Significance
RNAs move within cells and often reside at specific locations. The need to detect those movements and locales across the transcriptome is pressing. We report an approach in living cells that provides a record of localization across the entire collection of RNAs a cell contains. It relies on covalently marking the RNA when it directly encounters a specific site. Using this approach, we identify a conserved battery of RNAs that are identified at more than one location in yeast and human cells, noncanonical modes of localization, and RNAs undergoing processing or surveillance at discrete subcellular locations. Our findings provide an entrée to the histories of individual RNA molecules through covalent marks.
Keywords: RNA localization, RNA regulation, localized RNA records
Abstract
RNA movements and localization pervade biology, from embryonic development to disease. To identify RNAs at specific locations, we developed a strategy in which a uridine-adding enzyme is anchored to subcellular sites, where it directly marks RNAs with 3′ terminal uridines. This localized RNA recording approach yields a record of RNA locations, and is validated through identification of RNAs localized selectively to the endoplasmic reticulum (ER) or mitochondria. We identify a broad dual localization pattern conserved from yeast to human cells, in which the same battery of mRNAs encounter both ER and mitochondria in both species, and include an mRNA encoding a key stress sensor. Subunits of many multiprotein complexes localize to both the ER and mitochondria, suggesting coordinated assembly. Noncoding RNAs in the course of RNA surveillance and processing encounter both organelles. By providing a record of RNA locations over time, the approach complements those that capture snapshots of instantaneous positions.
Localization of specific RNAs to discrete subcellular locations was first observed in striking examples during early development (1–6) and in yeast (7). We now know RNA localization is widespread and critical in secretion, patterning, cell fate determination, and neurobiology (8, 9). Many messenger RNAs (mRNAs) in embryos and mammalian cells exhibit discrete patterns of localization, emphasizing its breadth. Localization hinges on the interplay between sequences in the RNAs, RNA binding proteins, molecular motors, and subcellular structures, such as the endoplasmic reticulum (ER) or cytoskeleton (10–13). Advancements in FISH (14–17), live imaging (8, 18, 19), and sequencing-based methods, including proximity-specific ribosome profiling (20, 21), APEX-RIP (22), and APEX-seq (23, 24), are very powerful, but typically provide snapshots of localized RNA, with a time scale of minutes, since cells do not survive the required treatments. They also often require either custom oligonucleotide probes or sophisticated equipment. Approaches are needed to identify RNAs at any subcellular location across the entire transcriptome and to do so in living cells.
We developed a strategy that provides a record of RNAs as they interact with a cellular site in vivo. In this approach, termed “localized RNA recording,” a U-adding enzyme is anchored to a specific subcellular location. The anchored enzyme marks RNA molecules it encounters in vivo, which are identified through deep sequencing. Since cells live during RNA recording, the number of Us added to a given molecule likely mirrors the cumulative time spent at that location. The approach identifies RNAs that encountered specific sites, as evidenced by our analysis of the ER and mitochondria in the yeast, Saccharomyces cerevisiae). We identify RNAs recorded at both locations, including noncoding (nc) RNAs and mRNAs, RNAs undergoing processing and surveillance, and mRNAs recorded at different locations that encode subunits of the same multiprotein complex.
Results
Broad-Specificity Recording.
We first designed a protein construct intended to mark, or “record,” its interaction with most or all cellular RNAs, and so provide a baseline for comparison. We selected Caenorhabditis elegans PUP-2 (25) as the recording agent since it adds uridines to RNA 3′ ends, lacks RNA-binding domains, and has been used to identify RNAs bound to specific proteins in living cells (26–28) (SI Appendix, Fig. S1A). To facilitate identification of most RNAs in the cytoplasm, we linked PUP-2 to the RNA-recognition motifs (RRMs) of yeast poly(A)-binding protein, generating a construct here termed “PUP alone (+PAB).” A control chimera, “PUP alone (−PAB),” was constructed that lacked the PAB RRMs (SI Appendix, Fig. S1A). Both proteins were expressed under control of the SEC63 promoter (20) (used later to enable direct comparison to RNAs at the ER).
To identify recorded RNAs and the number of Us they received, we prepared polyadenylated RNA via oligo(dT) selection and ribosomal RNA depletion (26) (Materials and Methods). RNAs then were reverse-transcribed using a primer designed to enrich uridylated RNAs (SI Appendix, Fig. S1B). The resulting DNA libraries were analyzed on an Illumina sequencer using paired-end sequencing (SI Appendix, Fig. S1B). Sequencing data were processed using a computational pipeline (26) that identified all RNAs that had received U residues, as well as the number of reads obtained, and the number of Us added (SI Appendix, Fig. S1 C and D).
The two PUP alone proteins added Us to cellular RNAs with very different efficiencies. PUP alone (+PAB) yielded three to four orders of magnitude more recorded reads per million across all U-tail lengths (SI Appendix, Fig. S2A). PUP alone (+PAB) was more reproducible (SI Appendix, Fig. S2 B and C), and yielded more recorded species (SI Appendix, Fig. S2D and Datasets S1 and S2). The number of Us added was correlated with RNA abundance with both proteins (SI Appendix, Fig. S3A and Dataset S3). Recorded RNAs were ranked based on the number of Us added and the number of reads obtained (SI Appendix, Fig. S3B), which revealed the dramatic differences with and without the PAB RRMs. We adopted the protein with RRMs for subsequent experiments due to its efficiency and ability to tag most cellular RNAs. We refer to it simply as “PUP alone” hereafter.
ER-Localized RNA Recording.
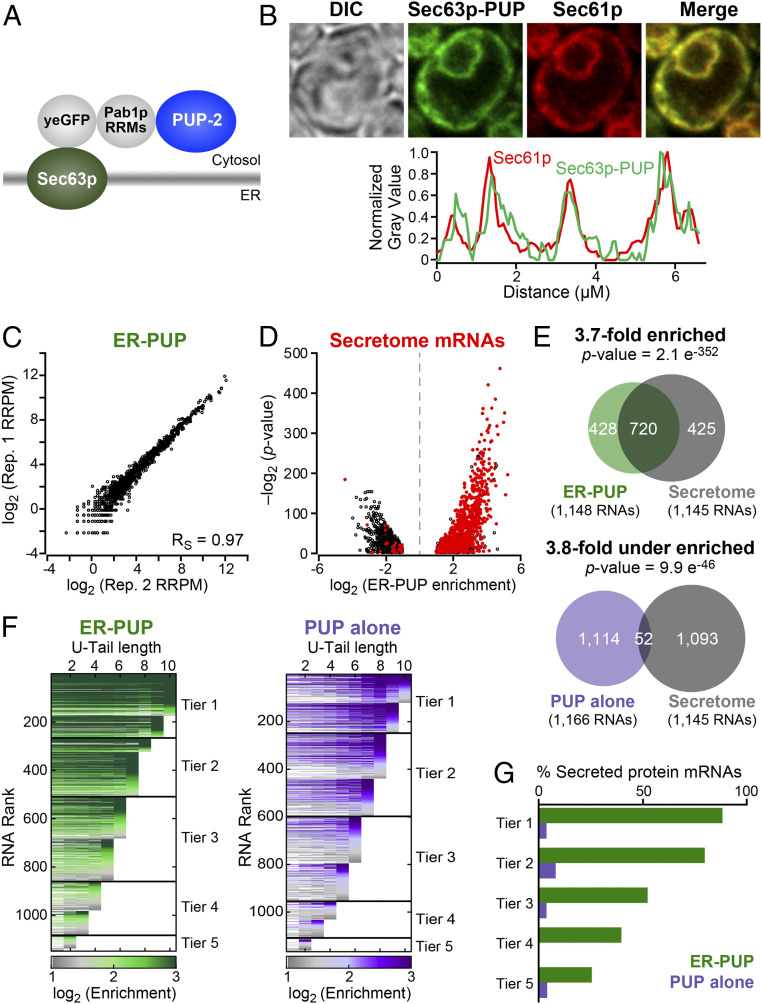
To detect RNAs that encounter the ER, we fused the PUP alone chimera to the C terminus of Sec63p, a protein embedded in the ER (Fig. 1A), and expressed the chimeric protein from the endogenous SEC63 locus. The Sec63p chimera, termed “ER-PUP,” is predicted to be cotranslationally embedded into the ER membrane by three transmembrane segments of Sec63p, and place the C-terminal PUP-2 domain in the adjacent cytosol (29). As predicted, GFP fluorescence from ER-PUP mirrored the pattern reported for Sec63p, and colocalized with signal from the Sec61p-mCherry ER-marker (30, 31) (Fig. 1B and SI Appendix, Fig. S4A), indicating that ER-PUP was anchored to the ER membrane.
