Abstract
Most active non-LTR (long terminal repeat) retrotransposons carry two open reading frames (ORFs) encoding ORF1p and ORF2p proteins. The ORF2p proteins are relatively well studied and are known to contain endonuclease/reverse transcriptase domains. At the same time, the biological function of ORF1p proteins remains poorly understood, except in that they nonspecifically bind single-stranded mRNA/DNA molecules. CR1-like elements form the most widely distributed clade/superfamily of non-LTR retrotransposons. We found that ORF1p proteins encoded by diverse CR1-like elements contain conserved esterase domain (ES) or plant homeodomain (PHD). This indicates that CR1-like ORF1p proteins are either lipolytic enzymes or are involved in protein-protein interactions related to chromatin remodeling. Sequence conservation of ES suggests that interaction with cellular membranes is an important phase in life circles of CR1-like elements. Presumably such interaction helps in penetrating host cells. As a consequence, the presence of multiple young CR1 families characterized by ∼10% intrafamily and 40% interfamily identities may be explained by a relatively frequent horizontal transfer of these CR1-like elements. Unexpectedly, ES links together non-LTR retrotransposons and single-stranded RNA viruses like influenza C and coronaviruses, which are known to depend on their own ES.
Keywords: non-LTR retrotransposon, CR1 clade, ORF1p, esterase, PHD homeodomain
Introduction
Genomes of all known eukaryotes are populated by transposable elements (TEs) capable of intragenomic multiplication or transposition (Berg and Howe 1987). For example, recognizable fossils of TEs constitute approximately 45% and 12% of the Homo sapiens (Lander et al. 2001) and Arabidospis thaliana (Kapitonov and Jurka 1999) genomes, respectively. Eukaryotic TEs can be divided into the following four classes: endogenous retroviruses and long terminal repeat (LTR) retrotransposons (Coffin, Hughes, and Varmus 1997), non-LTR retrotransposons, including so-called LINEs, SINEs, and processed pseudogenes (Malik, Burke, and Eickbush 1999; Weiner 2000), cut-and-paste DNA transposons (Craig 1995; Capy et al. 1998), and rolling-circle DNA transposons (Kapitonov and Jurka 2001). Duplication of a retrotransposon depends on reverse transcription and endonucleolytic cleavage, both of which are catalyzed by a reverse transcriptase (RT) and endonuclease domains of a polyprotein encoded by itself or by other retrotransposons. Primed by an endonucleolytic nick at the host DNA, an mRNA molecule, expressed during transcription of the retrotransposon DNA, is reverse transcribed and inserted in the genome. At present, known non-LTR retrotransposons belong to ∼10 superfamilies or clades identified on the basis of phylogenetic studies of their protein sequences (Malik, Burke, and Eickbush 1999). Non-LTR retrotransposons form a clade if they share a common ancestor that is not shared by any other non-LTR retrotransposons outside the clade. Most non-LTR retrotransposons carry two long open reading frames, ORF1 and ORF2, which encode ORF1p and ORF2p proteins, respectively. ORF2p includes the RT, apurinic/apyrimidinic endonuclease (APE) or restriction-enzyme-like endonuclease domains. In some retroelements, ORF2p also includes a ribonuclease H domain. Whereas both the structure and the function of ORF2p are relatively well understood, properties of ORF1p remain obscure, in part because of the lack of significant similarity between ORF1p and proteins with known enzymatic functions. To date, the only structural elements discovered in different ORF1p proteins are the nonspecific zinc finger, leucine zipper, and coiled coil motifs (Holmes, Singer, and Swergold 1992; Dawson et al. 1997; Haas et al. 1997; Kajikawa, Ohshima, and Okada 1997; Poulter, Butler, and Ormandy 1999). Experimental data also suggest (Dawson et al. 1997; Hohjoh and Singer 1997; Kolosha and Martin 1997) that ORF1p proteins from the L1 and I clades bind single-stranded RNA–DNA. Overall, the role of ORF1 proteins in non-LTR retrotransposons is uncertain, although there are indications (Dawson et al. 1997; Hohjoh and Singer 1997; Kolosha and Martin 1997; Martin and Bushman 2001) linking ORF1p to retroviral nucleocapsid proteins involved in packaging retroviral RNA and in other important steps of a retroviral “life cycle” (Coffin, Hughes, and Varmus 1997).
CR1 is one of the most abundant and widely distributed clades of non-LTR retrotransposons (Malik, Burke, and Eickbush 1999). Most CR1 elements are severely truncated at their 5′ ends. Therefore, it was found only recently that they are non-LTR retrotransposons populating genomes of birds, amphibians, and fishes (Burch, Davis, and Haas 1993); lizards and turtles (Vandergon and Reitman 1994; Kajikawa, Ohshima, and Okada 1997); mammals (Smit 1996; Jurka and Kapitonov 1999a); and invertebrates (Drew and Brindley 1997).
In this paper we report new full-length CR1-like elements from zebrafish, medaka, and fruit fly. We show that ORF1-encoded proteins in various CR1-like non-LTR retrotransposons include conserved plant homeodomain (PHD) and esterase domains. Given the conservation of the PHD and esterase domains in highly divergent CR1-like retrotransposons from different species, including those split several hundred million years ago, we assume that the PHD and esterase activities of the ORF1-encoded proteins were necessary for survival of these retrotransposons. Interestingly, as for the CR1-like non-LTR retrotransposons, the life cycle of enveloped negative-stranded and positive-stranded RNA viruses in birds and mammals depends on their own esterase.
Materials and Methods
All non-LTR retroelements reported here were found by using various methods of computational analysis. Starting with a compilation of known transposable elements collected in Repbase Update (Jurka 2000) at www.girinst.org/Repbase_Update, we identified their copies in DNA sequences deposited in GenBank. The identification began with comparing DNA and protein sequences of known TEs against the sequenced portion of the Danio rerio and Drosophila melanogaster genomes by using CENSOR (Jurka et al. 1996) and TBLASTN (Altschul et al. 1997), respectively.
