Abstract
Clonal hematopoiesis is associated with various age-related morbidities. Error-corrected sequencing (ECS) of human blood samples, with a limit of detection of ≥0.0001, has demonstrated that nearly every healthy individual >50 years old harbors rare hematopoietic clones below the detection limit of standard high-throughput sequencing. If these rare mutations confer survival or proliferation advantages, then the clone(s) could expand after a selective pressure such as chemotherapy, radiotherapy, or chronic immunosuppression. Given these observations and the lack of quantitative data regarding clonal hematopoiesis in adolescents and young adults, who are more likely to serve as unrelated hematopoietic stem cell donors, we completed this pilot study to determine whether younger adults harbored hematopoietic clones with pathogenic mutations, how often those clones were transferred to recipients, and what happened to these clones over time after transplantation. We performed ECS on 125 blood and marrow samples from 25 matched unrelated donors and recipients. Clonal mutations, with a median variant allele frequency of 0.00247, were found in 11 donors (44%; median, 36 years old). Of the mutated clones, 84.2% of mutations were predicted to be molecularly pathogenic and 100% engrafted in recipients. Recipients also demonstrated de novo clonal expansion within the first 100 days after hematopoietic stem cell transplant (HSCT). Given this pilot demonstration that rare, pathogenic clonal mutations are far more prevalent in younger adults than previously appreciated, and they engraft in recipients and persist over time, larger studies with longer follow-up are necessary to correlate clonal engraftment with post-HSCT morbidity.
INTRODUCTION
Matched, unrelated allogeneic hematopoietic stem cell transplantation (HSCT) is a curative therapy for a variety of nonmalignant β-globinopathies (1), constitutional enzyme deficiencies, and hematologic malignancies (2). However, HSCT recipients often suffer multiple early and late post-HSCT morbidities (3). These range from relatively common conditions such as cardiac dysfunction, coronary artery disease (4), graft-versus-host disease (GvHD) (5), immune dysfunction/infection, cytopenias, and myelodysplasia to very rare events such as donor cell leukemia (6). Many of these common morbidities have been anecdotally attributed to donor clone(s) with pathogenic mutations in a discrete panel of candidate genes (5, 7, 8). These anecdotal clones would qualify as clonal hematopoiesis of indeterminate potential [CHIP; with ≥2% variant allele frequency (VAF)] in an otherwise healthy person (9), and about 5% of healthy individuals older than 50 years harbor CHIP clones (10-12). However, this definition of CHIP is primarily based on the limit of detection of standard next-generation sequencing (NGS), hence the age-related prevalence because it takes decades of selection for some clones to expand to the level of this detection. In contrast, error-corrected sequencing (ECS) has a limit of detection of 0.0001 and has revealed that nearly everyone older than 50 years harbors hematopoietic clones with mutations associated with acute myeloid leukemia (AML) and atherosclerosis (13, 14), and there are very few differences in clonal variability and frequency between those who stay healthy and those who actually develop AML (15). The clinical relevance of hematopoietic clones with <2% VAF was recently demonstrated in AML prediction (16) and mutation clearance after allogeneic HSCT for myelodysplastic syndrome (17), where clones as rare as 0.005 VAF were clinically relevant for disease progression. Recently, Frick and colleagues (5) studied common clonal mutations in the context of CHIP from older, matched, related HSCT donors (>55 years old), where about 5 to 10% of this population would be expected to harbor CHIP clones based on prior studies (10-12). This study found that the presence of CHIP correlated with the development of chronic GvHD. However, the study was limited by only examining older, related donors and mutations above 0.02 VAF. Unlike older related HSCT donors who are expected to have CHIP, 86% of eligible unrelated donors are adolescents and young adults (AYA) aged 18 to 44 years, an age group where CHIP is virtually nondetectable (10-12), but recipient morbidity generally exceeds that seen in related HSCT. Despite not having CHIP, it has been hypothesized that the AYA population harbors hematopoietic somatic mutations of low VAF, undetectable via standard NGS (18), and these mutations could serve as a reservoir for future disease development when relevant selective pressure is present (19). Hence, the appro-priate way to study these low VAF mutations in the AYA group and the effects thereof in HSCT recipients is via ultrasensitive sequenc-ing techniques, such as ECS, that could circumvent the error rate of standard NGS (14).
In addition, the genes frequently mutated in AYA leukemia (20) differ substantially from leukemia in older adults (21), suggesting that the AYA population may harbor a different clonal hematopoietic mutation spectrum than that seen in the CHIP literature. However, the physiologic prevalence and mutation spectrum of hematopoietic clones with mutations <0.02 VAF in the AYA population has not been quantitatively characterized. Thus, our 80-gene targeted panel included genes that are frequently mutated in both pediatric/AYA and older adult AML.
In summary, this caused us to hypothesize that (i) unrelated, AYA HSCT donors may harbor hematopoietic clones with mutations <0.02 VAF in genes other than those associated with CHIP, and (ii) these mutations may confer a growth or survival advantage and may there-fore be selected and engrafted in recipients. In this model, prior and ongoing chemotherapy, radiotherapy, and immunosuppression can act as potent selective pressures on any cell with a survival or proliferation advantage. ECS has previously demonstrated a comparable process in therapy-related AML (t-AML) (13), where preexisting TP53-mutated hematopoietic progenitors, as rare as 0.0003 VAF, are selected by treatment of the primary malignancy and result in t-AML months to years later. To interrogate this hypothesis, our primary goal was to find retrospectively banked, matched unrelated donor:recipient samples with as many longitudinal time points as possible. For each pair, five samples were evaluated: donor pre-HSCT, recipient pre-HSCT, and recipient at 30 (D30), 100 (D100), and 365 days (D365) after HSCT. We asked the following questions: (i) What is the clonal hematopoietic spectrum in younger, healthy donors? (ii) How many donor clones are typically transferred to recipients? (iii) What happens to these clones longitudinally in re-cipients? Given that the presence of clonal hematopoiesis is un-expected in this donor age group and there may have been little to no clonal transfer to recipients, this study was not designed to cor-relate clinical outcomes with donor clonal hematopoiesis, but the results indicate that such a study is warranted.
