Abstract
Fig4 is a phosphoinositide phosphatase that converts PI3,5P2 to PI3P. Paradoxically, mutation of Fig4 results in lower PI3,5P2, indicating that Fig4 is also required for PI3,5P2 production. Fig4 promotes elevation of PI3,5P2, in part, through stabilization of a protein complex that includes its opposing lipid kinase, Fab1, and the scaffold protein Vac14. Here we show that multiple regions of Fig4 contribute to its roles in the elevation of PI3,5P2: its catalytic site, an N-terminal disease-related surface, and a C-terminal region. We show that mutation of the Fig4 catalytic site enhances the formation of the Fab1-Vac14-Fig4 complex, and reduces the ability to elevate PI3,5P2. This suggests that independent of its lipid phosphatase function, the active site plays a role in the Fab1-Vac14-Fig4 complex. We also show that the N-terminal disease-related surface contributes to the elevation of PI3,5P2 and promotes Fig4 association with Vac14 in a manner that requires the Fig4 C-terminus. We find that the Fig4 C-terminus alone interacts with Vac14 in vivo and retains some functions of full-length Fig4. Thus, a subset of Fig4 functions are independent of its phosphatase domain and at least three regions of Fig4 play roles in the function of the Fab1-Vac14-Fig4 complex.
INTRODUCTION
Fig4, a Sac1-related phosphoinositide phosphatase (PI-PTPase), belongs to a diverse family of cysteine-dependent phosphatases that includes protein tyrosine phosphatases. These enzymes are highly divergent in sequence, but share an essential catalytic cysteine within a core CX5R catalytic motif (Alonso et al., 2016). PI-PTPases modulate signaling cascades in response to stimuli through localized dephosphorylation of phosphoinositide lipids on specific membrane domains (Vicinanza et al., 2008; Waugh, 2015). Mutations in PI-PTPases are associated with diseases including cancer and neurodegeneration (Liu and Bankaitis, 2010; Pulido et al., 2013; Billcliff and Lowe, 2014; Waugh, 2015). A common assumption is that pathologies caused by mutations in PI-PTPases arise from failure to dephosphorylate lipid substrates. However, catalytically impaired mutants of cysteine-dependent PI-PTPases frequently exhibit phenotypes distinct from their null-mutations, which suggests that these enzymes possess functions independent of their lipid phosphatase activity (Amoasii et al., 2012; Krebs et al., 2013; Quadri et al., 2013; Dong et al., 2015; Bharadwaj et al., 2016; Lenk et al., 2016).
Fig4 is a phosphatidylinositol 3,5-bisphosphate (PI3,5P2) 5-phosphatase (Rudge et al., 2004) and is composed of an N-terminal Sac1-type phosphatase domain and a C-terminal domain specific to Fig4. Fig4 is proposed to be the primary enzyme responsible for the turnover of PI3,5P2 (Gary et al., 2002; Rudge et al., 2004; Duex et al., 2006a). Thus, loss of this enzyme is predicted to elevate levels of PI3,5P2. However, knockout of Fig4 paradoxically leads to lower levels of PI3,5P2 in yeast (Duex et al., 2006a) and mammals (Chow et al., 2007). Moreover, Fig4 has a positive role in the hyperosmotic shock–induced increase in PI3,5P2 (Duex et al., 2006a). Consistent with a conserved role for Fig4 in PI3,5P2 synthesis, the loss of Fig4 results in enlarged lysosomes/vacuoles in yeast, flies, and mammals (Rudge et al., 2004; Duex et al., 2006b; Chow et al., 2007; Bharadwaj et al., 2016), a phenotype associated with lower than normal levels of PI3,5P2 (Gary et al., 1998; Ikonomov et al., 2001; Rudge et al., 2004; Nicot et al., 2006). Notably, mutations in yeast Fig4 that correspond to human Fig4 mutations associated with Charcot–Marie–Tooth syndrome Type 4J (CMT4J) and amyotrophic lateral sclerosis (ALS) result in defects in the stimulus-induced elevation of PI3,5P2 (Chow et al., 2007 , 2009). These findings suggest that mutations in Fig4 that underlie some neurological diseases impair Fig4-dependent elevation of PI3,5P2.
Fig4 is proposed to enhance PI3,5P2 synthesis independent of its lipid phosphatase function through stabilization of the protein complex harboring Fab1/PIKfyve (Sbrissa et al., 2007, 2008; Botelho et al., 2008; Jin et al., 2008), the sole enzyme responsible for PI3,5P2 production (Gary et al., 1998; Ikonomov et al., 2001; Nicot et al., 2006; Rusten et al., 2006). This is supported by findings that catalytically impaired Fig4 rescues the enlarged lysosome phenotype observed in Fig4 knockout flies (Bharadwaj et al., 2016) and partially restores osmotic shock–induced elevation of PI3,5P2 in yeast (Duex et al., 2006b). Catalytically impaired Fig4 also partially rescues neonatal abnormalities in Fig4 null mice (Lenk et al., 2016). The failure of catalytically impaired Fig4 to fully support elevation of PI3,5P2 following hyperosmotic shock in yeast (Duex et al., 2006b) suggests that the Fig4 active site may contribute to Fab1 activation.
Here we show that formation of the yeast Fab1-Vac14-Fig4 complex is modulated by at least three regions of Fig4: the Fig4 catalytic site, a conserved N-terminal surface mutated in some neurological diseases, and the C-terminus. Mutation of the catalytic cysteine of yeast or mammalian Fig4 enhances formation of the Fab1/PIKfyve-Vac14-Fig4 complex in both yeast and human cells. In addition, we show that mutation of a disease-related N-terminal surface impairs the ability of Fig4 to interact with Vac14. Furthermore, we show that expression of the Fig4 C-terminus alone supports some in vivo functions of Fig4 and that this region interacts with Vac14 in yeast lysates. Moreover, the N-terminus promotes Fig4 association with Vac14 in a manner that also requires the Fig4 C-terminus. Together, the presence of three contact sites in Fig4 raises the possibility that the catalytic region in coordination with the N- and C-termini changes the conformation of Fig4, which in turn regulates the assembly and/or activity of the Fab1-Vac14-Fig4 complex.
