Abstract
Astrocytes perform multiple essential functions in the developing and mature brain, including regulation of synapse formation, control of neurotransmitter release and uptake, and maintenance of extracellular ion balance. As a result, astrocytes have been implicated in the progression of neurodegenerative disorders such as Alzheimer's disease, Huntington's disease, and Parkinson's disease. Despite these critical functions, the study of human astrocytes can be difficult because standard differentiation protocols are time-consuming and technically challenging, but a differentiation protocol recently developed in our laboratory enables the efficient derivation of astrocytes from human embryonic stem cells. We used this protocol along with microarrays, luciferase assays, electrophoretic mobility shift assays, and ChIP assays to explore the genes involved in astrocyte differentiation. We demonstrate that paired-like homeodomain transcription factor 1 (PITX1) is critical for astrocyte differentiation. PITX1 overexpression induced early differentiation of astrocytes, and its knockdown blocked astrocyte differentiation. PITX1 overexpression also increased and PITX1 knockdown decreased expression of sex-determining region Y box 9 (SOX9), known initiator of gliogenesis, during early astrocyte differentiation. Moreover, we determined that PITX1 activates the SOX9 promoter through a unique binding motif. Taken together, these findings indicate that PITX1 drives astrocyte differentiation by sustaining activation of the SOX9 promoter.
Keywords: astrocyte, gliogenesis, human embryonic stem cell, transcription factor, paired-like homeodomain transcription factor 1 (PITX1), sex-determining region Y box 9 (SOX9), glial cell, neural stem cell (NSC), cell differentiation
Astrocytes are among the most abundant cells in the brain (1) and contribute to a multitude of complex and essential functions. For example, they regulate the local microcirculation by signaling to vascular endothelial cells, guide synapse formation, modulate synaptoplastic function, provide nutrients to neurons, control neurotransmitter release and uptake, and maintain extracellular ion balance (2). Therefore, disruption of these functions will have deleterious effects on neural function; indeed, astrocytes have been implicated in the progression of several acute and chronic neurodegenerative diseases (3). For instance, astrocytes contribute to Aβ-induced death of primary rat neurons, suggesting contribution to Alzheimer's disease pathology (4). In addition, astrocytes in two mouse models of Huntington's disease express dysfunctional Kir4.1 channels, resulting in reduced extracellular K+ level (5). In Parkinson's disease model mice, astrocytes accumulate α-synuclein and induce microglial activation, resulting in a significant loss of dopaminergic and motor neurons (6). Consequently, human astrocyte models are essential for in vitro studies of human neurodegenerative diseases.
Despite vital contributions to healthy brain function and pathology, the study of human astrocytes is difficult because the standard differentiation methods are time-consuming and technically challenging. One of first protocols developed to differentiate astrocytes from human embryonic stem cells (hESCs) require 180 days to generate a pure astrocyte population (7). Alternatively, following our protocol, we can obtain pure cultures of mature astrocytes from neural progenitor cells (NPCs) in <4 weeks as confirmed by expressions of markers such as glial fibrillary acidic protein (GFAP), S100 calcium-binding protein β (S100β), and CD44; reaction to inflammation; and promotion of synaptogenesis (8).
Paired-like homeodomain transcription factor 1 (PITX1) regulates the expressions of many developmental genes including genes involved in early tissue patterning. The homedomain of PITX1 includes a lysine residue at the recognition helix that interacts with the DNA major groove and recognizes the CC doublet of the target gene promoter. PITX1 also recognizes the bicoid subfamily of homeoboxes (9), which control the expressions of genes implicated in axis formation and are essential for the development of anterior structures in Drosophila (10, 11). Further, PITX1 regulates genes involved in the early stages of brain development. Lanctôt et al. (12) reported that PITX1 was up-regulated during mouse and chick embryogenesis. PITX1 expression was also detected in the ectoderm-derived stomodeum of mouse embryo at E8, where it contributes to head formation. In addition, PITX1 protein binds a cis-acting element of the pituitary hormone gene promoter and binds to the TAAT/GCC core motif. (13), thereby contributing to pituitary lineage specification (including specification of gonadotropes, thyrotropes, somatotropes, lactotropes, and corticotropes), pituitary morphogenesis, and formation of Rathke's pouch (11, 13, 14). Gene inactivation experiments in mice and chicks have also demonstrated important contributions to hindlimb development. PITX1 knockout mice showed reduced hindlimb size and complete loss of the ilium and patella, whereas PITX1 induction initiated hindlimb outgrowth through regulation of the transcription factor TBX4. PITX1 acts as a regulator of hindlimb bone and soft tissue morphology (15, 16).
Sex-determining region Y box 9 (SOX9) is a member of the SOX family and has a conserved high mobility group box. SOX9 recognizes the consensus DNA sequence AACAAT, through which it regulates various differentiation processes (17). The transcriptional function of SOX9 is associated with chondrogenesis and gliogenesis; indeed, SOX9 is a well-known chondrogenic marker (18). SOX9 is also expressed on astrocyte progenitor cells. Conditional deletion of SOX9 prolongs neurogenesis and delays gliogenesis (19). A complex of SOX9 and NFIA co-regulates genes induced after glial initiation (20).
To date, no studies have examined the functions of PITX1 in astrocyte differentiation or function. In this study, we demonstrate that PITX1 expression increased during human astrocyte differentiation and that activation of the SOX9 promoter induced by PITX1 is critical for this process.
Results
PITX1 expression increases during astrocyte differentiation
We obtained mature astrocytes from hESCs using the multistep differentiation method developed in our laboratory (21, 22). Briefly, NPCs were derived from hESCs through sequential formation of embryonic bodies, rosettes, and neurospheres (Fig. 1A, panels a–e), which finally differentiated into star-shaped astrocytes (Fig. 1A, panels f–h). Astrocyte differentiation was confirmed by immunostaining for GFAP (Fig. 1A, panels i and j), an intermediate filament protein that helps to maintain the mechanical strength of astrocytes (23) and is highly expressed on reactive astrocytes (24, 25), as well as by co-immunostaining for GFAP and S100β (Fig. S1), a neurotrophic protein that acts as an autocrine factor promoting astrocyte proliferation (26, 27).
Figure 1.
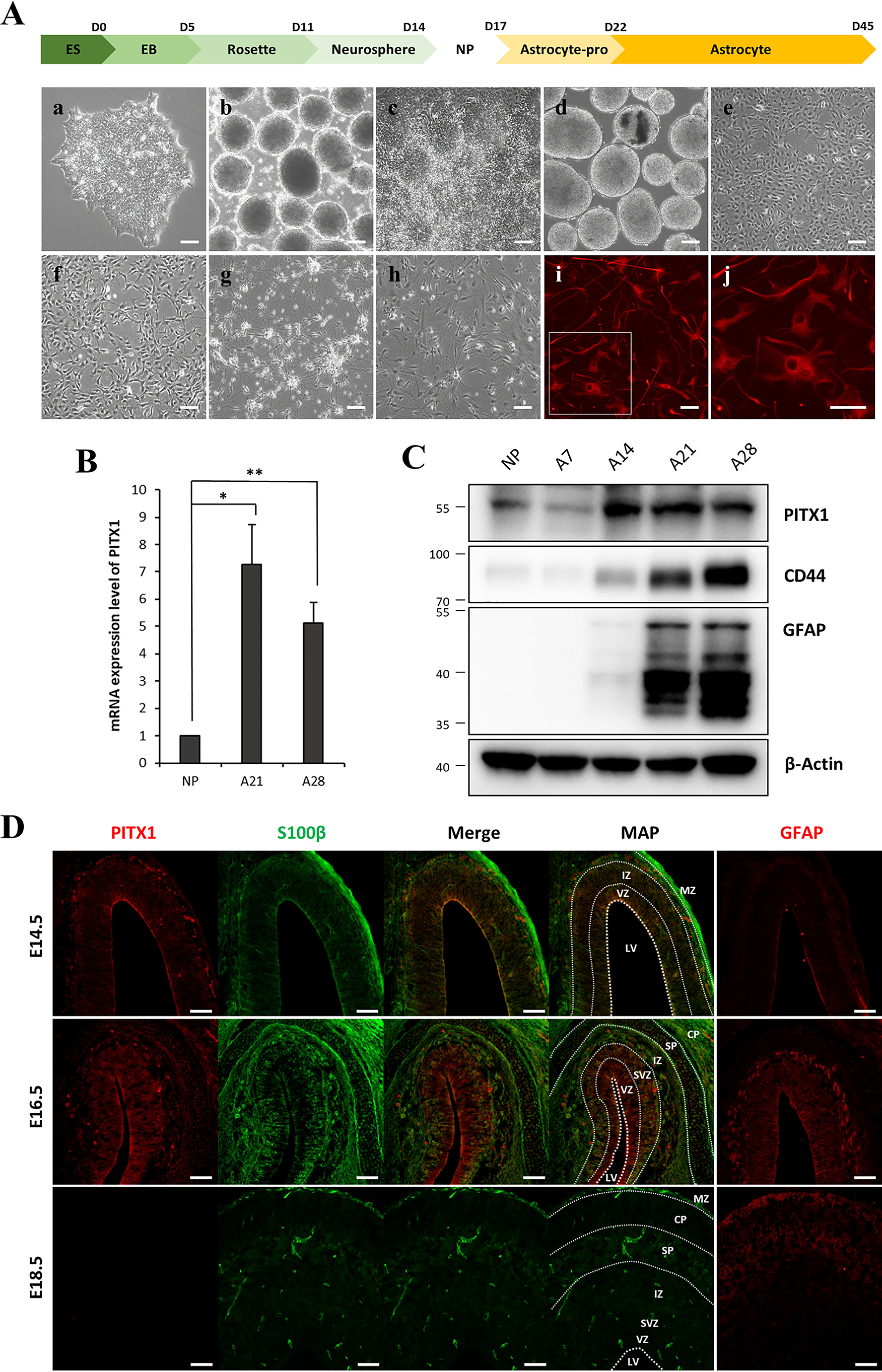
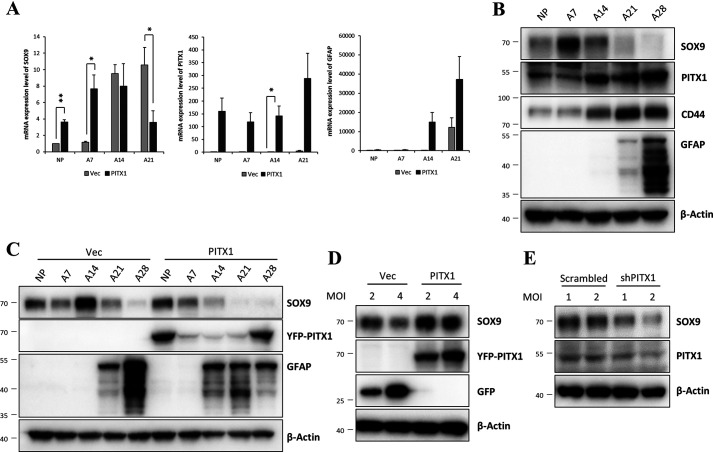
Expression of PITX1 increased during astrocyte differentiation. A, schematic showing the process of astrocyte differentiation from hESCs. Panels show light microscopic images of from hESCs (panel a), EBs (panel b), rosettes (panel c), neurospheres (panel d), NPCs (panel e), astrocyte progenitors on day 5 (panel f), astrocytes on day 14 (panel g), and astrocytes on day 28 of differentiation (panel h), as well as immunofluorescence images of the mature astrocyte marker GFAP (red) on day 28 at low magnification (panel i) and high magnification (panel j). B, relative PITX1 mRNA expression on different days of astrocyte differentiation according to real-time PCR. mRNA expression levels were normalized to β-actin expression level. The bars represent the fold change relative to (undifferentiated) NPCs. C, Western blotting showing the protein levels of PITX1, CD44, GFAP, and β-actin on different days of astrocyte differentiation. The data are expressed as the means ± S.E. of three independent experiments. Statistical significance was determined by independent sample t test. *, p < 0.05; **, p < 0.01. D, immunoexpression pattern of differentiating astrocytes in vivo. Rat embryonic brains (E14.5, E16.5, and E18.5) were immunostained for PITX1 (red), S100β (green), and GFAP (red). Map panels show the structure of brain layers, and dashed lines indicate layer boundaries. PITX1 and S100β were co-stained, and GFAP was stained separately in other sections. Scale bars, 100 μm in A and 75 μm in D.
