Abstract
The evolutionarily conserved multiprotein Mediator complex (MED) serves as an interface between DNA-bound transcription factors (TFs) and the RNA Pol II machinery. It has been proposed that each TF interacts with a dedicated MED subunit to induce specific transcriptional responses. But are these binary partnerships sufficient to mediate TF functions? We have previously established that the Med1 Mediator subunit serves as a cofactor of GATA TFs in Drosophila, as shown in mammals. Here, we observe mutant phenotype similarities between another subunit, Med19, and the Drosophila GATA TF Pannier (Pnr), suggesting functional interaction. We further show that Med19 physically interacts with the Drosophila GATA TFs, Pnr and Serpent (Srp), in vivo and in vitro through their conserved C-zinc finger domains. Moreover, Med19 loss of function experiments in vivo or in cellulo indicate that it is required for Pnr- and Srp-dependent gene expression, suggesting general GATA cofactor functions. Interestingly, Med19 but not Med1 is critical for the regulation of all tested GATA target genes, implying shared or differential use of MED subunits by GATAs depending on the target gene. Lastly, we show a direct interaction between Med19 and Med1 by GST pulldown experiments indicating privileged contacts between these two subunits of the MED middle module. Together, these findings identify Med19/Med1 as a composite GATA TF interface and suggest that binary MED subunit–TF partnerships are probably oversimplified models. We propose several mechanisms to account for the transcriptional regulation of GATA-targeted genes.
Keywords: GATA transcription factors, Serpent, Srp, Pannier, Pnr, mediator complex, Med19, Med1, cofactor, Drosophila development, thoracic closure, DNA-binding domain, Activation domain, GATA transcription factor, gene regulation, transcription coregulator, transcription regulation, drosophila genetics
Transcription, the first stage of gene expression, is a fundamental cellular process governed by the binding of sequence-specific transcription factors (TFs) at gene enhancers, inducing the recruitment/activation of the general RNA Polymerase II (Pol II) machinery at gene promoters. In eukaryotes, TFs do not bind directly the Pol II enzyme but instead contact a multisubunit complex called Mediator (MED), serving as a physical and functional interface between DNA-bound TFs and PolII (for review see Refs. 1–3). Although TF DNA-binding specificity has been largely decoded, how TFs interact with the Mediator complex has been less extensively studied, and it is not clear whether each TF binds a specific MED subunit or whether TF–MED interactions obey more complex rules.
Mediator is an evolutionarily conserved complex composed of 25 to 30 distinct proteins distributed in four modules: Head, middle, and tail forming the core MED, and a separable regulatory Cdk8 kinase module (CKM) (1). Despite a general role of the Mediator complex in regulating transcription, some MED subunits display striking functional specificities, as exemplified by their differential requirements for cell viability (4, 5), their involvement in specific human diseases (6, 7), or their roles in given developmental processes (8–10). It has been proposed that MED subunit specificity comes from their ability to contact specific transcription factors and mediate their regulatory activity (11, 12). For example, specific interactions have been demonstrated between Med15 and SMAD transcription factors in Xenopus (13), Med23 and RUNX2 in mice (14), Med12 and Gli3 in mammalian cells (15), Med19 and REST in mammals and Med19 and HOX developmental regulators in Drosophila (16, 17), or also between Med1 and hormone nuclear receptors or GATA TF families in mammalian cells (18, 19).
GATA transcription factors represent a good model to analyze interaction between TFs and Mediator subunits. The mammalian GATA TF family comprises six members (GATA1–6), shown to specifically interact with the Med1 Mediator subunit (20). They have conserved homologs between both vertebrates and invertebrates (21) and contain two highly conserved zinc finger (ZF) domains. The C-terminal one (C-ZF) is both necessary and sufficient for sequence-specific DNA binding at WGATAR genomic sites, whereas the N-terminal ZF (N-ZF) appears only to modulate DNA-binding affinity (22) and has been involved in direct interactions with GATA cofactors (23–26). Mammalian GATAs are key regulators of developmental processes: GATA1, -2, and -3 are crucial hematopoietic TFs whereas GATA4, -5, and -6 control cardiac development, among other functions (21). Interestingly, among the five GATA TFs encoded by the Drosophila genome, only Serpent (Srp), is a bona fide hematopoietic GATA factor, whereas Pannier (Pnr) is involved in cardiac development (27). Pnr activity is also crucial during central thorax patterning and dorsocentral (DC) mechanosensory bristle formation, and it has been studied in depth in this context (28–30). Within the wing imaginal disc, the Pnr TF directly activates proneural genes of the achaete-scute complex in the dorsocentral cluster, which gives rise to the DC bristles (28). In addition, Pnr activates the wingless gene in a strip of cells of the presumptive thorax (31).
In a genome-wide RNAi screen in Drosophila cultured cells we identified a set of MED subunits as modulators of GATA/Serpent–induced transactivation, among which were Med12, Med13, Med1, and Med19 (32). This work further showed that Med12 and Med13 subunits are required in vivo for Srp-driven developmental processes, but we were unable to detect direct physical interaction with Srp in vitro, suggesting that GATA/Srp may recruit the Mediator complex by contacting other MED subunits. Indeed, we recently showed that Med1 mediates GATA TFs function in Drosophila (33). Med1 does interact physically with both Pnr and Srp GATA TFs, through their conserved zinc finger region. Furthermore, in vivo experiments showed that Med1 is involved in Srp-driven hematopoiesis and Pnr-driven thorax differentiation and is required for Srp and Pnr target gene expression in the corresponding tissues. These data established that the Med1 GATA cofactor activity is evolutionarily conserved and involves the GATA N- and C-zinc finger domains in both mammals and Drosophila. Nevertheless, we also showed that Drosophila Med1 is not critical for wingless-induced transactivation by Pnr, raising the possibility that other MED subunits could mediate some GATA TFs functions.
Here, we reveal that another MED subunit, Med19, also acts as a GATA coactivator. Med19 mutants phenocopy pnr loss-of-function and extinguish the expression of both Pnr target genes achaete and wg, whereas Med1 mutants were previously shown to affect only achaete expression. Using immunoprecipitation, pulldown, and bimolecular fluorescence complementation (BiFC) techniques, we establish that Med19 physically interacts with Pnr in cellulo, in vivo, and in vitro through its C-ZF domain. Med19 also interacts physically with GATA/Srp, suggesting that Med19 acts as a generic GATA cofactor. Moreover, we show that both Med1 and Med19 jointly regulate a series of Srp target genes in Drosophila cultured cells. Finally, in vitro experiments revealed that Med1 and Med19 physically interact through the Med1 domain which is conserved throughout eukaryotes. Taken together, our results show that GATA-driven regulatory functions in Drosophila require two MED complex subunits, Med19 in all tested cases and Med1 in a majority. The evolutionary conservation of Med19 and GATA interacting domains suggests that Med19 may play a conserved GATA cofactor function in mammals.
Results
Drosophila Med19 is required for notum morphogenesis, bristle development, and GATA/pannier target gene activation
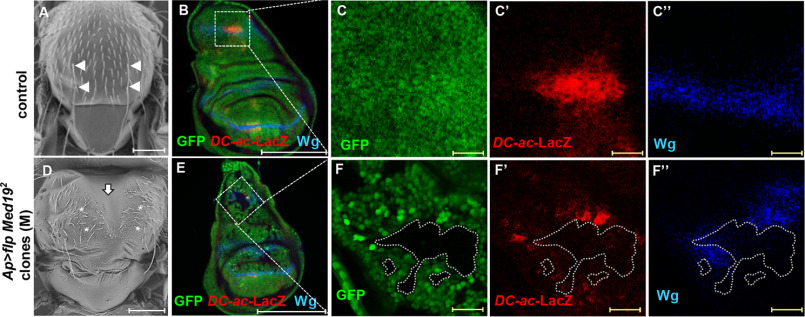
Our whole-genome dsRNA screen in Drosophila cultured cells identified Med19 as one MED subunit capable of modulating Srp TF-induced transactivation ex vivo (32). This led us to ask whether and how Med19 could interact with GATA TFs in vivo. To this end, we generated Med19 mutant clones in the larval wing imaginal disc, which gives rise to adult thoracic structures whose proper development depends on GATA/Pnr activity. Flies bearing Med19- clones displayed specific phenotypes in the thorax, including thoracic cleft and loss of DC mechanosensory macrochaetes (Fig. 1, A and D), typical of pnr loss-of-function (29, 30). We observed similar phenotypes upon expression of RNAi against Med19 in the apterous (ap) domain encompassing all the presumptive notum (Fig. S1, A and B). To investigate the functional relationship between Med19 and Pnr, we first examined pnr gene expression in Med19-deficient wing discs by FISH and observed that pnr is expressed in Med19-depleted wing discs (Fig. S1, C–F). Thus, Med19 mutant phenotypes cannot be explained by a loss of pnr expression. To further analyze the functional relationship between Med19 and Pnr, we then examined GATA/Pnr TF activity in Med19 loss-of-function clones by analyzing the expression of known Pnr target genes. Compared with WT cells shown in Fig. 1, B–C′′, we observed that both wingless (wg) and achaete (ac) expression was cell autonomously lost in Med19−/− cells (Fig. 1, E–F′′), indicating that Med19 is required for the expression of both Pnr target genes. Note that ac expression has been visualized by a DC-ac-lacZ reporter gene which is directly activated upon Pnr binding to the DC ac enhancer (28).
Figure 1.
