Significance
Strong evidence supports the tumor immune landscape as a determinant of patient responses to immunotherapy. Readily available therapies, including radiation, are being investigated as modifying agents with immune checkpoint blockade. However, little is known regarding radiotherapy’s impact within the tumor microenvironment and intratumoral T cell repertoires of patients, leaving critical gaps in the guided design of clinical protocols. In this study, samples from renal cell carcinoma (RCC) patients underwent high-throughput analysis to reveal transcriptional immune activation and increased clonality in irradiated tumors. Analysis across longitudinal blood samples showed that tumor-enriched clonotypes undergo phases of peripheral expansion and contraction following radiation. Collectively, these findings demonstrate that radiotherapy remodels intratumoral T cell responses and support refined sequencing of combination strategies in RCC.
Keywords: radiation, T cell repertoire, RNA sequencing, renal cell carcinoma
Abstract
Clinical studies combining radiation and immunotherapy have shown promising response rates, strengthening efforts to sensitize tumors to immune-mediated attack. Thus, there is an ongoing surge in trials using preconditioning regimens with immunotherapy. Yet, due to the scarcity of resected tumors treated in situ with radiotherapy, there has been little investigation of radiation’s sole contributions to local and systemic antitumor immunity in patients. Without this access, translational studies have been limited to evaluating circulating immune subsets and systemic remodeling of peripheral T cell receptor repertoires. This constraint has left gaps in how radiation impacts intratumoral responses and whether tumor-resident T cell clones are amplified following treatment. Therefore, to interrogate the immune impact of radiation on the tumor microenvironment and test the hypothesis that radiation initiates local and systemic expansion of tumor-resident clones, we analyzed renal cell carcinomas from patients treated with stereotactic body radiation therapy. Transcriptomic comparisons were evaluated by bulk RNA sequencing. T cell receptor sequencing monitored repertoires during treatment. Pathway analysis showed radiation-specific enrichment of immune-related processes, and T cell receptor sequencing revealed increased clonality in radiation-treated tumors. The frequency of identified, tumor-enriched clonotypes was tracked across serial blood samples. We observed increased abundance of tumor-enriched clonotypes at 2 wk postradiation compared with pretreatment levels; however, this expansion was not sustained, and levels contracted toward baseline by 4 wk posttreatment. Taken together, these results indicate robust intratumoral immune remodeling and a window of tumor-resident T cell expansion following radiation that may be leveraged for the rational design of combinatorial strategies.
In-depth preclinical studies have shown that, in addition to radiation’s blunt capacity to reduce tumor burden, this hallmark cancer therapy can also enhance T cell-mediated cytotoxicity (1–3). Historically, reports of rare abscopal responses in cancer patients with solid tumors have offered the most promising correlative clinical evidence of the immunostimulatory properties of radiation (4, 5). More recent strategies combining radiation and immunotherapy with impressive objective response rates and prolonged survival provide more compelling support for radiation’s role in enhancing endogenous human antitumor immunity (6–8).
The advent of targeted forms of radiation, including stereotactic body radiation therapy (SBRT), has broadened radiation’s application across a range of tumor histologies. In particular, this precise delivery method has been applied to renal cell carcinoma (RCC) (9) — a tumor type previously classified as radioresistant due to insensitivity to low-dose treatment (10, 11). Several studies have demonstrated the ability of high-dose radiation delivered with SBRT to control RCC growth (12–16). In addition, clinical trials that combined SBRT with IL-2, immune checkpoint blockade (ICB), or tyrosine kinase inhibition have shown promise in treating RCC (6, 7, 17).
Given these findings and compelling evidence in other tumor types (8, 18, 19), more than 100 open clinical trials are investigating radiation as a tumor-modifying agent in combination with immunotherapy for a variety of malignancies (20). However, due to restricted access of tissues, little is known about on-treatment changes to the tumor immune landscape following radiation. The field’s unfamiliarity with patient tumors following radiation stands in direct contrast to the intense investigation of tumor-infiltrating lymphocytes and the T cell repertoire prior to immunotherapy. These studies have evaluated both intratumoral T cell lymphocyte populations and remodeling of the T cell receptor repertoire after immunotherapy as predictors of patient response to immunotherapy (21, 22). These findings have sparked further interest in the use of available tumor-modulating agents, including radiation, to sensitize patients to immunotherapy. With mounting questions surrounding the synergy between immune therapies and SBRT (6, 17), there is an urgent need to understand how radiation impacts the tumor immune landscape in patients.
To ascertain radiation’s effects on the tumor microenvironment and resident T cell populations, we analyzed tumors resected from RCC patients treated with SBRT. Analysis of resected SBRT-treated RCC revealed enrichment of immune response pathways within SBRT samples, including those indicative of T cell activation and signaling. T cell receptor (TCR) sequencing of patient tumors identified increased clonality in post-SBRT RCC, suggesting accumulation of T cells with similar specificities (23). Evaluation of serial blood samples pinpointed dynamic remodeling of the circulating T cell repertoire following treatment. These translational findings detailing the kinetics of expansion and contraction of tumor-enriched clonotypes provide an immediate rationale for sequencing of SBRT with immunotherapy combinations.
Results
Transcriptional Analysis of RCC Reflects Broad T Cell Activation in SBRT-Treated Patients.
