ABSTRACT
Background
Adipose tissue plays important roles in health and disease. Given the unique association of visceral adipose tissue with obesity-related metabolic diseases, the distribution of lipids between the major fat depots located in subcutaneous and visceral regions may shed new light on adipose tissue–specific roles in systemic metabolic perturbations.
Objective
We sought to characterize the lipid networks and unveil differences in the metabolic infrastructure of the 2 adipose tissues that may have functional and nutritional implications.
Methods
Paired visceral and subcutaneous adipose tissue samples were obtained from 17 overweight patients undergoing elective abdominal surgery. Ultra-performance LC-MS was used to measure 18,640 adipose-derived features; 520 were putatively identified. A stem cell model for adipogenesis was used to study the functional implications of the differences found.
Results
Our analyses resulted in detailed lipid metabolic maps of the 2 major adipose tissues. They point to a higher accumulation of phosphatidylcholines, triacylglycerols, and diacylglycerols, although lower ceramide concentrations, in subcutaneous tissue. The degree of unsaturation was lower in visceral adipose tissue (VAT) phospholipids, indicating lower unsaturated fatty acid incorporation into adipose tissue. The differential abundance of phosphatidylcholines we found can be attributed at least partially to higher expression of phosphatidylethanolamine methyl transferase (PEMT). PEMT-deficient embryonic stem cells showed a dramatic decrease in adipogenesis, and the resulting adipocytes exhibited lower accumulation of lipid droplets, in line with the lower concentrations of glycerolipids in VAT. Ceramides may inhibit the expression of PEMT by increased insulin resistance, thus potentially suggesting a functional pathway that integrates ceramide, PEMT, and glycerolipid biosynthetic pathways.
Conclusions
Our work unveils differential infrastructure of the lipid networks in visceral and subcutaneous adipose tissues and suggests an integrative pathway, with a discriminative flux between adipose tissues.
Keywords: lipidomics, adipose tissues, phosphatidylcholines, sphingolipids, ceramides, PEMT, triacylglycerols, diacylglycerols, degree of unsaturation
Introduction
It is largely accepted today that fat depots represent key secretory organs, releasing multiple bioactive molecules, including lipid mediators. Increased adipose tissue mass is associated with chronic low-grade inflammation, systemic metabolic dysfunction, and obesity-linked disorders (1–3). Accumulating evidence indicates that adipose tissue distribution plays specific roles in health and disease. Subcutaneous and visceral adipose tissues (SAT and VAT, respectively) are white fat depots that are anatomically distinct: while the former is situated under the skin, the latter resides within the abdominal cavity.
The anatomical distinction has important implications, as VAT drains directly through the portal circulation to the liver (4). Functional differences between the 2 tissues are reflected in the stronger association of VAT accumulation with systemic low-grade inflammation and obesity-related cardiometabolic complications such as insulin resistance, type 2 diabetes, hyperlipidemia, and atherosclerosis (5–10), suggesting that VAT is a “unique, pathogenic fat depot” (6). In fact, increased visceral adiposity is a pivotal component of the metabolic syndrome (11). The association between abdominal obesity and the cardiometabolic comorbidities of obesity has led to the hypothesis that the concentrations of signaling molecules produced by the adipocytes in the 2 tissues vary. In line with this notion, recent work provides evidence for differences in lipid composition between VAT and SAT. SAT (but not VAT) from diabetic individuals contain elevated concentrations of sphingolipids (SLs) compared with samples from nondiabetic patients, which play a role in the regulation of SAT adipose browning, inflammation, and metabolism (12). SAT and VAT demonstrated differential accumulation of arachidonic acid concomitant to prostaglandin synthesis–related enzymes, suggesting their association with colorectal tumor stage (8). A recent pioneering attempt of a lipidomics analysis of the 2 adipose tissues focused on the 50% more abundant fraction of lipids, 97% of which were glycerolipids (13). The authors then resorted to a targeted analysis, demonstrating a differential cholesterol epoxide metabolism. The full characterization of the lipid network of VAT and SAT is therefore still needed.
Here we sought to provide a comprehensive characterization of the distinct lipid network in VAT and SAT. We therefore did not limit our analyses to abundant lipids. This study represents the first attempt, to the best of our knowledge, of a lipid network-based analysis of adipose tissues. Our analyses pointed to alterations in the concentrations of unsaturated lipids with known nutritional and health implications. The potential functional implications of our analyses were demonstrated by the downregulation of adipogenesis and lipid droplet formation in phosphatidylethanolamine methyl transferase (PEMT)–deficient differentiating pluripotent stem cells.
Methods
Study population
Participants were recruited in Soroka University Medical Center (SUMC), Beer-Sheva, Israel, before undergoing elective abdominal surgery (primarily bariatric surgery and elective cholecystectomy), after providing written informed consent. Tissue collection and handling procedures were detailed in multiple prior publications (14, 15). In brief, paired SAT and VAT biopsies were obtained during surgery and immediately delivered to the laboratory where they were rinsed, frozen in liquid nitrogen, and stored at -80°C until they were processed for lipidomics or mRNA expression analyses (herein). Basic mean (±SD) characteristics of this cohort are as follows: age, 46 ± 13.9 y; 33% males; and BMI (kg/m2), 34 ± 5.2. All procedures were approved, in advance, by the ethics committee of the SUMC, and were conducted in accordance with Declaration of Helsinki guidelines. For a lipidomics validation data set, an additional set of 5 subcutaneous and 8 visceral tissues were obtained. Clinical characteristics of patients are provided in Table 1.
TABLE 1.
Characteristics of participants in the study1
| Characteristics | Values |
|---|---|
| Age, y | 46 ± 13.9 |
| Gender, % males | 33 |
| BMI, kg/m2 | 34.0 ± 5.2 |
| Diabetes, number of patients with glycated hemoglobin ≥6.5% | 1 |
| Insulin fasting concentration, pmol/L | 14.7 ± 6.6 |
| Total cholesterol, mg/dL (mg%) | 187.5 ± 47.9 |
| TAGs, mg/dL (mg%) | 149.6 ± 82.7 |
1Values are means ± SDs unless otherwise indicated. TAG, triacylglycerol.
