To the Editor:
Global coronavirus (CoV) disease (COVID-19) diagnoses have eclipsed 18 million, with nearly 700,000 deaths (1). Beyond optimized supportive and ventilatory care for acute respiratory distress syndrome (2), no targeted therapies have been shown to improve patient survival (3). Although numerous antiviral agents are under active investigation, only remdesivir has improved clinically meaningful outcomes, resulting in an urgent need for alternate interventions.
Investigating means to manipulate innate lung defenses to protect against infectious pneumonias, our group has identified lung epithelial cells as potent effectors of therapeutically inducible antimicrobial responses. Once regarded as inert airflow conduits or passive gas-exchange filters, the airway and alveolar epithelia supplement lung defenses by undergoing profound structural and functional changes upon encountering pathogens (4). In addition to conferring a physical barrier, these cells modulate lung leukocyte responses and release microbicidal molecules, including antimicrobial polypeptides and reactive oxygen species (5, 6). Harnessing this defensive capacity, we found that stimulation of lung epithelial cells promotes pathogen killing and host survival. This inducible epithelial resistance develops within hours and extends to bacteria, fungi, and, notably, viruses, including influenza A and parainfluenza viruses (7–9). We have shown that these inducible epithelial responses protect against antibiotic-resistant organisms and bioterror pathogens, and we have hypothesized that such pathogen-agnostic mechanisms would similarly protect against emerging infections (10).
CoVs are enveloped positive-sense RNA viruses with characteristic surface spikes and large genomes (11). Severe acute respiratory syndrome CoV 2 (SARS-CoV-2) is the seventh CoV known to infect humans, with SARS-CoV and Middle East respiratory syndrome CoV (MERS-CoV) also capable of causing severe disease (12) in immunocompetent individuals. These three CoVs share substantial sequence homology, and both SARS-CoV and SARS-CoV-2 enter the cell via binding of their spike glycoprotein to human angiotensin converting enzyme-2 (ACE2) (13).
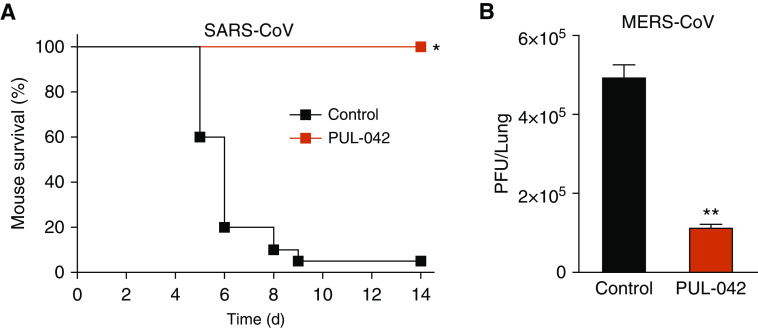
Given the hypothesis that inducible epithelial resistance could protect against emerging infections, we tested whether our lead compound, PUL-042, could protect against the other highly pathogenic CoVs even before the first report of COVID-19. PUL-042 is a combination therapeutic composed of the TLR2/6 ligand Pam2CSK4 (4 μM) and the TLR9 ligand ODN M362 (1 μM). Wild-type BALB/c mice and hDPP4 transgenic mice (14) were intranasally challenged with five times the median lethal dose of mouse-adapted SARS-CoV or MERS-CoV, respectively, 24 hours after intranasal instillation of PBS (sham) or PUL-042. As shown in Figure 1, a single PUL-042 treatment significantly improved survival of otherwise lethal SARS-CoV infection and significantly reduced the lung MERS-CoV burden 3 days after challenge, congruent with our prior work demonstrating a consistent correlation between survival advantage and reduced pathogen burden. Of note, the relationship between host survival and pathogen burden in respiratory viral infections can be very steep, with a 1.6-fold increase in the viral load resulting in a decrease in mouse survival from 80% to 20% in one model (15).
Figure 1.
PUL-042 treatment induces resistance against CoV pneumonia. (A) Survival of mice challenged with SARS-CoV. N = 20 mice/group. (B) Lung viral burden of mice 3 days after challenge with MERS-CoV. N = 5 mice/group. *P < 0.00001 versus sham treated. **P < 0.0001 versus sham treated. CoV = coronavirus; MERS-CoV = Middle East respiratory syndrome CoV; PFU = plaque-forming unit; SARS-CoV = severe acute respiratory syndrome CoV.