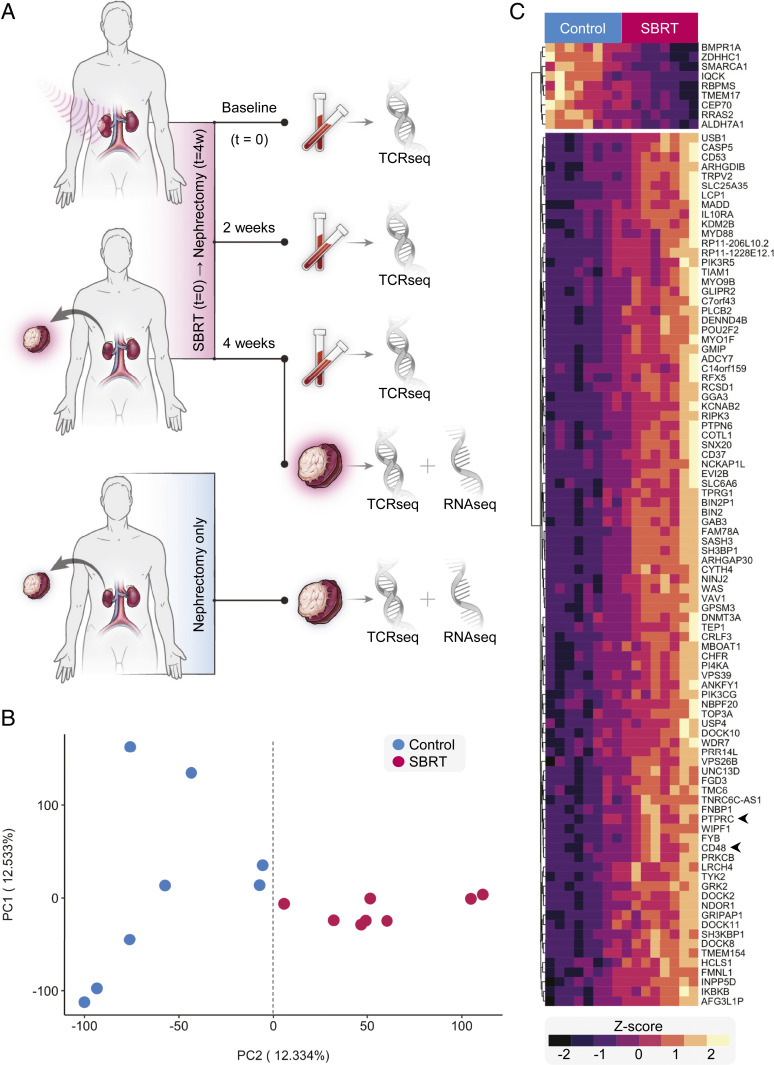
To determine the effects of in situ radiation therapy on human tumors, we analyzed samples from a completed clinical trial (NCT01892930) that treated patients with 15 Gy SBRT to primary RCC followed 4 wk later by nephrectomy. Details of this trial’s protocol have been described previously (24). Transcriptomes were sequenced from SBRT-treated as well as nephrectomy-only RCC tumors, which served as controls (Fig. 1A and SI Appendix, Tables S1 and S2). Principal component analysis (PCA) of expression data revealed separation of tumor transcriptomes by SBRT treatment along PC2 (Fig. 1 B and C and SI Appendix, Fig. S1A). Major contributors to PC2 included the immune cell markers PTPRC, which encodes CD45, and CD48 (Fig. 1C). Pathway enrichment of the top 100 contributors to PC2 revealed several immune response pathways (SI Appendix, Fig. S1B). A similar analysis of PC1 identified deubiquitination as the sole significantly enriched pathway (SI Appendix, Fig. S1B). PCA of control samples did not reveal clusters, and subsequent examination of contributors to PC1 and PC2 did not uncover thematic pathway enrichment (SI Appendix, Fig. S1 C and D).
Fig. 1.
Transcriptional signature of RCC following SBRT treatment. (A) Primary tumors from RCC patients after nephrectomy-only (control) and SBRT plus nephrectomy (SBRT) specimens were processed and later analyzed by bulk RNA sequencing. (B) PCA plot of bulk RNA sequencing data from patient RCC tumors. (C) Heat map of the top-100 contributors to PC2. Genes (rows) clustered by normalized expression among patient samples. Samples (columns) are arranged according to distribution along PC2.
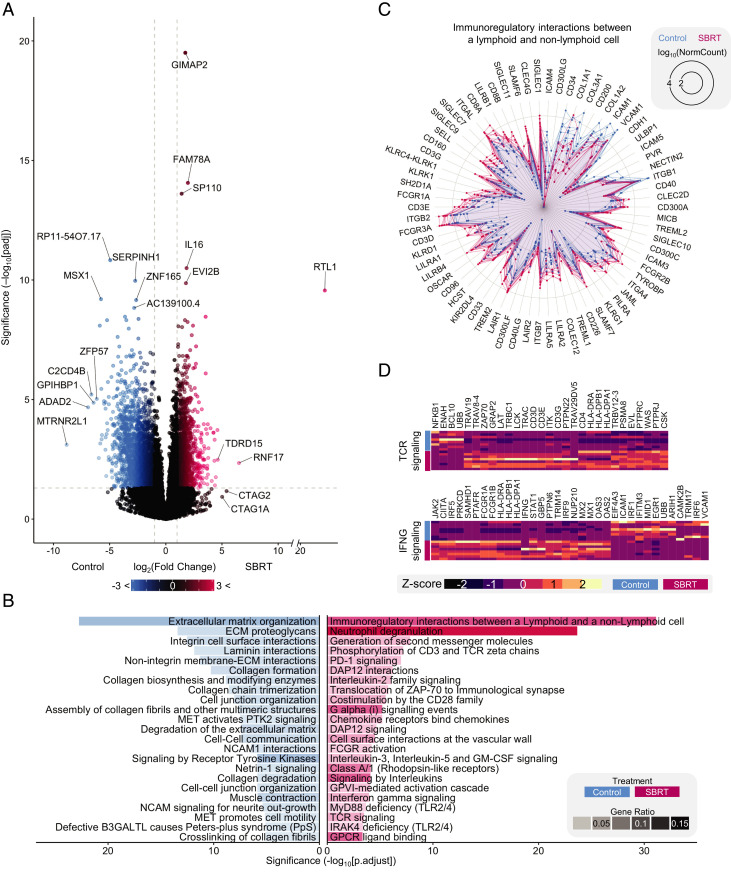
Evaluation of the 23,946 identified transcripts showed increased expression of 1,471 genes within SBRT-treated RCC and 2,547 transcripts that were lower in SBRT-treated compared with nephrectomy-only RCC (Fig. 2A and Dataset S1). Among the most significant SBRT-increased genes was IL16, encoding a T cell chemoattractant (Fig. 2A). Although just shy of significance following P value correction (Fig. 2A), expression of the tumor antigen gene CTAG1A was specific to SBRT-treated samples (Dataset S1), in line with our previous study that identified heightened postradiation NY-ESO-1 protein levels by flow cytometry (24).
Fig. 2.
Postirradiated RCC gene expression is associated with changes to immune pathways. (A) Volcano plot showing differential expression of genes in SBRT vs. control. Significance threshold is |log2(fold change)| >1 (vertical dashed lines) and Padj < 0.05 (horizontal dashed line). Genes in black did not pass the threshold. (B) GSEA of control DE genes (Left, blue) and SBRT DE genes (Right, red). The top-23 most significant are shown. Opacity is the ratio of DE genes in the pathway to total DE genes. (C) Radar plot of immune interactions between lymphoid and nonlymphoid cells. Lines represent individual patient values for indicated DE genes. (D) Heatmaps of DE genes for TCR signaling and IFNG signaling pathways.