We examined changes in the expression levels of differentiation-associated genes during astrocyte differentiation using microarrays. These analyses identified 243 genes showing a more than 10-fold change between the NPC and mature astrocyte stages (days 21 and 28). Among these genes, we focused on 76 genes encoding transcriptional factors. Heat maps (Fig. S2A) showed clustering between NPC and mature astrocyte expression groups. Among these genes, 13 up-regulated genes were selected for confirmation by real-time PCR, none of which have been studied for association with astrocyte differentiation or function (Fig. S2B). Finally, we selected the PITX1 gene because it was markedly up-regulated at both mRNA (Fig. 1B) and protein levels (Fig. 1C). We also examined CD44 and GFAP protein expression levels by Western blotting to confirm astrocyte differentiation. Sequential Western blotting during differentiation revealed CD44 expression prior to detectable GFAP expression, indicating that CD44 is expressed by astrocyte precursors, consistent with the findings of previous studies (28, 29). Specifically, differentiating astrocytes expressed CD44 on day 14, and expression levels of both GFAP and CD44 gradually increased from days 21 to 28. Therefore, we used CD44 as an astrocyte precursor marker and GFAP as a mature astrocyte marker.
Next, to confirm this expression pattern in vivo, we examined PITX1 expression levels at different stages of astrocyte differentiation in rat embryonic brain. During rat brain development, cell layers are distinguishable on embryonic day 10.5 (E10.5). In this process, the single cortical ventricular zone (VZ) is divided into the subventricular zone (SVZ), intermediate zone (IZ), subplate, cortical plate (CP), and marginal zone (30). On E18.5, most of these layers are clearly distinguished as in the adult brain (31). Astrocyte development begins with the emergence of radial glial cells in VZ on E12.5. As brain development progresses and distinct layers form, radial glial cells migrate from VZ to SVZ and IZ and concomitantly differentiate into glial progenitor cells before finally differentiating into mature astrocytes in the cerebral cortex (32). To examine the temporal and spatial profiles of PITX1 expression in differentiating astrocytes, we prepared sections of rat embryonic brain and conducted co-immunofluorescence staining for S100β as an astrocyte progenitor marker and GFAP as a mature astrocyte marker. S100β expression was previously demonstrated in mouse astrocyte progenitor cells in VZ (33). Similarly, PITX1 was weakly expressed in the innermost VZ on E14.5, but little GFAP expression was detected. On E16.5, VZ was divided into layers as revealed by the S100β staining pattern, and S100β-positive cells in VZ, SVZ, and IZ co-expressed both PITX1 and GFAP, indicating that astrocyte progenitor cells and mature astrocytes co-exist in these regions. On E18.5, as brain development progressed, layer size further increased, and the expression of PITX1 disappeared completely, whereas the expression of S100β weakened. Conversely, GFAP was robustly expressed in SP and CP, indicating that mature astrocytes had migrated to the upper part of the brain. In summary, we confirmed that PITX1 is expressed by astrocyte progenitor cells in the early stages of glial cell development (E14.5–16.5) but is lost in mature astrocyte at E18.5, suggesting PITX1 plays an essential role in the early stages of astrocyte differentiation during brain development.
Overexpression of PITX1 promotes astrocyte differentiation in vitro
To directly examine if PITX1 regulates astrocyte differentiation, we generated stably YFP-PITX1–overexpressing NPC lines using pLV-Venus-PITX1 lentivirus and compared astrocyte marker patterns and morphology during astrocyte differentiation to YFP-Vector–overexpressing NPC lines. First, human hESCs overexpressing YFP-PITX1 or YFP-Vector were selected using FACS. The fluorescence of sorted cells expressing YFP-PITX1 was weaker than those of sorted cells expressing YFP-Vector alone, but only cells with weak YFP-PITX1 fluorescence could survive and differentiate into NPCs (Fig. S3). The derived NPCs were checked for expression of the neural pluripotency marker Nestin and SRY-box transcription factor 2 (34, 35). In addition, PITX1 overexpression was confirmed through immunofluorescence staining with anti-PITX1 antibodies (Fig. S4).
These stable NPC lines expressing YFP-Vec (YFP-vector stable cells) or YFP-PITX1 (YFP-PITX1 stable cells) were subjected to our astrocyte differentiation protocol over 28 days, and we monitored YFP fluorescence and morphology throughout this process (Fig. 2A and Fig. S5). In addition, the expression levels of GFAP and PITX1 were monitored by Western blotting and immunofluorescence staining. Western blotting confirmed that GFAP protein appeared earlier in the YFP-PITX1 line compared with the YFP-Vector line. Expression of GFAP appeared on day 14 in the YFP-PITX1 line cells and was greater than that in YFP-Vector line on day 21 (Fig. 2. B and C). Next, using immunofluorescence, we counted the number of GFAP-positive cells (red) of the total population of cells (stained with DAPI, blue). In accordance with the Western blotting results, the number of GFAP-positive cells increased rapidly in the YFP-PITX1 line compared with control the YFP-Vector line, again with GFAP-positive cells first appearing on day 14, reaching higher numbers than in the YFP-Vector line on day 21 (Fig. 2, D and E).
Figure 2.
