Abstract
Since the advent of protein crystallography, atomic-level macromolecular structures have provided a basis to understand biological function. Enzymologists use detailed structural insights on ligand coordination, interatomic distances, and positioning of catalytic amino acids to rationalize the underlying electronic reaction mechanisms. Often the proteins in question catalyze redox reactions using metal cofactors that are explicitly intertwined with their function. In these cases, the exact nature of the coordination sphere and the oxidation state of the metal is of utmost importance. Unfortunately, the redox-active nature of metal cofactors makes them especially susceptible to photoreduction, meaning that information obtained by photoreducing X-ray sources about the environment of the cofactor is the least trustworthy part of the structure. In this work we directly compare the kinetics of photoreduction of six different heme protein crystal species by X-ray radiation. We show that a dose of ∼40 kilograys already yields 50% ferrous iron in a heme protein crystal. We also demonstrate that the kinetics of photoreduction are completely independent from variables unique to the different samples tested. The photoreduction-induced structural rearrangements around the metal cofactors have to be considered when biochemical data of ferric proteins are rationalized by constraints derived from crystal structures of reduced enzymes.
Keywords: protein crystallography, radiation damage, heme proteins, heme, enzyme structure, oxidation-reduction (redox), X-ray crystallography, protein crystallization
Enzyme-mediated transfer of electrons is an integral component of numerous fundamental biological reactions, such as respiration, photosynthesis, catabolic and anabolic transformations in metabolism, molecular signaling, or cellular defense. Any catalytic reaction mediated by redox-active enzymes is principally dependent on the nature and electronic state of the respective transition metals, which are modulated by inner- and outer-sphere ligands and solvent exposure of the active site. To elucidate the reaction mechanism of a given redox enzyme, X-ray crystallography has been the principal means of high-resolution structural characterization of these enzymes and their intermediate states. Here, the main focus lies on the metal coordination and ligation state, together with inter- and intramolecular distances, which depend primarily on the oxidation state of the metal center itself. However, X-rays cause rapid reduction of metal centers, thereby inducing subsequent changes in their stereochemistry (1). Consequently, care must be taken when interpreting X-ray crystal structures obtained from single-crystal data sets through routine X-ray diffraction experiments, here nominally defined as a 360° rotation data set collected at 100 K, with a total mean diffraction weighted dose (calculated by RADDOSE-3D version 4.0) of more than 1 MGy.
X-rays induce both site-specific and global damage in protein crystals (2, 3). Of chief interest in the case of metalloproteins is the site-specific damage inflicted by the formation of photoelectrons and free radical species created by the ionizing effects of X-ray radiation (1). Cryo-cooling the crystals to 100 K or lower has been shown to alleviate the effect of free radicals by reducing their diffusion through the solvent channels in the crystal (4). On the other hand, the rate of formation of photoelectrons is dose-dependent. These electrons can travel freely through the protein crystal, even at cryo-temperatures, and will reduce the metal centers of proteins (5). Redox-active proteins have been optimized by evolution to efficiently channel electrons into an oxidized active site (6). Heme iron was aptly described by Beitlich and co-workers as a “vacuum cleaner” for sucking in electrons (7), emphasizing concerns already stated by Berglund et al. (6) in 2002 about the actual oxidation state of many redox proteins in the Protein Data Bank.
To overcome this problem, protein crystallographers have developed methods to lower the overall dose absorbed per crystal used in X-ray diffraction experiments. This can effectively be done by using a multicrystal methodology (8, 9) and has already been applied to obtain ferric cryo-structures of three heme proteins. Nonreduced X-ray structures of the cytochrome c peroxidase intermediate Compound I were reported by two groups independently. These two structures, estimated to be 10% reduced, were derived from either 10 or 19 crystals, with each crystal receiving a dose of 20 or 35 kGy, respectively (8, 9). A study on horseradish peroxidase obtained a ferric structure from seven crystals with 15° rotational data collected per crystal. However, direct comparison is difficult, because no final dose absorbed by the crystals was reported (6). Crucially, an in-depth study, using UV-visible and online Resonance Raman microspectroscopy, also investigated radiation-induced reduction of ferric metmyoglobin crystals and found that reduction of the ferric heme iron occurs almost immediately upon exposure to the X-ray beam. The authors observed the reductive influence of X-rays already after an absorbed dose of 5.3 kGy. They calculated a dose of 38–110 kGy for 50% reduction of ferric metmyoglobin crystals (10). A study on an A-class dye-decolorizing peroxidase from Streptomyces lividans also measured dose-dependent reduction and found the dose required for half-maximal reduction to be 3 kGy (11). These investigations were all conducted at 100 K and collectively report a wide dose range to achieve a 50% reduction in their respective heme proteins. The cause of the broad range (3–175 kGy) remains unclear because both the investigated proteins and the employed methods differ in individual studies.
The use of X-ray free-electron laser (XFEL) light sources with pulses on the order of 10–20 fs has promised the possibility of outrunning radiation damage. XFEL sources have already been employed to investigate metalloproteins (5, 12–16), capturing the nonreduced redox center for the first time (13, 17). However, XFEL experiments are not routine, and beamtime is scarce. Similarly, the use of nonreducing neutron sources is limited not only by beamtime availability but additionally by the requirement for large, deuterated crystals, which is a severe limitation (18). Therefore, both methods are unlikely to replace synchrotrons as the main radiation source for structural biology studies in the near future.
Here, we present a study of the pre-steady-state reduction kinetics of X-ray induced photoreduction of six diverse heme proteins to determine the rate constant for reduction (kred) and a general dose limit for the collection of nonreduced data sets for redox-active metalloproteins at 100 K. Radiation-induced reduction follows a reaction in the form of “Aox + e− → Ared” and could potentially depend on sample-derived variables (secondary and tertiary structure of the protein, crystallization conditions and crystal morphology, type of cofactor) as well as radiation-derived variables (dose and dose rate, wavelength). The results show that the reduction kinetics are solely dependent upon dose and that sample-derived variables for dose calculations can be neglected. A protein-independent dose limit of ∼40 kGy that corresponds to a 50% reduction of the initial redox state can therefore be deduced. Furthermore, we used a method for collecting ultra-low dose (<3 kGy/crystal) rotation data at synchrotron light sources at 100 K as part of a dose series from 5 to 100% Fe(II) reduction. Structures of B-class dye-decolorizing peroxidase from Klebsiella pneumoniae (KpDyP) in water and cyanide ligation were obtained by merging data from ∼700 crystals. Structural differences in the active site are observed between the ferric and photoreduced structures. This study strongly highlights the need to critically scrutinize active-site structures of metalloproteins that were derived from non-low-dose data collection strategies.
Results
Selection of metalloproteins
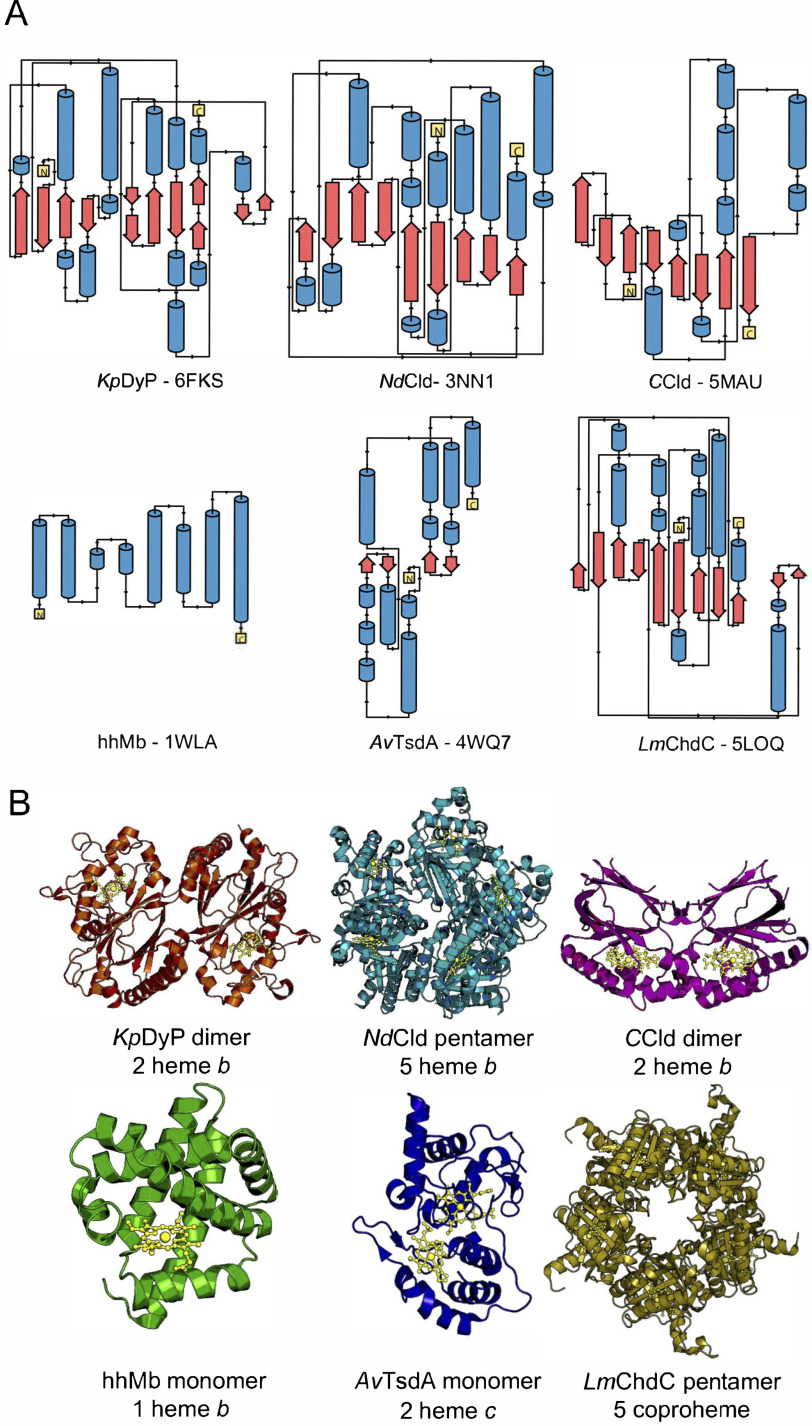
Reduction caused by X-ray radiation is a major problem in correlating the structure and function of metalloproteins. Several independent studies have set out to study the reduction of metal centers as well as possible ways to mitigate their reduction (e.g. by the use of scavengers, variation of wavelength, lower temperatures, or composite data collection strategies). For heme proteins, there are essentially three reported cases: myoglobin (10, 19–21), cytochrome c peroxidase, and horseradish peroxidase (8, 9). For a more general approach to study reduction kinetics, a set of six diverse heme proteins was selected; metmyoglobin from horse heart (hhMb), a diheme c protein, thiosulfate dehydrogenase from Allochromatium vinosum (AvTsdA) (22–25), a B-class dye-decolorizing peroxidase (KpDyP) (26), chlorite dismutases from Nitrospira defluvii (NdCld) (27) and from Cyanothece sp. PCC7425 (CCld) (28), and a coproheme decarboxylase from Listeria monocytogenes (LmChdC) (29). Of the six proteins, two are monomeric (hhMb and AvTsdA), two are dimeric (KpDyP and CCld), and two are pentameric (NdCld and LmChdC). Four of the six proteins (KpDyP, NdCld, CCld, and hhMb) have one heme b cofactor per subunit. LmChdC incorporates an iron coproporphyrin III (coproheme), a heme b precursor with four propionate groups. Except for AvTsdA, all the heme groups are pentacoordinated with a histidine as the proximal ligand. AvTsdA is a diheme cytochrome carrying two heme c cofactors (heme c is a thiolated heme variant that is covalently bound via thioether linkages to two cysteine residues). The set of model proteins also encompasses a variety of secondary structure compositions and folds, different crystal morphologies, and crystallization conditions. Approximate distributions of secondary structure elements calculated from the available crystal structures are shown in Fig. 1 and Table 1.
