Significance
In Arabidopsis, MAC5, a component of the conserved MOS4-associated complex, is required for development and immunity. This study shows that MAC5 interacts with Serrate (SE) and the stem loop of pri-miRNAs, facilitates pri-miRNA processing, and protects pri-miRNAs from SE-dependent 5′-to-3′ exoribonuclease activities. Consequently, this study provides insights into the mechanism regulating miRNA biogenesis. Since MAC5 is a conserved protein, this study may have a broader impact.
Keywords: miRNA biogenesis, MAC5A, Serrate, pri-miRNA stablity
Abstract
MAC5 is a component of the conserved MOS4-associated complex. It plays critical roles in development and immunity. Here we report that MAC5 is required for microRNA (miRNA) biogenesis. MAC5 interacts with Serrate (SE), which is a core component of the microprocessor that processes primary miRNA transcripts (pri-miRNAs) into miRNAs and binds the stem-loop region of pri-miRNAs. MAC5 is essential for both the efficient processing and the stability of pri-miRNAs. Interestingly, the reduction of pri-miRNA levels in mac5 is partially caused by XRN2/XRN3, the nuclear-localized 5′-to-3′ exoribonucleases, and depends on SE. These results reveal that MAC5 plays a dual role in promoting pri-miRNA processing and stability through its interaction with SE and/or pri-miRNAs. This study also uncovers that pri-miRNAs need to be protected from nuclear RNA decay machinery, which is connected to the microprocessor.
In plants, microRNAs (miRNAs) are short (∼21 nt) endogenous small RNAs that play essential roles in various biological processes (1–6). miRNAs are first transcribed as primary miRNA transcripts (pri-miRNAs) by the DNA-dependent RNA polymerase II (Pol II). In plants, the DICER-LIKE 1 (DCL1, an ribonuclease III enzyme) complex, which is composed of SERRATE (SE, a zinc-finger protein), HYPONASTIC LEAVES 1 (HYL1, a double-strand RNA [dsRNA]-binding protein), cotranscriptionally processes the stem-loop region of pri-miRNAs in the nucleus to generate miRNAs (7–9), which are subsequently incorporated into the effector protein ARGONAUTE1 (AGO1) to repress gene expression through target mRNA cleavage or translational inhibition (10–12). The efficient loading of miRNA into AGO1 requires HEAT SHOCK PROTEIN90 and CYCLOPHILIN40 (13, 14).
Plant miRNA generation is controlled through pri-miRNA transcription and processing. In Arabidopsis, the transcription factors, including the mediator complex, CYCLIN-DEPENDENT KINASE F;1, CYCLIN-DEPENDENT KINASE D, Negative on TATA less 2 (NOT2), CELL DIVISION CYCLE 5 (CDC5, a DNA-binding protein), the Elongator complex, and SNC1 positively regulate pri-miRNA transcription through modulating Pol II activity (15–20). In addition, many protein factors regulate pri-miRNA processing through modulating the DCL1 complex-pri-miRNA interaction and/or the activity/assembly of the DCL1 complex. These proteins include CBP20/80, DDL, NOT2, CDC5, PRL1, RACK1, MOS2 (MODIFIER OF SNC1,2), PINP1 (a DEAH-box helicase), GRP7 (a glycine-rich RNA-binding protein), STV1 (a ribosomal protein), MAC3, MAC7, and CHR2 (18, 19, 21–33). Furthermore, the levels or activities of DCL1 and HYL1 can be transcriptionally, posttranscriptionally, and/or posttranslationally modulated to control miRNA biogenesis (34–43). For instance, HYL1 phosphorylation and dephosphorylation play crucial roles in maintaining proper miRNA accumulation (34, 40).
Pri-miRNA stability is another important factor contributing to miRNA accumulation. Most pri-miRNAs have similar structural and sequence characteristics of mRNAs (10, 44, 45). However, they often contain premature stop codons, which are known to active the degradation of mRNAs (46–48). This fact raises interesting questions as to whether pri-miRNAs are the targets of the nuclear quality surveillance machinery, and if so, how they are protected from degradation during processing Moreover, increased pri-miRNA degradation reduces miRNA accumulation and causes defects in development and physiology, revealing that pri-miRNA degradation is a potential mechanism regulating miRNA biogenesis (24–26, 49). In fact, it has been shown that a chloroplast-to-nucleus signal can potentially promote miRNA accumulation through inhibiting pri-miRNA degradation (49). Currently, the mechanisms stabilizing pri-miRNAs, and the mechanisms degrading pri-miRNAs when protecting factors are defective, are poorly understood.
The MOS4-associated complex (MAC) from plants is a conserved complex in eukaryotes. Its orthologous complexes including the CDC5-SNEVPrp19-Pso4 complex (PRP19) in human and the Nineteen complex (NTC) in yeast are associated with spliceosome and participate in splicing (50, 51). MAC is composed of five core components and ∼13 accessory components. It plays critical roles in plant development and immunity (52). However, related mechanisms are not clear as the effect of MAC on splicing is moderate (27, 52, 53). Several MAC components such as MAC3, MAC7, CDC5, and PRL1, associate with the DCL1 complex to promote miRNA biogenesis through modulating pri-miRNA transcription and processing (19, 26, 27, 29). Among MAC accessory components, MAC5A and MAC5B are orthologs of CWC2 from yeast and the RNA-binding motif protein 22 (RBM22) from human (53). MAC5A and MAC5B display unequal genetic redundancy in a dosage-dependent manner and MAC5A is the dominant contributor (53). Interestingly, the loss-of-function mac5a mac5b double mutant is embryo lethal, suggesting that MAC5 is critical for plant development.
Here we report that MAC5A and MAC5B display a dosage-dependent effect on the accumulation of miRNAs. MAC5 is an RNA-binding protein and binds the stem-loop region of pri-miRNAs. In addition, MAC5 interacts with SE and promotes pri-miRNA processing. Interestingly, we show that a loss-of-function mac5a mutation reduces pri-miRNA levels without impacting their transcription. Moreover, lack of XRN2 or XRN3, the nuclear localized 5′-to-3′ exoribonuclease, partially recovers the accumulation of pri-miRNA in mac5a. More interestingly, the reduction of pri-miRNA levels in mac5a depends on SE, but not HYL1. These results reveal that MAC5 protects pri-miRNAs from SE-dependent exoribonuclease activities. Taken together, our study uncovers a dual role of MAC5 in processing and stabilizing pri-miRNAs through its interaction with SE and/or pri-miRNAs.
Results
MAC5 Is Required for miRNA Accumulation.
