Abstract
The hippocampus is a plastic region and highly susceptible to ageing and dementia. Previous studies explicitly imposed a priori models of hippocampus when investigating ageing and dementia-specific atrophy but led to inconsistent results. Consequently, the basic question of whether macrostructural changes follow a cytoarchitectonic or functional organization across the adult lifespan and in age-related neurodegenerative disease remained open. The aim of this cross-sectional study was to identify the spatial pattern of hippocampus differentiation based on structural covariance with a data-driven approach across structural MRI data of large cohorts (n = 2594). We examined the pattern of structural covariance of hippocampus voxels in young, middle-aged, elderly, mild cognitive impairment and dementia disease samples by applying a clustering algorithm revealing differentiation in structural covariance within the hippocampus. In all the healthy and in the mild cognitive impaired participants, the hippocampus was robustly divided into anterior, lateral and medial subregions reminiscent of cytoarchitectonic division. In contrast, in dementia patients, the pattern of subdivision was closer to known functional differentiation into an anterior, body and tail subregions. These results not only contribute to a better understanding of co-plasticity and co-atrophy in the hippocampus across the lifespan and in dementia, but also provide robust data-driven spatial representations (i.e. maps) for structural studies.
Keywords: dementia, temporal lobe, structural covariance, parcellation, elderly
Plachti et al. report that in the healthy brain, hippocampal macrostructure can be divided into anterior, lateral and medial subregions reminiscent of the cytoarchitecture. In patients with dementia, however, patterns of hippocampal co-atrophy follow functional subdivisions between anterior, body and tail subregions.
Some of the data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Introduction
The hippocampus is a notable brain region from its lifelong plasticity potential (Moreno-Jiménez et al., 2019), which can be observed with microstructural and molecular investigations but also at the macrostructural level using morphological measurements of structural MRI. From macrostructural studies, the plasticity of the hippocampus seems to relate to experience and more particularly to cognitive training (Maguire et al., 2006; Boyke et al., 2008). Relatedly, morphological measurements of the hippocampus across individuals suggest an important inter-individual variability (Van Petten, 2004; Fleming Beattie et al., 2017; Llera et al., 2019).
As ageing and Alzheimer’s disease atrophy patterns resemble each other, in particular, showing important atrophy in temporal lobes (Fjell et al., 2014), several authors suggested that dementia simply represents a more severe or accelerated ageing process. It has been frequently pointed out that clinically normal individuals demonstrate an accumulation of amyloid-β and tau pathologies in the hippocampus and entorhinal cortex suggesting that neurobiological features associated with Alzheimer’s disease can also be found in apparently healthy elderly populations (Sperling et al., 2019; Ziontz et al., 2019). Thus, the neurobiological relationship between healthy ageing and dementia and in particular the hypothesis of dementia as a form of increased ageing process remains controversial and poorly understood.
Most research studies have focused on hippocampal atrophy assessed at the macrostructural level and as representing the most straightforward non-invasive estimates of age-related structural changes. In other words, investigations have aimed to identify a specific pattern of atrophy across hippocampus’s organization. Two different models of hippocampus organization were referred to: the subfield model (based on cytoarchitecture features), and the tripartite model, differentiating regions along the longitudinal axis such as the head-body and tail (based on functional and large-scale connectivity features). Since subfields and subregions are suggested to be characterized by different neurobiological features, they are likely to be differently affected by ageing and pathological processes. Despite several studies having investigated this question, no convergence towards individual subfields and subregions as being specifically affected by atrophy has emerged hindering our understanding of the underlying mechanisms.
In sum, our fundamental understanding of structural changes in the human hippocampus across the adult lifespan and in dementia remain fairly limited, but several issues should be pointed out to account for the current state of art. First, as described above, most studies were based on an a priori model of hippocampus organization while it is unclear which model is the most appropriate. On the one hand, one could expect macrostructural changes to be constrained by the topology defined by cytoarchitecture, but on the other hand, as plasticity has been related to behavioural function, one could expect macrostructural changes to follow the functional organization of the human hippocampus along the longitudinal axis. Second, partly related to the first conundrum, the question of whether the pattern of structural changes in ageing and dementia follow a similar topological pattern remains as an open question.
In this study, we probed morphological changes across large datasets of structural MRI in healthy subjects and patients with dementia applying a data-driven approach to reveal latent patterns of differentiation in the hippocampus. Using the pattern of covariance with other brain regions across individuals to guide the clustering, importantly, allows the integration of interrelationships between the hippocampus and the whole brain hence revealing a more systemic pattern of change.
To implement the aforementioned objectives practically, we used a parcellation approach applied on hippocampus structural covariance in five different age and disease groups: young, middle-aged, elderly adults, mild cognitive impairment patients (MCI) and patients with dementia coming from independent datasets. We use the term ‘covariance’ to refer to healthy lifespan changes in structural covariation, which are assumed to be driven mainly by co-plasticity (e.g. regions developing together) and partly by co-atrophy, especially in older adults (e.g. regions degenerating together). In contrast, in dementia, we expect covariation to be primarily driven by co-degeneration of brain regions. Accordingly, we use the term ‘co-atrophy’ in the context of dementia patients (even though technically, the same ‘structural covariance’ measure was applied across age and disease groups).
In this framework, a data-driven approach of structural covariance offers a bottom-up examination of the topological patterns of co-plasticity/covariation in the first adult life periods and co-atrophy in the elderly and in dementia. Importantly, we examined the stability of the patterns across datasets by using split-half cross-validation and robustness across groups with bootstrapping approaches. We explored the possible mechanisms explaining these patterns by examining the similarity of these topological patterns with the pattern of functional organization of the hippocampus, and investigated the structural networks that underlie the different hippocampus subregions. Finally, we characterized these structural networks with regards to behavioural functions and compared these structural networks with functional networks.
Materials and methods
Datasets, cohort samples and age-phenotypical groups
We included six different datasets: Human Connectome Project (HCP) (http://www.humanconnectome.org), Enhanced Nathan Kline Institute-Rockland Sample (eNKI) (http://fcon_1000.projects.nitrc.org/indi/enhanced/), Cambridge Centre for Ageing and Neuroscience (CamCAN) (https://www.cam-can.org/) (Shafto et al., 2014; Taylor et al., 2017), 1000BRAINS from Forschungszentrum Juelich (Caspers et al., 2014), Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu/) and Open Access Series of Imaging Studies (OASIS3) (https://www.oasis-brains.org/). From these datasets, we formed five cohort samples: young, middle-aged, elderly, MCI and dementia participants. The age range of the group of young adults was set to 20–35 years. In turn, the age range of the middle-aged group was 35–55 years and for the elderly, we set a conservative age range of 60–80 years. MCI and Alzheimer’s disease patients were selected within the same age range as the elderly group. For the dementia group we included patients with probable Alzheimer’s type pathology by selecting Alzheimer’s disease patients from the OASIS3 dataset and ADNI dataset, as well as the late cognitive impaired individuals from the ADNI dataset who are considered as patients at the early stage of Alzheimer’s disease (Qiu et al., 2014). The MCI group was formed by the participants with the diagnosis ‘early MCI’ (ADNI dataset) and by participants with a Clinical Dementia Rating (CDR) score of 0.5 from the OASIS3 dataset. The demographic data of each study samples and groups are reported in Tables 1 and 2. The analyses of these data were approved by the ethical committee of the Heinrich Heine University Düsseldorf.
Table 1.