RESULTS AND DISCUSSION
The Fig4 phosphatase active site is required for full activation of Fab1
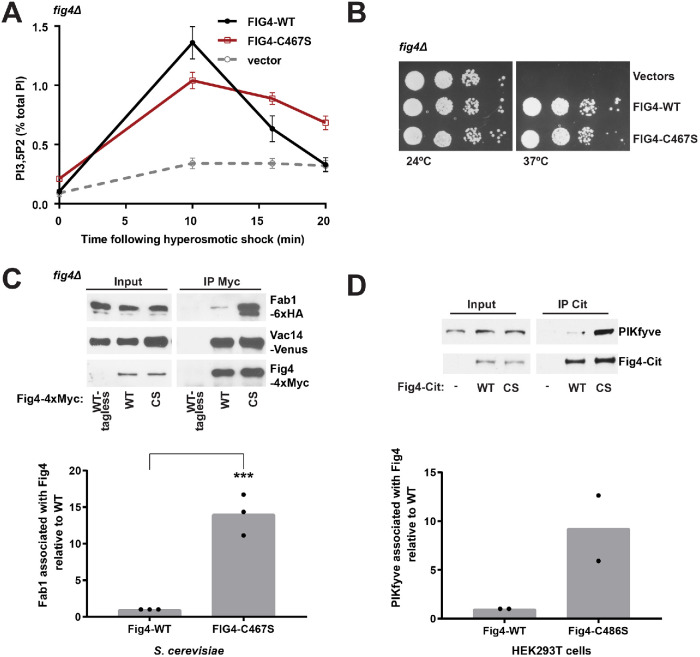
To investigate a potential role of Fig4 catalytic activity in Fab1 activation, we substituted the catalytic cysteine for serine (Fig4-C467S) to impair Fig4 catalytic function (Flint et al., 1997; Rohde et al., 2003; Berger et al., 2006). Notably, Fig4-C467S displays a 33% decrease in the elevation of PI3,5P2 following hyperosmotic shock (Figure 1A). This suggests that the Fig4 catalytic cysteine is necessary for the full activation of Fab1. As previously shown (Rudge et al., 2004), Fig4 functions as a lipid phosphatase. In further support of this hypothesis, expression of Fig4-C467S as the sole copy of Fig4 in a fig4Δ yeast strain resulted in a twofold elevation of basal PI3,5P2 levels relative to wild type (Fig4-WT) and an impaired rate of PI3,5P2 turnover following hyperosmotic shock (Figure 1A).
FIGURE 1:
Catalytically impaired Fig4 supports partial Fig4 function. (A) PI3,5P2 was analyzed by HPLC from fig4Δ cells labeled with [3H]inositol. NaCl (0.9 M) was added to the media at 0 time. The hyperosmotic shock–induced rise in PI3,5P2 (10 min levels minus basal levels) is decreased by 33 ± 1% in Fig4-C467S compared with Fig4-WT (p = 0.0022, two-tailed t test). Basal levels of PI3,5P2 are 2.1± 0.4-fold higher in Fig4-C467S relative to wild type (p = 0.0082, two-tailed t test). Mean of four independent experiments. Error bars: SD. (B) Catalytically impaired Fig4 substitutes for wild-type Fig4 to support growth at 37°C in a fig4Δ Vac14-Venus Fab1-TAP strain. Cells expressing no Fig4, Fig4-wild-type (WT), or catalytically impaired Fig4 (C467S) from a plasmid were grown on selective SC–leu plates at 24° and 37°C. (C) Western blot of proteins immunoprecipitated with anti-Myc antibody from a fig4Δ strain expressing Fig4-4xMyc wild-type (WT-Myc), C467S (CS-Myc), or untagged Fig4 wild-type (WT-tagless) from a plasmid with native 5′ and 3′ UTR. Vac14-Venus and Fab1-6xHA expressed from endogenous loci. (D) Western blot of proteins immunoprecipitated with anti-Citrine antibody from HEK293T cells transfected with Fig4-Citrine, Fig4-C486S-Citrine, or Citrine alone. Bar graphs show quantification of Western blots in C and D. In yeast, Fab1 association with Fig4 C467S is 14.06 ± 2.81 times higher than wild-type Fig4, normalized to Fig4. Data points and mean of three independent experiments (** p = 0.0006, two-tailed t test). In HEK293T cells, PIKfyve association with Fig4 C486S is 9.27 ± 4.75-fold higher relative to wild-type Fig4. Data points and mean from two independent experiments.
The Fig4-C467S mutant does not impair yeast viability, which indicates that some functions of Fig4 do not require catalytic activity. In a yeast strain where Vac14 and Fab1 are C-terminally tagged (fig4Δ Vac14-Venus Fab1-TAP), Fig4 expression is required for growth at temperatures higher than 30°C. Using this strain, we find that expression of Fig4-C467S supports growth similarly to WT (Figure 1B). This fits with studies in mice (Lenk et al., 2016) and Drosophila (Bharadwaj et al., 2016), where catalytically impaired Fig4 partially rescues Fig4 knockout mutants.The Fig4 active site modulates its association with Fab1.
The defect in Fab1 activation due to mutation of the Fig4 catalytic cysteine could result from partial misfolding and impaired association with the Fab1-Vac14-Fig4 complex. To test this possibility, we performed pull-down experiments of Fig4 in yeast expressing Fig4-WT versus Fig4-C467S. Fig4-C467S is expressed at levels similar to those in wild-type Fig4, yet strikingly, the Fig4-C467S mutant resulted in an ∼14-fold increase in Fab1 association with Fig4 (Figure 1C). Two other catalytically impaired mutants, Fig4-D469A and Fig4-Y538A, do not show the same degree of enhanced binding (Supplemental Figure S1, A–D). This implies that enhanced association of Fig4-C467S with Fab1 is not a consequence of loss of Fig4 catalytic function but is directly mediated by contacts with the mutant Fig4-C467S active site. Association with Fab1 is not a result of altered localization of Fig4-C467S in yeast cells, as Fig4-C467S-Envy and Fig4-Y538A-Envy show similar localization (Supplemental Figure S1E). Moreover, Fab1-3xGFP shows similar localization in the presence of Fig4-WT, -C467S, or -Y538A (Supplemental Figure S1F). Some, but not all, mutations of catalytic residues in protein phosphatases lead to enhanced binding to their protein substrates (Flint et al., 1997; Blanchetot et al., 2005). It is possible that enhanced binding of Fig4 to Fab1 reflects protein targets of Fig4 phosphatase function within the Fab1-Vac14-Fig4 complex. Alternatively the Fig4 active site may modulate conformational rearrangements in the Fab1/PIKfyve-Vac14-Fig4 complex that alter Fab1 binding. Notably, enhanced binding of Fig4-C467S to Fab1 requires prior assembly of the Fab1-Vac14-Fig4 complex as binding of this mutant to Fab1 is not detected in the absence of Vac14 (Supplemental Figure S2). The mechanism of enhanced association of the Fig4-C467S mutant with Fab1 is likely conserved. In HEK293T cells that overexpress human Fig4 with a mutated catalytic cysteine, Fig4-C486S, we observed an approximately ninefold increase in association with the mammalian Fab1 homologue, PIKfyve (Figure 1D).