Med19 is required for GATA/Pannier target gene expression. A and D, Drosophila thoraces: (A) WT or (D) displaying Med19−/− clones. Arrowheads point to DC bristles in A and asterisks to their expected position in D. The vertical arrow points to the thoracic cleft. Scale bars, 200 μm. B–F, expression of Pannier target genes in the dorsal compartment of control wing discs (B and C) or in Med19−/− mitotic clones (GFP-) (E and F). Scale bar, 200 μm. The expression of DC-ac-LacZ reporter and Wg protein are revealed with anti-βgal (red, C′, F′), and anti-Wg (blue, C′′, F′′) antibodies. C and F, magnifications of the DC region are shown. B and E, white scale bars, 200 μm. C and C′′ and F and F′′, yellow scale bars (in), 20 μm.
These data show that Med19 is cell autonomously required for Pnr activity but not for Pnr expression, suggesting that it could act as a GATA/Pnr cofactor.
Drosophila Med19 interacts physically with the pannier GATA TF
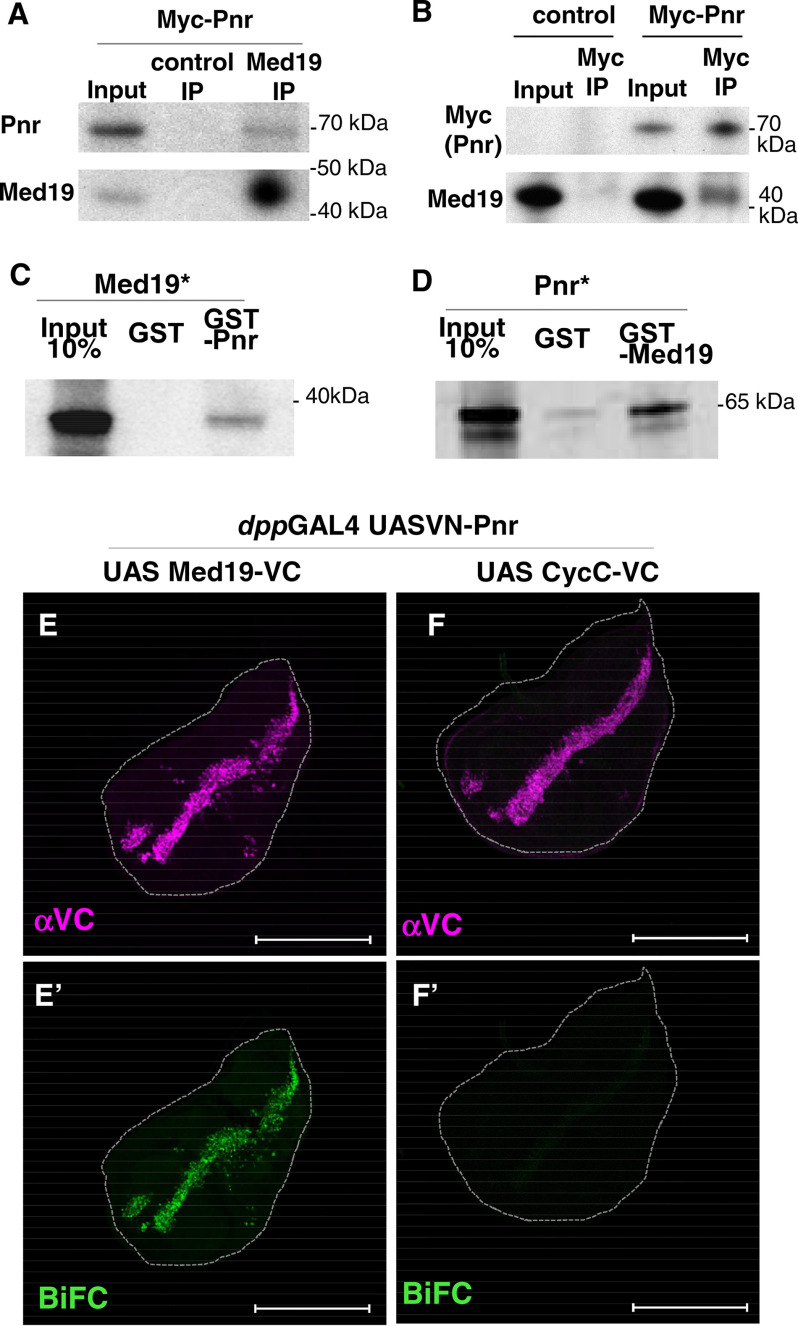
We investigated whether GATA/Pnr transcription factor and Med19 physically interact by using three independent experimental approaches: Co-immunoprecipitation from cultured cells, in vitro pulldown and in vivo Bimolecular Fluorescence Complementation (BiFC) interaction tests. We first tested whether Pnr-MED complexes actually form within Drosophila cells by performing co-immunoprecipitations experiments on total protein extracts from cultured cells expressing a functional Myc-tagged Pnr form. We observed that Pnr co-precipitated with endogenous Drosophila Med19 (Fig. 2A). In the reverse experiment, endogenous Med19 protein co-precipitated with Myc-tagged Pnr protein (Fig. 2B). These data provide complementary evidence for the formation of Med19-GATA complexes in Drosophila cells.
Figure 2.
Med19 physically interacts with GATA/Pnr in cellulo, in vitro, and in vivo. A and B, co-IP experiments from S2 cells transfected with pAct-Myc-Pnr using anti Med19 antibody or preimmune serum (control IP) (A). The reverse experiment (B) was performed using anti-Myc beads from control cells or cells transfected with pAct-Myc-Pnr. Western blot assays using αMed19, αPnr, or αMyc antibodies are shown. C and D, autoradiographs from GST pulldown assays between GST-Pnr and 35S-labeled (asterisk) in vitro–translated Med19 (C), and between GST-Med19 and in vitro–translated 35S-Pnr (D). E–F′, BiFC assays using VN-Pnr and either Med19-VC (E) or CycC-VC (F). Expression of fusion proteins along the A/P boundary of the wing disc is under the control of dppGAL4 driver. Immunostaining shows expression of VC constructs (magenta, E and F), and BiFC signals are shown in green (E′ and F′). Scale bars, 200 μm.
To investigate whether Med19–Pnr interaction is direct, we tested the ability of Med19 and Pnr proteins to bind each other physically in vitro through pulldown assays with GSH S-transferase (GST) fusion proteins. In vitro–produced Med19 readily bound full-length recombinant GST-Pnr (Fig. 2C), and vice versa (Fig. 2D). These results show that Med19 and Pnr can interact physically in the absence of any other Drosophila MED subunits.
We then used BiFC (16, 34) to analyze Med19–GATA interaction in vivo. Based on fusing N- and C-terminal portions (VN and VC) of the GFP-variant Venus protein with two proteins of interest respectively, this technique allows the reconstitution of a fluorescent Venus protein if the two candidate proteins are close enough within the cell. We used the dppGAL4 driver (Gal4/UAS system (35)) to co-express VN-Pnr with either Med19-VC or another MED subunit fusion, CycC-VC, along the antero/posterior frontier of the wing imaginal disc (Fig. 2, E and F). The co-expression of VN-Pnr and Med19-VC resulted in a clear BiFC signal, whereas the control VN-Pnr/CycC-VC combination gave a very low signal (Fig. 2, E′ and F′), even though CycC-VC and Med19-VC proteins were expressed at similar levels (Fig. 2, E and F). These data indicate that the BiFC technique discriminates specific interactions between different subunits within the MED complex and that Med19 and Pnr are in close proximity in the nucleus of living cells. Of note, the BiFC signal was observed in the entire dppGAL4 expression domain, including the wing pouch where endogenous GATA/Pnr is not expressed, showing that the Pnr-Med19 interaction can occur at ectopic locations independently of tissue-specific Pnr partners, thus providing further support for a direct molecular interaction in vivo.
Collectively, in cellulo, in vitro, and in vivo data support a direct physical interaction between the GATA/Pnr transcription factor and the Med19 Mediator subunit. Together with our previous results (33), these data suggest that the Pnr TF can interact with the entire MED complex via a direct molecular contact with the Med19 subunit in addition to or in place of the Med1 subunit.
Med19 core and HIM domains bind the C-zinc finger domain of GATA/pnr
We previously showed that Med1 directly interacts with the dual zinc finger domains of Pnr (33). We therefore decided to characterize interacting domains within Pnr and Med19 to determine whether Med19 and Med1 interact with the same Pnr domain.