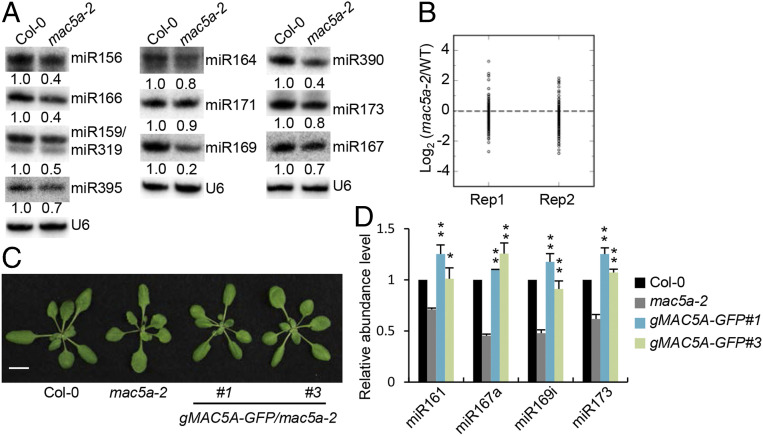
Given the function of several MAC components in promoting miRNA accumulation, we reasoned that the putative RNA-binding protein MAC5 might also have a role in miRNA biogenesis. To test this possibility, we examined miRNA accumulation in mac5a using Northern blot. All examined miRNAs were reduced in abundance in mac5a relative to Col (wild-type plants [WT]) (Fig. 1A). However, several of them, such as miR164 and miR167, showed only slight reduction. This is likely due to the redundant function of MAC5B with MAC5A. Indeed, the accumulation of miR164 and miR167 was strongly reduced in the mac5a MAC5B/mac5b mutant that has stronger developmental defects than mac5a (SI Appendix, Fig. S1 A–C). To identify the global effects of mac5a on miRNA accumulation, we compared the miRNA profile in mac5a with that in Col through Illumina deep sequencing analyses. The results revealed that most miRNAs displayed reduced accumulation in mac5a (Fig. 1B and Datasets S1 and S2). In addition, the expression of a wild-type copy of MAC5A gene under the control of its native promoter (gMAC5A-GFP) fully recovered the developmental and miRNA defects in mac5a (Fig. 1 C and D and SI Appendix, Fig. S2 A and B). These results demonstrate that MAC5 is required for miRNA accumulation.
Fig. 1.
MAC5A is required for miRNA abundance. (A) Northern blot analyses of miRNA levels in various genotypes. U6 RNA serves as the loading control. The numbers shown below the pictures indicate the relative amounts of miRNAs (the values in Col-0 were set as 1). (B) Small RNA sequencing analysis in wild type (Col-0) and mac5a-2. The miRNA abundance was calculated as reads per million, and a Log2-transformed ratio of mac5a-2/Col-0 was plotted. Each circle represents one miRNA. Two replicates are displayed. (C) Phenotypes of transgenic plants harboring genomic sequence of MAC5A. The MAC5A genomic sequence was fused to the N terminus of GFP. (Scale bar: 1 cm.) (D) qRT-PCR analysis of miRNA abundance in various genotypes (the values in Col-0 were set as 1). *P < 0.05; **P < 0.01 by Student’s t test compared with mac5a-2 value.
We next asked the effect of mac5a on the expression levels of miRNA targets. qRT-PCR analyses revealed the transcript levels of several examined mRNAs, including APS3, ARF6, CUC1/CUC2, MYB33, SCL6, and SPL9, which are targeted by miR395, miR167, miR164, miR159, miR171, and miR156, respectively, were increased (SI Appendix, Fig. S1D), indicating that miRNA activities are impaired in mac5a. In addition, the levels of miRNA targets were much higher in mac5a MAC5B/5b than those in mac5a or mac5b single mutants (SI Appendix, Fig. S1E), consistent with the redundant role of MAC5A and MAC5B in miRNA biogenesis.
MAC5 Positively Contributes to Pri-miRNA Accumulation without Affecting MIR Promoter Activities.
Next, we asked how MAC5 affects miRNA accumulation. We first examined the influence of MAC5A on the transcript levels of several genes, including CBP20, CBP80, DCL1, DDL, HEN1, HYL1, and SE, which act in miRNA biogenesis. The transcript levels of these genes did not show significant change in mac5a relative to Col (SI Appendix, Fig. S3A). Furthermore, mac5a did not change the protein levels of SE, HYL1, and DCL1 (SI Appendix, Fig. S3B). These results suggest that MAC5A may not regulate the expression of genes involved in miRNA biogenesis.
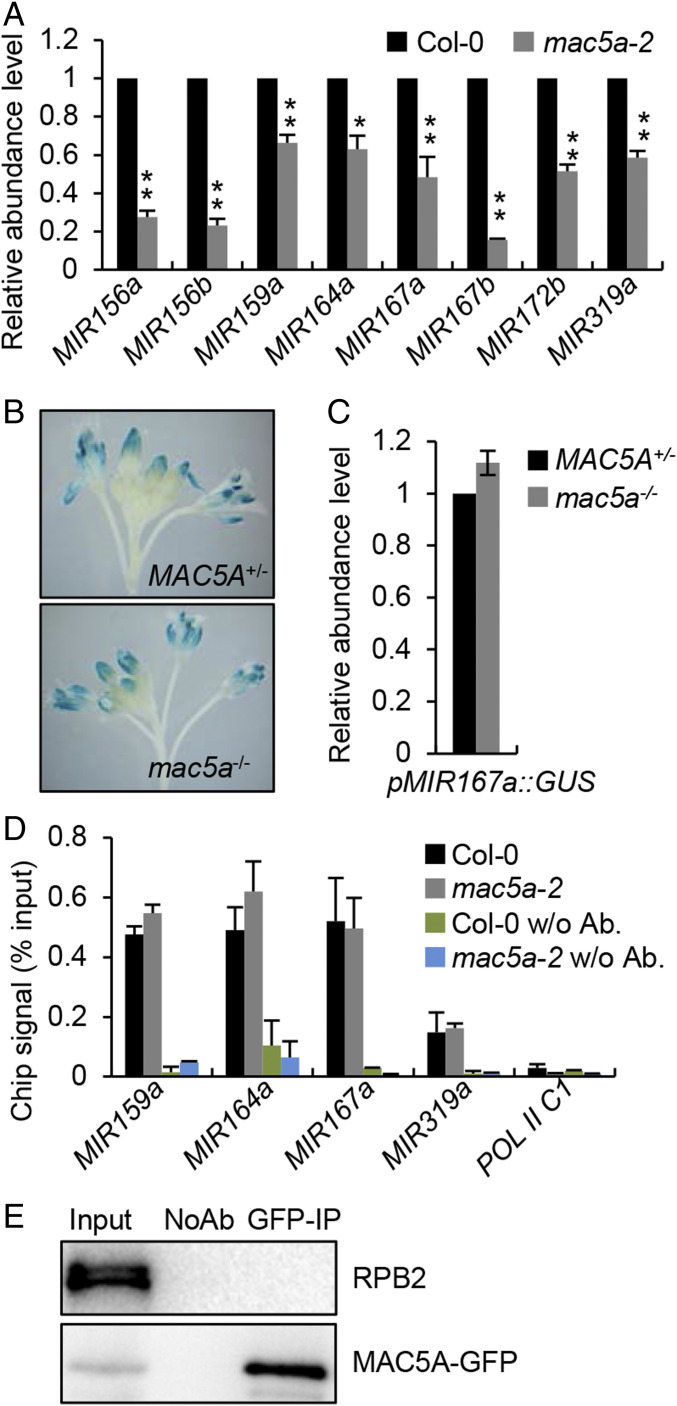
We next tested the impact of MAC5 on pri-miRNA accumulation that is one of the major factors determining miRNA levels. qRT-PCR analyses showed all eight examined pri-miRNA were reduced in abundance in mac5a compared with Col (Fig. 2A). In addition, pri-miRNA levels were further reduced in mac5a MAC5B/mac5b relative to mac5a (SI Appendix, Fig. S1F). These results demonstrate that MAC5 promotes pri-miRNA accumulation. Since pri-miRNA levels are partially controlled by transcription, we next asked if MAC5 plays a role in regulating MIR promoter activity using a GUS report gene assay as previously described (26). In this assay, a pMIR167a::GUS transgenic line was crossed into mac5a. In an F2 population, mac5a and MAC5A+ plants (Col and MAC5A/mac5a) containing the GUS transgene were isolated through PCR genotyping. GUS histochemical staining and qRT-PCR showed that the protein and transcript levels of GUS in mac5a were comparable to those in MAC5A+ (Fig. 2 B and C), suggesting that mac5a may not change MIR promoter activities. To confirm this result, we examined the occupancy of Pol II at MIR promoters in mac5a and Col through chromatin immunoprecipitation (ChIP) assay using antibodies against the second largest subunit of Pol II (RPB2). mac5a did not affect Pol II occupancy at the MIR promoters (Fig. 2D). We also examined the interaction of MAC5A with the RPB2 in the complementation line harboring the gMAC5A-GFP transgene through the coimmunoprecipitation (Co-IP) assay. RPB2 was not detected in MAC5A-GFP immunoprecipitates (Fig. 2E). These results reveal that MAC5 may not affect MIR transcription.