A conserved, disease-related N-terminal surface of the Fig4 phosphatase domain modulates its association with Fab1 and Vac14
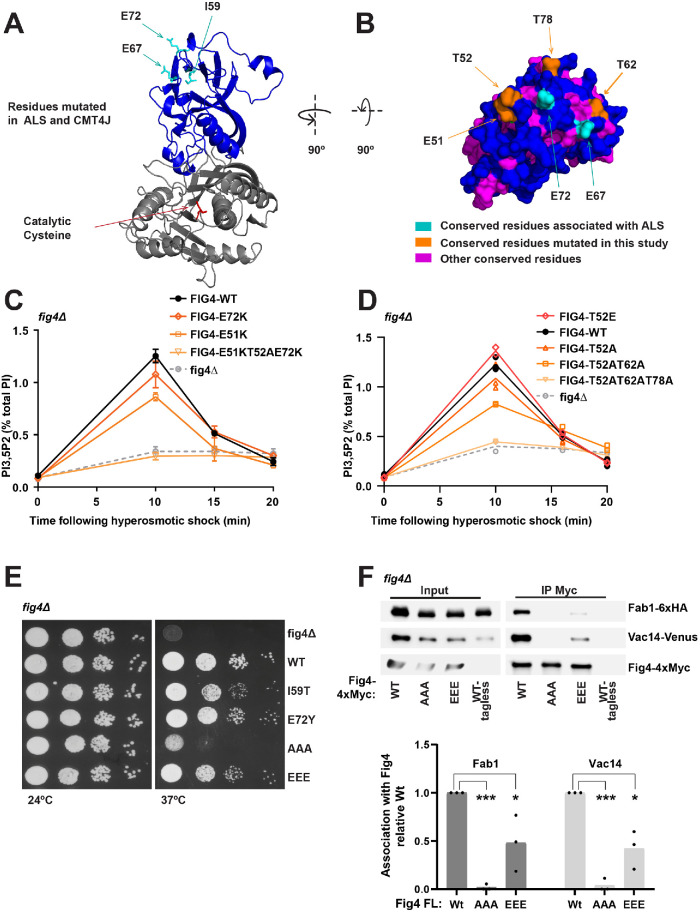
In addition to the catalytic site, we investigated the role of a conserved, disease-related surface of Fig4. Previous studies raised the possibility that this region may be critical to Fig4 function. The CMT4J disease mutation I49T (I59T in yeast) (Chow et al., 2007) is predicted to be buried within the Fig4 N-terminus (Manford et al., 2010; Lenk et al., 2011; Figure 2A). This mutation disrupts association of human Fig4 with human Vac14. In addition, yeast Fig4-I59T exhibits impaired interactions with yeast Vac14, as measured in yeast two-hybrid assays as well as in pull-down assays (Lenk et al., 2011). Importantly, yeast Fig4-I59T results in a >50% defect in elevation of PI3,5P2 following hyperosmotic shock (Chow et al., 2007). However, because the Fig4-I59T mutation is likely a buried residue, it is difficult to conclude whether the associated defects are due to overall misfolding of the protein, or whether they are due to some specific function of the nearby N-terminal surface.
FIGURE 2:
A conserved N-terminal surface of Fig4 is required for the activation of Fab1 and for Fig4 binding to Fab1 and Vac14. (A, B) Predicted structure of the Fig4 phosphatase domain (amino acids 32–529) using Phyre2 (Kelley et al., 2015), based on homology with Sac1 (Manford et al., 2010). (A) Cartoon representation of the N-terminal lobe (blue) and catalytic region (gray) of the Fig4 phosphatase domain. ALS and CMT4J mutations (cyan) and the catalytic cysteine (red) are shown as sticks. I59 corresponds to human CMT4J mutation I41T. E67 and E72 correspond to ALS mutations D48G and D53Y, respectively. (B) Surface representation of a top-down view of the predicted Fig4 N-terminal surface. (C, D) PI3,5P2 was analyzed by HPLC from cells labeled with [3H]inositol. NaCl (0.9 M) was added to the media at 0 time. (C) Mean of three independent experiments. The 10 min level decreased relative to WT in E51K (p = 0.0008, two-tailed t test) and T52AE51KE72K (p = 0.0003, two-tailed t test). The change was not statistically significant for E72K (p = 0.0986, two-tailed t test). (D) Mean of two independent experiments with individual data points shown. (E) Growth defects in Fig4 mutants in fig4Δ Vac14-Venus Fab1-TAP at 37°C correlate with corresponding defects in elevation of PI3,5P2. Cells expressing no Fig4, Fig4-wild-type (WT), or indicated Fig4 mutants from a plasmid were grown on selective Sc–leu plates at 24° and 37°C. (F) Western blot of proteins immunoprecipitated with anti-Myc antibody from a fig4Δ strain expressing Fig4-4xMyc from a plasmid with native 5′ and 3′ UTR, and Vac14-Venus and Fab1-6xHA expressed from their corresponding endogenous loci. Bar graph shows quantification of band densities in Western blot, relative to wild type, normalized to Fig4-Myc, with data points and mean from three independent experiments (* p < 0.01, *** p < 0.0001, by two-tailed t test). Fig4 mutants analyzed: E72K, E72Y, E51K, triple mutant T52AE51KE72K, T52E, T52A, double mutant T52AT62A, triple mutant T52ET62ET72E (EEE), and triple mutant T52AT62AT72A (AAA).
We hypothesized that the ALS-related mutations D48G or D53Y and surrounding surface area would likely provide a better means to study the functional significance of the N-terminal surface. These mutations are on a predicted surface in close proximity to I49 (Manford et al., 2010; Cai et al., 2014). Mutation of the corresponding residues in yeast, E67G and E72Y, results in enlarged vacuole morphology, suggesting that these mutations are impaired in the activation of Fab1 (Chow et al., 2009).
Notably, using a structural model of Fig4 (Manford et al., 2010, and Figure 2A), we found that this N-terminal surface of Fig4 is highly conserved in Fig4 proteins from diverse species (Figure 2B). This surface includes several negatively charged residues, including E51 and the residues corresponding to the ALS mutations, E67 and E72. This surface also harbors three highly conserved threonines, T52, T62, and T78 (Figure 2B). Charge reversal mutations of acidic residues and alanine mutations of the threonines, singly or in combination (Figure 2, C and D), did not display significant defects in PI3,5P2 turnover, but resulted in partial defects in the elevation of PI3,5P2 following hyperosmotic shock. The triple mutants FIG4-E51KT52AE72K and FIG4-T52AT62AT78A have large defects in PI3,5P2 synthesis and turnover and phenocopy the defects in PI3,5P2 levels observed in a fig4Δ mutant, which suggests that this region is critical to formation of the Fab1-Vac14-Fig4 complex. Consistent with the importance of acidic residues on this surface, mutation of one of the threonines to glutamate (T52E) slightly enhanced Fab1 activation following hyperosmotic shock (Figure 2D). In addition, growth defects imparted by the disease-related mutations correlate with corresponding defects in PI3,5P2 elevation. FIG4-E72Y and FIG4-I59T result in a minor growth defect relative to WT at 37°C. In addition, whereas FIG4-T52ET62ET78E displays a minor growth defect, FIG4-T52AT62AT78A has a severe growth defect (Figure 2E). These data further support the hypothesis that the N-terminal surface of Fig4 plays a role in the activation of Fab1.