Figure 1.
Illustration of the diversity of secondary, tertiary and quaternary structure elements of the investigated heme proteins. A, secondary structure elements. α helices are shown in blue, β sheets are in red, and loop regions are shown as black lines. N and C termini are shown as yellow boxes. B, crystal structures of the investigated proteins. KpDyP (orange) and CCld (pink) form dimeric structures with one heme b moiety (gold) per subunit and a dimeric or truncated α-β ferredoxin fold. NdCld (turquoise) and LmChdC (yellow) form pentamers with one heme b or coproheme moiety per subunit and an α/β ferredoxin fold. hhMb (green) is a monomeric α-helical protein with one heme b cofactor. AvTsdA (blue) is a monomeric protein with two heme c cofactors and a predominantly α-helical structure. The protein backbone and cofactors are depicted as cartoon and ball-and-stick representations, respectively.
Table 1.
Biochemical and biophysical properties of the studied heme proteins
Overview of the respective redox potentials E°′ (Fe(III)/Fe(II)), amino acid composition, and overall secondary structure.
| Protein | Redox potential E°′ (Fe(II)/Fe(III)) | No. of amino acids (multimer) | Secondary structure elements (%) |
||
|---|---|---|---|---|---|
| β-Strands | Helix | Others | |||
| KpDyP | −350 ± 1 mV (26) | 299 (598) | 23.2 | 32.6 | 44 |
| NdCld | −113 ± 1 mV (30) | 264 (1320) | 26.9 | 42.0 | 31 |
| CCld | −126 ± 1.9 mV (31) | 182 (364) | 35.8 | 28.9 | 35.3 |
| hhMb | 28 ± 5 mV (32) | 154 (154) | 0 | 73.9 | 26.1 |
| AvTsdA | Heme 1 (His/Cys ligation): −185 mV (25) | 270 (270) | 4.8 | 39.5 | 55.7 |
| Heme 2 (His/Lys ligation): −129 mV (25) | |||||
| Heme 2 (His/Met ligation): +266 mV (25) | |||||
| LmChdC | −205 ± 3 mV (33) | 251 (1255) | 31 | 37.9 | 31 |
Crystal morphology and composition
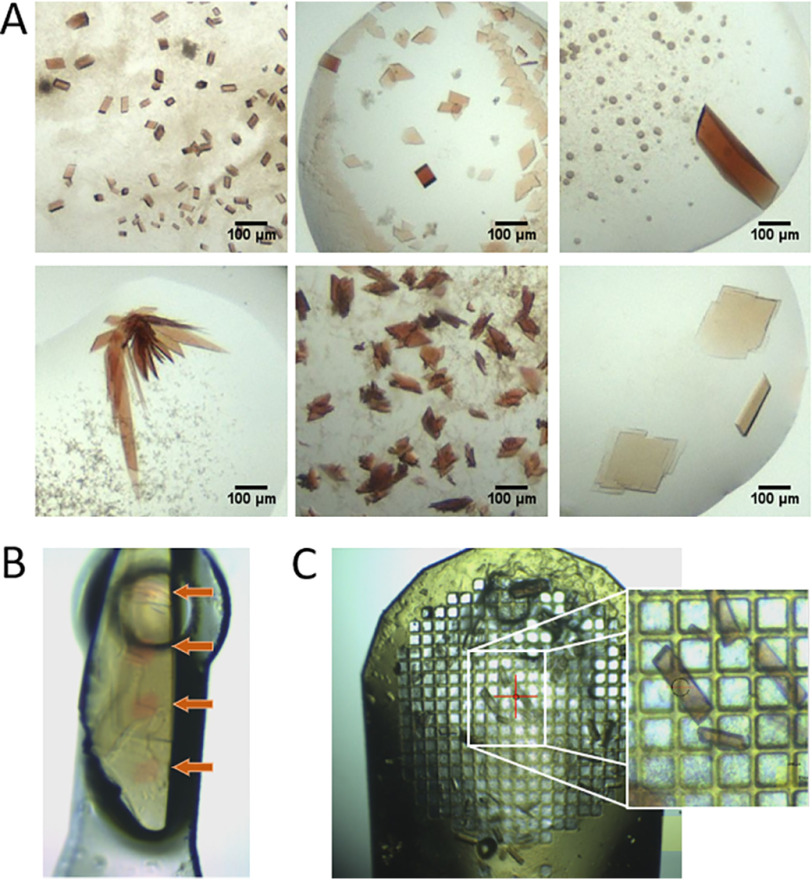
The diversity of shapes that protein crystals can form is well-known and difficult to influence. The crystal volume exposed to X-ray radiation (and thus the amount of iron exposed to the beam) differs, depending on the crystal form and its thickness along the beam direction. NdCld and LmChdC crystallized as plates of rhomboid or irregular shape with a thickness of 5–20 or 5–10 μm. The hhMb rosetta-shaped clusters of thin, platelike crystals were described previously (approximate thickness of up to 5 μm) (21). AvTsdA formed clusters of rhomboid plates from which the most platelike with an approximate thickness of 5 μm were selected. KpDyP crystals are cuboids with an average thickness of 15 μm. CCld formed the largest crystals, which were either triangular or rhomboid in shape and had an approximate thickness of 20 μm (Fig. 2A and Fig. S1).
Figure 2.
Representative crystal forms and sizes of the investigated heme proteins. A, crystal forms and sizes of KpDyP (top, left), NdCld (top, middle), CCld (top, right), hhMb (bottom, left), AvTsdA (bottom, middle), and LmChdC (bottom, right). B, visualized impact of X-ray–induced reduction as seen on a mounted crystal of LmChdC. Red dots (indicated by the orange arrows) show areas hit by the X-ray beam. C, 750-μm mesh loaded with crystals from KpDyP and close up. Mesh cryo-cooling allows the rapid manual collection of up to 30–50 crystals without the need for time-consuming sample changing.
Theoretically, a combination of crystal packing and crystal size determines the amount of reducible Fe(III) in a crystal. Therefore, the approximate radiation-exposed crystal volume, assuming a beam size of 20 × 20 μm, Fe(III) concentration, and total Fe(III) amount available for reduction (Table 2) were inferred for all protein crystals. The Fe(III) concentration was found to be similar for all proteins (29 µmol/cm3 on average) for the UV-visible experiment. Taking the exposed crystal volume into account, the absolute Fe(III) amount exposed to X-ray radiation was calculated to be between 0.2 and 200 fmol (Table 2). Irradiated and reduced spots on an initially ferric heme crystal are easily distinguishable by color (Fig. 2B).
Table 2.
Calculation of iron content of protein crystals used in this study
| Protein | Approximate crystal volume in X-ray beam | Space group | Unit cell volume (× 10−20) | Fe(III)/unit cell | Fe(III) concentration | Fe(III) amount |
|---|---|---|---|---|---|---|
| μm3 | cm3 | μmol/cm3 | fmol | |||
| KpDyP (6FKS) | 6200 | P 1 21 1 | 27.6 | 4 | 24.1 | 17.1 |
| NdCld (3NN1) | 1570–6200 | P 32 2 1 | 250 | 30 | 19.9 | 39.3–155.3 |
| CCld (5MAU) | 7850–15,700 | P 1 | 127.7 | 2 | 2.6 | 100.2–200.5 |
| hhMb (1WLA) | 314 | P 1 21 1 | 6.4 | 2 | 52.0 | 0.2 |
| AvTsdA (4WQ7) | 314–1570 | C 1 2 1 | 24.8 | 8 | 53.5 | 0.8–3.9 |
| LmChdC (6FXJ) | 1570–3100 | P 1 21 1 | 75.4 | 10 | 22.0 | 11.8–23.4 |
Crystallization conditions were shown to influence global radiation damage, because specific chemicals containing atoms with large X-ray absorption cross-section can boost the radiation-induced damage (10, 34–36). Our set of model proteins crystallized in a total of 10 different conditions. NdCld crystallized in over 50% of the conditions from the PACT screen (Molecular Dimensions), and crystals from five representative conditions were tested (Fig. S1).
Photoreduction is independent of sample-derived variables
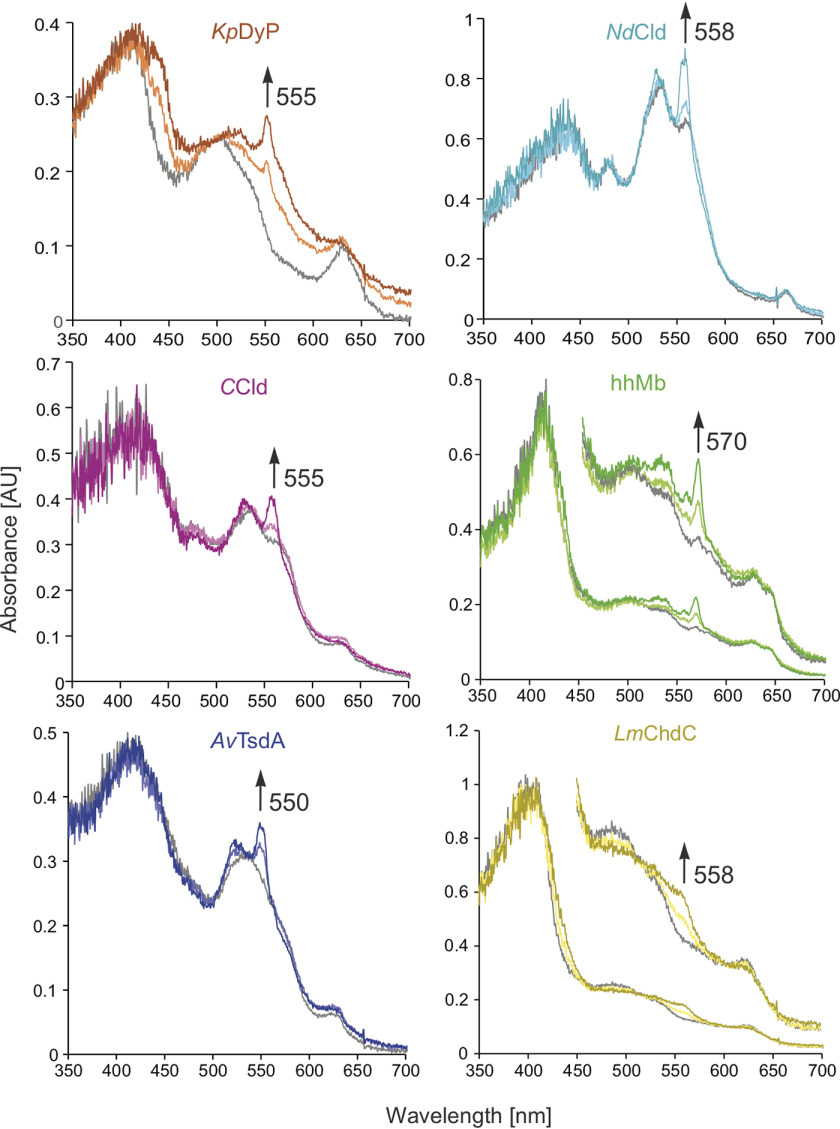
The absorption spectra of the heme protein crystal test set were investigated using the online, off-axis microspectrometer available at I24, Diamond Light Source (DLS) (Didcot, UK). Initially, the resting state spectral characteristics of each protein crystal in its respective cryo-conditions at 100 K (Fig. 3, gray spectra) were recorded. Half of the model proteins (KpDyP, hhMb, and LmChdC) exhibited spectra corresponding to their high-spin (HS) ferric resting state in solution. CCld and NdCld showed maxima corresponding to their low-spin (LS) OH-ligated ferric resting states (26, 31, 33, 37). The spectral characteristics of AvTsdA correspond to the LS state due to hexacoordination of the heme c cofactors by histidine and cysteine or methionine (22).