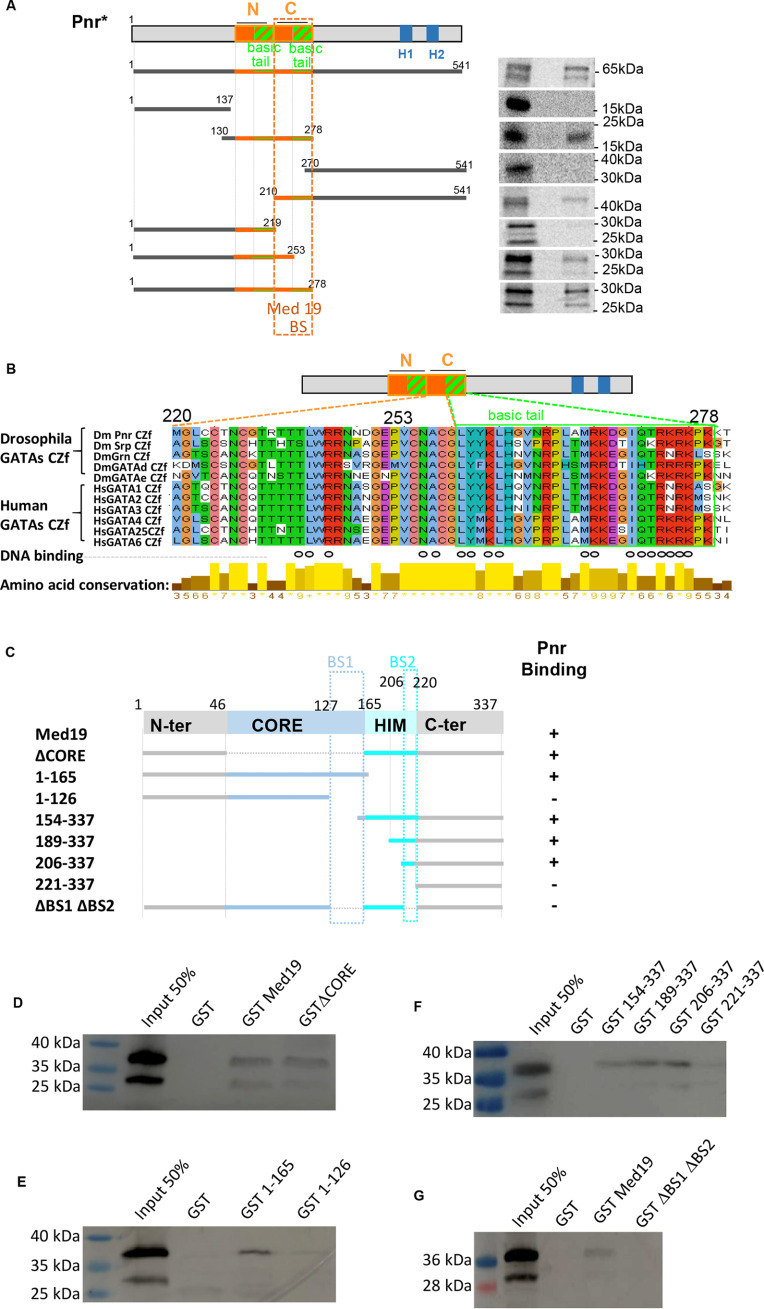
We first looked for the Med19-interacting domain(s) within the GATA/Pnr protein using full-length GST-Med19 as a bait (Fig. 3A). Pnr was split into three parts: The poorly evolutionarily conserved N-terminal region (amino acids (aa) 1–137), the strongly conserved central region spanning the two zinc fingers, N- and C-ZF, and the divergent C-terminal region containing two amphipathic α helices, H1 and H2. Only the ZF-containing region (aa 130–278) displayed significant binding. When cutting full-length Pnr into two halves separating the two zinc finger domains, binding was observed only with the C-ZF–containing part (Fig. 3A), suggesting that C-ZF mediates binding of Pnr to Med19. Consistently, the ability of the N-ZF–containing half of Pnr to bind Med19 was recovered when we added back the C-ZF proper (aa 220–253) containing the four zinc-chelating cysteines forming the finger structure. Interestingly, binding was increased when the C-ZF proper was extended by its neighboring C-terminal 25 amino acids (basic tail motif, aa 253–278) (Fig. 3B). Sequence alignment of Drosophila and mammalian GATAs indicates that the C-ZF basic tail has been strongly conserved during evolution, especially at positions shown to participate in DNA binding (open circles in Fig. 3B) (36, 37). Together, these experiments indicate that the entire Pnr C-ZF domain, zinc-finger proper and adjacent basic tail, is necessary for optimal Med19 binding.
Figure 3.
The GATA/Pnr C-zinc finger domain interacts with two conserved Med19 domains. A, GST pulldown assays delimitating Med19 interacting domains within Pnr. Left, schematic representation of GATA/Pnr and Pnr fragments generated to probe for binding to full-length GST-Med19. N and C show proper zinc fingers (orange boxes) and their basic tails (hatched green and orange boxes). H1 and H2 show amphipathic α helices. Right, corresponding autoradiographs from GST pulldown experiments. The critical domain for strong binding is narrowed down to Pnr amino acids 220–278 (Med19 BS dotted rectangle) comprising C-ZF proper zinc finger and its basic tail. B, sequence alignment shows that this domain is highly conserved in Drosophila GATA factors (Srp, Grn, GATAd, and GATAe) as well as human GATA factors (GATA1–6). Open circles denote residues participating in DNA binding (1, 2). Level of each amino acid conservation is represented underneath. C, schematic representation of the full-length Med19 protein and the Med19 subdomains generated as GST fusions. Pluses and minuses summarize Pnr GST pulldown results, based on HA-Pnr1-291 detection on Western blots shown in (D–G). The two distinct domains of Med19 which mediate binding of Pnr are squared in blue and denoted BS1 and BS2 in C.
In the reciprocal experiment, we identified the GATA/Pnr interacting domain within Med19. Our prior analysis of Drosophila Med19 function and evolutionary conservation within the eukaryotic kingdom (16, 38) allowed us to define four structural domains: A conserved MED-anchoring “CORE” region, an animal-specific basic HOX homeodomain–interacting motif (HIM) and two less well-conserved N- and C-terminal regions. To investigate which protein domain(s) is (are) required for Pnr binding, we tested the ability of in vitro translated Pnr1–291 to bind a series of GST-Med19 truncated forms (Fig. 3, C–G). A Med19 protein deleted for its evolutionarily conserved CORE domain (ΔCORE) still bound Pnr1–291 (Fig. 3D). Binding was also retained after truncating both C-terminal and HIM domains but was abolished if the deletion included the C-terminal end of the CORE domain (aa 126–165) (Fig. 3E). Deletions starting from the Med19 N terminus indicated that a truncated protein containing HIM and C-terminal domains also interacts with Pnr 1-291 (Fig. 3F). Further deletions revealed that one fragment of HIM from aa 206–220 was critical for Pnr binding in the absence of the CORE domain. Taken together, our data suggest the presence of two Pnr binding sites within Med19, aa 126–165 of the CORE (BS1), and aa 206–220 of the HIM domain (BS2) (Fig. 3C). To further assess their implication, we deleted both BS1 and BS2 in an otherwise full-length Med19 protein. As shown in Fig. 3G, the ΔBS1-BS2 mutant no longer bound GATA/Pnr, indicating that the presence of either BS1 or BS2 is essential for interacting with Pnr.
In conclusion, our in vitro binding assays indicate that the GATA/Pnr C-zinc finger domain, including its basic tail, binds two separate domains within the evolutionarily conserved Med19 CORE and HIM regions. Med19 appears only to bind the C-ZF, whereas Med1 has been shown to bind both GATA/Pnr C-ZF or N-ZF domains in vitro (33).
Med19 also physically interacts with GATA/Srp
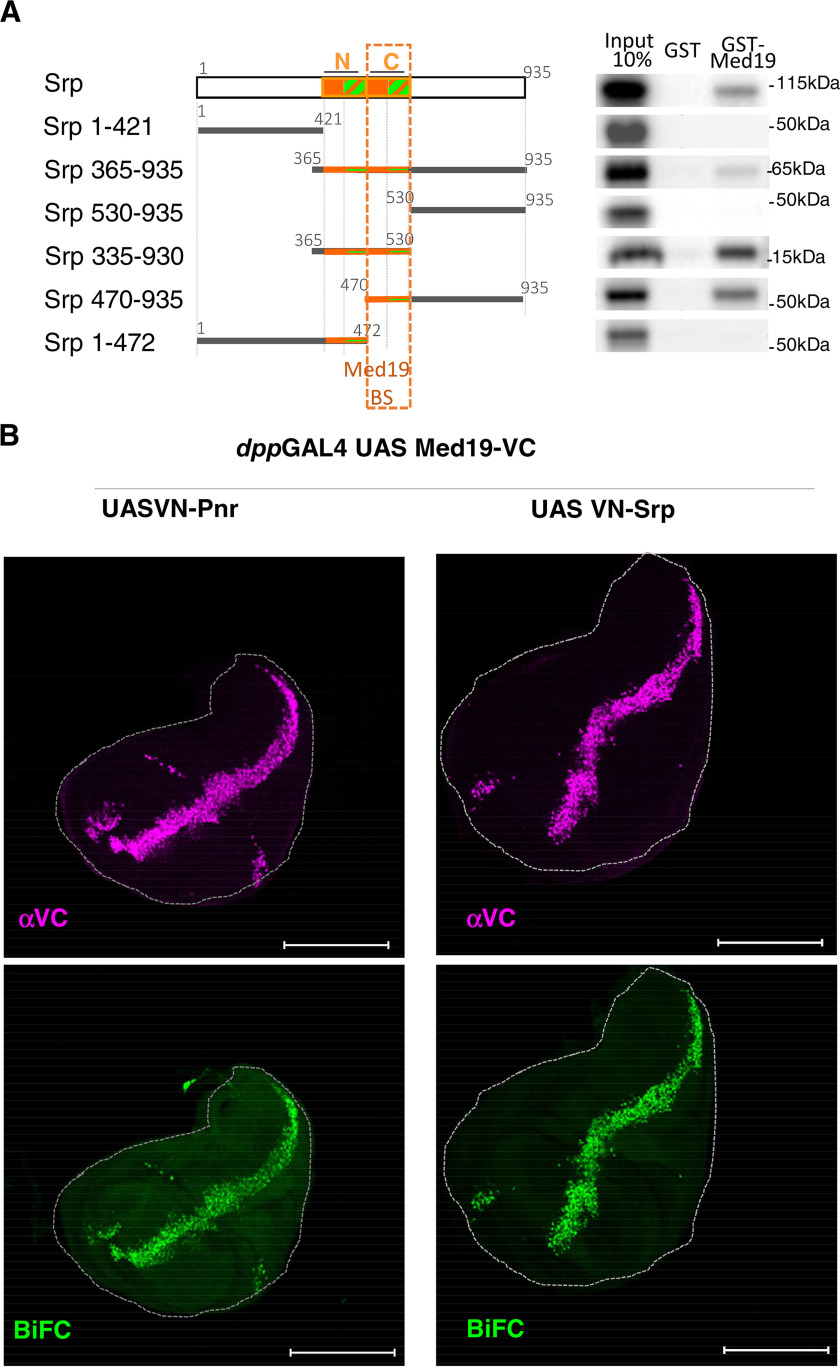
To investigate whether Med19 is a general GATA cofactor, we tested if it is able to interact with Srp, another Drosophila GATA TF family member. First, we used similar GST pulldown assays (Fig. 4A). They showed that recombinant GST-Med19 protein bound in vitro–translated full-length GATA/Srp protein. As previously shown for GATA/Pnr, when assaying Srp truncated forms, binding was only retained with the ZF-containing middle part. Splitting the Srp protein in two halves and separating both zinc finger domains indicated that only the C-ZF is involved in binding Med19 (Fig. 4A), as it is also the case for GATA/Pnr (Fig. 3E).
