Abstract
The ClpAP complex is a conserved bacterial protease that unfolds and degrades proteins targeted for destruction. Two ClpA AAA+ hexamer rings power substrate unfolding and translocation into the ClpP proteolytic chamber. Here, we determined high-resolution structures of wild-type Escherichia coli ClpAP undergoing active unfolding and proteolysis. A spiral of pore loop-substrate contacts spans both ClpA AAA+ domains. Protomers at the spiral seam undergo nucleotide-specific rearrangements supporting substrate translocation. IGL loops extend flexibly to bind the planar, heptameric ClpP surface with the empty, symmetry-mismatched IGL pocket maintained at the seam. Three different structures identify a binding-pocket switch by the IGL loop of the lowest-positioned protomer, involving release and re-engagement with the clockwise pocket. This switch is coupled to a ClpA rotation and a network of conformational changes across the seam, suggesting that ClpA can rotate around the ClpP apical surface during processive steps of translocation and proteolysis.
Introduction
The Hsp100 (Clp) AAA+ family of proteins, widely present in bacteria and eukaryotes, function as protein unfoldases and disaggregases1,2. Conserved members ClpX and ClpP assemble into large proteolytic machines with the serine protease ClpP and serve critical roles in targeted protein degradation and quality control3-7. Proteolysis requires substrate recognition and ATP hydrolysis-driven unfolding by the AAA+ machine, which unfolds and translocates the substrate into the proteolytic chamber of ClpP8-12. The ClpP chamber is formed by a double ring of heptamers13,14, which partner with 1-2 ClpX or ClpA AAA+ hexamers in bacteria, assembling into single and double-capped complexes15-17. To promote client degradation, ClpXP and ClpAP are aided by SspB18,19 and ClpS20,21, specificity adaptors that promote recognition of substrates including those containing the ssrA degron22,23 and N-end rule substrates24, respectively. Other substrates, such as the RepA DNA-binding protein, recognized by ClpA, are remodeled or degraded in support of specific cellular functions3,25.
Hsp100 interactions with ClpP involve a hexamer-heptamer symmetry-mismatch, which is a conserved feature among some proteolytic machines such as the 26S and PAN proteasomes3,6. Contacts are mediated by IGF/L-motif loops in ClpX or ClpA and hydrophobic binding pockets on the apical surface of ClpP6,26. Engagement of these loops triggers an open-gate conformational change of adjacent N-terminal loops on ClpP, facilitating substrate transfer to proteolytic sites27-29. Indeed, the acyldepsipeptide class of antibiotics (ADEPs) compete for binding to these pockets and stabilize an open-gate conformation, thereby converting ClpP to an uncontrolled, general protease30-33. How these Hsp100-ClpP interactions are coordinated during active unfolding and translocation is unknown.
ClpA contains two nucleotide-binding AAA+ domains (D1 and D2) per protomer which power unfolding34. Structures of related disaggregases, Hsp104 and ClpB, identify the substrate-bound hexamer adopts a right-handed spiral in which conserved, Tyr-bearing pore loops across both AAA+ domains contact and stabilize the polypeptide substrate via backbone interactions spaced every two amino acids35-38. Distinct substrate-bound states reveal a ratchet-like mechanism defined by the spiral arrangement, in which the ATP hydrolysis cycle drives substrate release at the lower position and rebinding to the topmost position along the substrate1,36,39. A similar spiral architecture and array of substrate contacts has now been identified for many AAA+ machines, supporting a universal rotary translocation mechanism40-43. However, for this Hsp100 family it is unclear how the dynamic substrate translocation steps are coupled to proteolysis, or how interactions are maintained at the hexamer:heptamer interface during processive steps of unfolding.
Here, we sought to determine the structural basis for coupled protein unfolding and proteolysis by the ClpAP complex. Using ATP and a RepA-tagged GFP substrate we determined cryo-EM structures of intact, wildtype ClpAP from E. coli to ~3.0 Å resolution which reveal three distinct substrate translocation states. Comparison of these state reveals the ClpP-connecting IGL loop of the protomer in lowest substrate-bound position undergoes release and rebinding to the clockwise pocket on ClpP. This IGL switch movement coincides with a ClpA rotation that is supported by conformational plasticity of 5 IGL loops which are bound to the apical surface of ClpP. Nucleotide-specific rearrangements in the AAA+ domains are identified which support a two amino acid-step translocation cycle. Together, these results reveal a model in which IGL-loop rearrangements enable ClpA to rotate its position on ClpP consecutively with substrate translocation steps thereby coupling substrate unfolding with ClpP activity.
Results
Architecture of Active, Substrate-Bound ClpAP
Structures of wildtype ClpAP undergoing active substrate unfolding and proteolysis were desired in order to capture functional states. RepA-GFP constructs are proteolyzed by ClpAP and can be used to monitor unfolding by ClpA10,44,45. Therefore, RepA-GFP containing the first 25 residues of RepA (RepA1-25-GFP) was tested for proteolysis and complex formation (Extended Data Fig. 1a-c). While the slowly-hydrolysable analog, ATPγS, enables stable formation of AAA+ complexes containing translocated substrates36,37, the reduced hydrolysis impairs function10 and may limit the ClpAP conformational cycle. Indeed, substantial degradation of RepA1-25-GFP occurs within 15 minutes in the presence of saturating (10 mM) ATP while little degradation is observed with ATPγS (Extended Data Fig. 1a). Therefore, in order to achieve active ClpAP for cryo-EM, incubations were carried out initially with ATPγS to promote assembly then 10mM ATP was added to initiate unfolding prior to vitrification. Assembly with ATPγS and mixtures with ATP have been previously established to support ClpA function44,46 and we identify robust degradation occurs under these ATPγS-ATP conditions, indicating ClpAP is active prior to vitrification (Extended Data Fig. 1b,c).
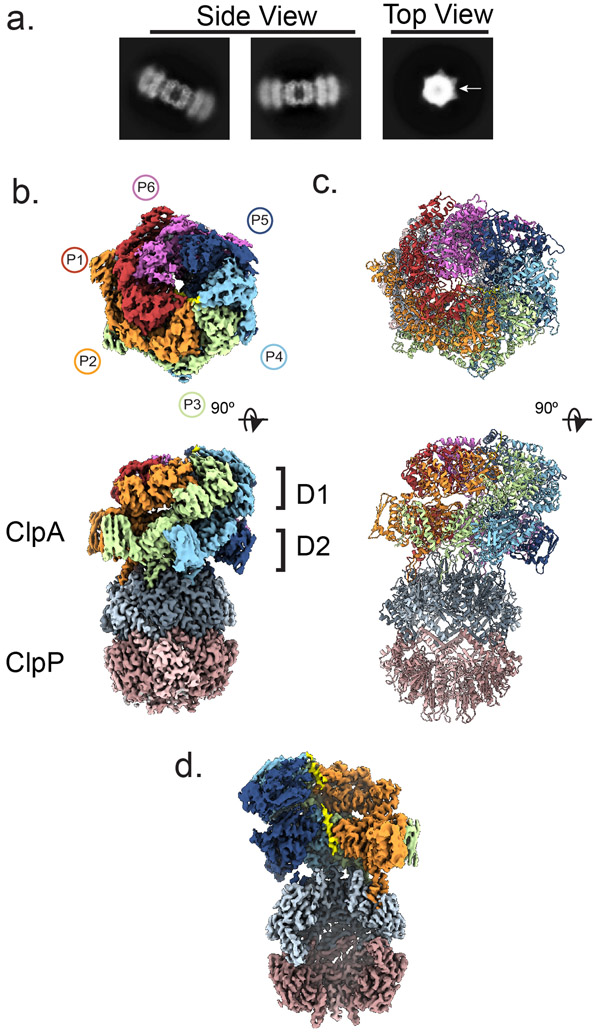
In reference-free 2D class averages, side and top views of ClpP particles double-capped with ClpA predominate (Fig. 1a and Extended Data Fig. 1d). Typically one ClpA hexamer of the double-capped complex showed well-resolved features, indicating preferred alignment likely due to flexibility across the double-capped complex. 3D classification yielded three distinct ClpAP conformations which refined to high-resolution (2.7-3.2 Å), hereafter referred to as the Engaged-1 (ClpAPEng1), Disengaged (ClpAPDis) and Engaged-2 (ClpAPEng2) states based on the binding states of the IGL loops (Extended Data Fig. 1e, f). As with 2D analysis, one ClpA hexamer showed improved features over the other. Therefore, the final models included one ClpA hexamer and two ClpP heptamers. In all states the D1 and D2 AAA+ rings of the ClpA hexamer adopt a right-handed spiral with the D2 ring contacting the planar, heptameric surface of ClpP via the IGL loops (residues 611-623) (Fig. 1b). ClpA is comprised of protomers P1-P6 with P1 at the lowest and P5 at the highest position of the spiral, while P6 is asymmetric and positioned at the seam interface (Fig. 1b). This architecture is similar to related ClpB and Hsp104 double-ring disaggregases in their substrate-bound states35-37. Resolution is the highest for ClpP (~2.5 Å), while ClpA is more variable (~2.5-4.5 Å for ClpAPEng1, ~3-6 Å for ClpAPDis, and ~3-6 Å for ClpAPEng2), with the spiral seam protomers (P1, P5 and P6) at lower resolutions due to their flexibility (Extended Data Fig. 1g-i). The high-resolution of the maps permitted accurate atomic models to be built for ClpAP (Fig. 1c, Extended Data Fig. 1j-k; Table 1). Density for the flexible N-terminal (NT) domain of ClpA (residues 1-168) was not well-resolved, and thus was not modeled.
