Abstract
ABSTRACT
Salmonella enterica
serovar Typhi is the etiologic agent of typhoid fever, a major public health problem in the developing world. Moving toward and adhering to the intestinal epithelium represents key initial steps of infection by S. Typhi. We examined the role of the S. Typhi yrbE gene, which encodes an inner membrane phospholipid transporter, in these interactions with epithelial cells. Disruption of yrbE resulted in elevated expression of flagellin and a hypermotile phenotype. It also significantly reduced the ability of S. Typhi to adhere to the HeLa epithelial cell line and to polarized primary epithelial cells derived from human ileal organoids. Interestingly, the yrbE-deficient strain of S. Typhi induced higher production of interleukin-8 from the primary human ileal epithelial cell monolayers compared to the wild-type bacteria. Deletion of the flagellin gene (fliC) in the yrbE-deficient S. Typhi inhibited motility and attenuated interleukin-8 production, but it did not correct the defect in adhesion. We also disrupted yrbE in S. Typhimurium. In contrast to the results in S. Typhi, the deficiency of yrbE in S. Typhimurium had no significant effect on flagellin expression, motility or adhesion to HeLa cells. Correspondingly, the lack of yrbE also had no effect on association with the intestine or the severity of intestinal inflammation in the mouse model of S. Typhimurium infection. Thus, our results point to an important and serovar-specific role played by yrbE in the early stages of intestinal infection by S. Typhi.
KEYWORDS: Salmonella, typhoid, epithelial cells, adhesion, inflammation
Introduction
Typhoid fever is a systemic febrile disease caused by infection with the human-restricted Gram-negative enteropathogen Salmonella enterica serovar Typhi.1,2 It is a major public health problem in the developing world: the risk-adjusted burden of typhoid has been estimated to be 12 million illnesses and 130,000 deaths annually in low- and middle-income countries, with most of the cases occurring in children less than 5 y of age.3 Diagnosis of the condition can be difficult, and treatment is increasingly complicated by the emergence of multidrug-resistant (MDR) and even extensively drug-resistant (XDR) strains of S. Typhi.1,4,5 Thus, there is an urgent need for new strategies to deal with the infection therapeutically and prophylactically. The development of such strategies depends on detailed understanding of typhoid pathogenesis.
Since S. Typhi infection occurs following the ingestion of contaminated food or water, the early steps in pathogenesis involve adhesion to and invasion of the intestinal epithelium, particularly the follicle-associated epithelium overlying Peyer’s patches in the ileum.6,7 Because S. Typhi infects only humans, much of what we know about Salmonella–host interactions has been learnt from the study of S. Typhimurium, a broad host range serovar that infects a variety of animals, including laboratory mice.8 Although there are important differences between S. Typhi and S. Typhimurium with respect to genomic structure, host range, virulence factors and disease outcomes, the general mechanisms involved in infection are similar in the two pathogens.6,7,9–11 Following ingestion and transit through the proximal gut, Salmonella moves from the intestinal lumen to the epithelium using flagella-mediated motility and then adheres loosely to the epithelial cells by means of a variety of surface adhesins.12–16 Insertion of the Salmonella pathogenicity island 1 (SPI1)-encoded type III secretion system (TTSS) into the apical membrane of the epithelial cell follows, resulting in firm adhesion or docking.16,17 A number of bacterial effector proteins are then injected through the syringe-like SPI1 TTSS into the epithelial cytoplasm, leading to actin polymerization-mediated protrusions of the plasma membrane that ultimately engulf the bacteria within a membrane-bound vacuole.18 The bacteria are then translocated across the epithelium into the lamina propria and, in the case of S. Typhi, are disseminated to systemic tissues.19 S. Typhimurium infection in humans is usually confined to the gastrointestinal tract, where it causes an acute inflammatory response that is responsible for the associated diarrhea, vomiting and abdominal pain.1,11 This response is attenuated by various mechanisms during S. Typhi infection, which may facilitate systemic spread of the pathogen and help to explain the relatively minor intestinal symptoms of typhoid.1
The bacterial cell envelope, which encompasses the outer and inner membranes in the case of Gram-negative organisms, helps to maintain cellular integrity and also constitutes the interface with the environment, including the tissues and cells of infected hosts. In the latter capacity, the various components of the envelope are involved in sensing a number of environmental factors and inducing appropriate responses via changes in gene expression and other mechanisms.20,21 In addition, virulence determinants such as adhesins, the TTSS and flagella are embedded in the cell envelope in association with the outer and/or inner membranes.14,22,23 An important characteristic of the Gram-negative outer membrane is the asymmetric composition of its two leaflets, with enrichment of lipopolysaccharide in the outer leaflet and phospholipids in the inner leaflet. This asymmetry is maintained in part by a conserved ATP-binding cassette (ABC) transport system, which removes phospholipids from the outer leaflet of the outer membrane and is referred to as the Mla pathway in E. coli and the Yrb pathway in several other Gram-negative bacteria.24,25 Mutations in this pathway have been associated with a variety of phenotypic abnormalities, including increased sensitivity to detergent and EDTA in E. coli, increased formation of outer membrane vesicles in Haemophilus influenzae and Vibrio cholerae, decreased serum resistance in H. influenzae, and decreased intercellular spread in Shigella flexneri.24–27 The Yrb system has received little attention in either S. Typhimurium or S. Typhi, the only insights being provided by a recent study of the latter organism indicating that deficiency of YrbE, the inner membrane permease that is believed to transport phospholipids, leads to inappropriately elevated expression of the typhoid toxin in the cytosol of infected cells.28 Given the paucity of information on Yrb function in Salmonella, we decided to investigate this issue in S. Typhi by generating a mutant strain with a disruption in the yrbE gene. We found that the YrbE-deficient strain of S. Typhi had abnormalities of flagellin expression, motility, adhesion to epithelial cells and the induction of intestinal inflammatory responses. Surprisingly, disruption of yrbE had little or no effect on these characteristics in S. Typhimurium.