Fig. 1.
ER-localized recording provides an in vivo record. (A) ER-PUP chimera designed to tag ER-proximal RNAs. PUP(+PAB) was fused to the C terminus of Sec63p, which tethers the chimera to the cytosolic surface of the ER (29). (B) Subcellular distribution: ER-PUP GFP fluorescence (green) vs. an ER marker (30, 31), Sec61p-mCherry (red); merged (yellow). Fluorescence intensities for ER-PUP (green) and the ER marker (red) across a representative cell. The cells were imaged using a 63× numerical aperture 1.4 objective lens with oil immersion. (C) Reproducibility of ER-PUP recording across biological replicates. (D) Distribution of mRNAs that encode secreted proteins (“secretome”) among the ER-PUP–recorded mRNAs. (E) Secretome mRNAs in recording by ER-PUP and PUP alone. (F) Distribution of relative enrichment (ER-PUP vs. PUP alone) across each U-tail length. RNA species that had significant (adjusted P < 0.05) difference [log2(∆ recorded reads) ≥ 1] in recording efficiency (enrichment) in any of 10 U-tail lengths (1U-10Us) were isolated and plotted as horizontal lines. Each row is one mRNA. Lines are segmented, and segments colored in accord with enrichment values: most enriched denoted in green (ER-PUP) or purple (PUP) and the lowest gray (both). RNAs were binned into five groups (“tiers”), and tiers ranked by the highest tag length (e.g., tier 1 RNAs had a minimum of 9 to 10 U’s; tier 5 had a minimum of 1 to 2 Us). (G) Proportion (%) secretome mRNAs in each tier of ER-PUP-recorded (green) or PUP alone-recorded (purple) tier.
To identify RNAs recorded by ER-PUP, we compared RNAs identified with and without the Sec63p anchor, using the analytical tool DESeq2 (32), a method that identifies the statistical strengths of observed differences in RNA populations (SI Appendix, Fig. S4 B–E). The data were highly reproducible (Fig. 1C and SI Appendix, Fig. S4F). The ER-anchored PUP selectively recorded 1,148 RNAs (Dataset S4), which we refer to as “ER-enriched,” while the unanchored PUP preferentially recorded 1,166 (“ER-depleted”) (Dataset S5). Many mRNAs identified by ER-PUP encoded secreted proteins (33) (“secretome mRNAs”, P = 2.1 e−352) and had ER-related gene ontology (GO) associations (34, 35), which were reduced or missing among RNAs recorded less efficiently at the ER (Fig. 1 D and E, SI Appendix, Fig. S4G, and Dataset S6). Thus, the Sec63p anchor RNAs recorded RNAs that encountered the ER in vivo.
To aid in further analyses, we grouped RNAs recorded by ER-PUP and the control, PUP alone, into five tiers based on U-tail lengths. RNAs with the longest U-tails were grouped in tier 1 and those with the shortest tails in tier 5 (Fig. 1F). Within a tier, each RNA was ranked by the fold-enrichments in that dataset relative to the other. With both ER-PUP and PUP alone, RNAs with the longest U-tails generally had the highest enrichment (Fig. 1F). Among RNAs identified by ER-PUP, the fraction of secretome mRNAs was highest in tiers 1 and 2 (88% and 79%) and declined progressively to tier 5 (25%) (Fig. 1G). The control, PUP alone, yielded little enrichment or correlation with tiers (Fig. 1 F and G), demonstrating that ER-PUP preferentially recorded RNAs with ER-related functions.
To assess the relationship between ER-PUP enrichment and mRNA abundance, we binned all yeast RNAs into five tiers based on RNA sequencing (RNA-seq) (26) (fragments per kilobase of transcript per million mapped reads [FPKM]), from most (tier 1) to least abundant (tier 5) (SI Appendix, Fig. S5 A and B and Dataset S3). The distribution of abundances of RNAs identified by ER-PUP was much more similar across tiers as compared to mRNA abundances in the cell (SI Appendix, Fig. S5B). Indeed, highly abundant RNAs (abundance tiers 1 and 2) with secretome association were dramatically enriched by ER-PUP (4.6-fold enriched, P = 3.98e−233), while ones that lack secretome association were depleted (2.9-fold depleted, P = 1.8e−74) (SI Appendix, Fig. S5C). Furthermore, the gamut of RNA abundances was represented across all ER-PUP tiers, while PUP-alone tiers primarily contained the most abundant species (SI Appendix, Fig. S5D). Finally, the fraction of secretome mRNAs among ER-PUP recorded mRNAs dramatically exceeds that seen with PUP alone in tiers 1, 2, and 3 (Fig. 1 F and G and SI Appendix, Fig. S5E), and is highest for the best-recorded RNAs in every abundance tier. Thus, the primary driver of modification by ER-localized PUP is localization rather than abundance.
Many but Not all ER-Enriched RNAs Are Bound by ER-Proximal Ribosomes.