Fig. 1: Architecture of the substrate-bound ClpAP complex.
a, Side and top view 2D class averages of double-capped ClpAP. Rings corresponding to ClpA (arrow) and ClpP rings identified in top views. b, Top and side views of the final map and c, model of ClpAPEng. ClpA is colored by individual protomer, as indicated. d, Channel view showing substrate peptide bound to ClpA (yellow).
Table 1.
Cryo-EM data collection, refinement and validation statistics of ClpAPEng-1, ClpAPDis, and ClpAPEng-2
| ClpAPEng1 | ClpAPDis | ClpAPEng2 | Focus ClpAPEng1 |
Focus ClpAPDis |
Focus ClpAPEng2 |
|
|---|---|---|---|---|---|---|
| EMDB 21519 PDB 6W1Z |
EMDB 21520 PDB 6W20 |
EMDB 21521 PDB 6W21 |
EMDB 21522 PDB 6W22 |
EMDB 21523 PDB 6W23 |
EMDB 21524 PDB 6W24 |
|
| Data collection and processing | ||||||
| Microscope | Titan Krios | |||||
| Camera | K3 | |||||
| Magnification | 58,600 | |||||
| Voltage (kV) | 300 | |||||
| Data acquisition software | Serial EM | |||||
| Exposure navigation | Image Shift | |||||
| Electron exposure (e–/Å2) | 68 | |||||
| Defocus range (μm) | .8-1.2 | |||||
| Pixel size (Å) | .853 | |||||
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 739,000 | 739,000 | 739,000 | 739,000 | 739,000 | 739,000 |
| Final particle images (no.) | 176,232 | 57,848 | 39,177 | 176,232 | 57,848 | 39,177 |
| Map resolution (Å) | 2.7 | 3.0 | 3.2 | 3.0 | 3.1 | 3.4 |
| FSC threshold | .143 | .143 | .143 | .143 | .143 | .143 |
| Map resolution range (Å) | ||||||
| Refinement | ||||||
| Model resolution (Å) | 3.0 | 3.1 | 3.2 | 2.8 | 3.0 | 3.3 |
| FSC threshold | .143 | .143 | .143 | .143 | .143 | .143 |
| Map sharpening B factor (Å2) | −73.0 | −68.5 | −60.8 | −80.3 | −65.6 | −55.8 |
| Model composition | ||||||
| Non-hydrogen atom | 48,556 | 48,252 | 48,346 | 27,542 | 27,238 | 27,332 |
| Protein residues | 6,180 | 6,134 | 6,147 | 3,492 | 3,446 | 3,459 |
| Ligands | 12 | 12 | 12 | 12 | 12 | 12 |
| B factors (Å2) | ||||||
| Protein | 33.4 | 125.4 | 177.4 | 0.5 | 153.0 | 210.8 |
| Ligand | 20.0 | 151.4 | 210.6 | 20.0 | 151.4 | 210.6 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | .03 | .01 | .011 | .026 | .014 | .013 |
| Bond angles (°) | 1.76 | .60 | 0.58 | 1.81 | .95 | 0.61 |
| Validation | ||||||
| MolProbity score | 1.72 | 1.96 | 1.78 | 1.62 | 2.26 | 1.9 |
| Clashscore | 9.43 | 13.39 | 13.92 | 8.42 | 15.9 | 14.4 |
| Poor rotamers (%) | .90 | 0.20 | .10 | 0.55 | .03 | .1 |
Density corresponding to an unfolded polypeptide substrate is identified spanning the D1 and D2 domains in all three structures and modeled as a 24-residue poly-Ala chain (Fig. 1d, 2a-c). Substrate is not observed in the ClpP pore or chamber potentially due to flexibility and the absence of substrate-interacting residues. In low-passed filtered maps of the final reconstruction, globular density at the entrance to the ClpA channel is visible at a reduced threshold that approximately corresponds to a GFP molecule (Extended Data Fig. 1j). These data together with SEC and proteolysis analysis above indicate that these ClpAP structures, determined under active conditions using ATP, contain RepA-GFP substrate and likely represent conformational states associated with processive translocation and proteolysis.
Fig. 2: Three distinct structures of ClpAP showing IGL loop rearrangement.

a, ClpAPEng-1, b, ClpAPDis, and c, ClpAPEng-2 cryo-EM maps showing degree offset (arrow) of the ClpA channel axis (solid line) and substrate position (yellow density) compared to the ClpP pore and proteolytic chamber (dashed line). Schematic (lower left) shows occupancy of the ClpA IGL-loops (circles, colored and numbered by protomer) around the ClpA hexamer, with the empty IGL pockets (white circles) and ClpA protomers indicated (letters) for the different states. Schematic (lower right) shows top view of ClpP with ClpA as a hexagon overlay (red: current state, black: previous state), and colored cylinders indicating substrate positions (red: current state). d, Cryo-EM density of the ClpA-P interface showing IGL loop interaction with ClpP in ClpAPEng-1 (left), ClpAPDis(center), and ClpAPEng-2(right).
Structures Reveal ClpA IGL Loop Switches to Engage the ClpP Symmetry-Mismatched Pocket
Following multiple rounds of 3D classification three distinct conformations of substrate-bound ClpAP refined to high resolution (Fig. 2a-c, Extended Data Fig. 1e-f). The major conformational differences involve ClpA and include changes in substrate interactions and nucleotide states (discussed below), and changes in the IGL loops and orientation across the ClpA-P interface. No substantial conformational differences are identified for ClpP between the different states (RMSD < 1 Å). In the ClpAPEng1 structure, well-resolved density for the IGL loops from all 6 ClpA protomers is identified in the pockets around the ClpP apical surface (Fig. 2a, d). One remaining empty pocket on ClpP, which results from the symmetry mismatch of the heptamer, is positioned at the ClpA spiral seam between protomers P1 and P6 (Fig. 2a, d). In the ClpAPDis structure, density for the IGL loop of protomer P1, which is at the lowest position along the substrate, is no longer observed in the ClpP pocket, resulting in two neighboring empty pockets at the ClpA seam (Fig. 2b, d). Remarkably, in the ClpAPEng2 structure, density for the P1 IGL loop is instead observed in the clockwise adjacent pocket, revealing that the loop has switched position in comparison with ClpAPEng1, and the empty, symmetry-mismatched pocket now resides between protomers P1 and P2 compared to P6 and P1 for the ClpAPEng1 (Figure 2c, d). Difference maps of the ClpA-P interface region further validate the position of the P1 IGL loop in these structures (Extended Data Fig. 2). If these structures represented a mix of states then the difference maps would show positive density in both IGL pockets. Importantly, however, positive density for the IGL loop only appears from P1 in the correct ClpP pocket corresponding to the ClpAPEng1 or ClpAPEng2 states, thereby verifying that these structures represent distinct states of the P1 IGL loop. Notably, for ClpAPDis, density for the P1 IGL loop is not observed in either pocket, indicating this loop is indeed unbound from ClpP and in an intermediate state (Extended Data Fig. 2).
