ABSTRACT
Background
Klebsiella pneumoniae
(hereafter, Kp) is a major public health threat responsible for high levels of multidrug resistant (MDR) human infections. Besides, Kp also causes severe infections in the community, especially in Asia and Africa. Although most Kp infections are caused by endogenous intestinal carriage, little is known about the prevalence and microbiological characteristics of Kp in asymptomatic human carriage, and attached risk factors including environmental sources exposure.
Methods
Here, 911 pregnant women from communities in Madagascar, Cambodia, and Senegal were screened for gut colonization by Kp. Characteristics of Kp strains (antimicrobial susceptibility, genomic diversity, virulence, and resistance genes) were defined, and associated risk factors were investigated.
Results
Kp carriage rate was 55.9%, and Kp populations were highly heterogeneous (6 phylogroups, 325 sequence types, Simpson index 99.6%). One third of Kp isolates had acquired antimicrobial resistance genes. MDR-Kp (11.7% to 39.7%) and extended spectrum beta-lactamase (ESBL)-producing Kp (0.7% to 14.7%) varied among countries. Isolates with virulence genes were detected (14.5%). Environmental exposure factors including food, animal contacts, or hospitalization of household members were associated with carriage of Kp, antimicrobial resistance and hypervirulence. However, risk factors were country-specific and Kp subpopulation-specific.
Conclusion
This large-scale multicenter study uncovers the huge diversity of Kp in human gut carriage, demonstrates that antimicrobial resistance is widespread in communities of three low-income countries, and underlines the challenges posed by Kp colonization to the control of antimicrobial resistance.
KEYWORDS: Klebsiella pneumoniae, carriage, antibiotic resistance, genomic diversity, community, low-income countries
Background
Klebsiella pneumoniae (Kp) represents an increasing threat to public health. Kp strains that produce extended spectrum beta-lactamases (ESBLs) and carbapenemases often exhibit cross-resistance to other available agents, and pan-resistant Klebsiella are increasingly described.1 In low-income countries (LICs), Kp is responsible for a large part of early neonatal infections and for severe community-acquired infections.2 The latter can occur in previously healthy persons and are caused by Kp strains that are regarded as hypervirulent (HV-Kp).1 In LICs, multidrug resistant (MDR)-Kp is one important driver of unfavorable outcome in infections because appropriate therapy is often not available nor affordable. Worryingly, the convergence of MDR and HV phenotypes in single Kp strains is being increasingly reported.3
The taxonomy of Kp has been recently updated, highlighting the existence of five different species distributed in seven phylogroups (Kp1 to Kp7): K. pneumoniae sensu stricto (phylogroup Kp1), K. quasipneumoniae [subsp. quasipneumoniae (Kp2) and subsp. similipneumoniae (Kp4)], K. variicola [subsp. variicola (Kp3) and subsp. tropica (Kp5)], ‘K. quasivariicola’ (Kp6) and K. africana (Kp7).4 In the manuscript, ‘Kp’ abbreviation refers to the K. pneumoniae species complex (‘sensu lato’, including the five species/seven phylogroups).
Kp is a well-established bacteria of the gut microbiota, and most Kp infections are caused by endogenous strains that previously colonize the host asymptomatically.5,6 However, Kp carriage has been mostly analyzed within health-care facilities and with a focus on antimicrobial resistant strains. Currently, little is known about naturally occurring human carriage Kp populations, and on the prevalence and diversity of MDR-Kp or HV-Kp among these.
Kp is a ubiquitous bacterium present in animals, environmental sources, and food.7 Therefore, environmental exposures or behavioral factors may impact individual Kp carriage. Understanding the determinants of Kp carriage may lead to novel strategies to control the spread of this pathogen. To our knowledge, risk factors associated with Kp carriage have not been investigated in the community, nor in LICs.
The objectives of this study were to define the prevalence, antimicrobial resistance, genomic diversity, and virulence potential of Kp isolates carried by pregnant women from three LICs, and to identify risk factors associated with colonization of Kp and its resistant or virulent subpopulations.
Methods
Study design
This cross-sectional study was nested within the BIRDY program.8 Briefly, it consists of a cohort of pregnant women and their newborns in Madagascar, Senegal, and Cambodia with an urban and rural site in each country (Figure S1). Pregnant women enrolled between January 2015 and December 2016 were invited to participate in the present Klebsiella carriage study. A maternal stool sample was taken at delivery or, in case of home delivery, shortly after delivery. Data on maternal socio-demographic characteristics, medical history, and exposures, possibly related to Kp and its resistant or virulent subpopulations carriage, including dietary habits, antibiotic consumption, and contacts with animals, were collected.
Klebsiella pneumoniae isolation
Fecal samples were obtained using endorectal swabs or stool samples, inoculated in a Luria-Bertani broth with amoxicillin (10 mg/L final concentration) and incubated 18 h at 37°C.9 The enrichment was performed with amoxicillin because Kp isolates carry a constitutively expressed class A beta-lactamase (of blaSHV, blaOKP or blaLEN type) that confers intrinsic resistant to aminopenicillins (such as ampicillin and amoxicillin), whereas many other bacteria including E. coli (the most abundant potential competitor) are susceptible to amoxicillin. 100 microliters of the enrichment culture were plated onto selective Simmons citrate with inositol medium agar plates.10 Large, yellow, glossy colonies suspected of being Klebsiella were identified using either API20E or mass spectrometry (MALDI-ToF, Bruker). The first isolate identified as either Klebsiella pneumoniae or Klebsiella variicola was kept. For 7 carriers, two isolates genetically distinct based on 7-gene MLST were retained.11
Antimicrobial susceptibility testing
The susceptibility of Kp isolates to 16 antimicrobial agents (amoxicillin + clavulanic acid, ticarcillin + clavulanic acid, piperacillin + tazobactam, cephalothin, cefotaxime, ceftazidime, cefepime, aztreonam, imipenem, gentamicin, amikacin, tobramycin, nalidixic acid, ciprofloxacin, tetracycline, and trimethoprim + sulfamethoxazole) was defined using the agar disk diffusion method and interpreted according to EUCAST 2015 guidelines, except for cephalothin, nalidixic acid and tetracycline (CLSI M100-S25). ESBL isolates were identified using the double-disk synergy test (DDST).