Fig. 2.
mac5a reduces pri-miRNA accumulation without affecting transcription. (A) qRT-PCR analysis of pri-miRNA levels in Col-0 and mac5a-2. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with Col-0 values). (B) Histochemical staining of GUS in inflorescence of wild-type and mac5a-2 harboring pMIR167a::GUS transgene. Fifteen plants for each genotype were analyzed and a picture is shown. (C) Detection of relative levels of GUS transcript in the indicated plants (the values in Col-0 were set as 1). Error bar indicates SD of three replicates. (D) Pol II occupancy at MIR promoter in the indicated plants detected by ChIP followed by qPCR. The intergenic region between At2g17470 and At2G17460 (POL C1) was used as a negative control. (E) Co-IP between RPB2 and MAC5A. Protein extracts from inflorescences of gMAC5A-GFP transgenic plants were performed with anti-GFP antibodies. The proteins were detected by anti-GFP or anti-RRB2 antibodies. Input indicates total proteins before immunoprecipitation; NoAb indicates immunoprecipitation using agarose beads without antibodies.
MAC5 Interacts with SE, but Not DCL1 and HYL1.
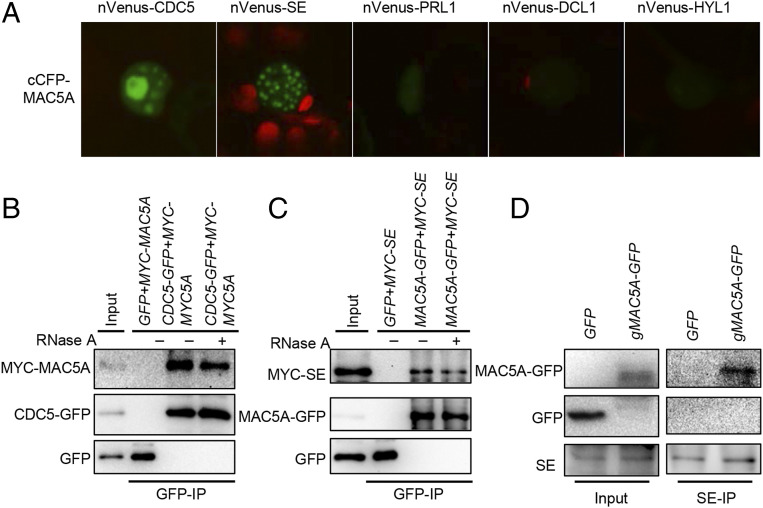
Given the fact that CDC5 and PRL1, two core components of MAC interact with the DCL1 complex, it is possible that MAC5 also associates with the DCL1 complex as an accessory component. We thus tested the interactions of MAC5A with DCL1, HYL1, and SE through a bimolecular fluorescence complementation (BiFC) assay. In this assay, MAC5A fused with a C-terminal fragment of CFP at its N terminal (cCFP-MAC5A) was coexpressed with DCL1, HYL1, or SE containing an N-terminal fragment of Venus (nVenus) at their N terminus in Nicotiana benthamiana. We also included CDC5, which was known to interact with MAC5A (53), as a positive control. Coexpression of MAC5A with SE and CDC5, but not other proteins, generated yellow florescence protein signal (YFP) (Fig. 3A; shown in green), indicating that MAC5A may interact with SE and CDC5, but not HYL1 and DCL1.
Fig. 3.
MAC5A interacts with SE. (A) The interactions of MAC5A with various proteins detected by BiFC. The MAC5A-CDC5 interaction is used as a positive control. Paired cCFP- and nVenus-fusion proteins were transiently coexpressed in N. benthamiana leaves. Green color indicates the BiFC signal (originally yellow fluorescence) detected by confocal microscopy at 48 h after infiltration. One hundred nuclei were examined for each pair and a picture is shown. Red: autofluorescence of chlorophyll. (B and C) Co-IP analyses of the MAC5A-CDC5 (B), and the MAC5A-SE interactions (C). CDC5-GFP or MAC5A-GFP/GFP were coexpressed with MYC-MAC5A or MYC-SE in N. benthamiana, respectively. IP was performed using anti-GFP antibodies. MYC-MYC5A, MYC-SE, CDC5-GFP, MYC5A-GFP, and GFP were detected by immunoblot. (D) The interaction between MAC5A and SE detected by Co-IP in Arabidopsis. Total proteins were extracted in transgenic plants harboring p35S::GFP or gMAC5A-GFP. IP was performed using anti-SE antibody. After IP, GFP and MAC5A-GFP proteins were detected by immunoblot using anti-GFP antibody. Inputs in B–D show the total protein before IP. RNase A was used to digest RNA stands.
Next, we performed Co-IP to test the interactions of MAC5A with SE and CDC5 (positive control) in N. benthamiana transiently expressing MYC-MAC5A/CDC5-GFP or MYC-SE/MAC5A-GFP. After IP with GFP antibodies, MAC5A and SE were detected in the CDC5 IPs and in the MAC5A IPs, respectively (Fig. 3 B and C). RNase A treatment did not abolish the MAC5A-SE and MAC5A-CDC5 interactions. Moreover, we also detected MAC5A in SE IPs and SE in MAC5A IPs from the stable transgenic mac5a harboring gMAC5A-GFP, respectively (Fig. 3D and SI Appendix, Fig. S4). Taken together, these results demonstrate that MAC5A interacts with SE and the interaction is independent of RNAs.
MAC5A Interacts with Pri-miRNAs In Vivo.
