Abstract
Staphylococcal peptidoglycan is characterized by pentaglycine cross-bridges that are cross-linked between adjacent wall peptides by penicillin-binding proteins to confer robustness and flexibility. In Staphylococcus aureus, pentaglycine cross-bridges are synthesized by three proteins: FemX adds the first glycine, and the homodimers FemA and FemB sequentially add two Gly-Gly dipeptides. Occasionally, serine residues are also incorporated into the cross-bridges by enzymes that have heretofore not been identified. Here, we show that the FemA/FemB homologues FmhA and FmhC pair with FemA and FemB to incorporate Gly-Ser dipeptides into cross-bridges and to confer resistance to lysostaphin, a secreted bacteriocin that cleaves the pentaglycine cross-bridge. FmhA incorporates serine residues at positions 3 and 5 of the cross-bridge. In contrast, FmhC incorporates a single serine at position 5. Serine incorporation also lowers resistance toward oxacillin, an antibiotic that targets penicillin-binding proteins, in both methicillin-sensitive and methicillin-resistant strains of S. aureus. FmhC is encoded by a gene immediately adjacent to lytN, which specifies a hydrolase that cleaves the bond between the fifth glycine of cross-bridges and the alanine of the adjacent stem peptide. In this manner, LytN facilitates the separation of daughter cells. Cell wall damage induced upon lytN overexpression can be alleviated by overexpression of fmhC. Together, these observations suggest that FmhA and FmhC generate peptidoglycan cross-bridges with unique serine patterns that provide protection from endogenous murein hydrolases governing cell division and from bacteriocins produced by microbial competitors.
Keywords: peptidoglycan, biosynthesis, cell division, antibiotic resistance, bacteriocin, Staphylococcus aureus (S. aureus)
Peptidoglycan is a large macromolecule that surrounds bacteria to support cell integrity and shape (1–3). In Staphylococcus aureus, the monomeric unit of peptidoglycan is the disaccharide [4(N-acetylmuramic acid-β(1–4)-GlcNAc-β)1]n, abbreviated to NAM-NAG, with the attached wall peptide [l-Ala-d-iGln-l-Lys-(NH2-Gly5)-d-Ala-d-Ala] (4–8); the pentaglycine cross-bridge (NH2-Gly5) is linked to the ε amino group of l-Lys within the wall peptide (9). During the transpeptidation reaction, penicillin-binding proteins (PBPs) cleave the amide bond between d-Ala-d-Ala of wall peptides to form an acyl enzyme intermediate that is resolved upon nucleophilic attack from the amino group (NH2) of pentaglycine cross-bridges. In this manner, d-Ala at position 4 of wall peptides is linked to the cross-bridges of adjacent wall peptides (10, 11).
Methicillin-resistant S. aureus (MRSA) uses penicillin-binding protein PBP2a, the product of the mecA gene, to synthesize cross-linked peptidoglycan in the presence of β-lactam antibiotics (12, 13). Berger-Bächi (14) isolated MRSA fem mutants (factors essential for expression of methicillin resistance) with insertional lesions that abrogate methicillin resistance. Strains harboring mutations in femA or femB synthesize altered cell wall cross-bridges with either one or three glycyl residues, respectively, instead of five (15–19). Efforts to elucidate staphylococcal cell wall synthesis in the late 1960s used S. aureus crude enzyme preparations and demonstrated the transfer of glycine from glycyl-tRNA to the peptidoglycan biosynthetic intermediate lipid II [C55-(PO4)2-NAM(l-Ala-d-iGln-l-Lys-d-Ala-d-Ala)-NAG] (20–24). Edman degradation of these reaction products revealed that the cross-bridge is synthesized by the sequential addition of glycine from glycyl-tRNA to the ε amino group of l-Lys, demonstrating that peptidyl-transfer occurs at the NH2 terminus in a ribosome-independent manner (25). Based on these data, it was concluded that femA and femB must encode the factors that utilize tRNA-Gly for the sequential addition of Gly2-3 and Gly4-5, respectively, thereby generating mature lipid II product with the pentaglycine cross-bridge [C55-(PO4)2-NAM(l-Ala-d-iGln-(NH2-Gly5)l-Lys-d-Ala-d-Ala)-NAG] (15–19). This mature product is the preferred substrate for extracellular transglycosylation and transpeptidation reactions (4, 25, 26). The new model also implied that addition of Gly1 to the ε amino group of the stem peptide lysine required a third peptidyltransferase dubbed FemX (27). Homology searches of S. aureus genome sequence identified three femAB-like genes: fmhA, fmhB (femX), and fmhC (28, 29). Unlike fmhB, deletion of fmhA or fmhC does not affect S. aureus growth or methicillin resistance (30). Genetic depletion of fmhB causes the accumulation of wall peptides lacking glycine, indicating that fmhB encodes FemX, the catalyst adding the first glycyl residue to the side chain of lysine in the wall peptide (30).
The pentaglycine cross-bridge of S. aureus is the target of bacteriocins, products of bacteria competing for the same replication niche (31). Staphylococcus simulans biovar staphylolyticus secretes lysostaphin and Staphylococcus capitis EPK1 secretes Ale-1, which are endopeptidases that cleave pentaglycine cross-bridges to lyse S. aureus (31–33). To protect themselves from the lytic activity of lysostaphin and Ale-1, S. simulans and S. capitis produce the immunity factors Lif (lysostaphin immunity factor) and Epr (endopeptidase resistance), respectively (34–36), which exhibit sequence similarity with FemA and FemB. When expressed in S. aureus, Lif and Epr insert serine residues into peptidoglycan cross-bridges with the structure Gly-Gly-Ser-Gly-Ser (36, 37). Serine-modified cross-bridges are substrates for PBP-catalyzed transpeptidation reactions and provide increased resistance to lysostaphin (36, 38). Earlier work also described the relationship between lysostaphin resistance and serine content in peptidoglycan cross-bridges of S. aureus and Staphylococcus epidermidis (39, 40). In S. aureus, only 5% of peptidoglycan harbors serine residues; however, the abundance of Gly4-Ser cross-bridges is dramatically increased in a femAB promoter mutant (19, 27, 41).
In this study, we investigated whether FmhA and FmhC facilitate the incorporation of serine into the peptidoglycan of S. aureus. fmhA is conserved across all staphylococcal genomes analyzed to date. fmhC occurs in a more limited subset of staphylococci and is located immediately adjacent to lytN, whose product functions as a cysteine histidine-dependent amidohydrolases/peptidase murein hydrolase with d-Ala-Gly endopeptidase activity in cross-wall peptidoglycan (42). We report that FmhA and FmhC function to incorporate serine into peptidoglycan cross-bridges and interact with FemA and FemB to generate heterodimers that are the likely catalysts for serine incorporation into peptidoglycan, thereby affecting staphylococcal resistance to lysostaphin, LytN, and β-lactam antibiotics.
Results
fmhA and fmhC are dispensable for growth under laboratory conditions
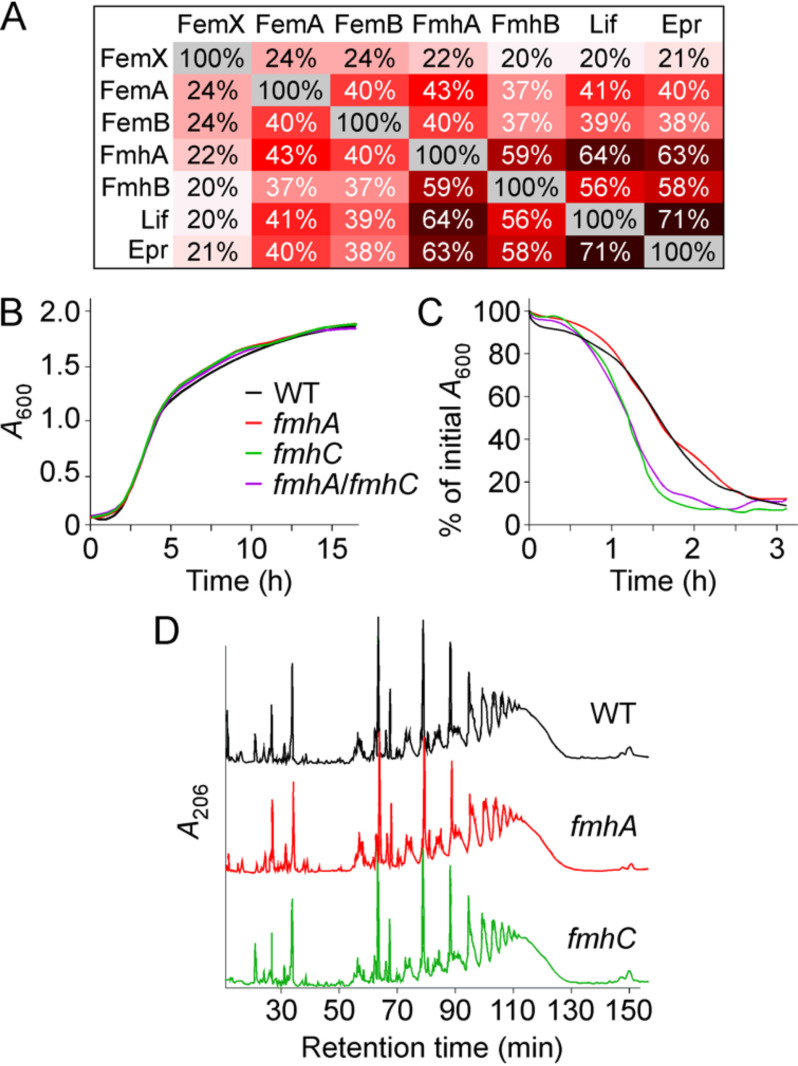
FemX, FemA, and FemB are involved in the synthesis of the pentaglycine cross-bridge in S. aureus (15–19). FmhA and FmhC are Fem-like factors and, as of yet, possess no assigned activity or function. Lif and Epr have been shown to share sequence similarity with FemA and FemB, albeit that these proteins have been proposed to incorporate serine instead of glycine in the peptidoglycan cross-bridges (36, 43). Sequence alignments between Fem and Fmh factors of S. aureus strain Newman and Lif and Epr demonstrate that FmhA and FmhC share greater sequence identity with Lif and Epr than FemA or FemB and that FemX is distinct from the other six factors (Fig. 1A). Next, mutants lacking fmhA, fmhC, or both genes were generated. Unlike fmhB encoding FemX and the femAB operon, fmhA and fmhC are dispensable for growth in S. aureus strain 8325 (28, 30, 44). In agreement with this notion, bursa aurealis transposon insertions were tolerated in both the fmhA and fmhC genes of strain Newman (45). To generate a double mutant, a deletion in the fmhC gene was obtained by allelic replacement using plasmid pKOR1 (46). Strain ΦΝΞ02665 with bursa aurealis insertion at nucleotide 554 of fmhA (45) was used to generate a lysate with bacteriophage ϕ85 for transduction using the WT strain Newman and isogenic fmhC variant to yield single fmhA and double fmhA/fmhC mutants. The growth rates of these strains monitored as absorbance of cultures over time at 600 nm (A600) revealed no discernible difference between WT and fmhA, fmhC, or fmhA/fmhC mutants (Fig. 1B). Differences in cross-bridge structure led to lysostaphin resistance, i.e. reduced rates of bacterial lysis. This can be measured by monitoring the reduction in A600 over time following addition of lysostaphin to bacterial cultures. Inactivation of fmhC but not fmhA results in a small increase in lysostaphin-mediated lysis (Fig. 1C). To further investigate differences in the cell wall structure, murein sacculi of WT and mutant strains were isolated and subjected to acid and enzymatic treatments to remove teichoic acids, proteins, and polysaccharides. Next, peptidoglycan preparations were treated with mutanolysin, an N-acetylmuramidase that cleaves the glycan strands and leaves cross-bridges intact. Such peptidoglycan fragments can be separated by reverse-phase HPLC based on cross-linking: monomers elute first, whereas highly cross-linked materials elute later (47). The HPLC elution profiles of mutanolysin digests of peptidoglycan preparations from strain Newman and isogenic variants fmhA and fmhC reveal comparable degrees of low (eluting between 20 and 40 min) and highly cross-linked materials (eluting between 50 and 130 min) (Fig. 1D). In summary, loss of neither fmhA nor fmhC results in altered growth, but strains lacking fmhC are more susceptible to lysostaphin. Loss of lysostaphin resistance occurs through subtle change(s) in peptidoglycan structure that cannot be revealed through simple HPLC analysis.