Genomic sequencing and bioinformatics analyses
NextSeq-500 sequencing of Nextera XT 2 × 150 nt libraries was performed. Genomic assemblies (Table S1) were obtained using SPAdes v3.9.12 Multilocus sequence typing (MLST) was performed using the BIGSdb-Kp database (https://bigsdb.pasteur.fr).11,13 This resource was also used for detecting quinolone target genes mutations, and variants of the wzi and intrinsic class A beta-lactamases.13,14 Novel variants were submitted to BIGSdb (for blaSHV, blaOKP, and blaLEN variants) and to NCBI beta-lactamases database (blaSHV-206 to blaSHV-228). Other beta-lactamases and antimicrobial resistance gene families were detected using Kleborate (https://github.com/katholt/Kleborate). Isolates that carried only the intrinsic blaSHV (or blaOKP or blaLEN), fosA and oqxAB genes were considered as having an ancestral resistome. Other antimicrobial resistance genes were considered acquired. To evaluate the correlation between the antimicrobial resistance phenotype and genotype, and to depict co-resistance networks, we constructed a correlation matrix for binary variables (in the case of resistance genes: 1, presence; 0, absence; in the case of antimicrobial drugs: 1, resistant/intermediate; 0, susceptible) using the ‘corr.test’ function (Pearson method, which for a pair of binary variables equates to the Phi coefficient) from the ‘corrplot’ R package. Significant correlations were visualized utilizing the ‘corrplot’ function from the same package.
Plasmid replicons were detected using PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/).15 Kleborate (https://github.com/katholt/Kleborate) was used to search for yersiniabactin, colibactin, aerobactin, and salmochelin clusters, and to predict the capsular types.16,17 BIGSdb-Kp was used to define the presence of other virulence-associated genes (Table S1).18 Isolates harboring at least one of the genes rmpA and rmpA2, and/or at least one complete gene cluster among iucABCD-iutA (aerobactin), iroBCDN (salmochelin), ybtAEPQSTUX-fuyA-irp1-irp2 (yersiniabactin), clbAB-CDEFGHIJKLMNOPQR (colibactin) and allABCDRS (allantoinase) were considered virulent. Hypervirulent Kp were defined as isolates harboring at least one of the genes rmpA and rmpA2, and/or at least one complete gene cluster among iucABCD-iutA (aerobactin) and iroBCDN (salmochelin). We defined ‘high-risk clones’ as clones harboring 7-gene MLST sequence types (ST) represented at least 10 times in NCBI genomes (April 2019: 6,258 genomes available) and mentioned in the title or abstract of at least five publications in NCBI PubMed (“Klebsiella” + “pneumoniae” + “STxxx”, April 2019).
Phylogenetic and statistical analyses
Genome phylogeny was derived from MASH distances using JolyTree (https://gitlab.pasteur.fr/GIPhy/JolyTree) and visualized using iTOL (https://itol.embl.de/).19 Unique strains were defined with a ≤ 10 allelic mismatch cutoff within allelic variation recorded using BIGSdb genome comparator function based on the 5,073 protein-coding genes of strain Kp616 (GCA_003076555.1; 90% identity and 90% length coverage).
Statistical analyses were performed using Stata version 15 (Stata Corp., College Station, TX). We defined multidrug resistant (MDR) strains as being resistant to at least one antimicrobial agent in at least 3 antibacterial categories. Chi square or Fisher exact test was used to compare the prevalence of Kp subsets (all, MDR, ESBL) and antibiotic resistance patterns among countries. Risk factor analysis was performed separately for each country given the inter-country variation of the pregnant women’s characteristics. We constructed univariate analysis to examine associations between women’s environmental exposures, and overall Kp, MDR-, ESBL-, and virulent Kp carriage. All variables with a univariate p-value ≤0.2 were included in multivariate analysis. We used a logistic regression model for all, MDR, and ESBL-Kp subsets. We conducted backwards stepwise elimination of non-significant parameters (p < .05). All multivariate models were adjusted for site (urban/rural). For the seven women from whom 2 Kp were isolated, both were kept in the multivariate analysis; sensitivity analyses performed with and without these duplicates led to identical results.4,20–22
Ethics
The study was approved by the ethics committees of Madagascar (068-MSANP/CE), Senegal (SEN 14–20), Cambodia (132 NEHCR) and was authorized by the Institut Pasteur in Paris, France. Written informed consent was given by all participants.
Results
Kp carriage
A total of 911 pregnant women from Madagascar (n = 423), Cambodia (n = 152), and Senegal (n = 336) were enrolled. For 874 (95.9%) of these, both epidemiological data and a fecal sample were obtained (Figure S2). Women enrolled in the urban sites represented 32.6%, 55.5%, and 35.2% of the total in Madagascar, Cambodia, and Senegal, respectively. The study population (Table 1) was young (mean 27 years; SD 6.4) and underprivileged, as a large majority (79.3%-88.9%) did not complete secondary school and an important fraction did not have toilets within the housing (Madagascar: 93.1%; Cambodia: 49.3%; and Senegal: 29.9%).