Results
We used the method of Datsenko and Wanner to disrupt the yrbE gene in the wild-type (WT) S. Typhi strain Ty2.29 PCR of genomic DNA from the mutant strain, designated as Ty2ΔyrbE, was used to confirm the disruption of the yrbE gene and final excision of the chloramphenicol resistance cassette used for the disruption. We also complemented Ty2ΔyrbE with a plasmid expressing yrbE to generate the strain Ty2ΔyrbE/pyrbE+. We used quantitative reverse transcription PCR (qRT-PCR) to compare yrbE expression in Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ and found it to be essentially undetectable in Ty2ΔyrbE and about 11-fold higher than WT in Ty2ΔyrbE/pyrbE+ (Supplementary Figure 1A). We also examined the expression of yrbB and yrbC, genes that are downstream of yrbE in the operon, and found that the disruption of yrbE did not significantly alter their expression, confirming the absence of polar effects (Supplementary Figure 2A).24 yrbE did not have a significant effect on growth since the Ty2 and Ty2ΔyrbE strains grew at essentially identical rates (Supplementary Figure 3A). The Ty2ΔyrbE/pyrbE+ strain had a minor, non-significant lag in growth compared to the other two strains during the early phase of culture but by 7 h it had achieved a similar density (Supplementary Figure 3A).
Since yrbE was listed as one of the genes affecting motility in a genome-wide screen carried out in E. coli, we compared swimming and swarming motility in Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+.30 We found that yrbE deficiency resulted in a hypermotile phenotype in the swimming assay, while complementation with the yrbE-expression plasmid corrected this abnormality (Figure. 1(A,B)). There was no consistent effect of the yrbE disruption on swarming motility (data not shown). These results were somewhat surprising because the E. coli screen indicated that a mutation in yrbE strongly repressed swarming but did not significantly affect swimming.30 Nevertheless, in keeping with the increased motility of Ty2ΔyrbE, this strain expressed significantly higher levels of fliC, the sole flagellin gene in most S. Typhi isolates, relative to Ty2 (Figure 1(C)).31,32 As in the motility assay, plasmid complementation restored expression of fliC to levels that were not significantly different from WT (Figure 1(C)).
Figure 1.
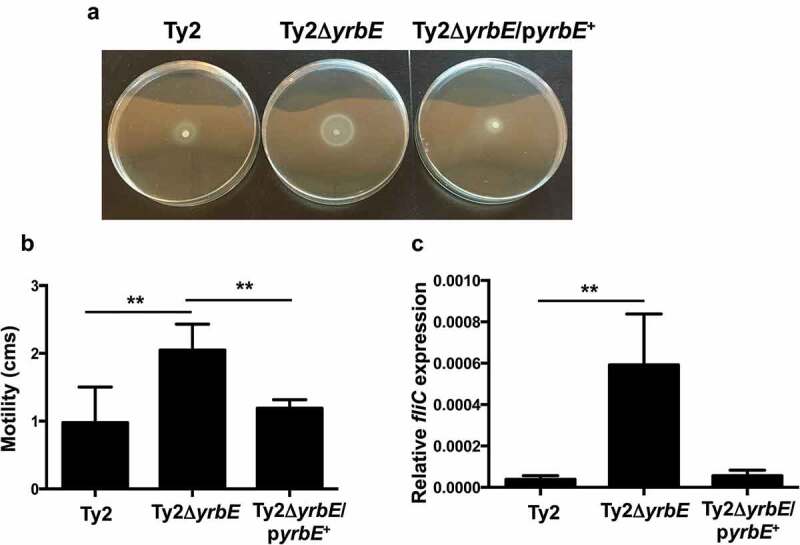
Effect of yrbE on motility and flagellin expression of S. Typhi. Overnight cultures of Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ were spotted onto LB/0.25% agar plates. After 8 h at 37°C, the plates were (A) photographed, and (B) motility diameters measured. **p < .01, n = 6 or 7 per group. (C) fliC expression in overnight cultures of Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ was measured by qRT-PCR. **p < .01, n = 4 or 6 per group.
We then proceeded to examine the effect of yrbE deficiency on adhesion of S. Typhi to the HeLa cervical epithelial cell line. As described by others, the adhesion assay was carried out on ice in order to prevent internalization of the bacteria.13 The bacteria were centrifuged onto the cells to correct for differences in motility. As shown in Figure 2, yrbE deficiency resulted in significantly reduced adhesion of S. Typhi to HeLa cells (about 1.5 log lower), whereas complementation of the mutant strain with plasmid-expressed yrbE restored WT adhesion levels. The effects of yrbE on adhesion could not be explained by differences in the numbers of Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ added to the cells. The actual infecting doses of the three bacterial strains were determined by plating and were found to be similar in each experiment.
Figure 2.
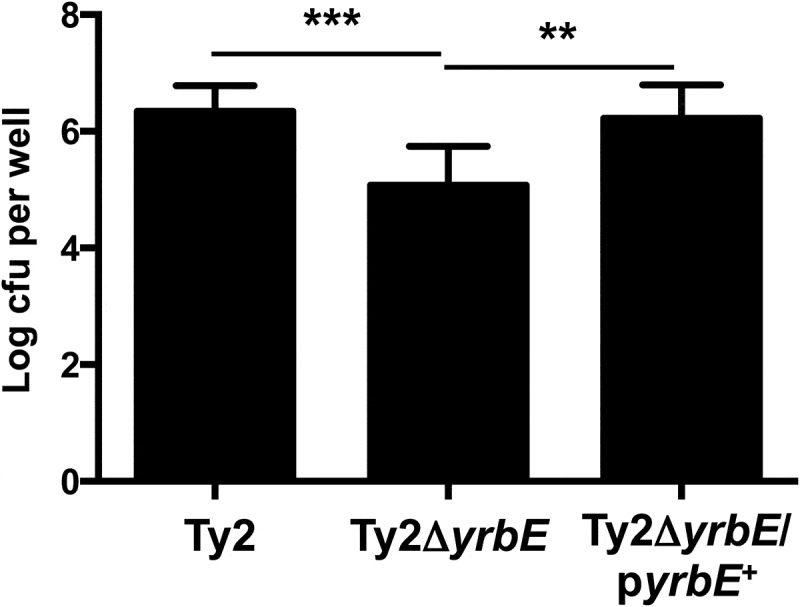
Effects of yrbE on S. Typhi adhesion to HeLa cells. Adhesion of Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ to HeLa cells was measured as described in the text. **p < .01, ***p < .001, n = 12–14 per group.