Figure 1.
Phenotypic characterization of strain variants lacking fmhA and fmhC. A, shared amino acid identities between Fem and Fmh factors of S. aureus, S. simulans Lif, and S. capitis Epr. B, the growth rate of S. aureus Newman (WT, WT) and isogenic variants fmhA, fmhC or fmhA/fmhC was monitored by recording absorbance at A600 over 16 h at 37 °C. C, overnight cultures of the S. aureus strains were treated with lysostaphin, and lysis was measured as a decline in A600 over time. Each curve represents the average of three technical repeats (error bars were omitted) and is representative of three biological repeats. D, peptidoglycan was prepared from S. aureus Newman (WT), and isogenic variants fmhA and fmhC cultures were grown to A600 0.8–1.0 and treated with mutanolysin. Solubilized products were resolved by HPLC over a C18 column by measuring absorbance at 206 nm over time.
FmhA and FmhC incorporate serine into the cross-bridge of S. aureus peptidoglycan
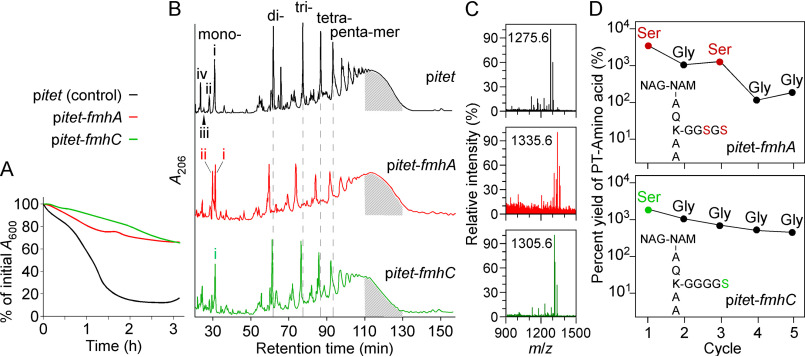
Lif and Epr activities were previously characterized by overexpressing the corresponding genes in heterologous hosts (36, 43, 48). In a similar approach, fmhA and fmhC were cloned under the anhydrotetracycline-inducible promoter of vector pMF312, herein referred to as pitet (49). Next, normalized cultures of strain Newman carrying plasmids pitet-fmhA or pitet-fmhC, or vector control pitet were incubated with lysostaphin. Overexpression of either fmhA or fmhC reduced the rate of lysostaphin-mediated cell lysis relative to vector control (Fig. 2A). In the case of fmhC, resistance was observed even in the absence of anhydrotetracycline inducer because of some leaky expression from pitet (42). Murein sacculi were extracted from cultures grown in the presence of anhydrotetracycline, and peptidoglycan was purified and subjected to mutanolysin treatment. Solubilized products were separated over a C18 column using HPLC (Fig. 2B). Reduced cross-linking material was observed for fmhA and fmhC overexpressing samples. Specifically, the abundance of di-, tri-, tetra-, and pentamer species was reduced upon fmhA overexpression, and elution of these species occurred ∼2 min earlier as compared with the control (Fig. 2B; dashed lines). Higher order cross-linking was reduced upon fmhC overexpression, as suggested by the reduced absorbance of material eluting between 110 and 130 min (Fig. 2B). Measurement of the area under the curve between 110 and 130 min (indicated by the area shaded in gray) suggests that material with higher cross-linking is reduced by ∼10 and 30% upon fmhA and fmhC overexpression, respectively, relative to WT. A peak unique to pitet-fmhA was observed with a retention time of 29 min corresponding to peptidoglycan monomers (Fig. 2B; red trace, peak ii). Fractions eluting between 26–32 min, containing peaks i–iv, were subjected to MALDI-TOF MS (Fig. 2C and Table 1). For the pitet-fmhC sample, peak i was found to contain a major product with m/z value consistent with a cross-bridge of four glycines and one serine; m/z values for products containing more than one serine were not observed (Table 1). This product was also found in peak i of the pitet-fmhA sample (cross-bridge of four glycines and one serine), along with another product with an m/z value consistent with a cross-bridge of two serines and three glycines (Fig. 2B and Table 1). Peak ii of the pitet-fmhA sample contained heterogenous fragments of truncated cross-bridges with up to two serines. These truncated cross-bridges may have been incorporated into the mature peptidoglycan or may be the product of cell wall hydrolytic activity during cell growth. The increased heterogeneity of pitet-fmhA peptidoglycan fragments is re-flected by the broader peaks in the chromatogram relative to pitet and pitet-fmhC (Fig. 2B). All m/z values for the vector control sample corresponded to glycine-containing cross-bridges (Table 1). To ascertain the amino acid sequence of cross-bridges in the monomeric fragments (peak i, Fig. 2, B and C), samples were subjected to Edman degradation (Fig. 2D). Consistent with observed m/z values, two phenylthiohydantoin-Ser were released in cycles one and three of Edman degradation of the pitet-fmhA sample. One phenylthiohydantoin-Ser was released in cycle one of Edman degradation of the pitet-fmhC sample. These results suggest that following fmhA overexpression, the cross-bridges contain serine residues at positions 3 and 5 (position 1 being the most proximal to the stem peptide lysine). Overexpression of fmhC results in the formation of cross-bridges with one serine at position 5 (Fig. 2D).
Figure 2.
Phenotypic characterization of strains overexpressing fmhA and fmhC. A, overnight cultures of WT strain Newman carrying control vector pitet, pitet-fmhA, or pitet-fmhC were treated with lysostaphin, and lysis was recorded as a decline in A600 over time as described for Fig. 1C. B, peptidoglycan prepared from the same strains grown to A600 0.8–1.0 was treated with mutanolysin, and products were resolved by HPLC as described for Fig. 1D. Peaks denoted with Roman numerals (i–iv) were subjected to MALDI-TOF analysis and m/z values of the corresponding species are listed in Table 1. Shaded area under curves between 110–130 min highlights higher order cross-linking of peptidoglycan fragments (47). C, MALDI-TOF spectra of species eluting in peak i shown in panel C. D, Edman degradation profile of the major peptidoglycan fragment in peak i of the samples overexpressing fmhA and fmhC. The yield of released phenylthiohydantoin (PT)-modified amino acids is shown for five cleavage cycles.
Table 1.
MALDI-TOF mass spectrometry of mutanolysin-digested peptidoglycan fragments
| Compound1 |
m/z2 |
Δ (Observed and calculated) | Predicted structure | |
|---|---|---|---|---|
| Observed | Calculated | |||
| pitet | ||||
| iv | 990.4475 | 990.0698 | 0.3777 | (NAG-NAM)-AQKAA |
| iii | 1033.4456 | 1033.1198 | 0.3258 | (NAG-NAM)-AQK(G2)A |
| ii | 1204.5112 | 1204.3298 | 0.1814 | (NAG-NAM)-AQK(G5)A |
| ii | 1261.5270 | 1261.3998 | 0.1272 | (NAG-NAM)-AQK(G5)A(G1)3 |
| ii | 1318.5313 | 1318.4698 | 0.0615 | (NAG-NAM)-AQK(G5)A(G2)3 |
| ii | 1375.5273 | 1375.5398 | 0.0125 | (NAG-NAM)-AQK(G5)A(G3)3 |
| ii | 1432.5298 | 1432.6098 | 0.0800 | (NAG-NAM)-AQK(G5)A(G4)3 |
| i | 1104.5363 | 1104.2098 | 0.3265 | (NAG-NAM)-AQK(G2)AA |
| i | 1161.5384 | 1161.2798 | 0.2586 | (NAG-NAM)-AQK(G3)AA |
| i | 1218.5429 | 1218.3498 | 0.1931 | (NAG-NAM)-AQK(G4)AA |
| i | 1275.5616 | 1275.4198 | 0.1418 | (NAG-NAM)-AQK(G5)AA |
| pitet-fmhA | ||||
| ii | 1005.5708 | 1006.0828 | 0.5120 | (NAG-NAM)-AQKA(S1)3 |
| ii | 1047.5819 | 1047.1398 | 0.4421 | (NAG-NAM)-AQK(G1)AA |
| ii | 1062.5872 | 1063.1528 | -0.5656 | (NAG-NAM)-AQK(G1)A(S1)3 |
| ii | 1149.6054 | 1150.2558 | -0.6504 | (NAG-NAM)-AQKA(G1S2)3 |
| ii | 1191.6037 | 1191.3128 | 0.2909 | (NAG-NAM)-AQK(G2S1)AA |
| i | 1263.6192 | 1264.3896 | -0.7704 | (NAG-NAM)-AQK(G3S2)A |
| i | 1293.6184 | 1294.4226 | -0.8042 | (NAG-NAM)-AQK(G2S3)4 |
| i | 1305.6181 | 1305.4528 | 0.1653 | (NAG-NAM)-AQK(G4S1)AA |
| i | 1335.6164 | 1335.4831 | 0.1333 | (NAG-NAM)-AQK(G3S2)AA |
| pitet-fmhC | ||||
| i | 1104.9077 | 1104.2098 | 0.6979 | (NAG-NAM)-AQK(G2)AA |
| i | 1161.8864 | 1161.2798 | 0.6066 | (NAG-NAM)-AQK(G3)AA |
| i | 1218.9403 | 1218.3498 | 0.5905 | (NAG-NAM)-AQK(G4)AA |
| i | 1305.9793 | 1305.4528 | 0.5265 | (NAG-NAM)-AQK(G4S1)AA |
1See Fig. 2 for compounds, i.e. absorption peaks of reverse-phase HPLC of mutanolysin-treated S. aureus peptidoglycan.
2Observed and calculated m/z values are for sodiated ions.
3These structures include cross-bridges from two cross-linked disaccharide units; amino acids from a cross-linked unit are listed after the fourth residue of the stem peptide from the first unit.