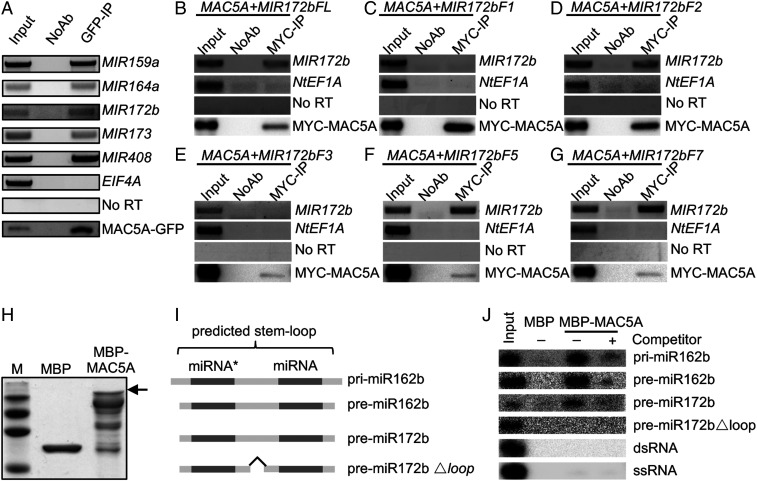
Because MAC5A is a putative RNA-binding protein, we asked if MAC5A binds pri-miRNAs in vivo. We performed an RNA immunoprecipitation assay (RIP) on the seedlings of mac5a harboring the gMAC5A-GFP transgene. Following cross-linking, nuclear isolation, and IP, RT-PCR was performed to detect the enrichment of pri-miR159a, pri-miR164a, pri-miR172b, pri-miR173, and pri-miR408 in the MAC5A-GFP immunoprecipitates (IPs), but not in the no-antibody control (Fig. 4A). Moreover, the negative control EIF4A was not detected in the MAC5A-GFP IPs (Fig. 4A). These results demonstrate that MAC5A binds pri-miRNAs in vivo.
Fig. 4.
MAC5A binds the stem-loop region of pri-miRNAs. (A) The association of MAC5A with pri-miRNAs in vivo detected by RIP assay. RIP assay was performed on the transgenic mac5a plants harboring gMAC5A-GFP using anti-GFP antibodies. After RIP, RNA was extracted and detected by RT-PCR. NoAb means no antibody control. EIF4A was used as negative control. (B–G) The interactions of MAC5A with full-length and truncated MIR172b fragments. MYC-MAC5A and various MIR172b RNAs were transiently coexpressed in N. benthamiana as indicated. IP was performed with anti-MYC antibodies. NtEF1A was used as negative control. (H) Purified MBP and MBP-MAC5A detected by SDS/PAGE. (I) The diagram of RNAs used for in vitro binding assay. (J) The in vitro interaction of MAC5A with the stem-loop region of pri-/premiRNAs, dsRNAs, and ssRNAs. The RNAs were generated through in vitro transcription.
MAC5A Binds the Stem-Loop Region of the Pri-miRNAs and Requires the Loop for Its Binding.
Next, we asked the potential MAC5A-binding region within pri-miRNAs using an in vivo pri-miRNA-binding assay that has been used to analyze STV1-pri-miRNA interaction (28). In this assay, we first examined the interaction of MYC-MAC5A with MIR172b (Fig. 4B and SI Appendix, Fig. S5 A–C) in N. benthamiana transiently expressing p35S::MYC-MAC5A and p35S::MIR172b. p35S::MIR172b produced a MIR172b transcript that comprises an ∼287-nt upstream arm, a stem loop, and a 417-nt downstream arm, flanked by an ∼120-nt upstream vector sequence and a ∼354-nt downstream vector sequence (MIR172bFL). As expected, MYC-MAC5A binds MIR172bFL, but not the endogenous control NtEF1A RNA from N. benthamiana (Fig. 4B). We further tested the interaction of MYC-MAC5A with the 5′ arm (MIR172bF1; ∼287 nt), MIR172bF2 containing a 5′ arm (∼287 nt) and a 6-nt 3′ arm, the 3′ arm (MIR172bF3; ∼417 nt), MIR172F5 containing a 39-nt 5′ arm and a 6-nt 3′ arm, and MIR172F7 containing a 10-nt 5′ arm and a 6-nt arm (SI Appendix, Fig. S5C). MAC5A was able to bind MIR172bF2, MIR172bF4, and MIR172bF7, but not MIR172bF1 and MIR172bF3 (Fig. 4 C–G). These results suggest that MAC5A may bind the stem-loop region of pri-miRNAs and requires the loop region for its binding.
To confirm the result, we next tested if the recombinant MAC5A protein fused with a MBP tag (MBP-MAC5A) can directly bind a short form pri-miR162b, which contains the stem loop with 6-nt arms at each end, premiR162b and premiR172b, both of which have a 2-nt 3′ overhang (Fig. 4I) (26). After purification with amylose resin, MBP and MBP-MAC5A (Fig. 4H) were incubated with radioactive-labeled pri-miR162b, premiR162b, or premiR172b. MBP-MAC5A, but not MBP, retained pri-miR162b, premiR162b, and premiR172b (Fig. 4J). These results confirmed that MAC5A is able to bind the stem-loop region of pri-miRNAs. We also examined whether MAC5A binds dsRNA or single-strand RNA (ssRNA) (Fig. 4I). However, MAC5A does not bind either dsRNA or ssRNA, suggesting that MAC5A may recognize the stem-loop structure within pri-miRNAs. Consistent with this notion, deletion of the loop region of premiR172b blocked the MAC5A binding (Fig. 4 I and J).
MAC5A Protects Pri-miRNAs from XRN2 or XRN3 Activity.