Materials and reagents
Acetonitrile, methanol (both ultra-LC-MS grade), chloroform, and water (HPLC-MS grade) were supplied by JT Baker; isopropanol (HPLC-MS grade) from ChemSolute; and formic acid (HPLC-MS grade) by TCI. Ammonium fluoride (>99%) was supplied by Sigma-Aldrich. Internal standard EquiSPLASH LIPIDOMIX (MS-quantitative grade) mix was obtained from Avanti Polar Lipids, Inc.
Sample preparation for lipidomics analysis
Adipose tissues were kept at −80°C until sample preparation. The samples were prepared for lipidomics analysis using a modified Bligh and Dyer protocol, following our preliminary studies. Tissues (∼20 mg) were extracted in 800 µL of ice cold extraction solvent. Extraction solvent was composed of the following: 85% methanol, 13% water, and 2% formic acid and contained an EquiSPLASH LIPIDOMIX mixture of 13 deuterated lipid internal standards [15:0–18:1 (d7) phosphatidylcholine (PC); 18:1(d7) Lyso-PC; 15:0–18:1 (d7) phosphatidylethanolamine (PE); 18:1 (d7) Lyso-PE; 15:0–18:1 (d7) phosphatidylglycerol; 15:0–18:1 (d7) phosphatidylinositol (NH4 salt); 15:0–18:1 (d7) phosphatidylserine; 15:0–18:1 (d7)-15:0 triacylglycerol (TAG); 15:0–18:1 (d7) diacylglycerol (DAG); 18:1 (d7) monoacylglycerol; 18:1 (d7) cholesteryl ester; d18:1–18:1 (d9) sphingomyelin; C15 ceramide-d7], at a concentration of 0.1 µg/mL in each sample. Samples went through 3 freeze-thaw cycles, and were then ground using a magnetic bead homogenizer (Bullet Blender® Storm; Next Advance, USA). For the extraction of lipids, samples were vortexed for 30 s and ultra-sonicated (Bioruptor Plus: Diagenode, USA) for 30 s × 5 cycles at 4°C. The homogenate was transferred to clean glass tubes, and half a volume of chloroform was added. Samples were vortexed once again and water was added at the same volume as chloroform. Samples were incubated on ice for 20 min, followed by another 5-cycle round of ultra-sonication. Following centrifugation (770 × g, 10 min, 4°C), the hydrophilic upper phase was discarded, and the lipophilic lower phase of samples was transferred to clean glass tubes. Solvents were evaporated in a SC210A SpeedVac concentrator (Thermo Fisher Scientific) at 30°C, and dry samples kept at −80°C until analysis. For the LC-MS run, samples were reconstituted in 95% acetonitrile, 5% water, and 0.1% formic acid.
Ultra-high-performance LC–quadrupole time-of-flight MS
LC-MS analysis was performed using a Waters Acquity UPLC H-Class and Xevo X2-XS Q-ToF high resolution, High Mass Accuracy Q-ToF (Waters). Electrospray ionization was used in positive (ES+) and negative (ES−) modes in separate acquisitions for preliminary studies. Following preliminary experiments, positive mode was used for the main data set. A UPLC CSH C18 column (100 mm × 2.1 mm, 1.7 μm; Waters) was used for the separation of metabolites. The mobile phase consisted of 0.1% formic acid in water (phase A) and 0.1% formic acid (vol:vol) in acetonitrile (phase B). The gradient program was as follows: 60% mobile phase A (0.1% formic acid in water) and 40% mobile phase B for 1 min. Mobile phase B proportion was increased to 70% (vol:vol) in 5 min. From 5 to 8 min, mobile phase consisted of 24% A, 40% B, and 36% C (isopropanol); from 8 to 9 min, 20% A, 35% B, and 45% C; from 9 to 12 min, 18.4% A, 33% B, and 48.6% C; then from 12 to 17 min, 12% A, 25% B, and 63% C. Then, up to 25 min, 0.4% A, 10.5% B, and 89.1% C. From 25.5 to 35 min the system was allowed to re-equilibrate to initial conditions. Following preliminary experiments, features eluted at 0–1 min were excluded, and only features eluted at the retention time of 1.0–25.0 min were used for analysis. The following settings were applied during the LC-MS runs: the flow rate was 0.4 mL/min and the column temperature was kept at 60°C. Capillary spray was maintained at 3.0 kV, cone voltage at 40 eV, collision energy was 40–65 eV. Full-scan and MSE mass spectra were acquired from all masses between 30 and 2000 Da. Argon was used as the collision gas for collision-induced dissociation. The mass spectrometer was calibrated using sodium formate, and leucine enkephalin was used as the lock mass (m/z 556.2771, 200 pg mL−1) and continuously infused at 6 µL/min. MassLynx software version 4.1 (Waters) was used to control the instrument and calculate accurate masses. Post-column derivatization was used with ammonium fluoride to improve the yields of the neutral charged lipids in the ES+ mode as [M + NH4]+. Ammonium fluoride 1 mM in 50:50 methanol:water was automatically continuously injected into the mass spectrometer together for post-column derivatization, to improve the yields of neutral charged lipids. The exact mass, fragmentation pattern, and isotope pattern were compared against 18 libraries compatible with Progenesis QI for putative identifications. The fragmentation pattern of lipids was further validated against theoretical mass fragments and the literature. Retention time was evaluated vis-à-vis our data from previous analyses and standards from the internal standard mixture.
Gene expression mining
The list of enzymes involved in lipid synthesis was extracted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) publicly available database (16). We focused on 3 KEGG maps: glycerophospholipids, glycerolipids, and SLs. Only genes expressed in humans were included. We used the GTEx publicly available database (17) to compare the expression of lipid transcripts in VAT and SAT. The expression data were then incorporated into KEGG maps and visualized using Microsoft PowerPoint and Adobe Photoshop.