RESULTS
Engraftment of pathogenic somatic variants of donor origin
Given that the prevalence of hematopoietic clones at <0.02 VAF in the healthy AYA population has not been quantified, we first characterized the prevalence and genetic spectrum of clonal hematopoietic mutations in donors before transplantation. Because clonal hematopoiesis is associated with multiple complex health problems and all-cause mortality (10), we were not solely interested in mutations associated with hematologic malignancies, but rather any mutation that would confer a growth or survival advantage to a cell due to altered molecular functions.
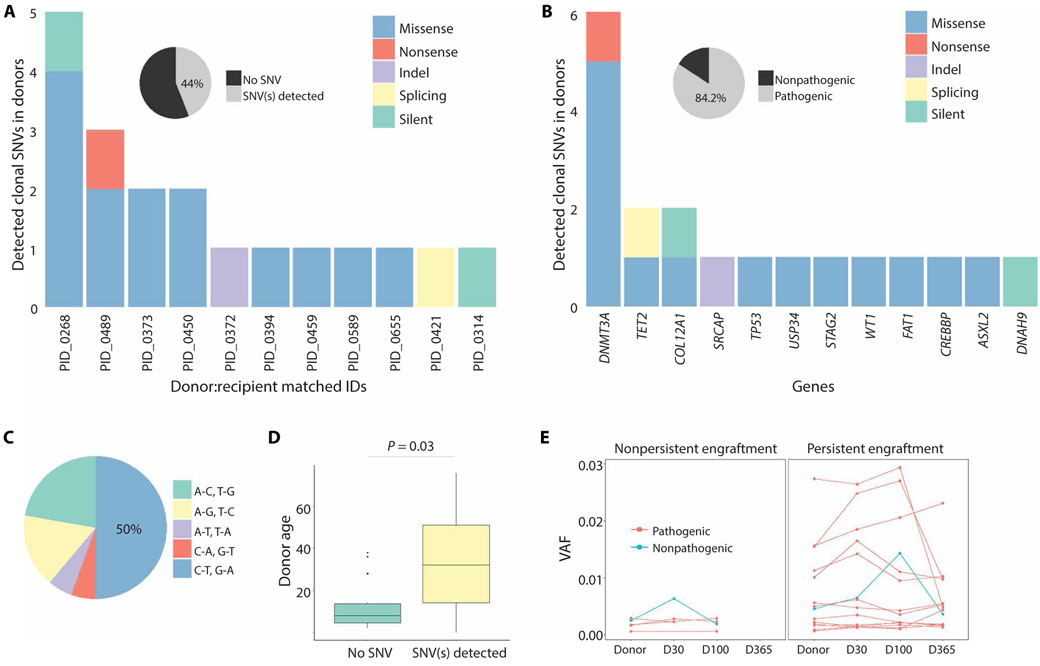
The donor pool consisted of 25 individuals with a median age of 26 years (range, 20 to 58). Only one donor, aged 23 (4% of donors), harbored a CHIP clone >0.02 VAF [SRCAP frame shift insertion-deletion (indel)]. In total, we identified 19 somatic mutations in 11 donors, aged 20 to 58 (44% of donors) (Fig. 1A and data file S1). The median VAF of these somatic mutations was 0.00247 (an order of magnitude more rare than the definition of CHIP) with a range of 0.00058 to 0.0274. Fourteen donors had no clonal mutations in the 80 target genes. Consistent with previous studies, despite a younger cohort, donors had mutations most frequently in DNMT3A and TET2 (Fig. 1B). None of the mutations detected in donors were observed in the pre-HSCT samples of recipients. Each mutation was annotated using the combined annotation-dependent depletion (CADD) scoring system. Mutations with a scaled CADD score ≥20 represent the top 1% of mutations expected to be most pathogenic to any cellular func-tion (22) and were, thus, labeled as “pathogenic” mutations in this study. We found that 84.2% of the detected mutations were patho-genic (Fig. 1B and Table 1), and 100% of detected somatic mutations engrafted in recipients. The most common mutations were cytosine-to- thymine transitions (Fig. 1C), as previously seen in healthy, elderly adults (14). The median ages for the donors with clonal hematopoiesis and those without were 36 and 24, respectively, which was a signif-icant difference (P = 0.03; two-sided Wilcoxon rank-sum test; Fig. 1D).
Fig. 1. Mutation burden and spectrum in unrelated donors.
(A) Number and types of somatic mutations detected in donors. Of the donor cohort, 44% (11 of 25) harbored at least one somatic mutation. (B) Mutation spectrum of detected mutations in donors. (C) Types of nucleotide changes. (D) Age of donors with and without detected SNV(s). Boxes show the 25th and 75th percentiles, as well as median (P = 0.03, two-sided Wilcoxon rank-sum test). (E) Clonal dynamics of engrafted mutations in recipients after HSCT.
Table 1.
Somatic mutations detected in donors that were predicted to be pathogenic according to CADD.
| Gene | Type | Amino acid change | CADD | COSMIC | Engrafted |
|---|---|---|---|---|---|
| COL12A1 | Missense | p.I530L | 22.1 | COSM271996 | Yes |
| CREBBP | Missense | p.T1242I | 25.5 | – | Yes |
| DNMT3A | Nonsense | p.W288X | 40 | C0SM1130818 | Yes |
| DNMT3A | Missense | p.R174S | 26.5 | – | Yes |
| DNMT3A | Missense | p.G398R | 30 | C0SM256035* | Yes |
| DNMT3A | Missense | p.I158M | 23.6 | – | Yes |
| DNMT3A | Missense | p.Q222P | 26.1 | – | Yes |
| DNMT3A | Missense | p.H669P | 23.3 | – | Yes |
| FAT1 | Missense | p.D1554N | 25.8 | COSM1429043 | Yes |
| SRCAP | Indel | T:TGCTTCGCC | 29 | – | Yes |
| STAG2 | Missense | p.Y188D | 26.3 | – | Yes |
| TET2 | Splicing | c.3954 + 1G > A | 34 | C0SM87141* | Yes |
| TET2 | Missense | p.Y1345C | 32 | – | Yes |
| TP53 | Missense | p.R150W | 25.7 | COSM99925* | Yes |
| USP34 | Missense | p.H1874R | 22.5 | – | Yes |
| WT1 | Missense | p.R74W | 28.7 | – | Yes |
Six mutations were found to be associated with various malignancies, and three were specifically associated with hematologic malignancies.
