Abstract
Current preclinical studies in drug development utilize high-throughput in vitro screens to identify drug leads, followed by both in vitro and in vivo models to predict lead candidates' pharmacokinetic and pharmacodynamic properties. The goal of these studies is to reduce the number of lead drug candidates down to the most likely to succeed in later human clinical trials. However, only 1 in 10 drug candidates that emerge from preclinical studies will succeed and become an approved therapeutic. Lack of efficacy or undetected toxicity represents roughly 75% of the causes for these failures, despite these parameters being the primary exclusion criteria in preclinical studies. Recently, advances in both biology and engineering have created new tools for constructing new preclinical models. These models can complement those used in current preclinical studies by helping to create more realistic representations of human tissues in vitro and in vivo. In this review, we describe current preclinical models to identify their value and limitations and then discuss select areas of research where improvements in preclinical models are particularly needed to advance drug development. Following this, we discuss design considerations for constructing preclinical models and then highlight recent advances in these efforts. Taken together, we aim to review the advances as of 2020 surrounding the prospect of biological and engineering tools for adding enhanced biological relevance to preclinical studies to aid in the challenges of failed drug candidates and the burden this poses on the drug development enterprise and thus healthcare.
Keywords: Bioengineering, Drug development, Preclinical studies, Disease modeling, Microphysiological systems, Stem cell engineering, Tissue engineering
Abbreviations: HCT, human clinical trials; 2D, two-dimensional; 3D, three-dimensional; HTS, high-throughput screen; PK, drug pharmacokinetics; PD, drug pharmacodynamics; DMPK, drug metabolism and pharmacokinetics studies; ADME, absorption, distribution, metabolism, and excretion studies; PKPD, Pharmacokinetic-pharmacodynamic studies; GEM, genetically engineered mouse; FDA, United States Food and Drug Administration; NTD, neural tube closure defect; BBB, blood-brain barrier; hPSC, human pluripotent stem cell; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; ECM, extracellular matrix; MPS, microphysiological system; NIH, United States National Institutes of Health; DARPA, United States Defense Advanced Research Projects Agency
Graphical abstract
1. Introduction
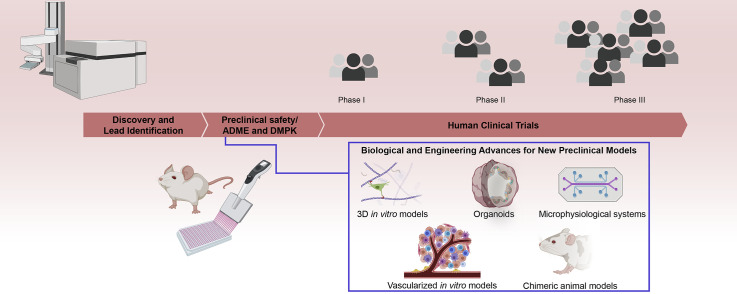
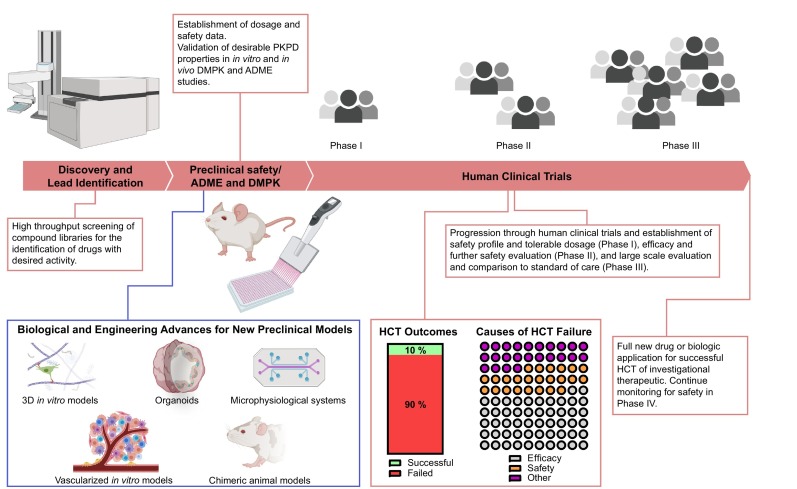
Drug development is a long and expensive process that follows a series of sequential steps (Fig. 1 ) [[1], [2], [3]]. Initially, researchers screen candidate pharmacological agents for desired activity and specificity and then establish potential ranges of safe and therapeutic dosages in preparation for human clinical trials (HCT) [1,3,4]. Upon regulatory authorization, an investigational new drug is tested in additional sequential stages of HCTs to validate and establish drug efficacy and safety in preparation for a new drug or biologics license application [3]. At the preclinical stage, researchers use in vitro cell culture models primarily for the initial characterization of a drug candidate's activity and specificity [1,4] and rely on in vivo non-human animal models to confirm the efficacy and establish a therapeutic index, or the range between the median effective and toxic doses, in preparation for HCTs (Fig. 1) [2,4]. While in vitro and non-human in vivo preclinical models have long been an integral component in the history of drug development [4], they frequently fail to accurately predict a drug candidate's performance in subsequent HCTs in terms of efficacy, specificity, toxicity, or a combination of all the above (Fig. 1) [5]. Currently, the drug development enterprise is facing increasing challenges in the successful production of new therapeutics [6]. These challenges are due, in part, to the increasing costs associated with these drug candidates that passed preclinical studies but failed in subsequent HCTs [6,7]. As a result, there is a critical need to make advancements in the models available for preclinical studies.
Fig. 1.
Drug development: current pipeline, challenges, and areas for opportunity. The current drug development pipeline is comprised of the preclinical and clinical stages. At the preclinical stage, drug discovery and preclinical safety, ADME, and DMPK studies take place to identify and validate drug candidates for human clinical trials (HCTs). The lack of efficacy or concerns of safety represent 75% of all failed HCTs, and with fewer than 1 in 10 drug candidates that emerge from preclinical studies succeeding in a new drug or biologics application, there is a need for new models for preclinical studies. Recent advances in biology and engineering, such as new 3D culture strategies, generation of and access to disease-relevant cell types, methods for creating perfusable and vascular tissues and tissue systems in vitro, and methods for improving in vivo models of human tissues, offer numerous opportunities for facilitating improvements in the preclinical stage of drug development. These advancements can particularly augment the validation stage of preclinical studies after the identification of lead candidates in discovery.
1.1. Specific rationale for new preclinical models
Approximately 75% of drugs that emerge from preclinical studies go on to fail in phase II or phase III HCTs due to lack of efficacy or safety (Fig. 1) [5,8]. The reliance on non-human animal models in preclinical studies is a significant contributor to this failure. There are fundamental biological differences between small animals, such as mice, and humans, and this frequently causes a failure to predict a potential drug's efficacy and toxicity [7,9]. Example differences between humans and small animals that impact drug development include the structure, size, and regenerative capacity of organs and tissues, as well as physiological differences in metabolism, immunology, and drug transport [7,10]. Large animal models, such as pigs, dogs, and non-human primates, can improve the predictive value of preclinical models by introducing anatomies and physiologies that are more similar to humans [[11], [12], [13]]. However, large animal models introduce a significant burden of cost, time, and increased ethical considerations. Furthermore, even with the improved predictive power of large animals, molecular, genetic, cellular, anatomical, and physiological differences persist [7,9]. As a result, there is a significant demand for preclinical models based on human tissues.
In vitro, cell-based assays are critical tools in drug development, as they offer simple-to-assemble experimental platforms with clear and precise outputs and easy incorporation of human cells and tissues [14,15]. Also, they are less costly and faster to perform than in vivo studies [16,17]. Currently, researchers regularly utilize two-dimensional (2D) in vitro cultures of established human cell lines in high-throughput screens (HTS) of large drug libraries for potential efficacy in treating monogenic and cell-autonomous diseases [4,18]. Monogenic forms of muscular dystrophy [19], spinal muscular atrophy [20], and hereditable forms of neurodegenerative disorders [21] are representative diseases where 2D HTS drug discovery efforts are common. These screens are also useful for human cancers that result from well-characterized and common genetic mutations [22,23]. However, these in vitro assays are overly simplistic and often lack much relevance to in vivo human biology [24]. Indeed, fewer than 1 in 10 drug leads emerging from in vitro preclinical studies results in a successful clinical trial (Fig. 1) [25]. As a result, there is a significant demand for a new generation of preclinical models that bridge the gap between the in vivo relevance of non-human preclinical animal models and in vitro models comprised of human tissues.
1.2. Scope of this review
Recent biological and engineering advances have greatly expanded the tools available to researchers for designing new and improved models of human tissues for preclinical studies. These advances include improvements in the in vitro isolation, derivation, culture, characterization, and ultimately utilization of primary- and stem cell-derived human cells, as well as the development of new biomaterials, microfabrication techniques, and tissue engineering approaches to create vascularized three-dimensional (3D) in vitro tissues. By combining these biological and engineering advances, researchers are now better able to create new preclinical models that recapitulate the structural organization and integrated dynamics of human tissues and organ systems in vitro. In addition, advances in biological and genetic tools, combined with increased access to disease-relevant cell types, can improve in vivo preclinical models. The proposition for integrating these new models into preclinical studies is that bridging the gap between HTS discovery and in vitro/in vivo validation will improve the predictive value of preclinical studies as well as provide relevant insight into disease mechanisms. Additionally, the development of new preclinical models can potentially provide entirely new platforms for drug discovery, validation, and the study of human diseases for which there are no current suitable models. However, the advances described here are best considered as an augmentation rather than a replacement for existing preclinical models. In this review, we will discuss the current values, limitations, and missed opportunities in drug development incurred by current preclinical models as well as select research areas where improvements might afford much-needed advances in drug development. Following this, we will discuss the design considerations for implementing new biological and engineering tools into novel preclinical models, and then highlight recent strategies to create in vitro and in vivo tissues that better reflect the structures and physiology of human tissues and organ systems. Taken together, we aim to review and discuss how new preclinical models comprised of more sophisticated in vitro and in vivo representations of human tissues and tissue systems can be developed to augment existing preclinical studies and advance drug development.
2. Current preclinical models: uses, limitations, and missed opportunities
Traditionally, preclinical models have prioritized simple, high-throughput, parallelizable in vitro assays and small animal in vivo models. Choosing these approaches over complex models is due to the need to screen millions of potential compounds and the lack of ability to feasibly recreate sophisticated features of human tissues in vitro at sufficient scale [24]. As a result, findings from these models are often of limited relevance to human biology and thus expected human in vivo outcomes [10,[26], [27], [28], [29]]. Compounding the limitations of oversimplicity are abnormal genetic, epigenetic, and cellular phenotypes of the human cell lines frequently used in in vitro models [30]. Furthermore, there is a need for advances in in vitro or in vivo models for non-cell-autonomous processes, such as failed regeneration after injury, sporadic neurodegenerative disorders, and spontaneous conversions from benign to metastatic phenotypes of cancers [31].
2.1. Value and limitations of current preclinical models
The ability of a drug to produce the intended therapeutic effect in the absence of adverse events is heavily dependent on drug pharmacokinetics (PK) and pharmacodynamics (PD) [4]. As a result, drug metabolism and pharmacokinetics (DMPK) studies are an integral component of preclinical studies [18]. Following computational prediction of physicochemical properties, DMPK studies use HTS to assist lead drug identification through validation and optimization [18,32,33]. This process increases the predictive capacity of in vitro preclinical studies to identify lead drugs with desirable PK properties [33,34]. Additionally, these studies test drug metabolism parameters in the effort to detect potential adverse events, such as drug-drug interactions [18]. After these studies, researchers test promising leads for their absorption, distribution, metabolism, and excretion (ADME) properties in both in vitro and in vivo preclinical models [34]. Understanding ADME is critical for the next stage of using a pharmacokinetic-pharmacodynamic (PKPD) relationship to establish a dosing regimen in animal studies. Overall, lead selection based on desirable drug properties in DMPK and ADME studies has improved the success of predicting drug safety in later HCTs [18].
Despite the ability of DMPK and ADME to improve the predictive value of preclinical studies, lack of efficacy and the emergence of previously undetected toxicity represents a large fraction of failed HCTs (50% and 25%, respectively) [5,8]. This high failure rate illustrates that the predictive value of human outcomes in drug performance is still a major limitation of current models used for preclinical studies [35]. Failure to adequately predict drug efficacy in preclinical studies is due to a combination of the use of simplistic 2D in vitro models for drug lead identification and subsequent validation in non-human animal models [36]. To overcome this limitation, new preclinical in vitro and in vivo models comprised of human tissues that better represent human biology are needed. Improved preclinical models comprised of human tissues may also aid identification of otherwise undetected, untoward effects and prevent the pursuit of drug leads that ultimately will fail for toxicity [37], as animal models are only of limited value for predicting efficacy or the likelihood of adverse events in humans [38]. This limitation is particularly true for metabolic, blood and lymphatic, and renal disorders [38].
2.2. Understanding of disease mechanisms and drug discovery
A poor understanding of underlying disease mechanisms can also contribute to drug candidate failure [39,40]. For example, gaps in knowledge of which cells within a tissue are the initiator of a disease, the cellular and tissue progression of the disease, and whether a disease is cell-autonomous or non-autonomous could all result in the development and use of preclinical models that poorly recapitulate the true nature of a human disease. Specifically, these gaps in knowledge of disease etiology may increase the likelihood of preclinical models selecting lead drugs that are unlikely to provide a meaningful therapeutic benefit. For example, an in vitro model that focuses on a singular and specific target of a disease, absent any other characteristics of the tissue or disease, may ultimately be prioritizing lead drug identification for a short-lived or non-druggable target in vivo. The outcome of such a preclinical model would be lead candidate drugs that are ineffective in vivo, despite showing efficacy and specificity in vitro. As a result, limited understanding of disease mechanisms inherently contributes to preclinical models' poor predictive value for drug efficacy.
There are also missed opportunities in drug development incurred by simplistic in vitro or inappropriate in vivo animal models. As a straightforward example of this, some drugs within given dosing regimens are found to be ineffective or toxic in animal models but would be effective and safe in humans. Aspirin is illustrative of this, as it is toxic in several animal models at doses well-tolerated and effective in humans [41,42]. Additionally, simplistic preclinical models that insufficiently capture human disease mechanisms can hinder more complex forms of drug discovery. For example, the discovery of new tumor antigens, as well as inhibitors of tumor metastases, are both of significant interest in drug development for cancer immunotherapies [43,44]. In addition, combinations of both strategies may be particularly effective in new cancer treatments [43,45]. Unfortunately, modeling cancer metastasis in animal models either requires the use of animal tumors or human tumor xenografts in immunocompromised animals, reducing the ability to examine combination therapy approaches [46]. A new in vitro model that can recreate a human neoplasm within a complete tumor microenvironment containing human immune cells would allow screening for such combination therapies.
3. Specific preclinical model limitations in different research areas
Having described the general value, limitations, and missed opportunities with current preclinical models, here we will discuss specific, representative research areas where new models of human tissues may offer a benefit for advancing drug development. Within these distinct areas, we will discuss current preclinical model features and key opportunities that new preclinical models of human tissues might afford. Both in vitro and in vivo models may be relevant in these distinct research areas, with varying relevance. These examples are not meant to be exhaustive, but to provide a sense of the range of potential approaches.
3.1. Modeling of human cancers
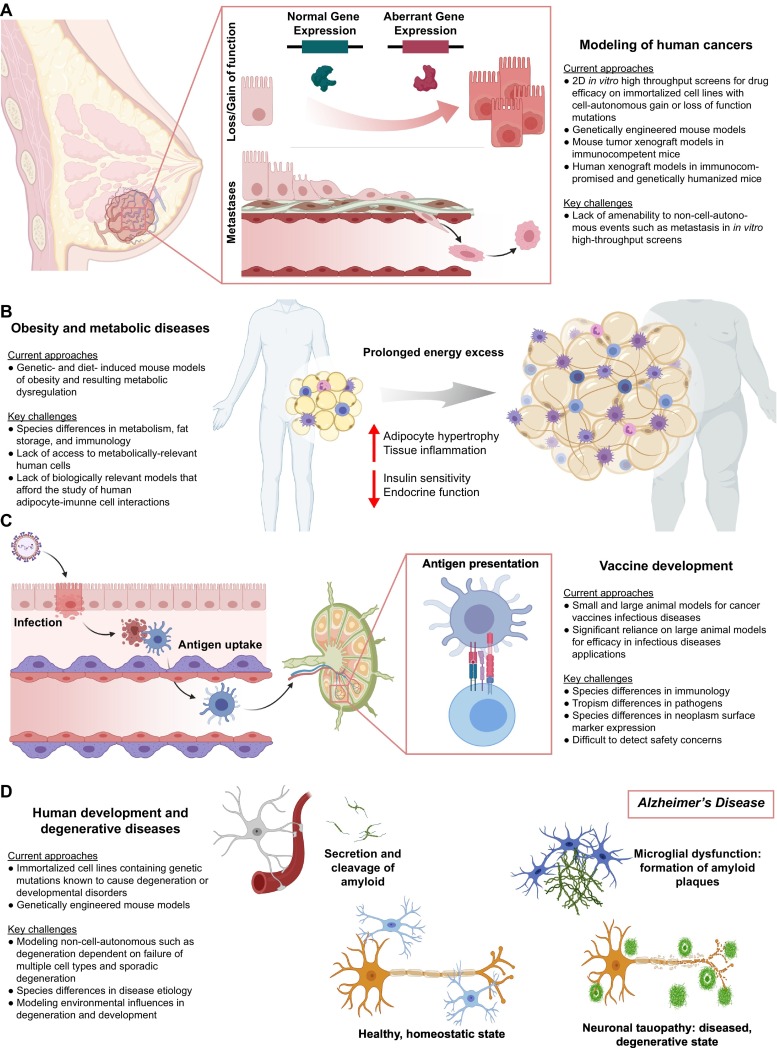
The value of preclinical cancer models depends on the ability to recreate an accurate representation of a human malignancy. Key aspects of this representation include the genetic basis for the malignancy, the capacity and propensity for metastases, and increasingly the interactions of the malignancy with the immune system [[47], [48], [49]]. Additionally, the model should ideally be anatomically and physiologically similar to the human neoplasm and progress through in vivo-like stages of development [48,49]. In vitro, immortalized cell lines with specific genetic perturbations facilitate HTS systems for the identification of drug leads that modulate cell-autonomous neoplasm outcomes, such as dysregulated proliferation and response to pathway inhibition (Fig. 2A) [50]. Genetically engineered mouse (GEM) models are an emerging and promising example of in vivo preclinical models for cancer [51]. In GEMs, targeted key genetic perturbations result in a reproducible malignancy in an immunocompetent animal. The enhanced feasibility for creating these genetic perturbations with new technologies such as CRISPR/Cas9 is significantly increasing the variety and availability of GEMs [52].