Figure 3.
Online microspectroscopy. Overlay of single-crystal UV-visible spectra obtained from online microspectroscopy (10% transmission) of the ferric (gray), ∼50% reduced (lightly shaded), and final (fully reduced, dark shaded) crystals of KpDyP (orange), NdCld (turquoise), CCld (pink), hhMb (green), AvTsdA (blue), and LmChdC (yellow). Arrows indicate the distinct increase in absorbance at the wavelength that was used to monitor the progress of photoreduction.
The X-ray–induced spectral changes to the resting state spectra were then obtained using a flux of 4.2 × 1011 photons s−1 (10% transmission at I24, DLS) as presented in Fig. 3 All radiation-induced spectral changes in the crystals were generated using a 20 × 20 μm focus (full-width half-maximum) with a Gaussian beam profile. The reduction-mediated spectral transitions in crystallo from ferric to ferrous states are also observed in solution at pH 7.0 (Fig. S2); ratios of HS and LS species in NdCld and CCld depend on pH and buffer conditions (28, 38). In solution, the reduction of the ferric resting state is typically characterized by absorption changes in the visible range (400–700 nm). In KpDyP, the intensity of the absorption peaks at 507 nm with shoulders at 540 (Q-bands) and 630 nm (charge transfer (CT) bands) decreased upon X-ray radiation, and a new band at 550 nm, corresponding to the HS ferrous iron appeared. In NdCld, the band at 535 nm showed a slight shift toward 530 nm, whereas the band at 560 nm exhibited a marked increase in absorbance in combination with a shift to 558 nm. A similar shift was observed for CCld; bands at 535 and 570 nm exhibited the same spectral shifts toward 530 and 558 nm, respectively, and increased in absorbance. The spectrum of hhMb by comparison showed five bands at 500, 535, 570, 630, and 645 nm. The X-ray radiation results in the formation of two split Q-bands at 530/540 and 558/568 nm, which are in good agreement with previously reported spectral changes (7, 20, 35, 39). In AvTsdA, a shift from the LS band at 540 (shoulder at 575 nm) to a split Q-band with maxima at 527/550 nm was observed, whereas the CT-band at 630 remained. Finally, LmChdC showed bands at 490, 530, and 630 nm that decreased in absorbance upon X-ray radiation, whereas a new band appeared at 560 nm.
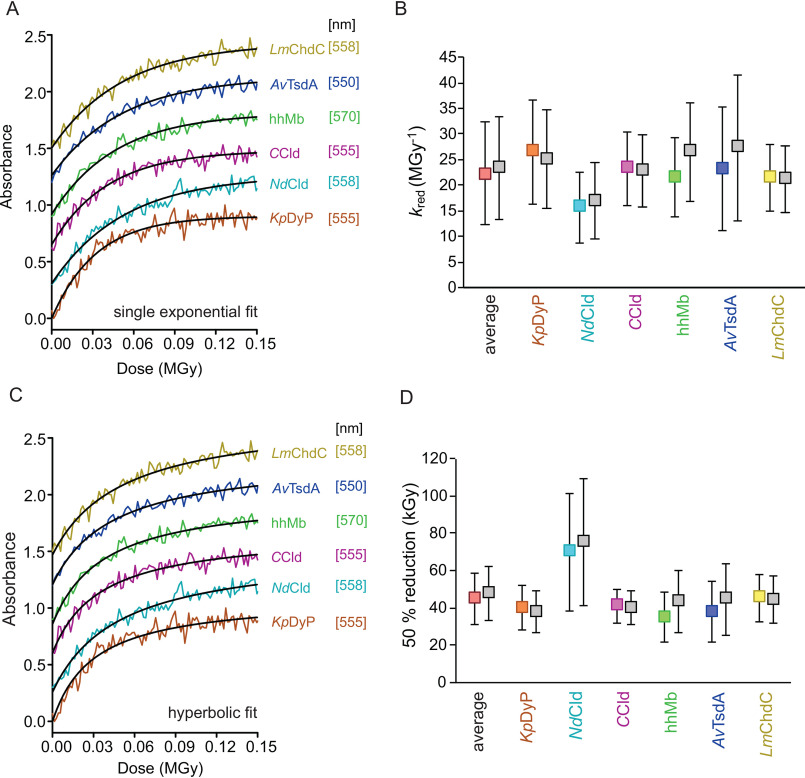
Logically, full reduction at the site of radiation must be reached once no absorption changes at the wavelength indicative of the ferrous species is observed. However, even after full reduction at the radiation site, both ferric and ferrous species will be present within one crystal. Mixed spectral characteristics will be therefore expected, as the UV-visible radiation passes through a partially different volume of the crystal than the irradiated volume. This is mainly due to the off-axis positioning of the UV-visible microspectrophotometer relative to the X-ray beam and differences in size and shape of the protein crystals (Fig. S3). Depending on crystal size, shape, and positioning, the amount of residual ferric spectral signatures varies, hampering interpretation of the reduction progress. To imply spectroscopic/kinetic formalisms, we followed the absorbance change at distinct wavelengths for each protein, where the highest reduction-dependent change in absorbance was observed over the accumulated absorbed dose (in MGy). This allows for the determination of the rate of reduction by single-exponential fits of the dose traces (kred values in MGy−1). A single-exponential increase in absorbance upon X-ray radiation was observed in all cases that led to saturation after an accumulated absorbed dose of 0.1 MGy (settings of depicted dose traces in Fig. 4 were 5% transmission, flux: 2.1 × 1011 photons s−1) and was markedly similar in all tested proteins (Fig. 4A).
Figure 4.
Kinetics of radiation-mediated reduction of Fe(III) to Fe(II). A, representative dose traces (at 5% transmission) with single-exponential fits to derive the rate of reduction (kred). B, rates of reduction (kred) for all proteins in the respective color and the calculated average for all six proteins (in red). C, representative dose-dependent changes of absorption at protein-specific wavelength (see Fig. 3) at 5% transmission (flux: 2.1 × 1011 photons s−1) with hyperbolic fits to derive the dose of half-maximal reduction. D, doses needed to reduce the heme iron in the protein crystals to obtain 50% ferric and ferrous heme (average value for all proteins in red). Dose traces in A and C were lifted by multiples of 0.3 for each sample for better visibility. Plotted values in B and D calculated using RADDOSE-3D version 4.0 with individual input for all different samples are shown in the respective color for each sample. Calculations using RADDOSE-3D version 4.0 input from a generic heme protein crystal are shown in gray beside the respective individual data points (KpDyP in orange, NdCld in turquoise, CCld in pink, hhMb in green, AvTsdA in blue, and LmChdC in yellow). Error bars, S.D.
To assess the kinetics of X-ray–induced photoreduction, it was assumed that the observed reduction was independent of the Fe(III) concentration. This was based on the idea that the amount of Fe(III) (0.2–200 fmol) within the crystal is rapidly overtaken by the secondary electrons produced through the photoelectric effect. It has been shown that, whereas radical species generated during secondary damage are immobile at cryo-temperatures, electrons easily traverse via the protein backbone to seek out the most electrophilic center (40). At 1% transmission (the lowest flux measured), the photon flux amounts to 4.2 × 1010 photons s−1, or 4.2 × 108 photons/image (0.1° oscillation, 10 ms). Up to 500 electrons/absorbed photon can be produced based on the assumption that of the roughly 2% interacting incident photons of a 12.1-keV X-ray beam, 84% are annihilated in production of photoelectrons, 8% are scattered by the Compton process, and 8% are Bragg scattered (34, 41). The “concentration” of electrons produced in the crystal during acquisition of one image is therefore at least 10-fold higher than the concentration of Fe(III). Thus, pseudo-first-order reaction settings are nominally guaranteed. The dose-dependent rate constant, kred (MGy−1), was directly derived from the single-exponential fits of dose traces from reduction experiments in the X-ray beam using different transmissions (1–20%, Fig. 4A). The rates of reduction were found to be similar in all cases: KpDyP, 26.5 ± 10.2 MGy−1; NdCld, 15.7 ± 6.9 MGy−1; CCld, 23.2 ± 7.2 MGy−1; hhMb, 21.4 ± 7.8 MGy−1; AvTsdA, 23.2 ± 12.0 MGy−1; LmChdC, 21.5 ± 6.6 MGy−1; average of all samples, 21.9 ± 9.3 MGy−1 (Fig. 4B). Initially, all dose calculations were performed with RADDOSE-3D version 4.0 (42, 43) using input parameters individually specified for each protein crystal (see supporting information D1–D8).
To simplify the workflow of dose calculations, we decided to test a more general approach using a generic heme crystal as input. Parameters for the generic heme protein crystal are averages of all heme protein structures deposited in the Protein Data Bank (PDB) (44–46) (parameters: crystal size, unit cell, number of monomers, number of residues, number of metal atoms in the protein, concentration/presence of heavy atoms in the cryo-solution (e.g. Br−), and average solvent fraction; supporting information D9 and D10). We then performed a dose calculation for all samples with this set of general input parameters. No significant differences were observed compared with the individual-calculation approach; KpDyP, 25.0 ± 9.7 MGy−1; NdCld, 16.9 ± 7.5 MGy−1; CCld, 22.7 ± 7.0 MGy−1; hhMb, 26.5 ± 9.6 MGy−1; AvTsdA, 27.4 ± 14.2 MGy−1; LmChdC, 21.1 ± 6.5 MGy−1; average of all samples, 23.3 ± 10.1 MGy−1 (Fig. 4B).
To investigate the dose required for 50% reduction of the heme protein crystals, absorbance spectra were recorded for each protein crystal at least in triplicate at six difference incident X-ray fluxes: 4.2 × 1010, 8.4 × 1010, 2.1 × 1011, 4.2 × 1011, 6.3 × 1011, and 8.4 × 1011 photons s−1 (these correspond to transmissions of 1, 2, 5, 10, 15, and 20%, respectively at I24, DLS). Here, a flux of 4.2 × 1010 photons s−1 corresponds to an average diffraction weighted dose (calculated by RADDOSE-3D version 4.0) (42, 43, 47) of 0.0151 MGy s−1 (for the generic heme protein crystal), 0.0143 MGy s−1 (KpDyP), 0.0163 MGy s−1 (NdCld), 0.0148 MGy s−1 (CCld), 0.0188 MGy s−1 (hhMb), 0.0179 MGy s−1 (AvTsdA), and 0.0149 MGy s−1 (LmChdC). The average diffraction weighted dose to reach the half-maximal increase in absorbance at different beam intensities was obtained from hyperbolic fits (Fig. 4C) of all dose traces for each heme protein. In all cases, the required absorbed dose was between 20 and 100 kGy (error-inclusive), the average being 47.7 ± 14.6 kGy (individual dose calculations) and 45.0 ± 13.4 kGy (generic dose calculations) (Fig. 4D).