Demographic data of all collected samples
| Samples | Sample size, n | Mean age (SD; range) | % female | Education (SD; range) | CDR (SD; range) | MMSE (SD; range) |
|---|---|---|---|---|---|---|
| Young_HCP | 304 | 27.8 (3.55; 22–34) | 50.6 | SSAGA_Educ: 14.8 (1.75; 11–17); NAs: 0 | – | 29.0 (SD =1.07; 23-30); NAs: 0 |
| Young_eNKI | 140 | 24.8 (3.85; 20–34) | 50 | SES–Adult Education code, 5.4 0.8; 4–7; 14.8 (1.6; 11–18); NAs: 0 | – | – |
| MiddleAged_eNKI | 72 | 43.6 (5.7; 35–54) | 52.7 | SES–Adult Education code, 5.5 (1; 3–7); 15.1 (2.3; 10–21); missing n = 2 | – | – |
| Old_eNKI | 76 | 68.3 (5.5; 60–79) | 51.3 | SES-Adult Education code, 6.0 (1.0 4–7); 16.1 (2.4; 12–24); NAs: 0 | – | – |
| Young_ CamCAN | 94 | 28.4 (3.97; 20–34) | 50 | Education scoring: 6.2 (1.7; 2–8); missing n = 21 | – | 29.4 (1.17, 25–30); NAs: 0 |
| MiddleAged_CamCAN | 207 | 44.3 (5.78; 35–54) | 50.7 | 5.7 (1.8, 1–8); missing n = 35 | – | 29.1 (1.17, 26–30); NAs: 0 |
| Old_CamCAN | 213 | 69.8 (5.96; 60–79) | 50.2 | 5.0 (2.2, 1–8); missing n = 65 | – | 28.4 (1.47, 25–30); missing n = 1 |
| Old_1000BRAINS | 492 | 66.9 (4.24; 60–75) | 50 | Education years: 13.7 (3.7, 3–27); missing n = 1 | – | Demtec: 15.2 (2.3, 8–18); missing n = 9 |
| Old_ADNI | 139 | 71.6 (4.65; 61–79) | 51.7 | Education years: 16.6 (2.6, 12–20); NAs: 0 | CDR sum of boxes: 0.02 (0.11, 0–0.5) NAs: 0 | 28.9 (1.24, 24–30); NAs: 0 |
| MCI_ADNI | 213 | 69.2 (5.05; 60–79) | 50.2 | Education years: 15.9 (2.6, 10–20); NAs: 0 | CDR sum of boxes: 1.22 (0.76, 0.5–4) NAs: 0 | 28.4 (1.5, 23–30); NAs: 0 |
| AD_ADNI | 219 | 71.0 (5.42; 60–79) | 51.1 | Education years: 16.1 (2.6, 11–20); NAs: 0 | CDR sum of boxes: 2.9 (1.8, 0.5–10) NAs: 0 | 25.8 (3.0, 19–30); NAs: 0 |
| Old_OASIS3 | 298 | 70.3 (4.42; 60–79) | 50 | Education years: 16.0 (2.7, 8–24); NAs: 0 | 0 (0, 0–0) NAs: 0 | 28.8 (1.8, 9–30); missing n =2 |
| MCI_OASIS3 | 74 | 70.9 (4.58; 61–79) | 50 | Education years: 15.3 (2.7, 8–20); NAs: 0 | 0.5 (0, 0.5–0.5) NAs: 0 | 26.7 (3.4, 13–30); missing n = 2 |
| AD_OASIS3 | 53 | 69.9 (5.58; 60–79) | 47.2 | Education years: 15.1 (2.8, 11–20); NAs: 0 | 0.92 (0.57, 0–2) NAs: 0 | 23.5 (4.8, 10–30); missing n = 1 |
CDR = Clinical Dementia Rating; MMSE = Mini-Mental State Examination; NAs = not available, n; SD = standard deviation.
Table 2.
Demographic data of the age and disease groups created from independent samples
| Phenotypical group | Size, n | Mean age (SD; age range) | % females |
|---|---|---|---|
| Young | 538 | 27.1 (3.95; 20–34) | 50.5 |
| Middle aged | 279 | 44.0 (5.77; 35–54) | 51.0 |
| Elderly | 1218 | 68.9 (5.07; 60–79) | 50.2 |
| MCI | 287 | 69.7 (4.98; 60–79) | 50.2 |
| Dementia | 272 | 70.7 (5.46; 60–79) | 50.4 |
Structural MRI acquisition, preprocessing and structural covariance computation
Only 3 T MRI anatomical scans were included in this study, acquired with different scanning parameters (Table 3). All images were preprocessed with SPM12 and the CAT12 toolbox, running on MATLAB R2016a. The normalization was performed with the DARTEL algorithm to the ICBM-152 template using both affine and non-linear spatial normalization. The MRI images were bias-field corrected and segmented into grey, white matter, and CSF tissues. The grey matter segments were then modulated for non-linear transformations only and subsequently smoothed with an isotropic Gaussian kernel (full-width at half-maximum = 8).
Table 3.
Sequence parameters of the different datasets
| Datasets | Sequence parameters |
|---|---|
| HCP | T1 (3D-MPRAGE), Siemens Skyra, 256 slices, TR = 2400 ms, TE = 2.14 ms, TI = 1000 ms, FoV = 224 x 224 mm2, flip angle = 8°, voxel size = 0.7 × 0.7 × 0.7 mm3 |
| eNKI | Cross Sectional Lifespan Connectomics and Longitudinal Developmental Connectomics study: T1 (3D-MPRAGE), Tim Trio, 176, TR = 1900 ms, TE = 2.52 ms, TI = 900 ms, FoV = 250 × 250 mm2, flip angle = 9°, voxel size = 1 × 1 × 1 mm3; Neurofeedback study: T1 (3D-MPRAGE), Tim Trio, 192 slices, TR = 2600 ms, TE = 3.02 ms, TI = 900 ms, flip angle = 8°, voxel size = 1 × 1 × 1 mm3 |
| CamCAN | T1 (3D-MPRAGE), Tim Trio, 192, TR = 2250 ms, TE = 2.98 ms, TI = 900 ms, FoV = 256 × 256 mm2, flip angle = 9°, voxel size = 1 × 1 × 1 mm3 |
| 1000BRAINS | T1 (3D-MPRAGE), Tim-TRIO, 176 slices, TR = 2.25 s, TE = 3.03 ms, TI = 900 ms, FoV = 256 × 256 mm2, flip angle = 9°, voxel resolution = 1 × 1 × 1 mm3 |
| ADNI | ADNI1: T1 (3D-MPRAGE), TR = 0.65 s, TE = min full, FoV = 256 × 256 mm2, flip angle = 8°, voxel resolution = 1.2 mm3; ADNIGO/2: T1 (3D-MPRAGE), TR = 0.4 s, TE = min full, FoV = 256 × 256 mm2, flip angle = 11°, voxel size = 1.2 mm3; ADNI3: T1 (3D-MPRAGE), TR = 2300 ms, TE = min full echo, TI = 900 ms, FoV = 256 mm, resolution = 1 × 1 × 1 mm3 |
| OASIS3 | T1 (3D-MPRAGE), Tim Trio, TR = 2400 ms, TE = 3.08 ms, TI = 1, FoV = 256 × 256 mm2, flip angle = 8°, voxel size = 1 × 1 × 1 mm3 |
FoV = field of view; TE = echo time; TI = inversion time; TR = repetition time.
We used a mask of the human hippocampus created in a previous study (Plachti et al., 2019) from macro-anatomical atlas and cytoarchitecture maps. Structural covariance was computed by correlating hippocampal voxels with all other grey matter voxels using Pearson's correlation, which were z-transformed. For each dataset, hundreds of bootstrap samples (corresponding to the size of the dataset) were created and a respective structural covariance matrix was computed for each bootstrap sample (Supplementary material).