Using the majority rule applied to the corresponding set of multiple aligned copies of retrotransposons, we built their consensus sequences. Copies of TEs not produced directly by transpositions, such as those created by chromosomal duplications or redundant sequencing, were discarded based on the similarities between their flanking regions.
Distantly related proteins were identified using PsiBLAST (Altschul et al. 1997). Multiple alignments of protein sequences were created by CLUSTAL-W (Thompson, Higgins, and Gibson 1994). Alignments of DNA sequences were performed using the VMALN2 and PALN2 programs developed at the Genetic Information research Institute, Mountain View, California (GIRI). Phylogenetic analysis was conducted using MEGA 2.1 (Kumar et al. 2001). Protein domains described in this article were identified using the Family Pairwise Search (FPS) algorithm (Bailey and Grundy 1999; http://fps.sdsc.edu/) and the SUPERFAMILY protein assignments server (Gough et al. 2001; http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/). Scoring of the protein sequences by FPS was performed against Pfam, a collection of protein family alignments reconstructed using hidden Markov models (Bateman et al. 2002). Assignment by SUPERFAMILY has been performed using SCOPE, a library of protein superfamilies (Murzin et al. 1995).
Sequences of retrotransposons reported here were deposited in the Repbase Update in the sections designated for fruit fly, zebrafish, human, vertebrates and invertebrates.
Results
CR1 Elements from Zebrafish
Screening reverse transcriptase-like sequences in the publicly available DNA sequences, covering ∼1% of the D. rerio genome revealed multiple copies of non-LTR elements that belong to the CR1 clade. Cluster analysis of these sequences indicates that CR1-like elements in the D. rerio genome belong to over 10 young and diverse families characterized by ∼5% intrafamily and ∼35% interfamily nucleotide divergence (unpublished data). We assembled three consensus sequences that belong to families named CR1-1_DR, CR1-2_DR, and CR1-3_DR (fig. 1).
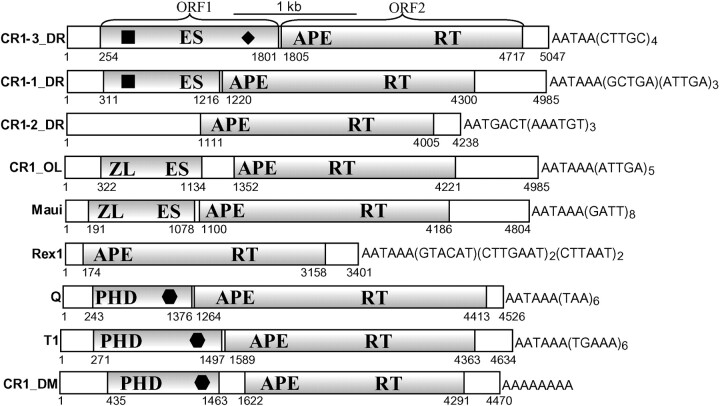
Fig. 1.
Schematic structure of complete CR1-like retrotransposons from fishes and insects. CR1-1_DR, CR1-2_DR and CR1-3_DR—are consensus sequences of retrotransposons that belong to the three families of retrotransposons identified in the Danio rerio genome. Maui and Rex1 are the consensus sequences of two retrotransposons from CR1-like families present in the Fugu rubripes genome. CR1_OL is a slightly damaged element identified in the Oryzias latipes genome. Horizontally shaded boxes mark ORF1s and ORF2s. ORF2s encode proteins composed of the apurinic/apyrimidinic endonuclease (APE) and reverse transcriptase (RT) domains. Proteins encoded by ORF1s are composed of putative zinc finger/leucine zipper (ZL) motifs, the plant homeodomain (PHD) and the esterase (ES) domains. Black squares, diamonds and hexagons indicate different unclassified domains. The 3′ termini of all retrotransposons, excluding CR1-2_DR and CR1_DM, are shown starting from the polyadenylation signal, followed by terminal microsatellite repeats composed of different 4–7-bp units repeated 2–8 times. The average number of the repetitions is shown as a subscript index
The 4985-bp CR1-1_DR consensus sequence was built from 10 copies and it is ∼98% identical with them. One partially truncated CR1-1_DR copy has been reported recently as the CR1DR1 element (Jekosch 2002). The CR1-1_DR consensus sequence harbors two ORFs encoding 300-aa CR1-1DR1p and 1000-aa CR1-1DR2p proteins (fig. 1).
The second family of zebrafish CR1-like elements is represented by a 4238-bp CR1-2_DR consensus sequence (fig. 1) assembled from another 10 copies that are also ∼98% identical to the consensus. Originally, a 600-bp fragment, 98% identical to a portion of the CR1-2_DR consensus sequence (positions 3422–4062), was reported as a LINE element (Okada et al. 1997). Recently, a ∼2900-bp CR1-2_DR copy was deposited in Repbase Update as the CR1DR2 element (Jekosch 2002), which is 98% identical to the coding region of the CR1-2_DR consensus sequence (positions 1111–4008). Surprisingly, the CR1-2_DR consensus sequence includes a long 1110-bp 5′ UTR region corresponding to ORF1 in various CR1-like elements. The only ORF in CR1-2_DR encodes a 965-aa CR1-2_DRp protein composed of the APE and RT domains.
Finally, the 5047-bp CR1-3_DR consensus sequence was built from seven different copies; they are ∼95% identical with the consensus. CR1-3_DR carries ORF1 (positions 254-1801 ) and ORF2 (positions 1805–4717) encoding, respectively, a 516-aa CR1-3_DR1p protein and a 971-aa CR1-3_DR2p protein. As expected, CR1-3_DR2p is composed of the APE and RT domains.
