017 using primers 223 (ACACCCTCTTTATTTTGCAG) and 224 (AGAAGATAATGAAAACAGGAAG) [42]. These primers were designed based on intron 2 and the 3ʹUTR to avoid the amplification of PRNP pseudogene, with an expected amplicon size of 788 bp [42]. In addition to these primers, internal primers for Sanger sequencing, PRNP-IF (ATGCTGGGAAGTGCCATGA) and PRNP-IR (CATGGCACTTCCCAGCAT), were designed using the software Primer3 (http://primer3.ut.ee/) [60]. PCR used 0.4 µM final concentration of each oligonucleotide primer in 1.5 mM MgCl2, 200 µM of each of the dNTPs (Promega Corporation, U1515), and 1X Colourless GoTaq Flexi Buffer with 0.08 units/µl final concentration of GoTaq Flexi DNA Polymerase (Promega Corporation, M8296) in 25 µl volume reaction. PCR consisted of an initial 95°C for 2 minutes; with cycles of 30 seconds denaturing at 95°C, followed by 30 seconds of annealing at 58°C (five cycles); 56°C (five cycles); or 54°C (30 cycles), followed by 1 minute extension at 72°C; with a final extension of 5 minutes at 72°C. After confirming the amplification with a 1% agarose gel with ethidium bromide under ultraviolet, we removed unincorporated primers and dNTPs from the PCR amplicons with exonuclease I (New England Biolabs, B0293S) and shrimp alkaline phosphatase (New England Biolabs, M0371S) [61]. The purified products were submitted to the Core DNA Sequencing Facility of the University of Illinois at Urbana-Champaign for Sanger sequencing analyses where samples were cycle sequenced using the BigDye Terminator v3.1 and resolved on ABI 3730XL capillary sequencer. The software Sequencher 5.1 (Gene Codes Corporation) was used to edit chromatograms, assemble contigs for each amplicon.
PRNP polymorphisms analyses
Haplotype phase was inferred using PHASE [62], which assumes Hardy-Weinberg equilibrium and uses a coalescent-based Bayesian method, implemented in DnaSP version 5.10.1 (http://www.ub.edu/dnasp/) [63] with 10,000 iterations and 100 burn-in iterations, using the available sequence data accumulated (n = 2899 deer). Each inferred haplotype was then translated to a protein sequence using the Translate tool of ExPASy (https://web.expasy.org/translate/) [64]. Sequence and protein variant nomenclature follows Sequence Variant Nomenclature (https://varnomen.hgvs.org/).
Haplotype diversity and nucleotide diversity were calculated using the software DnaSP version 5.10.1 [63]. To test the associations between the (translated) PrP variants and CWD susceptibility, Fisher’s exact test was conducted using R version 3.4.0 [65] in RStudio version 1.1.423 [66]. The year CWD spread to each county was obtained from the Illinois Department of Natural Resources [11] and the Wisconsin Department of Natural Resources [67]. The p-values by multiple comparisons were adjusted using the Benjamini-Hochberg procedure (https://alexandercoppock.com/statistical_comparisons.html) [68].
We tested whether the frequency of PrP variant A changed over time using the data from counties that had more than five years of CWD. A generalized linear mixed-effects model was implemented in lme4 package version 1.1–21 [69] treating the years after CWD had expanded to each county as an explanatory variable, frequency of PrP variant as a response variable, and county harvested as random effects. We also compared the frequency of PrP variant A within five years and after five years after CWD had spread to each county using the Cochran-Mantel-Haenszel test in R [65] treating counties as strata. The Cochran-Mantel-Haenszel test assumes the homogeneity of odds ratios across strata; thus the Woolf test was conducted using vcd package version 1.4–7 [70,71] in R, ahead of the Cochran-Mantel-Haenszel test to determine if it is applicable.
Supplementary Material
Acknowledgments
We thank the Illinois and Wisconsin Department of Natural Resources for collecting samples, and the hunting communities for their collaboration and engagement with the CWD surveillance and control programmes. We thank W. M. Brown for assistance with data management, technicians and undergraduate students in the Novakofski and Mateus-Pinilla laboratory for assistance with the genetic samples.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the U.S. Fish and Wildlife Service [Federal Aid in Wildlife Restoration Project (W-146-R)]; Illinois Natural History Survey – Prairie Research Institute; Office of the Vice Chancellor for Research, University of Illinois at Urbana-Champaign (US)..
Disclosure statement
The authors declare there is no conflict of interest.
Supplementary material
Supplemental data of this article can be accessed here.