To gain insight into the mechanistic basis for the impaired adhesion caused by yrbE deficiency, we considered the possibility that the hypermotility of Ty2ΔyrbE might interfere with the ability to form normal adhesive interactions with the plasma membrane of mammalian cells. To investigate this idea, we deleted the fliC gene in Ty2ΔyrbE to generate the double mutant strain Ty2ΔyrbEΔfliC. Deletion of fliC completely abolished the hypermotility of Ty2ΔyrbE (Figure 3(A)) but did not increase the adhesion of this strain to HeLa cells (Figure 3(B)). Since flagella have been implicated in initial, nonspecific adherence to host cells, it was possible that disruption of fliC could have affected adhesion independently of motility.33 Therefore, to substantiate our findings, we disrupted the flagellar motor gene motB in Ty2ΔyrbE to generate the strain Ty2ΔyrbEΔmotB. motB is part of the stator complex of the flagellar motor and couples proton flux to rotation of the rotor-flagellar filament.34 Mutations of motB result in a paralyzed phenotype in which flagella are assembled but cannot rotate.35 In keeping with these earlier observations, disruption of motB in Ty2ΔyrbE inhibited motility (Figure 3(C)). However, it did not improve adhesion to HeLa cells (Figure 3(D)). Taken together, our results indicate that the increased flagellin expression and hypermotility of Ty2ΔyrbE do not explain the impaired adhesion of this strain.
Figure 3.
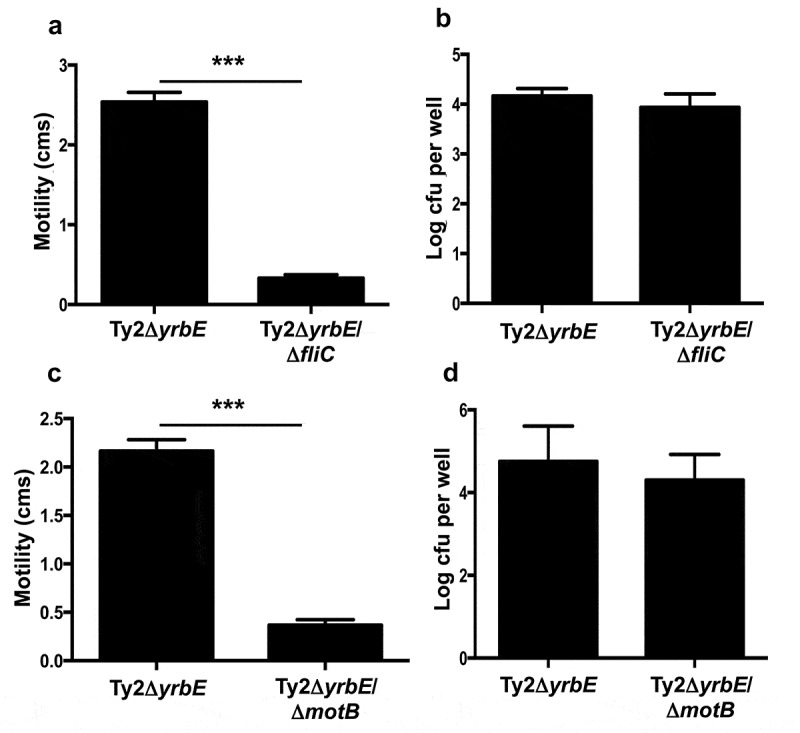
Effects of fliC and motB deletion on motility and adhesion of Ty2ΔyrbE. (A) Swimming motility of Ty2ΔyrbE and Ty2ΔyrbEΔfliC. ***p < .001, n = 5 per group. (B) Adhesion of Ty2ΔyrbE and Ty2ΔyrbEΔfliC to HeLa cells. The difference between groups is not significant, n = 6 per group. (C) Swimming motility of Ty2ΔyrbE and Ty2ΔyrbEΔmotB. ***p < .001, n = 3 per group. (D) Adhesion of Ty2ΔyrbE and Ty2ΔyrbEΔmotB to HeLa cells. The difference between groups is not significant, n = 6 per group.
Although the HeLa cell line is widely used in studies of Salmonella-epithelial interactions, it is derived from a cell type that is not the natural target of S. Typhi. Accordingly, we also used polarized monolayers of primary intestinal epithelial cells generated from human ileal organoids to examine the effects of yrbE deficiency. Short of infecting human volunteers, the organoid-derived monolayers represent one of the most physiologically relevant experimental systems for studying interactions between S. Typhi and the human intestinal epithelium.36 Polarized monolayers of the primary ileal epithelial cells were infected apically with Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ and adhesion was assessed as before. Similar to the results with HeLa cells, we found that disruption of yrbE significantly reduced the adhesion of S. Typhi to the polarized gut epithelial monolayers and that this abnormality could be corrected by plasmid complementation (Figure 4(A)). We also examined the effect of the yrbE deficiency on the ability of the bacteria to induce secretion of the pro-inflammatory cytokine interleukin-8 (IL-8) from the basolateral aspect of the monolayers. We found that Ty2ΔyrbE induced a higher amount of IL-8 relative to the WT strain (although not to the level of statistical significance), and that complementation of Ty2ΔyrbE with the yrbE expression plasmid significantly reduced this amount (Figure 4(B)). The increase in IL-8 induction by Ty2ΔyrbE was probably related to its higher expression of flagellin since the disruption of fliC in this strain significantly reduced production of the cytokine (Supplementary Figure 4), consistent with earlier demonstrations of the essential role played by Salmonella flagellin in activating inflammatory responses in intestinal epithelial cells.37–39
Figure 4.
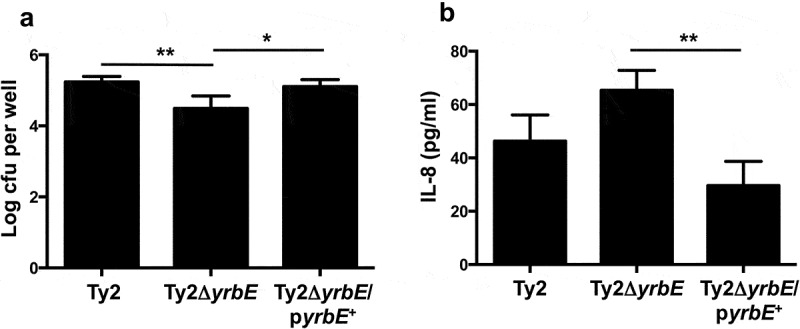
Effects of yrbE on interactions between S. Typhi and human ileum organoid-derived polarized primary epithelial monolayers. (A) Adhesion of Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ to the monolayers was measured as described in the text. *p < .05, **p < .001, n = 6 or 7 per group. (B) Induction of basolateral IL-8 secretion from the monolayers in response to Ty2, Ty2ΔyrbE and Ty2ΔyrbE/pyrbE+ was measured by ELISA. **p < .01, n = 3 per group.