CR1 Retrotransposon from Fruit Fly
A TBLASTN-based screening of CR1-like reverse transcriptases encoded by the Drosophila melanogaster genome revealed a rather abundant family of CR1-like non-LTR retrotransposons, hereafter named CR1_DM. The 4470-bp CR1_DM consensus sequence was constructed from 20 copies that were ∼10% divergent from one another. It contains ORF1 and ORF2 respectively encoding a 355-aa CR1_DM1p protein and a 964-aa CR1_DM2 protein (fig. 1). Approximately 100 copies of CR1_DM are present in a sequenced portion of the D. melanogaster genome that covers mainly euchromatin regions representing ∼70% of the genome. Multiple subfamilies of CR1_DM are present in the genome (unpublished data).
CR1 Elements from Medaka and Blood Fluke
We also characterized a full-length 4985-bp copy of a CR1-like element in the Oryzias latipes (medaka fish) genome, called CR1-1_OL (fig. 1). Its 5′ and 3′ boundaries (GenBank AB054295, positions 330–5314) are labeled by ∼300-bp direct repeats composed of an 18-bp minisatellite unit. The CR1-1_OL element has been inserted into the genome relatively recently. Its ORF1 encodes a 271-aa CR1-1_OL1p protein (positions 322-1134), and ORF2 (positions 1352–4221) is corrupted by only two false frame shifts and one false stop codon.
Using genome survey sequences (GSS) from GenBank, we built a 3032-bp consensus sequence of the Schistosoma mansoni SR1 retrotransposon that is 700-bp longer than its sequence reported previously (Drew and Brindley 1997). The consensus sequence encodes a 950-aa SR1p protein (positions 36–2885) composed of the APE and RT domains. The extended region encodes APE. Available sequence data do not permit obtaining of any further 5′-extension of the SR1 consensus sequence, and we cannot prove or disprove the existence of ORF1p encoded by this element.
Diversity of the 3′ Tails
Studies of DNA sequences flanking CR1-like elements presented in this article have revealed characteristics similar to those of known CR1-like elements reported previously (Haas et al. 1997; Kajikawa, Ohshima, and Okada 1997; Poulter, Butler, and Ormandy 1999). These elements do not generate target site duplications, and their 3′ tails are composed of microsatellites (fig. 1). It appears that different families of CR1-elements, even those that populate the same genome, are characterized by different microsatellites that are specific for the each family. For example, the 3′ termini of CR1-1_DR elements are composed of (ATTGA)n which follows GCTTGA and the polyadenylation signal. The 3′ termini of CR1-2_DRs contain (AAATGT)n and they do not have any polyadenylation signal. In contrast, the 3′ termini of CR1-3_DR elements are composed of the polyadenylation signal followed by (CTTGC)n.
It has not yet been proved, however, whether 3′ microsatellite tails of CR1-like retrotransposons are their real termini or genomic microsatellites that served as targets during insertions of the retrotransposons. To resolve this question, we identified several CR1-like elements inserted into copies of other known TEs (unpublished data) that do not contain the microsatellites at positions targeted by the insertions. This observation suggests that the 3′ microsatellites have been inserted into the genome together with CR1-like elements, and they can be considered to be distinctive hallmarks or signatures of different families. Presumably, these signatures depend on slightly different family-specific enzymatic activities encoded by the CR1-like elements. It is likely that generation of microsatellites at the 3′ ends of CR1-like elements is a result of nontemplated additions by CR1-like reverse transcriptases, as shown experimentally for the I and R2 non-LTR retrotransposons (Chaboissier, Finnegan, and Bucheton 2000; Eickbush, Luan, and Eickbush 2000).
Phylogenetic Analysis
Figure 2 shows a phylogeny of ORF2p proteins encoded by CR1-like non-LTR retrotransposons and several other elements that belong to non-CR1 clades. The phylogenetic analysis strongly suggests that the CR1 clade is composed of three major subclades.
Fig. 2.
Phylogeny of the CR1-like non-LTR retrotransposons based on their endonuclease and reverse transcriptase domains. The phylogenetic tree also includes several retrotransposons from the Jockey, LOA, I, and L1 clades. Numbers next to each node indicate bootstrap values calculated as percentages of similar topologies out of 1,000 replicas for the neighbor-joining method. The names of non-LTR retrotransposons families and their host species are shown adjacent to the tree nodes. A scale of distances between the protein sequences is indicated. Solid triangles denote retrotransposons whose ORF1s code for the esterase. GenBank proteins identification numbers are as follows: Jockey (134083), Juan-C (1079026), Doc (8823), Lian (7511795), I (903726), CR1_BF (17529698), CR1 (2331059), CR1_PS (6576738), Q (11359829), T1 (159644), L1 (2072977). Sequences of the remaining retrotransposons have been deposited in the following sections of Repbase Update: humrep.ref (L2 and L3), dmrep.ref (CR1_DM, BAGGINS1, IVK), fugrep.ref (Maui, REX1), zebrep.ref (CR1-1_DR, CR1-2_DR, CR1-3_DR) and invrep.ref (SR1)
The first CR1 subclade, called CR1-I, includes CR1 and CR1_PS from chicken and turtle, L3 from mammals, and SR1 and CR1_BF from blood fluke and lancelet, respectively. Given the tree topology, distances, and bootstrap values, it is highly likely that, pending additional sequence data, this subclade will be split into at least three minor subclades. If so, SR1 and CR1_BF represent the potential minor subclades.
The second major CR1 subclade, called CR1-II, includes CR1-elements identified in insects only: Q and T1 from the African malaria mosquito, and CR1_DM from the fruit fly genome. Actually, T1 element was the first element from the CR1 clade identified as a non-LTR retrotransposon (Besansky 1990). CR1 replaced T1 as the name of the clade after classification introduced by Malik, Burke, and Eickbush (1999).