Table 1.
Characteristics of the inclusion population.
| n (%) or mean (SD) |
||||
|---|---|---|---|---|
| Madagascar N = 405 | Cambodia N = 146 | Senegal N = 323 | p-value | |
| Urban Site | 132 (32.6) | 81 (55.5) | 105 (35.2) | <0.001 |
| Age (years) | 25.9 (6.4) | 27 (5.7) | 28 (6.71) | 0.08 |
| Gestity | ||||
| 1 | 121 (29.9) | 48 (32.9) | 70 (21.7) | 0.04 |
| >1 | 284 (70.1) | 119 (67.1) | 253 (78.3) | |
| Education | ||||
| Absence/primary school | 90 (22.2) | 78 (53.4) | 227 (70.3) | <0.001 |
| Partial secondary school | 231 (57.0) | 50 (34.2) | 62 (19.2) | |
| Complete secondary or higher | 84 (20.7) | 18 (12.3) | 34 (11.1) | |
| Occupancy | ||||
| Unemployed | 263 (64.9) | 50 (34.3) | 258 (79.9) | <0.001 |
| Manual job | 129 (31.9) | 90 (61.6) | 55 (17.0) | |
| Clerical job | 13 (3.2) | 6 (4.1) | 10 (3.1) | |
| House characteristics | ||||
| Individual housing | 135 (33.3) | 93 (64.1) | 210 (65.0) | <0.001 |
| Housing within a compound | 133 (32.8) | 30 (20.7) | 73 (22.6) | |
| Accommodation shared with other families | 137 (33.8) | 22 (15.2) | 40 (12.4) | |
| Electricity | 299 (73.8) | 145 (99.3) | 307 (95.6) | <0.001 |
| Toilets | ||||
| Inside the house | 28 (6.9) | 74 (50.6) | 225 (70.2) | <0.001 |
| Outside the house | 377 (93.1) | 72 (49.3) | 96 (29.9) | |
| Pregnancy follow-up | ||||
| Skilled health-care workers | 384 (97.2) | 142 (97.3) | 322 (99.7) | 0.02 |
Kp colonization was detected in 489 of 874 (55.9%) women. Carriage rate per country (Figure 1a) was significantly lower in Senegal (40.2%, 95% CI [36.3–46.9]; p < .001) compared to Madagascar (64.7%, CI [59.9–69.2]) and Cambodia (66.4%, [58.3–73.7]). Carriage rate was lower in the urban area than in the rural area (Figure 1a) in Senegal (29.5% versus 45.4%, p = .006) but not in Madagascar (63.6% versus 65.2%, p = .7) and Cambodia (61.5% versus 70.4%, p = .3).
Figure 1.
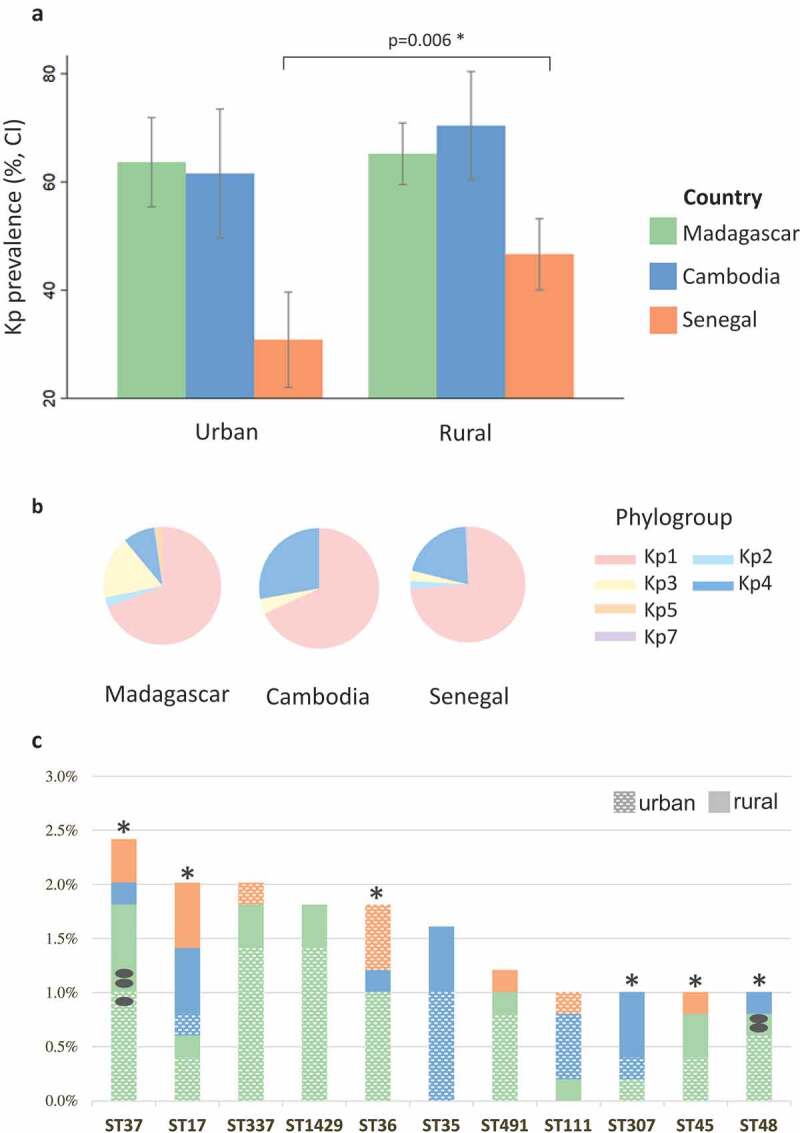
Rates of Kp carriage and genotype diversity.