Gene set enrichment analysis (GSEA) of differentially expressed (DE) genes in nephrectomy-only RCC revealed significant enrichment of multiple extracellular matrix pathways (Fig. 2B, SI Appendix, Fig. S2A, and Dataset S2). This was consistent with a previous study demonstrating decreased expression of extracellular matrix genes in tissues treated with radiation (25). We identified significant up-regulation of immune-related pathways in SBRT-treated RCC (Fig. 2B and Dataset S3). Of particular interest, among the SBRT GSEA set were multiple pathways involved in lymphocyte activity (Fig. 2 B–D). The most significant of these pathways—immunoregulatory interactions between lymphoid and nonlymphoid cells—included the expression of genes encoding hallmark T cell surface markers: CD3D, CD3E, CD3G, CD8A, and CD8B (Fig. 2C). Heightened expression of CD4 was also detected in SBRT-treated tumors (Dataset S1). Recent findings in patients treated with SBRT and ICB show a correlation between the expression of IFN-γ–associated genes within postirradiated lesions and a decreased volume of out-of-field, unirradiated solid tumors (19). We found increased expression of three of the four genes evaluated in that study (LCK, TLR8, and GPR171) in SBRT-treated RCC (Dataset S1), as well as up-regulation of IFNG and components of the IFN-γ signaling pathway (Fig. 2 B and D). Previous reports identified the postradiation production of CXCL10 as vital for the recruitment of CXCR3+ T cells (2, 26). In line with this, our experiment identified increased expression of IFN-inducible chemokines CXCL9, CXCL10, and CXCL11 and the corresponding chemokine receptor CXCR3 in SBRT-treated tumors (SI Appendix, Fig. S2B and Dataset S1). Additional enriched lymphocyte-related pathways included phosphorylation of CD3 as well as TCR signaling (Fig. 2 B and D). Collectively, these transcriptomic analyses suggest enhanced immunogenicity within SBRT-treated RCC, including enriched genes and pathways indicative of T cell activity.
TCR Sequencing Reveals Increased Repertoire Clonality in SBRT-Treated RCC.
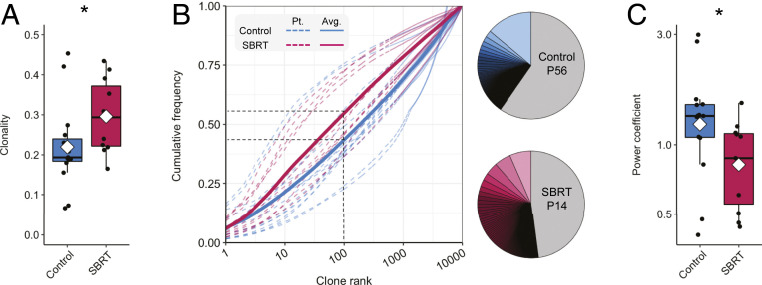
Clear cell RCC is one of the most heavily immune infiltrated solid tumor types with T cells, rather than macrophages, being the dominant immune subset (27). The composition of the T cell repertoire within cancer patients has been an area of intense focus (8, 21). Based on our transcriptional data showing broad immune activation in SBRT-treated RCC (Fig. 2), we hypothesized that intratumoral T cell clonality in RCC would be altered by radiation, and that T cell clones present within the tumor microenvironment would expand systemically. To test this hypothesis, we performed TCRβ sequencing of RCC patient tumors and peripheral blood. This high-throughput technique was used in the analysis of 53 samples, including 26 tumors and 27 peripheral blood samples, and identified 4,016,671 unique CDR3 nucleotide sequence variants encoding 2,658,974 unique CDR3 amino acid sequences. Exploration of overlap between individual amino acid sequences in patient tumors revealed that most sequences were unique to a given patient regardless of treatment (SI Appendix, Fig. S3). Evaluation of repertoires revealed higher clonality in SBRT-treated RCC compared with control tumors (Fig. 3A).
Fig. 3.
Analysis of the intratumoral T cell repertoire of nephrectomy and SBRT-treated RCC. (A) Boxplot showing T cell repertoire clonality in control and SBRT patient tumors. Per-patient values were calculated with the immunoSEQ analyzer. Dots are individual patient values; a diamond indicates the mean. (B) Cumulative frequency distributions calculated for the control and SBRT groups (control regression: 0.171 * log10rank1.193; SBRT regression: 0.283 * log10rank0.858). Black dashed lines indicate the average cumulative frequencies of the top-100 clones for SBRT and control tumor T cell repertoires. Pie charts show representative cumulative frequencies of the top-100 clonotypes in indicated patients. (C) Boxplot of nonlinear regression power coefficients by patient. *P < 0.05, Mann–Whitney–Wilcoxon two-tailed test.
The increased clonality observed within SBRT-treated tumors is further supported by cumulative frequency analysis of the top 10,000 clonotypes showing that a smaller number of the most abundant clonotypes represented a greater proportion of the total T cell repertoire (Fig. 3B). On average, the top-100 most abundant clonotypes represented 43% of the T cell repertoire within nephrectomy tumors and 54% of that within SBRT-treated RCC (P < 0.05, two-tailed Mann–Whitney test) (Fig. 3B, Dataset S4, and SI Appendix, Fig. S4). Power model regression revealed SBRT samples had on average lower power coefficients than control (Fig. 3C and SI Appendix, Fig. S5), consistent with SBRT samples being closer to a high clonality model (23).