The ClpA channel and bound polypeptide substrate are offset between ~14° and 16°, from the ClpP pore in the different structures (Fig. 2a-c). Upon alignment of the structures, ClpA is identified to be in three distinct positions relative to ClpP. These differences appear to occur through a pivot across ClpP and clockwise twist around the substrate channel axis which coincides with the binding site-switch of the P1 IGL loop (Fig. 2a-c, Supplementary Video 1). Going from the Engaged-1 to Disengaged states, ClpA pivots towards the P5-P6 side of the hexamer, shifting by approximately 10 Å across ClpP. From the Disengaged to Engaged-2 states ClpA twists clockwise, resetting the orientation of the channel relative to ClpA but with an overall rotation of ~10° compared to ClpAPEng1. The ClpA rotation is visualized in a morph between these states, revealing how protomers P4-P6 tilt towards ClpP, compressing the interface in this region and then expand through a clockwise rotation around the axial channel in the ClpAPEng2 state (Supplementary Video 1).
In addition to these structures, we determined structures of ATPγS-stabilized ClpAP bound to RepA1-25-GFP in which ATP was not added prior to vitrification (Extended Data Fig. 3). Following similar data classification and refinement procedures, we determined two ClpAP structures at 3.0 and 3.1 Å resolution which match the ClpAPEng1 and ClpAPDis states described above (Extended Data Fig. 3a-g and Table 2). Notably, the ClpAPEng2 state was unable to be classified as a distinct conformation despite similar-sized datasets. This could be due to changes in the conformational equilibrium resulting from the ATPγS-stabilized conditions compared to active conditions with ATP. Nonetheless, these structures further establish that P1 IGL loop undergoes engaged and disengaged conformational changes under conditions in which substrate binding and processing occurs.
Table 2.
Cryo-EM data collection, refinement and validation statistics of ATPγS-ClpAP
| Disengaged State |
Engaged State |
|
|---|---|---|
| EMDB 20845 PDB 6UQE |
EMDB 20851 PDB 6UQO |
|
| Data collection and processing | ||
| Microscope | Titan Krios | |
| Camera | K3 | |
| Magnification | 58,600 | |
| Voltage (kV) | 300 | |
| Data acquisition software | SerialEM | |
| Exposure navigation | Image Shift | |
| Electron exposure (e–/Å2) | 68 | |
| Defocus range (μm) | 1.2-2 | |
| Pixel size (Å) | .853 | |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 1,800,000 | 1,800,000 |
| Final particle images (no.) | 314,000 | 169,000 |
| Map resolution (Å) | 3.0 | 3.1 |
| FSC threshold | .143 | .143 |
| Map resolution range (Å) | ||
| Refinement | ||
| Model resolution (Å) | 3.1 | 3.1 |
| FSC threshold | .143 | .143 |
| Model resolution range (Å) | ||
| Map sharpening B factor (Å2) | ||
| Model composition | −112.9 | −103.2 |
| Non-hydrogen atom | 48,402 | 48,522 |
| Protein residues | 6,161 | 6,174 |
| Ligands | 12 | 12 |
| B factors (Å2) | ||
| Protein | 33.7 | 33.4 |
| Ligand | 44.1 | 20.0 |
| R.m.s. deviations | ||
| Bond lengths (Å) | .025 | .026 |
| Bond angles (°) | 1.82 | 1.84 |
| Validation | ||
| MolProbity score | 1.37 | 1.25 |
| Clashscore | 3.52 | 2.58 |
| Poor rotamers (%) | .37 | .08 |
IGL-Loop Plasticity Enables ClpP Engagement by the ClpA Spiral
Previous crystal structures of ClpA were unable to resolve the IGL loops due to flexibility, but biochemical data for ClpX IGF loops suggest that they make static interactions with ClpP and all 6 IGF loops are required for optimal activity47. In the ClpAP structures, density for the ClpA IGL loops is well-defined, enabling atomic modeling for nearly all loop residues in each pocket (Extended Data Fig. 4a). The IGL-loop region extends from residues N606 and T637 in the base of the D2 large subdomain as two short α-helices. Residues 616-620 form the flexible loop, which extends into the hydrophobic binding pocket on ClpP, resulting in ~600 Å2 of buried surface area compared to the empty pocket (Fig. 3a, left). The IGL-loop binding pocket is formed by the interface of two ClpP protomers and includes α-helices B and C from one protomer and a 3-strand β-sheet (strands 1, 2, and 3) and the C-terminal (CT) strand from the adjacent protomer (Fig. 3a). The loop residues I617, G618, L619, and I620 bind a hydrophobic region in the pocket comprised of A52, L48, F49, and F82 in α-helices of one protomer and L23, Y60, Y62, I90, M92, F112, L114, L189 in the adjacent protomer (Fig. 3a, middle). Additional electrostatic contacts likely stabilize the loop as well, including R192 in the CT strand, and E26, which appear to interact with H621 and R614 and Q622, respectively (Fig. 3a, right).
Fig. 3: IGL loop interactions and conformational flexibility.

a, Representative view of a bound IGL loop (orange, ribbon view) positioned in the binding pocket of ClpP shown in surface view with hydrophobic residues colored in yellow (left), and shown in ribbon views with hydrophobic interactions (middle) and electrostatic contacts (right) labeled. b, Overlay of IGL loops (colored by protomer) of ClpAPEng-1 (left), ClpAPDis (middle) and ClpAPEng-2 (right). IGL loops are aligned to connecting residues 638-649. Dotted line represents missing residues not present in the density. c, Map and model showing P1 IGL-loop density extends into the IGL pocket for ClpAPEng-1 (left) and ClpAPEng-2 (right) but is disengaged for ClpAPDis (middle), contacting the adjacent apical ClpP surface (right). The distances between ClpP E67 and ClpA-P1 S625 in the three states are shown to indicate the shift in the position of the P1 IGL loop relative to ClpP. d, Overlay of IGL loops of P1 for ClpAPEng-1 (red) and ClpAPEng-2 (grey). e, Map and model of the P5 IGL-loop for ClpAPEng-1 (left), ClpAPDis (middle) and ClpAPEng-2 (right) showing extended and compact conformations, respectively, based on distance measurements between loop residues 605-619 and 633-619 (red dots).
While the IGL loops all make identical contacts with ClpP, flexibility of the connecting helices (residues 608-615 and 624-635) enables the loops to extend from ClpA in a number of orientations around the hexamer and between the different states (Extended Data Fig. 4b). The largest changes occur with the P1 loop, which switches binding pockets on ClpP between the three states, as discussed above (Fig. 3b-d, Supplementary Video 2). The loop is largely well-resolved in the ClpAEng1 and ClpAEng2 states, however residues 609-624 were unable to be modeled for ClpADis due to weak density in the unbound, disengaged conformation. By comparison of the P1 IGL loop position in the different states, the binding-pocket switch is identified to result from two changes: an overall clockwise rotation of ClpA hexamer (Fig. 2a-c) and a large, 80° rotation of the loop around residues T604 and T637 in the connecting helices (Fig. 3c,d). Surprisingly, the P5 IGL loop is also identified to contract and extend between the states through a partial unfolding of both connecting helices (residues 609-613 and 614-629) (Fig. 3e and Supplementary Video 2). In the Engaged-1 state, the loop is extended by ~5 Å compared to the Disengaged state, whereas in the Engaged-2 state the P5 loop is partially extended by ~ 3 Å. Notably, this loop extension is only observed at the P5 position and appears to correlate with the orientation of ClpA in the different states. Overall, these results reveal a remarkable conformational plasticity of the IGL loops which likely functions to support consistent interactions with ClpP around the variable hexamer-heptamer interface during substrate translocation and enable the binding-pocket switch movement of the P1 loop.