Kp phylogenetic diversity
Genomic sequence-based phylogenetic relationships (Figure 2) revealed the presence of six deep lineages, corresponding to K. pneumoniae phylogroups Kp1 (70.1%), Kp2 (2%), Kp3 (10.7%), Kp4 (15.7%) and Kp5 (2.3%) and a novel phylogroup we named Kp7, described elsewhere as K. africana.4 Kp1 was the most frequent phylogroup in each country (68–73.7%), but phylogroups distribution differed among countries (p < .001) (Figure 1b): Kp4 was the second most common group in Cambodia and Senegal (27.8% and 20.4%, respectively) but not in Madagascar (Kp3: 17.2%; Kp4: 8.4%).
Figure 2.
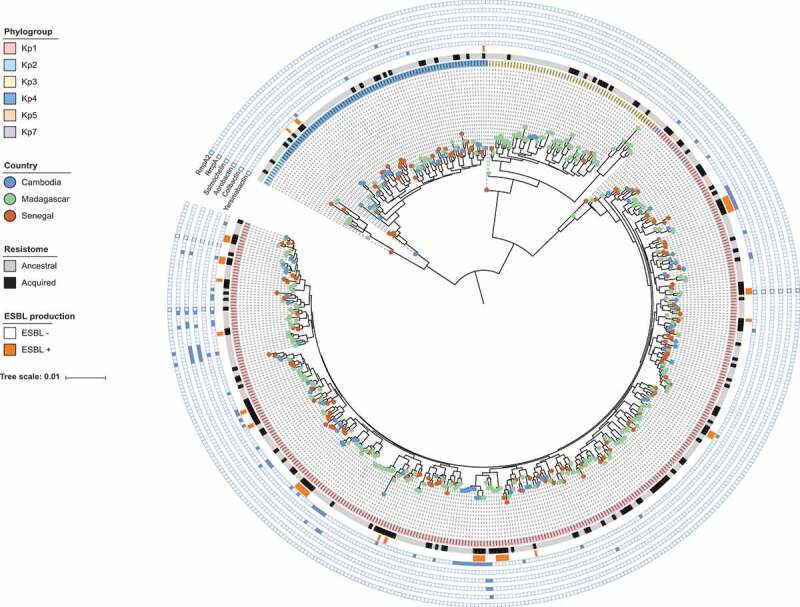
Phylogenetic relationships. The scale bar corresponds to 0.01 substitutions per site. iTOL (https://itol.embl.de/) was used to visualize country of origin (circles at branch tips), phylogroups (background color in first circle comprising the isolates names), the ancestral or acquired character of isolates’ resistome (second circle), ESBL phenotype (third circle), and virulence genes as indicated in front of each external circle.
A remarkably high genotypic diversity was found based on MLST. There were 325 STs, a majority of which (242, 74.5%) were represented by a single isolate (Figure S3). The genotype distribution into multiple low-frequency STs was reflected by a very high (99.6%) Simpson diversity index, even within individual countries or sites (range, 98.7–99.7%). High-risk clones (Table S2) represented 70 (14.1%) isolates (Table S3). Among 11 STs with ≥5 isolates, six were high-risk clones and nine were isolated in two or three countries (Figure 1c). However, based on the number of allelic differences between strains within these clonal groups, only two strains were observed in more than one carrier (Figure 1c; Table S1).
Antimicrobial resistance
Antimicrobial resistance rates differed among the three countries (Figure 3a; Table S4). Low levels of resistance were observed for imipenem (0.8%) and amikacin (0.4%). Resistance rates to quinolones (15.5%), other aminoglycosides (17.5%), and third-generation cephalosporins (14.4%) were higher (p < .001) in Cambodia compared to Madagascar (2.7%, 8%, and 10%, respectively) and Senegal (~3% each). ESBL-Kp were almost absent in Senegal (0.7%) but they were frequent in Cambodia (14.4%) and Madagascar (8.4%). MDR-Kp rates were higher in Madagascar (39.7%) compared to Cambodia (25.8%) and Senegal (11.7%; p < .001). Multidrug-resistance was higher in the urban site compared to the rural site only in Cambodia (42.5% vs 14%, p = .002; Table S4). In Cambodia, 10.3% of the isolates were resistant to at least 8 categories of antibiotics. As expected, higher resistance levels were observed for older antibiotics (amoxicillin and ticarcillin with clavulanate, tetracycline, and trimethoprim/sulfamethoxazole).
Figure 3.