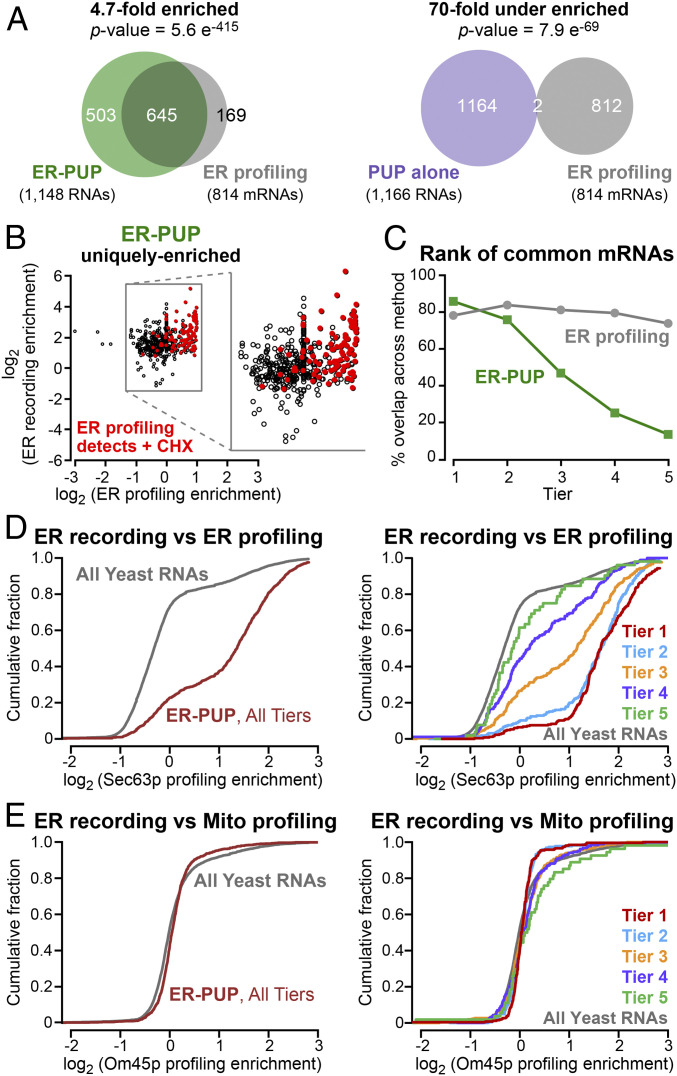
mRNAs recorded by ER-PUP are predicted to include ones translated at or near the ER. We compared mRNAs identified in ER-recording with those detected in proximity-specific ribosome profiling experiments that had used the same Sec63p anchor (20) (termed “ER profiling”) (Dataset S7). ER-profiling identifies ribosome-bound mRNAs near an ER-anchored biotin ligase that biotinylates Avi-tagged ribosomes (20). Of mRNAs associated with Sec63-proximal ribosomes, 79% were preferentially identified by ER-PUP (4.7-fold enriched, P = 5.6 e−415), and only 0.2% were preferentially identified by PUP alone vs. ER-PUP (70-fold depleted, P = 7.9 e−69) (Fig. 2A). However, a sizable fraction (43%) of ER-PUP-enriched mRNAs were not identified via ER ribosome profiling (Fig. 2A). In contrast, ER-PUP–enriched mRNAs were depleted in mRNAs identified by mitochondria-specific ribosome profiling (21) (1.7-fold depleted, P = 7.3 e−6) (SI Appendix, Fig. S6A and Dataset S8). Recording also uniquely identified 453 mRNAs and 50 noncoding at the ER, while profiling detected 169 mRNAs not detected by our approach (Fig. 2 A, Left, and SI Appendix, Fig. S6B). Thus, many but not all recorded RNAs were detected as translated at the ER under normal conditions.
Fig. 2.
Many but not all ER-enriched RNAs are bound by ER proximal ribosomes. (A) mRNAs enriched by ER-PUP or PUP alone vs. positional profiling at the ER (20). (B) Enrichment of RNAs uniquely enriched [log2(∆ recorded reads) ≥ 1] by ER-PUP, and their enrichment in ER profiling. Red dots mark RNAs whose enrichment increases above the cutoff in profiling after arresting elongation with cycloheximide (20). (C) Rank of the commonly enriched RNAs in recording (green) and profiling (gray). (D) Cumulative fraction distribution of ER-enriched mRNAs vs. ER profiling; per tier analysis is shown Right. (E) Cumulative fraction distribution of ER-enriched mRNAs vs. mitochondrial profiling (21), with per tier analysis (Right).
ER-recording detected RNAs seen in profiling only when ribosomes are arrested. Cycloheximide blocks translation elongation and so likely increases the time that ribosome–nascent chain complexes are near the ER-anchored biotin ligase in profiling (20, 36). mRNAs enriched by ER-recording include those that are enriched by profiling only when ribosomes are trapped by cycloheximide treatment (Fig. 2B). We suspect that recording detects ribosome-bound mRNAs that have only brief interactions with the ER, and so can be detected independent of elongation arrest (Fig. 2B).
We next analyzed the relationship between U-tail length and ribosome profiling. mRNAs detected by Sec63p-mediated profiling (20) were grouped into five tiers using k-means clustering, from highest ribosome association (tier 1) to least (tier 5) (SI Appendix, Fig. S6C and Dataset S7). mRNAs with the longest U-tails were more likely to associate with ER-proximal ribosomes (tiers 1 to 3, P = 1.2e−40), and mRNAs were recorded regardless of their rank in profiling (Fig. 2C). The highest-ranking mRNAs identified at the ER (tiers 1 and 2) were more engaged with ribosomes, as inferred from profiling; poorly ranked RNAs progressively decreased in their associations with ribosomes (20) (Fig. 2D). As expected, ER-recorded RNAs did not exhibit this correlation with RNAs translated by mitochondria-proximal ribosomes, as inferred from Om45p-mediated profiling (21) (Fig. 2E).
Taken together, these findings indicate that recording identified RNAs with ER-ribosome association, as well as RNAs that do not. Thus, localized recording and profiling yield overlapping but nonidentical sets of RNAs, and together provide a more complete view of RNAs that encounter the ER than does either approach alone.
Mitochondria-Localized RNA Recording.
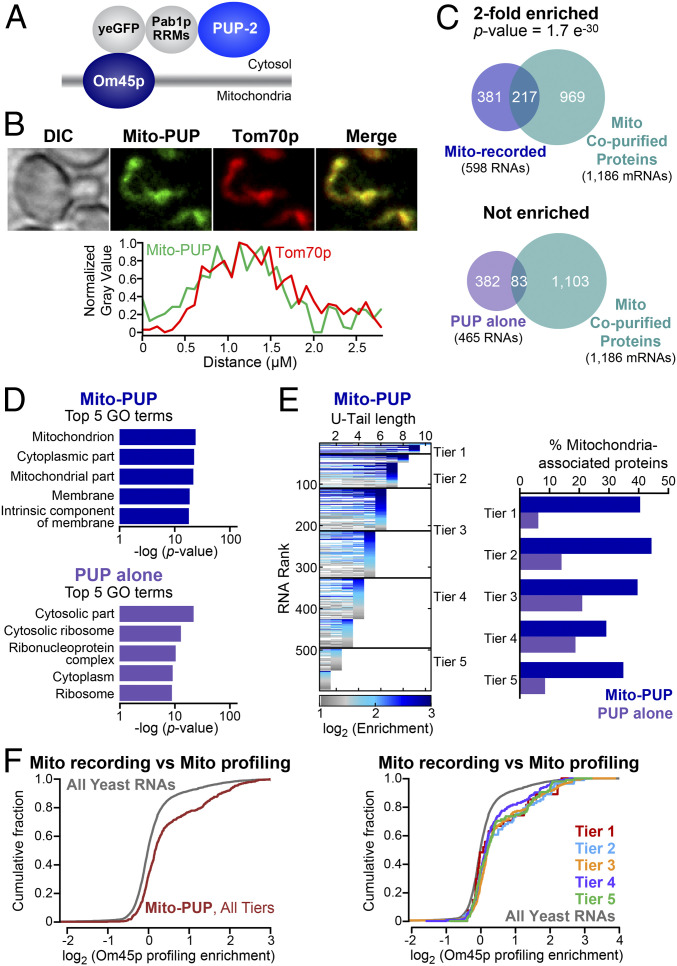
To detect RNAs near the mitochondrial outer membrane, we inserted PUP-2 downstream of the OM45 gene, resulting in an Om45p–PUP-2 chimera, which we refer to as “Mito-PUP” hereafter. Om45p is predicted to be cotranslationally inserted into the mitochondrial membrane with its C-terminal PUP-2 domain in the cytosol (37–42) (Fig. 3A). GFP from Mito-PUP colocalized with the Tom70p-mCherry mitochondrial marker in yeast and both proteins yielded fluorescence patterns comparable to those of the endogenous proteins fused to GFP (31) (Fig. 3B).