Fig. 2.
Representative research areas where new models of human tissues may offer a benefit for advancing drug development: current models and limitations. (a) Breast cancer as a representation of cancer. Key model features should include cell-autonomous events such as the genetic gain or loss of function that disrupts the cell cycle, as well as non-cell-autonomous events such as tumor site initiation and metastasis. Human cancer-immune system interactions are additional desirable features not readily available in many current preclinical cancer models. (b) Obesity and metabolic diseases are the product of prolonged energy excess and result in an expansion of inflamed adipose tissue. Current models consist of genetically- and diet-induced systems in small animals. However, the species differences in immunology, metabolism, eating behaviors, and interactions between these systems are substantial limitations in the current models. The use of metabolically relevant human cells and allowing for adipocyte-immune cell interactions can aid in the drug development efforts for these diseases. (c) Establishing humoral immunity involves a complex cascade of antigen-presenting cell activation, antigen uptake and processing, cellular trafficking to lymph nodes, and antigen presentation. Current models recreate this process primarily in small and large animal models, with a very high reliance on large animals in infectious disease vaccine development. Immunology differences between humans and animal models with respect to this cascade, as well as restrictive tropisms of pathogens and neoplasm surface marker expression, significantly hinder vaccine development. Models capable of recreating humoral immunity establishment with human cells are highly desirable. (d) Human development and degenerative diseases are particularly difficult to model for drug development due to lack of access to disease-relevant cell types as well as the non-cell-autonomous nature of many degenerative diseases such as sporadic Alzheimer's Disease. Challenges in modeling developmental and degenerative diseases include species differences in these processes and recreating the environmental conditions that drive these disorders in disease-relevant human cells. Models utilizing disease-relevant human cells and culture systems that allow for the recreation of developing tissues can help to overcome these challenges.
Despite the advantages of current all-murine preclinical cancer models, the lack of a human biology component ultimately means that these models provide information that is more relevant to mouse biology. To add the human biology component in mouse studies, preclinical cancer models traditionally include human tissue xenografts [46]. In this approach, transplantation of human tumor cells into an immunocompromised mouse allows researchers to study the formation, progression, and potential tumor metastases, as well as test the efficacy of potential drug candidates to inhibit any of these stages (Fig. 2A) [53]. Additionally, xenograft approaches allow for the capture of genetic and epigenetic complexity of human malignancies [54,55]. However, the recent appreciation for the role of the immune system in the development of human cancers and as a potential target for therapeutics is bringing increased attention to the limitations of using immunocompromised mice [54,55]. Refining existing 3D culture models and improving the representation of human tissues in in vivo models can improve the predictive capacity of cancer therapeutic efficacy in preclinical studies.
3.2. Obesity and metabolic diseases
Rising obesity is a significant challenge in global health, as the resulting metabolic dysfunction leads to numerous comorbidities, such as type-2 diabetes, dyslipidemia, non-alcoholic fatty liver disease, hypertension, stroke, coronary artery disease, and several gastrointestinal cancers [56,57]. Type-2 diabetes and the associated comorbidities alone are the 7th leading cause of mortality in the United States and contribute to $113 billion in medical costs annually [58]. Central to the pathological state of obesity (Fig. 2B), a positive energy balance from excess nutrients results in a shift from adipogenesis to adipocyte hypertrophy to accommodate increased lipid storage [59]. This persistent hypertrophic state coincides with tissue inflammation, loss of adipocyte insulin sensitivity, and adipocyte endocrine dysfunction via currently unknown mechanisms [59]. This dysfunctional adipose tissue results in changes in adipokine and free fatty acid secretions that are thought to lead to insulin resistance in distal tissues, such as skeletal muscle and liver, as well as impairment of β-cell function [60].
Current models for studying obesity and its comorbidities include diet- and genetically-induced obesity and insulin resistance in mice [61]. While these models provide a reproducible system with complete physiology to study metabolism in the context of obesity- and type 2 diabetes-like phenotypes, non-human mammals, including rodents, fail to recapitulate several dynamics of human physiology and metabolism [[61], [62], [63]]. In a specific example of the disparity between humans and mice, omental and mesenteric adipose tissue depots are the largest visceral fat deposits in the human body, while in contrast, these are relatively small tissues in mice compared to epididymal fat depots [64]. As a consequence, epididymal fat pads are the most frequently studied tissue in mice but are only visceral-like in comparison to omental or mesenteric fat. This is particularly concerning, as obesity-related metabolic disorders in humans are most highly correlated with hypertrophic visceral adipose tissue [65,66]. Given the disparity in adipose tissue distribution and metabolism across species and the central role of adipose tissue dysregulation and pathological obesity in humans, it is critical to develop models that allow us to study human adipocyte behavior in physiologically relevant environments.
3.3. Vaccine development
There is a need for improving the vaccine development pipeline [67], particularly in light of the COVID-19 pandemic [68]. Unfortunately, lack of suitable in vitro models combined with the substantial differences between human and mouse immunology hinders this endeavor [10,67]. These limitations necessitate the use of non-human primate models to develop vaccines with the potential to establish humoral immunity [69]. However, challenges with pathogen tropism and species differences in receptors that regulate immune response persist even in large animals, creating a very lengthy vaccine development pipeline with high failure rates [67]. Generally, vaccine performance is dependent on the ability to elicit a specific immune response that is distinct from tolerance [70,71]. Antigen selection is key to promote a specific immune response, while the magnitude and robustness of the initial response is impacted by adjuvants, with both being key to durable prophylaxis [72,73]. This is true both of the vaccines against infectious agents and emerging cancer vaccines [70,71].
There are also unique safety concerns in vaccine development, as certain vaccines established as effective in non-human animal models have resulted in worsening the outcomes of vaccinated patients that were later exposed to the pathogen [74,75]. For example, children vaccinated with an inactivated respiratory syntactical virus, shown to be safe and efficacious in rodents and non-human primates, later suffered from enhanced respiratory syntactical virus disease when exposed to the actual virus [74,75]. Recent findings suggest that in vitro co-culture models of human immune cells can be potentially predictive of vaccine safety for respiratory syntactical virus vaccine candidates [76]. However, simple co-cultures do not recapitulate the spatio-temporal cascade of cellular and molecular events in antigen processing. Vaccine component interactions within antigen processing events where resident antigen-presenting cells, particularly dendritic cells, take up and present antigens to naïve T cells within nearby lymph nodes (Fig. 2C) can impact both vaccine efficacy and safety [73]. The limitations of current models for recreating the complexities of this cascade with human cells, combined with increased applications beyond infectious diseases, such as cancer vaccines, motivate the creation of new models for vaccine development.
3.4. Human development and degenerative diseases
As of 2016, developmental disorders affect 6.99% of all live births in the United States, representing an increasing historical trend [77]. Beyond the often devastating human impact, birth defects cost the US healthcare system $22.9 billion annually as of 2013 [78]. Globally, six select developmental disorders affected 52.9 million births in 2016 [79]. While there are many potential causes of developmental disorders, the identification of teratogenic effects of pharmacological compounds is an important component of birth defect prevention [80]. Drugs with unknown human fetal effects are classified as Class B and C compounds by the US Food and Drug Administration (FDA) [81]. As of 2006, 78% of pregnant women were exposed to a Class B or C compound during gestation [82]. Thalidomide, an anti-nausea medication originally prescribed to pregnant women for morning sickness, is a classic case study of the potential harm of drugs with unknown fetal interactions. Thalidomide was recalled after this compound was established to result in limb deformations in the offspring of exposed mothers [83]. However, this recall occurred after four years of epidemiological analysis and an estimated negative effect on 10,000 embryos [83]. In addition to pharmacologic compounds, pregnant women are also regularly exposed to potentially teratogenic industrial and agricultural substances, most of which have no toxicological assessment on human development [80,84]. As an example, only 6.7% of chemical additives permitted in food have human reproductive toxicology data in the FDA database [84]. The ethical limits concerning randomized clinical trials on pregnant women and a lack of suitable models of human development make detecting developmental and reproductive toxic effects of drug and drug-related compounds a difficult challenge to address.
In addition to a need for improved models for fetal drug toxicology, there is a need for systems that model human developmental disorders for therapeutic drug development. The specific utility of such models is that they would allow the study of the underlying genetic, environmental, or combined mechanisms for pathology as an efficacy screening platform for compounds or methods of correction for disorders of developing human tissues. Prenatal nutraceuticals represent one example of such opportunities in drug development. As an illustrative example, neural tube closure defects (NTDs) affect approximately 300,000 live births globally and result in 88,000 deaths each year [85,86]. Fortunately, folic acid fortification of our food supply has transformed the rate of NTDs, and this current number, while still large, is at a historical low [85,86]. However, it took decades of observational human population studies to support and propose a clinical trial to confirm the therapeutic benefit of folic acid supplementation [87], and there are likely many more nutrient or environmental factors that influence NTDs and other developmental disorders. Overall, there is a need for new preclinical models that capture the underlying biological processes of human development [88]. In addition to the screening for teratogenic effects of pharmacologic compounds, advancements in models of human development are needed that can provide a discovery platform for identifying potential drugs or drug-like compounds that can potentially mitigate developmental disorders. The drug development opportunity of such models is that increasing confidence in extrapolating in vitro toxicological and therapeutic preclinical testing to clinical outcomes can potentially reduce the costly burden of teratogenic effects and developmental disorders [89].
In addition to disorders affecting the development of human tissues, human tissue degeneration, often associated with advanced age, can benefit from improvements in preclinical models [90]. This is particularly true for neurodegenerative diseases such as Alzheimer's, Multiple Sclerosis, and Parkinson's Disease. Alzheimer's Disease alone affects 50 million people worldwide and results in $800 billion in annual healthcare costs [91]. Current challenges with modeling human tissue degeneration include accessing disease-relevant cell types, the ability to introduce both genetic and environmental perturbations that lead to tissue degeneration, and modeling of sporadic forms of degenerative diseases. For neurodegenerative diseases, additional unique drug transport issues arise from the blood-brain barrier (BBB) [92,93]. Degenerative diseases are often highly complex and non-cell-autonomous (Fig. 2D), additional elements that result in a decrease in the likelihood that simple single-cell 2D assays will be able to recapitulate the disease phenotypes in a meaningful way for drug development. The non-cell-autonomous nature also increases the likelihood that animal models will be poor representations of the disease, as the pathologic degeneration is dependent on the failure of multiple cell types all in a manner that is similar to human biology. For example, mouse models of Alzheimer's Disease readily form amyloid plaques, a hypothesized combined failure of brain endothelium and microglia [94,95], but fail to progress to the neuronal tauopathies observed in human patients [96]. Putative new models for degenerative diseases require access to disease-relevant human cell types and the ability to study them in the complex multicellular environments in which these diseases occur.
4. Design considerations for new in vitro preclinical models
Recently, advances in biology and engineering are affording new tools that provide increasingly accurate recreations of human tissues in vitro and in vivo. As these advances emerge, there are several design considerations that are important to acknowledge, so that researchers' implementation of these tools can best increase the relevance of new preclinical models to human biology and physiology. These considerations include the source of the biological materials, implementation of 3D culture models, degree of in vivo complexity captured, the introduction of dynamics and transport phenomena that influence drug PKPD, and balancing complexity with throughput and feasibility.
4.1. Considering the starting cellular material
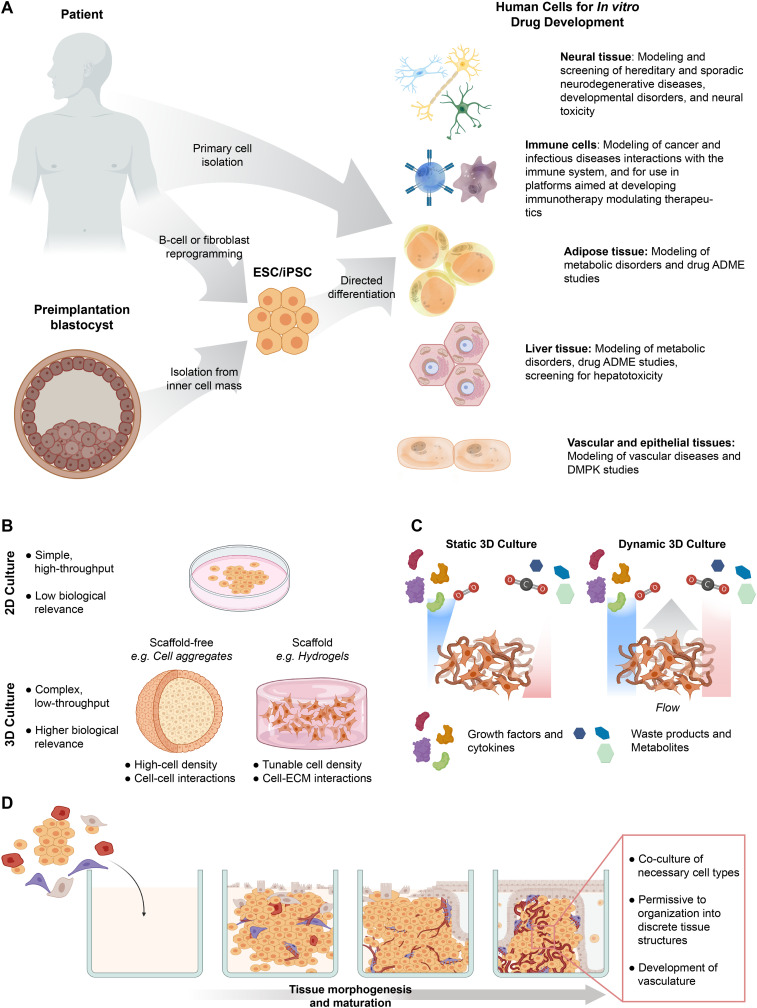
The source of human cellular material is an early and critical design consideration for a preclinical model. Broadly, the potential sources available are immortalized human cell lines, primary human cells, or an increasingly common option over the past decade, adult or human pluripotent stem cell (hPSC)-derived cells [97,98]. This early consideration is important, as the cell source often predetermines which questions concerning drug efficacy or toxicity researchers can ask with a given model. As an example, in determining the efficacy of a drug targeting a specific cell-autonomous disease, the source cell material must either contain or be amenable to the introduction of the required genetic aberration. For drug toxicity, the sourced cells must have and maintain a phenotype that is relevant to the potentially sensitive tissues. In essence, the key biological phenomena under investigation for a particular step in drug development should inform the choice of source for the cellular material.
Immortalized cells have genetically disrupted key cell-cycle checkpoint pathways and therefore permit cells to divide in culture longer than primary cells [99]. These cells are extensively used in drug development as they are relatively easy to culture and can divide indefinitely in vitro, providing a reliable and consistent cell source [97,99]. Also, these cells are often relatively easy to genetically manipulate, and there is a robust body of data with which to compare new studies [16,100]. However, there are several inherent limitations associated with these cells. For example, there are well-documented changes in cellular phenotype and epigenetics that come with immortalization [30,99], and long-term culture of these immortalized cell lines results in frequent and extensive karyotypic abnormalities [101]. Additionally, cell lines immortalized from a single source intrinsically introduce genetic homogeneity, a potential problem given that variance across populations is one factor missing from many current preclinical models [102,103]. Lastly, the contamination of one immortalized cell line with cells from other sources is a significant concern [104].
In contrast to immortalized cells, primary cells are those taken directly from human patients or fetal tissues that are not immortalized by transgenic manipulation or prolonged culture (Fig. 3A) [105]. Primary human cell types with potential use for preclinical studies include hepatocytes [106], primary endothelial cells [107], adipocytes [108], and various cells of the immune system [109,110]. A major advantage of using these cells for preclinical models, relative to immortalized lines, is their biological relevance concerning healthy and diseased phenotypes [105]. Challenges associated with the use of primary cells include lack of standardization of sourced cellular material due to patient-to-patient variability, limited availability of many specific cell types, and cost [98]. Further compounding these limitations, primary cells often have limited proliferative capacity and quickly lose their mature phenotype or expression of key receptors in in vitro culture [111,112]. These combined limitations make scaling of a single, standardized cell source for industry particularly challenging.
Fig. 3.
Design considerations for developing in vitro preclinical models. (a) More biologically relevant sources of primary human cells for drug development are available directly from patients or via differentiation from embryonic or induced pluripotent stem cells (ESC/iPSC). Human cells have relevant utility in numerous facets of drug development. (b) Transitioning from 2D to 3D culture can increase the biological relevance of in vitro models, but often decreases throughput. There are numerous strategies for creating 3D models, which can be scaffold or scaffold-free. (c) 3D in vitro models introduce complexities related to transport, such as poor diffusion of oxygen and growth factors into the tissue, as well as carbon dioxide, metabolites, and waste products out of the tissues. The introduction of flow can aid in overcoming this complexity and create dynamic cultures with transport that are more biologically relevant for drug development models. (d) In vitro morphogenesis can better mimic the architecture of native tissues in models for drug development. Engineering strategies to assemble cells into 3D tissues should be permissive to and, if possible, promote this morphogenesis to allow for mature tissue features to develop and emerge.