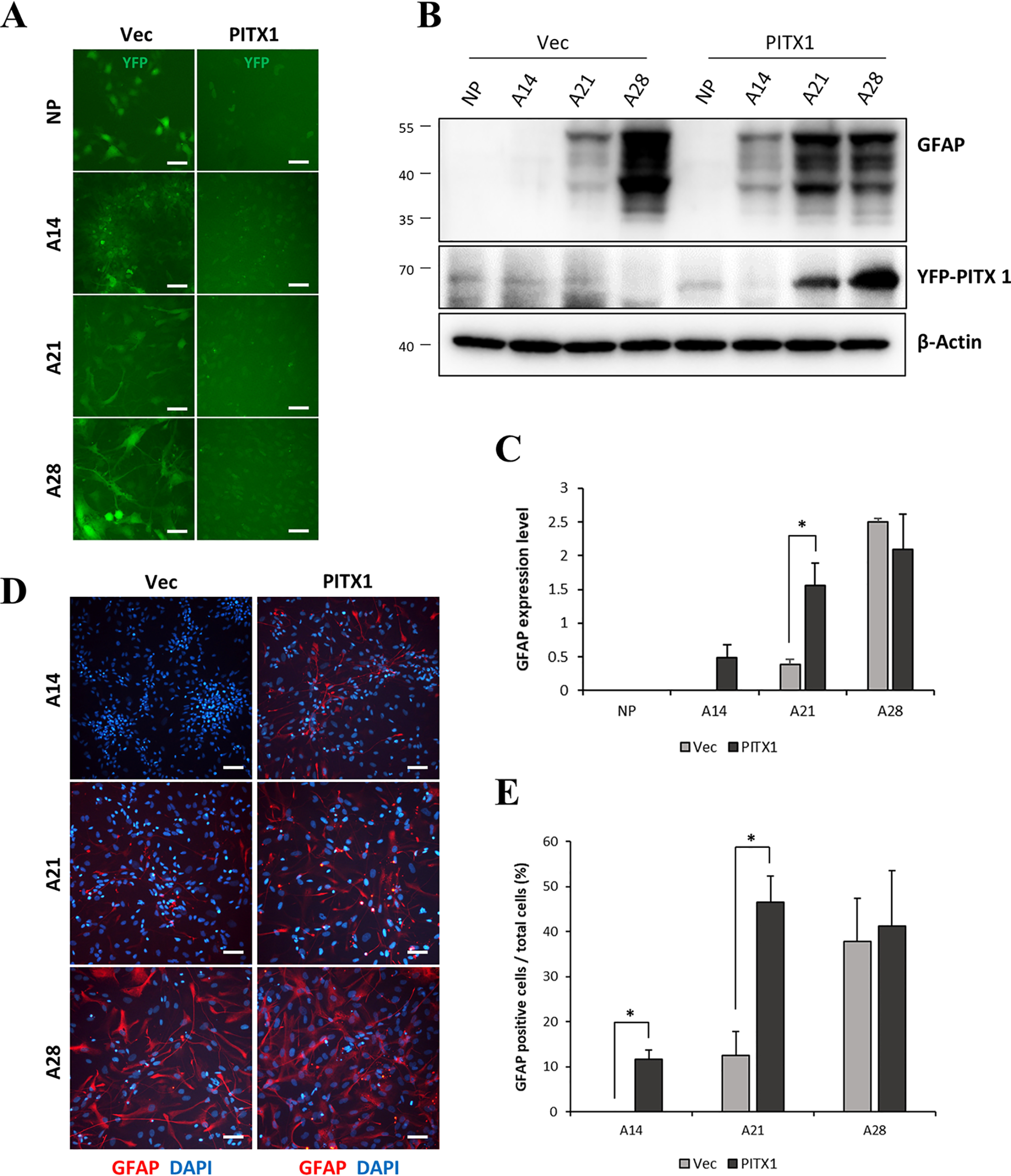
Overexpression of PITX1-accelerated astrocyte differentiation. A, schematic showing live cells during astrocyte differentiation (days 14, 21, and 28) from NPCs. Cells overexpressed YFP-Vector (Vec) or YFP-PITX1 (both emitting green fluorescence, PITX1). B, Western blotting showing GFAP, YFP-PITX1, and β-actin protein expression levels by YFP-Vector and YFP-PITX1 NPC lines on different days during astrocyte differentiation. C, quantification of Western blots (as in B) showing accelerated expression of GFAP (normalized to β-actin expression) in YFP-PITX1 NPC lines. D, immunofluorescence images of astrocytes derived from YFP-Vector and YFP-PITX1 NPC lines during astrocyte differentiation. The cells were stained with antibodies against GFAP (red) and with DAPI (blue). E, proportion of GFAP-positive cells relative to total cells (DAPI-positive). The data are expressed as the means ± S.E. of three independent experiments. Statistical significance was determined by independent sample t test. *, p < 0.05; **, p < 0.01; ***, p < 0.005. Scale bars, 100 μm in A and D.
Knockdown of PITX1 blocks astrocyte differentiation
To provide further evidence that PITX1 promotes astrocyte differentiation, we prepared PITX1-knockdown NPCs using a lentiviral vector encoding a targeted shRNA and compared astrocyte marker expression and morphology with those of NPCs expressing a scrambled shRNA. Astrocytes expressing scrambled shRNA had long and thin bodies, whereas PITX1-knockdown cells had wide and flat bodies on day 28 (the final day of astrocyte differentiation) (Fig. 3A). Western blotting indicated that GFAP expression was eliminated in PITX1-knockdown cells but was up-regulated in cells transfected with scrambled shRNA during astrocyte differentiation (Fig. 3, B and C). Immunofluorescence staining revealed reduced GFAP-positive cell numbers (red) among all DAPI-stained cells (blue) in PITX1 knockdown cells compared with scrambled shRNA-infected cells on days 21 and 28 (Fig. 3, D and E). Taken together, these findings of PITX1-overxpressing and PITX1-knockdown cells demonstrate that PITX1 is essential for astrocyte differentiation from NPCs.
Figure 3.

Knockdown of PITX1 blocked astrocyte differentiation. A, schematic showing live cells during astrocyte differentiation (days 14, 21, and 28) from PITX1-knockdown NPCs and control NPCs expressing scrambled shRNA. B, Western blotting showing the protein levels of GFAP, PITX1, and β-actin on different days of differentiation. C, graph showing greatly reduced expression of GFAP during astrocyte differentiation from PITX1-knockdown cells. D, pictures showing GFAP immunofluorescence (red) and DAPI (blue) on days 21 and 28 of astrocyte differentiation. E, the proportion of GFAP-positive cells relative to total cells. The data are expressed as the means ± S.E. of three independent experiments. Means were compared for statistical significance by independent sample t test. *, p < 0.05; **, p < 0.01; ***, p < 0.005. Scale bars, 100 μm in A and D.
SOX9 protein levels are regulated with PITX1 during early astrocyte differentiation
The level of PITX1 protein gradually increased during astrocyte differentiation (Fig. 1, B and C), and knockdown of PITX1 completely blocked astrocyte differentiation (Fig. 3, B and D), strongly suggesting that PITX1 is involved in the initiation of astrocyte differentiation. To identify the downstream of PITX1 for astrocyte differentiation, we selected several known targets and examined whether expression levels were altered by stable PITX1 overexpression (Fig. S6). We focused specifically on the relationship between PITX1 and SOX9 because SOX9 is an important transcription factor in gliogenesis and is a key factor governing the neuron–glial switch during brain development (19, 20). Consistent with PITX1-dependent regulation, YFP-PITX1 NPCs resulted in earlier expression pattern of SOX9 mRNA (days 7–14) compared with in YFP-Vector NPCs (days 14–21) (Fig. 4A). Next, we examined the protein levels of PITX1 and SOX9 during astrocyte differentiation. Expression of PITX1 protein gradually increased from the beginning of differentiation, whereas that of SOX9 increased markedly from day 7 to 14, after which it decreased (Fig. 4B). Further, in accord with real-time PCR results, SOX9 protein increased faster in YFP-PITX1-NPCs compared with in YFP-Vector NPCs (Fig. 4C). SOX9 protein level peaked at day 14 in YFP-Vector NPCs but was highest at day 0 (before differentiation) in YFP-PITX1-NPCs.
Figure 4.
Association between PITX1 and SOX9 expression levels during early astrocyte differentiation. A, relative mRNA expression levels of GFAP, SOX9, and PITX1 by YFP-Vector (Vec) and YFP-PITX1 lines at multiple astrocyte differentiation time points according to real-time PCR. mRNA expression level was normalized to β-actin expression level. The bars represent the fold change relative to control NPCs. B, Western blotting showing the protein expression levels of PITX1, SOX9, CD44, GFAP, and β-actin after the indicated astrocyte differentiation duration. C, Western blotting showing the protein expression levels of SOX9, YFP-PITX1, GFAP, and β-actin in YFP-Vector and YFP-PITX1 lines after the indicated astrocyte differentiation duration. D and E, Western blotting analysis showing protein levels of SOX9, PITX1, GFP, and β-actin in PITX1-overexpressing and -knockdown cells during early astrocyte differentiation. Differentiating astrocytes were infected by lentivirus on day 6 and harvested on day 9. The data are expressed as the means ± S.E. of three independent experiments. Statistical significance was determined by independent sample t test. *, p < 0.05; **, p < 0.01.
Next, we investigated whether SOX9 and PITX1 expression levels were associated during early astrocyte differentiation following PIXT1 overexpression or knockdown. Differentiating astrocytes were infected with lentivirus on day 6 and harvested on day 9 for the measurement of protein expression by Western blotting. In addition, the influence of PITX1 on SOX9 expression was examined by controlling the transfection multiplicity of infection (MOI). Overexpression of PITX1 increased SOX9 expression during early astrocyte differentiation (Fig. 4D), whereas its knockdown reduced the levels of SOX9 protein compared with in control. Moreover, we demonstrated this association more precisely by manipulating the magnitude of transfection (2 or 4 MOI). Again, reduction in SOX9 expression paralleled the reduction in PITX1 protein expression (Fig. 4E). Therefore, we conclude that PITX1 induces SOX9 protein expression during early astrocyte differentiation.
PITX1 regulates the SOX9 promoter through a unique binding motif
PITX1 activates transcription of genes by binding to the sequence TAATCC within the promoter (11, 36). Therefore, we investigated the SOX9 promoter to determine whether it contains this PITX1-binding motif. The SOX9 promoter contains binding sites for multiple transcription factors, most of which are located in the proximal promoter region within 300 bp of the transcription start site (TSS). The proximal promoter region was highly conserved among chicks, mice, and humans (37). The SOX9 promoter has two expected PITX1-binding motifs between −1015 bp and TSS, a TAATCC sequence at −578 bp, and a CAATCC sequence (similar to the PITX1 motif) at −129 bp. We first conducted luciferase assay to confirm that PITX1 regulates the SOX9 promoter directly. The luciferase reporter gene was inserted after the SOX9 promoter (−1015 bp to +14 bp) and co-transfected with PITX1 into BE(2)C cells. Luciferase activity was found to increase in parallel with PITX1 concentration (Fig. 5A), consistent with direct induction of SOX9 by PIXT1 binding to the SOX9 promoter.