Fig. 3.
RNA contacts with the mitochondrial outer membrane. (A) Architecture of Mito-PUP. PUP alone was fused to the cytosolic C-terminal end of the mitochondrial membrane protein, Om45p (37–42). (B) Confocal localization of Mito-PUP fluorescence (green) and the mitochondrial marker, Tom70p-mCherry (31) (red), including fluorescence intensity across a line that bisects the cell (below). Images were collected with an oil-immersed 63× numerical aperture 1.4 objective lens. (C) Overlaps of Mito-PUP- and PUP alone-enriched mRNAs, and those that encode proteins that copurify with biochemically isolated mitochondria (43, 44). (D) Top five GO associations for Mito-PUP (Upper) and PUP alone (Lower). (E) Rank of Mito-PUP–enriched RNAs plotted across each of 10 U-tail lengths (Left). The fraction of the RNAs in each Mito-PUP (blue bars) and PUP-alone tier (purple bars) that encode mitochondrial proteins is plotted (Right). (F) Cumulative fraction of all (Left) and per tier (Right) Mito-PUP–enriched mRNAs with a given degree of mitochondrial profiling (21) enrichment.
Mito-PUP preferentially recorded mRNAs that encode proteins physically associated with mitochondria and mitochondrial functions. Compared to PUP alone, Mito-PUP recorded 598 RNAs at least twofold more efficiently (mitochondria-enriched) (Dataset S9), and 465 RNAs less efficiently (mitochondria-depleted) (Fig. 3C, SI Appendix, Fig. S7, and Dataset S10). The entire set of RNAs that were preferentially recorded by Mito-PUP had a statistically significant association with mitochondria-related GO terms (34, 35), and for mRNAs that encode mitochondria-associated proteins (43, 44) (twofold enriched, P = 1.7e−30) (Fig. 3 C and D and Dataset S11). mRNAs depleted from mitochondria by our DESeq2 analyses did not have these associations (Fig. 3 C and D). RNAs in the highest recording tiers were more likely to encode mitochondrial proteins than those in lower tiers (Fig. 3E and SI Appendix, Fig. S8A). The number of U residues added at mitochondria also correlated with mitochondrial translation at that location, as judged by ribosome profiling (21) (threefold enriched, P = 1.8 e−34) (Fig. 3F and SI Appendix, Fig. S8B). Together, these analyses strongly suggest that Mito-PUP preferentially records mRNAs near mitochondria.
Despite the overlap between recording and profiling, each method identified unique mRNAs. The two methods detected 133 mRNAs in common, and these were enriched for mitochondrial functions by GO analysis (Dataset S12). The rank of commonly detected RNAs greatly differed between methods; for example, they trended toward longer uridine tags (tiers 1 to 3) but recording rank was reduced among RNAs detected most efficiently in ribosome profiling (tier 1) (SI Appendix, Fig. S8 C and D). Instead, the common RNAs were distributed nearly evenly across the midrange ribosome profiling tiers (tiers 2 to 4), which likely indicates that differences in detection requirements influence the rank for each method (SI Appendix, Fig. S8 C and D).
Mito-PUP recorded hundreds of RNAs that were not identified by ribosome profiling. Of these, 457 were mRNAs and 8 were ncRNAs (SI Appendix, Fig. S8E). Conversely, profiling identified 325 mRNAs that were not detected by our approach, but these were mostly lower abundance RNAs (SI Appendix, Fig. S8 E and F). Of the RNAs uniquely identified by each method, those unique to recording were associated with ion transport processes, and those unique to profiling were associated with transfer RNAs (tRNAs) and respiration (Datasets S13 and S14). Thus, recording and ribosome profiling yield unique but complementary results.
mRNAs localized to the outer periphery of mitochondria fall into two classes: Class 1, which require Puf3p for localization, and class II, whose localization is independent of Puf3p (45). A third group of mRNAs, termed class 3, are translated in the cytoplasm, not near mitochondria (45). Mito-PUP identified mRNAs localized near mitochondria, whether they require Puf3p for localization (class 1, 3.4-fold enriched, P = 6.1e−24) or not (class 2, 3.5-fold enriched, P = 3.1e−22) (Dataset S15). In contrast, MitoPUP identified class 3 mRNAs poorly, indicating those mRNAs are too far from the Om45p anchor to be marked by the Mito-PUP (Dataset S15). Thus, localized recording specifically discriminated groups of RNAs whose proteins are destined for the same organelle, but traffic there differently.
RNAs Detected by Both ER-PUP and Mito-PUP.
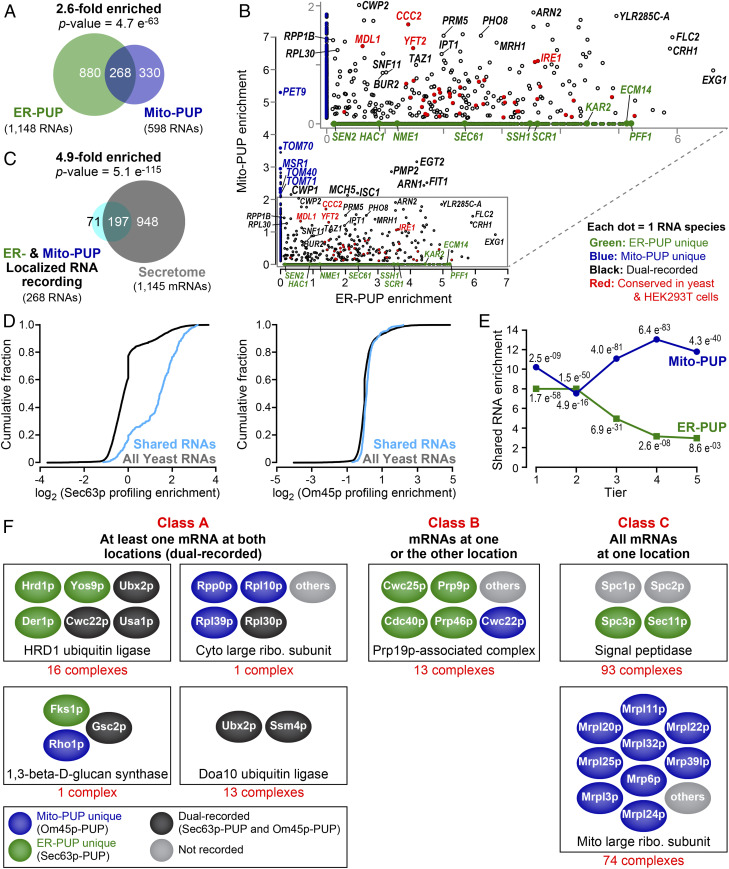
Most RNAs recorded by ER-PUP or Mito-PUP (Fig. 4A) yielded GO enrichments anticipated for that organelle (Datasets S16 and S17). For example, several components of the TOM (Tom70p, Tom40p. and Tom71p) and TIM (Tim18p, Tim50p, Tim22p, Tim44p, and Tim54p) protein import complexes exhibit high recording only by Mito-PUP, while those that encode certain secreted proteins (e.g., Ecm14p and Pff1p), ER-resident chaperones and translocon components (e.g., Kar2p, Ssh1p, and Sec63p), and the ncRNA of the signal recognition particle (SRP), SCR1 (46), were recorded only at the ER (Fig. 4B).