Of the 19 engrafted mutations, 14 (74%) clones persisted through D365 after HSCT, and 13 of these had pathogenic mutations (Fig. 1E and fig. S1). The likelihood of persistent engraftment was not depen-dent on the initial VAF in donors (P = 0.105; two-sided Wilcoxon rank-sum test). Despite an initially low VAF, three recipients (12%) had engrafted clones that expanded beyond the defined CHIP threshold of ≥0.02 VAF after HSCT at D100 and D365 (fig. S2). All mutations that expanded to ≥0.02 VAF were scored as pathogenic, and the mutated genes were TP53 p.R150W [Catalogue of Somatic Mutations in Cancer (COSMIC) ID: COSM99925; CADD = 25.7], DNMT3A p.Q222P (CADD = 26.1), and CREBBP p.R445X (COSMIC ID: COSM255965; CADD = 38).
Presence of de novo pathogenic somatic mutations in recipients after HSCT
Next, we examined longitudinal differences in the mutational spectrum of engrafted clones. By comparing the recipient’s clonal profile before HSCT and after HSCT, we accounted for residual physiologic hematopoietic clones and residual primary disease (data files S2 and S3). These recipient clones were filtered out accordingly.
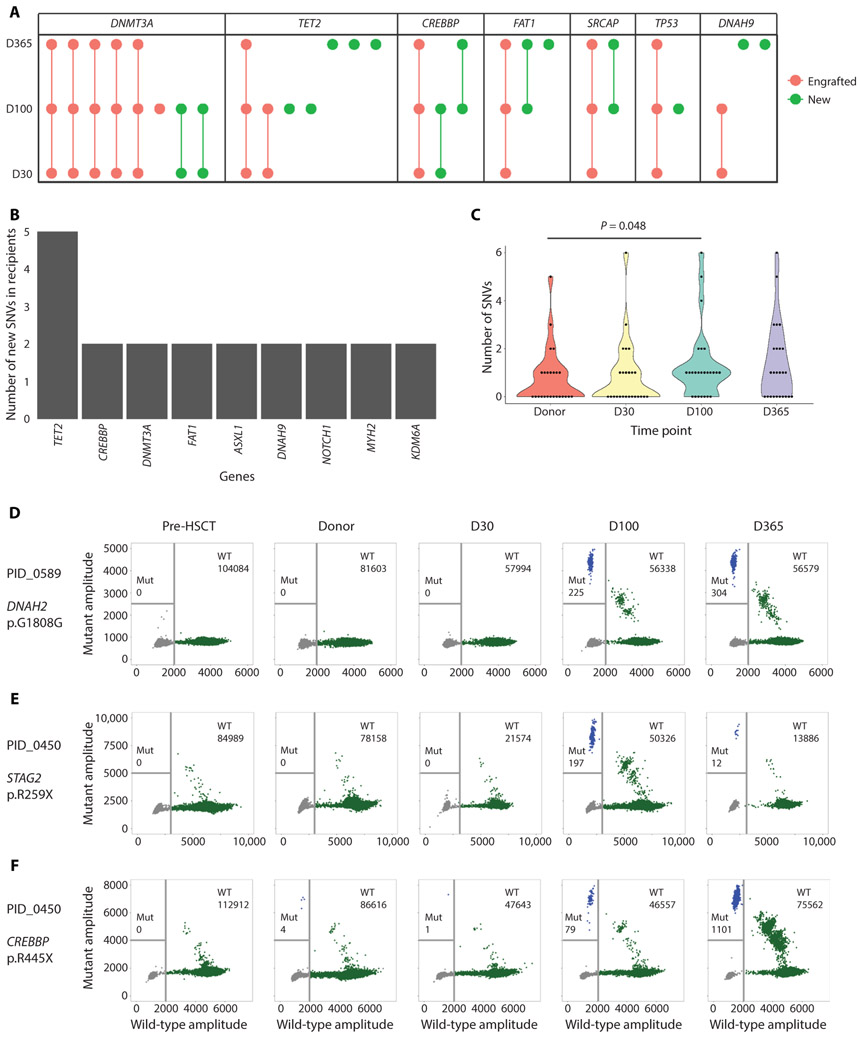
As expected, given their high prevalence, DNMT3A mutations were most commonly observed after HSCT across all time points (Fig. 2A), and most of these were engrafted from donors. In addition, 30 (61.2%) of the total detected unique mutations in recipients after HSCT were new mutations not previously observed in donors. Of these 30 mutations, 9 were observed at two different time points within the same recipients after HSCT. These newly detected mutations were called in different genes from those observed in donors and in previous CHIP studies (10-12). For instance, TET2, CREBBP, and FAT1 were more commonly mutated in recipients after HSCT than in donors before HSCT [mutations observed only in donor- derived cells in recipients after HSCT (Fig. 2B); mutations observed only in donors before HSCT (Fig. 1B)]. The most common type of nucleotide change was cytosine to thymine (fig. S3). We also found that the mutation burden across the entire cohort significantly in-creased from pre-HSCT (19 total somatic mutations) in donors to D100 (33 total somatic mutations) (P = 0.048, one-sided Wilcoxon rank-sum test; Fig. 2C). The presence of these mutations was not due to differences in sequencing metrics (fig. S4). In addition, when comparing the presence of these mutations in recipients who were transplanted from donors with (n = 11) or without (n = 14) detectable clonal mutations, we found no difference in this observation (not significant, P = 0.44, two-sided Wilcoxon rank-sum test; data file S2 and fig. S5).
Fig. 2. Mutation burden and spectrum of donor clonal mutations engrafted in the recipients.
(A) Mutation spectrum in recipients at different time points after HCST. “Engrafted” mutations are known to be from the donor, and “New” mutations were not observed before HSCT in either donor or recipient specimens. Genes with ≥1 engrafted and ≥ 1 new mutations are shown. (B) Mutation spectrum of new mutations detected in recipients after HSCT. Genes with ≥2 new mutations were represented. (C) Violin plots showing mutation burden at different time points after HSCT. The P value was calculated using one-sided Wilcoxon rank-sum test. (D to F) Representative plots showing ddPCR experiment results. The blue dots in the panels indicate positive mutant droplets, green dots indicate positive wild-type (WT) droplets, and gray dots indicate empty droplets. (D) ddPCR results demonstrating the presence of a de novo, persistent mutation in DNAH2 in donor-derived cells engrafted in the recipient (PID_0589). The mutation was not detected in the donor before HSCT. (E) ddPCR results demonstrating the presence of a de novo, transient mutation in STAG2 in donor-derived cells engrafted in the recipient (PID_0450). The mutation was not detected in the donor before HSCT, and it reduced in VAF by 1 year after HSCT. (F) ddPCR results demonstrating the presence of an engrafted donor-derived mutation in CREBBP that was detected at an extremely low VAF in the donor (PID_0450). This mutation increased in VAF at D100 and 1 year after HSCT in the recipient.
