Abstract
Mechanical stimulation of fibroblasts induces changes in the actin cytoskeleton including stress fiber (SF) reinforcement and realignment. Here we characterize the nuclear response to mechanical stimulation (uniaxial cyclic stretch). Using fluorescence microscopy and quantitative image analysis we find that stretch-induced nuclear elongation and alignment perpendicular to the stretch vector are dependent on formin-regulated actin polymerization. The mechanosensitive transcription factors Yes-associated protein/Transcriptional coactivator with PDZ domain (YAP/TAZ) and myocardin-related transcription factor (MRTF-A, also known as MKL1 and MAL1) accumulate in the nucleus and activate their target genes in response to uniaxial cyclic stretch. We show that transmembrane actin nuclear (TAN) lines are induced by stretch stimulation and nuclear envelope (NE) proteins including nesprins, SUN2, and lamins form Linkers of the Nucleoskeleton and Cytoskeleton (LINC) complexes aligned with actin SFs. These NE structures are altered by pharmacological treatments (Cytochalasin D and Jasplakinolide) or genetic disruption (zyxin gene deletion) that alter actin, and their persistence requires maintenance of stretch stimulation. Nuclear pore complexes (NPCs) accumulate at TAN lines providing a potential mechanism for linking mechanical cues to NPC function.
INTRODUCTION
Although artists’ illustrations of the prototypical diploid mammalian cells show a centrally located, radially symmetric nucleus, the position of the cell nucleus is known to vary based on developmental timing, differentiation state, motile behavior, and other physiological factors (Gundersen and Worman, 2013). The shape and orientation of the cell nucleus is also modulated in response to a variety of developmental signals. One of the most dramatic examples of nuclear sculpting occurs during spermatogenesis when cell nuclei are profoundly compacted and elongated (Kierszenbaum et al., 2007), in some cases achieving axial ratios in which nuclear length exceeds width by a factor of 10.
Nuclear shape is dynamic and can be influenced by both cytoplasmic and nucleoplasmic factors. Both microtubules and the actin cytoskeleton have been shown to influence nuclear shape determination (Kierszenbaum et al., 2007; Xue et al., 2013). Actomyosin-dependent contractility has also been shown to be involved in rotation of cell nuclei (Kumar, Maitra, et al., 2014). In the nucleoplasm, lamins play a critical role in shaping of cell nuclei from the inside out, with lamin A/C playing a key role in regulating nuclear rigidity (Lammerding et al., 2004; Swift, Ivanovska, et al., 2013).
In recent years, nuclear shape has been shown to influence cell physiology and behavior, clearly reinforcing the interconnection between nuclear form and function. Experimental manipulation of nuclear shape by plating cells on microfabricated substrata results in alterations in gene expression (Thomas et al., 2002; Talwar et al., 2014). When human lamin genes are mutated, as occurs in laminopathies, nuclear shape is often altered in association with changes in gene expression that have pathological consequences (Davidson and Lammerding, 2014).
Nuclear remodeling also occurs during cell migration under normal and pathophysiological conditions. Neuroblast nuclei undergo dramatic elongation during extensive cortical migrations in development (Tsai et al., 2005; Pajerowski et al., 2007). Likewise, tumor metastasis often requires that cells traverse through very tight interstitial spaces, putting pressure on the relatively rigid nuclear compartment. Abnormal nuclear morphology and nuclear envelope (NE) changes are evident in cancer cells (Chow et al., 2012; Bell and Lammerding 2016). Nuclear lamin stiffness is a barrier to the migration of cells within complex 3D environments that exhibit confining micropores (Harada et al., 2014; Thiam et al., 2016). Indeed, when nuclei face severe geometric challenges during cell migration, nuclear rupture leading to genome instability can occur (Denais, Gilbert, Isermann, et al., 2016; Shah et al., 2017; Hatch, 2018).
A growing body of evidence suggests that mechanical stress influences nuclear structure and function. Classic experiments have shown that tugging on integrin adhesion receptors results in distortion of the nucleus, illustrating a physical linkage between the cell surface and the nucleus (Maniotis et al., 1997). Recent studies have revealed that mechanical stress promotes the nuclear accumulation of the transcriptional coactivators Yes-associated protein (YAP)/Transcriptional coactivator with PDZ domain (TAZ) (Dupont et al., 2011; Aragona et al., 2013). One way in which mechanical signals influence YAP/TAZ distribution and transcriptional output is via down-regulation of the MST1/2 (Hippo in Drosophila) signaling pathway, a kinase cascade whose output is exquisitely tuned in response to mechanical cues (Dupont et al., 2011; Halder and Johnson, 2011; Wada et al., 2011; Halder et al., 2012, Aragona et al., 2013; Thomasy et al., 2013; Yu et al., 2013; Meng et al., 2016). Activation of the MST1/2 kinase cascade results in phosphorylation of YAP/TAZ and its sequestration in the cytoplasm, whereas dephosphorylation of YAP/TAZ leads to nuclear localization and activation. In addition, the actin cytoskeleton has been identified as an MST 1/2 (Hippo)-independent regulator of YAP/TAZ nuclear function (Dupont et al., 2011; Aragona et al., 2013; Feng et al., 2014; Gaspar et al., 2015), but the mechanism by which actin stress fibers (SFs) influence the nuclear response to mechanical stress is not understood. Myocardin-related transcription factor (MRTF-A) is another mechanosensitive transcription factor that is regulated by actin and is responsive to nuclear rigidity (Talwar et al., 2014; Finch-Edmondson and Sudol, 2016).
To probe the links among mechanical stress, the actin cytoskeleton, and the nucleus, we have applied controlled mechanical stimulation to cultured cell populations and describe the impact on nuclear orientation, shape, and NE molecular organization. We have uncovered changes in nuclear morphology and membrane organization that occur under conditions in which transcription factors YAP/TAZ and MRTF-A accumulate within cell nuclei. We report that uniaxial cyclic stretch induces nuclear translocation of transcription factors, gene expression changes, the reorientation and elongation of cell nuclei, establishment of linear arrays of Linker of the Nucleoskeleton and Cytoskeleton (LINC) proteins into robust transmembrane actin-associated nuclear (TAN) lines, and accumulation of nuclear pore complexes (NPCs) in association with this dynamically organized domain of the NE. Our work details a reliable experimental approach to study the effects of directional mechanical stress on nuclear structure and function, providing a cell biological description of the response of cell nuclei to cyclic stretch and a foundation for future mechanistic studies.
RESULTS
Uniaxial cyclic stretch promotes YAP/TAZ and MRTF-A nuclear accumulation
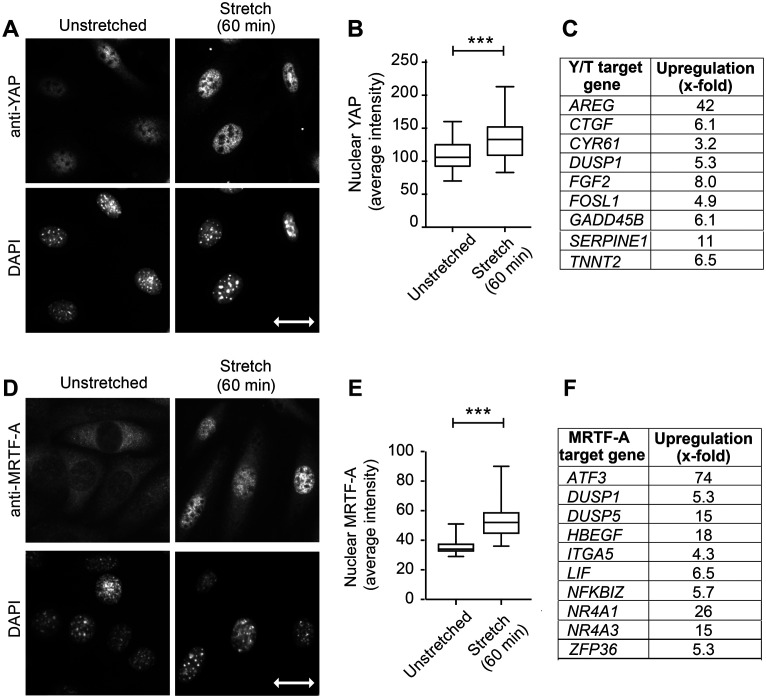
We previously reported that cells respond to uniaxial cyclic stretch stimulation by reinforcement of actin SFs and realignment of these cytoskeletal elements perpendicular to the stretch vector (Yoshigi, Hoffman, et al., 2005; Hoffman et al., 2006). To probe how this mechanical input impacts nuclear function, we examined the subcellular distribution of mechanosensitive transcription factors YAP/TAZ and MRTF-A (Finch-Edmondson and Sudol, 2016) in cells exposed to uniaxial cyclic stretch. It was previously reported that dynamic stretch applied to mesenchymal stem cells induced a nuclear translocation of YAP (Driscoll et al., 2015). We applied uniaxial cyclic stretch (15%) delivered at 0.5 Hz for 60 min to mouse fibroblasts with our previously described custom-made device (Yoshigi, Hoffman, et al., 2005). Although the relatively stiff substrate used results in significant nuclear YAP at baseline condition, mechanical stimulation results in further accumulation of YAP within cell nuclei as evidenced by visual inspection of cells labeled with anti-YAP antibody by indirect immunofluorescence compared with DAPI (Figure 1A) and by quantitative image analysis of nuclear signal (Figure 1B). Moreover, RNA sequencing analysis of mRNAs from unstretched and stretch-stimulated cells provides evidence that YAP/TAZ-dependent transcriptional programs are activated by uniaxial cyclic stretch (Figure 1C). We find a number of previously reported YAP/TAZ transcriptional target genes (Zhang et al., 2009; Cordenonsi et al., 2011; Zanconato et al., 2015), including AREG, CTGF, CYR61, DUSP1, FGF2, FOSL1, GADD45B, SERPINE1, and TNNT2, are up-regulated in response to uniaxial cyclic stretch (Supplemental Figure S1). These findings validate our uniaxial cyclic stretch regimen as a way to activate mechanically sensitive changes in gene expression and nuclear response.