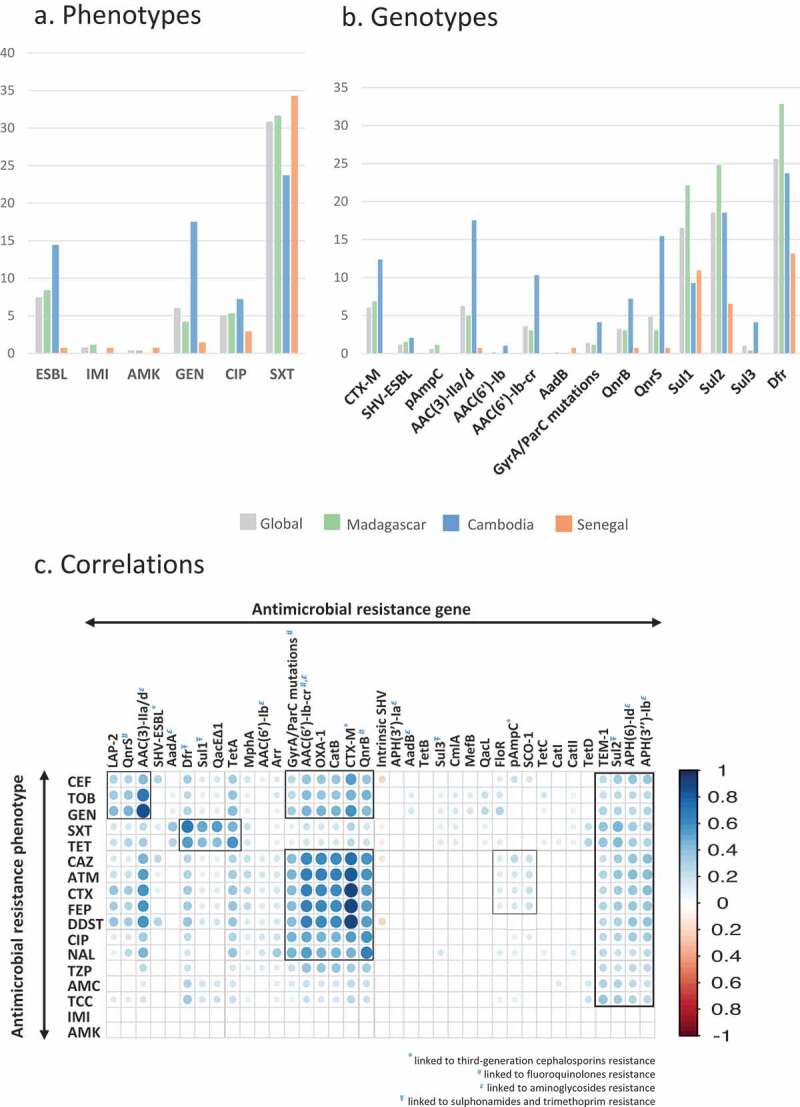
Antimicrobial resistance.
Comparisons among phylogroups showed differences only in Cambodia, with higher resistance rates against gentamicin and tetracycline in Kp1 (p = .04 and p = .006, respectively; Table S3).
Antimicrobial resistance genes, plasmids and sublineages
As expected, oqxAB, fosA, and the chromosomal beta-lactamase gene were ubiquitous (Table S3; Figure 3b); 117 variants of the latter were found, with blaSHV-11 (n = 145, 29.7%) being the most frequent (Figure S4). Remarkably, 334 (67.4%) of isolates had no acquired resistance gene nor any GyrA or ParC alterations, thus corresponding to the ancestral (“wild-type”) resistome type. The median number of acquired resistance genes was 3 (similar in each country), ranging from 9 to 11 among ESBL producers. Of the 37 ESBL enzymes, CTX-M-15 was the most common (n = 22, 60%). Resistance genes were strongly associated with phenotypic resistance (Figure 3c), and some gene associations were depicted, reflecting their genetic linkage on common genetic platforms (plasmids, transposons, integrons) and resulting in co-resistance phenotypes. For example, blaOXA-1, aac(6’)-Ib-cr, catB, and qnrB were associated with blaCTX-M-15 gene, whereas blaLAP-2, qnrS, and aac(3)-IId were linked to blaCTX-M-14/-27; detection rates for both combinations were higher in Cambodia (Table S5).
Plasmid replicons were found in 70.6%, 73.9%, and 55.5% of Kp from Madagascar, Cambodia, and Senegal, respectively. Twenty-two replicon types were detected (Table S2), of which IncFIB(K) (36.1%), Col440I (27.4%), IncFII (26.4%), IncR (15.9%), and IncFIA(HI1) (15.1%) were the most frequent. However, the predominant type in Senegal was Col440I (31.4%), and plasmid distribution differed among countries (p < .001).
Seventy isolates (14.1%) belonged to widespread genotypes commonly associated with ESBL or carbapenemase production (Table S3). Furthermore, among ESBL-Kp, 19 (51.4%) belonged to previously recognized MDR STs. Although ST307 is emerging as a major carbapenemase-producing sublineage,23,24 our ST307 carriage isolates did not harbor carbapenemase genes.
Virulence genes and convergence with resistance
Seventy-two (14.5%) virulent isolates were found (Figure 1), with yersiniabactin being the most frequent virulence factor (62 isolates, 12.5%; Figure S5; TableS5). The regulator of mucoid phenotype genes rmpA and rmpA2 were present in six isolates (1.2%), whereas aerobactin and salmochelin clusters were present in 14 (2.8%) and 11 (2.2%) isolates, respectively; these hypervirulence-associated clusters were observed mostly in Madagascar and were associated with STs of capsular serotype K2 previously described as hypervirulent:25,26 ST25, ST65, ST375, and ST380 (Table S3). In contrast, no isolate of the liver abscess-associated serotype K1 ST23 was found.
Virulence and antibiotic resistance elements were generally observed in distinct isolates (Table S3; Figure S6). In particular, aerobactin and salmochelin hypervirulence clusters were associated with ancestral resistome isolates (in Madagascar, p = .05 and p = .06, respectively). However, in one urban Cambodian isolate (SB5663, ST17), a convergence of MDR (blaCTX-M-27, blaLAP-2, dfrA, sul2, tetA, floR) and hypervirulence (rmpA, yersiniabactin, salmochelin/ICEKp1) was observed (Table S3; Figure S6).
