Abstract
Liquid–liquid phase separation (LLPS) of biomolecules drives the formation of subcellular compartments with distinct physicochemical properties. These compartments, free of lipid bilayers and therefore called membraneless organelles, include nucleoli, centrosomes, heterochromatin, and centromeres. These have emerged as a new paradigm to account for subcellular organization and cell fate decisions. Here we summarize recent studies linking LLPS to mitotic spindle, heterochromatin, and centromere assembly and their plasticity controls in the context of the cell division cycle, highlighting a functional role for phase behavior and material properties of proteins assembled onto heterochromatin, centromeres, and central spindles via LLPS. The techniques and tools for visualizing and harnessing membraneless organelle dynamics and plasticity in mitosis are also discussed, as is the potential for these discoveries to promote new research directions for investigating chromosome dynamics, plasticity, and interchromosome interactions in the decision-making process during mitosis.
Keywords: membraneless organelle, chromosome compartments, centromere, bubristatin, organoids, centrosome, chemical biology, chromatin, chromosomes, subcellular organelle, molecular motor, molecular imaging, mitotic spindle, mitosis, CENP-E, syntelin
Interest in the cell nucleus and its structures dates back to the late 19th century, when Friedrich Miescher discovered nucleic acids (1) and Walther Flemming coined the term “chromatin” during his study of mitosis (2). Mitosis is a process of cell duplication by which one cell gives rise to two genetically identical daughter cells through a series of orchestrated movements by dynamic interactions between chromosomes and spindles (Fig. 1A) (3). The process of chromosome movements in mitosis is defined as prophase, prometaphase, metaphase, anaphase, and telophase, resulting in equal distribution of duplicated genomes to two daughter cells (3). Although decades of study might leave the impression that most of the important mechanisms of mitotic regulation have already been discovered, many fundamental questions remain. For example, stem cells undergo an asymmetric division in which cellular constituents are preferentially segregated into only one of the two daughter cells, thus endowing the two daughter cells with different fates (4). Two remaining concerns in stem cell biology are how stem cells segregate asymmetrically and what mechanisms govern this decision-making process during the metaphase-anaphase transition (Fig. 1, B and C).
Figure 1.
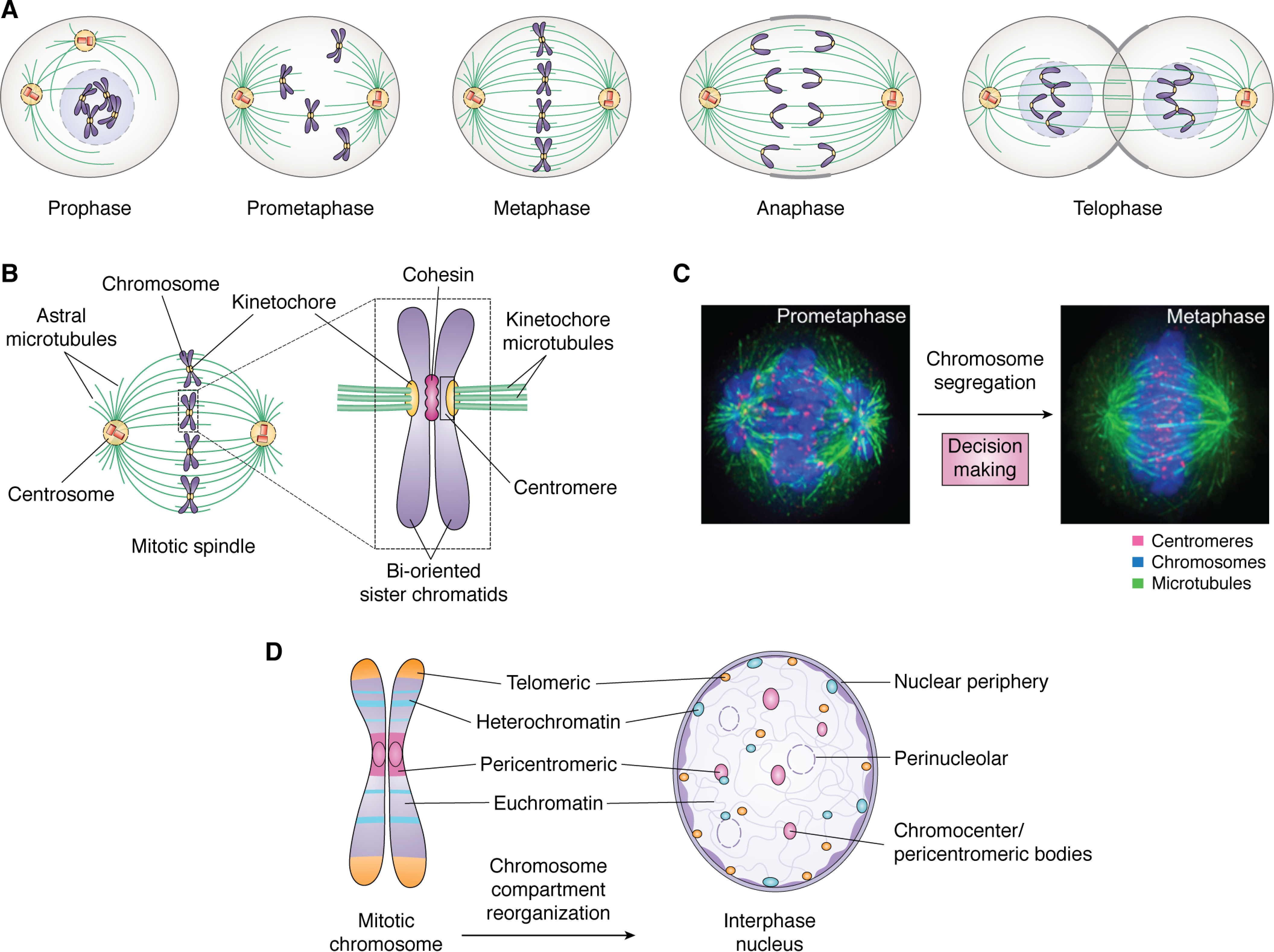
Mitosis is a model system to study membraneless organelle dynamics during cell fate decision. A, schematic illustration of dynamic reorganization of various membraneless organelles, including the mitotic spindle, chromosomes, centromere, central spindles, and midbody, during the process of mitosis. The structure and morphological changes are suitable for a two-dimensional image-based phenotypic screen. B, anatomy of a mitotic spindle that is comprised of several membraneless compartments, including spindle microtubules, astral microtubules, chromosomes, and centrosomes. Mitotic chromosomes contain functional subcompartments, including the telomere, centromere, and heterochromatin. The molecular composition of these compartments is dynamically regulated by cell-cycle machinery, and the assembly of each compartment is driven by LLPS under spatiotemporal cues. Adapted from Ref. 38. C, immunofluorescence images of prometaphase and metaphase spindles from mitotic HeLa cells. The transition from prometaphase to metaphase involves chromosome alignment and spindle assembly checkpoint signaling, which determines the cell fate after mitosis and whether the division is asymmetric or symmetric. A concern is how asymmetric or symmetric division is determined and how LLPS drives the process. Red, centromeres; green, microtubules; blue, chromosomes. Adapted from Ref. 8. D, schematic drawing of the dynamic change of compartments in mitotic chromosomes and the interphase nucleus. Chromosomes contain functional subcompartments, including telomeres, centromeres, and heterochromatin. These compartments are under the control of cell-cycle machinery, and the assembly of those membraneless compartments is driven by LLPS. Current interests in the field are how reversible LLPS is regulated by cell-cycle machinery and the spatiotemporal order of the assembly/disassembly.
Chromosomes are thread-like structures made of proteins and DNA organized in a compact manner to allow the accurate transmission of genetic material to daughter cells in mitosis. Centromeres, chromatins, and telomeres are compartments of all eukaryotic chromosomes (Fig. 1D) (5–9). Centromeres were serendipitously identified by study of antisera from patients with scleroderma (10). The centromere is a chromosomal locus for spindle organization; the telomere provides stability to the chromosome during mitosis (3, 7). The two major types of chromatin, euchromatin and heterochromatin, bear different transcriptional activities and spatial distribution. Heterochromatin is mainly localized at the interphase nuclear periphery and the region surrounding the nucleolus; euchromatin is localized in the interior of the nucleus of interphase cells (Fig. 1D) (6, 11). A challenging question in cell biology is how chromosomal compartments are reorganized during cell division.
Early studies aiming to delineate the mechanisms of action in mitosis involved classical genetic approaches to identify genes involved in the cell cycle and biochemical approaches to characterize proteins necessary for mitosis (12–15). These studies revealed that cyclin-dependent kinase signaling and post-translational modifications regulate cell division. Although biochemical reconstitution and phenotypic characterization have identified parts of the molecular machinery that drives cell division, it remains unclear how the mitotic machinery is assembled and disassembled (16). In addition, functional proteomics, biochemical studies, and computational analyses indicate that a large portion of spindle and chromosome proteins exhibit intrinsic disorder (17), which presents a roadblock for structural biological approaches to dissect the machinery for decision making in cell division.
Liquid–liquid phase separation (LLPS) of macromolecules that possess low structural complexity or intrinsic disorder has recently emerged as a phenomenon governing the formation of distinct intracellular compartments with context-dependent functions (18, 19). Membraneless condensates, spontaneously formed by LLPS, drive the assembly of both cytoplasmic structures and nuclear speckles (20). Thus, phase separation is a means to generate functional compartments and scaffolding for specific biochemical catalysis (21–23), force generation (24), cell growth control, and beyond (21, 25–37).
In this review, we begin by outlining the key features of membraneless organelles and chromosome compartments during cell division. We then address why and how mitotic processes exploit the cooperativity and efficiency of phase separation, and phase transitions more broadly, during cell division. We next discuss how LLPS and post-translational modifications orchestrate the chromosome dynamics and plasticity during mitosis. Finally, we discuss future directions, applying three-dimensional model systems to unravel the dynamics of membraneless organelles and phase transition in the control of cell division.