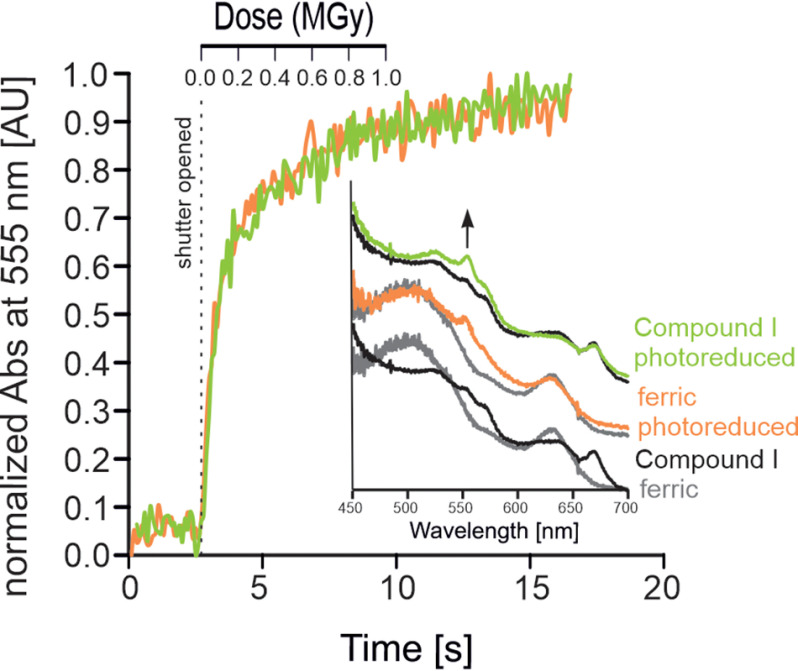
Next, the effect of different redox potentials (E°′) of the given redox couples (e.g. Fe(III)/Fe(II)), which in principle dictate reduction behavior on photoreduction, was investigated. To cover the entire range of redox potentials (approximately –400 to +1000 mV), we directly compared the redox couples of Fe(III)/Fe(II) and the Compound I/Fe(II) in KpDyP. The Compound I intermediate (i.e. oxoiron(IV) porphyryl radical) is formed by the two-electron oxidation of ferric KpDyP mediated by hydrogen peroxide. Compound I formation leads to altered vibrational characteristics of the chromophore due to the heme ligand (oxoiron) characterized by absorption bands in the visible region at 530, 575, 615, and 648 nm at room temperature and 525, 572, 615, and 640 nm under cryo-conditions with an additional low temperature–induced maximum at 552 nm and a band at 670 nm, most probably indicating a certain fraction of degraded heme (48, 49) (Fig. 5). This intermediate represents the highest oxidation state possible in heme proteins (E°′ ∼1000 mV) (50, 51). Compound I formation was achieved in crystallo by soaking ferric KpDyP crystals with H2O2. The rates of reduction between the ferric KpDyP and Compound I crystals at 100 K were compared by following the absorption change over the accumulated absorbed dose at the low-temperature maximum of 555 nm. The similar rates of reduction observed for the ferric KpDyP (kred = 21.4 MGy−1) and Compound I (kred = 24.8 MGy−1) suggest that the same kinetics are applicable for the formation of ferrous KpDyP (Fig. 5). Moreover, the appearance of a band at 555 nm indicates that Compound I undergoes a three-electron reduction to HS ferrous KpDyP.
Figure 5.
Kinetics of radiation-mediated reduction of Compound I to Fe(II) in KpDyP. Shown are time- and dose-dependent changes in absorption at 555 nm of the ferric resting state (orange) and the Compound I intermediate (green) of KpDyP upon photoreduction (10% transmission, flux = 4.2 × 1011 photons s−1). Inset (bottom), overlay of single-crystal UV-visible spectra of the ferric (gray) resting state and the Compound I intermediate (black) obtained by soaking with H2O2. Inset (middle), ferric resting state before (gray) and after (orange) exposure. Inset (top), Compound I intermediate before (black) and after (green) exposure.
Influence of photoreduction on resting state and ligated iron centers can be resolved using a dose-dependent method
The results from the spectroscopic analysis of the heme protein test set suggest that 75–100 kGy of absorbed X-ray dose is sufficient for complete reduction of Fe(III) to Fe(II) in a heme protein crystal at 100 K. This is in good agreement with other spectroscopic investigations (10, 11). However, as described previously, studies using mainly structural indicators (i.e. interatomic distances between the heme iron and a ligand molecule) to estimate dose limits have routinely arrived at higher acceptable absorbed doses (20–35 kGy) (8, 9), where no significant structural effects could be observed. To correlate the spectroscopically observed reduction with structural changes to the heme environment, a dose series of structures was determined for the model protein KpDyP at 100 K. Merged data sets corresponding to 5, 10, 25, 50, and 100% Fe(II) were created (see Table 3, structures not shown), allowing a stepwise visualization of the structural changes induced by Fe(III) reduction.
Table 3.
Data collection and refinement statistics
Values for the highest-resolution shell are shown in parentheses. RMSD, root mean square deviation.
| Time/dose-resolved water-bound heme | Cyanide-bound heme | |||||||
|---|---|---|---|---|---|---|---|---|
| Beamline | I04 DLS | ID23-1 ESRF | ||||||
| Oscillation (degrees) | 0.1 | 0.1 | ||||||
| Exposure/image (s) | 0.04 | 0.1 | 0.1 | |||||
| Beam size (μm) | 26 × 15 | 20 × 20 | 20 × 20 | |||||
| Images/wedge | 100 | 100 | 3600 | |||||
| Wavelength (Å) | 0.9795 | 0.9919 | 0.9919 | |||||
| Flux (photon s−1) | 6.99 × 1010 | 1.89 × 1010 | 1.89 × 1010 | |||||
| Scaled wedges | 723 | 645 | 1 | |||||
| Image range from wedge | 1–4 | 5–8 | 17–20 | 35–38 | 71–74 | 96–99 | 1–5 | 1–3600 |
| Dose (kGy) | 2.15 | 4.3 | 10.8 | 20.4 | 40.3 | 53.6 | 2.3 | 1590.0 |
| Total exposed time (s) | 0.12 | 0.32 | 0.8 | 1.52 | 2.96 | 4 | 0.5 | 360 |
| Fe(II) % reduced | 5.2 | 10.5 | 26.4 | 50 | 98.7 | 100 | 5.7 | 100 |
| Space group | P 1 21 1 | P 1 21 1 | P 1 21 1 | |||||
| Unit cell (Å) | a = 50.89 | a = 50.89 | a = 50.57 | |||||
| b = 76.65 | b = 76.59 | b = 75.74 | ||||||
| c = 76.58 | c = 76.31 | c = 75.71 | ||||||
| Unit cell (degrees) | α = γ = 90.00 | α = γ = 90.00 | α = γ = 90.00 | |||||
| β = 108.01 | β = 107.87 | β = 107.85 | ||||||
| Resolution range (Å) | 48.40–1.90 | 48.40–1.90 | 48.40–1.90 | 47.67–1.90 | 47.67–1.90 | 47.67–1.90 | 36.31–1.80 | 36.03–1.52 |
| Rmerge | 0.18 (0.48) | 0.20 (0.54) | 0.20 (0.54) | 0.20 (0.57) | 0.20 (0.58) | 0.21 (0.60) | 0.21 (0.75) | 0.06 (0.85) |
| Rmeas | 0.22 (0.59) | 0.23 (0.64) | 0.24 (0.64) | 0.24 (0.68) | 0.24 (0.70) | 0.25 (0.74) | 0.25 (0.90) | 0.07 (0.10) |
| Rpim | 0.12 (0.33) | 0.12 (0.34) | 0.12 (0.34) | 0.12 (0.36) | 0.13 (0.37) | 0.14 (0.41) | 0.13 (0.49) | 0.04 (0.57) |
| All observations | 185,150 (11,453) | 238,147 (14,639) | 241,170 (14,925) | 241,538 (14,758) | 235,160 (14,352) | 202,833 (12,376) | 267,452 (15,345) | 246,785 (24,852) |
| Unique observations | 42,694 (2727) | 43,499 (2791) | 43,564 (2794) | 43,584 (2795) | 43,552 (2789) | 43,298 (2778) | 51,235 (3003) | 81,528 (8142) |
| Mean (I/σI) | 5.4 (3.0) | 6.2 (3.4) | 6.1 (3.4) | 6.2 (3.4) | 6.0 (3.3) | 5.5 (2.9) | 6.1 (2.7) | 3.0 (3.1) |
| CC½ | 0.96 (0.65) | 0.96 (0.37) | 0.96 (0.47) | 0.96 (0.39) | 0.95 (0.33) | 0.95 (0.41) | 0.97 (0.70) | 0.99 (0.50) |
| Completeness (%) | 96.8 (96.6) | 98.6 (98.8) | 98.7 (98.9) | 98.8 (98.9) | 98.7 (98.7) | 98.1 (98.3) | 99.2 (99.0) | 97.8 (97.4) |
| Multiplicity | 4.3 (4.2) | 5.5 (5.2) | 5.5 (5.3) | 5.5 (5.3) | 5.4 (5.1) | 4.7 (4.5) | 5.2 (5.1) | 3.0 (3.1) |
| Rwork (%) | 17.2 | 17 | 16.9 | 18.4 | 18.1 | 17.2 | 17.2 | 16.7 |
| Rfree (%) | 19.9 | 19.1 | 18.1 | 20.1 | 19.8 | 19 | 19.7 | 18.2 |
| Refined residues | 599 | 599 | 599 | 599 | 599 | 599 | 597 | 597 |
| Refined H2O | 671 | 634 | 655 | 644 | 621 | 669 | 785 | 560 |
| Average B-factor (Å2) | 12 | 12 | 12 | 12 | 12 | 12 | 10.34 | 26.26 |
| RMSD bond length (Å) | 0.023 | 0.022 | 0.024 | 0.018 | 0.017 | 0.015 | 0.013 | 0.015 |
| RMSD bond angle (degrees) | 1.54 | 1.52 | 1.59 | 1.42 | 1.36 | 1.24 | 1.15 | 1.66 |
| Ramachandran favored (%) | 98.15 | 97.98 | 97.64 | 98.15 | 97.98 | 97.82 | 98.65 | 98.48 |
| Rotamer outliers (%) | 0.6 | 1.18 | 1.19 | 0.8 | 1.01 | 0.4 | 1.38 | 1.2 |
| Molprobity overall (percentile) | 100 | 98 | 99 | 100 | 100 | 100 | 99 | 100 |
| PDB accession code | 6RQY | 6RR1 | 6RR4 | 6RR5 | 6RR6 | 6RR8 | 6RPE | 6RPD |
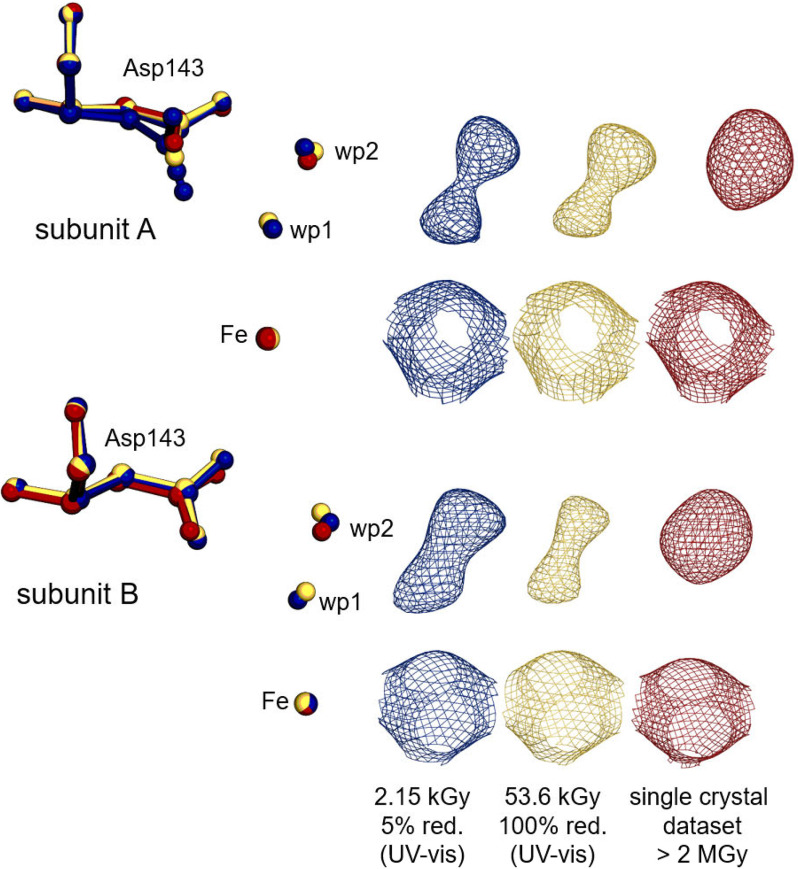
Dose series were collected from KpDyP in complex with two small-molecule ligands of peroxidases, water and cyanide. These ligands bind on the distal heme side (site of catalysis) and show differences in their attractive propensities to Fe(II) and Fe(III). Cyanide binds to both Fe(II) and Fe(III) but with significantly reduced affinity to Fe(II), whereas distal ligated water is typically released during reduction. Molecular dynamics simulations of ferric and ferrous state clearly indicate two water positions in the active site susceptible to the oxidation state. Water position 1 (wp1) is occupied only in the ferric state, and wp2 is occupied in both states (30, 52). Previous studies assumed that a stepwise reduction of the metal center should be reflected by a gradual increase of the distance between the iron ion and the distal ligand (5). However, reduction of ferric iron to its ferrous state is a one-electron step. Consequently, only two possible ligation states exist in the crystal. Therefore, we expect to observe a gradual loss of ligand density at the ferric ligand site upon reduction to the ferrous state throughout the dose series. Gradual increase of the distance (movement) of the ligand is impossible, as one iron cannot be partially reduced.