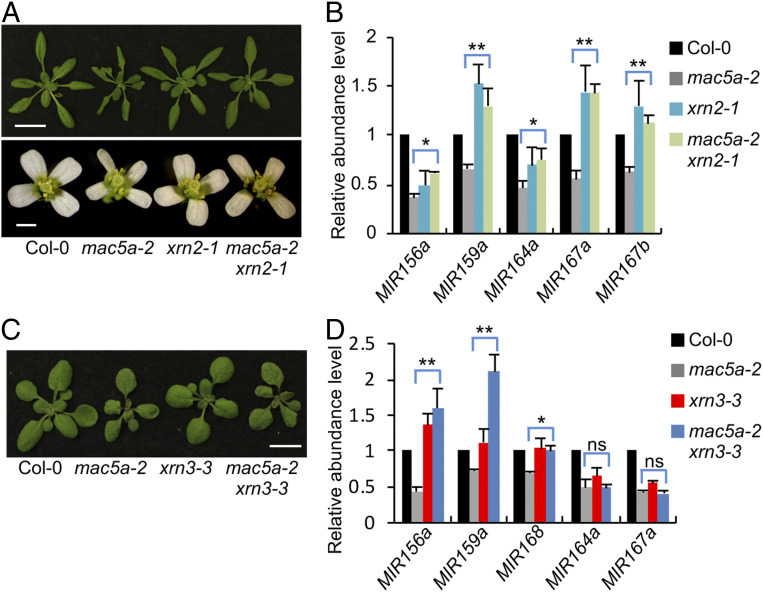
The facts that MAC5A binds the stem loop of pri-miRNAs and mac5a reduces the abundance of pri-miRNAs without effects on MIR transcription suggest that MAC5A may protect pri-miRNA from degradation by ribonucleases. In plants, the nuclear-localized exosome that degrades RNAs from 3′-to-5′ and the 5′-to-3′ exoribonucleases XRN2 and XRN3 are known to degrade aberrant RNAs. We thus examined if they could degrade pri-miRNAs when MAC5A is lacking. We first used artificial miRNA to target RRP44, an essential component of nuclear-localized exosome. We did not detect the effect of RRP44 on pri-miRNA accumulation in mac5a (SI Appendix, Fig. S6A). We also generated a mac5a-2 xrn2-1 double mutant and a mac5a-2 xrn3-3 double mutant. If XRN2/3 degrades pri-miRNAs in mac5a, we would expect that the amounts of pri-miRNAs in the double mutants should be similar to that in xrn2/3 single mutants. Introducing the xrn2 mutation into mac5a partially rescued the developmental defects of mac5a (Fig. 5A and SI Appendix, Fig. S6 B–E). Agreeing with this observation, the amounts of several examined pri-miRNAs and miRNAs were higher in mac5a xrn2 than those in mac5a and were similar to those in xrn2 (Fig. 5B and SI Appendix, Fig. S6F). Moreover, the examined miRNA target levels were decreased in mac5a xrn2 relative to mac5a (SI Appendix, Fig. S6G). Although the xrn3 mutation did not rescue the growth deficiency of mac5a (Fig. 5C), it did increase the amount of pri-miR156a, pri-miR159a, and pri-miR168 among several examined pri-miRNAs (Fig. 5D). We also generated a mac5a xrn4 double mutant to test the effect of cytoplasm-localized XRN4 on pri-miRNAs in mac5a. xrn4 did not rescue the pri-miRNA levels in mac5a or the growth defects of mac5a (SI Appendix, Fig. S6 H and I). These results reveal that nuclear-localized XRN2 and XRN3 are enzymes degrading pri-miRNAs in mac5a. To confirm that MAC5 indeed protects pri-miRNAs from XRN2-mediated degradation, we analyzed the half-lives of pri-miRNAs using cordycepin as a transcriptional inhibitor in WT, mac5a, xrn2, and mac5a xrn2. The half-lives of examined pri-miRNAs in mac5a were similar to those in WT. This result supports that MAC5 stabilizes pri-miRNAs (SI Appendix, Fig. S7A), because less efficient pri-miRNA processing in mac5a (see below) shall increase pri-miRNA half-life. Moreover, the half-life of pri-miRNAs is longer in mac5a xrn2 than that in WT, mac5a, and xrn2 (SI Appendix, Fig. S7A), further supporting the dual function of MAC5, promoting pri-miRNA processing and stabilizing pri-miRNAs.
Fig. 5.
MAC5A protects pri-miRNAs from 5′-to-3′ degradation. (A) Phenotypes of 4-wk-old plants (Top) and flowers (Bottom) of the four indicated genotypes. (Scale bars: 2 cm, Top and 1 mm, Bottom.) (B) pri-miRNA levels detected by qRT-PCR. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with mac5a-2 value). (C) Phenotypes of 3-wk-old plants of the four indicated genotypes. (Scale bar: 1 cm.) (D) pri-miRNA levels detected by qRT-PCR. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with mac5a-2 value. ns indicates no significant difference).
It should be noted that not all examined pri-miRNA are affected by XRN2 or XRN3. Since this observation might be caused by the potential redundancy between XRN2 and XRN3, we generated a mac3a xrn2 xrn3 mutant. However, we did not observe the additive impacts of XRN2 and XRN3 on pri-miRNA accumulation in mac5a (SI Appendix, Fig. S8), suggesting that other enzymes may also be involved in pri-miRNA degradation in mac5a.
MAC5A and SE Act Additively on Pri-miRNA Processing.
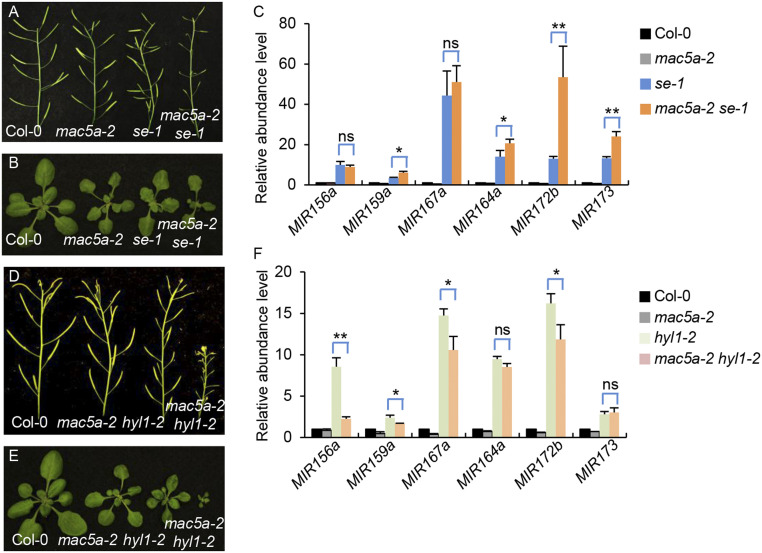
The physical interaction between MAC5A and SE promoted us to test their genetic interaction by double mutant analysis. We generated a mac5a-2 se-1 double mutant by crossing mac5a-2 into se-1. mac5a se showed more severe growth and fertility defects than either mac5a-2 or se-1, suggesting that MAC5A and SE function additionally in regulating growth and development (Fig. 6 A and B). Consistent with these observations, the miRNA levels in double mutants were further reduced compared with mac5a-2 or se-1 (SI Appendix, Fig. S9A), suggesting that MAC5A functions synergistically with SE in controlling miRNA levels. The reduction of miRNA levels in mac5a-2 se-1 relative to mac5a-2 and se-1 has at least two nonmutually exclusive explanations. MAC5A and SE may contribute differently on DCL1 activities. Alternatively, mac5a may impair the accumulation of pri-miRNAs in se. We first examined the effect of mac5a and se on premiRNA processing using an in vitro assay (9, 54). The radioactive labeled premiR162b was incubated with the protein extracts from young flower buds of Col, mac5a, se, or mac5a se. Both mac5a and se reduced the processing of premiR162b (SI Appendix, Fig. S9 B and C). Notably, the processing efficiency of premiR162b was lower in mac5a se than that in either mac5a or se single mutants (SI Appendix, Fig. S9 B and C), suggesting that MAC5A and SE contribute additively to miRNA processing.
Fig. 6.
pri-miRNA degradation in mac5a depends on SE, but not HYL1. (A and B) Phenotypes of siliques (A) and 3-wk-old plants (B) in Col-0, mac5a-2, se-1, and mac5a-2 se-1. (B) pri-miRNA levels detected by qRT-PCR. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with se-1 value. ns indicates no significant difference). (D and E) Phenotypes of siliques (D) and 3-wk-old plants (E) in Col-0, mac5a-2, hyl1-2, and mac5a-2 hyl1-2. (F) pri-miRNA levels detected by qRT-PCR in Col-0, mac5a-2, hyl1-2, and mac5a-2 hyl1-2. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with hyl1-2 value. ns indicates no significant difference).
Pri-miRNA Degradation in mac5a Depends on SE.
