ABSTRACT
Immunotherapy using immune-checkpoint inhibitors is revolutionizing oncotherapy. However, the application of immunotherapy may be restricted because of the lack of proper biomarkers in a portion of cancer patients. Recently, emerging evidence has revealed that gut commensal bacteria can impact the therapeutic efficacy of immune-checkpoint inhibitors in several cancer models. In addition, testing the composition of gut bacteria provides context for prediction of the efficacy and toxicity of immunotherapy. In this review, we discuss the impacts of gut commensal bacteria on the tumoral immune milieu, highlighting some typical bacteria and their associations with immunotherapy.
KEYWORDS: Commensal bacteria, gut immunity, tumor immune milieu, immune-checkpoint inhibitor, immunotherapy biomarker
Introduction
Immune-checkpoint inhibitors (ICIs) have opened a new era of oncotherapy. Currently, numerous clinical trials investigating ICI efficacy are ongoing across a large range of cancer types.1 Accordingly, first-generation ICI drugs targeting CTLA-4, PD-1, or PD-L1 have been approved for fourteen indications.2 In general, the response rates to one of these drugs used alone vary from 20% to 40%.3,4 More strikingly, a portion of patients can achieve complete remission of tumors after receiving this therapy.5
Although ICI therapy tinctures offer cancer patients hope of a cure, the priority task is determining the biomarkers with high specificity and sensitivity to predict the candidates who can benefit from ICI therapy with more precision. Currently, the available biomarkers for selecting patients receiving ICI therapy include PD-L1,6 tumor mutational burden (TMB),7 high microsatellite instability (MSI-H), and deficient mismatch repair (dMMR).8 On this basis of MSI-H or dMMR, the drug approved by the FDA is pembrolizumab, regardless of cancer type.9 Nevertheless, different types of cancer have express biomarkers useful for selecting candidates for ICI therapy. For example, PD-L1 positivity and a high TMB value in patients with non-small cell lung cancer (NSCLC) and presentation with MSI-H or dMMR by colorectal cancer patients have been well-established markers.8,10 In addition to such biomarkers, several other ICI-therapy-associated biomarkers are being explored.11,12
As revealed by the molecular characteristics common across cancers, immune-associated biomarkers are also being discovered.13–15 Hence, intestinal commensal bacteria have attracted public interest because it is believed that intestinal commensal bacteria shape human immunity,16,17 presenting that disturbed homeostasis of bacterial ecosystem enables host immunity to be abnormal.18 This imbalance can be translated into ICI therapy. Emerging data have supported the idea that evaluating intestinal bacteria is a new route by which to predict the therapeutic response to and toxicity of ICI drugs.11 For example, commensal bacteria that can synergize with ICI therapy in tumoricidal processes include Bifidobacterium longum,19 Collinsella aerofaciens,19 Enterococcus faecium,19 Faecalibacterium genus bacteria20 and Akkermansia muciniphila in humans.21 In addition, poor efficacy of ICI therapy appears to be associated with increased frequency of Bacteroides in the gut.20,22 Referring to ICI-associated toxicity, it has been revealed that patients with a higher fecal level of Bacteroidales commonly exhibit lower incidences of colitis than those with lower levels.23 Conversely, cancer patients who have used antibiotics long term have not only a poor response to ICI therapy but also an increased incidence of ICI-related toxicity.24,25 It has been shown that antibiotics are able to reduce the total number and diversity of commensal bacteria in the gut.25 In this regard, homeostasis in the ecosystem of commensal bacteria is critical for ensuring the effectiveness of ICI therapy. Thus, a prevailing proposal has been presented suggesting that the richer of diversity of the commensal bacteria, the better response to ICI therapy is likely to be.19 Currently, several clinical trials concerning oral supplementation of commensal bacteria for improving the therapeutic efficacies of ICI therapy are ongoing (NCT03772899 and NCT03686202). However, the mechanisms by which gut commensal bacteria mediate the efficacy of ICI therapy are indeed complicated because bacteria-primed immune processes that favor ICI therapy have not been established for certain species. Instead, a community of bacteria elicits the tumoricidal response.26 In this review, we discuss the role of commensal bacteria in regulating host immunity and their influence on tumoral immune milieu formation, thus providing a rationale for commensal bacteria in guiding ICI therapy.
Composition and physiological functions of gut commensal bacteria
The gut contains a microbial world. As estimated, 3.8 × 1013 bacteria exist in the lumen,27 and most are commensal. Among these bacteria, the number of species ranges from 500 to 1000.28 Approximately 98% of these bacteria belong to Bacteroidetes and Firmicutes phyla, whereas Fusobacteria, Actinobacteria, Proteobacteria, Verrucomicrobia and Cyanobacteria account for a only a minor portion.28–30 A healthy intestine provides an environment that favors the growth of anaerobes.31,32 However, significant differences characterize bacterial diversity among individuals.33 Therefore, dietary habits and living environment are factors influencing the bacterial diversity in gut.34,35
At steady state, gut commensal bacteria have beneficial effects on the hosts in several respects, including maintaining epithelial homeostasis and a barrier function,36,37 facilitating food digestion and nutrient absorption, synthesizing bioactive substances in favor of cell metabolites, and shaping the host immunity.38,39 In terms of host immunity, gut commensal bacteria control the polarization of several T cell subsets, such as Th1, Th2, Th17 and Treg cells, thus enabling the host to defend against foreign stimuli while sustaining an immunotolerant milieu.40 During human evolution, the diversity of commensal bacteria was reduced in the gut constantly,35 causing abnormal colonization of commensal bacteria, sluggishness in the physical functions of the bacteria, and abnormal immunity.41 Consequently, hosts are prone to increased susceptibility to autoimmune diseases or even cancer.18 This notion has been typically translated into gastric and colorectal cancer, in which collective commensal bacterial dysbiosis is an intrinsic factor that induces carcinogenesis.42,43 In fact, reduced diversity represents a paradigm of bacterial dysbiosis.
Gut commensal bacteria in the regulation of host immunity
Gut commensal bacteria shape host immunity. In general, gut bacteria use antigens and metabolites to induce immune cell commitment.44 In contrast, the composition of the gut bacteria is maintained at a physical level by the substances provided by the gut immune and epithelial cells.45 Therefore, IgA secreted by gut B cells or IgM secreted by plasma cells is critical in defending against the outgrowth of pathogenic bacteria in the gut.46 In parallel with these secretions, mature epithelial cells, especially Paneth cells, can produce various anti-microbial peptides to sustain the bacterial ecosystem in the gut, enabling a moderate level of fixed bacteria.45 On this basis, this bacterial community is helpful in protecting against the occurrence of immune abnormity.