References
- [1].Richards B, Walsh D, Russell RE. Chronic wasting disease overview. Reston, Virginia: USGS National Wildlife Health Center; [cited 2020. January 6]. Available from: https://www.usgs.gov/centers/nwhc/science/chronic-wasting-disease?qt-science_center_objects=0 - qt-science_center_objects
- [2].Mathiason CK. Chapter Twelve - Scrapie, CWD, and transmissible mink encephalopathy. In: Legname G, Vanni S, editors. Progress in molecular biology and translational science. Vol. 150. Cambridge (MA): Academic Press; 2017. p. 267–292. [DOI] [PubMed] [Google Scholar]
- [3].New 5-year report shows 101.6 million Americans participated in hunting, fishing & wildlife activities [Internet]. Washington DC: U.S. Department of the Interior; [Cited 2017. September 7]. Available from: https://www.doi.gov/pressreleases/new-5-year-report-shows-1016-million-americans-participated-hunting-fishing-wildlife [Google Scholar]
- [4].Zhou Z, Xiao G.. Conformational conversion of prion protein in prion diseases. Acta Biochim Biophys Sin (Shanghai). 2013;45(6):465–476. [DOI] [PubMed] [Google Scholar]
- [5].Williams ES, Young S. Chronic wasting disease of captive mule deer - spongiform encephalopathy. J Wildl Dis. 1980;16(1):89–98. [DOI] [PubMed] [Google Scholar]
- [6].Moreno JA, Telling GC . Molecular mechanisms of chronic wasting disease prion propagation. Cold Spring Harb Perspect Med. 2018;8(6):a024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haley NJ, Hoover EA. Chronic wasting disease of cervids: current knowledge and future perspectives. Annu Rev Anim Biosci. 2015;3:305–325. [DOI] [PubMed] [Google Scholar]
- [8].Centers for Disease Control and Prevention . Chronic wasting disease (CWD): occurrence: Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of High-Consequence Pathogens and Pathology (DHCPP); 2019. [updated 2019 Mar 13; cited 2019 June 4]. Available from: https://www.cdc.gov/prions/cwd/occurrence.html
- [9].Rivera NA, Brandt AL, Novakofski JE, et al. Chronic wasting disease in Cervids: prevalence, impact and management strategies. Vet Med (Auckl). 2019;10:123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mateus-Pinilla N, Weng H-Y, Ruiz MO, et al. Evaluation of a wild white-tailed deer population management program for controlling chronic wasting disease in Illinois, 2003–2008. Prev Vet Med. 2013. July 1;110(3–4):541–548. [DOI] [PubMed] [Google Scholar]
- [11].Dufford D, McDonald P. Illinois chronic wasting disease: 2017-2018 surveillance and management report. CWD Annual Reports: Wildlife Disease Program, Illinois Department of Natural Resources; 2018. p. 1–16.
- [12].Manjerovic MB, Green ML, Mateus-Pinilla N, et al. The importance of localized culling in stabilizing chronic wasting disease prevalence in white-tailed deer populations. Prev Vet Med. 2014. January 1;113(1):139–145. [DOI] [PubMed] [Google Scholar]
- [13].Miller MW, Williams ES, Hobbs NT, et al. Environmental sources of prion transmission in mule deer. Emerg Infect Dis. 2004. June;10(6):1003–1006. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Plummer IH, Johnson CJ, Chesney AR, et al. Mineral licks as environmental reservoirs of chronic wasting disease prions. PLoS One. 2018;13(5):e0196745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sigurdson CJ, Williams ES, Miller MW, et al. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J Gen Virol. 1999;80(10):2757–2764. [DOI] [PubMed] [Google Scholar]
- [16].Hoover CE, Davenport KA, Henderson DM, et al. Pathways of prion spread during early chronic wasting disease in deer. J Virol. 2017;91(10):e00077–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Plummer IH, Wright SD, Johnson CJ, et al. Temporal patterns of chronic wasting disease prion excretion in three cervid species. J Gen Virol. 2017;98(7):1932–1942. [DOI] [PubMed] [Google Scholar]
- [18].Tamguney G, Richt JA, Hamir AN, et al. Salivary prions in sheep and deer. Prion. 2012. Jan-Mar;6(1):52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Haley NJ, Mathiason CK, Carver S, et al. Detection of chronic wasting disease prions in salivary, urinary, and intestinal tissues of deer: potential mechanisms of prion shedding and transmission. J Virol. 2011. July;85(13):6309–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Henderson DM, Denkers ND, Hoover CE, et al. Longitudinal detection of prion shedding in saliva and urine by chronic wasting disease-infected deer by real-time quaking-induced conversion. J Virol. 2015;89(18):9338–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mathiason CK, Powers JG, Dahmes SJ, et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006;314(5796):133. [DOI] [PubMed] [Google Scholar]