FIGURE 1:
Linking YAP/TAZ and MRTF-A to uniaxial cyclic stretch response. (A) YAP immunolocalization to fibroblast nuclei (DAPI staining) on unstretched membranes and following uniaxial cyclic stretch (15% 0.5Hz 60 min). (B) Quantitation of nuclear YAP signal (region of interest selected from DAPI image; n = 61 nuclei in unstretched condition and n = 76 nuclei following stretch stimulation). (C) YAP/TAZ target genes identified as up-regulated in RNA seq analysis comparing four pairs of unstretched and stretch-stimulated (15% 0.5 Hz 6 h) cell mRNAs are presented as increased-fold change. (D) MRTF-A immunolocalization and DAPI-stained nuclei are shown for unstretched and stretch-stimulated cells. (E) Quantitation of nuclear MRTF-A signal (region of interest selected from DAPI image; n = 108 nuclei in unstretched condition and n = 110 nuclei following stretch stimulation). (F) MRTF-A target genes up-regulated by stretch stimulation in RNA seq analysis described above; 10 highest responders are shown here and all 25 target genes are shown in Supplemental Figure S1. Stretch vector is shown as a double-headed arrow of 30 micron scale on the DAPI image. Graphs of nuclear signals are box and whisker plots with median shown and minimum to maximum range. Unpaired t tests were used to determine p values ***p < 0.0001. RNA sequence gene descriptions with log twofold changes, p values, and references that identify the target genes are included as Supplemental Figure S1.
We next evaluated the response of MRTF-A (also known as MAL1 and MKL1), an actin-regulated mechanosensitive transcription factor (Miralles et al., 2003; Finch-Edmondson and Sudol, 2016), to uniaxial cyclic stretch. Unstimulated cells display a cytoplasmic distribution of MRTF-A and uniaxial cyclic stretch stimulation induces a nuclear accumulation of MRTF-A detected by immunofluorescence microscopy (Figure 1D). Quantitative image analysis of MRTF-A nuclear signal across cell populations confirms the stretch-stimulated nuclear translocation (Figure 1E). Analysis of the RNA sequence data set reveals 25 MRTF-A target genes (Yu et al., 2016; Foster et al., 2017; Gegenfurtner et al., 2018) are significantly up-regulated by stretch stimulation. The top 10 MRTF-A target genes include ATF3, DUSP1, DUSP5, HBEGF, ITGA5, LIF, NFKBIZ, NR4A1, NR4A3, and ZFP36 (Figure 1F). Additional information about the YAP/TAZ and MRTF-A target genes, their p values, the references for the target genes, and a summary of overall stretch-stimulated pathways are included in the supplement (Supplemental Figure S1). Together these data provide evidence that uniaxial cyclic stretch stimulation elicits a functional nuclear response that includes transcription factor nuclear import and activation of transcriptional programs within nuclei.
Nuclear shape and orientation are influenced by mechanical stimulation in a formin-dependent manner
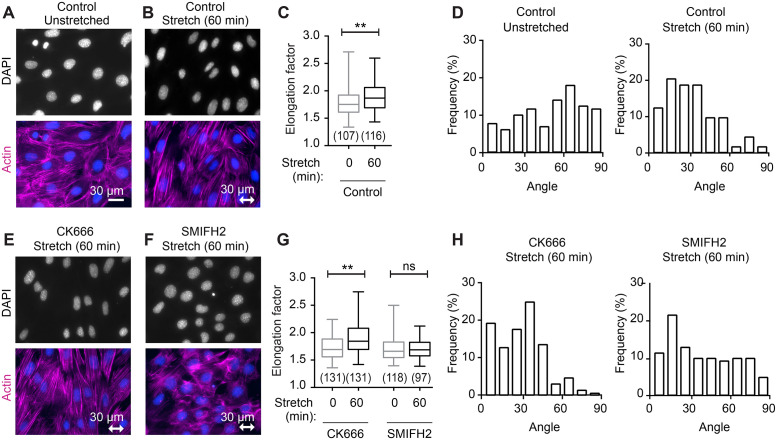
To explore the impact of stretch on nuclear morphology, we compared the nuclei from control cells and cells exposed to uniaxial cyclic stretch. Nuclei in unstretched cells appear rounded or slightly elongated (Figure 2A), whereas nuclei following stretch stimulation appear more oval-shaped and elongated (Figure 2B). Quantitative evaluation of nuclear shape was obtained by calculating the ratio of the long axis to short axis for nuclei from unstretched and stretch-stimulated cells. Exposure of cells to a regimen of uniaxial cyclic stretch for 60 min results in a statistically significant increase in the axial ratio (“elongation factor”) of cell nuclei (Figure 2C). Nuclear alignment relative to the horizontal stretch vector was evaluated for 127 unstretched cell nuclei and 112 stretch-stimulated cell nuclei. The nuclei were placed into 10o bins between 0 and ±90o based on their relative orientation, with the position perpendicular to the stretch vector set at 0o (Figure 2D). The orientation of nuclei from unstretched cells is random and covers the full 180o range from –90 to +90o. On exposure to uniaxial cyclic stretch, nuclear reorientation occurs with more than 70% of the cell nuclei oriented within 40o perpendicular to the stretch axis.
FIGURE 2:
Stretch-induced change in nuclear shape and positioning requires formin-dependent actin cytoskeleton. (A) Fluorescence microscopy of DAPI-stained nuclei (blue) and F-actin (phalloidin, magenta) on unstretched cells and (B) in cells subjected to stretch-stimulation (uniaxial cyclic stretch 15% 0.5 Hz 60 min). (C) Quantitation of nuclear elongation factor for unstretched (107 nuclei) and stretch-stimulated (116 nuclei) cells. (D) Graph of nuclear alignment relative to the horizontal axis shows nuclei binned into 10o increments with respect to perpendicular (0o) alignment. Unstretched nuclei (n = 127) in relatively random alignment shifted to perpendicular alignment in stretch-stimulated nuclei (n = 112), with 71% distributed to within 40o. DAPI (blue) and F-actin (magenta) staining of stretch-stimulated cells in the presence of inhibitors (E) CK666 (100 µm) Arp2/3 inhibitor or (F) SMIFH2 (15 µm) formin inhibitor. (G) Quantitation of nuclear elongation factor for cells stretch-stimulated in the presence of CK666 (n = 131, 131) or SMIFH2 (n = 118, 97), presented as median with minimum to maximum range. (H) Graph of nuclear alignment relative to the horizontal axis for cells stretch-stimulated in the presence of CK666 (n = 123) or SMIFH2 (n = 140). Box and whisker plots are presented as median with minimum to maximum range, and p values were determined by unpaired t tests. **p < 0.001 and n.s. is not statistically significant. The double-headed arrow indicates stretch vector and scale of 30 µm.
To determine whether the stretch-stimulated elongation or reorientation was dependent on the actin cytoskeleton, we used pharmacologic inhibitors of the two major classes of actin polymerization regulators: Arp2/3, which nucleates branched actin filaments (Swaney and Li, 2016), and formins, which promote nucleation and elongation of unbranched actin filaments (Goode and Eck, 2007). Treatment of cells with the Arp2/3 inhibitor CK666 (Yang, Zhang, et al., 2012) does not compromise the response of cells to uniaxial cyclic stretch. Actin SFs are evident in the stretched cells (Figure 2E), cell nuclei elongated (Figure 2G), and cell nuclei realigned perpendicular to the stretch vector (Figure 2H), illustrating that Arp2/3-dependent actin remodeling is not required for the stretch response. In contrast, treatment with the formin inhibitor, small molecule inhibitor of formin homology 2 domains (SMIFH2) (Rizvi, Neidt, et al., 2009), resulted in a compromised response to cyclic stretch. Cells treated with the formin inhibitor display diminished transcellular SFs (Figure 2F), lack of nuclear elongation (Figure 2G), and a failure of nuclear alignment relative to the stretch vector (Figure 2H), suggesting a role for formin-mediated actin assembly in the response of nuclei to cyclic stretch.
Uniaxial cyclic stretch promotes assembly of LINC complexes into nuclear lines
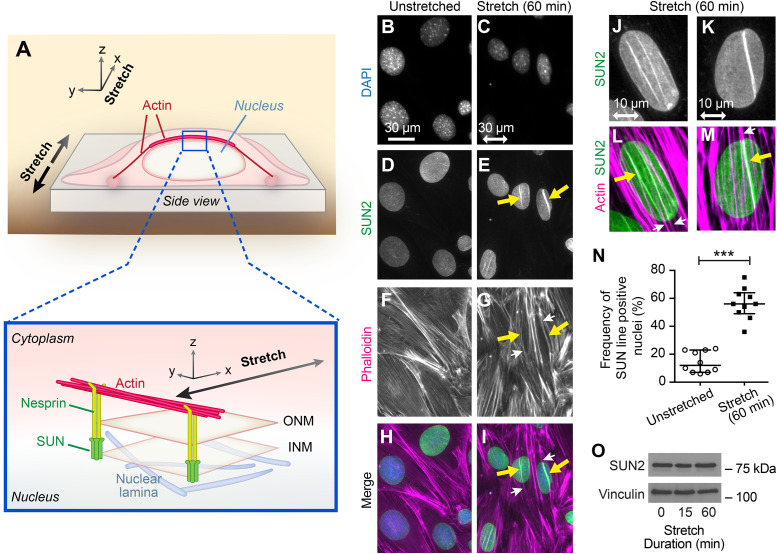
Pioneering work identified a role for LINC complexes in the establishment of a direct connection between the NE and the cytoskeleton (Crisp et al., 2006). The core LINC complex is comprised of outer nuclear membrane-resident Klarsicht, ANC-1, and Syne homology (KASH) domain proteins, such as nesprins, and inner nuclear membrane-resident Sad1 and UNC-84 (SUN) domain proteins, such as SUN1 and SUN2 (Haque et al., 2010; Mellad et al., 2011). A direct interaction of nesprin and SUN proteins creates a bridge across the inner and outer nuclear membranes, which is further linked to lamins in the nucleoplasm and cytoskeletal elements within the cytoplasm (Figure 3A) (Dahl et al., 2008; Chang et al., 2015). It was previously established that the association of particular LINC complex proteins with cytoplasmic actin SFs results in the formation of perinuclear actin caps that regulate nuclear morphology and protect nuclei from mechanical stress (Khatau et al., 2009; Kim et al., 2017). Dorsal actin SFs that anchor to the NE through LINC complexes during cell migration are called Transmembrane Actin Nuclear (TAN) lines (Luxton, Gomes, et al., 2010). Perinuclear actin caps and TAN lines share similar organizing principles. For simplicity, we refer to structures that display colinear accumulations of actin and LINC proteins as nuclear lines.