Fig. 4.
Dual recording at both ER and mitochondria. (A) mRNAs recorded by ER-PUP and Mito-PUP. (B) Comparison of RNAs uniquely enriched by the ER- (green dots) or Mito-PUP (blue dots), and those that were common to both (black, and red). Larger circles represent mRNAs that encode proteins typical of each organelle, while the red denote mRNAs that are detected at the ER (ER-PUP unique) and mitochondria (Mito-PUP unique) by independent, APEX-seq studies in HEK293T cells (24). (C) Dual-recorded RNAs vs. secretome mRNAs. (D) Cumulative fraction of common RNAs that have ER-profiling (20) (Left) and mitochondrial profiling (21) (Right) enrichment. (E) Distribution of common RNAs among ER- (green line) and Mito-PUP (blue line) rank. (F) mRNAs that encode subunits of multiprotein complexes exhibit three localization patterns. Representative complexes depicted for each class; physical interactions between proteins not implied. For a complete list, see Dataset S24.
Strikingly, however, a substantial fraction (ER: 23%, Mito: 44%) of all recorded mRNAs were detected by both ER-PUP and Mito-PUP (P = 4.7 e−63) (Fig. 4A and Dataset S18). Detection by both Om45p and Sec63p anchors suggests these mRNAs come near both the ER and mitochondrial outer membrane. As a whole, these “dual-recorded” mRNAs encoded primarily secreted proteins (4.9-fold enriched, P = 5.1e−115) (Fig. 4C and Dataset S19), and were translated (Fig. 4D) and received longer tails at the ER (Fig. 4E), suggesting a longer cumulative time at that location. We note that OSM1 mRNA was detected by both ER and mitochondrial profiling (21); in recording, it was enriched only by the ER-anchored PUP (Datasets S4 and S9). However, OSM1 was detected by mitochondrial ribosome profiling only after translational arrest with cycloheximide (21).
A second group of 45 shared RNAs were recorded roughly equally by ER- and mitochondria-anchored proteins (normalized to PUP alone in each location) (Fig. 4B and Dataset S20). High-ranking, shared mRNAs of this type include ones associated with lipid biosynthesis (ISC1, IPT1, YFT2, and TAZ1), ion transport (MDL1, MCH5, and CCC2), RNA polymerase II transcription (SNF11 and BUR2), a plasma membrane-associated proteolipid (PMP2), and a glycosylphosphatidylinositol (GPI)-anchored cell wall endonuclease (EGT2) (Fig. 4B and Dataset S20). These commonly enriched mRNAs may reside where the two organelles are in close proximity (47, 48) or move from one location to the other (Discussion).
Localization of specific RNAs to the proximity of both the ER and the outer mitochondrial membrane is conserved. We compared our recording results to data recently reported from human embryonic kidney (HEK293T) cells using APEX-seq (24). The high fraction of RNAs recorded at both locations in yeast (ER: 268 of 1,148; mitochondria: 268 of 598) (Fig. 4 A and B) was mirrored in HEK293T cells (24), as was the identity of many of the RNAs (ER membrane: 50%; outer mitochondrial membrane: 67%) (SI Appendix, Fig. S9 and Datasets S18, S21, and S22). Among the dual-recorded RNAs detected in both yeast (Fig. 4B) and human cells (SI Appendix, Fig. S9C) were ones that encode functions linked to ERMES (ER-mitochondria encounter structure) (48), formed where ER and mitochondria are in close proximity. These included mRNAs that encode proteins and functions that are associated with MAMs (ER-mitochondrial associated membranes) (47, 49), including transmembrane transporters (Fig. 4B and SI Appendix, Fig. S9C and Dataset S23), and proteins involved in lipid (CAX4, LAC1, TGL1, and ALE1) or glycoprotein (ROT2, CAX4, OST6) metabolism. Strikingly, the ER stress sensor, IRE1/ERN1 (50–56), was detected at both organelles, consistent with the presence of Ire1p protein at both organelles (49), and the requirement for Ire1p for the increase of mitochondrial respiration during the UPR (57) (Fig. 4B and SI Appendix, Fig. S9C and Dataset S23). Conservation of many shared targets suggests the importance of their presence at both locales, particularly given the substantial differences between the two techniques.
mRNAs that encode components of multiprotein complexes (identified through GO annotations) (34, 35) comprise three classes based on the range of locations between the ER and mitochondria (Fig. 4F and Dataset S24). Thirty-one complexes fell into class A, and included dual recorded mRNAs (Fig. 4F and Dataset S24) (e.g., HRD1 ubiquitin ligase complex). Most strikingly, 13 complexes fell into class B, with mixtures of mRNAs recorded at one or the other location (Fig. 4F) (e.g., Prp19 complex). Ninety-three complexes were uniquely enriched by ER-PUP (e.g., the signal peptidase) and 74 by Mito-PUP (e.g., the 54S mitochondria ribosomal subunit) (class C in Fig. 4F and Dataset S24). The recording of mRNAs for subunits in different locales suggests coordination to assemble the complexes, or that certain of the proteins have other roles.
Regulatory Sites and RNA-Binding Proteins.
RNA localization often is controlled by RNA-binding proteins (RBPs). We, therefore, identified sequence motifs in the 3′UTRs of RNAs identified by localized recording using MEME (58), and compared those motifs to known RNA-binding specificities (26, 59).
The top motif among RNAs identified by Mito-PUP was a degenerate form of the Puf3p binding site (SI Appendix, Fig. S10A). Puf3p binds and controls nuclear-encoded mRNAs with mitochondrial functions, and participates in their localization near mitochondria (26, 27, 45, 60–63). The proportion of Mito-recorded RNAs with such elements was similar across all tiers (SI Appendix, Fig. S10B, dark blue line). This prompted reanalysis of Om45p-mediated profiling data (21), which revealed the same phenomenon (SI Appendix, Fig. S10 A and B, light blue line). Our findings suggest that Puf3p promotes mitochondrial localization of only certain mRNAs, and that others arrive there through other mechanisms (45).
ER-recorded RNAs were enriched for Bfr1p targets (P = 1.9 e−280) (Dataset S25), consistent with the role of Bfr1p in the secretory pathway and RNA metabolism (26, 64–68); similarly, RNAs recorded at mitochondria were enriched for mRNAs bound by Puf3p (P = 1.2 e−26) (SI Appendix, Fig. S10C and Dataset S26). RNAs identified at the ER were moderately enriched among RNAs that bind Pub1p (P = 2.8e−36), Mrn1p (P = 4.5e−16), and Scp160p (P = 4.6e−11); of these, only Scp160p is known to localize to the ER (69, 70) (SI Appendix, Fig. S10D). Sec63p-recorded mRNAs revealed AU- or U-rich motifs (SI Appendix, Fig. S10E). These analyses point to proteins likely involved in the control of these mRNAs.
ncRNAs and RNA Metabolism.
Our analyses revealed connections between RNA metabolism and cell biology. ncRNAs from 53 genes were significantly enriched at mitochondria or the ER relative to the PUP-alone control (SI Appendix, Fig. S11, three representative paired reads are shown for each type of RNA, and Dataset S27). SCR1, the RNA component of the SRP (46), was the highest-ranking ncRNA identified by ER-PUP (Fig. 4B and SI Appendix, Fig. S11A). HAC1 mRNA, which is critical in the UPR (50–56), also was detected (Fig. 4B and SI Appendix, Fig. S11A).