RNA extraction and quantitative real-time PCR for adipose tissues
Total RNA from human SAT and VAT biopsies was extracted using the RNeasy lipid tissue mini-kit (Qiagen) and concentrations of the total RNA were quantified using nanodrop. Total RNA (2000 ng) was reverse-transcribed with a high-capacity cDNA reverse transcriptase kit (Applied Biosystems). The Universal Probe Library with specific primers (Roche) was used to quantify the expression of selected genes (Supplemental Table 1). Relative gene expression was obtained after normalization to housekeeping control genes [peptidylprolyl isomerase A (PPIA) and phosphoglycerate kinase 1 (PGK1), as recommended in (18)].
Embryonic stem cell maintenance and differentiation
V6.5 mouse embryonic stem cells (mESCs) were maintained as previously described (19, 20). Differentiation was initiated by generation of embryoid bodies (EBs), formed from hanging drops containing 103 mESCs in 20 μL of cultivation medium. After 2 d, EBs were treated with 10−7 M all-trans retinoic acid for 3 d and then transferred to gelatin-coated plates in differentiation medium composed of the cultivation medium supplemented with 85 nM insulin, 2 nM triiodothyronine, and 1 μM rosiglitazone. Differentiation was evaluated at day 24 of differentiation.
Assessment of adipocyte differentiation
Lipid droplets were visualized first under light microscopy and then cells were fixed and stained with Oil Red-O as previously described (21), and with 4′,6-diamidino-2-phenylindole (DAPI) to image nuclei. Images were recorded under confocal microscopy. Expression of adipogenic gene markers: Total RNA was purified on RNeasy columns (Qiagen) and real-time PCR assays were run on a StepOnePlus system (Applied Biosystems, Life Technologies). Transcript expression levels of fatty acid binding protein 4 (FABP4) and adiponectin, C1Q and collagen domain containing (AdipoQ) were evaluated using comparative Ct (2-ΔΔCT). ΔΔCTvalues were expressed relative to acidic ribosomal phosphoprotein P0 (36B4) for sample normalization.
Statistical analyses
Lipidomics analyses
Progenesis QI (Nonlinear Dynamics) was used for spectra deconvolution, alignment, normalization, and identification of lipid species. Masses with minimum intensity cutoff of 100 m/z, lowest mean abundance in blank (solvents that went through sample preparation, but contained no sample), and fold-change >100 from blank were used for analysis. Data were exported to EZInfo 3.0 (Umetrics, v2.0) and Metaboanalyst 4.0 (22) for multivariate statistical analysis. Following quantile normalization of data, a partial least-squares discriminant analysis (PLS-DA) model was generated. Ten-fold cross-validation was applied (Q2 and R2 are presented in Figure 1). The robustness of the class separation was assessed by permutation testing (1000 permutations).
FIGURE 1.
VAT and SAT present differential lipid profiles that cannot be attributed to a specific lipid pathway. The lipid composition of paired VAT and SAT samples is partially separated in a PCA (A) and a PLS-DA (B). R2, 0.86; Q2, 0.41. To address a possible overfit in PLS-DA, a permutation test was performed with 1000 permutations, suggesting prediction accuracy during training of an empirical P value (P = 0.01). This separation is further demonstrated by a heat map (C). (D–F) PLS-DA of the lipid species of the main differential lipid groups: (D) Phospholipids, R2, 0.64; Q2, 0.47; and P = 0.01 significance for 1000 permutations; (E) Sphingolipids, R2, 0.62; Q2, 0.35; and P = 0.001 significance for 1000 permutations. (F) The model for glycerolipids showed especially low predictive power: R2, 0.46; Q2, −0.14; and P = 0.006 significance for 1000 permutations. PC, principal component; PCA, principal component analysis; PLS-DA, partial least-squares discriminant analysis; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
Univariate analyses
Mass intensity was normalized to corresponding internal standards (see section on sample preparation). Following normalization to corresponding internal standards, the accumulative abundances of lipid subclasses were calculated. Paired t-test analyses were performed for univariate data.
Results
To study the alterations in lipid metabolism between SAT and VAT, we integrated lipidomics analyses with gene expression data mining and measurements. We analyzed a cohort of 17 paired samples from patients with elevated BMI (>25; 34 altogether, “main dataset”). To validate the reproducibility of the data, we performed a second analysis of the cohort independently of the original analysis, and carried out a third analysis of a separate unpaired cohort. The data presented are taken exclusively from the main dataset.
A lipidomics analysis demonstrates a differential lipidome in VAT and SAT
Our lipidomics analysis resulted in the detection of 64,538 features. Of these, we filtered out features with masses that match those found in blank samples and may have originated from solvents and plasticizers (see Methods). This filtering out resulted in 18,640 adipose-originated features. A total of 4595 features were assigned identifications by Progenesis QI after comparison to 18 MS databases. The lipidome of the 2 tissues (as represented by 18,640 features) could be distinguished by a principal component analysis (PCA; Figure 1A). A PLS-DA of the adipose tissue–derived features (Figure 1B) further suggested a separation of the lipidome of VAT and SAT, and a heat map (Figure 1C) corroborated this observation. The identification of 520 lipids was further validated by their exact mass (mass accuracy, 5 ppm), retention time, isotope pattern, and fragmentation pattern. The identified 520 lipids were used for univariate tests. These included 93 glycerolipids, 213 phospholipids (PLs), and 214 SLs. In an attempt to unveil the lipid class responsible for the separation of the lipidome of VAT and SAT, we carried out PLS-DA analyses of the identified lipids of the 3 major lipid classes. We found a mild separation of the PLs (Figure 1D) and SLs (Figure 1E), which was validated by a permutation test. The predictive power of the model for glycerolipids was especially low (Figure 1F; R2 = 0.46, Q2 = −0.14). Interestingly, the predictive power of the model of specific lipid subclasses was not higher than the one of the global lipidome, suggesting that VAT/SAT differential lipid metabolism should be studied across lipid subclasses. We reasoned that a network-based analysis would provide means to integrate the distinct lipid alterations between the tissues. We used the KEGG database for the list of enzymes that comprise the 3 major pathways found in our lipidomics analysis (“glycerophospholipid,” “sphingolipid,” and “glycerolipid”). We then mined the NIH GTEx open-data resource (17) for data on the expression of the genes of the lipid pathways in 442 SAT and 355 VAT samples. For visualization of the lipid networks in the 2 tissues, we incorporated the GTEx gene expression data as well as our lipidomics data in each of the tissues into the relevant KEGG maps. The abundance of specific lipid groups, as found in our lipidomics study, and the expression of individual genes, as found by quantitative reverse transcriptase–PCR (RT-PCR) experiments, are presented below the metabolic maps.