Figure 5.
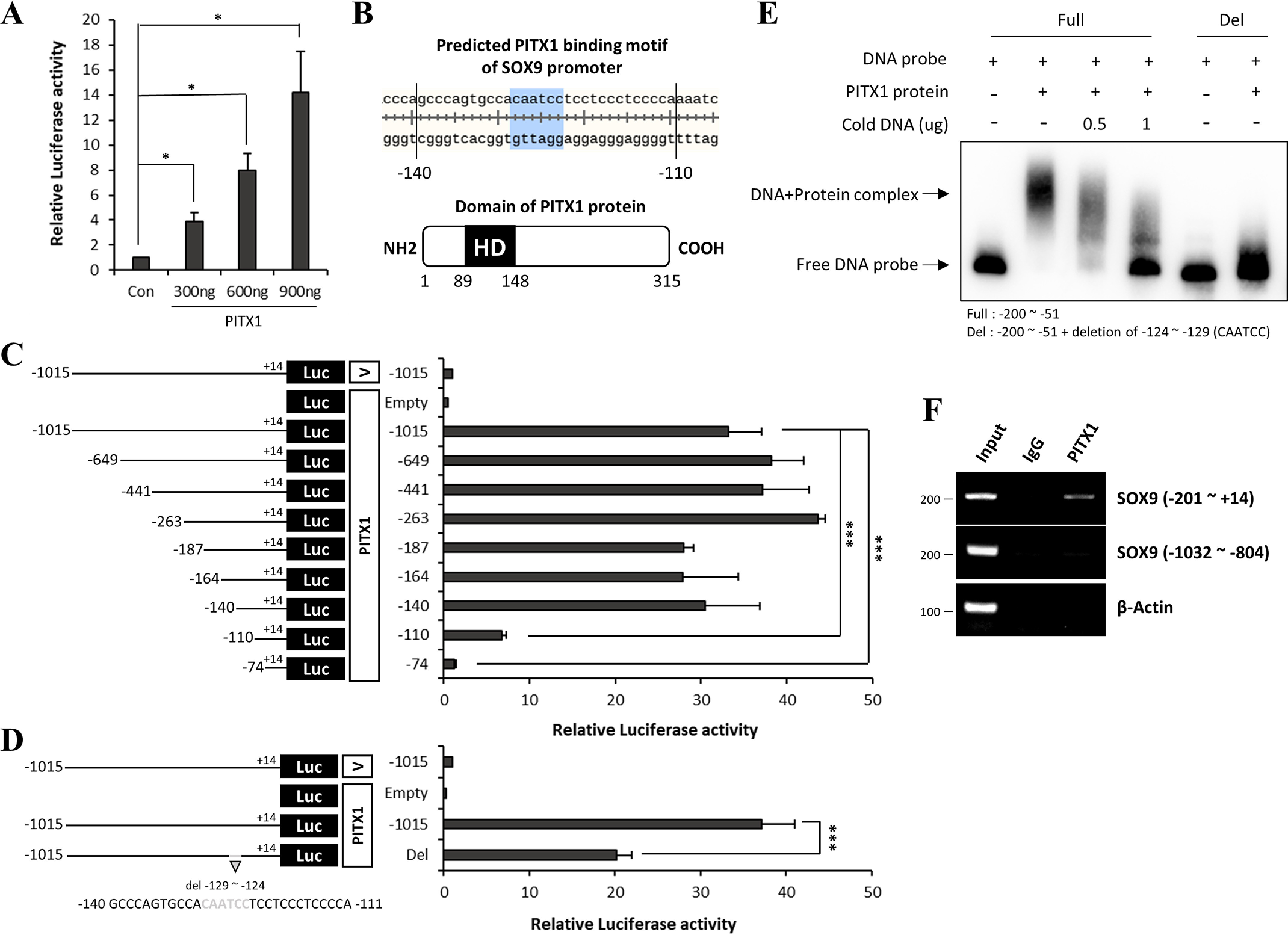
PITX1 activated the SOX9 promoter through a unique binding motif. A, luciferase activity of the SOX9 promoter with increasing PITX1 levels in BE(2)C cells. Luciferase activity was normalized to β-gal expression level. The bars represent the fold change relative to the control sample (Con). B, schematic showing predicted binding motifs within the SOX9 promoter (upper panel) and Homeodomain (HD) of the PITX1 protein (lower panel). C and D, schematic on the left illustrates SOX9 promoter deletion constructs. Graph on the right shows luciferase activity of the associated construct. The black line indicates the length of the SOX9 promoter (distance upstream of the TSS indicated by − and downstream by +). The specific deletion site is marked in gray. Each construct was fused to a luciferase reporter gene (black box). Expression genes of the Vector and PITX1 protein are depicted as white boxes. BE(2)C cells were co-transfected with the PITX1 expression gene, SOX9 promoter reporter gene, and β-gal expression gene. Luciferase activity was normalized to β-gal expression. The bars represent the fold change relative to the vector-transfected (control) sample. E, electrophoretic mobility shift assay demonstrating direct binding of the SOX9 promoter (probe) to the PITX1 homeodomain. The DNA probe was 5′- and 3′-biotin–labeled. The length of the full-form probe was −200 to −51 bp. Deletion forms of the probe were identical to the full-form probe except the PITX1-binding motifs. The concentration of biotinylated probe was 1 ng, and that of purified PITX1 homeodomain protein was 500 ng. Unlabeled probe was not biotin-labeled and was added at 500 or 1000 ng. The presence (+) or absence (−) of specific probes is indicated. F, ChIP assay showing interaction of the SOX9 promoter with the PITX1 protein. PIXT1 was pulled down with interacting DNAs from astrocytes on differentiation day 14 using a PITX1 antibody. PCR was performed using primers for the human SOX9 promoter. Primers of the SOX9 promoter were designed to amplify region −201 to +25 and −1032 to −804 bp. Region −201 to +25 bp included the PITX1-binding motif. The data are expressed as the means ± S.E. of three independent experiments. Statistical significance was determined by independent sample t test. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
We next identified the SOX9 promoter-binding site by serial deletion of promoter sequences. A series of 5′-deletions of the −1015 to +14-bp fragments were created in the luciferase reporter gene, and BE(2)C cells were co-transfected with these reporter constructs along with the PITX1 gene. Luciferase activity was greatly reduced by removing a region including the predicted PITX1-binding CAATCC motif at −129 bp, but it was not changed by deleting a region including the other tentative PITX1-binding motif TAATCC at −578 bp (Fig. 5C). The PITX1-binding sequence in the SOX9 promoter is shown in Fig. 5B. Compared with the full-length SOX9 promoter (−1015 bp to +14 bp), luciferase activity was decreased by half when the predicted PITX1-binding motif CAATCC at −129 bp was removed (Fig. 5D). These findings indicate that PITX1 activates the SOX9 promoter via a unique PITX1-binding motif CAATCC at −129 bp.
We next conducted electrophoretic mobility shift assay (EMSA) to confirm the interaction of PITX1 protein with the predicted PITX1-binding motif CAATCC at −129 bp in the SOX9 promoter. PITX1 contains a 7-kDa homeodomain on N-terminal (Fig. 5B) known to mediate both protein–DNA and protein–protein interactions (38, 39). For efficient EMSA, we purified the homeodomain through thrombin digestion and ion-exchange chromatography (Fig. S7). We then designed 150-bp biotinylated DNA probes with the predicted PITX1-binding motif of SOX9 (the full form). The addition of the PITX1 homeodomain shifted the probe DNA gel band to a higher than the free probe, and this gel mobility shift disappeared upon addition of unlabeled DNA probe. In contrast to the full probe, a probe with deletion of the predicted PITX1-binding motif (CAATCC) did not show band shift after addition of the PITX1 homeodomain (Fig. 5E). We then performed ChIP assay to determine whether PITX1 binds to this specific sequence of the SOX9 promoter in differentiating astrocytes (day 14). PITX1 was precipitated from cell lysates using anti-PITX1 antibodies and agarose beads. The genes bound by PITX1 were isolated through a series of procedures. The presence of the SOX9 promoter, including the predicted PITX1-binding motif, was confirmed by PCR. β-Actin and SOX9 promoter fragments without the predicted PITX1-binding motif were used as controls. Only the SOX9 promoter fragments containing the predicted PITX1-binding motif were immunoprecipitated by PITX1 (Fig. 5F). We further investigated binding of the SOX9 promoter to PITX1 protein during astrocyte differentiation using ChIP–real-time PCR, which revealed that PITX1 bound to the SOX9 promoter during early astrocyte differentiation (days 7 and 14). However, this interaction weakened in the later stages (days 21 and 28) (Fig. S8). We conclude that PITX1 activates the SOX9 gene through binding to a unique sequence (CAATCC) within the SOX9 promoter during early astrocyte differentiation.
Discussion
Astrocytes are essential for proper neuronal development, synapse formation, maintenance of extracellular ion concentration, regulation of synaptic transmission (40), inflammatory responses, detoxification pathways, and wound healing (41). Therefore, astrocyte dysfunction is strongly implicated in various brain disorders involving neuroinflammation and transmitter imbalances among other pathogenic processes. For instance, reactive astrocytes participate in clearance of Aβ in Alzheimer's disease (42), and astrocytic neuroinflammatory signaling may contribute to the progression of Alzheimer's disease and Huntington's disease (3).