Membraneless organelles in mitosis and chromosome compartmentalization
In mitosis, accurate chromosome segregation requires formation of a bipolar spindle and chromosome movement along the spindle, which is comprised of spindle microtubules, astral microtubules, centrosomes, and chromosomes (Fig. 1B) (3, 38). Microtubules are fibrous polymers that are dynamically regulated by GTP hydrolysis and proteins at the microtubule plus- and minus-ends (39). During cell division, chromosomes capture spindle microtubules through a search-and-capture mechanism by kinetochores, the supramolecular complexes assembled at the centromere (3, 40). The microtubule attachment to kinetochores and plus-end growth is necessary for chromosome stability (41–43). The mitotic spindle is a dynamic and complex membraneless organelle regulated by the Ran GTPase gradient (44–47). It undergoes structural changes to the central spindle during the metaphase-anaphase transition. The mitotic spindle is then reorganized into an interphase astral network during mitotic exit (Fig. 1D). The metaphase-anaphase transition is involved in decision making, as it determines whether a symmetric or an asymmetric division program will be executed (4).
Chromatin plasticity and LLPS
Chromatin is contained within compartments that are regulated spatiotemporally by cell-cycle machinery and extracellular cues such as growth factors (6, 48, 49). Heterochromatin, situated between the pericentromere and telomeres of mitotic chromosomes, is essential for maintaining genome stability (6). It is rich with repetitive sequences and undergoes reorganization and compartmentalization as the nuclear DNA condenses into chromosomes during mitosis (Fig. 1D). Heterochromatin is involved in nuclear processes ranging from gene repression to chromosome segregation. The diversified functions of heterochromatin depend on the capacity of these structures to interact with the underlying DNA and to orchestrate various types of stimulation signals in a context-dependent manner during the cell division cycle (48, 49).
However, the molecular mechanisms regulating the intrinsic properties of chromatin and interchromosomal interactions are poorly illustrated (3, 50, 51). A recent cryogenic EM study of reconstituted chromatin fibers revealed a double helix twisted by tetranucleosomal units, which provides mechanistic insights into how nucleosomes are compacted into higher-order chromatin fibers (52). It was postulated that gene silencing by heterochromatin is due to the compaction by HP1 proteins that recruit diverse interacting proteins. Recent observations suggest that the HP1α protein possesses liquid droplet–like properties (33, 35). Unmodified HP1α is soluble, but either phosphorylation of its N-terminal extension or DNA binding promotes the formation of phase-separated droplets (33, 35), demonstrating a context-dependent change in physiochemical properties. Studies of plants revealed that, in Arabidopsis, ADCP1 acts as a multivalent H3K9me reader (53). Similar to human HP1α and fly HP1α, ADCP1 mediates heterochromatin LLPS. As determined by use of embryonic stem cell renewal as a model system, the LLPS-driven physicochemical property of TRIM66, which contains a PHD-bromo domain at its C terminus, regulates chromatin plasticity via binding to unmodified H3R2K4 and acetylated H3K56 (54). Thus, phase separation of the HP1α-H3K9me3 complex illustrates how the process and product of phase separation have distinct but coupled roles: LLPS provides the selectivity and context-dependent recognition, and the resulting compartment builds a foundation for establishment of a functional centromere and, during mitosis, accelerates specific biochemical reactions at heterochromatin and the centromere.
To elucidate the molecular mechanisms underlying control of chromatin plasticity by LLPS, Rosen and colleagues (55) used 12-mer nucleosome arrays containing fluorescence-labeled histone octamers to demonstrate that physiologic concentrations of monovalent and divalent cations induce the formation of reversible LLPS chromatin droplets. Interestingly, acetylation of histone tails removes the positive charges and causes dissolution of chromatin droplets, suggesting that epigenetic “readers” modulate the formation of local genomic compartments via protein-protein interactions elicited by post-translational modifications, demonstrating how the valence of molecular interactions drives LLPS (21, 56). It would be of interest to visualize the LLPS and chromosome segregation during live cell division.
Centromere plasticity and LLPS
The centromere is situated at the interface of the chromosome and the kinetochore that connects to spindle microtubules during mitosis (3, 40). As it contains structurally disordered histone tails (B1), the centromere exhibits characteristics of macromolecule phase separation. Various weak interactions, including hydrophobic and electrostatic contacts between macromolecules, are involved in forming and interacting with neighboring nucleosomes (18, 57).
Structurally, the centromere is comprised of three layers: an inner centromere that consists of a specialized layer of chromatin with kinase activity, an outer plate that consists of tightly packed centromeric nucleosomes, and an outermost corona containing the mitotic motor, CENP-E, and other proteins (3, 9, 40). The inner centromere, which is comprised of Aurora B, INCENP, borealin, and survivin, is a specialized region that promotes cohesion, the regulation of kinetochores, and the assembly of specialized chromatin and mechano-sensation of spindle-pulling forces (49, 58, 59). Inner centromere kinase activity is driven by LLPS via a molecular condensate scaffold built by the chromosome passenger complex (60). It is likely that this specialized compartment of the inner centromere enables the formation of a chromatin body with biochemical activities to regulate protein acetylation, methylation, and phosphorylation in a synergistic manner (45, 61). The recently identified TIP60-Aurora B acetylation-phosphorylation signaling cascade is an example of how chromatin bodies compartmentalize spatiotemporally regulated biochemical reactions (45, 50). It is also possible that coacervates formed by Aurora B and its accessory proteins at the inner centromere transduce physical force across the sister centromeres to fine-tune Aurora B kinase activity in situ during the chromosome segregation process (61). Given the role of inner centromere activity in sensing mechanical force generated by kinetochore microtubule pulling and coacervate-driven regulation of Aurora A kinase activity (62, 63), it would be important to track Aurora B kinase with a FRET-based optical sensor and determine whether Aurora B activity in the inner coacervates senses mechanical force across the sister centromeres (i.e. the pulling by spindle microtubules from two opposing poles).
Accurate chromosome segregation depends on stable interactions of the mitotic spindle with the centromere and with the spindle matrix. BuGZ, initially identified as an outer plate centromere protein Bub3-interacting protein, is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment (64, 65). BuGZ promotes Aurora A activation via zinc finger binding to the kinase domain, which promotes substrate MCAK phosphorylation (63). The coacervates formed by BuGZ and Aurora A are reminiscent of the functional regulation of coacervates formed by Aurora B, INCENP, borealin, and survivin in the inner centromere (60). In the future, it would be important to evaluate how the Aurora A kinase gradient responds to BuGZ in vivo by use of the previously established FRET-based activity reporter and relate its LLPS property to the spatiotemporal dynamics of Aurora A distribution (61, 66).
Kinetochore plasticity, reorganization, and LLPS
The kinetochore, the site for spindle microtubule-centromere association, functions as a molecular machine to power chromosome movements and serves as a signaling device governing chromosome segregation during mitosis (3, 40, 50). As mitosis progresses, kinetochores undergo structural and morphological changes (67), resulting in kinetochore disassembly and reorganization into the spindle midzone (68). Immunoelectron microscopy demonstrates the dynamic translocation of the kinetochore motor, CENP-E, during the metaphase-anaphase transition (68). During the transition, a subset of these central microtubules overlap in an anti-parallel orientation (Fig. 2A, bottom, red arrowheads), thought to be initiated by relocation of CENP-E, as visualized by the use of 10-nm gold particles (Fig. 2A) (68). However, the mechanism underlying the transition from the mitotic spindle to the central spindle before anaphase onset remains elusive. In addition, the physicochemical properties of kinetochore remodeling and biogenesis of the midzone have not been addressed despite the electron microscopic analysis of CENP-E dynamics during the metaphase-anaphase transition (68, 69) and a microinjection experiment suggesting the function of CENP-E in anaphase (70).
Figure 2.