A network-based analysis demonstrates tissue-specific wiring of the glycerophospholipid pathway in VAT and SAT
The map of glycerophospholipid metabolism in VAT and SAT (Figure 2A) reveals alterations in the expression of PL metabolism genes (as mined in the GTEx cohort) and concomitant differences in the relative accumulation of PL subclasses (as found by our lipidomics analysis). We found elevated accumulation of phosphatidylcholines (PCs) (Figure 2B), but not phosphatidylethanolamines (PEs) (Figure 2C) in SAT compared with VAT. We found no differences in the accumulation of phosphatidylserines (Figure 2D), nor phosphatidylglycerols (Figure 2E). The abundance of phosphatidylinositols was higher in SAT (Figure 2F), and so was that of ether lipids (Figure 2G). For a validation of the GTEx gene expression data, we carried out targeted expression profiling experiments by RT-PCR in the same samples used for the lipidomics analysis. We first studied the expression of the genes of the primary pathway responsible for the biosynthesis of PLs in mammalian cells: the cytidine diphosphate–choline (CDP-choline, or “Kennedy”) pathway. Consistent with the GTEx expression data (Figure 2A), the expression of choline kinase ɑ (CHKA), the first enzyme of the pathway, was higher in SAT (Figure 2H), as was the expression of the second enzyme of the pathway, CTP:phosphocholine cytidylyltransferase (CCT, also known as PCYT1A) (Figure 2I). In contrast, the expression of choline/ethanolamine phosphotransferase (CEPT) 1 (CEPT1) was lower in SAT (Figure 2J). The lack of consistency with GTEx data for this enzyme may arise from a potential sexual dimorphism, as higher CEPT expression only characterizes females but not males (Figure 2K, L). The expression of choline phosphotransferase 1 (CHPT1), a paralog of CEPT1, showed no difference between the tissues (Figure 2M). Together, enzymes of the CDP-choline pathway showed indecisive differences in their relative expression in VAT and SAT.
FIGURE 2.
Phosphatidylcholines constitute a major metabolic signature of VAT versus SAT. (A) A metabolic map of the glycerophospholipid pathway [modified from the KEGG pathway (23)]. The gene expression of enzymes of the pathway, as found in the GTEx database, is shown as rectangles and the relative lipid accumulation found in our lipidomics study is shown as circles. (B–O) The accumulation of phospholipids found by our lipidomics study, and key enzymes for differential lipid groups found by qRT-PCR experiments. Blue lines represent a change of ≥10% in the SAT:VAT ratio. Red lines represent a change of ≤ −10% in the SAT:VAT ratio. Black lines represent a similar accumulation ±10%. n = 17; error bars represent SEMs; 1-tailed paired t test. AGPS, alkyldihydroxyacetonephosphate synthase; CDP, cytidine diphosphate; CEPT, choline/ethanolamine phosphotransferase; CHKA, choline kinase alpha; CHPT1, choline phosphotransferase 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCYT1A,phosphate cytidylyltransferase 1, choline, alpha;PEMT, phosphatidylethanolamine N-methyltransferase; qRT-PCR, quantitative reverse transcriptase–PCR; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
On the other hand, in line with the GTEx data (Figure 2A), our RT-qPCR analysis of the samples used for the lipidomics analyses revealed that the expression of PEMT was uniformly (among all tested paired samples) higher in SAT (Figure 2N), providing a likely explanation for the higher PC abundance. We examined the expression of alkyldihydroxyacetonephosphate synthase (AGPS), which catalyzes the exchange of an acyl for a long-chain alkyl group and the formation of the ether bond in the biosynthesis of ether PLs, and found it highly variable with a nonsignificant difference (Figure 2O).
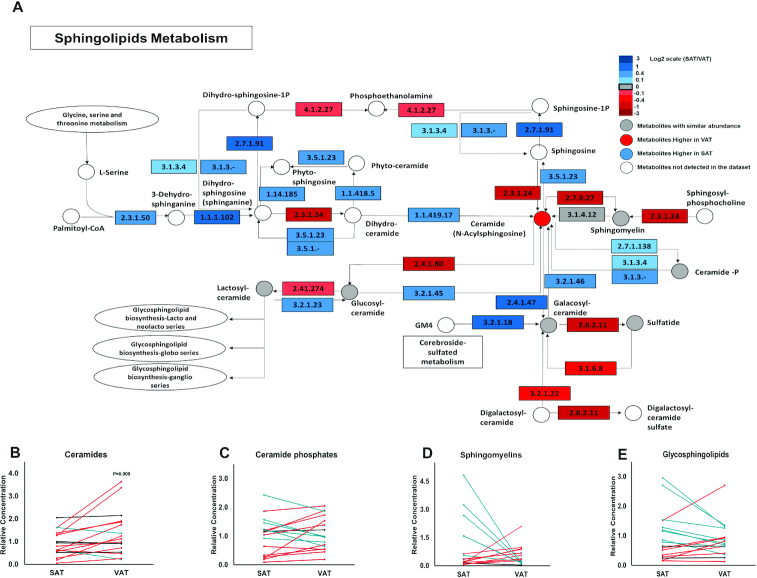
A network-based analysis demonstrates a differential ceramide accumulation in VAT and SAT
We subsequently studied the SL and GL metabolic networks using the approach described above. The alterations in SL metabolism between VAT and SAT are depicted in Figure 3A. We found a higher accumulation of nonglycosylated SLs (“ceramides”) in VAT (Figure 3B), whereas no significant changes in the abundance of ceramide phosphates (Figure 3C), sphingomyelins (Figure 3D) or the glycosylated forms of SLs (Figure 3E) were observed.