4Unexpected structure with three serines that could result from aberrant fmhA overexpression.
fmhA and fmhC overexpression impacts oxacillin resistance
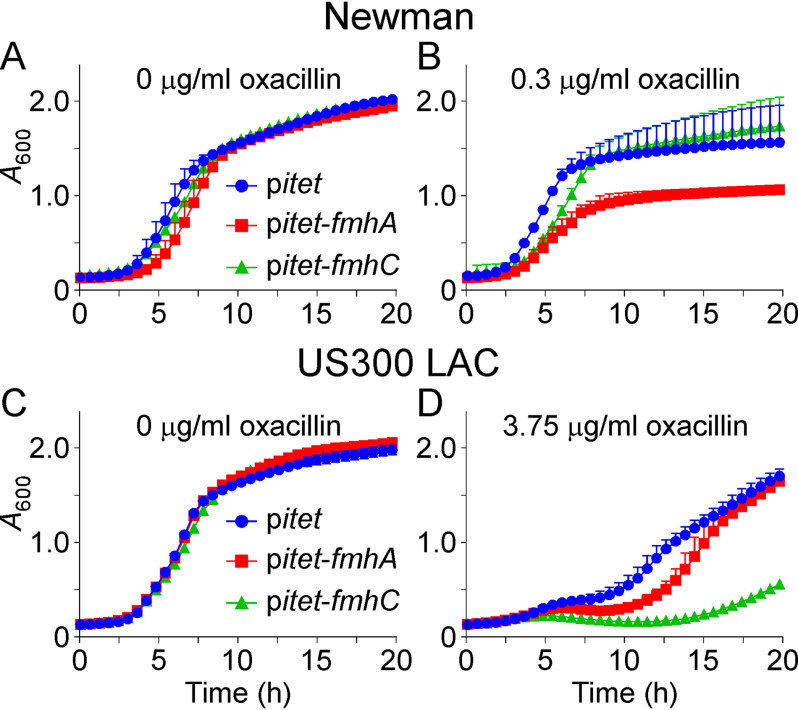
Oxacillin is a β-lactam that covalently inactivates sensitive PBPs. As PBPs catalyze the transpeptidation reaction between the N terminus of cross-bridges and stem peptide alanine, we wondered what impact, if any, cell wall serine content would have on oxacillin susceptibility. S. aureus Newman is an oxacillin-sensitive strain that does not possess an oxacillin-resistant PBP. Newman strains carrying pitet, pitet-fmhA, or pitet-fmhC were grown without addition of oxacillin to the culture medium (Fig. 3A). Overexpression of fmhA had little impact on growth, causing only a small growth delay (Fig. 3A). When grown in the presence of a sub-inhibitory concentration of oxacillin, overexpression of fmhA further exacerbated, the growth delay and the culture failed to reach a density comparable with that of the WT pitet control (Fig. 3B). fmhC overexpression did not affect bacterial growth significantly (Fig. 3B). Thus, cross-linking activity mediated by PBPs is more susceptible to oxacillin when cross-bridges are enriched for serine at positions 3 and 5. We wondered whether cross-bridge composition may have a similar impact on MRSA. MRSA strains encode an additional PBP2, PBP2a, which is insensitive to β-lactams and becomes active following β-lactam exposure to allow the cell to continue the transpeptidation reaction necessary for peptidoglycan biosynthesis. The MRSA strain USA300 was transformed with pitet, pitet-fmhA, or pitet-fmhC, and the corresponding bacterial cultures were grown with and without the addition of oxacillin. All cultures grew to the same absorbance over time in the absence of oxacillin (Fig. 3C). In contrast to the methicillin-sensitive strain Newman, addition of oxacillin to the growth medium altered the replication of all three strains (Fig. 3D). Overexpression of fmhA delayed bacterial replication slightly, whereas overexpression of fmhC resulted in a clear growth defect (Fig. 3D). These results suggest that FmhA and FmhC products are poor substrates for PBP2 and PBP2a; thus, the presence of serine in the cross-bridges reduces the resistance toward oxacillin in both methicillin-sensitive and methicillin-resistant strains of S. aureus.
Figure 3.
Impact of fmhA and fmhB overexpression on oxacillin susceptibility. Cultures of S. aureus Newman (A and B) and USA300 LAC* (C and D) carrying control vector pitet, pitet-fmhA, or pitet-fmhC, were grown without (A and C) or with (B and D) oxacillin. Growth was measured as increased A600 over time. The data are shown as an average of technical triplicates with error bars indicating the standard deviation and are representative of three biological replicates.
FmhC facilitates daughter cell separation by functioning as a LytN-immunity factor
fmhC is encoded directly downstream of lytN. LytN cleaves peptidoglycan between Gly5 of the cross-bridge and alanine of the adjacent stem peptide (50). LytN harbors a signal sequence with the conserved amino acid motif YSIRK-G/S for secretion at the cross-wall, the site of peptidoglycan biosynthesis during cell division (Fig. 4A) (51). Because of this localization, LytN has been proposed to cleave the newly synthesized cross-wall, thereby disrupting cross-linking and facilitating daughter cell separation (Fig. 4A) (42). It has been surmised that this activity is subject to regulation because both plasmid-driven overexpression and genetic inactivation of lytN precipitate damage to the cross-wall (42). We wondered whether FmhC might regulate LytN activity by generating LytN-resistant substrates. To test this hypothesis, we first used a genetic approach. The integrative plasmid pCL55 (52) was used to recombine lytN placed under the anhydrotetracycline-inducible promotor at the geh locus on the chromosome of strain Newman (pCL55-lytN). As reported before, this additional copy of lytN conferred a growth defect relative to the control (Fig. 4B; compare black and blue growth curves) (42). To examine whether overexpression of fmhA or fmhC would reverse this growth defect, the genes were cloned on the compatible replicative plasmid pJK4 (53). Plasmid expression of fmhC but not fmhA compensated for lytN overexpression (Fig. 4B). In fact, when compared with the control strain (pCL55-lytN pJK4), fmhC overexpression (pCL55-lytN pJK4-fmhC) appeared to accelerate entry into the exponential phase of growth (Fig. 4B). These results suggest that FmhC-mediated cross-linking of peptidoglycan slows down degradation by LytN. To test this possibility directly, HPLC dimeric fragments obtained from pitet and pitet-fmhC samples as described in Fig. 2B were incubated with either buffer or recombinant LytN (rLytN). When incubated with buffer, dimers eluted as a single peak (peak i) with the expected retention time of ∼65 min on HPLC. MALDI-TOF analysis of these peaks revealed m/z values for the expected dimers containing pentaglycine cross-bridges for the WT sample carrying pitet (Table 2 and Fig. 4Ca). For the pitet-fmhC sample, two dimeric fragments were observed; the predominant fragment was consistent with the presence of two Gly4-Ser1 cross-bridges, whereas the additional fragment was consistent with the presence of one Gly4-Ser1 cross-bridge and one pentaglycine cross-bridge (Table 2 and Fig. 4Cb). The presence of some pentaglycine cross-bridges in the pitet-fmhC peptidoglycan is not unexpected, because FemA and FemB are produced in this background. When dimers were incubated with rLytN and separated once more by HPLC, new peaks (peaks ii–iv) were eluted at ∼30–35 min (Fig. 4, Cc and Cd). MALDI-TOF analysis of peaks ii revealed an m/z value of 1,275, corresponding to monomeric peptidoglycan with a pentaglycine cross-bridge (Table 2 and Fig. 4, Cc and Cd). This is in agreement with the reported endopeptidase activity of LytN (Fig. 4A) (42). The other major product (peaks iii) corresponds to a monomeric fragment with a pentaglycine cross-bridge and a tetrapeptide stem lacking the terminal d-alanine (Table 2 and Fig. 4, Cc and Cd). Minor peak iv was only observed for the WT sample (pitet) and corresponded to a fragment with a truncated cross-bridge and a fragment lacking NAG, presumably from the action of endogenous glucosaminidases (Table 2 and Fig. 4Cc). A second fragment with a truncated cross-bridge was also found in fraction iii of WT. Additionally, a small peak interpreted as containing a Gly4-Ser1 cross-bridge was found in fraction ii of WT. Presumably, the abundance of these fragments was too low to be detected prior to digestion with rLytN and re-purification by HPLC. Importantly, dimeric substrates from the pitet sample were much better substrates for rLytN than dimers prepared from pitet-fmhC (compare intensities of peak i in Fig. 4, Cc and Cd). Consequently, products ii and iii were much more abundant when dimers from the pitet sample were incubated with rLytN (Fig. 4, Cc and Cd). Thus, the presence of Ser5 in the cross-bridge provides resistance to cleavage by LytN in vivo and in vitro, indicating that FmhC functions to protect the peptidoglycan from digestion by LytN during daughter cell separation (Fig. 4A).
Figure 4.
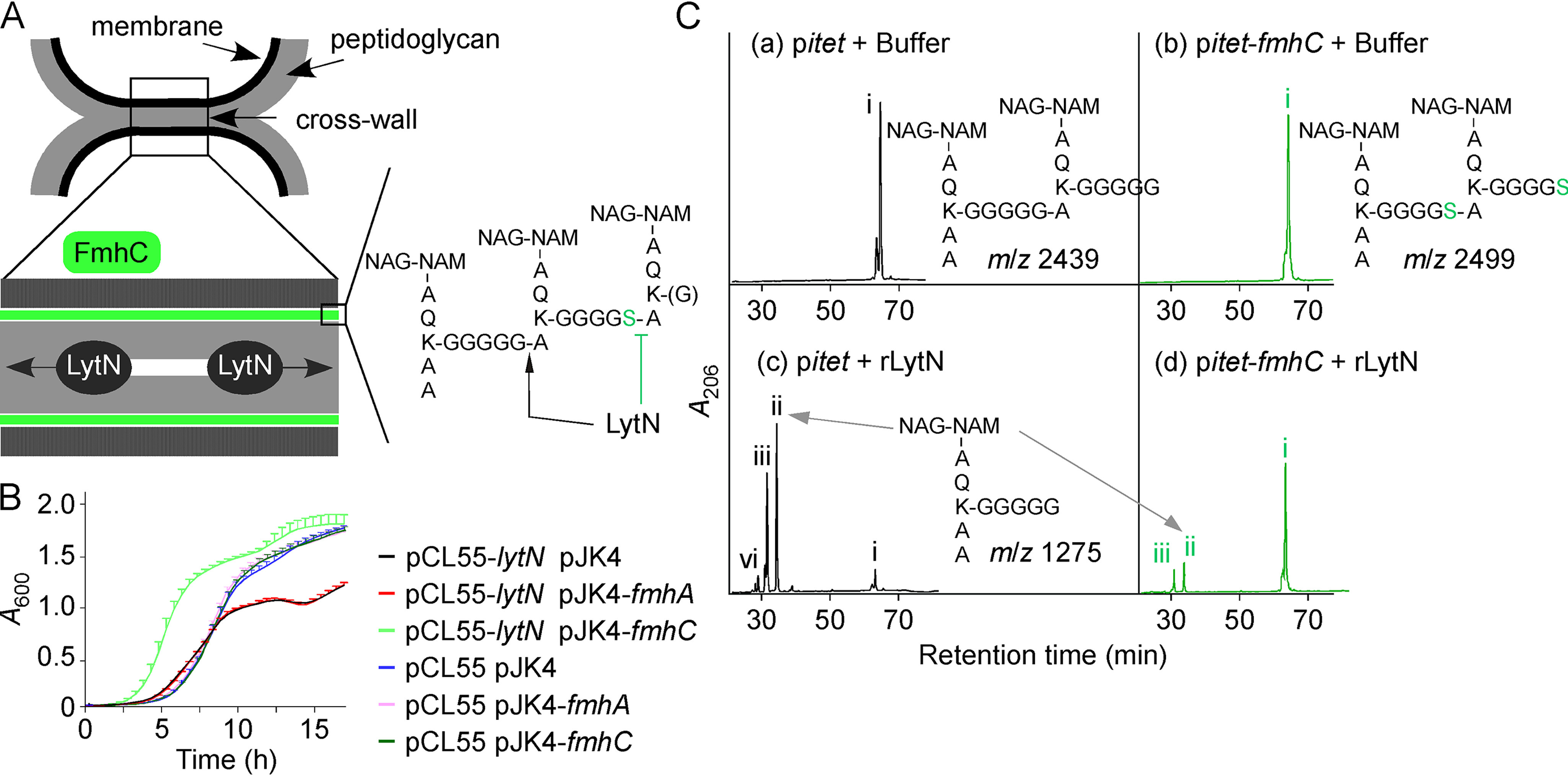
FmhC acts as an immunity factor for LytN. A, schematic of dividing daughter cells illustrating the location of the cell wall envelope, membrane (black), and peptidoglycan (gray). The cross-wall structure has been expanded to show LytN cleaving peptidoglycan. The model postulates that FmhC synthesizes peptidoglycan (shaded in green) that is resistant to LytN hydrolysis. The proposed structure of this cross-linked peptidoglycan is shown with sites cleaved by LytN (black arrow) or refractory to LytN cleavage (green inhibition sign). Green and blue hexagons: GlcNAc and N-acetylmuramic acid, respectively. B, the growth rate of S. aureus Newman pCL55 and Newman pCL55-lytN carrying pJK4, pJK4-fmhA, or pJK4-fmhC was interrogated by monitoring changes in A600 over 16 h at 37 °C. Cultures were grown in the presence of anhydrotetracycline and IPTG inducers. C, purified dimeric peptidoglycan fragments obtained from Newman carrying control vector pitet or pitet-fmhC, as shown in Fig. 2B, were treated with buffer (insets a and b) or rLytN (insets c and d). Reaction products were separated by HPLC. Peaks denoted with Roman numerals (i–iv) were subjected to MALDI-TOF analysis and the corresponding m/z values are listed in Table 2. Predicted structures of peptidoglycan fragments i (panels a and b) and ii (panels c–d) are shown on the figure.