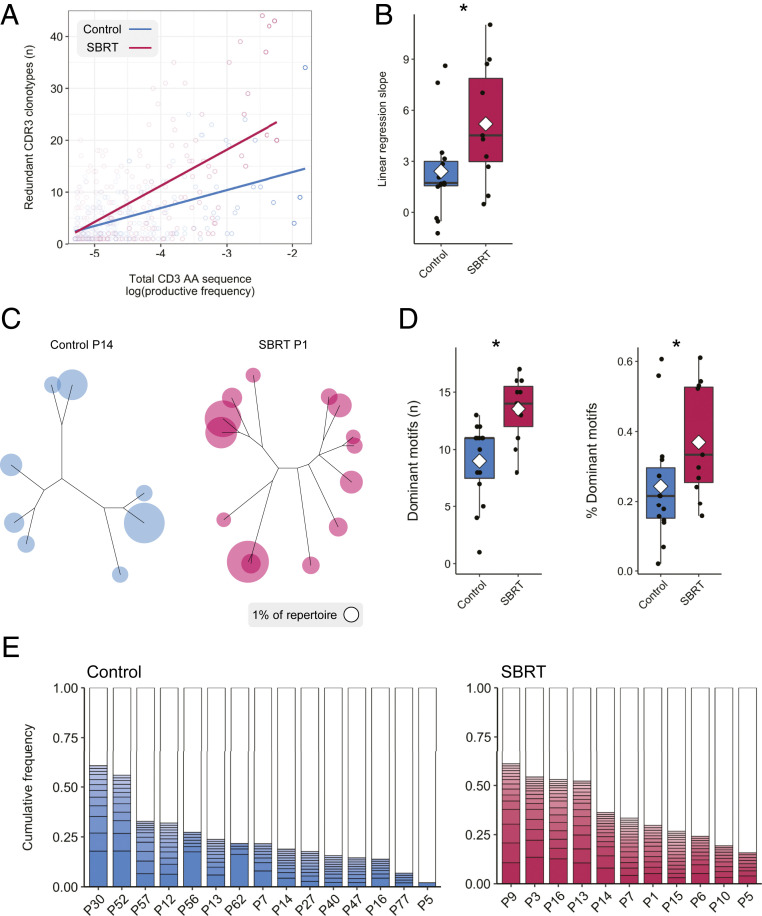
The increased clonality observed in SBRT-treated tumors led us to ask whether TCRs with similar amino acid sequences contribute to changes in the T cell repertoire. Although previous preclinical studies have shown that radiation of murine tumors resulted in increased frequency of longer CDR3 amino acid length (23), we did not observe this phenomenon in our patient samples (SI Appendix, Fig. S6). We examined sets of intrapatient clonotypes with different nucleotide sequences encoding identical amino acid sequences and found that as the combined frequency of an amino acid sequence increased within a repertoire, so did the number of redundant clonotypes per amino acid sequence (Fig. 4A). Linear regression revealed that this effect was more pronounced in SBRT-treated RCC (Fig. 4B).
Fig. 4.
SBRT influences the cumulative frequency of TCR repertoires occupied by clonotypes with similar specificities. (A) Evaluation of redundant templates within RCC tumors (control regression: 3.46 * log10frequency + 20.8; SBRT regression: 6.97 * log10frequency + 39.1). (B) Boxplot of linear regression slope coefficient by patient. (C) ImmunoMap dominant motif analysis performed on intratumoral TCR sequences to identify clusters of homologous sequences. (D) Boxplots of the number of dominant motifs and cumulative frequencies of dominant motifs (each representing ≥1% of the repertoire) shown for individual patient tumors. (E) Stacked bar graphs showing the number of dominant motifs and the percentage of the T cell repertoire occupied by dominant motifs. Dots represent individual patient values; a diamond denotes the mean. *P < 0.05, Mann–Whitney–Wilcoxon two-tailed test.
We and others have previously shown that radiation treatment of tumors can increase expression of tumor antigens (24, 28), possibly leading to clonotypes with similar but not necessarily identical specificities to respond to these antigens (29). To address whether RCC from patients treated with radiation had an increased frequency of lymphocytes with similar TCR specificities, we performed dominant motif analysis of intratumoral repertoires (23). Sets of CDR3 amino acid sequences that met a homology threshold and collectively accounted for 1% of the repertoire were designated dominant motifs. Dominant motifs were represented as dendrograms with amino acid sequence homology used to determine branch lengths (Fig. 4C). SBRT-treated tumors contained more dominant motifs than control, nephrectomy-only tumors (Fig. 4D). When evaluating the accumulation of motifs within samples, dominant motifs represented a larger proportion of the repertoire within SBRT-treated samples (Fig. 4 D and E). Collectively, our evaluation of intratumoral T cell repertoires indicates repertoire remodeling in SBRT-treated RCC through higher clonality, as well as an increased number and frequency of dominant motifs.
Tumor-Enriched Clonotypes Tracked in Peripheral Blood after SBRT Treatment.
To address questions regarding spatial and temporal differences in patient T cell populations, we evaluated repertoires in peripheral blood samples from SBRT-treated patients. For each patient, we considered only those T cell clonotypes that were among the top 10,000 in at least one patient sample. Consistent with reports evaluating breast cancer and melanoma patient samples (30, 31), we observed substantial sharing of clonotypes between a given patient’s tumor and peripheral blood; 42 ± 9% of clonotypes present in tumors were also detected in baseline peripheral blood (Fig. 5A and SI Appendix, Fig. S7A). Portions of each patient’s repertoire were identified as tumor-enriched clonotypes (TECs) and pretreatment blood-enriched clonotypes (BECs) with high relative abundance (|log2(ratio)| ≥5) (Fig. 5A and SI Appendix, Fig. S7A, filled squares), including novel tumor clonotypes that were not present in baseline blood samples (tumor frequency, ≥5 × 10−5; baseline frequency, 0) (Fig. 5A, left of vertical line). Because the patient baseline blood and tumor samples were from separate tissues and collected at different time points (Figs. 1A and 5 A and B), this initial observation raised the question of when TECs had arisen in the patient’s blood, as well as broader questions on reshaping of the T cell repertoire over the course of treatment.
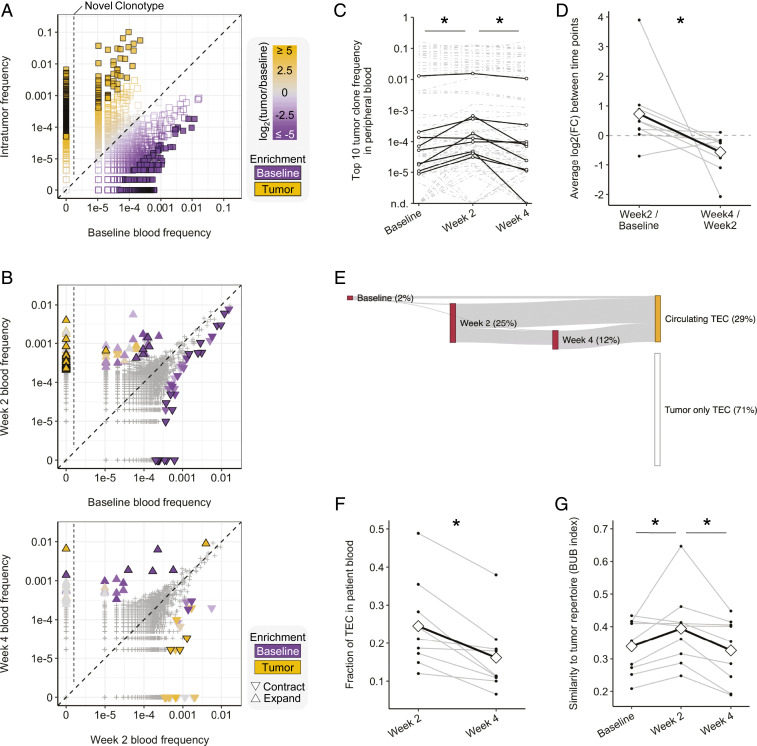
Fig. 5.
