Artemisinin-based combination therapy (ACT) is the first-line antimalarial regimen in Indonesia. Susceptibility of Plasmodium falciparum to artemisinin is falling in the Greater Mekong subregion, but it is not known whether the efficacy of current combinations is also threatened in nearby Sumatera. We evaluated the genetic loci pfcrt, pfmdr1, and pfk13, considered to be under selection by artemisinin combination therapy, among 404 P. falciparum infections identified by PCR detection in a cross-sectional survey of 3,731 residents of three regencies.
KEYWORDS: Indonesia, antimalarial agents, drug resistance mechanisms
ABSTRACT
Artemisinin-based combination therapy (ACT) is the first-line antimalarial regimen in Indonesia. Susceptibility of Plasmodium falciparum to artemisinin is falling in the Greater Mekong subregion, but it is not known whether the efficacy of current combinations is also threatened in nearby Sumatera. We evaluated the genetic loci pfcrt, pfmdr1, and pfk13, considered to be under selection by artemisinin combination therapy, among 404 P. falciparum infections identified by PCR detection in a cross-sectional survey of 3,731 residents of three regencies. The pfcrt haplotype SVMNT (codons 72 to 76) was the most prevalent and displayed significant linkage disequilibrium with the pfmdr1 haplotype YY (codons 86 and 184) (odds ratio [OR] 26.7; 95% confidence interval [CI], 5.96 to 239.4; P < 0.001). This contrasts with Mekong countries, where the CVIET haplotype of pfcrt predominates. Among 231 evaluable isolates, only 9 (3.9%) showed any evidence of nonsynonymous gene variants in the propeller domain of pfk13. The Thr474Ala variant was seen in six individuals, and Cys580Tyr was identified with low confidence in only a single isolate from an asymptomatic individual. Among a subset of 117 symptomatic P. falciparum-infected individuals randomized to receive either dihydroartemisinin-piperaquine or artemether-lumefantrine, the treatment outcome was not associated with pretreatment genotype. However, submicroscopic persistent parasites at day 28 or day 42 of follow-up were significantly more likely to harbor the pfmdr1 haplotype NF (codons 86 and 184) than were pretreatment isolates (P < 0.001 for both treatment groups). Current ACT regimens appear to be effective in Sumatera, but evidence of persistent submicroscopic infection in some patients suggests further detailed studies of drug susceptibility should be undertaken.
INTRODUCTION
Successful strategies for the elimination of malaria require effective first-line chemotherapies. Failure of the antimalarials chloroquine and sulfadoxine-pyrimethamine compromised malaria control strategies in many endemic countries and contributed to a significant increase in morbidity and mortality through the 1990s (1, 2). WHO currently recommends the use of artemisinin-based combination therapy (ACT) for the treatment of uncomplicated Plasmodium falciparum infection, a strategy which has contributed to reductions in malaria mortality in the last 2 decades (3). Nevertheless, decreased susceptibility of P. falciparum parasites to artemisinin and partner drugs has emerged in the Greater Mekong subregion (GMS), as evidenced by slow parasite clearance and an increased frequency of recrudescence in patients treated with the ACT dihydroartemisinin-piperaquine (DP) (4, 5). The continued progression of clinically relevant parasite resistance in this region may be slowed or prevented by deploying a more flexible treatment policy, informed by regular monitoring of candidate resistance-associated alleles of key genes in P. falciparum parasites, to identify genotypes with a selective advantage in parasites exposed to antimalarial drugs.
The marked reduction in in vivo parasite susceptibility to artemisinins was first observed in the GMS over a decade ago (6). This is caused by mutations in the P. falciparum gene pfk13 which affect the propeller domain of the kelch-13 protein (7, 8). Amplification of plasmepsin II gene copy number is linked to piperaquine resistance in the same region (9). Resistance to aminoquinolines is known to be mediated by the putative transporter pfcrt (10), with specific haplotypes at codons 72 to 76 associated with resistance to chloroquine (CVIET) and amodiaquine (SVMNT) (11, 12). The degree of resistance to aminoquinolones and to artemisinin is further modulated by additional variation in other genes, including pfmdr1, encoding P-glycoprotein H1. Polymorphisms in pfmdr1 have been associated with differential susceptibility to lumefantrine and amodiaquine (13). In vitro studies show that the codon 86 Tyr variant (86Y), which developed under aminoquinolone pressure in previous decades, has greater in vitro susceptibility to artemisinin than the wild-type 86N (14, 15). Further, the haplotype NFD at codons 86, 184, and 1246 of this locus is associated with parasite persistence in ACT-treated African patients (16, 17). Thus, understanding genetic changes in parasite populations where resistance is emerging can provide timely warning of threats to current therapies.
ACT has been used in Indonesia since 2004 after efficacy of chloroquine was severely reduced by the spread of parasites harboring the CVIET and SVMNT haplotypes of pfcrt (18–20). Two combinations were initially deployed, artesunate-amodiaquine (ASAQ) for western Indonesia and DP for eastern Indonesia (21). However, treatment failures with ASAQ were frequently documented, which led to further drug policy change in 2012, putting in place countrywide deployment of DP. In vivo studies using ASAQ for falciparum malaria have consistently demonstrated unsatisfactory clinical efficacy in Central Java, Papua, and Sumatera (22–25), with PCR-corrected efficacy as low as 80% in one study conducted prior to the adoption of ASAQ as the national recommendation (22). An explanation for the observed poor drug efficacy is hindered by a lack of information on parasite polymorphisms in this study. Also of great concern is that artemisinin-resistant parasites harboring pfk13 mutants have now spread across Southeast Asia, and so, with their proximity to the Mekong and a history of lower parasite susceptibility to ACT treatment, genetic markers of ACT resistance in P. falciparum parasites in western Indonesia urgently require investigation.