In general, T cell polarization in the gut induced by commensal bacteria can present in either a dendritic cell (DC)-dependent or DC-independent manner.47 DCs are professional antigen-presenting cells in humans.47 Especially in a bacterial antigen-enriched milieu, the pattern recognition receptors (PRRs) and damage-associated molecular patterns (DAMPs) are crucial molecules that mediate DCs in recognizing and subsequently responding to antigens.48,49 Thereafter, DCs process such antigens into adaptive immune cells by manipulating their survival and function through DC-produced cytokines under given conditions.50 After this process, commensal bacteria such as Lactobacillus sakei and Bifidobacteria are reported to have the capacity to upregulate MHC-II expression by DCs while recruiting them into the gut.51,52 Functionally, the antigens presented by MHC-II molecules are able to activate naïve T cells while priming their differentiation into other T cell subsets (Figure 1). Independent of DCs, gut B cells, macrophages, or other innate immune cells, such as NK cells and innate lymphoid cells, can also be activated by lumen bacteria presenting immunoregulatory phenotypes (Figure 1).53 For example, in gut Peyer′s patches (PPs), bacterial antigen presentation by epithelial M cells serves as a route for inducing B cell activation, allowing for IgA, IgM and IgG to clear pathogens.54 In this situation, the complex of commensal IgG can be cleared by residual macrophages.55 In response, the macrophages increase the production of IL-1β along with stimulating neutrophils and Th17 cells to clear the intestinal infection.55 Similar effects can be induced by NK cells as well. For example, Bacteroides fragilis and Salmonella can directly activate NK cells by interacting with TLR4 or TLR9, thus increasing NK cell cytotoxic activity.53,56
Figure 1.
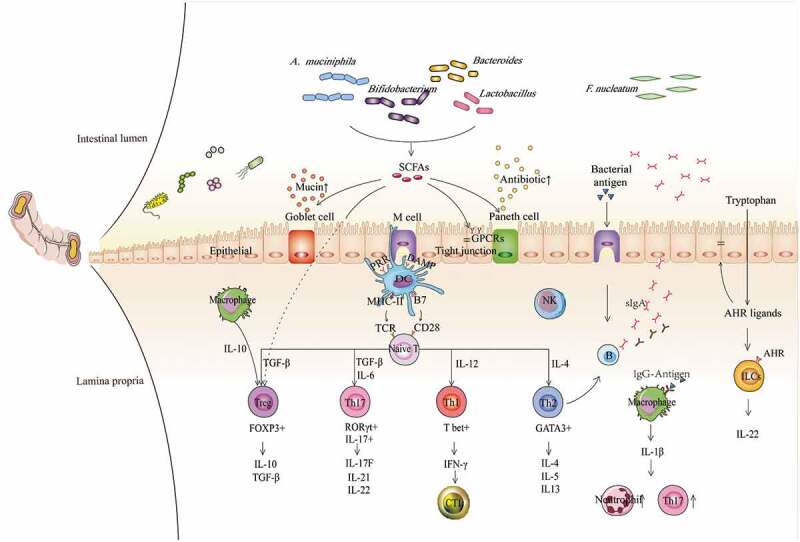
Contributions of gut commensal bacteria and their metabolites to the host immune system. Based on the function of antigen-presenting cells, such as DCs, NK cells, and macrophages, commensal bacteria mediate the differentiation of naïve CD4 + T cells into different subgroups, such as T-bet+ Th1 cells, GATA3+ Th2 cells, RORγt+ Th17 cells, and FOXP3+ Tregs, which further contribute to different immune modulation responses, and the production of various cytokines, such as TGF-β, IFN-γ and ILs. Immune regulation can be mediated not only by bacteria but also by their metabolites, especially SCFAs and AHR ligands, exerting functions by binding GPCRs and AHR on the surface of epithelial cells and immune cells, respectively, which subsequently contribute to augmented epithelial barrier function and improved gut immune tolerance. Conversely, some immune cells and epithelial cells can also mediate the balance of bacteria by secreting antibacterial substances, such as B cells secreting IgA, Goblet cells secreting mucins and Paneth cells secreting antimicrobial peptides, etc. Overall, microbiota-immune cross-talk contributes to gut homeostasis by forming a relatively stable feedback loop. SCFAs, short-chain fatty acids; AHR, aromatic hydrocarbon receptor; GPCRs, G protein-coupled receptors; TCR, T cell receptor; PRR, pattern recognition receptor; DAMP, damage-associated molecular pattern; MHC-II, major histocompatibility complex II; B7, B7.1(CD80)/B7.2(CD86); DC, dendritic cell; NK, natural killer cell; NE, neutrophil; Treg, regulatory T cell; CTL, cytotoxic T lymphocyte; ILC, innate lymphoid cell; IFN-γ, interferon-γ; TGF-β, transforming growth factor; and IL, interleukin.
Similar to bacterial antigens, the metabolites processed by commensal bacteria are critical in immunity homeostasis.44 For example, the most critical substances are short-chain fatty acids (SCFAs) (Figure 1). SCFAs can be generated by gut commensal bacteria such as Lactobacillus, Bacteroides, Bifidobacterium, and Akkermansia muciniphila after they conduct glycolysis of food fibers.57,58 SCFAs are able to sense immune cells, including DCs, T cells and B cells, with respect to increasing the gut numbers of Treg cells and DCs with a tolerogenic phenotype, minimizing Th2 cell-associated immune responses and improving the secretion of IgA by gut B cells.59 Alternatively, Lactobacillus spp. are able to utilize tryptophan to produce indole-3-aldehyde, which interacts with the aromatic hydrocarbon receptor (AHR) on innate immune cells to induce their secretion of IL-22 (Figure 1).60 In addition, SCFAs can induce epithelial cells to upregulate the production of anti-microbial peptides.59,61 In summary, gut commensal bacteria are essential for gut immunity.