Table 2.
MALDI-TOF mass spectrometry of rLytN-digested dimeric peptidoglycan fragments
| Compound1 |
m/z2 |
Δ (observed and calculated) | Predicted structure | |
|---|---|---|---|---|
| Observed | Calculated | |||
| pitet + Buffer | ||||
| i | 2416.57† | 2415.67 | 0.90 | (NAG-NAM)-AQK(G5)A-(NAG-NAM)-AQK(G5)AA |
| pitet-fmhC + Buffer | ||||
| i | 2446.53† | 2445.70 | 0.83 | (NAG-NAM)-AQK(G5)A-(NAG-NAM)-AQK(G4S1)AA |
| i | 2476.55† | 2475.74 | 0.81 | (NAG-NAM)-AQK(G4S1)A-(NAG-NAM)-AQK(G4S1)AA |
| pitet + rLytN | ||||
| ii | 1275.50 | 1275.42 | 0.08 | (NAG-NAM)-AQK(G5)AA |
| ii | 1233.49 | 1234.42 | 0.07 | (NAG-NAM)-AQK(G4S1)A |
| iii | 1204.49 | 1204.33 | 0.16 | (NAG-NAM)-AQK(G5)A |
| iii | 1162.48 | 1161.28 | 1.20 | (NAG-NAM)-AQK(G3)AA |
| iv | 1261.51 | 1261.40 | 0.11 | (NAG-NAM)-AQK(G5)A(G1) |
| iv | 1001.45 | 1001.12 | 0.33 | (NAM)-AQK(G5)A |
| pitet-fmhC + rLytN | ||||
| ii | 1275.50 | 1275.42 | 0.08 | (NAG-NAM)-AQK(G5)AA |
| iii | 1234.50 | 1234.42 | 0.08 | (NAG-NAM)-AQK(G4S1)A |
| iii | 1204.49 | 1204.33 | 0.16 | (NAG-NAM)-AQK(G5)A |
1See Fig. 3C for compounds i.e. absorption peaks of reverse-phase HPLC of mutanolysin-treated S. aureus peptidoglycan.
2Observed and calculated m/z values are for sodiated ions recorded in reflectron-positive mode, with the exception of † which were analyzed reflectron negative mode and therefore are [−H+].
FmhA mediates the insertion of serine into the peptidoglycan cross-bridge of strain BB308
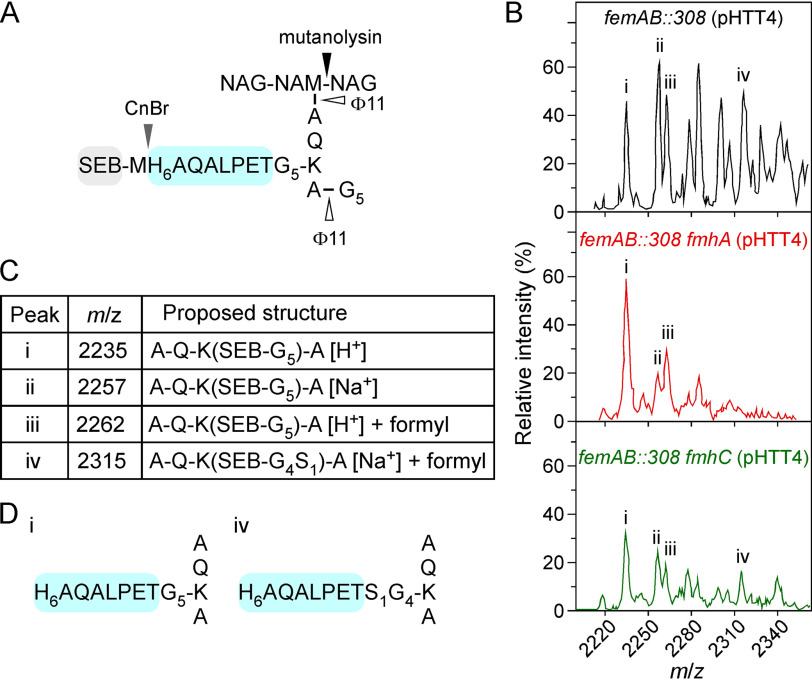
We previously reported that S. aureus strain BB308, which carries a transposon insertion in the promotor region of the femAB operon, elaborates tetraglycyl-monoseryl cross-bridges (41, 54). We reasoned that this could be exploited to identify the genetic factor(s) responsible for the enrichment of serine in the cross-bridges because WT strains such as Newman do not accumulate sufficient amounts of serine in their envelope to determine the contribution of the fmhA and fmhC genes. Phage-mediated transduction was used to transfer the erythromycin transposon of strain BB308 into strain Newman lacking either fmhA or fmhC to generate mutants femAB::308 fmhA and femAB::308 fmhC, respectively. Next, mutant strains were transformed with plasmid pHTT4, which encodes the protein hybrid SEB-MH6-CWS composed of staphylococcal enterotoxin B (SEB) carrying an N-terminal signal peptide and C-terminally fused cell-wall sorting signal (CWS) of staphylococcal protein A; a methionine followed by six histidines (MH6) is inserted at the fusion site between SEB and CWS (41). When produced in S. aureus, this protein hybrid is secreted and linked to peptidoglycan (41). After peptidoglycan solubilization with mutanolysin and ϕ11 hydrolase, SEB-MH6-CWS was purified over nickel-charged (Ni-NTA) affinity resin and cleaved at methionyl with cyanogen bromide (Fig. 5A). The resulting C-terminal anchor peptide was purified once more over Ni-NTA resin, desalted, and analyzed by MALDI-TOF spectrometry (Fig. 5B). The mass spectra for strains femAB::308 and femAB::308 fmhC showed similar profiles and ions with m/z 2,235, 2,257, 2,262, and 2,315 (Fig. 5, A and B). The presence of ions with m/z 2,235 and 2,315 is consistent with the calculated mass of anchor peptides harboring pentaglycine and tetraglycyl-monoseryl cross-bridges, respectively (Fig. 5, C and D). This result is in agreement with data reported for strain BB308 (41), suggesting that the phenotype observed in strain Newman femAB::308 is fully recapitulated and the increased serine content is caused by transposon insertion in the femAB promoter region. Additional ions were also observed; ions with m/z 2278 and sodiated adduct m/z 2,300 were consistent with the calculated mass of anchor peptide harboring a Gly3-Ser1 cross-bridge (Table 3). Serine-containing cross-bridge fragments were not observed in the femAB::308 fmhA sample (Fig. 5 and Table 3). Therefore, these data implicate FmhA as the catalyst of serine insertion in strains that carry a transposon insertion in the femAB promoter and corroborate our findings using strains that overexpress fmhA.
Figure 5.
fmhA mediates serine insertion in a femAB depleted background. A, the peptidoglycan of femAB pHTT4, femAB fmhA pHTT4, or femAB fmhC pHTT4 was isolated and treated with mutanolysin, ϕ11 hydrolase, and cyanogen bromide (CnBr. B, MALDI-TOF spectra of purified H6-CWS anchor peptides. C, the major m/z values are listed; see Table 3 for detailed m/z. D, diagrams of the assigned structures for peak i and peak iv. Of note, S1G4 in structure iv depicts the ratio between glycyl and seryl residues; the linkage unit (N to C terminus) could be GGSGG or SGGGG.
Table 3.
MALDI-TOF mass spectrometry of SEB-purified peptidoglycan fragments
|
m/z observed1 |
m/z calculated | Δ (observed and calculated) |
Predicted structure2 | ||||
|---|---|---|---|---|---|---|---|
| femAB::308 | femAB::308 fmhA | femAB::308 fmhC | femAB::308 | femAB::308 fmhA | femAB::308 fmhC | ||
| 2234.88 | 2234.59 | 2234.87 | 2236.35 | 1.47 | 1.76 | 1.48 | [NH2-AQK-(NH2-H6AQALPET-G5)-A-COOH] H+ |
| 2257.20 | 2256.97 | 2257.20 | 2258.34 | 1.14 | 1.37 | 1.14 | 2236.35 |
| 2262.78 | 2263.12 | 2262.65 | 2264.34 | 1.56 | 1.22 | 1.69 | 2236.35 |
| 2278.34 | 2278.32 | 2280.44 | 2.10 | 2.12 | [NH2- AQK-(NH2-H6AQALPET-G3S1)-AA-COOH] H+ | ||
| 2284.82 | 2285.62 | 2285.10 | 2286.33 | 1.51 | 0.71 | 1.23 | 2258.34 formylated |
| 2300.70 | 2300.0032 | 2302.42 | 1.72 | 2.42 | 2280.44 sodiated | ||
| 2306.39 | 2307.28021 | 2305.776 | 2307.44 | 1.05 | 0.16 | 1.66 | [NH2-AQK-(NH2-H6AQALPET-G5)-A A-COOH] H+ |
| 2316.26 | 2315.9331 | 2316.42 | 0.16 | 0.49 | [NH2-AQK-(NH2-H6AQALPET-G4S1)-A-COOH] Na+ formylated | ||
| 2341.44 | 2341.06 | 2343.42 | 1.99 | 2.37 | [NH2-AQK-(NH2-H6AQALPET-G5)-Ala-[G1]-COOH] Na+ formylated | ||
1See Fig. 5 for the corresponding mass spectra.