Parcellation: clustering of hippocampus voxels based on structural covariance
Clustering
To identify patterns of similar and different structural covariance among hippocampus voxels, we used an unsupervised clustering approach extensively applied in the field of brain parcellation. More precisely, for each voxel within the hippocampus, an individual structural covariance profile to all other brain voxels across subjects was computed. In the next step, hippocampus voxels were clustered based on the similarity/dissimilarity of their profiles. As a clustering algorithm we applied the k-means++ algorithm in MATLAB identifying two to seven parcels. We used 255 iteration and 500 repetition parameters in line with Plachti et al. (2019) to allow comparison with previous parcellations.
Split-half cross-validation as stability measure
To identify which cluster solution best summarized similarity and dissimilarity in the pattern of structural covariance of hippocampus voxels, we used split-half cross-validation to estimate the stability of differentiations. We divided each sample into halves 10 000 times (splits) and compared with the adjusted Rand Index the convergence between the two halves. The adjusted Rand Index estimates the consistency of two clusterings and is adjusted for chance. It can have values between 0 (not similar at all) and 1 (identical). A higher convergence reflects a higher consistency of the clusterings indicating high stability. In order to quantify statistically the stability of the different cluster solutions, we performed an ANOVA.
Cross-dataset group parcellation
After clustering, we merged the parcellation results from different datasets corresponding to the same age and disease group, in order to obtain robust patterns of structural covariance parcellation in each age/disease group. This procedure aimed to extract patterns that captured the relevant features under investigation (e.g. ageing or dementia effects) rather than dataset-specific effects (Jockwitz et al., 2019). First, the clustering approach was applied on structural covariance profiles of hippocampus voxels within each sample and age group, resulting in sample-group-specific matrices. We then concatenated the solution matrix of one sample (e.g. Young_HCP) with all the other samples (e.g. Young_eNKI, Young_CamCAN) belonging to the same age or disease group (e.g. Young) and applied bootstrapping (10 000 resampling) on the ‘merged’ solution matrix across bootstrap samples (Supplementary Fig. 1).
Covariance network of clusters and their relationship to functional large-scale networks
To identify the pattern of structural covariance underlying the clustering in each age/disease group (n = 2584), we examined the network of structural covariance more specifically associated to each cluster. To do so, we used the general linear model as implemented in SPM, hence at the voxel level. Accordingly, at each voxel, the linear relationship with the average grey matter value of the cluster of interest is tested. This procedure provided some insight into the individual pattern of structural covariance of the different subregions of the hippocampus that have driven the clustering. As the clustering is not performed on any thresholded values but based on the full pattern of structural covariance, we here examined the map of structural covariance of each cluster across the whole brain at an uncorrected level of P < 0.001 with a threshold of T = 1. Nevertheless, we additionally corrected for multiple comparisons using family wise error (FWE) rate at the significance level of P < 0.05 to examine the brain patterns that survived at a strict statistical threshold (Supplementary Fig. 7).
To test whether structural covariance networks in dementia follow functional coactivation networks, we examined the functional connectivity of the subregions derived in dementia but in a sample of healthy participants. Our underlying hypothesis was that the pattern of co-atrophy in dementia could mirror functional connectivity patterns observed in late life (but before dementia). To explore this question, we performed a similar general linear model analysis using resting-state functional MRI time-series in the group of healthy elderly (n = 428 in 1000BRAINS; EPI, 36 slices, repetition time = 2.2 s, echo time = 30 ms, field of view = 200 × 200 mm2, flip angel = 90°, voxel resolution = 3.1 × 3.1 × 3.1 mm3) for the hippocampus subregions derived from the dementia group. Preprocessing included movement correction by affine two-pass registration and alignment of the images to the first volume and to the mean of the volumes. The six motion parameters and their first derivatives from the realignment step were regressed out. Spatial normalization was performed to the MNI-152 template for the average EPI scans for each subject using the unified segmentation approach. Images were band-pass filtered with cut-off values of 0.01–0.08 Hz and smoothed with the isotropic Gaussian kernel (full-width at half-maximum = 5 mm). Denoising was performed using white matter and CSF signal regression.
For each grey matter voxel, a linear relationship with the average blood oxygen level-dependent (BOLD) response of the cluster of interest was computed. In this way, we obtained the functional connectivity network of each individual cluster and contrasted it against the whole brain pattern of association of other clusters.
Covariance network of clusters and their behavioural associations
After having identified the structural covariance network for each cluster, we characterized those networks in terms of associated behavioural functions using the NeuroSynth database (https://neurosynth.org/) and its cognitive decoding tool with >1300 terms included. For the most frequent terms reported in the literature (such as ‘episodic memory’), NeuroSynth provides meta-analytic maps of the most frequently associated voxels in activation studies. It therefore offers the possibility to compare any given brain pattern, such as the whole brain structural covariation patterns in the present study, to the collection of maps related to each term using the cognitive decoding tool. Accordingly, we used the uncorrected whole-brain maps of each cluster and ran Pearson correlations between our structural covariance maps and the meta-analytic maps of NeuroSynth. As our objective here was not to identify specific behavioural functions associated to a specific network but rather to identify the broad pattern of behavioural associations of cluster’s network, we included all correlations for associated terms >0.1, we excluded non-behavioural terms (e.g. hippocampus, dementia) and summarized similar lexical terms into a summary label (i.e. ‘emotions’, ‘affect’, ‘happy’, ‘fear’ → emotion). The pattern of associated behavioural terms, which could differ in number depending on the spatial extent of the cluster’s covariance pattern, was then interpreted qualitatively rather than with regards to magnitude of association.
Data availability
The data that support the findings of this study are available from open science initiatives reported and cited above. Code can be shared upon reasonable request from the corresponding author. The derived clusters are available at (http://anima.fz-juelich.de/) as ROI in .nii format.
Results
Stable clustering level
We used split-half cross-validation (10 000 splits) to identify the most stable cluster solution based on similarity across splits as measured by the adjusted Rand Index. We performed a 6 (datasets: HCP, eNKI, CamCAN, 1000BRAINS, ADNI, OASIS3) × 6 (cluster solution: 2–7) ANOVA with the adjusted Rand Index as dependent variable. The ANOVAs were performed separately for each hemisphere.
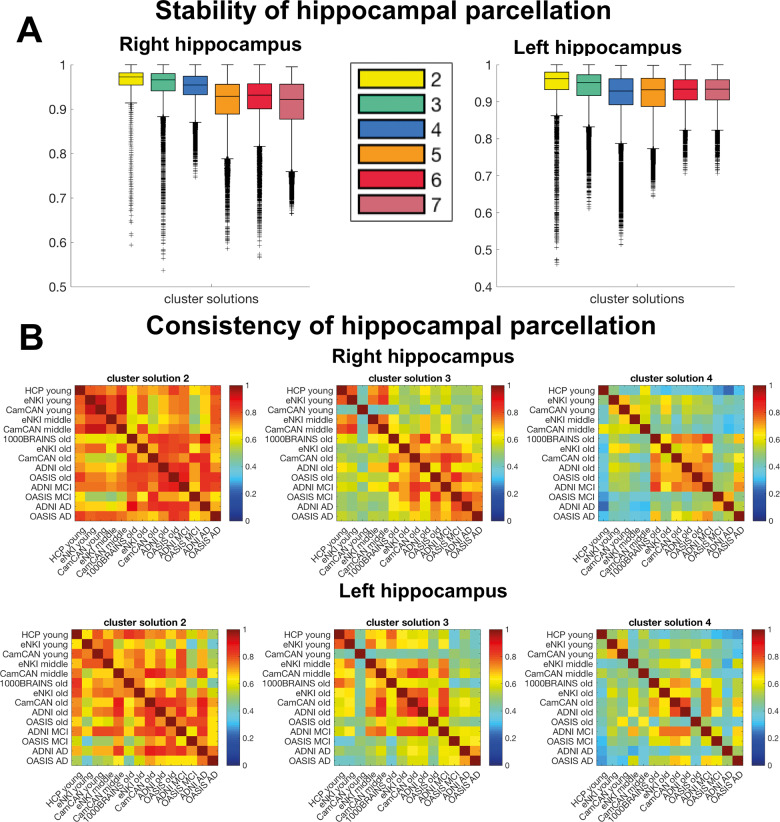
Overall, examining cluster solutions main effect [F(5,839964) = 32365.18, P < 0.001], in the right hippocampus, parcellations into two and three clusters were the most stable solutions even though the differences between all cluster solutions were marginal: two (mean = 0.97), three (mean = 0.96), four (mean = 0.95) (Fig. 1A). For the left hippocampus, cluster solutions two and three were also the most stable: two (mean = 0.97), three (mean = 0.96), four (mean = 0.94) [F(5,839964) = 25194.75, P < 0.001] (Fig. 1A). The significant interaction effects in right and left hippocampi indicated that the stability of parcellations was dependent on the dataset, F(25,839964) = 2006.7, P < 0.001, F(25,839964) = 4884.36, P < 0.001 (Supplementary Fig. 2).