FIGURE 3.
Ceramides show a discriminative accumulation in VAT. (A) A metabolic map of the sphingolipid pathway [modified from the KEGG pathway (24)]. The gene expression of enzymes of the pathway, as found in the GTEx database, is shown as rectangles and the relative lipid accumulation in the main dataset is shown as circles. (B–E) The accumulation of sphingolipid groups found by our lipidomics study. Blue lines represent a change of ≥10% in the SAT:VAT ratio. Red lines represent a change of ≤ −10% in the SAT:VAT ratio. Black lines represent a similar accumulation ±10%. n = 17; error bars represent SEMs; 1-tailed paired t test. KEGG, Kyoto Encyclopedia of Genes and Genomes; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
TAGs and DAGs are highly abundant in SAT
We next studied the network of glycerolipids (Figure 4A). We observed a higher accumulation of TAGs (Figure 4B) and DAGs (Figure 4C) in SAT. No significant changes were seen in the abundance of phosphatidic acids, essential substrates for enzymes involved in the synthesis of TAGs (Figure 4D).
FIGURE 4.
High levels of TAGs and DAGs found in SAT compared to VAT. (A) A metabolic map of the glycerolipid pathway (25). The gene expression of enzymes of the pathway as found in the GTEx database is shown as rectangles and the relative lipid accumulation found by our lipidomics study in the main data set is shown as circles. (B–D) The accumulation of glycerolipid groups found by our lipidomics study. Blue lines represent a change of ≥10% in the SAT:VAT ratio. Red lines represent a change of ≤ −10% in the SAT:VAT ratio. Black lines represent a similar accumulation ±10%. n = 17; error bars represent SEMs; 1-tailed paired t test. DAG, diacylglycerol; KEGG, Kyoto Encyclopedia of Genes and Genomes; SAT, subcutaneous adipose tissue; TAG, triacylglycerol; VAT, visceral adipose tissue.
The analyses of the validation data sets revealed relative abundances (no internal standards were added) and were used to confirm the trends observed in the main dataset. They corroborated a different distribution of PCs in comparison to PEs in the adipose tissues (implying differential PEMT expression), with a lower accumulation of ceramides although a higher accumulation of TAGs in SAT.
We proceeded to examine the possible implications of the alterations between SAT and VAT lipid networks on the distribution of lipids of nutritional value. Recent work demonstrated that PLs were the most accurate and sensitive lipids for assessing fatty acid incorporation into adipose tissue (26). We therefore quantified SFAs, MUFAs, and PUFAs in PLs from the 2 tissues. We found considerable differences in the distribution of unsaturated lipids: both MUFAs (Figure 5A) and PUFAs (Figure 5B) showed higher concentrations in SAT, whereas no differences were found in the concentrations of SFAs (Figure 5C). The lower concentration of MUFAs and PUFAs in VAT is also depicted by the lower degree of unsaturation (Figure 5D).
FIGURE 5.

The concentration of unsaturated fatty acids in phospholipids is lower in VAT. The dispersion of fatty acids in phospholipids is regarded as an accurate and sensitive measure for assessing fatty acid incorporation into adipose tissue (26). The concentrations of MUFAs and PUFAs were quantified in phospholipids from SAT or VAT as the cumulative concentrations of the corresponding phospholipid species. n = 17; error bars represent SEMs; 1-tailed paired t test. SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
We quantified the concentrations of 61 lipids that were previously associated with specific nutritional sources (Supplemental Table 2). Notably, the concentrations of PCs in this analysis were higher in SAT in 9 of the 16 PCs (q < 0.05), 3 were higher in VAT, and 4 with no significant change. Intriguingly, all of the PCs with higher concentrations in VAT were very recently associated with seafood consumption (27). Our lipidomics study demonstrates a differential distribution of PCs versus PEs in SAT and VAT, and higher concentrations of glycerolipids in SAT, whereas a lower abundance of ceramides. Our transcript analyses point to a robustly altered expression of PEMT, corroborated by an independent large cohort study (GTEx). Previous reports suggest that PEMT expression is induced during differentiation of 3T3-L1 adipocytes, and that PEMT-knockout mice are resistant to high-fat-diet–induced fat cell hypertrophy (28, 29). We therefore set out to determine the influence of PEMT expression on adipogenic differentiation of embryonic stem cells (ESCs). Given the seeming correlation between PEMT expression and the abundance of TAGs and DAGs, we studied the accumulation of lipid droplets in the differentiated ESCs. We generated a PEMT shRNA ESC line (Figure 6A, B), and tested their differentiation to adipocytes by quantifying the expression of adipocyte markers.
FIGURE 6.