Finally, the third major subclade, called CR1_III, includes L2 from mammals, Maui from pufferfish, and the CR1-1_DR, CR1-2_DR, and CR1-3_DR families from zebrafish. Interestingly, L2 and CR1-2_DR form a distinctive group separated from the other members of the third subclade by the 100% bootstrap value, and neither L2 nor CR1-2_DR encodes ORF1p-like proteins. In addition, the REX1 element (Volff, Korting, and Schartl 2000; Smit 2001) may also represent a major subclade. It was suggested recently that L2-like and REX1-like elements form two novel clades of non-LTR retrotransposons called L2 and REX1, respectively (Lovsin, Gubensek, and Kordi 2001). We do not think that the introduction of these two clades is strongly supported by available data. Moreover, we report here (see below) that the esterase domain is conserved in ORF1p encoded by different elements that belong to the CR1-I and CR1-III (L2) major subclades (fig. 2).
The PHD Domain
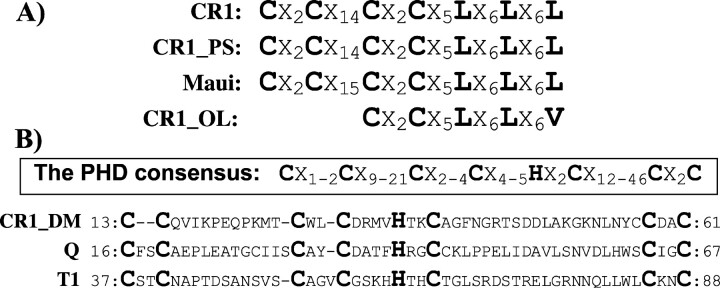
Computational analysis of the OFR1 proteins encoded by the zebrafish CR1-1_DR, CR1-2_DR, and CR1-3_DR elements failed to identify any zinc finger/leucine zipper motifs (ZL) similar to those present in the CR1, CR1_PS, and Maui retrotransposons from the chicken, turtle, and pufferfish genomes, respectively (Kajikawa, Ohshima, and Okada 1997; Poulter, Butler, and Ormandy 1999; Haas et al. 2001). However, ORF1p in CR1_OL from medaka fish harbors one motif distantly similar to ZL (fig. 3). Because stop codons and frame shifts that distort ORF2 encoded by the only available CR1_OL copy are present, it is likely that the originally intact ZL has also been damaged by mutations.
Fig. 3.
Zinc finger motifs in proteins encoded by ORF1s of different CR1-like non-LTR retrotransposons. C denotes cysteine; L, leucine; H, histidine; X, any residue; the subscript index indicates the number of the amino acid residues marked by it. A, Putative zinc finger/leucine zipper domains in CR1 (Haas et al. 2001), CR1_PS (Kajikawa, Ohshima, and Okada 1997), Maui (Poulter, Butler, and Ormandy 1999) and CR1_OL from chicken, turtle, pufferfish, and medaka, respectively. B, ORF1 proteins encoded by the fruit fly CR1_DM and the malaria mosquito Q1 and T non-LTR retrotransposons that harbor the PHD domain. Conserved cysteine and histidine residues matching the PHD consensus sequence are highlighted. Numbers at the beginning and the end of the amino acid sequences indicate positions of the corresponding amino acid residues in the protein sequences deposited in GenBank and Repbase Update
Surprisingly, N-terminal portions of the ORF1 proteins encoded by the fly CR1_DM, and mosquito Q (Besansky, Bedell, and Mukabayire 1994) and T1 elements include motifs (fig. 3) that fit the consensus sequence of a unique zinc finger domain called the PHD (plant homeodomain) or LAP (leukemia-associated protein) domain (Aasland, Gibson, and Stewart 1995; Saha et al. 1995). The PHD domain has been identified in proteins primarily associated with chromatin and involved in chromatin-mediated transcription control (Aasland, Gibson, and Stewart 1995; Capili et al. 2001). As indicated by a PsiBLAST search, all three ORF1p proteins from CR1_DM, T1, and Q are similar to one another over a span of ∼300 aa, although the overall sequence identity is only ∼20%. Additionally, presence of the PHD domain in the ORF1 proteins encoded by CR1_DM, T1, and Q was confirmed by other computational methods. Given an E-value < 0.01, PHD was the only domain identified in these proteins by the Family Pairwise Search algorithm (Bailey and Grundy 1999) and SUPERFAMILY (Gough et al. 2001). Therefore, the presence of PHD in the highly divergent N-terminal portions of the ORF1p proteins encoded by insect CR1-like elements should be considered to be a strong indication that this domain is important for their life cycle.
The exact function of the PHD domain is not yet known, but it is thought to be involved in protein–protein interactions and to be of importance for the assembly or activity of multicomponent complexes involved in transcriptional activation or repression (Aasland, Gibson, and Stewart 1995; Saha et al. 1995; Capili et al. 2001). Multiple lines of evidence suggest that PHD domain proteins can be targeted to DNA only indirectly via protein–protein interactions (Gibbons et al. 1997; Kehle et al. 1998; Koipally et al. 1999; Lyngso et al. 2000; Yochum and Ayer 2001). Therefore, it is unlikely that the zinc fingers encoded by ORF1s in CR1-like elements are involved directly in DNA or RNA binding, as proposed earlier for the putative zinc finger/leucine zipper domains in CR1_PS (Kajikawa, Ohshima, and Okada 1997).