Figure 1.
Stability and consistency of hippocampal parcellations. (A) Stable organizational patterns were found for right and left hippocampus for cluster solutions 2–4 estimated with split-half cross-validation. All clusterings reached very high stability >0.9 adjusted Rand Index. (B) Cross-sample consistency of lower cluster solutions measured with the adjusted Rand Index. Despite overall high stability, the simple parcellation schemes 2–4 were also very consistent >0.6 across datasets and within age/disease-specific groups (e.g. young, elderly) suggesting biological relevance in those differentiations. Cluster solution 3 was exceptionally useful to study age and disease-related patterns, because this scheme demonstrated not only high within age/disease similarity but to some extent also across age/disease groups indicating relatedness, which did not apply for cluster solution 4. In contrast cluster solution 2 showed very high similarity independent of age/disease and dataset suggesting on the one hand a robust biological differentiation, but on the other hand a less flexible scheme to represent lifespan and pathological alterations. Box plots with median, 1.5 interquartile range (IQR), min. Q1–1.5×IQR, max. Q3 + 1.5 × IQR.
In line with previous clustering studies, our first exploration showed a relatively linear decrease in the stability as the number of cluster increases, suggesting that the simpler, more parsimonious models are the most robust ones (additionally supported by silhouette plots in the Supplementary Fig. 3). In particular here, the 2- and 3-cluster solutions were the most stable levels of differentiation. Fig. 3
Similarity/consistency of hippocampal differentiation
To ensure that the stability of cluster solutions 2–4 was driven by intrinsic properties of the structural covariance pattern rather than by intrinsic properties of the dataset, we examined the pattern of similarity (measured by the adjusted Rand Index) between the different cohort samples (Fig. 1B).
The inspection of the similarity matrices revealed that cluster solution 2 showed a general pattern of high similarity, whatever the dataset or age group. This suggested a global differentiation being robust across data and age/disease group (Fig. 1B). The three-cluster solution mainly and remarkably showed a high within-group (age and disease) and between-group consistency suggesting a differentiation pattern driven by intrinsic features of the age/disease groups rather than by the intrinsic features of the dataset. This suggests that neurobiological rather than technical factors specific to the dataset guided the parcellation.
In contrast, the four-cluster solution showed high within-age group consistency only for the healthy elderly group in the right hippocampus, questioning its usability to study lifespan and disease-related changes. Finally, the higher clustering levels (five, six and seven-cluster solutions) showed relatively low similarity between samples (Supplementary Fig 2). Thus, the investigations of consistency/similarity between samples supported the focus on the three-cluster solution as the most stable and most likely biologically relevant pattern of differentiation of hippocampus voxels.
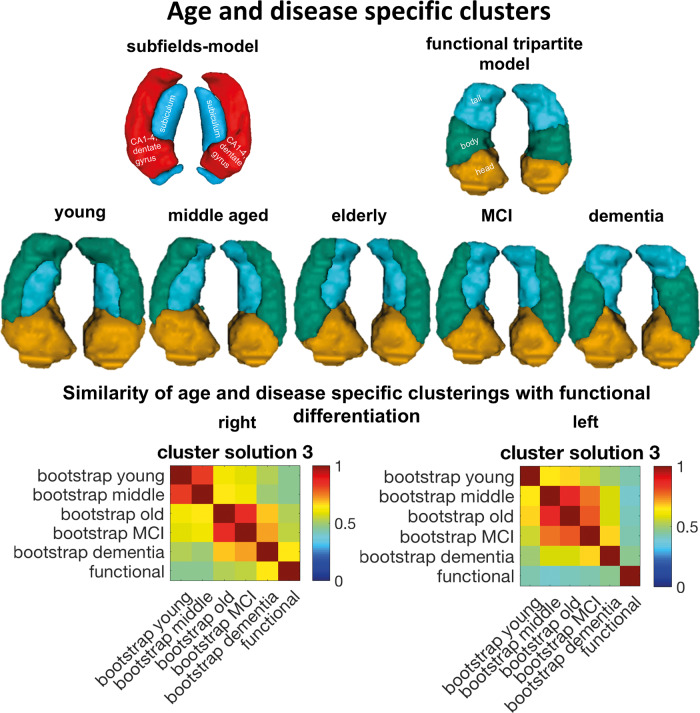
Figure 2.
Age and disease-specific clusterings of the hippocampus and its similarity to functional differentiation into head, body and tail parcellation. In younger age the hippocampal differentiation was reminiscent of the differentiation between subiculum versus cornu ammonis (CA) 1–4 and dentate gyrus subfields. With increasing age, the lateral subregion decreased from the tail, whereas the differentiation in dementia was reminiscent of the functional differentiation into head, body and tail also suggested by the similarity estimation. Clusterings were compared using adjusted Rand Index.
In summary, our first ‘bottom-up’ examination of the differentiation of the hippocampus based on structural covariance across different datasets suggested that a three-cluster solution could represent the data in a stable manner. Furthermore, our examination of consistency within age and disease group suggested that this high stability is not primarily driven by characteristics that were intrinsic to the dataset but rather by characteristics that were intrinsic to the population group and hence driven by neurobiological factors. Thus, altogether, hippocampus voxels within different age/disease groups could be optimally summarized with a three-cluster solution ideally applicable to study lifespan and disease alterations. Importantly, such a parsimonious three-partition model also meets previous theories on hippocampus organization.
Even though cluster solutions 2 and 4 displayed high stability and consistency compared to higher differentiations, they were either less informative, as in the case of cluster solution 2 (Supplementary Fig. 5), or demonstrated qualitatively divergent parcellations less comparable across age/disease group, as in the case of cluster solution 4 (Supplementary Fig. 5). Building on these explorations of the data and previous knowledge, we then focused on the three-cluster solution pattern.
Cross-dataset age and disease group parcellation
After deriving parcellations in each cohort sample, we merged them to obtain a robust pattern of differentiation of hippocampus voxels for five different age and disease groups: young, middle-aged, elderly, MCI and dementia patients using a bootstrapping approach to promote stability. This aggregation was done separately for the left and right hippocampi. Nevertheless, a very symmetrical pattern of differentiation could be observed across hemispheres. For both hippocampi, our maps (Fig. 2) showed a very similar pattern for the young, middle-aged, elderly and the MCI group. This pattern highlighted a division in the medial-lateral dimension of the hippocampus body and to some extent, of the tail while the head appeared as a relatively homogeneous region. This pattern replicated the findings from our previous parcellation work in the hippocampus performed in a sample of young participants from the HCP dataset (Plachti et al., 2019), and as already highlighted in our previous study, is reminiscent of the medial-lateral differentiation between cornu ammonis and subiculum subfields known from cytoarchitecture. Of note, it seemed that with increasing age the head cluster decreased slightly in size, while the medial (blue) cluster expanded into the tail area and the lateral (green) cluster expanded into the anterior direction (Fig. 2).