As an alternative, adult stem cells and pluripotent stem cells can afford the convenience of a highly expandable and standardized cell source without the need for viral immortalization. Researchers can readily derive adult stem cells from several human tissues [[113], [114], [115]] and pluripotent embryonic stem cells (hESC) from the inner cell mass of a fertilized blastocyst [116]. Additionally, overexpression of transcription factors Oct4, Klf4, Sox-2, c-Myc, NANOG, and Lin28 in adult somatic cells can provide induced pluripotent stem cells (hiPSC) [[117], [118], [119]]. This cellular reprogramming approach can afford a combination of isogenic hPSC- and primary cell-derived tissues [98]. With their ability to generate a nearly inexhaustible source of human cells and tissues, both adult and pluripotent stem cells have dramatically enhanced the range of cell types available to researchers to develop preclinical models for drug development (Fig. 2) [98]. However, a significant limitation is that current approaches for deriving specific cells and tissue from stem cells often result in tissues of an immature or fetal-like phenotype, which are of potentially limited value for preclinical studies of diseases that affect adults [120,121].
4.2. Complexities introduced by three-dimensionality of culture models
Native tissues are complex 3D environments with a hierarchical structural organization [[122], [123], [124]]. In vivo, the spatial arrangement of cells within tissues and the interactions of different groupings of cells within organized patterns of extracellular matrix (ECM) all communicate together in 3D to orchestrate the normal function of a tissue [125]. The lack of this organization and communication in simple 2D in vitro cultures likely contributes to the poor predictive value of in vivo outcomes by current preclinical studies. In appreciation of this, researchers have developed a wide variety of in vitro 3D culture strategies and technologies to better mimic the cell-cell and cell-ECM interactions, gradients of oxygen and nutrients, and the mechanical signaling parameters of in vivo tissues [1]. These recent advances have made 3D cultures more sophisticated, and their more common usage can improve the in vitro representations of human tissues for preclinical studies [126,127]. However, numerous design considerations arise when transitioning to 3D culture models.
3D culture strategies emerged early in the 20th century, so there are now many strategies and tools used for the implementation of three-dimensionality in cell culture [127]. Example strategies include scaffold-free approaches, such as cell aggregation into spheroids [128], scaffold approaches via encapsulation of cells in synthetic or natural ECM hydrogels (Fig. 3B) [129], combinations of these two approaches, and use of specialized micro manufactured devices that allow for placement of cells in precise 3D configurations. Given the range of tools, the particular tissue or organ system under study should motivate which approach researchers take for model design. Scaffold-free based systems may be appropriate where cell-cell or cell density interactions are of most importance to disease outcomes [122,128], while scaffold-based systems can model dynamics where mechanical cues and cell-ECM interactions are the primary parameters of interest [130,131]. Furthermore, specialized transwell or micro manufactured devices may be required for studies where barrier properties, drug transport, and drug partitioning are the most relevant factors [132].
In addition to choosing the appropriate 3D model, three-dimensionality introduces new variables that researchers should consider. These variables include porosity and cell density of the 3D construct, adhesion ligand identity and density, degradation, and remodeling capacity of the 3D microenvironment [130,133]. Often these parameters can fundamentally regulate how cells respond to signals in 3D. Furthermore, researchers should consider that principles established in 2D cultures may not directly translate to 3D. To illustrate this, it has been demonstrated that mechanical cues from a culture substrate can regulate cell phenotype and fate in 2D [134], and studies in 3D cultures later contextualized these substrate mechanical cues [135,136]. For example, the ability for a cell to deform the surrounding ECM in 3D culture was found to regulate the dependency of cell fate on substrate stiffness [136]. Lastly, three-dimensionality and the method for introducing it can intrinsically influence cell phenotypes. For example, intrinsic factors such as the rate of aggregation and centrifugation have been found to influence gene expression and bias cell fate decisions made by hPSC aggregates [137].
The source of biological materials can return as a consideration in the transition to many 3D models. For scaffold-based strategies, materials choices include natural, synthetic, or hybrid sources [133]. Natural ECM scaffolds offer the advantage of innate biological compatibility without the need for significant modification [138]. However, they are less tailorable for specific properties than their synthetic counterparts, often have a high degree of batch-to-batch variation due to their biological origin, and can contain poorly defined sets of biological molecules that can introduce latent variability into experimentation [138,139]. In contrast to natural ECMs, synthetic ECMs usually require chemical functionalization for biological compatibility, increasing complexity and potentially cost. However, synthetic ECMs also afford researchers precise tunability of important ECM properties, such as ECM stiffness, porosity, cell-adhesion ligand identity and density, as well as degradability and how the material responds to cells over time [140]. Synthetic ECMs comprised of hydrophilic polyol polymers that confer a bioinert backbone combined with chemical moieties for specific modification are frequently utilized [141]. The choice of ECM origin and composition will influence how microenvironmental parameters influence a 3D in vitro preclinical model.
4.3. Tissue engineering and considering the degree of morphogenesis
Tissue engineering has emerged over the past 30 years as a discipline focused on the creation of de novo biological structures that resemble human tissues for a range of applications [142,143]. These applications include the translation of engineered tissue for therapeutic purposes, the study of the fundamental biology of tissue function and development, and also a potential new platform for preclinical studies [142]. Utilizing tissue engineering to recreate tissue- and organ-level complexity in vitro for preclinical studies begins with considering which cells are needed to reconstitute a tissue, devising a strategy that is permissive for these cells to make connections and interact in ways reminiscent of the native tissue, and establishing clear and precise definitions of their phenotypes [144]. Additional considerations in tissue engineering approaches may include design or selection of specialized culture devices that help to spatially configure cells in 3D, whether terminally differentiated functional cell types or progenitors are most appropriate, and how to validate that the engineered tissue sufficiently represents the native tissue.
Most human tissues are highly vascularized and possess essential functional cells of the tissue or organ, as well as stromal support cells, resident immune cells, and cells of the nervous system [145,146]. Increasingly, research underscores that all of these cells exist in constant communication with each other to perform tissue functions [147,148]. Furthermore, in the course of the disease, tissue and organ dysfunction often involves a complex cascade of disruption of this communication [149]. Therefore, to improve preclinical models of human diseases for drug development, it may often be necessary to provide the right co-culture of cells to make the model more organ- or tissue-like (Fig. 3D). There are a number of strategies and advances in culture devices ranging from simple to complex to accomplish this goal, so the selection of the right culture strategy is an additional consideration in creating a tissue-engineered preclinical model. In addition, there are challenges associated with long-term culture stability within these devices and the ability to establish a shared medium between the different cell types that require consideration [150]. Lastly, tissue engineering is often not as simple as merely placing these cells together in a 3D culture. The chosen cells must be capable of and the culture devices permissive to allowing for sufficient tissue morphogenesis to occur in order to adequately represent the intended tissue architecture, physiology, and cellular communication.
As tissue morphogenesis is central to the tissue engineering process, design considerations should include validation strategies for determining to what extent the putative preclinical model sufficiently represents an in vivo human tissue with respect to the intended tissue biology and physiology. Relevant considerations for validation strategies aimed at determining the degree of tissue morphogenesis include selecting appropriate characterization and functional assays. For example, assayed features for tissue vascularization might look for immunohistochemical characterization of specific cadherins and tight junction proteins in the correct locations or functional assays such as barrier permeability or response to cytokines. Increasingly, cellular “omic” characterizations such as single-cell RNA and epigenetic sequencing methods are options for assaying morphogenesis in in vitro cultures [151,152]. Data dimensional reduction and comparison of the in vitro tissue to primary cells or tissue samples can help determine the degree of morphogenesis and maturation. However, there are caveats with this approach, as the application of single-cell analysis of the transcriptome and epigenome have illustrated that the act of merely isolating primary cells alters their transcriptome substantially [153]. Additionally, the penultimate validation requires that the tissue recreates the function of native tissues that are required for the specific modeling application at hand.
4.4. Space-and time-variable properties of dynamic cultures
Tissues of the human body are not static. In vivo, cells and tissues experience and respond to dynamic mechanical and chemical signals that vary over time and region of the tissues. The signals exist on the local cellular and tissue level during tissue homeostasis, repair and regeneration, and disease. Chemical cues that change over time and space include the gradients, production, consumption, and accumulation of metabolites, nutrients, and growth factors and cytokines. However, classic in vitro systems are static systems at steady-state and do not recapitulate the dynamics of these chemical cues in in vivo organs and tissue systems. Examples of mechanical signals include the local forces that result from cellular contraction and migration within the tissue extracellular matrix (ECM) and shear forces introduced by the pulsatile fluid flow through endothelial and interstitial spaces. At the tissue- or organ-level, forces from expanding or growing tissues, as in the case of development, injury and repair, and the growth of malignancies, are highly dynamic over time and can serve as mechanical cues [154]. How cells and tissues respond to drug candidates in vivo can be widely contextualized by how these chemical and mechanical cues change over time. Furthermore, changes in cell shape, force transmission, chemical cue concentrations, and gradients in 3D are all potent regulators and directors of tissue morphogenesis [155]. As a result, how properties of in vitro cultures are permitted or induced to change over time is an additional consideration for designing preclinical models.
Drug uptake across transport barriers, the degree of tissue vascularization and perfusion, the microscale organization and structure of tissues, and organismal level interactions between different tissue systems that influence drug metabolism all impact PKPD [156,157]. These drug interactions within human tissues and physiology that impact DMPK are all inherently dynamic processes with respect to their occurrence in space and time throughout the body. However, simplistic in vitro models often lack this dynamic nature, and non-human animal models often do not faithfully recapitulate human physiology. To address this, design strategies for in vitro preclinical models should account for fluid flow and transport across the appropriate endothelial and epithelial barriers, as well as drug accumulation, uptake, metabolism, and clearance in human tissues. In the event the intended target is intracellular, the cellular uptake is another PK parameter to consider [158,159]. For prodrugs, design considerations include the rate of drug metabolism and metabolite distribution into different tissue systems, and models that link together different human organ systems may be required. Given the importance of drug PKPD properties on drug performance, and that a combination of PKPD properties may result in an unacceptably narrow therapeutic index [8], design strategies that include PKPD-defining features of human biology and physiology in vitro will likely increase a model's predictive value [4].
To further improve the biological relevance of preclinical models, design strategies often necessitate increasingly complex and larger in vitro tissue cultures. With this development, important considerations concerning the dynamic aspects and transport challenges of large 3D cultures emerge. Of particular note, the dynamic oxygen and nutrient demands of metabolic tissues are increasingly difficult to regulate in large 3D culture settings [131]. The drug development enterprise often employs engineering tools such as bioreactors and liquid handlers that can allow for the introduction of dynamic culture features such as fluid flow to address this issue (Fig. 3C) [160]. However, implementing these tools requires precise optimization of flow and oxygenation to successfully avoid the buildup of toxic waste products and the formation of necrosis [161]. Additionally, whether it is necessary and how to incorporate transport epithelial or endothelial barriers in large 3D cultures is an issue that should be considered. When utilizing models comprising multiple large 3D cultures of different tissues, another design consideration is how to connect these systems. Overall, increasingly large, complex, 3D culture models that better represent the physiological scales of human tissues may aid drug development [150,[162], [163], [164]]. However, improving the viability, introducing transport barriers, and interconnecting different, large 3D in vitro tissue cultures may be required to develop these models.
4.5. Model feasibility and likelihood of adoption
In designing new preclinical models, their feasibility and ease of adoption are also important. For example, the cell source needs to be readily available, scalable, and standardizable. This is an important consideration, as the evaluation of drug candidate performance in a preclinical model is often determined by comparing the outcome of a singular study to a robust, preexisting body of literature [100]. While the lack of preexisting data in the literature is a challenge in the development of new preclinical models, no such body can ever exist if the model is ultimately not widely adopted. Any input materials and culture devices also need to be standardized and readily available. For example, novel, poorly standardized chemistries for synthetic ECMs or natural ECM modifications, culture devices requiring complex assembly, or highly customized machines can also contribute to the generation of models that are unlikely to be widely adopted or utilized in drug development. Lastly, contemporary drug development workflows employ a combination of model systems, not a singular “best” model. Engineered models should consider how they can augment and complement existing models used in the drug development process.
Inherently, preclinical animal models are lengthy, low throughput, and often difficult to analyze, so this is an area where engineered in vitro solutions already have an advantage. However, a caveat with increasing the sophistication and biological relevance of in vitro models is that it frequently results in a decrease in ease-of-assembly and throughput [165]. As a result, practical concerns such as model complexity, availability, and reproducibility are important considerations to balance with other design aspects, such as a model's predictive capacity. Thus, an overall balance between the two must be considered to determine which model features should be given the highest priorities. As an example, if changes in endothelial barrier function are the critical parameter for disease, a transwell assay might be sufficient, but attempting to develop a de novo in vitro vasculature might be required for studies particularly focused on angiogenic events. In general, complexity for the sake of complexity is not inherently useful in designing new preclinical models for drug development, and the preclinical application itself should motivate the model design.
The ability to make accurate measurements and readily collect outputs contributes to the throughput of an assay applied to a given in vitro model. Similar to inputs, the outputs typically decrease with the increasing complexity of the model. For example, in a cell aggregate 3D tumor model, significant variance results from forming cell aggregates via different methods, leading to challenges in making simple measurements such as cell viability in 3D [166]. Additionally, current in vitro preclinical models increasingly utilize genetically engineered cellular readouts, such as fluorescence or luminescence expression in response to biological changes, to make assay measurements in a high-throughput manner. However, while immortalized cells are relatively easy to genetically manipulate, primary cells are often quite challenging to do so in a controlled and standardized manner. Devising strategies to incorporate reporter systems in disease-relevant phenotypes is another feasibility consideration in the design of preclinical models.
5. Biological advances: improving access to disease-relevant human cells and difficult to model tissues
As discussed in 4.1, the selection of cell sources is a critical early design consideration for constructing an in vitro preclinical model. Recent advances in biology that improve access to more disease-relevant human cell types and tissues can greatly improve in vitro preclinical models as well as enable the creation of new models for human disorders that have been traditionally very difficult to recreate in vitro, such as human development. In this section, we will discuss recent strategies to both derive and better utilize disease-relevant human cells and tissues from hPSC and primary sources. In addition, we will discuss unique hPSC properties that enable the creation of highly sophisticated in vitro models of human organs and development.
5.1. Advances in producing disease-relevant cell types
Numerous advances in materials, engineering, and biology have resulted in improved production and utilization of primary- and stem cell-derived cells, substantially increasing researchers' access to disease-relevant cell types for constructing preclinical models (Fig. 3A). hPSCs, in particular, have afforded access to a virtually limitless supply of human cells and tissues that have not been available for preclinical in vitro models for the majority of the history of drug development. Illustrative recent advances relevant to research areas described in Section 4 include successfully derived microglia-like cells [167], type 1 diabetes hiPSC-derived β-cells [168], adipocytes [169], T cells [170], and germ cells [171]. In addition, the derivation of these cell types from hiPSC or primary progenitors can aid in understanding patient- and disease-specific responses to drugs, unlike models constructed of cell lines. For example, the use of in vitro adipose tissues derived from various patients' adipose stromal cells revealed genetic variations that predict the variable patient response to the thiazolidinedione PPARγ agonist, Rosiglitazone [172]. Additionally, recent studies have demonstrated the capacity for more precise specification of the phenotype of hPSC-derived cell types. For example, optimizing the timing of retinoic acid exposure during hPSC differentiation can produce motor neurons with Hox gene expression patterns specific to different regions of the spinal cord [173] as well as specify atrial or ventral cardiomyocyte identity [174].
In addition to the improved availability of biologically relevant cell types, there have been recent advances in strategies to improve their biological relevance concerning phenotypic maturity. Lack of maturity and a fetal-like phenotype is a persistent challenge for the use of hPSCs for disease modeling and therapeutics [120,121]. Several studies have shown that the implantation of hPSC-derived models increased their maturation [[175], [176], [177]]. However, ex vivo strategies that can increase the phenotypic maturity of hPSC-derived cell types will both improve the biological relevance of in vitro models and simplify maturation approaches. Recent research has identified influential parameters and strategies that regulate and promote hPSC-derived cell maturation. For example, cellular metabolism can regulate the phenotype of hPSC-derived cells, and glycolysis-to-oxidative phosphorylation mitochondrial switching enhances the maturity of hPSC-derived endothelial cells [178]. Similar metabolic switching via inhibition of hypoxia-inducible factor 1-alpha and its downstream target lactate dehydrogenase A mediated maturation of hPSC-derived cardiomyocytes [179], suggesting that cellular metabolism may be an important manipulatable factor for regulating the biological relevance of hPSC-derived cells for drug development.
The access to a standardizable and scalable source of primary human cells with increased biological relevance can improve the predictive capacity of in vitro models, relative to current models comprised of current cell lines. For example, the BBB is a unique endothelial barrier with tight regulation that prevents many candidate drug compounds for neurological indications from reaching the central nervous system [180]. Disorders of the central nervous system are particularly challenging for developing therapeutics and result in a higher rate of failed drug candidates than found in other diseases [39]. In vitro models of this DMPK-influencing physiological barrier have improved with recent advances that provide access to more relevant biological materials; however, primary human sources of relevant endothelial cells are limited and difficult to standardize [181]. In contrast, the development of BBB-like endothelial cells from hPSCs can yield up to 100-fold higher degree barrier properties, as measured by transendothelial electrical resistance, relative to primary endothelial cells [182], commonly used in barrier function assays. Derivation and addition of hPSC-derived pericytes to cultures of hPSC-derived BBB ECs further enhanced their barrier properties [183].