Potential explanations for the presence of these mutations were either that they (i) were present in donors before transplant with a VAF below the limit of ECS detection or (ii) arose de novo after engraftment. To distinguish between these two possibilities, we performed droplet digital polymerase chain reaction (ddPCR) on a subset of mutations in all five samples from matched pairs. We found that these mutations were a mixture of extremely rare donor mutations that engrafted in recipients and underwent clonal expansion and de novo mutations that appeared post-HSCT (Fig. 2, D to F, and fig. S6). Some de novo mutations persisted or expanded (Fig. 2D) over time, whereas some were transient and vanished by later time points (Fig. 2E). With respect to exceedingly rare preexisting clones, one recipient (PID_0450) was found to have a CREBBP nonsense mutation, which was not detected in the donor pre-HSCT sample but was detected at D365 via ECS. By ddPCR, the same mutation was detected in the donor pre-HSCT (Fig. 2F) and underwent an approximate 500-fold expansion with an increase in VAF from 0.000046 to 0.027 by D365 after HSCT. The prevalence of these mutations was associated with gene length (P < 0.00001; Pearson correlation = 0.5136; fig. S7), suggesting a stochastic mechanism of mutation.
Persistent engraftment of donor-derived mutations and clinical descriptors
Although this study was not designed or powered to establish clinical correlations to clonal hematopoiesis, we nevertheless examined the relationships between engrafted donor-derived mutations and clinical outcomes as a descriptive and exploratory pilot analysis. We were particularly interested in chronic GvHD, which was recently associated with CHIP clones engrafted from older, related donors (5). Be-cause young, unrelated donors with CHIP are rare (we detected CHIP in one donor), we examined the effect of persistent engraftment (up to 1 year) of donor-derived mutations. We found that 75% of recipients who had at least one persistently engrafted, pathogenic mutation developed chronic GvHD versus about 50% of those without any persistently engrafted mutated clones. However, given the sample size, the difference was not statistically significant (P = 0.17, Gray’s test; figs. S8 and S9). Descriptive results for other clinical outcome measures for donors with and without clonal mutations (as well as pathogenic or nonpathogenic) are provided in data file S4.
DISCUSSION
In this pilot study intended to quantify the presence of rare hematopoietic clones in the healthy AYA population and observe the dynamics of these clones over time in an unrelated allogeneic HSCT context, we have made five observations that address several outstanding questions. First, we showed that clonal hematopoietic mutations ≥0.0005 VAF are common (44%) in the AYA population—an age group where CHIP was virtually nondetectable in previous studies (10-12) but which constitutes 86% of eligible unrelated HSPC donors. Although not demonstrated here, previous data suggest that these mutations, which were present at 10-fold lesser VAF than CHIP, are likely to occur in hematopoietic progenitors due to their presence in myeloid and lymphoid lineages in comparable frequencies, as well as their persistent nature over time (14, 23). A substantial proportion of these clones harbor mutations that could confer a survival or proliferative advantage upon selective pressures. If we only examined common mutations at or above the defined CHIP threshold of 0.02 VAF without considering rare clones, we would miss most, if not all, of these mutations in unrelated donors that might have as yet unknown clinical impacts, as acknowledged by Frick and col-leagues (5). Given the many indications for unrelated, allogeneic HSCT and recent associations of clonal hematopoiesis with risks for developing leukemia (16), atherosclerosis (24), and chronic GvHD after HSCT (5), and given that under selective pressures these pre-existing clones can emerge to clinical relevance years after their se-lection (13), it is crucial to understand how putatively pathogenic clones in this age group can be transferred from healthy donors to recipients who have undergone combinations of radiation, chemo-therapy, and immunosuppression.
Second, we find that donor hematopoietic clones harbor mutations that are mostly pathogenic (84.2%) and have a seemingly strong predilection for engraftment (100% in this cohort). Third, rare clones with pathogenic mutations were likely to persist/expand for at least 1 year after HSCT, regardless of initial VAF. These two observations support the hypothesis that pathogenic mutations confer a variable fitness advantage to the donor cells (25) and would also explain why these engrafted rare, pathogenic mutations persist/expand in recipients after HSCT. Fourth, the fact that there was no difference in the pre-HSCT VAF of clones with and without persistent engraftment argues for quantifying the presence of rare clones with mutations conferring a strong effect over time and against recent reports attributing clinical relevance solely to “clone size” (26). An example of this is the recipient with a rare donor-derived CREBBP-mutated clone expanding 500-fold in the recipient 1 year after HSCT. CREBBP mutations have been shown to adversely affect hematopoietic development and are associated with malignant lymphoid stem-like properties (27). Thus, in the appropriate context, rare clones with mutations conferring a strong effect size or selective advantage can expand relatively rapidly regardless of their initial VAF.
Fifth, we found that the clonal hematopoietic spectrum of recipients after HSCT transiently changes over time, revealing mutations within the first year after HSCT that are less commonly seen in physiologic CHIP and appear to develop from de novo mutations gained after HSCT. The positive association between post-HSCT mutations and gene length suggests clonal drift. Under this scenario, the rapid proliferation of donor hematopoietic progenitors would introduce stochastic mutations across the genome, and only clones with an advantage would persist over time. In light of this, we suggest that there may be many rare hematopoietic progenitors with pathogenic mutations in unrelated, otherwise-healthy AYA donors that are otherwise neutral in the donor, due to a lack of selective pressure, but could undergo preferential expansion in recipients as a result of the selective pressures previously mentioned.