Indeed, a change in the electron density of distal site waters was observed throughout the dose series and ultimately led to a complete disappearance of one ligated water at wp1 (26). Fig. 6 shows the electron densities of active-site water molecules at wp1 and wp2 of subunits A and B of KpDyP together with the conformation of the catalytic aspartate of the first (5% reduced) and the final (100% reduced) structure, compared with the model derived from a single-crystal data collection, in which only wp2 is occupied. Calculated omit maps of the three selected structures support the interpretation of molecular rearrangements during photoreduction (Fig. S4A). Analysis of the dose series, representing photoreduction, shows a minor, but visually evident, decrease in electron densities for wp1 toward the 100% (53.6 kGy) reduced final wedge (Fig. S5), whereas no change of the electron density was observed for the carboxylate, as expected for the low absorbed dose. However, extra restraints on the wp1–iron and wp1–wp2 distances were required to maintain the water locations during refinement. Therefore, the modeled distances, in contrast to the measured electron densities, are subject to experimenter bias and cannot be considered accurate. Linear extrapolation to infer noncovalent interatomic distances, as done in a previous study (5) is not an appropriate tool for analysis of the active site over the dose series in the given case of KpDyP, because we observe a change in occupancies of the two waters rather than a movement of one molecule.
Figure 6.
Structural changes upon reduction of ferric high-spin KpDyP. Asp-143 (sticks), distal active-site water molecules, and the heme iron (sphere) are represented from KpDyP subunit A (top) and B (bottom) for the 5% reduced (blue, 6RQY), the 100% reduced (gold, 6RR8) structure, based on UV-visible spectroscopy, from the multicrystal approach and for the completely reduced routine single-crystal data set (red, 6FKS). 2mFo − DFc (contoured at 1 σ) electron density maps for the represented molecules are shown in the respective colors on the right.
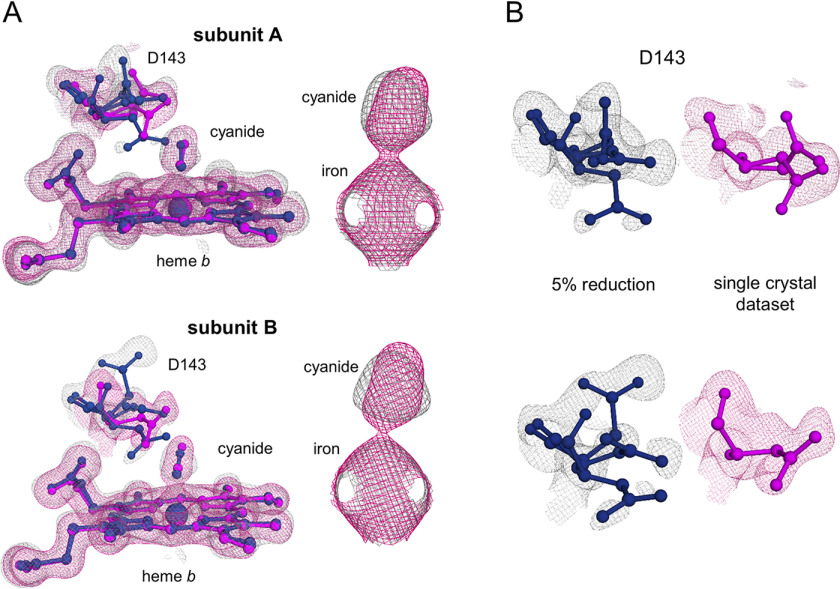
The cyanide ions, like the water molecules from the heme-water dose series, had to be restrained during refinement, preventing unequivocal confirmation of lengthening of the cyanide-iron distance during reduction. We therefore turned to examining the electron densities directly. Fig. 7A compares the cyanide complex structure at the beginning of the series (5% reduction) with the single-crystal data set (0.1° oscillation, 0.1-s exposure, 360° total oscillation, 1.59 MGy). There is no distinct change in either subunit for the heme iron–cyanide density apart from a minor elongation of the cyanide density in the standard rotation data set. A change in the distance between the iron and the cyanide density can be seen in the calculated omit maps of subunit B (Fig. S4B).
Figure 7.
Structural changes upon reduction of ferric cyanide bound low-spin KpDyP. A, heme b, Asp-143, and cyanide (sticks) are represented from KpDyP subunit A (top) and subunit B (bottom) for the 5% reduced (blue, gray mesh, 6RPE) structure derived from the multicrystal approach and for the completely reduced routine single-crystal data set (purple, pink mesh, 6RPD). 2mFo − DFc electron density maps are represented for all modeled structural elements (contoured at 1.5 σ (cyanide alone) or 1 σ (rest)). B, flexibility of Asp-143 in subunit A (top) and subunit B (bottom) in the same colors as in A, representing the same states of reduction.
Interestingly, significant differences were observed for the Asp-143 side-chain conformation when comparing the 5% reduced and the ferrous single-crystal structure (Fig. 7B). This aspartate is critical for catalytic activity in KpDyP and other dye-decolorizing peroxidases and has primarily been observed in a single “flat” conformation in photoreduced structures, approximately perpendicular to the peptide backbone. In the ferric structure, the Asp-143 side chain was observed in three alternative conformations (occupancies ranging from 0.25 to 0.50) leading to generally poorer electron densities, indicating multiple geometries and high flexibility. In addition to the “flat” conformation, we therefore modeled a downward and an upward (subunit B) or a tilted (subunit A) conformation (Fig. 7B). Although less pronounced, a movement of the Asp-143 carboxylic group is also visible in the heme-water structures. Here, a change in water coordination leads to a tilt and rotation of Asp-143 (Fig. 6).
Discussion
The observer effect, well-known in quantum mechanics, is the effect that the observation of a phenomenon changes the phenomenon itself. This applies equally to structure determination, where the incident beam used to collect structural information interacts with the observed specimen. This was recognized early on with studies focusing on damage to the crystal, protein, or redox-sensitive cofactors (4, 6, 7, 34, 35, 53–57). However, the tested parameters were limited to a handful of model proteins. Here, a comprehensive study on the pre-steady-state kinetics of the radiation-dependent photoreduction of six different heme proteins has been provided. The aim was to investigate and differentiate the parameters influencing photoreduction, namely beam characteristics versus sample-derived (protein and cofactor characteristics, crystal geometry, dimension, and composition) variables. These are the main factors for mathematically calculating a dose limit.
Pre-steady-state kinetics of the model proteins clearly demonstrate that in all cases, the criteria for pseudo-first-order reaction conditions, namely the dependence of the reduction velocity on only one variable (dose), were met. Global radiation damage was previously observed to be solely dose-dependent and dose rate–independent (58). Whereas it cannot be completely excluded that dose rate-dependent effects would be observed with a faster measurement technique, within the tested photon flux density range (0.42–8.4 × 1011 photons/s), the specific damage (i.e. photoreduction) is most probably solely dose-dependent.
The dose is determined by beam characteristics and sample-derived variables. As the beam characteristics were kept constant throughout the experiments shown in Fig. 4, any differences in the reduction kinetics and calculated dose can be attributed to sample-derived variables: protein and cofactor characteristics, crystal geometry, dimension, and composition. No changes in kred values and the absorbed dose at half-maximal reduction were observed.
We suggest, because the calculated dose limit for all investigated samples (i.e. systems) does not depend on the sample-derived variables, there is no need to empirically determine a new limit for each new system. All observed variations, resulting from different dose calculations (generic versus individual), are not significant (Fig. 4). Although it is in principle less accurate to use generic crystal parameters for dose calculations, empirical data presented here show that, in practice, it is a valid proxy (Fig. 4). To show this, we define a model heme crystal based on average parameters derived from the PDB. Data presented in Fig. 4 emphasize that this general, simplified approach for dose calculations using RADDOSE-3D is legitimate.
Therefore, these data suggest that whereas global radiation damage depends on the composition of the crystal, reduction of the metal center does not. In all cases, the reduction kinetics were only dependent on the absorbed dose, irrespective of sample variation. A dose limit of 3 kGy for 5% reduction is proposed that would be applicable to all heme proteins measured at synchrotron light sources at cryo-temperatures.
Employing the described multicrystal data collection strategy, we examined two test cases, namely the change in water and cyanide coordination in KpDyP at 100 K. The complete disappearance of water from wp1 in the single-cryo-crystal data set, compared with the (according to UV-visible spectroscopy) 5 and 100% Fe(II) reduced structures, was striking. This disappearance was also observed at room-temperature in the serial structures presented by Ebrahim and co-workers (5) and served as a striking reminder that cryo-temperatures do not mitigate the effects of radiation damage. The hydrogen-bonding networks of metalloproteins are often conserved access routes for small ligands, such as hydrogen peroxide. In the KpDyP test case, the ligated water is within hydrogen-bonding distance of two catalytically relevant residues: the aforementioned Asp-143 and the structurally important Arg-232 (26). Conformational changes were observed in the Asp-143 carboxylic group in response to Fe reduction in both test cases. In the resting state, these changes likely reflect either an altered hydrogen-bonding network of the active site or the charge-screening effect of the dipolar water molecule. Additionally, the high flexibility of Asp-143 side chain, reflected in the low occupancy of all modeled conformations in the cyanide-bound structures, might serve as an explanation for the relatively low affinity of cyanide to KpDyP (KD = 0.9 mm) (26). We speculate that the negatively charged cyanide anion, which displaces the water in the ferric resting state, has to break the hydrogen-bonding network within the active site and displaces the equally negatively charged Asp-143. In the ferrous cyanide-bound structure, derived from the single-crystal data set, the orientation of the Asp-143 side chain, which is the catalytically most important residue in the active site of KpDyP (26, 49), is well-resolved. However, the conformation does not correspond to the real situation in the ferric resting state (Fig. 7). This could possibly lead to incorrect mechanistic conclusions, based on the well-resolved but misleading Asp-143 side chain orientation in the single-crystal data set.
Our data also indicate that the distance and the angle of cyanide relative to the heme cofactor is only influenced to a small extent by the oxidation state of the heme iron. This may be due to the obtained resolution (1.5–1.7 Å), which was not high enough to resolve such small changes.
The importance of the heme iron redox state for ligand binding is aptly shown in studies of aldoxime dehydratase (59), which is only active in its ferrous state (60). The binding mode of the aldoxime molecule (R-CH=N-OH) changes significantly in response to the redox state of the heme iron (ferric, O-ligated; ferrous, N-ligated). Sawai and co-workers (61) used X-ray–induced photoreduction during data collection to obtain a catalytically relevant Michaelis complex structure of ferrous aldoxime dehydratase from crystals of the inactive ferric aldoxime complex. This is an example of major rearrangement of a ligand in response to photoreduction and how X-ray–induced photoreduction can be used as a tool to obtain catalytically important conformations.