- [22].Caughey B. Prion protein conversions: insight into mechanisms, TSE transmission barriers and strains. Br Med Bull. 2003;66(1):109–120. [DOI] [PubMed] [Google Scholar]
- [23].Pal LR, Yu C-H, Mount SM, et al. Insights from GWAS: emerging landscape of mechanisms underlying complex trait disease. BMC Genomics. 2015;16(Suppl8):S4–S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cortez LM, Sim VL. Implications of prion polymorphisms. Prion. 2013. Jul-Aug;7(4):276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Duque Velásquez C, Kim C, Herbst A, et al. Deer prion proteins modulate the emergence and adaptation of chronic wasting disease strains. J Virol. 2015;89(24):12362–12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res. 2008;Jul-Aug;39(4):30. [DOI] [PubMed] [Google Scholar]
- [27].Mead S, Whitfield J, Poulter M, et al. Genetic susceptibility, evolution and the kuru epidemic. Philos Trans R Soc Lond B Biol Sci. 2008. November 27;363(1510):3741–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mead S, Stumpf MP, Whitfield J, et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003. April 25;300(5619):640–643. [DOI] [PubMed] [Google Scholar]
- [29].Mead S, Poulter M, Uphill J, et al. Genetic risk factors for variant Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 2009. January;8(1):57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hizume M, Kobayashi A, Teruya K, et al. Human prion protein (PrP) 219K is converted to PrPSc but shows heterozygous inhibition in variant Creutzfeldt-Jakob disease infection. J Biol Chem. 2009. February 6;284(6):3603–3609. [DOI] [PubMed] [Google Scholar]
- [31].Shibuya S, Higuchi J, Shin R-W, et al. Codon 219 lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1998;43(6):826–828. [DOI] [PubMed] [Google Scholar]
- [32].Diack AB, Head MW, McCutcheon S, et al. Variant CJD. Prion. 2014. July 04;8(4):286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kobayashi A, Teruya K, Matsuura Y, et al. The influence of PRNP polymorphisms on human prion disease susceptibility: an update. Acta Neuropathol. 2015. August 01;130(2):159–170. [DOI] [PubMed] [Google Scholar]
- [34].Animal and Plant Health Inspection Service, USDA. Scrapie in sheep and goats: 84 FR 11170. 2019:11170–11196. Available from: https://www.federalregister.gov/documents/2019/03/25/2019-05430/scrapie-in-sheep-and-goats
- [35].Robinson SJ, Samuel MD, O’Rourke KI, et al. The role of genetics in chronic wasting disease of North American cervids. Prion. 2012. April 01;6(2):153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kurt TD, Sigurdson CJ. Cross-species transmission of CWD prions. Prion. 2016;10(1):83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Brandt AL, Green ML, Ishida Y, et al. Influence of the geographic distribution of prion protein gene sequence variation on patterns of chronic wasting disease spread in white-tailed deer (Odocoileus virginianus). Prion. 2018. July 04;12(3–4):204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brandt AL, Kelly AC, Green ML, et al. Prion protein gene sequence and chronic wasting disease susceptibility in white-tailed deer (Odocoileus virginianus). Prion. 2015. November 2;9(6):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Johnson C, Johnson J, Clayton M, et al. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J Wildl Dis. 2003. July;39(3):576–581. [DOI] [PubMed] [Google Scholar]
- [40].Johnson C, Johnson J, Vanderloo JP, et al. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006;87:2109–2114. [DOI] [PubMed] [Google Scholar]
- [41].Kelly AC, Mateus-Pinilla NE, Diffendorfer J, et al. Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus). Prion. 2008. Jan-Mar;2(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].O’Rourke KI, Spraker TR, Hamburg LK, et al. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004. May;85(Pt 5):1339–1346. [DOI] [PubMed] [Google Scholar]
- [43].Otero A, Duque Velásquez C, Johnson C, et al. Prion protein polymorphisms associated with reduced CWD susceptibility limit peripheral PrP(CWD) deposition in orally infected white-tailed deer. BMC Vet Res. 2019;15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].An P, Kirk GD, Limou S, et al. . Impact of APOL1 genetic variants on HIV-1 infection and disease progression. Front Immunol. 2019;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Meade-White K, Race B, Trifilo M, et al. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol. 2007;81(9):4533–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Race B, Meade-White K, Miller MW, et al. In vivo comparison of chronic wasting disease infectivity from deer with variation at prion protein residue 96. J Virol. 2011;85(17):9235–9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Miller MW, Wolfe LL, Sirochman TM, et al. Survival patterns in white-tailed and mule deer after oral inoculation with a standardized, conspecific prion dose. J Wildl Dis. 2012. April;48(2):526–529. [DOI] [PubMed] [Google Scholar]