Beyond the derivation and maturation of newly available cell types from hPSCs, engineering advances can improve the expansion and, therefore, the utilization and standardization of primary cells. For example, recent biomaterials advances offer new methods to expand antigen-specific primary human T cells [184]. Ex vivo expansion of antigen-specific primary T cells is a challenge to advancing T cell-based therapeutics, as this requires activation of the T cell receptor, and antigen-specific T cells require the appropriate presentation of antigens [184]. The leading commercial approaches [185] use antibodies conjugated to plastic beads that target the epsilon subunit of the T cell receptor but do not provide specificity of the expanded T Cell population. Recently, engineered antigen-presenting cell-mimicking scaffolds that combine sustained release of proliferation cues, such as Interleukin-2 with peptide-loaded recombinant major histocompatibility complex I and anti CD28 antibodies, have demonstrated the capacity to stimulate and expand antigen-specific cytotoxic T cells [184,[186], [187], [188]].
5.2. Modeling human development in vitro
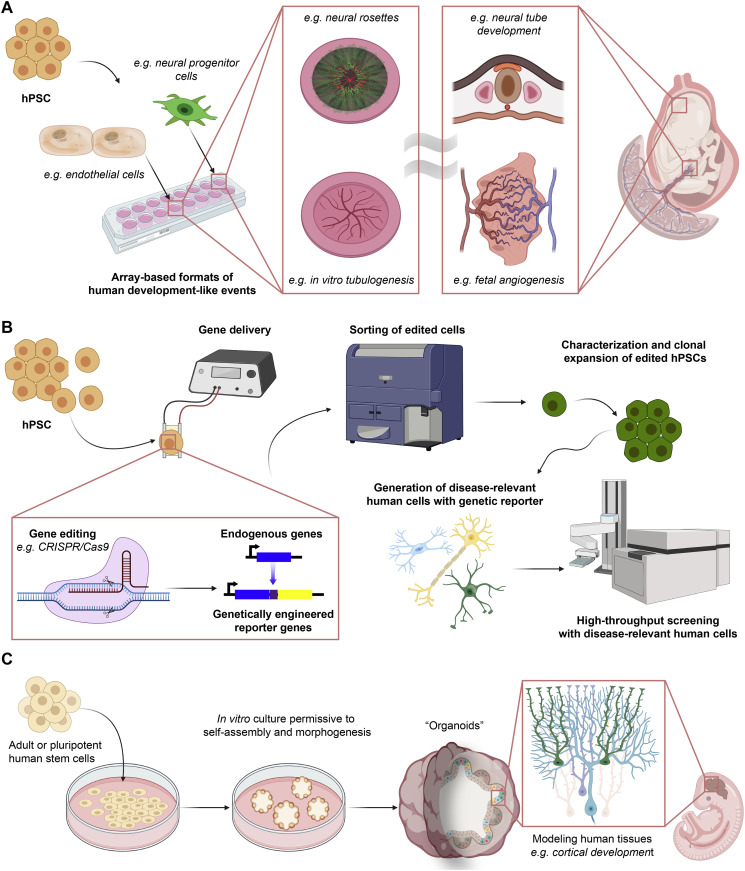
Fetal like hPSC-derived tissues may be particularly useful for modeling human development. In fact, they may hold a specific advantage over the use of primary cells and may have particular relevance to areas such as developmental toxicology. As an example, hPSCs-derived neural tissues have demonstrated the capacity to form neural rosettes, structures reminiscent of early neural tube patterning [189,190]. Demonstrating the potential utility of this phenomenon, rosettes generated from congenital Spina Bifida patient-derived hiPSCs correctly predicted folic acid as an NTD preventative factor [191]. Using mesendoderm specification as an output metric from hPSC differentiation allowed for the correct identification of the teratogenicity of 71 drug-like compounds with 92% accuracy [192]. Additionally, this approach was amenable to screening of environmental toxins and 300 pharmaceutical compounds [192] as well as correctly predicting thalidomide teratogenicity [193].
Additional strategies, such as array-based formats of hPSC-derived rosettes [194], can afford enhanced throughput screening platforms for identifying additional nutrients or potential toxins that mediate developmental disorders such as NTDs. Similarly, enhanced throughput tubulogenesis models can serve as platforms for detecting potential teratogens that perturb fetal angiogenesis. For example, an array-based tubulogenesis model on synthetic hydrogels, using both primary and hPSC-derived endothelial cells, afforded accurate identification of developmental toxins in a blinded screen [195]. Of note, both of these models exploit the self-directed tissue morphogenic capacity inherent to these cells to form the neural tube-like structures or vascular networks, illustrating the importance of the appropriate cell source selection. Additionally, the clear, array-based outputs demonstrate how proper consideration of model design feasibility, as discussed in Section 5.5, can afford increased physiological relevance of a preclinical model without the commensurate increase in model complexity hindering throughput.
In addition to modeling developmental events, the ease of implementing genetic tools in hPSCs can facilitate drug screening in disease-relevant human cells (Fig. 4B). As an example, after demonstrating that genetic correction of deficits in KCC2 expression in hiPSC-derived neurons from Rett Syndrome patients rescued normal neuronal function [196], hPSCs-with a luciferase reporter in the endogenous KCC2 locus were generated [197]. Using neurons derived from this reporter hPSC line, an unbiased drug screen identified pharmacological interventions that rescued neuronal activity and behavioral deficits in a murine Rett Syndrome model without the need for genetic correction [197]. In another example, normal hESC- and hiPSC-derived neurons generated from SHANK3 Haploinsufficiency Syndrome patients enabled identification of lithium and valproic acid, two approved therapeutics for other indications, as potential modulators of the autism disorder phenotype caused by the dose-dependent loss of SHANK3 [198].
Fig. 4.
Biology advances for improving in vitro preclinical models. (a) hPSCs possess properties that allow for mimicking developmental processes in vitro. Examples include the in vitro formation of neural rosettes and endothelial tubules that are reminiscent of the embryonic neural tube and fetal angiogenesis, respectively. Moreover, these processes can be developed into array-based formats that allow for toxin and nutrient screening for developmental disorders. (b) hPSCs can provide a highly efficient route for producing genetic readouts into disease-relevant human cells for HTS assays. Gene editing is straightforward and efficient in hPSCs, and edited cells can be sorted, expanded, and characterized before differentiating into the disease-relevant cells of interest. (c) hPSCs possess the capacity to undergo self-assembly and morphogenesis into complex human tissues called “organoids” that are otherwise difficult to recreate. Cerebral organoids are such an example, as they self-organize into structures that resemble the neuronal spatial arrangement and layering of the developing cortex. These self-contained tissues can also be used in screening for toxins and nutrient screening for developmental disorders as well as modeling non-cell-autonomous degenerative conditions such as sporadic Alzheimer's Disease.
5.3. Self-driven tissue morphogenesis for creating in vitro human organ-like tissues
In addition to providing access to more disease-relevant cell types, adult and pluripotent stem cells recently emerged as biological tools to recreate complex human tissues in vitro. When placed in permissive 3D microenvironments, these stem cells undergo a process of self-driven morphogenesis to form 3D microtissues called “organoids” (Fig. 4C). In an early demonstration, Lgr5+ stem cells isolated from intestinal crypts self-organized into crypt-villus like structures in vitro [115]. Later, hPSC-derived neural precursors showed the remarkable self-directed morphogenesis capacity with the spontaneous in vitro formation of the optic cup [199] and cerebral organoids containing multiple layers of the cortex as well as mid- and hindbrain structures [200,201]. A significant advantage of these organoids for drug development applications is that they recreate organ-specific hierarchical structures resembling native human tissues, which have been otherwise difficult to accomplish [202,203].
Since the seminal organoid discoveries, researchers have derived additional patient- and hPSC-derived organoids from other tissues, including the intestines, liver [204], pancreas [205], and kidney [206]. As organoids of these tissues form, either from adult stem cells [115,205] or hPSCs [199,201], the processes by which they self-organize can allow for the further study of human tissue development in vitro [203]. For example, physically confining cerebral organoids revealed that the forces from physical confinement at the membrane boundary and individual cellular contractility interacted to determine the overall organoid shape, folding, and cellular migration positioning at the microtissue scale [207]. As the neuronal migration and cortical folding mechanisms underpinning fetal cerebral gyration are relevant to disorders such as viral infection-mediated microcephaly, organoids that recreate these complex morphogenic events in vitro could serve as potential platforms in drug development for areas like infectious diseases.
In addition to the in vitro study of morphogenesis of native tissues, organoids have afforded researchers insights into disease mechanisms and provided models to improve cancer therapeutics. For example, a long-term in vitro culture of patient-derived organoid models of non-small cell lung carcinoma demonstrated similar molecular and histopathological phenotypes to the patient-derived xenografts and the primary tumor [208]. The model helped to correctly identify that KRAS mutant tumors would respond to MEK inhibition, which matched established patient outcomes, as well as predict that a combination of FGFR and MEK inhibition would be effective in treatment [208]. Self-assembled tissues, such as kidney-derived organoids, also offer an opportunity to screen for cancer-treatment associated toxicity [206]. Tumor organoids may also afford a neoantigen discovery platform for personalized immunotherapies [209].
6. Engineering advances: assembling advanced in vitro preclinical models for increased biological relevance
In the preceding section, we discussed biological advances that are improving access to disease-relevant human cells and tissues. Here, we will highlight recent engineering advances that enable improved in vitro modeling of human tissues.
6.1. Advances in recreating the extracellular matrix
As discussed in 5.2, the choice of natural vs. synthetic ECM, as well as the chemical functionalization, crosslinking, and degradation strategy, can influence how a 3D in vitro model can facilitate preclinical studies. In the case of natural ECMs, chemical functionalization can allow for the investigation of how different ECM components (Fig. 5A) contribute to disease. For example, using methacrylated gelatin, hyaluronic acid, and chondroitin sulfate, and systematically varying the composition of the natural ECM glycosaminoglycan components helped to elucidate the role of ECM moieties in the deposition of oxidized lipoproteins during the progression of calcific aortic valve disease [210]. In studying breast cancer, varying the collagen concentration in a methacrylated gelatin background scaffold demonstrated the impact of collagen fiber thickness and density on tumor metastases [211]. These examples illustrate how natural ECM modification approaches can be useful for improving the understanding of disease mechanisms and identifying targetable biomolecules for drug development.
Fig. 5.
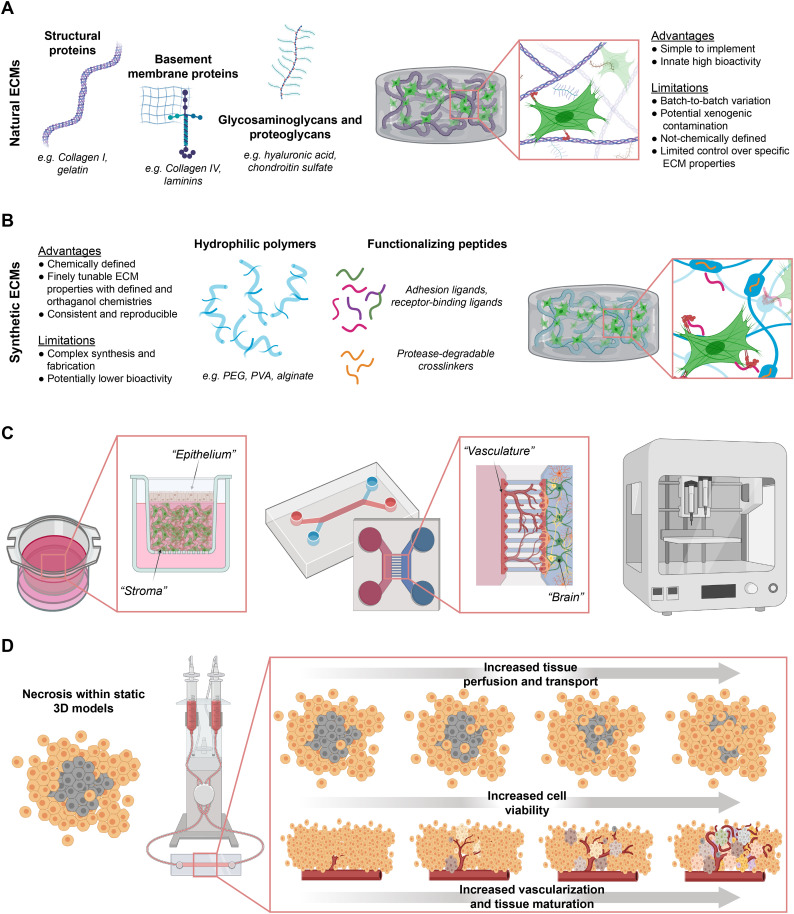
Engineering advances for improving in vitro preclinical models. (a) Natural ECMs can be utilized to create 3D scaffold-based tissue models. Example ECMs include structural and basement membrane proteins. Additional ECM components such as glycosaminoglycans and proteoglycans can also be incorporated. Natural ECMs are innately bioactive and offer a straightforward strategy to create biocompatible 3D culture systems. (b) Synthetic ECMs are alternatives to natural ECMS for 3D scaffold-based culture models. These synthetic ECMs are often based on biologically inert hydrophilic polymers decorated with functionalizing peptides to allow for cell adhesion and cell-mediated degradation and remodeling. While often requiring more complex synthesis and fabrication than their natural counterparts, synthetic ECM strategies allow for more fine control and tuning of ECM properties and therefore facilitate understanding of the relationship of ECM properties to disease mechanisms. Additionally, the more chemically defined nature of synthetic ECMs can reduce untoward effects from latent variables in natural ECMs, potentially improving reproducibility in drug development platforms. (c) Specialized culture devices and technologies such as transwell (left), microfluidic (middle), and bioprinting (right) allow for increased control over cellular spatial arrangement in 3D cultures. Examples of possible arrangements include 3D stroma-epithelial cultures (left) and the blood-brain barrier (middle). (d) As 3D cultures introduce challenges with nutrient transport, strategies that incorporate microfluidics with fluid flow and perfusion can increase transport to overcome these challenges. The introduction of vasculature can also improve transport and be utilized to introduce transport barrier properties in tissues or to provide morphogenic cues that drive further tissue maturation.
Relative to natural ECMs, synthetic ECMs allow for more precise control of 3D and studying how cells respond to physical matrix properties (Fig. 5B) [141]. For example, combining chemical and ionic crosslinking of alginate hydrogels with both natural and synthetic adhesion ligands demonstrated that both the stiffness and composition of an ECM act in concert to induce malignancy of mammary epithelium in vitro [212]. Combining polyethylene glycol and alginate to create a hybrid synthetic ECM allowed for independently varying the stress relaxation from the initial elastic modulus and demonstrating how ECM deformations resulting from cellular forces can induce changes in gene expression that promote cancer metastases [213]. Synthetic ECMs also offer strategies to recreate complex 3D niches to study disease in vitro. For example, an alginate foam that mimicked the 3D microporosity of the bone marrow microenvironment enhanced differentiation of myeloid precursors from a leukemia cell line and improved prediction of chemotherapeutic resistance, as compared to a 2D culture [214]. Overall, the tailorability of these synthetic ECM approaches enables understanding of the role that mechanobiology plays in disease progression, offering insights into disease mechanisms and potential drug development platforms.
Cellular-ECM interactions in 3D cultures can promote tissue morphogenesis to form complex human tissues in vitro [215,216]. For example, cellular traction and ECM density can influence vascular morphogenesis and capillary stability in vitro [217], and the exposure of hPSCs to proteolytically degradable ECM can promote epithelial-to-mesenchymal transition gene activation and thus enhance mesoderm specification [218]. While naturally-derived ECMs such as Matrigel™ are common scaffolds used to promote morphogenesis, synthetic ECMs can be employed to isolate ECM properties that promote and regulate in vitro morphogenesis [141]. By combining customizable synthetic ECMs with techniques such as single-cell RNA and ATAC sequencing, researchers may be able to isolate the morphogenetic cues of 3D ECM further. This information may enable researchers to utilize epigenetic and transcriptional reprogramming tools [219,220] to manipulate 2D cultures into better reflecting 3D cultures or in vivo tissues. This approach would potentially afford the production of more biologically relevant cells in a format more compatible with high-throughput and parallelizable 2D assays for drug development.
6.2. Technologies for assembling cells into 3D tissues
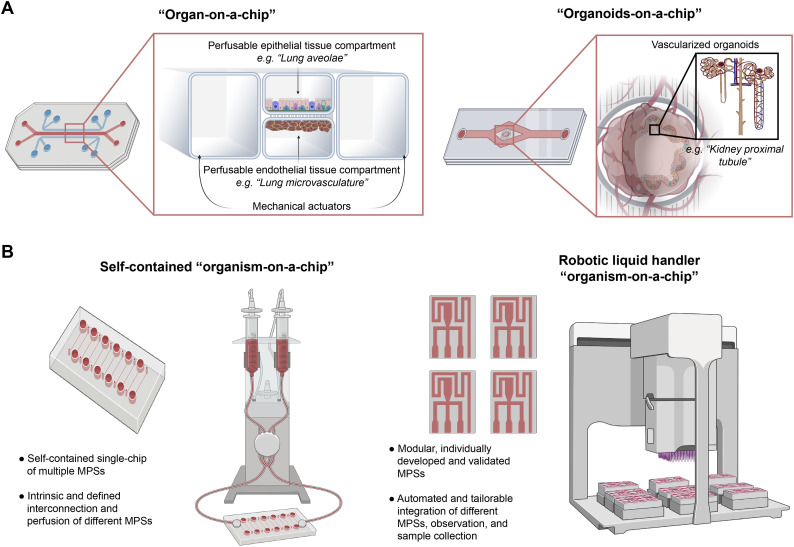
Assembly of 3D in vitro co-cultures into distinct configurations can offer distinct advantages that facilitate drug development (Fig. 5C). In vitro 3D models for vaccine development are illustrative of this potential, particularly in the early stages of immunodominant antigen identification, where the risk of candidate failure is highest [67]. This early-stage failure in vaccine development often occurs, in part, due to antigen selection in rodents and the differences between species in the mechanisms of establishing protective immunity [67]. In addition, improper immunodominant antigen selection may increase disease virulence of a pathogen, adding substantial safety concerns [67]. In vitro models comprised of human cells arranged in a manner that is permissive to initial infection, antigen-presenting cell activation, antigen uptake, antigen presentation, and subsequent T cell and B cell activation and maturation could provide a useful alternative. For example, an in vitro 3D human cell culture system comprised of a stromal compartment with an epithelial barrier allowed for modeling of monocyte extravasation, and then dendritic cell activation, antigen uptake, and presentation of antigens to activate T cells following viral infection [221]. In addition, the model detected differences in how elderly and young human patients' cells respond to an existing vaccine antigen, which matched known patient outcomes [221].