Twenty-one tRNA-related transcripts were recorded at the ER. These lacked CCA, contained 3′ extensions beyond the 3′ end of the mature tRNA, and possessed poly(A) tails upstream of the uridines added by ER-PUP (SI Appendix, Fig. S11B). Poly(A) is added to some ncRNAs as part of nuclear RNA surveillance, mediated by the TRAMP complex and exosome (71–74). We obtained no reads upstream of anticodons, which may be due to the presence of modified bases that arrest reverse transcriptase (75–77) or RNA cleavage events that may leave tRNA halves (75, 78). Among proteins that participate in tRNA splicing (79), mRNAs encoding Sen2p, a subunit of the endonuclease, were detected at the ER; while those encoding Tom70p, an enhancer of tRNA splicing in vivo (80), was detected at mitochondria, where it also is part of a translocase (Fig. 4B).
Twenty-five small nuclear RNA (snRNA)- and small nucleolar RNA (snoRNA)-related RNAs were recorded by either ER- or Mito-PUPs. These included snRN7-L, which was identified at both organelles (ER: Tier 3, Mito: Tier 4), and snR7-S and snR6, which were only detected at the ER (SI Appendix, Fig. S11C). Yeast snRNAs can shuttle to the cytoplasm as part of their maturation, perhaps to help prevent the inclusion of misprocessed snRNAs in the spliceosome (81). Some snoRNAs were also recorded at one or both sites, including NME1, the RNA component of mitochondrial RNA processing (MRP) that catalyzes RNA cleavage events (82) (Fig. 4B and SI Appendix, Fig. S11D). While snoRNAs are thought to be restricted to the nucleus, failure to properly process their guanosine cap can cause their accumulation in the cytoplasm (83). Together, the data on ncRNAs suggest recording captures mature ncRNAs as well as ones undergoing maturation and surveillance.
Discussion
Localized RNA recording identifies RNAs located to specific sites transcriptome-wide and is independent of hybridization, affinity purification, fractionation, cross-linking, or chemical treatments. RNAs recorded using Sec63p or Om45p displayed distinct properties consistent with each anchor. The number of uridines added to an individual RNA molecule likely reflects the integrated time it was at that location. mRNAs with longer U-tails were enriched for biological functions consistent with their location and association with local ribosomes. These findings mirror analyses of RNA–protein interactions using tagging, in which the number of uridines added to an individual RNA molecule correlates with the consensus match to the RNA binding site of the protein, and with in vitro assays (26, 84).
Localized recording provides a new entrée into RNA movements, and raises key issues likely to be resolved via in vivo imaging. The hypothesis that the number of Us added likely reflects time spent in a specific location has not been tested independently, but emerges from parallels to analysis of RBP binding to their mRNAs targets, as monitored by U addition (26). There, the number of Us added parallels the apparent affinity of binding sites and, likely, occupancy. Indeed, localized RNA recording is even more stringent as background labeling is subtracted via the PUP-alone control with DeSeq2 (32). Since locations are dependent on the distribution of the anchoring proteins, imaging the movements of RNAs we report will be of great interest.
In principle, uridine addition could affect mRNAs by altering their translation, stability, or movement. In rich fermentable media, the approximate growth rate of recording strains grown to midlog phase was 10% slower that of wild-type cells, perhaps implying effects of U-addition (Materials and Methods and SI Appendix, Fig. S12A). This effect is seen with both PUP alone as well as localized strains, suggesting it is due to U-addition per se, and not to localization of the activity. Similarly, after addition of DTT to induce the UPR, ER-PUP cells grew ∼27% more slowly than wild-type (SI Appendix, Fig. S12B); and in nonfermentable media, the mito-PUP strain grew ∼36% more slowly (Materials and Methods and SI Appendix, Fig. S12C). However, certain potential artifacts are circumvented through PUP-alone controls. For example, computational analyses have prompted the suggestion that oligouridine tracts cause mRNAs to adhere to membranes (85, 86), which in principle could attract RNAs tagged elsewhere to be recorded at the ER. However, such mRNAs would be tagged in the PUP-alone strain, and so would be removed during computational analysis (SI Appendix, Fig. S4 D and E).
Certain RNAs recorded at a given site were not enriched by proximity ribosome profiling, suggesting they may be subject to translational regulation, consistent with the observation that some mRNAs associate with membranes in a ribosome-independent manner (87). Other variables, such as the structure of specific RNAs, RNA compartmentalization, or 3′ bound factors may also affect recording efficiency.
Specific RNAs were readily detected by RNA-seq (26), yet not recorded in the PUP-alone strain. Several technical differences likely underlie this difference, including substantial differences in library preparation and data processing (Materials and Methods and SI Appendix, Fig. S1 B and C). Preparation of recording libraries also includes GI-tailing and poly(A) enrichment procedures. Finally, 56% of the “missed RNAs” were recorded, but in fewer than three replicates, and so did not satisfy our cut-offs (SI Appendix, Figs. S2 B and D and S12D). Deeper sequencing may also reveal the missed RNAs, as they tend to be low abundance (SI Appendix, Fig. S12E). Of the missing RNAs, 49% were nonpolyadenylated RNAs (mitochondria mRNAs and ncRNAs), which are depleted in our library preparation, and only 10% were ORFs of known function (SI Appendix, Fig. S12F).
Localized recording likely provides a cumulative “record” of an RNA’s movements, while the APEX-seq (24) approach yields a snapshot, a “registry” of RNA locations at a specific time. The two approaches are complementary and provide a fuller view of RNA movements than either alone. Similarly, localized recording and global studies of RBPs are synergistic (e.g., SI Appendix, Fig. S10). For example, Puf3p binds to and controls expression and localization of nuclear-encoded mitochondrial mRNAs (26, 27, 45, 60–63). RNAs identified by the mitochondrially localized PUP often contained poor or no Puf3p binding sites, consistent with the existence of Puf3p-indeendent mechanisms for mitochondrial localization. Comparison with profiling suggests other roles for optimal binding, perhaps including its ability to enhance rather than repress translation (88) (SI Appendix, Fig. S10 B and C). RNAs recorded at the ER contained novel motifs, some of which may be involved in the SRP-independent localization of mRNAs to the ER (89) or may bind RBPs, such as Bfr1p (26, 68). Indeed, Bfr1p is implicated in the secretory pathway, copurifies with secretory messenger ribonucleoproteins (mRNPs) and mRNAs, and localizes to the ER in a manner that requires its RNA binding activity (26, 64–68).
A specific subset of RNAs were recorded using both ER (Sec63p) and mitochondrial (Om45p) anchors. These “dual-recorded” RNAs might arise in several ways. First, they may be located where the two organelles are very close, perhaps as promoted by ERMES (48, 90). Second, a single RNA molecule may move between organelles. Certain dual-recorded RNAs are low-ranking at one or both locations, suggesting transient interactions. Third, individual RNA molecules may go from the nucleus to either one or the other location, rather than between the two organelles. The mechanism of dual localization we detect appears to be largely independent of translation since dual localization is very rare among mRNAs detected by yeast ribosome profiling at these two organelles (20, 21). However, mRNAs detected by localized recording might be translated, but in a manner that escapes detection in profiling, which requires ribosomes with their exit tunnels near the membrane (20, 21). Finally, since our approach hinges on the distribution of the anchor proteins, dual recording could stem from an exchange of protein- or RNA-containing lipid particles between locales. This could explain otherwise enigmatic localization patterns (e.g., enrichment of ribosomal protein mRNAs at both locales, and of the nuclear export heterodimer components MTR2 and MEX67 at the ER and mitochondria, respectively). In addition, it has been suggested that organelles may associate broadly with diverse RNA populations (87). Underlying mechanisms may be identified through the use of multiple recording devices in the same cell, live imaging (8, 18, 19, 91), or strategies that track localization and translation simultaneously (92, 93).