Remarkably, the differentiation of the hippocampus in the dementia group deviated from the pattern that was observed in the healthy population across adult age. Despite the anterior subregion also appearing as a relatively homogeneous region, the lateral (green) cluster was focused on the hippocampus body while the medial (blue) cluster appeared more prominent in the tail. As illustrated in Fig. 2, this pattern was reminiscent of the functional differentiation along the anterior-posterior dimension (and hence ‘head-body-tail’ tripartite model) observed in parcellations using large-scale functional connectivity. To further quantitatively evaluate these apparent divergences and resemblances, we compared the similarity of the age and disease groups among each other and with the functional map of the hippocampus derived in healthy adult functional MRI data (Plachti et al., 2019) using the adjusted Rand Index.
Strikingly, the highest similarity with the hippocampus functional map was found for the parcellation pattern obtained in dementia. This finding suggested that over time, the structural changes in the hippocampus in the pathological condition of dementia followed the large-scale functional organization of the hippocampus. Interestingly, this tendency was higher for the right than for the left hippocampus. Finally, it is worth noting that the pattern in participants with MCI was more similar to the healthy middle-aged and elderly participants than to the pattern observed in dementia.
Whole brain structural covariance patterns of each cluster
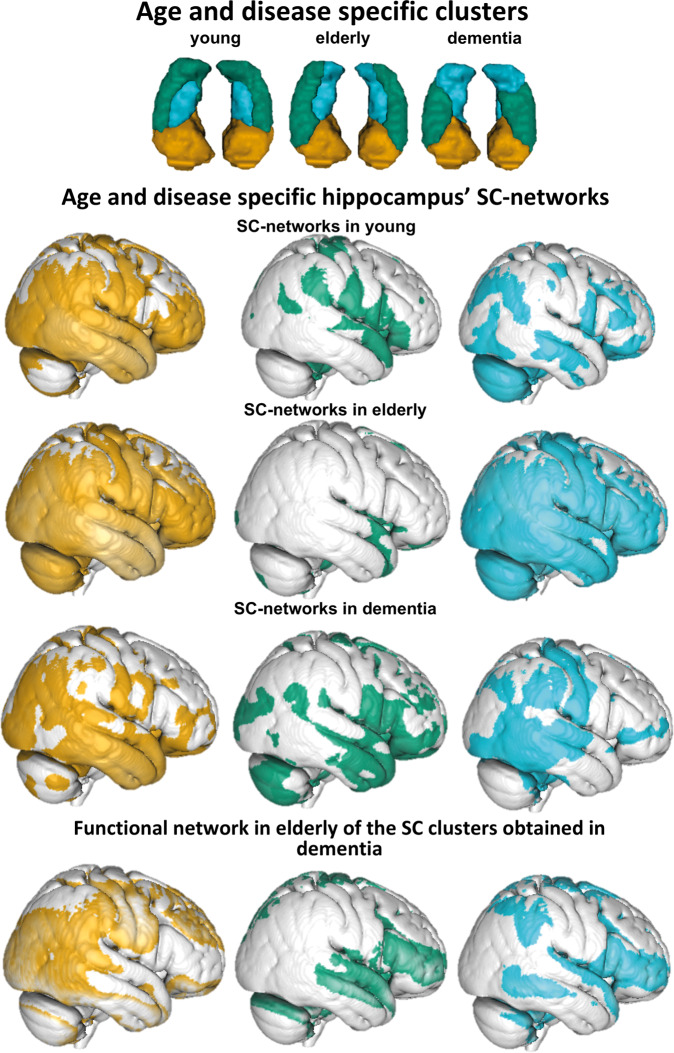
To better understand the structural covariance patterns that drove the differentiation among hippocampus voxels in each age/disease group, we examined the specific structural covariance pattern of each cluster and this, separately in each age/disease group. The structural covariance networks for young, elderly adults and dementia patients are presented below while the results obtained in middle aged and MCI participants (that were in line with other non-demented groups) are presented in Supplementary Fig. 6.
In young participants, the anterior cluster was associated with wide frontotemporal and parietal networks including frontal medial cortex, superior frontal gyrus, orbitofrontal cortex, cingulate cortex, temporal lobe, parahippocampal gyrus, (pre-)cuneal cortex, calcarine cortex, lingual gyrus and occipital pole. In addition, the putamen, pallidum, amygdala, insular cortex belonged to this network. A similar pattern was found in healthy elderly participants despite a slight expansion, additionally covering the inferior frontal gyrus, the whole cerebellum, pre- and postcentral gyri (Fig. 3).
Figure 3.
Patterns of structural covariance of each hippocampus subregion in young, elderly and dementia groups. Relative resting state functional connectivity networks of dementia hippocampus in healthy elderly resembled structural covariation (SC) networks of dementia hippocampus in dementia group. Uncorrected (P < 0.001), thresholded T = 1.
The lateral (green) cluster in the young group was mainly associated with subcortical structures such as putamen, pallidum, nucleus caudatus, thalamus but also with the cingulate gyrus, lingual gyrus, precuneous cortex and intracalcarine/supracalcarine cortex. Additionally, frontal and temporal brain regions were included such as frontal orbital cortex, frontal operculum cortex, inferior frontal gyrus, pars opercularis and superior temporal gyrus. In the older group, this network mainly reduced to the parieto-occipital (posterior cingulate cortex, precuneous, lingual and intracalcarine gyrus) and frontal medial (frontal medial cortex, subcallocal cortex, frontal pole) brain regions reminiscent of the default mode network.
The blue medial cluster in the group of young adults was mostly related to middle frontal, middle temporal gyri, cerebellum and lateral occipital cortex. Subcortical regions such as the caudate and thalamus, but also the insula, were included. Interestingly, the (blue) medial cluster showed in the group of healthy elderly a very broad pattern of structural covariation (Fig. 3), especially in the posterior brain regions (e.g. parietal, occipital lobes and motor related regions: cerebellum, pre-postcentral gyrus, thalamus, putamen, but also occipital gyrus, superior parietal lobule, and temporal gyri). Some smaller associated regions were also found in the inferior frontal and middle frontal cortex.
In contrast, in the group of patients with dementia, the pattern of structural covariance of each cluster was less spatially extended compared to all the other groups (Fig. 3). Furthermore, the pattern was also qualitatively different when compared to the patterns of the three clusters in the other age/disease groups confirming that the differentiation into subregions within the hippocampus itself is qualitatively different and did not follow the known pattern of healthy ageing. Hence, the (green) lateral-body cluster was not associated with posterior subcortical structures as the lateral (green) cluster in other groups but rather was more specifically associated with structures in the frontal (inferior frontal gyrus pars opercularis, frontal pole, opercular gyrus), temporal (middle temporal gyrus, Heschl’s gyrus) and occipital brain regions (Fig. 3). In contrast, the (blue) tail cluster was more associated with posterior brain regions [posterior parts of the temporal lobe, postcentral gyrus and (pre)cuneous, angular gyrus] while the anterior cluster was more associated with temporal, temporo-occipital fusiform cortex, and parietal regions losing mainly its covariation with frontal regions compared to younger healthy adults.
Because of apparent similarity between structural differentiation of the hippocampus in the dementia group with the functional organization model of the hippocampus known from previous studies in the healthy population, we further explored the relationship between functional and structural networks. More concretely, we investigated the pattern of resting state functional connectivity in the later life period of healthy participants (i.e. in healthy older adults) of the hippocampus cluster derived in dementia patients. This exploratory analysis suggested that the functional networks of the anterior and the lateral clusters that can be observed in an ageing population are very similar to their structural networks observed in patients with dementia hence further supporting the hypothesis of an influence of large-scale functional interaction in the co-atrophy pattern in dementia.