2Adventitious formylation of peptides in the presence of TFA has been previously noted.
FmhA and FmhC purify with FemA and FemB
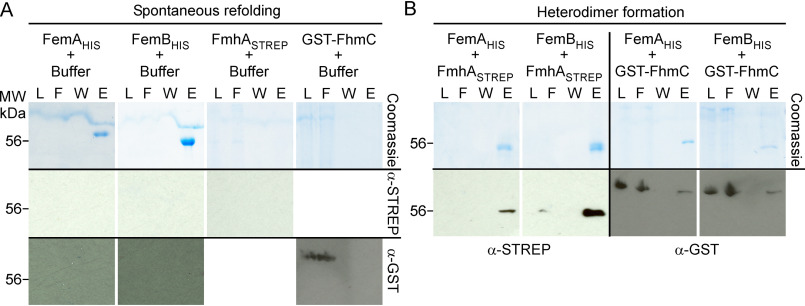
Using a bacterial two-hybrid system, Rohrer and Berger-Bächi (55) observed that S. aureus FemA, FemB, and FemX form homodimers and one heterodimer composed of FemA-FemB. These interactions were confirmed using protein pulldown assays and size exclusion chromatography (55). Genetic experiments supported a model whereby Lif and Epr heterodimerize with FemA and FemB, because production of neither Lif nor Epr alone is sufficient to restore the length of the shortened cross-bridges of femAB::308 mutants (48). We wondered whether FmhA and FmhC may interact with FemA and FemB. Recombinant FemA and FemB with a C-terminal six-histidine tag (FemAHis, FemBHis) were purified from E. coli extracts using Ni-NTA resin. As previously reported, both proteins were highly soluble (55). A STREP tag was appended at the C-terminal of FmhA, FmhASTREP, whereas FmhC was produced as a translational hybrid fused to GSH S-transferase, GST-FmhC. Both proteins were found to sediment following centrifugation at 100,000 × g, suggesting the formation of aggregates. To examine whether these proteins could refold spontaneously with or without Fem factors, materials in these sediments were suspended in 6 m guanidine hydrochloride (GdnHCl) and cleared once more by centrifugation at 100,000 × g. Similarly, purified FemAHis and FemBHis were also incubated with 6 m GdnHCl to disrupt existing homodimers and assess the ability of these factors to refold spontaneously. Refolding was initiated by 10-fold dilution in buffer lacking GdnHCl for each individual protein (Fig. 6A) or for pairwise combinations of Fem/Fmh proteins (Fig. 6B). Insoluble materials were removed by centrifugation at 100,000 × g, and soluble fractions were loaded over Ni-NTA (Fig. 6). Aliquots of proteins in loaded, flow-through, wash, and imidazole eluate were separated on SDS-PAGE for visualization using Coomassie Brilliant Blue or Western blotting (Fig. 6). FemAHis and FemBHis proteins were clearly visible in eluate fractions, suggesting that both proteins could refold spontaneously and interacted with Ni-NTA (because of the large dilution during renaturation, proteins could not be detected in loaded fractions). Anti-STREP antibodies (α-STREP) failed to detect FmhASTREP in the loaded fraction, indicating that FmhASTREP is prone to aggregation following dilution in buffer and cannot refold spontaneously (Fig. 6A). Anti-GST antibodies (α-GST) identified GST-FmhC in the loaded and flow-through fractions, indicating that the protein can refold spontaneously and does not bind to Ni-NTA resin in a nonspecific manner (Fig. 6A). When FmhASTREP was refolded in the presence of FemAHis or FemBHis, the protein formed a soluble complex that could be co-purified over Ni-NTA (Fig. 6B). A greater FmhASTREP yield was observed upon incubation with FemBHis, as documented by the increased intensity of the STREP immune signal (Fig. 6B). Similarly, a small amount of GST-FmhC was found to remain bound to FemAHis or FemBHis (Fig. 6B). In summary, FmhA and FmhC are insoluble when produced in E. coli. Refolding with FemA or FemB increases their solubility, dramatically so in the case of FmhA, and demonstrates heterodimerization between Fmh and Fem factors.
Figure 6.
Spontaneous refolding and heterodimer formation of Fem and Fmh proteins. A, purification over Ni-NTA following refolding of denatured proteins. Recombinant FemAHis, FemBHIS, FmhASTREP, and GST-FmhC were extracted from E. coli in the presence of 6 m guanidine hydrochloride. Protein preparations were diluted 10-fold in column buffer, spun at 100,000 × g to remove insoluble precipitates, and purified by gravity flow over Ni-NTA. B, heterodimer formation. Recombinant FmhASTREP and GST-FmhC prepared in 6 m guanidine hydrochloride were mixed in a pairwise fashion with either FemAHIS or FemBHIS prior to renaturation and purification over Ni-NTA as described above. Aliquots of the load (L), flow-through (F), wash (W) and eluates (E) collected during Ni-NTA purification were separated by SDS-PAGE and gels were stained with Coomassie Brilliant Blue or transferred to membranes for immunoblot analyses with α-STREP or α-GST antibodies. Numbers to the left of gels and blot indicate the position of the 56 kDa molecular weight (MW) marker.
Discussion
The cross-bridge of S. aureus peptidoglycan is composed of glycine residues with the infrequent occurrence of serine and alanine residues (9, 39). FemX, FemA, and FemB are responsible for the sequential addition of glycine residues and are essential for growth (30, 44). The genetic determinants of nonglycine amino acid insertion into cross-bridges of S. aureus have heretofore not been ascertained. Fem-like factors Lif and Epr of S. simulans and S. capitis incorporate serine residues in cross-bridges for protection against endogenous secreted glycylglycine endopeptidases. Lif and Epr share sequence homology with S. aureus FmhA and FmhC. We report here that the cross-bridge of S. aureus peptidoglycan is enriched in serine residues upon overproduction of either FmhA or FmhC. Specifically, FmhA mediates serine incorporation in positions 3 and 5 of cross-bridges whereas FmhC mediates serine incorporation in position 5 exclusively. The observed m/z values of peptidoglycan fragments extracted from FmhA-overproducing staphylococci indicate that cross-bridges may contain one or two serine residues, the former presumably with sequences Gly1-2-Ser3-Gly4-5 or Gly1-4-Ser5. Truncated cross-bridges with m/z values consistent with sequence Gly1-2-Ser3 were also observed and may account for the increased oxacillin susceptibility of the methicillin-sensitive strain Newman upon fmhA overexpression. In USA300, a methicillin-resistant strain that encodes the oxacillin-resistant PBP2a, overexpression of fmhC reduced the resistance toward oxacillin. This may reflect a difference in substrate recognition between PBP2 and PBP2a, with PBP2a being less likely to use Gly1-4-Ser5 as a substrate. Alternatively, although overexpression was achieved using the same plasmids, we cannot rule out that FmhA and FmhC were not produced to similar extent in the two strains.
S. epidermidis and other coagulase-negative species incorporate serine in positions 3 and 5 of peptidoglycan much more frequently than S. aureus (9, 56). Although the genetic requirement for cross-bridge synthesis has not been examined in S. epidermidis, we note the presence of conserved genes coding for FemA, FemB, FemX, and FmhA in S. epidermidis ATCC 12228 but not FmhC. Thus, we can postulate that FmhA is solely responsible for the incorporation of serine residues in the cross-bridge. The reason for the lower serine content in S. aureus is thus unclear. The pool of seryl-tRNA may be limited in S. aureus, or fmhA may be transcribed at a very low level. It has been noted that fmhA transcripts are down-regulated in response to stress, biofilm formation (57), and following inactivation of the cell wall stress regulator NsaRS (58), suggesting that a basal level of transcription does occur. Nonetheless, our data indicate that FmhA is responsible for the increased occurrence of serine in peptidoglycan cross-bridges when expression of femAB becomes limiting, such as in the BB308 strain. Only one serine was observed in these cross-bridges. This could be because depletion of femAB hinders the formation of FmhA heterodimers with FemA or FemB and thus impairs the catalysis of cross-bridge serine insertion. Alternatively, because peptidoglycan analysis was performed using SEB fused to a cell wall sorting signal, the data may indicate that cross-bridges with more than one serine cannot serve as nucleophiles for sortase-mediated anchoring.
In S. aureus, fmhC is encoded immediately downstream of lytN, a hydrolase that is secreted at the cross-wall of dividing cells and cleaves the bond between Gly5 of the cross-bridge and alanine of the stem peptide. Of note, S. epidermidis ATCC 12228 lacks both the fmhC and lytN genes. We show that fmhC overexpression leads to increased incorporation of Ser5 in the cross-bridges and increased resistance toward lysostaphin. Further, the corresponding peptidoglycan preparations are no longer substrates of LytN activity in vitro. In agreement with the notion that FmhC protects cells from LytN-mediated lysis, overexpression of fmhC, but not fmhA, mitigates the toxicity associated with lytN overexpression. There are several possibilities regarding the inability of FmhA to afford protection to LytN-mediated toxicity in this assay. First, overproduction of FmhA results in the accumulation of truncated cross-bridges, which might negate any LytN-resistance effect. Second, FmhA products include cross-bridges with a single serine at position 3, which presumably remain sensitive to LytN cleavage. Third, FmhA may be excluded from septal locations. Although our experiments cannot completely exclude FmhA, we favor a model whereby FmhC acts to safeguard the cell wall integrity of recently separated daughter cells from LytN.
The proposed activity of Fem/Fmh factors is based on comparative analyses of peptidoglycan composition using isogenic strains lacking or overexpressing candidate fem-like genes. The model for cross-bridge assembly is based on observing labeled glycine incorporation using crude enzyme and lipid preparations from S. aureus and precedes the identification of Fem/Fmh factors by two decades (20–24). Following the identification of FemA, FemB, and FemX, Schneider et al. (26) reexamined the model for single-step addition of glycine residues using histidine-tagged FemA, FemB, and FemX, purified staphylococcal tRNAs, and Gly-tRNA-synthetase. The use of purified components allowed these authors to establish that lipid II (not lipid I or soluble cell wall substrates) serves as a substrate for FemX and to demonstrate that nonproteinogenic Gly-tRNAs serve as glycine donors, resolving earlier conflicting reports (21, 25). S. aureus encodes distinct Gly-tRNAs for protein synthesis (proteinogenic) and peptidoglycan synthesis (nonproteinogenic), a concept that has led to the untested hypothesis that each Fem factor may use a cognate Gly-tRNA (26, 59). Schneider and colleagues (26) could also show that addition of FemX alone leads to the rapid formation of lipid II-Gly whereas addition of FemX and FemA together yields the lipid II-Gly3 product; lipid II charged with two glycines was not observed. A plausible model postulates that FemA homodimers add two glycines per cycle without releasing the lipid II intermediate. This notion is supported by bacterial two-hybrid studies suggesting that FemA is a homodimer whereas FemX behaves as a monomer. Here, we find that recombinant FmhA and FmhC form protein aggregates in E. coli but can be refolded in the presence of FemA and FemB. Aggregation likely results from high copy number, plasmid-borne overexpression of fmhA and fmhC genes, and the lack of binding partner in E. coli. In S. aureus, plasmid expression of fmhA and fmhC does not offset this fine balance, as manifested by the formation of serine-containing cross-bridges. As expected, FemA and FemB, which form homodimers, were found to be soluble in E. coli. We propose a model whereby FmhA incorporates Ser3 and Ser5 after heterodimeric association with FemA and FemB, respectively, whereas FmhC interacts solely with FemB in vivo, thereby providing a mechanism for the selective insertion of serine at position 5 of cross-bridges. Although a weak interaction was observed between GST-FmhC and FemAHis upon refolding of denatured monomers, it is reasonable to assume that in vivo the FemA homodimer is extremely stable and is not displaced by FmhC. In summary, this study defines a function for two previously uncharacterized staphylococcal factors, FmhA and FmhC. Although FmhA may act minimally in S. aureus, FmhC appears to act as the LytN-immunity factor, facilitating safe daughter cell separation during bacterial growth.