Dual localization of specific mRNAs is conserved between yeast and human cells and may reflect close proximity of organelles. Proximity to MAMs may help integrate events between ER and mitochondria. IRE1 mRNA is exemplary as it is detected at both locales in both yeast and human cells. Its dual localization may integrate inputs from both organelles and facilitate coordinated responses, such as the Ire1p-dependent rise in respiration after ER stress in yeast (57) or sustained UPR-induced apoptosis in mammalian cells (94).
ncRNAs were readily detected in localized recording without modifying the approach to specifically detect them, and include SCR1 and NME1, the RNA components of SRP (46) and MRP (82). Indeed, SCR1 was the most highly ranked ncRNA at the ER. RNAs related to tRNAs, snRNAs, and snoRNAs appear to have been caught during their maturation or surveillance. Recorded tRNAs were 3′-extended and polyadenylated upstream of their U-tails. Their polyadenylation suggests the action of the nuclear TRAMP complex before their encounter with the recording enzyme. The 3′-extended tRNAs may subsequently be processed by the endonuclease Trz1p (95), which is both nuclear and mitochondrial (31, 95, 96). On the other hand, the poly(A) tail on these presumably cytoplasmic ncRNAs may be a protective feature for the processing of these molecules in the cytoplasm, as with the 5′ caps on exported intron-containing tRNA precursors (97). Indeed, the Mex67/Mtr2 mRNA nuclear export machinery can export tRNA precursors to the cytoplasm (98). However, the putative polyadenylation protective mechanism may not be involved in tRNA splicing as we did not observe reads mapped to introns. The presence of recorded snRNAs supports the recent finding that yeast snRNAs are shuttled to the cytoplasm as part of their maturation to prevent the inclusion of misprocessed snRNAs in the spliceosome (81). Furthermore, the recorded snoRNAs may be of the same type that have been observed leaving the nucleus in yeast heterokaryons (99), or may arise through a different form of surveillance, as failure to properly process the guanosine cap of snoRNAs can lead to their accumulation into the cytoplasm (83).
Localized recording can be performed in living organisms, as is true of “TRIBE,” in which adenosine deaminases acting on RNA (ADAR)-catalyzed deaminations mark the binding of specific proteins (100, 101). APEX-seq, as currently configured, is not yet suitable in intact organisms. Conversely, in cells with endogenous uridylation activities, the application of the recording strategy would be simplified through enzymes with different nucleotide specificities (102) or reduction of endogenous uridylation activities.
The use of localized devices to covalently mark RNAs provides an entrée to creating histories of individual RNA molecules. Combinations of localized RNA recording, APEX-seq, and TRIBE, performed in the same cell, should yield biographies of individual RNAs, in which their movements and protein encounters are written in unique combinations of modifications and tail sequences.
Materials and Methods
Uridilyated RNAs were extracted from cells, prepared for sequencing, and reads processed and mapped to the genome as previously reported (26) (see detailed protocol in SI Appendix, Supplementary Methods). Please see the SI Appendix, Supplementary Methods for description of imaging and novel motif discovery protocols, how published datasets were obtained and processed, a list of tools used, and for a description yeast strains, plasmids, and primers.
Supplementary Material
Acknowledgments
We thank the M.W. and Kimble laboratories, and members of the University of Wisconsin–Madison community, particularly Judith Kimble and Dave Brow, for helpful advice and discussions; and Eric Phizicky and Elizabeth Grayhack (University of Rochester) and Scott Aoki (Indiana University) for their insights; Sarah Crittenden for help with confocal microscopy; Kim Haupt and Elle Kielar Grevstad (University of Wisconsin–Madison Optical Core) for assistance with image analyses; Aaron Hoskins, Tucker Carrocci, and Ian Norden for help with fluorescent proteins; Joshua R. Hyman, Molly Zeller, Amanda Maegli and Mike Sussman (University of Wisconsin–Madison Biotechnology Center) for help with sequencing; Laura Vanderploeg and the Biochemistry Media Laboratory for help preparing figures; and Carol Pfeffer for her help with the correspondence during the submission of this work. This work was supported by NIH Grant GM50942 (to M.W.); and an E.W. Hopkins fellowship and an NIH diversity supplement (to H.C.M.-M.). While at the University of Wisconsin–Madison, C.P.L. was supported by Wharton and Biochemistry Scholar Fellowships, and D.F.P. was supported by T32 GM008349 (NIH) and a Wisconsin Alumni Research Foundation (WARF) Scholarship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921408117/-/DCSupplemental.
Data Availability.
The data reported in this work have been deposited to the Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra/ (study no. SRP253075, BioProject accession PRJNA613017).