To test whether these conserved threonines on the N-terminal surface of Fig4 are regulated by phosphorylation, we performed affinity purification of FLAG-tagged Fig4 and subjected tryptic digests of copurified proteins to phosphopeptide enrichment by immobilized metal affinity chromatography. We did not detect phosphopeptides derived from the N-terminal domain of Fig4, despite the fact that many peptides from Fig4 were resolved (56% coverage of the total protein), including unmodified peptides covering T62 and T78 (Supplemental Table 1). This analysis does not rule out N-terminal regulation by phosphorylation—particularly at residues that could not be resolved by tryptic digest such as T52.
To test whether this N-terminal surface mediates binding of Fig4 to the Fab1-Vac14-Fig4 complex, we performed immunoprecipitation assays with a subset of the Fig4 point mutants (Figure 2F). The mutants were stably expressed. The triple mutant FIG4-T52AT62AT78A was severely impaired in its association with Vac14 and Fab1. The triple mutant FIG4-T52ET62ET78E was also partially deleterious to Fig4 association with Vac14 and Fab1. Together, these data indicate that the N-terminal surface of Fig4 is critical for the integrity of the Fab1-Vac14-Fig4 complex.
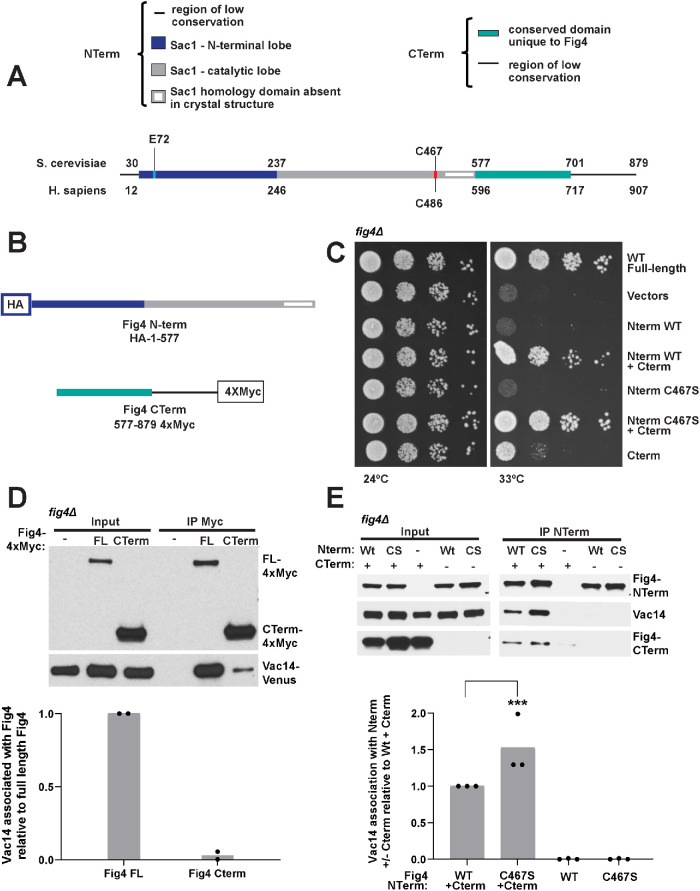
The Fig4 C-terminus mediates contact of the N-terminal phosphatase domain with the Fab1-Vac14-Fig4 complex
Fig4 is composed of two primary regions (Figure 3A), its N-terminal Sac1 phosphatase domain, which includes the catalytic site, and a C-terminal region specific to Fig4. To test the contributions of these domains to PI3,5P2 elevation independent of each other, we expressed Fig4 as two separate peptides: the N-terminal phosphatase domain (Fig4-Nterm, a.a. 1–577) and the C-terminus (Fig4-Cterm, a.a. 577–end[879]) (Figure 3B) and tested their ability to support Fig4-dependent growth. We found that the Fig4 C-terminus alone, but not the Fig4 N-terminus, partially restored growth. When coexpressed, the Fig4 N-terminus and Fig4 C-terminus complemented growth comparably to full-length Fig4 (Figure 3C). Together, these data indicate that the C-terminus of Fig4 retains some critical functions of the full-length protein, further supporting assertions that some Fig4 functions are entirely independent of its catalytic activity. Moreover, that the Fig4 N-terminus and Fig4 C-terminus support growth when expressed as separate polypeptides enabled us to perform further experiments to test the independent roles of these domains.
FIGURE 3:
Synergistic function of the Fig4 C- and N-termini depends on association of the Fig4 C-terminus with Vac14. (A) Linear representation of the Fig4 primary sequence. (B) N-terminal HA-tagged Fig4 phosphatase domain (Fig4-NTerm) (amino acids 1–577) and 4xMyc-tagged Fig4 C-terminal tail (Fig4-CTerm) (amino acids 577–879). (C) The Fig4-Cterm, but not the Fig4-Nterm, supports partial growth in fig4Δ Vac14-Venus Fab1-TAP at 33°C. The Fig4 Nterm contributes to and complements full Fig4 function only in the presence of the Fig4-Cterm. Cells expressing no Fig4, Fig4-wild-type (WT), or overexpressing the indicated Nterm and Cterm constructs from plasmids were grown on selective Sc–his–leu plates at 24° and 33°C. (D, E) Western blot of proteins immunoprecipitated with anti-Myc antibody (D) or anti-HA beads (E) from a fig4Δ strain expressing Vac14-Venus from its endogenous locus and expressing either no Fig4, Fig4-4xMyc from a plasmid with native 5′ and 3′ UTR, or overexpression (via an ADH promoter) of Fig4-Nterm and/or Fig4-CTerm. Bar graph shows quantification of band densities in Western blot (D) relative to full-length Fig4, normalized to Fig4-Myc, with data points and mean from two independent experiments and (E) relative to Nterm-WT+Cterm, normalized to NTerm, with data points and mean from three independent experiments (*** p < 0.0001, by two-tailed t test).
Fig4 binds Vac14 (Botelho et al., 2008; Ikonomov et al., 2009), and immunoprecipation studies with mammalian Fig4 and genetic analysis in yeast suggested that this interaction occurs through the Fig4 C-terminus (Botelho et al., 2008; Ikonomov et al., 2009). By immunoprecipitation, we found that the yeast Fig4 C-terminus binds Vac14 (Figure 3D). However, much less Vac14 is immunoprecipitated than is observed with full-length Fig4. On the basis of the observation that mutation of residues on the N-terminal surface disrupts Fig4 association with Vac14 (Figure 2F), we tested whether expression of the Fig4 N-terminal domain alone could pull down Vac14, but found that it could not (Figure 3E). However, when the Fig4 N-terminus and C-terminus were coexpressed, pulling down on the Fig4 N-terminus coprecipitated Vac14 (Figure 3E). These data suggest that when expressed as separate polypeptides, the Fig4 N-terminus interacts with either Vac14, Fab1 and/or the Fig4 C-terminus. Note that Fab1 is present in the yeast strains used in these pull-down studies and could be playing a role in the association of the Fig4 N-terminus with the C-terminus. They also suggest that, while the C-terminus is necessary and sufficient for Vac14 binding, in the context of full-length Fig4 the N-terminus contributes to binding interactions with the Fab1-Vac14-Fig4 complex.