In this study, we report the prevalence of polymorphisms of interest in the pfk13, pfcrt, and pfmdr1 genes of P. falciparum isolates from a large cross-sectional survey in three regencies in North Sumatera Province, Indonesia (26). We determined the alleles carried by P. falciparum isolates from a subset of survey participants enrolled in a randomized comparison of antimalarial efficacy of two ACTs, artemether-lumefantrine (AL) and DP (27), and tested for evidence of an association between variants of these three loci and treatment outcomes.
RESULTS
Population prevalence was estimated for each gene variant of interest by genotyping DNA from P. falciparum infections previously identified in our cross-sectional survey. PCR was positive for 304 tested individuals, of which 201 were identified as submicroscopic, low-density parasitemia (26). Resistance-associated loci were amplified from among these 304 isolates.
Polymorphisms in Pfcrt.
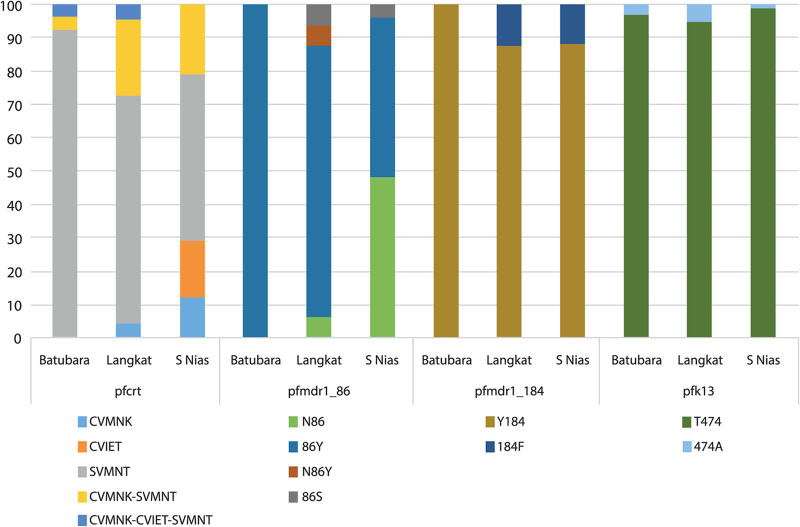
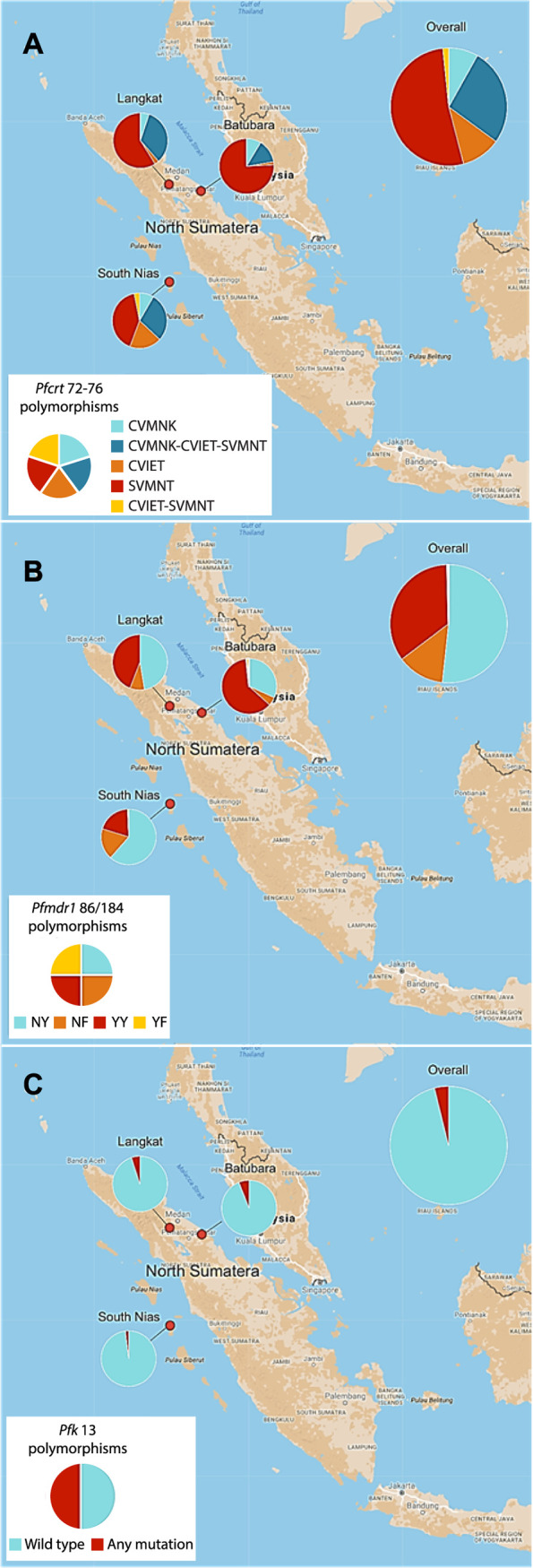
Pfcrt genotyping at codons 72 to 76 was successful for 183 isolates (60.2%). We observed the pfcrt-SVMNT haplotype as the dominant allele, being present in 140 of these (76.5% of evaluable isolates), either alone (68.6% of these) or mixed with CVMNK or CVIET haplotypes (31.4%) (Fig. 1A). The prevalence of parasites harboring the wild-type haplotype CVMNK, alone or mixed, was 34.9%. CVIET occurred in 20.2% of isolates. Parasites carrying the SVMNT haplotype, alone or mixed, were the most prevalent in each of the three sites, comprising 42/49 in Batubara regency (85.7%), 33/39 in Langkat regency (84.6%), and 65/95 in South Nias regency (68.4%). In South Nias, the CVIET haplotype was observed more commonly than in the other regencies, occurring in 28/95 of isolates (29.5%).