Commensal bacteria, tumoral immune milieu and effectiveness of ICI therapy
Immune deficiency serves as a critical factor for cancer pathogenesis.62 Mechanically, an action called immunoediting represents cancer cell clone evolution over time after the host immune clearance, thus enabling the clones with low immunogenicity to be preserved.63 In this setting, commensal bacteria can cross-talk with the residual cancer cells directly or indirectly to facilitate their aggressive behaviors.64 For example, intestinal bacterial dysbiosis is a state of early gut carcinogenesis, while intestinal bacterial dysbiosis persists during cancer progression.65 For example, Fusobacterium nucleatum (F. nucleatum) are regarded as ‘Oncobacteria’ of colorectal cancer because they can self-localize into tumors to facilitate their growth, induce chemoresistance and conduct immunosuppression.66 In another paradigm, an event called ‘bacterial succession’ suggests that one type of bacterium, followed by the others, continues to perform its oncogenic functions during different periods.67 As an example of this notion, Helicobacter pylori plays a pioneering role in inducing early lesions of gastric cancer,68 and other bacteria, such as F. nucleatum, continue to perform their function in promoting tumor progression.69 Similar to it effect in colorectal cancer, intestinal bacterial dysbiosis also influences pancreatic cancer pathogenesis.42,70 For example, antibiotics can be used to support the management of pancreatic cancer when the tumoral neoantigens share similarity with the antigens on bacteria.71 In this situation, tumoricidal T cells potentially recognize bacterial antigens, thus misleadingly clearing bacteria instead of tumor cells. In this regard, immunosuppression in tumors naturally requires bacterial participation. In fact, recent data support this notion, suggesting that intestinal bacteria influence the tumoral immune milieu mainly by altering the tumoral density of immune cells and changing their cytokine production.72 In general, commensal bacteria have discriminating roles in this event. By using the syngeneic melanoma or sarcoma transplantable tumor models, it was revealed that supplying mice with feces of human ICI responders can significantly enhance tumor remission after anti-PD-1 or -PD-L1 therapy.19–21 For example, such feces were found to upregulate PD-L1 expression in the tumor microenviroment of melanoma mice.20 By contrast, feces from non-ICI-responders can even promote tumor progression.20 Therefore, commensal bacteria are able to distort the effectiveness of ICI drugs.72 In the following section, we introduce the contributions of some commensal bacteria in altering the tumoral immune milieu and analyze their roles in improving or distorting the efficacy of ICI drugs (Figure 2).
Figure 2.
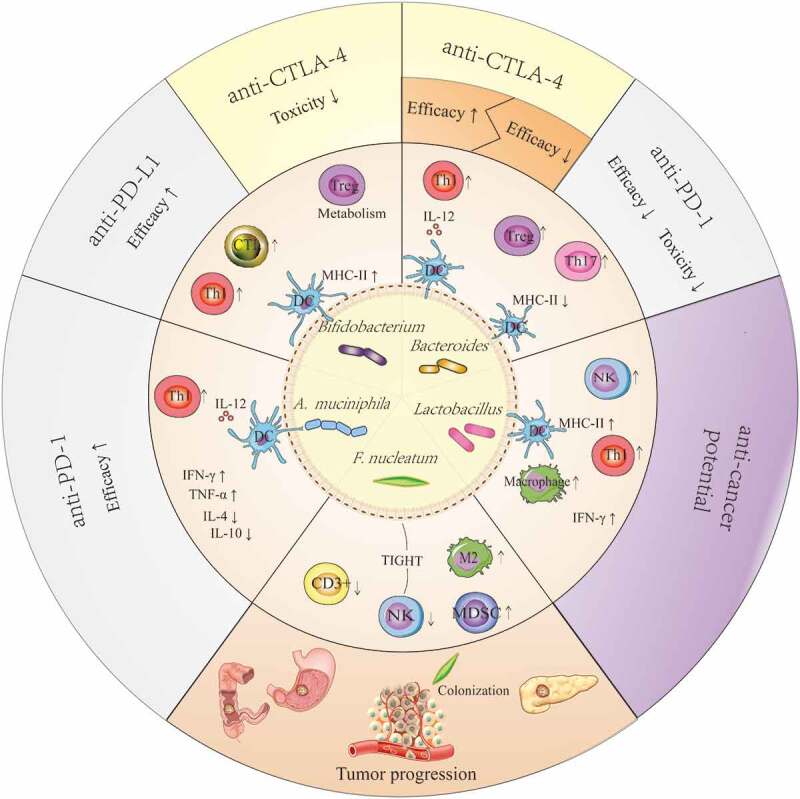
Potential mechanism that explains the anticancer or pro-cancer effects of some candidate ICI-therapy-associated bacteria by shaping the host immune status. Lactobacilli may upregulate the expression of MHC-II on DCs, enhance the activity of NK cells and macrophages, and improve Th1-mediated immune responses as well as increase the production of IFN-γ in tumors. The effects described above facilitate the anticancer potential of Lactobacilli. Bifidobacteria can improve the efficacy of anti-PD-L1 therapy by upregulating the expression of MHC-II on DCs, promoting Th1 polarization and CTL accumulation in the tumor microenvironments, and reducing the toxicity of anti-CTLA-4 therapy through enhanced Treg cell metabolism. Akkermansia muciniphila can enhance the efficacy of anti-PD-1 therapy in a manner dependent on the enhanced IL-12-dependent Th1-related immune response, along with increased levels of IFN-γ and TNF-α and decreased levels of IL-4 and IL-10. The results from a preclinical trial confirmed that Bacteroidetes can restore anti-CTLA-4 treatment efficacy by enhancing the IL-12-dependent Th1-related immune response, whereas these bacteria are associated with poor clinical outcomes of anti-CTLA-4 or anti-PD-1 therapy in human clinical trials. Fusobacterium nucleatum can promote tumor progression in gastrointestinal cancer and pancreatic cancer, and it is verified to be associated with reduced density of CD3 + T cells, exhaustion of NK cells, and augmentation of M2 polarization along with accumulation of MDSCs in tumor microenvironments. These effects may be closely linked with their colonization in tumors.
Lactobacilli
Lactobacilli are the most common probiotics distributed in the gut. They belong to the Firmicutes phylum. At the genus level, Lactobacillus includes more than 150 species, and most of them are facultative anaerobic bacteria. Among them, Lactobacillus reuteri (L. reuteri), Lactobacillus acidophilus (L. acidophilus) and Lactobacillus casei (L. casei) serve as common bacteria corresponding to intestinal health and disease.73
At steady state, several strains of L. reuteri can exert their functions, including promotion of Treg cell development and reduction in pro-inflammatory cytokine production, thus protecting against the exacerbated inflammation in gut.73 In fact, independent of Treg cells, L. reuteri also exhibits potency in inhibiting Th1- or Th2-related immune responses because L. reuteri are capable of restoring the levels of plasma inosine, which are dedicated to reducing IFN-γ and IL-4 along by inhibiting Th1/Th2 polarization via inosine–adenosine A2A receptor interactions (Table 1).74 Similarly, the supernatant from in vitro cultured L. reuteri is able to reduce TNF-α production by human myeloid cells, suggesting that the metabolites of L. reuteri also exert an anti–inflammatory effect.75 In addition to the aforementioned intrinsic roles, tryptophan catabolites generated by L. reuteri can eventually promote the proliferation of CD8+CD4+ double-positive intraepithelial lymphocytes (Table 1).76 After this proliferation is initiated, tryptophan catabolites act as ligands of AHR. Upon the AHR-ligand interactions, the production of IL-22 by innate lymphoid cells is improved.77 Functionally, IL-22 can protect the intestine against infection synergistically with IL-17 through the induction of enterocyte expansion and antibacterial peptide production. However, in colorectal cancer, IL-22 is able to promote tumor progression.78 In the latter situation, administration of the L. casei BL23 strain to mice bearing colorectal cancer reportedly reduced the tumoral level of IL-22; thus L. casei BL23 serves as a candidate treatment against colorectal cancer progression (Table 1).79
Table 1.