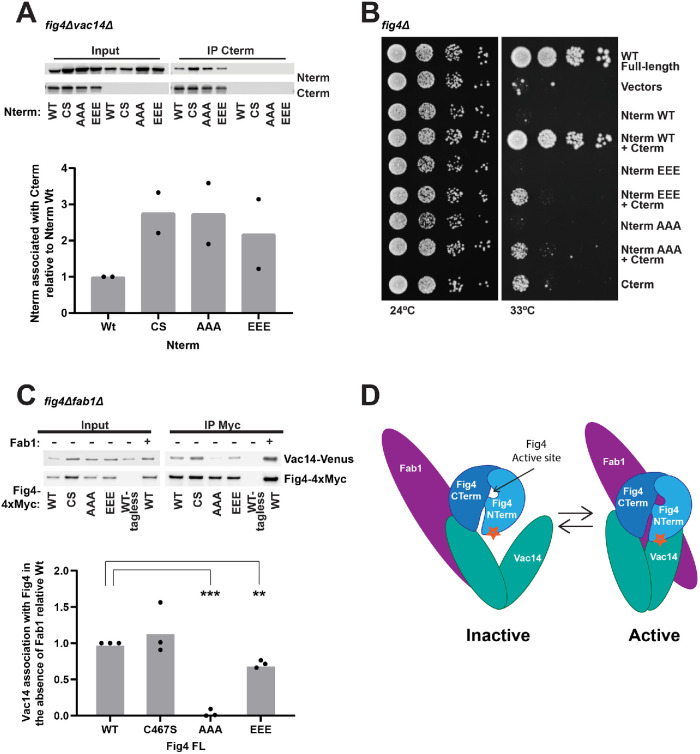
The Fig4 C-terminus interacts with the N-terminal phosphatase domain
To determine whether the Fig4 N-terminus binds the Fig4 C-terminus, we tested whether these regions stably interact in the absence of Vac14. Indeed, when expressed in fig4Δvac14Δ yeast, the Fig4 N-terminus coimmunoprecipitated with the Fig4 C-terminus (Figure 4A). Moreover, the N-terminal constructs mutated on the disease surface, T52AT62AT78A or T52ET62ET78E, retain the ability to bind the Fig4 C-terminus (Figure 4A).
FIGURE 4:
The Fig4 N-terminal and C-terminal domains mediate Fab1-Vac14-Fig4 complex assembly and function through contacts on Vac14. (A) Western blot of proteins immunoprecipitated with anti-Myc antibody from a fig4Δvac14Δ strain overexpressing the Fig4 C-terminus (CTerm-4xMyc) and/or the wild-type (WT), C467S (CS), T52AT62AT78A (AAA), or T52ET62ET78E (EEE) Fig4 phosphatase domain (HA-N-term) via an ADH promoter. Bar graph shows quantification of band densities in Western blot, relative to wild type, normalized to Cterm-Myc with data points and mean from two independent experiments. (B) In contrast to wild type, Fig4-Nterm AAA and Nterm-EEE do not support growth in the presence of the Fig4-Cterm in fig4Δ Vac14-Venus Fab1-TAP at 33°C. Cells expressing no Fig4, Fig4-wild-type (WT), or overexpressing the indicated Nterm and Cterm constructs from plasmids were grown on selective Sc–his–leu plates at 24° and 33°C. (C) Western blot of proteins immunoprecipitated with anti-Myc antibody from a fig4Δfab1Δ expressing Fig4-WT, Fig4-CS, Fig4-AAA, or Fig4-EEE. Vac14-Venus expressed from endogenous locus. Bar graph shows quantification of band densities in Western blot, relative to wild type, normalized to Fig4-Myc, with data points and mean from three independent experiments (*** p < 0.0001, ** p < 0.001, two-tailed t test). (D) Model for regulation of the Fab1-Vac14-Fig4 complex. The Fig4 C-terminus and an N-terminal surface (star) both contribute to its association with Vac14 within the Fab1-Vac14-Fig4 complex. The Fig4 catalytic site is also required for the full activation of Fab1.
To further test the mutations on the surface of the Fig4 N-terminus, we tested the ability of the T52AT62AT78A or T52ET62ET78E mutants to support yeast growth when coexpressed with the Fig4 C-terminus. However, we found that only the wild-type Fig4 N-terminus when coexpressed with the Fig4 C-terminus supported growth to the same extent as full-length wild-type Fig4 (Figure 4B). The Fig4 N-terminus with surface mutations made no further contribution to the ability of the Fig4 C-terminus alone to support yeast growth. This result was somewhat surprising given that full-length Fig4-T52ET62ET78E supported growth to the same extent as Fig4 wild type (Figure 2E). Thus, in the next study of the N-terminal surface, we utilized full-length Fig4.
The Fig4 N-terminal surface contributes to stable binding of Fig4 to Vac14
Fig4 binds Vac14 independent of Fab1 (Botelho et al., 2008; Ikonomov et al., 2009). To test whether the disease-related surface of Fig4 contributes to its binding to Vac14, we used immunoprecipitation assays and tested the ability of full-length Fig4 bearing N-terminal surface mutations to bind Vac14 in the absence of Fab1. Threonine to alanine mutations, FIG4-T52AT62AT78A, on the N-terminal disease-related surface impaired binding to Vac14 (Figure 4C). Mutation of these threonines to glutamate, FIG4-T52ET62ET78E, impaired binding to a lesser extent. These data suggest that, in the context of full-length Fig4, the disease-related N-terminal surface contributes to the interaction of Fig4 with Vac14. It is also possible that this surface contacts an as yet unidentified subunit in the complex.
Model for dynamic regulation of Fab1 by Fig4
Together, our data suggest a model (Figure 4D) whereby Fig4 contributes to the function of the Fab1-Vac14-Fig4 complex through both active site–dependent and active site–independent mechanisms. We show that three regions of Fig4 mediate the association and function of the Fab1-Vac14-Fig4 complex: the active site, an N-terminal disease-related surface, and the C-terminus. We find that the Fig4 active site influences the stability and function of the complex. Our data also suggest that Fig4 stabilizes the Fab1-Vac14-Fig4 complex through two contacts with Vac14, the disease-related N-terminal surface on Fig4 and the Fig4 C-terminus. Interactions involving the N-terminus are enhanced by mutation of the catalytic cysteine to serine (Figures 3E and 4A), suggesting that the active site also directly contacts the complex. Given that the Fig4 active site resides between two distal contact sites on Fig4, it is tempting to speculate that conformational changes that originate in the active site could impact the conformation of the Fab1-Vac14-Fig4 complex via their interactions with the Fig4 N-terminal disease-related surface and the C-terminus. These putative conformational changes may underlie the activation of Fab1 by Fig4.