FIGURE 3:
Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes and associated actin SFs are stimulated by mechanical stress. (A) General model of stretch-stimulated cell with NE LINC complexes (nesprin and SUN proteins) shown in relation to the nuclear lamina (below) and the cytoplasmic actin cytoskeleton (above). Nesprin proteins in the outer nuclear membrane (ONM) and SUN proteins in the inner nuclear membrane (INM) complete the molecular chain from cytoplasmic actin to nuclear lamina. (B–I) Widefield fluorescent microscopy of cells in unstretched and stretch-stimulated conditions shows nuclei (DAPI, B and C), LINCs (SUN2 immunolocalization-Abcam antibody, D and E), and F-actin (Phalloidin, F and G). Merge overlay (H and I) shows actin SFs (magenta, white arrows) colocalized with SUN2 (green, yellow arrows) nuclear lines in stretch-stimulated cells. The stretch vector is shown in the horizontal plane with a double-headed arrow of scale 30 microns. (J–M) Widefield fluorescent microscopy of stretch-stimulated nuclei showing two examples of SUN2 immunolocalization (green) and F-actin (Phalloidin, magenta) codistribution (yellow arrows). The stretch vector is shown in the horizontal plane with double-headed arrow of scale 10 microns. (N) Nuclei from unstretched and stretch-stimulated cells were scored for nuclear lines and displayed as a percentage of cells positive for SUN2 nuclear lines. Graph (mean with SD) shows a compilation of population percentages from 10 experiments of paired unstretched nuclei (n = 845 nuclei) and stretch-stimulated nuclei (n = 1244 nuclei) revealing an average nuclear line detection of 14% in the unstretched population that increased to 56% in the stretched population. ***p < 0.0001 was determined by Student’s unpaired t test. (O) Western immunoblot analysis of SUN2 levels in cell lysates (15 µg) from unstretched and stretch-stimulated (uniaxial cyclic stretch 15% 0.5 Hz for 15 and 60 min) cells using vinculin as loading control.
To explore whether mechanical stimulation affects LINC complex organization and/or TAN line assembly, we exposed cells to uniaxial cyclic stretch under conditions (15% 0.5 Hz 60 min) that promoted nuclear remodeling and reorientation and evaluated F-actin and SUN2 distributions by indirect immunofluorescence microscopy. In response to stretch stimulation, the nuclei reorient and elongate (Figure 3, B and C) as seen with DAPI staining. Linear arrays of the NE protein SUN2 are prominent in the stretch-activated cells (Figure 3E, yellow arrows) relative to unstretched cells (Figure 3D), although an occasional SUN2 line is observed in a field of unstretched cells. These SUN2 arrays are coincident with the location of reinforced actin SFs visualized by fluorescent-Phalloidin staining (Figure 3, F–I), indicative of nuclear line formation. Apical and basal focal planes (Supplemental Figure S2) on nuclei from stretch-stimulated cells reveal that the linear arrays of SUN2 are coincident with the location of actin SFs associated with the apical or dorsal surface of the nucleus, consistent with perinuclear actin caps (Khatau et al., 2009), TAN lines (Luxton, Gomes, et al., 2010), and the model shown in Figure 3A.
The linear arrays of SUN2 are coincident with the position of actin SFs; however, not all actin SFs that overlie the nucleus exhibit codistributed SUN2 protein (compare Figure 3, J and L with K and M, for example). We have also observed that individual nuclear lines display variable intensity of SUN2 labeling (see Figure 3, E, J, and K and Supplemental Figure S3, for examples). Individual nuclei of cells stretched for 60 min may display up to five discrete lines, though the vast majority (77%) have one (31%) or two (46%) SUN2-rich lines (Supplemental Figure S3). Quantitation of populations across multiple stretch experiments shows SUN2-rich lines associated with an actin SF are present in 14.4 ± 2.4% of unstimulated cells compared with 56.4 ± 3.5% of stretched cells (Figure 3N). No concomitant increase in the expression of SUN2 protein is observed (Figure 3O), suggesting that the accumulation at linear arrays is due to protein redistribution rather than increased protein expression.
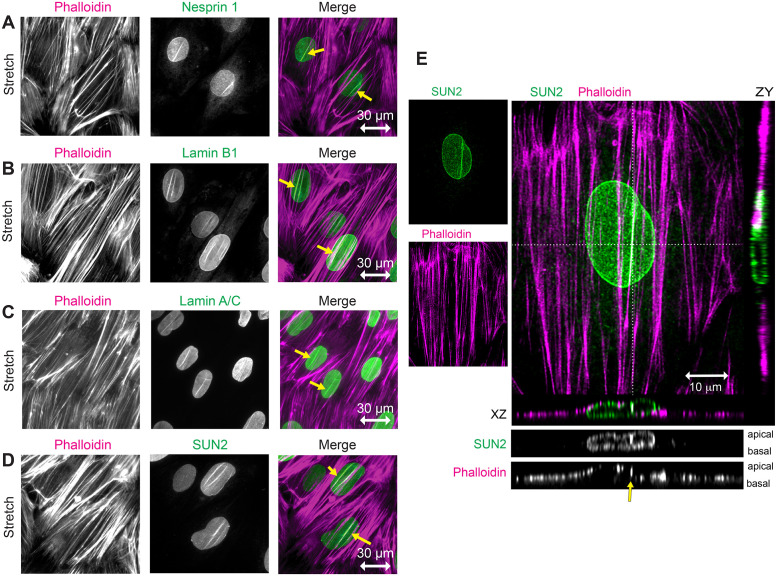
The stretch-induced nuclear lines display the full complement of core proteins that extend from the nuclear lamina to the actin cytoskeleton as evidenced by the presence of Nesprin 1 (Figure 4A), Lamin B1 (Figure 4B), and Lamin A/C (Figure 4C), as well as SUN2 (Figure 4D) in linear arrays coincident with actin SFs. We further determined that SUN2 and nesprins are colocalized in these nuclear lines by immunolocalization of SUN2 in stretch-stimulated cells programmed to express GFP-tagged mini-Nesprin (Luxton, Gomes, et al., 2010) (Supplemental Figure S4).
FIGURE 4:
Stretch-stimulated LINC components include Nesprin, SUN, and Lamin proteins. (A–D) Widefield fluorescence microscopy of F-actin (phalloidin, magenta) and LINC components (green) including Nesprin 1, Lamin B1, Lamin A/C, and SUN2 in stretch-stimulated cells. Merged images show codistribution of actin and LINC proteins (yellow arrows) in nuclei aligned perpendicular to the stretch vector (indicated by a horizontal double-headed arrow of 30 micron scale). (E) Confocal microscopy of stretch-stimulated fibroblast stained with SUN2-specific antibody (green, Burke #3.1E mab) and Phalloidin (magenta), shown as maximum intensity projections with 10 micron scale bar. Orthogonal z sections are designated by dashed lines in the x (below merge) and y (right of merge) planes. Grayscale z sections are shown with apical surface on top and basal surface on bottom. SUN2 is distributed throughout the NE and F-actin is distributed across the top of nucleus and along the flat cell edges. SUN2 localizes with one prominent actin SF (yellow arrow) but not with a neighboring SF crossing the nucleus; see associated Supplemental Videos S1–S3.
Video S1.
Confocal stack of SUN2 images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure 4E).
Video S2.
Confocal stack of Phalloidin/F-actin images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure 4E).
Video S3.
Merged images of SUN2 (green) and phalloidin (magenta) from a stretch-stimulated cell (Figure 4E) with 3D rotation for spatial resolution.
The codistribution of actin SFs with several NE proteins suggested the formation of stretch-stimulated nuclear line structures. However since it was possible that a deep groove in a nuclear membrane might lead to an apparent nuclear line signal when visualized in the xy dimensions by widefield microscopy, the stretch-stimulated actin and SUN2 nuclear lines were next resolved in the z dimension by confocal microscopy. This confocal data clarified the spatial nature of these flat fibroblasts, which are generally around 2–4 microns z depth in cells up to 100 microns in the xy dimensions (Figure 4E; Supplemental Videos S1–S3). The actin SFs are in close apposition to the nuclear membrane, sometimes forming actin and SUN2 nuclear lines as seen with orthogonal sections cut horizontally through the nucleus and viewed end-on (Figure 4E). The nuclei appear to be sandwiched between the overlying actin SFs and the substratum. We can distinguish the apical and basal boundaries of the NE and can distinguish ventral SFs from those that overlay the nucleus. We only detect SUN2 lines associated with dorsal or apical actin SFs, not with ventral or basal SFs. Notably, not all SFs are associated with SUN2 lines. For instance, a SUN2 nuclear line is evident along the actin SF right of center (Figure 4E, yellow arrow) but not along the actin SF crossing the center of the cell, which appears to have a similar actin fluorescence intensity. Actin indentation of the nucleus coincides with increased SUN2 signal in only one of these actin SFs, as seen in the xz orthogonal section.