The Esterase Domain
On the basis of a BLASTP search, we identified only three GenBank proteins similar to CR1-3_DR1p (E < 0.01). They are the ORF1 proteins from the chicken CR1 (Haas et al. 2001), turtle CR1_PS (Kajikawa, Ohshima, and Okada 1997), and pufferfish Maui (Poulter, Butler, and Ormandy 1999) retrotransposons. Because only a central portion of CR1-3_DR1p (positions 168–327) is similar to the ORF1p proteins (22% to 32% identity), we used it as a separate query for a PsiBLAST search (E < 0.005). After several iterations, the central portion converged with ∼150 eukaryotic and prokaryotic proteins from the esterase/acetylhydrolase superfamily (Drablos and Petersen 1997; Arpigny and Jaeger 1999). The same classification of CR1-3_DRp1 (E < 10−10) was also supported by the SUPERFAMILY genome assignments server (Gough et al. 2001). Figure 4 shows a multiple alignment of ORF1p proteins from CR1 retrotransposons and several prokaryotic and eukaryotic esterases. Two esterases included in the alignment were comprehensively studied experimentally: PAF-AH, a brain acetylhydrolase from cow (Ho et al. 1997), and RGAE, a rhamnogalacturonan acetylesterase from fungi (Molgaard, Kauppinen, and Larsen 2000). The most conserved structural hallmark of esterases is a catalytic triad composed of properly arranged serine, histidine, and aspartic acid residues (Drablos and Petersen 1997; Arpigny and Jaeger 1999; Molgaard, Kauppinen, and Larsen 2000). Different order and spacing of amino acid residues from the catalytic triad define several families of esterases (Dalrymple et al. 1997; Gough et al. 2001). Presumably, the esterase domain (ES) encoded by the CR1-like ORF1 proteins belongs to a specific family called GDSL (Arpigny and Jaeger 1999), SGNH (Molgaard, Kauppinen, and Larsen 2000), or the rhamnogalacturonan acetylesterase family (Gough et al. 2001). This family is characterized by GDS, GXND, and DXXH conserved motifs (Dalrymple et al. 1997). It has been shown experimentally (Ho et al. 1997) that serine from the first motif and aspartic acid plus histidine from the third motif belong to the catalytic triad. Strikingly, all three motifs and the catalytic triad are perfectly conserved in the highly divergent ORF1p encoded by CR1-elements from the chicken, turtle, medaka, pufferfish, and zebrafish genomes (fig. 4). The alignment also includes ES found in the ancient L3 retrotransposon fossilized in the human genome (see next section). Additionally, we found ES conserved in putative ORF1p proteins encoded by CR1-like elements fossilized in the crocodile, frog, and salmon genomes (unpublished data).
Fig. 4.

Multiple sequence alignment of the putative conserved esterase domains encoded by ORF1s in CR1-like non-LTR retrotransposons and other esterases. Solid arrowheads mark the catalytic serine-asparate-histidine triad. Ambiguous amino acids are denoted by Xs. GenBank protein identification numbers are as follows: CR1_PS (6576737), CR1 (2331058), Maui (4378024), NeuA (13876786, CMP-N-acetylneuramic acid synthetase from Streptococcus agalactiae), TesA (267107, Acyl-CoA thioesterase I from Escherichia coli), RGAE (7766904), rhamnogalacturonan acetylesterase from Aspergillus aculeatus), PAF-AH (2624421, platelet-activating factor acetylhydrolase from Bos taurus). Amino acid sequences of ORF1p proteins encoded by CR1-1_DR, CR1-2_DR, CR1-3_DR, CR1-1_TN, CR1_OL and L3 are deposited in Repbase Update
L3, the Most Ancient Transposable Element Ever Reconstructed In Silico
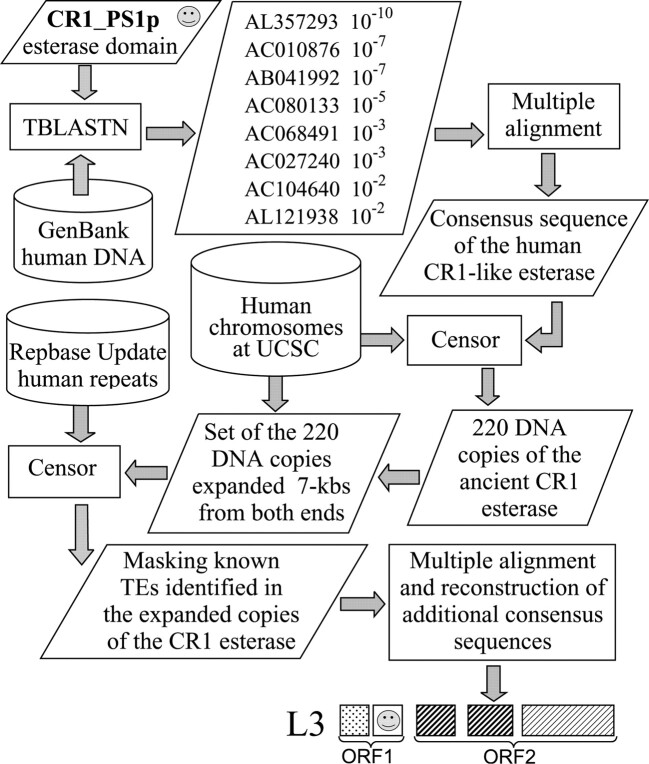
Using TBLASTN, we found that the 169-aa CR1_PS esterase domain matches eight proteins (10−10 ≤ E ≤ 10−2) encoded by human GenBank sequences (fig. 5). Apparently none of these proteins are functional because of multiple stop codons. DNA sequences encoding these proteins were extracted from the corresponding GenBank sequences. Based on a multiple alignment of the extracted fragments, a 700-bp consensus sequence was assembled, which was ∼73% identical to the fragments. Remarkably, a 120-aa protein encoded by the consensus sequence was much more similar to the CR1_PS esterase domain than any of the eight ORFs (E = 10−33, 52% identity). Moreover, the consensus sequence ORF was not interrupted by stop codons. Applying the BLASTN search with the consensus sequence as a query, we identified 13 fragments in the human genome similar to the consensus, including the original eight fragments. However, screening of assembled human chromosomes by CENSOR revealed ∼300 copies similar to the consensus sequence. Each copy has been expanded up to 7 kb in both directions. After pairwise alignment of the expanded sequences with each other, we eliminated chromosomal duplications more than 80% identical to each other. Subsequently, the final set of 220 sequences was screened for known TEs using CENSOR and Repbase Update. As we expected, there was a striking correlation between the extracted esterase-like fragments and L3. The CR1-like esterase domain was followed by remnants of L3 in 53 sequences, and both the esterase and L3 were in the same orientation. This is consistent with the fact that the L3 element is an ancient CR1-like non-LTR retrotransposon (Jurka and Kapitonov 1999a) whose reverse transcriptase is closest to CR1_PS2p. However, L3 is so old that only its relatively short 3′ portion was recovered previously as the ∼1.8-kb L3 consensus sequence (Jurka and Kapitonov 1999a; Smit 2000). Using the set of 220 masked sequences, we iteratively built three consensus sequences that represent missing parts of the ancient L3 (fig. 5). They encode domains closest to those present in CR1_PS, including the very beginning of ORF1p, esterase, endonuclease, and a middle portion of ORF2p. These domains are also present in rodents and other mammals (unpublished data).