Associations between environmental exposures and specific characteristics of Kp
Risk factors of Kp, MDR, ESBL, and virulent Kp carriage were investigated (Table 2; Table S7). The use of antibiotics during pregnancy was associated with a higher risk of Kp carriage (in Madagascar, adjusted odds ratio (aOR) = 2.1; p = .03), as was dry fish consumption (in Cambodia: aOR = 2; p = .05) and contact with chicken (in Senegal: aOR = 1.9; p = .008). In contrast, hands washing after using toilets (in Madagascar: aOR = 0.5; p = .02), but also manipulating animal excrement (in Madagascar, aOR = 0.5; p = .008), were associated with a reduced risk of Kp carriage.
Table 2.
Environmental exposures and colonization with Kp, MDR Kp and virulent Kp.
| Kpn + | Kpn - | Multivariate analysis* | MDR Kp |
non MDR Kp | Multivariate analysis* | Virulent Kp | non virulent Kp | Multivariate analysis* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Madagascar | ||||||||||||||
| N = 262 (n, %) |
N = 143 (n, %) |
aOR [95%CI] | p | N = 104 | N = 158 | aOR | p | N = 34 | N = 228 | aOR | p | |||
| Antibiotherapy during pregnancy | 42 (16.0) | 15(10.5) | 2.1 [1.1–4.1] |
0.03 | Hand washing after toilets use | 65(70.7) | 107(81.1) | 0.4 [0.2–0.9] |
0.02 | Rabbit meat consumption $$ | 7(20.6) | 15(6.6) | 3.6 [1.3–9.8] |
0.01 |
| Hand washing after toilets use | 172(76.9) | 115(86.4) | 0.5 [0.3–0.9] |
0.02 | Dry meat consumption $$ | 12(11.9) | 32(20.9) | 0.4 [0.2–0.8] |
0.01 | Hospitalization of a household member | 7(20.6) | 13(5.8) | 4.0 [1.4–11.2] |
0.009 |
| Manipulation of animal excrement | 35(13.4)) | 31(21.7) | 0.5 [0.2–0.8] |
0.008 | Raw vegetables consumption$ | 91 (88.4) | 128 (82.1) | 2.4 [1.1–5.5] |
0.03 | |||||
| Cambodia | ||||||||||||||
| N = 97 | N = 49 | aOR | p | |||||||||||
| Dry fish consumption $ | 51(52.6) | 17(34.7) | 2.0 [1.0–4.1] |
0.05 | ||||||||||
| Senegal | ||||||||||||||
| N = 137 | N = 193 | aOR | p | N = 16 | N = 121 | aOR | p | N = 29 | N = 108 | aOR | p | |||
| Contact with chicken | 82(59.9) | 78(40.4) | 1.9 [1.2–3.1] |
0.008 | Contact with chicken | 5(31.3) | 77(63.6) | 0.2 [0.06–0.7] |
0.01 | Fish consumption $ | 23 (79.3) | 101(93.5.) | 0.2 [0.1–0.8] |
0.02 |
| Use of fecal material as fertilizer | 8(27.6) | 11(10.2) | 3.7 [1.7–11.0] |
0.02 | ||||||||||
* All multivariate analyses are adjusted on site. Only significant parameters are shown
$ Consumption frequency was categorized as “at least once per week” versus “less than once per week”
$$ Consumption frequency was categorized as “ever” versus “never”
Among Kp-colonized women, those who reported washing their hands after using toilets (in Madagascar: aOR = 0.4; p = .02), to eat dried meat (in Madagascar: aOR = 0.4; p = .01), or to be in contact with chicken (in Senegal: aOR = 0.2; p = .01) were less at risk of being MDR-Kp colonized. In contrast, women who reported eating raw vegetables at least once per week (in Madagascar: aOR = 2.4; p = .03) were more at risk of being MDR-Kp colonized.
Regarding virulent Kp carriage, hospitalization of a household member in the previous year (in Madagascar: aOR = 4.0; p = .001), consumption of rabbit meat (in Madagascar: aOR = 3.6; p = .01), and use of fecal material as fertilizer (Senegal: 3.7, p = .02) were risk factors. In contrast, fish consumption was associated with decreased virulent Kp colonization (Senegal: OR = 0.2; p = .02).
No risk factor for MDR and virulent Kp carriage in Cambodia and for ESBL Kp carriage in the 3 countries were identified.
Discussion
Despite the importance of Kp carriage in the ecology and spread of this pathogen, knowledge on Kp colonization is scarce, especially in LIC communities. In previous carriage studies, other Enterobacteriaceae including E. coli were considered together with Kp, which represented a minority of studied isolates.27,28 In addition, previous work focused on ESBL- or carbapenemase-producing Enterobacteriaceae carriage, overlooking the natural diversity and ecology of susceptible populations from which resistant strains evolve.
The design of our community-based multicentric study is unique in that the naturally colonizing population of Kp was investigated irrespective of antimicrobial resistance phenotypes, providing insights into the natural ecology of Kp in human carriage. We found that Kp carriage rate per country ranged from 40% to 66%. Kp community carriage studies in high-income countries found carriage rates ranging from 6% to 35%; rates were much higher in one study of Chinese volunteers living in Asian countries.5–7,29 Although hospitalization is known to increase Kp carriage, our estimates are even higher than the prevalence of Kp carriage reported in hospital settings, typically ranging from 20 to 35%.5,6,27 Kp abundance in the gut microbiota is low; combining an enrichment step with a Klebsiella selective medium may have contributed to recover Kp at higher rates here than in previous studies.30 Of concern, pregnant women can transmit to their children during delivery or after birth, potentially leading to neonatal sepsis.31
Our data demonstrate that Kp1 is the most prevalent phylogroup in all sites. Currently, recognized high-risk clones all belong to Kp1, and this phylogroup is the most frequent in human infections.26,32 Our results suggest that the clinical predominance of Kp1 might be a direct reflection of the ecological dominance of this group in carriage, rather than to a higher infectious potential compared to other phylogroups. Consistent with this hypothesis, the high rate of Kp4 carriage in Cambodia is mirrored by common Kp4 bloodstream infections in this country.3
A striking inter-carrier genetic diversity of Kp was found, with almost three quarters of isolates having a unique ST. Further, genomic sequencing ruled out in all cases except two, the occurrence of recent transmission. Human Kp carriage populations thus appear structured into individual carriers acting as rarely connected islands, favoring evolutionary diversification into a multitude of genotypes.