PEMT-deficient ESCs show lower expression of adipocyte markers and decreased accumulation of lipid droplets following differentiation. (A) PEMT-deficient cells were generated from V6.5 ESCs by shRNA-mediated gene silencing. The PEMT-deficient ESCs demonstrate ∼90% silencing (B). Adipogenesis was then induced in mock-transfected and PEMT-deficient ESCs. (C–E) Twenty-four days following the induction of differentiation, cells were tested for adipocyte gene expression (C) or observed under microscopy (D, E). (C) The expression of FABP4 and AdipoQwas analyzed by real-time polymerase chain reaction, and is represented as the mean ± SEM. (D, E) Cells were fixed and stained with Oil red O for lipid droplets and DAPI for nuclei. Phase (D) or color (E) images were taken using confocal microscopy (E). n = 4; error bars represent SEMs; 1-tailed paired t test. *, P<0.05; ***, P<0.001. AdipoQ, adiponectin, C1Q, and collagen domain containing; DAPI, 4′,6-diamidino-2-phenylindole; ESC, embryonic stem cell; FABP4, fatty acid binding protein 4; PEMT, phosphatidylethanolamine N-methyltransferase; WT, wild-type.
Following differentiation of ESCs to adipocytes, the expression of the adipocyte markers FABP4 and AdipoQ was 10-fold lower in the PEMT shRNA ESC-derived adipocytes (Figure 6C), implying a dramatic decrease in adipogenesis following PEMT attenuation. In line with our lipidomics data, following differentiation, PEMT-deficient adipocytes presented a lower accumulation of lipid droplets (Figure 6D, E).
Discussion
The differences between VAT and SAT tissues have drawn attention, as they may point to the etiology and mechanisms underlying the unique association between excess VAT and metabolic diseases. Our integrated analysis provides comprehensive lipid metabolic maps of VAT and SAT in overweight patients. The study design addressed patient-specific variance with paired-sample collection of different adipose tissue compartments. The functional implications of key findings were then tested by a stem cell model.
Our data demonstrate differences in the distribution of lipids across lipid subclasses, and suggest differential wiring of lipid networks in the 2 tissues. Our lipidomics analysis pointed to differential accumulation of PCs in adipose tissues. Given the indecisive differences in the expression of enzymes of the Kennedy pathway, we assumed that the differences in PC biosynthesis may be mediated via the PEMT pathway. Aligned with this notion, PEMT was previously suggested to be differentially expressed according to waist-to-hip ratio, especially in females (30), and to exhibit a differential gene expression in VAT and SAT (13, 31). In our data set, the differential expression of PEMT was cross-gender. We found higher accumulation of TAGs and DAGs in SAT tissues, which is concordant with the expression of corresponding genes as found in GTEx. The higher concentrations of glycerolipids may be explained by the PEMT-dependent accumulation of lipid droplets we found in our stem cell model. The lower expression of PEMT we found in VAT and the lower accumulation of lipid droplets in PEMT-deficient adipocytes are also in agreement with the findings of several groups that reported a (somewhat counterintuitive) lower adipocyte size in VAT (32, 33) and its association with insulin resistance (34). Further studies demonstrate that PEMT expression is regulated by insulin (35, 36), and that ceramides induce insulin resistance (37–39). Taken together with these studies, our network-based analysis and our stem cell model suggest that PEMT forms a major metabolic junction that bridges a few biosynthetic pathways into a functional pathway in adipose tissues. Together with previous work, our study therefore suggests a model for how alterations in lipid network may impact fat depot–specific adipogenesis, lipid droplet accumulation, and adipocyte size. Higher ceramide accumulation in VAT, leading to insulin resistance and lower expression of PEMT, was followed by downregulated lipogenesis, and lower accumulation of glycerolipids and lipid droplets (Figure 7).
FIGURE 7.

(A, B) Scheme of a suggested pathway that integrates the differential accumulation of ceramides and phospholipids in VAT and SAT to the lower adipogenesis and lipid droplet accumulation in VAT. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PEMT, phosphatidylethanolamine N-methyltransferase; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
Although many previous studies have focused on secreted proteins (adipokines, cytokines) in VAT, the interest in lipid mediators (sometimes termed “lipokines”) is growing. In fact, lipids provide a plethora of signaling molecules and their precursors, and are central to cell function (40, 41). Differences in the lipid composition of the 2 fat depots may therefore lead to shifts in the signaling pathways. Indeed, VAT adipocytes are more metabolically active, and have greater capacity to release free fatty acids, features that were proposed to relate to their greater tendency to exhibit insulin resistance than SAT adipocytes (4). Of note, our data suggest a differential distribution of unsaturated fatty acids in SAT and VAT. Given the great volume of literature that demonstrates beneficial effects of unsaturated lipids, the low concentrations of unsaturated fatty acid lipids in VAT (or the higher ratio of saturated to unsaturated lipids) may be linked to the association between VAT and metabolic diseases. Our data on lipids that were associated with specific nutritional sources suggest that PCs from all sources but seafood are highly accumulated in SAT. Given that seafood consumption is supposedly low in the Israeli population examined, there may be an alternative source for these lipids.
PEMT offers an alternative pathway to the canonical CDP-choline pathway, the predominant mechanism by which mammalian cells synthesize PCs. The expression of PEMT was thought to be liver specific (42); however, multiple reports strongly suggest that it is functionally expressed in other tissues, and specifically in adipocytes and fat tissue (13, 43). Furthermore, PEMT was shown to be upregulated in adipogenic-differentiated cells while being downregulated in dedifferentiated cells (44). Upregulated choline metabolism, and specifically PEMT expression, was implicated in lipid droplets formation and stability and TAG concentration (28), offering a possible explanation to our finding in both human adipose samples and in stem cell–derived adipocytes. The regulation of TAG concentration in adipocytes by PEMT expression may be explained by the higher conversion of PEMT-derived PCs to TAGs (45). The shift in the PL composition following changes in the expression of PEMT may have clinical implications and is upregulated by insulin signaling (35).
It would be important to further study the interactions of clinical parameters and different phenotypes on the distribution of lipid subclasses in adipose tissues.