Experimental procedures
Bacterial growth and reagents
Strains of S. aureus were grown in tryptic soy broth or on tryptic soy agar plates supplemented with appropriate antibiotics. Erythromycin, kanamycin, and chloramphenicol were used at a concentration of 10 μg/ml. Oxacillin was used at 0.3 and 3.75 μg/ml for strains Newman and USA300 LAC* (a variant of the original clone of the epidemic community-acquired MRSA USA LAC strain (60) that has lost plasmid pUSA03 encoding ermC (61)), respectively. Strains of E. coli were grown in Lysogeny (62) or Terrific (pdb.rec085894, Cold Spring Harbor Protocols) broths or on Luria broth agar plates supplemented with either ampicillin (100 μg/ml) or kanamycin (50 μg/ml). When indicated, the inducers anhydrotetracycline and isopropyl β-d-thiogalactopyranoside (IPTG) were used at concentrations of 200 ng/ml and 1 mm, respectively. Lysostaphin was purchased from AMBI Products (AMBI Products, Lawrence, NY) and mutanolysin from Sigma-Aldrich. The stock solution of mutanolysin was treated with 1 mm PMSF to remove residual protease contamination. Most other chemicals used were purchased from either Sigma-Aldrich or Thermo Fisher Scientific.
To examine bacterial growth, overnight cultures were normalized to absorbance 3 at 600 nm (A600 = 3) and diluted 1:50 into 200 µl of fresh medium in 96-well plates. To assess oxacillin susceptibility, overnight cultures were diluted 1:1,000 into medium containing or lacking oxacillin. Next, plates were incubated with shaking at 37 °C in a Synergy HT plate reader (BioTek). Growth was monitored by recording A600 every 30 min for up to 16 h. Growth curves were recorded in triplicates and each experiment was reproduced three times. To assess the relative resistance toward lysostaphin, overnight cultures (1 ml) were washed twice with 50 mm Tris-HCl, pH 7.5, and cells were suspended in 650 μl of the same buffer. 90 μl of this suspension was aliquoted in two technical triplicates for each strain into a 96-well plate. One set received 10 μl of buffer (50 mm Tris, pH 8.0; control) and the second, 10 μl of lysostaphin (20 μg/ml). Decline in A600 was monitored every 5 min at 37 °C with agitation using the Synergy HT plate reader (BioTek). The change in cell density, expressed as a percentage of the input for each well, was normalized to the control samples for each time point.
Strains, vectors, and plasmids
S. aureus parent strains RN4220 (63), Newman (64), BB308 (54), and USA300 LAC* were used for this study. S. aureus Newman variant carrying a bursa aurealis insertion in fmhA was obtained from the Phoenix library (45). Allelic recombination with plasmid pKOR1 was used to delete the fmhC gene as described (46). For cloning into pKOR1, two 1-kb DNA fragments upstream and downstream of the fmhC gene were amplified from the chromosome of strain Newman with primers SW169/SW170 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGTATTCGCGAAATCAG-3′/5′-CATACTTTATAATTAAACCTTAGTTGAAAATTTCATATTTCAATGTCC-3′) and primers SW171/SW172 (5′-TGAAATATGAAATTTTCAACTAAGGTTTAATTATAAAGTATGTTGG-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCTTGGATACGTAATAAATTACC-3′), respectively. The two flanking regions were fused in a subsequent PCR, and the final PCR product was cloned into pKOR1 using the BP Clonase II kit (Invitrogen) and recombined on the chromosome of S. aureus as described (46). Allelic replacement of the fmhC gene on the chromosome was verified by DNA sequencing of PCR amplified products using genomic DNA of candidate strains and primer pair SW185/SW186 (5′-AACACGTTTAGTTGGTCCGAACTGTC-3′/5′-AATGACTAAATTATCTGCCAATGTCATTTCC-3′). Strains Newman fmhA and fmhA/fmhC were generated by transducing the fmhA::erm allele using bacteriophage ϕ85 from strain ΦΝΞ02665 (45). Successful transduction was confirmed by DNA sequencing of PCR amplified products using genomic DNA of candidate strains and primer pair SW187/SW188 (5′-TAAATCTAAATAGTGAACAACAACATGCG-3′/5′-ATAAGTGCCTTTAAATTCTGTCGAGC-3′). Deletion of fmhA in the femAB::308 strain was performed using the pKOR1 allelic recombination system as described above for fmhC. Fragments upstream and downstream of fmhA were amplified using primers SW165/SW166 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTTTTGTTAACCATTCTTTATTTTC-3′/5′-CATAATCTTGGAGCGATATTTTTATCCCATCCCTCTCTG3′ and SW167/SW168 (5'-GAGAGGGATGGGATAAAAATATCGCTCCAAGATTATGAC-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTAGGATTTATTTTAGTGGCTG-3′). For overexpression studies in S. aureus, the fmhA and fmhC genes were cloned in the replicative vectors, pMF312 and JK4 (49, 53). pMF312 is a modified pOS1 plasmid (pEC194-based) carrying the anhydrotetracycline-inducible promoter (itet) and chloramphenicol resistance (49). pJK4 carries the IPTG inducible spac promoter and kanamycin resistance (36). For cloning in pMF312, the fmhA and fmhC genes were amplified with primers SW159/SW160 (5′-GGCCTAGGAGGAGGACAGCTATGAACTTTGTAACGTTGACTTC-3′/5′-GATCCCGCGGTTAACGACGTTTATGATTTAAGTACG-3′) for fmhA and primers fmhCF/fmhCR (5′-GGCCTAGGAGGAGGACAGCTATGAAATTTTCAACTTTAAGTGAAGAAGAAT-3′/5′-GATCCCGCGGTTAAACCTTATAAATAAGTTTTGCAAATTTATATAGAA-3′) for fmhC. For cloning in pJK4, the fmhA and fmhC genes were amplified with primer pairs SW206/SW207 (5′-GATCTCTAGAATGAACTTTGTAACGTTGAC-3′/5′-GATCGGTACCTTAACGACGTTTATGATTTAAG-3′) for fmhA and primers SW208/SW209 (5′-GATCTCTAGAATGAAATTTTCAACTTTAAG-3′/5′-GATCGGTACCTTAAACCTTATAAATAAGTTTTG-3′) for fmhC. Chromosomal induction of the lytN gene was achieved by taking advantage of the integrative vector pCL55 (52) modified with the itet promoter (65). lytN was amplified using primers lytNf/lytNR (5′-GGCCTAGGAGGAGGACAGCTATGTTTTTATATTATTGTAAGGAGTGTTTCATC-3′/5′-GATCCCGCGGTTATGCTTTTTTAAATGGTCTAATAAAAATC-3′). The PCR product and vector were cut with AvrII and SacII for ligation. The resulting plasmid was integrated into the geh lipase gene of strain Newman as described (52), yielding the strain referred to as pCL55-lytN. For a control, pCL55 without insert was also integrated in the genome of strain Newman, yielding the strain referred to as pCL55. All plasmids carried the E. coli ColE1 replicon for cloning purposes. All clones were sequenced to confirm error-free constructs. Plasmids extracted from E. coli were first transformed in S. aureus RN4220, followed by electroporation into WT S. aureus Newman, USA300 LAC*, and isogenic variants.
Escherichia coli K12, DH5α, and BL21 were used for cloning and production of recombinant proteins. E. coli recombinant clones used for the purification of proteins were generated as follows. The femA and femB genes were amplified by PCR using primers SW210/SW211 (5′-GATCGCTAGCATGAAGTTTACAAATTTAAC-3′/5′-GATCCTCGAGAAAAATTCTGTCTTTAAC-3′) for femA and primers SW212/SW213 (5′-GATCGCTAGCATGAAATTTACAGAGTTAACTG-3′/5′-GATCCTCGAGTTTCTTTAATTTTTTACGTAATTTATC-3′) for femB and cloned into the pET24b vector thus generating C-terminal histidyl-6–tagged proteins, FemAHis and FemBHis, for affinity chromatography over Ni-NTA. For purification of recombinant FmhA, the corresponding gene was amplified by PCR using primers 5′-GATCCCATGGCGAACTTTGTAACG-3′/5′-GATCGGATCCTTATTTTTCGAATTGAGGATGTGACCAACGACGTTTATGATTTAAGTACGTTTG-3′ into pET15b vector to generate FmhA with a C-terminal STREP tag, FmhASTREP. fmhC was cloned into the pGEX-2TK vector to generate a translational hybrid with the N-terminal GSH S-transferase (GST-FmhC). Primers SW240/SW241 (5′-CCGGATCCATGAAATTTTCAACTTTAAGTGAAGAAGAAT-3′/5′-CCGAATTCTTAAACCTTATAAATAAGTTTTGCAAATTTATATAG-3′) were used to generate this clone. rLytN was produced from a clone lacking the signal peptide as previously described (42). All plasmids were analyzed by DNA sequencing and transformed into E. coli BL21(DE3) for production of recombinant proteins.