In addition, NE proteins that are not known to be part of a NE–cytoskeletal interaction, such as SERCA2 (Sarco(endo)plasmic reticulum Ca2+ -ATPase) (Hossain and Clarke 2019) or Sec61B (Rapoport 2007), do not accumulate in robust nuclear lines in response to uniaxial cyclic stretch, as compared with SUN2 or other nuclear line constituents described in this paper. Immunolocalization and microscopy in the xyz dimensions were consistent with endoplasmic reticulum and NE distributions of SERCA2 (Supplemental Figure S5, A–C; Supplemental Videos S4–S7) and Sec61B (Supplemental Figure S5, D–F; Supplemental Videos S8–S11) in stretch-stimulated cells, with minimal nuclear line signals along actin SFs. Thus, the robust accumulation of SUN2 and other LINC proteins on nuclear lines, though sometimes clearly occurring at sites of NE indentation, is not fully explained by nuclear indentation since other NE constituents do not similarly accumulate.
Video S4.
Confocal stack of SUN2 images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5A).
Video S5.
Confocal stack of phalloidin images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5A)..
Video S6.
Confocal stack of SERCA2 images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5A).
Video S7.
Confocal stack of merged images (green-SUN2, magenta-actin, yellow-SERCA2) from a stretch-stimulated cell (Figure S5A) with 3D rotation for spatial resolution.
Video S8.
Confocal stack of SUN2 images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5D).
Video S9.
Confocal stack of phalloidin images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5D)..
Video S10.
Confocal stack of Sec61B images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S5D).
Video S11.
Confocal stack of merged images (green-SUN2, magenta-actin, yellow-Sec61B) from a stretch-stimulated cell (Figure S5D) with 3D rotation for spatial resolution.
Nuclear line assembly and maintenance depend on sustained mechanical stimulation
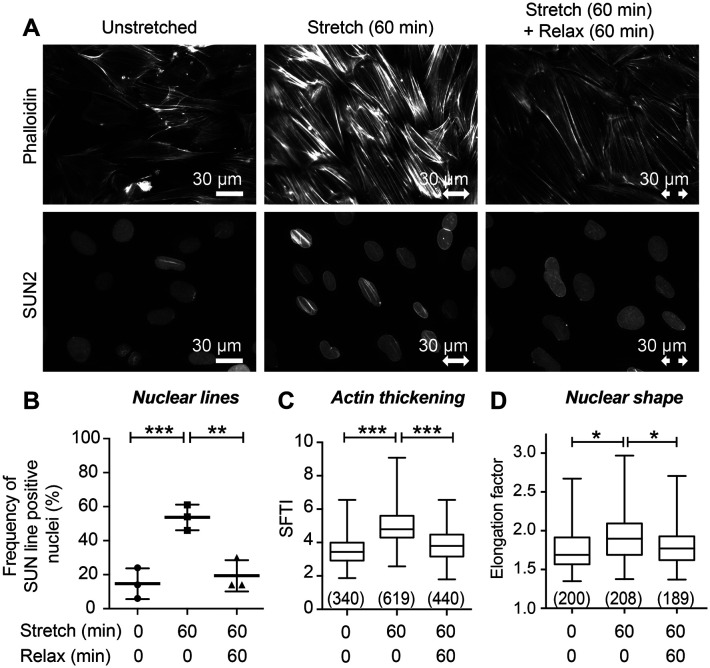
An important innovation of this work is the demonstration that uniaxial mechanical stimulation of fibroblasts promotes the establishment of nuclear lines that orient perpendicular to the stretch vector and contain the full complement of LINC proteins that bridge from the cytoskeleton to the nucleoskeleton. To address whether maintenance of mechanical signaling is required to sustain these structures, we conducted an experiment in which cells were subjected to a stretch regimen followed by a relaxation period. Under these conditions, we observe actin reinforcement and robust nuclear line assembly as evidenced by linear SUN2 distribution in stretched cells and a concomitant loss of actin reinforcement and SUN2 lines on relaxation (Figure 5A). Quantitative analysis revealed that 15% of unstretched cells display SUN2 nuclear lines, which increases to 54% on stretch, and returns to 20% with subsequent relaxation (Figure 5B).
FIGURE 5:
Actin and SUN2 nuclear line structures require sustained mechanical stimulation. (A) Widefield fluorescent microscopy of F-actin (phalloidin) and SUN2 immunolocalization on cells in unstretched condition prior to fixation, or subjected to stretch then fixed immediately, or subjected to stretch then allowed 60 min without stretch (“relaxation”) prior to fixation. Stretch vector is shown by a double-headed arrow of 30 micron scale. (B) Quantitation of cell population with SUN2 nuclear lines shows unstretched baseline of 15% of nuclei (n = 298 nuclei scored) increased to 54% of nuclei (n = 372 nuclei scored) with stretch-stimulation and decreased to 20% (n = 303 nuclei scored) following 60 min without stimulation. Graph is mean with SD of summary data points shown from three independent experiments. (C) SFTI analysis shows unstretched SF thickness (n = 340 measurements) increased with stretch stimulation (n = 619 measurements) and returned to baseline following 60 min without stretch (n = 440 measurements). (D) Nuclear Elongation factor (measured by Feret’s diameter analysis) of unstretched cells (n = 200) increased with 60 min of stretch (n = 208) and decreased following 60 min relaxation (n = 189). Graphs C and D are box and whisker plots showing median with minimum to maximum range and p values (*p < 0.05, **p < 0.001, ***p < 0.0001) determined by unpaired t tests.
We previously reported that uniaxial cyclic stretch induces an actin SF reinforcement response that can be quantitated using an analytical tool that assesses SF thickness (Yoshigi, Hoffman, et al., 2005; Hoffman et al., 2006). Analysis of the SF Thickness Index (SFTI) of unstretched, stretched, and relaxed cells revealed that SF reinforcement returns to a level comparable to unstretched cells when stretch stimulation is ceased and cells are allowed to recover for 60 min (Figure 5C). Similarly, when cells are allowed to relax after exposure to a stretch regimen, the nuclear elongation factor returns to baseline levels found in unstretched cells (Figure 5D). Thus, sustained physical stimulation is required to maintain nuclear lines, actin reinforcement, and nuclear elongation.
Disruption of actin SF reinforcement or integrity limits nuclear line formation
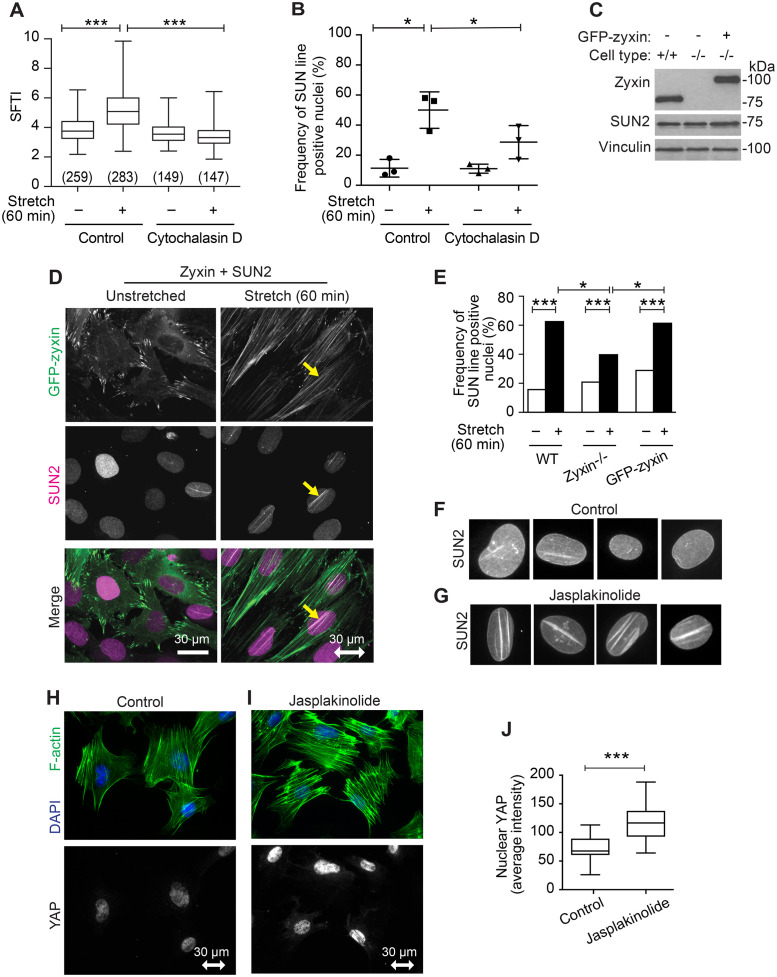
The observation that stretch-induced reinforcement of the actin cytoskeleton is correlated with the assembly of LINC complexes and TAN lines raised the possibility that the actin reinforcement promoted by mechanical stimulation contributes to these organizational events in the NE. If actin polymerization were required for stretch-induced reorganization of LINC proteins into TAN lines, we reasoned that inhibition of actin assembly would abrogate the formation of linear SUN2 arrays in response to uniaxial cyclic stretch. The mycotoxin Cytochalasin D inhibits actin polymerization by blocking the addition of actin monomers to the ends of actin filaments. Exposure of cells to a concentration of Cytochalasin D that eliminates evidence of F-actin perturbs cell adhesion making it impossible to study the stretch response. We therefore identified conditions (250 nM for 1 h prior to stretch, then for 1 h stretch duration) in which cells treated with Cytochalasin D retained actin SFs and cell adhesion, but fail to reinforce actin SFs in response to uniaxial cyclic stretch (Figure 6A). Although the induction of SUN2 nuclear line assembly is not completely abrogated by Cytochalasin D treatment under the conditions of our experiment, cells display a significant reduction of SUN2 nuclear line frequency compared with control, stretched cells (Figure 6B), illustrating a role for stretch-induced actin reinforcement in the assembly of SUN2-rich nuclear lines.