Dynamic peripheral changes in T cell clonotype frequencies. (A and B) Representative scatterplots (patient P16) showing the distribution of clonotypes between samples. (A) Baseline peripheral blood vs. tumor. Colors indicate the relative frequency in the tumor (orange) or baseline blood (purple); sample enriched clonotypes are filled squares, left of the vertical line are novel tumor clonotypes. (B) Comparisons between longitudinal peripheral blood samples. The up-triangle indicates expansion, the down-triangle indicates contraction, and color indicates sample enrichment. (C) Frequency of the top-10 most abundant tumor clones in peripheral blood. Black lines indicate representative clones per patient. (D) Average fold change per patient of the clones shown in C. Diamonds and the black line track the mean. (E) Representative Sankey plot (patient P6) showing emergence and detectability of TECs in peripheral blood samples. (F) Line graph showing percentage of TECs detected in postradiation patient blood. Diamonds and the black line track the mean. (G) Baroni-Urbani and Buser index showing the degree of patient blood sample similarity to patient tumor. Diamonds and the black line track the mean. *P < 0.05, Mann–Whitney–Wilcoxon paired two-tailed test.
To investigate the dynamic changes to the patient T cell repertoire after radiation, we extended our analysis to longitudinal peripheral blood samples. Recent high-impact reports have identified clonal T cell expansion and contraction in peripheral blood as predictors of response to radiation and ICB (8). Comparison of the top-10 tumor clonotypes in patient blood between baseline and week 2 revealed an increase in frequency over that interval (Fig. 5C and SI Appendix, Fig. S7B); the top-10 baseline clones decreased in frequency over that same interval (SI Appendix, Fig. S7C). We also noted that between weeks 2 and 4 posttreatment, the top-10 tumor clones showed significant decreases in frequency (Fig. 5C and SI Appendix, Fig. S7B). An analysis of the average fold change of each patient’s top-10 tumor clones across blood samples showed a positive fold change between baseline and week 2, followed by a negative fold change between weeks 2 and 4, consistent with a phasic increase and then decrease in peripheral blood frequency of those highly abundant tumor clones (Fig. 5D). In contrast, when evaluating baseline to week 4 changes (Fig. 5C) no significant differences were detected, highlighting the importance of timing of sample collection when evaluating repertoire dynamics.
This finding led us to analyze the dynamics of repertoire similarity between patient tumors and blood samples over time. Gross comparisons between patient tumors and serial peripheral blood samples showed the highest overlap at 2 wk (48% ± 10%). Differential abundance analysis of compartment-enriched clonotypes expanding between baseline and week 2 revealed these to be largely TECs, while characterization of contracted clonotypes between weeks 2 and 4 showed more TECs (22%) than BECs (1%) (SI Appendix, Fig. S7D). Furthermore, tracking of TECs in posttreatment blood samples revealed that even though the final blood draw and nephrectomy both occurred at 4 wk, the majority of circulating TECs were first detected at 2 wk (Fig. 5 E and F and SI Appendix, Fig. S8). While on average 25% of all TECs were detected at 2 wk, only 16% were detected at 4 wk; moreover, the majority of TECs detected at 4 wk were previously present at 2 wk (Fig. 5 E and F and SI Appendix, Fig. S8). This convergence of patient tumor- infiltrated repertoire with the 2 wk posttreatment blood sample was further supported by evaluating the Baroni-Urbani and Buser sample similarity index (32, 33). This analysis showed the highest intrapatient, intercompartment similarity between patient tumor and 2-wk blood T cell repertoires (Fig. 5G). Collectively, these findings suggest that expansion of tumor-resident clones occurs within the first 2 wk after SBRT, but that this expansion of tumor-associated T cells is transient, and their frequency is reduced at 4 wk posttreatment.
Discussion
Restricted access to patient tumors following radiotherapy has limited our understanding of how this pillar of cancer treatment influences intratumoral T cell responses. Unlike many murine models, human solid tumors do not constitutively express identifiable tumor antigens; thus, analysis of relevant T cell responses in the periphery is especially hindered when tumor samples are unavailable. Consequently, tumor immunologists have described the expansion and contraction of T cell clones by sequencing peripheral T cell receptor repertoires, leaving gaps in knowledge of the dynamic changes to clones present within tumors. Here we leveraged access to patient tumors following treatment to provide a snapshot of the intratumoral transcriptome and address the hypothesis that radiation promotes peripheral expansion of tumor-resident T cell clones.
Our findings within the tumor microenvironment show that RCC patients treated with SBRT exhibit broad transcriptional immune activation and increased clonality with an underlying heightened proportion of dominant motifs. Analysis of peripheral blood samples reveal a dynamic reshaping of the peripheral T cell repertoire wherein tumor-enriched T cell clonotypes expand within the first 2 wk following radiation. This initial burst of observed tumor clonotypes in the periphery is followed by a phase of contraction. Interestingly, global expansion and contraction of peripheral blood T cell clones has been shown to be among the strongest predictors of patient responses to radiation combined with ICB (8). Formenti et al. (8) compared T cell repertoires between baseline and day 22 of treatment. In our study, inclusion of a 14-d time point was crucial for observing differences in expansion and contraction. This time frame is similar to that of peripheral T cell activation detected weeks after treatment with PD-1 blockade (34, 35). Intriguingly, in one of these studies, two patients had received radiation therapy within 4 wk of ICB initiation and had high baseline levels of proliferating T cells, and ultimately both responded to anti–PD-1 (35).