Protein purification
Recombinant proteins were purified from E. coli BL21(DE3) with the exception of the ϕ11 hydrolase, which was purified from E. coli BL21(DE3) harboring pHTT2 and pLysS as previously described (66). Purified ϕ11 hydrolase was dialyzed against 50% glycerol, 50 mm sodium phosphate, 10 mm MgCl2, and 2 mm DTT, pH 6.8. For purification of Fem and Fmh factors, following IPTG induction of cultures, cells were sedimented (10,000 × g, 10 min), washed, and suspended in 15 ml of buffer A (100 mm Tris-HCl, pH 8.0, and 150 mm NaCl) and lysed by three passages in a French press at 14,000 lb/in2. Unbroken cells were removed by centrifugation (5,000 × g for 15 min) and crude lysates were subjected to ultracentrifugation (100,000 × g for 1 h at 4 °C). Soluble recombinant proteins FemAHis and FemBHis were subjected via gravity flow to chromatography on Ni-NTA resin (Qiagen) with a packed volume of 1 ml pre-equilibrated with buffer A containing 20 mm imidazole. Columns were washed with 20-bed volumes of buffer A and eluted with a step gradient of imidazole (20–500 mm) in buffer containing or lacking 6 m guanidine hydrochloride. FmhASTREP and GST-FmhC were found in the pellet fraction following ultracentrifugation at 100,000 × g. Pellets were suspended in buffer A containing 6 m guanidine hydrochloride at room temperature for 60 min. The samples were subjected to ultracentrifugation (100,000 × g for 1 h at 4 °C). 1 ml of soluble sample was mixed with 1 ml of either buffer A containing 6 m guanidine hydrochloride, or with FmhASTREP or GST-FmhC suspended in the same buffer. All samples were diluted 10-fold in buffer A containing 20 mm imidazole and spun at 100,000 × g for 1 h at 4 °C to remove insoluble proteins before purification over Ni-NTA as described above. Purification of rLytN was performed as described previously (42). Concentration of proteins was determined with the bicinchoninic acid assay (Pierce). Aliquots of samples loaded on the column, flow-through, wash, and eluted fractions were mixed with an equal volume of sample buffer and separated on 12 or 15% SDS-PAGE. Proteins in gels were visualized by staining with Coomassie Brilliant Blue or electro-transferred to a polyvinylidene difluoride membrane (Millipore). All antibody dilutions were performed in PBS with 5% milk. The mAb against the STREP tag (α-STREP) was obtained from IBA Lifesciences and used at a dilution of 1:5,000 with HRP-conjugated anti-mouse antibody used at a dilution of 1:10,000. Polyclonal antibodies against GST (α-GST, Abcam) and HRP-conjugated anti-rabbit antibody were used at dilutions of 1:10,000.
Peptidoglycan purification
Staphylococci from cultures grown to A600 between 0.8–1.0 were suspended in 50 ml 4% SDS buffered with 100 mm Tris-HCl, pH 6.8, and boiled for 30 min. Cells were washed five times in water and lysed via bead-beating (MP Biomedicals). Cellular material was collected by centrifugation (7,500 × g for 10 min), washed two times with water, and suspended in 50 mm Tris-HCl (pH 7.5), 10 mm CaCl2, and 20 mm MgCl2 for digestion with DNase (10 μg/ml), and RNase (50 μg/ml) for 2 h at 37 °C and then with trypsin (100 μg/ml) for 16 h at 37 °C. The cell wall material was sedimented (3,300 × g for 15 min), suspended in 100 mm Tris-HCL, pH 6.8, containing 1% SDS, boiled for 10 min to inactivate enzymes, and then washed twice with water, once with 8 m LiCl, once with 100 mm EDTA, twice with water, once with acetone, and twice with water. Sacculi were suspended in 5 ml of 47% hydrofluoric acid for 48 h at 4 °C. Peptidoglycan was recovered by centrifugation (33,000 × g for 45 min) and washed twice with water, twice with 100 mm Tris-HCl (pH 7.5), and twice with water. Sacculi were dried under vacuum, the dry weight was recorded, and the samples were suspended to 50 mg/ml with sterile water and stored at −20 °C.
Biochemical characterization of peptidoglycan preparations
Peptidoglycan (5 mg) was incubated for 16 h at 37 °C with 10 μl of enzyme: mutanolysin (0.5 unit) or rLytN (300 μg/ml). Peptidoglycan samples were incubated in 12.5 mm and 50 mm phosphate buffer, pH 5.5, respectively. Enzymatic reactions were ended by heat treatment (95 °C, 10 min) and insoluble materials were removed by centrifugation. The solubilized materials were dried under vacuum and reduced via the addition of 0.5 m sodium borate (100 μl) and 3–5 mg of sodium borohydride. After 15 min of incubation, reactions were inactivated by the addition of 20% phosphoric acid to reach pH < 4.0. Samples were centrifuged (23,000 × g for 10 min) to remove any precipitates before separation onto reverse-phase HPLC using a Waters 2695 Alliance system. 100 μl of sample was applied to a 250 × 4.6-mm reverse-phase C18 column (ODS-Hypersil, 3 µm; Thermo Fisher Scientific) via automated injection. The column was eluted at a flow rate of 0.5 ml/min with a linear gradient starting 5 min after injection of 5% (v/v) methanol in 100 mm NaH2PO4, (pH 2.5) to 30% (v/v) methanol in 100 mm NaH2PO4 (pH 2.8) for 150 min. Column temperature was maintained at 52 °C. The eluted compounds were detected by absorption at 206 nm (A206). Where indicated, area under the curves was determined with ImageJ for a rough estimate of muropeptide amounts. For MALDI-TOF MS, selected HPLC fractions were dried under vacuum, suspended in 50 μl of 0.1% TFA, and desalted into 30% acetonitrile 0.1% TFA using ZipTips (Millipore) according to the manufacturer's instructions. 0.5 μl of desalted fraction was spotted with 0.5 μl of matrix, α-cyano-4-hydroxycinnamic acid, at 10 mg/ml in 50% acetonitrile–0.1% TFA. The samples were subjected to MALDI-TOF MS using an Autoflex Speed Bruker MALDI instrument. The ions were detected in reflectron-positive mode unless otherwise stated. For automated Edman degradation analysis, desalted fractions were sent to the Protein Structure Core Facility at the University of Nebraska, where they were analyzed using a Shimadzu PPSQ-33A system.
Purification of anchor peptides
Overnight cultures (40 ml) of Newman variants femAB::308, femAB::308 fmhA, and femAB::308 fmhC harboring pHTT4 were used to inoculate 2 liters of tryptic soy broth supplemented with 10 µg/ml chloramphenicol. The cultures were grown with shaking for 5 h. The cells were collected by centrifugation, washed, suspended in 100 ml of water, extracted with 100 ml of ethanol-acetone (1:1), and incubated for 30 min on ice. The cells were collected by centrifugation, washed with 300 ml of ice-cold water, and suspended in 30 ml of 0.1 m Tris-HCl, pH 7.5, for incubation with mutanolysin (333 units/ml) for 16 h, followed by incubation with ϕ11 hydrolase (250 µg) for 16 h with rotation at 37 °C. Digested samples were centrifuged at 40,000 × g for 30 min, and the supernatant was subjected to Ni-NTA affinity chromatography as described for recombinant proteins above. Purified SEB-MH6-CWS was incubated in the dark overnight with a crystal of cyanogen bromide and re-purified by Ni-NTA affinity chromatography as described (41). Eluate was desalted using a C18 matrix cartridge (Waters) (Waters Corporation, Milford, MA) and dried under vacuum, as described (41). Dried peptides were resuspended in 20 μl of 50% CH3CN 0.1% TFA. 1 μl of sample was co-spotted with 1 μl of α-cyano-4-hydroxycinnamic acid (10 mg/ml in 50% CH3CN 0.1% TFA) and allowed to dry before analysis on a MALDI-TOF instrument (Bruker) in linear positive mode.
Data availability
All data described in the article are contained within the article. Strains and plasmids described in this manuscript are available upon request to the corresponding author.
Acknowledgments
We thank Chloe Schneewind, Blake Sanders, and members of our laboratory for suggestions and careful reading of the manuscript.
Author contributions—S. W., E. D., O. S., and D. M. conceptualization; S. W., E. D., O. S., and D. M. formal analysis; S. W., O. S., and D. M. validation; S. W., E. D., O. S., and D. M. investigation; S. W., O. S., and D. M. visualization; S. W., O. S., and D. M. methodology; S. W., O. S., and D. M. writing-original draft; O. S. and D. M. supervision; O. S. and D. M. funding acquisition; O. S. and D. M. project administration.
Funding and additional information—This research was supported by the National Institute of Allergy and Infectious Diseases, Infectious Disease Branch Grant AI038897 (to O. S. and D. M.). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PBP
- penicillin-binding protein
- MRSA
- methicillin-resistant S. aureus
- fem
- factors essential for expression of methicillin resistance
- NAG
- N-acetylglucosamine
- NAM
- N-acetylmuramic acid
- Lif
- lysostaphin immunity factor
- Epr
- endopeptidase resistance
- pitet
- anhydrotetracycline-inducible promoter of vector pMF312
- SEB
- staphylococcal enterotoxin B
- CWS
- C-terminally fused cell wall sorting signal
- Ni-NTA
- nickel-nitrilotriacetic acid
- Gdn-HCl
- guanidine hydrochloride
- IPTG
- isopropyl β-d-thiogalactopyranoside.