Creating 3D tissues can facilitate more physiologically relevant in vitro modeling of PK. In vitro PK modeling related to transport barriers, such as gut and lung epithelium or endothelial membranes, has traditionally relied on 2D monolayers on semipermeable membranes [132,222]. For example, oral bioavailability, a critical mode of absorption and drug PK for most therapeutics, is often predicted with a static model using the Caco-2 rectal epithelial cell line as a monolayer on transwell membrane to model drug gut uptake [223]. However, the use of more biologically relevant cell types, co-cultures with supporting cell types, and more realistic configurations than 2D monolayers on semipermeable membranes can improve the representation of these PK barriers in vitro. For example, a two-compartment transwell co-culture of hPSC-derived neurons, astrocytes, and BBB endothelial cells where the neurons and astrocytes were below the endothelial layer increased the barrier function properties in this model over 5-fold as measured by transendothelial electrical resistance [224].
Specialized culture devices provide the spatial control and placement of distinct cell populations into specific 3D configurations, increasing the organizational complexity of in vitro tissues. Microfluidic devices (Fig. 5C), often composed of molded, gas-permeable silicone polymers or micromachined plastic, allow for the spatial arrangement of cells and microtissues into perfusable arrangements at the micron scale of precision [[225], [226], [227], [228], [229], [230]]. These devices take advantage of the predominance of laminar flow at microscales, which allows for predictable fluid flow perfusion and spatial patterning [231]. In 2016, the US National Center for Advancing Translational Sciences spearheaded funding for the development of Tissue Chip Testing Centers to further develop and validate tissue chip technology for drug development in regulatory, academic, and industry settings [232]. In recent years, researchers have used the tissue assembly approach in microfluidic devices to create a variety of tissue constructs relevant for drug development, such as bile ducts [233] required for liver metabolism and adipose chips for ADME studies [234]. Table 1 summarizes recent representative advances in tissue chips relevant for drug development, as well as other chips for modeling human tissue physiology that are applicable to disease areas discussed in this review.
Table 1.
Selected recent studies developing “organ-on-a-chip” models that are relevant to preclinical studies in drug development.
|
Human tissue chip models | ||||
|---|---|---|---|---|
| Tissue | Type of device | Summary of drug development application | Cell source | Ref. |
| Cardiovascular | Static two-channel microfluidic device perfused with an actuator for cyclic mechanical stimulation | Platform development of mechanical heart stimulation for drug dose studies for cardiotoxicity | hPSC-derived cardiomyocytes | [274] |
| 3D printed microfabricated tissues on electrically coupled cantilevers | Platform development of electrically coupled heart stimulation for drug dose studies for cardiotoxicity | hPSC-derived cardiomyocytes | [275] | |
| Endothelial tubule microfluidic device perfused with a two-channel peristaltic pump | The study of endothelial transport barriers and angiogenesis in response to stimulus | Primary and immortalized endothelial cells | [276] | |
| Hepatic | Bioprinted cells encapsulated in a crosslinked scaffold within a bioreactor | Screening drug compounds for hepatotoxic effects | Immortalized hepatocytes | [277] |
| Single-channel microfluidic device perfused with a peristaltic pump | Platform development for enhancing 3D hepatocyte culture for DMPK and toxicity studies | Primary hPSC-derived hepatocytes | [278] | |
| Self-contained perfused bioreactor with 3D culture inserts | Platform development for utilizing primary 3D hepatocyte cultures to screen for patient variability in DMPK | Primary hepatocytes | [279] | |
| Renal | Two-channel microfluidic device perfused with a syringe pump | Replicating secretory function of kidney glomerulus for in vitro ADME and screening platform for nephrotoxicity | hPSC-derived podocytes | [280] |
| Kidney proximal tubule microfluidic device perfused with a two-channel peristaltic pump | Replicating the uptake and secretory functions of the kidney for in vitro ADME | Primary proximal tubular epithelial cells | [281] | |
| Neural | Two-channel passively pumped microfluidic device incorporated into a well-plate | The study of receptor-mediated transport of antibodies across the blood-brain barrier | Immortalized astrocytes, brain microvascular cells, and pericytes | [282] |
| Three-channel microfluidic device perfused with a syringe pump with organoid channels | The study of prenatal nicotine exposure using brain organoids as a model of fetal development | hPSC derived brain organoids | [283] | |
| Two-channel microfluidic device perfused with a syringe pump with a static culture well | The study of metabolic coupling between endothelial cells and neural cells of the neurovascular unit as well as modeling drug-induced blood-brain barrier permeability | Primary astrocytes, brain microvascular cells, pericytes, fetal stem cell-derived neurons | [284] | |
| Adipose and endocrine | Two-channel microfluidic device perfused with a syringe pump with a static culture channel | The study of lipid metabolism and adipocyte drug response for obesity | Primary adipocytes | [285] |
| Single-channel passively pumped microfluidic device with capture wells for cell aggregates | The study of insulin kinetics in 3D pancreatic islets exposed to flow for type 2 diabetes | Immortalized pancreatic cells | [286] | |
| Three-channel microfluidic device perfused with a syringe pump with a central static culture channel | The incorporation of flow and immune cells into 3D adipose tissue cultures for modeling insulin sensitivity for type 2 diabetes | Combination of immortalized and primary adipose and immune cells | [237] | |
| Cancer | Two-channel microfluidic device perfused with a peristaltic pump and vacuum channel with an actuator for mechanical stimulation | In vitro modeling of non-cell-autonomous cancer events such as microenvironmental and mechanical parameters regulating tumor site initiation, growth, and metastasis | Primary lung alveolar epithelial and microvascular cells as well as a cancer cell line | [287] |
| Microfluidic device containing a spheroid chamber and perfused with a peristaltic pump | The study of nano-particle transport accumulation with tumors exposed to fluid flow | Cancer cell line | [288] | |
| Passively pumped and gradient generating microfluidic device containing a spheroid chamber | The study of drug efficacy response to chemotherapeutics for glioblastoma | Cancer cell line | [289] | |
The precise control and orientation of cells within scaffolds in 3D tissue chips also allows for more physiologically relevant studies of cellular migration. For example, a polymeric silicone microfluidic device with parallel hollow lumens embedded in collagen helped to elucidate the signaling pathways that promote pancreatic cancer metastases [235]. Specifically, the model helped to show that adenocarcinoma metastases relied on pancreatic ductal endothelial ablation that was dependent on the ALK7 pathway of TGF-β signaling and that broad-spectrum inhibitors of TGF -β signaling could limit this effect [235]. Similarly, microfluidic devices containing primary endothelial and breast cancer cells demonstrated that signaling resulting from endothelium disruption promotes breast tumor metastases [236]. For inflammatory diseases, tissue chips can allow interactions of immune cells with tissues to be studied in a 3D in vitro setting. For example, in a microfluidic chip for long term culture of human adipocytes, with on-demand introduction of an immortalized human macrophage cell line [237], macrophage introduction resulted in decreased insulin sensitivity, a hallmark of obesity-induced type 2 diabetes [237]. Importantly, this in vitro platform allowed for on-chip assays such as cytokine production, glucose uptake, and microscopy [237], analysis not easily achievable in current in vivo models for studying obesity and type 2 diabetes.
6.3. Vascularizing in vitro tissues and introducing flow to 3D cultures
Dynamic cultures created via perfusable networks that allow for fluid flow and circulation improve the in vitro representation of in vivo tissues [238]. The use of microfluidic devices with combinations of passive or active pumping modalities represent the most common approaches for the implementation of dynamic cultures in vitro. Based on the concept that at small scales, viscous forces dominate over inertial forces [239], these devices allow for the simple introduction of controlled, laminar flow of fluids. Additionally, as microfabrication techniques allow for precise patterning microscale features such as channels and lumens, these technologies allow researchers to recreate a variety of culture models with perfusable vascular networks and endothelial barriers to study drug-development related transport phenomena in vitro. For example, microfluidic channels lined with confluent endothelium and superficially with kidney epithelium within a permeable ECM showed selective reabsorption of solutes, such as glucose, and showed a response to known pharmacological inhibition [240].
Introducing flow to 3D tissues can improve the utilization of primary cells for in vitro models (Fig. 5D) [161]. For example, primary hepatocytes used in drug development for hepatotoxicity screening need to be mature, metabolically active cells capable of metabolizing drugs [241]. However, their availability and cost significantly hinder the utilization of primary hepatocytes [161,241]. Compounding this limitation is the rapid dedifferentiation of hepatocytes that occurs in vitro [112]. As a result, researchers frequently assess hepatic drug metabolism using S9 fractions of primary liver homogenates [242]. While this approach affords drug metabolism prediction by key CYP proteins, the activities of these enzymes are only 20–25% of intact hepatocyte microsomes where membrane-bound drug metabolism occurs [243], and this approach offers no insights into hepatoxicity. Additionally, the intrinsic nature of S9 fractions as single-use liver homogenates renders this approach similarly restricted by the limited availability of primary tissues. Engineered perfusion bioreactors that regulate oxygen tension and nutrient delivery in 3D cultures offer strategies to improve the utilization of primary cells such as hepatocytes [161,162]. For example, a bioreactor with fluid flow computationally optimized to consider oxygen consumption helped to extend the viability of 3D primary hepatocytes cultures [162].
In addition to providing a mechanism for tissue perfusion combined with recreating cross-barrier transport, the implementation of vasculature into in vitro tissues can increase tissue development and maturation (Fig. 5D) [244]. For instance, exposing aggregates of metanephric mesenchyme derived from hPSCs to specific rates of flow in a perfusable millifluidic device resulted in enhanced formation of glomerular vascular networks and induced morphogenesis of in vitro kidney organoids to mature states [244]. Additionally, incorporation of primary human parenchymal and endothelial cells into 3D hPSC-derived hepatocyte aggregates increased CYP450 activity as well as albumin and urea production relative to 2D monoculture of hepatocytes [245]. These studies help to illustrate that the endothelium represents an important functional niche that regulates maturation in normal and diseased tissues in ways that increase the biological relevance of in vitro models for drug development.
Lastly, 3D printing has emerged as an alternative route for creating complex, vascularized tissues. This technology (reviewed here [246]) allows for the precise patterning of cells within 3D microenvironments. In particular, 3D printing of sacrificial networks followed by seeding of endothelial cells allows for direct 3D patterning of increasingly complex vascular tissues [246]. For example, 3D printing of sacrificial polysaccharide “carbohydrate glass” within a crosslinked polyethylene glycol hydrogel allowed for seeding primary endothelial cells into a perfusable, hierarchical vascular network within a 3D culture of stromal cells [247]. Combinations of 3D printing of endothelial cells with cell spheroids [248], as well as 3D printing and patterning of hPSC derived tissues [249], are additional avenues to create more complex and biologically relevant vascularized tissues in vitro. Further advances in 3D printing, such as techniques for printing composite biocompatible and dynamically responsive materials [250,251], will facilitate drug development by allowing for more complex preclinical models of human tissues in vitro, particularly vascularized tissues [248].
6.4. Integration of advances to create microphysiological systems and model whole organisms
Microphysiological systems (MPSs) are in vitro models of simple, representative, functional units of organs that seek to recreate key features of in vivo tissue (Fig. 6A) [163,222,252,253]. In 2010, a lung-on-a-chip MPS concept, in which a semipermeable polymeric silicone membrane lined with lung epithelium and endothelium connected to air channels, recapitulated whole-organ inflammatory responses to mechanical stretch and cytokines [254]. To support the development of MPS technology, the US National Institutes of Health (NIH) and FDA jointly created a program to advance MPSs as in vitro models for testing the efficacy and safety of drugs for more improved efficiency in developing therapies, followed by an additional initiative between the NIH, FDA, and the US Defense Advanced Research Projects Agency (DARPA) to expand this approach futher [229]. Since 2012, numerous examples of MPSs have emerged and led to the proliferation of “organ-on-a-chip” and “organoid-on-a-chip” devices (Fig. 6A-B) [2]. A key advantage of these organ-on-a-chip systems as preclinical models is that they can combine organ-like tissue architecture, function, intact endothelium, and flow with the reductionist and streamlined approach of a self-contained and modular tissue chip that allows for enhanced throughput in vitro modeling of drug PKPD and efficacy in complex human tissues [255]. For example, retina organoids on a perfusable chip allowed for the recreation of known drug-induced retinopathies [256], suggesting such an approach could be a useful addition to ADME studies to identify off-target drug toxicity.
Fig. 6.
Microphysiological systems and engineering tools for creating “organisms-on-a-dish.” (a) Representations of “organ-on-a-chip” and “organoid-on-a-chip” models such as Lung [254] and Kidney [244] chip systems. (b) Representations of “organism-on-a-chip” designs. Self-contained single-chip models afford simplicity in integration and perfusion of the different MPSs but lack modularity and present engineering challenges in liquid handling. Modular designs combined with automated robotic liquid handling strategies afford rapid and simple diversification of MPSs integration but require more specialized engineering resources.
While individual MPSs can mimic in vivo tissues, in vitro PKPD studies often require multi-organ or organismal interactions, interconnected via a perfusable vasculature. For example, some drugs may exhibit no direct toxicity in tissues but do after hepatic metabolism. In another circumstance, the cellular response to a drug might be contextually dependent on paracrine factors within a tissue or endocrine factors from multiple distal tissues [257,258]. Interconnected MPSs representing different tissues offer a strategy to model these complexities in vitro. For example, in a self-contained and interconnected gut-liver MPS, inflammatory signals from the gut modulated hepatic drug metabolism [259]. Expanding on this concept, combining a quantitative systems pharmacology approach with ten interconnected different tissue systems comprised of both primary and immortalized cell lines link allowed for the creation of in vitro PKPD models and characterization of interactions between different tissue systems to predict human outcomes [222]. This strategy of interconnecting different MPSs (Fig. 6B) has led to the establishment of “organisms-on-a-chip” culture systems and represents promising opportunities to create representations of organismal physiologies for modeling human DMPK and ADME in vitro [260]. Table 2 summarizes examples of these organism-on-a-chip approaches for recapitulating tissue and organ function in vitro.
Table 2.
Selected recent studies developing “organism-on-a-chip” models that are relevant to preclinical studies in drug development.
|
Human “organism-on-a-chip” models | ||||
|---|---|---|---|---|
| Tissue systems | Type of device | Summary of drug development application | Cell source | Ref. |
| Lung-liver-heart | Microfabricated individual devices (bioprinted cardiac, cross-linked scaffold liver, transwell lung epithelium) interconnected and perfused with a peristaltic pump | Modeling drug toxicity dependency on multi-tissue system interactions | Primary cells | [37] |
| Gut-liver | Self-contained, two-chamber, microfluidic device with transwell inserts and integrated pumping manifold | Modeling drug PK in vitro and examining the effects of gut inflammation on hepatic drug metabolism | Combination of immortalized and primary cells | [259,290] |
| Intestine-liver-skin-kidney | Self-contained, four-chamber, microfluidic device perfused with a peristaltic pump | Replicating multi-tissue system drug and ADME in vitro | Combination of immortalized and primary cells | [291] |
| Heart-liver, heart-liver-cancer | Modular design with individual microfluidic organ-on-a-chip models sequentially interconnected and perfused via a peristaltic pump and integrated small molecule sensors | Modeling liver and heart functionality in drug response studies and chemotherapeutic efficacy studies for liver cancer | Combination of primary, hPSC-derived, and cancer cells | [292] |
| Heart-skeletal muscle-liver-neuronal | Self-contained four-chamber microfluidic device with gravity-driven passive pumping | Modeling of cardio-, muscular- hepato-, and neuronal drug toxicity in vitro | Combination of immortalized, primary, and hPSC-derived cells | [293] |
| Gut-liver-kidney, bone marrow-liver-kidney | Modular design with individual microfluidic organ-on-a-chip models interconnected via a robotic liquid handler | Integrated components for modeling first-pass drug metabolism in in vitro DMPK studies (gut) as well as kidney drug toxicity screening (bone marrow) | Combination of immortalized and primary cells | [263] |
| Liver-pancreas-intestine-lung-heart-muscle-brain-endometrium-skin-kidney | Self-contained, ten-chamber, microfluidic device with transwell inserts and integrated pumping manifold | Integrated “human-on-a-chip” for performing in vitro PKPD studies in combination with systems biology | Combination of immortalized, primary, and hPSC-derived cells (pancreas cell-non human) | [222] |
Interconnected MPSs can enable better representation of human organ and organ systems in vitro; however, new challenges emerge with the use of this technology, including increased complexity concerning assembly, reproducibility, and complications with data collection [252]. For example, the exchange, collection, and analyses of media from multi MPSs with μL-scale volumes present analytical and engineering challenges [252]. In addition, the small dimensions of MPSs introduce concerns with the validation of the scaling parameters used in the computational prediction of PKPD models from MPS-generated data [261]. As an alternative approach to self-contained interconnected MPSs systems described above (Fig. 6B), advancements in robotics and automated liquid handlings and microscopy can be used to connect different modular organ-on-a-chip systems [262]. For example, a programmable robotic culture platform afforded the automated perfusion, observation, and sample collection in three interconnected different modular tissue systems that had been previously validated to quantitatively predict their tissue-specific drug PK properties [263]. Continued engineering advances to connect MPSs, combined with quantitative data that shows computational modeling outcomes from MPS data scales to the meso- and macro-scale of human physiology [259], will lead to further refinement of the organism-on-a-chip approach and improve the predictive value of in vitro PKPD, DMPK, and ADME studies of new drugs.