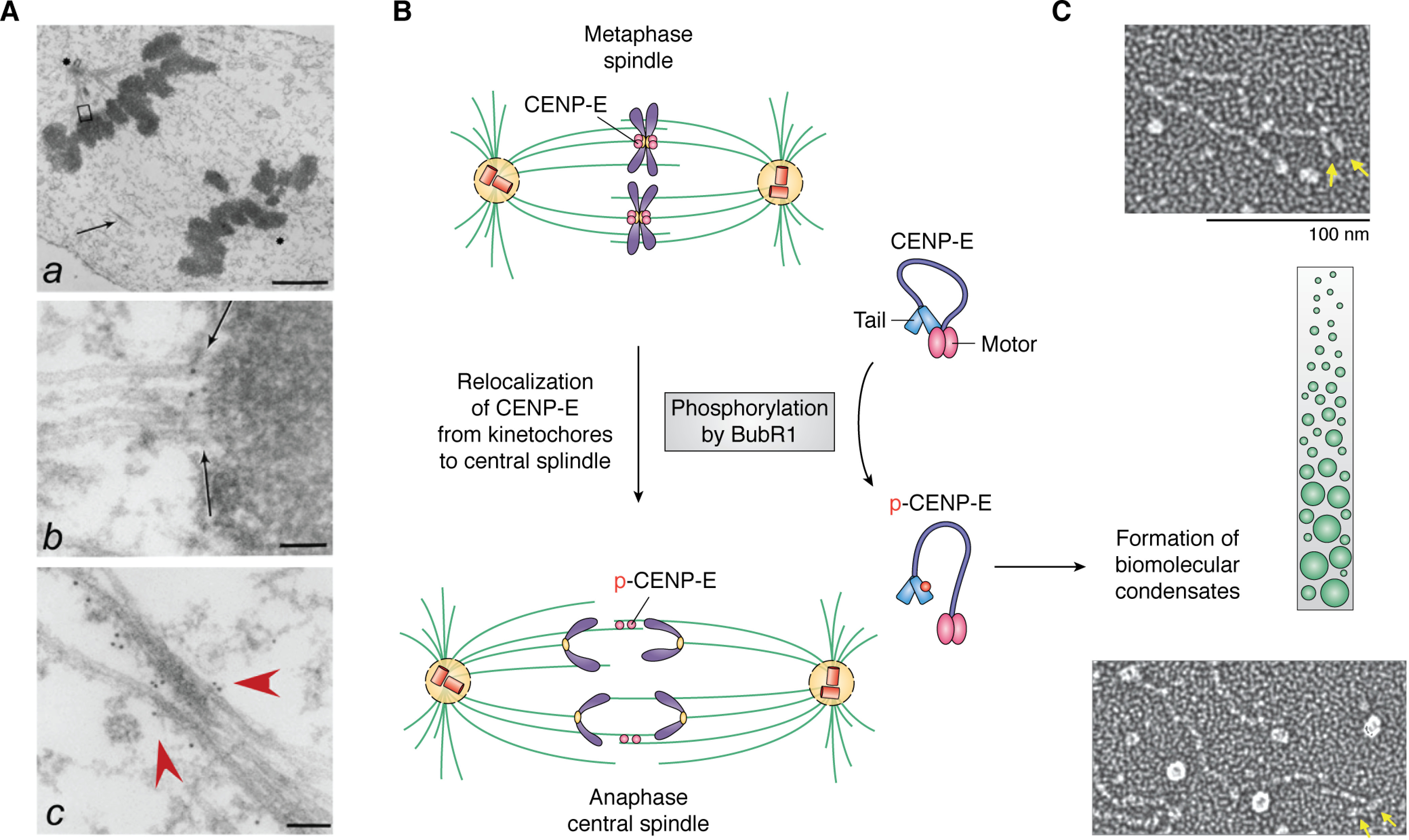
Membraneless organelle reorganization in mitosis is driven by LLPS and phosphorylation. A, electron microscopic image of an anaphase spindle showing membraneless organelles, including the centromere and the central spindle as well as the ultrastructure of chromatin. a, low-magnification view of a late anaphase HeLa cell showing elongated spindle poles, labeled with asterisks. One is apparent; another is in a different section. Interzonal microtubules are readily seen (arrow). b, magnified view of the upper boxed region in a, showing that CENP-E is located between a kinetochore and its associated spindle microtubules (arrow). c, magnified view of the area indicated by the arrow in a. Some CENP-E is now localized to the interzonal microtubules (red arrowheads). Bars, 2 μm (a), 70 nm (b), and 90 nm (c). 10-nm gold particles annotate the CENP-E molecules. Adapted from Ref. 68. B, schematic illustration of the reorganization of the metaphase spindle into an anaphase central spindle. Phosphorylation of CENP-E by BubR1 leads to intramolecular interactions of CENP-E and drives assembly of the central spindle. Unresolved questions are how LLPS drives coacervate formation and how BubR1 kinase activity is regulated in the coacervates. Adapted from Ref. 74. C, single-molecule electron microscopic analysis of CENP-E exhibiting its conformational changes. Adapted from Ref. 76. The green illustrates the crowding effect, whereas the yellow arrows indicate the motor domain of CENP-E. It remains to be examined whether BubR1 phosphorylation increases CENP-E molecule crowding during metaphase-anaphase transition.
To define the assembly order of spindle midzone architecture, two groups have identified chemical inhibitors of CENP-E using an enzymatic assay and a phenotype-based screen, respectively (71, 72). By use of the CENP-E inhibitor syntelin, Ding et al. (71) discovered that inhibition of CENP-E arrested cells in the prometaphase, with the chromosome syntelically attached to the spindle microtubules. Further delineation of CENP-E function in epithelial cell division in 3D organoids led the authors to uncover the role of CENP-E in central spindle organization (73). Surprisingly, the relocation of CENP-E from the kinetochore to the central spindle is regulated by BubR1 phosphorylation (Fig. 2B) (74). Mechanistically, CENP-E contains an N-terminal motor domain followed by a long stretch of highly disordered region with a property to form biomolecular condensates via multivalent interactions with PRC1, CLASP, and other proteins (75). Correlative single-molecule electron microscopic analyses and in vivo real-time studies suggest that CENP-E exhibits structural changes elicited by BubR1 phosphorylation, which drives the formation of biomolecular condensates at the metaphase-anaphase transition (Fig. 2C) (74, 76). As discussed here, the LLPS-mediated assembly of the central spindle is based on in vitro and correlative studies. A pressing issue is to establish whether formation and regulation of the LLPS-mediated midzone condensate are indeed operating in living cells. The use of bubristatin, a BubR1 inhibitor, combined with an optical reporter for LLPS property readout will shed light on LLPS-driven compartmentalization and regulation in mitotic control (74). It would also be important to delineate how the phase property of CENP-E is regulated in the central spindle and to determine whether it regulates asymmetric division.
Spindle plasticity and membraneless organelle dynamics in cell division
Centrosomes, also known as microtubule organization centers, replicate in the S phase and organize the mitotic spindle through anchoring of microtubule minus-ends (77). Hyman and colleagues (31, 78), who analyzed the centrosome proteome, found that the coiled-coil protein SPD-5 assembles into spherical condensates in the presence of crowding agents. Various centrosomal proteins, including microtubule-associated proteins, partition into these condensates via interactions with SPD-5. Thus, the SPD-5–based scaffold acts as a selective membraneless compartment for centrosome organization and spindle establishment (78).
Mitotic spindle orientation is controlled by a centrosomal network and a conserved molecular cascade involving Gαi, LGN, and the NuMA tripartite complex, which guides accurate positioning of the mitotic cleavage plane (79). Early studies demonstrated that, during metaphase, a ring-like F-actin structure surrounding the mitotic spindle is formed temporally (80). This cytoplasmic F-actin structure is relatively isotropic and less dynamic. Computational modeling of the spindle-positioning process suggests a mechanism by which the ring-like F-actin structure regulates astral microtubule dynamics and mitotic spindle orientation. Yu et al. (81) found that NDP52, a membrane organelle regulator, regulates spindle orientation by remodeling the polar F-actin structure. Mechanistically, NDP52 binds to phosphatidic acid–containing vesicles to enable NDP52-containing vesicles to anchor to the actin assembly factor N-WASP via a physical interaction and thereby shorten actin filaments. Because NDP52 binds with microtubule plus-ends, it is desirable to delineate the nature of NDP52-containing compartments in mitotic cells and evaluate how the LLPS activity of NDP52 underlines the aforementioned regulation of spindle position.
The microtubule plus-end tracking proteins (EB1 and its associated proteins, including TIP150 and SKAP) are essential for spindle assembly and orientation (Fig. 3A) (39, 43). In cells, EB1 and its associated proteins appear as dynamic, micrometer-long coacervates and ride on polymerizing microtubules (39). Xia et al. (42) attempted to define how the EB1 coacervates track growing microtubules and showed that mitosis-dependent acetylation of EB1 guides multivalent interactions among EB1, TIP150, and MCAK via regulation of the hydrophobic cavity located at its C terminus (Fig. 3B). They further developed a photoactivatable, complementary fluorescent protein procedure to image EB1 interactions in live cells at 35 nm resolution (Fig. 3C) (82). By use of this superresolution imaging tool, they identified the lysine residues necessary for EB1 tracking of growing microtubule plus-ends (82) and developed a new paradigm by which EB1 coacervates control the dynamics of microtubule plus-ends in space and time (Fig. 3D). Future studies should determine how EB1 coacervates guide the decision making in cell division, as mutations of the tumor suppressor APC (adenomatous polyposis coli) impair EB1 binding activity and compromise chromosome stability during division of gastrointestinal epithelial cells (83).
Figure 3.
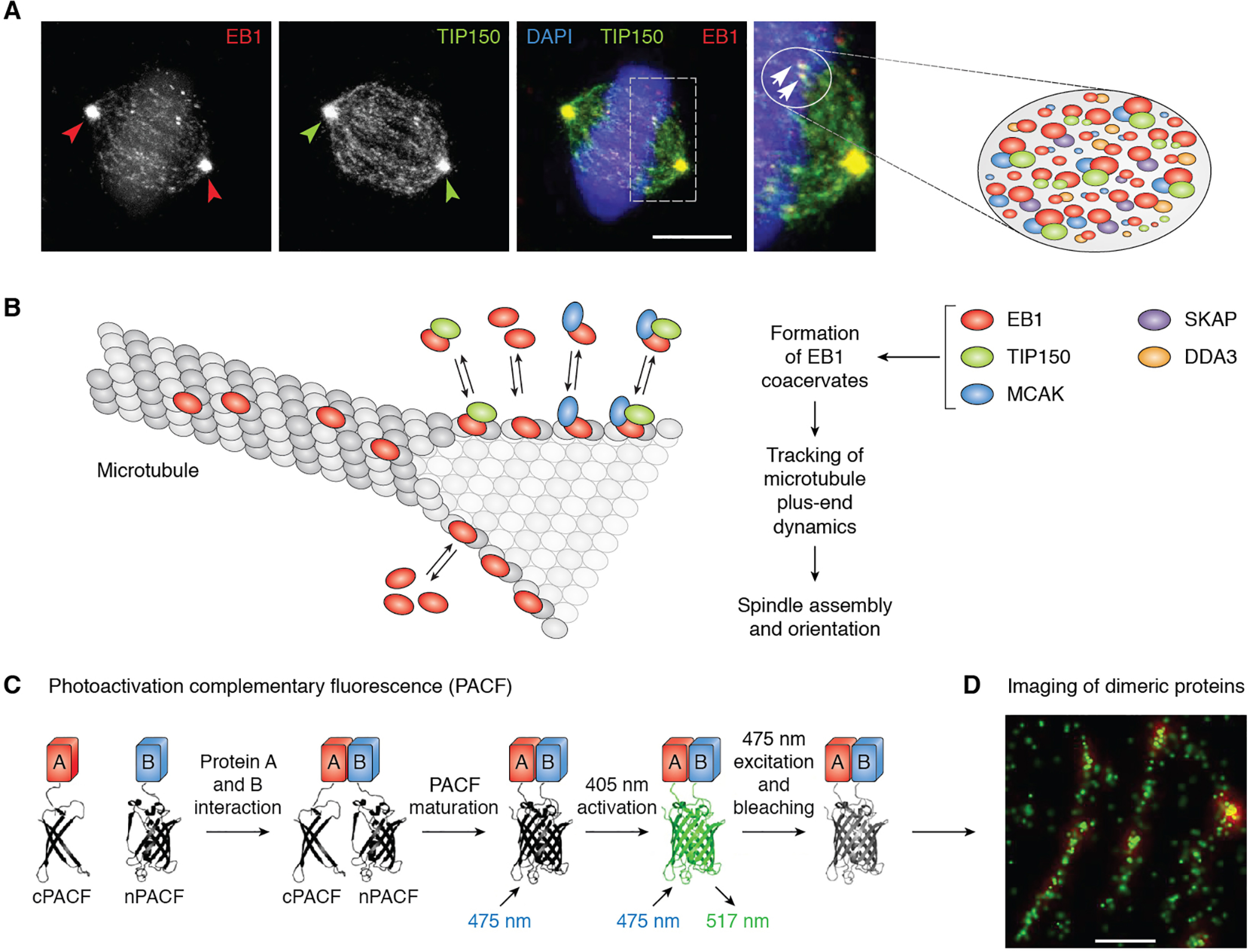
Superresolution imaging of microtubule plus-end coacervates and mapping of amino acids necessary for LLPS. A, subcellular distribution of EB1 and TIP150 in metaphase HeLa cells. EB1 comet-like coacervates (circles and arrowheads) are driven by LLPS from multiple components, including EB1, TIP150, MCAK, and SKAP. An unresolved question is how the multivalent interactions are orchestrated to exhibit context-dependent microtubule dynamics during cell division. Adapted from Refs. 43 and 109). B, schematic illustration of LLPS-driven coacervates formed by EB1 and its binding proteins, including TIP150 and MCAK, which account for the function of EB1 coacervates in tracking microtubule plus-end dynamics. C, schematic design of photoactivation complementary fluorescence (PACF) localization microscopy for molecular imaging of dimeric proteins at high resolution. Adapted from Ref. 82. D, example of superresolution imaging of EB1 dimerization at the single-molecule level in which microtubule plus-end tracking coacervates are organized by dimeric EB1 molecules in close proximity. Bar, 100 nm.