The YrbE transporter in S. Typhimurium is identical in protein sequence to the S. Typhi version. However, given the differences in pathogenicity of the two organisms, we were interested in determining whether yrbE played the same roles in S. Typhimurium as it did in S. Typhi. Accordingly, we disrupted the yrbE gene in the WT S. Typhimurium strain SL1344 to generate the strain SLΔyrbE, as well as the plasmid complemented version of this strain, SLΔyrbE/pyrbE+. We confirmed the absence of yrbE expression in SLΔyrbE and its marked increase in SLΔyrbE/pyrbE+ (Supplementary Figure 1B), as well as the lack of any polar effects of the yrbE disruption on the downstream genes yrbB and yrbC (Supplementary Figure 2B). As in the case of S. Typhi, yrbE did not have significant effects on growth of S. Typhimurium (Supplementary Figure 3B). We compared the swimming motility of SL1344, SLΔyrbE and SLΔyrbE/pyrbE+. As noted previously by others, S. Typhimurium is more motile in this assay than S. Typhi.40,41 Somewhat to our surprise, the lack of yrbE had no significant effect on either motility or flagellin expression in S. Typhimurium (Figure 5(A,B)). Note that the fliC primers used for qRT-PCR amplify both of the flagellin transcripts expressed by S. Typhimurium, fliC and fljB. Similar results were obtained with fljB-specific primers (data not shown). Interestingly, the plasmid complemented strain had significantly reduced motility and flagellin expression (Figure 5(A,B)), uggesting that yrbE was capable of influencing these characteristics in S. Typhimurium when over-expressed. In a further distinction from S. Typhi, yrbE had no significant effects on adhesion of S. Typhimurium to HeLa cells (Figure 6). Finally, we made use of the streptomycin pre-treatment mouse model of S. Typhimurium infection to examine the effects of yrbE in vivo.42 Here also, we found no significant differences between SL1344, SLΔyrbE and SLΔyrbE/pyrbE+ with respect to indicators of the severity of intestinal inflammation – gross appearance, length, histology or TNFα transcript levels of the cecum (Supplementary Figure 5A, Figure 7(A–C)) – or in the numbers of the bacteria present in the stool or associated with the cecal wall (Supplementary Figure 5B, Figure 7(D)). Thus, based on our findings, yrbE does not appear to have any effects on interactions between S. Typhimurium and the intestinal epithelium, a clear difference from its role in S. Typhi.
Figure 5.
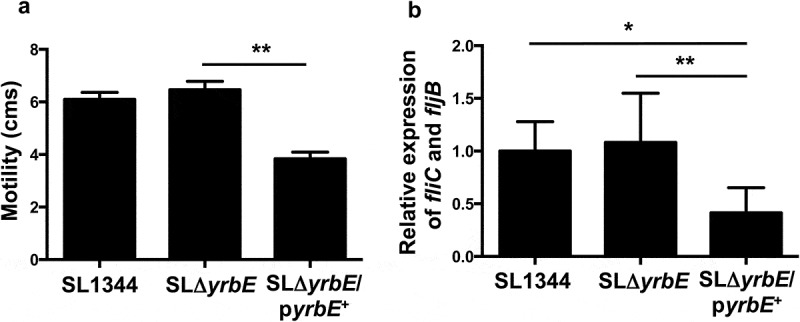
Effect of yrbE on motility and flagellin expression of S. Typhimurium. (A) Overnight cultures of SL1344, SLΔyrbE and SLΔyrbE/pyrbE+ were spotted onto LB/0.25% agar plates. After 8 h at 37°C, the motility diameters were measured. **p < .01, n = 5 per group. (B) Flagellin expression in overnight cultures of SL1344, SLΔyrbE and SLΔyrbE/pyrbE+ was measured by qRT-PCR using fliC primers (which amplify both fliC and fljB transcripts). *p < .05 **p < .01, n = 7–9 per group.
Figure 6.
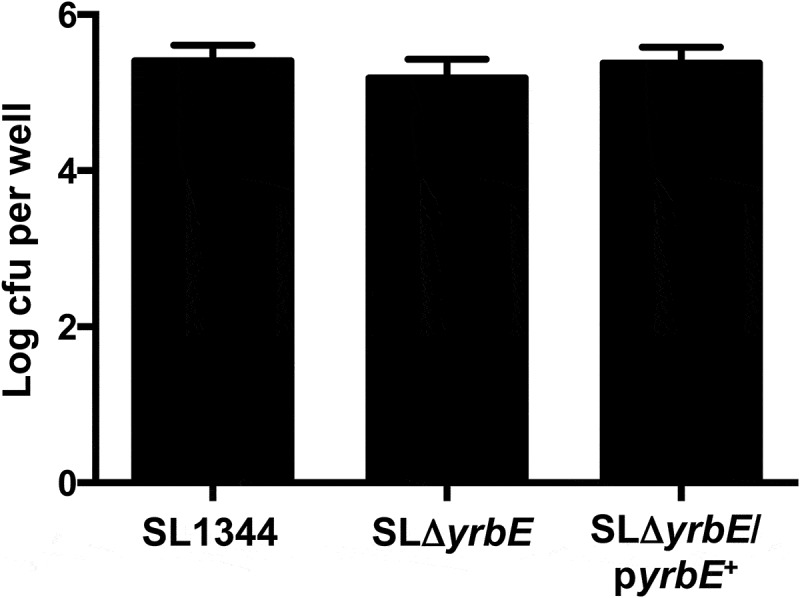
Effect of yrbE on S. Typhimurium adhesion to HeLa cells. Adhesion of SL1344, SLΔyrbE and SLΔyrbE/pyrbE+ to HeLa cells was measured as described in the text. The differences between groups are not significant, n = 9 per group.
Figure 7.