7. Looking forward: improving the relevance of in vivo models utilizing human tissues
While there are ample opportunities to improve preclinical studies with advances in in vitro representations of human tissues, in vivo animal models will undoubtedly continue to serve an integral role in drug development. As such, advances in biology and engineering can and should be applied to augment existing animal models as an additional strategy for improving preclinical studies. Currently, transplantation of human xenografts into immunocompromised animals is the gold standard for studying human tissues in vivo. However, the recent appreciation of immunology in a range of human diseases, as well as various immune cells being potential targets for therapeutic strategies, indicates that these models may be critically deficient for many avenues of emerging drug development. Contrary to conventional xenografts, transplantation of healthy tissues that can repopulate the human immune system in vivo may afford the study of human diseases in a more immunologically relevant setting. For example, engraftment of bone marrow, liver, and thymus tissues can allow for modeling HIV infection in mice and be used in research towards vaccines for HIV [264]. However, this model depends on fetal tissue, raising significant ethical and regulatory concerns for their widespread adoption.
The use of hPSC-derived tissues combined with humanized animals may lead to improved xenograft models. Genetic animal humanization, which involves the introduction of specific human- and removal of non-human cytokines and surface receptors, may improve the engraftment of human xenografts from non-fetal sources (Fig. 7A-B) [265]. For example, injection of human myeloid precursor into a humanized mouse expressing human cytokines for CSF1, IL3, SCF, and GM-SCF led to the development and maturation of human microglia in a mouse brain [266,267], albeit still in an immunocompromised animal. Given the newly appreciated role that microglia play in the development of many neurodegenerative diseases such as Alzheimer's [94], Parkinson's [268], and multiple sclerosis [269], this model may be particularly useful for the study of human microglia in vivo (Fig. 7B). Another promising humanization strategy involves chimeric mice generated via grafting of human hepatocytes as well as the transgenic introduction of human p450 [270] genes into mice. Given the importance of the predictive value of ADME- and hepatoxicity-screening data on drug efficacy and safety, these platforms have substantial potential value for improving preclinical studies.
Fig. 7.
Employing biological tools to advance future in vivo models. (a) Conventional xenograft models involving the delivery of non-fetal primary human cells to immunocompromised animals result in poor engraftment and limited utility as in vivo models of human tissues. (b) Delivering progenitors derived from hPSCs into genetically humanized mice results in improved engraftment and representation of human tissues in vivo after the progenitors differentiate. (c) Not being amenable to embryonic-chimeras, human-interspecies chimeras are achievable with in utero delivery of hPSC-derived progenitors that are matched to the developmental timing of the host embryo. (d) Conventional cancer xenografts resulting from the delivery of human tumor cells to immunocompromised mice do not allow for immunotherapy drug development with human tissues in vivo. Cancer models resulting from human-chimeras generated in immunocompetent mice may afford the potential to model cancer-immune system interactions in vivo for immunotherapy development.
Human-nonhuman species chimeras represent an alternative to engraftment of human tissue in humanized mice. This principle of interspecies chimeras depends on the convergence of developmental timing amongst vertebrates and the ability to derive and deliver developmentally appropriate human progenitor cells (Fig. 7C) [271,272]. For example, injection of developmentally matched hPSC-derived neural crest progenitors into E8.5 mouse embryos resulted in the contribution of human pigmented epithelium to mouse hair follicles in immunocompetent animals without immunosuppression [271]. Later, injection of hPSC-derived neural crest cells with a doxycycline-inducible MYCN + ALK F1174L oncogene mutation formed neuroblastomas in immunocompetent mice using this chimeric approach [273]. Impressively, the human tumors in this model more closely reflected the tumor tissue architecture and immune response of tumors in human patients. Overall, improvements in the ability to create human chimeras and increase the contribution of normal and diseased human tissues in immunocompetent animals (Fig. 7D) could be a powerful advance in preclinical models. However, complexities relating to the physiological crosstalk between human and nonhuman tissue systems in these types of models will likely result in new challenges [270].
8. Conclusions
Despite decades of advances in DMPK and ADME validation in drug development, it is empirically evident that preclinical studies are still of limited value in predicting drug performance in HCTs. At the intersection between biology and engineering, new tools and advances are creating a unique opportunity to both produce and configure human cells and tissues in such a way to increase the relevance of preclinical models to native human tissues and organs. Utilizing these tools requires careful considerations of the process from design to implementation that are required to promote the greatest benefit to drug development. Further, the biology and engineering research enterprises will continue to make advancements and provide new tools, so the material discussed here can be considered as a representative framework for synthesizing demands of different research areas with available biological and engineering tools. Increased interactions between academic researchers and those in the drug development industry can greatly facilitate this goal through continued refinement of this framework. Ultimately, new tools arising from advances in biology and engineering will likely improve preclinical studies in the drug development enterprise by providing disease-relevant human cells and tissues and improved in vitro and in vivo preclinical models.
Acknowledgments
A.S.K. would like to acknowledge funding from the US National Institutes of Health (Grant: T32 EB016652) and David G. Belair for helpful discussions regarding industry-relevant considerations in developing models for preclinical studies. D.J.M. would like to acknowledge funding from the US National Institutes of Health (Grants: 5U01CA214369, 1R01CA223255, and 1R01FD006589). R.J. would like to acknowledge funding from the US National Institutes of Health (Grants: R37HD045022, 1R01-NS088538, and 5R01-MH104610). Figures were generated using BioRender.com and Adobe Illustrator, both of which are licensed through the Whitehead Institute for Biomedical Research.
References
- 1.Langhans S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018;9:1–14. doi: 10.3389/fphar.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelm J.M., Lal-Nag M., Sittampalam G.S., Ferrer M. Translational in vitro research: integrating 3D drug discovery and development processes into the drug development pipeline. Drug Discov. Today. 2019;24:26–30. doi: 10.1016/j.drudis.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Tonkens R. An overview of the drug development process. Phys. Exec. 2005:48–52. [PubMed] [Google Scholar]
- 4.Tuntland T., et al. Implementation of pharmacokinetic and pharmacodynamic strategies in early research phases of drug discovery and development at Novartis institute of biomedical research. Front. Pharmacol. 2014;5:1–16. doi: 10.3389/fphar.2014.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waring M.J., et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- 6.Yildirim O., Gottwald M., Schüler P., Michel M.C. Opportunities and challenges for drug development : public – private partnerships , adaptive designs and big data. Front. Pharmacol. 2016;7:1–13. doi: 10.3389/fphar.2016.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pound P., Ritskes-Hoitinga M. Is it possible to overcome issues of external validity in preclinical animal research ? Why most animal models are bound to fail. J. Transl. Med. 2018;16:1–8. doi: 10.1186/s12967-018-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison R.K. Phase II and phase III failures : 2013–2015. Nat. Rev. Drug Discov. 2016;15:817–818. doi: 10.1038/nrd.2016.184. [DOI] [PubMed] [Google Scholar]
- 9.Epstein S.E., Luger D., Lipinski M.J. Large animal model efficacy testing is needed prior to launch of a stem cell clinical trial an evidence-lacking conclusion based on conjecture. Circ. Res. 2017;121:496–498. doi: 10.1161/CIRCRESAHA.117.311562. [DOI] [PubMed] [Google Scholar]
- 10.Mestas J., Hughes C.C.W. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 11.Whitelaw C.B.A., Sheets T.P., Lillico S.G., Telugu B.P. Engineering large animal models of human disease. J. Pathol. 2016;238:247–256. doi: 10.1002/path.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler A., Gonzalez L., Blikslager A. Large animal models: the key to translational discovery. Cell. Mol. Gastroenterol. Hepatol. 2016;2:716–724. doi: 10.1016/j.jcmgh.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein W.R., Griebel P.J. A road less travelled : large animal models in immunological research. Nat. Rev. Immunol. 2003;3:79–85. doi: 10.1038/nri977. [DOI] [PubMed] [Google Scholar]
- 14.MacGregor J.T., et al. In vitro human tissue models in risk assessment: report of a consensus-building workshop. Toxicol. Sci. 2001;59:17–36. doi: 10.1093/toxsci/59.1.17. [DOI] [PubMed] [Google Scholar]
- 15.Coleman R., Human A. Tissue in the evaluation of safety and efficacy of new medicines : a viable alternative to animal models? Int. Sch. Res. Netw. 2011;2011:1–8. doi: 10.5402/2011/806789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu N., Wang L. In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics. 2015;16:273–285. doi: 10.2217/pgs.14.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moleiro A.F., Conceição G., Leite-Moreira A.F., Rocha-Sousa A. Review article a critical analysis of the available in vitro and ex vivo methods to study retinal angiogenesis. Hindawi. 2017;2017:1–19. doi: 10.1155/2017/3034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni A., et al. Proposing advancement criteria for efficient DMPK triage of new chemical entities. Future Med. Chem. 2014;6:131–139. doi: 10.4155/fmc.13.190. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera P.V., et al. Vol. 287. 2012. High Throughput Screening for Compounds That Alter Muscle Cell Glycosylation Identifies New Role for N -Glycans in Regulating Sarcolemmal Protein Abundance and Laminin; pp. 22759–22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarecki J., et al. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening : early leads towards a therapeutic for spinal muscular atrophy. Hum. Mol. Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 21.Seyb K.I., et al. Cytotoxicity through a cell-based high-throughput screening platform. J. Biomol. Screen. 2008;13:870–878. doi: 10.1177/1087057108323909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Booth N.L., et al. A cell-based high-throughput screen to identify synergistic TRAIL sensitizers. Cancer Immunol. Immunother. 2009;58:1229–1244. doi: 10.1007/s00262-008-0637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iljin K., et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin. Cancer Res. 2009;15:6070–6079. doi: 10.1158/1078-0432.CCR-09-1035. [DOI] [PubMed] [Google Scholar]
- 24.Van Der Meer A.D., Van Den Berg A. Organs-on-chips : breaking the in vitro impasse integrative biology. Integr. Biol. 2012;4:461–470. doi: 10.1039/c2ib00176d. [DOI] [PubMed] [Google Scholar]
- 25.Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 26.Demetrius L. Of mice and men. EMBO Rep. 2005;6:S39–S44. doi: 10.1038/sj.embor.7400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight A. Animal experiments scrutinised: systematic reviews demonstrate poor human clinical and toxicological utility. ATLA- Altern. Lab. Anim. 2007;35:641–659. doi: 10.1177/026119290703500610. [DOI] [PubMed] [Google Scholar]
- 28.Edmondson R., Broglie J.J., Adcock A.F., Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birgersdotter A., Sandberg R., Ernberg I. Gene expression perturbation in vitro - a growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005;15:405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Lorsch J.R., Collins F.S., Lippincott-Schwartz J. Fixing problems with cell lines. Science. 2014;80(346):1452–1453. doi: 10.1126/science.1259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher E.M.C., Lana-Elola E., Watson S.D., Vassiliou G., Tybulewicz V.L.J. New approaches for modelling sporadic genetic disease in the mouse. Dis. Model. Mech. 2009;2:446–453. doi: 10.1242/dmm.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., et al. Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B. 2019;9:1113–1144. doi: 10.1016/j.apsb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szymański P., Markowicz M., Mikiciuk-Olasik E. Adaptation of high-throughput screening in drug discovery-toxicological screening tests. Int. J. Mol. Sci. 2012;13:427–452. doi: 10.3390/ijms13010427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang T.J., et al. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern. Med. 2016;176:1826–1833. doi: 10.1001/jamainternmed.2016.6008. [DOI] [PubMed] [Google Scholar]
- 36.Weaver R.J., Valentin J. Today’ s challenges to de-risk and predict drug safety in human “ mind-the-gap ”. Soc. Toxicol. 2019;167:307–321. doi: 10.1093/toxsci/kfy270. [DOI] [PubMed] [Google Scholar]
- 37.Skardal A., et al. Multi-tissue interactions in an integrated three-tissue organ-on-a- chip platform. Sci. Rep. 2017;7:1–16. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark M., Steger-Hartmann T. A big data approach to the concordance of the toxicity of pharmaceuticals in animals and humans. Regul. Toxicol. Pharmacol. 2018;96:94–105. doi: 10.1016/j.yrtph.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Gribkoff V.K., Kaczmarek L.K. The need for new approaches in CNS drug discovery: why drugs have failed, and what can be done to improve outcomes. Neuropharmacology. 2017;120:11–19. doi: 10.1016/j.neuropharm.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes P.J., et al. Barriers to new drug development in respiratory disease. Eur. Respir. J. 2015:1197–1207. doi: 10.1183/09031936.00007915. [DOI] [PubMed] [Google Scholar]
- 41.Boyd E.M. The acute oral toxicity of acetylsalicylic acid. Toxicol. Appl. Pharmacol. 1959;1:229–239. doi: 10.1016/0041-008x(59)90107-3. [DOI] [PubMed] [Google Scholar]
- 42.Hucker H.B. Species differences in drug metabolism. Annu. Rev. Pharmacol. 1970;10:99–118. doi: 10.1146/annurev.pa.10.040170.000531. [DOI] [PubMed] [Google Scholar]
- 43.Fontebasso Y., Dubinett S.M. Drug development for metastasis prevention. Crit. Rev. Oncog. 2015;20:449–474. doi: 10.1615/CritRevOncog.v20.i5-6.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun D.A., Wu C.J. Antigen discovery and therapeutic targeting in hematologic malignancies. Cancer J. 2017;23:115–124. doi: 10.1097/PPO.0000000000000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu C., et al. Combination of immunotherapy with targeted therapy: theory and practice in metastatic melanoma. Front. Immunol. 2019;10:1–19. doi: 10.3389/fimmu.2019.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi Y., et al. Studying cancer immunotherapy using patient-derived xenografts (PDXs ) in humanized mice. Exp. Mol. Med. 2018;50:1–9. doi: 10.1038/s12276-018-0115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khanna C., Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26:513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 48.Guerin M.V., Finisguerra V., Van Den Eynde B.J. Preclinical murine tumor models: a structural and functional perspective. Elife. 2020;9:1–24. doi: 10.7554/eLife.50740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day C., Merlino G., Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163:39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerby B., et al. High-throughput screening in niche-based assay identifies compounds to target preleukemic stem cells. J. Clin. Invest. 2016;126:4569–4584. doi: 10.1172/JCI86489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gopinathan A., Tuveson D.A. The use of GEM models for experimental cancer therapeutics the use of xenografts for preclinical testing. Dis. Model. Mech. 2008;1:83–86. doi: 10.1242/dmm.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J., Rho J., Devkota S., Sung Y.H., Lee H. Developing genetically engineered mouse models using engineered nucleases: current status , challenges , and the way forward. Drug Discov. Today Dis. Model. 2016;20:13–20. [Google Scholar]
- 53.Go L., Tracey N., Ma R., Qian B., Brunton V.G. Mouse models of metastasis: progress and prospects. Dis. Model. Mech. 2017:1061–1074. doi: 10.1242/dmm.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richmond A., Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. 2Dis. Mod. Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-david U., Beroukhim R., Golub T.R. Genomic evolution of cancer models: perils and opportunities. Nat. Rev. Cancer. 2019;19:97–109. doi: 10.1038/s41568-018-0095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bays H.E., Chapman R.H., Grandy S. The relationship of body mass index to diabetes mellitus, hypertension and dyslipidaemia: comparison of data from two national surveys. Int. J. Clin. Pract. 2007;61:737–747. doi: 10.1111/j.1742-1241.2007.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khaodhiar L., McCowen K.C., Blackburn G.L. Obesity and its comorbid conditions. Clin. Cornerstone. 1999;2:17–31. doi: 10.1016/s1098-3597(99)90002-9. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen T.H., Nguyen T.-N., Fischer T., Ha W., Tran T.V. Type 2 diabetes among Asian Americans: prevalence and prevention. World J. Diabetes. 2015;6:543–547. doi: 10.4239/wjd.v6.i4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorisky A. Effect of high glucose levels on white adipose cells and adipokines—fuel for the fire. Int. J. Mol. Sci. 2017;18:1–6. doi: 10.3390/ijms18050944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunmore S.J., Brown J.E.P. The role of adipokines in β-cell failure of type 2 diabetes. J. Endocrinol. 2013;216:T37–T45. doi: 10.1530/JOE-12-0278. [DOI] [PubMed] [Google Scholar]
- 61.King A.J.F. The use of animal models in diabetes research. Br. J. Pharmacol. 2012;166:877–894. doi: 10.1111/j.1476-5381.2012.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jansen H.T., et al. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun. Biol. 2019;2:1–10. doi: 10.1038/s42003-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hellgren E.C. Physiology of hibernation in bears author. A Sel. Pap.Tenth Int. Conf. Bear Res. Manag. 1998;10:467–477. [Google Scholar]
- 64.Meza-Perez S., Randall T.D. Immunological functions of the omentum. Trends Immunol. 2017;38:526–536. doi: 10.1016/j.it.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banerji M.A., Faridi N., Atluri R., Chaiken R.L., Lebovitz H.E. Body composition, visceral fat, leptin, and insulin resistance in Asian Indian men*. J. Clin. Endocrinol. Metab. 1999;84:137–144. doi: 10.1210/jcem.84.1.5371. [DOI] [PubMed] [Google Scholar]
- 66.Fukuda T., et al. Ratio of visceral-to-subcutaneous fat area predicts cardiovascular events in patients with type 2 diabetes. J. Diab. Investig. 2018;9:396–402. doi: 10.1111/jdi.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oyston P., Robinson K. The current challenges for vaccine development. J. Med. Microbiol. 2012:889–894. doi: 10.1099/jmm.0.039180-0. [DOI] [PubMed] [Google Scholar]
- 68.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020:1–5. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 69.Gerdts V., et al. Large animal models for vaccine development and testing. ILAR J. 2015;56:53–62. doi: 10.1093/ilar/ilv009. [DOI] [PubMed] [Google Scholar]