Alternatively, donor cells may experience a transient hyper-mutative phase upon encountering an unfamiliar microenvironment. Transient hypermutation of cellular subpopulations has been shown to give rise to adaptive mutations that allow new cellular phenotypes to emerge (28, 29), and the process selectively mutates epigenetic modifier genes because they promote cell phenotypic heterogeneity (30). Such a hypothesis would be consistent with the observed in-crease in clonal mutation burden as a function of time after HSCT, as well as with the observation that some de novo mutations dis-appear and some expand by D365, suggesting that only the clones with a selective advantage persist. In addition, most DNMT3A mutations observed in recipients were engrafted from donors, supporting the hypothesis that DNMT3A-mutated clones, or, more broadly, clones with mutations in epigenetic modifiers such as CREBBP or TET2, harbor a competitive advantage (31, 32).
In summary, we have shown that extremely rare, preexisting clones with pathogenic mutations engrafted the recipients regardless of their initial VAFs. Our sample size and only 1 year of post-HSCT follow-up prevented us from establishing clinical correlations. It would stand to reason that our demonstration of engraftment of clones at 10-fold lower VAF than CHIP would require a longer time for manifestation of clinical consequences. Thus, this pilot study interrogating the prevalence of rare clonal hematopoiesis in the AYA population and examining what happens to these clones in unrelated HSCT recipients merits a much larger study with longer follow-up to correlate post-HSCT morbidities with transfer and persistence of donor clones. Such correlations could enable clinicians to survey the clonal hematopoietic profile of potential donors to improve post-HSCT surveillance and mitigate potential long-term morbidity.
MATERIALS AND METHODS
Study design
This retrospective pilot study was designed to interrogate donor- derived clonal dynamics after HSCT. All patients provided informed consent for research. The Human Research Protection Office at Washington University approved the study. From the adult AML specimen repository at Washington University, we initially identified a total of 30 patients who had banked samples before transplant and at days 30, 100, and 365 after HSCT. There were no other selection criteria. From that group, the Center for International Blood and Marrow Transplant Research (CIBMTR) was able to provide donor pre-HSCT specimens for 25 of 30 recipients, again without any additional selection.
Sample collection
Four longitudinally collected peripheral blood and/or bone marrow samples per recipient were acquired for 25 recipients with primary hematological malignancies who had undergone matched, unrelated donor allogeneic HSCT at Barnes-Jewish Hospital/Siteman Cancer Center/Washington University School of Medicine (Table 2). Of the patients, 64% were transplanted for myeloid malignancies. For each patient, samples were collected before HSCT conditioning (pre-HSCT), 30 days (D30), 100 days (D100), and 1 year after HSCT (D365). In addition, aliquots from 25 corresponding unrelated do-nor leukocyte samples collected before HSCT were obtained from the CIBMTR repository. In total, 125 unique samples (100 patient samples from four time points and 25 donor samples) were processed and analyzed. An independent replicate for each sample was then prepared and deep sequenced to confirm the variants identified.
Table 2.
Demographic information of recipients and the corresponding matched donors in relation to engraftment of donor-derived mutations.
| Characteristic | Category | No donor mutation (n = 14) |
Mutation engrafted (n = 11) |
P | Test performed |
|---|---|---|---|---|---|
| Donor age | Median (range) | 24 (21 to 39) | 36 (20 to 58) | 0.03 | Wilcoxon rank-sum test |
| Donor gender | Male | 10 (71.4%) | 8 (72.7%) | 0.99 | Fisher’s exact test |
| Female | 4 (28.6%) | 3 (27.3%) | |||
| Recipient age | Median (range) | 51 (27 to 65) | 55 (19 to 69) | 0.66 | Wilcoxon rank-sum test |
| Recipient gender | Male | 13 (92.9%) | 7 (63.6%) | 0.13 | Fisher’s exact test |
| Female | 1 (7.1%) | 4 (36.4%) | |||
| Primary disease | AML/MDS | 7 (50%) | 9 (81.8%) | 0.21 | Fisher’s exact test |
| Non-AML | 7 (50%) | 2 (18.2%) | |||
| Disease status prior to transplant | CR | 7 (50%) | 5 (45.4%) | ||
| Non-CR | 7 (50%) | 5 (45.4%) | 0.99 | Fisher’s exact test | |
| Unknown | 0 (0%) | 1 (9.1%) | |||
| Conditioning | MAC | 8 (57.1%) | 7 (63.6%) | 0.99 | Fisher’s exact test |
| Non-MAC | 6 (42.9%) | 4 (36.4%) | |||
| HLA mismatch | No mismatch | 13 (92.9%) | 9 (81.8%) | 0.56 | Fisher’s exact test |
| Mismatch | 1 (7.1%) | 2 (18.2%) |
CR, complete remission; MAC, myeloablative conditioning.
ECS and mutation analysis
Genomic DNA was extracted from the blood/marrow samples using the DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer’s recommendations. The final DNA elution volume was 50 ml. The concentration of the extracted DNA was determined with the Qubit dsDNA HS Assay (Life Technologies). After quantification of DNA concentration, 200 to 250 ng of DNA per sample was used to make ultradeep ECS libraries. For this study, we generated a custom Illumina TruSight enrichment assay including a total of 1063 amplicons enriching for some exons or full length of 80 frequently mutated genes in pediatric/AYA and adult AML (data file S5). Adult AML genes were previously included in the Illumina TruSight Myeloid Assay, and pediatric AML genes were identified from the TARGET project (20). Details of the library preparation are comprehensively documented in two previously published papers (33, 34). Briefly, amplicon oligos were hybridized onto the genomic DNA following the Illumina TruSight’s protocol. After hybridization, unbound oligos were removed, and extension-ligation of the amplicons of interest was performed. After extension-ligation, the libraries were amplified for six cycles using the Q5 High-Fidelity 2× Master Mix (New England BioLabs) in a 75-ml reaction: 37.5 ml of Q5 master mix, 6 ml of 10 mM redesigned i5 adapters, 6 ml of 10 mM i7 adapters, and 22 ml of the extension-ligation solution. The rede-signed i5 adapters contain a string of 16 random nucleotides (16N) that replaces the original eight-nucleotide index sequence. The 16N serve as unique molecular indexes (UMIs) that are essential for error correction after sequencing. The redesigned i5 adapters can be ordered through Integrated DNA Technologies using the following oligo sequence: AATGATACGGCGACCACCGAGATCTA-CAC(N1:25252525)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)ACACTCTTTCCCTACACGAC-GCTCTTCCGATCT. The initial six-cycle amplification allows for tagging of molecules in the reaction with the UMIs. After the initial amplification, the libraries were cleaned using AMPure XP magnetic beads (Agencourt). The number of UMI-tagged molecules in the cleaned libraries was quantified using the QX200 ddPCR platform with EvaGreen (Bio-Rad). After ddPCR quantification, each library was normalized to 6.3 million UMI-tagged molecules, and a second round of PCR (14 cycles) was performed in a 50-ml reaction: 25 ml of Q5 master mix, 2 ml of P5 primer (1 mM), 2 ml of P7 primer (1 mM), and 21 ml of DNA molecules. After that, the amplified libraries were purified, and the libraries were normalized. Six purified libraries were pooled and sequenced per lane in an Illumina HiSeq 4000 instrument with the following settings: 2 × 144 paired-end, 8 cycles Index 1, 16 cycles Index 2 (account for 16N random bases used as UMI). For each sample, a technical replicate library was prepared via the same protocol. In total, 125 samples were processed.