There are several other examples of cysteine-dependent PI-PTPases that have both active site–dependent and –independent roles. In mice, catalytically dead mutants of the PI-PTPase Mtm1 partially rescue phenotypes caused by absence of the wild-type protein (Amoasii et al., 2012; Lenk et al., 2016). Moreover, the wild-type alleles of a number of physiologically important PI-PTPase–related enzymes, including almost half of the myotubularin family (Robinson and Dixon, 2006) as well as 10% of all cysteine-dependent protein tyrosine phosphatases, are predicted to be catalytically inactive because they lack the amino acids essential for phosphatase function in their otherwise conserved catalytic motifs (Moorhead et al., 2009).
PI-PTPases are activated and inactivated in response to specific stimuli (Yu and Zhang, 2018). Conformational changes associated with activation and inactivation have been demonstrated for PTEN (Raftopoulou et al., 2004; Odriozola et al., 2007; Rahdar et al., 2009; Bolduc et al., 2013; Masson et al., 2016) and are predicted for Sac1 (Manford et al., 2010; Zhong et al., 2012) and Mtm1 (Schaletzky et al., 2003). Moreover, catalytically impaired mutants have been shown to display altered noncatalytic behavior relative to the wild-type enzyme. For instance, inactive PTEN causes dominant negative phenotypes; mice bearing one wild-type and one phosphatase dead allele are more prone to tumor development than mice that have one wild-type and one null PTEN allele (Papa et al., 2014). In addition, Sac1 (Rohde et al., 2003; Papa et al., 2014; Yu et al., 2014; Dong et al., 2015) displays altered protein–protein interactions. Similarly, we find that the Fig4-C467S mutant exhibits an enhanced interaction with Fab1. Conformational changes within PI-PTPases may have the potential to influence physically associated proteins, analogous to regulatory GTPases (Clague et al., 2004). Together, these studies along with previously published findings reveal that PI-PTPases perform critical cellular functions that are independent of their catalytic activity, of which a subset likely involve conformational changes that are mediated via the PI-PTPase active site.
MATERIALS AND METHODS
Yeast strains and plasmids
Strains and plasmids used in this study are listed in Table 1. Yeast were grown in yeast extract–peptone–dextrose (1% yeast extract, 2% peptone, 2% dextrose; YEPD) or synthetic complete (SC) minimal medium without selective amino acids at 24°C. SC medium contained 2% dextrose. For plate assays, a fig4Δ Vac14-Venus Fab1-TAP yeast strain was transformed with plasmids expressing full-length Fig4 or Fig4 Nterm and Cterm. Transformed cells were grown overnight in Sc-Leu media (full-length Fig4) or Sc-His-Leu media (Fig4 Nterm and Cterm) to mid–log phase. Cultures were diluted to ∼5 × 106 cells per ml and 10 μl of a 10-fold serial dilution was plated on Sc-His or Sc-His-Leu plates and grown at 24° and 33°C for 3 d or 37°C for 2 d.
TABLE 1:
Strains and plasmids used in this study.
| Strain/plasmid | Genotype/description | Source |
|---|---|---|
| LWY6474 | MATa, leu2, 3-112, ura3-52, his3-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1 | Duex et al., 2006b |
| LWY6538 | MATα, leu2,3-112, ura3-52, his-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1, vac14Δ::TRP1 | Duex et al., 2006b |
| LWY8640 | MATa, leu2,3-112, ura3-52, his-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1, VAC14-Venus::KAN | Jin et al., 2008 |
| LWY8812 | MATa, leu2,3-112, ura3-52, his-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1, FAB1-3XGFP::KAN | Jin et al., 2008 |
| LWY14459 | MATa, leu2,3-112, ura3-52, his-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1, VAC14-Venus::KAN, FAB1-TAP::KAN | This study |
| LWY15782 | MATa, leu2,3-112, ura3-52, his-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, fig4Δ::TRP1, VAC14-Venus::KAN, FAB1-6xHA::NAT | This study |
| pRS415-Fig4 | Yeast centromere plasmid; LEU2 | Jin et al., 2008 |
| pRS415-Fig4-C467S | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-Y538A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-E72K | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-E51K | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-E51KT52AE72K | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T52A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T62A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T78A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T52AT62A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T52AT62AT78A | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T52E | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-T52ET62ET78E | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-4xMyc | Yeast centromere plasmid; LEU2 | Jin et al., 2008 |
| pRS415-Fig4-4xMyc-C467S | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-4xMyc-EEE | Yeast centromere plasmid; LEU2 | This study |
| pRS415-Fig4-4xMyc-AAA | Yeast centromere plasmid; LEU2 | This study |
| pRS415ADH | Yeast centromere plasmid; LEU2 | Mumberg et al., 1995 |
| pRS415ADH-Fig4-CTerm | Yeast centromere plasmid; LEU2 | This study |
| pRS413ADH | Yeast centromere plasmid; HIS3 | Mumberg et al., 1995 |
| pRS413ADH-Fig4-NTerm | Yeast centromere plasmid; HIS3 | This study |
| pRS413ADH-Fig4-NTerm-C467S | Yeast centromere plasmid; HIS3 | This study |
| pRS413ADH-Fig4-NTerm-EEE | Yeast centromere plasmid; HIS3 | This study |
| pRS413ADH-Fig4-NTerm-AAA | Yeast centromere plasmid; HIS3 | This study |
| pRS413-Fig4-Envy | Yeast centromere plasmid; HIS3 | This study |
| pRS413-Fig4-Envy-C467S | Yeast centromere plasmid; HIS3 | This study |
| pRS413-Fig4-Envy-Y538A | Yeast centromere plasmid; HIS3 | This study |
| pRS416 Fab1-3xGFP | Yeast centromere plasmid; URA3 | Jin et al., 2008 |
Phosphoinositide lipid labeling and quantification
Yeast [3H]inositol labeling and total cellular phosphatidylinositol extraction, deacylation, and measurements were performed as previously described (Bonangelino et al., 2002; Duex et al., 2006a). fig4Δ yeast with plasmids expressing wild-type Fig4 or Fig4-C467S from the native Fig4 5′ and 3′ UTR (or no Fig4) were labeled with [3H]inositol and treated with 0.9 M NaCl at 0 time. Cells were grown in the appropriate SC medium to an OD600 of 0.5. Cells (0.2 OD600 U) were washed with SC medium lacking inositol and used to inoculate 5 ml of medium lacking inositol and containing 50 µCi of myo-[3H]inositol. Cells were labeled 16 h with shaking at 24°C, harvested by centrifugation, and resuspended in 100 µl of inositol-free medium. For hyperosmotic shock, 100 µl of inositol-free medium with 1.8 M NaCl was added to the 100 μl of sample and then stopped via the addition of 800 µl ice-cold 4.5% perchloric acid. For basal conditions, 100 µl of inositol-free medium was added, followed by the addition of ice-cold 4.5% perchloric acid. Cells were lysed on a Beadbeater (Biospec) for 2 min at room temperature, followed by resting 2 min on ice. This was performed three times. Cell extracts were centrifuged at 14,000 rpm for 15 min at room temperature. Pellets were washed with 100 mM EDTA, pH 8.0, and resuspended in 50 µl of distilled deionized water. The lipids were deacylated by treatment with methylamine. Samples were dried in a SpeedVac. Pellets were resuspended in 300 µl of distilled deionized water. The samples were then mixed with 300 µl of butanol/ethyl ether/formic acid ethyl ester (20:4:1), vortexed, and centrifuged at 14,000 rpm for 5 min. The aqueous phase was transferred to a fresh microcentrifuge tube. This sample extraction was performed twice. Samples were then dried on a SpeedVac, resuspended in 60 µl of distilled deionized water, and analyzed by high-performance liquid chromatography (HPLC) using an anion exchange, PartisphereSAX (Whatman), column, as previously described (Bonangelino et al., 2002).To compare phosphatidylinositol polyphosphate (PI) levels, the raw counts in each peak were expressed as a percentage of total phosphatidylinositol calculated from summation of the counts of the five glycerol-inositol phosphate peaks (PI, PI3P, PI4P, PI3,5P2, and PI4,5P2).