- 70.Farkas A.M., Marvel D.M., Finn O.J. Antigen choice determines vaccine-induced generation of immunogenic versus tolerogenic DC that are marked by differential expression of pancreatic enzymes. J. Immunol. 2013;190:3319–3327. doi: 10.4049/jimmunol.1203321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makkouk A., Weiner G.J. Cancer immunotherapy and breaking immune tolerance: new approaches to an old challenge. Cancer Res. 2015;75:5–11. doi: 10.1158/0008-5472.CAN-14-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fucikova J., Palova-jelinkova L., Bartunkova J., Spisek R. Induction of tolerance and immunity by dendritic cells: mechanisms and clinical applications. Front. Immunol. 2019;10:1–17. doi: 10.3389/fimmu.2019.02393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beverley P.C.L. Immunology of vaccination. Br. Med. Bull. 2002:15–28. doi: 10.1093/bmb/62.1.15. [DOI] [PubMed] [Google Scholar]
- 74.Acosta P.L., Caballero M.T., Polack F.P. Brief history and characterization of enhanced respiratory syncytial virus disease. Clin. Vaccine Immunol. 2016;23:189–195. doi: 10.1128/CVI.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muralidharan A., Li C., Wang L., Li X. Expert review of vaccines immunopathogenesis associated with formaldehyde-inactivated RSV vaccine in preclinical and clinical studies preclinical and clinical studies. Exp. Rev. Vacc. 2017;16:351–360. doi: 10.1080/14760584.2017.1260452. [DOI] [PubMed] [Google Scholar]
- 76.Chirkova T., et al. In vitro model for the assessment of human immune responses to subunit RSV vaccines. PLoS One. 2020:1–19. doi: 10.1371/journal.pone.0229660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zablotsky B., Black L.I., Blumberg S.J. Estimated prevalence of children with diagnosed developmental disabilities in the United States, 2014-2016. NCHS Data Brief. 2017;291:1–8. [PubMed] [Google Scholar]
- 78.Arth A.C., et al. Inpatient hospitalization costs associated with birth defects among persons of all ages — United States , 2013. CDC Morb. Mortal. Wkly. Rep. 2017;66:43–65. doi: 10.15585/mmwr.mm6602a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olusanya B.O., et al. Developmental disabilities among children younger than 5 years in 195 countries and territories, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Glob. Health. 2018;6:e1100–e1121. doi: 10.1016/S2214-109X(18)30309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grason H.A., Misra D.P. Reducing exposure to environmental toxicants before birth: moving from risk perception to risk reduction. Public Health Rep. 2009;124:629–641. doi: 10.1177/003335490912400505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boland M.R., Polubriaginof F., Tatonetti N.P. Development of a machine learning algorithm to classify drugs of unknown fetal effect. Sci. Rep. 2017;7:1–15. doi: 10.1038/s41598-017-12943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ayad M. Epidemiology of medications use in pregnancy Martina. Semin. Perinatol. 2018;39:508–511. doi: 10.1053/j.semperi.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller M.T. Thalidomide embryopathy: a model for the study of congenital incomitant horizontal strabismus. Trans. Am. Ophthalmol. Soc. 1991;89:623–674. [PMC free article] [PubMed] [Google Scholar]
- 84.Neltner T.G., Alger H.M., Leonard J.E., Maffini M.V. Data gaps in toxicity testing of chemicals allowed in food in the United States. Reprod. Toxicol. 2013;42:85–94. doi: 10.1016/j.reprotox.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 85.Zaganjor I., et al. Describing the prevalence of neural tube defects worldwide: a systematic literature review. PLoS One. 2016;11:1–31. doi: 10.1371/journal.pone.0151586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitchell L.E. Epidemiology of neural tube defects. Am. J. Med. Genet. - Semin. Med. Genet. 2005;135C:88–94. doi: 10.1002/ajmg.c.30057. [DOI] [PubMed] [Google Scholar]
- 87.Crider K.S., Bailey L.B., Berry R.J. Folic acid food fortification—its history, effect, concerns, and future directions. Nutrients. 2011;3:370–384. doi: 10.3390/nu3030370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turner M.A., Kenny L., Alfirevic Z. Challenges in designing clinical trials to test new drugs in the pregnant woman and fetus. Clin. Perinatol. 2019;46:399–416. doi: 10.1016/j.clp.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 89.Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 90.Gitler A.D., Dhillon P., Shorter J. Neurodegenerative disease: models , mechanisms , and a new hope. Dis. Model. Mech. 2017;10:499–502. doi: 10.1242/dmm.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henstridge C.M., Spires-jones T.L. Modeling Alzheimer’s disease brains in vitro. Nat. Neurosci. 2018;21:897–902. doi: 10.1038/s41593-018-0177-2. [DOI] [PubMed] [Google Scholar]
- 92.Alavijeh M.S., Chishty M., Qaiser M.Z., Palmer A.M. Drug metabolism and pharmacokinetics , the blood-brain barrier , and central nervous system drug discovery. NeuroRX. 2005;2:554–571. doi: 10.1602/neurorx.2.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ugarte A., Corbacho D., Aymerich M.S., García-Osta A., Cuadrado-Tejedor M. Impact of neurodegenerative diseases on drug binding to brain tissues: from animal models to Human samples. Neurotherapeutics. 2018;15:742–750. doi: 10.1007/s13311-018-0624-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hemonnot A.-L., Hua J., Ulmann L., Hirbec H. Microglia in Alzheimer disease: well-known targets and new opportunities. Front. Aging Neurosci. 2019;11:1–20. doi: 10.3389/fnagi.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parodi-Rullan R., Sone J.Y., Fossati S. Endothelial mitochondrial dysfunction in cerebral amyloid angiopathy and Alzheimer’s disease. J. Alzheimers Dis. 2019;72:1019–1039. doi: 10.3233/JAD-190357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drummond E., Wisniewski T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol. 2017;133:155–175. doi: 10.1007/s00401-016-1662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eglen R., Reisine T. Primary cells and stem cells in drug discovery: emerging tools for high-throughput screening. Assay. 2011:108–124. doi: 10.1089/adt.2010.0305. [DOI] [PubMed] [Google Scholar]
- 98.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maqsood M.I., Matin M.M., Bahrami A.R., Ghasroldasht M.M. Immortality of cell lines: challenges and advantages of establishment. Cell Biol. Int. 2013;37:1038–1045. doi: 10.1002/cbin.10137. [DOI] [PubMed] [Google Scholar]
- 100.Jack J., Rotroff D., Motsinger-Reif A. Cell lines models of drug response: successes and lessons from this pharmacogenomic model. Curr. Mol. Med. 2014;14:833–840. doi: 10.2174/1566524014666140811113946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wenger S.L., et al. Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci. Rep. 2005;24:631–639. doi: 10.1007/s10540-005-2797-5. [DOI] [PubMed] [Google Scholar]
- 102.Dornbos P., LaPres J.J. Incorporating population-level genetic variability within laboratory models in toxicology: from the individual to the population. Toxicology. 2018:1–8. doi: 10.1016/j.tox.2017.12.007. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soldatow V.Y., Lecluyse E.L., Griffith L.G., Rusyn I. In vitro models for liver toxicity testing. Toxicol. Res. (Camb.) 2013;2:23–39. doi: 10.1039/C2TX20051A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hughes P., Marshall D., Reid Y., Parkes H., Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007;43:575–586. doi: 10.2144/000112598. [DOI] [PubMed] [Google Scholar]
- 105.Li Z. Comprehensive Biotechnology. Second ed. vol. 5. 2011. In vitro micro-tissue and -organ models for toxicity testing; pp. 551–563. [Google Scholar]
- 106.Bell C.C., et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y., et al. Effects of the combination of TRC105 and bevacizumab on endothelial cell biology. Investig. New Drugs. 2014;32:851–859. doi: 10.1007/s10637-014-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klingelhutz A.J., et al. Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-19024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ha M.K., et al. Mass cytometric study on the heterogeneity in cellular association and cytotoxicity of silver nanoparticles in primary human immune cells. Environ. Sci. Nano. 2020;7:1102–1114. [Google Scholar]
- 110.Ogese M.O., et al. Characterization of drug-specific signaling between primary human hepatocytes and immune cells. Soc. Toxicol. 2017;158:76–89. doi: 10.1093/toxsci/kfx069. [DOI] [PubMed] [Google Scholar]
- 111.Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics. 2009;8:443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mehta A., et al. Intrinsic hepatocyte dedifferentiation is accompanied by upregulation of mesenchymal markers , protein sialylation and core alpha 1,6 linked fucosylation. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aust L., et al. Yield of human adipose-derived adult stem cells from liposuction aspirates. Cytotherapy. 2004;6:7–14. doi: 10.1080/14653240310004539. [DOI] [PubMed] [Google Scholar]
- 114.Yu H., et al. Isolation of a novel population of multipotent adult stem cells from human hair follicles. Am. J. Pathol. 2006;168:1879–1888. doi: 10.2353/ajpath.2006.051170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sato T., et al. Single Lgr5 stem cells build crypt – villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–266. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 116.Thomson J.A., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;80(282):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 117.Takahashi K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 118.Yu J., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;80(318):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 119.Meissner A., Wernig M., Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 120.Baxter M., et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ribeiro M.C., et al. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro - correlation between contraction force and electrophysiology. Biomaterials. 2015;51:138–150. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 122.Achilli T.-M., Meyer J., Morgan J.R. Advances in the formation, use and understanding of multi-cellular spheroids. Expert. Opin. Biol. Ther. 2012;12:1347–1360. doi: 10.1517/14712598.2012.707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yamada K.M., Cukierman E. Modeling tissue morphogenesis and Cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 124.Shamir E.R., Ewald A.J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014;15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pampaloni F., Reynaud E.G., Stelzer E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 126.Nelson C.M., Bissell M.J. Modeling dynamic reciprocity: engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin. Cancer Biol. 2005;15:342–352. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simian M., Bissell M.J. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 129.Leong B.P., Chan K.W. Scaffolding in tissue engineering : general approaches and tissue-specific considerations. Eur. Spine J. 2008;17:S467–S479. doi: 10.1007/s00586-008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wade R.J., Burdick J.A. Engineering ECM signals into biomaterials. Mater. Today. 2012;15:454–459. [Google Scholar]
- 131.Khalil A.S., Xie A.W., Murphy W.L. Context clues: the importance of stem cell-material interactions. ACS Chem. Biol. 2013 doi: 10.1021/cb400801m. SI, A-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ghaffarian R., Muro S. Models and methods to evaluate transport of drug delivery systems across cellular barriers. J. Vis. Exp. JOVE. 2013;80:1–12. doi: 10.3791/50638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Drury J.L., Mooney D.J. Hydrogels for tissue engineering : scaffold design variables and applications. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 134.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 135.Huebsch N., et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chaudhuri O., et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xie A.W., et al. Controlled self-assembly of stem cell aggregates instructs pluripotency and lineage bias. Sci. Rep. 2017;7:1–15. doi: 10.1038/s41598-017-14325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Catoira M.C., Fusaro L., Di D., Martina F., Francesca R. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci. Mater. Med. 2019;30:1–10. doi: 10.1007/s10856-019-6318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 140.Lutolf M.P., Hubbell J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 141.Aisenbrey E.A., Murphy W.L. Synthetic alternatives to matrigel. Nat. Rev. Mater. 2020:1–13. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shafiee A., Atala A. Tissue engineering: toward a new era of medicine. Annu. Rev. Med. 2017;68:29–40. doi: 10.1146/annurev-med-102715-092331. [DOI] [PubMed] [Google Scholar]
- 143.Langer R., Vacanti J.P. Tissue engineering. Science. 1993;80(260):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 144.Yamada K.M., et al. Extracellular matrix dynamics in cell migration, invasion and tissue morphogenesis. Int. J. Exp. Pathol. 2019;100:144–152. doi: 10.1111/iep.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Auger F.A., Gibot L., Lacroix D. The pivotal role of vascularization in tissue engineering franc. Annu. Rev. Biomed. Eng. 2013;15:177–202. doi: 10.1146/annurev-bioeng-071812-152428. [DOI] [PubMed] [Google Scholar]
- 146.Das S., et al. Innervation: the missing link for biofabricated tissues and organs. NPJ Regen. Med. 2020;5:1–19. doi: 10.1038/s41536-020-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rossello R.A., Kohn D.H. Cell communication and tissue engineering. Commun. Integr. Biol. 2010;106:53–56. doi: 10.4161/cib.3.1.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Giladi A., et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 2020;38:629–637. doi: 10.1038/s41587-020-0442-2. [DOI] [PubMed] [Google Scholar]
- 149.Garden G.A., La Spada A.R. Intercellular (Mis)communication in neurodegenerative disease. Neuron. 2012;73:886–901. doi: 10.1016/j.neuron.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sarkar U., et al. Metabolite profiling and pharmacokinetic evaluation of hydrocortisone in a perfused three-dimensional human liver bioreactor. Drug Metab. Dispos. 2015;43:1091–1099. doi: 10.1124/dmd.115.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ren X., et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020;0:1–16. doi: 10.1038/s41422-020-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xie H., et al. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci. Adv. 2020;6:1–17. doi: 10.1126/sciadv.aay5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van Velthoven C.T.J., de Morree A., Egner I.M., Brett J.O., Rando T.A. Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 2017;21:1994–2004. doi: 10.1016/j.celrep.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mammoto T., Ingber D.E. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lecuit T., Lenne P.-F., Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- 156.Jones H.M., Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. Pharm. Syst. Pharmacol. 2013;2:1–12. doi: 10.1038/psp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Negus S.S., Banks M.L. Pharmacokinetic—Pharmacodynamic (PKPD) analysis with drug discrimination. Curr. Top. Behav. Neurosci. 2018;39:245–259. doi: 10.1007/7854_2016_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides ☆. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 159.Mateus A., Treyer A., Wegler C., Karlgren M., Matsson P. Intracellular drug bioavailability: a new predictor of system dependent drug disposition. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sailon A.M., et al. A novel flow-perfusion bioreactor supports 3D dynamic cell culture. J. Biomed. Biotechnol. 2009;2009:1–7. doi: 10.1155/2009/873816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ebrahimkhani M.R., Neiman J.A.S., Raredon M.S.B., Hughes D.J., Griffith L.G. Bioreactor technologies to support liver function in vitro. Adv. Drug Deliv. Rev. 2014;69–70:132–157. doi: 10.1016/j.addr.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Domansky K., et al. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2014;10:51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu J., et al. Quantitative systems pharmacology approaches applied to microphysiological systems (MPS): data interpretation and multi-MPS integration. CPT Pharmacometrics Syst. Pharmacol. 2015;4:585–594. doi: 10.1002/psp4.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.D’Costa K., et al. Biomaterials and culture systems for development of organoid and organ-on-a-chip models. Ann. Biomed. Eng. 2020:1–26. doi: 10.1007/s10439-020-02498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Varma H., Lo D.C., Stockwell B.R. High throughput screening for Neurodegeneration and complex disease phenotypes. Comb. Chem. High Throughput Screen. 2008;11:238–248. doi: 10.2174/138620708783877753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zanoni M., et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Nat. Publ. Gr. 2016;6:1–11. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muffat J., et al. Efficient derivation of microglia-like cells from human pluripotent stem cells HHS public access author manuscript. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Millman J.R., et al. Generation of stem cell-derived b-cells from patients with type 1 diabetes. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ahfeldt T., et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell Biol. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Montel-Hagen A., et al. Organoid-induced differentiation of conventional T cells from Human pluripotent stem cells. Cell Stem Cell. 2019;24:1–14. doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhou Y., et al. Overexpression of GATA2 enhances development and maintenance of human embryonic stem cell-derived hematopoietic stem cell-like progenitors. Stem Cell Rep. 2019;13:31–47. doi: 10.1016/j.stemcr.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hu W., et al. Patient adipose stem cell-derived adipocytes reveal genetic variation that predicts antidiabetic drug response. Cell Stem Cell. 2019;24:299–308. doi: 10.1016/j.stem.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lippmann E.S., et al. Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Rep. 2015;4:632–644. doi: 10.1016/j.stemcr.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Stem Cells. 2017;21:179–194. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 175.Kadota S., Pabon L., Reinecke H., Murry C.E. In vivo maturation of human induced pluripotent stem cell-derived cardiomyocytes in neonatal and adult rat hearts. Stem Cell Rep. 2017;8:278–289. doi: 10.1016/j.stemcr.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pepper A.R., et al. Posttransplant characterization of long-term functional hESC-derived pancreatic endoderm grafts. Diabetes. 2019;68:953–962. doi: 10.2337/db18-0788. [DOI] [PubMed] [Google Scholar]
- 177.Rezania A., et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tiemeier G.L., et al. Closing the mitochondrial permeability transition pore in hiPSC-derived endothelial cells induces glycocalyx formation and functional maturation. Stem Cell Rep. 2019;13:803–816. doi: 10.1016/j.stemcr.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hu D., et al. Metabolic maturation of human pluripotent stem cell derived cardiomyocytes by inhibition of HIF1α and LDHA. Circ. Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Begley D.J. Delivery of therapeutic agents to the central nervous system: the problems and the possibilities. Pharmacol. Ther. 2004;104:29–45. doi: 10.1016/j.pharmthera.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 181.Onat D., Brillon D., Colombo P.C., Schmidt A.M. Human vascular endothelial cells: a model system for studying vascular inflammation in diabetes and atherosclerosis. Curr. Diab.Rep. 2011;11:193–202. doi: 10.1007/s11892-011-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Qian T., et al. Directed differentiation of human pluripotent stem cells to podocytes under defined conditions. Sci. Adv. 2017;3:1–12. doi: 10.1038/s41598-019-39504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Stebbins M.J., et al. Human pluripotent stem cell–derived brain pericyte–like cells induce blood-brain barrier properties. Sci. Adv. 2019;5:1–16. doi: 10.1126/sciadv.aau7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cheung A.S., Zhang D.K.Y., Koshy S.T., Mooney D.J. Scaffolds that mimic antigen-presenting cells enable ex vivo expansion of primary T-cellsexpansion of primary T-cells. Nat. Biotechnol. 2018;36:160–169. doi: 10.1038/nbt.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Smith C., et al. Ex vivo expansion of human T cells for adoptive immunotherapy using the novel Xeno-free CTS immune cell serum replacement. Clin. Transl. Immunol. 2015;4:1–10. doi: 10.1038/cti.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Steenblock E.R., Fahmy T.M. A comprehensive platform for ex vivo T-cell expansion based on biodegradable polymeric artificial antigen-presenting cells. Mol. Ther. 2008;16:765–772. doi: 10.1038/mt.2008.11. [DOI] [PubMed] [Google Scholar]