Deep sequencing was performed on the Illumina HiSeq 4000 at the McDonnell Genome Institute of Washington University. A mini-mum of three raw reads sharing the same UMI were processed to give error-corrected consensus sequence (ECCS). Each library was deep sequenced to an average ECCS depth of 9200× (fig. S4). The raw sequencing data in fastq format were first demultiplexed into corresponding samples using a custom script. The demultiplexed reads were subsequently processed using an UMI-aware custom script. First, the first 30 nucleotides of each read were hard clipped to remove oligo sequences. Next, reads sharing the same UMIs were aligned to one another to form read families. Each read family was required to have three reads or more for deduplication and error correction, which would output a consensus read for each read family. The consensus reads were aligned locally to hg19 using Bowtie2 with local alignment setting. The bam files were realigned using GATK’s Indel Realigner. Next, the aligned reads were processed with Mpileup using the following parameters: –BQ0 –d 10,000,000,000,000 to remove coverage thresholds to ensure a proper pileup output. The output was filtered to include bases with ≥700× consensus read coverage and within the target regions of the Illumina TruSight panel that are not common variants (≥0.01 minor allele fraction) identified by the 1000 Genomes Project. For single-nucleotide polymorphisms, a position-specific binomial background error model was implemented in variant calling. Each genomic position was modeled independently by compiling the background error rate of all samples for that specific genomic position (sum of all variant bases relative to the sum of reference bases). A sample with a number of variant bases at a genomic position that was significantly different from the background error rate based on binomial distribution after Bonferroni correction was considered a positive for that position. Typically, the P value (after Bonferroni correction) for calling a variant as positive was <0.00000001. After variant calling, several other filters were applied to remove artifacts and to obtain high-confidence variants: (i) variants that were only called in one technical sequencing replicate but not in the other were removed, (ii) variants called due to sequencing batch effect were removed, (iii) nonhotspot variants identified in more than one donor-recipient matched pair were removed, (iv) variants with <0.001 VAF were removed unless the variants were observed at multiple time points in the matched sample set, and (v) variants that had a coefficient of variation >15% between 3-read and 5-read error corrections were removed. After applying the filters, we retained a set of high-confidence variants by removing false-positive calls and common variants that are observed in the general population. Indels were identified using VarScan2 with the mpileup2indel setting after error correction into a consensus read sequence (35).
Two independent replicate sequencing libraries were made and sequenced separately (DNA was extracted from different aliquots of leukocytes from the same sample). Variants that passed the established filters in all available libraries for that sample were retained for further analysis. Variants present in pre-HSCT recipient samples represented the clonal hematopoietic profile of the recipient and, potentially, any remaining primary leukemia. These pre-HSCT germ-line variants in recipients were used to evaluate the degree of mixed chimerism in the recipient after HSCT. Engraftment of donor hematopoietic clone(s) was evaluated on the basis of the presence of variants from donor pre-HSCT observed in recipient samples after HSCT. Persistent engraftment was further defined as having donor-derived mutation(s) that persist through 1 year (D365) after HSCT.
Validation of observed mutations via ddPCR and triplicate sequencing
For validation of called mutations, we performed ddPCR using the Bio-Rad QX200 platform or triplicate ECS with independently pre-pared and sequenced libraries on these observed variants. For ddPCR, a primer/probe set specific to the variant of interest was designed by Bio-Rad according to MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines for quantitative PCR (data file S6). Probes targeted both reference and mutated nucleotides at the same genomic positions via different fluorophores. All ddPCRs were performed in accordance with the manufacturer’s recommendations using “ddPCR Supermix for Probe (no dUTP).” For triplicate sequencing, we considered only those variants observed in all three independent sequencing runs to be true positives.
Statistical analysis of clinical correlates
Categorical variables [donor gender, recipient gender, primary dis-ease = AML/multidimensional scaling (MDS) or others, disease status before transplant = remission, conditioning = myeloablative or reduced intensity, and human leukocyte antigen (HLA) mismatch = no] were compared using Fisher’s exact test. A nonparametric Wilcoxon rank-sum test was used to compare continuous, non-Gaussian variables (duration of cytopenia, age of donor, and age of recipient). Cytopenia was defined as white blood cell count <2 × 109/liter, hemoglobin <10 g/dl, and platelets <100 × 109/liter. Because several patients died without chronic GvHD, the cumulative incidence of chronic GvHD was accessed using the Fine-Gray subdistribution hazard model to account for death as a competing risk for this endpoint. The start time for chronic GvHD was defined as after D100 after transplant. Leukemia-free survival was compared using a Kaplan- Meier model. Mixed chimerism was assessed repeatedly as a presence/absence, and it was compared using a repeated-measures logistic regression. The analysis was intended to be exploratory, so no attempt was made to adjust the P values for multiple tests.
Supplementary Material
Fig. S1. Engrafted donor mutations in recipients.
Fig. S2. Clonal expansion of mutations reaching the threshold for CHIP (≥0.02 VAF) in three patients after HSCT.
Fig. S3. Types of somatic substitutions in recipients after HSCT.
Fig. S4. The sequencing depth of each ECS library at all time points.
Fig. S5. New mutations detected in recipients after HSCT.
Fig. S6. ECS calls validated by ddPCR.