Immunoprecipitations from yeast
fig4Δ yeast strains (fig4ΔVac14-Venus, fig4ΔVac14-VenusFab1-6xHA, or fig4Δvac14Δ) were transformed with plasmids expressing 4xMyc-tagged or HA-tagged Fig4 or empty vector. Log-phase cells (25 OD600 U) were harvested and lysed in immunoprecipitation (IP) buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 10 mM EDTA, 1 mM ethylene glycol tetraacetic acid [EGTA], 5 mM 2-glycerophosphate, 1 × Roche Complete inhibitor cocktail, and 1x Sigma Protease Inhibitor Cocktail) for use with fungal and yeast extracts supplemented with 3 mM benzamidine, 3 µg/ml leupeptin, 5 µg/ml aprotonin, and 18 µg/ml chymostatin. All steps were carried out at 4°C. Debris was removed by centrifugation for 5 min at 500 × g. The supernatant was mixed with 5% octyl-glucoside (Sigma) in lysis buffer for a final concentration of 0.5% octyl-glucoside and incubated for 30 min. Octyl-glucoside–solubilized lysate was cleared by spinning at 13,000 × g for 10 min. For Fig4-4xMyc and Vac14-Venus or Fab1-GFP pull downs, 1 µl per 100 µl lysate mouse anti-Myc (9E10; Fisher) or mouse anti-GFP (Roche) was added, respectively. Supernatants were incubated with antibodies 1 h, and then ∼100 µl lysate was applied to 10 µl equilibrated Protein-G Sepharose beads (Sigma) or SureBeads Protein-G magnetic beads (Bio-Rad) and incubated with rocking for 1 h. For HA-Fig4 pull downs, supernatants were incubated with rocking for 1 h with 20 µl of goat anti-HA agarose beads (Bethel). Protein-G or anti-HA beads were washed three times with 500 µl of IP buffer containing 0.5% octyl-glucoside. Bound protein was eluted by heating immunoglobulin G beads with 25 μl of sample buffer (1% SDS, 8 M urea, 10 mM Tris, pH 6.8, 10 mM EDTA, 0.01% bromophenol blue) at 75°C for 10 min. SDS–PAGE and Western blot analysis were used to detect Fab1-6xHA, HA-Fig4 fragments (rabbit anti-HA; Cell Signalling), Fig4-4xMyc full-length and fragments (rabbit anti-Myc; Cell Signalling), and Vac14-Venus (mouse anti-GFP-Roche). Western blots were quantified using ImageJ. Fab1 bands normalized to Fig4 in corresponding pull down.
Immunoprecipitations from HEK293T cells
Each 10 cm dish of HEK293T cells was transfected with 8 μg of the indicated pCMV-based plasmid, Fig4-Citrine Wild-type, Fig4-C486S-Citrine, or Citrine alone using lipofectamine 2000 (Invitrogen). Forty-two hours posttransfection, cells were lysed on ice in 500 µl binding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 1x Roche EDTA free Complete protease inhibitors) with 1% Brij. All subsequent steps were performed at 4°C. Lysates were cleared by centrifugation for 15 min at 13,000 rpm and supernatants were applied to 20 µl protein-G agarose beads (Sigma) prebound with 1 µl anti-GFP antibody in 100 µl lysis buffer and incubated with rocking for 2 h. Beads were washed three times with 1 ml binding buffer with 0.1% Brij and eluted with 30 μl sample buffer (see Immunoprecipitations from yeast) at 80°C for 5 min. SDS–PAGE and Western blot analysis were used to detect Fig4-Citrine (mouse anti-GFP; Roche) and endogenous PIKfyve (anti-PIKfyve antibody clone R159.4.3C9; Developmental Studies Hybridoma Bank, University of Iowa). Western blots quantified using ImageJ. Pikfyve bands normalized to Fig4 in corresponding pull down.
Fluorescence microscopy
Yeast were grown in the appropriate SC medium to an OD600 of 0.5 and then were labeled with FM4-64 (Vida and Emr, 1995). Images were acquired using the DeltaVision RT Restoration Microscopy System (Applied Precision, Issaquah, WA).
Mass spectrometry analysis of Fig4
Cells expressing Fig4-FLAG were grown to mid–log phase (OD600 ∼ 0.7), and affinity purification was performed as previously described (MacGurn et al., 2011; Lee et al., 2017). Briefly, cells were pelleted, washed with Tris-EDTA, pH 8.0, and frozen as dry pellets. Cell lysates were prepared by bead beating in lysis buffer (50 mM Tris, 0.2% NP-40, 150 mM NaCl, 5 mM EDTA, 2X Complete EDTA-Free protease inhibitor tablets, 1X PhosStop). M2 flag beads (Sigma) were added to the lysates and rotated for 2 h at 4°C. The beads were washed and then eluted in elution buffer (100 mM Tris, 1% SDS) at 95°C for 5 min. Eluates were reduced by addition of 10 mM dithiothreitol and then alkylated by addition of 20 mM iodoacetimide. Samples were then precipitated, resuspended (50 mM Tris, 8 M urea), and digested with 1 μg of trypsin overnight at 37°C. Phosphopeptide enrichment by immobilized metal affinity chromatography was performed as previously described (Albuquerque et al., 2008). Peptide samples were dried, reconstituted in 0.1% trifluoroacetic acid, and analyzed by liquid chromatography with tandem mass spectrometry using an Orbitrap XL mass spectrometer. Raw data were analyzed using MaxQuant software.