References
- 1.Driever W., Nüsslein-Volhard C., The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Berleth T. et al., The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749–1756 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ephrussi A., Dickinson L. K., Lehmann R., Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37–50 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Kim-Ha J., Smith J. L., Macdonald P. M., Oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66, 23–35 (1991). [DOI] [PubMed] [Google Scholar]
- 5.Melton D. A., Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature 328, 80–82 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Johnstone O., Lasko P., Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365–406 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Long R. M. et al., Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Tutucci E., Livingston N. M., Singer R. H., Wu B., Imaging mRNA in vivo, from birth to death. Annu. Rev. Biophys. 47, 85–106 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Das S., Singer R. H., Yoon Y. J., The travels of mRNAs in neurons: Do they know where they are going? Curr. Opin. Neurobiol. 57, 110–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin A., Lécuyer E., RNA localization: Making its way to the center stage. Biochim. Biophys. Acta, Gen. Subj. 1861, 2956–2970 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Martin K. C., Ephrussi A., mRNA localization: Gene expression in the spatial dimension. Cell 136, 719–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipshitz H. D., Smibert C. A., Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 10, 476–488 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Besse F., Ephrussi A., Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971–980 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., Zhuang X., RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J. H. et al., Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Femino A. M., Fay F. S., Fogarty K., Singer R. H., Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Bertrand E. et al., Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Tutucci E. et al., An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 15, 81–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jan C. H., Williams C. C., Weissman J. S., Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams C. C., Jan C. H., Weissman J. S., Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaewsapsak P., Shechner D. M., Mallard W., Rinn J. L., Ting A. Y., Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife 6, e29224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padrón A., Iwasaki S., Ingolia N. T., Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fazal F. M. et al., Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak J. E., Wickens M., A family of poly(U) polymerases. RNA 13, 860–867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapointe C. P., Wilinski D., Saunders H. A., Wickens M., Protein-RNA networks revealed through covalent RNA marks. Nat. Methods 12, 1163–1170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapointe C. P. et al., Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 6, 125–135.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapointe C. P. et al., Architecture and dynamics of overlapped RNA regulatory networks. RNA 23, 1636–1647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldheim D., Rothblatt J., Schekman R., Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12, 3288–3296 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finke K. et al., A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 15, 1482–1494 (1996). [PMC free article] [PubMed] [Google Scholar]
- 31.Huh W. K. et al., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ast T., Cohen G., Schuldiner M., A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Balakrishnan R. et al., YeastMine—An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012, bar062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong E. L. et al., Gene ontology annotations at SGD: New data sources and annotation methods. Nucleic Acids Res. 36, D577–D581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogg S. C., Walter P., SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81, 1075–1084 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Yaffe M. P., Jensen R. E., Guido E. C., The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J. Biol. Chem. 264, 21091–21096 (1989). [PubMed] [Google Scholar]
- 38.Sesaki H., Jensen R. E., UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123–1134 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dukanovic J., Rapaport D., Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim. Biophys. Acta 1808, 971–980 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Riezman H. et al., The outer membrane of yeast mitochondria: Isolation of outside-out sealed vesicles. EMBO J. 2, 1105–1111 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapaport D., The mitochondrial protein OM45 is exposed to the cytosol. J. Biol. Chem. 287, 27415–27416, author reply 27416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauffer S. et al., Reply to Rapaport: The mitochondrial protein OM45 is exposed to the cytosol. J. Biol. Chem. 287, 27416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinders J., Zahedi R. P., Pfanner N., Meisinger C., Sickmann A., Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern M. et al., Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saint-Georges Y. et al., Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One 3, e2293 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hann B. C., Walter P., The signal recognition particle in S. cerevisiae. Cell 67, 131–144 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Vance J. E., Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 (1990). [PubMed] [Google Scholar]
- 48.Kornmann B. et al., An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori T., Hayashi T., Hayashi E., Su T. P., Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8, e76941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T., Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1, 803–817 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Cox J. S., Shamu C. E., Walter P., Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Cox J. S., Walter P., A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Mori K., Ma W., Gething M. J., Sambrook J., A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743–756 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Nikawa J., Akiyoshi M., Hirata S., Fukuda T., Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 24, 4222–4226 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidrauski C., Cox J. S., Walter P., tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Sidrauski C., Walter P., The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Knupp J., Arvan P., Chang A., Increased mitochondrial respiration promotes survival from endoplasmic reticulum stress. Cell Death Differ. 26, 487–501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey T. L., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- 59.Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O., Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, e255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson J. S. Jr., Houshmandi S. S., Lopez Leban F., Olivas W. M., Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 10, 1625–1636 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerber A. P., Herschlag D., Brown P. O., Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gadir N., Haim-Vilmovsky L., Kraut-Cohen J., Gerst J. E., Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 17, 1551–1565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu D., Stumpf C. R., Krahn J. M., Wickens M., Hall T. M., A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 20192–20197 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson C. L., Képès F., BFR1, a multicopy suppressor of brefeldin A-induced lethality, is implicated in secretion and nuclear segregation in Saccharomyces cerevisiae. Genetics 137, 423–437 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trautwein M., Dengjel J., Schirle M., Spang A., Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 5021–5037 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lang B. D., Li Am, Black-Brewster H. D., Fridovich-Keil J. L., The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res. 29, 2567–2574 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidner J., Wang C., Prescianotto-Baschong C., Estrada A. F., Spang A., The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J. Cell Sci. 127, 1992–2004 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Manchalu S., Mittal N., Spang A., Jansen R. P., Local translation of yeast ERG4 mRNA at the endoplasmic reticulum requires the brefeldin A resistance protein Bfr1. RNA 25, 1661–1672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wintersberger U., Kühne C., Karwan A., Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast 11, 929–944 (1995). [DOI] [PubMed] [Google Scholar]
- 70.Frey S., Pool M., Seedorf M., Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 276, 15905–15912 (2001). [DOI] [PubMed] [Google Scholar]
- 71.LaCava J. et al., RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Wyers F. et al., Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Vanácová S. et al., A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houseley J., LaCava J., Tollervey D., RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Motorin Y., Muller S., Behm-Ansmant I., Branlant C., Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 425, 21–53 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Machnicka M. A. et al., MODOMICS: A database of RNA modification pathways–2013 update. Nucleic Acids Res. 41, D262–D267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hrabeta-Robinson E., Marcus E., Cozen A. E., Phizicky E. M., Lowe T. M., High-throughput small RNA sequencing enhanced by AlkB-facilitated RNA de-methylation (ARM-Seq). Methods Mol. Biol. 1562, 231–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson D. M., Parker R., The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185, 43–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trotta C. R. et al., The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858 (1997). [DOI] [PubMed] [Google Scholar]
- 80.Wan Y., Hopper A. K., From powerhouse to processing plant: Conserved roles of mitochondrial outer membrane proteins in tRNA splicing. Genes Dev. 32, 1309–1314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Becker D. et al., Nuclear pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Rep. 27, 3199–3214.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Schmitt M. E., Clayton D. A., Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 6, 1975–1985 (1992). [DOI] [PubMed] [Google Scholar]
- 83.Grzechnik P. et al., Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage. Nat. Commun. 9, 1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webster M. W., Stowell J. A., Passmore L. A., RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not. eLife 8, e40670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prilusky J., Bibi E., Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 6662–6666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kraut-Cohen J., Gerst J. E., Addressing mRNAs to the ER: cis sequences act up!. Trends Biochem. Sci. 35, 459–469 (2010). [DOI] [PubMed] [Google Scholar]
- 87.Hoffman A. M., Chen Q., Zheng T., Nicchitta C. V., Heterogeneous translational landscape of the endoplasmic reticulum revealed by ribosome proximity labeling and transcriptome analysis. J. Biol. Chem. 294, 8942–8958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee C. D., Tu B. P., Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 11, 1638–1650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kraut-Cohen J. et al., Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 24, 3069–3084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kornmann B., Walter P., ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 123, 1389–1393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tutucci E., Livingston N. M., Singer R. H., Wu B., Imaging mRNA in vivo, from birth to death. Annu. Rev. Biophys. 47, 85–106 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Pichon X. et al., Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 214, 769–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee B. H., Bae S. W., Shim J. J., Park S. Y., Park H. Y., Imaging single-mRNA localization and translation in live neurons. Mol. Cells 39, 841–846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rutkowski D. T., Kaufman R. J., A trip to the ER: Coping with stress. Trends Cell Biol. 14, 20–28 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Skowronek E., Grzechnik P., Späth B., Marchfelder A., Kufel J., tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20, 115–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hazbun T. R. et al., Assigning function to yeast proteins by integration of technologies. Mol. Cell 12, 1353–1365 (2003). [DOI] [PubMed] [Google Scholar]
- 97.Ohira T., Suzuki T., Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol. 12, 648–655 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Chatterjee K. et al., Sharing the load: Mex67-Mtr2 cofunctions with Los1 in primary tRNA nuclear export. Genes Dev. 31, 2186–2198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olson B. L., Siliciano P. G., A diverse set of nuclear RNAs transfer between nuclei of yeast heterokaryons. Yeast 20, 893–903 (2003). [DOI] [PubMed] [Google Scholar]
- 100.McMahon A. C. et al., TRIBE: Hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahman R., Xu W., Jin H., Rosbash M., Identification of RNA-binding protein targets with HyperTRIBE. Nat. Protoc. 13, 1829–1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Preston M. A. et al., Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this work have been deposited to the Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra/ (study no. SRP253075, BioProject accession PRJNA613017).