ClpP Structure and N-terminal Gating
The flexible N-terminal loop residues of ClpP (1-18) form a pore on the apical surface that functions as a substrate gate which is allosterically controlled by engagement of the adjacent IGF/L-binding pockets by ClpX/A or ADEP compounds33. In all three ClpAP structures, the ClpP NT loops from each protomer are well-resolved and adopt an extended configuration resulting in an open gate conformation that is positioned adjacent the ClpA translocation channel, ~30 Å away from where substrate is resolved (Fig. 4a and Extended Data Fig. 5a,b). This is distinct from crystal structures showing the NT loops adopt an asymmetric open-gate arrangement48, but similar to ADEP-bound structures where all the loops are in an extended conformation31,33. Additionally, no contact is observed between the NT loops and ClpA (Fig. 4a), which may be distinct compared to ClpXP, in which NT loops have been identified to contact the ClpX pore-2 loops47.
Fig. 4: Structure of ClpP and NT gating loops.

a, Channel view of ClpAP highlighting the ClpP NT gating loops (red) relative to substrate density (yellow). b, (left) top view of ClpP NT loops with ClpA IGL loops (colored by protomer). c, Expanded view of an NT loop pair with cis (E8-K25) and trans (R15-E14) salt-bridge contacts. d, Side-view map of single-capped ClpAP complex. e, Expanded views of the ClpP pore for the ClpA-bound, and -unbound surfaces showing open- and closed-gate conformations, respectively. The open-gate conformation was modeled into both sites to show differences compared to the closed-gate density. f, Top views showing ClpP pore diameter for the (top) open- and (bottom) closed-gate conformations.
We identify two specific interactions: one across the ClpP NT loops and one with an adjacent helix A in the IGF/L pocket, which have not been previously characterized and appear to stabilize the open gate conformation (Fig. 4b). A salt-bridge contact between residues R15 in one loop and E14 in the clockwise loop is identified in each protomer (Fig. 4b and Extended Data Fig. 5c). Additionally, a potential salt-bridge contact involving E8 and K25 is also observed which may additionally stabilize the loop orientation (Fig. 4b and Extended Data Fig. 5d). Notably, K25 is located in a helix that comprises part of the hydrophobic, IGL-binding pocket (Fig. 4b). Thus, this interaction may be involved in the allosteric gating mechanism.
For the three structures both ClpP pores (top and bottom) adopt an open gate conformation due to the double-capped configuration of the complex. However, in an initial dataset of ATPγS-stabilized ClpAP, we identified a population of single-capped complexes which resolved into one 3D class (Fig. 4c and Extended Data Fig. 5e,f), enabling us to characterize the open and closed-gate conformations in one structure. While the resolution of the NT loops was not sufficient to model the closed conformation, at lower threshold values, density for the loops on the unbound end of ClpP appears to extend ~8 Å from ClpP, while density for the ClpA-bound end NT loops extends ~16 Å (Fig. 4d). Additionally, the pore diameter is identified to be ~25 Å for the ClpA-bound end of ClpP, which is substantially wider compared to the unbound end, which is ~15 Å (Fig. 4e). Thus, we identify the NT loop gating mechanism is specifically triggered by engagement of the cis-bound ClpA IGL loops, which may allosterically regulate the NT loops potentially through specific salt bridges which stabilize the extended loop arrangement.
ClpA Substrate Contacts and Translocation States
To improve the resolution of the ClpA pore loop interactions and the seam protomers, particle subtraction and focused refinement of the ClpA hexamer was performed (Extended Data Fig. 6a). This resulted in an estimated resolution of 3.0Å and 3.1Å and 3.4Å for the ClpAEng-1, ClpADis and ClpAEng-2 focused maps, respectively (Extended Data Fig. 6b). While the overall resolution did not increase compared to the full map containing ClpP, improvements in the map density for the seam protomers and substrate contacts is observed, particularly for the Engaged-1 state (Extended Data Fig. 6 c-e). Nonetheless, the seam protomers remain at a lower resolution (~3.5-6 Å) compared to the rest of the map, due to their flexibility. Models were further refined using these maps in order to characterize the substrate interactions and conformational changes between the states. Similar to other AAA+ structures, the conserved Tyr-pore loops in the D1 and D2 of ClpA extend into the channel and form a double spiral of substrate interactions spaced every two amino acids along a 24 amino acid-long polypeptide (Fig. 5a). For all states, the D1 stabilizes a 9-residue segment through direct contact by Y259 from protomers P1-P4, which intercalates between the substrate side chains and contacts the backbone (Fig. 5b, Extended Data Fig. 6f). The conserved flanking residues, K258 and R260, extend laterally to make electrostatic contacts with the upper and lower adjacent pore loops (D262 and E264), similar to ClpB D137,38. Notably, in the ClpAPEng-1 structure the P5 and P6 D1 pore loops are disconnected from the substrate, with Y259 positioned ~18 Å and ~17 Å away, respectively (Fig. 5a and Extended Data Fig. 6g and Supplemental Video 3). This 4-bound, 2-unbound configuration of the D1 pore loops is distinct from previous structures of ClpB and Hsp10436-38. The D2 similarly shows well-defined pore loop-substrate contacts for protomers P1-P4 in both states (Fig. 5c, Extended Data Fig. 6f). These interactions stabilize a longer, 11 residue polypeptide segment and are primarily mediated by Y540 and V541, which form a Y-shaped clamp around the substrate backbone. Additional, pore-2 loops49,50, conserved in ClpB and Hsp10436-38, are present in both the D1 (residues: 292-302) and D2 (residues: 613-625), and line the channel, likely making additional contributions to stabilizing the polypeptide. Notably, residues E526, R527 and H528 from protomers P1-P5 contact the substrate and together form an “exit pore” which is adjacent the ClpP gating loops and thus may serve to facilitate transfer to the ClpP chamber (Extended Data Fig. 6h,i).
Fig. 5: ClpA pore loop-substrate contacts and translocation states.

a, Segmented map and model of the substrate-bound P1-P6 pore loops, colored by protomer, with substrate (yellow) for ClpAPEng-1. Distances shown indicating length of substrate interactions for the the D1 and D2. Model of the b, D1 and c, D2 pore loops and substrate for ClpAPEng-2 (colored by protomer) and overplayed with ClpAPEng-1 (grey). Substrate-contacting residues are indicated and shifts in the position of the pore loops protomers, P5 and P6 between states are shown. d, ClpAPEng-2 model is displayed showing alpha carbon RMSD between the three states, determined by alignment to protomer P3. Large changes >7 Å) are indicated in red with wider tubes, intermediate changes (~6.0 Å) are colored in white, and small/no changes are colored in blue. e, Individual seam protomers shown with alpha carbon rmsd mapped as in d for P5(left), P6 (middle), and P1(right).
As with previous Hsp100 structures36-38, protomers P2-P4 show no substantial conformational changes between the states. Therefore, in order to compare conformational changes of the seam protomers (P1, P5 and P6), protomer P3 was used for alignments of the ClpA hexamer. The largest changes occur for these protomers between ClpAPEng-1 and ClpAPDis, and between ClpAPEng-1 and ClpAPEng-2, (RMSD ≈ 5.1 Å and 3.5 Å, respectively), while changes between ClpAPDis and ClpAPEng-2 are more modest (RMSD ≈ 2.3 Å). For simplification, comparisons between ClpAPEng-1 and ClpAPEng-2 are shown (Fig. 5b, c). Overall, the pore loops for P5 and P6 shift closer to the polypeptide substrate and move up the channel axis going from ClpAPEng-1 to the ClpAPDis and ClpAPEng-2 states (Fig. 5b, c and Supplementary Video 3). Notably, the P5 pore loop moves up by ~4 Å and towards the substrate by ~8 Å (Extended Data Fig. 6g). This positions P5 Y259 adjacent the substrate two residues above the P4 Y259 position supporting the two-amino acid translocation step, however direct contact is not identified. The largest changes occur with D2 pore loop of protomer P6, which moves up the channel axis by ~7 Å, corresponding to a two-residue shift in the substrate position, but remains unbound to substrate in all three states (Fig. 5c). Together these changes reveal protomer movements up channel axis and appear on-path to a translocation step through engagement of the next contact site along the substrate by the D1 in protomer P5 (Extended Data Fig. 6j). In order to identify how these changes are connected to the IGL loop movement, the C-α deviation between the three states was mapped onto the hexamer model (Figure 5d). As expected, the IGL loops of the seam protomers show the greatest variability while protomers P2-P4 show little change. Remarkably, connected regions of variability are identified at the spiral seam across the subdomains and protomer interfaces, revealing a path of conformational changes that extend from the C- to N-termini for P1, P6 and P5, respectively. The greatest variability occurs in the IGL loop and D2 small subdomain of P1, the D2 large subdomain of P6 and the D1 large subdomain of P5 (Fig. 5e and Supplementary Video 4). Remarkably, these changes reveal an 80 Å-long allosteric communication network which appears to connect IGL-loop movement in P1 to translocation steps that occur in P5 and P6.