FIGURE 6:
SUN2 nuclear lines require robust actin filaments. (A) SFTI for cells in unstretched (n = 259 measurements) and stretch-stimulated (n = 283) conditions, or in the presence of 250 nM actin inhibitor Cytochalasin D (unstretched n = 149, stretch-stimulated n = 147) are presented as box and whisker plots of median with minimum to maximum range of measurements and p values are determined by unpaired Student’s t test. (B) Graph of mean with SD of SUN2 nuclear line-positive cells in three populations of unstretched and stretch-stimulated (n = 495 nuclei) cells and in cells stimulated in the presence of Cytochalasin D (n = 349 nuclei). The population percentages of cells positive for nuclear lines are presented from three independent experiments; p values determined by paired Student’s t tests. (C) Western immunoblot analysis for zyxin and SUN2 levels in WT, zyxin-null, and zyxin-null cells engineered to express GFP-zyxin; vinculin serves as a loading control (load 20 µg). (D) SUN2 immunolocalization in unstretched and stretch-stimulated cells expressing GFP-zyxin shows stretch-stimulated zyxin distribution along entire actin SFs and codistribution with SUN2 nuclear lines (yellow arrow). Stretch vector is shown as a double-headed arrow in the horizontal plane of a 30 micron scale. (E) Graph of nuclei (n > 200 nuclei per set) with SUN2-positive nuclear lines in unstretched and stretch-stimulated WT cells, zyxin-null cells, and zyxin-null cells expressing GFP-zyxin. Data were analyzed using Chi-square test (Fisher’s exact test) to determine p values. (F) Four examples of SUN2 immunolocalization in control nuclei not treated with inhibitor. (G) Four examples of SUN2 immunolocalization in nuclei from cells treated with Jasplakinolide (50 nM, 2 h). F-actin (phalloidin, green) and DAPI (blue) shown for untreated cells (H) and for cells treated with 50 nM Jasplakinolide for 2 h (I). YAP immunolocalization is shown for the same fields with scale bar of 30 microns. (J) Using a region of interest outlined by the DAPI signal, the nuclear YAP signal was identified and quantitated for control cells (n = 30 nuclei) and Jasplakinolide-treated cells (n = 32 nuclei) and presented as median with minimum to maximum range. *p < 0.05, ***p < 0.0001.
We previously reported (Yoshigi, Hoffman, et al., 2005) that the LIM protein zyxin is required for stretch-induced actin reinforcement. Therefore, we explored whether zyxin is required for SUN2 line formation in stretched cells using fibroblasts isolated from wild-type (WT) mice, zyxin –/– mice, or zyxin-null cells reconstituted by expression of GFP-zyxin (Figure 6C). Zyxin is typically found at focal adhesions, sites where actin SFs are anchored to the cytoplasmic membrane through transmembrane integrin receptors, as is seen in unstretched conditions (Figure 6D). In cells subjected to uniaxial cyclic stretch, GFP-zyxin distributes along actin SFs and colocalizes with SUN2 nuclear lines (Figure 6D). Codistribution of zyxin and SUN2 along nuclear lines was also observed using confocal microscopy of immunolocalized proteins in stretch-stimulated WT cells. Zyxin distributes along entire actin SFs, including the region over the nucleus associated with SUN2 (Supplemental Figure S6; Supplemental Videos S12–S15). WT cells assemble SUN2 lines in response to stretch stimulation, but the SUN2 nuclear line formation is decreased in cells lacking zyxin (Figure 6E). Zyxin re-expression (GFP-zyxin in zyxin-null cells) restores the stretch-stimulated SUN2 line formation (Figure 6E), illustrating a positive contribution of zyxin to establishment of TAN lines.
Video S12.
Confocal stack of SUN2 images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S6).
Video S13.
Confocal stack of zyxin images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S6).
Video S14.
Confocal stack of F-actin images from a stretch-stimulated cell going from top of the cell to the bottom at 0.35 micron steps (Figure S6).
Video S15.
Confocal stack of merged images (green-SUN2, magenta-zyxin) from a stretch-stimulated cell (Figure S6) with 3D rotation for spatial resolution.
Collectively these results illustrate that the assembly of linear SUN2 arrays is enhanced by actin reinforcement downstream of uniaxial cyclic stretch. We next explored whether pharmacological induction of actin reinforcement is sufficient to induce the assembly of SUN2-rich TAN lines. The marine toxin Jasplakinolide is an F-actin promoting agent (Bubb et al., 1994), induces SF thickening independent of mechanical stimulation (Hoffman et al., 2006) and promotes robust assembly of randomly oriented SUN2-rich nuclear lines (compare Figure 6, F and G) independent of stretch stimulation. Jasplakinolide treatment promotes nuclear accumulation of YAP/TAZ in these mouse fibroblasts, as can be seen in Figure 6, H and I and quantitated in Figure 6J, consistent with a report for nuclear YAP in HeLa cells (Reddy et al., 2013). We also find that Jasplakinolide-stimulated actin assembly promotes nuclear translocation of MRTF-A in these fibroblasts (Supplemental Figure S7), consistent with a previous report in HeLa cells (Iyer et al., 2012). These findings illustrate that pharmacological induction of actin SF reinforcement is sufficient to trigger nuclear line formation as well as nuclear accumulation of mechanically sensitive transcription factors. To determine whether the stretch-stimulated transcription factor translocation follows or precedes the establishment of nuclear lines, we conducted a kinetic analysis and found that MRTF-A displays relatively rapid nuclear accumulation in cells exposed to cyclic stretch (peak at 5–15 min), preceding nuclear line formation (peak at 1–6 h) (Supplemental Figure S7). While SUN2 nuclear lines and MRTF-A nuclear translocation are both stimulated by Jasplakinolide and cyclic stretch, the nuclear translocation precedes the peak timing of nuclear line formation.
NPCs accumulate at TAN lines in stretch-stimulated cells
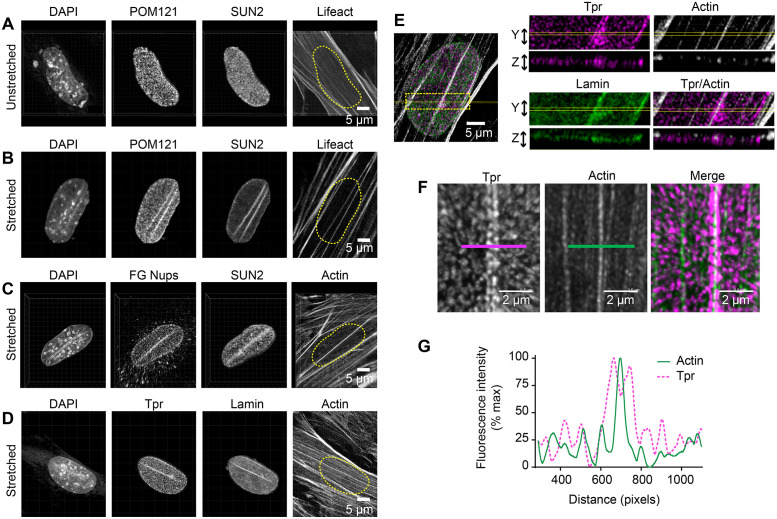
Transmission of mechanical signals between the cytoskeleton and the nucleus is thought to involve both LINC complexes and NPCs (Lee and Burke, 2018). Both LINC proteins and nuclear pores display a punctate, monodisperse distribution in the NEs of unstimulated fibroblasts (Figure 7A). However, given prior reports that at least one member of the SUN protein family, SUN1, associates directly with a nuclear pore constituent (Liu et al., 2007), we postulated that uniaxial cyclic stretch might trigger the redistribution of NPCs coincident with TAN line formation. Indeed, we found that both the NPC marker pore membrane protein of 121 kDa (POM121) and SUN2 are colocalized in cells that have been exposed to uniaxial cyclic stretch (Figure 7B). POM121-GFP distribution faithfully recapitulates the POM121-specific antibody signal, showing that at least one component of NPCs colocalizes with SUN2 in nuclear lines (Supplemental Figure S8). If functional NPCs accumulated at the nuclear lines of stretched cells, we would expect other nuclear pore constituents to be present at these sites following mechanical stimulation. Indeed, nucleoporins detected with a monoclonal antibody (mAb414) specific for a phenylalanine–glycine repeat (FG)-rich domain found in several nucleoporins (Figure 7C) and the nucleoporin, translocated promoter region (TPR) (Figure 7D), also colocalize with TAN lines in cells exposed to uniaxial cyclic stretch. In some cells, we observe a linear concentration of DAPI staining adjacent to the sites of nuclear pore protein accumulation (Figure 7, B and C), indicating concomitant changes in chromatin organization at these sites. Optical sectioning (orthogonal slice = 0.25 microns) to visualize the relative distributions of TPR, actin and lamin A/C in a single stretch-stimulated cell revealed that the three proteins were observed in a single optical section (Figure 7E). High resolution 3D images and fluorescence intensity line scans reveal two rows of nuclear pore constituent TPR flanking the actin SF (Figure 7, F and G). These data provide evidence that NPCs are colocalized with LINC proteins and the actin cytoskeleton in mechanically stimulated cells.