Our findings, within the constraints of 15-Gy radiation treatment of RCC, indicate that expansion of TECs occurs within the first 2 wk post-SBRT, followed by a separate phase of contraction. The emergence of novel clonotypes at week 2 that are also present within resected patient tumors but undetected in baseline blood samples merits further inquiry. However, interpretation of this observation (Fig. 5B and SI Appendix, Fig. S7A) is obscured due to lack of pretreatment biopsy specimens in which to analyze the RCC microenvironment before treatment. Therefore, we could not determine the proportion of these tumor clonotypes that were present in RCC at baseline. This limitation hinders the ability to delineate the relative contributions of SBRT-induced T cell activation and peripheral expansion against tumor antigens from mobilization and retention of tumor-specific T cell clones within the tumor microenvironment. Given evidence that responses to ICB may rely on replenishment of clonotypes from the periphery (22), along with findings in RCC patient tumors demonstrating the importance of a stem-like T cell pool that gives rise to effector CD 8 T cells (36), both of these mechanisms merit further investigation.
The unique nature of this clinical trial allowed for investigation into the transcriptional changes in RCC after treatment. A previous clinical study provided evidence that SBRT may prime patient responses to systemic IL-2 therapy (6). Our pathway analysis identified enrichment of IL-2 family signaling in the SBRT-treated gene set (Fig. 2B and Dataset S3). Similarly, gene expression of IL-2 receptor subunits (IL2RB and IL2RG) and major upstream components of IL-2 signal transduction (JAK3, PIK3CD, and GRB2) were increased in SBRT-treated tumors (Dataset S1). While a clear immune signal differentiated treated samples from control samples (Figs. 1B, PC2 and 2), few inferences could be drawn from the variation among control samples (Fig. 1B, PC1 and SI Appendix, Fig. S1). The comparatively low noise among SBRT-treated samples vs. control samples suggests that radiation and the subsequent immune response may have harmonized those tumor transcriptomes. Although transcriptional data may shed light on certain avenues of inquiry, other questions remain unanswered owing to limitations of the study. The single-armed design left us unable to address the impact of radiation dose and scheduling on the RCC transcriptome and TCR repertoire. This fundamental issue must be resolved for optimization of strategies designed to capitalize on increased immunogenicity following radiation. It is also likely that dose and scheduling for improved immune and patient responses are disease- and treatment-specific (8, 37), reflecting current recommendations for curtailing tumor growth that are dependent on the targeted tissue and disease. Available tissue was restricted to small regions within patient tumors (∼1 g), and thus we were unable to capture the well-characterized tumor heterogeneity within RCC and other tumor types (38, 39). In addition, while we did observe peripheral changes to T cell repertoires following treatment, the duration of contraction and tumor-enriched clone frequency was not evaluated past 4 wk due to the nature of the study.
Collectively, the data presented here showing the dynamics of tumor-enriched clone expansion and contraction provide justification for single-dose radiation as an immune-sensitizing agent in RCC. Transcriptional findings of increased IFN responses and associated chemotactic signatures have been linked to radiation-induced immunogenicity (2, 26). The specific observations of T cell expansion and contraction within distinct time points of observation offer a rationale for the timing of combination strategies that leverage endogenous T cell responses for improved patient outcomes.
Materials and Methods
RCC Patient Samples.
All subjects in this study presented with clear cell pathology (SI Appendix, Tables S1 and S2). Deidentified SBRT-treated samples for this study (SI Appendix, Table S1) were from a previously described single-center, Institutional Review Board (IRB)-approved pilot study (ClinicalTrials.gov identifier NCT01892930) (24). Patient primary tumors received 15 Gy SBRT as described previously (24, 40).
Deidentified control RCC tumor samples from treatment-naïve RCC patients who underwent cytoreductive nephrectomy and gave written consent for research use on resected specimens (SI Appendix, Table S2) were released according to an IRB-approved protocol. Samples used for comparative RNA-seq were from six females and two males, resected between 2012 and 2014; the median age of these patients was 61 y (range, 50 to 74 y) (SI Appendix, Table S2). Samples used for TCR sequencing were from four females and 11 males, resected between 2011 and 2014; the median patient age was 57 y (range, 40 to 82 y) (SI Appendix, Table S2). All RCC patient tumors were processed by mechanical separation, incubation with a collagenase/hyaluronidase mixture (Stem Cell Technology) with end-on-end rotation at 37 °C, followed by separation with gentleMACS C tubes (Miltenyi Biotec) and stored in liquid nitrogen for later analysis. Only samples that passed all quality control checks were included in these analyses.
RNA Sample Preparation and Sequencing.
Thawed cells were immediately lysed and resuspended in 700 μL of QIAzol Lysis Reagent (Qiagen) for 15 min at room temperature. RNA was isolated by chloroform extraction and ethanol precipitation. Samples were then run through an RNeasy Mini spin column with on-column DNase treatment (Qiagen). Quantitative assessment of the purified total RNA was then accomplished using a Qubit Broad-Range RNA Kit (Thermo Fisher Scientific). RNA was further evaluated qualitatively with the Agilent 4200 TapeStation system.
Sequencing libraries were prepared with the Illumina TruSeq Stranded mRNA Kit purified with AMPure XP beads (Beckman Coulter) and validated for appropriate size on an Agilent 4200 TapeStation D1000 Screentape. DNA libraries were quantitated using a Kapa Biosystems qPCR Kit and pooled together in an equimolar fashion. Each pool was denatured and diluted to 2.4 pM with 1% PhiX control library added. The resulting pool was loaded into the appropriate NextSeq Reagent cartridge for 75 cycle single-read sequencing and sequenced on a NextSeq500 (Illumina) following the manufacturer’s recommended protocol.
Samples were aligned to the human genome build GRCh38.p7 and corresponding GENCODE annotation version 25 (March 2016) (41, 42). Fastqc was used for quality control of raw reads (43). TopHat2 was used to align reads to reference genome (44), allowing a maximum of one mismatch per read. Quality control of alignment was performed with RseQC (45). Mapped reads were quantified at the gene level using Subread (46) with fracOverlap = 1.
TCR Sequencing Sample Preparation and Sequencing.
DNA was prepared using the QIAamp DNA Blood Mini Kit (Qiagen). Samples were resuspended in 200 µL of cold 1× PBS, pH 7.4 (Thermo Fisher Scientific). Then 20 µL of Qiagen Protease was added to the sample, which was homogenized with the addition of 200 µL of Buffer AL, followed by vortexing for 15 s (Scientific Industries). Sample was centrifuged (VWR International) and incubated at 56 °C for 10 min in a water bath (Thermo Fisher Scientific). Then 200 µL of 200-proof ethanol (Pharmco-Aaper) was added, followed by further homogenization by vortexing. The sample was run through the QIAamp Mini spin column, and quantitative assessment of the purified DNA was performed with the Qubit Broad-Range DNA kit (Thermo Fisher Scientific).