Kp isolates causing hypervirulent infections belong to recognized sublineages of capsular serotypes K1 and K2, but the source of these infections is poorly known. K1-ST23 was common among Asian healthy carriers.33 Here, several isolates belonging to hypervirulent K2 sublineages were found, suggesting that the human gut may act as a reservoir of these strains.
Antimicrobial resistance differed among countries, likely reflecting the different levels of antibiotic use, hygiene, and infection control between the 3 countries. The proportion of MDR and ESBL-producing isolates was high in Madagascar and Cambodia. These findings are worrisome, as the gut is a well-known reservoir of infections by these strains.5,6 Besides, the role of Kp as a source of ESBL genes and other resistance determinants that can be transmitted horizontally to other pathogens is a clear cause of concern.34 We did not find any carbapenemase-producing Kp, indicating that its spread in the community is still limited in these countries, consistent with low carbapenem consumption.35
We were able to investigate for the first time, risk factors associated with natural carriage of Kp and its genetic or phenotypic subpopulations. We found that women who washed their hands after toilet use were less likely to be colonized with Kp or MDR Kp in Madagascar. Hand washing is effective in controlling the transmission of fecal-oral infection in the community, but is often neither properly done nor practiced at key times.36 Our finding supports that promotion of a simple hygienic behavior could contribute to the control of the spread of antimicrobial resistance in the community.
Consumption of food or contact with some animals were found associated with Kp, MDR, or virulent Kp carriage. Raw vegetables or rabbit meat (although rabbit meat consumption was low) were associated with a higher risk of being colonized with MDR-Kp or virulent Kp, respectively, suggesting a possible food origin for these isolates. Bacteria can persist on products that are inadequately cooked prior to consumption. However, at which step of the food supply chain these products are contaminated remains an open question. This study calls for more focused investigations of the presence of Kp on food. We also found that manipulating animal excrement and having animal contacts appear protective of Kp and MDR-Kp colonization. One explanation could lie in negative ecological associations between Kp and other bacteria during animal hosts colonization. Future studies on interactions of Kp with the gut microbiota are warranted.
We acknowledge several limitations to this study. Due to the cross-sectional design, the interpretation of the risk factors must be cautious, in particular due to recall bias. Our sample size was not very large, specifically in Cambodia, which might explain that we found few risk factors associated with MDR or virulent Kp. Although the majority of the samples consisted in a stool collection (85%), endorectal swabbing was performed instead when stool was not available. However, previous studies showed that bacterial communities defined from endorectal swabbing and stool specimens are similar.37,38 In addition, we used an enrichment step that allows the recovery of Kp isolates even when the amount of starting material is low, which could potentially be the case when using rectal swabs.
Human-to-human contact was not investigated in depth here. Characteristics reflecting a potential human transmission (number of household members) were collected and were not significantly associated with Kp carriage. We performed our carriage study in pregnant women, who represent a homogeneous and generally healthy population, with limited socioeconomic status bias. However, the immune system is modified during pregnancy, impacting the gut microbiota.39 Whether pregnancy has an impact on Kp carriage would require dedicated studies. Finally, food or environmental sampling performed in parallel to human screening would have allowed comparing Kp populations in these niches, possibly leading to their association with human carriage, but these studies will be particularly challenging in light of the huge diversity of Kp revealed herein.40
Conclusions
To conclude, this multicentric study provides the most detailed picture to date of Kp colonization in human LICs communities. One important outcome is the high prevalence of Kp carriage, and high levels of antimicrobial resistance among carriage Kp isolates. Our findings also illustrate the challenges of understanding the transmission dynamics of ubiquitous pathogens. As Kp carriage risk factors were country-dependent, major drivers could not be pointed out, probably reflecting the influence of a complex combination of antibiotic use, hygienic and environmental conditions in the given local context. Consequently, our results suggest that there is “no one size fits all” intervention to tackle the Kp community burden and that public health strategies to control Kp spread may need to be defined based on local specificities.
Supplementary Material
Acknowledgments
We thank all collaborators of the BIRDY program: Rasoanaivo Fanjalalaina Vololonirina, Andrianonimiadana Lova Maminirina, Rabearitiana Lydos, Rasoloson Dimitri, Randrianirina Frédérique, Ratsima Hariniaina Elisoa, Volahasina Antsa Tanjona, Volahasina Antso Fenitra, Randriamamonjiarison Aina Nirina, Andrianirina Zafitsara Zo, Andriatahina Todisoa, Vincent Richard, Awa Ndir, Amy Gassama Sow, Balla Sy, Marguerite Diatta, Pape Samba Dieye, Jean Baptiste Diouf, Joseph Faye, Abibatou Ndiaye, Bouya Ndao, Thida Chon, Veronique Ngo, Arnaud Tarantola, Sok Touch, Patrice Piola, Sophie Goyet, Siyin Lach, Elsa Kermorvant-Duchemin, Michael Padget, Laurence Watier, and Abdou Armya Youssouf.