Considerable attention has been paid recently to attenuating the host response in COVID-19, with an intent to improve patient survival by reducing immunopathology. Here, we show an example of how inducing early, effective, proinflammatory responses can protect against pathogenic CoV pneumonia. The similarity of the pathogenic CoVs, together with our demonstrations of protection against other respiratory viruses, suggest this approach can also protect against SARS-CoV-2. As such, PUL-042 is under investigation in two U.S. Food and Drug Administration–approved randomized multicenter clinical trials to reduce pneumonia severity and mortality in patients with early COVID-19 (NCT 04312997) or in COVID-19–exposed healthcare workers and personal contacts (NCT 04313023).
The urgent, ongoing need to test interventions against SARS-CoV-2 precludes our immediate ability to do additional studies of SARS-CoV or MERS-CoV; thus, these results must be interpreted in view of this limitation. Nonetheless, these data suggest this host-focused strategy can be used in patients with suspected COVID-19 without requiring delays for confirmatory testing, as it is well tolerated by humans (NCT 02124278) and has broad activity against other infectious agents. Although it is suspected that a second pandemic wave may arise concurrently with seasonal influenza, a patient with a virus-like illness can be preemptively treated with PUL-042 with the expectation that she or he will appreciate a benefit, whether the syndrome results from SARS-CoV-2, influenza A, both viruses, or another pathogen. CoVs possess the capacity to jump species boundaries, making it highly likely that more pathogenic CoVs will emerge. However, this same strategy of manipulating lung epithelial responses can be anticipated to also protect against future emergent viruses.
Supplementary Material
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2020-0247LE on July 24, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.World Health Organization Coronavirus disease (COVID-2019) situation reports 2020[accessed 2020 Aug 6]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 2.Poston JT, Patel BK, Davis AM. Management of critically ill adults with COVID-19. JAMA. doi: 10.1001/jama.2020.4914. [online ahead of print] 26 Mar 2020; DOI: 10.1001/jama.2020.4914. [DOI] [PubMed] [Google Scholar]
- 3.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. doi: 10.1001/jama.2020.6019. [online ahead of print] 13 Apr 2020; DOI: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 4.Leiva-Juárez MM, Kolls JK, Evans SE. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 2018;11:21–34. doi: 10.1038/mi.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bals R, Hiemstra PS. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur Respir J. 2004;23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett JA, Fischer AJ, McCray PBJ., Jr Innate immune functions of the airway epithelium. Contrib Microbiol. 2008;15:147–163. doi: 10.1159/000136349. [DOI] [PubMed] [Google Scholar]
- 7.Evans SE, Scott BL, Clement CG, Larson DT, Kontoyiannis D, Lewis RE, et al. Stimulated innate resistance of lung epithelium protects mice broadly against bacteria and fungi. Am J Respir Cell Mol Biol. 2010;42:40–50. doi: 10.1165/rcmb.2008-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkpatrick CT, Wang Y, Leiva Juarez MM, Shivshankar P, Pantaleón García J, Plumer AK, et al. Inducible lung epithelial resistance requires multisource reactive oxygen species generation to protect against viral infections. MBio. 2018;9:e00696-18. doi: 10.1128/mBio.00696-18. [Published erratum appears in MBio 10:e00496-19.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggan JM, You D, Cleaver JO, Larson DT, Garza RJ, Guzmán Pruneda FA, et al. Synergistic interactions of TLR2/6 and TLR9 induce a high level of resistance to lung infection in mice. J Immunol. 2011;186:5916–5926. doi: 10.4049/jimmunol.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans SE, Xu Y, Tuvim MJ, Dickey BF. Inducible innate resistance of lung epithelium to infection. Annu Rev Physiol. 2010;72:413–435. doi: 10.1146/annurev-physiol-021909-135909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr AR, Stanley P. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng CT, Huang C, Newman P, Wang N, Narayanan K, Watts DM, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldblatt DL, Flores JR, Valverde Ha G, Jaramillo AM, Tkachman S, Kirkpatrick CT, et al. Inducible epithelial resistance against acute Sendai virus infection prevents chronic asthma-like lung disease in mice. Br J Pharmacol. 2020;177:2256–2273. doi: 10.1111/bph.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.