Sequencing libraries were generated using the ImmunoSEQ kit (Adaptive Biotechnologies) according to the manufacturer’s recommendations. The first round of PCR was accomplished using the ImmunoSEQ proprietary PCR primer mix. A positive control reaction, provided in the kit, and a negative control reaction were included with each sample batch. The first-round PCR was purified using the PCR Cleanup beads provided. A second round of PCR was performed to generate uniquely barcoded sequencing libraries using the barcode primer plate included in the kit. Final library purification was done using PCR Cleanup beads. Final libraries were validated on an Agilent 4200 TapeStation D1000 Screentape. Samples were pooled volumetrically, and the final pool was quantitated using a Kapa Biosystems qPCR kit. The pool was denatured and diluted to 1.0 pM with 20% PhiX control library added. The pooled samples were loaded into an Illumina NextSeq500 MidOutput 150-cycle reagent cartridge and sequenced on a NextSeq500 per the manufacturer’s recommended protocol with 156 Read1 cycles and 15 Index1 cycles using the custom primers included in the ImmunoSEQ kit.
For all analyses of changes and differences in T cell clone abundance, frequency was normalized to the top-10,000 most abundant clones. Public T cell clones (47), which on average accounted for fewer than five clones per sample per patient after initial normalization, were also removed before the present analysis.
Comparative Transcriptomics.
High-dimensional comparisons of tumor transcriptomes were performed by PCA in R (48). A heatmap of the top-100 contributors to the principal components was generated in the R package pheatmap (49). Differential gene expression was performed using the Bioconductor package DESeq2 (50); differential expression was defined as |log2(fold change)| ≥ 1 (±2 fold) with adjusted P value < 0.05. Pathway enrichment of DE genes was performed using the Reactome pathway database Bioconductor package ReactomePA (51, 52).
Unless stated otherwise, this and all subsequent data visualization was performed in the R package ggplot2 (53).
Intratumoral TCR Repertoire Analysis.
Processed sequencing data were retrieved from Adaptive Biotechnologies and analyzed in R. Herein, a clonotype is defined as a set of all T cells with the same CDR3 nucleotide sequence. Clonality represents the distribution of T cell clones, with low clonality indicative of an equal distribution of T cells across all clonotypes and higher clonality indicative of a larger fraction of the repertoire occupied by fewer clonotypes. For boxplots, the maximum whisker length is 1.5 times the interquartile range. Statistical comparisons between treatment group repertoires was performed with the two-tailed wilcox.test function in R.
Redundant clonotype analysis was performed with a custom R script. In brief, redundant clonotypes were defined as having degenerate CDR3 DNA sequences encoding for identical CDR3 amino acid sequences. Redundant clonotypes were plotted by their total log10(normalized frequency) and number. Linear regression was performed in R.
Dominant motif analysis was performed as described previously (23) using the ImmunoMap tool (54). In brief, sequence distance was determined with the PAM10 scoring matrix (gap penalty = 30, homology threshold = 0.3). Sequence clusters below the homology threshold were classified as a motif. Only motifs with cumulative frequency ≥0.01 were classified as dominant motifs. Visualization of dominant motifs was performed in ImmunoMap; for clarity, only amino acids in a motif were used for visualization. Statistical comparisons between treatment group dominant motif number and % motifs were performed with the two-tail wilcox.test function in R.
Serial Peripheral Blood TCR Repertoire Analysis.
Differential abundance analysis was calculated using the beta-binomial model in the Adaptive Biotechnologies’ differential abundance tool. In brief, expanded and contracted clonotypes are defined as having a significant increase or decrease in clonotype frequency between early and later time points, respectively. The applied model reduces noise from normal peripheral blood T cell repertoire variance over 2-wk and 4-wk time points (55).
Statistical comparisons of clonotype frequency between time points for top tumor and baseline peripheral blood clonotypes was performed with the two-tailed paired wilcox.test function in R.
Tumor-enriched vs. baseline peripheral blood-enriched clones with a ≥32-fold difference in abundance were determined as described previously (30).
Expanded and contracted tumor and peripheral blood enriched clonotypes were tallied with a custom R script. Statistical comparisons were performed with the two-tailed paired wilcox.test function in R.
TECs in peripheral blood samples for generating a Sankey plot analysis were tallied with a custom R script. In brief, TECs were determined for each intrapatient blood sample, and a Sankey plot was generated using the networkDC package in R (56). Statistical comparisons of the number of novel TECs in posttreatment blood and total detected TECs in posttreatment blood were performed with the two-tailed paired wilcox.test function in R.
Calculation of Baroni-Urbani and Buser coefficient for sample similarity between tumor and serial peripheral blood sample T cell clonotypes was performed as described previously (32, 33).
Supplementary Material
Acknowledgments
The research reported in this paper was supported by the NIH, National Center for Advancing Translational Sciences to the University at Buffalo (UL1TR001412). Funding was also provided by NIH Grant R01 CA172105 (to S.I.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Support was also provided by the National Cancer Institute (Grant P30CA016056 supporting use of the Roswell Park Comprehensive Cancer Center’s Genomic and Biomedical Data Science Shared Resources), Roswell Park Friends of Urology, the Elsa Kreiner Memorial Fund, and the Fraternal Order of Eagles.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001933117/-/DCSupplemental.
Data Availability.
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE153262). Immunosequencing data can be accessed at the Adaptive Biotechnologies immuneACCESS site (https://clients.adaptivebiotech.com/pub/chow-2020-pnas).