Figure 4.
Med19 physically interacts with GATA/Srp. A, left, schematic representation of GATA/Srp and the multiple Srp fragments generated to probe for binding to full-length GST-Med19. Autoradiographs from GST pulldown experiments are shown on the right. Again, the critical binding domain is restricted to the GATA C-ZF domain containing the zinc finger proper (orange square) and its basic tail (hatched green and orange boxes). B, BiFC assays using expression of Med19-VC with either VN-Pnr or VN-Srp under the control of the dppGAL4 driver. Immunostaining shows similar expression of VC constructs (magenta), and strong BiFC signals (green) with Pnr and also with Srp. Scale bars, 200 μm.
To test whether Med19-Srp interaction also occurs in vivo, we used again the BiFC experimental approach. Upon expression of Med19-VC with VN-tagged Srp in the dpp expression domain, we observed a strong BiFC signal, similar to what we obtained with Pnr-VN (Fig. 4B), indicating that Med19 and GATA Srp indeed interact in vivo.
Altogether, these results show that, Med19 interacts in vivo and in vitro with both Pnr and Srp, suggesting that Med19 is a general GATA cofactor. This interactions occurs via the GATA family–defining C-zinc finger domain.
Med19 shares GATA-cofactor functions with the Med1 mediator subunit
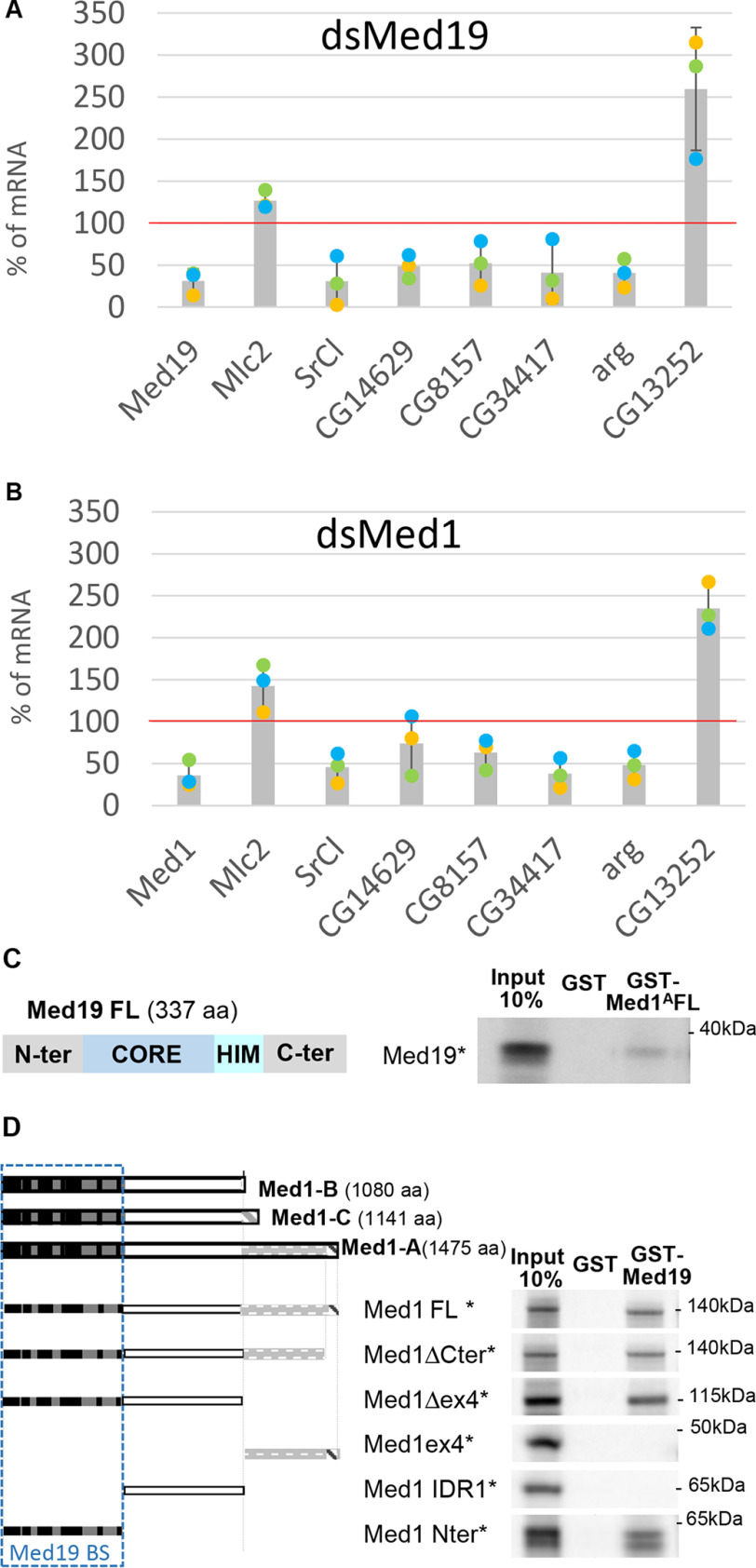
We previously showed that Med1, another subunit of the MED middle module, is required for Pnr and Srp TF activities in vivo and interacts directly with Srp and Pnr, in this case through both their N- and C-zinc finger domains (33). Our new data showing that Med19 can also act as GATA cofactor thus raises the question of the respective roles of Med19 and Med1 in GATA-driven transcriptional regulation. Concerning Pnr-dependent transcriptional activity, we have shown that Med19 is cell autonomously required for DC-ac-lacZ reporter expression like Med1, whereas wg expression requires Med19, but not Med1. This prompted us to consider each GATA target gene as a particular case that could involve interaction with both MED subunits or with Med19, or possibly Med1 alone. Kuuluvainen et al. (39) identified a set of Srp target genes in Drosophila S2 cells, which can be used to test the impact of Med1 or Med19 mRNA depletion. Here, we quantified the expression of six Srp target genes: SrCl, CG14629, CG8157, arg, and CG34417, which are activated (positive targets), and CG13252, which is repressed by Srp, using real-time quantitative PCR (RT-qPCR) in control, Med1-, or Med19-mRNA–depleted S2 cells. Quantification of mRNAs coding for the myosin light chain (Mlc2) served as a control for housekeeping transcription. As shown in Fig. 5A, in cells depleted for Med19 mRNA, expression of the five activated Srp target genes SrCl, CG14629, CG8157, arg, and CG34417 was significantly down-regulated and the Srp-repressed target gene CG13252 was instead up-regulated, indicating that Med19 is required in cellulo for GATA/Srp transcriptional activity. In cells depleted for the Med1 transcript (Fig. 5B), expression of Srp target genes followed the same trend than after Med19 mRNA depletion, although less efficiently.
Figure 5.
Med19 and Med1 are both required for GATA/Srp target genes expression and interact physically. A and B, real-time quantitative PCR analysis of mRNA expression in Med19-depleted cells (dsMed19, A), or Med1-depleted cells (dsMed1, B). The control cultured cells treated with dsGFP were used as reference (100%), and gapdh1 as an internal normalization gene. Each dot represents the result of one of three independent biological experiment (yellow, green, and blue dots). Bar graph indicates the mean and S.D. The experiment was also reproduced with a second dsRNA against Med1 and Med19. C, GST pulldown with GST-Med1A and full-length Med19 reveal direct interaction between the two MED subunits. D, Left, schematic representation of the three Med1 protein isoforms (Med1-A, -B, and -C) and Med1 fragments generated to probe for binding to full-length GST-Med19. The N-terminal region (darker gray rectangle) comprises short evolutionarily conserved motifs (black boxes) and correspond essentially to the yeast Med1 orthologue (38). The middle (white) and C-terminal (light gray) regions emphasizes the divergent long metazoan-specific extensions except for a conserved C-terminal a helix (hatched box). Right, GST-pulldown assays narrow the Med19-binding domain to the highly conserved N-terminal portion of Med1 proteins.
Several conclusions can be drawn from these experiments: (i) Contrary to other MED components, Med1 and Med19 are not required for general PolII-dependent transcription given that some genes are unchanged or even up-regulated. (ii) Med19 and Med1 are both required for Srp-mediated gene regulation in cultured cells, seemingly on the same target genes and (iii) both for activation and repression.