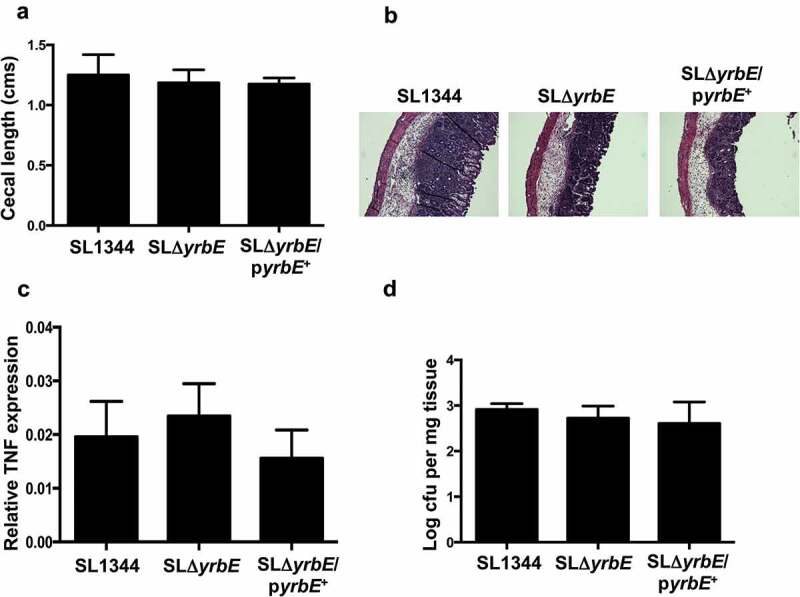
Effects of yrbE in the in vivo streptomycin pre-treatment mouse model of S. Typhimurium infection. Groups of C57BL/6J mice were infected orally with SL1344, SLΔyrbE and SLΔyrbE/pyrbE+ after streptomycin pre-treatment and euthanized 48 h after infection. (A) Cecal lengths (n = 4–8 per group). (B) Formaldehyde-fixed sections of the ceca were stained with hematoxylin and eosin and visualized with a 10X objective. (C) Total RNA prepared from segments of the ceca was used for qRT-PCR analysis of TNFα expression (n = 4–7 per group). (D) Weighed portions of the ceca were homogenized in 1% Triton X-100 and serial dilutions plated to determine the numbers of intestine-associated Salmonella (n = 3–4 per group). The differences between groups are not significant.
Discussion
The results presented here provide novel insights into the functions of the YrbE transporter in S. Typhi. Our findings indicate that YrbE plays an important role in controlling the expression of flagellin, and that it influences key aspects of pathogenesis, viz., motility, and adhesion to epithelial cells. The hypermotility associated with disruption of the yrbE gene in the Ty2ΔyrbE strain results from increased expression of flagellin, while the impaired adhesion of this strain is not the result of the increases in flagellin or motility. The exact mechanism by which yrbE influences adhesion remains to be elucidated. The YrbE transporter is involved in maintaining the normal phospholipid composition of the outer bacterial membrane.24–26 Although a very recent study has raised some questions about the direction in which phospholipids are moved by the transporter, disruption of yrbE is expected to cause changes in outer membrane phospholipid composition.43 Such alterations could affect the biophysical properties of the membrane, which in turn could affect adhesion of the bacteria to host cells. Further work will be required to investigate and substantiate this idea.
Interestingly, the elevated levels of flagellin expression that result from disruption of yrbE appear to enhance the inflammatory effects of S. Typhi at the gut epithelium, presumably as a result of increased activation of Toll-like receptor 5 (TLR5), a major receptor for flagellin that is expressed on the basolateral surface of intestinal epithelial cells.37–39 Thus, yrbE may contribute to the attenuation of intestinal inflammatory responses that facilitates extra-intestinal spread of S. Typhi.10 Absence of yrbE-dependent control of fliC expression is also likely to affect the course of S. Typhi infection at systemic sites. Flagellin sensors such as TLR5 and the NLRC4 inflammasome that are expressed in macrophages and dendritic cells play important roles in restricting bacterial growth.44 Correspondingly, Salmonella mutants that over-express flagellin have been shown to be attenuated in vivo.45
Together with earlier experiments showing that yrbE inhibits the inappropriate expression of the typhoid toxin in the cytosol of infected cells, our observations suggest that a major function of this transporter is to regulate S. Typhi gene expression, presumably as a result of changes in outer membrane phospholipid composition.28 Interestingly, a recent study showed that deficiency of the Neisseria gonorrhoeae mlaA gene, which acts in the same phospholipid transport pathway as yrbE, resulted in abnormally elevated expression of some adhesins.46 This finding provides further support for the idea that YrbE-dependent regulation of outer membrane phospholipids is required for normal gene regulation. This function is likely to be a reflection of the outer membrane’s role in detecting and responding to environmental cues. Based on this idea, it seems reasonable to speculate that an abnormality of outer membrane phospholipid composition caused by yrbE deficiency would interfere with the sensing and/or signal-transducing properties of other molecules located in this structure. Further work will be required to fill in the details of this model. The involvement of yrbE in gene regulation does not rule out its more traditional role in protection against membrane disrupting agents, nor its more recently described activity in membrane vesiculation.24–26 All these functions probably arise from the requirement for yrbE in maintaining the structural integrity of the outer membrane.
It was surprising to discover that disruption of yrbE had little or no effect on flagellin expression, motility, adhesion and the induction of intestinal inflammatory responses in S. Typhimurium. This finding suggests that the function of yrbE is serovar-specific and raises the intriguing possibility that this gene might contribute to the differences in types of disease caused by S. Typhi and S. Typhimurium. Further characterization of yrbE function in the two serovars will help to shed light on this issue. Meanwhile, it may be pertinent that a recent report identified unusual trehalose phospholipid molecules that are present at higher levels in the cell envelope of S. Typhi compared to S. Typhimurium.47 If YrbE is involved in the transport of such phospholipids, disrupting the yrbE gene could have functional consequences in S. Typhimurium that differ from those in S. Typhi. This possibility merits further investigation.
In summary, our findings indicate that yrbE plays an important and serovar-specific role in the early interactions between S. Typhi and the intestinal epithelium. Further characterization of its function could provide novel insights into typhoid pathogenesis and suggest new ways to prevent and treat this important public health problem.
Materials and methods
Growth of bacteria and generation of gene disruptions
The WT S. Typhi strain Ty2 was obtained from the American Type Culture Collection, Manassas, VA and was cultured at 37°C in lysogeny broth (LB). To carry out infections, a colony from a freshly streaked plate was inoculated into 2 ml of LB and incubated in a 12 ml culture tube with a loose cap at 37°C with shaking at 220 rpm for 8 h. Ten microliters of the culture was then back-diluted 1/1000 into 10 ml of fresh LB in a tightly capped 12 ml culture tube and incubated overnight at 37°C under static conditions (i.e., without shaking).