FIG 1.
Prevalence of genotypes of interest in pfcrt, pfmdr1, and pfk13 in a cross-sectional community sample in 3 regencies. Genotypes are shown for pfcrt at codons 72 to 76 (A), codons 86/184 of pfmdr1 gene (B), and the pfk13 propeller domain (C) in three study sites in North Sumatera province. Pfcrt haplotypes were identified by multiplex qPCR; pfmdr1 and pfk13 genotypes were established by direct sequencing of PCR products (see Materials and Methods). Denominators are n = 183 (n = 49 for Batubara, n = 39 for Langkat, and n = 95 for South Nias) in panel A, n = 261 (n = 59 for Batubara, n = 57 for Langkat, and n = 145 for South Nias) in panel B, and n = 232 (n = 66 for Batubara, n = 60 for Langkat, and n = 106 for South Nias) in panel C.
Polymorphisms in Pfmdr1.
Codons 25 to 201 of pfmdr1 were successfully amplified and sequenced for 267 isolates (66.1%). The prevalence of the Pfmdr1 N86 (Asn) wild-type allele was predominant overall (174/267, 65.2%) but varied among sites 37% to 79%. The 86Y (Tyr) variant, associated with chloroquine and amodiaquine resistance, occurred in 93/267 (34.8%), and two rare mutations, 86F (Phe) and 86S (Ser), were also observed, each in two individuals. The wild-type Y184 was highly prevalent, occurring in more than 90% of isolates in Batubara regency and over 80% in Langkat and South Nias regencies (Fig. 2). We did not observe any mutation in the Pfmdr1 codons 1034, 1042, or 1246 alleles among 73, 74, and 69 evaluable sequences, respectively, and no further analysis of these codons was conducted.
FIG 2.
Pretreatment prevalence of variants in codons of interest in the pfcrt, pfmdr1, and pfkelch13 genes by regency. Allele-specific qPCR (pfcrt only) or direct sequencing of nested PCR products was used to enumerate P. falciparum alleles of interest present among pretreatment samples from prospective trial participants (n = 117). These alleles were at the following codons: 72 to 76 (pfcrt), 86 and 184 (pfmdr1), and 474 (pfkelch13) (propeller domain).
The combined haplotype at pfmdr1 codons 86 and 184 was determined for each isolate. The NF haplotype is known to be selected by artemether-lumefantrine, while the YY haplotype is selected by amodiaquine (13). We also included samples with mixed alleles at only one of the two positions, such that two haplotypes could be unambiguously assumed to occur in that isolate. We noted the haplotype YY (91/261, 34.9%) was almost three times more prevalent in the population than the parasites carrying haplotype NF (34/261, 13.0%). However, this ratio differed by site, with YY predominant over NF in Batubara and Langkat but equally distributed in South Nias (Fig. 1B).
Population prevalence of polymorphisms in Pfk13.
The Kelch13 propeller domain sequence was determined on at least one DNA strand for P. falciparum isolates from 231 participants, with the wild-type genotype present in the majority. Previous surveys of allele prevalence at this locus have sampled among clinical malaria cases, whereas the majority of our 231 sequences came from asymptomatic individuals tested as part of our cross-sectional survey (26). Parasite densities were therefore usually low, and sequencing quality was not always adequate to confirm genotypes on both DNA strands of the pfk13 amplicon. Nine isolates were considered to harbor nonsynonymous polymorphisms with low, moderate, or high confidence (Table 1). The previously described amino acid substitution T474A was the most prevalent, occurring in six individuals and at least once in each regency, and the C580Y substitution was identified at low confidence in a single isolate from South Nias. The other common Southeast Asian mutant alleles R539T and F446I were not observed among our isolates (28). Although K13 polymorphisms occurred in all 3 sites, the prevalence was uniformly low, with 4 of 66 in Batubara, 3 of 60 in Langkat, and 2 of 106 in South Nias (Fig. 1C).
TABLE 1.