Med19 and Med1 can interact directly
Given the functional implication of both Mediator subunits in mediating GATA activity, we lastly asked whether Med1 and Med19 proteins are able to interact physically using GST pulldown assays. As shown in Fig. 5C, a GST fusion of Med1 largest isoform A (Med1A) bound in vitro–translated full-length Med19. In the reverse experiment, purified GST-Med19 also bound in vitro–translated Med1A (Fig. 5D), showing that both proteins indeed bind to each other in vitro, in absence of other MED subunits.
We next sought to identify Med19-interacting domain(s) within the Med1 isoforms by analyzing truncated proteins. As shown in Fig. 5D, the Med19 interacting domain lies within the evolutionarily conserved Med1 N-terminal part, which has been proposed to be required for its incorporation within the MED complex (40) and is shared by the three Med1 isoforms. Conversely, Med1 isoform-specific parts are not required for Med19 interaction.
Taken together, GST pulldown data reveal a direct interaction between Med1 and Med19 which could not be anticipated from structural data (see Fig. 6) given that Med1 and Med19 have been proposed to lie in opposite parts of the Mediator complex middle module (41).
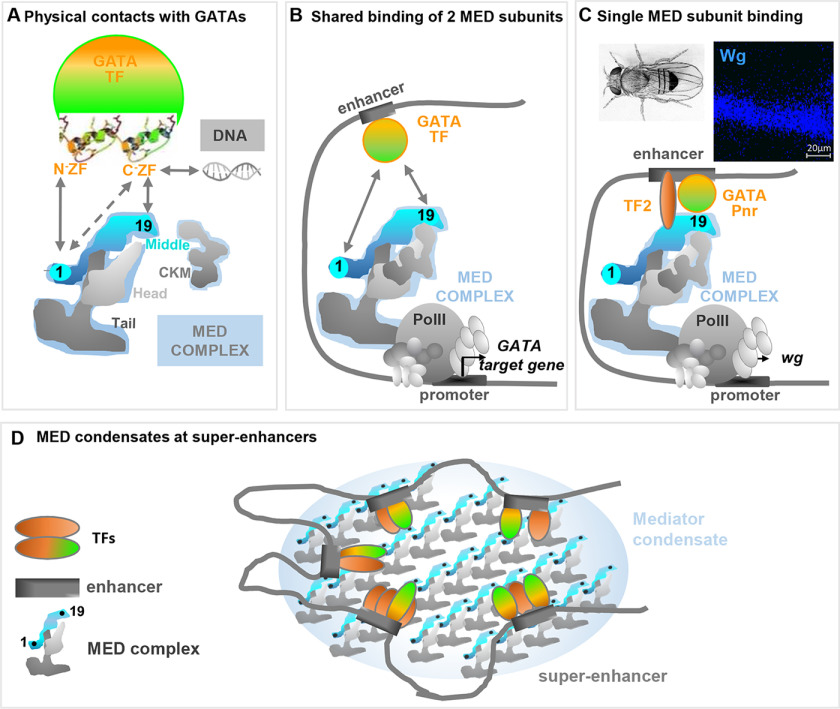
Figure 6.
Models of MED-GATA interaction involving both Med19 and Med1. A, in addition to Med1, another Mediator subunit belonging to the middle module, Med19, directly interacts with GATA factors. Whereas Med1 interacts with both the GATA N and C-ZF domains, Med19 only interacts with the GATA C-ZF that serves both as transactivation domain and DNA-binding domain. B, given that both Med19 and Med1 are required for the expression of most tested GATA Pnr and GATA Srp target genes, we propose that DNA-bound GATAs recruit the Mediator complex through direct contacts with both Med19 and Med1, thus enabling PolII machinery recruitment at GATA target genes. C, in some cases such as Pnr-dependent wg expression in Drosophila wing discs, Med1 is dispensable. We propose a model in which additional contacts between MED subunits and other TFs or GATA cofactors would render GATA-Med19-interaction sufficient to recruit the Mediator complex at GATA target genes. D, the binding affinity observed in vitro between Med1 and Med19 MED subunits, located at distant positions according to MED structural analyses, could reflect inter-MED instead of intra-MED interactions. Such interactions could promote the formation of MED complex condensates at phase-separated nuclear foci, previously shown to boost transcription at TF-bound super-enhancers.
Discussion
In this work, using molecular, cellular, and genetic analyses, we establish that Drosophila GATA factors' transcriptional activity depends on the Mediator complex subunit Med19, in addition to the previously characterized Med1 cofactor (33). Four main conclusions that are discussed below can be drawn from our results (i) Med19 interacts with the GATA C-ZF domain which also serves as the GATA DNA-binding domain. (ii) Med19 and GATA interacting domains are evolutionarily conserved, suggesting conserved Med19 cofactor functions in higher metazoans. (iii) Comparative analysis of Med19 and Med1 depletion indicates that Med19 but not Med1 is systematically required for GATA target gene expression, suggesting a differential use of MED subunits by GATAs depending on the target gene. (iv) Med1 and Med19 interact in vitro. Taken together our data allow us to propose new models of Mediator complex mechanism of action.
Overlapping DNA-binding and activation domains of GATA TFs
TFs minimally contain two domains: The DNA binding domains (DBDs), which have been extensively studied and allowed to define different TF families, and transcriptional activation domains, which link TFs to the RNA polymerase II machinery, and whose structure and characteristics are less well defined. GATA TFs are characterized by the presence of two ZFs which were, so far, thought to play distinct roles. While the C-ZF appeared to be dedicated to DNA binding, the N-ZF was shown to bind co-activators such as dLMO (42) and FOG (43). Our present data show that Med19 interacts specifically with the Pnr C-ZF (Fig. 6A). Full interaction requires both the zinc finger and its adjacent basic tail, which also contributes to DNA binding (36, 37). It is the first evidence that the Drosophila GATA C-ZF may play a dual role, in DNA binding and as an interface with MED subunit(s). Interestingly, the analysis of GATA ZF evolutionary conservation indicates that N- and C-ZF domains come from a duplication event of the C-ZF with its basic tail (44). Thus, this transactivation function of GATAs' DBD might represent an ancestral GATA function allowing minimal primitive GATAs, essentially composed of the DBD, to connect the MED complex and thus recruit the transcriptional machinery to regulate its target genes. They provide rationale why slightly extended GATA ZF domains are in some cases sufficient for transcriptional activities in vivo (45).
This dual activity of DBD is not restricted to GATA factors. We have previously shown that HOX TFs also contact Med19 through their DNA-binding homeodomain (16). Our data also corroborate results from a recent high-throughput approach, looking for transactivation domains of Drosophila transcription factors. This work shows that transactivation domains of several ZF and basic helix-loop-helix TFs overlap structured DNA-binding domains (46). Altogether, these results identify a novel class of TF characterized by overlapping transcriptional activation domains and DBD and suggest an emerging Med19 property as a dedicated cofactor directly connecting these TFs DNA-binding domains to the general PolII transcriptional machinery.
How is this dual function of DBDs achieved? Do DNA binding and transactivation functions use distinct or shared molecular determinants? Recent improvements of EM analyses could allow characterizing GATA molecular residues involved in MED versus DNA binding to try to separate the GATA DNA binding from GATA transactivation functions.
Evolutionarily conserved GATA-coactivator functions of Med19
Whereas Med1 is a known GATA cofactor both in mammals and in Drosophila (19, 33), the role of Med19 in mediating GATA transcription regulatory properties had never been investigated until now. Here we show that Drosophila Med19 binds GATA factors, via motifs lying within the evolutionarily conserved Med19 CORE and HIM domains (16). Both of these domains bind to the C-ZF domain of GATAs, which is a hallmark of GATA TF family suggesting that interaction with Med19 is likely to be conserved in mammals. Yet, Med1 depletion experiments in mammalian cultured cells induces defects in only a subset of GATA1-activated genes and does not prevent GATA1-dependent repression (47, 48). Furthermore, in studies of the different blood cell types produced by conditional Med1 knockout mice, Med1 appears to be critical for erythroid lineages which depend upon GATA1 function but is dispensable for hematopoietic stem cell production and T-cell development which require GATA2 and GATA3, respectively (49). Thus, despite being capable of binding all GATA factors in vitro, Med1 is not critical for all GATA functions, which suggests that (an)other MED subunit(s) also bind(s) GATAs to relay their regulatory signals to the PolII machinery. Considering the evolutionary conservation of interaction motifs within both GATAs and Med19, we argue that Med19 is a strong candidate as a GATA cofactor in mammals.
New models for GATA–MED interactions
Our data show that most Drosophila GATA target genes require both Med19 and Med1. How does this work? We showed that Med19 only interacts with the C-ZF domain, but Med1 can bind both GATA zinc finger domains (33), suggesting that Med1 and Med19 can simultaneously bind GATA factors (Fig. 6A). We thus propose that in the majority of cases where GATA-driven gene expression requires both Med19 and Med1, (Fig. 6B), enhancer-bound GATAs must directly contact both Med1 and Med19 subunits to recruit the Mediator complex and thus the PolII machinery at GATA target genes.
Some genes (e.g. wingless) require Med19 but not Med1. How does this kind of gene specificity occur mechanistically? We hypothesize that for these Med1-independant genes, other transcription factors might be involved in recruiting the MED (through other subunits) and hence overcome the necessity for Med1-GATA interaction (see Fig. 6C).