Fig. S7. Number of detected mutations in genes according to gene length.
Fig. S8. Leukemia-free survival of recipients with or without persistent engraftment of donor-derived mutations.
Fig. S9. Cumulative incidence of chronic GvHD in recipients with or without persistent engraftment of donor-derived mutations.
Data file S1. Detected somatic mutations in donors.
Data file S2. Detected somatic mutations in recipients after HSCT after removing recipient’s own hematopoietic clones.
Data file S3. Shared variants in pre-HSCT and post-HSCT recipient samples due to incomplete clearance of recipient’s hematopoietic clones after HSCT.
Data file S4. Analysis of recipient clinical outcomes in relation to engraftment of donor-derived mutations.
Data file S5. Recurrently mutated genes in adult and pediatric AML.
Data file S6. ddPCR probe sequences.
Acknowledgments:
We thank the leadership at the CIBMTR and Siteman Cancer Center for providing the donor and recipient samples, respectively. We also thank G. Challen, J. Welch, M. Jacoby, and M. Ferris for the helpful and insightful discussions. E. Martin and B. Koebbe at the Edison Family Center for Genome Sciences and System Biology provided IT and computational infrastructure support. We also thank the McDonnell Genome Institute for the NGS resources.
Funding: The genomic portion of this study was supported by NCI R01CA211711 to T.E.D., the Hyundai Quantum Award to T.E.D., the Leukemia and Lymphoma Society Scholar Award to T.E.D., the Eli Seth Matthews Leukemia Foundation to T.E.D., and the Kellsie’s Hope Foundation to T.E.D. The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement 5U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); Grant/Cooperative Agreement 1U24HL138660 from NHLBI and NCI; contract HHSH250201700006C with Health Resources and Services Administration (HRSA/DHHS); and three grants (N00014-17-1-2388, N00014-17-1-2850, and N00014-18-1-2045) from the Office of Naval Research. J.R.B. is supported by a UKRI future leaders fellowship and by a CRUK Cambridge Centre Early Detection Programme group leader grant. Author contributions: W.H.W. and T.E.D. formulated the initial concept for this study with S.B., I.P., K.E., G.E.S., D.L.C., J.D., M.A.P., N.N.S., J.S., and B.E.S. I.P., K.E., and J.D. curated the recipient samples before and after HSCT, while G.E.S., D.L.C., M.A.P., N.N.S., J.S., and B.E.S. curated the donor samples.
Footnotes
W.H.W. and N.M. prepared the ECS libraries. W.H.W. and A.B. performed ddPCR validations. Bioinformatics analyses were performed by W.H.W. with guidance from J.R.B., clinical correlation analysis by S.B., and statistical analysis by F.W. and K.T. The manuscript was written by W.H.W. and T.E.D., with input and comments from all coauthors.
Competing interests: The Washington University Office of Technology Management has filed patent application #62/106,967 for “Ultra-rare Variant Detection from Next-generation Sequencing,” which has been licensed by Canopy Biosciences as RareSeq. T.E.D. is a coinventor on this patent. Canopy Biosciences was not involved in the generation of the data presented herein. T.E.D. has ownership and employment by ArcherDX Inc. and serves as the chief medical officer for this molecular cancer diagnostics company. ArcherDX and its products were not involved in the generation or preparation of any data in this report. All other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Shenoy S, Boelens JJ, Advances in unrelated and alternative donor hematopoietic cell transplantation for nonmaglinant disorders. Curr. Opin. Pediatr 27, 9–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyurkocza B, Rezvani A, Storb RF, Allogeneic hematopoietic cell transplantation: The state of the art. Expert. Rev. Hematol 3, 285–299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afessa B, Peters SG, Major complications following hematopoietic stem cell transplantation. Semin. Respir. Crit. Care Med 27, 297–309 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Scott JM, Armenian S, Giralt S, Moslehi J, Wang T, Jones LW, Cardiovascular disease following hematopoietic stem cell transplantation: Pathogenesis, detection, and the cardioprotective role of aerobic training. Crit. Rev. Oncol. Hematol 98, 222–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frick M, Chan W, Arends CM, Hablesreiter R, Halik A, Heuser M, Michonneau D, Blau O, Hoyer K, Christen F, Galan-Sousa J, Noerenberg D, Wais V, Stadler M, Yoshida K, Schetelig J, Schuler E, Thol F, Clappier E, Christopeit M, Ayuk F, Bornhäuser M, Blau IW, Ogawa S, Zemojtel T, Gerbitz A, Wagner EM, Spriewald BM, Schrezenmeier H, Kuchenbauer F, Kobbe G, Wiesneth M, Koldehoff M, Socié G, Kroeger N, Bullinger L, Thiede C, Damm F, Role of donor clonal hematopoiesis in allogeneic hematopoietic stem-cell transplantation. J. Clin. Oncol 37, 375–385 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Kato M, Yamashita T, Suzuki R, Matsumoto K, Nishimori H, Takahashi S, Iwato K, Nakaseko C, Kondo T, Imada K, Kimura F, Ichinohe T, Hashii Y, Kato K, Atsuta Y, Taniguchi S, Fukuda T, Donor cell-derived hematological malignancy: A survey by the Japan Society for Hematopoietic Cell Transplantation. Leukemia 30, 1742–1745 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Gibson CJ, Kennedy JA, Nikiforow S, Kuo FC, Alyea EP, Ho V, Ritz J, Soiffer R, Antin JH, Lindsley RC, Donor-engrafted CHIP is common among stem cell transplant recipients with unexplained cytopenias. Blood 130, 91–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jian J, Hao H, Yuan C, Donor-cell-derived myelodysplastic syndrome involving U2af1 mutation developing 8 years after matched unrelated bone marrow transplantation for acute leukemia and literature review (2017); www.eajournals.org.