Effects of potential immune-associated bacteria on the host immune system.
| Bacteria | Model | Effects on the immune system | Refs |
|---|---|---|---|
| Lactobacillus reuteri | Treg-deficient mice | Decreased levels of IFN-γ and IL-4; Inhibition of Th1- or Th2- responses. |
74 |
| Germ-free mice and mice without CD8+CD4+ double-positive intraepithelial lymphocytes | Induction of CD8+CD4+ double-positive intraepithelial lymphocytes cells. | 76 | |
| Breast cancer-bearing mice | Induction of CD4+CD25+ Treg cells; Suppression of mammary tumorigenesis. |
109 | |
| Lactobacillus acidophilus | Breast cancer-bearing mice | Increased levels of IFN-γ, IL-4 and TGF-β; Establishment of a Th1 protective pattern. |
80 |
| Lactobacillus brevis | Breast cancer-bearing mice | Increased levels of IFN-γ, TNF-α, IL-2, IL-17; Increased cytotoxic activity of NK cells. |
81,82 |
| Lactobacillus casei | Colorectal cancer-bearing mice | Reduced level of IL-22; Increased levels of IL-6, IL-17, IL-10, TGF-β; Mediation of Treg-to-Th17-biased immune response; Protection against dysbacteriosis. |
79,110 |
| Lactobacillus rhamnosus | Mice with depletion of EGFR in intestinal epithelial cells | Promotion of epithelial cell proliferation, differentiation and tight junction formation; Regulation of the differentiation of T helper cells, Treg cells, and B cells. |
111 |
| Bifidobacterium spp. | Melanoma-bearing mice and melanoma-bearing Foxp3-DTR mice; Anti-CTLA-4 mAb |
Amelioration of intestinal immunopathology without distorting antitumor effects of anti-CTLA-4 mAb; Decreased levels of IL-6. | 88 |
| Bifidobacterium spp. | Melanoma-bearing mice; Anti-PD-L1 mAb |
Upregulation of MHC-II by DCs; Accumulation of CD8 + T cells in tumors; Increased levels of IFN-γ in tumors. |
52 |
| Bifidobacterium longum | Healthy adult mice | Upregulation of IL-4, IL-6, IL-10, IFN-γ and TGF-β in ileal Peyer’s patches | 112 |
| Mice with gliadin-induced enteropathy | Reduction in CD4+Foxp3+ cells. Increases in CD8+ T cell populations; Upregulation of NF-kB and IL-10 expression; Decrease in TNF-α production. |
113 | |
| Healthy infant mice | Increase in IFN-γ-secreting cells and ratio of IFN-γ-secreting to IL-4-secreting cells; Strengthened Th1-related immune response. |
85 | |
| Bifidobacterium adolescentis | Healthy conventional mice and germ-free mice | Increased number of Th17 cells in the small intestine. | 86 |
| Akkermansia muciniphila | Epithelial tumor patients Sarcoma-bearing mice Anti-PD-1 mAb |
Beneficial clinical responses to anti-PD-1 mAb in patients; IL-12-dependent recruitment of Th1 cells into tumor beds; Increased levels of IFN-γ and TNF-α and decreased levels of IL-4 and IL-10. |
21 |
| Bacteroidetes | Melanoma patients Melanoma-bearing mice Anti-PD-1 mAb |
High number of Tregs and MDSCs; Blunted cytokine response in melanoma patients; Increased frequency of Th17 cells and Tregs in model mice. |
20 |
| Melanoma patients Anti-CTLA-4 mb |
Poor response to anti-CTLA-4 mAb; High number of α4+β7+ T cells in the periphery. |
22 | |
| Bacteroides Fragilis/Bacteroides thetaiotaomicron | Sarcoma-bearing mice Anti-CTLA-4 mAb |
Accumulation of CD11b+ DCs in the lamina propria IL-12-dependent Th1 immune response in tumor-draining lymph nodes. |
103 |
| Fusobacterium nucleatum | Colorectal cancer patients | Inverse correlation with CD3+ T cell density in tumors. | 104 |
| ApcMin/+ mice | Accumulation of MDSCs and M2-like TAMs. | 107 |
Concerning L. acidophilus, previously reported data suggested that oral administration of L. acidophilus to breast cancer-bearing mice could activate NK cells while enhancing Th1-mediated immune responses and increasing the production of IFN-γ in tumors, thus eliciting antitumoral immunity (Table 1).80 In parallel with L. acidophilus, L. plantarum and L. brevis were found to generate effective immune responses against tumors in breast cancer-bearing mice because administration of these bacteria cause increases of the production of IFN-γ, IL-2, and TNF-α by activating T cells and macrophages and causes increased cytotoxic activity of NK cells (Table 1).81,82 In this respect, tumoral upregulation of IFN-γ is believed to be a biomarker that can be used to predict the response of a tumor to ICI therapy. In this regard, despite the lack of valid evidence suggesting the synergistic effect of Lactobacillus in ICI therapy, it is reasonable to speculate that maintaining the gut frequency of Lactobacillus at a stable level assists in gut immune homeostasis, thereby minimizing the distortion of ICI drugs.