We thank all physicians, laboratory staff, field interviewers, and community workers involved in the project. We thank Leonardo Panunzi for assistance with genomic analyses of resistance genes, and the “Plateforme de Microbiologie Mutualisée” of Institut Pasteur for genomic sequencing. We are grateful to all women participating in the BIRDY program.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the Institut Pasteur programme Action Concertee Inter-Pasteurienne (Grant A-14-2014) and by the French government’s Investissement d’Avenir program Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant ANR-10-LABX-62-IBEID). Support to the BIRDY project was provided by the Total Foundation, MSDAVENIR and the Monaco Department of International Cooperation. CR was supported financially by the MedVetKlebs project, a component of European Joint Programme One Health EJP, which has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830. The funders had no role in study design, data collection and analysis, interpretation, or writing of the manuscript.
Accession numbers
The genomic sequencing data were deposited in the NCBI/ENA/DDBJ databases and are accessible via the BioProject PRJEB29143.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Woodford N, Turton JF, Livermore DM.. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. [DOI] [PubMed] [Google Scholar]
- 2.Zaidi AK, Thaver D, Ali SA, Khan TA.. Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect J. 2009;28:S10–8. [DOI] [PubMed] [Google Scholar]
- 3.Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchêne S, Jenney A, Holt KE. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:e1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues C, Passet V, Rakotondrasoa A, Diallo TA, Criscuolo A, Brisse S. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol. 2019;170:165–170. [DOI] [PubMed] [Google Scholar]
- 5.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Pilcher DV, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65:208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, Malani PN, Rao K, Bachman MA. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere [Internet] 2016; 1. http://www.ncbi.nlm.nih.gov/pubmed/27777984 [DOI] [PMC free article] [PubMed]
- 7.Brisse S, Grimont F, Grimont PA, Schleifer K-H, Stackebrandt E. The Genus Klebsiella. In: Dworkin M, Falkow S, Rosenberg E, editors. The Prokaryotes. A handbook on the biology of bacteria. New York: Springer; 2006. [Google Scholar]
- 8.Huynh B-T, Kermorvant-Duchemin E, Herindrainy P, Padget M, Rakotoarimanana FMJ, Feno H, Hariniaina-Ratsima E, Raheliarivao T, Ndir A, Goyet S, et al. Bacterial infections in neonates, madagascar, 2012-2014. Emerg Infect Dis. 2018;24:710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeggman S, Löfdahl S, Paauw A, Verhoef J, Brisse S. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:2400–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Kregten E, Westerdaal NA, Willers JM. New, simple medium for selective recovery of Klebsiella pneumoniae and Klebsiella oxytoca from human feces. J Clin Microbiol. 1984;20:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: bIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51:4073–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genomics. 2016;2:e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bojer MS, Struve C, Ingmer H, Hansen DS, Krogfelt KA. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS One. 2010;5:e15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blin C, Passet V, Touchon M, Rocha EPC, Brisse S. Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Env Microbiol. 2017;19:1881–1898. [DOI] [PubMed] [Google Scholar]
- 21.Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol. 2001;51:915–924. [DOI] [PubMed] [Google Scholar]
- 22.Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T, Olsen RJ. Whole-genome sequencing of a human clinical isolate of the Novel Species Klebsiella quasivariicola sp. nov. Genome Announc. 2017;5:e01057–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, Endimiani A, Mammina C, Ocampo AM, Jimenez JN, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genomics. 2017;3:e000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, Hamidian M, Howden BP, Löhr IH, Holt KE. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother. 2019;74:577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U A. 2015;112:E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luvsansharav UO, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y. Prevalence of and risk factors associated with faecal carriage of CTX-M beta-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother. 2012;67:1769–1774. [DOI] [PubMed] [Google Scholar]
- 28.Chereau F, Herindrainy P, Garin B, Huynh BT, Randrianirina F, Padget M, Piola P, Guillemot D, Delarocque-Astagneau E. Colonization of extended-spectrum-beta-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother. 2015;59:3652–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y-T, Siu LK, Lin J-C, Chen T-L, Tseng C-P, Yeh K-M, Chang F-Y, Fung C-P. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy chinese and overseas chinese adults in Asian countries. BMC Microbiol. 2012;12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One. 2012;7:e47075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan GJ, Lee AC, Baqui AH, Tan J, Black RE. Risk of early-onset neonatal infection with maternal infection or colonization: a global systematic review and meta-analysis. PLoS Med. 2013;10:e1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brisse S, van Himbergen T, Kusters K, Verhoef J. Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. Clin Microbiol Infect. 2004;10:942–945. [DOI] [PubMed] [Google Scholar]
- 33.Chung DR, Lee H, Park MH, Jung SI, Chang HH, Kim YS, Son JS, Moon C, Kwon KT, Ryu SY, et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis. 2012;31:481–486. [DOI] [PubMed] [Google Scholar]
- 34.Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139. [DOI] [PubMed] [Google Scholar]
- 35.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018;115:E3463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halder AK, Tronchet C, Akhter S, Bhuiya A, Johnston R, Luby SP. Observed hand cleanliness and other measures of handwashing behavior in rural Bangladesh. BMC Public Health. 2010;10:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, Hayden MK. CDC Prevention Epicenters Program. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyman M, van Houten MA, Arp K, Sanders EAM, Bogaert D. Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S-rRNA sequencing. Sci Rep. 2019;9:16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Kling Bäckhed H, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the Gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanage WP. Two health or not two health? That is the question. mBio. 2019;10(2):e00550-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, Malani PN, Rao K, Bachman MA. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere [Internet] 2016; 1. http://www.ncbi.nlm.nih.gov/pubmed/27777984 [DOI] [PMC free article] [PubMed]