Aberrant LLPS and chromosome instability
Error-free mitosis depends on accurate chromosome attachment to spindle microtubules, congression of those chromosomes, segregation in anaphase, and assembly of a spindle midzone upon mitotic exit (3, 50). Recent studies of the Cancer Genome Atlas (83) show that gastric tumorigenesis is a result of perturbation of chromosome stability during cell division. Bub1B, which encodes a mitotic kinase, BubR1, and its mutants are implicated in gastric tumorigenesis (84). Although previously proposed to be a pseudokinase, Huang et al. (74) solved the crystal structure of the kinase domain of Drosophila melanogaster BubR1, which is predicted to be catalytically active based on its folding conformation. As discussed earlier, BubR1 is a bona fide kinase that phosphorylates CENP-E, causing a switch from a laterally attached microtubule motor to a plus-end microtubule tip tracker. Inhibition of CENP-E phosphorylation by inhibiting BubR1 with bubristatin prevents proper microtubule capture at kinetochores and proper assembly of the central spindle at mitotic exit (74). Thus, BubR1-mediated CENP-E phosphorylation is a temporal switch that enables transition from lateral to end-on microtubule capture and organization of microtubules into stable midzone arrays (Fig. 4A). The availability of BubR1 chemical probes will enable us to determine how LLPS-mediated formation of central spindle condensate controls chromosome stability in mitosis. Because BubR1 mutations are implicated in chromosome stability and tumorigenesis in gastrointestinal tracts (83, 85), it would be of interest to see whether and how the coacervates formed by CENP-E and BubR1 are modulated by disease-associated mutations.
Figure 4.
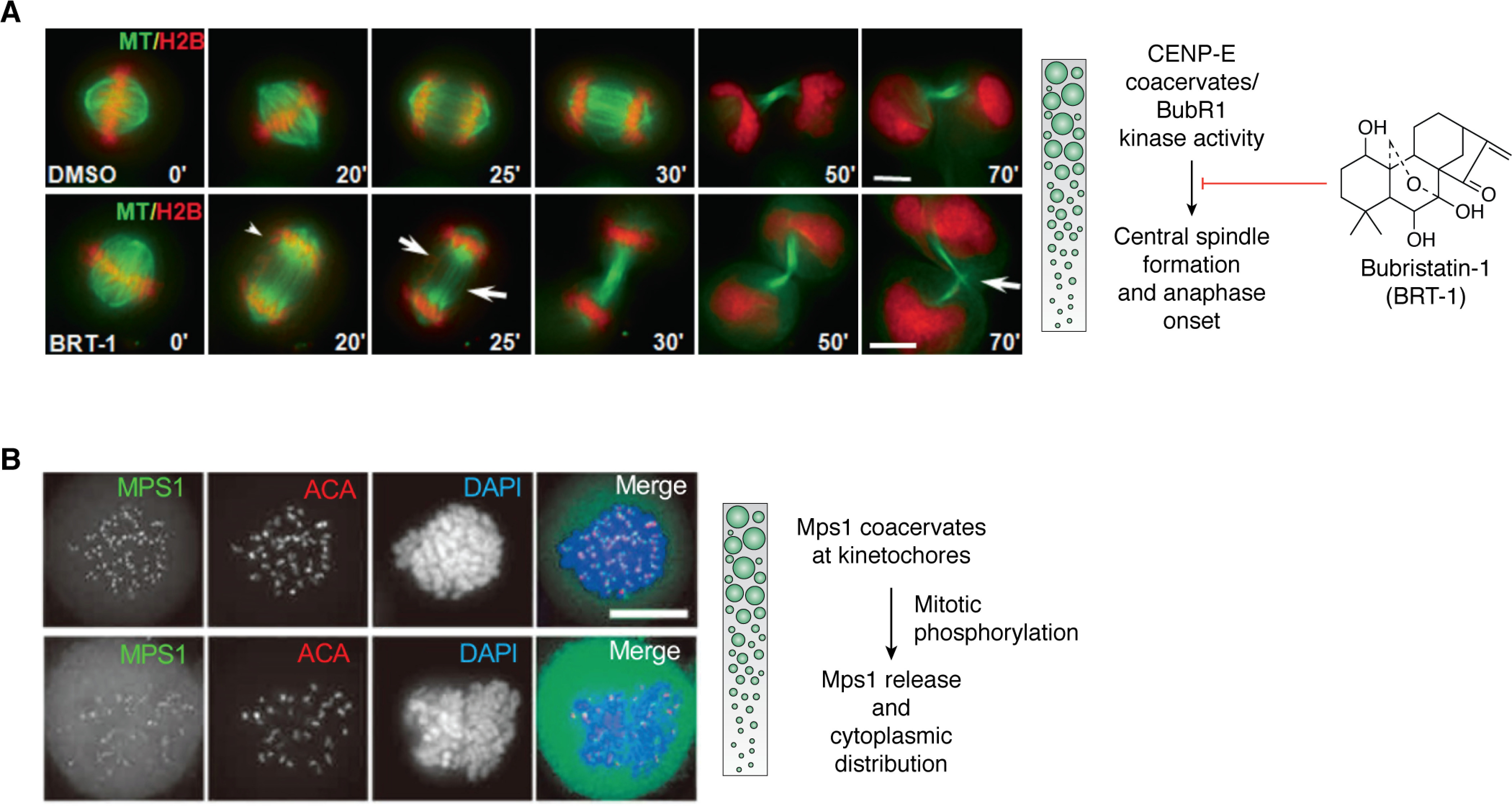
Chromosome compartmentalization dynamics and cell fate decision during mitosis. A, LLPS-driven central spindle assembly by membraneless organelles requires CENP-E and BubR1 kinase. Representative phenotypes of the metaphase-anaphase transition in HeLa cells treated with bubristatin-1 (BRT-1). HeLa cells expressing GFP-tubulin and mCherry-H2B were synchronized at metaphase with MG132. Once released from metaphase arrest, cells were treated with BRT-1 or DMSO followed by real-time imaging with deconvolution microscopy. A current interest is to visualize how CENP-E-BubR1 coacervates regulate BubR1 kinase activity in the central spindle. Scale bar, 10 μm. Adapted from Ref. 74. B, LLPS-driven centromere kinase Mps1 (monopolar spindle 1) coacervates are essential for stable kinetochore localization in mitosis. HeLa cells expressing LAP-Mps1 were exposed to DMSO (bottom) or reversine (an Mps1 inhibitor; top) in the presence of MG132 and nocodazole for synchronization, before being fixed and counterstained for anti-centromere antibody (ACA; shown as red in merged images) and DNA (blue). It was apparent that Mps1 kinase coacervates are stable in the presence of reversine, an inhibitor for preventing Mps1 from utilizing ATP, indicating that Mps1 autophosphorylation liberates Mps1 coacervates from kinetochores. A current interest is to determine whether coacervate dissolution is regulated by Mps1 or its downstream effectors. Scale bar, 10 μm. Adapted from Refs. 89 and 110.
Cancer cell genomes are frequently characterized by numerical and structural chromosomal abnormalities (83). However, the mechanisms remain elusive. Cleveland and colleagues (86) have recently integrated a centromere-specific inactivation approach with selection for a conditionally essential gene, a strategy termed CEN-SELECT. They showed that a single-chromosome missegregation into a micronucleus triggers a broad spectrum of genomic rearrangements, suggesting that individual chromosome segregation errors during mitotic cell division are sufficient to drive extensive structural variations that recapitulate genomic features commonly associated with human disease (86). In fact, perturbations in heterochromatin structure or interchromosomal interactions result in chromosome segregation errors and chromothripsis (or chromosome shattering) (87). However, it remains to be established whether the perturbation is related to alteration of phase properties at the centromere and kinetochore (3, 6, 87).
TTK/Mps1, a kinetochore-associated dual kinase initially identified in breast cancer as an amplified oncogene (88), exhibits low structural complexity and coacervates to stabilize its localization at the kinetochore (Fig. 4B) (88, 89). Mitotic phosphorylation liberates TTK/Mps1 coacervates from the kinetochore and promotes its cytoplasmic distribution. Because amplification of TTK/Mps1 promotes breast cancer progression, and inhibition of TTK/Mps1 kinase activity leads to improved outcomes in clinical oncology (90), it would be of interest to determine whether TTK/Mps1 forms coacervates with its chemical inhibitor at the kinetochore and whether coacervates at the kinetochore are involved in the underlying mechanism for interrogation of breast cancer progression (91, 92). Pelkmans and colleagues (93) showed that another mitotic dual kinase, DYRK3, regulates membraneless organelles by preventing unmixing of the cytoplasm into aberrant organelles during mitosis, indicating that, during cell division, the kinase activities of Mps1 and DYRK3 control the LLPS. It is now desirable to discover how DYRK3 activity is regulated by mitotic kinase(s) and to determine whether there is cross-talk between TTK/Mps1 and DYRK3 signaling (74, 93).