Behavioural characterization of structural covariance networks of clusters
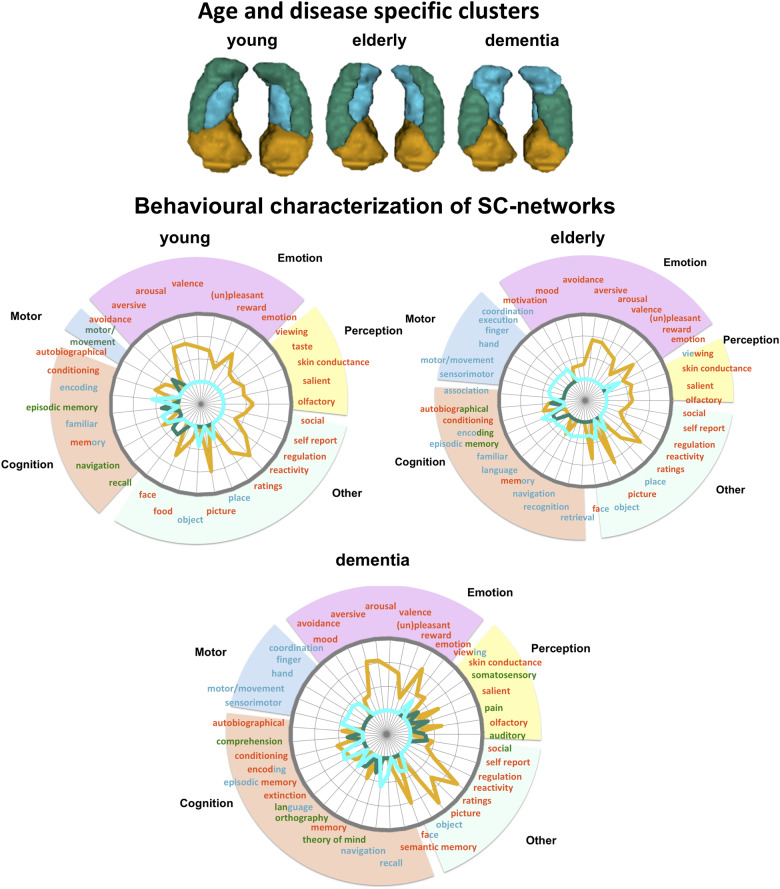
To explore whether the structural covariance patterns of each cluster could reflect functional networks subserving specific behavioural functions, we characterized the spatial pattern of each cluster’s covariance network with regards to behavioural terms with NeuroSynth. Results of middle-aged and MCI patients are presented in Supplementary Fig. 10, while we here focused on the associations in the young, elderly and the dementia group, as showing a slightly different pattern.
Overall, the spatial pattern of the anterior cluster was primarily associated with emotional, perceptual (olfactory, viewing) and self-related (autobiographical) terms, but also with other less ontologically defined terms such as faces, ratings and reactivity (Fig. 4). Overall, this behavioural pattern pointed to an automatic and more perceptual-emotional processing and integration of information into self-related internal states. This behavioural profile of the anterior subregion was even preserved in dementia pathology. In contrast, the pattern of the lateral (green) and the medial (blue) clusters diverged depending on the age and disease group. Whereas the medial blue cluster networks in the group of healthy young adults were associated with visual processing of objects and places, in the group of elderly and dementia patients, however, it was behaviourally additionally associated with motor/movement and orientation (Fig. 4).
Figure 4.
Behavioural characterization of cluster covariance networks in age and disease groups using NeuroSynth. Behavioural profiles of anterior clusters covariance network remained relatively stable across the lifespan and in disease playing a major role in automatic perceptual-emotional approach-behaviour in learning, establishing self-related memories. Across the lifespan the medial (blue) subregion’s network changed from being associated with visual processing in younger years to being also motor-related in older age. The lateral body (green) subregion in the group of dementia was behaviourally associated with language and theory of mind processing while the lateral subregion did not show a clear behavioural specificity in the second half of lifespan compared to the anterior subregion.
Most changes in structural covariation and behaviour were observed for the lateral (green) cluster. In the group of young healthy adults, the network was associated with motor-related behaviour (e.g. motor, navigation), whereas in the elderly, the behavioural association suggested an involvement of storing self-related information (e.g. autobiographic memory, episodic memory). In the group of dementia patients, on the other hand, the network was primarily associated with communication and social cognition, both of its own internal states (e.g. pain) as well as external information (e.g. comprehension, theory of mind). Overall, these results suggested that the changes in the patterns of structural covariation of the medial and lateral clusters over the lifespan and in pathology could be related to associations with different behavioural functions.
Discussion
The hippocampus is susceptible to senescence and neurodegenerative processes but the patterns of structural changes at the macroscale revealed inconsistencies across studies. Observed changes in grey matter volume could be either constrained by micro-anatomical organization of the cytoarchitecture or follow an organization determined by lifelong functional large-scale networks.
In a previous study, we used a parcellation approach to study human hippocampus organization with a multimodal parcellation approach. We hence examined the pattern of structural covariance in the human hippocampus in healthy young adults and found a topology that mimics both medial-lateral differentiation from cytoarchitecture and anterior-posterior differentiation shown by functional connectivity profiles (Plachti et al., 2019). A similar pattern was found in a recent study using a similar population but different parcellation approaches (Ge et al., 2019), and was reproduced again in this study, hence suggesting that this pattern reflects a robust pattern of co-plasticity in young adults.
Here we investigated if structural changes represented in covariations in older age and dementia follow or deviate from the patterns of co-plasticity observed in young adults. Our results indicated that during ageing, the overall pattern of structural covariance follows the pattern of structural covariance observed in young adult age with some small differences discussed below. However, in participants with probable dementia disease, the pattern of co-atrophy in the hippocampus deviates from what was observed in these healthy populations. In patients with dementia, the co-atrophy seems to follow the functional large-scale networks with a pattern that resembles more than the functional model of hippocampus organization than what was observed in other groups. Overall, the most prominent differences between groups in the differentiation patterns of the hippocampus were found in the body and tail whereas the head always appears as a uniform region. Group differences were shown not only in the topological pattern within the hippocampus, but also in the whole brain structural covariance pattern that drove the clustering and their associated behavioural associations.
Consistent pattern of head differentiation in hippocampus structural covariance across the lifespan
Independent of age and disease, the head of the hippocampus emerged consistently as one homogeneous subregion, except for some minor reductions with higher age and ongoing pathology. But the actual underlying covariance pattern of the anterior hippocampal subregion changed across age/disease groups. In young adulthood the anterior hippocampal covariation pattern was characterized by a broad network extending across frontal, temporal and occipital lobes as well as (inferior) parietal regions. In accordance with the large spatial distribution of this network, behavioural associations showed a relatively broader spectrum including emotional, cognitive and perceptual processes. These results could suggest that the hippocampus head is a plastic region (based, for example, on cell proliferation in the dentate gyrus during the lifespan) (van Praag et al., 2005), the structure of which is modulated by rich functional interaction with large-scale brain networks subserving various behavioural functions. The structural covariance networks of the hippocampus head in early and late adulthood demonstrated that the anterior hippocampus covaried with the same brain regions in both halves of healthy lifespan suggesting a perseverance of co-plasticity and resilience. However, in dementia, the structural covariance network of the anterior subregion decreased mainly to the temporal lobe suggesting a loss of network.