Nonsynonymous single-nucleotide polymorphisms in the Pfk13 propeller domain of nine isolates in the community sample among 231 sequenced
| Regency | Identifier | Codon | Coverage | Evidencea |
|---|---|---|---|---|
| Batubara | BB02030 | Mixedb T474A | Both strands | High confidence |
| BB02033 | Mixed T474A | Both strands | Moderate confidence | |
| BB13019 | Unmixed T535A, C542R | One strand | Low confidence | |
| BB22036 | Unmixed N523S, T535A, T593A | One strand | Low confidence | |
| Langkat | LK01061 | Mixed T474A, mutant peak low | Both strands | Moderate confidence |
| LK06042 | Mixed T474A | Both strands | Moderate confidence | |
| LK10083 | Mixed T474A | Both strands | Moderate confidence | |
| South Nias | NS23031 | Mixed T474A | Both strands | Moderate confidence |
| NS27031 | Mixed E461G, C580Y; mixed synon a→g codon 521c | One strand | Low confidence |
Only polymorphisms confirmed on all available DNA strand sequence reads are presented. Equivocal sequences, or polymorphisms observed on only one of two strands, were not considered to have been verified and were scored as wild type. For isolates BB13019, BB22036, and NS 27031, only a single strand was available, and so, the results are presented as low confidence.
“Mixed” denotes the presence of two different DNA sequences at the codon named in the isolate, indicative of a multiclonal infection.
Nucleotide change at codon 521 did not alter the amino acid encoded.
Associations between Pfcrt, Pfmdr1, and Pfk13 polymorphisms in the P. falciparum population.
We investigated any evidence of linkage disequilibrium between the pfcrt and pfmdr1 polymorphisms among isolates in our cross-sectional survey. Isolates carrying the SVMNT pfcrt haplotype were significantly more likely to carry the pfmdr1 YY haplotype (odds ratio [OR], 26.7; 95% confidence interval [CI], 5.96 to 239.4; P < 0.001). Conversely, only 8 of 116 isolates harboring pfcrt SVMNT also carried the pfmdr1 haplotype NF (7.0%), compared to 11 of 26 harboring other pfcrt genotypes (OR, 0.101; 95% CI, 0.031 to 0.333; P < 0.001). We observed that pfk13 propeller domain variant alleles were present in a background of pfcrt SVMNT (all four evaluable) and pfmdr1 YY or NY (four and three evaluable, respectively), but it was not possible to test these associations statistically, as we had too few isolates successfully typed at all three loci.
Pfcrt, Pfmdr1, and Pfk13 polymorphisms in parasites before and after ACT treatment.
A subset of individuals with symptomatic P. falciparum infections were enrolled in a prospective treatment efficacy study, randomized to receive either AL or DP (27). We observed an unexpectedly high proportion of ACT-treated patients with persisting subpatent P. falciparum parasites, and so, we explored whether pfcrt, pfmdr1, and pfk13 genotypes in the pretreatment parasite population contributed to trial outcomes. Among 71 evaluable PCR-confirmed P. falciparum isolates with remaining DNA samples available, the amodiaquine-resistant SVMNT haplotype of pfcrt (at codons 72 to 76) dominated in both treatment groups (28/34 in the DP group [82.4%]; 35/37 in the AL group [94.6%]) (Fig. 2). The chloroquine-resistant CVIET and drug-sensitive CVMNK pfcrt haplotypes were both less common, together accounting for 11/34 and 8/37 of pretreatment isolates in the DP and AL treatment groups, respectively, including a number of mixed infections in which SVMNT was also present. The relative proportions of SVMNT differed according to site, with the highest in Batubara and the lowest in South Nias (Fig. 2).
For pfmdr1, the YY haplotype at codons 86 and 184 was predominant in the pretreatment population for both ACT groups (32/49, 65.3% for DP; 36/47, 76.6% for AL), reflecting the high prevalence of this haplotype observed in the cross-sectional population survey (Fig. S1). The rare 86S allele was also identified in two individuals (Fig. 2). The 86N allele was common only in South Nias and rare in pretreatment isolates from the other 2 regencies. For pfk13, wild-type genotypes (96%, 72/75) dominated in the propeller domain. The T474A polymorphism was detected in 3 (4.0%) pretreatment isolates in the AL group, in each case, mixed with the wild-type sequence. All parasite isolates harboring pfk13 mutations also carried the SVMNT haplotype of pfcrt. We found no evidence of slow clearance by quantitative PCR (qPCR) during the first 72h following treatment with either ACT, except in a single DP-treated patient who exhibited PCR-confirmed early treatment failure (27).