Nucleotide States Support Hydrolysis-Driven Translocation
Similar to Hsp104 and ClpB, ATP hydrolysis activity in D1 and D2 is required for ClpA substrate translocation steps51. All three structures show well resolved nucleotide pockets and the nucleotide state of each pocket was assessed based on the density for ATP and the position of the trans-activating Arg-finger residues (R339-R340 in the D1 and R643 in D2) (Extended Data Fig. 7). For the substrate bound protomers P2, P3 and P4, the D1 and D2 nucleotide pockets are largely identical across the three states and in an ATP, active configuration (Fig. 6a and Extended Data Fig. 7a). The D2 of protomer P2 is an exception and appears to be bound to ADP and in a post-hydrolysis state in ClpAPEng2. For the seam protomers P1, P5 and P6 the nucleotide states vary, but are similar to previous Hsp100 structures, and thus support models for consecutive hydrolysis during processive translocation previously described36-38 (Fig. 6a and Extended Data Fig. 7b). The P5-D1 appears to switch from an ADP state in ClpAEng-1 to an ATP state in ClpADis and ClpAEng-2, indicating nucleotide exchange may occur between these states. Notably, this coincides with the conformational changes that bring the P5-D1 pore-loop towards with the next contact position along the substrate after P4 (Figure 5b), supporting models proposing that the translocation step occurs upon ATP re-binding1. Conversely, the P5-D2 is in an ATP state and bound to substrate in all three structures (Fig. 6a and Extended Data Fig. 7b). Protomer P6, which is at the spiral seam and unbound to substrate, is in a post-hydrolysis, ADP state for both the D1 and D2 across all three structures. For protomer P1, which is at the lowest substrate-contact position and undergoes IGL-loop switching between the states, the D1 appears bound to ATP in ClpAEng-1 and bound to ADP in ClpADis and ClpAEng-2, indicating hydrolysis likely occurs between these states. However, the P1-D2 appears bound to ADP and inactive based on the distal position of Arg finger all states.
Fig. 6: Nucleotide states and ClpA rotation model for processive unfolding and proteolysis.

a, Schematic showing nucleotide states and substrate contact for the D1 and D2 of ClpAPEng-1 (left), ClpAPDis (middle), and ClpAPEng-2 (right) determined based on difference maps (Extended Data Fig. 7). Protomer nucleotide states are denoted by colored circles (green for ATP and red for ADP). b, Model for ClpAP processive substrate translocation cycle. Two translocation steps are depicted and coupled to IGL-loop disengagement (step 1 and 4) and engagement to the next clockwise IGL pocket on ClpP (step 2 and 5), indicated by arrows. Top view schematics show rotary cycle of substrate binding by ClpA (left) and occupancy ClpP IGL pockets (right). The protomer at the lowest substrate contact site, which releases the IGL loop is indicated (*).
Together, the changes in nucleotide states between the three structures indicate ATP hydrolysis occurs at the spiral seam and likely proceeds counter-clockwise around the hexamer, supporting the rotary substrate translocation cycle in which protomers toward low position in the spiral (P1 and P2) undergo ATP hydrolysis and substrate release, then re-bind substrate at the top position (P5) with ATP binding1 (Fig. 6a and Extended Data Fig. 7a). Based on the different D1-D2 nucleotide states within protomers P1, P2 and P5, hydrolysis may be asynchronous, and possibly initiate in the D2 ring based on the ATP-ADP change identified for P2 between the ClpADis and ClpAEng-2 structures. This finding is similar to what is identified for ClpB37 and indicates the D1 and D2 regulate distinct steps of translocation and coordination with ClpP. Surprisingly, certain conformational changes, including release of substrate, movement of the P1 IGL loop, and changes in P6, do not appear to directly correlate with changes in the cis nucleotide pocket. Allosteric communication and distinct functional roles have been described for the D1 and D2 of ClpB52,53. Thus, hydrolysis at adjacent sites, either across the D1 and D2 or between protomers connected by the Arg finger, may allosterically drive the conformational changes identified in the different structures. Indeed, the P1 IGL loop switching may be supported by hydrolysis at P1-D1 during disengagement (ClpAEng-1 to ClpADis) and at P2-D2 during engagement of the next pocket (ClpADis to ClpAEng-2) (Fig. 6a and Extended Data Fig. 7b).
Discussion
For the conserved class of AAA+ protease complexes such as ClpXP and ClpAP, it has been unclear how dynamic steps of ATP hydrolysis-driven substrate translocation could occur in coordination with the attached heptameric protease. The structures presented here reveal a dynamic ClpA-P interface, in which the connecting IGL loops undergo large conformational changes that may enable the ClpA hexamer to rotate on the ClpP apical surface during processive translocation steps (Fig. 2). Most notably, the IGL loop of the protomer in the lowest substrate-bound site (P1) is observed in three different positions that together reveal a clockwise binding-pocket switch movement. This IGL loop movement appears coordinated with conformational changes associated with the substrate translocation steps, based on the large allosteric communication path we identify across the seam protomers which connects ClpP interactions with the pore loop-substrate contacts (Fig. 5d).
The conformational differences between the three structures suggest a model for substrate translocation by ClpAP in which hexamer-heptamer symmetry mismatch is continually maintained with an empty IGL binding pocket aligned at the spiral seam of ClpA. During consecutive translocation steps, the IGL loop of the adjacent protomer (P1) at the lowest substrate contact site, disengages from ClpP (ClpAPEng-1 to ClpAPDis, step 1) then re-binds to the clockwise empty pocket (ClpAPDis to ClpAPEng2, step 2) in a manner that is regulated by ATP hydrolysis and conformational changes associated with substrate release and re-binding (Fig. 6b and Supplementary Video 5). Based on the path of conformational variability (Fig. 5d) and changes in nucleotide state (Fig 6a) across the seam protomers, ATP hydrolysis and translocation movements by neighboring protomers likely regulate the P1-IGL loop movement, coordinating the binding-pocket switch with translocation steps.
The nucleotide states of the protomers and pore-loop spacing along the substrate are consistent with a rotary, two amino-acid step translocation mechanism proposed in previous studies35-37,43. This step size is smaller than what has been reported for ClpA and ClpX by single molecule54,55 and transient state kinetic methods56. However, recent single molecule studies of ClpB identify rapid modes of consecutive translocation in which 6-7Å steps could occur but are not resolvable due to the high translocation rate57. For a processive cycle2,58,59, we propose that these steps could continue with IGL-loop switching at each translocation step, rotating the position of empty IGL pocket around ClpP with the spiral seam (Fig. 6b). This rotation of ClpA relative to ClpP would enable the hexamer to shift by one clockwise binding position on the ClpP apical surface per 6 substrate translocation steps down the axial channel. Other models involving larger translocation step sizes55 or alternate hydrolysis mechanisms60 would likely confer different coordination with IGL-loop switching. Nonetheless, we suggest that the functional significance of the hexamer:heptamer mismatch is that the 7th binding pocket on ClpP is available for the IGL loops to sequentially switch position with the substrate translocation steps, allowing processivity by ClpA without altering contact with ClpP. Additionally, this rotation may be substrate-specific and perhaps more critical for proteolysis of stable, folded substrates compared to labile structures.
While other mechanisms may support substrate translocation and proteolysis by ClpAP, we note that IGL-loop switching between the same sites, stochastically or counterclockwise would result in an offset between the empty IGL pocket and the spiral seam of ClpA hexamer. None of these potential configurations of ClpAP were observed in any of the 3D classes for our ATPγS and ATP datasets. Additionally, recent structures of the related ClpXP complex bound to substrate identify conformations which are similar to the Engaged-1 and Disengaged states determined here and a complimentary rotary mechanism is proposed61. The additional Engaged-2 state structure determined here further supports these models by identifying that the P1 IGL loop indeed switches position and ClpA rotates clockwise relative to ClpP with an apparent substrate translocation step. The discovery of this additional state in our study may have resulted from the use of WT enzyme and ATP, allowing an additional active state of substrate translocation to be captured.