Consistent pattern of medial-lateral differentiation in hippocampus structural covariance
Across different age groups of the healthy population, we found a consistent differentiation pattern along the medial-lateral dimension of the hippocampus dividing it into a lateral and a medial subregion. This pattern replicated previous findings and seemed to follow the cytoarchitectonic differentiation between the cornu ammonis and subiculum subfields (Plachti et al., 2019). Importantly, this pattern, like the head subregion, appeared to remain stable across the whole adult lifespan suggesting a very strong and robust scheme of structural covariance that should be referred to when studying structural changes with MRI in adults. This scheme was even further retained when subdividing the hippocampus into four subregions in healthy adults and MCI patients (Supplementary Fig. 5), even if one additional cluster appeared either in the anterior or posterior-lateral region depending on the age/disease group. Even though the differentiation into a lateral and medial parcel was preserved over the lifespan, the lateral cluster decreased posteriorly with age and the medial cluster expanded into the tail. This change in the cluster pattern was reflected both in the associated structural pattern and the related behavioural associations.
The medial hippocampal subdivision showed a covariation pattern with occipito-parietal, temporal (middle temporal gyrus), and frontal (inferior and middle frontal gyri) brain regions. Furthermore, the network included subcortical brain regions such as thalamus, caudate, and insula. With increasing age, the covariance network expanded highly in size, especially covering posterior brain regions. The shift from mostly anteriorly associated brain regions in younger years to posteriorly associated regions in elderly is not unusual for the hippocampus. It has already been reported in functional connectivity (Blum et al., 2014; Stark et al., 2019), in structural covariance studies (Li et al., 2018), and for anatomical connectivity with strengthened connections to medial occipital regions (Maller et al., 2019), which was in line with our results, even though the responsible mechanisms remain to be elucidated.
These alterations were also mirrored in the behavioural association patterns. While in younger adults visual cognition (e.g. object, place, encoding, familiarity) was prominent, in the elderly, however, the behavioural spectrum expanded to language processing as well as to motor-related (learning) behaviour. Both, structural covariation networks and behavioural profiling suggest that brain regions connected by the inferior longitudinal fasciculus (ILF) covary more likely with the medial subregion of the hippocampus. The ILF is an occipito-temporal association tract with close relationships to the occipital radiations and hippocampus through the tapetum (Herbet et al., 2018). The ILF is behaviourally associated with visual object and face recognition, reading as well as lexical and semantic processing (Herbet et al., 2018), which is in accordance with our behavioural profiling of the medial subregion across the lifespan.
While the medial cluster expanded into the tail during healthy ageing, the lateral cluster decreased from the tail. The lateral subregion’s covariance network in young adulthood yielded primarily associations with subcortical regions (e.g. thalamus, caudate nuclei) and additionally with the parieto-occipital fissure. Anatomically those associated brain regions were reminiscent to some extent to the grey matter regions around the dorsal hippocampal commissure, being connected with posterior cingulum, tapetum, and fornix (Postans et al., 2019). The dorsal hippocampal commissure is associated with learning, memory and recently also with recognition (Postans et al., 2019). The fornix is the white matter output of the hippocampus through the tail (Amaral et al., 2018) whereas the tapetum transfers information between hemispheres. The hippocampus is connected via the fornix with limbic structures (i.e. hypothalamus, thalamus, nucleus accumbens) (Douet and Chang, 2015), and has been suggested to play a major role in transferring information from short-term to long-term memory via the Papez circuit and is accordingly, involved in long-term memory encoding and retrieval (Eichenbaum et al., 2007; Douet and Chang, 2015; Foster et al., 2019).
Structural covariance pattern in the hippocampus in dementia resembles functional organization
In the healthy population, structural covariance across the brain is assumed to reflect maturational, developmental and experience-based co-plasticity (Alexander-Bloch et al., 2013; Geng et al., 2017). In patients with neurodegenerative disorders, structural covariance across the brain could be expected to mainly reflect brain structure co-atrophy. The moderate-to-high convergence between structural covariance and task-(un)related functional connectivity (Reid et al., 2016; Kotkowski et al., 2018; Paquola et al., 2018; Shah et al., 2018) suggests that abnormalities in structural and functional network topology is predictive of brain disorders (Seeley et al., 2009; Goodkind et al., 2015) and weaker cognitive performance (Spreng and Turner, 2013; McTeague et al., 2016; Montembeault et al., 2016). However, the question remains whether structural atrophy changes functional BOLD response (He et al., 2007) or the other way around (Chang et al., 2018). From a neuropathological standpoint, Alzheimer’s pathology is assumed to follow a specific topological pattern distributed along large-scale networks (Braak and Braak, 1991; Corder et al., 2000; Montembeault et al., 2016). For example, amyloid plaque distribution in the brain seems to follow functional organization mirrored in the default mode network (Klunk et al., 2004; Buckner et al., 2005; Montembeault et al., 2016). Similarly, the spreading of tau neurofibrillary tangles seems to follow a functional pattern, which is not explained by spatial proximity (Franzmeier et al., 2019). In other words, brain regions that are more likely to be functionally coupled together share a stronger tau covariance, which is not explained by pure spatial neighbourhood. This apparent convergence between spatial distribution of pathology markers and the spatial organization of functional networks may be explained by the fact that synchronous neuronal firing establishes a network-based synaptogenesis (Katz and Shatz, 1996; Bi and Poo, 1999), which can then be assumed to be vulnerable to pathological processes.
Linking these neuropathological considerations to the pattern of differentiation based on structural covariance found in the hippocampus of patients with probable Alzheimer’s disease in this study, we can hypothesize that the pattern of co-atrophy in these patients followed the pattern of functional organization subserving broad behavioural functions. In this regard, we can note that the pattern of structural covariance networks of the hippocampal body in dementia patients in this study was associated with temporal and frontal regions in turn associated with comprehension, language, orthography and theory of mind. We hypothesize that the structural covariance network of the hippocampus body reflects a functional network of higher cognitive functions of social cognition additionally supported by the functional coactivation pattern of the lateral body subregion when applied to healthy elderly. It therefore emphasizes that the hippocampal differentiation based on structural covariance in dementia follows functional differentiation. Overall our findings point to the necessity of accounting for hippocampus functional organization related to large-scale networks subserving broad behavioural functions when studying hippocampus structural changes at the macroscale in dementia.
Supplementary Material
Acknowledgements
Data collection and sharing for this project was provided by the Cambridge Centre for Ageing and Neuroscience (CamCAN).
Funding
Clinical data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc; Cogstate; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; EuroImmun; F Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research and Development, LLC; Johnson and Johnson Pharmaceutical Research and Development LLC; Lumosity; Lundbeck; Merck and Co, Inc; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc; Piramal Imageng; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. CamCAN funding was provided by the UK Biotechnology and Biological Sciences Research Council (grant number BB/H008217/1), together with support from the UK Medical Research Council and University of Cambridge, UK. This study was supported by Deutsche Forschungsgemeinschaft (GE 2835/1–1), Helmholtz-Gemeinschaft (Helmholtz Portfolio Theme ‘Supercomputing and Modelling for the Human Brain’), Horizon 2020 Framework Programme [720270 (HBP SGA1)], Horizon 2020 Framework Programme [785907 (HBP SGA2)], and Deutsche Forschungsgemeinschaft (EI 816/4–1).
Competing interests
The authors report no competing interests.