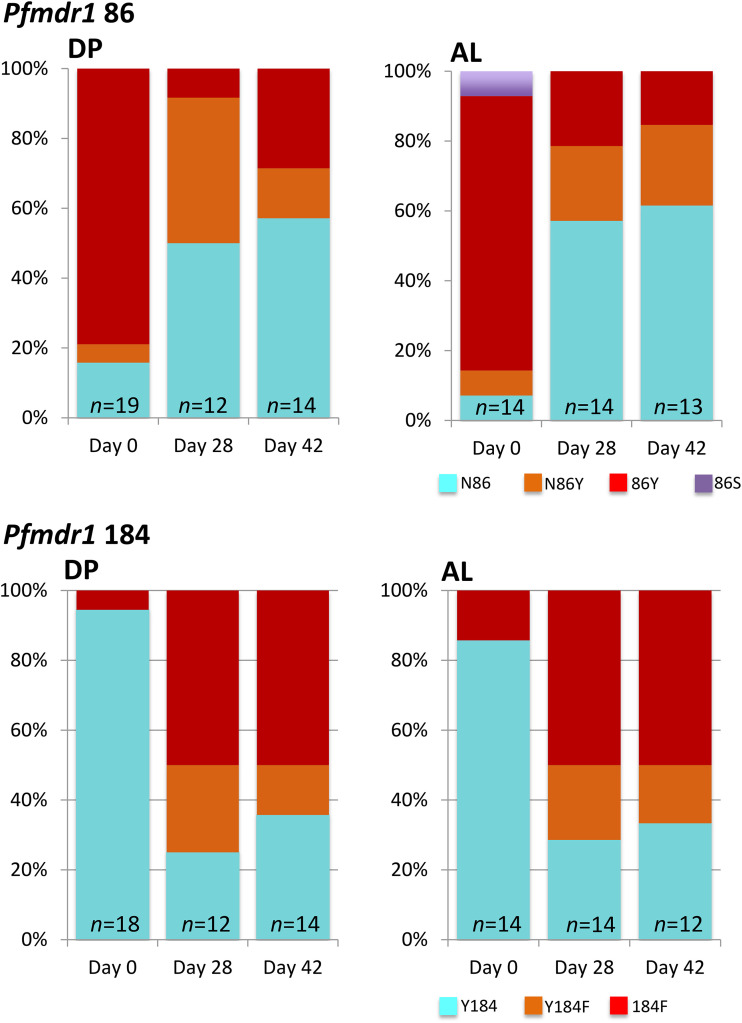
An unexpected finding of our clinical study was that a significant number of persistent PCR-detectable P. falciparum infections remained 28 or 42 days after treatment (27). We therefore attempted to genotype pfmdr1 in these recurrent isolates and compare them to those of the baseline isolates. Successful amplification of the pfmdr1 amplicon containing codons 86 and 184 was achieved for 31 and 30 samples at days 28 and 42, respectively. We observed a significant selection for N86 and 184F at days 28 and 42 in both treatment arms but found no evidence that the presence of the NF haplotype before treatment was associated with persistent parasitemia in follow-up (P = 0.62). The proportion of patients in the DP and AL groups carrying the pfmdr1 haplotype NF increased from 6.1% and 4.6% at baseline to 58.8% (10/17) and 50.0% (7/14) at day 28 (OR, 21.9; 95% CI, 4.0 to 143.8; P < 0.001 and OR, 21.0; 95% CI, 2.9 to 227.5; P < 0.001 for DP and AL, respectively). Corresponding figures for day 42 were 42.1% (8/19) and 53.3% (8/15) (OR, 11.2; 95%, CI 2.1 to 72.5; P = 0.0003 and OR, 24; 95% CI, 3.5 to 254.7; P < 0.001, respectively). Paired analysis of pre- and posttreatment pfmdr1 genotypes by McNemar’s test of asymmetry confirmed directional selection favoring the pfmdr1 NF haplotype at both day 28 (21 evaluable participants pooled across DP and AL groups; P < 0.001) and day 42 (23 evaluable participants; P = 0.002) (Fig. 3). There were insufficient data to stratify this analysis by treatment group.
FIG 3.
Prevalence of pfmdr1 alleles in 15 and 13 individuals randomized to the DP and AL treatment groups, respectively, with PCR-detectable P. falciparum at days 28 or 42 during follow-up. “Baseline” denotes the pretreatment isolates in the same individuals evaluated at days 28 and 42. Pale blue color denotes the wild-type allele, red indicates the mutant allele associated with aminoquinoline resistance, and orange represents a mixture of both alleles present simultaneously.
Unfortunately, parasite densities were very low in the subpatent parasite infections at day 28 and day 42, and insufficient material was available to perform qPCR-based genotyping of pfcrt or direct sequencing of pfk13 amplicons in this group of isolates.
DISCUSSION
We performed a survey of antimalarial drug resistance markers in northwestern Indonesia to identify genetic polymorphisms present in the P. falciparum parasite population. We found that pfk13 variants, although rare, were present in parasites harboring the SVMNT genotype at codons 72 to 76 of pfcrt, which is the predominant haplotype in our three study sites. This contrasts with P. falciparum in the GMS, where pfk13 variant parasites carry the CVIET pfcrt allele at codons 72 to 76 (29), together with additional acquired mutations associated with piperaquine resistance at other pfcrt codons (30). Decreased piperaquine susceptibility is associated with the C350R pfcrt polymorphism in French Guiana, where it occurs with the SVMNT haplotype at codons 72 to 76, although this is not linked to artemisinin resistance (31). Among a subset of symptomatic participants randomized to receive the ACT regimens DP or AL, we found strong evidence of directional selection on pfmdr1. In both drug arms, the NF haplotype at codons 86 and 184 was much more abundant in persistent subpatent parasites identified at day 28 or day 42 of follow-up than in the pretreatment population. We identified only nine pfk13 propeller domain-variant alleles with moderate to high confidence in the cross-sectional survey, six of which encoded the Thr to Ala change at codon 474.