FIGURE 7:
LINCs and NPCs codistribute following uniaxial cyclic stretch. Multichannel imaging of nuclei in unstretched condition (A) or following uniaxial cyclic stretch (B) includes DAPI (DNA), POM121-GFP (NPC component), SUN2 (inner nuclear membrane LINC), and Lifeact-apple (cytoplasmic F-actin). Nuclear position is outlined (yellow dashed line) on Lifeact image. (C) Multichannel imaging of stretch-stimulated nucleus stained for DAPI, FG Nucleoporins (mAb414), SUN2, and F-actin. (D) Multichannel imaging of stretch-stimulated nucleus stained for DAPI, TPR (NPC component), Lamin, and F-actin. Scale bar is 5 microns. (E) Maximum intensity projection of confocal images of TPR (magenta), F-actin (white), and Lamin A/C (green). Images were sampled (yellow box) in z plane (0.25 microns) to show the distribution of signals in a single plane. Scale bar 5 microns. (F) Confocal microscopy of NPC component TPR (magenta) and F-actin (green) resolves the NPCs aligned adjacent to the actin SF. Scale bar 2 microns. (G) Line scans of TPR (dashed magenta line) and F-actin (solid green line) images. Images presented are maximum intensity projections from a confocal stack of images.
DISCUSSION
Mechanical stimulation of cells in culture leads to a dramatic reorganization of the actin cytoskeleton including the promotion of actin SF assembly (Kaunas et al., 2005; Yoshigi, Hoffman, et al., 2005; Hoffman et al., 2012). Here we have applied uniaxial cyclic stretch to a population of cultured fibroblasts and monitored the accompanying changes in nuclear morphology and function. Fibroblast nuclei elongate and align perpendicular to the orientation of the stretch vector and the mechanosensitive transcription factors YAP/TAZ and MRTF-A are translocated into the nucleus. In response to exposure to uniaxial cyclic stretch, cells display changes in both nuclear shape and position relative to the stretch vector. These nuclear changes are disrupted by pharmacological and genetic manipulation of actin cytoskeletal assembly and dynamics, illustrating the central role of the actin cytoskeleton in the shaping of nuclear architecture in response to mechanical cues. Coordinate with the stretch-induced changes in the actin cytoskeleton, NE-resident LINC components form robust nuclear lines aligned perpendicular to the stretch vector and coincident with actin SFs. Previous work has demonstrated that TAN line-resident proteins, such as the outer nuclear membrane protein nesprin, display reduced lateral mobility relative to protein that was not proximal to a TAN line (Luxton, Gomes, et al., 2010); this finding is consistent with their anchorage to stable cellular structures. Data presented here demonstrate that continuous mechanical stimulation is required to maintain the nuclear lines and actin SFs, illustrating their dynamic responsiveness to mechanical cues. The presence and orientation of nuclear lines is directly correlated with the directionality and maintenance of the stretch signal, thus the presence of a nuclear line with the actin cytoskeleton can serve as a surrogate marker indicative of the mechanical environment of the cell. In addition to providing a morphological indication of a cell’s mechanical environment, TAN lines also provide a potential mechanism for conveyance of mechanical signals across the NE. Our experimental approach provides a reproducible system for the activation of mechanically sensitive gene expression coupled with the ability to characterize phenotypic changes that accompany the transcriptional response at the level of cellular architecture with a high degree of spatial resolution.
We also report the novel observations that NPCs accumulate at TAN lines. Whenever a SUN2 line is observed, both actin and NPCs are colocalized with the SUN2 line. Likewise, we have observed no SUN2 lines that do not also display accumulation of NPCs. We show the NPCs appear as beads on a string flanking actin SFs that display accumulation of SUN2. Only SFs that overlay the nucleus are associated with SUN2 and NPCs, but not all dorsal SFs are colocalized with SUN2 and NPCs. The development of SUN2 lines appears to be highly dependent on the actin cytoskeleton, since agents that compromise actin assembly or elimination of zyxin, which is known to promote actin SF reinforcement (Hoffman et al., 2006), result in a reduction in nuclear lines. Reciprocally, the actin-stabilizing agent, Jasplakinolide, promotes the establishment of SUN2 lines. Many questions remain to be answered about how these nuclear lines are established in response to uniaxial mechanical stimulation, including what is the temporal order in which the components come together, which associations are constitutive versus stretch stimulated, and what is the half-life of the nuclear line complex, etc.? Live cell imaging will be required to decipher the dynamic behavior of these components in response to mechanical cues.
To our knowledge, this is the first report of NPCs associated with the actin cytoskeleton. Colocalization of NPCs with TAN lines raises the intriguing possibility that the actomyosin-dependent contractility could influence physically proximal NPCs. The possibility that mechanical force might promote widening of nuclear pore channels to alter transport of cargo has been considered (Wang et al., 2009; Kirby and Lammerding 2018; Kassianidou et al., 2019) and it was recently demonstrated that the nuclear import of YAP is responsive to substrate rigidity and experimentally delivered nuclear compression (Elosegui-Artola et al., 2017). Plating of cells on a rigid substratum induces widening of nuclear pore diameter from ∼58 nm to ∼75 nm, as evidenced by morphometric analysis of NPCs by Transmission Electron Microscopy, and promotes YAP nuclear import (Elosegui-Artola et al., 2017). Although additional experimental work will be required to validate if and how NPC conductance might be influenced by uniaxial cyclic stretch, it is intriguing to consider the possibility that NPCs associated with TAN lines might be positioned to respond to SF contractility, providing a mechanism to elicit stretch-induced changes in gene expression via regulated nuclear transport of proteins and/or RNA.
Initially viewed as passive conduits for the passage of molecules across the NE, there has been much speculation about the role of nuclear pores in the active regulation of gene expression (Burns and Wente, 2014; Ben-Yishay et al., 2016; Raices and D’Angelo, 2017). As early as 1985, Günter Blobel and colleagues proposed the “gene gating” hypothesis in which gene transcription was envisioned to be functionally coupled to efficient mRNA export via nuclear pores (Blobel, 1985). More recently, it has been proposed that chromatin connections may influence NPC-dependent cargo import and export, and that NPCs may influence gene regulation via the functional nuclear organization, positioning active chromatin domains adjacent to nuclear pores (Cremer et al., 2015; Ibarra and Hetzer, 2015; D’Angelo, 2018). Adding another level of potential control, mechanical cues can also impact the epigenetic regulation of chromatin to influence gene expression (Discher et al., 2017; Miroshnikova et al., 2017; Uhler and Shivashankar, 2017; Heo et al., 2018; Stephens et al., 2018).
A growing number of examples show that mechanical signals influence gene expression by promoting nuclear accumulation of mechanosensitive transcriptional regulators such as YAP/TAZ and MRTF-A (Finch-Edmondson and Sudol, 2016). Previous work has highlighted an important role for the actin cytoskeleton in activating these transcription factors, but by distinct molecular mechanisms. The nuclear translocation of MRTF-A (also known as MAL or MKL1) is regulated through a well-defined actin-dependent mechanism (Miralles et al., 2003). On exposure to stimuli that promote F-actin assembly, cytoplasmic MRTF-A loses its G-actin interaction and binds to importins for transport through the nuclear pore (Nakamura, Hayashi, et al., 2010; Pawlowski et al., 2010). MRTF-A nuclear export is regulated by Crm1-dependent nuclear pore export (Hayashi and Morita, 2013). Studies in mammalian cells and in Drosophila have revealed that YAP/TAZ can be regulated independent of Hippo signaling by a mechanically stimulated actin cytoskeletal pathway that depends on zyxin to promote nuclear accumulation of the transcriptional regulators (Dupont et al., 2011; Aragona et al., 2013; Gaspar et al., 2015; Harvey 2015). Still, the precise mechanism by which filamentous actin stimulates the nuclear accumulation of YAP/TAZ has remained a mystery. The promotion of NPC arrays in association with TAN lines provides a potential mechanism by which the actin cytoskeleton might directly influence NPC function and transcription factor subcellular distribution via mechanical gating of nuclear pores. However, our kinetic studies (Supplemental Figure S7) show that the establishment of robust NPC-associated TAN lines lags the nuclear accumulation of MRTF-A in stretched cells. Available reagents did not allow rigorous assessment for YAP/TAZ stretch kinetics. Thus, while a role for mechanical regulation of NPCs in association with LINC complexes cannot be ruled out, at least for MRTF it does not appear that nuclear import depends on TAN line formation.
It has been reported that actin SFs compress and indent the NE, and that LINC complexes and nuclear lamina accumulate at these nuclear indentations (Li et al., 2014; Versaevel et al., 2014; Kim et al., 2017). Consistent with this, we report here that stretch-stimulated cells develop actin SFs that compress nuclei and result in nuclear lines of LINC proteins and NPCs. We conclude the stretch-stimulated nuclear lines of SUN2 and other proteins, which often occur at sites of nuclear indentations, are not fully explained by the nuclear membrane in the groove because the indentations are not deep enough to generate sufficient nuclear membrane signal, the nuclear lines are not present with all actin SF indentations, and not all NE constituents generate such robust nuclear lines.
LINC proteins play important roles in structural and functional connections between the cytoplasm and the nucleus. Experimental disruption of components of the LINC complex compromises perinuclear actin cap formation (Khatau et al., 2009), cellular alignment (Chancellor et al., 2010), nuclear mechanical stiffness (Stewart-Hutchinson et al., 2008), nuclear deformation (Lombardi et al., 2011), nuclear morphology (Kim et al., 2017), gene expression (Alam et al., 2016), and cell migration (Kim et al., 2014; Chang et al., 2015). We show here that uniaxial cyclic stretch induces the formation of robust nuclear lines in which LINC proteins are aligned with cytoplasmic actin SFs and intranuclear lamin proteins, and that NPCs also accumulate at these sites. The demonstration that it is possible to induce robust nuclear line formation by exposure of cells to uniaxial cyclic stretch should facilitate the future study of these intriguing structures, including assembly and disassembly dynamics, relationship to NPC structure and function, and potential influence on chromatin organization and epigenetic regulation.