In the past, most studies on astrocyte physiology and their contribution to disease were conducted on cells isolated from the developing brains of animals because it is not feasible to extract healthy astrocytes from the human brain. However, recent advances in human pluripotent stem cell differentiation can provide researchers with supply of human glial cells for basic mechanistic studies, as well as preclinical investigations on potential clinical applications in regenerative medicine (43, 44). Indeed, transplantation of human astrocytes has been shown to protect spinal cord neurons following injury (45). Moreover, transplantation of glial-restricted precursors survived in diseased tissue, differentiated efficiently into astrocytes, and attenuated motor neuron loss in rat spinal cord (46). Given the important contributions of astrocytes to brain physiology and pathology, as well as the clinical potential of astrocyte transplantation, it is vital to establish improved methods for isolation of human astrocytes. However, this has been hampered by low efficiency. In fact, using traditional methods, the differentiation of astrocytes from induced pluripotent stem cells requires >120 (47) and >60 days from hESCs (21, 22). We developed a method for differentiation of astrocytes from hESCs that requires only 45 days (efficiency: 80%) (8). This rapid and efficient astrocyte differentiation protocol provides a robust platform for human-specific pathology and cell therapy studies. In the current study, we investigated the molecular factors contributing to astrocyte differentiation using this protocol.
Expression of PITX1 increased during astrocyte differentiation as revealed by microarrays, real-time PCR, and Western blotting. Further, PITX1 expression level was strongly associated with astrocyte differentiation because exogenous PITX1 overexpression accelerated and its knockdown reduced the emergence of mature astrocyte markers. In this study, we newly observed that PITX1 is critical for astrocyte differentiation.
SOX transcription factors, including SOX9, have important functions during astrocyte and oligodendrocyte development. In glial precursor cells, SOX9 function was suppressed under the influence of SOX3, but SOX9 induced astrocyte differentiation when SOX3 was removed (48). SOX9 is known to induce gliogenesis (19) through cooperation with NFIA (20). We screened for transcription factors regulated by PITX1 and found that PITX1 overexpression both enhanced SOX9 expression in YFP-PITX1 NPCs and shifted the peak of SOX9 expression during astrocyte differentiation to an earlier period. Furthermore, we wanted to confirm the effect of PITX1 about SOX9 expression in astrocyte precursor cells, not NPCs. Endogenous PITX1 was only weakly expressed on NPCs, but its expression increased substantially by day 7 of astrocyte differentiation. Therefore, we investigated SOX9 expression level depending on PITX1 overexpression or knockdown in an early stage of astrocyte differentiation (Fig. 4, D and E). In particular, the patterns of SOX9 induced by knockdown of PITX1 were carefully identified, because SOX9 protein increased in early astrocyte differentiation. The overexpression of PITX1 induced marked increase of SOX9, and knockdown of PITX1 induced gradual decrease of SOX9 as MOI increased. As a result, the stepwise increase and decrease of PITX1 can gradationally change SOX9 expression in the early stage of astrocyte differentiation. It can be expected that PITX1 strongly interacted with SOX9. In addition, luciferase assay confirmed that PITX1 directly regulates the SOX9 promoter (Fig. 5, A, C, and D). Next, we identified two putative PITX1-binding motifs in the SOX9 promoter within 1 kilobase pair of TSS, a TAATCC sequence at −578 bp and a CAATCC at −129 bp, but only deletion of the latter reduced PITX1-driven luciferase activity. Thus, PITX1 directly activates the SOX9 promoter through a novel CAATCC-binding motif; this was further confirmed by sequence deletion assay, EMSA, and ChIP assay.
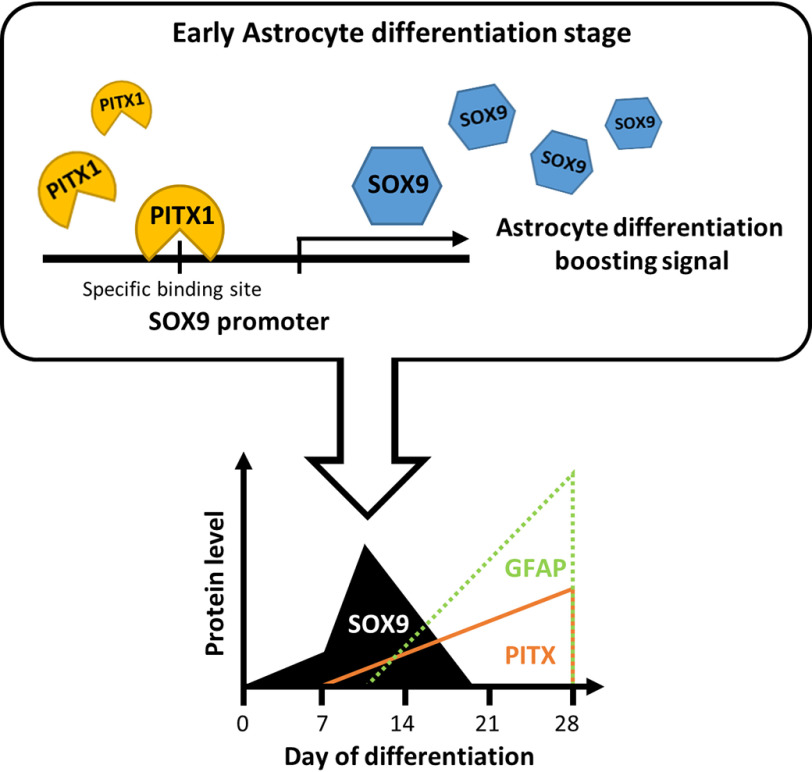
The correlation of PITX1 and SOX9 proteins was shown in the diagram (Fig. 6). At the start of differentiation, SOX9 expression level increases independently of PITX1, presumably under the influence of other signaling and transcription factors. A previous study reported that Sonic Hedgehog stimulated the generation of neural stem cells by inducing SOX9 expression (49). Moreover, Notch1 led to up-regulation of Sox9 expression during neuroectodermal differentiation and induced astrocyte differentiation (50). To derive astrocytes from precursors, we used NPCs; these have similar lineage capacities as neural stem cells, so they may share similar pathways regulating SOX9 aside from PITX1. After the initiation of astrocyte differentiation, PITX1 expression level gradually increased in parallel with the increase in SOX9 expression, but SOX9 disappeared completely during the late stage of differentiation. We found that PITX1 and SOX9 interacted through a specific motif within the SOX9 promoter during the early stage of astrocyte differentiation. However, the expression levels of PITX1 and SOX9 do not match during late stage of differentiation because transcription of SOX9 may not be simply influenced by one protein. During male gonadogenesis, Sf1 and SRY up-regulated SOX9 cooperatively, and then SOX9 replaced SRY (51). In another study, β-catenin interacted with TCF and activated SOX9 transcription, which was then autorepressed through an interaction with β-catenin (52). In the early stage of astrocyte differentiation, we hypothesized that SOX9 transcription is initiated by other transcription factors and that PITX1 acts as a secondary transcription factor to maintain SOX9 expression. In our knockdown experiments, SOX9 expression decreased when PITX1 was suppressed and increased when PITX1 was overexpressed (Fig. 4, D and E). Subsequently, when astrocyte differentiation progressed, SOX9 expression decreased. In fact, astrocyte differentiation was completely blocked in SOX9-overexpressing cells lines (Fig. S9). We believe that SOX9 expression decreases because of down-regulation of the transcription factor that first initiates expression or because of an increase in transcriptional repressors. In summary, PITX1 does not increase SOX9 transcription alone but rather maintains expression with other factors. We suggest that maintaining SOX9 expression in the early stage of differentiation is important for sustained progression of astrocyte differentiation. Once differentiation is complete (or nearly complete), SOX9 expression falls even in the continued presence of PITX1 because of the influence of other factors.
Figure 6.
Schematic depicting the interaction of PITX1 protein with the SOX9 promoter during the early stage of astrocyte differentiation.
In the adult brain, astrocytes regulate inflammation, neuronal excitability, transmitter signaling, metabolic status, and neurogenesis. Therefore, numerous etiologically distinct brain disorders may stem from astrocyte dysfunction. We report that PITX1 directly up-regulates SOX9, a transcription factor that drives the expressions of other genes required for astrocyte differentiation, and this occurs through a newly discovered PITX1-binding motif in the SOX9 promoter. These findings may provide mechanistic insight into disorders stemming from the disruption of astrocyte differentiation and function, as well as aid in human cell therapies requiring differentiation and/or transplantation of human astrocytes.
Experimental procedures
Culture of human embryonic stem cell line
The hESC line H9 was cultured on Matrigel® GFR basement membrane-coated dishes (Corning) in mTeSR™1 basal medium (STEMCELL Technologies) with mTeSR™1 supplement (STEMCELL Technologies) and 25 ng/ml basic fibroblast growth factor (bFGF; Gibco). The cells were transferred into new culture dishes every 5 days and provided with fresh medium daily. Cultured cells were maintained in a humidified incubator under 5% CO2 at 37 °C. All procedures were conducted according the principles outlined in the Declaration of Helsinki.
Differentiation of NPCs
Embryoid bodies (EBs) were generated by splitting H9 cells into 500-μm-sized colonies using needles and deplating using collagenase 4 (Worthington Biochemical). EBs were maintained on Petri dishes with EB medium–DMEM/F12 supplemented with 20% KnockOut™ serum replacement (Gibco), 100 nm LDN193189 (SelleckChem), 10 μm SB525334 (SelleckChem), and 2 mm l-glutamine (Gibco). The EB medium was changed daily. After 5 days, EBs were replated on dished coated with Matrigel® matrix basement membrane (Corning) in rosette medium–DMEM/F12 containing 1× N2 (Gibco), 1× B27 (Gibco), l-glutamine, and 40 ng/ml bFGF. Regarding generation of neural tube-like rosettes, the cells were maintained for 5 days, and fresh rosette medium was replaced every alternate day. Formed rosettes were selected using stretched Pasteur pipettes and maintained on Petri dishes with rosette medium to generate neurospheres. Fresh rosette medium was replaced every alternate day. After 3 days, the neurospheres were dissociated into single NPCs using Accutage® (Innovative Cell Technologies). These single NPCs were seeded on Matrigel®-coated dishes in STEMdiff neural progenitor basal medium (STEMCELL Technologies) containing STEMdiff neural progenitor supplement A and B (STEMCELL Technologies). All cultured cells were maintained in a humidified incubator under 5% CO2 at 37 °C.