We observed a high proportion of parasite genotypes associated with amodiaquine and chloroquine resistance in our samples, with 76.5% carrying the pfcrt haplotype SVMNT and 20.2% the CVIET haplotype. Despite the discontinuation of chloroquine in 2004 and subsequent introduction of ACT, the proportion of mutant 76T in this region remains above 90%, similar to pre-2004 data (19, 20), likely due to the use of ASAQ. This contrasts with data from East Africa, where wild-type pfcrt has recovered to high prevalence following the widespread deployment of AL (17). Evidence of treatment failure with ASAQ triggered a recent change in recommendations for treating P. falciparum infection in Indonesia (22–25). DP is now the approved first-line regimen, with AL licensed and widely available in the private sector. Recently, evidence has accumulated of decreased DP efficacy in western Cambodia, and the phenotype has been associated with an increased copy number of the plasmepsin II gene and other emerging gene variants (9, 30, 32). This leads to concern that Indonesian parasites may also develop piperaquine resistance, and studies of polymorphisms known to be associated with piperaquine susceptibility are now needed. We found PCR-based evidence of submicroscopic parasite persistence at D28 and/or D42 in both drug arms (30% of evaluable patients in the AL arm, 40% in the DP arm) (27), as has previously been observed in imported P. falciparum malaria cases in France (33).
The pfmdr1 86Y allele was formerly common in the Southeast Asian region, but it significantly decreased in frequency, consistent with the abandonment of chloroquine and amodiaquine (34). A similar dramatic fall in the prevalence of 86Y was also observed in Nias, from 100% in 2003 (20) to 31.4% in 2005 (35). Nevertheless, this was not concomitant with an increase in abundance of wild-type pfcrt. Our findings are consistent with these data, as Pfmdr1 86Y is at moderate prevalence but accompanied by a high prevalence of mutant Pfcrt 76T (Fig. 2). The 184F allele has also slowly disappeared in mainland Southeast Asia, possibly driven by pressure from mefloquine, except in western Cambodia and eastern Thailand (36), but as mefloquine is not available in Indonesia, this cannot explain the relatively low prevalence of 184F in Sumatera. It is important to also recognize compelling evidence in the literature that artemisinins themselves directly select for the NF haplotype of pfmdr1, both in vivo (16) and in genome editing experiments in vitro (15).
We show a strong association between the pfcrt SVMNT and the pfmdr1 YY haplotypes among our parasite populations. Both alleles have been associated with amodiaquine resistance (12, 13, 34). The SVMNT haplotype is distributed across Indonesia, Papua New Guinea, East Timor, South Asia, and, as an allele with an independent origin, in South America (18, 37, 38). However, these high-grade amodiaquine-resistant parasites remain uncommon in most parts of mainland Southeast Asia and are absent from Africa, where CVIET predominates and amodiaquine may still be effective (36). The ongoing presence of these gene mutations in our study sites is likely the result of extended drug pressure from amodiaquine as the partner drug in the previously recommended ASAQ regimen and the continuing access to chloroquine in the private sector. This occurrence of SVMNT alleles may therefore explain the low clinical efficacy of ASAQ for treatment of P. falciparum infection observed in Indonesian efficacy studies (22–25).
Pfk13 propeller domain polymorphisms have been linked to reduced sensitivity to artemisinin in Southeast Asia and are thought to have emerged independently in Cambodia and Myanmar. The mutants C580Y, R539T, and M446I associated with slow clearance of P. falciparum after artesunate monotherapy or ACT are the most frequent and geographically specific in mainland Southeast Asia. In eastern Indonesia, this trend has not been seen, as only 0.9% of 106 samples from Sumba harbored the pfk13 allele G497V (28), and no pfk13 mutation was detected among 65 samples from southern Papua (39). In our study sites, 6 of 9 variant isolates harbored the T474A propeller domain polymorphism, which is not prevalent in the GMS, although a T474I variant has been described (28). Codon 474 variants have not been associated with reduced susceptibility to artemisinin to date. We were unable to evaluate the impact of this genotype on parasite clearance, and phenotypic studies of these mutants are now needed to assess their significance.
We observed diversity in the P. falciparum genetic signature among the three study sites, which is in line with differences in transmission intensity, treatment-seeking behavior, access to health care, and antimalarial use in these communities. However, our study was not designed to scrutinize the factors contributing to these differences in genetic profiles, and so, their importance remains unclear. A limitation of our study was the difficulty of obtaining high-quality genotypes from multiple loci in these parasite isolates, the majority of which were low-density asymptomatic infections. Even among patients with clinical malaria enrolled in our prospective study, posttreatment isolates were difficult to analyze at all the loci of interest, even when evidence of persisting P. falciparum was obtained from at least one gene amplification. Another limitation of our study is the use of a convenience sampling approach (26), and this may have introduced bias in the proportion of drug resistance markers presented. Nevertheless, new evidence of mutations in the Pfk13 propeller domain in western Indonesia was found. The lack of information on the associated phenotypic profiles warrants future studies to measure artemisinin susceptibility of these parasites in vivo and in vitro. We have also confirmed that selective impact of ACT favoring the pfmdr1 haplotype NF (codons 86 and184), originally described in African studies, is also clearly evident in Sumatera.
In summary, our study provides new information on the genetic profiles of P. falciparum parasites in western Indonesia. We provide evidence of selective pressure from ASAQ in the recent past, including linkage disequilibrium between certain alleles of pfcrt and pfmdr1, and evidence of more recent counterselection by current regimens on the pfmdr1 locus in particular. This can guide antimalarial policy for ACT use in the country. We found no evidence that artemisinin-resistant parasites had spread from the nearby GMS. The presence of some Pfk13 mutations among the sampled parasite population is of potential concern and demonstrates the need to further evaluate artemisinin susceptibility of parasites from western Indonesia. DP and AL currently appear to be effective treatment options for P. falciparum infection in North Sumatera, but further efficacy studies are needed.