The IGL-loop interactions with the ClpP hydrophobic pockets are identical at all positions, while flexibility of the helices that connect the loops to the D2 base of ClpA enables substantial variability in the ClpA position relative to ClpP. This flexibility is likely critical for maintaining ClpP binding during ratcheting conformational changes associated with substrate translocation and the rotations in ClpA between the different states (Fig. 2a-c). Furthermore, the extension and unfolding of the P5 IGL-loop helices in the Engaged-1 state is striking and may also provide energetic constraints that could facilitate release and clockwise switch of the P1 IGL loop during the conformational change to the Disengaged and Engaged-2 states (Fig. 3c).
Binding by IGF/L loops is well-understood to trigger gate-opening in ClpP27-29 and the conformational plasticity and asymmetric binding interactions we identify reveal new insight into how these loops facilitate allosteric regulation between ClpA and ClpP56,62. A number of proteolytic machines, including the 26S proteasome, operate as hexamer:heptamer assemblies3,4. Notably, assembly of the eukaryotic Rpt and archaeal PAN AAA+ with its respective 20S core, involves interaction with flexible C-terminal HbYX motifs and gate-opening of the 20S63,64. While the HbYX interactions are distinct and likely operate differently during translocation, recent structures reveal a conserved spiral staircase arrangement of 26S41,65 and PAN66 bound to substrates and a sequential rotation of the ATPase ring has been proposed for PAN66. For the Clp protease system the symmetry mismatch and IGF/L loop binding pocket switch likely serves a critical role in processivity by coordinating the rotary ATPase cycle and directional translocation steps with substrate transfer and proteolysis by ClpP.
Online Methods
Purification and analysis of ClpA, ClpP and RepA(1-25)-GFP
ClpA and ClpP were purified as previously described1,2. RepA 1-25 protein was expressed with a C-terminal His6-tag construct from the pDS56/RBSII plasmid. Transformed BL21 cells were inoculated in LB media with 100 ug/mL Ampicillin and grown at 37°C to OD600nm = ~0.6–0.8. The cell culture was induced with 1 mM IPTG for ~4 h at 30°C. Cell pellet was resuspended in 40 mM HEPES pH 7.4, 2 mM β-mercaptoethanol, and 10% glycerol with protease inhibitors (EDTA-free) (Roche) and then lysed by sonication. Following centrifugation (16,000 x g, 20 min, 30°C), the supernatant was applied to a Ni-NTA column (GE Healthcare) followed by a gradient elution from 20 mM imidazole to 500 mM imidazole. Purity was verified by SDS-PAGE and fractions were combined and concentrated into a storage buffer (40 mM HEPES pH 7.4, 500 mM KCl, 20 mM MgCl2, 10% glycerol (v/v), and 2 mM β-mercaptoethanol).
The RepA(1-25)-GFP degradation assay (Extended Data Fig. 1a-b) was performed in triplicate and consisted of 6 μM ClpA, 7 μM ClpP or ClpP-S98A, 1 μM RepA(1-25)-GFP and 2 mM nucleotide incubated in buffer at 20° for15 min containing 50 mM Tris-HCl pH 7.5, 150 mM KCl, 10 mM MgCl2, and 1 mM DTT. For the assay with spiked nucleotide, 10 mM nucleotide was added after the initial incubation. Aliquots of the reaction were separated from the reaction at the specified time points and quenched in 2% SDS buffer, heated for 10 min and ran onto an acrylamide gel. The bands were visualized using silver staining (Sigma-Aldrich). Size exclusion chromatography (SEC) analysis and purification was performed by incubating 36 μM ClpA, 42 μM ClpP, 30 μM RepA(1-25)-GFP and 2 mM ATPγS in 50 mM Tris-HCl pH 7.5, 150 mM KCl, 10 mM MgCl2, 1 mM DTT for 15 minutes at 20°. The complex incubation reaction was then injected onto a Superose 6 Increase 3.2/300 column (GE Healthcare) and the eluted peaks were analyzed using SDS-PAGE.
Cryo-EM Data Collection and Processing
The fraction corresponding to the largest molecular weight complex from SEC (Extended Data Fig. 1b) was isolated and incubated with 1 mM ATPγS. Before freezing, proper dilutions were made and 10 mM ATP was added to the dilution. After a 30 s. incubation, a 3.5 uL drop was applied to glow discharged holey carbon grid (R 1.2/1.3; Quantifoil), in which sample was then blotted for 2.5 s. at 4° and 100% humidity with Whatman No. 1 filter paper before being plunge frozen liquid ethane using a vitrobot (Thermo Fischer Scientific). The sample was then imaged on a Titan Krios TEM (Thermo Fischer Scientific) operated at 300 keV and equipped with a Gatan BioQuantum imaging energy filter using a 20eV zero loss energy slit (Gatan Inc). Movies were acquired in super-resolution mode on a K3 direct electron detector (Gatan Inc.) at a calibrated magnification of 58,600X corresponding to a pixel size of 0.4265 Å/pixel. A defocus range of .8 to 1.2 μm was used with a total exposure time of 2 seconds fractionated into 0.2s subframes for a total dose of 68 e-/Å2 at a dose rate of 25 e-/pixel/s. Movies were subsequently corrected for drift using MotionCor2 (10.1038/nmeth.4193) and were Fourier-cropped by a factor of 2 to a final pixel size of 0.853 Å/pixel.
A total of ~18,000 micrographs were collected over two different datasets. The two datasets were processed separately and then were combined at the end. All the data-processing was performed in cryosparc23. For particle picking, templates were generated from 100 particles, in which only side-views were selected. After inspecting the particles picked, approximately 1.6 million particles were extracted. Two rounds of 2D classification were performed to remove contamination and junk particles, which amounted to ~54% of the dataset. A five-class ab-initio reconstruction was performed from the particle set and was used for initial classification steps.
To identify different conformations, heterogenous refinement was performed with 4 different classes (Extended Data Fig. 1f). Following this first round, maps showing high resolution features, which accounted for ~54% of the 739,000 particles going into 3D, were kept and grouped together. Another round of heterogenous refinement with 5 different classes was then performed. Following this second round, two unique states, ClpAPEng1 (24%, ~176,000 particles) and ClpAPDis (24%, ~176,000 particles), were identified. The ClpAPDis particles underwent another 5 class heterogenous refinement to further identify any more conformations. Following this third round, two unique states, ClpAPDis (8%, 58,000 particles) and ClpAPEng2 (5%, 40,000 particles), were identified. Particles associated with each unique class were combined and homogenous refinement was performed separately on each state. To better improve the resolution of the mobile protomers following Non-Uniform refinement, the particles from each state underwent particle subtraction. Particle subtraction was performed in which the bottom half of ClpP was subtracted. A local-refinement was then performed, in which the fulcrum position was set to the center of ClpA. The same procedure was completed on all the states.
The final resolution of ClpAPEng1 was 2.8Å, ClpAPDis was 3.2Å, and ClpAPEng2 was 3.4Å (Extended Data Fig. 6b). After completing local CTF refinement on of the final refinement runs the resolutions were improved to 2.7Å for ClpAPEng1, 3.0 Å for ClpAPDis and 3.2 Å for ClpAPEng2 (Extended Data Fig. 1e).
Molecular Modeling
An initial model for ClpA was obtain by using a ClpB structure (PDB 5ofo)4 and generated in SWISS-MODEL5 and the initial model for ClpP was taken directly from a ClpP crystal structure (PDB 1yg6)6 previously solved. Both initial models were docked into the EM maps using the UCSF chimera’s function fit in map7. Initial refinement was performed using Phenix8 with 1 round of simulated annealing and morphing and 5 rounds of real-space refinement that included minimization_global, rigid_body, adp, local_grid_search, secondary structural restraints and non-crystallographic symmetry (NCS) restraints. The resulting model then underwent real space refinement in Coot9. Nucleotides were added in manually using Coot and real space refinement using cif files generated for ADP and ATPγS in Phenix eLBOW10.