Fig. 5.
Flowchart of the identification of ORF1p encoded by the ancient L3 retrotransposon fossilized in the human genome. Arrows indicate information flow directions. Rectangles illustrate different computational processes indicated by corresponding program names. Parallelograms indicate specific sets of data. Cans symbolize databases. GenBank accession numbers of sequences containing DNA regions which encode protein sequences TBLASTN-similar to CR1_PS1p are indicated together with corresponding E values. The “smiley face” marks the esterase domain found in different CR1-like elements
The L3 consensus sequence reported here is composed of four separate segments ∼65% identical to L3 copies. Given this diversity and the age (over 200 Myr), we can recover only the most conserved parts of L3s. Our data indicate that the esterase domain was functional in CR1-like retrotransposons that had been multiplied in ancestors of all mammals. It should be pointed out that many L3 copies are interrupted by L2 elements inserted randomly at different positions (unpublished data). Therefore, L3 elements are older than L2 elements. As the oldest transposable element identified in the human genome, L3 can be an extremely useful reference sequence in evolutionary studies.
Discussion
Despite the abundance and variety of non-LTR retrotransposons, many aspects of their life cycles and evolution are not known. Overall, they are viewed quite mechanistically as genomic parasites vertically transmitted during evolution of eukaryotic genomes (Malik, Burke, and Eickbush 1999). Our functional understanding of non-LTR retrotransposons is definitely lagging behind what we know about endogenous retroviruses/LTR retrotransposons (1997). In particular, we know little about the function of proteins encoded by ORF1 in different families of non-LTR retrotransposons. Our discovery of two specific and conserved domains, PHD (plant homeodomain) and ES (esterase, a lipolytic acetylhydrolase) may represent a breakthrough in this respect. Based on a classification by SCOP (Gough et al. 2001), the esterase/acetylhydrolase superfamily is composed of four families: (I) esterase, (II) esterase domain of hemagglutinin glycoprotein HEF1, (III) acetylhydrolase, and (IV) rhamnogalacturonan acetylesterase. Presumably, families III and IV can be joined together as the so-called GDSL or SGNH families (Dalrymple et al. 1997; Arpigny and Jaeger 1999). GDSL includes secreted and outer membrane–bound esterases, acetylhydrolases, and arylesterases (Dalrymple et al. 1997; Arpigny and Jaeger 1999; Molgaard, Kauppinen, and Larsen 2000). Usually, these enzymes remove acetyl or fatty acids from complex polysaccharides, viral glycoproteins, and cellular proteins interacting with membranes and involved in cell signaling or the regulation of the immune system.
Esterase is important for the life cycles of enveloped negative-stranded and positive-stranded RNA viruses infecting birds and mammals. For example, esterase domains are included in membrane glycoproteins, so called hemagglutinin-esterase or the HEF1 proteins, which are present on the surfaces of influenza C (Herrler et al. 1985; Rosenthal et al. 1998) and coronaviruses and toroviruses (Wurzer, Obojes, and Vlasak 2002). During the infection process, hemagglutinins interact with sialic acid molecules bound to the cell receptors. This interaction is followed by entrance into the cell of virus particles that cannot be efficient unless ester bonds formed between the hemagglutinin glycoproteins and the receptor sialic acids are cut by esterase (Herrler et al. 1985).
It is known that esterases perform enzymatic depalmitoylation of viral glycoproteins and various cellular proteins. As a result, fatty acids (usually palmitate) covalently attached to cysteines near C-termini of palmitoylated proteins are cleaved off. It is thought (Dunphy and Linder 1998) that palmitoylation can affect a protein's affinity for membranes, subcellular localization, and interactions with membrane proteins. Rhamnogalacturonan acetylesterase from fungi (RGAE, fig. 4) catalyzes degradation of polysaccharides that constitute a cell wall in the plant host (Molgaard, Kauppinen, and Larsen 2000).
The esterase domain is conserved in the highly divergent ORF1p proteins of CR1-like elements from the chicken, turtle, fish, and human genomes (and putatively from the frog and crocodile genomes). This underscores its functional importance for life cycles of CR1-like elements. Surprisingly, its function is not linked directly to any known stages of a non-LTR retrotransposon life cycle. It is really difficult to understand why the esterase was preserved by non-LTR retrotransposons whose evolution is thought to follow, usually, a “vertical transmission” model (Malik, Burke, and Eickbush 1999). However, a regular horizontal transfer/transmission of CR1-like elements would favor esterases involved in penetration of cell membranes. Interestingly, the chicken and zebrafish genomes harbor multiple CR1-like families of approximately the same age. Six of them have been identified in the chicken genome (Vandergon and Reitman 1994). Three families of CR1-like elements from the zebrafish are reported in this article. All three have been retrotransposed relatively recently because of a low ∼5% to 10% nucleotide divergence between elements that belong to the same family. However, there is an enormous ∼40% divergence between elements that belong to any of different families residing in the same genome. It is conceivable that these families have invaded the host independently, and most of their diversity was acquired in some other hosts.