The S. Typhi yrbE gene was disrupted following the protocol of Datsenko and Wanner.29 Briefly, Ty2 was transformed by electroporation with the temperature-sensitive plasmid pKD46 encoding the Lambda Red recombinase. The pKD46 transformant was then transformed with a DNA fragment containing a chloramphenicol resistance marker flanked by regions of yrbE homology that was generated by PCR amplification with the Phusion DNA polymerase (New England Biolabs, Ipswich, MA) using the primers STyYrbEP1 and STyYrbEP2 and the plasmid pKD3 as the template (sequences of all primers are provided in Supplementary Table 1). Transformants were selected on LB with 10 µg/ml chloramphenicol at 37°C to eliminate pKD46. Genomic DNA was prepared from several transformants and used for PCR amplification with the primers STyYrbEUp and STyYrbEDn to confirm the disruption of yrbE by insertion of the chloramphenicol resistance cassette. A transformant with the confirmed disruption was transformed with the temperature-sensitive plasmid pCP20 encoding the FLP recombinase in order to excise the chloramphenicol resistance cassette. Following curing of the plasmid by growth at 37°C in the absence of any antibiotic selection, the excision was confirmed by PCR amplification of genomic DNA with STyYrbEUp and STyYrbEDn. The yrbE-disrupted strain was designated Ty2ΔyrbE.
To complement Ty2ΔyrbE, the WT yrbE open reading frame was PCR amplified from Ty2 genomic DNA using the primers STyYrbER1 containing a flanking EcoR1 site and STyYrbEN1 containing a flanking Not1 site. The EcoR1-Not1-digested PCR product was cloned into the multi-copy plasmid pBH and the resultant product sequenced to confirm that the cloned yrbE retained the WT sequence.38 The plasmid was transformed into Ty2ΔyrbE to generate the Ty2ΔyrbE/pyrbE+ strain.
The disruptions of the fliC and motB genes in Ty2ΔyrbE were carried out and confirmed as described above for yrbE. The corresponding primers were: STyFliCP1, STyFliCP2, STyFliCUp, STyFliCDn, STyMtBP1, STyMtBP2, STyMtBUp, STyMtBDn. The fliC-disrupted strain was designated Ty2ΔyrbEΔfliC, while the motB disrupted strain was designated Ty2ΔyrbEΔmotB.
The WT S. Typhimurium strain SL1344 was originally obtained from Dr. Beth McCormick, University of Massachusetts Medical Center, Worcester, MA. It was grown similarly to S. Typhi and the disruption of the yrbE gene carried out exactly as described above using the same sets of primers (which target identical nucleotide sequences in the S. Typhimurium genome). The yrbE-disrupted strain – SLΔyrbE – was transformed with the pBHyrbE expression plasmid to obtain the complemented strain SLΔyrbE/pyrbE+
Growth assays were carried out by inoculating 8 µl of saturated overnight cultures of the S. Typhi and S. Typhimurium strains into quadruplicate 800 µl aliquots of LB (with ampicillin added to 100 µg/ml in the case of the plasmid-complemented strains) in a 48-well tissue culture plate. The plate was incubated at 37°C with shaking at 220 rpm. One hundred µl aliquots of the cultures were removed every hour and the absorbance at 650 nm measured on a microplate reader using LB as the blank.
Bacterial motility assays
Motility of bacterial strains was analyzed essentially as described by Kalai Chelvam et al. by spotting 2 µl of the overnight cultures onto the center of triplicate LB plates made with either 0.25% agar (for swimming) or 0.5% agar (swarming).41 The plates were incubated at 37°C for 8 h (swimming) or overnight (swarming) and the maximum diameter of the motility circle was measured.
Analysis of bacterial gene expression
Total RNA was prepared from 1.2 ml aliquots of triplicate overnight static cultures of the bacteria using the RNeasy Mini kit (Qiagen, Germantown, MD) and following the manufacturer’s guidelines. The RNA (0.5–1 µg per sample) was treated with RNAse-free DNAse to eliminate any contaminating genomic DNA, reverse transcribed and amplified using the iScript cDNA synthesis kit and the iQ SYBR Green Supermix kit, respectively (both kits from Bio-Rad, Hercules, CA). Amplification was carried out using primers specific for S. enterica fliC and S. Typhimurium fljB, as well as bacterial 16S rRNA, and was monitored using the Bio-Rad CFX Connect Real-Time System. The specificity of the amplification was confirmed based on the generation of a single, sharp melting curve. Relative expression was calculated using the 2−ΔCt method, with normalization to 16S rRNA. The sequences of the primers for Salmonella fliC and universal bacterial 16S rRNA have been published previously.48,49 The primers used for amplication of S. Typhimurium fljB were STmFljbF and STmFljbR (all primer sequences are provided in Supplementary Table 1). Note that because of the marked sequence similarity between fliC and fljB, it was not possible to design primers specific for only the former. Thus, the fliC primers that we used amplify both transcripts in S. Typhimurium. The fljB primers amplify only the fljB transcript. The expression of the yrbB and yrbC genes was analyzed by qRT-PCR in the same way, using the primers STyYrbBF and STyYrbBR, and STyYrbCF and STyYrbCR, respectively.
Growth of HeLa cells
HeLa cells were maintained in a 37°C tissue culture incubator in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA) with 10% heat-inactivated fetal bovine serum (iFBS) and penicillin-streptomycin, with trypsinization and passage every 2–3 d.
Adhesion assays
On the day before infection, 1.75 × 105 HeLa cells in 0.5 ml of medium were seeded in triplicate in the wells of a 24-well tissue culture plate and allowed to adhere overnight. The next morning, the wells were typically 80-90% confluent. The medium was aspirated from the wells and the cells were washed 3 times with PBS and placed in antibiotic-free DMEM with 10% iFBS (infection medium) prior to infections. The tissue culture plate with the cells was incubated on ice starting about 30 min before the addition of bacteria.
Overnight static cultures of the various bacterial strains were harvested by centrifugation, washed with PBS and suspended in infection medium at a calculated density of 4 × 109 cfu/ml. Twenty-five microliters of the appropriate bacterial suspension (approximately 108 cfu) was added to the corresponding wells. The actual inoculum of each bacterial strain was determined by plating serial dilutions of the suspensions used for infection. The inocula of the WT, yrbE-disrupted and plasmid-complemented strains were similar in any given experiment.