MATERIALS AND METHODS
Study sites, sample collection, and patient recruitment.
As previously described, we conducted a parasitological survey between January and June 2015 in Batubara, Langkat, and South Nias regencies in North Sumatera province, Indonesia (26). A total of 3,731 participants were screened for Plasmodium species infection by microscopy and post hoc nested PCR. All microscopy-positive participants were treated with the standard 3-dose DP or 6-dose AL regimens, and those meeting inclusion criteria for a prospective efficacy trial of AL versus DP, and who gave consent, were followed up for 42 days as described elsewhere (27).
The study was approved by the Research Ethics Committees of the University of Sumatera Utara, Indonesia (401/KOMET/FK USU/2014) and the London School of Hygiene and Tropical Medicine, United Kingdom (8504-01).
Parasite genotyping for resistance markers.
Parasite DNA was extracted from dried blood spots as described (26). We performed genotyping of pfcrt, pfmdr1, and the pfkelch13 propeller domain using established methods with minor modifications. Polymorphisms at codons 72 to 76 in pfcrt were determined using multiplex qPCR (40). Polymorphisms at codons 86, 184, 1034, 1042, and 1246 in pfmdr1 were identified by direct sequencing (13). Pfk13 polymorphisms were identified by nested amplification and direct sequencing of PCR products (7, 28). The prevalence of each polymorphism in the evaluated genes was estimated. Samples yielding mixed alleles contributed to the prevalence of both alleles.
Treatment outcomes.
For 117 symptomatic participants with PCR-confirmed P. falciparum infections, randomized to receive AL or DP, pgmet qPCR positivity at day (D) 3 and pfmdr1 nested PCR positivity at D28 or D42 were indicators of unsuccessful treatment (27).
Statistical analysis.
Statistical analyses were performed in the Stata 11 package. Binary variables were compared across categories by estimating odds ratios (ORs) with 95% confidence intervals (CIs), and significance was determined using the X2 distribution. Linkage disequilibrium between loci was examined in 2 by 2 contingency tables.
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.White NJ, Nosten F, Looareesuwan S, Watkins WM, Marsh K, Snow RW, Kokwaro G, Ouma J, Hien TT, Molyneux ME, Taylor TE, Newbold CI, Ruebush TK, Danis M, Greenwood BM, Anderson RM, Olliaro P. 1999. Averting a malaria disaster. Lancet 353:1965–1967. doi: 10.1016/s0140-6736(98)07367-x. [DOI] [PubMed] [Google Scholar]
- 2.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. 2002. Epidemiology of drug-resistant malaria. Lancet Infect Dis 2:209–218. doi: 10.1016/S1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2015. Guidelines for the treatment of malaria, 3rd ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium . 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 7.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. 2015. K13-propellor mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 11.Sidhu ABS, Verdier-Pinard D, Fidock DA. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, Hallett RL. 2010. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother 54:3714–3716. doi: 10.1128/AAC.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, Hallett RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. 2000. The tyrosine-86 allele of pfmdr1 gene of the Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol 108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 15.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, Martin RE, Lehane AM, Fidock DA. 2016. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, van Schalkwyk DA, Sawa P, Omar SA, Clark TG, Bousema T, Sutherland CJ. 2014. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis 210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, Walakira A, Nankabirwa J, Yeka A, Staedke SG, Greenhouse B, Nsobya SL, Kamya MR, Dorsey G, Rosenthal PJ. 2016. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 207:631–635. doi: 10.1093/infdis/jiw614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huaman MC, Yoshinaga K, Suryanatha A, Suarsana N, Kanbara H. 2004. Short report: polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok, Indonesia. Am J Trop Med Hyg 71:40–42. doi: 10.4269/ajtmh.2004.71.40. [DOI] [PubMed] [Google Scholar]
- 19.Syafruddin D, Asih PBS, Aggarwal SL, Shankar AH. 2003. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Purworejo District, Central Java Province, Indonesia. Am J Trop Med Hyg 69:614–620. doi: 10.4269/ajtmh.2003.69.614. [DOI] [PubMed] [Google Scholar]
- 20.Syafruddin D, Asih PBS, Casey GJ, Maguire JD, Baird JK, Nagesha HS, Cowman AF, Reeder JC. 2005. Molecular epidemiology of Plasmodium falciparum resistance to antimalarial drugs in Indonesia. Am J Trop Med Hyg 72:174–181. doi: 10.4269/ajtmh.2005.72.174. [DOI] [PubMed] [Google Scholar]
- 21.Elyazar IR, Hay SI, Baird JK. 2011. Malaria distribution, prevalence, drug resistance and control in Indonesia. Adv Parasitol 74:41–175. doi: 10.1016/B978-0-12-385897-9.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djatmiko W. 2005. Uji efikasi terapi kombinasi artesunate+amodiaquine pada malaria falciparum tanpa komplikasi di Banjarnegara Propinsi Jawa Tengah (in Indonesian). Masters thesis. Universitas Diponegoro, Semarang, Indonesia. (In Indonesian.) [Google Scholar]