- [48].Haley NJ, Merrett K, Buros Stein A, et al. Estimating relative CWD susceptibility and disease progression in farmed white-tailed deer with rare PRNP alleles. Plos One. 2019;14(12):e0224342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Johnson CJ, Aiken JM, McKenzie D, et al. Highly efficient amplification of chronic wasting disease agent by protein misfolding cyclic amplification with beads (PMCAb). PloS One. 2012;7(4):e35383–e35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Griffiths PC, Spiropoulos J, Lockey R, et al. Characterization of atypical scrapie cases from Great Britain in transgenic ovine PrP mice. J Gen Virol. 2010;91(8):2132–2138. [DOI] [PubMed] [Google Scholar]
- [51].Götte DR, Benestad SL, Laude H, et al. Atypical scrapie isolates involve a uniform prion species with a complex molecular signature. Plos One. 2011;6(11):e27510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miller MW, Wild MA, Williams ES. Epidemiology of chronic wasting disease in captive Rocky Mountain elk. J Wildl Dis. 1998; 34(3): 532–538. [DOI] [PubMed] [Google Scholar]
- [53].Henderson DM, Denkers ND, Hoover CE, et al. Progression of chronic wasting disease in white-tailed deer analyzed by serial biopsy RT-QuIC and immunohistochemistry. Plos One. 2020;15(2):e0228327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Johnson CJ, Herbst A, Duque-Velasquez C, et al. Prion protein polymorphisms affect chronic wasting disease progression. Plos One. 2011. 18;6(3):Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Robinson SJ, Samuel MD, Johnson CJ, et al. Emerging prion disease drives host selection in a wildlife population. Ecol Appl. 2012;22(3):1050–1059. [DOI] [PubMed] [Google Scholar]
- [56].Johnson CJ, Phillips KE, Schramm PT, et al. Prions adhere to soil minerals and remain infectious. PLoS Pathog. 2006. April;2(4):e32–e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kuznetsova A, McKenzie D, Banser P, et al. Potential role of soil properties in the spread of CWD in western Canada. Prion. 2014. January 1;8(1):92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nichols TA, Pulford B, Wyckoff AC, et al. Detection of protease-resistant cervid prion protein in water from a CWD-endemic area. Prion. 2009. Jul-Sep;3(3):171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lavelle MJ, Phillips GE, Fischer JW, et al. Mineral licks: motivational factors for visitation and accompanying disease risk at communal use sites of elk and deer [journal article]. Environ Geochem Health. 2014. December 01;36(6):1049–1061. [DOI] [PubMed] [Google Scholar]
- [60].Kõressaar T, Lepamets M, Kaplinski L, et al. Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics. 2018;34(11):1937–1938. [DOI] [PubMed] [Google Scholar]
- [61].Hanke M, Wink M. Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primer and dNTPs. BioTechniques. 1994;17(5):858. [PubMed] [Google Scholar]
- [62].Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003. November 01;73(5):1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- [64].Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].R Core Team . R: a language and environment for statistical computing. 3.0.0. R Foundation for Statistical Computing; 2016.
- [66].Team R RStudio: integrated development environment for R 1.1.423. 2016.
- [67].Wisconsin Department of Natural Resources . CWD deer testing results by county Madison, Wisconsin: Wisconsin Department of Natural Resources; 2019. [cited 2019 October29]. Available from: https://dnr.wi.gov/wmcwd/Summary/County [Google Scholar]
- [68].Coppock A Multiple comparison calculator 2019 [cited 2019 October30]. Available from: https://alexandercoppock.com/statistical_comparisons.html
- [69].Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4 [sparse matrix methods; linear mixed models; penalized least squares; Cholesky decomposition]. 2015. October 7;67(1):48. [Google Scholar]
- [70].Meyer D, Zeileis A, Hornik K. The strucplot framework: visualizing multi-way contingency tables with vcd. J Stat Softw. 2006;17(3):1–48. [Google Scholar]
- [71].Meyer D, Zeileis A, Hornik K. vcd: visualizing categorical data. R package version 1.4-7. 2020
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