References
- 1. Weidel W., Frank H., and Martin H. H. (1960) The rigid layer of the cell wall of Escherichia coli strain B. J. Gen. Microbiol 22, 158–166 10.1099/00221287-22-1-158 [DOI] [PubMed] [Google Scholar]
- 2. Salton M. R. J. (1952) Cell wall of Micrococcus lysodeikticus as the substrate of lysozyme. Nature 170, 746–747 10.1038/170746a0 [DOI] [PubMed] [Google Scholar]
- 3. Giesbrecht P., Kersten T., Maidhof H., and Wecke J. (1998) Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62, 1371–1414 10.1128/MMBR.62.4.1371-1414.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strominger J. L., Izaki K., Matsuhashi M., and Tipper D. J. (1967) Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 26, 9–18 [PubMed] [Google Scholar]
- 5. Ghuysen J.-M., and Strominger J. L. (1963) Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of the disaccharides. Biochemistry 2, 1119–1125 10.1021/bi00905a036 [DOI] [PubMed] [Google Scholar]
- 6. Ghuysen J.-M., Tipper D. J., Birge C. H., and Strominger J. L. (1965) Structure of the cell wall of Staphylococcus aureus strain Copenhagen. VI. The soluble glycopeptide and its sequential degradation by peptidases. Biochemistry 4, 2245–2254 10.1021/bi00886a043 [DOI] [PubMed] [Google Scholar]
- 7. Tipper D. J., Ghuysen J.-M., and Strominger J. L. (1965) Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. III. Further studies of the disaccharides. Biochemistry 4, 468–473 10.1021/bi00879a015 [DOI] [PubMed] [Google Scholar]
- 8. Tipper D. J. (1968) Alkali-catalyzed elimination of d-lactic acid from muramic acid and its derivatives and the determination of muramic acid. Biochemistry 7, 1441–1449 10.1021/bi00844a029 [DOI] [Google Scholar]
- 9. Tipper D. J., and Berman M. F. (1969) Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureus Copenhagen. I. Chain length and average sequence of cross-bridge peptides. Biochemistry 8, 2183–2191 10.1021/bi00833a060 [DOI] [PubMed] [Google Scholar]
- 10. Tipper D. J., and Strominger J. L. (1965) Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl. Acad. Sci. U. S. A. 54, 1133–1141 10.1073/pnas.54.4.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yocum R. R., Waxman D. J., Rasmussen J. R., and Strominger J. L. (1979) Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial d-alanine carboxypeptidases. Proc. Natl. Acad. Sci. U. S. A. 76, 2730–2734 10.1073/pnas.76.6.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hartman B. J., and Tomasz A. (1984) Low affinity penicillin binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158, 513–516 10.1128/JB.158.2.513-516.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsuhashi M., Song M. D., Ishino F., Wachi M., Doi M., Inoue M., Ubukata K., Yamashita N., and Konno M. (1986) Molecular cloning of the gene for penicillin-binding protein supposed to cause high resistance to β-lactamase antibiotics in Staphylococcus aureus. J. Bacteriol. 167, 975–980 10.1128/jb.167.3.975-980.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berger-Bächi B. (1994) Expression of resistance to methicillin. Trends Microbiol. 2, 389–393 10.1016/0966-842X(94)90617-3 [DOI] [PubMed] [Google Scholar]
- 15. Ehlert K., Schröder W., and Labischinski H. (1997) Specificities of FemA and FemB for different glycine residues: FemB cannot substitute for FemA in staphylococcal peptidoglycan pentaglycine side chain formation. J. Bacteriol. 179, 7573–7576 10.1128/jb.179.23.7573-7576.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strandén A., Ehlert K., Labischinski H., and Berger-Bächi B. (1997) Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179, 9–16 10.1128/JB.179.1.9-16.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Jonge B. L. M., Sidow T., Chang Y. S., Labischinski H., Berger-Bächi B., Gage D. A., and Tomasz A. (1993) Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J. Bacteriol. 175, 2779–2782 10.1128/jb.175.9.2779-2782.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henze U., Sidow T., Wecke J., Labischinski H., and Berger-Bächi B. (1993) Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175, 1612–1620 10.1128/jb.175.6.1612-1620.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maidhof H., Reinicke B., Blümel P., Berger-Bächi B., and Labischinski H. (1991) femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin susceptible Staphylococcus aureus strains. J. Bacteriol. 173, 3507–3513 10.1128/jb.173.11.3507-3513.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chatterjee A. N., and Park J. T. (1964) Biosynthesis of cell wall mucopeptide by a particulate fraction from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 51, 9–16 10.1073/pnas.51.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuhashi M., Dietrich C. P., and Strominger J. L. (1965) Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: Role of sRNA and lipid intermediates. Proc. Natl. Acad. Sci. U. S. A. 54, 587–594 10.1073/pnas.54.2.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katz W., Matsuhashi M., Dietrich C. P., and Strominger J. L. (1967) Biosynthesis of the peptidoglycan of bacterial cell walls. IV. Incorporation of glycine in Micrococcus lysodeikticus. J. Biol. Chem. 242, 3207–3217 [PubMed] [Google Scholar]
- 23. Higashi Y., Strominger J. L., and Sweeley C. C. (1967) Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of C55 isoprenoid alcohol. Proc. Natl. Acad. Sci. U. S. A. 57, 1878–1884 10.1073/pnas.57.6.1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higashi Y., Strominger J. L., and Sweeley C. C. (1970) Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus. J. Biol. Chem. 245, 3697–3702 [PubMed] [Google Scholar]
- 25. Kamiryo T., and Matsuhashi M. (1972) The biosynthesis of the cross-linking peptides in the cell wall peptidoglycan of Staphylococcus aureus. J. Bacteriol. 247, 6306–6311 [PubMed] [Google Scholar]
- 26. Schneider T., Senn M. M., Berger-Bächi B., Tossi A., Sahl H. G., and Wiedemann I. (2004) In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53, 675–685 10.1111/j.1365-2958.2004.04149.x [DOI] [PubMed] [Google Scholar]
- 27. Kopp U., Roos M., Wecke J., and Labischinski H. (1996) Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target?. Microb. Drug Resist. 2, 29–41 10.1089/mdr.1996.2.29 [DOI] [PubMed] [Google Scholar]
- 28. Tschierske M., Mori C., Rohrer S., Ehlert K., Shaw K. J., and Berger-Bächi B. (1999) Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol. Lett. 171, 97–102 10.1111/j.1574-6968.1999.tb13417.x [DOI] [PubMed] [Google Scholar]
- 29. Sugai M., Fujiwara T., Komatsuzawa H., and Suginaka H. (1998) Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224, 67–75 10.1016/S0378-1119(98)00508-3 [DOI] [PubMed] [Google Scholar]
- 30. Rohrer S., Ehlert K., Tschierske M., Labischinski H., and Berger-Bächi B. (1999) The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. U. S. A. 96, 9351–9356 10.1073/pnas.96.16.9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schindler C. A., and Schuhardt V. T. (1964) Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. U. S. A. 51, 414–421 10.1073/pnas.51.3.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Browder H. P., Zygmunt W. A., Young J. R., and Tavormina P. A. (1965) Lysostaphin: enzymatic mode of action. Biochem. Biophys. Res. Com. 19, 383–389 10.1016/0006-291X(65)90473-0 [DOI] [PubMed] [Google Scholar]
- 33. Sugai M., Fujiwara T., Akiyama T., Ohara M., Komatsuzawa H., Inoue S., and Suginaka H. (1997) Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. J. Bacteriol. 179, 1193–1202 10.1128/jb.179.4.1193-1202.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dehart H. P., Heath H. E., Heath L. S., Leblanc P. A., and Sloan G. L. (1995) The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 61, 1475–1479 10.1128/AEM.61.4.1475-1479.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heath H. E., Heath L. S., Nitterauer J. D., Rose K. E., and Sloan G. L. (1989) Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem. Biophys. Res. Com. 160, 1106–1109 10.1016/S0006-291X(89)80117-2 [DOI] [PubMed] [Google Scholar]
- 36. Sugai M., Fujiwara T., Ohta K., Komatsuzawa H., Ohara M., and Suginaka H. (1997) epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J. Bacteriol. 179, 4311–4318 10.1128/jb.179.13.4311-4318.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tschierske M., Ehlert K., Strandén A. M., and Berger-Bächi B. (1997) Lif, the lysostaphin immunity factor, complements FemB in staphylococcal peptidoglycan interpeptide bridge formation. FEMS Microbiol. Lett. 153, 261–264 10.1111/j.1574-6968.1997.tb12583.x [DOI] [PubMed] [Google Scholar]
- 38. Robinson J. M., Hardman J. K., and Sloan G. L. (1979) Relationship between lysostaphin endopeptidase production and cell wall composition in Staphylococcus staphylolyticus. J. Bacteriol. 137, 1158–1164 10.1128/JB.137.3.1158-1164.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Browder H. P., Tavormina P. A., and Zygmunt W. A. (1968) Optical configuration of staphylococcal cell wall serine. J. Bacteriol. 96, 1452–1453 10.1128/JB.96.4.1452-1453.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tipper D. J. (1969) Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureus Copenhagen. II. Structure of neutral and basic peptides from hydrolysis with the Myxobacter al-1 peptidase. Biochemistry 8, 2192–2202 10.1021/bi00833a061 [DOI] [PubMed] [Google Scholar]
- 41. Ton-That H., Labischinski H., Berger-Bächi B., and Schneewind O. (1998) Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 273, 29143–29149 10.1074/jbc.273.44.29143 [DOI] [PubMed] [Google Scholar]
- 42. Frankel M. B., Hendrickx A. P., Missiakas D. M., and Schneewind O. (2011) LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J. Biol. Chem. 286, 32593–32605 10.1074/jbc.M111.258863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thumm G., and Götz F. (1997) Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 23, 1251–1265 10.1046/j.1365-2958.1997.2911657.x [DOI] [PubMed] [Google Scholar]
- 44. Monteiro J. M., Covas G., Rausch D., Filipe S. R., Schneider T., Sahl H. G., and Pinho M. G. (2019) The pentaglycine bridges of Staphylococcus aureus peptidoglycan are essential for cell integrity. Sci. Rep. 9, 5010 10.1038/s41598-019-41461-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bae T., Banger A. K., Wallace A., Glass E. M., Aslund F., Schneewind O., and Missiakas D. M. (2004) Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U. S. A. 101, 12312–12317 10.1073/pnas.0404728101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bae T., and Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 47. Roos M., Pittenauer E., Schmid E., Beyer M., Reinike B., Allmaier G., and Labischinski H. (1998) Improved high-performance liquid chromatographic separation of peptidoglycan isolated from various Staphylococcus aureus strains for mass spectrometric characterization. J. Chromatogr. B Biomed. Sci. Appl. 705, 183–192 10.1016/S0378-4347(97)00506-9 [DOI] [PubMed] [Google Scholar]
- 48. Ehlert K., Tschierske M., Mori C., Schröder W., and Berger-Bächi B. (2000) Site-specific serine incorporation by Lif and Epr into positions 3 and 5 of the staphylococcal peptidoglycan interpeptide bridge. J. Bacteriol. 182, 2635–2638 10.1128/jb.182.9.2635-2638.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Frankel M. B., Wojcik B. M., DeDent A. C., Missiakas D. M., and Schneewind O. (2010) ABI-domain containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol. Microbiol. 78, 238–252 10.1111/j.1365-2958.2010.07334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeDent A., Bae T., Missiakas D. M., and Schneewind O. (2008) Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 27, 2656–2668 10.1038/emboj.2008.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pinho M. G., and Errington J. (2003) Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50, 871–881 10.1046/j.1365-2958.2003.03719.x [DOI] [PubMed] [Google Scholar]
- 52. Lee C. Y., Buranen S. L., and Ye Z.-H. (1991) Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103, 101–105 10.1016/0378-1119(91)90399-V [DOI] [PubMed] [Google Scholar]
- 53. Kern J., Ryan C., Faull K., and Schneewind O. (2010) Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J. Mol. Biol. 401, 757–775 10.1016/j.jmb.2010.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berger-Bächi B. (1983) Insertional inactivation of staphylococcal methicillin resistance by Tn551. J. Bacteriol. 154, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rohrer S., and Berger-Bächi B. (2003) Application of a bacterial two-hybrid system for the analysis of protein-protein interactions between FemABX family proteins. Microbiology 149, 2733–2738 10.1099/mic.0.26315-0 [DOI] [PubMed] [Google Scholar]
- 56. Salton M. R., and Pavlik J. G. (1960) Studies of the bacterial cell wall. VI. Wall composition and sensitivity to lysozyme. Biochim. Biophys. Acta 39, 398–407 10.1016/0006-3002(60)90191-8 [DOI] [PubMed] [Google Scholar]
- 57. Scherr T. D., Roux C. M., Hanke M. L., Angle A., Dunman P. M., and Kielian T. (2013) Global transcriptome analysis of Staphylococcus aureus biofilms in response to innate immune cells. Infect. Immun. 81, 4363–4376 10.1128/IAI.00819-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kolar S. L., Nagarajan V., Oszmiana A., Rivera F. E., Miller H. K., Davenport J. E., Riordan J. T., Potempa J., Barber D. S., Koziel J., Elasri M. O., and Shaw L. N. (2011) NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157, 2206–2219 10.1099/mic.0.049692-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Green C. I., and Vold B. S. (1993) Staphylococcus aureus has clustered tRNA genes. J. Bacteriol. 175, 5091–5096 10.1128/jb.175.16.5091-5096.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A., Mongodin E. F., Sensabaugh G. F., and Perdreau-Remington F. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 61. Burts M. L., DeDent A. C., and Missiakas D. M. (2008) EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69, 736–746 10.1111/j.1365-2958.2008.06324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bertani G. (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186, 595–600 10.1128/jb.186.3.595-600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., and Novick R. P. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 64. Baba T., Bae T., Schneewind O., Takeuchi F., and Hiramatsu K. (2008) Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacterial. 190, 300–310 10.1128/JB.01000-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gründling A., and Schneewind O. (2007) Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J. Bacteriol. 189, 2521–2530 10.1128/JB.01683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ton-That H., Faull K. F., and Schneewind O. (1997) Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272, 22285–22292 10.1074/jbc.272.35.22285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in the article are contained within the article. Strains and plasmids described in this manuscript are available upon request to the corresponding author.