After the addition of the bacteria, the plate was centrifuged at 500 g for 10 min at 4°C to deposit the bacteria onto the cells. The plate was incubated on ice for 1 h, as described by others.13 The cells were then washed three times with ice-cold PBS and lysed by the addition of sterile 1% Triton X-100 in water. After incubation on ice with gentle agitation for 15 min, serial dilutions of the lysates were made and plated on LB agar to enumerate recovered bacteria.
Human ileum-derived organoid monolayers
Polarized monolayers of primary human intestinal epithelial cells were prepared as previously described.50,51 In brief, biopsies of the terminal ileum were obtained from individuals in good general health and without chronic medical conditions who were undergoing diagnostic colonoscopy. Human subject recruitment and use of the biopsies were carried out with informed consent and with the approval of the Massachusetts General Hospital/Partners Healthcare Institutional Review Board (protocol number 2014P002001). Biopsies were transported immediately to the laboratory and processed to obtain crypt fractions. Crypts were cultured to generate organoids as described in detail in ref.50 The organoids were trypsinized and the cells seeded on Transwell inserts in 24-well plates (Corning Life Sciences, Tewksbury, MA) and cultured until the formation of monolayers. Forty-eight hours before use, the monolayers were treated apically with N-[2S-(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester-glycine (DAPT, MilliporeSigma, Burlington, MA) and the stem cell factors were reduced to induce differentiation. Monolayer integrity and polarization were assessed by the measurement of trans-epithelial electrical resistance. About an hour before infection, the medium was aspirated from the apical and basolateral compartments, the cells were washed 3 times with PBS and placed in antibiotic-free DMEM with 10% iFBS. Bacterial adhesion to the organoid-derived polarized monolayers of primary intestinal epithelial cells was quantified in the same way as for the HeLa cells, with the bacteria being added to the apical chamber and volumes of medium and PBS adjusted for the size of the Transwell compartments. For the measurement of IL-8 secretion, the organoid monolayers were infected on the apical side for 1 h at 37°C. The cells were then washed and incubated in medium containing 100 µg/ml of gentamicin to kill extracellular bacteria. The basolateral supernatants were collected 4–5 h later and IL-8 concentration measured by enzyme-linked immunosorbent assay (ELISA) as described previously.39
Mouse infections
Groups of 6-week-old male WT C57BL/6J mice (Jackson Laboratory) were treated with streptomycin, 20 mg per mouse, by oral gavage and then infected 24 h later with approximately 5 × 108 cfu of SL1344, SLΔyrbE or SLΔyrbE/pyrbE+ by oral gavage, as described in the published protocol.42 The mice were euthanized by controlled flow carbon dioxide asphyxia 48 h after infection. At necropsy, the cecum, which is the site of maximal inflammation in this model, was excised, photographed and its length recorded. Segments of the cecum were removed for fixation and processing for hematoxylin and eosin staining, preparation of RNA, and quantitation of tissue pathogen burden by homogenization in sterile 1% Triton X-100 and plating of serial dilutions of the homogenate on MacConkey agar containing 50 µg/ml streptomycin. A sample of stool was collected aseptically from the colon and homogenized and plated similarly. qRT-PCR for cecal TNFα transcript levels was carried out as described previously.52 The animal experiments were approved by the Institutional Animal Care and Use Committee under protocol number 2008N000061 and were performed in accordance with all relevant guidelines.
Statistical analysis
Each assay was performed in triplicate or quadruplicate and was carried out at least twice. The results from representative or multiple experiments are presented as the mean ± standard deviation. The Student’s t-test was used to assess significance in experiments involving comparison of just two groups while the Kruskal–Wallis test with post-hoc Dunn’s test was used for experiments involving multiple comparisons. A p value < .05 was considered to be significant. All statistical analyses were carried out using Prism 6 (GraphPad Software, San Diego, CA). The p values are indicated in the figures, with * representing p < .05, ** representing p < .01 and *** representing p < .001. The number of replicates (n) is provided in each figure legend.
Supplementary Material
Funding Statement
This work was supported by National Institutes of Health (NIH) grant [R01 AI089700] to BJC. Support for the Intestinal Organoid Core was provided by NIH grant [U19 AI082655]. EH was supported by NIH grant [R25 DK103579] through the MGHfC Digestive Disease Summer Research Program.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Gibani MM, Britto C, Pollard AJ.. Typhoid and paratyphoid fever: a call to action. Curr Opin Infect Dis. 2018;31:440–448. doi: 10.1097/QCO.0000000000000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiyoshi H, Tiffany CR, Bronner DN, Baumler AJ. Typhoidal Salmonella serovars: ecological opportunity and the evolution of a new pathovar. FEMS Microbiol Rev. 2018;42:527–541. doi: 10.1093/femsre/fuy024. [DOI] [PubMed] [Google Scholar]
- 3.Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E, Kim YE, Park JK, Wierzba TF. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk factor adjustment. Lancet Glob Health. 2014;2:e570–e580. doi: 10.1016/S2214-109X(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 4.Britto CD, Wong VK, Dougan G, Pollard AJ. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl Trop Dis. 2018;12:e0006779. doi: 10.1371/journal.pntd.0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:pii: e00105-18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog. 2012;8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R, Mylona E, Frankel G. Typhoidal Salmonella: distinctive virulence factors and pathogenesis. Cell Microbiol. 2018;20:e12939. doi: 10.1111/cmi.12939. [DOI] [PubMed] [Google Scholar]
- 8.Higginson EE, Simon R, Tennant SM. Animal models for salmonellosis: applications in vaccine research. Clin Vaccine Immunol. 2016;23:746–756. doi: 10.1128/CVI.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305:1–13. doi: 10.1111/fml.2010.305.issue-1. [DOI] [PubMed] [Google Scholar]
- 10.Keestra-Gounder AM, Tsolis RM, Baumler AJ. Now you see me, now you don’t: the interaction of Salmonella with innate immune receptors. Nat Rev Microbiol. 2015;13:206–216. doi: 10.1038/nrmicro3428. [DOI] [PubMed] [Google Scholar]
- 11.Wotzka SY, Nguyen BD, Hardt W-D. Salmonella Typhimurium diarrhea reveals basic principles of enteropathogen infection and disease-promoted DNA exchange. Cell Host Microbe. 2017;21:443–454. doi: 10.1016/j.chom.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop A, House D, Perkins T, Baker S, Kingsley RA, Dougan G. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: roles of surface structures in adhesion and invasion. Microbiology. 2008;154:1914–1926. doi: 10.1099/mic.0.2008/016998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Wagner C, Hensel M. Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol. 2011;715:17–34. [DOI] [PubMed] [Google Scholar]