Together, our data provide comprehensive metabolic maps of lipid networks in the 2 major mammalian fat depots, underscore the metabolic differences between SAT and VAT, and suggest potential nutritional and functional implications for these alterations, implying an integrated lipid regulatory pathway that crosses a few traditional biosynthetic pathways and regulates adipogenesis and lipid accumulation. The roles of this putative functional pathway should be further investigated.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—AZ, AS, and RS: analyzed data and prepared the figures; DS: analyzed data; CTM: analyzed data and helped revise the manuscript; NMK: participated in methods development and performed preliminary analysis; AZ: prepared samples for lipidomics analyses and generated PEMT-deficient ESCs; RB-H: generated PEMT-deficient ESCs; NG and YH: performed RT-qPCR experiments; CD: induced and examined differentiation to adipocytes; AR: designed research and drafted the manuscript; AM: conceived the study, designed research and all experiments, analyzed and interpreted data, drafted and prepared the manuscript; and all authors: read and approved the final manuscript. The authors report no conflicts of interest.
Notes
This research was funded in part by grants from the DFG (German Research Foundation) – Projektnummer 516 209933838 – SFB 1052 (projects B2), and the Israel Science Foundation (2176/19).
Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Data described in the manuscript will be made available upon request.
Abbreviations used: CDP-choline, cytidine diphosphate–choline; CEPT, choline/ethanolamine phosphotransferase; DAG, diacylglycerol; EB, embryoid body; ESC, embryonic stem cell; KEGG, Kyoto Encyclopedia of Genes and Genomes; mESC, mouse embryonic stem cell; PC; phosphatidylcholine; PCA, principal component analysis; PE, phosphatidylethanolamine; PEMT, phosphatidylethanolamine N-methyltransferase; PL, phospholipid; PLS-DA, partial least-squares regression discriminant analysis; SL, sphingolipid; RT-PCR, reverse transcriptase–PCR; SAT, subcutaneous adipose tissue; SUMC, Soroka University Medical Center; TAG, triacylglycerol; VAT, visceral adipose tissue.
Contributor Information
Anish Zacharia, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Daniel Saidemberg, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Chanchal Thomas Mannully, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Natalya M Kogan, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Alaa Shehadeh, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Reut Sinai, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Avigail Zucker, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Reut Bruck-Haimson, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
Nir Goldstein, Department of Clinical Biochemistry and Pharmacology and The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
Yulia Haim, Department of Clinical Biochemistry and Pharmacology and The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
Christian Dani, University Côte d'Azur, CNRS, INSERM, iBV, Faculté de Medicine, Nice, France.
Assaf Rudich, Department of Clinical Biochemistry and Pharmacology and The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
Arieh Moussaieff, The Institute for Drug Research, Hebrew University of Jerusalem, Jerusalem, Israel.
References
- 1. Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjorntorp P. “Portal” adipose tissue as a generator of risk factors for cardiovascular disease and diabetes. Arteriosclerosis. 1990;10(4):493–6. [PubMed] [Google Scholar]
- 3. Bays HE. “Sick fat,” metabolic disease, and atherosclerosis. Am J Med. 2009;122(1 Suppl):S26–37. [DOI] [PubMed] [Google Scholar]
- 4. Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev. 2010;11(1):11–18. [DOI] [PubMed] [Google Scholar]
- 5. Dusserre E, Moulin P, Vidal H. Differences in mRNA expression of the proteins secreted by the adipocytes in human subcutaneous and visceral adipose tissues. Biochim Biophys Acta. 2000;1500(1):88–96. [DOI] [PubMed] [Google Scholar]
- 6. Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, Vasan RS, Murabito JM, Meigs JB, Cupples LA et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116(1):39–48. [DOI] [PubMed] [Google Scholar]
- 7. Goodpaster BH, Thaete FL, Simoneau JA, Kelley DE. Subcutaneous abdominal fat and thigh muscle composition predict insulin sensitivity independently of visceral fat. Diabetes. 1997;46(10):1579–85. [DOI] [PubMed] [Google Scholar]
- 8. Liesenfeld DB, Grapov D, Fahrmann JF, Salou M, Scherer D, Toth R, Habermann N, Bohm J, Schrotz-King P, Gigic B et al. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare study. Am J Clin Nutr. 2015;102(2):433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol. 2017;13(11):633–43. [DOI] [PubMed] [Google Scholar]
- 10. Linder K, Arner P, Flores-Morales A, Tollet-Egnell P, Norstedt G. Differentially expressed genes in visceral or subcutaneous adipose tissue of obese men and women. J Lipid Res. 2004;45(1):148–54. [DOI] [PubMed] [Google Scholar]
- 11. Einstein FH, Atzmon G, Yang XM, Ma XH, Rincon M, Rudin E, Muzumdar R, Barzilai N. Differential responses of visceral and subcutaneous fat depots to nutrients. Diabetes. 2005;54(3):672–8. [DOI] [PubMed] [Google Scholar]
- 12. Chaurasia B, Kaddai VA, Lancaster GI, Henstridge DC, Sriram S, Galam DL, Gopalan V, Prakash KN, Velan SS, Bulchand S et al. Adipocyte ceramides regulate subcutaneous adipose browning, inflammation, and metabolism. Cell Metab. 2016;24(6):820–34. [DOI] [PubMed] [Google Scholar]
- 13. Jove M, Moreno-Navarrete JM, Pamplona R, Ricart W, Portero-Otin M, Fernandez-Real JM. Human omental and subcutaneous adipose tissue exhibit specific lipidomic signatures. FASEB J. 2014;28(3):1071–81. [DOI] [PubMed] [Google Scholar]