New technology to unravel membraneless organelle dynamics
The LLPS-mediated organelle formations discussed here are largely based on in vitro experimentation and reconstitution. A pressing objective is to establish whether LLPS-mediated membraneless organelle formation and regulation are involved in live cell division as modeled by 2D and 3D culture systems. Several emerging technologies are suitable to delineate the molecular and cellular mechanisms that control this LLPS-driven membraneless organelle dynamics in mitosis. Advances in molecular imaging, optical activity reporters, chemical probes, and 3D organoid models will facilitate our understanding of the role of LLPS-driven membraneless organelle dynamics in cell division, renewal homeostasis, and disease processes.
Molecular imaging and membraneless organelle dynamics in 2D cell cultures
In eukaryotic cells, membrane-bound and membraneless organelles undergo dynamic yet organized interactions that orchestrate complex cellular functions. A better understanding of organelle contacts and trajectory relies on real-time visualization of live cells. Total internal reflection fluorescence microscopy (TIRFM) provides high-resolution and high-speed imaging but is limited by the z-depth of samples as it can image only within ∼100 nm from the basal membrane (82), which is too small a distance to span many organelles and their contact sites. To overcome this depth limitation of TIRFM, Li and colleagues (94) developed grazing incidence structured illumination microscopy (GI-SIM) by enabling the illumination to enter the objective rear pupil inside the critical angle for TIRFM, which creates an illumination field parallel to the substrate that is comparable in thickness with the objective depth-of-focus for organelle interactions in live cells. GI-SIM captures organelle dynamics at 97-nm resolution (x-y) and 266 frames/s, which allows precise measurements of microtubule growth or shrinkage events and therefore distinguishes models of microtubule dynamic instability in interphase cells (94). A challenge ahead is to capture organelle interactions during mitosis, as GI-SIM has an optimal sample thickness without compromising imaging resolution and speed.
Fluorescent proteins and chemical probes are valuable tools for studying dynamic processes within living cells. A systems-level analysis of multiple organelle interactions has been developed using a multi-spectral image acquisition method that overcomes the challenge of spectral overlap in the fluorescent protein palette (95). This spectral imaging protocol achieves, for a single live cell, simultaneous measurements of five different organelles, their numbers, volumes, speeds, positions, and dynamic interorganelle contacts (Fig. 5A). Because the size and dynamics of membraneless organelles are similar to those of membrane organelles, the spectral imaging is applicable to study the spatiotemporal dynamics of compartmentalization in the same or different chromosomes during the entire process of mitosis.
Figure 5.
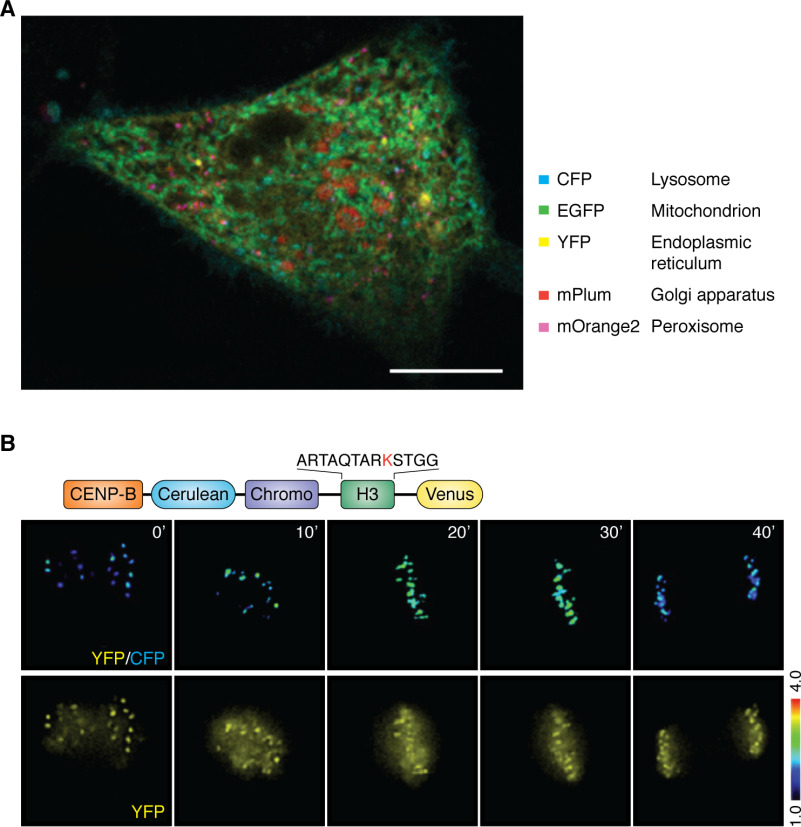
Emerging molecular imaging tools for delineating molecular interactions in membraneless organelle dynamics. A, example of spectral imaging of five organelles in an interphase HeLa cell, which provides a proof-of-principle for imaging membraneless organelles during cell division. The protocol has been optimized for imaging LLPS-driven centromere assembly dynamics. Scale bar, 10 μm (X. Liu and X. Yao, unpublished observations). B, reversible assembly dynamics of membraneless organelles are driven by post-translational modifications. Those modifications modulate the nature of multivalent interactions and provide a scaffold for organizing multivalent interactions. The figure shows the molecular design of a FRET-based methylation sensor located in the centromere and an example of the methylation gradient in live mitotic cells. The rational design of this sensor is based on the selective and reversible interaction between a chromodomain and trimethylated Lys-9 in histone 3. Adapted from Ref. 66. A similar design can be used to visualize and quantify the concentration gradients of phosphorylation and acetylation on different membraneless organelles.
Recent studies in cellular and molecular biophysics, as well as biochemical reconstitutions, have revealed that mitotic phosphorylation, acetylation, and methylation of kinetochore proteins have roles in kinetochore organization (96–98). FRET-based biosensors can be used to measure localized post-translational modification dynamics. In general, the FRET-based sensor reports changes in intramolecular FRET between cyan and yellow fluorescent proteins (61, 66). Fig. 5B is an example of a FRET-based methylation sensor based on the selective binding between the chromodomains and methylated H3K9 in vitro or in living cells (61). As seen in Fig. 5B, methylation at the centromere peaks in metaphase cells (30') is followed by a gradual decay at anaphase onset and reaches a low at telophase (40'), which projects a cell cycle–dependent methylation profile. It would be of interest to accomplish simultaneous visualization of phosphorylation and methylation gradients during chromosome segregation with parallel measurements of chromatin elasticity. This would enable us to consolidate the post-translational modification dynamics into a working model in which LLPS-driven compartmentalization controls chromatin plasticity and decision making in mitosis.
Modeling membrane organelle dynamics and cell fate in organoids
To date, studies of LLPS have relied on the use of well-documented experimental models, including primary or transformed cell lines and model systems, such as Caenorhabditis elegans. Such approaches have elucidated details of LLPS but have limited clarification of the physiological relevance of membraneless organelles to human health. Thus, the mechanisms underlying membraneless organelle spatiotemporal regulation and perturbation of its dynamics in pathogenesis of diseases remain poorly understood. Although chronic perturbation of LLPS may result in profound pathological afflictions, including ALS (99) and enteric infections such as Helicobacter pylori and SARS-CoV-2 (100, 101), the dynamics of pathogenesis and mechanisms of action have not been fully characterized. The development of 3D human organoids from various contexts therefore provides a powerful tool to study, in real time, epithelial development, stem cellular dynamics, and the host cell response to infection and other ecological perturbations (73, 102, 103).
Light-sheet fluorescence microscopy allows rapid acquisition of three-dimensional images over large fields of view and over long durations (103). Three-dimensional images are compiled from successive light-sheet illuminations of thin, two-dimensional optical sections. The high spatial and temporal resolution of the imaging enables tracking of intracellular components in relatively thick samples, such as single chromosome trajectories in 3D gastric organoids (Fig. 6A) (73). At a higher magnification, compartments in a single chromosome can be visualized (Fig. 6B). This platform provides a toolkit for real-time imaging of LLPS-driven compartmental dynamics of mitotic cells in 3D organoids. Future studies, likely using patient-derived organoids combined with spectral imaging, will afford insights into the molecular mechanisms underlying LLPS and will identify small molecules for regulating LLPS in 3D cultures (Fig. 6C).
Figure 6.