- 16.Misselwitz B, Kreibich SK, Rout S, Stecher B, Periaswamy B, Hardt W-D. Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect Immun. 2011;79:330–341. doi: 10.1128/IAI.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lara-Tejero M, Galan JE. Salmonella enterica serovar Typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRock DL, Chaudhary A, Miller SI. Salmonella interactions with host processes. Nat Rev Microbiol. 2015;13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson KG, Holden DW. Dynamics of growth and dissemination of Salmonella in vivo. Cell Microbiol. 2010;12:1389–1397. doi: 10.1111/j.1462-5822.2010.01511.x. [DOI] [PubMed] [Google Scholar]
- 20.Laloux G, Collet JF. Major Tom to ground control: how lipoproteins communicate extracytoplasmic stress to the decision center of the cell. J Bacteriol. 2017;199:pii: e00216-17. doi: 10.1128/JB.00216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Nisco NJ, Rivera-Cancel G, Orth K. The biochemistry of sensing: enteric pathogens regulate type III secretion in response to environmental and host cues. MBio. 2018;9:pii: e02122-17. doi: 10.1128/mBio.02122-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apel D, Surette MG. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim Biophys Acta. 2008;1778:1851–1858. doi: 10.1016/j.bbamem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Hu B, Lara-Tejero M, Kong Q, Galan JE, Liu J. In situ molecular architecture of the Salmonella type III secretion machine. Cell. 2017;168:1065–1074. doi: 10.1016/j.cell.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinverni JC, Silvahy TJ. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun. 2016;7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 2011;7:e1001247. doi: 10.1371/journal.ppat.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun. 2014;82:660–669. doi: 10.1128/IAI.01057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fowler CC, Galan JE. Decoding a Salmonella Typhi regulatory network that controls typhoid toxin expression within human cells. Cell Host Microbe. 2018;23:65–76. doi: 10.1016/j.chom.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol. 2007;189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moshitch S, Doll L, Rubinfeld BZ, Stocker BA, Schoolnik GK, Gafni Y, Frankel G. Mono- and biphasic Salmonella Typhi: genetic homogeneity and distinguishing characteristics. Mol Microbiol. 1992;6:2589–2597. doi: 10.1111/j.1365-2958.1992.tb01436.x. [DOI] [PubMed] [Google Scholar]
- 32.Baker S, Hardy J, Sanderson KE, Quail M, Goodhead I, Kingsley RA, Parkhill J, Stocker B, Dougan G. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 2007;3:e59. doi: 10.1371/journal.ppat.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. Bacterial flagella: twist and stick or dodge across the kingdoms. PLoS Pathog. 2015;11:e1004483. doi: 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blair DF, Kim DY, Berg HC. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickerson KP, Senger S, Zhang Y, Lima R, Patel S, Ingano L, Flavahan WA, Kumar DKV, Fraser CM, Faherty CS, et al. Salmonella Typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EbioMedicine. 2018;31:92–109. doi: 10.1016/j.ebiom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 38.Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, DiDonato JA. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-κB and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 2004;4:33. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang FC, Werne A, Li Q, Galyov EE, Walker WA, Cherayil BJ. Cooperative interactions between flagellin and SopE2 in the epithelial interleukin-8 response to Salmonella enterica serovar Typhimurium infection. Infect Immun. 2004;72:5052–5062. doi: 10.1128/IAI.72.9.5052-5062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forbes SJ, Eschmann M, Mantis NJ. Inhibition of Salmonella enterica serovar Typhimurium motility and entry into epithelial cells by a protective anti-lipopolysaccharide immunoglobulin A antibody. Infect Immun. 2008;76:4137–4144. doi: 10.1128/IAI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalai Chelvam K, Chai LC, Thong KL. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog. 2014;6:2. doi: 10.1186/1757-4749-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hapfelmeier S, Hardt WD. A mouse model for S. Typhimurium-induced enterocolitis. Trends Microbiol. 2005;13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol. 2019;4:1692–1705. doi: 10.1038/s41564-019-0481-y. [DOI] [PubMed] [Google Scholar]
- 44.Broz P. Recognition of intracellular bacteria by inflammasomes. Microbiol Spectr. 2019;7. doi: 10.1128/microbiolspec.BAI-0003-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai MA, Quarles EK, Lopez-Yglesias AH, Zhao X, Hajjar AM, Smith KD. Innate immune detection of flagellin positively and negatively regulates Salmonella infection. PLoS One. 2013;19:e72047. doi: 10.1371/journal.pone.0072047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baarda BI, Zielke RA, Le Van A, Jerse AE, Sikora AE. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog. 2019;15:e1007385. doi: 10.1371/journal.ppat.1007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinink P, Buter J, Mishra VK, Ishikawa E, Cheng TY, Willemsen PTJ, Porwollik S, Brennan PJ, Heinz E, Mayfield JA, et al. Discovery of Salmonella trehalose phospholipids reveals functional convergence with mycobacteria. J Exp Med. 2019;216:757–771. doi: 10.1084/jem.20181812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Russmann H, Baumler AJ. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in motility. Mol Microbiol. 2009;74:175–193. doi: 10.1111/j.1365-2958.2009.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shanmugam NK, Trebicka E, Fu LL, Shi HN, Cherayil BJ. Intestinal inflammation modulates expression of the iron-regulating hormone hepcidin depending on erythropoietic activity and the commensal microbiota. J Immunol. 2014;193:1398–1407. doi: 10.4049/jimmunol.1400278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 51.Senger S, Ingano L, Freire R, Anselmo A, Zhu W, Sadreyev R, Walker WA, Fasano A. Human fetal-derived enterospheres provide insights on intestinal development and a novel model to study necrotizing enterocolitis (NEC). Cell Mol Gastroenterol Hepatol. 2018;5:549–568. doi: 10.1016/j.jcmgh.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Johnson EE, Shi HN, Walker WA, Wessling-Resnick M, Cherayil BJ. Attenuated inflammatory responses in hemochromatosis reveal a role for iron in the regulation of macrophage cytokine translation. J Immunol. 2008;181:2723–2731. doi: 10.4049/jimmunol.181.4.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