- 9.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL, Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL, Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landén M, Höglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Grönberg H, Hultman CM, McCarroll SA, Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L, Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, Heath S, Baty JD, Klco JM, Ding L, Mardis ER, Westervelt P, Dipersio JF, Walter MJ, Graubert TA, Ley TJ, Druley TE, Link DC, Wilson RK, Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 518, 552–555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young AL, Challen GA, Birmann BM, Druley TE, Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun 7, 12484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young AL, Tong RS, Birmann BM, Druley TE, Clonal haematopoiesis and risk of acute myeloid leukemia. Haematologica 104, 2410–2417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, Barda, Zuzarte PC, Heisler L, Sundaravadanam Y, Luben R, Hayat S, Wang TT, Zhao Z, Cirlan I, Pugh TJ, Soave D, Ng K, Latimer C, Hardy C, Raine K, Jones D, Hoult D, Britten A, McPherson JD, Johansson M, Mbabaali F, Eagles J, Miller JK, Pasternack D, Timms L, Krzyzanowski P, Awadalla P, Costa R, Segal E, Bratman SV, Beer P, Behjati S, Martincorena I, Wang JCY, Bowles KM, Quirós JR, Karakatsani A, La Vecchia C, Trichopoulou A, Salamanca-Fernández E, Huerta JM, Barricarte A, Travis RC, Tumino R, Masala G, Boeing H, Panico S, Kaaks R, Krämer A, Sieri S, Riboli E, Vineis P, Foll M, McKay J, Polidoro S, Sala N, Khaw K-T, Vermeulen R, Campbell PJ, Papaemmanuil E, Minden MD, Tanay A, Balicer RD, Wareham NJ, Gerstung M, Dick JE, Brennan P, Vassiliou GS, Shlush LI, Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncavage EJ, Jacoby MA, Chang GS, Miller CA, Edwin N, Shao J, Elliott K, Robinson J, Abel H, Fulton RS, Fronick CC, O’Laughlin M, Heath SE, Brendel K, Saba R, Wartman LD, Christopher MJ, Pusic I, Welch JS, Uy GL, Link DC, DiPersio JF, Westervelt P, Ley TJ, Trinkaus K, Graubert TA, Walter MJ, Mutation clearance after transplantation for myelodysplastic syndrome. N. Engl. J. Med 379, 1028–1041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson CJ, Papula A, Poon YPG, Wong WH, Young AL, Druley TE, Fisher DS, Blundell JR, The evolutionary dynamics and fitness landscape of clonal haematopoiesis. bioRxiv, doi: 10.1101/569566 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Bowman RL, Busque L, Levine RL, Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, Alonzo TA, Guidry Auvil JM, Davidsen TM, Gesuwan P, Hermida L, Muzny DM, Dewal N, Rustagi N, Lewis LR, Gamis AS, Wheeler DA, Smith MA, Gerhard DS, Meshinchi S, Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res. 76, 2197–2205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M, CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arends CM, Galan-Sousa J, Hoyer K, Chan W, Jäger M, Yoshida K, Seemann R, Noerenberg D, Waldhueter N, Fleischer-Notter H, Christen F, Schmitt CA, Dörken B, Pelzer U, Sinn M, Zemojtel T, Ogawa S, Märdian S, Schreiber A, Kunitz A, Krüger U, Bullinger L, Mylonas E, Frick M, Damm F, Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 32, 1908–1919 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu C-L, Sano S, Muralidharan S, Rius C, Vuong J, Jacob S, Muralidhar V, Robertson AAB, Cooper MA, Andrés V, Hirschi KK, Martin KA, Walsh K, Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlesworth B, The effects of deleterious mutations on evolution at linked sites. Genetics 190, 5–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, Schmid T, Brüne B, Wagner S, Serve H, Hoffmann J, Seeger F, Dimmeler S, Zeiher AM, Rieger MA, Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 4, 25–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton SJ, Giotopoulos G, Yun H, Vohra S, Sheppard O, Bashford-Rogers R, Rashid M, Clipson A, Chan W-I, Sasca D, Yiangou L, Osaki H, Basheer F, Gallipoli P, Burrows N, Erdem A, Sybirna A, Foerster S, Zhao W, Sustic T, Petrunkina Harrison A, Laurenti E, Okosun J, Hodson D, Wright P, Smith KG, Maxwell P, Fitzgibbon J, Du MQ, Adams DJ, Huntly BJP, Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat. Cell Biol 19, 1093–1104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosche WA, Foster PL, The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A 96, 6862–6867 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galhardo RS, Hastings PJ, Rosenberg SM, Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol 42, 399–435 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feinberg AP, Koldobskiy MA, Göndör A, Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet 17, 284–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao H, Wang L-M, Luo Y, Lai X, Li C, Shi J, Tan Y, Fu S, Wang Y, Zhu N, He J, Zheng W, Yu X, Cai Z, Huang H, Mutations in epigenetic regulators are involved in acute lymphoblastic leukemia relapse following allogeneic hematopoietic stem cell transplantation. Oncotarget 7, 2696–2708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong M, Park HJ, Celik H, Ostrander EL, Reyes JM, Guzman A, Rodriguez B, Lei Y, Lee Y, Ding L, Guryanova OA, Li W, Goodell MA, Challen GA, Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 23, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young AL, Wong TN, Hughes AEO, Heath SE, Ley TJ, Link DC, Druley TE, Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia 29, 1608–1611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong WH, Tong RS, Young AL, Druley TE, Rare event detection using error-corrected DNA and RNA sequencing. J. Vis. Exp, 10.3791/57509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK, VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Engrafted donor mutations in recipients.
Fig. S2. Clonal expansion of mutations reaching the threshold for CHIP (≥0.02 VAF) in three patients after HSCT.
Fig. S3. Types of somatic substitutions in recipients after HSCT.
Fig. S4. The sequencing depth of each ECS library at all time points.
Fig. S5. New mutations detected in recipients after HSCT.
Fig. S6. ECS calls validated by ddPCR.
Fig. S7. Number of detected mutations in genes according to gene length.
Fig. S8. Leukemia-free survival of recipients with or without persistent engraftment of donor-derived mutations.
Fig. S9. Cumulative incidence of chronic GvHD in recipients with or without persistent engraftment of donor-derived mutations.
Data file S1. Detected somatic mutations in donors.
Data file S2. Detected somatic mutations in recipients after HSCT after removing recipient’s own hematopoietic clones.
Data file S3. Shared variants in pre-HSCT and post-HSCT recipient samples due to incomplete clearance of recipient’s hematopoietic clones after HSCT.
Data file S4. Analysis of recipient clinical outcomes in relation to engraftment of donor-derived mutations.
Data file S5. Recurrently mutated genes in adult and pediatric AML.
Data file S6. ddPCR probe sequences.