The PHD domain is another specific domain that we identified in the ORF1p proteins encoded by the CR1_DM, T1, and Q non-LTR retrotransposons from fruit fly and African malaria mosquito, respectively (fig. 3). These elements form the only well-defined CR1 sub-clade whose members do not code for esterase (fig. 2). As for esterase, the PHD domain is conserved in highly divergent proteins, and its function is not related to DNA/RNA binding. The PHD domain is thought to be involved in protein–protein interactions related to chromatin remodeling (Aasland, Gibson, and Stewart 1995; Kehle et al. 1998; Yochum and Ayer 2001). Therefore, it is possible that the PHD domain in CR1-like retrotransposons is necessary for both efficient retrotransposition and minimization of potentially harmful insertions of retrotransposons into the host genome by providing dynamic regulatory feedback between chromatin structure, expression of reverse transcriptase/endonuclease by retrotransposons, and their target-specificity. Interestingly, T1 and Q elements are most abundant in paracentromeric heterochromatin (Mukabayire and Besansky 1996). Similar abundance of different TEs in paracentromeric heterochromatin has been observed in other species (Kapitonov and Jurka 1999). It is possible that most of the TEs inserted accidentally into paracentromeric heterochromatin were fixed, whereas most of their relatives inserted originally into euchromatin have been lost. It is also possible, however, that insertion of some TEs can be channeled to heterochromatin regions by PHD-like regulatory elements, which may suppress transcription of retrotransposons at stages when most of the euchromatin is open (Jurka and Kapitonov 1999b). It is striking that some gypsy-like LTR retrotransposons have acquired “chromodomain” (Aasland and Stewart 1995; Malik and Eickbush 1999) which, like to PHD, is also involved in chromatin remodeling (Aasland, Gibson, and Stewart 1995; Aasland and Stewart 1995).
Interestingly, the PHD domain was acquired by Kaposi's sarcoma–associated herpesvirus (Coscoy, Sanchez, and Ganem 2001). The N-terminal PHD domain in MIR proteins encoded by the herpesvirus is directly involved in recruiting cellular proteins that regulate endocytosis of host immune recognition proteins (Coscoy, Sanchez, and Ganem 2001). As for the herpesvirus, CR1-like elements might have recruited the PHD domain to evade the host defense. Such evasion may be potentially important if these elements regularly trespass host cells.
One may design other interesting models employing function of PHD and ES in ORF1p proteins. However, our main goal was to identify new domains in the ORF1p proteins and to underscore the complexity of the life cycle of non-LTR retrotransposons concealed by the popular “vertical transmission” model.
Acknowledgments
We are grateful to Jolanta Walichiewicz and Michael Jurka for help with illustrations, and for editing the manuscript. We thank reviewers of the manuscript for useful comments. This work was supported by National Institutes of Health grant 2 P41 LM06252-04A1.
Footnotes
E-mail: jurka@girinst.org or vladimir@girinst.org.
Literature Cited
- Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci 20:56-59. [DOI] [PubMed] [Google Scholar]
- Aasland, R., and A. F. Stewart. 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res 23:3168-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- Bailey, T. L., and W. N. Grundy. 1999. Classifying proteins by family using the product of correlated p-values, pp. 10–14. in P. Istrail, P. Pevzner, and M. Waterman, eds. Proceedings of the Third International Conference on Computational Molecular Biology (RECOMB99). ACM, New York.
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, D. E., and M. H. Howe. 1987. Mobile DNA. American Society for Microbiology Press, Washington, DC.
- Besansky, N. J. 1990. A retrotransposable element from the mosquito Anopheles gambiae Mol. Cell. Biol 10:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky, N. J., J. A. Bedell, and O. Mukabayire. 1994. Q: a new retrotransposon from the mosquito Anopheles gambiae Insect Mol. Biol 3:49-56. [DOI] [PubMed] [Google Scholar]
- Burch, J. B., D. L. Davis, and N. B. Haas. 1993. Chicken repeat 1 elements contain a pol-like open reading frame and belong to the non-long terminal repeat class of retrotransposons. Proc. Natl. Acad. Sci. USA 90:8199-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy, P., C. Bazin, D. Higuet, and T. Langin. 1998. Dynamics and evolution of transposable elements. Chapman & Hall, New York.