We measured the reduction rate of the Fe(III) spectroscopically and thereby defined an acceptable limit of 3 kGy at 100 K to obtain structures with only 5% reduced heme iron. However, as stated previously, several studies have used consecutive data truncation to monitor reduction via redox-sensitive interatomic distances (8, 9). In all these cases, the allowed dose limit at which the data set was truncated lies significantly higher than the limit proposed here. In analyzing the consecutive wedges obtained during our data collection, we can show in a time-resolved fashion that whereas Fe(III) reduction occurs nearly instantaneously according to online UV-visible spectroscopy, the actual Fe(III) reduction–related structural alterations of the coordinating atoms are observed only at a later stage. We propose that this is due to the cryo-temperature, which significantly slows down molecular movements and diffusion. This conclusion is supported by data obtained at room temperature, where larger structural changes were observed relative to cryo-temperature (5). Theoretically, cryo-cooling during data collection would therefore allow the collection of more wedges, as full reduction of the metal centers is reached at a significantly earlier time point than the slowed down structural effects are taking hold. However, cryogenically slowed down molecular movements and diffusion might depend on additional factors (e.g. investigated protein or ligand, the interaction of this ligand with its surroundings within the crystal, the shape of the active-site, temperature). It therefore remains difficult to set a general dose limit for this approach to data collection.
In summary, our spectroscopic data show that X-ray–induced reduction of Fe(III) in a heme group happens at a much lower dose than previously shown. We propose a dose of 40 kGy to be sufficient for half-maximal reduction. We did not observe significant sample-dependent differences. The 3-kGy dose limit used in this study guarantees a nearly nonreduced ferric structure and is a low-risk approach.
Apart from general crystal-derived variables, the main physical property of metal centers is their respective redox potential of the metal cofactor couples (e.g. the Fe(III)/Fe(II) couple). In this study, the maximal redox potential range of metalloproteins is covered (E°′ of Fe(III)/Fe(II) ranging from −400 mV to +50 mV and E°′ of Compound I/Fe(III) being ∼1000 mV) (50, 51). This range also includes enzymes such as high-redox potential copper enzymes (E°′ (Cu(II)/Cu(I)) up to +750 mV) (62, 63). Here, the investigated proteins cover approximately half of the maximal redox potential range of all metalloproteins (−400 to +28 mV). Further, we included a direct comparison of the kred values of the reduction kinetics of KpDyP crystals in their ferric and Compound I state that were identical, despite a difference of ∼1200 mV of the redox potential of the respective redox couples. Therefore, the susceptibility of the metal cofactor to X-ray–induced reduction does not depend on its redox potential (E°′). This limit is hence applicable to any heme protein in any oxidation state without the need for time-consuming individual analysis.
It remains to be shown whether this limit also applies to other metals commonly found in metalloproteins and if radiation-induced reduction kinetics of different metals are equally disconnected from their respective redox and protein properties. In line with the results of our experiments, previous studies on photosystem II have shown that Mn(III) and Mn(IV) reduction is uncoupled from global radiation damage (64). Furthermore, it is recommended to collect the diffraction data far from the absorption edge of the investigated metal, because its absorption cross-section is largest at the wavelength corresponding to Δf′. We employed a simple method to obtain a nonreduced multicrystal data set under cryo-conditions within a routine macromolecular crystallography experiment that will allow reliable structural characterization of the resting state of metalloproteins.
Experimental procedures
Protein preparation and crystallization conditions
hhMb was purchased from Sigma–Aldrich (catalog no. 100684-32-0). CCld was purified as described previously (31). NdCld was purified as described by Hofbauer et al. (37). KpDyP was purified as described by Pfanzagl et al. (26). AvTsdA was purified as described by Denkmann et al. (65). LmChdC was purified as described by Hofbauer et al.(66). All proteins were subjected to size-exclusion chromatography using a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) and pre-equilibrated with 50 mm phosphate buffer, pH 7.0, prior to crystallization.
Crystallization experiments were performed using a 48-well MRC Maxi crystallization plate (AvTsdA) or SWISSCI MRC three-well crystallization plates (Molecular Dimensions, Newmarket, UK) adopting the vapor diffusion method. Crystallization drops were set by hand (for AvTsdA) or using a Mosquito LCP (TTP Labtech, Melbourn Science Park, Melbourn, UK). hhMb crystalized in 3.2 m (NH4)2SO4, 0.1 m Tris-HCl, pH 8.5. CCld crystallized in 0.15 m MgSO4, 0.1 m MES, pH 6.5, 22% PEG 3350, 5% glycerol. NdCld crystallized in 50% of all conditions in the PACT screen from Molecular Dimensions. Crystals from five different conditions were used: Condition 1 (well A4), 20% (w/v) PEG 1500, 0.1 m SPG, pH 7.0; condition 2 (well A5), 20% (w/v) PEG 1500, 0.1 m SPG, pH 8.0; condition 3 (well E2), 20% (w/v) PEG 3350, 0.2 m NaBr; condition 4 (well E8), 20% (w/v) PEG 3350, 0.2 m NaSO4; condition 5 (well G2), 20% (w/v) PEG 3350, 0.1 m BisTris propane, pH 7.5, 0.2 m NaBr. AvTsdA crystallized in 23.5% (w/v) PEG 3350, 0.2 m (NH4)2SO4, 0.1 m BisTris, pH 6.5, 0.1 m NaI. KpDyP crystallized in 23% (w/v) PEG 3350, 0.1 m MgCl2, 0.1 m Tris-HCl, pH 8.5. LmChdC crystallized in 10% (w/v) PEG 6000, 0.1 m Bicine, pH 9.0. For cryoprotection, glycerol was added to a final concentration of 25% to the crystallization mother liquor (diluting the mother liquor accordingly) and soaked for 5 min. For cyanide-ligated KpDyP, crystals were soaked for 1 min in cryoprotectant with 250 mm NaCN.
Online UV-visible spectroscopy and beamline settings
For UV-visible absorption spectroscopy, crystals were fished locally and directly placed in the cryostream and kept at 100 K. Spectra were measured using an Andor Shamrock 303 imaging spectrograph controlled through the Andor Solis software and linked to an air-cooled Newton EM-CCD camera and an Ocean optics deuterium tungsten halogen lamp. The microspectrometer was mounted on the diffractometer of I24 (DLS) to allow the collection of time series during X-ray exposure. The focus of the microspectrometer was 20 × 20 μm to match the diameter of the incoming X-ray beam (20 × 20 μm) to ensure that the whole X-ray–exposed area of the crystal was sampled by the light beam. To collect time series, spectra were recorded with a temporal resolution of 0.02 s. After starting spectral data collection, the shutters were opened manually to start X-ray radiation. The incident X-ray energy was 12.1 keV with a flux of 4.2 × 1012 photons/s and Gaussian beam characteristics. Time series were recorded at 1, 2, 5, 10, 15, and 20% transmission, with four independent measurements per transmission. Time series were converted to diffraction-weighted dose series using RADDOSE 3D (43) with the following parameters for the generic crystal: cuboid, 20 20 20, 0.5, RD3D, 80 95 120, 2.5 (one calculation with 2, one with 3), 350, Fe 1 S 13, P 100, 0.43, Gaussian, 4.2e12, 20 20, 12.1, 100 100, 0 10, 1, 0.1 (the inputs for the individual calculations are given in the supporting information). It must be noted that the generic crystal parameters are not based on averages of the crystals used in this study, but on averages obtained from all deposited heme proteins to the PDB. Parameters for individual dose calculations are given in the supporting information (supporting information D1–D10). For data analysis and fitting, kinetic slices at 550 nm (AvTsdA), 555 nm (KpDyP, CCld), 558 nm (NdCld, LmChdC), or 570 nm (hhMb) were baseline-corrected and scaled to an absolute absorbance range of 1.0 to allow visual comparison. All kinetic slices were fitted using SigmaPlot 13.0 from the starting point of radiation until saturation using a single-exponential equation f = a*(1 − exp(−b*x)) to derive kred (MGy−1), and the time until half-maximal reduction was determined using hyperbolic fits f = a*x/(b + x).
UV-visible spectroscopy of ferric and ferrous heme proteins in solution
Spectra of WT KpDyP, NdCld, CCld, hhMb, AvTsdA, and LmChdC were recorded between 350 and 700 nm with a Cary 60 scanning photometer (Agilent Technologies). Conditions were 50 mm phosphate buffer, pH 7.0. 20 mg ml−1 sodium dithionite from a freshly prepared stock solution was used to reduce the sample.
X-ray data collection and structure determination
Diffraction data of the reduced cyanide-ligated KpDyP structure were collected at beamline ID23-1 at the European Synchrotron Radiation Facility (Grenoble, France) at 100 K using an PILATUS3 6 M detector. Diffraction data of the multidata sets were obtained by manually applying native or NaCN-soaked crystals on 700/25 MicroMeshes (MiTeGen) and removing excess cryoprotectant with wicks (Fig. 2C). Meshes were then flash-cooled in liquid nitrogen. From each crystal, 10° (equivalent to 100 images in 0.1° wedges) were measured with a total photon flux of 109 photons/image. Diffraction data of the native KpDyP were collected from 750 crystals at beamline I04 of the DLS at 100 K using a PILATUS3 6 M detector. Diffraction data of 700 crystals of the NaCN-soaked KpDyP were collected at beamline ID23-1 at the European Synchrotron Radiation Facility (Grenoble, France) at 100 K using a PILATUS3 6 M detector. For all experiments, the detector distance was set to collect 1.5 Å resolution.
Data reduction
Individual wedges for the multicrystal data were indexed and integrated using XDS (67). The wedges were then combined into a single mtz file using POINTLESS (68), and those data sets with unit cell vectors that did not conform to the majority (here defined as a TOLERANCE of 2 in POINTLESS) were manually excluded. The remaining data sets were collectively scaled together in AIMLESS (69) using the following settings: SCALES ROTATION SPACING, 5; SECONDARY, 0; BFACTOR ON BROTATION SPACING, 10. This created, for the oxygen- and cyanide-bound heme structures, a single scaled data set that comprised 723 and 645 wedges, respectively. To create the sub-data sets that corresponded to different proportions of Fe(II) (5, 10, 25, and 50% and 2 × 100%) based on the UV-visible spectroscopy and RADDOSE-3D calculations, these single scaled data sets were then fed back into AIMLESS, and selected scaled images were merged together. For example, to create a Ferric-O structure with a diffraction-weighted absorbed dose of 2.15 kGy and a time resolution of 160 ms, the first 1–4 scaled images from the 723 wedges were merged together. This process could then be repeated for the other time/dose points of interest (see Table 3). As few images as was feasible were used to make these sliced data sets. The principle criteria used were CC½, completeness, and Rpim. CTRUNCATE (70) was then used to convert the intensities into structure factors and apply the same Rfree flag to each individual data set. The ferrous-CN data set was processed with XDS using xia2 as part of the CCP4 version 7.0 program suite (70).
Phasing and model building
The phases for each data set were solved using molecular replacement, using the WT KpDyP structure (PDB code 6FKS) (26) as a search model in PHASER-MR (71). PHENIX.AUTO-BUILD (72) was then run to minimize any potential model bias. Models were then manually built using COOT (73) and refined using PHENIX.REFINE (74) and BUSTER (75). Angles were applied to the water and cyanide carbon atoms such that a 90° angle was maintained between the heme Fe and the Fe-coordinating nitrogen atoms. The cyanide also required angle restraints to maintain the 180° angle between the Fe cyanide carbon and nitrogen. Distance restraints were placed on the heme-bound waters and cyanide molecule to ensure the molecules refined in the observed density. For example, the distance restraints used in the 5% Fe(II)-reduced Ferric-O structure were 2.50, 2.00, and 2.95 Å, for the heme Fe–wp1, wp1–wp2, and wp2–Ser-234 O bonds, respectively. A σ of 10 was used for all restraints to allow for some flexibility in their application. MOLPROBITY (76) was used to validate the structures and guide the refinement of problem areas. Figures were prepared with PyMOL (77).