A future challenge will be to test these models by site-directed mutagenesis in vivo to assess the functional contribution of each GATA-MED contact. Nevertheless, this task is complicated because of overlapping DNA- and MED- interacting domains within GATAs (see above). On the other hand, Med19 CORE domain has also a dual function of MED anchorage and interaction with GATA. It thus requires prior structural analysis of molecular contacts to specifically target GATA-MED interaction without affecting essential DNA-binding activity of GATAs or Med19 ability to incorporate the MED-complex.
Another interpretation of our results could be that other subunits necessary for GATA target gene expression fall off from the complex when Med19 or Med1 are deleted or knocked down. However, structural analyses of MED complexes from yeast and mammalian cells lacking Med19 or Med1 indicate that global MED organization is unchanged (4, 50), and we therefore considered it very unlikely. Other lines of evidence indicate that complexes missing only Med19 can be isolated from Med19-depleted mammalian or yeast cells (51, 52). Altogether, these results suggest that Drosophila MED subunit loss is unlikely in Med19 or Med1 depletion conditions. Because we found a direct physical interaction between GATAs and Med1/Med19, we consider that the simplest explanation for our results is that the loss of either of these subunits is enough to abolish GATA regulatory signals.
Previous models of core MED structure-function analysis suggested that the middle and head modules contact the PolII enzyme and associated general transcription factors while the tail module interacts with sequence-specific TFs (2, 3). Our data show that two MED subunits of the middle module, Med1 and Med19, are able to bind GATA factors and are required for their function. They emphasize that MED should be viewed as a much more complex interface using multiple MED subunits to contact different TF combinations, thus mediating specific transcriptional responses.
Unexpected direct interaction between Med1 and Med19 middle module subunits
Modelization of MED spatial organization indicates that Med1 and Med19 are most likely located at opposite ends of the middle module, Med1 near the tail module and Med19 within the so-called hook domain proposed to anchor the separable CDK8 module (CKM) (4, 53) (Fig. 6A). Nevertheless, our data indicate that Med1 and Med19 interact in vitro. Furthermore, this interaction occurs via the highly conserved, N-terminal, MED-addressing domain of Med1, suggesting an evolutionary conservation. How, then, to reconcile the proposed MED architecture with our results showing a direct interaction between Med1 and Med19 subunits in vitro?
We propose two nonexclusive hypotheses: First, MED complexes could adopt different conformations, which would differ from the “canonical” architecture of the MED complex in isolation. This is supported by observations that the MED complex changes its overall shape when engaged in interactions with either TF, CKM, or PolII (50). Perhaps when MED is recruited by GATA, Med1-Med19 contacts within the MED complex could stabilize one of these “alternative” conformations.
A second possibility is that Med1-Med19 interactions do not occur within but between MED complexes and could thus stabilize “multi-MED” structures. It has been shown that master TFs control gene expression programs by establishing clusters of enhancers called super-enhancers, at genes with prominent roles in cell identity (54). Recent studies have revealed that, at super-enhancers, master TFs and the Mediator coactivator form phase-separated condensates, which compartmentalize and concentrate the PolII machinery to specific nuclear foci to ensure high level of transcription (55–57). Interestingly, mammalian Med1 can form such phase-separated droplets that concentrate the transcription machinery at super-enhancers (56). Bringing together several MED complexes associated with TFs via Med1-Med19 trans-interaction might thus help phase-separated droplet formation at clustered gene enhancers and ensure high transcriptional level (Fig. 6D).
In conclusion, our work shows that two MED subunits physically bind GATAs and are required to relay the regulatory signals from common TFs. This argues against the generally admitted view of binary interaction between one MED subunit and one TF, which appears as an oversimplified model for MED action. The Mediator should be viewed as a complex interface allowing fine-tuned gene regulation by TFs through specific contacts with different MED subunit combinations. This study highlights the unexpected role of Drosophila Med19 as a GATA cofactor and Med1 interactor. This work sheds new light on the GATA-MED paradigm and suggests novel means by which several MED subunits might collaborate to regulate gene transcription.
Experimental Procedures
Drosophila stocks, genetic mosaics, and phenotypic analyses
No vertebrate animals were used in this study. No applicable regulations are available for Drosophila but this animal study followed guidelines from the Animal Ethics Committee of CNRS. Each experiment with Drosophila was performed with at least two independent replicates. There were no predetermined exclusion criteria for animal work.
Stocks and crosses were raised at 25°C or 22°C and 12-h:12-h light–dark cycle on standard yeast-agar-cornmeal medium (Bloomington Drosophila Stock Center). Mitotic clones were generated using the Flp-FRT system with the Med192 null allele FRT80B chromosome (16). The Flp recombinase was expressed in the dorsal part of the wing using the ap-GAL4 driver recombined with UAS-Flp. The following stocks were used: apGAL4 UASFlp/Cyo; Med192 FRT2A/TM6B, UbGFP M FRT2A/TM6B; apGAL4 (MD544) UASGFP/CyO, UAS dsRNA Med19 (no. 27559); and dppGAL4 from BDSC, UAS VN-AbdA and UAS VN-Srp kindly provided by S. Merabet, DC-ac-lacZ kindly provided by P. Heitzler UAS-Med19VC (16), and UAS-Med1AVC (33). Phenotypes of ∼20 adults/genotype were analyzed by scanning EM (Hitachi TM-1000 Tabletop model) of frozen adults.
Immunofluorescence and FISH of wing discs
Approximately 30 third-instar larval imaginal discs were prepared and stained using standard procedures. Antibodies used were rabbit anti–β-gal (Cappel, 1:2500) for both DC-ac-LacZ and UAS-lacZ detection; mouse anti-Wg (1:200) (4D4 antibody from Developmental Studies Hybridoma Bank, Iowa University). For BiFC experiments, we used mouse anti-GFP from Roche (1:200), which only recognizes the C-terminal half of the GFP variant Venus (ΔVC).
For FISH, DIG-U–labeled antisense RNA probes against pannier, spanning the whole Pnr ORF were used. Fast-RED revelation was optimal when carried out at 4°C overnight. Image acquisition was performed on a Leica SP5 confocal microscope. Z-stacks were generated using ImageJ-related FiJi software.
BiFC assay
This technique is based on expressing in vivo two-candidate partner proteins with the N- and C-terminal portions (VN and VC) of the Venus protein to test the reconstitution of a functional fluorescent protein. UAS-Pnr-VN, UAS-VN-Pnr, and UAS CycC-VC lines were generated by inserting PnrA or CycC ORF in phase with VN173 (aa 1–172) or VC155 (aa 155–238) ORF in a recipient pUAST-attB plasmid, allowing site-specific insertion. attP-carrying embryos expressing PhiC31 integrase in the presumptive germline were injected with these plasmids: Pnr constructions were inserted on the X chromosome (attP ZH-2A), and CycC constructions on the second chromosome (attP 51D) to ensure identical expression of the different VN lines, and easy combination of VN and VC lines. Note that the BiFC constructions were functionally validated for their ability to rescue mutant lethality for Med19-VC, Med1-VC, and CycC-VC or to produce typical gain-of-function phenotypes for VN-Pnr.
Crosses were carried out at 22°C for interaction tests to express candidate proteins at homogenous and relatively low levels to avoid nonspecific signal. We used the dppGAL4 driver (Gal4/UAS system) to direct co-expression of fusion proteins at the anteroposterior frontier of the wing imaginal disc both in the thorax where pnr is normally expressed and in the wing pouch region that does not express pnr.
Image acquisition was performed on a Leica SP5 using the same settings and number of z slices for the different genetic contexts. BiFC fluorescence was quantified using ImageJ software.
Co-immunoprecipitation experiments
Cultured Drosophila S2 cells were grown in 10% serum containing Schneider's medium at 25°C, and transfected using FuGENE HD transfection reagent (Roche) following manufacturer recommendations. Transfections of 18 × 106 cells per plate were carried out with pActin-Myc-Pnr (3 µg) and pActin-GAL4 (2 µg) plasmids. The cell harvest, protein extraction, and IP were performed as described in Ref. 33 with the following modifications: The buffer used for protein extraction and subsequent IP contained 0.1% Nonidet P-40 instead of 0.5% to increase purification of large complexes such as MED; 1 mg of total protein extract was used for each IP instead of 1.5mg. Anti-Med19 and nonrelevant IPs were performed with 5 µl of decomplemented serum from a Med19-immunized guinea pig (16) or 5 µl of the same animal's pre–immune serum, respectively. We used 10 µl G protein–coupled Sepharose beads per IP (Sigma, P3296). Anti-Myc IPs were performed with 10 µl anti-Myc-agarose bead (Sigma, A7470).
Med19 was revealed using a home-made polyclonal serum from guinea-pig (diluted 1:500) immunized with a full-length Med19-GST fusion protein (16). Antibody specificity has been assessed by immunodetection from Med19-depleted (see Fig. S1) or Med19 overexpressing tissues; Myc-Pnr was detected with rabbit anti-Pnr (kind gift from G. Morata, diluted 1:1000). We used Lumi-Light PLUS Western Blotting Substrate (Roche, 12015196001) and high-performance chemiluminescence film (Amersham Biosciences HyperfilmTM ECL, GE Healthcare, 28906837) for revelation.