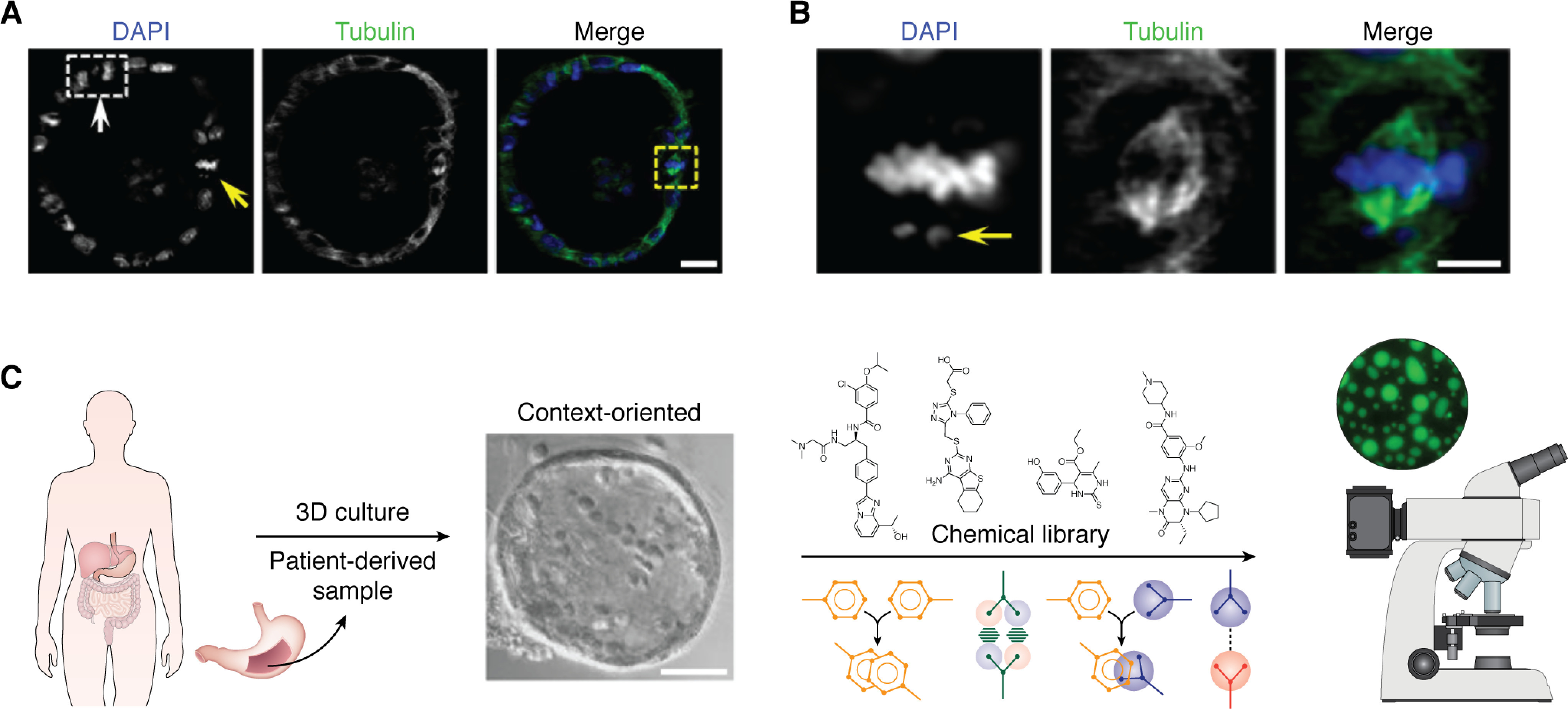
Three-dimensional organoids for modeling spatiotemporal dynamics of membraneless organelles in healthy and disease contexts. A, mouse gastric organoids were fixed for immunocytochemical staining of α-tubulin (green) and DNA staining with 4′,6-diamidino-2-phenylindole (DAPI) (blue). The light-sheet micrograph shows two mitotic cells, indicated by arrows, which contain one anaphase cell (white box, white arrow) and three metaphase cells (yellow arrow). Visualization of chromosome segregation in gastric organoids is used to study chromosome stability and establishment of polarity during cell renewal and progression of tumor cells in response to extracellular cues such as cytokines. Light-sheet micrography of gastric epithelial and stem cells can be used to study their responses to various extracellular cues or therapeutic agents. For example, organoids derived from gastric cancer cells of patients might be used to identify the most effective treatments. Real-time spectral imaging could be used to study the effects of various combinations of agents (adapted from Ref. 73). Scale bar, 20 μm. B, magnified image from A. This mitotic metaphase cell has a lagging chromosome and spindle orientation error (arrow). This image demonstrates that a combination of light-sheet microscopy with 3D organoids allows high-resolution imaging of single chromosome dynamics in a 3D context (adapted from Ref. 73). Scale bar, 10 μm. C, schematic illustration showing how human patient–derived samples can be used to visualize membraneless organelle dynamics and formation of coacervates and a chemical biological screen for compounds and/or regimens tailored to regulate LLPS for precision and combination therapeutics of cancers without targeted therapy (adapted from Ref. 73).
Perspectives
Combinations of advanced optical imaging protocols, such as lattice light-sheet microscopy with adaptive optics and photoactivatable complementary fluorescence, spectral imaging analyses, and correlative light and cryo-electron microscopic tomography (104), would increase our understanding of membraneless organelle dynamics and organelle communications. The newly developed lattice light-sheet microscopy with adaptive optics has enabled, for live organoids, noninvasive, aberration-free imaging of subcellular processes, including organelle contacts and remodeling during mitosis. This technology reveals the phenotypic diversity within cells from various organisms and developmental stages and could be useful for determining, in real-time and at the organelle level, how host cells adapt to a physiological stimulus or in response to SARS-CoV-2 infection (101). In this context, inclusion of the expanding collection of gene-edited organoids will allow modeling of chromosome interactions and LLPS regulation underlying control of cell division.
The large size and complex architecture of the centromere, which contains numerous proteins possessing low-complexity regions, are linked to its regulatory mechanisms involving post-translational modifications and interchangeable complex subunits between compartments in mitosis (9). Thus, the centromere must possess intrinsic self-control mechanisms. According to proteomic and bioinformatics analyses, the centromere is composed of perhaps 15 scaffolding proteins with a total of more than 150 proteins (3, 9, 17). Future work will describe the spatiotemporal dynamics and physicochemical properties of these low-complexity kinetochore proteins driven by LLPS during cell division in 3D organoids.
Finally, recent clinical and translational studies support our early rationale that genetic variation affects protein post-translational modifications and is involved in rewiring biological pathways to generate asymmetric division of tumor cells (17, 105). These studies demonstrate how mutations near phosphoresidues create molecular switches from its cognate kinase to a new kinase that rewires cell signaling networks and reveals oncogenic switch properties in a lung cancer bearing an epidermal growth factor receptor mutant (105). A future challenge will be to integrate a FRET-based sensor to illuminate, in live cells, how post-translational modifications and an oncogenic switch regulate the LLPS of chromosome compartments. The goal is to break the barrier of sample thickness for illumination and adapt a context-dependent physiology model system, such as 3D organoids, for molecular imaging of live organelle interactions at a superresolution level.
Despite the progress, over the past century, in understanding the function and mechanism of centromere assembly and plasticity control, much remains to be explored (16, 106). We still have a minimal understanding of the LLPS regulation underlying centromere assembly/disassembly during the cell cycle. Advances will require cryoelectron tomographic analyses of the different states of the centromeres of mitotic cells in situ. Currently, it is unclear to what extent LLPS-mediated assembly affinities compare with the allosteric structure changes elicited by post-translational modification. In addition, it remains elusive how individual centromere assembly is regulated and how this is subjected to interchromosomal interactions in cell-cycle regulation. LLPS-mediated chromosomal compartmentalization is also involved in regulating telomere assembly and plasticity (107). Thus, the challenges ahead are to delineate and distinguish the characteristics of membraneless organelles that are driven by LLPS from those independent of LLPS (108).
Overall, we anticipate that advances in our understanding of the molecular language of membraneless organelle communication will enable us to amalgamate the LLPS-driven communications in 3D organoids into a working model for decision making in cell division and targeted interrogation for aberrant LLPS-driven pathogeneses.
Acknowledgments
We thank Drs. Sandra Harris-Hooker, Peter MacLeish and Gianluca Tosini for support, Dr. Mingjie Zhang for input on the manuscript, and Dr. Phil Yao for insightful discussion during the revision.
Funding and additional information—This work was supported in part by National Natural Science Foundation of China Grants 31621002, 21922706, 81630080, 31430054, 91854203, 91853115, and 31671405; National Key Research and Development Program of China Grants 2017YFA0503600 and 2016YFA0100500; Ministry of Education Grants IRT_17R102 and 20113402130010; Strategic Priority Research Program of Chinese Academy of Sciences Grant XDB19000000; Fundamental Research Funds for the Central Universities Grants WK2340000066, WK2070000194, KB2070000023, and YD2070006001; and National Institutes of Health Grants CA164133, DK115812, MD000101, and DK56292. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- LLPS
- liquid–liquid phase separation
- H3
- histone H3
- 2D
- two-dimensional
- 3D
- three-dimensional
- TIRFM
- total internal reflection fluorescence microscopy
- GI-SIM
- grazing incidence structured illumination microscopy.