Data availability
Structural data were deposited to the RCSB Protein Data Bank under the following accession codes: 6RPD, 6RPE, 6RQY, 6RR1, 6RR4, 6RR5, 6RR6, 6RR8. All other data are stored on the network space of the Department of Chemistry at the University of Natural Resources and Life Sciences, Vienna, and partially at the Synchrotron facilities, at which diffraction data were collected for this work. Data will be shared upon request to the corresponding authors.
Supplementary Material
Acknowledgments
We thank Elspeth Garman for correspondence and help concerning radiation damage and dose calculations. We thank Ben Siegl for fruitful discussions. We thank Graeme Winter, James Beilsten-Edmunds, David Waterman, Danny Axford, and Richard Gildea for guidance, assistance, and productive conversations toward the data reduction of large small-wedge data sets. We thank Margarida Archer and Jose A. Brito from the ITQB (Oeiras, Portugal) for providing AvTsdA protein.
This article contains supporting information.
Author contributions—V. P., K. D.-C., and S. H. conceptualization; V. P., J. H. B., H. M., D. S., T. G., and S. H. data curation; V. P., J. H. B., H. M., D. S., T. G., and S. H. formal analysis; V. P. and S. H. supervision; V. P., J. H. B., and S. H. validation; V. P. and S. H. visualization; V. P., J. H. B., K. D.-C., and S. H. methodology; V. P.,J. H. B., K. D.-C., and S. H. writing-original draft; V. P. and S. H. project administration; V. P., J. H. B., C. O., K. D.-C., and S. H. writing-review and editing; J. H. B. software; C. O. and K. D.-C. resources; C. O., K. D.-C., and S. H. funding acquisition.
Funding and additional information—This project was supported by the Austrian Science Fund, FWF (Doctoral Program BioToP - Molecular Technology of Proteins (W1224) and the project P29099), Diamond Light Source Proposal MX19036, ESRF Proposal MX1998, and the iNEXT program (PID: 4482). The work of K. D.-C. was supported by Austrian Science Fund (FWF) Projects I525, I1593, P22276, P19060, and W1221; Wellcome Trust Collaborative Award 201543/Z/16/Z; by COST action BM1405—Non-globular proteins–from sequence to structure, function, and application in molecular physiopathology (NGP-NET); WWTF (Vienna Science and Technology Fund) Chemical Biology Project LS17-008; the Austrian Federal Ministry of Science, Research, and Economy; and the National Foundation for Research, Technology, and Development through funding the Christian Doppler Laboratory for High-Content Structural Biology and Biotechnology, Austria.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- Gy
- gray(s)
- AvTsdA
- thiosulfate dehydrogenase from Allochromatium vinosum
- CCld
- chlorite dismutase from Cyanothece sp. PCC7425
- hhMb
- horse heart myoglobin
- KpDyP
- dye-decolorizing peroxidase from Klebsiella pneumoniae
- LmChdC
- coproheme decarboxylase from Listeria monocytogenes
- HS
- high-spin
- LS
- low-spin
- NdCld
- chlorite dismutase from Nitrospira defluvii
- PDB
- Protein Data Bank
- XFEL
- X-ray free-electron laser
- DLS
- Diamond Light Source
- wp
- water position
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
References
- 1. Hough M. A., Antonyuk S. V., Strange R. W., Eady R. R., and Hasnain S. S. (2008) Crystallography with online optical and X-ray absorption spectroscopies demonstrates an ordered mechanism in copper nitrite reductase. J. Mol. Biol. 378, 353–361 10.1016/j.jmb.2008.01.097 [DOI] [PubMed] [Google Scholar]
- 2. Holton J. M. (2009) A beginner's guide to radiation damage. J. Synchrotron Radiat. 16, 133–142 10.1107/S0909049509004361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carugo O., and Djinović Carugo K. (2005) When X-rays modify the protein structure: radiation damage at work. Trends Biochem. Sci. 30, 213–219 10.1016/j.tibs.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 4. Corbett M. C., Latimer M. J., Poulos T. L., Sevrioukova I. F., Hodgson K. O., and Hedman B. (2007) Photoreduction of the active site of the metalloprotein putidaredoxin by synchrotron radiation. Acta Crystallogr. D Biol. Crystallogr. 63, 951–960 10.1107/S0907444907035160 [DOI] [PubMed] [Google Scholar]
- 5. Ebrahim A., Moreno-Chicano T., Appleby M. V., Chaplin A. K., Beale J. H., Sherrell D. A., Duyvesteyn H. M. E., Owada S., Tono K., Sugimoto H., Strange R. W., Worrall J. A. R., Axford D., Owen R. L., and Hough M. A. (2019) Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins. IUCrJ 6, 543–551 10.1107/S2052252519003956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berglund G. I., Carlsson G. H., Smith A. T., Szöke H., Henriksen A., and Hajdu J. (2002) The catalytic pathway of horseradish peroxidase at high resolution. Nature 417, 463–468 10.1038/417463a [DOI] [PubMed] [Google Scholar]
- 7. Beitlich T., Kühnel K., Schulze-Briese C., Shoeman R. L., and Schlichting I. (2007) Cryoradiolytic reduction of crystalline heme proteins: analysis by UV-Vis spectroscopy and X-ray crystallography. J. Synchrotron Radiat. 14, 11–23 10.1107/S0909049506049806 [DOI] [PubMed] [Google Scholar]
- 8. Gumiero A., Metcalfe C. L., Pearson A. R., Raven E. L., and Moody P. C. (2011) Nature of the ferryl heme in compounds I and II. J. Biol. Chem. 286, 1260–1268 10.1074/jbc.M110.183483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meharenna Y. T., Doukov T., Li H., Soltis S. M., and Poulos T. L. (2010) Crystallographic and single-crystal spectral analysis of the peroxidase ferryl intermediate. Biochemistry 49, 2984–2986 10.1021/bi100238r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hersleth H. P., and Andersson K. K. (2011) How different oxidation states of crystalline myoglobin are influenced by X-rays. Biochim. Biophys. Acta 1814, 785–796 10.1016/j.bbapap.2010.07.019 [DOI] [PubMed] [Google Scholar]
- 11. Kekilli D., Moreno-Chicano T., Chaplin A. K., Horrell S., Dworkowski F. S. N., Worrall J. A. R., Strange R. W., and Hough M. A. (2017) Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction. IUCrJ 4, 263–270 10.1107/S2052252517002159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halsted T. P., Yamashita K., Hirata K., Ago H., Ueno G., Tosha T., Eady R. R., Antonyuk S. V., Yamamoto M., and Hasnain S. S. (2018) An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase. IUCrJ 5, 22–31 10.1107/S2052252517016128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fukuda Y., Tse K. M., Nakane T., Nakatsu T., Suzuki M., Sugahara M., Inoue S., Masuda T., Yumoto F., Matsugaki N., Nango E., Tono K., Joti Y., Kameshima T., Song C., et al. (2016) Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc. Natl. Acad. Sci. U. S. A. 113, 2928–2933 10.1073/pnas.1517770113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tosha T., Nomura T., Nishida T., Saeki N., Okubayashi K., Yamagiwa R., Sugahara M., Nakane T., Yamashita K., Hirata K., Ueno G., Kimura T., Hisano T., Muramoto K., Sawai H., et al. (2017) Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate. Nat. Commun. 8, 1585 10.1038/s41467-017-01702-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimada A., Kubo M., Baba S., Yamashita K., Hirata K., Ueno G., Nomura T., Kimura T., Shinzawa-Itoh K., Baba J., Hatano K., Eto Y., Miyamoto A., Murakami H., Kumasaka T., et al. (2017) A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase. Sci. Adv. 3, e1603042 10.1126/sciadv.1603042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chreifi G., Baxter E. L., Doukov T., Cohen A. E., McPhillips S. E., Song J., Meharenna Y. T., Soltis S. M., and Poulos T. L. (2016) Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer. Proc. Natl. Acad. Sci. U. S. A. 113, 1226–1231 10.1073/pnas.1521664113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johansson L. C., Arnlund D., Katona G., White T. A., Barty A., DePonte D. P., Shoeman R. L., Wickstrand C., Sharma A., Williams G. J., Aquila A., Bogan M. J., Caleman C., Davidsson J., Doak R. B., et al. (2013) Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nat. Commun. 4, 2911 10.1038/ncomms3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blakeley M. P., Langan P., Niimura N., and Podjarny A. (2008) Neutron crystallography: opportunities, challenges, and limitations. Curr. Opin. Struct. Biol. 18, 593–600 10.1016/j.sbi.2008.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hersleth H. P., Hsiao Y. W., Ryde U., Görbitz C. H., and Andersson K. K. (2008) The crystal structure of peroxymyoglobin generated through cryoradiolytic reduction of myoglobin compound III during data collection. Biochem. J. 412, 257–264 10.1042/BJ20070921 [DOI] [PubMed] [Google Scholar]
- 20. Hersleth H. P., Hsiao Y. W., Ryde U., Görbitz C. H., and Andersson K. K. (2008) The influence of X-rays on the structural studies of peroxide-derived myoglobin intermediates. Chem. Biodivers. 5, 2067–2089 10.1002/cbdv.200890189 [DOI] [PubMed] [Google Scholar]
- 21. Hersleth H. P., Uchida T., Røhr A. K., Teschner T., Schünemann V., Kitagawa T., Trautwein A. X., Görbitz C. H., and Andersson K. K. (2007) Crystallographic and spectroscopic studies of peroxide-derived myoglobin compound II and occurrence of protonated FeIV O. J. Biol. Chem. 282, 23372–23386 10.1074/jbc.M701948200 [DOI] [PubMed] [Google Scholar]
- 22. Kurth J. M., Brito J. A., Reuter J., Flegler A., Koch T., Franke T., Klein E. M., Rowe S. F., Butt J. N., Denkmann K., Pereira I. A., Archer M., and Dahl C. (2016) Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J. Biol. Chem. 291, 24804–24818 10.1074/jbc.M116.753863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brito J. A., Gutierres A., Denkmann K., Dahl C., and Archer M. (2014) Production, crystallization and preliminary crystallographic analysis of Allochromatium vinosum thiosulfate dehydrogenase TsdA, an unusual acidophilic c-type cytochrome. Acta Crystallogr. F Struct. Biol. Commun. 70, 1424–1427 10.1107/S2053230X14019384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brito J. A., Denkmann K., Pereira I. A., Archer M., and Dahl C. (2015) Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238 10.1074/jbc.M114.623397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenner L. P., Kurth J. M., van Helmont S., Sokol K. P., Reisner E., Dahl C., Bradley J. M., Butt J. N., and Cheesman M. R. (2019) Heme ligation and redox chemistry in two bacterial thiosulfate dehydrogenase (TsdA) enzymes. J. Biol. Chem. 294, 18002–18014 10.1074/jbc.RA119.010084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfanzagl V., Nys K., Bellei M., Michlits H., Mlynek G., Battistuzzi G., Djinovic-Carugo K., Van Doorslaer S., Furtmüller P. G., Hofbauer S., and Obinger C. (2018) Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage. J. Biol. Chem. 293, 14823–14838 10.1074/jbc.RA118.004773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostan J., Sjöblom B., Maixner F., Mlynek G., Furtmüller P. G., Obinger C., Wagner M., Daims H., and Djinović-Carugo K. (2010) Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: identification of a catalytically important amino acid residue. J. Struct. Biol. 172, 331–342 10.1016/j.jsb.2010.06.014 [DOI] [PubMed] [Google Scholar]