Astrocyte differentiation
NPCs were seeded at 30% confluence on Matrigel®-coated dishes. The next day, the medium was replaced with astrocyte progenitor medium–DMEM/F12 containing 1× N2, 1× B27, l-glutamine, 20 ng/ml bFGF, 1× NEAA (Gibco), 5 units/ml heparin (Sigma), and 20 ng/ml EGF (Millipore). The medium was changed every alternate day. After 5 days, the cells were transferred onto Matrigel®-coated dishes with astrocyte medium–Stempro® hESC SFM containing 10 ng/ml Activin A (Gibco), 10 ng/ml Neuroregulin 1 (Sino Biological), 200 ng/ml insulin-like growth factor 1 (Gibco), 10 ng/ml ciliary neurotrophic factor (Peprotech), and 8 ng/ml bFGF. The cells were differentiated for 23 days, and astrocyte medium was changed every alternate day. All cultured cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
Generation of stable cell lines and viral transduction of astrocytes
Stable cell lines were generated by infecting hESC (H9) colonies with 2 MOI of viral supernatant (lentiviral vector; pLV-Venus-Vector (53), pLV-Venus-PITX1) and LentiBOOST™ (Sirion Biotech). After 5 h, the infection medium was replaced with the original maintenance medium. After 2 days, stably transduced cells were selected by YFP fluorescence using FACSAriaTM (BD).
Differentiating astrocytes on day 6 were plated at a density of 1 × 105 cells/well in 6-well plates. The following day, the cells were transfected with 2 MOI of viral supernatant (lentiviral vector; pLV-Venus-Vector, pLV-Venus-PITX1) and LentiBOOST™ (Sirion Biotech). After 5 h, the infection medium was replaced with the original maintenance medium. After 2 days, differentiated astrocytes of day 9 were harvested for analysis.
Microarrays
Total RNA was isolated from NPCs and differentiating astrocytes (days 21 and 28) using TRIzol reagent (Ambion). The differentiating astrocytes were sorted using FITC anti-human CD44 antibody (BioLegend; 338803). Three independent RNA samples were acquired to minimize experimental variation. RNA was amplified and labeled using Agilent's low RNA input linear amplification kit PLUS. Amplified and labeled cRNA was purified on the cRNA cleanup module (Agilent Technology) according to the manufacturer's protocol. Labeled cRNA target was quantified using the ND-1000 spectrophotometer (NanoDrop Technologies). After checking labeling efficiency, hybridization was performed using the Agilent gene expression hybridization kit. Fragmentation of cRNA was performed by adding 10× blocking agent and 25× fragmentation buffer and incubating at 60 °C for 30 min. The fragmented cRNA was resuspended in 2× hybridization buffer and directly pipetted onto the Agilent human oligo microarray (44K). The array was hybridized at 65 °C for 17 h in the Agilent hybridization oven (Agilent Technology). The hybridized microarrays were washed according to the manufacturer's protocol (Agilent Technology). Hybridized chips were scanned using the Agilent DNA microarray Scanner and quantified using the Agilent feature extraction software. All data normalization and further analysis were performed using GeneSpringGX 7.3 (Agilent Technology). All processing of microarrays was performed by Ebiogen.
Real-time PCR
RNA was isolated from cells using TRIzol reagent (Ambion) and the RNeasy kit (Qiagen) following the manufacturers' protocols. cDNA was synthesized from 2 μg of RNA using Moloney murine leukemia virus reverse transcriptase (Promega). Quantitative real-time PCR was performed with TB Green™Premix Ex Taq™ (TaKaRa) on the CFX Connect real-time PCR detection system (Bio-Rad). Relative expression levels were normalized to the β-actin gene expression level. The primer sequences used were as follows: β-actin, forward, 5′-GAGCACAGAGCCTCGCCTTT-3′, and reverse, 5′-ACATGCCGGAGCCGTTGTC-3′; GFAP, forward, 5′-GGCCCGCCACTTGCAGGAGT-3′, and reverse, 5′-CTTCTGCTCGGGCCCCTCAT-3′; PITX1, forward, 5′-GACCCAGCCAAGAAGAAGAA-3′, and reverse, 5′-AACTGCTGGCTTGTGAAGTG-3′; and SOX9, forward, 5′-GAGCTGAGCAGCGACGTCAT-3′, and reverse, 5′-CGTAGCTGCCCGTGTAGGTG-3′
Immunofluorescence
Cultured cells were rinsed with PBS and fixed in 4% paraformaldehyde (T&I) diluted in PBS for 10 min at room temperature. After washing with PBS, the fixed cells were permeabilized and blocked with 0.3% Triton® X-100 (Sigma) diluted in 1× Western blocking reagent solution (Roche) for 1 h at room temperature. Cells were incubated with rabbit anti-GFAP antibody (Abcam; ab7260) and mouse anti-S100β (Abcam; ab11178) diluted in 1× Western blocking reagent solution at 4 °C overnight. After washing with PBS, the cells were stained with goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 594 (Invitrogen; A11012) and goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 488 (Invitrogen; A11001) for 1 h at room temperature. After washing with PBS, the cells were mounted using ProLong® Gold antifade reagent with DAPI solution (Invitrogen). The images were acquired with the Leica DM IL LED inverted laboratory microscope (Leica).
Histology
The embryos of Sprague–Dawley rats were dissected on E14.5, E16.5, or E18.5 and fixed by immersion in 4% paraformaldehyde (T&I) at 4 °C overnight. Then fixed embryos were soaked in 30% sucrose solution overnight at 4 °C and embedded in FSC 22® clear frozen section compound (Leica). The brains were cryosectioned at 10 μm (E16.5 and E18.5) or 20 μm (E14.5) with a vibratome (VT1000S, Leica).
Regarding immunostaining, the brain sections were washed with PBS and permeabilized with 0.3% Triton® X-100 (Sigma) for 30 min. Next, sections were blocked with 1× Western blocking reagent solution (Roche) for 1 h at room temperature. Sections were then incubated at 4 °C overnight with antibodies against GFAP (Abcam; ab7260), S100β (Abcam; ab11178), and PITX1 (Novus; NBP1-88644) diluted in 1× Western blocking reagent solution. After washing with PBS, the sections were stained with goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 594 (Invitrogen; A11012) and goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 488 (Invitrogen; A11001) for 1 h at room temperature. After washing with PBS, the sections were mounted using ProLong® Gold antifade reagent with DAPI solution (Invitrogen). The images were acquired using a Leica DM IL LED inverted laboratory microscope. All animal experiments were performed in accordance with relevant guidelines and regulations, approved by the ethics committees of the Korea Research Institute of Bioscience and Biotechnology.
Western blotting
The cells were rinsed with PBS, harvested, and lysed using Nonidet P-40 lysis buffer containing protease inhibitors (cOmplete, Mini, EDTA-free; Roche) for 30 min on ice. The cell lysates were centrifuged at 14,000 rpm for 20 min. The protein concentration of the supernatants was measured using the 1× Bio-Rad protein assay dye reagent concentrate (Bio-Rad Laboratories). The proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore). The membranes were blocked with 5% (w/v) dry skim milk diluted in TBS (pH 7.5) with 0.05% Tween® 20 (TBS-T; Sigma) for 1 h. After washing with TBS-T, the membranes were incubated overnight at 4 °C with primary antibody diluted in TBS-T with the addition of 5% BSA (GenDEPOT) on a rocking platform. After washing with TBS-T, the membranes were incubated with horseradish peroxidase–conjugated secondary antibody diluted in TBS-T for 1 h. After washing with TBS-T, the membranes were developed using Immobilon Western chemiluminescent horseradish peroxidase substrate (Merck) and visualized using the Fusion Solo 6X system (Vilber). The primary antibodies used were as follows: rabbit anti-GFAP (Abcam; ab7260), mouse anti-CD44 (Novusbio; NBP1-47386), mouse anti-GFP (Santa Cruz; sc-9996), mouse anti–β-actin (Sigma; A5441), rabbit anti-PITX1 (Abcam; ab70273), and rabbit anti-SOX9 (Cell Signaling; 82630).
Luciferase assay
BE(2)C cells were plated at a density of 1.5 × 105 cells/well in 24-well plates. The following day, the cells were transfected using Lipofectamine 3000 (Invitrogen) with 0.5 μg of DNA (pGL3 basic reporter genes, 200 ng; pLV-Venus-PITX1 expression gene, 100 ng; and RSV–β-Gal normalized gene, 200 ng). After 24 h of incubation, the medium was changed to DMEM with 10% fetal bovine serum (Gibco). After 48 h of transfection, luciferase assay was performed using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol. A portion of the cell lysate was used for measurement of β-gal activity. Ortho-nitrophenyl-β-D-galactopyranoside (Sigma) solution was added to a portion of the cell lysate, and the solution was incubated at 37 °C for 15 min. Measurements of luciferase and β-gal activities were conducted using the VICTORTM × 3 plate reader (PerkinElmer).