MATERIALS AND METHODS
Antibodies
The following antibodies and staining reagents were used: Primary antibodies: lamin A/C (#4777), YAP (#14074XP), and YAP/TAZ (#8418) from Cell Signaling Technology; lamin B1(#16048), Nesprin 1 (#24742), nucleoporins FG414 (#50008), and SUN2 (#124916) from Abcam; MRTF-A (G8 sc-390324) from Santa Cruz Biotechnology; POM121 (GTX-102128) from GeneTex; TPR (IHC-00099) from Bethyl Labs; vinculin (v-9131) from Sigma; SUN2 (#3.1E) from Brian Burke; and zyxin (Beckerle B71; ABC1463) from EMD Millipore. Alexa Fluor secondary antibodies, phalloidin, and DAPI were from Molecular Probes/Invitrogen.
Cell culture
WT mouse fibroblast cells were isolated and cultured as described previously (Hoffman et al., 2006), as were cells isolated from zyxin-null mice and expressing a GFP-zyxin transgene (Hoffman et al., 2006). Cells were cultured in high-glucose DMEM supplemented with pyruvate, glutamine, penicillin/streptomycin (Invitrogen), and 10% fetal bovine serum (FBS; Hyclone Labs, Logan, UT). POM121 DNA (Transomic Technologies) was tagged with GFP using Gateway cloning, the LifeAct sequence was tagged using Gateway cloning, and the DNAs were transiently transfected into WT fibroblasts using Lipofectamine 2000 (Invitrogen).
Uniaxial cyclic stretch
Cells were stretch stimulated using a custom-designed system previously described (Yoshigi, Hoffman, et al., 2005). Briefly, cells were seeded onto precoated (25 µg/ml Collagen I and 2 µg/ml Fibronectin) silicone membranes (2.2 million cells onto three 26 × 33 mm membranes in a 10-cm plate) and grown to confluence overnight. Cells were subjected to uniaxial cyclic stretch (15%, 0.5 Hz, 0.25, 1, and 6 h as designated in the figure legends), then either lysed and proteins harvested for immunoblot analysis or cells were fixed (3.7% formaldehyde 15 min) for cell staining and microscopy (Yoshigi, Hoffman, et al., 2005; Hoffman et al., 2012). For stretch experiments with inhibitors (Cytochalasin D at 250 nM, CK666 at 100 µm, SMIFH2 at 15 and 30 µm), cells on membranes were preincubated with 250 nM Cytochalasin D for 1 h prior to stretch, or with 100 µm CK666 or 15-30 uM SMIFH2 for 2 hr prior to stretch. The stretch stimulation and inhibitor experiments were performed in complete DMEM media with fresh inhibitor added at the beginning of stretch stimulation.
SF thickening, nuclear alignment, and elongation analysis
SFTI analysis of Phalloidin-stained cells utilized a custom erosion/brightness decay software written in LabView (National Instruments) and previously described (Yoshigi, Hoffman, et al., 2005). Multiple SFs in multiple cells in >10 microscopic fields were measured (regions of interest, ROI) for relative SFTI within single experiments, in at least three independent stretch experiments. SF alignment analysis was performed as previously described (Yoshigi et al., 2003, 2005). For source code, contact M. Yoshigi (masaaki.yoshigi@hsc.utah.edu). Briefly, nuclear alignment was calculated by Sobel filter algorithm, and orientation histograms were obtained from >100 nuclei as described in the figure legends. Distribution kurtosis of orientation histogram was designated as Alignment Index. Nuclei were binned into 10o increments, with perpendicular to stretch vector set as 0o and parallel with stretch vector (horizontal) set as –90o and +90o (Yoshigi et al., 2003; Hoffman et al., 2017).
Immunofluorescence microscopy
Cells were seeded onto glass coverslips (25,000 cells/well in 12-well dish, grown overnight DMEMc + 10% FBS), then treated for 2 h with 250 nM Cytochalasin D, 100 nM Jasplakinolide, or DMSO control. Cells seeded onto Collagen I–Fibronectin-coated stretch-silicone membranes (2.2 million cells/10-cm dish) were grown overnight and then stretched as described above. For localizations, cells on coverslips or on silicone membranes were fixed (15 min in 3.7% formaldehyde/phosphate-buffered saline) and then permeabilized (5 min in 0.5% Triton X-100), blocked (1 h in 5% normal goat serum), and incubated with antibodies as recommended by suppliers. Widefield fluorescent cell images were captured with a Zeiss Axioskop 2 mot plus fluorescence widefield microscope (Zeiss 40× NeoFluor 0.75NA dry objective and Zeiss 63× ApoChromat 1.4NA oil objective), Zeiss AxioCamMRm camera, and Zeiss AxioVision version 4.8.1 software. Confocal images were captured on a Leica SP8 DMi8 inverted microscope with a Leica 63× oil objective (HC PL APO CS2 NA1.4) using PMT and hybrid detectors and LASX software (Leica). To remove out of focus light, Lightning software (Leica) was used to deconvolve the images. Images are presented as maximum intensity projections of a confocal stack of images. Unless otherwise noted, z-steps are 0.35 µm with a total z thickness ranging 2–5 microns for the mouse fibroblasts. Orthogonal sectioning and 3D imaging of a confocal stack of images were processed with LASX software.
Nuclear YAP, MRTF-A, and SUN2 nuclear line analysis
Nuclear signal was measured on cells with immunolocalized YAP (CST#14074) or YAP/TAZ (CST#8418) or MRTF-A (sc#390324) antibody. Red–green–blue images were captured (>10 fields each) and the nuclei were thresholded on the DAPI image. ROI created around those DAPI nuclei were transferred onto the YAP or MRTF-A image, and the average intensity in the nuclear regions was measured (Metamorph Imaging software). To score SUN2 nuclear lines, the SUN2 nuclear signals were captured (>10 fields/sample, Zeiss 40× objective) on unstretched and stretched cells using identical exposures and camera settings. The images were subsequently opened in Photoshop to visualize on screen and scored positive or negative for nuclear lines, and data were tallied for the percentage of nuclei positive for SUN2 lines.
Western immunoblot analysis
Protein concentration of cell lysates were measured (Pierce Coomassie Protein Assay) and proteins (7.5–30 µg/lane, described in the figure legends) were electrophoresed through denaturing 10% polyacrylamide gels (Bio-Rad) and transferred onto nitrocellulose filters. Antibodies were diluted as recommended by the manufacturers and detected by HRP-conjugated antibodies and enhanced chemiluminescence (GE Healthcare).
RNA sequence analysis
Mouse fibroblasts were stretch stimulated (uniaxial, 15%, 0.5 Hertz, 6 h) and then mRNA was prepared using RNAeasy mini spin columns according to the manufacturer’s direction (Qiagen). Four independent stretch experiments were performed to generate four pairs of unstretched and stretched samples for pairwise gene expression analysis. Illumina HiSeq RNA sequencing was performed at the Genomics core facility, with data analysis guided by the Bioinformatics core facility (University of Utah Shared Resources).
Statistical analysis
GraphPad-Prism version 7 was used for statistical analysis. Graphs with >90 data points are presented as box and whisker plots in quartiles with median in center and minimum to maximum range, with the number of measurements included below each box as described in the figure legends. Bar graph is presented as mean with SE mean. For data from multiple experiments where the data points are 3–10 experiments, the box and whisker plot of mean with minimum to maximum includes each experiment point. The horizontal lines above the graphs identify the two groups compared using unpaired Student’s t tests that assumed a Gaussian distribution and equal SD between populations. As described in the figure legend, Chi-square analysis (Fisher’s exact test) from a contingency table was used to determine statistical significance in Figure 6E; *p < 0.05, **p < 0.01, ***p < 0.001, comparisons that were not statistically significant are designated n.s. or are not shown.
Figure preparation
Graphs were constructed in Prism 7 (GraphPad, San Diego, CA), cell images were processed with Photoshop CC, and figures were assembled in Illustrator CC (Adobe, San Jose, CA). Videos of confocal stack of images were assembled with FIJI (National Institutes of Health ImageJ) and 3D rotations were made with LASX (Leica) software.
Resource Table
A resource table included as Supplemental Table 1.
Supplementary Material
Acknowledgments
The High Throughput Genomics and Bioinformatics Shared Resource of the University of Utah Health Sciences Center provided invaluable technical support and guidance for analysis of RNA sequencing data, with thanks to Brian Dalley and Chris Stubben. The University of Utah Fluorescence Microscopy Core Facility provided microscopes critical to this research (NCRR Shared Equipment Grant No. 1S10RR024671), with guidance from Michael Bridge. Gregg Gundersen provided mini-Nesprin2G DNA and Brian Burke supplied LINC-specific antibodies. Diana Lim contributed expert figure design and preparation. We acknowledge Hiro Yoshigi for software development and data analysis, Haley Halberg for image analysis, and Ioana Pop for gene expression analysis. We thank Doug Mackay and Dollie LaJoie for insights, reagents, and discussions. This work was supported by the National Institutes of Health (R01-GM50877 to M.C.B. including a supplemental equipment grant and R01-GM131052 to K.S.U.) and the Huntsman Cancer Foundation. The Cancer Center Support Grant (National Cancer Institute P30-CA042014) awarded to the Huntsman Cancer Institute at the University of Utah provided pilot project funds (CRR-180205), developmental funds, and supported shared resources critical to this project.
Abbreviations used:
- FBS
fetal bovine serum
- FG
phenylalanine–glycine repeat
- LINC
Linker of the Nucleoskeleton and Cytoskeleton
- MRTF-A
myocardin-related transcription factor, also known as MKL1 and MAL1
- NE
Nuclear Envelope
- NPC
nuclear pore complex
- POM121
pore membrane protein of 121 kDa
- SF
stress fiber
- SFTI
SF Thickness Index
- SMIFH2
small molecule inhibitor of formin-homology domain
- SUN
Sad-1 and UNC
- TAN
transmembrane actin-associated nuclear
- TAZ
Transcriptional coactivator with PDZ domain
- TPR
translocated promoter region
- YAP
Yes-associated protein
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-01-0027) on January 22, 2020.