Density for the ClpA focus refinement was higher quality than the full map, therefore was used to model individual protomers using Rosetta Comparitive Modeling (RosettaCM)11,12. The structures for ClpA (PDB 1r6b)13, Hsp104 (PDB 5d4w and 5vjh)14, ClpB BAP form (PDB 5og1)4 and PTEX (PDB 6e10)15 were determined as homology models with HHpred16 and used to constrain model refinement in Rosetta CM with template_weight=0 and the initial model with template_weight=1. The lowest energy models were examined by eye to ensure the model fit into the density, the protomer was placed into the context of the whole structure and the Rosetta Relax protocol was run on the full complex.
Rosetta Enumerative Sampling (Rosetta ES) was used to de novo build in the IGL loops and NT loops for each protomer17. The ClpA residues 612 to 628 were deleted from each protomer and Rosetta ES was run to rebuild the loops with a beamwidth of 32. The resulting model with rebuilt IGL loops was added into the full model and the Rosetta Relax protocol was run. Residues 16 to 32 from ClpP were deleted from each protomer and the same RosettaES parameters were used to build in the NT loops, followed by the Rosetta Relax protocol.
Extended Data
Extended Data Fig. 1. ClpAP complex formation with RepA(1-25)-GFP and cryoEM data analysis.

a RepA1-25-GFP degradation assay in the presence of either ATPγS or ATP along with ClpA and ClpP. The assay was performed at 20°. Arrow represents RepA degradation product. b Size exclusion chromatography (SEC) trace of the components and formed ClpAP complex following incubation with RepA1-25-GFP and ATPyS. The 280 absorbance traces are shown for ClpA alone (red, dashed), ClpA with RepA1-25-GFP (red, solid), ClpAP alone (black, dashed) and ClpAP with RepA1-25-GFP (black, solid). c RepA1-25-GFP degradation assay in the presence of ATPγS with both ClpP WT and ClpP_S98A. ATP was spiked into the reaction at 10 mM after the initial complex formation for 15 min was completed with ATPγS. The zero-time point is before spiking ATP into the reaction. The assay was performed at 20°. d Reference-free 2D class averages of ClpAP bound to RepA1-25-GFP. The scale bar equals 125 Å. e Gold standard FSC-curves for the final refinement of ClpAPEng-1(red), ClpAPDis(cyan), ClpAPEng-2(black) of the ClpAP-RepA(1-25)-GFP complex. f 3D classification scheme used to identify the two different states in the ClpAP-RepA1-25-GFP dataset. Green asterisk represents the classes in which the particles were pooled together for further classification and refinement. The local resolution map of ClpAPEng-1 (g), ClpAPDis (h) and ClpAPEng-2 (i). j Low-pass filtered map showing globular density docked with GFP (PDB 1GFL) and additional N-terminal ClpA density (NTD). k Map vs. Model FSC of ClpAPEng-1(red), ClpAPDis(cyan), ClpAPEng-2 (black) of the ClpAP-RepA(1-25)-GFP complex following atomic modeling in Rosetta. Uncropped gel images are available as source data online.
Extended Data Fig. 2. Difference maps of the ClpAP interface.

Difference maps of the cryo-EM maps of a ClpAPEng1 vs. ClpAPDis and ClpAPEng-2 , b ClpAPDis vs. ClpAPEng-1 and ClpAPEng-2, c ClpAPEng-2 vs. ClpAPDis and ClpAPEng-1. The IGL pockets are encompassed by red circle, open pocket (dashed) and occupied pocket (solid). Schematic (right) shows occupancy of the ClpA IGL-loops (circles, colored and numbered by protomer) around the ClpA hexamer, with the empty IGL pockets (white circles) and ClpA protomers indicated (letters) for the different states. Asterisk represents the IGL-loop that is engaging in that state.
Extended Data Fig. 3. ATPγS-ClpAP cryoEM data analysis.

a Reference-free 2D class averages of ClpAP-γS bound to RepA1-25-GFP. The scale bar equals 125 Å. b Gold standard FSC-curves for the final refinement of ATPγS-ClpAPEng (blue) and ATPγS-ClpAPDis (red) of the ClpAP-RepA(1-25)-GFP complex. ATPγS-ClpAPEng-1 (c) and ATPγS-ClpAPDis (d) cryo-EM maps showing degree offset (arrow) of the ClpA channel axis (solid line) and substrate position (yellow density) compared to the ClpP pore and proteolytic chamber (dashed line). Schematic (below,left) shows occupancy of the ClpA IGL-loops (circles, colored and numbered by protomer) around the ClpA hexamer, with the empty IGL pockets (white circles) and ClpA protomers indicated (letters) for the different states. e 3D classification scheme used to identify the two different states in the ATPγS-ClpAP-RepA1-25-GFP dataset. Dotted boxes represent the classes in which the particles were pooled together for further classification and refinement. The maps for ClpAPEng (red) and ClpAPDis (yellow) are colored accordingly. Map vs. Model FSC of ATPγS-ClpAPEng(f) and ATPγS-ClpAPDis(g) following atomic modeling in Rosetta.
Extended Data Fig. 4. Comparison of IGL loops between the different states.

a EM map and model of the IGL-loop in the hydrophobic pocket of P1 (top), P2-P4 (middle, top), P5 (middle, bottom) and P6 (bottom) for ClpAPEng-1(left), ClpAPDis (middle) and ClpAPEng-2 (right). b Overlay of IGL-loops of ClpAPEng-1 (colored by protomer) vs. ClpAPDis (black) vs. ClpAPEng-2 (grey) laid out after alignment to the residues (638-649) above the IGL-loop. The dotted loop in P1 represents the missing loop in ClpAPDis and ClpAPEng-2.
Extended Data Fig. 5. Single capped ClpAP structure and ClpP N-terminal loop interactions.

a Map of the ClpP N-terminal gating loops and the model for ClpA with substrate for ClpAPDis (b) ClpAPEng. Map and model view of ClpP residues E14 and R15 (c) and E8 and K25 (d). e Gold standard FSC curve and (f) 2D reference-free class averages of the single capped ClpAP structure.
Extended Data Fig. 6. Particle Subtraction and Focus Refinement of ClpAPEng-1, ClpAPEng-2 and ClpAPDis.

a EM map with mask (grey) used for particle subtraction of ClpA. Red dot represents the point in which particles were shifted to. b Gold standard FSC curve of both focus maps for ClpAPEng-1 (red), ClpAPDis (cyan), and ClpAPEng-2 (black). The local resolution map of ClpAPEng-1 (c), ClpAPDis (d) and ClpAPEng-2 (e). f EM map and model of each Tyr-containing pore loop in ClpAPEng-1 for both D1 (top) and D2 (bottom), the substrate channel density is colored yellow. g EM map and model of each Tyr-containing pore loop in P5 for ClpAPEng-1 (left), ClpAPDis (middle), and ClpAPEng-2 (right) for both D1 (top) and D2 (bottom), the substrate channel density is colored yellow. The distance between the Tyr and the substrate is represented by dotted line. h EM map and model of ClpAPEng-1 (colored by protomer) with the D2 secondary pore loops residues interacting with substrate. i ClpAPEng-1 EM map colored by protomer with D2 secondary pore loops (red) and ClpP NTD-loops (green). j Overlay of the seam protomers P5 (left), P1 (middle), and P6 (right) for ClpAPEng-1 (grey) and ClpAPEng-2 (colored) showing conformational shifts (arrows) supporting translocation step.
Extended Data Fig. 7. Nucleotide States of ClpAPEng-1, ClpAPEng-2 and ClpAPDis.

a Difference map density for P4 D1 and D2 ATP with Arg finger residues displayed in green. There are no differences between P3 and P4, therefore P3 ATP density is not shown. b Difference map density for P1, P2, P5 and P6 for both D1 and D2 and Arg finger residues colored green.
Supplementary Material
Acknowledgements
We thank K. Mack, Z. March, and R. Cupo, T. Pospiech and J. Braxton for feedback on the manuscript. We thank the UCSF BACEM Facility for assistance with data collection. This work was supported by an Alzheimer’s Association Research Fellowship (to J.B.L.), an NSF Graduate Research Fellowship DGE-0822 (to L.M.C.), a GAANN fellowship (to A.N.R.), NSF grant MCB-1412624 (to A.L.L.), and NIH grants R01GM099836 (to J.S.), and R01GM110001 (to D.R.S.).