PITX1 homeodomain purification
BL21(DE3) competent Escherichia coli were transfected with target DNA (pGEX-4T-1 GST-PITX1 homeodomain) and induced with 0.1 mm isopropyl β-d-thiogalactopyranoside at 18 °C overnight. The bacteria were resuspended in lysis buffer (50 mm Tris-HCl pH 7.5, 200 mm NaCl, and 3 mm β-mercaptoethanol), and the homeodomain was extracted with GST resin purification followed by thrombin digestion. The proteins were purified by ion-exchange chromatography and HitrapQ column size-exclusion chromatography. The purified proteins were separated by SDS-PAGE. The SDS-PAGE gel was fixed and stained in Sun-Gel staining solution (SPL solution) for 1 h with gentle agitation. Then the PAGE gel was washed with distilled water until the background of the gel was fully destained. Protein purification was conducted by Bonsu Ku laboratory. The amino acid sequence of PITX1 homeodomain is as follows: QRRQRTHFTSQQLQELEATFQRNRYPDMSMREEIAVWTNLTEPRVRVWFKNRRAKWRKRE.
EMSA
EMSA was performed using the LightShift® chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer's protocol. Biotinylated probe and unlabeled probe were made by Bioneer. 1 ng of biotinylated probe was incubated with 500 ng of purified PITX1 protein and 500 ng or 1000 ng of unlabeled probe for 20 min at room temperature. The reaction mixtures included 50 ng of poly(dI·dC). The reacted probes were separated on 6% polyacrylamide gels and transferred to Biodyne™ B nylon membranes (Thermo Scientific). Biotinylated probe was detected using the Fusion Solo 6X system (Vilber).
ChIP assay
Differentiating astrocytes were fixed in 1% formaldehyde (Sigma) for 10 min, washed with PBS, resuspended in lysis buffer, and centrifuged at 8000 rpm for 10 min. The nuclei were isolated with nuclei extraction buffer and sonicated using Ultrasonic Processors (Sonics Vibra-Cell™) at 20% amplitude for 3 min. The nuclei solution was centrifuged at 14,000 rpm for 20 min, and the concentration of supernatants was measured using Nanodrop One (Thermo Scientific). Then 50 ng of sample of the supernatant was used as input control, whereas 200 ng of sample was mixed with 2 μg of anti-PITX1 antibody (Abcam; ab70273) and rabbit IgG control (Santa Cruz; sc-2027) each, followed by incubation at 4 °C overnight. Protein A/G Plus agarose beads (Santa Cruz; sc-2003) were added to the samples and incubated at 4 °C for 3 h. Then the beads were washed twice with each of the following wash buffers (150 mm NaCl low-salt buffer, 500 mm NaCl high-salt buffer, 250 mm LiCl buffer, 10 mm Tris, and 1 mm EDTA buffer). The supernatant was removed, and the beads were resuspended in immunoprecipitation elution buffer (1% SDS, 1 mm EDTA, Tris-Hcl 10 mm). Additionally, input control samples were added with immunoprecipitation elution buffer. The bead mixture was then centrifuged at 14,000 rpm for 3 min, and the supernatants of bead and input samples were incubated at 65 °C overnight. 10 μl of proteinase K (Sigma) and 250 μl of Tris buffer were added to the samples and incubated at 42 °C for 2 h. 500 μl of phenol/chloroform/isoamyl alcohol (Sigma) was added to the sample and vortexed. The sample was centrifuged at 14,000 rpm at 4 °C for 10 min, and the supernatant was combined with 500 μl of chloroform/isoamylalcohol (Sigma) and vortexed. These samples were then centrifuged at 14,000 rpm at 4 °C for 10 min, and the supernatant was mixed with glycogen (Sigma) and sodium acetate (Sigma). Pure ethanol was added to the samples and vortexed. The samples were incubated at −20°C overnight. The samples were centrifuged at 14,000 rpm at 4 °C for 30 min, and the supernatant was removed. The precipitate was washed with 80% ethanol. The samples were then centrifuged at 14,000 rpm for 10 min at 4 °C, and the supernatant was removed. The pellet was dried for 10 min at room temperature and subsequently dissolved in Tris-EDTA buffer. The isolated DNA was analyzed by PCR assays using primers for the human SOX9 promoter and β-actin. The primer sequences were as follows: β-actin, forward, 5′-GAGCACAGAGCCTCGCCTTT-3′, and reverse, 5′-ACATGCCGGAGCCGTTGTC-3′; SOX9 (−201 to +14), forward, 5′-CTACCGTCCGCCCTTTG-3′, and reverse, 5′-GCTTTCGGCTCTCCAACTC-3′; and SOX9 (−1032 to −804), forward, 5′-GGAGCGTTTTGTCTGCGGT-3′, and reverse, 5′-GCCGACGCTCCTCAGTAA-3′.
Data availability
All data are contained within the article and the supporting information. The microarray array data were deposited into the NCBI-GEO database under accession number GSE153054.
Supplementary Material
This article contains supporting information.
Author contributions—J. S. B. data curation; J. S. B. validation; J. S. B., M. O., S. L., J.-E. G., Y. M., and B. K. investigation; J. S. B., W.-K. K., K.-J. O., E.-W. L., K.-H. B., S. C. L., and B.-S. H. writing-original draft; J. S. B. and B.-S. H. writing-review and editing; S. C. L. and B.-S. H. conceptualization; B.-S. H. project administration.
Funding and additional information—This work was partly supported by Korea Research Institute of Bioscience and Biotechnology Research Initiative Programme Grants KGM5321911 and KGM1121911 and the Korea National Research of Foundation Grants 2013M3A9A7046301 and 2017R1E1A1A01074745.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- hESC
- human embryonic stem cell
- EB
- embryoid body
- NPC
- neural progenitor cell
- PITX1
- paired-like homeodomain transcription factor 1
- SOX9
- sex-determining region Y box 9
- GFAP
- glial fibrillary acidic protein
- MOI
- multiplicity of infection
- bFGF
- basic fibroblast growth factor
- S100β
- S100 calcium-binding protein β
- VZ
- ventricular zone
- SVZ
- subventricular zone
- IZ
- intermediate zone
- HD
- homeodomain
- EMSA
- electrophoretic mobility shift assay
- En
- embryonic day n
- DAPI
- 4′,6-diamino-2-phenylindole
- TSS
- transcription start site.
References
- 1. von Bartheld C. S., Bahney J., and Herculano-Houzel S. (2016) The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J. Comp. Neurol. 524, 3865–3895 10.1002/cne.24040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sofroniew M. V., and Vinters H. V. (2010) Astrocytes: biology and pathology. Acta Neuropathol. 119, 7–35 10.1007/s00401-009-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phatnani H., and Maniatis T. (2015) Astrocytes in neurodegenerative disease. Cold Spring Harb. Perspect. Biol. 7, a020628 10.1101/cshperspect.a020628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garwood C. J., Pooler A. M., Atherton J., Hanger D. P., and Noble W. (2011) Astrocytes are important mediators of Aβ-induced neurotoxicity and Tau phosphorylation in primary culture. Cell Death Dis. 2, e167 10.1038/cddis.2011.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khakh B. S., and Sofroniew M. V. (2014) Astrocytes and Huntington's disease. ACS Chem. Neurosci. 5, 494–496 10.1021/cn500100r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu X. L., Long C. X., Sun L., Xie C., Lin X., and Cai H. (2010) Astrocytic expression of Parkinson's disease-related A53T α-synuclein causes neurodegeneration in mice. Mol. Brain 3, 12 10.1186/1756-6606-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krencik R., Weick J. P., Liu Y., Zhang Z. J., and Zhang S. C. (2011) Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 29, 528–534 10.1038/nbt.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Byun J. S., Lee C. O., Oh M., Cha D., Kim W. K., Oh K. J., Bae K. H., Lee S. C., and Han B. S. (2020) Rapid differentiation of astrocytes from human embryonic stem cells. Neurosci. Lett. 716, 134681 10.1016/j.neulet.2019.134681 [DOI] [PubMed] [Google Scholar]
- 9. Treisman J., Gönczy P., Vashishtha M., Harris E., and Desplan C. (1989) A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell 59, 553–562 10.1016/0092-8674(89)90038-X [DOI] [PubMed] [Google Scholar]
- 10. Driever W., and Nüsslein-Volhard C. (1988) The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104 10.1016/0092-8674(88)90183-3 [DOI] [PubMed] [Google Scholar]
- 11. Prince K. L., Walvoord E. C., and Rhodes S. J. (2011) The role of homeodomain transcription factors in heritable pituitary disease. Nat. Rev. Endocrinol. 7, 727–737 10.1038/nrendo.2011.119 [DOI] [PubMed] [Google Scholar]
- 12. Lanctôt C., Lamolet B., and Drouin J. (1997) The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development 124, 2807–2817 [DOI] [PubMed] [Google Scholar]
- 13. Lamonerie T., Tremblay J. J., Lanctôt C., Therrien M., Gauthier Y., and Drouin J. (1996) Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10, 1284–1295 10.1101/gad.10.10.1284 [DOI] [PubMed] [Google Scholar]