- 23.Hasugian AR, Purba HL, Kenangalem E, Wuwung RM, Ebsworth EP, Maristela R, Penttinen PM, Laihad F, Anstey NM, Tjitra E, Price RN. 2007. Dihydroartemisinin-piperaquine versus artesunate-amodiaquine: superior efficacy and posttreatment prophylaxis against multidrug-resistant Plasmodium falciparum and Plasmodium vivax malaria. Clin Infect Dis 44:1067–1074. doi: 10.1086/512677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoso 2011. Evaluasi penggunaan artesunat-amodiakuin (artesdiakuin) pada pengobatan malaria tanpa komplikasi di Puskesmas Penyandingan dan Tanjung Lengkayap Kabupaten Oku. Bull Penelit Kesehat 39:99–109. (In Indonesian.) [Google Scholar]
- 25.Faranita T, Pasaribu AP, Ali M, Lubis M, Pasaribu S. 2012. Efficacy of artemether-lumefantrine and artesunate-amodiaquine for treating uncomplicated falciparum malaria in children. Paediatr Indones 52:260–266. doi: 10.14238/pi52.5.2012.260-6. [DOI] [Google Scholar]
- 26.Lubis IND, Wijaya H, Lubis M, Lubis CP, Divis PCS, Beshir KB, Sutherland CJ. 2017. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J Infect Dis 215:1148–1155. doi: 10.1093/infdis/jix091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubis IND, Wijaya H, Lubis M, Lubis CP, Beshir KB, Staedke SG, Sutherland CJ. 2020. Recurrence of Plasmodium malariae and P falciparum following treatment of uncomplicated malaria in North Sumatera with dihydroartemisinin-piperaquine or artemether-lumefantrine. Open Forum Infect Dis, in press. doi: 10.1093/ofid/ofaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ménard D, Khim N, Beghain J, Adegnika AA, Amodu O, Barnadas C, Berry A, Boum Y, Bustos MDG, Cao J, Caridad J, Espino FE, Chen JH, Collet L, Cui L, Das Thakur G, Dieye A, Djallé DD, Dorkenoo MA, Eboumbou Moukoko C, Fall B, Fandeur T, Ferreira-da-Cruz MF, Fola AA, Fuehrer HP, Hassan AM, Herrera S, Hongvanthong B, Houzé S, Hua TL, Huy R, Jahirul Karim M, Jiang L, Kano S, Khan W, Khanthavong M, Kremsner PG, Lacerda M, Laminou IM, Leang R, Leelawong M, Lin K, Mazarati JB, Mei L, Menard S, Morlais I, Muhindo Mavoko H, Musset L, Na-Bangchang K, Nambozi M, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhingra SK, Gabryszewski SJ, Small-Saunders JL, Yeo T, Henrich PP, Mok S, Fidock DA. 2019. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio 10:e02731-18. doi: 10.1128/mBio.02731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kümpornsin K, Takala-Harrison S, Witkowski B, Fairhurst RM, Ariey F, Menard D, Fidock DA. 2018. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 9:3314. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA, Neafsey DE, Musset L. 2015. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci U S A 112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, Dhorda M, Amaratunga C, Woodrow CJ, Ashley EA, White NJ, Dondorp AM, Fairhurst RM, Ariey F, Menard D, Wirth DF, Volkman SK. 2018. Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 9:1769. doi: 10.1038/s41467-018-04104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamaliddin C, Joste V, Hubert V, Kendjo E, Argy N, Houze S. 2019. Evaluation of PCR to monitor Plasmodium falciparum treatment efficacy in a nonendemicity setting. J Clin Microbiol 58:e01080-19. doi: 10.1128/JCM.01080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A 106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syafruddin D, Asih PB, Wahid I, Dewi RM, Tuti S, Laowo I, Hulu W, Zendrato P, Laihad F, Shankar AH. 2007. Malaria prevalence in Nias District, North Sumatra Province, Indonesia. Malar J 6:116–123. doi: 10.1186/1475-2875-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srimuang K, Miotto O, Lim P, Fairhurst RM, Kwiatkowski DP, Woodrow CJ, Imwong M, Tracking Resistance to Artemisinin Collaboration . 2016. Analysis of anti-malarial resistance markers in pfmdr1 and pfcrt across Southeast Asia in the Tracking Resistance to Artemisinin Collaboration. Malar J 15:541. doi: 10.1186/s12936-016-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, McNamara DT, Bockarie MJ, Kazura JW, Kyle DE, Fidock DA, Zimmerman PA. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci U S A 98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen N, Baker J, Ezard N, Burns M, Edstein MD, Cheng Q. 2002. Short report: molecular evaluation of the efficacy of chloroquine treatment of uncomplicated Plasmodium falciparum malaria in East Timor. Am J Trop Med Hyg 67:64–66. doi: 10.4269/ajtmh.2002.67.64. [DOI] [PubMed] [Google Scholar]
- 39.Poespoprodjo JR, Kenangalem E, Wafom J, Chandrawati F, Puspitasari AM, Ley B, Trianty L, Korten Z, Surya A, Syafruddin D, Anstey NM, Marfurt J, Noviyanti R, Price RN. 2018. Therapeutic response to dihydroartemisinin-piperaquine for P falciparum and P vivax nine years after its introduction in Southern Papua, Indonesia. Am J Trop Med Hyg 98:677–682. doi: 10.4269/ajtmh.17-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadalla NB, Elzaki SE, Mukhtar E, Warhurst DC, El-Sayed B, Sutherland CJ. 2010. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malar J 9:74–83. doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.