Glossary
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative;
- MCI =
mild cognitive impairment
References
- Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci 2013; 33: 2889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral RSC, Park MTM, Devenyi GA, Lynn V, Pipitone J, Winterburn J, et al. Manual segmentation of the fornix, fimbria, and alveus on high-resolution 3T MRI: application via fully-automated mapping of the human memory circuit white and grey matter in healthy and pathological aging. Neuroimage 2018; 170: 132–50. [DOI] [PubMed] [Google Scholar]
- Bi G, Poo M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature 1999; 401: 792–6. [DOI] [PubMed] [Google Scholar]
- Blum S, Habeck C, Steffener J, Razlighi Q, Stern Y. Functional connectivity of the posterior hippocampus is more dominant as we age. Cogn Neurosci 2014; 5: 150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyke J, Driemeyer J, Gaser C, Buchel C, May A. Training-induced brain structure changes in the elderly. J Neurosci 2008; 28: 7031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–59. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, et al. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci 2005; 25: 7709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers S, Moebus S, Lux S, Pundt N, Schütz H, Mühleisen TW, et al. Studying variability in human brain aging in a population-based German cohort-rationale and design of 1000BRAINS. Front Aging Neurosci 2014; 6: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-T, Huang C-W, Chang W-N, Lee J-J, Chang C-C. Altered functional network affects amyloid and structural covariance in Alzheimer's disease. Biomed Res Int 2018; 2018: 8565620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Woodbury MA, Volkmann I, Madsen DK, Bogdanovic N, Winblad B. Density profiles of Alzheimer disease regional brain pathology for the huddinge brain bank: pattern recognition emulates and expands upon Braak staging. Exp Gerontol 2000; 35: 851–64. [DOI] [PubMed] [Google Scholar]
- Douet V, Chang L. Fornix as an imaging marker for episodic memory deficits in healthy aging and in various neurological disorders. Front Aging Neurosci 2015; 6: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, Yonelinas AP, Ranganath C. The medial temporal lobe and recognition memory. Annu Rev Neurosci 2007; 30: 123–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB. What is normal in normal aging? Effects of aging, amyloid and Alzheimer's disease on the cerebral cortex and the hippocampus. Prog Neurobiol 2014; 117: 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming Beattie J, Martin RC, Kana RK, Deshpande H, Lee S, Cure J, et al. Hippocampal dentation: structural variation and its association with episodic memory in healthy adults. Neuropsychologia 2017; 101: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CM, Kennedy KM, Hoagey DA, Rodrigue KM. The role of hippocampal subfield volume and fornix microstructure in episodic memory across the lifespan. Hippocampus 2019; 29: 1206–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmeier N, Rubinski A, Neitzel J, Kim Y, Damm A, Na DL, et al. Functional connectivity associated with tau levels in ageing, Alzheimer’s, and small vessel disease. Brain: a Journal of Neurology 2019; 142: 1093–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Kot P, Liu X, Lang DJ, Wang JZ, Honer WG, et al. Parcellation of the human hippocampus based on gray matter volume covariance: replicable results on healthy young adults. Hum Brain Mapp 2019; 40: 3738–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Li G, Lu Z, Gao W, Wang L, Shen D, et al. Structural and Maturational Covariance in Early Childhood Brain Development. Cereb Cortex 2017; 27: 1795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodkind M, Eickhoff SB, Oathes DJ, Jiang Y, Chang A, Jones-Hagata LB, et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry 2015; 72: 305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang L, Zang Y, Tian L, Zhang X, Li K, et al. Regional coherence changes in the early stages of Alzheimer's disease: a combined structural and resting-state functional MRI study. Neuroimage 2007; 35: 488–500. [DOI] [PubMed] [Google Scholar]
- Herbet G, Zemmoura I, Duffau H. Functional anatomy of the inferior longitudinal fasciculus: from historical reports to current hypotheses. Front Neuroanat 2018; 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockwitz C, Mérillat S, Liem F, Oschwald J, Amunts K, Caspers S, et al. Generalizing age effects on brain structure and cognition: a two-study comparison approach. Hum Brain Mapp 2019; 40: 2305–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science 1996; 274: 1133–8. [DOI] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol 2004; 55: 306–19. [DOI] [PubMed] [Google Scholar]
- Kotkowski E, Price LR, Mickle Fox P, Vanasse TJ, Fox PT. The hippocampal network model: a transdiagnostic metaconnectomic approach. Neuroimage Clin 2018; 18: 115–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li Q, Wang X, Li D, Li S. Differential age-related changes in structural covariance networks of human anterior and posterior hippocampus. Front Physiol 2018; 9: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llera A, Wolfers T, Mulders P, Beckmann CF. Inter-individual differences in human brain structure and morphology link to variation in demographics and behavior. eLife 2019; 8: e44443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire EA, Woollett K, Spiers HJ. London taxi drivers and bus drivers: a structural MRI and neuropsychological analysis. Hippocampus 2006; 16: 1091–101. [DOI] [PubMed] [Google Scholar]
- Maller JJ, Welton T, Middione M, Callaghan FM, Rosenfeld JV, Grieve SM. Revealing the hippocampal connectome through super-resolution 1150-direction diffusion MRI. Sci Rep 2019; 9: 2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTeague LM, Goodkind MS, Etkin A. Transdiagnostic impairment of cognitive control in mental illness. J Psychiatr Res 2016; 83: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembeault M, Rouleau I, Provost JS, Brambati SM. Altered gray matter structural covariance networks in early stages of Alzheimer's disease. Cereb Cortex 2016; 26: 2650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Jiménez EP, Flor-García M, Terreros-Roncal J, Rábano A, Cafini F, Pallas-Bazarra N, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med 2019; 25: 554–60. [DOI] [PubMed] [Google Scholar]
- Paquola C, Bennett MR, Lagopoulos J. Structural and functional connectivity underlying gray matter covariance: impact of developmental insult. Brain Connect 2018; 8: 299–310. [DOI] [PubMed] [Google Scholar]
- Plachti A, Eickhoff SB, Hoffstaedter F, Patil KR, Laird AR, Fox PT, et al. Multimodal parcellations and extensive behavioral profiling tackling the hippocampus gradient. Cereb Cortex 2019; 29: 4595–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postans M, Parker GD, Lundell H, Ptito M, Hamandi K, Gray WP, et al. Uncovering a role for the dorsal hippocampal commissure in recognition memory. Cerebl Cortex 2019; 30: 1001–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Li L, Zhou TY, Lu W. Alzheimer's disease progression model based on integrated biomarkers and clinical measures. Acta Pharmacol Sin 2014; 35: 1111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AT, Bzdok D, Langner R, Fox PT, Laird AR, Amunts K, et al. Multimodal connectivity mapping of the human left anterior and posterior lateral prefrontal cortex. Brain Struct Funct 2016; 221: 2589–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009; 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafto MA, Tyler LK, Dixon M, Taylor JR, Rowe JB, Cusack R, et al. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) study protocol: a cross-sectional, lifespan, multidisciplinary examination of healthy cognitive ageing. BMC Neurol 2014; 14: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Bassett DS, Wisse LEM, Detre JA, Stein JM, Yushkevich PA, et al. Mapping the structural and functional network architecture of the medial temporal lobe using 7T MRI. Hum Brain Mapp 2018; 39: 851–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Mormino EC, Schultz AP, Betensky RA, Papp KV, Amariglio RE, et al. The impact of amyloid-beta and tau on prospective cognitive decline in older individuals. Ann Neurol 2019; 85: 181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN, Turner GR. Structural covariance of the default network in healthy and pathological aging. J Neurosci 2013; 33: 15226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark SM, Frithsen A, Stark CEL. Age-related alterations in functional connectivity along the longitudinal axis of the hippocampus and its subfields. bioRxiv 2019; 577361. [DOI] [PMC free article] [PubMed]
- Taylor JR, Williams N, Cusack R, Auer T, Shafto MA, Dixon M, et al. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) data repository: structural and functional MRI, MEG, and cognitive data from a cross-sectional adult lifespan sample. Neuroimage 2017; 144 (Pt B): 262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Petten C. Relationship between hippocampal volume and memory ability in healthy individuals across the lifespan: review and meta-analysis. Neuropsychologia 2004; 42: 1394–413. [DOI] [PubMed] [Google Scholar]
- van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci 2005; 25: 8680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziontz J Bilgel M Shafer AT Moghekar A Elkins W Helphrey J, . et al. Tau pathology in cognitively normal older adults. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring 2019; 11: 637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from open science initiatives reported and cited above. Code can be shared upon reasonable request from the corresponding author. The derived clusters are available at (http://anima.fz-juelich.de/) as ROI in .nii format.