GST pulldown experiments
Preparation of GST fusion proteins, 35S methionine–labeled proteins and pulldown were performed essentially as described in Mojica et al. (2017) (58). Med1, Pannier, and Serpent proteins or subfragments have been produced from cDNA corresponding to PnrA (Ramain et al. (29)), SrpB (Waltzer et al. (2002)) (59), and Med1A (Immarigeon et al. (33)) by in vitro transcription/translation coupled reactions using rabbit reticulocyte extracts (TnT, Promega) isoforms, labeled. cDNA encoding full-length dMed19 and deletion derivatives were amplified by PCR using appropriated oligonucleotides and inserted into the BamHI/NotI site of the pGEX-6P1 vector (GE Healthcare). Pnr aa 1–291 fragment-encoding was amplified by PCR and cloned into pcDNA3 vector, with an HA-tag at the C terminus and a FLAG-tag at the N terminus. All clones were verified by sequencing. Primers sequences and complete clone sequences are available upon request. Bacterial expression vectors pGEX-6P1 were transformed in competent Escherichia coli strain BL21 (DE3). The transformed cells were plated in LB agar containing 50 µg/ml of ampicillin. A single colony was grown overnight in 25 ml of LB medium containing ampicillin on a rotary shaker (180 rpm) at 37°C. Overnight starter culture was diluted 1:30 and bacteria were grown in 150 ml of LB medium containing ampicillin at 37°C to an optical density of 0.8–0.9 at 600 nm, and expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) for 2 h at 37°C. Bacteria were pelleted by centrifugation and pellets were stored overnight at −20°C. Pellet was resuspended in 15 ml lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% w/v glycerol, 0.1% Nonidet P-40) including one cOmpleteTM EDTA-free protease inhibitor tablet and sonicated on ice. After centrifugation at 10 000 × g 45 min at 4°C, the supernatant was mixed 2 h at 4°C on a rotating platform with 2 ml GSH Sepharose 4B resin. Beads were washed four times with lysis buffer and stored at 4°C.
6 µl of tagged Pnr 1-291-HA in vitro translation product was mixed with 50 µl of GSH–agarose bead–GST Med19 derivatives in 200 µl of pulldown buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% w/v glycerol, 0.1% Nonidet P-40, 10 μm ZnSO4). The mixture was incubated for 2 h at 4°C and washed four times with 500 µl pulldown buffer. Protein complexes were eluted from the beads with 2 × Laemmli sample buffer, boiled 5 min, and separated by SDS-PAGE on Mini-PROTEAN® TGXTM precast gels (Bio-Rad). Bound Pnr 1–291 was identified by Western blot (1:5000 rabbit anti-HA polyclonal) using an ECL kit (Amersham Biosciences GE Healthcare Life Sciences) based on the manufacturer's recommendations. HRP-conjugated secondary antibodies were used at 1:5000 and were purchased from Amersham Biosciences GE Healthcare Life Sciences. All the membranes were scanned on an ImageQuant LAS 500 (GE Healthcare Life Sciences).
Sequence conservation analysis
C-ZF domains from the Drosophila and human GATA family members were extracted from the NCBI website (RRID:SCR_003257), aligned with the MAFFT software (RRID:SCR_011811), and using default parameters, and amino acid conservation were visualized with Jalview 2.10.5 version (RRID:SCR_006459) using Clustal coloring.
RT-qPCR
Two different dsRNA were used for Med1 or Med19 mRNA depletion, only one for control GFP mRNA. The indicated dsRNAs (see Table S1) is added at 2 µg/ml to exponentially growing S2 cells, in an orbital shaker, at 2 106 cell/ml in serum-free medium. After 40 min, serum is added. 24 h later a second addition of dsRNA is done at 1 µg/ml. Cells are collected 5 days after the first dsRNA treatment.
For mRNA quantification, mRNAs were purified by RNeasy Kit (Qiagen). Reverse transcription was done using SuperScriptTM II Reverse Transcriptase (Thermo Fisher Scientific) and cDNA were quantified by real-time qPCR (CFX Bio-Rad) with specific oligonucleotides (Table S1). Absolute quantification of each mRNA was normalized to GAPDH mRNA quantity in the same sample. mRNA measured in cells treated with a control dsRNA GFP was set at 100% to compare with cells treated with a dsRNA against Med1 or Med19.
Data Availability
All data are contained within this manuscript
Supplementary Material
Acknowledgments
We gratefully acknowledge P. Heitzler, S. Merabet, and BDSC for plasmids and fly stocks; D. Jullien, A. Vincent, and C. Monod for critical reading of the manuscript; V. Gobert and B. Augé for technical help; J. Favier, A. Destenabes, and V. Nicolas for fly facilities; and finally the Toulouse Rio Imaging platform.
This article contains supporting information.
Author contributions—C. I., A. V., D. M., V. V., H.-M. B., and M. B. conceptualization; C. I., V. V., and M. B. formal analysis; C. I., S. B.-F., E. G., A. V., E. P., M. A. B., A. P., M. C., and D. M. investigation; C. I., S. B.-F., E. G., E. P., M. A. B., A. P., M. C., and D. M. methodology; C. I. and M. B. writing-original draft; C. I., E. G., A. V., V. V., H.-M. B., and M. B. writing-review and editing; S. B.-F., E. G., H.-M. B., and M. B. supervision; A. V., V. V., H.-M. B., and M. B. funding acquisition; V. V., H.-M. B., and M. B. project administration.
Funding and additional information—This work was supported by French Ministère de l'Enseignement et de la Recherche (to C. I.), the Ligue Nationale contre le Cancer (to C. I. and A. P.), the Association pour la Recherche sur le Cancer Grant ARC PJA 20141201932, and the Agence Nationale de Recherche Grant ANR-16 CE12-0021-01. This work was also supported by the Centre National de Recherche Scientifique (CNRS) and Toulouse III University.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TF
- transcription factors
- MED
- Mediator
- CKM
- Cdk8 kinase module
- ZF
- zinc finger
- Srp
- Serpent
- Pnr
- Pannier
- DC
- dorsocentral
- BiFC
- bimolecular fluorescence complementation
- aa
- amino acid
- HIM
- HOX homeodomain–interacting motif
- DBD
- DNA binding domains
- IP
- immunoprecipitation
- ac
- achaete gene.
References
- 1. Soutourina J. (2018) Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 10.1038/nrm.2017.115 [DOI] [PubMed] [Google Scholar]
- 2. Jeronimo C., and Robert F. (2017) The Mediator complex: At the nexus of RNA polymerase II transcription. Trends Cell. Biol. 27, 765–783 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 3. Verger A., Monté D., and Villeret V. (2019) Twenty years of Mediator complex structural studies. Biochem. Soc. Trans. 47, 399–410 10.1042/BST20180608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El Khattabi L., Zhao H., Kalchschmidt J., Young N., Jung S., Van Blerkom P., Kieffer-Kwon P., Kieffer-Kwon K. R., Park S., Wang X., Krebs J., Tripathi S., Sakabe N., Sobreira D. R., Huang S. C., et al. (2019) A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158.e20 10.1016/j.cell.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boube M., Faucher C., Joulia L., Cribbs D. L., and Bourbon H. M. (2000) Drosophila homologs of transcriptional Mediator complex subunits are required for adult cell and segment identity specification. Genes Dev. 14, 2906–2917 10.1101/gad.17900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schiano C., Casamassimi A., Rienzo M., de Nigris F., Sommese L., and Napoli C. (2014) Involvement of Mediator complex in malignancy. Biochim. Biophys. Acta 1845, 66–83 10.1016/j.bbcan.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 7. Berk A. J. (2012) Yin and yang of mediator function revealed by human mutants. Proc. Natl. Acad. Sci. U. S. A. 109, 19519–19520 10.1073/pnas.1217267109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grants J. M., Goh G. Y. S., and Taubert S. (2015) The Mediator complex of Caenorhabditis elegans: Insights into the developmental and physiological roles of a conserved transcriptional coregulator. Nucleic Acids Res. 43, 2442–2453 10.1093/nar/gkv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loncle N., Boube M., Joulia L., Boschiero C., Werner M., Cribbs D. L., and Bourbon H. M. (2007) Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 26, 1045–1054 10.1038/sj.emboj.7601566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malik N., Agarwal P., and Tyagi A. (2017) Emerging functions of multi-protein complex Mediator with special emphasis on plants. Crit. Rev. Biochem. Mol. Biol. 52, 475–502 10.1080/10409238.2017.1325830 [DOI] [PubMed] [Google Scholar]
- 11. Borggrefe T., and Yue X. (2011) Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 22, 759–768 10.1016/j.semcdb.2011.07.022 [DOI] [PubMed] [Google Scholar]
- 12. Yin J. W., and Wang G. (2014) The Mediator complex: A master coordinator of transcription and cell lineage development. Development 141, 977–987 10.1242/dev.098392 [DOI] [PubMed] [Google Scholar]
- 13. Kato Y., Habas R., Katsuyama Y., Näär A. M., and He X. (2002) A component of the ARC/mediator complex required for TGFβ/nodal signalling. Nature 418, 641–646 10.1038/nature00969 [DOI] [PubMed] [Google Scholar]
- 14. Liu Z., Yao X., Yan G., Xu Y., Yan J., Zou W., and Wang G. (2016) Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun. 7, 1–11 10.1038/ncomms11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou H., Kim S., Ishii S., and Boyer T. G. (2006) Mediator modulates Gli3-dependent sonic hedgehog signaling. Mol. Cell Biol. 26, 8667–8682 10.1128/MCB.00443-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boube M., Hudry B., Immarigeon C., Carrier Y., Bernat-Fabre S., Merabet S., Graba Y., Bourbon H. M., and Cribbs D. L. (2014) Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of Mediator subunit Med19. PLoS Genet. 10, e1004303 10.1371/journal.pgen.1004303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crona F., Holmqvist P. H., Tang M., Singla B., Vakifahmetoglu-Norberg H., Fantur K., and Mannervik M. (2015) The Brakeless co-regulator can directly activate and repress transcription in early Drosophila embryos. Dev. Biol. 407, 173–181 10.1016/j.ydbio.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 18. Fondell J. D. (2013) The Mediator complex in thyroid hormone receptor action. Biochim. Biophys. Acta 1830, 3867–3875 10.1016/j.bbagen.2012.02.012 [DOI] [PubMed] [Google Scholar]