- Chaboissier, M. C., D. Finnegan, and A. Bucheton. 2000. Retrotransposition of the I factor, a non-long terminal repeat retrotransposon of Drosophila, generates tandem repeats at the 3′ end. Nucleic Acids Res 28:2467-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [PubMed]
- Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, N. L. 1995. Unity in transposition reactions. Science 270:253-254. [DOI] [PubMed] [Google Scholar]
- Dalrymple, B. P., D. H. Cybinski, I. Layton, C. S. McSweeney, G. P. Xue, Y. J. Swadling, and J. B. Lowry. 1997. Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology 143:2605-2614. [DOI] [PubMed] [Google Scholar]
- Dawson, A., E. Hartswood, T. Paterson, and D. J. Finnegan. 1997. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J 16:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drablos, F., and S. B. Petersen. 1997. Identification of conserved residues in family of esterase and lipase sequences. Methods Enzymol 284:28-61. [DOI] [PubMed] [Google Scholar]
- Drew, A. C., and P. J. Brindley. 1997. A retrotransposon of the non-long terminal repeat class from the human blood fluke Schistosoma mansoni Similarities to the chicken-repeat-1-like elements of vertebrates. Mol. Biol. Evol 14:602-610. [DOI] [PubMed] [Google Scholar]
- Dunphy, J. T., and M. E. Linder. 1998. Signalling functions of protein palmitoylation. Biochim. Biophys. Acta 1436:245-261. [DOI] [PubMed] [Google Scholar]
- Eickbush, D. G., D. D. Luan, and T. H. Eickbush. 2000. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster Mol. Cell. Biol 20:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, R. J., S. Bachoo, D. J. Picketts, et al 1997. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet 17:146-148. [DOI] [PubMed] [Google Scholar]
- Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol 313:903-919. [DOI] [PubMed] [Google Scholar]
- Haas, N. B., J. M. Grabowski, J. North, J. V. Moran, H. H. Kazazian, and J. B. Burch. 2001. Subfamilies of CR1 non-LTR retrotransposons have different 5′ UTR sequences but are otherwise conserved. Gene 265:175-183. [DOI] [PubMed] [Google Scholar]
- Haas, N. B., J. M. Grabowski, A. B. Sivitz, and J. B. Burch. 1997. Chicken repeat 1 (CR1) elements, which define an ancient family of vertebrate non-LTR retrotransposons, contain two closely spaced open reading frames. Gene 197:305-309. [DOI] [PubMed] [Google Scholar]
- Herrler, G., R. Rott, H. D. Klenk, H. P. Muller, A. K. Shukla, and R. Schauer. 1985. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J 4:1503-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y. S., L. Swenson, U. Derewenda, et al 1997. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature 385:89-93. [DOI] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J 16:6034-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, S. E., M. F. Singer, and G. D. Swergold. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem 267:19765-19768. [PubMed] [Google Scholar]
- Jekosch, K. 2002. CR1-like repeat from Danio rerio Repbase Reports 2:7-8 (http://girinst.org/Repbase_Reports). [Google Scholar]
- Jurka, J. 2000. Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet 16:418-420. [DOI] [PubMed] [Google Scholar]
- Jurka, J., and V. V. Kapitonov. 1999a. L3, humrep, Repbase Update (http://girinst.org/Repbase_Update).
- Jurka, J., and 1999b. Sectorial mutagenesis by transposable elements. Genetica 107:239-248. [PubMed] [Google Scholar]
- Jurka, J., P. Klonowski, V. Dagman, and P. Pelton. 1996. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem 20:119-121. [DOI] [PubMed] [Google Scholar]
- Kajikawa, M., K. Ohshima, and N. Okada. 1997. Determination of the entire sequence of turtle CR1: the first open reading frame of the turtle CR1 element encodes a protein with a novel zinc finger motif. Mol. Biol. Evol 14:1206-1217. [DOI] [PubMed] [Google Scholar]
- Kapitonov, V. V., and J. Jurka. 1999. Molecular paleontology of transposable elements from Arabidopsis thaliana Genetica 107:27-37. [PubMed] [Google Scholar]
- Kapitonov, V. V., and 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98:8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- Koipally, J., A. Renold, J. Kim, and K. Georgopoulos. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J 18:3090-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha, V. O., and S. L. Martin. 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94:10155-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, et al 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- Lovsin, N., F. Gubensek, and D. Kordi. 2001. Evolutionary dynamics in a novel L2 clade of non-LTR retrotransposons in Deuterostomia Mol. Biol. Evol 18:2213-2224. [DOI] [PubMed] [Google Scholar]
- Lyngso, C., G. Bouteiller, C. K. Damgaard, D. Ryom, S. Sanchez-Munoz, P. L. Norby, B. J. Bonven, and P. Jorgensen. 2000. Interaction between the transcription factor SPBP and the positive cofactor RNF4. An interplay between protein binding zinc fingers. J. Biol. Chem 275:26144-26149. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol 16:793-805. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol 73:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molgaard, A., S. Kauppinen, and S. Larsen. 2000. Rhamnogalacturonan acetylesterase elucidates the structureand function of a new family of hydrolases. Structure Fold Des 8:373-383. [DOI] [PubMed] [Google Scholar]
- Mukabayire, O., and N. J. Besansky. 1996. Distribution of T1, Q, Pegasus and mariner transposable elements on the polytene chromosomes of PEST, a standard strain of Anopheles gambiae Chromosoma 104:585-595. [DOI] [PubMed] [Google Scholar]
- Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol 247:536-540. [DOI] [PubMed] [Google Scholar]
- Okada, N., M. Hamada, I. Ogiwara, and K. Ohshima. 1997. SINEs and LINEs share common 3′ sequences: a review. Gene 205:229-243. [DOI] [PubMed] [Google Scholar]
- Poulter, R., M. Butler, and J. Ormandy. 1999. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarity to the CR1 family of non-LTR retrotransposons. Gene 227:169-179. [DOI] [PubMed] [Google Scholar]
- Rosenthal, P. B., X. Zhang, F. Formanowski, W. Fitz, C. H. Wong, H. Meier-Ewert, J. J. Skehel, and D. C. Wiley. 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92:9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev 6:743-748. [DOI] [PubMed] [Google Scholar]
- Smit, A. F. 2000. L3, humrep. Repbase Update (http://girinst.org/Repbase_Update).
- Smit, A. F. 2001. REX1_FURC. Repbase Update (fugrep.ref) (http://girinst.org/Repbase_Update).
- Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandergon, T. L., and M. Reitman. 1994. Evolution of chicken repeat 1 (CR1) elements: evidence for ancient subfamilies and multiple progenitors. Mol. Biol. Evol 11:886-898. [DOI] [PubMed] [Google Scholar]
- Volff, J. N., C. Korting, and M. Schartl. 2000. Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol. Biol. Evol 17:1673-1684. [DOI] [PubMed] [Google Scholar]
- Weiner, A. M. 2000. Do all SINEs lead to LINEs?. Nat. Genet 24:332-333. [DOI] [PubMed] [Google Scholar]
- Wurzer, W. J., K. Obojes, and R. Vlasak. 2002. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: a proposal to reorganize group 2 Coronaviridae J. Gen. Virol 83:395-402. [DOI] [PubMed] [Google Scholar]
- Yochum, G. S., and D. E. Ayer. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol 21:4110-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]