References
- 1.Lugade A. A. et al., Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. J. Immunol. 174, 7516–7523 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Vanpouille-Box C. et al., DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reits E. A. et al., Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 203, 1259–1271 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehlers G., Fridman M., Abscopal effect of radiation in papillary adenocarcinoma. Br. J. Radiol. 46, 220–222 (1973). [DOI] [PubMed] [Google Scholar]
- 5.Wersäll P. J. et al., Regression of non-irradiated metastases after extracranial stereotactic radiotherapy in metastatic renal cell carcinoma. Acta Oncol. 45, 493–497 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Seung S. K. et al., Phase 1 study of stereotactic body radiotherapy and interleukin-2: Tumor and immunological responses. Sci. Transl. Med. 4, 137ra74 (2012). [DOI] [PubMed] [Google Scholar]
- 7.He L. et al., Survival outcomes after adding stereotactic body radiotherapy to metastatic renal cell carcinoma patients treated with tyrosine kinase inhibitors. Am. J. Clin. Oncol. 43, 58–63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formenti S. C. et al., Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat. Med. 24, 1845–1851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siva S. et al., Radiotherapy for renal cell carcinoma: Renaissance of an overlooked approach. Nat. Rev. Urol. 14, 549–563 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Talley R. W., Moorhead E. L. 2nd, Tucker W. G., San Diego E. L., Brennan M. J., Treatment of metastatic hypernephroma. JAMA 207, 322–328 (1969). [PubMed] [Google Scholar]
- 11.Deschavanne P. J., Fertil B., A review of human cell radiosensitivity in vitro. Int. J. Radiat. Oncol. Biol. Phys. 34, 251–266 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Correa R. J. M. et al., The emerging role of stereotactic ablative radiotherapy for primary renal cell carcinoma: A systematic review and meta-analysis. Eur. Urol. Focus 5, 958–969 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Siska P. J., Beckermann K. E., Rathmell W. K., Haake S. M., Strategies to overcome therapeutic resistance in renal cell carcinoma. Urol. Oncol. 35, 102–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teh B. et al., The treatment of primary and metastatic renal cell carcinoma (RCC) with image-guided stereotactic body radiation therapy (SBRT). Biomed. Imaging Interv. J. 3, e6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beitler J. J., Makara D., Silverman P., Lederman G., Definitive, high-dose-per-fraction, conformal, stereotactic external radiation for renal cell carcinoma. Am. J. Clin. Oncol. 27, 646–648 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y. et al., Stereotactic ablative radiation therapy (SAbR) used to defer systemic therapy in oligometastatic renal cell cancer. Int. J. Radiat. Oncol. Biol. Phys. 105, 367–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dengina N. et al., Stereotactic body radiation therapy in combination with systemic therapy for metastatic renal cell carcinoma: A prospective multicentre study. ESMO Open 4, e000535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaverdian N. et al., Previous radiotherapy and the clinical activity and toxicity of pembrolizumab in the treatment of non-small-cell lung cancer: A secondary analysis of the KEYNOTE-001 phase 1 trial. Lancet Oncol. 18, 895–903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luke J. J. et al., Safety and clinical activity of pembrolizumab and multisite stereotactic body radiotherapy in patients with advanced solid tumors. J. Clin. Oncol. 36, 1611–1618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J. et al., Trial watch: The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 17, 854–855 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Riaz N. et al., Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yost K. E. et al., Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudqvist N. P. et al., Radiotherapy and CTLA-4 blockade shape the TCR repertoire of tumor-infiltrating T cells. Cancer Immunol. Res. 6, 139–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A. K. et al., A pilot study of stereotactic body radiation therapy combined with cytoreductive nephrectomy for metastatic renal cell carcinoma. Clin. Cancer Res. 23, 5055–5065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkelstein J. N., Johnston C. J., Baggs R., Rubin P., Early alterations in extracellular matrix and transforming growth factor beta gene expression in mouse lung indicative of late radiation fibrosis. Int. J. Radiat. Oncol. Biol. Phys. 28, 621–631 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Lim J. Y., Gerber S. A., Murphy S. P., Lord E. M., Type I interferons induced by radiation therapy mediate recruitment and effector function of CD8(+) T cells. Cancer Immunol. Immunother. 63, 259–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevrier S. et al., An immune atlas of clear cell renal cell carcinoma. Cell 169, 736–749.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A. et al., Gamma radiation promotes immunological recognition of cancer cells through increased expression of cancer-testis antigens in vitro and in vivo. PLoS One 6, e28217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derré L. et al., Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proc. Natl. Acad. Sci. U.S.A. 105, 15010–15015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beausang J. F. et al., T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc. Natl. Acad. Sci. U.S.A. 114, E10409–E10417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valpione S. et al., Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat. Can. 1, 210–221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheikh N. et al., Clonotypic diversification of intratumoral T cells following sipuleucel-T treatment in prostate cancer subjects. Cancer Res. 76, 3711–3718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baroni-Urbani C., Buser M. W., Similarity of binary data. Syst. Biol. 25, 251–259 (1976). [Google Scholar]
- 34.Huang A. C. et al., T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamphorst A. O. et al., Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl. Acad. Sci. U.S.A. 114, 4993–4998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen C. S. et al., An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voorwerk L. et al., Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: The TONIC trial. Nat. Med. 25, 920–928 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Kim K. T. et al., Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 17, 80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuzhakova D. V. et al., Measuring intratumoral heterogeneity of immune repertoires. Front. Oncol. 10, 512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanzly M. et al., Stereotactic body radiotherapy for the treatment of renal tumors. Urol. Case Rep. 2, 147–149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrow J. et al., GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derrien T. et al., The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews S., Fast QC: A quality control tool for high throughput sequence data (2010). www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 28 June 2018.
- 44.Kim D. et al., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Liao Y., Smyth G. K., Shi W., The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Britanova O. V. et al., Dynamics of individual T cell repertoires: From cord blood to centenarians. J. Immunol. 196, 5005–5013 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Team R. C., A Language and Environment for Statistical Computing (R Foundation for Statistical computing, Vienna, Austria, 2017). https://www.R-project.org/. Accessed 24 July 2020.
- 49.Kolde R., Package ‘pheatmap’ (Version 1.0.12, 2011). https://CRAN.R-project.org/package=pheatmap. Accessed 24 July 2020.
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G., He Q. Y., ReactomePA: An R/bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Jassal B. et al., The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer New York, 2009). [Google Scholar]
- 54.Sidhom J. W. et al., ImmunoMap: A bioinformatics tool for T-cell repertoire analysis. Cancer Immunol. Res. 6, 151–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rytlewski J. et al., Model to improve specificity for identification of clinically-relevant expanded T cells in peripheral blood. PLoS One 14, e0213684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allaire J. J., Package ‘networkD3’ (Version 0.4, 2017). https://CRAN.R-project.org/package=networkD3. Accessed 24 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE153262). Immunosequencing data can be accessed at the Adaptive Biotechnologies immuneACCESS site (https://clients.adaptivebiotech.com/pub/chow-2020-pnas).