References
- 1. Miescher F. (1871) Ueber die chemische. Zusammensetzung der Eiterzellen. Medicinisch-chemische Untersuchungen 4, 441–460 [Google Scholar]
- 2. Flemming W. (1882) Zellsubstanz, Kern und Zelltheilung, F. C. W. Vogel, Leipzig [Google Scholar]
- 3. Cleveland D. W., Mao Y., and Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- 4. Knoblich J. A. (2008) Mechanisms of asymmetric stem cell division. Cell 132, 583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. (2004) T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5, 323–329 10.1038/nrm1359 [DOI] [PubMed] [Google Scholar]
- 6. Janssen A., Colmenares S. U., and Karpen G. H. (2018) Heterochromatin: guardian of the genome. Annu. Rev. Cell Dev. Biol. 34, 265–288 10.1146/annurev-cellbio-100617-062653 [DOI] [PubMed] [Google Scholar]
- 7. Pluta A. F., Cooke C. A., and Earnshaw W. C. (1990) Structure of the human centromere at metaphase. Trends Biochem. Sci. 15, 181–185 10.1016/0968-0004(90)90158-8 [DOI] [PubMed] [Google Scholar]
- 8. Yao X., Abrieu A., Zheng Y., Sullivan K. F., and Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 10.1038/35019518 [DOI] [PubMed] [Google Scholar]
- 9. McKinley K. L., and Cheeseman I. M. (2016) The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Earnshaw W. C., and Rothfield N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- 11. Parada L., and Misteli T. (2002) Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12, 425–432 10.1016/S0962-8924(02)02351-6 [DOI] [PubMed] [Google Scholar]
- 12. Evans T., Rosenthal E. T., Youngblom J., Distel D., and Hunt T. (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33, 389–396 10.1016/0092-8674(83)90420-8 [DOI] [PubMed] [Google Scholar]
- 13. Hartwell L. H., Culotti J., Pringle J. R., and Reid B. J. (1974) Genetic control of the cell division cycle in yeast. Science 183, 46–51 10.1126/science.183.4120.46 [DOI] [PubMed] [Google Scholar]
- 14. Miake-Lye R., and Kirschner M. W. (1985) Induction of early mitotic events in a cell-free system. Cell 41, 165–175 10.1016/0092-8674(85)90071-6 [DOI] [PubMed] [Google Scholar]
- 15. Simanis V., and Nurse P. (1986) The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell 45, 261–268 10.1016/0092-8674(86)90390-9 [DOI] [PubMed] [Google Scholar]
- 16. Ong J. Y., and Torres J. Z. (2019) Dissecting the mechanisms of cell division. J. Biol. Chem. 294, 11382–11390 10.1074/jbc.AW119.008149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ren J., Liu Z., Gao X., Jin C., Ye M., Zou H., Wen L., Zhang Z., Xue Y., and Yao X. (2010) MiCroKit 3.0: an integrated database of midbody, centrosome and kinetochore. Nucleic Acids Res. 38, D155–D160 10.1093/nar/gkp784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banani S. F., Lee H. O., Hyman A. A., and Rosen M. K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 20. Mao Y. S., Zhang B., and Spector D. L. (2011) Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beutel O., Maraspini R., Pombo-Garcia K., Martin-Lemaitre C., and Honigmann A. (2019) Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11 10.1016/j.cell.2019.10.011 [DOI] [PubMed] [Google Scholar]
- 23. Zeng M., Shang Y., Araki Y., Guo T., Huganir R. L., and Zhang M. (2016) Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 166, 1163–1175.e12 10.1016/j.cell.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwayer C., Shamipour S., Pranjic-Ferscha K., Schauer A., Balda M., Tada M., Matter K., and Heisenberg C. P. (2019) Mechanosensation of tight junctions depends on ZO-1 phase separation and flow. Cell 179, 937–952.e18 10.1016/j.cell.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 25. Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B. T., Alberti S., Diez S., and Hyman A. A. (2017) Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saha S., Weber C. A., Nousch M., Adame-Arana O., Hoege C., Hein M. Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., Pozniakovski A., Eckmann C. R., Julicher F., and Hyman A. A. (2016) Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nott T. J., Craggs T. D., and Baldwin A. J. (2016) Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 8, 569–575 10.1038/nchem.2519 [DOI] [PubMed] [Google Scholar]
- 28. Su X., Ditlev J. A., Hui E., Xing W., Banjade S., Okrut J., King D. S., Taunton J., Rosen M. K., and Vale R. D. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du M., and Chen Z. J. (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freeman Rosenzweig E. S., Xu B., Kuhn Cuellar L., Martinez-Sanchez A., Schaffer M., Strauss M., Cartwright H. N., Ronceray P., Plitzko J. M., Forster F., Wingreen N. S., Engel B. D., Mackinder L. C. M., and Jonikas M. C. (2017) The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 10.1016/j.cell.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woodruff J. B., Ferreira Gomes B., Widlund P. O., Mahamid J., Honigmann A., and Hyman A. A. (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 32. Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., and Zheng Y. (2015) Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122 10.1016/j.cell.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., and Narlikar G. J. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., and Karpen G. H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bouchard J. J., Otero J. H., Scott D. C., Szulc E., Martin E. W., Sabri N., Granata D., Marzahn M. R., Lindorff-Larsen K., Salvatella X., Schulman B. A., and Mittag T. (2018) Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 10.1016/j.molcel.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., and Zhou Q. (2018) Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 10.1038/s41586-018-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pesenti M. E., Weir J. R., and Musacchio A. (2016) Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 37, 152–163 10.1016/j.sbi.2016.03.003 [DOI] [PubMed] [Google Scholar]
- 39. Akhmanova A., and Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- 40. Cheeseman I. M., and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 41. Ward T., Wang M., Liu X., Wang Z., Xia P., Chu Y., Wang X., Liu L., Jiang K., Yu H., Yan M., Wang J., Hill D. L., Huang Y., Zhu T., et al. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785 10.1074/jbc.M112.448886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., and Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U. S. A. 109, 16564–16569 10.1073/pnas.1202639109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang K., Wang J., Liu J., Ward T., Wordeman L., Davidson A., Wang F., and Yao X. (2009) TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865 10.1038/embor.2009.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bao X., Liu H., Liu X., Ruan K., Zhang Y., Zhang Z., Hu Q., Liu Y., Akram S., Zhang J., Gong Q., Wang W., Yuan X., Li J., Zhao L., et al. (2018) Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation. J. Mol. Cell Biol. 10, 18–32 10.1093/jmcb/mjx045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Y., and Yu H. (2018) Partner switching for Ran during the mitosis dance. J. Mol. Cell Biol. 10, 89–90 10.1093/jmcb/mjx048 [DOI] [PubMed] [Google Scholar]
- 46. Clarke P. R., and Zhang C. (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9, 464–477 10.1038/nrm2410 [DOI] [PubMed] [Google Scholar]
- 47. Zhang C., and Clarke P. R. (2000) Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288, 1429–1432 10.1126/science.288.5470.1429 [DOI] [PubMed] [Google Scholar]
- 48. Chu L., Huo Y., Liu X., Yao P., Thomas K., Jiang H., Zhu T., Zhang G., Chaudhry M., Adams G., Thompson W., Dou Z., Jin C., He P., and Yao X. (2014) The spatiotemporal dynamics of chromatin protein HP1α is essential for accurate chromosome segregation during cell division. J. Biol. Chem. 289, 26249–26262 10.1074/jbc.M114.581504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu X., Song Z., Huo Y., Zhang J., Zhu T., Wang J., Zhao X., Aikhionbare F., Zhang J., Duan H., Wu J., Dou Z., Shi Y., and Yao X. (2014) Chromatin protein HP1 interacts with the mitotic regulator borealin protein and specifies the centromere localization of the chromosomal passenger complex. J. Biol. Chem. 289, 20638–20649 10.1074/jbc.M114.572842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yao X., and Fang G. (2009) Visualization and orchestration of the dynamic molecular society in cells. Cell Res. 19, 152–155 10.1038/cr.2009.7 [DOI] [PubMed] [Google Scholar]
- 51. Maass P. G., Barutcu A. R., and Rinn J. L. (2019) Interchromosomal interactions: a genomic love story of kissing chromosomes. J. Cell Biol. 218, 27–38 10.1083/jcb.201806052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R. M., Zhu P., and Li G. (2014) Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 10.1126/science.1251413 [DOI] [PubMed] [Google Scholar]
- 53. Zhao S., Cheng L., Gao Y., Zhang B., Zheng X., Wang L., Li P., Sun Q., and Li H. (2019) Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation. Cell Res. 29, 54–66 10.1038/s41422-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen J., Wang Z., Guo X., Li F., Wei Q., Chen X., Gong D., Xu Y., Chen W., Liu Y., Kang J., and Shi Y. (2019) TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells. Nat. Commun. 10, 4273 10.1038/s41467-019-12126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gibson B. A., Doolittle L. K., Schneider M. W. G., Jensen L. E., Gamarra N., Henry L., Gerlich D. W., Redding S., and Rosen M. K. (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Choi J. M., Holehouse A. S., and Pappu R. V. (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 10.1146/annurev-biophys-121219-081629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., Zhang X., Yao X., Zhou Z. H., and Zang J. (2018) Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28, 374–378 10.1038/cr.2018.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mukherjee S., Sandri B. J., Tank D., McClellan M., Harasymiw L. A., Yang Q., Parker L. L., and Gardner M. K. (2019) A gradient in metaphase tension leads to a scaled cellular response in mitosis. Dev. Cell 49, 63–76.e10 10.1016/j.devcel.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akram S., Yang F., Li J., Adams G., Liu Y., Zhuang X., Chu L., Liu X., Emmett N., Thompson W., Mullen M., Muthusamy S., Wang W., Mo F., and Liu X. (2018) LRIF1 interacts with HP1α to coordinate accurate chromosome segregation during mitosis. J. Mol. Cell Biol. 10, 527–538 10.1093/jmcb/mjy040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trivedi P., Palomba F., Niedzialkowska E., Digman M. A., Gratton E., and Stukenberg P. T. (2019) The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol. 21, 1127–1137 10.1038/s41556-019-0376-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chu Y., Yao P. Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., and Yao X. (2011) Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 3, 260–267 10.1093/jmcb/mjq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu D., Vader G., Vromans M. J., Lampson M. A., and Lens S. M. (2009) Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang Y., Li T., Ems-McClung S. C., Walczak C. E., Prigent C., Zhu X., Zhang X., and Zheng Y. (2018) Aurora A activation in mitosis promoted by BuGZ. J. Cell Biol. 217, 107–116 10.1083/jcb.201706103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang H., He X., Wang S., Jia J., Wan Y., Wang Y., Zeng R., Yates J. 3rd, Zhu X., and Zheng Y. (2014) A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell 28, 268–281 10.1016/j.devcel.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Toledo C. M., Herman J. A., Olsen J. B., Ding Y., Corrin P., Girard E. J., Olson J. M., Emili A., DeLuca J. G., and Paddison P. J. (2014) BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell 28, 282–294 10.1016/j.devcel.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., and Yao X. (2012) SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340 10.1093/jmcb/mjs023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rieder C. L. (1982) The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79, 1–58 10.1016/s0074-7696(08)61672-1 [DOI] [PubMed] [Google Scholar]