Bifidobacteria
Bifidobacteria are also commonly believed to be probiotics in the human gut. They are anaerobic bacteria that belong to the Actinobacteria phyla.83 In general, Bifidobacteria exert beneficial effects on immunomodulatory effects by using their own bacterial components and various immune-related metabolites, and they maintain immune homeostasis through a cross-feeding mechanism.84 Typically, different species of Bifidobacteria facilitate Th1 and Th2 polarization and CTL accumulation.52,85,86 In addition, bacterial metabolites also play an important role in stimulating immune function. Another study confirmed that Bifidobacterium longum BB536 has positive effects on the early establishment of healthy intestinal bacteria and plays a significant role in enhancing the Th1 immune response (Table 1).85 Additionally, Bifidobacterium dentium, as desirable mucus-layer builders, have been shown to enhance the intestinal mucus layer through autophagy and calcium signaling pathways.87
In an experimental model, oral administration of Bifidobacterium to melanoma-bearing mice was found to synergize with anti-PD-L1 therapy to induce tumor shrinkage.52 The underlying mechanism is indicated by Bifidobacterium administration enhancements of the antigen-presentation function of DCs, thus facilitating CD8+ T cell activation and infiltration into tumors (Table 1).52 In addition to exerting a synergistic effect, Bifidobacteria were found to reduce the toxicity of anti-CTLA-4 therapy by modulating the metabolism of Treg cells rather than altering Treg cell density in the tumors, thus not distorting the efficacy of anti-CTLA-4 therapy (Table 1).88 Upon integrating the above data, Bifidobacteria can be regarded as beneficial for ICI therapy.89
Akkermansia muciniphila
Akkermansia muciniphila (A. muciniphila) are bacteria that are initialized in the early life of humans.90 They are gram-negative and colonize in the outer layer of the mucin covering the intestinal epithelium,91 In the gut, cecum has the largest number of A. muciniphila. However, they only account for 1 ~ 4% of all commensal bacteria.90 Despite being a small portion overall, they are regarded as promising probiotics. At steady state, A. muciniphila are capable of renewing the mucus layer by degrading mucins, providing a route to strengthened intestinal barrier function.92 Alternatively, the bacterial component Amuc-1100 can interact with TLR2-positive cells, thereby enhancing intestinal barrier function by upregulating tight-junction-associated proteins.93 Upon epithelial damage, A. muciniphila preferentially localize in the wound to elicit proliferation and migration of enterocytes to this site.94 Once at the wound, A. muciniphila can also increase the levels of IFN-γ and TNF-α while decreasing IL-4 and IL-10 production (Table 1).95 In this regard, A. muciniphila exhibits the capability to improve the effect of ICI therapy (Table 1).21 In fact, basic research has confirmed that A. muciniphila can promote Th1 polarization.96,97 To show this effect, A. muciniphila was administered to germ-free mice bearing sarcoma to increase the Th1-related immune response significantly, thus enhancing the efficacy of anti-PD-1 therapy in causing tumor shrinkage.21 Moreover, cancer patients with a high number of A. muciniphila in the gut commonly show a better response to ICI drugs than those with a low number of A. muciniphila.21 Therefore, A. muciniphila can be regarded as an ICI therapy-favored bacterium.
Bacteroidetes
In the healthy human gut, gram-negative Bacteroidetes are universally distributed in the colon.98 Herein, Bacteroidetes fragilis (B. fragilis) are the most prominent bacteria and have long been considered pathogens in humans.99 In fact, they are capable of inducing immune tolerance of the gut.100 In parallel with tolerance induction, it has also been revealed that nontoxigenic B. fragilis can counteract enterotoxigenic B. fragilis to protect against colitis and tumorigenesis in the gut.101
In summary, Bacteroidetes are able to induce cell polarization, including Th1, Th17 and Treg cells, in the gut.102,103 In the context of the administration of Bacteroidetes, the efficacy of anti-CTLA-4 therapy was restored in germ-free mice bearing melanoma or colon cancer (Table 1).103 Mechanically, in germ-free mice, pure colonization of Bacteroidetes promoted the maturation of IL-12-producing DCs in tumors and induced a Th1-related immune response.89,103 However, melanoma patients with a high fecal Bacteroidetes count have poor clinical outcomes after receiving anti-CTLA-4 therapy (Table 1).22 This outcome is translated into anti-PD-1 therapy.20 The underlying mechanism involves a reduced level of MHC-II molecules and a higher number of Treg cells and Th17 cells in nonresponders to anti-PD-1 therapy than are presented by responders (Table 1).20 In this context, different species of Bacteroidetes can have different impacts on the efficacy of ICI therapy.
Fusobacterium nucleatum
Fusobacterium nucleatum (F. nucleatum) has been well established as pathogens in the induction of colorectal cancer.43,66 They can infect CRC cells to induce robust proliferation of CRC cells. Immunosuppression is another hallmark of CRC tumors after F. nucleatum infection. In general, in CRC specimens, it was revealed that the tumoral density of CD3+ T cells inversely correlated with F. nucleatum number (Table 1).104 This result can be attributed to T cell loss, which is driven by the interaction between bacterial Fap2 and the TIGIT inhibitory receptor on T cells.105 This outcome also translates to NK cells. Thus, it was found that exhaustion of NK cells occurs in early gut carcinogenesis.105 After the initiation of these processes, F. nucleatum facilitates M2-like TAM polarization via IL-6/c-MYC/STAT3 axis activation.106 In addition, F. nucleatum has been shown to enhance the accumulation of myeloid-derived suppressive cells in CRC tumors (Table 1).107 Thus, the immunosuppressive milieu for CRC tumors is thus formed. Generally, CRC patients with a high number of F. nucleatum in their tumors commonly have poor clinical outcomes than those without F. nucleatum infection. Referring to its impact on ICI therapy, it was found that the presence of F. nucleatum is inversely related to tumoricidal infiltrates, even in MSI-H tumors,108 thus implying that F. nucleatum represents an ICI therapy-unfavorable bacteria.
Concerns related to the prediction of ICI efficacy by testing fecal bacteria
Above information has exemplified some typical bacteria that can impact the immune milieu in tumors. Yet, there are some concerns still existing in predicting the response of cancer patients to ICI therapy by testing the composition of their fecal bacteria. For example, albeit collecting data from melanoma patients, two separate research groups reported all the bacteria including Bifidobacterium longum,19 Collinsella aerofaciens,19 Enterococcus faecium,19 Faecalibacterium genus bacteria20 were highly related to clinical response of patients to ICI therapy. Moreover, high fecal frequency of A. muciniphila was associated with well response of NSCLC to ICI therapy.21 Although the specific roles of aforementioned bacteria in improving tumoral immune milieu have been well characterized in corresponding animal models,19–21 an open question is emerged as which will be the most reliable bacteria in predicting or comparing the therapeutic efficacies of ICI drugs within a certain type of cancer, or among different cancers? Remarkably, a basic study has confirmed that a consortium of eleven strains of bacteria function jointly in eliciting CD8+ cell accumulation in gut, whereas such an effect will be abated if in absence of one or more certain strains of bacteria.26 Otherwise, it should be asked whether the mice can support the colonization of all bacterial species from human feces.11 In these situations, it is conceivable that the response to ICI therapy should not be merely attributed to one single major species of bacteria that increase their frequency in cancer patient feces. Probably, other bacteria will assist in this process albeit they do not significantly alter their frequencies in gut. Thus, we believe that defining a group of bacteria may be more precise in predicting ICI response than a certain type of bacteria does.