We next analyzed the pri-miRNA levels in mac5a-2 se-1 using qRT-PCR analyses. As previously reported, se-1 increased the accumulation of pri-miRNAs (55). The levels of several examined pri-miRNAs in mac5a-2 se-1 were either increased or unaltered relative to se-1 (Fig. 6C and SI Appendix, Fig. S9D). This result further supports that the reduced miRNA levels are caused by the additive effect of SE and MAC5A on DCL1 activity. More importantly, this result shows that the degradation of pri-miRNAs in mac5a depends on a functional SE. Since SE is a key regulator of DCL1 activity, it is possible that the pri-miRNA degradation in mac5a may rely on pri-miRNA processing. To test this possibility, we generated a mac5a hyl1 double mutant. As expected, mac5a hyl1 had much stronger developmental defects and contained lower miRNAs than mac5a or hyl1 (Fig. 6 D and E and SI Appendix, Fig. S9 A and E). However, the pri-miRNA levels in mac5a hyl1 were lower than that in hyl1 (Fig. 6F), demonstrating that the degradation of pri-miRNAs in mac5a is independent of pri-miRNA processing. We also examined the half-lives of pri-miRNAs in WT, mac5a-2, se-1, and mac5a-2 se-1, hyl1-2, and mac5a-2 hyl1-2. We observed that the half-lives of pri-miRNAs in mac5a-2 se-1 were similar to those in se-1 (SI Appendix, Fig. S7B), agreeing with the observation of the steady pri-miRNA levels either elevated or unchanged in mac5a se relative to se. These data show that se is epistatic to mac5, demonstrating that the degradation of pri-miRNAs in mac5a depends on SE. The half-lives of pri-miRNAs in mac5a-2 hyl1 were slightly shorter than those in hyl1 (SI Appendix, Fig. S7C). It should be noted in hyl1, the half-lives of pri-miRNAs were longer than WT. One possible explanation is that the processing of pri-miRNAs is decreased in hyl1. Another nonmutually exclusive possibility is that HYL1 may also be able to stabilize pri-miRNAs, which may also explain the further reduction of pri-miRNAs in mac5a hyl1 relative to hyl1.
Since XRN2 is likely an enzyme degrading pri-miRNAs in mac5a, we next asked if SE interacts with XRN2. We transiently coexpressed XRN2-GFP and MYC-SE in N. benthamiana and performed IP with anti-GFP antibodies. We were able to detect MYC-SE in the XRN2-GFP IPs but not in GFP IPs (SI Appendix, Fig. S10), suggesting a potential SE-XRN2 interaction.
Discussion
MAC5 is a conserved protein in eukaryotes. Its orthologs CWC2 in yeast and RBM22 in human play critical roles in splicing and development as members of NTC and RPP19, respectively. In plants, they have crucial roles in development and immunity. However, MAC5 has only a minor effect on splicing (53). Consistent with this observation, lack of MAC3, PRL1, or MAC7 has a moderate effect on splicing at global levels, which does not correlate with gene expression (27). Here, we show that MAC5 is required for miRNA accumulation. Interestingly, miRNA levels in mac5a MAC5B/mac5b are lower than those in mac5a, suggesting that MAC5A and MAC5B may have a dosage-dependent impact on miRNA abundance. In addition, MAC5 interacts with SE and pri-miRNAs. These results support that MAC5 acts in miRNA biogenesis. Several other MAC components also play essential roles in miRNA biogenesis (19, 26, 27, 29), suggesting that MAC acts as a complex to promote miRNA biogenesis. However, the MAC components are diversified proteins such as transcription factors, ubiquitin ligases, RNA-binding proteins, RNA helicases, and others. Thus, it is likely that the individual MAC component has specific and coordinate functions in miRNA biogenesis as subunits of MAC.
MAC5 may enhance the DCL1 activity since the in vitro miR162 production is reduced in the mac5a protein extracts. However, MAC5 does not interact with DCL1, indicating that it may not directly modulate the DCL activity. There are several nonmutually exclusive possibilities by which MAC5 promotes pri-miRNA processing. It may function as an accessory protein of CDC5, which directly promotes the DCL1 activity (19, 26). It may enhance the association of MAC with the DCL1 complex through its interaction with pri-miRNAs. Agreeing with this notion, CWC2 in yeast links NTC to the spliceosome through its interaction with the U6 snRNA (56). It is also possible that MAC5 binding alters the stem-loop structure, and thereby facilitates pri-miRNA processing. Indeed, CWC5/RBM22 interaction induces an active conformation of the spliceosome’s catalytic RNA elements in human and yeast (57).
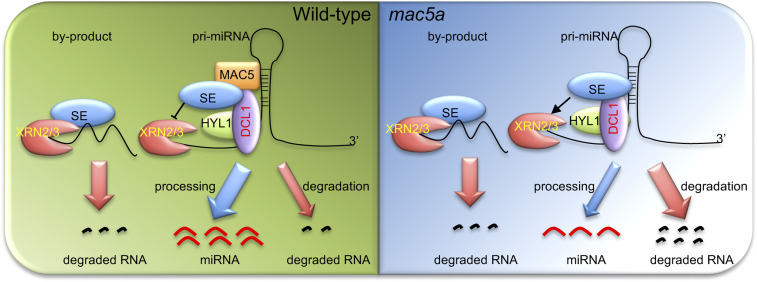
MAC5 is also required for the accumulation of pri-miRNAs. The reduced pri-miRNA accumulation in mac5a can be caused by impaired pri-miRNA transcription or stability. However, MAC5 does not affect pri-miRNA promoter activity and the occupancy of Pol II at pri-miRNA promoters, arguing against the first possibility. In fact, loss-of-function nuclear-localized XRN2 or XRN3, but not cytoplasm-localized XRN4, recovers pri-miRNA accumulation in mac5a, strongly supporting that MAC5 protects pri-miRNAs from the nuclear-localized ribonuclease activities. However, additional enzymes should also be involved in pri-miRNA degradation in mac5a as not all of the examined pri-miRNAs are affected by xrn2 or xrn3. Interestingly, pri-miRNA levels are either elevated or unchanged in mac5a se relative to se. This result shows that the reduction of pri-miRNA abundance in mac5a is dependent on SE. It is not due to the role of SE in pri-miRNA processing, given the fact that pri-miRNA levels are lower in mac5a hyl1 relative to hyl1. It is also unlikely caused by the role of SE in transcription, since SE mainly promotes the transcription of intron-less genes in Arabidopsis (58). We also detect the potential association between SE and XRN2. Thus, MAC5 likely protects pri-miRNAs from SE-dependent nuclease activities. How does MAC5 stabilize pri-miRNA? We suspect that MAC5 binding may prevent the attack of SE-dependent nuclease on pri-miRNAs (Fig. 7). It is also possible that MAC5 may prevent the nuclease activities through decoying SE (Fig. 7). Clearly, these possibilities need further investigation.
Fig. 7.
The proposed model for processing and degradation of pri-miRNAs. In wild-type plants, SE may link the RNA degradation enzymes including XRN2/3 and others to degrade the byproducts. MAC5 binds and protects pri-miRNAs from degradation. MAC5 may also decoy SE. MAC5 interacts with SE or pri-miRNAs to promote pri-miRNA processing. While in mac5a background, XRN2/3 and other enzymes will act on pri-miRNAs.