- 187.Meyer R.A., et al. Biodegradable Nanoellipsoidal artificial antigen presenting cells for antigen specific T-cell activation. Small. 2015;11:1519–1525. doi: 10.1002/smll.201402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fadel T.R., et al. A carbon nanotube-polymer composite for T-cell therapy. Nat. Nanotechnol. 2014;9:639–647. doi: 10.1038/nnano.2014.154. [DOI] [PubMed] [Google Scholar]
- 189.Fedorova V., et al. Differentiation of neural rosettes from human pluripotent stem cells in vitro is sequentially regulated on a molecular level and accomplished by the mechanism reminiscent of secondary neurulation. Stem Cell Res. 2019;40:1–10. doi: 10.1016/j.scr.2019.101563. [DOI] [PubMed] [Google Scholar]
- 190.Valensisi C., et al. Epigenomic landscapes of hESC-derived neural rosettes : modeling neural tube formation and diseases. Cell Rep. 2017;20:1448–1462. doi: 10.1016/j.celrep.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sahakyan V., et al. Folic acid exposure rescues spina bifida aperta phenotypes in human induced pluripotent stem cell model. Sci. Rep. 2018;8:1–19. doi: 10.1038/s41598-018-21103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kameoka S., Babiarz J., Kolaja K., Chiao E. A high-throughput screen for teratogens using Human pluripotent stem cells. Toxicol. Sci. 2013;137:76–90. doi: 10.1093/toxsci/kft239. [DOI] [PubMed] [Google Scholar]
- 193.Belair D.G., et al. Thalidomide inhibits human iPSC mesendoderm differentiation by modulating CRBN-dependent degradation of SALL4. Sci. Rep. 2020;10:1–18. doi: 10.1038/s41598-020-59542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Knight G.T., et al. Engineering induction of singular neural rosette emergence within hPSC-derived tissues. Elife. 2018;7:1–23. doi: 10.7554/eLife.37549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nguyen E.H., et al. Versatile synthetic alternatives to Matrigel for vascular toxicity screening and stem cell expansion. Nat. Biomed. Eng. 2017;1:1–34. doi: 10.1038/s41551-017-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tang X., et al. KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. PNAS. 2016;113:751–756. doi: 10.1073/pnas.1524013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tang X., et al. Pharmacological enhancement of KCC2 gene expression exerts therapeutic effects on human Rett syndrome neurons and Mecp2 mutant mice. Sci. Transl. Med. 2019:1–14. doi: 10.1126/scitranslmed.aau0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Darville H., et al. Human pluripotent stem cell-derived cortical neurons for high throughput medication screening in autism: a proof of concept study in SHANK3 haploinsufficiency syndrome. EBioMedicine. 2016;9:293–305. doi: 10.1016/j.ebiom.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Eiraku M., et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 200.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;80(345):283–293. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 201.Eiraku M., et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 202.Li M., Izpisua Belmonte J.C. Organoids — preclinical models of human disease. N. Engl. J. Med. 2019;380:569–579. doi: 10.1056/NEJMra1806175. [DOI] [PubMed] [Google Scholar]
- 203.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 204.Huch M., et al. Article long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Huch M., et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5 / R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Takasato M., et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564585. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 207.Karzbrun E., Kshirsagar A., Cohen S.R., Hanna J.H., Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shi R., et al. Organoid cultures as preclinical models of non – small cell lung cancer. Clin. Cancer Res. 2020;26:1162–1174. doi: 10.1158/1078-0432.CCR-19-1376. [DOI] [PubMed] [Google Scholar]
- 209.Sahin U. Studying tumor-reactive T cells: a personalized organoid model Ugur. Cell Stem Cell. 2018;23:318–319. doi: 10.1016/j.stem.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 210.Porras A.M., Westlund J.A., Evans A.D., Masters K.S. Creation of disease-inspired biomaterial environments to mimic pathological events in early calcific aortic valve disease. Proc. Natl. Acad. Sci. U. S. A. 2017:363–371. doi: 10.1073/pnas.1704637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Berger A.J., Linsmeier K., Kreeger P.K., Masters K.S. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin- methacrylate and collagen. Biomaterials. 2018;141:125–135. doi: 10.1016/j.biomaterials.2017.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chaudhuri O., et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 213.Nam S., Stowers R., Lou J., Xia Y., Chaudhuri O. Varying PEG density to control stress relaxation in alginate-PEG hydrogels for 3D cell culture studies. Biomaterials. 2019;200:15–24. doi: 10.1016/j.biomaterials.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Karimpoor M., et al. Alginate foam-based three-dimensional culture to investigate drug sensitivity in primary leukaemia cells. J. R. Soc. Interface. 2018;15:1–8. doi: 10.1098/rsif.2017.0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kleinman H.K., Philp D., Hoffman M.P. Role of the extracellular matrix in morphogenesis. Curr. Opin. Botechnol. 2003;14:526–532. doi: 10.1016/j.copbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 216.Daley W.P., Yamada K.M. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr. Opin. Genet. Dev. 2013;23:408–414. doi: 10.1016/j.gde.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kniazeva E., Putnam A.J. Endothelial cell traction and ECM density influence both capillary morphogenesis and maintenance in 3-D. Am. J. Cell Phsyiol. 2020;297:179–187. doi: 10.1152/ajpcell.00018.2009. [DOI] [PubMed] [Google Scholar]
- 218.Nguyen A.H., Wang Y., White D.E., Platt M.O., Mcdevitt T.C. Biomaterials MMP-mediated mesenchymal morphogenesis of pluripotent stem cell aggregates stimulated by gelatin methacrylate microparticle incorporation. Biomaterials. 2016;76:66–75. doi: 10.1016/j.biomaterials.2015.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu X.S., et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Perez-Pinera P., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sanchez-Schmitz G., et al. Microphysiologic human tissue constructs reproduce autologous age-specific BCG and HBV primary immunization in vitro. Front. Immunol. 2018;9:1–19. doi: 10.3389/fimmu.2018.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Edington C.D., et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 2018;8:1–18. doi: 10.1038/s41598-018-22749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hubatsch I., Ragnarsson E.G.E., Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat. Protoc. 2007;2:2111–2119. doi: 10.1038/nprot.2007.303. [DOI] [PubMed] [Google Scholar]
- 224.Canfield S.G., et al. An isogenic blood–brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J. Neurochem. 2017;140:874–888. doi: 10.1111/jnc.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.van Duinen V., et al. Perfused 3D angiogenic sprouting in a high-throughput in vitro platform. Angiogenesis. 2018:1–9. doi: 10.1007/s10456-018-9647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 227.Hui E.E., Bhatia S.N. Microscale control of cell contact and spacing via three-component surface patterning. Langmuir. 2007;23:4103–4107. doi: 10.1021/la0630559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sung K.E., et al. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials. 2009;30:4833–4841. doi: 10.1016/j.biomaterials.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Fabre K.M., Livingston C., Tagle D.A. Organs-on-chips (microphysiological systems): tools to expedite efficacy and toxicity testing in human tissue. Exp. Biol. Med. 2014;239:1073–1077. doi: 10.1177/1535370214538916. [DOI] [PubMed] [Google Scholar]
- 230.Ewart L., et al. Navigating tissue chips from development to dissemination: a pharmaceutical industry perspective. Exp. Biol. Med. 2017;242:1579–1585. doi: 10.1177/1535370217715441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Whitesides G.M. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 232.Livingston C.A., Fabre K.M., Tagle D.A. Facilitating the commercialization and use of organ platforms generated by the microphysiological systems (tissue chip) program through public – private partnerships. Comput. Struct. Biotechnol. J. 2016;14:207–210. doi: 10.1016/j.csbj.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Du Y., et al. A bile duct-on-a-chip with organ-level functions. Hepatology. 2020;71:1350–1363. doi: 10.1002/hep.30918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Loskill P., et al. WAT-on-a-chip: a physiologically relevant microfluidic system incorporating white adipose tissue. Lab Chip. 2018;17:1645–1654. doi: 10.1039/c6lc01590e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nguyen D.-H.T., et al. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019;5:1–9. doi: 10.1126/sciadv.aav6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jeon J.S., et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. PNAS. 2014;112:1–7. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu Y., et al. Adipose-on-a-chip: a dynamic microphysiological in vitro model of the human adipose for immune- metabolic analysis in type II diabetes. Lab Chip. 2019;19:241–253. doi: 10.1039/c8lc00481a. [DOI] [PubMed] [Google Scholar]
- 238.Ferrarini M., et al. Ex-vivo dynamic 3-D culture of human tissues in the RCCS TM bioreactor allows the study of multiple myeloma biology and response to therapy. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Convery N., Gadegaard N. 30 years of microfluidics. Micro Nano Eng. 2019;2:76–91. [Google Scholar]
- 240.Lin N.Y.C., et al. Renal reabsorption in 3D vascularized proximal tubule models. PNAS. 2019;116:5399–5404. doi: 10.1073/pnas.1815208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gómez-Lechón M.J., Tolosa L. Human hepatocytes derived from pluripotent stem cells: a promising cell model for drug hepatotoxicity screening. Arch. Toxicol. 2016;90:2049–2061. doi: 10.1007/s00204-016-1756-1. [DOI] [PubMed] [Google Scholar]
- 242.Richardson S.J., Bai A., Kulkarni A.A., Moghaddam M.F. Efficiency in drug discovery: liver S9 fraction assay as a screen for metabolic stability. Drug Metab. Lett. 2016;10:83–90. doi: 10.2174/1872312810666160223121836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jia L., Liu X. The conduct of drug metabolism studies considered good practice (II): in vitro experiments. Curr. Drug Metab. 2007;8:822–829. doi: 10.2174/138920007782798207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Homan K.A., et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ardalani H., et al. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes q. Acta Biomater. 2019;95:371–381. doi: 10.1016/j.actbio.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 246.Ma X., et al. 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling. Adv. Drug Deliv. Rev. 2018:235–251. doi: 10.1016/j.addr.2018.06.011. July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Miller J.S., et al. Rapid casting of patterned vascular networks for perfusable engineered 3D tissues. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sun W., et al. The bioprinting roadmap. Biofabrication. 2020;12:1–33. doi: 10.1088/1758-5090/ab5158. [DOI] [PubMed] [Google Scholar]
- 249.Skylar-Scott M.A., et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019;5:1–13. doi: 10.1126/sciadv.aaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Skylar-scott M.A., Mueller J., Visser C.W., Lewis J.A. Voxelated soft matter via multimaterial multinozzle 3D printing. Nature. 2019;575:330–349. doi: 10.1038/s41586-019-1736-8. [DOI] [PubMed] [Google Scholar]
- 251.Greco Song H.-H., Rumma R.T., Ozaki C.K., Edelman E.R., Chen C.S. Vascular tissue engineering: progress, challenges, and clinical promise. Cell Stem Cell. 2018;22:340–354. doi: 10.1016/j.stem.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wikswo J.P. The relevance and potential roles of microphysiological systems in biology and medicine. Exp. Biol. Med. 2014;239:1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Marx U., et al. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX. 2020;37:1–30. doi: 10.14573/altex.2001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Huh D., et al. Reconstituting organ-level lung functions on a chip. Science. 2010;80(328):1662–1669. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Park S.E., Georgescu A., Huh D. Organoids-on-a-chip. Science. 2019;80(365):960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Achberger K., et al. Merging organoid and organ-on-a-chip technology to generate complex multi- layer tissue models in a human retina-on- a-chip platform. Elife. 2019;8:1–26. doi: 10.7554/eLife.46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Smith M.P., et al. Targeting endothelin receptor signalling overcomes heterogeneity driven therapy failure. EMBO Mol. Med. 2017;9:1011–1029. doi: 10.15252/emmm.201607156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhou M., et al. Estrogen receptor beta enhances chemotherapy response of GBM cells by down regulating DNA damage response pathways. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-42313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tsamandouras N., et al. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS J. 2017;19:1499–1512. doi: 10.1208/s12248-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ronaldson-Bouchard K., Vunjak-Novakovic G. Review organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22:310–324. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wikswo J.P., et al. Scaling and systems biology for integrating multiple organs-on- a-chip. Lab Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Novak R., et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 2020;4:407–420. doi: 10.1038/s41551-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Herland A., et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020;4:421–436. doi: 10.1038/s41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vizcardo R., et al. Generation of tumor antigen-specific iPSC-derived thymic emigrants using a 3D Thymic culture system. Cell Rep. 2018;22:3175–3190. doi: 10.1016/j.celrep.2018.02.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pearson T., Greiner D.L., Shultz L.D. Creation of “humanized” mice to study human immunity. Curr. Protoc. Immunol. 2008;2 doi: 10.1002/0471142735.im1521s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Svoboda D.S., et al. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2019;116:25293–25303. doi: 10.1073/pnas.1913541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hasselmann J., et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019;103:1016–1033. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ferreira S.A., Romero-Ramos M. Microglia response during Parkinson’s disease: alpha-Synuclein intervention. Front. Cell. Neurosci. 2018;12:1–17. doi: 10.3389/fncel.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Guerrero B.L., Sicotte N.L. Microglia in multiple sclerosis: friend or foe ? Front. Cell. Neurosci. 2020;11:1–8. doi: 10.3389/fimmu.2020.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bissig K.-D., et al. P450-humanized and human liver chimeric mouse models for studying xenobiotic metabolism and toxicity. Drug Metab. Dispos. 2018;46:1734–1744. doi: 10.1124/dmd.118.083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cohen M.A., Markoulaki S., Jaenisch R. Matched developmental timing of donor cells with the host is crucial for chimera formation. Stem Cell Rep. 2018;10:1445–1452. doi: 10.1016/j.stemcr.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cohen M.A., et al. Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proc. Natl. Acad. Sci. U. S. A. 2015:1–6. doi: 10.1073/pnas.1525518113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cohen M.A., et al. Formation of human neuroblastoma in mouse-human neural crest chimeras. Cell Stem Cell. 2020;26:579–592. doi: 10.1016/j.stem.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Marsano A., et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip. 2016;16:599–610. doi: 10.1039/c5lc01356a. [DOI] [PubMed] [Google Scholar]
- 275.Yuan H., et al. Instrumented cardiac microphysiological devices via multi-material 3D printing. Nat. Mater. 2017;16:303–308. doi: 10.1038/nmat4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Tourovskaia A., Fauver M., Kramer G., Simonson S., Neumann T. Brief communication: tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med. 2015;239:1264–1271. doi: 10.1177/1535370214539228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Knowlton S., Tasoglu S. A bioprinted liver-on-a-chip for drug screening applications. Trends Biotechnol. 2016;34:681–682. doi: 10.1016/j.tibtech.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 278.Schepers A., Li C., Chhabra A., Tschudy Seney B., Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip. 2016;16:2644–2653. doi: 10.1039/c6lc00598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tsamandouras N., et al. Quantitative assessment of population variability in hepatic drug metabolism using a perfused three-dimensional human liver microphysiological system. J. Pharmacol. Exp. Ther. 2017;360:95–105. doi: 10.1124/jpet.116.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Derda R., et al. High-throughput discovery of synthetic surfaces that support proliferation of pluripotent cells. J. Am. Chem. Soc. 2010;132:1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Weber E.J., et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90:627–637. doi: 10.1016/j.kint.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wevers N.R., et al. A perfused human blood-brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS. 2018;15:1–12. doi: 10.1186/s12987-018-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wang Y., Wang L., Zhu Y., Qin J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip. 2018;18:851–860. doi: 10.1039/c7lc01084b. [DOI] [PubMed] [Google Scholar]
- 284.Maoz B.M., et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 2018;36:865–877. doi: 10.1038/nbt.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rogal J., et al. WAT-on-a-chip integrating human mature white adipocytes for mechanistic research and pharmaceutical applications. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-63710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zbinden A., et al. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 2020;85–86:205–220. doi: 10.1016/j.matbio.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 287.Hassell B.A., et al. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 288.Albanese A., Lam A.K., Sykes E.A., Rocheleau J.V., Chan W.C.W. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 2013:1–8. doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Akay M., et al. Drug screening of human GBM spheroids in brain cancer chip. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-33641-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Chen W.L.K., et al. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol. Bioeng. 2017;114:2648–2659. doi: 10.1002/bit.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Maschmeyer I., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 292.Shrike Y., et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 2017:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Oleaga C., et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016;6:1–17. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]