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
ClpAP cryo-EM maps and atomic coordinates have been deposited in the EMDB and PDB with accession codes EMDB-21519 and PDB-6W1Z for ClpAPEng1, EMDB-21520 and PDB-6W20 for ClpAPDis, EMDB-21521 and PDB-6W21 for ClpAPEng2, EMDB-21522 and PDB-6W22 for ClpAPEng1 focus, EMDB-21523 and PDB-6W23 for ClpAPDis focus, EMDB-21524 and PDB-6W24 for ClpAPEng2 focus, EMDB-20851 and PDB-6UQO for ATPγS-ClpAPEng, and EMDB-20845 and PDB-6UQE for ATPγS-ClpAPDis. The source data underlying Extended Data Fig. 1a and 1c are provided as a Source Data file. Other data are available from the corresponding author upon reasonable request.
References
- 1.Shorter J & Southworth DR Spiraling in Control: Structures and Mechanisms of the Hsp104 Disaggregase. Cold Spring Harb Perspect Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duran EC, Weaver CL & Lucius AL Comparative Analysis of the Structure and Function of AAA+ Motors ClpA, ClpB, and Hsp104: Common Threads and Disparate Functions. Front Mol Biosci 4, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivares AO, Baker TA & Sauer RT Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14, 33–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer RT & Baker TA AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Sauer RT et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119, 9–18 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessel M et al. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol 250, 587–94 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Sousa MC et al. Crystal and solution structures of an HslUV protease-chaperone complex. Cell 103, 633–43 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Baker TA & Sauer RT ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid BG, Fenton WA, Horwich AL & Weber-Ban EU ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci U S A 98, 3768–72 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins JR, Pak M, Maurizi MR & Wickner S The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad Sci U S A 95, 12135–40 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber-Ban EU, Reid BG, Miranker AD & Horwich AL Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–3 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa T et al. Translocation pathway of protein substrates in ClpAP protease. Proc Natl Acad Sci U S A 98, 4328–33 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Hartling JA & Flanagan JM The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–56 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Yu AY & Houry WA ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett 581, 3749–57 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Grimaud R, Kessel M, Beuron F, Steven AC & Maurizi MR Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem 273, 12476–81 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Ortega J, Lee HS, Maurizi MR & Steven AC ClpA and ClpX ATPases bind simultaneously to opposite ends of ClpP peptidase to form active hybrid complexes. J Struct Biol 146, 217–26 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ortega J, Singh SK, Ishikawa T, Maurizi MR & Steven AC Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol Cell 6, 1515–21 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Levchenko I, Seidel M, Sauer RT & Baker TA A specificity-enhancing factor for the ClpXP degradation machine. Science 289, 2354–6 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Bolon DN, Grant RA, Baker TA & Sauer RT Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell 16, 343–50 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Erbse A et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439, 753–6 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Wang KH, Roman-Hernandez G, Grant RA, Sauer RT & Baker TA The molecular basis of N-end rule recognition. Mol Cell 32, 406–14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keiler KC, Waller PR & Sauer RT Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–3 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Gottesman S, Roche E, Zhou Y & Sauer RT The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12, 1338–47 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk A, Schmidt R & Bukau B The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17, 165–72 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Wickner S et al. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A 91, 12218–22 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YI et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8, 230–3 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Effantin G, Maurizi MR & Steven AC Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase. J Biol Chem 285, 14834–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bewley MC, Graziano V, Griffin K & Flanagan JM Turned on for degradation: ATPase-independent degradation by ClpP. J Struct Biol 165, 118–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jennings LD, Bohon J, Chance MR & Licht S The ClpP N-terminus coordinates substrate access with protease active site reactivity. Biochemistry 47, 11031–40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brotz-Oesterhelt H et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med 11, 1082–7 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Lee BG et al. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat Struct Mol Biol 17, 471–8 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Gersch M et al. AAA+ chaperones and acyldepsipeptides activate the ClpP protease via conformational control. Nat Commun 6, 6320 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Li DH et al. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem Biol 17, 959–69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wendler P, Ciniawsky S, Kock M & Kube S Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823, 2–14 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Deville C et al. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv 3, e1701726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gates SN et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizo AN et al. Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase. Nat Commun 10, 2393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H et al. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc Natl Acad Sci U S A 115, E9560–E9569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gates SN & Martin A Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation. Protein Sci 29, 407–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H, Monroe N, Sundquist WI, Shen PS & Hill CP The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Pena AH, Goodall EA, Gates SN, Lander GC & Martin A Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho CM et al. Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puchades C et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoskins JR, Singh SK, Maurizi MR & Wickner S Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc Natl Acad Sci U S A 97, 8892–7 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoskins JR & Wickner S Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc Natl Acad Sci U S A 103, 909–14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JM & Lucius AL ATPgammaS competes with ATP for binding at Domain 1 but not Domain 2 during ClpA catalyzed polypeptide translocation. Biophys Chem 185, 58–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin A, Baker TA & Sauer RT Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell 27, 41–52 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bewley MC, Graziano V, Griffin K & Flanagan JM The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J Struct Biol 153, 113–28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW & Horwich AL Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–41 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Martin A, Baker TA & Sauer RT Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell 29, 441–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kress W, Mutschler H & Weber-Ban E Both ATPase domains of ClpA are critical for processing of stable protein structures. J Biol Chem 284, 31441–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogk A et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem 278, 17615–24 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Higuero JA et al. Allosteric communication between the nucleotide binding domains of caseinolytic peptidase B. J Biol Chem 286, 25547–55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olivares AO, Nager AR, Iosefson O, Sauer RT & Baker TA Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol 21, 871–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA & Lang MJ Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 145, 257–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JM, Lin J, Li T & Lucius ALE coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J Mol Biol 425, 2795–812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avellaneda MJ et al. Processive extrusion of polypeptide loops by a Hsp100 disaggregase. Nature (2020). [DOI] [PubMed] [Google Scholar]
- 58.Lee C, Schwartz MP, Prakash S, Iwakura M & Matouschek A ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell 7, 627–37 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Olivares AO, Kotamarthi HC, Stein BJ, Sauer RT & Baker TA Effect of directional pulling on mechanical protein degradation by ATP-dependent proteolytic machines. Proc Natl Acad Sci U S A (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cordova JC et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158, 647–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ripstein ZA, Vahidi S, Houry WA, Rubinstein JL & Kay LE A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. Elife 9(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajendar B & Lucius AL Molecular mechanism of polypeptide translocation catalyzed by the Escherichia coli ClpA protein translocase. J Mol Biol 399, 665–79 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Rabl J et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 30, 360–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith DM et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell 27, 731–44 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong Y et al. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majumder P et al. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc Natl Acad Sci U S A 116, 534–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 1.Miller JM, Lin J, Li T & Lucius ALE coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J Mol Biol 425, 2795–812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veronese PK, Stafford RP & Lucius AL The Escherichia coli ClpA molecular chaperone self-assembles into tetramers. Biochemistry 48, 9221–33 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Deville C et al. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv 3, e1701726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bewley MC, Graziano V, Griffin K & Flanagan JM The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J Struct Biol 153, 113–28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriarty NW, Grosse-Kunstleve RW & Adams PD electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65, 1074–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMaio F et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods 12, 361–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia D, Esser L, Singh SK, Guo F & Maurizi MR Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J Struct Biol 146, 166–79 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Gates SN et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho CM et al. Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann L et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Frenz B, Walls AC, Egelman EH, Veesler D & DiMaio F RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat Methods 14, 797–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ClpAP cryo-EM maps and atomic coordinates have been deposited in the EMDB and PDB with accession codes EMDB-21519 and PDB-6W1Z for ClpAPEng1, EMDB-21520 and PDB-6W20 for ClpAPDis, EMDB-21521 and PDB-6W21 for ClpAPEng2, EMDB-21522 and PDB-6W22 for ClpAPEng1 focus, EMDB-21523 and PDB-6W23 for ClpAPDis focus, EMDB-21524 and PDB-6W24 for ClpAPEng2 focus, EMDB-20851 and PDB-6UQO for ATPγS-ClpAPEng, and EMDB-20845 and PDB-6UQE for ATPγS-ClpAPDis. The source data underlying Extended Data Fig. 1a and 1c are provided as a Source Data file. Other data are available from the corresponding author upon reasonable request.