SE is a component of the CAP-binding complex. Its counterpart, ARS2, from human and yeast participates in the nuclear aberrant RNA decay through its interaction with the RNA surveillance machinery (59, 60). Thus, SE-dependent degradation of pri-miRNAs in mac5 suggests that SE connects RNA processing with degradation in plants (Fig. 7). Indeed, the degradation of pri-miRNA processing byproducts requires XRN2/3 (61, 62). The connection of SE, a component of the DCL1 complex, with the RNA surveillance machinery will ensure the quick removal of the processing byproducts, which are produced cotranscriptionally. Therefore, the finding that MAC5 protects pri-miRNAs from SE-dependent nucleases reveals that pri-miRNAs and the miRNA/miRNA* duplex-containing products needs to be protected from the RNA surveillance machinery during pri-miRNA transcription and processing.
Materials and Methods
Plant Materials and Growth Condition.
The mac5a-2 (Salk_142085), mac5b-1 (GK-419F11), se-1 (CS3257), xrn2-1 (Salk_041148), xrn3-3 (Sail_1172C07), and xrn4-5 (CS68822) mutants are in the Columbia (Col-0) background. These mutants were obtained from the Arabidopsis Biological Resources Center (ABRC). The transgenic line harboring the pMIR167a::GUS transgene was crossed to mac5a-2. In the F2 generation, MAC5A+ (MAC5A/MAC5A and MAC5A/mac5a-2) and mac5a-2 were isolated through PCR genotyping. Fifteen MAC5A+ or mac5a-2 plants were pooled for GUS transcript level analyses. The mac5a-2 se-1, mac5a-2 hyl1-2, mac5a-2 xrn2-1, mac5a-2 xrn3-3, and mac5a-2 xrn4-5 mutants were isolated by PCR genotyping with the primers listed in SI Appendix, Table S1. All plants were grown at 22 °C with 16-h light and 8-h dark cycles.
Plasmid Construction.
The MAC5A genomic sequence containing a 1,215-bp promoter and 2,231-bp coding regions was PCR amplified from Col-0 with the primers of proMAC5A-1F and MAC5Acds-1R. The resulted PCR product was cloned into pENTR/D-TOPO vector and subsequently cloned into pMDC204 binary vector to generate the proMAC5A::MAC5A-GFP (gMAC5A-GFP) plasmid. The MAC5A full-length cDNA was RT-PCR amplified with the primers of MAC5Acds-1F and MAC5Acds-1R, cloned into pENTR/D-TOPO vector, and subcloned into pEarleyGate203 and pMDC83 to generate the p35S::MYC-MAC5A and p35S::MAC5A-GFP constructs, respectively. To construct cCFP-MAC5A, MAC5A cDNA sequence was PCR amplified using the primer pair MAC5A-2F and MAC5A-2R and cloned into pSAT4-cCFP-C vector. Then, the p35S::cCFP-MAC5A fragment was released by I-SceI restriction enzyme digestion and cloned into pPZP-RCS2-ocs-bar-RI vector. The constructs nVenus-CDC5, nVenus-DCL1, nVenus-HYL1, nVenus-SE, and nVenus-PRL1 were described previously (9). To construct MBP-MAC5A, the MAC5A cDNA sequence was amplified with primer MAC5A-3F and MAC5A-3R and subsequently inserted into the pMAL-C5X vector. The XRN2 cDNA sequence was PCR amplified using primer XRN2cds-1F and XRN2cds-1R, cloned into pENTR/D-TOPO vector, and subsequently cloned into pMDC83. The primers are listed in SI Appendix, Table S1.
Plant Complementation.
The plasmids of gMAC5A-GFP and p35S::MYC-MAC5A were transformed into mac5a using the Agrobacterium-mediated floral dip method, respectively. The transgenic plants harboring gMAC5A-GFP were selected on MS (Murashige and Skoog) medium containing hygromycin (30 μg/mL). p35S::MYC-MAC5A transformants were selected by spraying seedlings with 120 mg/L BASTA (Glufosinate, ammonium salt) solution.
In Vitro Protein Expression and Purification.
Escherichia coli strain BL21 (DE3) containing MBP-MAC5A fusion construct was grown to OD600 = 0.5 and induced overnight at 16 °C with 0.5 mM IPTG (isopropyl β-d-1-thiogalactopyranoside). Bacterial cells were harvested and sonicated in extraction buffer {50 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 5% glycerol and 1 mM PMSF (phenylmethylsulfonyl fluoride)}. The expressed protein was purified with amylose resin (NEB).
Co-IP Assay.
The Co-IP assay was performed as previously described (9). To examine the interactions of MAC5A with CDC5 or SE, MYC-MAC5A and MAC5A-GFP were transiently coexpressed with CDC5-GFP and MYC-SE in N. benthamiana leaves, respectively. The interaction between XRN2 and SE was performed by coexpressing XRN2-GFP and MYC-SE in N. benthamiana. IP was performed on protein extracts using anti-YFP or anti-MYC antibodies coupled to protein G agarose beads. After IP, proteins were detected with Western blot using monoclonal antibodies against YFP or MYC. For the interaction of MAC5A and SE in Arabidopsis, IP was performed with anti-SE antibody in p35S::GFP and gMAC5A-GFP transgenic plants, respectively. After IP, proteins were detected with Western blot using anti-GFP or anti-SE antibodies.
BiFC Assay.
The BiFC assay was performed as described (19). Paired cCFP and nVenus fusion proteins were coexpressed in N. benthamiana leaves. After a 40-h expression, confocal microscopy (Fluoview 500 workstation; Olympus) was used to detect YFP and chlorophyll autofluorescence signals at 488 nm with a narrow barrier (505 to 525 nm, BA505-525; Olympus).
Northern Blot and qRT-PCR Analyses.
The Northern blot was performed as described (9). A total of ∼15 μg total RNAs extracted from inflorescences were resolved on 16% PAGE (polyacrylamide gel electrophoresis) gel and transferred to nylon membranes. 32P-labeled antisense DNA oligonucleotides were used to detect small RNAs. Radioactive signals were detected with a PhosphorImager and quantified with ImageQuant.
The levels of pri-miRNAs, miRNA target transcripts, and GUS mRNA were determined using qRT-PCR. A total of 1 μg total RNAs from inflorescences was used to generate cDNAs using the SuperScript III reverse transcriptase (Invitrogen) and an oligo dT18 primer. cDNAs were then used as templates for qPCR on an iCycler apparatus (Bio-Rad) with the SYBR green kit (Bio-Rad). The primers used for PCR are listed in SI Appendix, Table S1.
RIP Analyses.
RIP was performed according to refs. 9, 63. A total of ∼2 g seedlings of transgenic plants harboring pMAC5A::MAC5A-GFP (gMAC5A-GFP) construct were used to examine the RNA binding activity of MAC5A in vivo. After cross-linking with 1% formaldehyde for 10 min, glycine was added to quench the reaction for 10 min. Nuclei were extracted and lysed in the buffer {50 mM Tris⋅HCl pH 8.0, 10 mM EDTA (ethylenediaminetetraacetic acid), 1% SDS (sodium lauryl sulfate)} by sonication five times. After debris was removed by centrifuge at 16,000 × g for 10 min, equal amounts of proteins from various samples were diluted with RIP dilution buffer and incubated with anti-MYC antibodies conjugated to protein G agarose beads. The immunoprecipitates were then eluted with elution buffer (100 mM NaHCO3, 1% SDS) at 65 °C. Flowing reversing cross-linking with proteinase K (Invitrogen) and 200 mM NaCl at 65 °C, RNAs were extracted and used as templates for RT-PCR analyses.