- 28. Schaffner I., Mlynek G., Flego N., Pühringer D., Libiseller-Egger J., Coates L., Hofbauer S., Bellei M., Furtmüller P. G., Battistuzzi G., Smulevich G., Djinović-Carugo K., and Obinger C. (2017) Molecular mechanism of enzymatic chlorite detoxification: insights from structural and kinetic studies. ACS Catalysis 7, 7962–7976 10.1021/acscatal.7b01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofbauer S., Mlynek G., Milazzo L., Pühringer D., Maresch D., Schaffner I., Furtmüller P. G., Smulevich G., Djinović-Carugo K., and Obinger C. (2016) Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ—lessons from the first crystal structure and kinetic studies. FEBS J. 283, 4386–4401 10.1111/febs.13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hofbauer S., Bellei M., Sündermann A., Pirker K. F., Hagmüller A., Mlynek G., Kostan J., Daims H., Furtmüller P. G., Djinović-Carugo K., Oostenbrink C., Battistuzzi G., and Obinger C. (2012) Redox thermodynamics of high-spin and low-spin forms of chlorite dismutases with diverse subunit and oligomeric structures. Biochemistry 51, 9501–9512 10.1021/bi3013033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schaffner I., Hofbauer S., Krutzler M., Pirker K. F., Bellei M., Stadlmayr G., Mlynek G., Djinovic-Carugo K., Battistuzzi G., Furtmüller P. G., Daims H., and Obinger C. (2015) Dimeric chlorite dismutase from the nitrogen-fixing cyanobacterium Cyanothece sp. PCC7425. Mol. Microbiol. 96, 1053–1068 10.1111/mmi.12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Battistuzzi G., Bellei M., Casella L., Bortolotti C. A., Roncone R., Monzani E., and Sola M. (2007) Redox reactivity of the heme Fe3+/Fe2+ couple in native myoglobins and mutants with peroxidase-like activity. J. Biol. Inorg. Chem. 12, 951–958 10.1007/s00775-007-0267-1 [DOI] [PubMed] [Google Scholar]
- 33. Hofbauer S., Dalla Sega M., Scheiblbrandner S., Jandova Z., Schaffner I., Mlynek G., Djinović-Carugo K., Battistuzzi G., Furtmüller P. G., Oostenbrink C., and Obinger C. (2016) Chemistry and molecular dynamics simulations of heme b-HemQ and coproheme-HemQ. Biochemistry 55, 5398–5412 10.1021/acs.biochem.6b00701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garman E. F. (2010) Radiation damage in macromolecular crystallography: what is it and why should we care? Acta Crystallogr. D Biol. Crystallogr. 66, 339–351 10.1107/S0907444910008656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macedo S., Pechlaner M., Schmid W., Weik M., Sato K., Dennison C., and Djinović-Carugo K. (2009) Can soaked-in scavengers protect metalloprotein active sites from reduction during data collection? J. Synchrotron Radiat. 16, 191–204 10.1107/S0909049509003331 [DOI] [PubMed] [Google Scholar]
- 36. Southworth-Davies R. J., and Garman E. F. (2007) Radioprotectant screening for cryocrystallography. J. Synchrotron Radiat. 14, 73–83 10.1107/S0909049506044177 [DOI] [PubMed] [Google Scholar]
- 37. Hofbauer S., Gysel K., Bellei M., Hagmüller A., Schaffner I., Mlynek G., Kostan J., Pirker K. F., Daims H., Furtmüller P. G., Battistuzzi G., Djinović-Carugo K., and Obinger C. (2014) Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity. Biochemistry 53, 77–89 10.1021/bi401042z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hofbauer S., Gruber C., Pirker K. F., Sündermann A., Schaffner I., Jakopitsch C., Oostenbrink C., Furtmüller P. G., and Obinger C. (2014) Transiently produced hypochlorite is responsible for the irreversible inhibition of chlorite dismutase. Biochemistry 53, 3145–3157 10.1021/bi500401k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Denisov I. G., Victoria D. C., and Sligar S. G. (2007) Cryoradiolytic reduction of heme proteins: maximizing dose dependent yield. Radiat. Phys. Chem. Oxf. Engl. 1993 76, 714–721 10.1016/j.radphyschem.2006.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones G. D., Lea J. S., Symons M. C., and Taiwo F. A. (1987) Structure and mobility of electron gain and loss centres in proteins. Nature 330, 772–773 10.1038/330772a0 [DOI] [PubMed] [Google Scholar]
- 41. Spence J. C. H. (2017) Outrunning damage: electrons vs X-rays—timescales and mechanisms. Struct. Dyn. 4, 044027 10.1063/1.4984606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bury C. S., Brooks-Bartlett J. C., Walsh S. P., and Garman E. F. (2018) Estimate your dose: RADDOSE-3D. Protein Sci. 27, 217–228 10.1002/pro.3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeldin O. B., Gerstel M., and Garman E. F. (2013) Optimizing the spatial distribution of dose in X-ray macromolecular crystallography. J. Synchrotron Radiat. 20, 49–57 10.1107/S0909049512044706 [DOI] [PubMed] [Google Scholar]
- 44. Kantardjieff K. A., and Rupp B. (2003) Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 10.1110/ps.0350503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kozlowski L. P. (2017) Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 45, D1112–D1116 10.1093/nar/gkw978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 10.1016/0022-2836(68)90205-2 [DOI] [PubMed] [Google Scholar]
- 47. Zeldin O. B., Brockhauser S., Bremridge J., Holton J. M., and Garman E. F. (2013) Predicting the X-ray lifetime of protein crystals. Proc. Natl. Acad. Sci. U. S. A. 110, 20551–20556 10.1073/pnas.1315879110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jakopitsch C., Pirker K. F., Flemmig J., Hofbauer S., Schlorke D., Furtmüller P. G., Arnhold J., and Obinger C. (2014) Mechanism of reaction of chlorite with mammalian heme peroxidases. J. Inorg. Biochem. 135, 10–19 10.1016/j.jinorgbio.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chance B. (1949) The properties of the enzyme-substrate compounds of peroxidase and peroxides; the spectra of the primary and secondary complexes. Arch. Biochem. 21, 416–430 [PubMed] [Google Scholar]
- 50. Furtmüller P. G., Arnhold J., Jantschko W., Pichler H., and Obinger C. (2003) Redox properties of the couples compound I/compound II and compound II/native enzyme of human myeloperoxidase. Biochem. Biophys. Res. Commun. 301, 551–557 10.1016/S0006-291X(02)03075-9 [DOI] [PubMed] [Google Scholar]
- 51. Battistuzzi G., Bellei M., Bortolotti C. A., and Sola M. (2010) Redox properties of heme peroxidases. Arch. Biochem. Biophys. 500, 21–36 10.1016/j.abb.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 52. Pfanzagl V., Bellei M., Hofbauer S., Laurent C. V. F. P., Furtmüller P. G., Oostenbrink C., Battistuzzi G., and Obinger C. (2019) Redox thermodynamics of B-class dye-decolorizing peroxidases. J. Inorg. Biochem. 199, 110761 10.1016/j.jinorgbio.2019.110761 [DOI] [PubMed] [Google Scholar]
- 53. Henderson R. (1990) Cryo-protection of protein crystals against radiation-damage in electron and X-ray diffraction. Proc. R. Soc. B Biol. Sci. 241, 6–8 10.1098/rspb.1990.0057 [DOI] [Google Scholar]
- 54. Unno M., Chen H., Kusama S., Shaik S., and Ikeda-Saito M. (2007) Structural characterization of the fleeting ferric peroxo species in myoglobin: experiment and theory. J. Am. Chem. Soc. 129, 13394–13395 10.1021/ja076108x [DOI] [PubMed] [Google Scholar]
- 55. Haas D. J., and Rossmann M. G. (1970) Crystallographic studies on lactate dehydrogenase at −75 degrees C. Acta Crystallogr. B 26, 998–1004 10.1107/s0567740870003485 [DOI] [PubMed] [Google Scholar]
- 56. Petsko G. A. (1975) Protein crystallography at sub-zero temperatures: cryo-protective mother liquors for protein crystals. J. Mol. Biol. 96, 381–392 10.1016/0022-2836(75)90167-9 [DOI] [PubMed] [Google Scholar]
- 57. Hope H. (1988) Cryocrystallography of biological macromolecules: a generally applicable method. Acta Crystallogr. B 44, 22–26 10.1107/S0108768187008632 [DOI] [PubMed] [Google Scholar]
- 58. Sliz P., Harrison S. C., and Rosenbaum G. (2003) How does radiation damage in protein crystals depend on X-ray dose? Structure 11, 13–19 10.1016/S0969-2126(02)00910-3 [DOI] [PubMed] [Google Scholar]
- 59. Kato Y., Nakamura K., Sakiyama H., Mayhew S. G., and Asano Y. (2000) Novel heme-containing lyase, phenylacetaldoxime dehydratase from Bacillus sp. strain OxB-1: purification, characterization, and molecular cloning of the gene. Biochemistry 39, 800–809 10.1021/bi991598u [DOI] [PubMed] [Google Scholar]
- 60. Kobayashi K., Yoshioka S., Kato Y., Asano Y., and Aono S. (2005) Regulation of aldoxime dehydratase activity by redox-dependent change in the coordination structure of the aldoxime-heme complex. J. Biol. Chem. 280, 5486–5490 10.1074/jbc.M410474200 [DOI] [PubMed] [Google Scholar]
- 61. Sawai H., Sugimoto H., Kato Y., Asano Y., Shiro Y., and Aono S. (2009) X-ray crystal structure of Michaelis complex of aldoxime dehydratase. J. Biol. Chem. 284, 32089–32096 10.1074/jbc.M109.018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li K., Xu F., and Eriksson K. E. (1999) Comparison of fungal laccases and redox mediators in oxidation of a nonphenolic lignin model compound. Appl. Environ. Microbiol. 65, 2654–2660 10.1128/AEM.65.6.2654-2660.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marshall N. M., Garner D. K., Wilson T. D., Gao Y. G., Robinson H., Nilges M. J., and Lu Y. (2009) Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature 462, 113–116 10.1038/nature08551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yano J., Kern J., Irrgang K. D., Latimer M. J., Bergmann U., Glatzel P., Pushkar Y., Biesiadka J., Loll B., Sauer K., Messinger J., Zouni A., and Yachandra V. K. (2005) X-ray damage to the Mn4Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. U. S. A. 102, 12047–12052 10.1073/pnas.0505207102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Denkmann K., Grein F., Zigann R., Siemen A., Bergmann J., van Helmont S., Nicolai A., Pereira I. A., and Dahl C. (2012) Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688 10.1111/j.1462-2920.2012.02820.x [DOI] [PubMed] [Google Scholar]
- 66. Hofbauer S., Hagmüller A., Schaffner I., Mlynek G., Krutzler M., Stadlmayr G., Pirker K. F., Obinger C., Daims H., Djinović-Carugo K., and Furtmüller P. G. (2015) Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes. Arch. Biochem. Biophys. 574, 36–48 10.1016/j.abb.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Afonine P. V., Mustyakimov M., Grosse-Kunstleve R. W., Moriarty N. W., Langan P., and Adams P. D. (2010) Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 10.1107/S0907444910026582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., and Womack T. O. (2017) BUSTER, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 76. Chen V. B., Arendall W. B., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data were deposited to the RCSB Protein Data Bank under the following accession codes: 6RPD, 6RPE, 6RQY, 6RR1, 6RR4, 6RR5, 6RR6, 6RR8. All other data are stored on the network space of the Department of Chemistry at the University of Natural Resources and Life Sciences, Vienna, and partially at the Synchrotron facilities, at which diffraction data were collected for this work. Data will be shared upon request to the corresponding authors.