- 14. Szeto D. P., Ryan A. K., O'Connell S. M., and Rosenfeld M. G. (1996) P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc. Natl. Acad. Sci. U.S.A. 93, 7706–7710 10.1073/pnas.93.15.7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szeto D. P., Rodriguez-Esteban C., Ryan A. K., O'Connell S. M., Liu F., Kioussi C., Gleiberman A. S., Izpisúa-Belmonte J. C., and Rosenfeld M. G. (1999) Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 13, 484–494 10.1101/gad.13.4.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duboc V., and Logan M. P. (2011) Pitx1 is necessary for normal initiation of hindlimb outgrowth through regulation of Tbx4 expression and shapes hindlimb morphologies via targeted growth control. Development 138, 5301–5309 10.1242/dev.074153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song I., Jeong B. C., Choi Y. J., Chung Y. S., and Kim N. (2016) GATA4 negatively regulates bone sialoprotein expression in osteoblasts. BMB Rep. 49, 343–348 10.5483/bmbrep.2016.49.6.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao Z., Dou C., Li J., Tang X., Xiang J., Zhao C., Zhu L., Bai Y., Xiang Q., and Dong S. (2016) Cordycepin inhibits chondrocyte hypertrophy of mesenchymal stem cells through PI3K/Bapx1 and Notch signaling pathway. BMB Rep. 49, 548–553 10.5483/bmbrep.2016.49.10.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolt C. C., Lommes P., Sock E., Chaboissier M. C., Schedl A., and Wegner M. (2003) The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang P., Lee H. K., Glasgow S. M., Finley M., Donti T., Gaber Z. B., Graham B. H., Foster A. E., Novitch B. G., Gronostajski R. M., and Deneen B. (2012) Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74, 79–94 10.1016/j.neuron.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shaltouki A., Peng J., Liu Q., Rao M. S., and Zeng X. (2013) Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells 31, 941–952 10.1002/stem.1334 [DOI] [PubMed] [Google Scholar]
- 22. Swistowski A., Peng J., Han Y., Swistowska A. M., Rao M. S., and Zeng X. (2009) Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS One 4, e6233 10.1371/journal.pone.0006233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eng L. F., and Ghirnikar R. S. (1994) GFAP and astrogliosis. Brain Pathol. 4, 229–237 10.1111/j.1750-3639.1994.tb00838.x [DOI] [PubMed] [Google Scholar]
- 24. Pekny M., and Pekna M. (2004) Astrocyte intermediate filaments in CNS pathologies and regeneration. J. Pathol. 204, 428–437 10.1002/path.1645 [DOI] [PubMed] [Google Scholar]
- 25. Hol E. M., and Pekny M. (2015) Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell Biol. 32, 121–130 10.1016/j.ceb.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 26. Barger S. W., Wolchok S. R., and Van Eldik L. J. (1992) Disulfide-linked S100 β dimers and signal transduction. Biochim. Biophys. Acta 1160, 105–112 10.1016/0167-4838(92)90043-D [DOI] [PubMed] [Google Scholar]
- 27. Raponi E., Agenes F., Delphin C., Assard N., Baudier J., Legraverend C., and Deloulme J. C. (2007) S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia 55, 165–177 10.1002/glia.20445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai N., Kurachi M., Shibasaki K., Okano-Uchida T., and Ishizaki Y. (2012) CD44-positive cells are candidates for astrocyte precursor cells in developing mouse cerebellum. Cerebellum 11, 181–193 10.1007/s12311-011-0294-x [DOI] [PubMed] [Google Scholar]
- 29. Liu Y., Han S. S., Wu Y., Tuohy T. M., Xue H., Cai J., Back S. A., Sherman L. S., Fischer I., and Rao M. S. (2004) CD44 expression identifies astrocyte-restricted precursor cells. Dev. Biol. 276, 31–46 10.1016/j.ydbio.2004.08.018 [DOI] [PubMed] [Google Scholar]
- 30. Molyneaux B. J., Arlotta P., Menezes J. R., and Macklis J. D. (2007) Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437 10.1038/nrn2151 [DOI] [PubMed] [Google Scholar]
- 31. Del Río J. A., Martínez A., Auladell C., and Soriano E. (2000) Developmental history of the subplate and developing white matter in the murine neocortex: neuronal organization and relationship with the main afferent systems at embryonic and perinatal stages. Cereb. Cortex 10, 784–801 10.1093/cercor/10.8.784 [DOI] [PubMed] [Google Scholar]
- 32. Tabata H. (2015) Diverse subtypes of astrocytes and their development during corticogenesis. Front. Neurosci. 9, 114 10.3389/fnins.2015.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao X., Chen Y., Zhu Q., Huang H., Teng P., Zheng K., Hu X., Xie B., Zhang Z., Sander M., and Qiu M. (2014) Control of astrocyte progenitor specification, migration and maturation by Nkx6.1 homeodomain transcription factor. PLoS One 9, e109171 10.1371/journal.pone.0109171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park D., Xiang A. P., Mao F. F., Zhang L., Di C. G., Liu X. M., Shao Y., Ma B. F., Lee J. H., Ha K. S., Walton N., and Lahn B. T. (2010) Nestin is required for the proper self-renewal of neural stem cells. Stem Cells 28, 2162–2171 10.1002/stem.541 [DOI] [PubMed] [Google Scholar]
- 35. Marinaro C., Butti E., Bergamaschi A., Papale A., Furlan R., Comi G., Martino G., and Muzio L. (2011) In vivo fate analysis reveals the multipotent and self-renewal features of embryonic AspM expressing cells. PLoS One 6, e19419 10.1371/journal.pone.0019419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Infante C. R., Park S., Mihala A. G., Kingsley D. M., and Menke D. B. (2013) Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev. Biol. 374, 234–244 10.1016/j.ydbio.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ushita M., Saito T., Ikeda T., Yano F., Higashikawa A., Ogata N., Chung U., Nakamura K., and Kawaguchi H. (2009) Transcriptional induction of SOX9 by NF-κB family member RelA in chondrogenic cells. Osteoarthritis Cartilage 17, 1065–1075 10.1016/j.joca.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 38. Island M. L., Mesplede T., Darracq N., Bandu M. T., Christeff N., Djian P., Drouin J., and Navarro S. (2002) Repression by homeoprotein pitx1 of virus-induced interferon a promoters is mediated by physical interaction and trans repression of IRF3 and IRF7. Mol. Cell Biol. 22, 7120–7133 10.1128/mcb.22.20.7120-7133.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohira T., Kojima H., Kuroda Y., Aoki S., Inaoka D., Osaki M., Wanibuchi H., Okada F., Oshimura M., and Kugoh H. (2019) PITX1 protein interacts with ZCCHC10 to regulate hTERT mRNA transcription. PLoS One 14, e0217605 10.1371/journal.pone.0217605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farhy-Tselnicker I., and Allen N. J. (2018) Astrocytes, neurons, synapses: a tripartite view on cortical circuit development. Neural Dev. 13, 7 10.1186/s13064-018-0104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rolls A., Shechter R., and Schwartz M. (2009) The bright side of the glial scar in CNS repair. Nat. Rev. Neurosci. 10, 235–241 10.1038/nrn2591 [DOI] [PubMed] [Google Scholar]
- 42. Wyss-Coray T., Loike J. D., Brionne T. C., Lu E., Anankov R., Yan F., Silverstein S. C., and Husemann J. (2003) Adult mouse astrocytes degrade amyloid-β in vitro and in situ. Nat. Med. 9, 453–457 10.1038/nm838 [DOI] [PubMed] [Google Scholar]
- 43. Mandal A., Tipnis S., Pal R., Ravindran G., Bose B., Patki A., Rao M. S., and Khanna A. (2006) Characterization and in vitro differentiation potential of a new human embryonic stem cell line, ReliCellhES1. Differentiation 74, 81–90 10.1111/j.1432-0436.2006.00051.x [DOI] [PubMed] [Google Scholar]
- 44. Mertens J., Marchetto M. C., Bardy C., and Gage F. H. (2016) Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 17, 424–437 10.1038/nrn.2016.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davies S. J., Shih C. H., Noble M., Mayer-Proschel M., Davies J. E., and Proschel C. (2011) Transplantation of specific human astrocytes promotes functional recovery after spinal cord injury. PLoS One 6, e17328 10.1371/journal.pone.0017328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lepore A. C., Rauck B., Dejea C., Pardo A. C., Rao M. S., Rothstein J. D., and Maragakis N. J. (2008) Focal transplantation-based astrocyte replacement is neuroprotective in a model of motor neuron disease. Nat. Neurosci. 11, 1294–1301 10.1038/nn.2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krencik R., and Zhang S. C. (2011) Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 6, 1710–1717 10.1038/nprot.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klum S., Zaouter C., Alekseenko Z., Björklund A. K., Hagey D. W., Ericson J., Muhr J., and Bergsland M. (2018) Sequentially acting SOX proteins orchestrate astrocyte- and oligodendrocyte-specific gene expression. EMBO Rep. 19, e46635 10.15252/embr.201846635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott C. E., Wynn S. L., Sesay A., Cruz C., Cheung M., Gomez Gaviro M. V., Booth S., Gao B., Cheah K. S., Lovell-Badge R., and Briscoe J. (2010) SOX9 induces and maintains neural stem cells. Nat. Neurosci. 13, 1181–1189 10.1038/nn.2646 [DOI] [PubMed] [Google Scholar]
- 50. Martini S., Bernoth K., Main H., Ortega G. D., Lendahl U., Just U., and Schwanbeck R. (2013) A critical role for Sox9 in notch-induced astrogliogenesis and stem cell maintenance. Stem Cells 31, 741–751 10.1002/stem.1320 [DOI] [PubMed] [Google Scholar]
- 51. Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L. L., Mok J., Lee M. J., and Haydon R. C. (2014) The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 1, 149–161 10.1016/j.gendis.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prévostel C., and Blache P. (2017) The dose-dependent effect of SOX9 and its incidence in colorectal cancer. Eur. J. Cancer 86, 150–157 10.1016/j.ejca.2017.08.037 [DOI] [PubMed] [Google Scholar]
- 53. McCaffrey L. M., and Macara I. G. (2009) The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23, 1450–1460 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information. The microarray array data were deposited into the NCBI-GEO database under accession number GSE153054.