Another concern will be presented in the methodology. As we know, 16 S rRNA or metagenomic whole-genome shotgun sequencing can identify most of the abundant bacteria that impact the efficacy of ICI therapy.11 On this basis, we should note the rare bacteria in feces, such as those residing in the small intestine but with less frequencies in feces. As estimated, these bacteria may distort the efficacy of ICI therapy as well, but methods concerning culture, isolation, identification and functional testing for these bacteria are technically difficult.11 Thus, more advanced methods should be developed to fulfill this aim. As we have exemplified the bacteria that can differ their roles in priming immune cells even if they are from the same taxonomy; so in our opinion, more deep sequencing or identification work should be done in the future. For example, a basic study has revealed that the non-toxigenic strain and the enterotoxigenic strain of B. fragilis exhibit opposite effects on gut tumor progression.101 Collectively, all these concerns will provide new sparks for future research in this field.
Conclusion
Gut commensal bacteria can certainly influence the therapeutic effects of ICI drugs. Due to the predominance of gut bacteria in controlling host immune integrity, maintaining homeostasis of gut commensal bacteria appears to be the basis of ensuring the effectiveness of ICI therapy. Moreover, due to the certainty that some commensal bacteria impact the effectiveness of ICI therapy and are predictive of the response to ICI therapy in cancer patients, the composition of these bacteria provides context in the field of oncotherapy.
Funding Statement
This study is mainly supported by National Natural Science Foundation of China (Grant No. 81874254), and partially by National Natural Science Foundation of China (Grant No. 81773353) and Jilin Scientific and Technological Development Program (Grant No. 20190201204JC).
Disclosure of Potential Conflicts of Interest
Disclosure of Potential Conflicts of Interest
Contributions
PX and CP wrote this paper; PX and GC prepared the figures and tables of this review; CP and DL conceived the topic of this review, and CP designed the logic flow.
References
- 1.Xin Yu J, Hubbard-Lucey VM, Tang J.. Immuno-oncology drug development goes global. Nat Rev Drug Discov. 2019;18(12):899–900. PMID: 31780841. doi: 10.1038/d41573-019-00167-9. [DOI] [PubMed] [Google Scholar]
- 2.Tang J, Yu JX, Hubbard-Lucey VM, Neftelinov ST, Hodge JP, Lin Y. Trial watch: the clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat Rev Drug Discov. 2018;17(12):854–855. PMID: 30482962. doi: 10.1038/nrd.2018.210. [DOI] [PubMed] [Google Scholar]
- 3.Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, Lao CD, Schadendorf D, Wagstaff J, Dummer R, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19(11):1480–1492. PMID: 30361170. doi: 10.1016/s1470-2045(18)30700-9. [DOI] [PubMed] [Google Scholar]
- 4.Tunger A, Sommer U, Wehner R, Kubasch AS, Grimm MO, Bachmann MP, Platzbecker U, Bornhäuser M, Baretton G, Schmitz M. The evolving landscape of biomarkers for anti-PD-1 or anti-PD-L1 therapy. J Clin Med. 2019;8(10):1534. PMID: 31557787. doi: 10.3390/jcm8101534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24(1):26. PMID: 28376884. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters S, Gettinger S, Johnson ML, Janne PA, Garassino MC, Christoph D, Toh CK, Rizvi NA, Chaft JE, Carcereny Costa E, et al. Phase II trial of atezolizumab as first-line or subsequent therapy for patients with programmed death-ligand 1-selected advanced non-small-cell lung cancer (BIRCH). J Clin Oncol. 2017;35(24):2781–2789. PMID: 28609226. doi: 10.1200/jco.2016.71.9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. PMID: 29658845. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. PMID: 28734759. doi: 10.1016/s1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753–3758. PMID: 30787022. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- 10.Cyriac G, Gandhi L. Emerging biomarkers for immune checkpoint inhibition in lung cancer. Semin Cancer Biol. 2018;52:269–277. PMID: 29782924. doi: 10.1016/j.semcancer.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359(6382):1366–1370. PMID: 29567708. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 12.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. PMID: 30755690. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wistuba-Hamprecht K, Martens A, Heubach F, Romano E, Geukes Foppen M, Yuan J, Postow M, Wong P, Mallardo D, Schilling B, et al. Peripheral CD8 effector-memory type 1 T-cells correlate with outcome in ipilimumab-treated stage IV melanoma patients. Eur J Cancer. 2017;73:61–70. PMID: 28167454. doi: 10.1016/j.ejca.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daud AI, Loo K, Pauli ML, Sanchez-Rodriguez R, Sandoval PM, Taravati K, Tsai K, Nosrati A, Nardo L, Alvarado MD, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest. 2016;126(9):3447–3452. PMID: 27525433. doi: 10.1172/jci87324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balatoni T, Mohos A, Papp E, Sebestyen T, Liszkay G, Olah J, Varga A, Lengyel Z, Emri G, Gaudi I, et al. Tumor-infiltrating immune cells as potential biomarkers predicting response to treatment and survival in patients with metastatic melanoma receiving ipilimumab therapy. Cancer Immunol Immunother. 2018;67(1):141–151. PMID: 28988380. doi: 10.1007/s00262-017-2072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens B. Gut bacteria link to immunotherapy sparks interest. Nat Biotechnol. 2018;36(2):121. PMID: 29406499. doi: 10.1038/nbt0218-121. [DOI] [PubMed] [Google Scholar]
- 17.Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–130. PMID: 28625867. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. PMID: 28260787. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 19.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. PMID: 29302014. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. PMID: 29097493. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Routy B, Le Chatelier E, Derosa L, Duong CP, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. PMID: 29097494. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 22.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. PMID: 28368458. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 23.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7(1):10391. PMID: 26837003. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol. 2019;14(8):1378–1389. PMID: 31026576. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, Long H, Plodkowski AJ, Arbour KC, Chaft JE, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29(6):1437–1444. PMID: 29617710. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. PMID: 30675064. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 27.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. PMID: 27541692. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. PMID: 20664075. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 29.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27(4):626–638. PMID: 28167665. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Front Microbiol. 2011;2:93. PMID: 21747801. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinken A, Thiele I, Drake HL. Anoxic conditions promote species-specific mutualism between gut microbes in silico. Appl Environ Microbiol. 2015;81(12):4049–4061. PMID: 25841013. doi: 10.1128/AEM.00101-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362(6418):eaat9076. PMID: 30498100. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551(7681):507–511. PMID: 29143816. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 34.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Walter J, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25(6):789–802.e5. PMID: 31194939. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, Ochman H. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A. 2014;111(46):16431–16435. PMID: 25368157. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. PMID: 24566914. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 37.Cabinian A, Sinsimer D, Tang M, Jang Y, Choi B, Laouar Y, Laouar A. Gut symbiotic microbes imprint intestinal immune cells with the innate receptor SLAMF4 which contributes to gut immune protection against enteric pathogens. Gut. 2018;67(5):847–859. PMID: 28341747. doi: 10.1136/gutjnl-2016-313214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352(6285):535–538. PMID: 27126035. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, Isoherranen N, Vaishnava S. Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate Interleukin-22 activity and prevent microbial dysbiosis. Immunity. 2018;49(6):1103–15.e6. PMID: 30566883. doi: 10.1016/j.immuni.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159(2):122–127. PMID: 26141651. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Round JL, Palm NW. Causal effects of the microbiota on immune-mediated diseases. Sci Immunol. 2018;3(20):eaao1603. PMID: 29440265. doi: 10.1126/sciimmunol.aao1603. [DOI] [PubMed] [Google Scholar]
- 42.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–1911. PMID: 24316595. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah MA. Gastric cancer: the gastric microbiota - bacterial diversity and implications. Nat Rev Gastroenterol Hepatol. 2017;14(12):692–693. PMID: 29042691. doi: 10.1038/nrgastro.2017.140. [DOI] [PubMed] [Google Scholar]
- 44.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35:8–15. PMID: 27883933. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Brown EM, Kenny DJ, Xavier RJ. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37(1):599–624. PMID: 31026411. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 46.Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 2019. PMID: 31740744. doi: 10.1038/s41385-019-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko HJ, Chang SY. Regulation of intestinal immune system by dendritic cells. Immune Netw. 2015;15(1):1–8. PMID: 25713503. doi: 10.4110/in.2015.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua Y, Yang Y, Sun S, Lwanowycz S, Westwater C, Reizis B, Li Z, Liu B. Gut homeostasis and regulatory T cell induction depend on molecular chaperone gp96 in CD11c cells. Sci Rep. 2017;7(1):2171. PMID: 28526855. doi: 10.1038/s41598-017-02415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandolfi F, Altamura S, Frosali S, Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin Ther. 2016;38(5):1017–1028. PMID: 27021609. doi: 10.1016/j.clinthera.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 50.Kalinski P, Muthuswamy R, Urban J. Dendritic cells in cancer immunotherapy: vaccines and combination immunotherapies. Expert Rev Vaccines. 2013;12(3):285–295. PMID: 23496668. doi: 10.1586/erv.13.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zegarra-Ruiz DF, El Beidaq A, Iniguez AJ, Lubrano Di Ricco M, Manfredo Vieira S, Ruff WE, Mubiru D, Fine RL, Sterpka J, Greiling TM, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25(1):113–27.e6. PMID: 30581114. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. PMID: 26541606. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Tian Z. Gut-liver axis: gut microbiota in shaping hepatic innate immunity. Sci China Life Sci. 2017;60(11):1191–1196. PMID: 28840534. doi: 10.1007/s11427-017-9128-3. [DOI] [PubMed] [Google Scholar]
- 54.Komban RJ, Stromberg A, Biram A, Cervin J, Lebrero-Fernandez C, Mabbott N, Yrlid U, Shulman Z, Bemark M, Lycke N. Activated Peyer’s patch B cells sample antigen directly from M cells in the subepithelial dome. Nat Commun. 2019;10(1):2423. PMID: 31160559. doi: 10.1038/s41467-019-10144-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity. 2019;50(4):1099–114.e10. PMID: 30876876. doi: 10.1016/j.immuni.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marinelli L, Tenore GC, Novellino E. Probiotic species in the modulation of the anticancer immune response. Semin Cancer Biol. 2017;46:182–190. PMID: 28844794. doi: 10.1016/j.semcancer.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, Shi F, Gao J, Fu XL. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J Cell Physiol. 2019;234(10):17023–17049. PMID: 30888065. doi: 10.1002/jcp.28436. [DOI] [PubMed] [Google Scholar]
- 58.Cani PD. Microbiota and metabolites in metabolic diseases. Nat Rev Endocrinol. 2019;15(2):69–70. PMID: 30602737. doi: 10.1038/s41574-018-0143-9. [DOI] [PubMed] [Google Scholar]
- 59.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. PMID: 27231050. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng HY, Ning MX, Chen DK, Ma WT. Interactions between the gut microbiota and the host innate immune response against pathogens. Front Immunol. 2019;10:607. PMID: 30984184. doi: 10.3389/fimmu.2019.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. Circulating and tissue-resident CD4 T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153(5):1320–37.e16. PMID: 28782508. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayor PC, Eng KH, Singel KL, Abrams SI, Odunsi K, Moysich KB, Fuleihan R, Garabedian E, Lugar P, Ochs HD, et al. Cancer in primary immunodeficiency diseases: cancer incidence in the United States immune deficiency network registry. J Allergy Clin Immunol. 2018;141(3):1028–1035. PMID: 28606585. doi: 10.1016/j.jaci.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16(3):151–167. PMID: 30523282. doi: 10.1038/s41571-018-0142-8. [DOI] [PubMed] [Google Scholar]
- 64.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806.e12. PMID: 31398337. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6(1):8727. PMID: 26515465. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. PMID: 30546113. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8(1):158. PMID: 29317709. doi: 10.1038/s41598-017-18596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Powell SE, Betel D, Shah MA. The gastric microbiome and its influence on gastric carcinogenesis: current knowledge and ongoing research. Hematol Oncol Clin North Am. 2017;31(3):389–408. PMID: 28501083. doi: 10.1016/j.hoc.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Zhang C, Cleveland K, Schnoll-Sussman F, McClure B, Bigg M, Thakkar P, Schultz N, Shah MA, Betel D. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol. 2015;16(1):265. PMID: 26614063. doi: 10.1186/s13059-015-0821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Riquelme E, Maitra A, McAllister F. Immunotherapy for pancreatic cancer: more than just a gut feeling. Cancer Discov. 2018;8(4):386–388. PMID: 29610286. doi: 10.1158/2159-8290.Cd-18-0123. [DOI] [PubMed] [Google Scholar]