To identify the binding region of pri-miRNA by MAC5A protein, MYC-MAC5A and a series of truncated pri-miR172b were transiently coexpressed in N. benthamiana leaves and RIP assay was performed as previously reported (28).
For in vitro RNA-binding assay, DNA templates for RNA strands of pri-miR162b, premiR162b, premiR172b, premiR172bΔloop, dsRNA, and ssRNA were PCR amplified using the primers listed in SI Appendix, Table S1. Resulting DNA templates were used to synthesize RNA strands through in vitro transcription using T7 RNA polymerase (Promega) under the presence of [α-32P] UTP. The unlabeled competitors were generated in the absence of [α-32P] UTP. After transcription, the RNA transcripts were annealed in the annealing buffer (10 mM Tris pH 8.0, 20 mM NaCl, 1 mM EDTA). The RNA-binding assay was performed as previously report (9).
ChIP Assay.
ChIP was performed using 14-d-old seedlings from Col-0 and mac5a-2 as described by Kim et al. (17). Three biological replicates were performed. Anti-RPB2 antibody (Clontech) was used for immunoprecipitation. qPCR was performed on DNAs copurified with RPB2 antibody, using primers listed in SI Appendix, Table S1.
Small RNA Sequencing.
A small RNA library was prepared using total RNAs extracted from inflorescences of Col and mac5a. Two biological replicates were performed. After sequencing, miRNA analysis was performed according to Ren et al. (9). The total numbers of perfectly aligned reads were used for normalization (64). EdgeR with trimmed mean of M value (TMM) normalization method was used to compare miRNA abundance (65).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31872816 to S.L.), the NIH (GM127414 to B.Y.), and the NSF (MCB-1808182 to B.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008283117/-/DCSupplemental.
Data Availability.
All data generated by this study are included in the paper and SI Appendix. All sequencing data generated for this study have beendeposited to the National Center for Biotechnology Information Gene Expression Omnibus (Col accession nos. GSM2829820 and GSM2829821; mac 5a accession nos. GSM2829824 and GSM2829825).
References
- 1.Baulcombe D., RNA silencing in plants. Nature 431, 356–363 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Song X., Li Y., Cao X., Qi Y., MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Voinnet O., Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Achkar N. P., Cambiagno D. A., Manavella P. A., miRNA biogenesis: A dynamic pathway. Trends Plant Sci. 21, 1034–1044 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Li S., Castillo-Gonzalez C., Yu B., Zhang X., The functions of plant small RNAs in development and in stress responses. Plant J. 90, 654−670 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez R. E., Schommer C., Palatnik J. F., Control of cell proliferation by microRNAs in plants. Curr. Opin. Plant Biol. 34, 68–76 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kurihara Y., Watanabe Y., Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 101, 12753–12758 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Z., Han M. H., Fedoroff N., The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 9970–9975 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren G. et al., Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 12817–12821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z. et al., Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers K., Chen X., Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumberger N., Baulcombe D. C., Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 11928–11933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earley K. W., Poethig R. S., Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 286, 38184–38189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith M. R. et al., Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5424–5429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C., CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24, 1626–1642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X., Cui Y., Li Y., Qi Y., Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1, 15075 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Kim Y. J. et al., The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–822 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L. et al., NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25, 715–727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S., Xie M., Ren G., Yu B., CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 110, 17588–17593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Q. et al., The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 9, 5080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., A silencing safeguard: Links between RNA silencing and mRNA processing in Arabidopsis. Dev. Cell 14, 811–812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S. et al., Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 49, 1634–1644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laubinger S. et al., Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 8795–8800 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speth C., Willing E. M., Rausch S., Schneeberger K., Laubinger S, RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 76, 433−445 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Yu B. et al., The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 10073–10078 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S., Liu Y., Yu B., PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10, e1004841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia T. et al., The Arabidopsis MOS4-associated complex promotes MicroRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29, 2626–2643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S. et al., STV1, a ribosomal protein, binds primary microRNA transcripts to promote their interaction with the processing complex in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 1424–1429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S. et al., MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell 30, 481–494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köster T. et al., Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 42, 9925–9936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao Y., Shi J., Zhai Y., Hou Y., Ma W., Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U.S.A. 112, 5850–5855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X. et al., A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 23, 645–657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z. et al., SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Manavella P. A. et al., Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151, 859–870 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Ben Chaabane S. et al., STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 41, 1984–1997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang X., Shi Y., Lu X., Chen Z., Qi Y., CMA33/XCT regulates small RNA production through modulating the transcription of Dicer-like genes in Arabidopsis. Mol. Plant 8, 1227–1236 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Raghuram B., Sheikh A. H., Rustagi Y., Sinha A. K., MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 282, 521–536 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Yan J. et al., The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13, e1006753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho S. K., Ben Chaabane S., Shah P., Poulsen C. P., Yang S. W., COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5, 5867 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Su C. et al., The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev. Cell 41, 527–539.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Li S. et al., SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 46, 9148–9159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z. et al., Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genet. 14, e1007247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Achkar N. P. et al., A quick HYL1-dependent reactivation of MicroRNA production is required for a proper developmental response after extended periods of light deprivation. Dev. Cell 46, 236–247.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Cai X., Hagedorn C. H., Cullen B. R., Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y. et al., MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J., XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta 1829, 590–603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jopling C. L., Stop that nonsense!. eLife 3, e04300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lykke-Andersen S., Jensen T. H., Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Fang X. et al., Chloroplast-to-nucleus signaling regulates MicroRNA biogenesis in Arabidopsis. Dev. Cell 48, 371–382.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Deng X. et al., Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. U.S.A. 113, 5447–5452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palma K. et al., Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monaghan J. et al., Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monaghan J., Xu F., Xu S., Zhang Y., Li X., Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant Physiol. 154, 1783–1793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi Y., Denli A. M., Hannon G. J., Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19, 421–428 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Yang L., Liu Z., Lu F., Dong A., Huang H., SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850 (2006). [DOI] [PubMed] [Google Scholar]
- 56.McGrail J. C., Krause A., O’Keefe R. T., The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res. 37, 4205–4217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasche N. et al., Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J. 31, 1591–1604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speth C. et al., Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. eLife 7, e37078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugiyama T. et al., Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 61, 747–759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan J. et al., Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 36, 2870–2886 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gy I. et al., Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurihara Y. et al., Surveillance of 3′ noncoding transcripts requires FIERY1 and XRN3 in Arabidopsis. G3 (Bethesda) 2, 487–498 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wierzbicki A. T., Haag J. R., Pikaard C. S., Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nobuta K., McCormick K., Nakano M., Meyers B. C., Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol. Biol. 592, 89–106 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated by this study are included in the paper and SI Appendix. All sequencing data generated for this study have beendeposited to the National Center for Biotechnology Information Gene Expression Omnibus (Col accession nos. GSM2829820 and GSM2829821; mac 5a accession nos. GSM2829824 and GSM2829825).