REFERENCES
- Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R, Kc B, Aggarwal V, Shrestha S, Jones AL, et al (2016). The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep , 38063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell , 1047–1059. [DOI] [PubMed] [Google Scholar]
- Bell ES, Lammerding J. (2016). Causes and consequences of nuclear envelope alterations in tumour progression. Eur J Cell Biol , 449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yishay R, Ashkenazy AJ, Shav-Tal Y. (2016). Dynamic encounters of genes and transcripts with the nuclear pore. Trends Genet , 419–431. [DOI] [PubMed] [Google Scholar]
- Blobel G. (1985). Gene gating: a hypothesis. Proc Natl Acad Sci USA , 8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Senderowicz AM, Sausville EA, Duncan KL, Korn ED. (1994). Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J Biol Chem , 14869–14871. [PubMed] [Google Scholar]
- Burns LT, Wente SR. (2014). From hypothesis to mechanism: uncovering nuclear pore complex links to gene expression. Mol Cell Biol , 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor TJ, Lee J, Thodeti CK, Lele T. (2010). Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J , 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Antoku S, Ostlund C, Worman HJ, Gundersen GG. (2015). Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus , 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. (2015). Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol , 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KH, Factor RE, Ullman KS. (2012). The nuclear envelope environment and its cancer connections. Nat Rev Cancer , 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al (2011). The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell , 759–772. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Hubner B, Strickfaden H, Smeets D, Popken J, Sterr M, Markaki Y, Rippe K, Cremer C. (2015). The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett (20 Pt A), 2931–2943. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol , 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA. (2018). Nuclear pore complexes as hubs for gene regulation. Nucleus , 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJ, Lammerding J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ Res , 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. (2014). “ Broken nuclei–lamins, nuclear mechanics, and disease.” Trends Cell Biol , 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Smith L, Cho S, Colasurdo M, Garcia AJ, Safran S. (2017). Matrix Mechanosensing: From Scaling Concepts in ‘Omics Data to Mechanisms in the Nucleus, Regeneration, and Cancer. Annu Rev Biophys , 295–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll TP, Cosgrove BD, Heo SJ, Shurden ZE, Mauck RL. (2015). Cytoskeletal to nuclear strain transfer regulates YAP signaling in mesenchymal stem cells. Biophys J , 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al (2011). Role of YAP/TAZ in mechanotransduction. Nature , 179–183. [DOI] [PubMed] [Google Scholar]
- Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, et al (2017). Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell , 1397–1410.e1314. [DOI] [PubMed] [Google Scholar]
- Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, Zaidi MR, Ksander BR, Merlino G, Sodhi A, et al (2014). Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell , 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Edmondson M, Sudol M. (2016). Framework to function: mechanosensitive regulators of gene transcription. Cell Mol Biol Lett , 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CT, Gualdrini F, Treisman R. (2017). Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev , 2361–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Holder MV, Aerne BL, Janody F, Tapon N. (2015). Zyxin antagonizes the FERM protein expanded to couple F-actin and Yorkie-dependent organ growth. Curr Biol , 679–689. [DOI] [PubMed] [Google Scholar]
- Gegenfurtner FA, Jahn B, Wagner H, Ziegenhain C, Enard W, Geistlinger L, Radler JO, Vollmar AM, Zahler S. (2018). Micropatterning as a tool to identify regulatory triggers and kinetics of actin-mediated endothelial mechanosensing. J Cell Sci , jcs212886. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. (2007). Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem , 593–627. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. (2013). Nuclear positioning. Cell , 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. (2012). Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol , 591–600. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. (2011). Hippo signaling: growth control and beyond. Development , 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. (2010). Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem , 3487–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. (2014). “ Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival.” J Cell Biol , 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF. (2015). Growth control: re-examining Zyxin’s role in the hippo pathway. Curr Biol , R230–R231. [DOI] [PubMed] [Google Scholar]
- Hatch EM. (2018). Nuclear envelope rupture: little holes, big openings. Curr Opin Cell Biol , 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morita T. (2013). Differences in the nuclear export mechanism between myocardin and myocardin-related transcription factor A. J Biol Chem , 5743–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SJ, Cosgrove BD, Dai EN, Mauck RL. (2018). Mechano-adaptation of the stem cell nucleus. Nucleus , 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L, Jensen CC, Yoshigi M, Beckerle M. (2017). Mechanical signals activate p38 MAPK pathway-dependent reinforcement of actin via mechanosensitive HspB1. Mol Biol Cell , 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC. (2012). Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell , 1846–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. (2006). Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol , 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain KR, Clarke RJ. (2019). General and specific interactions of the phospholipid bilayer with P-type ATPases. Biophys Rev , 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Hetzer MW. (2015). Nuclear pore proteins and the control of genome functions. Genes Dev , 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer KV, Pulford S, Mogilner A, Shivashankar GV. (2012). Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys J , 1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Kalita J, Lim RYH. (2019). The role of nucleocytoplasmic transport in mechanotransduction. Exp Cell Res , 86–93. [DOI] [PubMed] [Google Scholar]
- Kaunas R, Nguyen P, Usami S, Chien S. (2005). Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci USA , 15895–15900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. (2009). A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA , 19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL. (2007). Molecular biology of sperm head shaping. Soc Reprod Fertil Suppl , 33–43. [PubMed] [Google Scholar]
- Kim DH, Cho S, Wirtz D. (2014). Tight coupling between nucleus and cell migration through the perinuclear actin cap. J Cell Sci (Pt 11), 2528–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Louhghalam A, Lee G, Schafer BW, Wirtz D, Kim DH. (2017). Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat Commun , 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, Lammerding J. (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol , 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Maitra A, Sumit M, Ramaswamy S, Shivashankar GV. (2014). Actomyosin contractility rotates the cell nucleus. Sci Rep , 3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest , 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YL, Burke B. (2018). LINC complexes and nuclear positioning. Semin Cell Dev Biol , 67–76. [DOI] [PubMed] [Google Scholar]
- Li Q, Kumar A, Makhija E, Shivashankar GV. (2014). The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials , 961–969. [DOI] [PubMed] [Google Scholar]
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. (2007). Functional association of Sun1 with nuclear pore complexes. J Cell Biol , 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem , 26743–26753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science , 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. (1997). Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA , 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellad JA, Warren DT, Shanahan CM. (2011). Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol , 47–54. [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. (2016). Mechanisms of Hippo pathway regulation. Genes Dev , 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. (2003). Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell , 329–342. [DOI] [PubMed] [Google Scholar]
- Miroshnikova YA, Nava MM, Wickstrom SA. (2017). Emerging roles of mechanical forces in chromatin regulation. J Cell Sci , 2243–2250. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Hayashi K, Iwasaki K, Fujioka T, Egusa H, Yatani H, Sobue K. (2010). Nuclear import mechanism for myocardin family members and their correlation with vascular smooth muscle cell phenotype. J Biol Chem , 37314–37323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. (2007). Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA , 15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski R, Rajakyla EK, Vartiainen MK, Treisman R. (2010). An actin-regulated importin alpha/beta-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J , 3448–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, D’Angelo MA. (2017). Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol , 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Deguchi M, Cheng Y, Hsueh AJ. (2013). Actin cytoskeleton regulates Hippo signaling. PLoS One , e73763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. (2009). Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol , 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Wolf K, Lammerding J. (2017). Bursting the bubble—nuclear envelope rupture as a path to genomic instability? Trends Cell Biol , 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. (2018). Separate roles for chromatin and lamins in nuclear mechanics. Nucleus , 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. (2008). Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res , 1892–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Li R. (2016). Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol , 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science , 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwar S, Jain N, Shivashankar GV. (2014). The regulation of gene expression during onset of differentiation by nuclear mechanical heterogeneity. Biomaterials , 2411–2419. [DOI] [PubMed] [Google Scholar]
- Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al (2016). Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun , 10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CH, Collier JH, Sfeir CS, Healy KE. (2002). Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA , 1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasy SM, Morgan JT, Wood JA, Murphy CJ, Russell P. (2013). Substratum stiffness and latrunculin B modulate the gene expression of the mechanotransducers YAP and TAZ in human trabecular meshwork cells. Exp Eye Res , 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. (2005). LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol , 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler C, Shivashankar GV. (2017). Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat Rev Mol Cell Biol , 717–727. [DOI] [PubMed] [Google Scholar]
- Versaevel M, Braquenier JB, Riaz M, Grevesse T, Lantoine J, Gabriele S. (2014). Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci Rep , 7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development , 3907–3914. [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. (2009). Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol , 75–82. [DOI] [PubMed] [Google Scholar]
- Xue JZ, Woo EM, Postow L, Chait BT, Funabiki H. (2013). Chromatin-bound Xenopus Dppa2 shapes the nucleus by locally inhibiting microtubule assembly. Dev Cell , 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zhang XF, Pollard TD, Forscher P. (2012). Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol , 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshigi M, Clark EB, Yost HJ. (2003). Quantification of stretch-induced cytoskeletal remodeling in vascular endothelial cells by image processing. Cytometry A , 109–118. [DOI] [PubMed] [Google Scholar]
- Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. (2005). Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol , 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhang Y, Park HW, Jewell JL, Chen Q, Deng Y, Pan D, Taylor SS, Lai ZC, Guan KL. (2013). Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev , 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu OM, Miyamoto S, Brown JH. (2016). Myocardin-Related Transcription Factor A and Yes-Associated Protein Exert Dual Control in G Protein-Coupled Receptor- and RhoA-Mediated Transcriptional Regulation and Cell Proliferation. Mol Cell Biol , 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol , 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. (2009). YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol , 1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.