- 71.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. PMID: 29567829. doi: 10.1158/2159-8290.Cd-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41–47. PMID: 30684593. doi: 10.1016/j.canlet.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 73.Mu Q, Tavella VJ, Luo XM. Role of lactobacillus reuteri in human health and diseases. Front Microbiol. 2018;9:757. PMID: 29725324. doi: 10.3389/fmicb.2018.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, Luo M, Tran DQ, Zhou J, Tatevian N, et al. Resetting microbiota by lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med. 2017;214(1):107–123. PMID: 27994068. doi: 10.1084/jem.20160961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao PH, Kuo WW, Hsieh DJ, Yeh YL, Day CH, Chen YH, Chang SH, Padma VV, Chen YH, Huang CY. Heat-killed lactobacillus reuteri GMNL-263 prevents epididymal fat accumulation and cardiac injury in high-calorie diet-fed rats. Int J Med Sci. 2016;13(8):569–577. PMID: 27499689. doi: 10.7150/ijms.15597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, et al. Lactobacillus reuteriinduces gut intraepithelial CD4+CD8αα+T cells. Science. 2017;357(6353):806–810. PMID: 28775213. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. PMID: 23973224. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 78.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210(5):917–931. PMID: 23589566. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacouton E, Chain F, Sokol H, Langella P, Bermudez-Humaran LG. Probiotic strain lactobacillus casei BL23 prevents colitis-associated colorectal cancer. Front Immunol. 2017;8:1553. PMID: 29209314. doi: 10.3389/fimmu.2017.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maroof H, Hassan ZM, Mobarez AM, Mohamadabadi MA. Lactobacillus acidophilus could modulate the immune response against breast cancer in murine model. J Clin Immunol. 2012;32(6):1353–1359. PMID: 22711009. doi: 10.1007/s10875-012-9708-x. [DOI] [PubMed] [Google Scholar]
- 81.Yazdi MH, Mahdavi M, Kheradmand E, Shahverdi AR. The preventive oral supplementation of a selenium nanoparticle-enriched probiotic increases the immune response and lifespan of 4T1 breast cancer bearing mice. Arzneimittelforschung. 2012;62(11):525–531. PMID: 22945771. doi: 10.1055/s-0032-1323700. [DOI] [PubMed] [Google Scholar]
- 82.Yazdi MH, Mahdavi M, Setayesh N, Esfandyar M, Shahverdi AR. Selenium nanoparticle-enriched lactobacillus brevis causes more efficient immune responses in vivo and reduces the liver metastasis in metastatic form of mouse breast cancer. Daru. 2013;21(1):33. PMID: 23631392. doi: 10.1186/2008-2231-21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. PMID: 27446020. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruiz L, Delgado S, Ruas-Madiedo P, Sanchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. 2017;8:2345. PMID: 29255450. doi: 10.3389/fmicb.2017.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu BB, Yang Y, Xu X, Wang WP. Effects of Bifidobacterium supplementation on intestinal microbiota composition and the immune response in healthy infants. World J Pediatr. 2016;12(2):177–182. PMID: 25846071. doi: 10.1007/s12519-015-0025-3. [DOI] [PubMed] [Google Scholar]
- 86.Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. 2016;113(50):E8141–e50. PMID: 27911839. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Engevik MA, Luk B, Chang-Graham AL, Hall A, Herrmann B, Ruan W, Endres BT, Shi Z, Garey KW, Hyser JM, et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. MBio. 2019;10(3). PMID: 31213556. doi: 10.1128/mBio.01087-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang F, Yin Q, Chen L, Davis MM. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc Natl Acad Sci U S A. 2018;115(1):157–161. PMID: 29255057. doi: 10.1073/pnas.1712901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15(8):465–478. PMID: 28529325. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 90.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74(5):1646–1648. PMID: 18083887. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Derrien M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(5):1469–1476. PMID: 15388697. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 92.Kosciow K, Deppenmeier U. Characterization of a phospholipid-regulated β-galactosidase from Akkermansia muciniphila involved in mucin degradation. Microbiologyopen. 2019;8:e796. PMID: 30729732. doi: 10.1002/mbo3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. PMID: 27892954. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 94.Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, Jones RM, Nusrat A, Neish AS. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016;1(2):15021. PMID: 27571978. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Chin J Nat Med. 2019;17:11. PMID: 31006995. doi: 10.1016/S1875-5364(19)30101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114(40):10713–10718. PMID: 28893978. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8(1):80–93. PMID: 24917457. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wexler AG, Goodman AL. An insider’s perspective: bacteroides as a window into the microbiome. Nat Microbiol. 2017;2(5):17026. PMID: 28440278. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rocha ER, Smith CJ. Ferritin-like family proteins in the anaerobe bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals. 2013;26(4):577–591. PMID: 23842847. doi: 10.1007/s10534-013-9650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. PMID: 27133167. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chan JL, Wu S, Geis AL, Chan GV, Gomes TAM, Beck SE, Wu X, Fan H, Tam AJ, Chung L, et al. Non-toxigenic bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019;12(1):164–177. PMID: 30279518. doi: 10.1038/s41385-018-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. PMID: 18854238. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. PMID: 26541610. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1(5):653–661. PMID: 26181352. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. PMID: 25680274. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen T, Li Q, Wu J, Wu Y, Peng W, Li H, Wang J, Tang X, Peng Y, Fu X. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol Immunother. 2018;67(10):1635–1646. PMID: 30121899. doi: 10.1007/s00262-018-2233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. PMID: 23954159. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, Kosumi K, Masugi Y, Twombly TS, Cao Y, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. 2018;6(11):1327–1336. PMID: 30228205. doi: 10.1158/2326-6066.Cir-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lakritz JR, Poutahidis T, Levkovich T, Varian BJ, Ibrahim YM, Chatzigiagkos A, Mirabal S, Alm EJ, Erdman SE. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int J Cancer. 2014;135(3):529–540. PMID: 24382758. doi: 10.1002/ijc.28702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lenoir M, Del Carmen S, Cortes-Perez NG, Lozano-Ojalvo D, Munoz-Provencio D, Chain F, Langella P, de Moreno de LeBlanc A, LeBlanc JG, Bermudez-Humaran LG. Lactobacillus casei BL23 regulates Treg and Th17 T-cell populations and reduces DMH-associated colorectal cancer. J Gastroenterol. 2016;51(9):862–873. PMID: 26749362. doi: 10.1007/s00535-015-1158-9. [DOI] [PubMed] [Google Scholar]
- 111.Shen X, Liu L, Peek RM, Acra SA, Moore DJ, Wilson KT, He F, Polk DB, Yan F. Supplementation of p40, a lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes. Mucosal Immunol. 2018;11(5):1316–1328. PMID: 29875401. doi: 10.1038/s41385-018-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Makioka Y, Tsukahara T, Ijichi T, Inoue R. Oral supplementation of bifidobacterium longum strain BR-108 alters cecal microbiota by stimulating gut immune system in mice irrespectively of viability. Biosci Biotechnol Biochem. 2018;82(7):1180–1187. PMID: 29557273. doi: 10.1080/09168451.2018.1451738. [DOI] [PubMed] [Google Scholar]
- 113.Laparra JM, Olivares M, Gallina O, Sanz Y, Leulier F. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One. 2012;7(2):e30744. PMID: 22348021. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]