- 19. Stumpf M., Waskow C., Krötschel M., Van Essen D., Rodriguez P., Zhang X., Guyot B., Roeder R. G., and Borggrefe T. (2006) The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl. Acad. Sci. U. S. A. 103, 18504–18509 10.1073/pnas.0604494103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crawford S. E., Qi C., Misra P., Stellmach V., Sambasiva Rao M., Enge J. D., Zhu Y., and Reddy J. K. (2002) Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 277, 3585–3592 10.1074/jbc.M107995200 [DOI] [PubMed] [Google Scholar]
- 21. Chlon T. M., and Crispino J. D. (2012) Combinatorial regulation of tissue specification by GATA and FOG factors. Development 139, 3905–3916 10.1242/dev.080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whyatt D. J., deBoer E., and Grosveld F. (1993) The two zinc finger-like domains of GATA-1 have different DNA binding specificities. EMBO J. 12, 4993–5005 10.1002/j.1460-2075.1993.tb06193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsang A. P., Visvader J. E., Turner C. A., Fujiwara Y., Yu C., Weiss M. J., Crossley M., and Orkin S. H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90, 109–119 10.1016/S0092-8674(00)80318-9 [DOI] [PubMed] [Google Scholar]
- 24. Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K. E., Guyot B., Heck A. J. R., Vyas P., De Boer E., Grosveld F., and Strouboulis J. (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24, 2354–2366 10.1038/sj.emboj.7600702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C. R., Gregory G. D., Hardison R. C., and Blobel G. A. (2009) SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113, 2191–2201 10.1182/blood-2008-07-169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilkinson-White L., Gamsjaeger R., Dastmalchi S., Wienert B., Stokes P. H., Crossley M., Mackay J. P., and Matthews J. M. (2011) Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc. Natl. Acad. Sci. U. S. A. 108, 14443–14448 10.1073/pnas.1105898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sorrentino R. P., Gajewski K. M., and Schulz R. A. (2005) GATA factors in Drosophila heart and blood cell development. Semin. Cell Dev. Biol. 16, 107–116 10.1016/j.semcdb.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 28. García-García M. J., Ramain P., Simpson P., and Modolell J. (1999) Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development 126, 3523–3532 [DOI] [PubMed] [Google Scholar]
- 29. Ramain P., Heitzler P., Haenlin M., and Simpson P. (1993) Pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development 119, 1277–1291 [DOI] [PubMed] [Google Scholar]
- 30. Heitzler P., Haenlin M., Ramain P., Calleja M., and Simpson P. (1996) A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143, 1271–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sato M., and Saigo K. (2000) Involvement of pannier and u-shaped in regulation of Decapentaplegic-dependent wingless expression in developing Drosophila notum. Mech. Dev. 93, 127–138 10.1016/S0925-4773(00)00282-3 [DOI] [PubMed] [Google Scholar]
- 32. Gobert V., Osman D., Bras S., Augé B., Boube M., Bourbon H.-M., Horn T., Boutros M., Haenlin M., and Waltzer L. (2010) A genome-wide RNA interference screen identifies a differential role of the Mediator CDK8 module subunits for GATA/RUNX-activated transcription in Drosophila. Mol. Cell Biol. 30, 2837–2848 10.1128/MCB.01625-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Immarigeon C., Bernat-Fabre S., Augé B., Faucher C., Gobert V., Haenlin M., Waltzer L., Payet A., Cribbs D. L., Bourbon H.-M. G., and Boube M. (2019) Drosophila Mediator subunit Med1 is required for GATA-dependent developmental processes: Divergent binding interfaces for conserved coactivator functions. Mol. Cell Biol. 39, 1–18 10.1128/MCB.00477-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu C.-D., Grinberg A. V., and Kerppola T. K. (2005) Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC). Anal. Curr. Protoc. Cell Biol. 29, 21.3.1–21.3.21 10.1002/0471143030.cb2103s29 [DOI] [PubMed] [Google Scholar]
- 35. Brand A. H., and Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 36. Ghirlando R., and Trainor C. D. (2003) Determinants of GATA-1 binding to DNA: The role of non-finger residues. J. Biol. Chem. 278, 45620–45628 10.1074/jbc.M306410200 [DOI] [PubMed] [Google Scholar]
- 37. Newton A., Mackay J., and Crossley M. (2001) The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem. 276, 35794–35801 10.1074/jbc.M106256200 [DOI] [PubMed] [Google Scholar]
- 38. Bourbon H. M. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 10.1093/nar/gkn349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuuluvainen E., Hakala H., Havula E., Estimé M. S., Rämet M., Hietakangas V., and Mäkelä T. P. (2014) Cyclin-dependent kinase 8 module expression profiling reveals requirement of Mediator subunits 12 and 13 for transcription of Serpent-dependent innate immunity genes in Drosophila. J. Biol. Chem. 289, 16252–16261 10.1074/jbc.M113.541904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ge K., Cho Y.-W., Guo H., Hong T. B., Guermah M., Ito M., Yu H., Kalkum M., and Roeder R. G. (2008) Alternative mechanisms by which Mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor -stimulated adipogenesis and target gene expression. Mol. Cell Biol. 28, 1081–1091 10.1128/MCB.00967-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soutourina J. (2019) Mammalian Mediator as a functional link between enhancers and promoters. Cell 178, 1036–1038 10.1016/j.cell.2019.07.040 [DOI] [PubMed] [Google Scholar]
- 42. Asmar J., Biryukova I., and Heitzler P. (2008) Drosophila dLMO-PA isoform acts as an early activator of achaete/scute proneural expression. Dev. Biol. 316, 487–497 10.1016/j.ydbio.2008.01.040 [DOI] [PubMed] [Google Scholar]
- 43. Haenlin M., Cubadda Y., Blondeau F., Heitzler P., Lutz Y., Simpson P., and Ramain P. (1997) Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11, 3096–3108 10.1101/gad.11.22.3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lowry J. A., and Atchley W. R. (2000) Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 50, 103–115 10.1007/s002399910012 [DOI] [PubMed] [Google Scholar]
- 45. Visvader J. E., Crossley M., Hill J., Orkin S. H., and Adams J. M. (1995) The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Mol. Cell Biol. 15, 634–641 10.1128/mcb.15.2.634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arnold C. D., Nemčko F., Woodfin A. R., Wienerroither S., Vlasova A., Schleiffer A., Pagani M., Rath M., and Stark A. (2018) A high-throughput method to identify trans-activation domains within transcription factor sequences. EMBO J. 37, e98896 10.15252/embj.201798896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pope N. J., and Bresnick E. H. (2013) Establishment of a cell-type-specific genetic network by the Mediator complex component Med1. Mol. Cell Biol. 33, 1938–1955 10.1128/MCB.00141-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pope N. J., and Bresnick E. H. (2010) Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res. 38, 2190–2200 10.1093/nar/gkp1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stumpf M., Yue X., Schmitz S., Luche H., Reddy J. K., and Borggrefe T. (2010) Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc. Natl. Acad. Sci. U. S. A. 107, 21541–21546 10.1073/pnas.1005794107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsai K. L., Tomomori-Sato C., Sato S., Conaway R. C., Conaway J. W., and Asturias F. J. (2014) Subunit architecture and functional modular rearrangements of the transcriptional Mediator complex. Cell 157, 1430–1444 10.1016/j.cell.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baidoobonso S. M., Guidi B. W., and Myers L. C. (2007) Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae Mediator complex. J. Biol. Chem. 282, 5551–5559 10.1074/jbc.M609484200 [DOI] [PubMed] [Google Scholar]
- 52. Ding N., Tomomori-Sato C., Sato S., Conaway R. C., Conaway J. W., and Boyer T. G. (2009) MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem. 284, 2648–2656 10.1074/jbc.M806514200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robinson P. J., Trnka M. J., Pellarin R., Greenberg C. H., Bushnell D. A., Davis R., Burlingame A. L., Sali A., and Kornberg R. D. (2015) Molecular architecture of the yeast Mediator complex. Elife e08719 10.7554/eLife.08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hnisz D., Abraham B., Lee T., Lau A., Saint-Andre V., Sigova A., Hoke H., and Young R. (2013) Transcriptional super-enhancers connected to cell identity and disease. Cell 155, 934–947 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boija A., Klein I. A., Sabari B. R., Dall'Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cho W. K., Spille J. H., Hecht M., Lee C., Li C., Grube V., and Cisse I. I. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mojica-Vázquez L. H., Benetah M. H., Baanannou A., Bernat-Fabre S., Deplancke B., Cribbs D. L., Bourbon H. M., and Boube M. (2017) Tissue-specific enhancer repression through molecular integration of cell signaling inputs. PLoS Genet. 13, e1006718 10.1371/journal.pgen.1006718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waltzer L., Bataillé L., Peyrefitte S., and Haenlin M. (2002) Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J. 21, 5477–5486 10.1093/emboj/cdf545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this manuscript