Supplementary Material
Acknowledgments
We thank Karin Reinisch and Nikit Kumar for thoughtful discussions and helpful suggestions. This work was supported by National Institutes of Health Grants R01-NS099340 and R01-NS064015 to L.S.W. B.S.S. was supported in part by a Jane Coffin Childs Memorial Fund Fellowship and National Institutes of Health Grant K99-GM-120511. N.S. was supported in part by National Institutes of Health grant T32 GM007315 and through a Rackham Warner Lambert Fellowship.
Abbreviations used:
- ALS
amyotrophic lateral sclerosis
- CMT4J
Charcot–Marie–Tooth syndrome Type 4J
- PI
phosphatidylinositol
- PI3,5P2
phosphatidylinositol (3,5)-bisphosphate; PTPase, phosphatase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0556) on June 17, 2020.
REFERENCES
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. (2008). A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics , 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Nunes-Xavier CE, Bayon Y, Pulido R. (2016). The extended family of protein tyrosine phosphatases. Methods Mol Biol , 1–23. [DOI] [PubMed] [Google Scholar]
- Amoasii L, Bertazzi DL, Tronchere H, Hnia K, Chicanne G, Rinaldi B, Cowling BS, Ferry A, Klaholz B, Payrastre B, et al. (2012). Phosphatase-dead myotubularin ameliorates X-linked centronuclear myopathy phenotypes in mice. PLoS Genet , e1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P, Berger I, Schaffitzel C, Tersar K, Volkmer B, Suter U. (2006). Multi-level regulation of myotubularin-related protein-2 phosphatase activity by myotubularin-related protein-13/set-binding factor-2. Hum Mol Genet , 569–579. [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Cunningham KM, Zhang K, Lloyd TE. (2016). FIG4 regulates lysosome membrane homeostasis independent of phosphatase function. Hum Mol Genet , 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billcliff PG, Lowe M. (2014). Inositol lipid phosphatases in membrane trafficking and human disease. Biochem J , 159–175. [DOI] [PubMed] [Google Scholar]
- Blanchetot C, Chagnon M, Dube N, Halle M, Tremblay ML. (2005). Substrate-trapping techniques in the identification of cellular PTP targets. Methods , 44–53. [DOI] [PubMed] [Google Scholar]
- Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, Devreotes P, Gabelli SB, Cole P. (2013). Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. eLife , e00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. (2002). Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol , 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD. (2008). Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell , 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Deng Y, Horenkamp F, Reinisch KM, Burd CG. (2014). Sac1-Vps74 structure reveals a mechanism to terminate phosphoinositide signaling in the Golgi apparatus. J Cell Biol , 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, et al. (2009). Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet , 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, et al. (2007). Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature , 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Dove SK, Barr FA. (2004). I-proteins — a proposed switch in myotubularin function. Trends Biochem Sci , 58–61. [DOI] [PubMed] [Google Scholar]
- Dong Y, Gou Y, Li Y, Liu Y, Bai J. (2015). Synaptojanin cooperates in vivo with endophilin through an unexpected mechanism. eLife , e05660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. (2006a). Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell , 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS. (2006b). The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol , 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK. (1997). Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA , 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD. (2002). Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell , 1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol , 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. (2009). PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem , 35794–35806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Shisheva A. (2001). Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem , 26141–26147. [DOI] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang YL, Park S, Duex JE, et al. (2008). VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P-2 in yeast and mouse. EMBO J , 3221–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CE, Karkheiran S, Powell JC, Cao M, Makarov V, Darvish H, Di Paolo G, Walker RH, Shahidi GA, Buxbaum JD, et al. (2013). The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum Mutat , 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Tumolo JM, Ehlinger AC, Jernigan KK, Qualls-Histed SJ, Hsu PC, McDonald WH, Chazin WJ, MacGurn JA. (2017). Ubiquitin turnover and endocytic trafficking in yeast are regulated by Ser57 phosphorylation of ubiquitin. eLife , e29176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk GM, Ferguson CJ, Chow CY, Jin N, Jones JM, Grant AE, Zolov SN, Winters JJ, Giger RJ, Dowling JJ, et al. (2011). Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet , e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk GM, Frei CM, Miller AC, Wallen RC, Mironova YA, Giger RJ, Meisler MH. (2016). Rescue of neurodegeneration in the Fig4 null mouse by a catalytically inactive FIG4 transgene. Hum Mol Genet , 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bankaitis VA. (2010). Phosphoinositide phosphatases in cell biology and disease. Prog Lipid Res , 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD. (2011). TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell , 1104–1117. [DOI] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, Mao Y. (2010). Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J , 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson GR, Perisic O, Burke JE, Williams RL. (2016). The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem J , 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead GB, De Wever V, Templeton G, Kerk D. (2009). Evolution of protein phosphatases in plants and animals. Biochem J , 401–409. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene , 119–122. [DOI] [PubMed] [Google Scholar]
- Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. (2006). The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell , 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odriozola L, Singh G, Hoang T, Chan AM. (2007). Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem , 23306–23315. [DOI] [PubMed] [Google Scholar]
- Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, et al. (2014). Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell , 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido R, Stoker AW, Hendriks WJ. (2013). PTPs emerge as PIPs: protein tyrosine phosphatases with lipid-phosphatase activities in human disease. Hum Mol Genet , R66–R76. [DOI] [PubMed] [Google Scholar]
- Quadri M, Fang M, Picillo M, Olgiati S, Breedveld GJ, Graafland J, Wu B, Xu F, Erro R, Amboni M, et al. (2013). Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum Mutat , 1208–1215. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. (2004). Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science , 1179–1181. [DOI] [PubMed] [Google Scholar]
- Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. (2009). A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA , 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE. (2006). Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol , 403–412. [DOI] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. (2003). The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem , 52689–52699. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Anderson DM, Emr SD. (2004). Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell , 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Rodahl LM, Pattni K, Englund C, Samakovlis C, Dove S, Brech A, Stenmark H. (2006). Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell , 3989–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. (2008). ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol , 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. (2007). Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem , 23878–23891. [DOI] [PubMed] [Google Scholar]
- Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. (2003). Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol , 504–509. [DOI] [PubMed] [Google Scholar]
- Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. (2008). Function and dysfunction of the PI system in membrane trafficking. EMBO J , 2457–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol , 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh MG. (2015). PIPs in neurological diseases. Biochim Biophys Acta , 1066–1082. [DOI] [PubMed] [Google Scholar]
- Yu ZH, Zhang RY, Walls CD, Chen L, Zhang S, Wu L, Liu S, Zhang ZY. (2014). Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry , 4136–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZH, Zhang ZY. (2018). Regulatory mechanisms and novel therapeutic targeting strategies for protein tyrosine phosphatases. Chem Rev , 1069–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Hsu F, Stefan CJ, Wu X, Patel A, Cosgrove MS, Mao Y. (2012). Allosteric activation of the phosphoinositide phosphatase Sac1 by anionic phospholipids. Biochemistry , 3170–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.