- 14. Bashan N, Dorfman K, Tarnovscki T, Harman-Boehm I, Liberty IF, Bluher M, Ovadia S, Maymon-Zilberstein T, Potashnik R, Stumvoll M et al. Mitogen-activated protein kinases, inhibitory-kappaB kinase, and insulin signaling in human omental versus subcutaneous adipose tissue in obesity. Endocrinology. 2007;148(6):2955–62. [DOI] [PubMed] [Google Scholar]
- 15. Haim Y, Bluher M, Konrad D, Goldstein N, Kloting N, Harman-Boehm I, Kirshtein B, Ginsberg D, Tarnovscki T, Gepner Y et al. ASK1 (MAP3K5) is transcriptionally upregulated by E2F1 in adipose tissue in obesity, molecularly defining a human dys-metabolic obese phenotype. Mol Metab. 2017;6(7):725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Available from: https://www.genome.jp/kegg. [Google Scholar]
- 17. Available from: https://gtexportal.org/home. [Google Scholar]
- 18. Neville MJ, Collins JM, Gloyn AL, McCarthy MI, Karpe F. Comprehensive human adipose tissue mRNA and microRNA endogenous control selection for quantitative real-time-PCR normalization. Obesity. 2011;19(4):888–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21(3):392–402. [DOI] [PubMed] [Google Scholar]
- 20. Dani C, Smith AG, Dessolin S, Leroy P, Staccini L, Villageois P, Darimont C, Ailhaud G. Differentiation of embryonic stem cells into adipocytes in vitro. J Cell Sci. 1997;110(Pt 11):1279–85. [DOI] [PubMed] [Google Scholar]
- 21. Wdziekonski B, Villageois P, Dani C. Development of adipocytes from differentiated ES cells. Methods Enzymol. 2003;365:268–77. [DOI] [PubMed] [Google Scholar]
- 22. Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Available from: https://www.genome.jp/kegg-bin/show_pathway?map00564. [Google Scholar]
- 24. Available from: https://www.genome.jp/kegg-bin/show_pathway?map00600. [Google Scholar]
- 25. Available from: https://www.genome.jp/kegg-bin/show_pathway?map=map00561&show_description = show. [Google Scholar]
- 26. Stanley EG, Jenkins BJ, Walker CG, Koulman A, Browning L, West AL, Calder PC, Jebb SA, Griffin JL. Lipidomics profiling of human adipose tissue identifies a pattern of lipids associated with fish oil supplementation. J Proteome Res. 2017;16(9):3168–79. [DOI] [PubMed] [Google Scholar]
- 27. Furtado JD, Beqari J, Campos H. Comparison of the utility of total plasma fatty acids versus those in cholesteryl ester, phospholipid, and triglyceride as biomarkers of fatty acid intake. Nutrients. 2019;11(9):2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horl G, Wagner A, Cole LK, Malli R, Reicher H, Kotzbeck P, Kofeler H, Hofler G, Frank S, Bogner-Strauss JG et al. Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo. J Biol Chem. 2011;286(19):17338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao X, van der Veen JN, Hermansson M, Ordonez M, Gomez-Munoz A, Vance DE, Jacobs RL. Decreased lipogenesis in white adipose tissue contributes to the resistance to high fat diet-induced obesity in phosphatidylethanolamine N-methyltransferase-deficient mice. Biochim Biophys Acta. 2015;1851(2):152–62. [DOI] [PubMed] [Google Scholar]
- 30. Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12(8):1217–22. [DOI] [PubMed] [Google Scholar]
- 32. Fang L, Guo F, Zhou L, Stahl R, Grams J. The cell size and distribution of adipocytes from subcutaneous and visceral fat is associated with type 2 diabetes mellitus in humans. Adipocyte. 2015;4(4):273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96(2):E268–77. [DOI] [PubMed] [Google Scholar]
- 34. Verboven K, Wouters K, Gaens K, Hansen D, Bijnen M, Wetzels S, Stehouwer CD, Goossens GH, Schalkwijk CG, Blaak EE et al. Abdominal subcutaneous and visceral adipocyte size, lipolysis and inflammation relate to insulin resistance in male obese humans. Sci Rep. 2018;8(1):4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kelly KL, Kiechle FL, Jarett L. Insulin stimulation of phospholipid methylation in isolated rat adipocyte plasma membranes. Proc Natl Acad Sci. 1984;81(4):1089–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiechle FL, Malinski H, Strandbergh DR, Artiss JD. Stimulation of phosphatidylcholine synthesis by insulin and ATP in isolated rat adipocyte plasma membranes. Biochem Biophys Res Commun. 1986;137(1):1–7. [DOI] [PubMed] [Google Scholar]
- 37. Huang S, Huang S, Wang X, Zhang Q, Liu J, Leng Y. Downregulation of lipin-1 induces insulin resistance by increasing intracellular ceramide accumulation in C2C12 myotubes. Int J Biol Sci. 2017;13(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petersen MC, Shulman GI. Roles of diacylglycerols and ceramides in hepatic insulin resistance. Trends Pharmacol Sci. 2017;38(7):649–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reali F, Morine MJ, Kahramanogullari O, Raichur S, Schneider HC, Crowther D, Priami C. Mechanistic interplay between ceramide and insulin resistance. Sci Rep. 2017;7:41231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veronika M, Welsch R, Ng A, Matsudaira P, Rajapakse JC. Correlation of cell membrane dynamics and cell motility. BMC Bioinformatics. 2011;12(Suppl 13):S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453(7194):475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Devlin AM, Singh R, Wade RE, Innis SM, Bottiglieri T, Lentz SR. Hypermethylation of Fads2 and altered hepatic fatty acid and phospholipid metabolism in mice with hyperhomocysteinemia. J Biol Chem. 2007;282(51):37082–90. [DOI] [PubMed] [Google Scholar]
- 43. Cole LK, Vance DE. A role for Sp1 in transcriptional regulation of phosphatidylethanolamine N-methyltransferase in liver and 3T3-L1 adipocytes. J Biol Chem. 2010;285(16):11880–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ullah M, Stich S, Notter M, Eucker J, Sittinger M, Ringe J. Transdifferentiation of mesenchymal stem cells-derived adipogenic-differentiated cells into osteogenic- or chondrogenic-differentiated cells proceeds via dedifferentiation and have a correlation with cell cycle arresting and driving genes. Differentiation. 2013;85(3):78–90. [DOI] [PubMed] [Google Scholar]
- 45. Vance DE. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim Biophys Acta. 2014;1838(6):1477–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.