- 68. Yao X., Anderson K. L., and Cleveland D. W. (1997) The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447 10.1083/jcb.139.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schaar B. T., Chan G. K., Maddox P., Salmon E. D., and Yen T. J. (1997) CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382 10.1083/jcb.139.6.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yen T. J., Compton D. A., Wise D., Zinkowski R. P., Brinkley B. R., Earnshaw W. C., and Cleveland D. W. (1991) CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10, 1245–1254 10.1002/j.1460-2075.1991.tb08066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ding X., Yan F., Yao P., Yang Z., Wan W., Wang X., Liu J., Gao X., Abrieu A., Zhu T., Zhang J., Dou Z., and Yao X. (2010) Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 20, 1386–1389 10.1038/cr.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wood K. W., Lad L., Luo L., Qian X., Knight S. D., Nevins N., Brejc K., Sutton D., Gilmartin A. G., Chua P. R., Desai R., Schauer S. P., McNulty D. E., Annan R. S., Belmont L. D., et al. (2010) Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. U. S. A. 107, 5839–5844 10.1073/pnas.0915068107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu X., Xu L., Li J., Yao P. Y., Wang W., Ismail H., Wang H., Liao B., Yang Z., Ward T., Ruan K., Zhang J., Wu Q., He P., Ding X., et al. (2019) Mitotic motor CENP-E cooperates with PRC1 in temporal control of central spindle assembly. J Mol. Cell Biol. 10.1093/jmcb/mjz051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang Y., Lin L., Liu X., Ye S., Yao P. Y., Wang W., Yang F., Gao X., Li J., Zhang Y., Zhang J., Yang Z., Liu X., Yang Z., Zang J., et al. (2019) BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res. 29, 562–578 10.1038/s41422-019-0178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu X., Shen J., Xie L., Wei Z., Wong C., Li Y., Zheng X., Li P., and Song Y. (2020) Mitotic implantation of the transcription factor prospero via phase separation drives terminal neuronal differentiation. Dev. Cell 52, 277–293.e8 10.1016/j.devcel.2019.11.019 [DOI] [PubMed] [Google Scholar]
- 76. Kim Y., Heuser J. E., Waterman C. M., and Cleveland D. W. (2008) CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J. Cell Biol. 181, 411–419 10.1083/jcb.200802189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nigg E. A., and Holland A. J. (2018) Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zwicker D., Decker M., Jaensch S., Hyman A. A., and Julicher F. (2014) Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. U. S. A. 111, E2636–E2645 10.1073/pnas.1404855111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu J., Wen W., Zheng Z., Shang Y., Wei Z., Xiao Z., Pan Z., Du Q., Wang W., and Zhang M. (2011) LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gαi/LGN/NuMA pathways. Mol. Cell 43, 418–431 10.1016/j.molcel.2011.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lu H., Zhao Q., Jiang H., Zhu T., Xia P., Seffens W., Aikhionbare F., Wang D., Dou Z., and Yao X. (2014) Characterization of ring-like F-actin structure as a mechanical partner for spindle positioning in mitosis. PLoS ONE 9, e102547 10.1371/journal.pone.0102547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yu H., Yang F., Dong P., Liao S., Liu W. R., Zhao G., Qin B., Dou Z., Liu Z., Liu W., Zang J., Lippincott-Schwartz J., Liu X., and Yao X. (2019) NDP52 tunes cortical actin interaction with astral microtubules for accurate spindle orientation. Cell Res. 29, 666–679 10.1038/s41422-019-0189-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xia P., Liu X., Wu B., Zhang S., Song X., Yao P. Y., Lippincott-Schwartz J., and Yao X. (2014) Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell 25, 4166–4173 10.1091/mbc.E14-06-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rio Frio T., Lavoie J., Hamel N., Geyer F. C., Kushner Y. B., Novak D. J., Wark L., Capelli C., Reis-Filho J. S., Mai S., Pastinen T., Tischkowitz M. D., Marcus V. A., and Foulkes W. D. (2010) Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N. Engl. J. Med. 363, 2628–2637 10.1056/NEJMoa1006565 [DOI] [PubMed] [Google Scholar]
- 85. Cahill D. P., Lengauer C., Yu J., Riggins G. J., Willson J. K., Markowitz S. D., Kinzler K. W., and Vogelstein B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature 392, 300–303 10.1038/32688 [DOI] [PubMed] [Google Scholar]
- 86. Ly P., Brunner S. F., Shoshani O., Kim D. H., Lan W., Pyntikova T., Flanagan A. M., Behjati S., Page D. C., Campbell P. J., and Cleveland D. W. (2019) Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 51, 705–715 10.1038/s41588-019-0360-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Umbreit N. T., Zhang C. Z., Lynch L. D., Blaine L. J., Cheng A. M., Tourdot R., Sun L., Almubarak H. F., Judge K., Mitchell T. J., Spektor A., and Pellman D. (2020) Mechanisms generating cancer genome complexity from a single cell division error. Science 368, eaba0712 10.1126/science.aba0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dou Z., Prifti D. K., Gui P., Liu X., Elowe S., and Yao X. (2019) Recent progress on the localization of the spindle assembly checkpoint machinery to kinetochores. Cells 8, 278 10.3390/cells8030278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gui P., Sedzro D. M., Yuan X., Liu S., Hei M., Tian W., Zohbi N., Wang F., Yao Y., Aikhionbare F. O., Gao X., Wang D., Yao X., and Dou Z. (2020) Mps1 dimerization and multisite interactions with Ndc80 complex enable responsive spindle assembly checkpoint signaling. J. Mol. Cell Biol. 10.1093/jmcb/mjaa006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chandler B. C., Moubadder L., Ritter C. L., Liu M., Cameron M., Wilder-Romans K., Zhang A., Pesch A. M., Michmerhuizen A. R., Hirsh N., Androsiglio M., Ward T., Olsen E., Niknafs Y. S., Merajver S., et al. (2020) TTK inhibition radiosensitizes basal-like breast cancer through impaired homologous recombination. J. Clin. Invest. 130, 958–973 10.1172/JCI130435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Song X., Liu W., Yuan X., Jiang J., Wang W., Mullen M., Zhao X., Zhang Y., Liu F., Du S., Rehman A., Tian R., Li J., Frost A., Song Z., et al. (2018) Acetylation of ACAP4 regulates CCL18-elicited breast cancer cell migration and invasion. J. Mol. Cell Biol. 10, 559–572 10.1093/jmcb/mjy058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen J., Yao Y., Gong C., Yu F., Su S., Chen J., Liu B., Deng H., Wang F., Lin L., Yao H., Su F., Anderson K. S., Liu Q., Ewen M. E., et al. (2011) CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555 10.1016/j.ccr.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rai A. K., Chen J. X., Selbach M., and Pelkmans L. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 10.1038/s41586-018-0279-8 [DOI] [PubMed] [Google Scholar]
- 94. Guo Y., Li D., Zhang S., Yang Y., Liu J. J., Wang X., Liu C., Milkie D. E., Moore R. P., Tulu U. S., Kiehart D. P., Hu J., Lippincott-Schwartz J., Betzig E., and Li D. (2018) Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175, 1430–1442.e17 10.1016/j.cell.2018.09.057 [DOI] [PubMed] [Google Scholar]
- 95. Valm A. M., Cohen S., Legant W. R., Melunis J., Hershberg U., Wait E., Cohen A. R., Davidson M. W., Betzig E., and Lippincott-Schwartz J. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 10.1038/nature22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Allan L. A., Camacho Reis M., Ciossani G., Huis In 't Veld P. J., Wohlgemuth S., Kops G. J., Musacchio A., and Saurin A. T. (2020) Cyclin B1 scaffolds MAD1 at the kinetochore corona to activate the mitotic checkpoint. EMBO J. 39, e103180 10.15252/embj.2019103180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yan K., Yang J., Zhang Z., McLaughlin S. H., Chang L., Fasci D., Ehrenhofer-Murray A. E., Heck A. J. R., and Barford D. (2019) Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282 10.1038/s41586-019-1609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mo F., Zhuang X., Liu X., Yao P. Y., Qin B., Su Z., Zang J., Wang Z., Zhang J., Dou Z., Tian C., Teng M., Niu L., Hill D. L., Fang G., et al. (2016) Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232 10.1038/nchembio.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gasset-Rosa F., Lu S., Yu H., Chen C., Melamed Z., Guo L., Shorter J., Da Cruz S., and Cleveland D. W. (2019) Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 10.1016/j.neuron.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Morey P., Pfannkuch L., Pang E., Boccellato F., Sigal M., Imai-Matsushima A., Dyer V., Koch M., Mollenkopf H. J., Schlaermann P., and Meyer T. F. (2018) Helicobacter pylori depletes cholesterol in gastric glands to prevent interferon γ signaling and escape the inflammatory response. Gastroenterology 154, 1391–1404.e9 10.1053/j.gastro.2017.12.008 [DOI] [PubMed] [Google Scholar]
- 101. Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., Ravelli R. B. G., Paul van Schayck J., Mykytyn A. Z., Duimel H. Q., van Donselaar E., Riesebosch S., Kuijpers H. J. H., Schipper D., van de Wetering W. J., et al. (2020) SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., and Clevers H. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 103. Yao X., and Smolka A. J. (2019) Gastric parietal cell physiology and Helicobacter pylori-induced disease. Gastroenterology 156, 2158–2173 10.1053/j.gastro.2019.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tao C. L., Liu Y. T., Sun R., Zhang B., Qi L., Shivakoti S., Tian C. L., Zhang P., Lau P. M., Zhou Z. H., and Bi G. Q. (2018) Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J. Neurosci. 38, 1493–1510 10.1523/JNEUROSCI.1548-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lundby A., Franciosa G., Emdal K. B., Refsgaard J. C., Gnosa S. P., Bekker-Jensen D. B., Secher A., Maurya S. R., Paul I., Mendez B. L., Kelstrup C. D., Francavilla C., Kveiborg M., Montoya G., Jensen L. J., et al. (2019) Oncogenic mutations rewire signaling pathways by switching protein recruitment to phosphotyrosine sites. Cell 179, 543–560.e26 10.1016/j.cell.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 106. Kops G., and Gassmann R. (2020) Crowning the kinetochore: the fibrous corona in chromosome segregation. Trends Cell Biol. 30, 653–667 10.1016/j.tcb.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 107. Zhang H., Zhao R., Tones J., Liu M., Dilley R., Chenoweth D. M., Greenberg R. A., and Lampson M. A. (2020) Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 31, 2048–2056 10.1091/mbc.e19-10-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. McSwiggen D. T., Mir M., Darzacq X., and Tjian R. (2019) Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 33, 1619–1634 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., Ding X., Huang Y., Wang D., and Yao X. (2012) Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287, 39380–39390 10.1074/jbc.M112.406652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dou Z., Liu X., Wang W., Zhu T., Wang X., Xu L., Abrieu A., Fu C., Hill D. L., and Yao X. (2015) Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl. Acad. Sci. U. S. A. 112, E4546–E4555 10.1073/pnas.1508791112 [DOI] [PMC free article] [PubMed] [Google Scholar]